SUMMARY
Axonal degeneration is an early and prominent feature of many neurological disorders. SARM1 is the central executioner of the axonal degeneration pathway that culminates in depletion of axonal NAD+; yet the identity of the underlying NAD+ depleting enzyme(s) is unknown. Here, in a series of experiments using purified proteins from mammalian cells, bacteria, and a cell-free protein translation system, we show that the SARM1-TIR domain itself has intrinsic NADase activity – cleaving NAD+ into ADP Ribose (ADPR), cyclic ADPR, and Nicotinamide, with Nicotinamide serving as a feedback inhibitor of the enzyme. Using traumatic and vincristine-induced injury models in neurons, we demonstrate that the NADase activity of full-length SARM1 is required in axons to promote axonal NAD+ depletion and axonal degeneration after injury. Hence, the SARM1 enzyme represents a novel therapeutic target for axonopathies. Moreover, the widely utilized TIR-domain is a protein motif that can possess enzymatic activity.
Keywords: Axonal Degeneration, NAD+, Enzyme, SARM1, Toll/Interleukin-1 Receptor Domain, Innate Immunity
eTOC-BLURB
Essuman et al. demonstrate that the TIR domain of SARM1 is an enzyme that depletes axonal NAD+ to induce pathological axon loss. This enzymatic activity represents a novel therapeutic target for many neurological conditions characterized by axonal degeneration. More broadly, this study shows that TIR domains can possess enzymatic function.
INTRODUCTION
Axonal degeneration is a hallmark of several neurological disorders including peripheral neuropathy, traumatic brain injury, and neurodegenerative diseases (Gerdts et al., 2016). In Parkinson’s disease and Amyotrophic Lateral Sclerosis, for example, axonal degeneration is an early event, preceding symptom onset and widespread neuronal loss (Kurowska et al., 2017; Fischer and Glass, 2007). Although these neurological conditions have unique underlying etiologies, blocking axonal degeneration in the early stages of these disorders may slow or perhaps halt their progression by preventing the loss of functional synapses, and maintaining neuronal connectivity. In order to develop effective therapies targeting pathological axonal degeneration, the underlying molecular mechanisms need to be defined and pharmacologically-inhibitable targets identified.
Damaged or unhealthy axons are eliminated via an intrinsic self-destruction program that is distinct from traditional cellular death pathways like apoptosis (Gerdts et al., 2016). This programmed subcellular destructive pathway is triggered in Wallerian degeneration, which was initially thought to be a passive ‘wasting’ process of the severed distal axon segment. However, the discovery of the naturally occurring Wallerian degeneration slow (Wlds) mutant mice (Lunn et al., 1989; Mack et al., 2001), whose distal axons degenerate in a delayed fashion after injury, challenged this notion of passive degeneration. Early studies of Wlds showed that the Nicotinamide Adenine Dinucleotide (NAD+) synthesis enzyme, Nicotinamide Mononucleotide Adenylyltransferase 1 (NMNAT1) (Figure 1A), was the functional moiety of the Wlds protein responsible for axonal protection (Araki et al., 2004; Mack et al., 2001). Later, its paralog NMNAT2, was identified as a short-lived axonal enzyme that is required for axon maintenance (Gilley and Coleman, 2010). While the gain-of-function Wlds mutation suggested that damaged axons activate a self-destructive program, the existence of a distinct pathway was secured by the discovery of genes that are required for axonal degeneration after injury (Gerdts et al., 2013; Miller et al., 2009; Osterloh et al., 2012; Walker et al., 2017; Xiong et al., 2012; Yang et al., 2015). Among these pro-degenerative genes, SARM1 (Sterile Alpha and Toll/Interleukin-1 Receptor motif-containing 1) is the central executioner of the degenerative program.
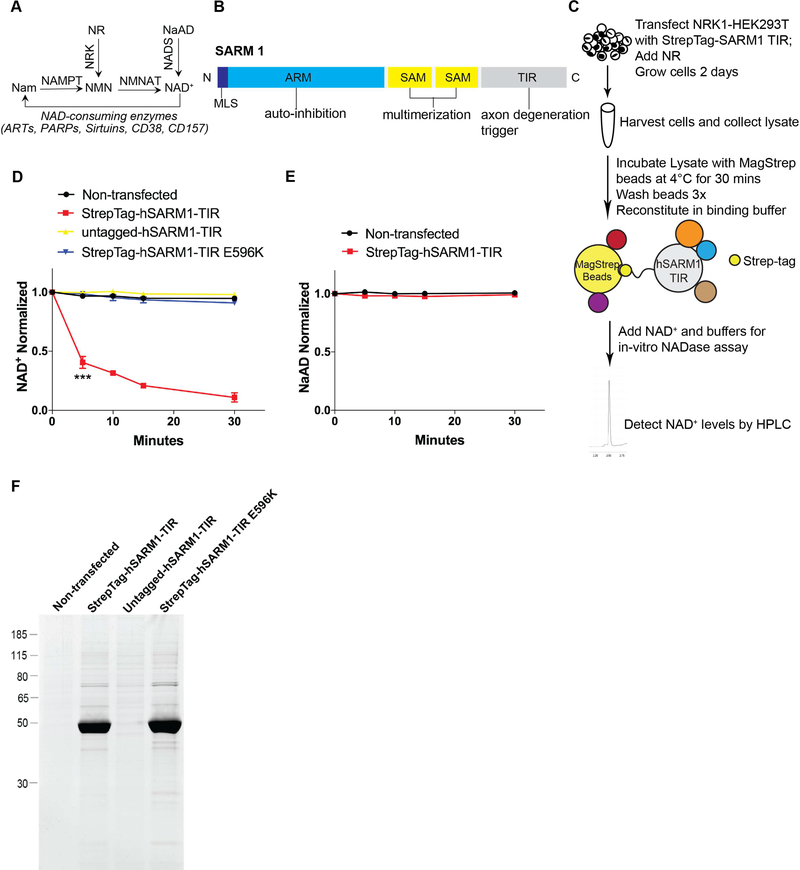
Figure 1: Native SARM1-TIR protein complex cleaves NAD+ in an in vitro assay.
A) Selected pathways of NAD+ synthesis and degradation. Nam–Nicotinamide; NMN–Nicotinamide Mononucleotide; NAD+–Nicotinamide Adenine Dinucleotide; NR–Nicotinamide Riboside; NaAD–Nicotinic Acid Adenine Dinucleotide; NAMPT–Nicotinamide Phosphoribosyltransferase; NRK–Nicotinamide Riboside Kinase; NMNAT–Nicotinamide Mononucleotide Adenylyltransferase; NADS– NAD+ synthetase; ART–ADP Ribosyltransferase; PARP–Poly ADP-Ribose Polymerase. B) SARM1 domains. MLS–Mitochondrial Localization Signal; ARM–Armadillo/HEAT Motifs; SAM–Sterile Alpha Motif; TIR–Toll/Interleukin 1 Receptor. C) Schematic illustrating the in vitro NADase assay.D) NAD+ cleavage reaction timecourse of human SARM1-TIR (wild type and mutant) laden beads in NADase assay (normalized to control at 0 min). E) NaAD reaction timecourse of human SARM1-TIR laden beads in NADase assay (normalized to control at 0 min). F) SYPRO Ruby gel of SARM1-TIR laden beads used in assay. Data for each time point was generated from three independent experiments using purified protein from three independent transfection experiments. Data are presented as mean ± SEM; Error bars: SEM; ***P<0.001 one-way ANOVA. See also Figure S1.
Loss of SARM1 blocks axonal degeneration for weeks after injury (Gerdts et al., 2013; Osterloh et al., 2012) and improves functional outcomes in mice after both traumatic brain injury (Henninger et al., 2016) and vincristine-induced peripheral neuropathy (Geisler et al., 2016). Axonal injury induces NAD+ loss (Wang et al., 2005), and SARM1 is required for this injury-induced NAD+ depletion both in vitro and in vivo (Gerdts et al., 2015; Sasaki et al., 2016). Moreover, activation of SARM1 signaling, via enforced dimerization of its TIR domain, is sufficient to induce axonal degeneration in the absence of injury due to a catastrophic depletion of axonal NAD+ (Gerdts et al., 2015).
NAD+ is a dinucleotide that is essential for many redox reactions, but it is also consumed by a variety of enzymes (e.g. PARPs, CD38, Sirtuins) where the resulting metabolites influence signaling pathways via their effects on calcium mobilization or protein parylation (Cantó et al., 2015; Fliegert et al., 2007). The identity of the NADase enzyme(s) responding to SARM1 activation and mediating NAD+ loss in injured axons has been unknown, although PARP1 and CD38 were previously eliminated as candidates (Gerdts et al., 2015; Sasaki et al., 2009). Furthermore, SARM1 is not known to have enzymatic activity, nor have TIR domains from any protein ever been associated with enzymatic activity. TIR domains are rather known for their scaffolding properties in Toll-like Receptor signaling, where they activate downstream enzymes to regulate pro-inflammatory and defense genes (O’Neill et al., 2013).
To further our understanding of this axon death pathway, we sought to identify the responsible NADase enzyme(s). Through a series of experiments, we now show that the TIR domain of SARM1 acts as an enzyme to cleave NAD+, and that SARM1 enzymatic activity is necessary to promote axonal NAD+ depletion and axon degeneration after both traumatic and vincristine induced axonal injuries. Our findings therefore identify SARM1 enzymatic activity as novel therapeutic target against diseases characterized by axonal degeneration including peripheral neuropathy, traumatic brain injury, and neurodegenerative diseases. More broadly, these findings show that TIR domains can possess intrinsic enzymatic activity.
RESULTS
SARM1-TIR complex purified from mammalian cells cleaves NAD+
SARM1 contains multiple conserved motifs including SAM domains, ARM/HEAT motifs and a Toll/Interleukin-1 Receptor (TIR) domain (Figure 1B) that mediate oligomerization and protein-protein interactions (O’Neill et al., 2013; Tewari et al., 2010; Qiao et al., 2005). Dimerization of SARM1-TIR domains in neurons is sufficient to induce axonal degeneration and to trigger rapid degradation of NAD+, demonstrating that the NADase activity is either associated with or induced by dimerized SARM1-TIR domains. TIR domains are common in signaling proteins functioning in innate immunity pathways where they serve as scaffolds for protein complexes (O’Neill et al., 2013). Therefore, we hypothesized that NAD+ depletion after SARM1 activation or enforced SARM1-TIR domain dimerization was mediated via an interaction with and activation of an NAD+ consuming enzyme like a member of the Sirtuin or Poly ADP Ribose Polymerase (PARP) families. To address this hypothesis, we reasoned that purification of the native SARM1-TIR complex in a manner that retained NADase activity would be a major step forward in identifying the responsible NAD+ consuming enzyme.
To perform these biochemical experiments, the human SARM1-TIR domain was engineered to include a tandem StrepTag II at the N-terminus and a Venus fluorescent tag at the C-terminus. This protein was expressed transiently in NRK1-HEK293T cells supplemented with Nicotinamide Riboside (NR) to augment cellular NAD+ levels and promote cell viability (Figure S1A). To maintain intact complexes, cell lysates were subsequently prepared by lysing cells under native conditions by sonication, and the recombinant SARM1-TIR protein complexes were affinity purified using MagStrep magnetic beads. We reasoned that extensive purification could disrupt interactions important for detecting NAD+ depletion, so we opted to test directly the magnetic beads loaded with complexes for activity. We incubated beads loaded with SARM1-TIR complexes with NAD+ (5 μM) for up to 30 minutes, extracted the metabolites, and measured NAD+ levels using HPLC (Figure 1C). We found that NAD+ levels dropped precipitously, within 5 minutes, when beads loaded with SARM1-TIR complexes were tested (Figure 1D). In contrast, no decrease in NAD+ was observed if beads exposed to lysates were prepared from either non-transfected NRK1-HEK293T cells or from NRK1-HEK293T cells expressing SARM1-TIR lacking the StrepTag II (Figure 1D). We also tested a non-functional TIR domain mutant [SARM1(E596K)] that we recently identified in a structure/function analysis of SARM1 (Summers et al., 2016). Magnetic beads loaded with complexes assembled on this SARM1-TIR(E596K) mutant failed to degrade NAD+ in this in vitro assay (Figure 1D). Another previously identified non-functional mutant SARM1(G601P), also failed to degrade NAD+ in the in vitro assay (Figure S1B and S1C). Finally, we examined the substrate specificity of the SARM1-TIR in vitro NADase reaction. We previously showed that Nicotinic Acid Adenine Dinucleotide (NaAD), a closely related analog of NAD+, was not cleaved after SARM1 activation (Gerdts et al., 2015). Using this in vitro assay, we found that wild type SARM1-TIR complexes do not degrade NaAD (Figure 1E). Together, these results show that the purified SARM1-TIR complex actively degrades NAD+ in a manner consistent with previous characterization of the axonal degeneration process.
We next asked if this enzymatic activity was unique to complexes associated with the SARM1-TIR domain or whether TIR domains from other proteins could also assemble complexes that exhibit NADase activity. We expressed and purified the TIR domains of TLR4, a Toll-like receptor, and MyD88, another member of the TIR adaptor family, from NRK1-HEK293T cells and tested them in the in vitro NAD+ depletion assay. Both TLR4 and MyD88 TIR containing complexes showed no NADase activity (Figure S1B and S1C). These results support the previously reported unique roles of SARM1 among TIR adaptor proteins (Gerdts et al., 2015; O’Neill et al., 2013, Summers et al., 2016) in promoting axonal degeneration and neuronal NAD+ depletion.
Multiple biochemical experiments fail to identify a bona fide NAD+ consuming enzyme in the purified SARM1-TIR complex
To identify SARM1-TIR associated proteins, we first used gel electrophoresis followed by protein staining with the highly sensitive fluorescent stain SYPRO Ruby. The beads loaded with wild type SARM1-TIR complexes isolated from NRK1-HEK293T cells revealed a prominent band corresponding to SARM1-TIR, and contained few co-purifying proteins (Figure 1F). Furthermore, all bands detected by SYPRO Ruby in wild type SARM1-TIR complex were present in similar abundance in complexes containing inactive SARM1-TIR mutant (E596K), a seemingly unlikely result if one of the bands represented an associated NAD+ consuming enzyme (Figure 1F). However, to rigorously explore this possibility, we pursued a proteomic strategy using mass spectrometry (LC-MS/MS) to identify potential NAD+ consuming proteins present in the purified SARM1-TIR complexes. To perform these studies, we further purified wild type and mutant SARM1-TIR complexes by tandem affinity purification (TAP). Wild type SARM1-TIR complexes still robustly degraded NAD+ and had very few co-purifying proteins detectable by SYPRO Ruby (Figure S2A and S2B). To identify associated proteins, we performed LC-MS/MS analysis on both wild type and mutant TAP complexes. Interestingly, we found no bona fide NAD+ consuming enzymes (e.g.,PARPs, Sirtuins, NUDIX hydrolases, ADP Ribosyl cyclases) in either complex (Table S1). Furthermore, no proteins were significantly enriched in wild type vs. mutant SARM1-TIR complexes, as would be expected if such a protein were responsible for the NADase activity (Table S1). In summary, these analyses did not identify a SARM1-TIR associated protein that is a likely source of the NADase activity.
SARM1-TIR domain possesses intrinsic NAD+ cleavage activity
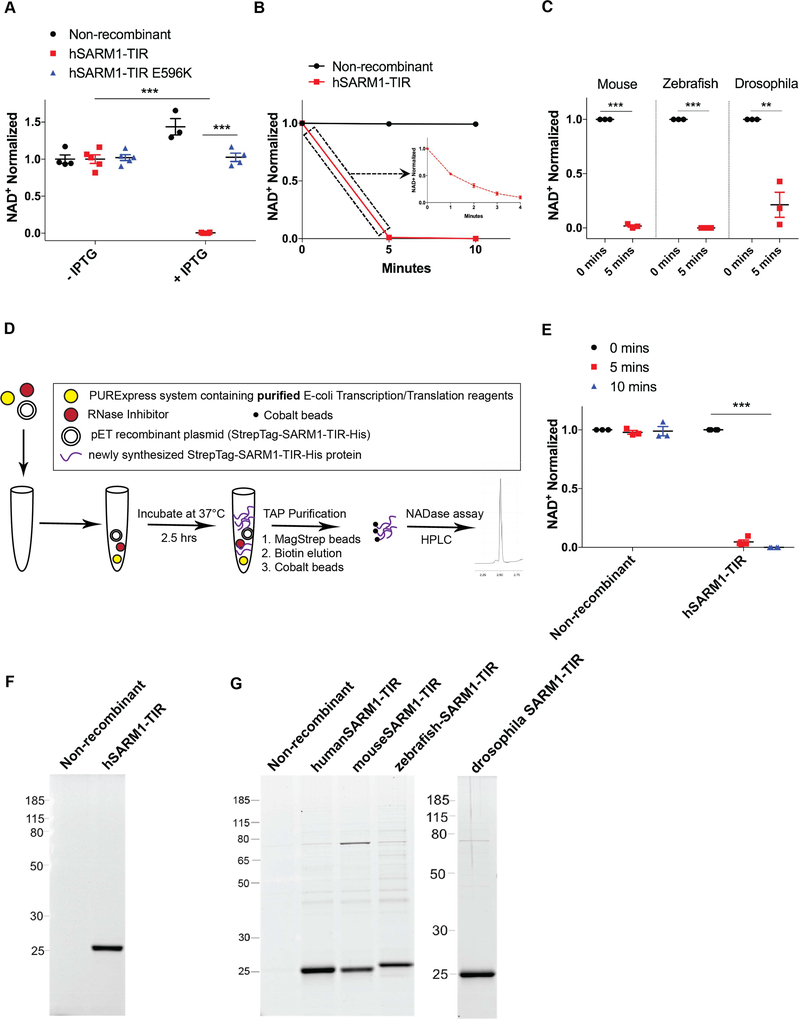
The failure to identify proteins specific to the wild type vs. mutant SARM1-TIR complexes as well as any bona fide NAD+ consuming enzymes, suggested that the SARM1-TIR domain itself might possess NADase enzymatic activity. To explore this possibility, we expressed human SARM1-TIR in E. coli so that proteins with NADase activity would not be co-purified. We first asked whether the bacteria expressing human SARM1-TIR protein had decreased levels of NAD+, as this would indicate that it was also cleaving endogenous bacterial NAD+ pools. SARM1-TIR expression in E. coli was induced by IPTG addition, endogenous metabolites were extracted, and NAD+ levels were assessed by HPLC. We found that bacteria producing wild type SARM1-TIR had remarkably low (almost undetectable) levels of endogenous NAD+ within 60 minutes after IPTG addition when compared to bacteria harboring non-recombinant vector. Further, bacteria harboring mutant SARM1-TIR (E596K) had NAD+ levels comparable to those harboring non-recombinant vector or to bacteria in which wild type SARM1 was not induced (Figure 2A). Next, we purified the bacterially expressed SARM1-TIR using TAP and tested for NADase activity. Consistent with our results using SARM1-TIR complexes isolated from mammalian cells, NAD+ was rapidly consumed by bacterially produced SARM1-TIR protein (Figure 2B). Although it is highly unlikely that human SARM1-TIR would associate with an E. coli NADase, we wished to test the intrinsic nature of the SARM1 NADase activity by stringently washing the SARM1 TIR purified complexes with either high salt or detergents to remove potential associated proteins. Using these washed SARM1 TIR beads, we found no decrease in NAD+ cleavage activity, indicating that SARM1 itself is the NADase (Figure S2Cand S2D).
Figure 2: NAD+ cleavage enzymatic activity is intrinsic to SARM1-TIR.
A) Endogenous NAD+ levels in bacteria after IPTG induction of human SARM1-TIR. B) In vitro NAD+ cleavage reaction by human SARM1-TIR protein expressed and purified from bacteria. C) Bacterially expressed mouse, zebrafish, and drosophila SARM1-TIR proteins cleave NAD+ in NADase assay. D) Schematic of cell-free protein expression system. E) Human SARM1-TIR purified from cell-free protein expression system cleaves NAD+ in NADase assay. F) SYPRO Ruby gel of SARM1-TIR laden beads purified from cell-free transcription/translation system. G) SYPRO Ruby gel of SARM1-TIR laden beads purified from bacteria. These cell-free and bacterially expressed proteins lack the Venus fluorescent tag and thus run at a different size than the proteins expressed in NRK1-HEK293T cells (compare to Figure 1). Data was generated from at least three independent reaction experiments using purified protein from at least three independent bacteria clones. Data are presented as mean ± SEM; Error bars: SEM; **P<0.01, ***P<0.001 unpaired two tailed Student’s t-test and one-way ANOVA for multiple comparisons. See also Figure S2.
Because the role of SARM1 in axonal degeneration is evolutionarily conserved (Gerdts et al., 2015; Osterloh et al., 2012), we reasoned that SARM1 NADase activity should also be conserved. We expressed and purified mouse, zebrafish, and Drosophila SARM1-TIR domains in E. coli and tested these purified proteins for their ability to cleave NAD+. Similar to the human SARM1-TIR domain, bacterially expressed mouse, zebrafish, and Drosophila SARM1-TIR domains also degrade NAD+ in vitro (Figure 2C and 2G).
To demonstrate definitively that SARM1-TIR itself possessed the enzymatic activity, we synthesized the human SARM1-TIR in a cell-free protein expression system that utilizes purified E-coli components for transcription and translation. None of the purified E-coli transcription/translation components are known NADases (Shimizu et al., 2001), and we experimentally confirmed that these purified components do not exhibit NADase activity (Figure S2E). To test if SARM1-TIR purified from this in vitro translation system could cleave NAD+, we first incubated human SARM1-TIR plasmid DNA with the purified transcription and translation reagents and RNase inhibitor for 2.5 hours at 37°C. Next, we purified the newly synthesized protein from the reaction by TAP, and tested for NADase activity in our assay (Figure 2D). The purified SARM1-TIR from this cell-free protein translation system rapidly cleaved NAD+, consistent with our prior findings with SARM1-TIR purified from both mammalian cells and bacteria (Figure 2E and 2F).
In summary, we find that the SARM1-TIR domain depletes endogenous NAD+ in bacteria, that bacterially synthesized SARM1-TIR from multiple species cleaves NAD+ in vitro, and that SARM1-TIR synthesized and purified from a cell-free protein translation system cleaves NAD+ in vitro. These results demonstrate that the SARM1-TIR domain has intrinsic NADase activity, and, hence, SARM1 itself is responsible for the NAD+ depletion observed after axon injury. Moreover, these findings reveal for the first time that a TIR domain, an evolutionary ancient domain previously demonstrated to function as a protein interaction domain, can also harbor enzymatic activity.
Characterization of the SARM1-TIR enzymatic reaction reveals both cyclase and glycohydrolase activities
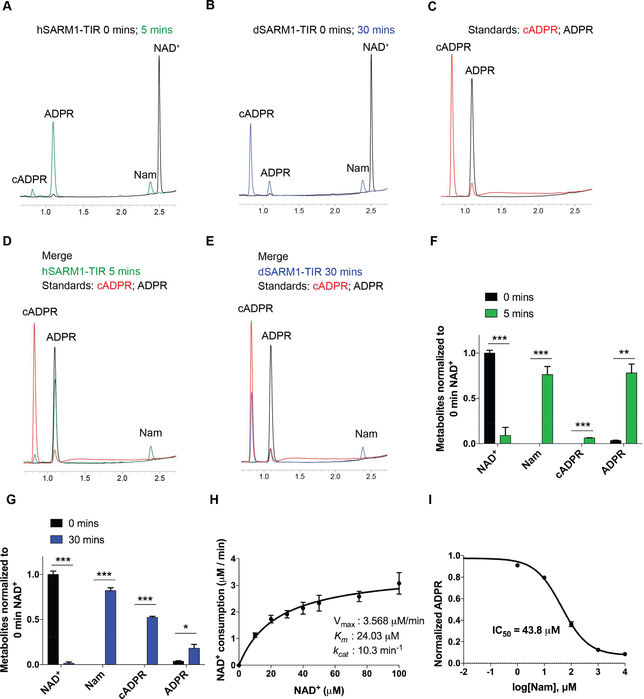
To further characterize the SARM1-TIR NADase activity, we sought to identify the NAD+ cleavage products of this enzymatic reaction as well as establish reaction parameters. Our previous study using cultured cells and C14-labelled NAD+ identified Nicotinamide (Nam) as a reaction product (Gerdts et al., 2015). However, we were unable to detect other products due to the position of the label on the NAD+ molecule. We therefore performed HPLC and LC-MS/MS analysis of the metabolites produced by human SARM1-TIR, and identified Nam and ADP Ribose (ADPR) as major products, and cyclic ADPR (cADPR) as a minor product (Figure 3A-G and Figure S3A-C). Interestingly, while the mouse and zebrafish orthologs generated a similar ratio of reaction products as the human enzyme (Figure S3D), the Drosophila SARM1-TIR purified either from bacteria or NRK1–293T cells generated more cADPR than ADPR (Figure 3A-G). This finding is similar to results with the ADP-Ribosyl cyclase family of NADases (Liu et al., 2009), in which the mammalian ADP Ribosyl Cyclase CD38 cleaves NAD+ to generate ADPR as the major product, with minor amounts of cADPR; while the ADP Ribosyl Cyclase isolated from the sea mollusk Aplysia californica cleaves NAD+ into cADPR (Liu et al., 2009). This difference in reaction products between the Drosophila and vertebrate SARM1-TIR NADase may provide insights into the divergent enzymatic activities of the ADP Ribosyl cyclase family of enzymes.
Figure 3: Characterization of the SARM1-TIR NAD+ cleavage reaction.
(A-E) HPLC chromatograms showing NAD+ cleavage products of human and drosophila SARM1-TIR. Retention time: Nam t~2.40 min; cADPR at t~0.85 min; ADPR at t~1.10 min. (F-G) Quantification of metabolites generated by human (F) and drosophila (G) SARM1-TIR as displayed in A-E (normalized to 0 min NAD+). H) Kinetic parameters for human SARM1-TIR cleavage reaction. Vmax, Km, kcat were determined by fitting the data to the Michaelis-Menten equation and are presented as mean ± SEM for three independent biological samples. I) Nam dose response inhibition of human SARM1-TIR enzymatic activity. Data was generated from three independent reaction experiments using purified protein from three independent bacteria clones. Data are presented as mean ± SEM; Error bars: SEM; *P<0.05; **P<0.01; ***P<0.001 unpaired two tailed Student’s t-test. See also Figure S3.
Next, we performed kinetic assays of the SARM1-TIR enzyme, which revealed saturation kinetics (Figure 3H), a distinguishing feature of enzyme catalysts, with an estimated Michaelis constant (Km) of 24 μM, maximum velocity (Vmax) of 3.6 μM/min, and turnover number (kcat) of 10.3 min−1 (Figure 3H). Although the estimated kcat is lower than the reported values for other ADP-Ribosyl cyclases and NAD+ glycohydrolases (Ghosh et al., 2010), the estimated Km values are similar (Cantó et al., 2015). In order to obtain information that could be helpful in developing SARM1 inhibitors, we tested whether either of the reaction products could inhibit the enzymatic activity of SARM1-TIR. ADPR did not inhibit SARM1-TIR NADase activity (Figure S3E), however Nam inhibits the enzymatic activity with an IC50 of 43.8 μM, which is about 9-fold higher than the starting reaction NAD+ concentration (Figure 3I). Interestingly, Nam also inhibits other NAD+ consuming enzymes such as Sirtuins and PARPs (Avalos et al., 2005; Gibson et al., 2012). Inhibitors of the SARM1-TIR domain modeled after nicotinamide could be useful in preventing the early stages of axonal degeneration (Gerdts et al., 2016; Wang et al., 2005). Ara-2’-F-NAD+, a slow binding inhibitor of CD38 (Berthelier et al., 1998), however did not inhibit SARM1-TIR NADase even at high doses (Figure S3F), so there is likely to be selectivity of inhibitors for different families of NADases.
Glutamic Acid 642 is a putative catalytic residue in the active site of the SARM1-TIR enzyme
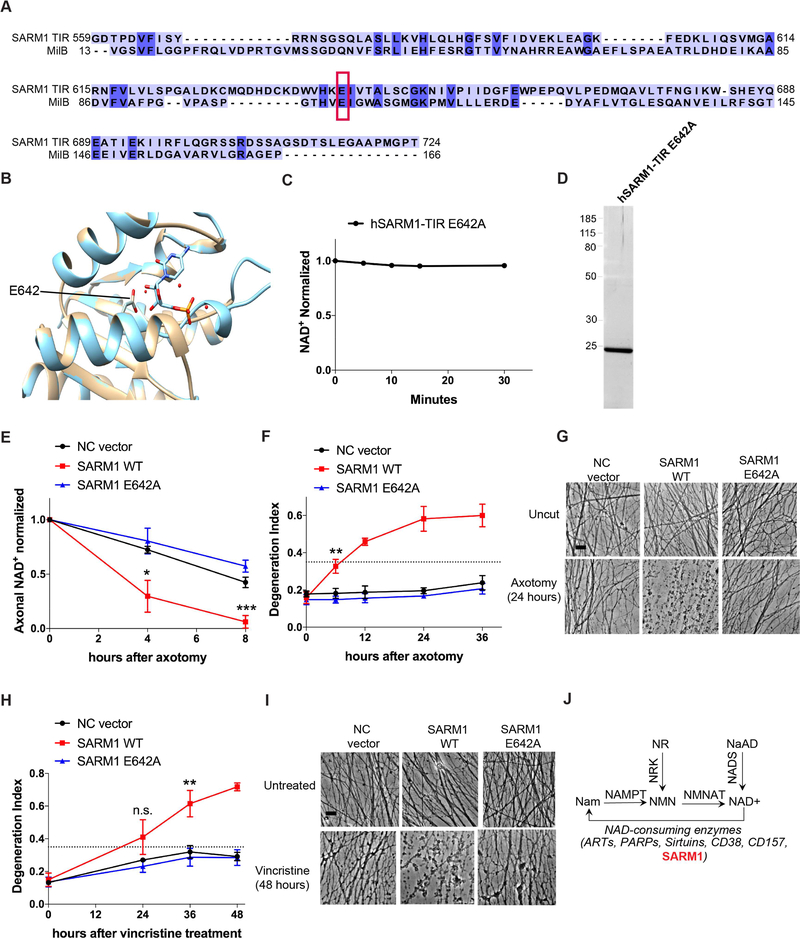
Next, we sought to gain some insight into the structural features enabling SARM1-TIR NAD+ cleavage. Since there is no reported crystal structure of the SARM1-TIR domain, we used an unbiased template-based prediction (Söding et al., 2005) to identify protein homologs of SARM1-TIR. Interestingly, a recent bioinformatics study showed that some TIR domains share strong structural similarity to nucleotide/nucleoside hydrolases (Burroughs et al., 2015). From our domain prediction analysis using SARM1-TIR, we identified other TIR domains as expected. However, in addition to these TIR domains, we also detected a number of nucleotide hydrolase/transferase enzymes (Table S2). For some of these enzymes, residues that contribute to catalytic activity have been established (Sikowitz et al., 2013; Armstrong et al., 1995). We therefore used structural modeling and sequence alignments to identify putative residues in the SARM1-TIR domain that might contribute to enzymatic activity (Figure 4A, 4B, S4A, S4B). The SARM1-TIR domain was modeled using the crystal structure of two enzymes identified from our prediction: MilB Cytidine 5’ monophosphate (CMP) Hydrolase (PDB: 4JEM) (Figure 4B) and Nucleoside 2-deoxyribosyltransferase (PDB: 1F8Y) (Figure S4B). Importantly, we found that a glutamic acid E642 in the SARM1-TIR domain aligned with both the key catalytic glutamic acid residue in CMP hydrolase (Sikowitz et al., 2013) and the proposed nucleophilic glutamic acid in the active site of nucleoside 2-deoxyribosyltransferase (Armstrong et al., 1995) (Figure 4A, 4B and S4C). Moreover, glutamic acid residues are also known catalytic residues in other NADases (Ghosh et al., 2010). To test if SARM1 TIR E642 had similar catalytic properties, we mutated this residue to an Alanine (E642A) in SARM1-TIR, purified the protein from the cell-free protein translation system, and tested it for NAD+ cleavage activity. We found that purified SARM1-TIR E642A failed to cleave NAD+ in the NADase assay (Figure 4C and 4D). Hence, we suggest that E642 in the SARM1-TIR domain is a key catalytic residue within the active site that is responsible for NAD+ cleavage.
Figure 4: SARM1 enzymatic activity functions in axons to promote pathological axonal degeneration.
A) Amino acid sequence alignment of SARM1-TIR with MilB Cytidine 5’ Monophosphate (CMP) Hydrolase. CMP catalytic glutamic acid is highlighted in red box and aligns to glutamic acid 642 in the SARM1-TIR domain. B) Modeling of the SARM1-TIR domain on the crystal structure of CMP Hydrolase bound to CMP. E642 aligns with a catalytic residue of CMP Hydrolase. C) NAD+ reaction timecourse of human SARM1-TIR E642A purified from cell-free protein translation system (normalized to control at 0 min). D) SYPRO Ruby gel of SARM1-TIR E642A purified from cell-free protein translation system. E) Axonal NAD+ levels after axotomy (normalized to control at 0 hr). NC vector, SARM1 WT, and SARM1 E642A constructs were expressed in SARM1−/− DRG neurons, and levels of NAD+ were obtained at indicated timepoints after axotomy. F) Axonal degeneration timecourse after axotomy, quantified as degeneration index (DI) where a DI of 0.35 (indicated by dotted line) or above represents degenerated axons. G) Bright-field micrographs of axons expressing indicated constructs represented in F. H) Axonal degeneration timecourse after vincristine treatment, quantified as DI. I) Bright-field micrographs of axons after vincristine treatment corresponding to selected groups in H. Scale bar, 5μm. Quantification data were generated from at least three independent biological experiments. Data are presented as mean ± SEM; Error bars: SEM.*P<0.05, **P<0.01, ***P<0.001 one-way ANOVA. J) Selected pathways of NAD+ synthesis and degradation including SARM1 as a NAD+ consuming enzyme. See also Figure S4.
SARM1 enzymatic activity functions in axons to promote pathological axonal degeneration
Having demonstrated that the SARM1 TIR domain is an enzyme, and now identified its putative catalytic residue, we next investigated whether this enzymatic activity and, in particular, the identified glutamate, were required for the pro-degenerative functions of full-length SARM1 in neurons. In wild type neurons, axotomy triggers rapid depletion of axonal NAD+ and axonal degeneration, while in SARM1-deficient neurons axonal degeneration is blocked and NAD+ levels remain significantly higher than in injured wild type axons (Gerdts et al., 2015). First, we tested whether SARM1 NADase activity is necessary for injury-induced axonal NAD+ depletion and subsequent axonal degeneration. We expressed either wild type (enzymatically active) full-length SARM1 or mutant (enzymatically disabled) SARM1(E642A) in cultured SARM1-deficient DRG neurons and found both were well expressed in axons (Figure S4D and S4E). Following axotomy, we measured axonal NAD+ levels and axonal degeneration. Expression of enzymatically active, wild type SARM1 in SARM1-deficient DRG neurons promotes both axonal NAD+ depletion and axonal degeneration after axotomy. In contrast to wild type SARM1, when the enzymatically disabled SARM1(E642A) mutant is expressed in these neurons, axotomy did not induce axonal degeneration or rapid NAD+ depletion (Figure 4E-G). We also tested the requirement for SARM1 enzyme activity in another injury model – vincristine-induced neurotoxicity. Cultured SARM1-deficient DRG axons are protected from vincristine-induced axonal degeneration (Gerdts et al., 2013). Moreover, we recently demonstrated in vivo that SARM1 is required in mice for the development of vincristine-induced peripheral neuropathy (Geisler et al., 2016). As with axotomy, we expressed either wild type (enzymatically active) full-length SARM1 or mutant (enzymatically disabled) SARM1(E642A) in cultured SARM1-deficient DRG neurons. Enzymatically active SARM1 mediates axon loss in response to the chemotherapeutic vincristine, while enzymatically disabled SARM1 does not promote axon loss following vincristine administration (Figure 4H, and 4I). Altogether, these findings demonstrate that the intrinsic NADase activity of SARM1 (Figure 4J) is necessary to promote axonal degeneration after both traumatic and neurotoxic injuries, and suggest that inhibitors of the SARM1 NADase could block pathological axonal degeneration.
DISCUSSION
Here we demonstrate that the TIR domain of SARM1 acts as an enzyme to cleave NAD+, and that this enzymatic activity is necessary to promote pathological axonal degeneration. These findings have a number of implications. First, our results describe the first enzymatic activity intrinsic to a TIR domain. Many proteins central to innate immune signaling contain TIR domains (O’Neill et al., 2013), and this finding therefore raises the possibility that these other TIR domains could also be enzymatically active. Second, the SARM1-TIR domain is the closest mammalian relative to the ancestral TIR domains in prokaryotes (Zhang et al., 2011), suggesting that this domain, as an enzyme cleaving NAD+, may be a component of an ancient cell death pathway. Third, the generation of cADPR and ADPR by SARM1-TIR may contribute to axonal degeneration. cADPR and ADPR are signals for intracellular calcium mobilization (Fliegert et al., 2007), and calcium is a major stimulus for axonal degeneration (Villegas et al., 2014). However, whether this is a primary mechanism for axonal demise or whether energetic failure secondary to NAD+/ATP depletion (Gerdts et al., 2015; Yang et al., 2015) and/or NAD+/NADH electron donor activities is most critical is unclear. Fourth, detailed structural studies with SARM1-TIR protein crystallized in complex with its substrate NAD+ and/or with analogs could provide useful information about residues within or outside of the active site that are important for enzymatic catalysis. Finally, NADase activity is integral to the conserved axon death program (Gerdts et al., 2015), and so the discovery that SARM1 is the axonal NADase now provides an identified target for the rational design of inhibitors that will serve as novel therapeutic candidates for the treatment of disorders characterized by axonal degeneration including peripheral neuropathies, traumatic brain injuries, and neurodegenerative diseases.
STAR METHODS
KEY RESOURCE TABLE
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jeffrey Milbrandt (jmilbrandt@wustl.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mammalian Cell Culture
HEK293T and NRK1-HEK293T cells were maintained in 10% FBS in DMEM, supplemented with penicillin/streptomycin and glutamine, and passaged by suspending in 0.05% trypsin. NRK1-HEK293T is a polyclonal cell line we developed that stably expresses Nicotinamide Riboside Kinase 1 (NRK1) so that supplementation with Nicotinamide Riboside (NR), an NAD+ biosynthetic precursor, during protein expression would significantly augment cellular NAD+ levels and maintain cell viability adequate for protein purification.
Bacterial Cultures for protein purification
The appropriate dual tag (StrepTag and HisTag) SARM1-TIR was cloned into a pET30a+ plasmid. These constructs as well as non-recombinant pET30a+ were transformed into Shuffle T7 Express Competent E-coli (New England BioLabs). Single colonies were grown overnight and the next day, cultures were diluted in LB media, grown at 30°C until they reached A600 = 0.4–0.8, when IPTG (0.5 mM final concentration) was added. The bacteria were grown for an additional 4 h, pelleted by centrifugation, washed with PBS and stored at −80° C for future protein purification.
Cell-free protein transcription/translation
In vitro cell-free protein transcription/translation was performed using the PURExpress In Vitro Protein Synthesis Kit (New England BioLabs Catalog # E6800S). For a total reaction volume of about 25μL, the reaction was assembled in the following order: 10μL of Solution A, 7.5μL of Solution B, 3μL of RNase inhibitior, water, and ~500ng of pET30a+ non-recombinant/recombinant DNA. Reaction was incubated at 37°C for 2.5 hours and subsequently placed on ice to stop the reaction. Tandem Affinity purification of proteins was subsequently performed as described in Method Details.
Mouse embryonic dorsal root ganglion (DRG) neuronal culture
DRG neurons were isolated from SARM1−/− E13.5 mouse embryos as previously described (Gerdts et al., 2015) and seeded on plates pre-coated with poly-D-Lysine (Sigma-Aldrich) and laminin (Life Technologies). DRG neurons were maintained in neurobasal medium supplemented with L-glutamine, 2% B27 (Gibco), 50ng/mL nerve growth factor (Envigo Bioproducts), and 1μM 5-fluoro-2’deoxyuridine plus 1μM uridine (Sigma-Aldrich). On DIV 1, neurons were transduced with lentiviral particles generated from HEK293T cells as previously described (Sasaki et al., 2009) expressing Venus alone or the indicated SARM1 construct fused to Venus at the C-terminus. Axons from SARM1−/−
DRGs expressing the indicated construct were severed with a razor blade or treated with 40 nM vincristine on DIV 7. SARM1−/− mice (C57/BL6) were housed (12 hr dark/light cycle and less than 5 mice per cage) and used under the direction of institutional animal study guidelines at Washington University in St. Louis.
METHOD DETAILS
Protein Expression and purification from NRK1-HEK293T stable line.
Approximately 10 million NRK1-HEK293T cells were plated and transfected the next day with 15 μg of StrepTag SARM1-TIR construct DNA using X-tremeGENE 9 reagent (Sigma). Nicotinamide Riboside (NR) was added at a final concentration of 1 mM to improve cell viability. After 2 days the cells were harvested and lysed by sonication in binding buffer (50 mM Sodium Phosphate buffer pH 8, 300 mM Sodium Chloride, 0.01% Tween-20) with EDTA-free protease inhibitors. For single step affinity purification, the whole cell lysates were incubated with 20 μL MagStrep (Strep-Tactin) type 3 XT beads suspension (IBA Lifesciences) for 30 min. The beads were then washed three times with binding buffer and resuspended in 100 μL of binding buffer for enzymatic assays and other downstream applications.
Tandem Affinity purification (TAP) from NRK1-HEK293T stable line.
Dual tagged (Strep-tag and His tag SARM1-TIR) proteins were first purified by Strep Tag affinity methods as described above. For tandem affinity purification, the proteins were then eluted from MagStrep type 3 XT beads with 22.5 mM biotin (Sigma, B4501) for 25 min. Supernatant containing the eluted protein was separated from MagStrep beads, and then incubated with 10 μL Co2+ Dynabead suspension (ThermoFisher) for 30 min to bind SARM1-TIR proteins via the His tag. The beads were then washed at least two times with binding buffer and resuspended in 100 μL of binding buffer for downstream applications.
Bacterial protein expression and Tandem Affinity Purification (TAP).
See ‘Experimental Model and Subject Details’ section for expression of proteins in bacteria. For protein purification, the frozen bacterial pellet was thawed on ice, resuspended in binding buffer (without protease inhibitors) and incubated with 100 μg/mL lysozyme for 15 min on ice. EDTA-free protease inhibitor cocktail was then added and the cells were lysed by sonication. Tandem affinity purification was carried out as described above. In instances where high salt (1M, 2M NaCl in binding buffer) or detergent (0.5% Triton X-100, 0.5% Tween-20 in binding buffer) washes were employed, protein laden beads were washed (x3) with these solutions after initial binding to MagStrep beads. Biotin elution and His Tag purification were subsequently performed as described above.
NADase assay and metabolite extraction.
Ten microliters of beads incubated with the indicated cell lysate was incubated with 5 μM NAD+ and reaction buffer (92.4 mM NaCl and 0.64X PBS), for a total reaction volume of 50 μL. Reactions were carried out at room temperature (25° C) for the indicated amount of time, and stopped by addition of 1M of perchloric acid (HClO4) and placing the tube on ice. NAD+ metabolites were extracted using HClO4/K2CO3 method and quantified by HPLC (see HPLC for metabolite measurement). For LC-MS/MS analysis, the extraction was performed using 50% Methanol in distilled water, and chloroform (see LC-MS/MS metabolite measurement for further details).
HPLC metabolite measurement.
Metabolites were isolated from enzyme reaction mixture by extracting with 1M HClO4, placing on ice for 10 min, neutralized with 3M K2CO3, placing on ice for 10 min, then followed by separation by centrifugation. The supernatant (90 μL) containing the extracted metabolites was mixed with 0.5M Potassium Phosphate buffer (10 μL) and metabolites were analyzed by HPLC (Nexera X2) with Kinetex (100 × 3 mm, 2.6 μm; Phenomenex) column. Internal standards for NAD+, Nicotinamide (Nam), Nicotinic Acid Adenine Dinucleotide (NaAD), ADP Ribose (ADPR), cyclic ADPR were used to generate standard curves for quantification of the respective compounds. The levels for each compound in each experimental sample were normalized to the 0 min time point that was analyzed concurrently.
LC-MS/MS metabolite measurement.
Samples were prepared by mixing the reactions with 50% methanol in distilled water. The samples were placed on ice, centrifuged, soluble metabolites in the supernatant were extracted with chloroform, and the aqueous phase was lyophilized and stored at −20° C until LC-MS/MS analysis. For LC-MS/MS, the metabolite samples were reconstituted with 5 mM ammonium formate, centrifuged 12,000 × g for 10 min, and the cleared supernatant was applied to the LC-MS/MS for metabolite identification and quantification. Liquid chromatography was performed by HPLC system (1290; Agilent) with Synergi Fusion-RP (4.6 × 150mm, 4 μm; Phenomenex) column. Samples (10 μl) were injected at a flow rate of 0.55 ml/min with 5 mM ammonium formate for mobile phase A and 100% methanol for mobile phase B and metabolites were eluted with gradients of 0–7 min, 0–70% B; 7–8 min, 70% B; 9–12 min, 0% B. The metabolites were detected with Triple Quad mass spectrometer (6460 MassHunter; Agilent) under positive ESI multiple reaction monitoring (MRM). Metabolites were quantified by MassHunter quantitative analysis tool (Agilent) with standard curves. Standard curves for each compound were generated by analyzing NAD+, cADPR, ADPR, and Nam reconstituted in 5 mM ammonium formate. The levels for each compound in each experimental sample were normalized to the 0 min time point that was analyzed concurrently. Sample identity was blinded to individual performing the experiment.
Endogenous bacterial and mammalian cell NAD+ quantification.
Overnight cultures of E. coli harboring a SARM1-TIR construct were diluted and grown at 30° C until they reached A600= 0.4–0.8. IPTG (0.1 mM final concentration) was added to induce protein expression and the cultures were harvested 60 min later. The cultures were normalized to A600= 0.5 ± 0.05 and the pellet from 500 μl of culture suspension was lysed by adding 0.5M HClO4. NAD+ metabolites were extracted using HClO4/K2CO3 method and measured by HPLC. Two hundred thousand NRK1-HEK293T cells grown in presence of NR were transfected with 1 μg SARM1-TIR expression construct. After two days, the NAD+ metabolites were extracted with 0.5M HClO4 and 3M K2CO3 and measured by HPLC.
SYPRO Ruby Gel Staining.
Purified bead-SARM1-TIR protein complexes were boiled in Laemmli buffer for 10 min and separated on a 10% Bis-Tris Plus gel. After electrophoresis, the gel was fixed in 50% Methanol/7% acetic acid for 30 min x 2, then incubated overnight in SYPRO Ruby Protein Gel stain (Thermo Fisher). The next day, the gel was washed with 10% methanol/7% acetic acid solution for 30 min, rinsed in distilled water for 5 minutes x 2, and stained proteins were visualized with a UV transilluminator.
Preparation of peptides for LC-MS.
Purified TAP complexes were eluted by boiling the cobalt magnetic beads for 15 min in Tris-HCl buffer (pH 7.6, 100 mM) (40 μL) containing 4% SDS and dithiothreitol (100 mM). The beads were spun at 16,000 × g for 5 min and the eluted proteins were mixed with 300 μL of Tris-HCl buffer (pH 8.5, 100 mM) containing 8M urea. The SDS was removed using a filter-aided-sample-preparation (FASP) method (Wisniewski et al., 2009). After buffer exchange, 100 μL of buffer (ammonium bicarbonate, pH 7.8, 50 mM) was pipetted into the Microcon® filtration unit (YM-30) and trypsin was added (1 μg in 1 μL). The digest was incubated for 4h at 37°C and then overnight in a humid chamber after the addition of another aliquot of trypsin. The digest was acidified (5 μL of neat formic acid) and the peptides were recovered by centrifugation to the lower chamber. The acidified peptides were treated with ethyl acetate as previously described (Erde et al., 2014). The peptides were desalted by solid phase extraction on a Beckman BioMek NxP robot with C4 and porous graphite carbon Nutips (Glygen) (Chen et al., 2012). The peptides that eluted with acetonitrile (60% in 1% formic acid) were combined, dried in a vacuum centrifuge, dissolved in acetonitrile/formic acid (1%/0.1%) (16 μL). An aliquot (2 μL) was taken for analysis using a fluorescent assay (ThermoFisher Scientific) and the remainder was pipetted into autosampler vials (SUN-SRi), concentrated by vacuum centrifugation and dissolved in aqueous TFA (0.1%) (0.6/μg) for LC-MS analysis (see below).
Protein Identification by LC-MS Analysis.
LC-ESI/MS/MS analysis was performed using a Q-Exactive™ Plus Hybrid Quadrupole-Orbitrap™ Plus mass spectrometer (ThermoFisher Scientific) coupled to an EASY-nanoLC 1000 system (ThermoFisher Scientific). The samples were loaded (2 μL) onto a 75 μm i.d. × 25 cm Acclaim® PepMap 100 RP column (Thermo-Fisher Scientific). The peptides were eluted at a flow rate of 300 nL/min with an acetonitrile gradient in aqueous formic acid (1%) as mobile phase A. After isocratic elution with A for 5 min the acetonitrile proportion was increased linearly to 30% with sovent B (100% ACN, 0.1% FA) over 180 min, followed by sequential increases in B to 45% in 25 min, 95% B over 5 min and an isocratic wash at 90% B for 7 min. Full-scan mass spectra were acquired by the Orbitrap™ mass analyzer in the mass-to-charge ratio (m/z) of 375 to 1400 and with a mass resolving power set to 70,000. Fifteen data-dependent high-energy collisional dissociations (HCD) were performed with a mass resolving power set to 35,000, a fixed first m/z 100, an isolation width of 0.7 m/z, and the normalized collision energy (NCE) setting of 32. The maximum injection time was 50 ms for parent-ion analysis and 105 ms for product-ion analysis. Target ions already selected for MS/MS were dynamically excluded for 30 sec. An automatic gain control (AGC) target value of 3e6 ions was used for full MS scans and 1e5 ions for MS/MS scans. Peptide ions with charge states of one or greater than six were excluded from MS/MS acquisition. The tandem mass spectra were processed using Matrix Science Distiller version 2.5 without charge state deconvolution and deisotoping. The processed files were used for protein database searches using Mascot (Matrix Science, London, UK; version 2.5.1). The database was UniProt Human Reference database (downloaded May 3, 2014, 69021 entries). A parent ion tolerance and MS2 fragment tolerance were set to 10 ppm and 0.05 Da, respectively. Carbamidomethyl of cysteine was specified as a fixed modification and oxidation of methionine was set as a variable modification. Protein identifications were performed using Scaffold, version_4.4.8 (Proteome Software Inc., Portland, OR) implementing the Protein and Peptide Prophet algorithms (Keller et al., 2002; Nesvizhskii et al., 2003). Peptide identifications were accepted with > 90.0% probability. Protein identifications were accepted if they could be established at greater than 95.0% probability and contained at least 2 peptides with unique sequences (a low cut-off to allow for the identification of any low abundant NAD+ consuming protein). Protein probabilities were assigned by the Protein Prophet algorithm. Proteins that contained similar peptides and could not be differentiated based on identification of unique peptide sequences were grouped to satisfy the principles of parsimony. Relative protein quantities were estimated using a normalized total spectrum count generated from two independent transfection experiment (Table S1). Spectral count data from TAP wild type and mutant complexes were each normalized to spectra count of wild type SARM1-TIR lacking Strep Tag II, to account for background non-specific binding. Peptide counts that had a negative value after subtraction of control untagged SARM1-TIR from tagged wild type or mutant SARM1, were assigned a value of 0.
Enzyme kinetics studies.
Vmax, Km, kcat were determined from the reaction velocity of NAD+ consumption in the first 60 seconds of reaction for increasing substrate (NAD+) concentration, and fitting the data to the Michaelis-Menten equation using nonlinear curve fit in GraphPad Prism 7. kcat was calculated per dimer of purified hSARM1-TIR. Data are presented as Mean ± SEM from three independents biological samples and reaction measurements. Enzyme concentration was determined via densitometry analysis on SYPRO Ruby gel of purified protein, with carbonic anhydrase used as a standard.
Enzyme inhibition studies.
Purified bacterial hSARM1-TIR was tested in the NADase assay with the addition of 1 mM Nam or 1mM ADPR in the reaction mixture. For dose-response inhibition experiments, varying concentrations of Nam or ara-2’-F-NAD+ were added to the reaction mixture. The reaction was stopped after 5 min and metabolites were extracted by the perchloric acid method and measured by HPLC as indicated above.
Axonal NAD+ measurement.
SARM1−/− DRGs were transduced with lentivirus as described above. Cells were supplemented with fresh media every 2 days. On DIV 7, axons were severed with a razor blade. At the indicated timepoint, cell bodies were removed then axonal NAD+ was extracted using perchloric acid/sodium carbonate method and separated with high performance liquid chromatography as previously described (Sasaki et al., 2009).
Modeling SARM1-TIR domain.
The human SARM1 TIR domain (aa559–724) was analyzed for structural homologs in the protein data bank (PDB) using HHpred (Söding et al., 2005) and PHYRE2 (Kelley et al 2015). Protein sequence alignments were generated by HHpred and formatted with JalView. Hits with an E-value greater than 0.1 and score below 40 have a reduced probability of accurate prediction and were excluded. PHYRE2 and SWISS-MODEL (Arnold et al., 2006) were used to generate 3D structural models of the SARM1 TIR domain using MilB CMP-glycosidase as a template (PDB: 4JEM) or nucleoside 2-deoxyribsoyltransrferase (PDB: 1F8Y). These structures were visualized and superimposed with Chimera (www.rbvi.ucsf.edu/chimera)
QUANTIFICATION AND STATISTICAL ANALYSIS
Number of n is indicated in each figure legend or appropriate method section. One-way analysis of variance (ANOVA) comparisons were performed for multiple groups and unpaired two-tailed t-tests were used for individual comparisons with an assumption of equal variance between groups. All error bars represent SEM. For quantification of Venus expression, DRGs were fixed in paraformaldyhyde and Venus fluorescence visualized by microscopy from multiple fields of axons for each experiment. DRGs were co-stained for beta tubulin (Mouse anti-beta3 tubulin (TUJ1); from Biolegend) to assess total axon area for each field. Axon degeneration was quantified in distal axons from brightfield images using an ImageJ macro (Sasaki et al., 2009) that measures the ratio of fragmented axon area to total axon area. For an individual experiment, six fields were analyzed from 2–3 wells per condition. Other data analyses were done with Graph Pad Prism 7, Image J macro, Microsoft Excel, Adobe Illustrator and Photoshop.
DATA AND SOFTWARE AVAILABILITY
Recombinant DNA sequences have been deposited in BankIt with Accession numbers: KY584388-KY584401. Table S1 contains the list of identified proteins from LC-MS/MS analysis of SARM1-TIR complex purified from mammalian cells.
Supplementary Material
HIGHLIGHTS.
SARM1-TIR cleaves NAD+ into Nam, ADPR, and cADPR
SARM1 NADase activity is necessary for pathological axon loss
SARM1 is the first member of a new class of NADase enzyme
TIR-domains can possess enzymatic activity
ACKNOWLEDGEMENTS:
This work was supported by the National Institutes of Health (Grant RO1NS065053 (A.D.), RO1AG013730 (J.M.), RO1NS087632 (J.M. and A.D.). K.E. is a Howard Hughes Medical Institute Medical Research Fellow. D.W.S. is supported by a Development Grant from the Muscular Dystrophy Association. The protein identifications and LC-MS analyses were generated at the Washington University Proteomics Shared Resource (WU-PSR). WU-PSR is supported by the WU Institute of Clinical and Translational Sciences (NCATS UL1 TR000448), the WU Mass Spectrometry Research Resource (NIGMS P41 GM103422) and the Siteman Comprehensive Cancer Center (NCI P30 CA091842). Washington University, J.M. and Y.S. may derive income from licensing of technology to ChromaDex. We thank T. Fahrner, and K. Simburger for technical assistance, and members of the Milbrandt and DiAntonio lab for fruitful discussions.
Footnotes
SUPPLEMENTAL INFORMATION: Supplemental Information includes four figures, and two tables (including Table S1 Excel table).
ACCESSION NUMBERS: The accession numbers for the recombinant DNA sequences used are BankIt: KY584388-KY584401.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES:
- Araki T, Sasaki Y, and Milbrandt J (2004). Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science 305, 1010–1013. [DOI] [PubMed] [Google Scholar]
- Armstrong SR, Cook WJ, Short SA, and Ealick SE (1996). Crystal structures of nucleoside 2-deoxyribosyltransferase in native and ligand-bound forms reveal architecture of the active site. Structure 4, 97–107. [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, and Schwede T (2006). The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201. [DOI] [PubMed] [Google Scholar]
- Avalos JL, Bever KM, and Wolberger C (2005). Mechanism of sirtuin inhibition by nicotinamide: altering the NAD(+) cosubstrate specificity of a Sir2 enzyme. Mol. Cell 17, 855–868. [DOI] [PubMed] [Google Scholar]
- Berthelier V, Tixier JM, Muller-Steffner H, Schuber F, and Deterre P (1998). Human CD38 is an authentic NAD(P)+ glycohydrolase. Biochem. J 330 ( Pt 3), 1383–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Zhang D, Schaffer DE, Iyer LM, and Aravind L (2015). Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling. Nucleic Acids Res. 43, 10633–10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C, Menzies KJ, and Auwerx J (2015). NAD(+) Metabolism and the Control of Energy Homeostasis: A Balancing Act between Mitochondria and the Nucleus. Cell Metab. 22, 31–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZW, Fuchs K, Sieghart W, Townsend RR, and Evers AS (2012). Deep amino acid sequencing of native brain GABAA receptors using high-resolution mass spectrometry. Mol. Cell. Proteomics 11, M111.011445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erde J, Loo RR, and Loo JA (2014). Enhanced FASP (eFASP) to increase proteome coverage and sample recovery for quantitative proteomic experiments. J. Proteome Res 13, 1885–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer LR, and Glass JD (2007). Axonal degeneration in motor neuron disease. Neurodegener. Dis 4, 431–442. [DOI] [PubMed] [Google Scholar]
- Fliegert R, Gasser A, and Guse AH (2007). Regulation of calcium signalling by adenine-based second messengers. Biochem. Soc. Trans 35, 109–114. [DOI] [PubMed] [Google Scholar]
- Geisler S, Doan RA, Strickland A, Huang X, Milbrandt J, and DiAntonio A (2016). Prevention of vincristine-induced peripheral neuropathy by genetic deletion of SARM1 in mice. Brain 139, 3092–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdts J, Brace EJ, Sasaki Y, DiAntonio A, and Milbrandt J (2015). SARM1 activation triggers axon degeneration locally via NAD(+) destruction. Science 348, 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdts J, Summers DW, Milbrandt J, and DiAntonio A (2016). Axon Self-Destruction: New Links among SARM1, MAPKs, and NAD+ Metabolism. Neuron 89, 449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdts J, Summers DW, Sasaki Y, DiAntonio A, and Milbrandt J (2013). Sarm1-mediated axon degeneration requires both SAM and TIR interactions. J. Neurosci 33, 13569–13580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh J, Anderson PJ, Chandrasekaran S, and Caparon MG (2010). Characterization of Streptococcus pyogenes beta-NAD+ glycohydrolase: re-evaluation of enzymatic properties associated with pathogenesis. J. Biol. Chem 285, 5683–5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson BA, and Kraus WL (2012). New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol 13, 411–424. [DOI] [PubMed] [Google Scholar]
- Gilley J, and Coleman MP (2010). Endogenous Nmnat2 is an essential survival factor for maintenance of healthy axons. PLoS Biol. 8, e1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henninger N, Bouley J, Sikoglu EM, An J, Moore CM, King JA, Bowser R, Freeman MR, and Brown RH Jr. (2016). Attenuated traumatic axonal injury and improved functional outcome after traumatic brain injury in mice lacking Sarm1. Brain 139, 1094–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, and Aebersold R (2002). Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem 74, 5383–5392. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, and Sternberg MJ (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc 10, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurowska Z, Kordower JH, Stoessl AJ, Burke RE, Brundin P, Yue Z, Brady ST, Milbrandt J, Trapp BD, Sherer TB, et al. (2016). Is Axonal Degeneration a Key Early Event in Parkinson’s Disease? J. Parkinsons Dis 6, 703–707. [DOI] [PubMed] [Google Scholar]
- Liu Q, Graeff R, Kriksunov IA, Jiang H, Zhang B, Oppenheimer N, Lin H, Potter BV, Lee HC, and Hao Q (2009). Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase. J. Biol. Chem 284, 27637–27645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn ER, Perry VH, Brown MC, Rosen H, and Gordon S (1989). Absence of Wallerian Degeneration does not Hinder Regeneration in Peripheral Nerve. Eur. J. Neurosci 1, 27–33. [DOI] [PubMed] [Google Scholar]
- Mack TG, Reiner M, Beirowski B, Mi W, Emanuelli M, Wagner D, Thomson D, Gillingwater T, Court F, Conforti L, et al. (2001). Wallerian degeneration of injured axons and synapses is delayed by a Ube4b/Nmnat chimeric gene. Nat. Neurosci 4, 1199–1206. [DOI] [PubMed] [Google Scholar]
- Miller BR, Press C, Daniels RW, Sasaki Y, Milbrandt J, and DiAntonio A (2009). A dual leucine kinase-dependent axon self-destruction program promotes Wallerian degeneration. Nat. Neurosci 12, 387–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, and Aebersold R (2003). A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem 75, 4646–4658. [DOI] [PubMed] [Google Scholar]
- O’Neill LA, Golenbock D, and Bowie AG (2013). The history of Toll-like receptors - redefining innate immunity. Nat. Rev. Immunol 13, 453–460. [DOI] [PubMed] [Google Scholar]
- Osterloh JM, Yang J, Rooney TM, Fox AN, Adalbert R, Powell EH, Sheehan AE, Avery MA, Hackett R, Logan MA, et al. (2012). dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science 337, 481–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao F, and Bowie JU (2005). The many faces of SAM. Sci. STKE 2005, re7. [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Nakagawa T, Mao X, DiAntonio A, and Milbrandt J (2016). NMNAT1 inhibits axon degeneration via blockade of SARM1-mediated NAD+ depletion. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Vohra BP, Lund FE, and Milbrandt J (2009). Nicotinamide mononucleotide adenylyl transferase-mediated axonal protection requires enzymatic activity but not increased levels of neuronal nicotinamide adenine dinucleotide. J. Neurosci 29, 5525–5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, and Ueda T (2001). Cell-free translation reconstituted with purified components. Nat. Biotechnol 19, 751–755. [DOI] [PubMed] [Google Scholar]
- Sikowitz MD, Cooper LE, Begley TP, Kaminski PA, and Ealick SE (2013). Reversal of the substrate specificity of CMP N-glycosidase to dCMP. Biochemistry 52, 4037–4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J, Biegert A, and Lupas AN (2005). The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers DW, Gibson DA, DiAntonio A, and Milbrandt J (2016). SARM1-specific motifs in the TIR domain enable NAD+ loss and regulate injury-induced SARM1 activation. Proc. Natl. Acad. Sci. USA 113, E6271–E6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari R, Bailes E, Bunting KA, and Coates JC (2010). Armadillo-repeat protein functions: questions for little creatures. Trends Cell Biol. 20, 470–481. [DOI] [PubMed] [Google Scholar]
- Villegas R, Martinez NW, Lillo J, Pihan P, Hernandez D, Twiss JL, and Court FA (2014). Calcium release from intra-axonal endoplasmic reticulum leads to axon degeneration through mitochondrial dysfunction. J. Neurosci 34, 7179–7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LJ, Summers DW, Sasaki Y, Brace EJ, Milbrandt J, and DiAntonio A (2017). MAPK signaling promotes axonal degeneration by speeding the turnover of the axonal maintenance factor NMNAT2. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhai Q, Chen Y, Lin E, Gu W, McBurney MW, and He Z (2005). A local mechanism mediates NAD-dependent protection of axon degeneration. J. Cell. Biol 170, 349–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, and Mann M (2009). Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362. [DOI] [PubMed] [Google Scholar]
- Xiong X, Hao Y, Sun K, Li J, Li X, Mishra B, Soppina P, Wu C, Hume RI, and Collins CA (2012). The Highwire ubiquitin ligase promotes axonal degeneration by tuning levels of Nmnat protein. PLoS Biol. 10, e1001440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Wu Z, Renier N, Simon DJ, Uryu K, Park DS, Greer PA, Tournier C, Davis RJ, and Tessier-Lavigne M (2015). Pathological axonal death through a MAPK cascade that triggers a local energy deficit. Cell 160, 161–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zmasek CM, Cai X, and Godzik A (2011). TIR domain-containing adaptor SARM is a late addition to the ongoing microbe-host dialog. Dev. Comp. Immunol 35, 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.