Summary
B lymphocytes, known as antibody producers, mediate tumor cell destruction in the manner of antibody‐dependent cell‐mediated cytotoxicity; however, their anti‐tumor function seems to be weakened during tumorigenesis, while the underlying mechanisms remain unclear. In this study, we found that IgG mediated anti‐tumor effects, but IgG‐producing B cells decreased in various tumors. Considering the underlying mechanism, glycometabolism was noteworthy. We found that tumor‐infiltrating B cells were glucose‐starved and accompanied by a deceleration of glycometabolism. Both inhibition of glycometabolism and deprivation of glucose through tumor cells, or glucose‐free treatment, reduced the differentiation of B cells into IgG‐producing cells. In this process, special AT‐rich sequence‐binding protein‐1 (SATB1) was significantly silenced in B cells. Down‐regulating SATB1 by inhibiting glycometabolism or RNA interference reduced the binding of signal transducer and activator of transcription 6 (STAT6) to the promoter of germline Cγ gene, subsequently resulting in fewer B cells producing IgG. Our findings provide the first evidence that glycometabolic inhibition by tumorigenesis suppresses differentiation of B cells into IgG‐producing cells, and altering glycometabolism may be promising in improving the anti‐tumor effect of B cells.
Keywords: B cells, metabolic disorder, tumor immunology
Abbreviations
- 2‐DG
2‐deoxy‐d‐glucose
- AOM
azoxymethane
- CRC
colorectal cancer
- DSS
dextran sodium sulfate
- FACS
fluorescence‐activated cell sorting
- IgG
immunoglobulin G
- LDH
lactate dehydrogenase
- PGK‐1
phosphoglycerate kinase 1
- PKM2
pyruvate kinase M2
- SATB1
special AT‐rich sequence‐binding protein‐1
- STAT6
transcription 6
Introduction
Intratumoral immune cells participate and act in all the stages of tumor progression,1 and among them, B lymphocytes have gained more attention in recent years.2, 3 Increasing evidence reveals that B cells make up a significant component of the lymphocytic infiltration in the tumor microenvironment,4, 5 and dense infiltration of B cells into the tumor is shown to correlate with the outcome of many tumor types including colorectal,6 breast,7 ovarian8 and pancreatic9 cancers. Since their discovery in the mid‐1960s, B lymphocytes were mainly known for their ability to produce antibodies.10, 11 The B‐cell‐derived antibody response has been reported to be critical to protect against microbial infections or mediate autoimmune diseases.12, 13, 14, 15 Although the role of B cells in tumors is still conflicting and controversial, due to their antibody‐producing capacity, tumor‐infiltrating B cells are recognized as anti‐tumor immune components.16, 17
Mature B cells from tumor patients were able to produce antibodies and mediate tumor cell cytotoxicity.18 Among them, IgG, especially IgG1, was identified as the most effective antibody type driving antibody‐dependent cell‐mediated cytotoxicity.19 In antibody‐dependent cell‐mediated cytotoxicity, IgG specially binds to the target cells and their FC region is combined by Fc‐γ receptors (FcγR or FCgR) on the surface of cytotoxic effector cells.20, 21 Emerging evidence shows that the tumor‐reactive G (IgG) produced by B cells is correlated with better outcome in patients.21, 22, 23 However, there is also evidence of a gradual reduction of anti‐tumor IgG antibodies and of an anti‐tumor response of B cells with disease progression.24, 25, 26 These indicate the antibody‐secreting capacity of B cells and their anti‐tumor protective functions may be impaired in tumors. However, knowledge of the mechanisms underlying is still lacking.
The metabolic pathways of immune cells and their effects on the overall immune response have become the concern of research in the emerging field of immunometabolism.27, 28 The central role of metabolism is to provide energy coupled with resources to fuel cellular function.27 A major source of cellular energy is glucose, which can be used through glycolysis and the tricarboxylic acid cycle.29 Different immune cells during their growth and proliferation adopt distinct levels of glycometabolism to balance their requirements for energy and biosynthesis. Those metabolic choices in immune cells play an important role in their fate and function.30, 31, 32 Previous studies have shown that the rates of glycometabolism increase in activated B cells in response to various stimuli in vitro.27, 33 Recent work suggests that the tumor microenvironment always has a shortage of glucose, and further studies on T cells indicate that tumor cells can outcompete T cells for glucose to mediate T‐cell hyporesponsiveness.34, 35 However, whether this lack of glucose in the tumor can influence the metabolism of B cells and then impede their anti‐tumor protective function remains unknown.
In the present study, we reported that IgG‐producing B cells decreased in various neoplastic tissues including human lung, esophageal and colorectal cancers. Using the murine model of primary colorectal cancer (CRC), we identified the anti‐tumor effect of IgG and confirmed the decrease of IgG‐producing B cells during tumor progression. Regarding the mechanisms, lack of glucose in the tumor microenvironment impeded B‐cell metabolism and repressed their differentiation to IgG‐producing B cells. Moreover, we demonstrated that special AT‐rich sequence‐binding protein‐1 (SATB1) was down‐regulated in the ‘glucose‐starved’ B cells and was involved in the regulation by affecting the binding of signal transducer and activator of transcription 6 (STAT6) into the germline Cγ gene promoter. Based on these findings, we demonstrated that the deceleration of glycometabolism caused by shortage of glucose in tumor impeded the differentiation of B cells into anti‐tumor subsets, and this may be a new target for improving B‐cell‐based anti‐tumor responses.
Materials and methods
Mice and samples
C57BL/6 mice at 8–12 weeks of age from the Shanghai SLAC Laboratory Animal Co. Ltd (Shanghai, China) were used for all experiments. All mice were housed in the animal facility of the Fudan University (Shanghai, China) under specific pathogen‐free conditions. All animal were cared for according to the Guidelines for the Care and Use of Laboratory Animals.
The adjacent normal and neoplastic tissues were collected from three individuals with lung adenocarcinoma (one man, 72 years old; two women, 65 and 69 years old), four individuals with esophageal squamous cancer (three men, 65, 58 and 54 years old; one woman, 63 years old), two individuals with rectal carcinoma (both men, 63 and 52 years old) and one individual with cancer of the descending colon (a 62‐year‐old woman). Informed consents for the use of samples were obtained from all patients. The study was approved by the Zhongshan Hospital Research Ethics Committee.
Mouse model of colorectal cancer
Murine colorectal carcinogenesis was established by azoxymethane (AOM; Sigma‐Aldrich, St. Louis, MO) plus dextran sodium sulfate (DSS, molecular weight 36 000–50 000 molecular weight; MP Biomedicals, Santa Ana, CA). Female C57BL/6 mice were injected (day 0) intraperitoneally with 9 mg/kg body weight of AOM, followed by ad libitum access to three DSS (at a final concentration of 2·5%)/water cycles (7 days of DSS and 14 days of water). Leukocytes from colon tissues, including intraepithelial lymphocytes and lamina propria lymphocytes, were isolated as previously reported.36 The animal protocol was approved by the institutional IACUC.
Polyclonal antibody preparation In the intravenously injected IgG experiment, the mouse IgG purified from the serum of the mice bearing CRC was used. The purified mouse IgG (2·5 μg/g) was intravenously injected into CRC mice once every 10 days. Blocking of FC receptor was performed using an anti‐CD16/CD32 monoclonal antibody (eBioscience, San Diego, CA). The anti‐CD16/CD32 monoclonal antibody (2·5 μg/g) was intravenously injected into CRC mice once every 10 days. Phosphate‐buffered saline was used as control.
Detection of secretory IgG
Titers of IgG1 in cell‐culture supernatants of in‐vitro‐stimulated B cells were measured using a specific enzyme‐linked immunosorbent assay (ELISA) kit (eBioscience) according to the manufacturer's protocol.
Mouse serum was collected from murine colorectal carcinogenesis in different stages of tumor development. The concentrations of IgG, IgG1, IgG2a, IgG2b, IgG2c and IgG3 were measured by ELISA kit.
Glucose assay
The glucose concentration in cell‐culture supernatants of in‐vitro‐stimulated B cells was measured using the Glucose Assay Kit (Eton Bioscience, San Diego, CA). For determining glucose levels in established mouse models, harvested colon tissues were weighed and pulped in fixed amounts of phosphate‐buffered saline. Ex vivo glucose concentration was quantified in accordance with the weight of tumors and the volume of collected supernatant.
Isolation and stimulation of mouse B cells
B cells isolated from naive C57BL/6 wild‐type mouse splenocytes were purified by negative selection using the EasySep™ mouse B‐cell isolation kit (Stem Cell Technologies, Vancouver, BC), and the purity was > 90% as judged by fluorescence‐activated cell sorting (FACS). Naive B cells were then cultured in RPMI 1640 (Gibco, Portland, OR) medium with 10% FBS (Gibco), 50 mm β‐mercaptoethanol (Sigma‐Aldrich) and 1× antibiotic mixture (Gibco) at 37° in 48‐well plates and stimulated with lipopolysaccharide (LPS; 10 μg/ml; Sigma‐Aldrich), interleukin‐4 (IL‐4; 20 ng/ml, PeproTech, Rocky Hill, NJ) for differentiation to IgG1. 2‐Deoxy‐d‐glucose (2‐DG) (Sigma‐Aldrich) and STAT6 inhibitor AS1517499 (AXON Medchem BV, Groningen, The Netherlands) were added at a final concentration of 0·5 mm and 100 nm, respectively.
Mass spectrum analysis of protein expression
The protein expression of in‐vitro‐stimulated B cells was analyzed using the Triple TOF 4600 (AB SCIEX, Framingham, MA). The manipulation was conducted in the technical platform of the Institute of Biomedical Sciences, Fudan University. Differentially expressed protein was identified through fold change filtering.
Transfection of small interfering RNAs
Small interfering RNAs were transfected into B cells using electroporation. Before electroporation, B cells were washed three times with Opti‐MEM (Gibco) and resuspended in Opti‐MEM medium. Then, indicated doses of diluted mRNA were mixed with B cells in 0·1 ml Opti‐MEM medium and electroporated in a 2‐mm cuvette using an ECM830 Electro Square Wave Porator (Harvard Apparatus BTX, San Diego, CA).
Flow cytometry analysis
Anti‐CD38, anti‐IgG, anti‐CD45, anti‐CD19, anti‐CD138, anti‐IgG1, anti‐IgG2a, anti‐B220, anti‐NK1.1, anti‐CD3, anti‐CD11b, anti‐Gr‐1, anti‐CD107a, anti‐tumor necrosis factor‐α (TNF‐α), Annexin V, 7‐AAD, anti‐Ki67, anti‐CXCR3, anti‐CXCR5 (all from eBioscience) and anti‐lactate dehydrogenase (LDH), anti‐PKM2, anti‐PGK‐1, anti‐SATB1 (all from Abcam, Burlingame, CA) were used for flow cytometry per the manufacturer's instructions. Cells were stained using fluorochrome‐conjugated antibodies according to the manufacturer's protocols. For the intranuclear staining of SATB1, cells were stained for surface markers; then, SATB1 was stained according to a Foxp3/Transcription Factor Buffer Staining Set (eBioscience). For detection of total IgG, cells were stained for both surface IgG and intracellular IgG. The intracellular IgG was strained by using an Intracellular Fixation and Permeabilization Buffer Set (eBioscience) according to the manufacturer's protocols.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as previously described elsewhere.37 The treated cells were cross‐linked using 1% formaldehyde, lysed and sonicated in a Covaris instrument (shearing time 30 min, 20% duty cycle, intensity 10, 200 cycles per burst, 30 seconds per cycle) in 2 ml. ChIP was performed using anti‐STAT6 (1:100, Cell Signaling Technology, Danvers, MA) antibody, following a standard protocol. Selected genes that contain STAT6‐binding elements were subsequently confirmed by polymerase chain reaction (PCR). The primer used was designed according to previous reports,38 and was synthesized by Sangon Biotech (Shanghai, China). Cγ1 promoter: Sense Primer (S): AGGCTTCCTACCTTCTCC, Antisense Primer (A): GTCCAAACCTGACTTAGATTAC.
Immunoprecipitation
Immunoprecipitation was performed as previously reported.39 The stimulated B cells were resuspended in lysis buffer and rotated at 4° for 20 min. The lysate was centrifuged at 12 000 g at 4° for 15 min, and supernatant was used for immunoprecipitation with the indicated antibody (SATB1, 1:100; Abcam, Cambridge, UK). Proteins were incubated overnight at 4° and subsequently with Protein G Dynabeads (Life Technologies, Gaithersburg, MD) for 1 hr. Beads were washed with NETN buffer (10mM Tris‐Cl, 100mM NaCl, 1mM EDTA, 0.5% Nonidet P‐40) three times, boiled in Laemmli sampling buffer, and subjected to Western blot.
Quantitative real‐time PCR and Western blotting
For the quantitative detection of mRNA, quantitative real‐time PCR (qRT‐PCR) or immunoblotting analysis was performed as previously reported.40 The primer used was synthesized by Sangon Biotech. Ighg1: sense primer (S): ACCGAAGGCTCCACAGGTGTAC, antisense primer (A): CCATTCCACTGCCACTCCACAG. The primary antibodies used in Western blotting were as follows: rabbit anti‐STAT6 (1 : 1000; Cell Signaling Technology), rabbit anti‐phospho‐STAT6 (1 : 1000; Cell Signaling Technology), rabbit anti‐SATB1 (1 : 1000; Abcam), mouse anti‐β‐actin (1 : 5000; ProteinTech Group, Chicago, IL).
Statistical analysis
Data were analyzed using the graphpad prism software (version 5; GraphPad Software Inc., La Jolla, CA) and were presented as the means ± standard error of the mean (SEM). The Student's unpaired t‐test or unpaired t‐test with Welch's correction was used to analyze intergroup differences for two groups, analysis of variance was used to analyze more than two groups, and the Pearson's correlation coefficient was used to analyze the correlation between groups.
Results
IgG‐producing B cells mediate anti‐tumor immune response, but decrease in neoplastic tissues during tumor progression
To investigate the IgG‐producing B cells and their IgG production during tumorigenesis, we first assessed whether their presence was linked to tumors. The results showed that the percentage of total IgG‐producing B cells in various neoplastic tissues from patients with lung, esophageal and colorectal cancers were significantly lower than that in the adjacent normal tissues (Fig. 1a). To further determine the dynamic changes of IgG‐producing B cells and their IgG production during tumorigenesis, a primary CRC mouse model induced by AOM plus DSS was established. Subsequently, B‐cell‐derived IgG production was detected during the tumorigenesis. Consistently, in the CRC model, compared with normal tissue, total IgG dramatically decreased in tumor tissue, and further reduced at the advanced stage of tumorigenesis (T3) compared with the early stage of tumor initiation (T1). Moreover, analysis of IgG subclasses showed that IgG1 and IgG2a decreased most dramatically (Fig. 1b). To determine whether the change of concentration was caused by the reduction of antibody‐producing B cells, IgG1‐producing B and IgG2a‐producing B‐cell subsets were detected in the CRC model. Results showed that the percentage of IgG1‐producing B cells and IgG2a‐producing B cells were both reduced in tumor tissues compared with adjacent normal tissues (Fig. 1c), and further down‐regulated during tumor progression, which was consistent with the changes of these antibodies (Fig. 1d).
Figure 1.
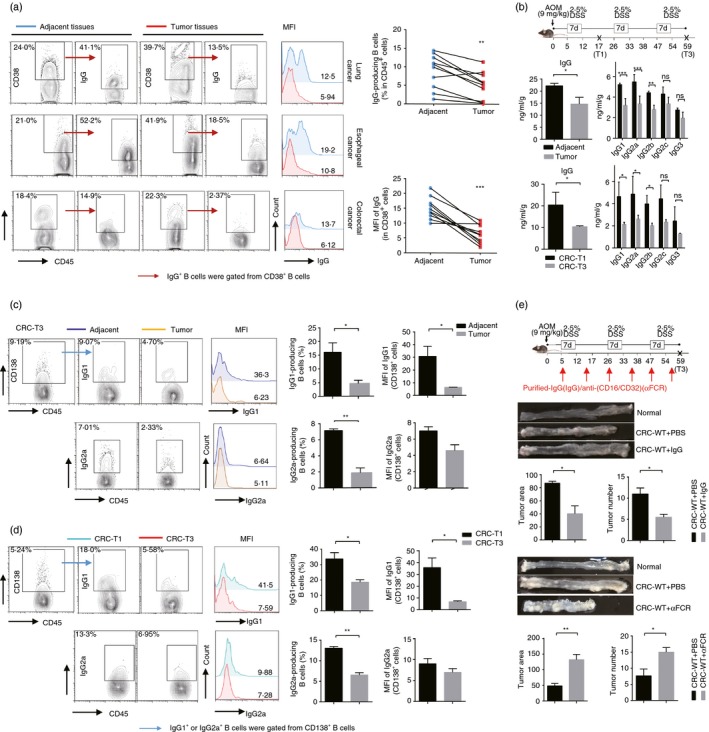
IgG‐producing B cells mediate anti‐tumor immune response, but decrease in neoplastic tissues during tumor progress. (a) The expression of total IgG including the surface and the intracellular IgG in CD38+ B cells in neoplastic and adjacent normal tissues of human lung, esophageal and colorectal cancers. (b) The concentration of IgG and its subclasses in the adjacent normal tissues and tumor tissues of colorectal cancer (CRC) mice at stage T3 (top), and in the colon tissues from CRC mice at stage T1 and T3 (bottom). (c) Fluorescence‐activated cell sorting (FACS) analysis of IgG1‐producing and IgG2a‐producing B cells, which all gated from CD138+ B cells in adjacent normal sites and tumor sites in the colon tissues from CRC mice at stage T3. (d) FACS analysis of IgG1‐producing and IgG2a‐producing B cells in colon tissues from CRC mice at stage T1 or stage T3. (e) Purified IgG or anti‐CD16/CD32 (α FCR) antibodies were intravenously injected into the C57BL/6 mice at the indicated time‐point, and the CRC model was constructed. Tumor numbers and volumes were detected at stage T3. Mice treated with phosphate‐buffered saline were served as the negative controls. Data shown in (b) to (e) are pooled from three independent experiments each with three or four mice and expressed as mean ± SEM. *P < 0·05, **P < 0·01, ***P < 0·001.
To determine the functional significance of IgG‐producing B cells in tumorigenesis, purified IgG was injected into CRC mice. The tumor growth was greatly decreased following administration of IgG, displaying fewer tumor nodes and smaller tumor volumes compared with the control mice (Fig. 1e). Consistent with this observation, FACS analysis also showed that the expression of CD107a in natural killer cells and TNF‐α secretion of neutrophils were increased in the IgG intravenously injected group (see Supplementary material, Fig. S1a). In addition, IgG purified from non‐tumor‐bearing mice was also injected into CRC mice, and the tumor progressed at the same rate compared with control mice (data not shown). It has been shown that murine IgG bound and activated effector cells to kill tumors through Fc receptors,41 so we used anti‐CD16/CD32 monoclonal antibody to block the FcγRII/III, and then detected the tumorigenesis. With FcγRII/III blocked, the AOM+DSS‐induced CRC mice had more aggressive tumor growth, presenting more tumor nodes and larger tumor volumes, with the lower expression of CD107a in natural killer cells and decreased TNF‐α secretion of neutrophils compared with the CRC‐control mice (Fig. 1g, and see Supplementary material, Fig. S1b). Furthermore, we found that the CD107a surface expression of natural killer cells, which reflected their degranulation process capacities decreased during CRC progression (see Supplementary material, Fig. S1c). Taken together, these data reveal that IgG is one of essential anti‐tumor factors; however, IgG‐producing B cells decrease during tumor progression.
Glycometabolism in tumor‐infiltrating B cells decelerates and correlates with the decrease of IgG‐producing B cells in tumor
Recent work has shown that activation and differentiation of effector immune cells depended on rapid synthesis of enormous amounts of energy through glycometabolism, and dysregulation of glycometabolism profoundly affected immune cell differentiation and function,30 so we further explored the effect of glycometabolism on the IgG production of B cells in tumors. The cellular glucose metabolic changes of B cells were examined in vivo by detecting several important glycometabolism‐associated enzymes, including lactate dehydrogenase (LDH), pyruvate kinase M2 (PKM2) and phosphoglycerate kinase 1 (PGK‐1). We observed that all of the enzymes were significantly diminished in B cells from tumor tissues compared with normal tissues (Fig. 2a). Consistent with this result, protein levels of those enzymes also decreased in B cells during the CRC progress (Fig. 2b). We further assessed the phenotypes of the tumor‐infiltrating B cells. And we found that compared with B cells that did not produce IgG (IgG−), those IgG‐producing (IgG+) B cells including IgG1+ and IgG2a+ B cells highly expressed those glucose‐associated enzymes, indicating that a much higher level of glycometabolism was required for IgG+ B cells compared with IgG− B cells (Fig. 2c).
Figure 2.
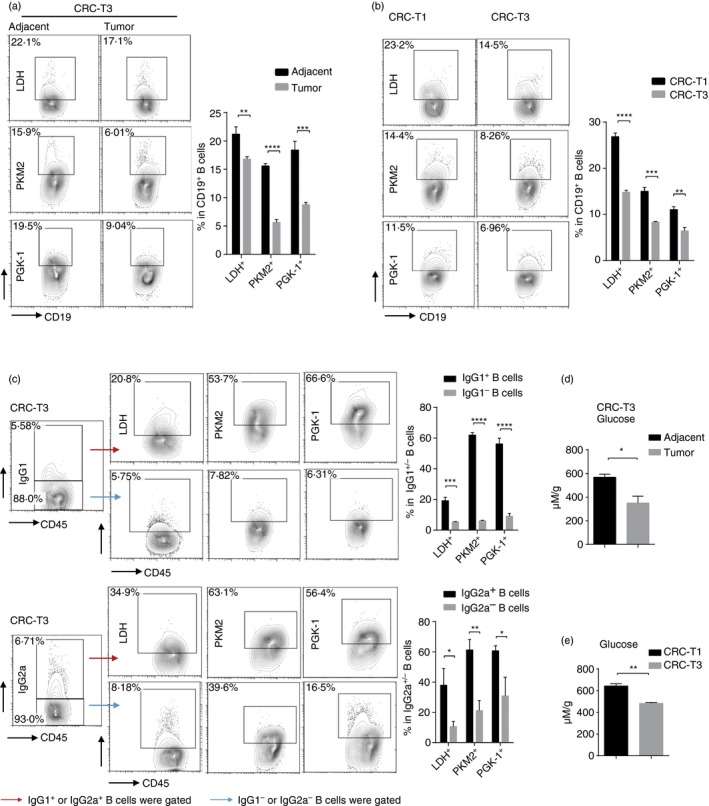
Glycometabolism in tumor‐infiltrating B cells decelerates and correlates with the decrease of IgG‐producing B cells in tumor. (a) Levels of LDH, PKM2 and PGK‐1 in CD19+ B cells in adjacent normal sites or tumor sites in the colon tissues of colorectal cancer (CRC) mice at stage T3. (b) FACS analysis of LDH, PKM2 and PGK‐1 levels in CD19+ B cells in colon tissues from CRC mice at stage T1 or T3. (c) FACS analysis of LDH, PKM2 and PGK‐1 levels in IgG‐producing (IgG1+) B cells/without IgG production (IgG1−), as well as in IgG2a+/IgG2a− B cells in the colon tissues from CRC mice at stage T3. (d) Glucose concentrations in adjacent normal sites and tumor sites in the colon tissues from CRC mice at stage T3. (e) Glucose concentrations in the colon tissues from CRC mice at stage T1 and stage T3. All the data shown are pooled from three independent experiments each with 3–4 mice and expressed as mean ± SEM. *P < 0·05, **P < 0·01, ***P < 0·001, ****P < 0·0001.
Given that the level of surrounding glucose is associated with the rate of cell metabolism, we wondered if the glucose in the tumor microenvironment contributed to the deceleration of glycometabolism in B cells. Therefore, we measured the glucose levels in the CRC model. Consistent with the decrease of glycometabolism‐associated enzymes, glucose levels in neoplastic tissues were lower than in normal tissues, and with the development of tumor, the glucose concentration further decreased in colon tissues (Fig. 2d,e). These data indicate that the glycolytic activity is reduced in B cells during tumor progression and associated with the decrease of IgG+ B cells, which may be caused by glucose‐restricted microenvironment.
Both inhibition of glycometabolism and deprivation of glucose by tumor cells reduce the transformation of B cells into IgG‐producing B cells
To clarify the role of glycometabolism in the generation of IgG+ B cells, differentiation of B cells into IgG1+ B cells was constructed in vitro by stimulation of LPS plus IL‐4. Under stimulation of LPS plus IL‐4, B cells differentiated to IgG1+ B cells, and the IgG1+ B cells showed ‘clock‐face’ nuclei and clear perinuclear regions in their cytoplasm (Fig. 3a). We next determined the change of glycometabolism in B cells in response to the stimulation. Results showed that the expression of glucose‐associated enzymes – LDH, PGK‐1 and PKM2 – in B cells increased in a time‐dependent manner following stimulation (Fig. 3b).
Figure 3.
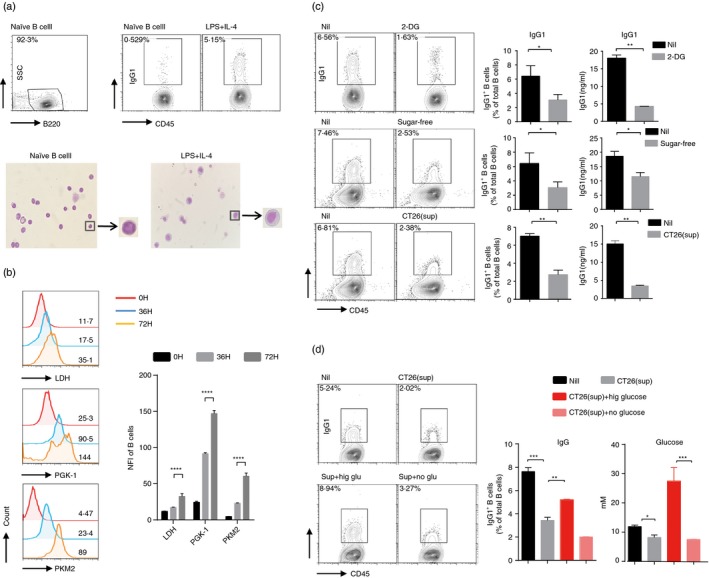
Both inhibition of glycometabolism and deprivation of glucose by tumor cells reduce the transformation of B cells into IgG‐producing B cells. (a) B cells were stimulated with lipopolysaccharide (LPS) plus interleukin‐4 (IL‐4) for 3 days. May–Grünwald–Giemsa staining of IgG1+/IgG1− (b) FACS analysis of LDH, PKM2 and PGK‐1 levels was detected during IgG1+ B‐cell differentiation. (c) B cells stimulated with LPS plus IL‐4 were cultured in medium with indicated doses of 2‐DG, sugar‐free medium or supernatant of CT26 cell line for 3 days. B cells cultured in medium with treatment of LPS and IL‐4 were used as control. Expression and secretion of IgG1 were detected by FACS and ELISA, respectively. (d) B cells stimulated with LPS and IL‐4 were cultured in supernatant of the CT26 cell line with high‐glucose medium or sugar‐free medium. The glucose concentration of each culture medium (right) was measured before stimulation. Expression of IgG1 (left) in B cells in each group was assessed at day 3. B cells cultured in supernatant of CT26 cell line with treatment of LPS and IL‐4 were used as control. The data are shown as the mean ± SEM from one experiment that was repeated three times, yielding similar results. *P < 0·05, **P < 0·01, ***P < 0·001, ****P < 0·0001.
To further understand whether increased glycolytic flux contributed to the generation of IgG1+ B cells, we used the glycolysis inhibitor 2‐DG to inhibit glycometabolism. 2‐DG treatment was non‐toxic to B cells (see Supplementary material, Fig. S2a), but sharply reduced the percentage of IgG1+ B cells and the antibody production. To confirm this result and test whether glucose was required for IgG1+ B‐cell generation, we cultured B cells in sugar‐free medium with stimulation of LPS and IL‐4. Compared with control groups, B cells that were cultured in sugar‐free medium displayed the low glycometabolism with the diminished expression of glucose‐associated enzymes (see Supplementary material, Fig. S2b), and showed a lower proportion of IgG1+ B cells and was associated with the decreased titers of IgG1, while cell vitality and apoptosis in response to stimulation were comparable between them (Fig. 3c, and see Supplementary material, Fig. S2c).
Given that the tumor microenvironment was shortage of glucose, we wondered if the shortage of glucose in tumor contributed to the decrease of IgG+ B cells. We used the CRC line CT26 cell‐culture supernatants to simulate the glucose‐restricted tumor microenvironment, and detected the contribution of glucose level to the generation of IgG1+ B cells. Results showed that stimulation of CT26 cell‐culture supernatants could also reduce LPS plus IL‐4 induced generation of IgG1+ B cells (Fig. 3c,d). When we added high‐glucose media to the supernatants to up‐regulate the amount of glucose, the percentage of IgG1+ B cells significantly increased, but only added sugar‐free media could not cause the above phenomenon (Fig. 3d, and see Supplementary material, Fig. S2d). Together these results suggest that the glycometabolism appears essential for B‐cell‐derived IgG production.
SATB1 is required for the glycometabolism‐triggered differentiation of B cells into IgG‐producing B cells
To understand how glycometabolism regulated the generation of IgG+ B cells, we compared the protein expression profiles of control and 2‐DG‐treated B cells using mass spectrometry. The results showed that 48 proteins were differentially expressed. Among them, we found the expression of DNA‐binding protein‐SATB1, which was reported to be associated with the immunoglobulin heavy‐chain gene expression, was reduced significantly and consistent with the expression of Cγ1 immunoglobulin heavy (immunoglobulin heavy constant γ1, Ighg1) (Fig. 4a). We further evaluated the SATB1 expression levels in B cells with 2‐DG treatment or cultured in sugar‐free medium under stimulation of LPS and IL‐4. Results showed that the expression of SATB1 in B cells was significantly down‐regulated under 2‐DG treatment or sugar‐free culture, which was in consonance with protein sequence analysis (Fig. 4b). Similar to the trends in vitro, in vivo data showed that the SATB1 expression of B cells also decreased during the CRC progression (Fig. 4c), whereas those IgG1+ B cells showed higher SATB1 expression than the IgG1– B cells (Fig. 4d), indicating that SATB1 might be required for the generation of IgG1+ B cells.
Figure 4.
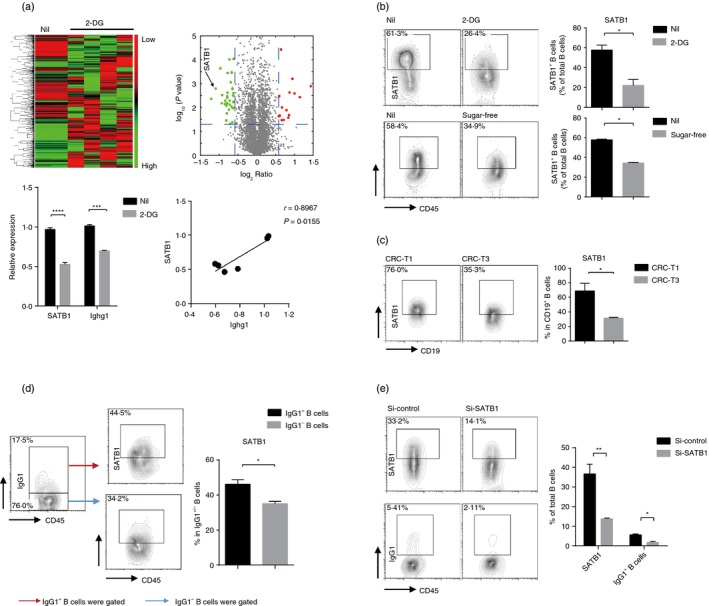
SATB1 is required for the glycometabolism‐triggered differentiation of B cells into IgG‐producing B cells. (a) B cells were stimulated with lipopolysaccharide (LPS) and interleukin‐4 (IL‐4) in the presence of 2‐DG or not for 3 days. Heat map and scattered plot display the protein expression relative to the control group mean on a log2 scale (top). Heat maps depicting the protein expressions of Ighg1 and SATB1, as well as the correlation between Ighg1 and SATB1(bottom). (b) B cells stimulated with LPS plus IL‐4 were cultured in medium with or without 2‐DG, or cultured in sugar‐free medium. FACS analysis of the protein expression of SATB1. (c) FACS analysis of SATB1 levels in CD19+ B cells in the colon tissues from colorectal cancer (CRC) mice at stage T1 and T3. (d) FACS analysis of SATB1 levels in IgG1+/IgG1− B cells in the colon tissues from CRC mice at stage T3. (e) B cells were stimulated with LPS plus IL‐4 for 3 days after transfection with short interfering RNAs of SATB1 (si‐SATB1) or control. The transfection efficiency of SATB1 and IgG1 expression in B cells. Data are pooled from three independent experiments and expressed as the mean ± SEM. *P < 0·05, **P < 0·01, ***P < 0·001, ****P < 0·0001.
To further verify the modulation of SATB1 in the generation of IgG+ B cells, we used the small interfering RNA to silence the SATB1 gene and examined the percentage of IgG+ B cells. Similar to 2‐DG treatment, interference of SATB1 in B cells reduced generation of IgG1+ B cells under stimulation by LPS and IL‐4 (Fig. 4e, and see Supplementary material, Fig. S3a). Overall, these results show that differentiation of B cells to IgG+ B cells is inhibited when glycometabolism is decelerated, probably due to the down‐regulation of SATB1.
STAT6 is the cofactor of SATB1 and mediates the regulation of IgG‐producing B cells by SATB1
The importance of SATB1 in regulating the generation of IgG+ B cells has been revealed, so we further studied the mechanisms underlying. As the primary transducer under IL‐4 stimulation, STAT6 was considered as the potential helper in this process. STAT6 has been demonstrated as a key mediator in IL‐4‐induced transcription of germline Cγ1 and switch recombination in B cells by binding to the promoter of Cγ1 gene. As expected, under the stimulation of LPS plus IL‐4, phosphorylation of STAT6 in B cells was dramatically up‐regulated, but not changed after decreasing SATB1 by 2‐DG treatment (Fig. 5a), which indicated that SATB1 did not affect IL‐4 induced STAT6 activation.
Figure 5.
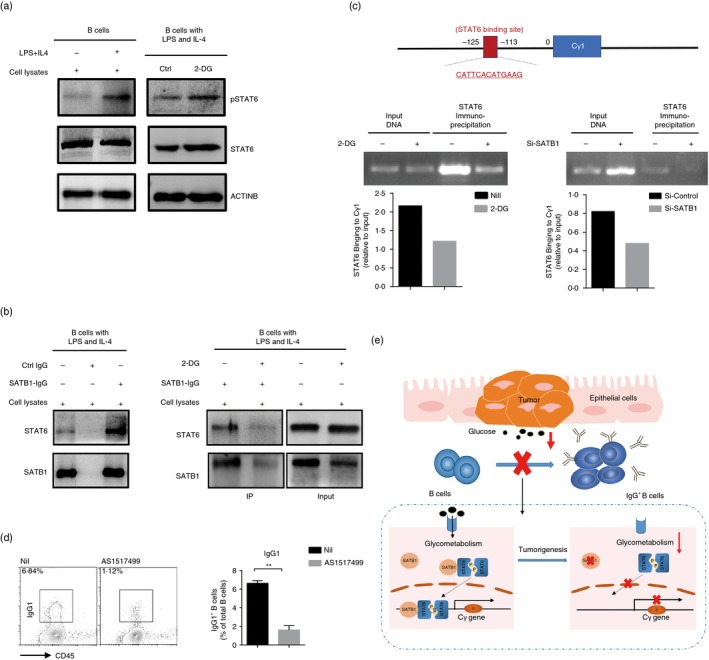
Signal transducer and activator of transcription 6 (STAT6) is the cofactor of SATB1 and mediates the regulation of IgG‐producing B cells by SATB1. B cells were stimulated with lipopolysaccharide (LPS) and interleukin‐4 (IL‐4) in the presence of 2‐DG or not for 3 days. (a) Protein levels of STAT6 and pSTAT6 in B cells with different treatments were detected by the Western blot. (b) Co‐immunoprecipitation of SATB1 with STAT6 was performed in B cells treated with 2‐DG or not. Protein levels of STAT6 and SATB1 were detected by Western blot. (c) ChIP assays of STAT6 on the Cγ1 promoter were performed in B cells in the presence of 2‐DG or not, or transfected with si‐STATB1 or control siRNA. STAT6‐binding sites on the Cγ1 promoter were detected by PCR. (d) Expression of IgG1 in B cells with indicated stimulation in the presence of STAT6 inhibitor or not for 3 days. (e) The regulatory mechanism of microenvironment in IgG‐producing B‐cell generation in tumor. The data are shown as the means ± SEM from one experiment that was repeated three times, yielding similar results. **P < 0·01.
Many reports indicated that SATB1 could recruit some transcription factors to SATB1‐binding status to regulate gene expression, so we further explored whether SATB1 could bind to the STAT6 and help it bind to the Cγ1 gene promoters. We performed immunoprecipitation of SATB1 in the LPS plus IL‐4 activated B cells with or without 2‐DG treatment. Immunoprecipitation of SATB1 in the activated B cells pulled down a protein of 100 000 molecular weight specifically detected by the anti‐STAT6 antibody, whereas the signal of STAT6 decreased when SATB1 was down‐regulated by 2‐DG treatment, implying that SATB1 might serve as cofactor of STAT6 in B cells (Fig. 5b). Next, we further performed a ChIP‐PCR assay to understand whether SATB1 affected STAT6 specifically to the Cγ1 gene promoter. Results showed that STAT6 could tightly bind to the Cγ1 gene promoter region; however, the binding was attenuated upon SATB1‐silence by 2‐DG treatment or small RNA interference (Fig. 5c). Consistent with the results, the real‐time PCR analysis showed that silence of SATB1 reduced Cγ1 germline mRNA expression in LPS‐ and IL‐4‐stimulated B cells (see Supplementary material, Fig. S3b). Moreover, we used the AS1517499, a widely used STAT6 inhibitor, to treat B cells 30 min before IL‐4 stimulation. As shown in Fig. 5(d), FACS analysis showed that AS1517499 reduced the generation of IgG1+ B cells. These data provide evidence of the interaction of SATB1 with STAT6, and reveal that STAT6 contributes to the regulation of IgG+ B cells by SATB1 (Fig. 5e).
Discussion
Growing clinical studies have revealed that tumor‐specific IgG was significantly associated with improved overall and recurrence‐free survival of cancer patients.23 There is evidence of an anti‐tumor effect of B cells being impaired and failed,26 but the mechanisms underlying the deficiency remain largely unknown. In this study, we found that the number of IgG‐producing B cells was significantly lower in both human and mouse tumor tissues compared with the normal tissues and further decreased during the tumor progression. Importantly, using a murine model of primary CRC, for the first time, we revealed that the glucose‐restricted tumor microenvironment could reduce the glycometabolism of B cells, resulting in repression of IgG‐producing B‐cell differentiation. In this process, SATB1 was identified as being required for glycometabolism‐triggered generation of IgG‐producing B cells, and STAT6 was the cofactor of SATB1. Therefore, altering glycometabolism may be promising in improving IgG‐producing B‐cell‐mediated anti‐tumor effect.
IgG was initially identified to be secreted by IgG+ B cells and had a protective role in infectious diseases.42 However, their tumor‐reactive functions remained under debate. In this study, we demonstrated that IgG could mediate tumor cell cytotoxicity. Consistently, Saito et al.26 reported that the decreased serum concentration of IgG was closely related to poor prognosis for patients with gastric cancer. This report strongly supports our hypothesis that IgG+ B cells have a protective function in the tumor. It should be pointed out that the circulating IgGs also had tumor reactivity in accordance with the IgGs produced by tumor‐infiltrated B cells (see Supplementary material, Fig. S4). Similar observations have been made by others.43 However, some studies also found that deletion of B cells could alleviate the progression of the tumor.44, 45 Hence, the anti‐tumor function of B cells is impaired in some tumors. We considered that this impairment seemed to be dependent on the impedance of the generation of IgG+ B cells and IgG production. Furthermore, we found that the IgG‐producing CD38+ B cells that might include both plasma cells and memory cells (see Supplementary material, Fig. S5) in human tumor tissues, and the IgG‐producing plasma B cells (IgG+ CD138+ B cells) in mouse tumor tissues were decreased compared with the normal tissues. In the future, it will be interesting to analyze which subset of IgG‐producing B cells can primarily lead to humoral immunity against the tumor.
The tumor microenvironment is an intricate network that includes cells, soluble factors and extracellular matrix molecule.46 This environment plays a pivotal role in modulating the function of immune cells. A previous report found that breast cancer cells produced leukotriene B4 to induce the generation of regulatory B cells.47 Our previous reports also demonstrated that the colon epithelial cells could secrete chemokines to recruit immunosuppressive B cells in CRC tissues.6 In addition, several cytokines, such as IL‐4, IL‐6 and transforming growth factor‐β, have been shown to participate in both the initiation and progression of tumor,48 those cytokines may play important roles in B‐cell development, or survival/proliferation of B cells.10, 48 However, little is known about the mechanisms by which the tumor microenvironment modulates the differentiation of tumor‐infiltrated B cells. In our report, we identified that the ‘glucose‐restricted’ microenvironment influenced the glycometabolism of B cells and then shaped the ability of B cells to perform in tumor. Moreover, we also discovered that the tumor microenvironment affected neither B‐cell proliferation nor cell apoptosis (see Supplementary material, Fig. S6a,b). There is a growing appreciation of the fact that under activation, B cells increase glucose uptake and induction of glycolysis to support their differentiation and biosynthesis.30 We envisage that the various states of B‐cell hyporesponsiveness that have been described in cancer may be induced by nutrition restriction. Our data further indicated that both inhibition of glycometabolism and deprivation of glucose limited the generation of IgG1+ B cells in vivo. Moreover, we supported that decreased SATB1 expression of B cells in glucose‐restricted microenvironment prevented the differentiation of B cells to IgG+ B cells.
SATB1 was recognized as a cell‐type‐specific ‘genomic organizer’, which could recruit chromatin remodeling complexes to the anchored sites and thereby regulate the gene expression.49 Cai et al.50 have reported that SATB1 was a necessary determinant for T helper type 2 cell activation, though inducing IL‐4, IL‐5, IL‐13 and c‐Maf expression, although its role in the differentiation of B cells has not been reported. In this report, we found that both inhibition of glycometabolism and deprivation of glucose reduced the expression of SATB1 in B cells. Using RNAi, we have shown that when SATB1 expression was reduced by > 50%, the differentiation of IgG1+ B cells did not induce as normal.
A previous study also reported that SATB1 involved in immunoglobulin heavy‐chain (IgH) gene expression,51 but the mechanisms underlying its regulation remain to be further investigated. We demonstrated that IgG1+ B‐cell‐specific transcriptional pathway‐STAT6 was regulated by SATB1. IgG1 switching is known to be induced by IL‐4 stimulation through the transcriptional regulator STAT6.52 Once phosphorylated, STAT6 forms homodimers and transfers to the nucleus, activates gene transcription through binding to the specific DNA domain. In our study, the immunoprecipitation analysis showed that SATB1 could bind to the STAT6, and 2‐DG treatment reduced their combination. We also found that using the RNAi to down‐regulate the expression of SATB1 could reduce binding of STAT6 with Ighg1 promoter. In this study, we first demonstrated that SATB1 probably helped STAT6 to bind Ighg1 promoter to direct B cells into IgG1+ B cells.
In conclusion, our results suggest that IgG+ B cells exert an anti‐tumor effect in neoplastic tissues. Although the ‘glucose‐restricted’ tumor microenvironment can inhibit the glycometabolism of B cells, resulting in the decrease of their differentiation into IgG‐producing cells. In this process, SATB1 in B cells is down‐regulated by the deceleration of the glycometabolism, which disrupts the transcriptional factors to bind to the germ‐line gene promoter. Based on these findings, future therapies may consider restoring the glucose supply to B cells to reinforce their anti‐tumor effect.
Disclosures
The authors have declared that no conflict of interest exists.
Author contributions
JL performed the research, discussed and analyzed the data, and wrote the paper. RL and YC designed the research, discussed and analyzed the data, and wrote the paper. YL, ZL, JG, YL, EH, ZW, HZ, LW, DZ and HX performed the research.
Supporting information
Figure S1. IgG mediates an anti‐tumor effect through antibody‐dependent cell‐mediated cytotoxicity in colorectal cancer progression.
Figure S2. Inhibition of glucose metabolism or glucose deprivation do not influence the viability of B cells.
Figure S3. Silence of SATB1 reduces Cγ1 mRNA expression in lipopolysaccharide and interleukin‐4 stimulated B cells.
Figure S4. IgGs produced by peripheral or tumor‐derived B cells, or circulating IgGs bind to tumor cells.
Figure S5. The phenotype of IgG‐producing B cells in human specimen.
Figure S6. The cell proliferation, apoptosis and chemokine receptor expression of B cells in the colon tissues during the colorectal cancer progression.
Acknowledgements
This research was supported by the National Science Foundation of China (Grant numbers: 81730045, 31570892 and 81601362); Science and Technology Commission of Shanghai Municipality (Grant number: 15JC1401200); National Science and Technology Major Project of China (Grant number: 2017ZX10203207); Shanghai Municipal Commission of Health and Family Planning (Grant numbers: 2018YQ16, 20184Y0089). Special thanks to Dr Lin Wang (The Affiliated Zhongshan Hospital of Fudan University, Shanghai, China) for collecting tissue samples and thanks to Mr Xing Gao and Mr Lei Zhang (Institute of Biomedical Sciences, Fudan University, Shanghai, China) for their support with the mass spectrometry analysis.
Contributor Information
Ronghua Liu, Email: ronghualiu@fudan.edu.cn.
Yiwei Chu, Email: yiweichu@fudan.edu.cn.
References
- 1. Erdag G, Schaefer JT, Smolkin ME, Deacon DH, Shea SM, Dengel LT et al Immunotype and immunohistologic characteristics of tumor‐infiltrating immune cells are associated with clinical outcome in metastatic melanoma. Cancer Res 2012; 72:1070–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation‐induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer 2013; 13:759–71. [DOI] [PubMed] [Google Scholar]
- 3. Sarvaria A, Madrigal JA, Saudemont A. B cell regulation in cancer and anti‐tumor immunity. Cell Mol Immunol 2017; 14:662–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nelson BH. CD20+ B cells: the other tumor‐infiltrating lymphocytes. J Immunol 2010; 185:4977–82. [DOI] [PubMed] [Google Scholar]
- 5. Chien CH, Chiang BL. Recent advances in regulatory T cells induced by B cells. Cell Mol Immunol 2017; 15:539–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu R, Lu Z, Gu J, Liu J, Huang E, Liu X et al MicroRNAs 15A and 16‐1 activate signaling pathways that mediate chemotaxis of immune regulatory B cells to colorectal tumors. Gastroenterology 2018; 154:637–51.e7. [DOI] [PubMed] [Google Scholar]
- 7. Schmidt M, Bohm D, von Torne C, Steiner E, Puhl A, Pilch H et al The humoral immune system has a key prognostic impact in node‐negative breast cancer. Cancer Res 2008; 68:5405–13. [DOI] [PubMed] [Google Scholar]
- 8. Lundgren S, Berntsson J, Nodin B, Micke P, Jirstrom K. Prognostic impact of tumour‐associated B cells and plasma cells in epithelial ovarian cancer. J Ovarian Res 2016; 9:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pylayeva‐Gupta Y, Das S, Handler JS, Hajdu CH, Coffre M, Koralov SB et al IL‐35‐producing B cells promote the development of pancreatic neoplasia. Cancer Discov 2015; 6:247–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vazquez MI, Catalan‐Dibene J, Zlotnik A. B cells responses and cytokine production are regulated by their immune microenvironment. Cytokine 2015; 74:318–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. LeBien TW, Tedder TF. B lymphocytes: how they develop and function. Blood 2008; 112:1570–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lu LL, Chung AW, Rosebrock TR, Ghebremichael M, Yu WH, Grace PS et al A functional role for antibodies in tuberculosis. Cell 2016; 167:433–43 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ruan S, Cai Y, Ramsay AJ, Welsh DA, Norris K, Shellito JE. B cell and antibody responses in mice induced by a putative cell surface peptidase of Pneumocystis murina protect against experimental infection. Vaccine 2017; 35:672–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim M, Kim CH. Regulation of humoral immunity by gut microbial products. Gut Microbes 2017; 8:392–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sakai J, Akkoyunlu M. The role of BAFF system molecules in host response to pathogens. Clin Microbiol Rev 2017; 30:991–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pavoni E, Monteriu G, Santapaola D, Petronzelli F, Anastasi AM, Pelliccia A et al Tumor‐infiltrating B lymphocytes as an efficient source of highly specific immunoglobulins recognizing tumor cells. BMC Biotechnol 2007; 7:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zitvogel L, Kroemer G. Cancer: antibodies regulate antitumour immunity. Nature 2015; 521:35–7. [DOI] [PubMed] [Google Scholar]
- 18. Wang SS, Liu W, Ly D, Xu H, Qu L, Zhang L. Tumor‐infiltrating B cells: their role and application in anti‐tumor immunity in lung cancer. Cell Mol Immunol 2018; 10.1038/s41423-018-0027-x. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kapur R, Einarsdottir HK, Vidarsson G. IgG‐effector functions: “the good, the bad and the ugly”. Immunol Lett 2014; 160:139–44. [DOI] [PubMed] [Google Scholar]
- 20. Steplewski Z, Sun LK, Shearman CW, Ghrayeb J, Daddona P, Koprowski H. Biological activity of human‐mouse IgG1, IgG2, IgG3, and IgG4 chimeric monoclonal antibodies with antitumor specificity. Proc Natl Acad Sci USA 1988; 85:4852–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gilbert AE, Karagiannis P, Dodev T, Koers A, Lacy K, Josephs DH et al Monitoring the systemic human memory B cell compartment of melanoma patients for anti‐tumor IgG antibodies. PLoS ONE 2011; 6:e19330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brunner SM, Itzel T, Rubner C, Kesselring R, Griesshammer E, Evert M et al Tumor‐infiltrating B cells producing antitumor active immunoglobulins in resected HCC prolong patient survival. Oncotarget 2017; 8:71002–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maletzki C, Jahnke A, Ostwald C, Klar E, Prall F, Linnebacher M. Ex‐vivo clonally expanded B lymphocytes infiltrating colorectal carcinoma are of mature immunophenotype and produce functional IgG. PLoS ONE 2012; 7:e32639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carpenter EL, Mick R, Rech AJ, Beatty GL, Colligon TA, Rosenfeld MR et al Collapse of the CD27+ B‐cell compartment associated with systemic plasmacytosis in patients with advanced melanoma and other cancers. Clin Cancer Res 2009; 15:4277–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chiaruttini G, Mele S, Opzoomer J, Crescioli S, Ilieva KM, Lacy KE et al B cells and the humoral response in melanoma: the overlooked players of the tumor microenvironment. Oncoimmunology 2017; 6:e1294296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saito H, Miyatani K, Kono Y, Murakami Y, Kuroda H, Matsunaga T et al Decreased serum concentration of total IgG is related to tumor progression in gastric cancer patients. Yonago Acta Med 2017; 60:119–25. [PMC free article] [PubMed] [Google Scholar]
- 27. Kunisawa J. Metabolic changes during B cell differentiation for the production of intestinal IgA antibody. Cell Mol Life Sci 2017; 74:1503–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mathis D, Shoelson SE. Immunometabolism: an emerging frontier. Nat Rev Immunol 2011; 11:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol 2011; 27:441–64. [DOI] [PubMed] [Google Scholar]
- 30. Pearce EL, Pearce EJ. Metabolic pathways in immune cell activation and quiescence. Immunity 2013; 38:633–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chang CH, Curtis JD, Maggi LB Jr, Faubert B, Villarino AV, O'Sullivan D et al Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 2013; 153:1239–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu G, Bi Y, Shen B, Yang H, Zhang Y, Wang X et al SIRT1 limits the function and fate of myeloid‐derived suppressor cells in tumors by orchestrating HIF‐1α‐dependent glycolysis. Cancer Res 2014; 74:727–37. [DOI] [PubMed] [Google Scholar]
- 33. Torigoe M, Iwata S, Nakayamada S, Sakata K, Zhang M, Hajime M et al Metabolic reprogramming commits differentiation of human CD27(+)IgD(+) B cells to plasmablasts or CD27(−)IgD(−) cells. J Immunol 2017; 199:425–34. [DOI] [PubMed] [Google Scholar]
- 34. Gupta S, Roy A, Dwarakanath BS. Metabolic cooperation and competition in the tumor microenvironment: implications for therapy. Front Oncol 2017; 7:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chang CH, Qiu J, O'Sullivan D, Buck MD, Noguchi T, Curtis JD et al Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell 2015; 162:1229–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang E, Liu R, Lu Z, Liu J, Liu X, Zhang D et al NKT cells mediate the recruitment of neutrophils by stimulating epithelial chemokine secretion during colitis. Biochem Biophys Res Commun 2016; 474:252–8. [DOI] [PubMed] [Google Scholar]
- 37. Hsu YA, Huang CC, Kung YJ, Lin HJ, Chang CY, Lee KR et al The anti‐proliferative effects of type I IFN involve STAT6‐mediated regulation of SP1 and BCL6. Cancer Lett 2016; 375:303–12. [DOI] [PubMed] [Google Scholar]
- 38. Zhuang B, Luo X, Rao H, Li Q, Liu X, Qi H. [Expression and significance of SATB1 and wnt/β‐catenin signaling molecule in the placenta of preeclampsia]. Zhonghua Fu Chan Ke Za Zhi 2015; 50:283–90. [PubMed] [Google Scholar]
- 39. Stephen TL, Payne KK, Chaurio RA, Allegrezza MJ, Zhu H, Perez‐Sanz J et al SATB1 expression governs epigenetic repression of PD‐1 in tumor‐reactive T cells. Immunity 2017; 46:51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lu Z, Liu J, Liu X, Huang E, Yang J, Qian J et al MicroRNA 15a/16‐1 suppresses aryl hydrocarbon receptor‐dependent interleukin‐22 secretion in CD4+ T cells and contributes to immune‐mediated organ injury. Hepatology 2018; 67:1027–40. [DOI] [PubMed] [Google Scholar]
- 41. Schwab I, Nimmerjahn F. Intravenous immunoglobulin therapy: how does IgG modulate the immune system? Nat Rev Immunol 2013; 13:176–89. [DOI] [PubMed] [Google Scholar]
- 42. Flynn NJ, Somasundaram R, Arnold KM, Sims‐Mourtada J. The multifaceted roles of B cells in solid tumors: emerging treatment opportunities. Target Oncol 2017; 12:139–52. [DOI] [PubMed] [Google Scholar]
- 43. Punt CJ, Barbuto JA, Zhang H, Grimes WJ, Hatch KD, Hersh EM. Anti‐tumor antibody produced by human tumor‐infiltrating and peripheral blood B lymphocytes. Cancer Immunol Immunother 1994; 38:225–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Balkwill F, Montfort A, Capasso M. B regulatory cells in cancer. Trends Immunol 2013; 34:169–73. [DOI] [PubMed] [Google Scholar]
- 45. Ganti SN, Albershardt TC, Iritani BM, Ruddell A. Regulatory B cells preferentially accumulate in tumor‐draining lymph nodes and promote tumor growth. Sci Rep 2015; 5:12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Romero‐Garcia S, Moreno‐Altamirano MM, Prado‐Garcia H, Sanchez‐Garcia FJ. Lactate contribution to the tumor microenvironment: mechanisms, effects on immune cells and therapeutic relevance. Front Immunol 2016; 7:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wejksza K, Lee‐Chang C, Bodogai M, Bonzo J, Gonzalez FJ, Lehrmann E et al Cancer‐produced metabolites of 5‐lipoxygenase induce tumor‐evoked regulatory B cells via peroxisome proliferator‐activated receptor α . J Immunol 2013; 190:2575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Landskron G, De la Fuente M, Thuwajit P, Thuwajit C, Hermoso MA. Chronic inflammation and cytokines in the tumor microenvironment. J Immunol Res 2014; 2014:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pavan Kumar P, Purbey PK, Sinha CK, Notani D, Limaye A, Jayani RS et al Phosphorylation of SATB1, a global gene regulator, acts as a molecular switch regulating its transcriptional activity in vivo . Mol Cell 2006; 22:231–43. [DOI] [PubMed] [Google Scholar]
- 50. Cai S, Lee CC, Kohwi‐Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet 2006; 38:1278–88. [DOI] [PubMed] [Google Scholar]
- 51. Oancea AE, Berru M, Shulman MJ. Expression of the (recombinant) endogenous immunoglobulin heavy‐chain locus requires the intronic matrix attachment regions. Mol Cell Biol 1997; 17:2658–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Turner ML, Corcoran LM, Brink R, Hodgkin PD. High‐affinity B cell receptor ligation by cognate antigen induces cytokine‐independent isotype switching. J Immunol 2010; 184:6592–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. IgG mediates an anti‐tumor effect through antibody‐dependent cell‐mediated cytotoxicity in colorectal cancer progression.
Figure S2. Inhibition of glucose metabolism or glucose deprivation do not influence the viability of B cells.
Figure S3. Silence of SATB1 reduces Cγ1 mRNA expression in lipopolysaccharide and interleukin‐4 stimulated B cells.
Figure S4. IgGs produced by peripheral or tumor‐derived B cells, or circulating IgGs bind to tumor cells.
Figure S5. The phenotype of IgG‐producing B cells in human specimen.
Figure S6. The cell proliferation, apoptosis and chemokine receptor expression of B cells in the colon tissues during the colorectal cancer progression.


