Abstract
Background: It is well established that maternal exercise during pregnancy improves metabolic outcomes associated with obesity in mothers and offspring, however, its effects on the gut microbiota of both mother and offspring, are unknown. Here, we investigated whether wheel running exercise prior to and during pregnancy and prolonged feeding of an obesogenic diet were associated with changes in the gut microbiomes of Sprague-Dawley rat dams and their offspring. Female rats were fed either chow or obesogenic diet, and half of each diet group were given access to a running wheel 10 days before mating until delivery, while others remained sedentary. 16S rRNA gene amplicon sequencing was used to assess gut microbial communities in dams and their male and female offspring around the time of weaning.
Results: Statistical analyses at the operational taxonomic unit (OTU) level revealed that maternal obesogenic diet decreased gut microbial alpha diversity and altered abundances of bacterial taxa previously associated with obesity such as Bacteroides and Blautia in dams, and their offspring of both sexes. Distance based linear modeling revealed that the relative abundances of Bacteroides OTUs were associated with adiposity measures in both dams and offspring. We identified no marked effects of maternal exercise on the gut microbiota of obesogenic diet dams or their offspring. In contrast, maternal exercise decreased gut microbial alpha diversity and altered the abundance of 88 microbial taxa in offspring of control dams. Thirty of these taxa were altered in a similar direction in offspring of sedentary obesogenic vs. control diet dams. In particular, the relative abundances of Oscillibacter OTUs were decreased in offspring of both exercised control dams and sedentary obesogenic diet dams, and associated with blood glucose concentrations and adiposity measures. Analyses of predicted bacterial metabolic pathways inferred decreased indole alkaloid biosynthesis in offspring of both obesogenic diet and exercised control dams.
Conclusions: Our data suggest that maternal exercise prior to and during pregnancy resulted in gut dysbiosis in offspring of control dams. Importantly, alterations in the maternal gut microbiota by obesogenic diet or obesity were transferred to their offspring.
Keywords: maternal obesity, voluntary exercise, programming, gut microbiome, adiposity, birthweight
Introduction
The worldwide incidence of overweight and obesity among children and adults has nearly tripled since 1975 (1). Overweight and obesity are usually described using the Body Mass Index (BMI); BMI scores ≥25 and ≥30 kg/m2 are classified as overweight and obese, respectively. The strong increase in obesity prevalence has affected as many as 50% of women of reproductive age in Western countries (2, 3). In addition to increasing the risk of several complications during pregnancy (4), maternal obesity can have long term detrimental consequences on offspring, including greater likelihood to develop childhood and adult obesity (5, 6). Physical exercise during pregnancy may help reduce the negative effects of maternal obesity on mothers and offspring (7, 8). Work from our laboratory and others has shown benefits of voluntary exercise before and during pregnancy on maternal and offspring metabolic outcomes associated with diet-induced obesity in rodents (9–11), but the underlying molecular mechanisms are not clear.
Over the last few decades, gut microbiota dysbiosis (perturbations in gut microbiota composition) has been implicated as an important factor contributing to various diseases including obesity, inflammatory bowel diseases, non-alcoholic fatty liver disease and gastrointestinal malignancies (12). Diet is intimately linked to obesity, and known to be a major factor influencing the human gut microbial composition (13). The infant gut is colonized by maternal gut microbes during gestation in-utero, and then further through delivery and lactation (14–17). Thus, it is likely that maternal gut microbiota dysbiosis during pregnancy and lactation constitutes a mechanism by which the effects of maternal obesity or over nutrition are conferred to the offspring. In humans, children born to women who were overweight or obese during pregnancy vs. those from normal-weight mothers showed significant differences in gut microbiota composition at 1 month, 6 months (18) and 2 years of age (19). Moreover, gut bacterial populations previously associated with obesity such as Bacteroides, Oscillibacter, and Blautia species were altered in newborns of mothers who reported a higher intake of fat during pregnancy (20). Similarly, across different animal models, consumption of a high-fat/western-style diet (HFD) during pregnancy and lactation has been associated with offspring gut microbiota dysbiosis (21–24). However, very few studies have analyzed gut microbial communities in both maternal and offspring specimens, essential to establish a direct link between microbial profiles.
Virtually no studies have investigated the impact of maternal diet and exercise prior to and during pregnancy on the gut microbiome of both mothers and their offspring. Recent studies in male rodents have explored the impacts of HFD and exercise on the gut microbiome. For instance, voluntary exercise by male mice could induce shifts in major bacterial phyla (Bacteroidetes and Firmicutes) and prevent weight gain and adiposity associated with HFD feeding (25). Voluntary or forced exercise altered the gut microbiota composition and these were orthogonal to alterations induced by HFD feeding in male mice (26, 27). But these studies were limited to the use of sequencing techniques such as denaturing gradient gel electrophoresis which only detect predominant bacterial members in the gut microbiome. In this study we sought to determine whether prolonged feeding of obesogenic diet and voluntary exercise prior to and during pregnancy were associated with changes in the gut microbiomes of Sprague-Dawley rat dams and their offspring. To investigate this, 16S rRNA gene sequencing was employed on feces collected from dams and their male and female offspring around the time of weaning, alongside detailed anthropometric and blood and tissue sampling for metabolic measures. We also investigated whether changes in the maternal and offspring gut microbiota composition were associated with metabolic measures at the time of sampling.
Results
Obesogenic diet had impacts on maternal gut microbiota α-diversity and composition
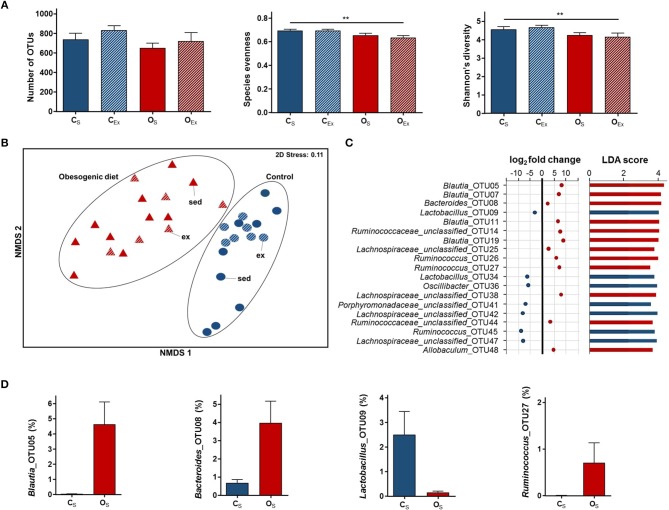
We previously reported that at 4 weeks post-partum, dams consuming obesogenic diet had higher plasma triglyceride and insulin concentrations, increased body weight and adiposity relative to those on control diet (10). There was no significant difference in blood glucose concentrations between diet groups (10). To analyse impacts of diet and exercise on the maternal gut microbiota, we performed 16S rRNA amplicon sequencing on fecal samples collected from dams at this time (control diet sedentary: CS, control diet exercised: CEx, obesogenic diet sedentary: OS, and obesogenic diet exercised: OEx). We first examined several α-diversity measures across maternal diet groups. Both species evenness [F(1, 28) = 11.1, p = 0.002; CS: 0.69 ± 0.01, CEx: 0.69 ± 0.01, OS: 0.65 ± 0.02, and OEx: 0.63 ± 0.02] and Shannon's diversity index [F(1, 28) = 8.1, p = 0.008; CS: 4.60 ± 0.13, CEx: 4.70 ± 0.10, OS: 4.20 ± 0.14, and OEx: 4.20 ± 0.20] were significantly lower in dams fed obesogenic diet relative to control dams (Figure 1A). There was no significant difference in the number of OTUs between diet groups [F(1, 28) = 2.6, p = 0.12; CS: 738.4 ± 63.5, CEx: 829.3 ± 47.2, OS: 649.2 ± 47.6, and OEx: 718.0 ± 88.1; Figure 1A).
Figure 1.
Impact of obesogenic diet on maternal gut microbiome. (A) Measures of α-diversity (number of operational taxonomic units (OTUs), species evenness, and Shannon's diversity index). Data are displayed as mean ± SEM and were analyzed by two-way ANOVA. Main effects are indicated on top of horizontal line; **p ≤ 0.01 maternal diet effect. (B) Non-metric multidimensional scaling (NMDS) plot following square root transformation and Bray-Curtis resemblance of relative abundance data at the OTU level. Maternal exercise is denoted as sed (sedentary) and ex (exercised). (C) Microbial taxa among top 50 OTUs identified to be significantly different in abundance between OS and CS dams by DESeq2 (p ≤ 0.05) and LEfSe (LDA Score>2.0, p ≤ 0.05) analyses. In DESeq2, negative log2 fold change value denotes decreased abundance and positive log2 fold change value denotes increased abundance in OS dams relative to CS dams. (D) Relative abundance of four microbial taxa identified to be differentially abundant between OS and CS dams by DESeq2 and LEfSe analyses. Data are displayed as mean ± SEM. CS (control diet sedentary; blue): n = 9, CEx (control diet exercised; striped blue): n = 7, OS (obesogenic diet sedentary; red): n = 10, and OEx (obesogenic diet exercised; striped red): n = 6.
To determine whether the overall gut microbial composition of obesogenic and control diet fed dams were different, we examined different β-diversity measures. Dams fed obesogenic diet clustered differently to control dams at the OTU level as observed by NMDS (Figure 1B). PERMANOVA analyses likewise confirmed significant differences in β-diversity between diet groups (t = 2.6, df = 1, 30, p = 0.001). Maternal diet did not alter sample dispersions (variation of Bray-Curtis similarities; PERMDISP: t = 1.0, df = 1, 30, p = 0.35).
We next examined sedentary dams to identify individual microbial taxa that differed between diet groups at the OTU level. LEfSe analysis identified 292 differentially abundant OTUs, while DESeq2 analysis identified 167 differentially abundant OTUs between diet groups. A total of 131 differentially abundant OTUs was consistent across both analyses, with 60 OTUs enriched and 71 OTUs decreased in OS dams when compared to CS dams. Several OTUs affiliated with genera Lactobacillus, Alistipes, and unclassified genera within Porphyromonadaceae and Lachnospiraceae families were decreased in OS dams. On the other hand, OTUs affiliated with Blautia, Ruminococcus and Bacteroides were more abundant in OS dams (Figures 1C,D). LEfSe analysis at the higher taxonomic levels identified Lactobacillus, Blautia and Bacteroides genera as differentially abundant between OS and CS dams (LDA score>4), which was consistent with the OTU results.
Maternal gut microbiota composition was associated with metabolic measures
Distance based linear models (DistLM) were used to identify associations between the overall maternal gut microbial composition and several metabolic parameters. The metabolic parameters tested were: fat mass, blood glucose concentrations, plasma leptin, insulin and triglyceride (TG) concentrations at endpoint, and change in body weight over the experiment. The analysis was performed between metabolic parameters and a Bray-Curtis resemblance matrix of the overall gut microbiome at the OTU level. All the considered parameters, except for blood glucose, were significantly associated with the overall gut microbial composition at the OTU level (Table 1).
Table 1.
Correlations between overall maternal gut microbial composition and metabolic parameters.
| Variable | Pseudo-F | p(perm) | res.df |
|---|---|---|---|
| Leptin | 3.3 | 0.001 | 30 |
| Insulin | 1.8 | 0.015 | 30 |
| Glucose | 0.69 | 0.94 | 30 |
| Triglycerides | 4.3 | 0.001 | 30 |
| Fat mass | 4.6 | 0.001 | 30 |
| Change in body weight | 3.2 | 0.002 | 30 |
| Distance run | 1.4 | 0.11 | 30 |
Distance based linear modeling (DistLM) was performed using a Bray-Curtis resemblance matrix of the overall gut microbiome composition at the operational taxonomic unit level with metabolic parameters: plasma leptin, insulin, triglyceride and blood glucose concentrations, fat mass, and change in body weight over the experiment as predictor variables. Fat mass was measured as the sum of retroperitoneal, visceral and inguinal fat pads. Distance run represented average kilometers run per day by each rat. A forward-stepping selection procedure and Akaike Information Criterion selection criterion was used. Pseudo-F and p-values were obtained using 999 permutations (perm) and bold values indicate significance, p ≤ 0.05.
res.df = residual degrees of freedom.
Further, we performed DistLM analyses to investigate whether specific microbial taxa were associated with these metabolic parameters. Here the analysis was performed between a Euclidean distance resemblance matrix of each parameter and the relative abundances of the top 150 OTUs. Several OTUs differentially abundant with diet were associated with the tested parameters. Lactobacillus_OTU34 (decreased 6-fold in OS dams), Blautia_OTU5 (enriched 8-fold in OS dams) and Bacteroides_OTU8 (enriched 3-fold in OS dams) were associated with all tested metabolic parameters except blood glucose and insulin. Ruminococcus_OTU27 (enriched more than 7-fold in OS dams) was associated with insulin and leptin. Only Lachnospiraceae unclassified_OTU47 (decreased 8-fold in OS dams) was associated with blood glucose.
Maternal exercise altered gut microbial composition in control dams
Dams had access to running wheels from 10 days prior to mating until delivery, and exercise levels did not differ between diet groups (10). At the time of sampling, maternal exercise had no discernible metabolic impacts in obesogenic or control diet dams at 4 weeks post-partum (10), however, it is possible that some impacts were present in the gut microbiota. We identified no significant effect of maternal exercise on α-diversity measures (species evenness, number of OTUs, or Shannon's diversity index; Figure 1A) in the dams.
To determine whether maternal exercise had an impact on the overall gut microbial composition, PERMANOVA analysis was conducted between exercised and sedentary dams in each diet group, separately (CEx vs. CS, OEx vs. OS). Pairwise PERMANOVA revealed CEx dams had significantly different gut microbial compositions compared to CS dams (t = 1.2, df = 14, p = 0.041), in contrast, there was no significant difference between OEx and OS dams (t = 0.98, df = 14, p = 0.50). The microbial communities of sedentary dams were more dispersed than those of exercised dams (PERMDISP: t = 3.2, df = 1, 30, p = 0.009). We also tested whether average distance run per day was associated with the gut microbial composition, and found no significant correlation (Table 1).
We then applied LEfSe and DESeq2 analyses between sedentary and exercised dams in each diet group, separately. Consistent across both analyses, one differentially abundant OTU was identified between CEx and CS dams, while no differentially abundant OTUs were identified between OEx and OS dams. Anaerostipes_OTU70 was decreased in CEx dams relative to CS dams (DESeq2: log2 fold change = −8.6, p = 0.0018; LEfSe: LDA>3, p = 0.018). At the higher taxonomic levels, LEfSe identified genera that were not affiliated with OTUs differentially abundant with maternal exercise such as Clostridium_XlVa (decreased in CEx relative to CS; LDA>4) and Clostridium_XVIII (decreased in OEx relative to OS; LDA>3).
Offspring gut microbiota α-diversity measures were impacted by maternal diet and exercise
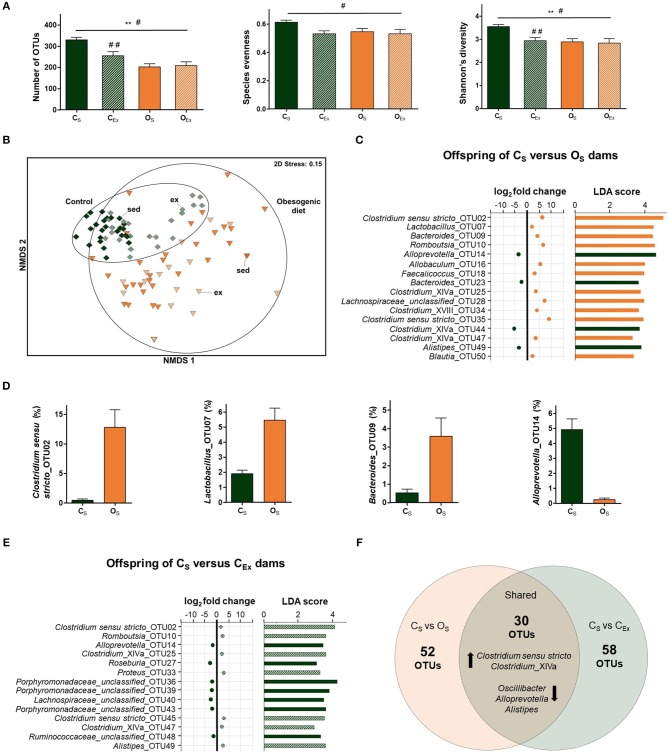
The gut microbiota of male and female offspring at postnatal day (PND) 19 was examined using 16S rRNA amplicon sequencing. Maternal obesogenic diet was associated with higher blood glucose concentrations, final body weight and adiposity in offspring at PND19 (10). Maternal diet and exercise had sex-specific effects on blood glucose concentrations, plasma insulin and TG concentrations in the offspring (10). In male offspring, maternal exercise was associated with significantly lower blood glucose concentrations (OEx vs. OS offspring). Insulin concentrations were lower in male offspring of both CEx and OEx dams, with no significant effect of maternal diet. In contrast, insulin concentrations were higher in female offspring of OS vs. CS dams, with no significant effect of maternal exercise. Plasma TG concentrations were significantly lower in male offspring of CEx vs. CS dams. There were no significant effects of maternal exercise on blood glucose concentrations, plasma insulin or TG concentrations in female offspring of obesogenic and control diet dams (10). Here, we found no sex-specific differences in α-diversity measures (three-way ANOVA: offspring sex, maternal diet and maternal exercise). Thus, data from both sexes was aggregated to examine differences in α-diversity across diet and exercise. A two-way ANOVA indicated a significant interaction between maternal diet and exercise on the number of OTUs [F(1, 89) = 7.4, p = 0.008; CS: 330.5 ± 10.3, CEx: 255.0 ± 19.3, OS: 203.0 ± 13.5, and OEx: 209.5 ± 15.8] and Shannon's diversity index [F(1, 89) = 4.6, p = 0.034; CS: 3.60 ± 0.08, CEx: 2.90 ± 0.13, OS: 2.90 ± 0.13, and OEx: 2.80 ± 0.18] in offspring. Simple main effects analysis indicated that maternal exercise only significantly decreased both α-diversity measures in offspring of control dams (p ≤ 0.001 for both), as shown in Figure 2A. Maternal exercise, across diet groups, significantly decreased species evenness [F(1, 89) = 5.8, p = 0.018; CS: 0.61 ± 0.01, CEx: 0.53 ± 0.02, OS: 0.55 ± 0.02, and OEx: 0.53 ± 0.03], with no significant effect of maternal diet (Figure 2A).
Figure 2.
Impacts of maternal obesogenic diet and voluntary exercise on offspring gut microbiome. (A) Measures of α-diversity (number of operational taxonomic units (OTUs), species evenness, and Shannon's diversity index). Data are displayed as mean±SEM and were analyzed by two-way ANOVA. Main effects indicated on top of horizontal line; **p ≤ 0.01 maternal diet effect; #p ≤ 0.05 maternal exercise effect. Simple main effects are indicated on top of bar; ##p ≤ 0.01 maternal exercise effect. (B) Non-metric multidimensional scaling (NMDS) plot following square root transformation and Bray-Curtis resemblance of relative abundance at the OTU level. Maternal exercise is denoted as sed (sedentary) and ex (exercised). (C) Microbial taxa among top 50 OTUs identified to be significantly different in abundance between offspring of OS and CS dams by DESeq2 (p ≤ 0.05) and LEfSe (LDA Score>2.0, p ≤ 0.05) analyses. In DESeq2, negative log2 fold change value denotes decreased abundance and positive log2 fold change value denotes increased abundance in offspring of OS relative to CS. (D) Relative abundance of four microbial taxa identified to be differentially abundant between offspring of OS and CS dams by DESeq2 and LEfSe analyses. Data are displayed as mean ± SEM. (E) Microbial taxa among top 50 OTUs identified to be significantly different in abundance between offspring of CEx and CS dams by DESeq2 (p ≤ 0.05) and LEfSe (LDA Score>2.0, p ≤ 0.05) analyses. In DESeq2, negative log2 fold change value denotes decreased abundance and positive log2 fold change value denotes increased abundance in offspring of CEx relative to CS. (F) Number of OTUs altered in a similar direction in offspring of both CEx (green) and OS (orange) dams relative to offspring of CS dams. Black arrows denote either increase or decrease in abundance of microbial taxa. Offspring from CS (control diet sedentary; green): n = 25, CEx (control diet exercised; striped green): n = 22, OS (obesogenic diet sedentary; orange): n = 29, and OEx (obesogenic diet exercised; striped orange): n = 17.
Maternal obesogenic diet altered offspring gut microbiota composition
No sex-specific differences in gut microbial composition were identified globally (PERMANOVA: t = 0.78, df = 1, 91, p = 0.66), within each sub-group (PERMANOVA: CS: t = 0.74, df = 1, 23, p = 0.72, CEx: t = 0.99, df = 1, 20, p = 0.39, OS: t = 0.84, df = 1, 27, p = 0.54, and OEx: t = 0.62, df = 1, 15, p = 0.80), or in abundance of individual microbial taxa within each sub-group (LEfSe and DESeq2 analyses) between male and female offspring. Thus, data from both sexes was aggregated to examine differences in β-diversity across diet and exercise. The microbial compositions of offspring from obesogenic diet dams differed significantly when compared to offspring from control dams, as visualized by NMDS (Figure 2B), and confirmed using PERMANOVA analyses (t = 3.5, df = 1, 91, p = 0.001). Microbial communities in offspring from control dams were less dispersed than in offspring from obesogenic diet dams (PERMDISP: t = 4.5, df = 1, 91, p = 0.001).
LEfSe and DESeq2 analyses were employed on microbial abundance data from offspring of sedentary dams across both diet groups. Of the 394 differentially abundant OTUs detected by LEfSe, 82 were consistent with DESeq2 analysis. More OTUs were decreased (53 OTUs) than enriched (29 OTUs) in OS relative to CS offspring. OTUs affiliated with Clostridium sensu stricto, Clostridium_XlVa, Bacteroides, Blautia and Lactobacillus were enriched, while Alistipes, Oscillibacter and Alloprevotella OTUs were decreased in OS offspring (Figures 2C,D). Consistent with the OTU results, LEfSe found Clostridium sensu stricto and Lactobacillus genera were enriched, while Alloprevotella was decreased in OS vs. CS offspring (LDA score>4).
Maternal exercise altered offspring gut microbiota composition in a maternal diet dependent manner
The impact of maternal exercise on β-diversity was calculated between offspring of exercised and sedentary dams in each diet group, separately. PERMANOVA analyses revealed significantly different gut microbial compositions in offspring from CEx vs. CS dams (t = 2.4, df = 45, p = 0.001). There was no significant difference in β-diversity between offspring from OEx and OS dams (t = 1.1, df = 44, p = 0.20). The dispersion of microbial communities was similar between offspring from exercised and sedentary dams (PERMDISP: t = 0.46, df = 1, 91, p = 0.67).
We then investigated whether maternal exercise altered the abundance of specific microbial taxa in the offspring. LEfSe and DESeq2 identified 88 differentially abundant OTUs between CEx and CS offspring. In contrast, no differentially abundant OTUs were identified between offspring of OEx and OS dams. OTUs affiliated with genera Romboutsia, Clostridium sensu stricto and Proteus were enriched, whereas OTUs affiliated with Roseburia, Alistipes and unclassified genera within Porphyromonadaceae and Lachnospiraceae families were decreased in CEx offspring (Figure 2E). Of the 88 differentially abundant OTUs between CEx and CS offspring, 30 were also differentially abundant between OS vs. CS offspring, as shown in Figure 2F. In both OS and CEx offspring, OTUs affiliated with Clostridium sensu stricto and Clostridium_XlVa were enriched, while OTUs affiliated with Alloprevotella, Alistipes and Oscillibacter were decreased relative to CS offspring (Figure 2F).
Offspring gut microbiota composition was associated with metabolic measures
DistLM was used to analyse the association between the overall gut microbial composition and metabolic parameters in the offspring. The analysis was performed against metabolic parameters: final body weight, visceral fat, and blood glucose concentrations, as well as plasma leptin, and insulin and TG concentrations. The overall offspring gut microbial composition (Bray-Curtis resemblance matrix) was significantly associated with final body weight, visceral fat, glucose and leptin (Table 2).
Table 2.
Correlations between overall offspring gut microbial composition and metabolic parameters.
| Variable | Pseudo-F | p(perm) | Res.df |
|---|---|---|---|
| Leptin | 4.1 | 0.001 | 87 |
| Insulin | 1.6 | 0.069 | 85 |
| Glucose | 2.9 | 0.005 | 91 |
| Triglycerides | 1.5 | 0.086 | 79 |
| Visceral fat | 10.6 | 0.001 | 91 |
| Final body weight | 10.6 | 0.001 | 91 |
Distance based linear modeling (DistLM) was performed using a Bray-Curtis resemblance matrix of the overall gut microbiome composition at the operational taxonomic unit level with metabolic parameters: plasma leptin, insulin, triglyceride and blood glucose concentrations, visceral fat, and final body weight as predictor variables. A forward-stepping selection procedure and Akaike Information Criterion selection criterion was used. Pseudo-F and p-values were obtained using 999 permutations (perm) and bold values indicate significance, p ≤ 0.05.
res.df = residual degrees of freedom.
The top 150 OTUs whose relative abundance was differentially affected by maternal diet and exercise were analyzed for associations with final body weight, visceral fat and blood glucose (Euclidean distance resemblance matrix) using DistLM. OTUs enriched in OS offspring such as Lactobacillus_OTU7, Blautia_OTU54 and Bacteroides_OTU9 were associated with all the tested metabolic parameters. We also identified associations for OTUs altered in both OS offspring (OS vs. CS) and CEx offspring (CEx vs. CS) with metabolic parameters. Clostridium sensu stricto_OTU2 and Clostridium XlVa_OTU25 (enriched in OS and CEx offspring) were associated with final body weight. Alloprevotella_OTU14, Alistipes_OTU49, and Oscillibacter_OTU55 (decreased in OS and CEx offspring) were associated with all the tested metabolic parameters.
Predicted functions of offspring gut microbiota were altered by maternal diet and exercise
In order to infer changes in microbial metabolic pathways in the offspring gut microbiota resulting from maternal diet or exercise, functional content was predicted from amplicon data using PICRUSt. All offspring samples had low nearest sequenced taxon index (NSTI) values (average NSTI: 0.058 ± 0.003) indicating high prediction accuracy. Several predicted pathways were significantly enriched in offspring from obesogenic vs. control diet dams (Bonferroni corrected ANOVA with maternal exercise status nested as subclass, p < 0.05), among them, fructose and mannose metabolism (Table 3). In contrast, pathways related to indole alkaloid biosynthesis, α-linolenic acid metabolism and carotenoid metabolism were significantly decreased (Table 3). No significantly different pathways were identified after FDR or Bonferroni correction for maternal exercise (ANOVA with maternal diet type nested as subclass). We then analyzed offspring from control diet dams separately (offspring of CEx vs. CS dams). Pathways related to vasopressin regulated water reabsorption (fold change = −3.0, p = 0.00064, q = 0.067), indole alkaloid biosynthesis (fold change = −2.2, p = 0.0011, q = 0.067), and betalain biosynthesis (fold change = −2.2, p = 0.0011, q = 0.067) were decreased, while those related to phosphotransferase system (fold change = 1.5, p = 0.00083, q = 0.067), and Staphylococcus aureus infection (fold change = 1.9, p = 0.0013, q = 0.067) were increased in CEx offspring.
Table 3.
Predicted bacterial pathways altered by maternal diet in the offspring gut microbiota.
| Pathway | Fold-change | Mean (C) | Mean (O) | P (ANOVA) | P(adj) |
|---|---|---|---|---|---|
| Alpha Linolenic acid metabolism | −2.016 | 4203.68 | 2084.87 | 0.00000014 | 0.000038 |
| Amyotrophic lateral sclerosis | −1.683 | 7158.94 | 4253.39 | 0.00000021 | 0.000057 |
| Carotenoid biosynthesis | −2.165 | 9428 | 4354.37 | 0.00000091 | 0.00025 |
| Vasopressin regulated water reabsorption | −5.581 | 8.98 | 1.61 | 0.0000014 | 0.00038 |
| Geraniol degradation | −1.724 | 20611.09 | 11953.09 | 0.0000023 | 0.00063 |
| Caprolactam degradation | −1.88 | 18865.79 | 10035.26 | 0.0000024 | 0.00065 |
| Lipopolysaccharide biosynthesis | −1.654 | 77024.87 | 46576.93 | 0.0000026 | 0.00071 |
| Lipopolysaccharide biosynthesis proteins | −1.547 | 119476.9 | 77230.09 | 0.0000033 | 0.0009 |
| Shigellosis | −2.196 | 3437.06 | 1564.85 | 0.0000045 | 0.0012 |
| Indole alkaloid biosynthesis | −16.09 | 6.3 | 0.39 | 0.0000046 | 0.0013 |
| Bladder cancer | −2.172 | 1734.15 | 798.35 | 0.0000054 | 0.0015 |
| Pertussis | −1.702 | 10961.15 | 6439.37 | 0.0000055 | 0.0015 |
| Prion diseases | −1.653 | 2211.87 | 1338.09 | 0.0000068 | 0.0018 |
| Bacterial invasion of epithelial cells | −2.125 | 5215.64 | 2454.93 | 0.0000073 | 0.002 |
| Betalain biosynthesis | −11.14 | 6.3 | 0.57 | 0.0000098 | 0.0027 |
| Metabolism of xenobiotics (cytochrome P450) | −1.705 | 12248.57 | 7182.28 | 0.00001 | 0.0027 |
| Chlorocyclohexane and chlorobenzene degradation | −1.707 | 9778.04 | 5726.59 | 0.000016 | 0.0044 |
| Drug metabolism (cytochrome P450) | −1.697 | 13039.94 | 7683.5 | 0.000019 | 0.0052 |
| Sporulation | 1.71 | 90806.6 | 155265.2 | 0.000044 | 0.012 |
| Fructose and mannose metabolism | 1.362 | 214300.3 | 291772.5 | 0.00015 | 0.041 |
| Inositol phosphate metabolism | −1.377 | 35218.09 | 25582.15 | 0.00017 | 0.046 |
| Restriction enzyme | 1.343 | 26422.96 | 35483.37 | 0.00018 | 0.049 |
Nested ANOVA was performed using KEGG level 3 pathway counts generated by PICRUSt. The main variable was diet with exercise nested within. P-values were adjusted (Padj) with Bonferroni correction and only pathways showing P(adjusted) < 0.05 are shown. Negative fold change value denotes decreased abundance and positive fold change value denotes increased abundance in OS dams relative to CS dams.
Discussion
Alterations in the maternal gut microbiota during critical periods of embryonic, fetal, and early postnatal development may have effects on the offspring gut microbiota, with lifelong consequences for susceptibility to disease (28). Our results indicate that chronic consumption of an obesogenic diet prior to and during gestation, and continuing during lactation, induced changes in the maternal and offspring gut microbiota. This affected the predicted profile of microbial metabolic pathways related to lipid and carbohydrate metabolism in the offspring. We identified no marked effects of maternal exercise on the gut microbiota of dams fed obesogenic diet or their offspring. In contrast, maternal exercise decreased α-diversity and altered the abundance of 88 microbial taxa in offspring of CEx vs. CS dams. Thirty of these taxa were altered in a similar direction in offspring of OS vs. CS dams and associated with several metabolic markers relevant to obesity. We postulate that this dysbiosis may be due to the impacts of catch-up growth in offspring of CEx dams.
Results of this study emphasize the importance of healthy dietary habits during pregnancy and lactation on maternal and offspring outcomes. We found that consumption of an obesogenic diet induced changes in the maternal gut microbiota and these were transferred to their offspring of both sexes. Our results are in accordance with a metagenomic study in primates that showed HFD feeding during pregnancy and lactation induced changes in the gut microbial composition of mothers and their offspring (22). OTUs affiliated with Bacteroides and Blautia were enriched in both dams fed obesogenic diet and their offspring. The relative abundances of these OTUs were associated with several measures of adiposity in both dams and offspring. In support of our results, Bacteroides was enriched in the gut microbiota of both overweight mothers and their infants at one and 6 months of age compared to infants from normal weight mothers (18). Other studies in humans have associated increased Bacteroides with overweight and obesity (29–31). However, decreased Bacteroides was reported in newborns of mothers reporting a greater intake of fat during pregnancy (20). Increased levels of Bacteroides have also been associated with weight loss in obese human subjects undergoing dietary or surgical interventions (32–34). Variations across these studies can be attributed to differences in sequencing methods, taxonomic level analyzed and diet. Our findings along with previous evidence suggest a role for Bacteroides in the modulation of host adiposity, however, a causal link is yet to be established.
We inferred the predicted metabolic functions of the offspring gut microbiota using PICRUSt. Our analyses revealed a clear difference in the predicted functional capability between the microbiota of offspring from obesogenic vs. control diet dams. Specifically, pathways related to fatty acid metabolism such α-linolenic acid metabolism were decreased in offspring from obesogenic diet dams. The metabolism of α-linolenic acid by gastrointestinal microbes is known to generate conjugated fatty acids, which have shown potential to reduce atherosclerosis and adiposity (35). Conversely, pathways involved in carbohydrate metabolism (specifically fructose and mannose metabolism) were more abundant in offspring from obesogenic diet dams. This has also been reported in juvenile primates exposed to maternal HFD during gestation and lactation (22). Further, an increased abundance of pathways related to carbohydrate metabolism is a consistent finding across previous studies from our laboratory (36, 37) and others (38, 39) in adult rodents fed HFD. However, these findings need to be confirmed experimentally.
We previously reported that maternal exercise prior to and during pregnancy limited the detrimental impacts of maternal obesity in offspring by reducing plasma insulin and glucose concentrations at PND19 (10). In the present study, maternal exercise had no significant impact on measures of α-diversity and microbial composition in dams fed obesogenic diet or their offspring. This may be due to the major impact of obesogenic diet on the gut microbiota and the moderate level of exercise achieved by the dams. More marked effects of exercise have been reported in previous studies comparing the effect of HFD and exercise on the gut microbiota in adult male rodents (25–40). This discrepancy may be related to differences in diet and exercise paradigms, and developmental stage that exercise was introduced, as here exercise was confined to pre-pregnancy and during gestation. Our results suggest that maternal exercise prior to and during pregnancy cannot rescue the detrimental effects of a maternal obesogenic diet on the maternal and offspring gut microbiota. The mechanism(s) by which maternal exercise induces metabolic benefits in obesogenic diet fed dams and their offspring warrants further investigation.
In contrast, voluntary maternal exercise via wheel running had significant effects on gut microbiota composition in control dams and their offspring. Though a different exercise regimen was used, our results are supported by a study which reported significant impacts of exercise on gut microbial composition in control male Wistar rats, while there was no impact in those fed HFD (41). Interestingly, we found that of the 88 OTUs altered by maternal exercise in offspring of control dams, 30 were altered in a similar direction in offspring of OS vs. CS dams. Of interest, Oscillibacter OTUs were decreased in offspring of both CEx and OS dams, and these OTUs were associated with blood glucose levels and several measures of adiposity. Other studies have also associated changes in the abundance of Oscillibacter with obesity and metabolic parameters. For instance, Oscillibacter was negatively correlated with BMI or postprandial glucose area under the curve in humans (42, 43). In monozygotic twins discordant for obesity in terms of BMI, members of Oscillibacter were more abundant in twins with lower BMI (44). However, very little is known about the physiological role of this bacterium. Our analyses also revealed similar alterations in microbial metabolic pathways in offspring of both obesogenic diet and control exercised dams. Notably, pathways related to indole alkaloid biosynthesis were decreased in both. The intestinal microbiota has been reported to metabolize tryptophan using tryptophanase to produce indole metabolites (45). It was recently reported that Lactobacillus murinus supplementation blunted high salt diet induced TH17 activation and ameliorated salt sensitive hypertension in mice, and this was associated with an increase in fecal indole metabolites (46). In weaning piglets, tryptophan supplementation enhanced indole alkaloid biosynthesis pathways and downregulated the expression of inflammatory cytokines and improved intestinal mucosal barrier function (47).
We postulate that gut dysbiosis in offspring of CEx dams could be a reflection of catch-up growth (accelerated postnatal growth). Maternal exercise by control dams was associated with lower birth weight in their offspring across both sexes and this effect was diminished at weaning (10). Despite evidence in human studies that catch-up growth can increase the risk of metabolic diseases in adulthood (48, 49), very few studies have investigated its effects on the gut microbiome. A recent study in mice identified changes in the gut microbial composition of pups experiencing catch-up growth, and this was associated with excessive adiposity and glucose intolerance in adulthood. Specifically, they found that members of the Lachnospiraceae family were decreased in pups experiencing catch-up growth at 4 weeks of age (50), which is similar to our observations in offspring of CEx dams. Future studies should investigate whether these changes in gut microbial composition in offspring persist into adulthood and increase their susceptibility to metabolic disease.
There are several limitations to our study worth noting. We ceased voluntary exercise after gestation in order to avoid any interruption to nursing behavior by dams during lactation. It is possible some impacts of exercise on the maternal and offspring gut microbiome abated over time or were overridden by diet. In addition, this study did not account for other maternal factors such as the vaginal, breast milk, and skin microbiome, which are also known to impact the offspring gut microbial composition (15, 51). Despite the fact that the analysis between dam and offspring was highly consistent, our use of different primer sets for 16S rRNA gene amplification in dams and offspring may have influenced the results. Studies have detected different changes in gut microbial composition data depending on the region of 16S rRNA gene sequenced (52, 53). Moreover, it is not clear whether our findings can be extended to humans, rodent pups have a very immature intestinal tract at birth and during lactation (day 0–21), whereas in human infants the intestinal tract is more mature at birth (54). Nevertheless, our data in rats is an essential starting point in elucidating the impact of maternal obesogenic diet and voluntary exercise on the gut microbiome, especially since it is difficult to undertake similar studies in humans in a controlled manner.
Conclusions
This study makes novel contributions to our understanding of diet and exercise as maternal factors influencing the maternal and offspring gut microbiome. Importantly, we showed that bacterial taxa previously associated with obesity were altered by obesogenic diet in dams and their offspring. Maternal exercise prior to and during pregnancy had major impacts on the gut microbiota of offspring from control dams, with reduced gut microbial diversity and alterations in microbial composition. Intriguingly, we found that 30 of the 88 microbial taxa altered in offspring of exercised control dams were also altered in a similar direction in offspring of sedentary obesogenic vs. control diet dams. These findings were likely driven by the effects of catch-up growth in offspring of exercised control dams, but further controlled animal studies are needed to interrogate this potential mechanism.
Materials and methods
Maternal obesity and exercise model
This study is based on a cohort of female Sprague-Dawley rats and their offspring previously generated in our laboratory (10). Briefly, 6-week old female rats were assigned either standard laboratory chow (11 kJ/g, 13% fat, 22% protein, and 65% carbohydrate by energy; Gordon's Stockfeeds, NSW, Australia) or obesogenic diet ad libitum: two types of high-fat pellet (SF03-020: 20 kJ/g, 43% fat, 16% protein, and 41% carbohydrate by energy, and SF03-002: 22.8 kJ/g, 59% fat, 14% protein, and 27% carbohydrate by energy; Specialty feeds, Australia) supplemented with a selection of three different western foods (selected from cakes, potato chips, biscuits, meat pie, pasta with lard, and oats mixed with condensed milk). After 6 weeks, half the rats from each diet group were randomly assigned to voluntary exercise through introduction of a running wheel, while the other half remained sedentary with a locked wheel. Female rats were mated (with males fed standard chow) after 10 days of exercise. After delivery, dams were transferred to cages with no running wheel in order to prevent any interference of exercise with nurturing of pups, and to better model likely post parturition behavior. Dams were maintained on their pre-pregnancy diet throughout pregnancy and lactation.
The average litter size and male to female ratios were not significantly different across the groups. At postnatal day (PND) 1, litters were adjusted to 12 pups per mother (10); pups were allocated to groups so that litters were equally represented. Feces from the distal colon were collected from pups and dams at 3 and 4 weeks postpartum, respectively, at the time of tissue sampling for anthropometric and blood analyses. Fecal samples were immediately flash frozen in liquid nitrogen following collection and stored at -80°C. For the purposes of this study a subset of dams and pups (range 2–4 pups per dam with equal male to female ratios) were used.
Fecal DNA extraction
DNA was extracted from the feces of dams (CS: n = 9, CEx: n = 7, OS: n = 10, and OEx: n = 7) using the PowerFecal DNA isolation kit (Catalog number: 12830-50, MO BIO Laboratories, Inc., Carlsbad, CA, USA), and offspring of both sexes (male: CS: n = 12, CEx: n = 11, OS: n = 13, and OEx: n = 11, and female: CS: n = 13, CEx: n = 11, OS: n = 16, and OEx: n = 10) using the PowerSoil DNA Isolation Kit (Catalog number: 12888-100, MO BIO Laboratories, Inc., Carlsbad, CA, USA). Extractions were performed according to the manufacturer's instruction. DNA concentration and quality was measured using a DeNovix DS-11 Spectrophotometer (DeNovix, Inc., Delaware, USA).
16S rRNA gene sequencing and taxonomic analysis
The composition of gut microbial communities was analyzed by Illumina amplicon sequencing of the 16S rRNA gene (performed by Ramaciotti Center for Genomics, UNSW Sydney) using DNA from dams (2 × 300 base pair MiSeq chemistry, V1-V3 region, 27F-519R primer pair) and offspring (2 × 250 base pair MiSeq chemistry, V4 region, 515F-806R primer pair). Sequence data were processed using Mothur, version 1.39.1 (55), which included removal of ambiguous bases and homopolymers longer than 15 base pairs, alignment with SILVA database, chimera checking with UCHIME, classification against the latest RDP Ribosomal Database training set (version 16_022016), and removal of singletons. Sequences were clustered into operational taxonomic units (OTU) at 97% nucleotide identity to generate an OTU table with the taxonomy and number of sequences per OTU in each sample. Commands were derived using the Mothur MiSeq SOP (56) and modified as required. The generated OTU table, combined with previously published anthropometric and biochemical measurements (10), were used as input in follow-up analyses.
Statistical analyses
Mother and offspring samples were rarefied down to 24,626 and 12,291 reads, respectively. OTU tables were standardized by dividing feature read counts by total number of reads in each sample. Standardized data were then square root transformed. All statistical analyses explored sex-specific differences in the offspring. One dam was identified as an outlier by Non-metric multidimensional scaling (NMDS). As 88% of the gut microbial composition in this dam was dominated by two OTUs (OTUs 1 and 2) from the genus Romboutsia, the dam and her offspring (2 male and 2 female offspring) were excluded from subsequent analyses.
Alpha diversity (number of OTUs, species evenness and Shannon's diversity index) analyses were performed using Calypso (57). Statistical analyses such as two and three-way ANOVA on diversity measures were performed using SPSS software (SPSS statistics version 22, SPSS Inc., an IBM Company). Simple main effects (with Bonferroni adjustment) was performed in the case of a significant interaction between variables on α-diversity. Results are expressed as mean ± SEM and were considered significant if p ≤ 0.05. GraphPad Prism 7 was used to plot diversity results. In bar graphs, symbols above horizontal line indicate main effects: **p ≤ 0.01 maternal diet effect and #p ≤ 0.05 maternal exercise effect, while symbols above bar indicate simple main effects: ##p ≤ 0.01 maternal exercise effect.
NMDS plots, Permutational Multivariate Analysis of Variance (PERMANOVA) and Permutational Analysis of Multivariate Dispersions (PERMDISP) were generated using a Bray-Curtis resemblance matrix on PRIMER (Primer-E Ltd., Plymouth, United Kingdom) (58). To identify OTUs differentially abundant between groups we used two different statistical analyses. Linear Discriminant Analysis (LDA) Effect Size (LEfSe) (59) was performed using the Galaxy web application (60) using default settings. The R package Phyloseq (61) was used for the negative binomial Wald test in DESeq2 (62). P-values were adjusted for multiple testing using Benjamini Hochberg false discovery rate correction in DESeq2. LEfSe was performed across all taxonomic levels (phylum to OTU level) whereas DESeq2 was performed at the OTU level. Statistical significance was defined as p ≤ 0.05 in both statistical methods. Only OTUs identified by both LEfSe and DESeq2, and those with relative abundances consistently represented across more than 50% of rats in a group were discussed.
Distance-based linear models (DistLM) analysis was performed using PRIMER to examine associations between the gut microbiota and metabolic parameters. When the analysis was performed between the overall gut microbial composition and metabolic parameters, a Bray-Curtis resemblance matrix of the overall gut microbial composition at the OTU level was used. The relative abundances of the top 150 OTUs and Euclidean distance resemblance matrix of each metabolic parameter was used to find associations between specific OTUs and metabolic parameters. In both cases DistLM was performed using a forward-stepping selection procedure and Akaike Information Criterion selection criterion (999 permutations). Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) was performed using Galaxy web to generate a profile of putative functions (through metagenomic prediction) from the 16S rRNA OTU data (63). Taxonomic classification was performed against Greengenes 13.5, and pathway counts were compared across groups using nested ANOVA with Bonferroni correction. Subgroup comparisons were conducted using Wilcoxon test with FDR (FDR corrected p-values represented as q).
Availability of data and material
The sequence data are available in the European Nucleotide Archive (ENA) under accession number PRJEB26886 (http://www.ebi.ac.uk/ena/data/view/PRJEB26886).
Ethics statement
This study was carried out in accordance with the recommendations of the Animal Research Act 1985 (NSW), the Animal Research Regulations 2010 (NSW), and the NHMRC Australian Code for the Care and Use of Animals for Scientific Purposes 2013 (8th edition). The protocol was approved by the University of New South Wales Animal Care and Ethics Committee (Approval No. 11/104B).
Author contributions
MM conceived and designed experiments. MM and NK supervised the project. MR, HB, and MM developed the rat cohort. SB, NK, and MM analyzed and interpreted data, and wrote the paper. MR and HB performed all anthropometric and metabolic measurements, and reviewed the paper.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We greatly acknowledge Miss. Sarah-Jane Leigh for her statistical guidance. We also thank members of the Morris lab for their assistance.
Glossary
Abbreviations
- BMI
Body mass index
- DNA
Deoxyribonucleic acid
- LEfSe
Linear discriminant analysis (LDA) effect size
- OTU
Operational taxonomic units
- rRNA
Ribosomal RNA gene
- PERMANOVA
Permutational multivariate analysis of variance
- PND
Postnatal day
- OEx
Obesogenic diet exercised
- OS
Obesogenic diet sedentary
- CEx
Control diet exercised
- CS
Control diet sedentary
- NMDS
Non-metric multidimensional scaling
- DistLM
Distance-based linear models
- SEM
Standard error of mean
- PERMDISP
Permutational Analysis of Multivariate Dispersions
- HFD
High-fat diet
- TG
Triglycerides
- perm
Permutations
- df
Degrees of freedom
- PICRUSt
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States.
Footnotes
Funding. The work was funded by National Health and Medical Research Council of Australia (NHMRC 568728, 1023073) to MM. MR was sponsored by Government of Madhya Pradesh State, India and the Australian Centre for Perinatal Science (ACPS). HB was sponsored by the Ministry of Higher Education Malaysia and Universiti Putra Malaysia. NK is supported by a Cancer Institute NSW Career Development Fellowship (15/CDF/1-11).
References
- 1.World Health Organization. Obesity and Overweight. (2016). Available online at: http://www.who.int/mediacentre/factsheets/fs311/en/. (Accessed April 9, 2018). [Google Scholar]
- 2.Flegal KM, Carroll MD, Kit BK, Ogden CL. Prevalence of obesity and trends in the distribution of body mass index among US adults, 1999-2010. JAMA (2012) 307:491–7. 10.1001/jama.2012.39 [DOI] [PubMed] [Google Scholar]
- 3.Ng M, Fleming T, Robinson M, Thomson B, Graetz N, Margono C, et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet (2014) 384:766–81. 10.1016/S0140-6736(14)60460-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Begum KS, Sachchithanantham K, De Somsubhra S. Maternal obesity and pregnancy outcome. Clin Exp Obstet Gynecol. (2011) 38:14–20. 10.1097/01.gco.0000045486.15021.C9 [DOI] [PubMed] [Google Scholar]
- 5.Morris MJ. Early life influences on obesity risk: maternal overnutrition and programming of obesity. Exp Rev Endocrinol Metab. (2009) 4:625–37. 10.1586/eem.09.45 [DOI] [PubMed] [Google Scholar]
- 6.Whitaker KL, Jarvis MJ, Beeken RJ, Boniface D, Wardle J. Comparing maternal and paternal intergenerational transmission of obesity risk in a large population-based sample. Am J Clin Nutr. (2010) 91:1560–7. 10.3945/ajcn.2009.28838 [DOI] [PubMed] [Google Scholar]
- 7.Weissgerber TL, Wolfe LA, Davies GA, Mottola MF. Exercise in the prevention and treatment of maternal–fetal disease: a review of the literature. Appl Physiol Nutr Metab. (2006) 31:661–674. 10.1139/h06-060 [DOI] [PubMed] [Google Scholar]
- 8.Mourtakos SP, Tambalis KD, Panagiotakos DB, Antonogeorgos G, Arnaoutis G, Karteroliotis K, et al. Maternal lifestyle characteristics during pregnancy, and the risk of obesity in the offspring: a study of 5,125 children. BMC Pregnancy Childbirth (2015) 15:66. 10.1186/s12884-015-0498-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carter LG, Lewis KN, Wilkerson DC, Tobia CM, Ngo Tenlep SY, Shridas P, et al. Perinatal exercise improves glucose homeostasis in adult offspring. Am J Physiol Endocrinol Metab. (2012) 303:E1061–8. 10.1152/ajpendo.00213.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raipuria M, Bahari H, Morris MJ. Effects of maternal diet and exercise during pregnancy on glucose metabolism in skeletal muscle and fat of weanling rats. PLoS ONE (2015) 10:e0120980. 10.1371/journal.pone.0120980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stanford KI, Lee MY, Getchell KM, So K, Hirshman MF, Goodyear LJ. Exercise before and during pregnancy prevents the deleterious effects of maternal high-fat feeding on metabolic health of male offspring. Diabetes (2015) 64:427–33. 10.2337/db13-1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parekh PJ, Balart LA, Johnson DA. The influence of the gut microbiome on obesity, metabolic syndrome and gastrointestinal disease. Clin Transl Gastroenterol. (2015) 6:e91. 10.1038/ctg.2015.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sonnenburg JL, Bäckhed F. Diet–microbiota interactions as moderators of human metabolism. Nature (2016) 535:56–64. 10.1038/nature18846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. (2010) 107:11971–5. 10.1073/pnas.1002601107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe (2015) 17:690–703. 10.1016/j.chom.2015.04.004 [DOI] [PubMed] [Google Scholar]
- 16.Planer JD, Peng Y, Kau AL, Blanton LV, Ndao IM, Tarr PI, et al. Development of the gut microbiota and mucosal IgA responses in twins and gnotobiotic mice. Nature (2016) 534:263–6. 10.1038/nature17940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prince AL, Ma J, Kannan PS, Alvarez M, Gisslen T, Harris RA, et al. The placental membrane microbiome is altered among subjects with spontaneous preterm birth with and without chorioamnionitis. Am J Obstet Gynecol. (2016) 214:627.e1–627.e16. 10.1016/j.ajog.2016.01.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collado MC, Isolauri E, Laitinen K, Salminen S. Effect of mother's weight on infant's microbiota acquisition, composition, and activity during early infancy: a prospective follow-up study initiated in early pregnancy. Am J Clin Nutr. (2010) 92:1023–30. 10.3945/ajcn.2010.29877 [DOI] [PubMed] [Google Scholar]
- 19.Galley JD, Bailey M, Dush CK, Schoppe-Sullivan S, Christian LM. Maternal obesity is associated with alterations in the gut microbiome in toddlers. PLoS ONE (2014) 9:e113026. 10.1371/journal.pone.0113026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu DM, Antony KM, Ma J, Prince AL, Showalter L, Moller M, et al. The early infant gut microbiome varies in association with a maternal high-fat diet. Genome Med. (2016) 8:77. 10.1186/s13073-016-0330-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myles IA, Fontecilla NM, Janelsins BM, Vithayathil PJ, Segre JA, Datta SK. Parental dietary fat intake alters offspring microbiome and immunity. J Immunol. (2013) 191:3200–9. 10.4049/jimmunol.1301057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma J, Prince AL, Bader D, Hu M, Ganu R, Baquero K, et al. High-fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate model. Nat Commun. (2014) 5:3889. 10.1038/ncomms4889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paul HA, Bomhof MR, Vogel HJ, Reimer RA. Diet-induced changes in maternal gut microbiota and metabolomic profiles influence programming of offspring obesity risk in rats. Sci Rep. (2016) 6:20683. 10.1038/srep20683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Val-Laillet D, Besson M, Guérin S, Coquery N, Randuineau G, Kanzari A, et al. A maternal Western diet during gestation and lactation modifies offspring's microbiota activity, blood lipid levels, cognitive responses, and hippocampal neurogenesis in Yucatan pigs. FASEB J. (2017) 31:2037–49. 10.1096/fj.201601015R [DOI] [PubMed] [Google Scholar]
- 25.Evans CC, LePard KJ, Kwak JW, Stancukas MC, Laskowski S, Dougherty J, et al. Exercise prevents weight gain and alters the gut microbiota in a mouse model of high fat diet-induced obesity. PLoS ONE (2014) 9:e92193. 10.1371/journal.pone.0092193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang SS, Jeraldo PR, Kurti A, Miller ME, Cook MD, Whitlock K, et al. Diet and exercise orthogonally alter the gut microbiome and reveal independent associations with anxiety and cognition. Mol Neurodegener. (2014) 9:36. 10.1186/1750-1326-9-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campbell SC, Wisniewski PJ, Noji M, McGuinness LR, Häggblom MM, Lightfoot SA, et al. The effect of diet and exercise on intestinal integrity and microbial diversity in mice. PLoS ONE (2016) 11:e0150502. 10.1371/journal.pone.0150502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gohir W, Ratcliffe EM, Sloboda DM. Of the bugs that shape us: maternal obesity, the gut microbiome, and long-term disease risk. Pediatr Res. (2014) 77:196–204. 10.1038/pr.2014.169 [DOI] [PubMed] [Google Scholar]
- 29.Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci USA. (2009) 106:2365–70. 10.1073/pnas.0812600106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwiertz A, Taras D, Schäfer K, Beijer S, Bos NA, Donus C, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (2010) 18:190–5. 10.1038/oby.2009.167 [DOI] [PubMed] [Google Scholar]
- 31.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, et al. Richness of human gut microbiome correlates with metabolic markers. Nature (2013) 500:541–6. 10.1038/nature12506 [DOI] [PubMed] [Google Scholar]
- 32.Nadal I, Santacruz A, Marcos A, Warnberg J, Garagorri JM, Moreno LA, et al. Shifts in clostridia, bacteroides and immunoglobulin-coating fecal bacteria associated with weight loss in obese adolescents. Int J Obes. (2009) 33:758–67. 10.1038/ijo.2008.260 [DOI] [PubMed] [Google Scholar]
- 33.Santacruz A, Marcos A, Wärnberg J, Martí A, Martin-Matillas M, Campoy C, et al. Interplay between weight loss and gut microbiota composition in overweight adolescents. Obesity (2009) 17:1906–15. 10.1038/oby.2009.112 [DOI] [PubMed] [Google Scholar]
- 34.Furet JP, Kong LC, Tap J, Poitou C, Basdevant A, Bouillot JL, et al. Differential adaptation of human gut microbiota to bariatric surgery–induced weight loss. Diabetes (2010) 59:3049–57. 10.2337/db10-0253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kishino S, Takeuchi M, Park SB, Hirata A, Kitamura N, Kunisawa J, et al. Polyunsaturated fatty acid saturation by gut lactic acid bacteria affecting host lipid composition. Proc Natl Acad Sci USA. (2013) 10:17808–13. 10.1073/pnas.1312937110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaakoush NO, Martire SI, Raipuria M, Mitchell HM, Nielsen S, Westbrook RF, et al. Alternating or continuous exposure to cafeteria diet leads to similar shifts in gut microbiota compared to chow diet. Mol Nutr Food Res. (2017) 61:1500815. 10.1002/mnfr.201500815 [DOI] [PubMed] [Google Scholar]
- 37.Beilharz J, Kaakoush NO, Maniam J, Morris MJ. Cafeteria diet and probiotic therapy: cross talk among memory, neuroplasticity, serotonin receptors and gut microbiota in the rat. Mol Psychiatr. (2017) 23:351–61. 10.1038/mp.2017.38 [DOI] [PubMed] [Google Scholar]
- 38.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature (2006) 444:1027–31. 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- 39.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science (2013) 341:1241214. 10.1126/science.1241214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petriz BA, Castro AP, Almeida JA, Gomes CP, Fernandes GR, Kruger RH, et al. Exercise induction of gut microbiota modifications in obese, non-obese and hypertensive rats. BMC Genom. (2014) 15:511. 10.1186/1471-2164-15-511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Batacan RB, Fenning AS, Dalbo VJ, Scanlan AT, Duncan MJ, Moore RJ, et al. A gut reaction: the combined influence of exercise and diet on gastrointestinal microbiota in rats. J Appl Microbiol. (2017) 122:1627–38. 10.1111/jam.13442 [DOI] [PubMed] [Google Scholar]
- 42.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science (2011) 334:105–8. 10.1126/science.1208344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martínez I, Lattimer JM, Hubach KL, Case JA, Yang J, Weber CG, et al. Gut microbiome composition is linked to whole grain-induced immunological improvements. ISME J. (2013) 7:269–80. 10.1038/ismej.2012.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tims S, Derom C, Jonkers DM, Vlietinck R, Saris WH, Kleerebezem M, et al. Microbiota conservation and BMI signatures in adult monozygotic twins. ISME J. (2013) 7:707–17. 10.1038/ismej.2012.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity (2013) 39:372–85. 10.1016/j.immuni.2013.08.003 [DOI] [PubMed] [Google Scholar]
- 46.Liang H, Dai Z, Liu N, Ji Y, Chen J, Zhang Y, et al. Dietary L-tryptophan modulates the structural and functional composition of the intestinal microbiome in weaned piglets. Front Microbiol. (2018) 9:1736. 10.3389/fmicb.2018.01736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilck N, Matus MG, Kearney SM, Olesen SW, Forslund K, Bartolomaeus H, et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature (2017) 551:585–9. 10.1038/nature24628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ong KK, Ahmed ML, Emmett PM, Preece MA, Dunger DB. Association between postnatal catch-up growth and obesity in childhood: prospective cohort study. BMJ (2000) 320:967–71. 10.1136/bmj.320.7240.967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jimenez-Chillaron JC, Hernandez-Valencia M, Lightner A, Faucette RR, Reamer C, Przybyla R, et al. Reductions in caloric intake and early postnatal growth prevent glucose intolerance and obesity associated with low birthweight. Diabetologia (2006) 49:1974–84. 10.1007/s00125-006-0311-7 [DOI] [PubMed] [Google Scholar]
- 50.Wang J, Tang H, Wang X, Zhang X, Zhang C, Zhang M, et al. The structural alteration of gut microbiota in low-birth-weight mice undergoing accelerated postnatal growth. Sci Rep. (2016) 6:27780. 10.1038/srep27780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pannaraj PS, Li F, Cerini C, Bender JM, Yang S, Rollie A, et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. (2017) 171:647–54. 10.1001/jamapediatrics.2017.0378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fouhy F, Clooney AG, Stanton C, Claesson MJ, Cotter PD. 16S rRNA gene sequencing of mock microbial populations-impact of DNA extraction method, primer choice and sequencing platform. BMC Microbiol. (2016) 16:123. 10.1186/s12866-016-0738-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rintala A, Pietilä S, Munukka E, Eerola E, Pursiheimo JP, Laiho A, et al. Gut microbiota analysis results are highly dependent on the 16S rRNA gene target region, whereas the impact of DNA extraction is minor. J Biomol Tech. (2017) 28:19–30. 10.7171/jbt.17-2801-003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Puiman P, Stoll B. Animal models to study neonatal nutrition in humans. Curr Opin Clin Nutr Metab Care (2008) 11:601–6. 10.1097/MCO.0b013e32830b5b15 [DOI] [PubMed] [Google Scholar]
- 55.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. J Appl Environ Microbiol. (2009) 75:7537–41. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. J Appl Environ Microbiol. (2013) 79:5112–20. 10.1128/AEM.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zakrzewski M, Proietti C, Ellis JJ, Hasan S, Brion MJ, Berger B, et al. Calypso: a user-friendly web-server for mining and visualizing microbiome–environment interactions. J Bioinform. (2016) 33:782–3. 10.1093/bioinformatics/btw725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clarke KR. Non-parametric multivariate analyses of changes in community structure. Austral Ecol. (1993) 18:117–43. 10.1111/j.1442-9993.1993.tb00438.x [DOI] [Google Scholar]
- 59.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. (2011) 12:R60. 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Afgan E, Baker D, Van den Beek M, Blankenberg D, Bouvier D, Cech M, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. (2016) 44:W3–10. 10.1093/nar/gkw343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE (2013) 8:e61217. 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. (2013) 31:814–21. 10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]