Abstract
Evidence indicates a complex link between microbiota, tumor characteristics, and host immunity in the tumor microenvironment. In experimental studies, bifidobacteria appear to modulate intestinal epithelial cell differentiation. Accumulating evidence suggests that bifidobacteria may enhance the antitumor immunity and efficacy of immunotherapy. We hypothesized that the amount of bifidobacteria in colorectal carcinoma tissue might be associated with tumor differentiation and higher immune response to colorectal cancer. Using a molecular pathologic epidemiology database of 1313 rectal and colon cancers, we measured the amount of Bifidobacterium DNA in carcinoma tissue by a quantitative PCR assay. The multivariable regression model was used to adjust for potential confounders, including microsatellite instability status, CpG island methylator phenotype, long-interspersed nucleotide element-1 methylation, and KRAS, BRAF, and PIK3CA mutations. Intratumor bifidobacteria were detected in 393 cases (30%). The amount of bifidobacteria was associated with the extent of signet ring cells (P = 0.002). Compared with Bifidobacterium-negative cases, multivariable odd ratios for the extent of signet ring cells were 1.29 (95% CI, 0.74–2.24) for Bifidobacterium-low cases and 1.87 (95% CI, 1.16–3.02) for Bifidobacterium-high cases (Ptrend = 0.01). The association between intratumor bifidobacteria and signet ring cells suggests a possible role of bifidobacteria in determining distinct tumor characteristics or as an indicator of dysfunctional mucosal barrier in colorectal cancer.
The human gut microbiome is composed of >30 trillion microorganisms, and it is under intense investigation because of its substantial role in intestinal tumorigenesis as well as immune modulation of the tumor microenvironment and response to immunotherapy.1, 2, 3, 4, 5, 6, 7, 8, 9 A growing body of evidence indicates relationships of Fusobacterium nucleatum in colorectal cancer tissue with distinct features, including high-level microsatellite instability (MSI), low-level CD3+ T-cell density, and worse survival.10, 11 Our incomplete knowledge of the interactions between microbes, distinctive tumor features, and the host immune system highlights the critical need for transdisciplinary integrated analyses of microorganisms and cancer.12, 13, 14
Colon and rectal cancers represent heterogeneous sets of neoplasms with differing combinations of genetic and epigenetic alterations, the accrual of which is influenced by complicated interplay between tumor cells, host cells, and microorganisms.14, 15, 16, 17, 18, 19, 20, 21 Accumulating evidence suggests that the presence of members of Bifidobacterium genus in the gut lumen may suppress colorectal carcinogenesis through prevention of enteropathogenic infection and inhibition of secondary bile acid production.22, 23, 24 In in vitro and in vivo experiments, bacteria, including bifidobacteria, appear to modulate intestinal epithelial cell differentiation factors.25, 26 Experimental studies have shown that bifidobacteria activate antitumor immunity and boost the efficacy of immunotherapy blocking CD274 (PDCD1 ligand 1).3, 27 Immune cells in the tumor microenvironment may be the key players in regulating tumor progression,13, 14, 28, 29, 30, 31, 32, 33 indicating that the assessment of microorganisms, host immunity, and cancer cells in the tumor microenvironment is increasingly significant in translational research and clinical practice for tumor classification. Therefore, we hypothesized that the amount of bifidobacteria in colorectal carcinoma tissue might be associated with tumor differentiation and higher immune response to colorectal cancer.
To test this hypothesis, a molecular pathologic epidemiology database of colorectal cancer cases within two large US prospective cohort studies was used, and the amount of Bifidobacterium DNA in relation to tumor differentiation and immune response in the tumor microenvironment were examined. In addition, as a secondary exploratory analysis, the prognostic association of the amount of bifidobacteria in colorectal carcinoma was assessed.
Materials and Methods
Study Population
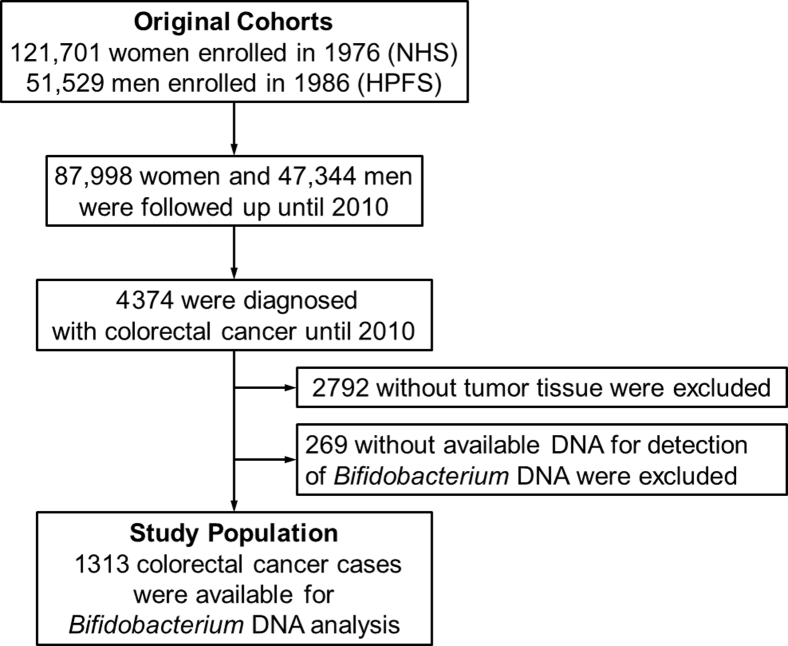
Data from two prospective cohort studies in the United States, the Nurses' Health Study (121,701 women aged 30 to 55 years, followed up since 1976) and the Health Professionals Follow-Up Study (51,529 men aged 40 to 75 years, followed up since 1986), were collected.34 Study participants have completed follow-up questionnaires to submit information on lifestyle factors and medical history, including colorectal cancer, every 2 years. The National Death Index was used to ascertain deaths of study participants and identify unreported lethal colorectal cancer cases. Participating physicians reviewed medical records to confirm diagnosis of colorectal cancer and to record tumor characteristics (eg, size, location, and the American Joint Committee on Cancer TNM classification) and causes of death for deceased participants. Formalin-fixed, paraffin-embedded tissue blocks were gathered from hospitals in which participants diagnosed with colorectal cancer had undergone tumor resection. A total of 1313 colorectal carcinoma patients with available tissue materials were included for measurement of the amount of intratumor Bifidobacterium DNA (Figure 1). Both colon and rectal carcinomas were included on the basis of the colorectal continuum model.35, 36 Patients were followed up until death or the end of follow-up (January 1, 2014, for the Health Professionals Follow-Up Study; May 31, 2014, for the Nurses' Health Study), whichever came first. Informed consent was obtained from every study participant. This study was approved by the institutional review boards at Harvard T.H. Chan School of Public Health and Brigham and Women's Hospital (Boston, MA).
Figure 1.
Flow diagram of study population in the Nurses' Health Study (NHS) and the Health Professionals Follow-Up Study (HPFS).
Histopathologic Analysis
A single pathologist (S.O.), blinded to other data, reviewed hematoxylin and eosin–stained tissue sections of all colorectal carcinoma cases and recorded pathologic features, including tumor differentiation, patterns and degrees of lymphocytic reactions, tumor growth pattern, and the extent of signet ring cells and extracellular mucin.37 Tumor differentiation was categorized as well to moderate or poor (>50% versus ≤50% glandular area, respectively). The proportions of signet ring cell component and extracellular mucinous component were recorded as percentage and categorized as 0%, 1% to 50%, or ≥51% of the tumor volume, as previously described.37 The cutoff value of 50% of signet ring cell or extracellular mucinous component was used, on the basis of the World Health Organization Classification of Tumors of the Digestive System, which defines signet ring cell carcinoma or mucinous carcinoma as carcinoma with >50% of signet ring cell or extracellular mucinous component, respectively.38 Any colorectal cancer can have signet ring cell or extracellular mucinous component to any extent (from 0% to 100%). Any colorectal cancer can have both signet ring cell component and mucinous component, either one of them, or neither of them.
Histopathologic lymphocytic reaction to tumor was evaluated, as previously described.31 Four components of lymphocytic reaction were examined, including tumor-infiltrating lymphocytes, intratumoral periglandular reaction, peritumoral lymphocytic reaction, and Crohn-like lymphoid reaction. Tumor-infiltrating lymphocytes were defined as lymphocytes on top of cancer cells. Intratumoral periglandular reaction was defined as lymphocytic reaction in tumor stroma within a tumor mass. Peritumoral lymphocytic reaction was defined as discrete lymphoid reaction surrounding a tumor mass. Crohn-like lymphoid reaction was defined as transmural lymphoid reaction. Each of the four components was graded as negative/low, intermediate, or high.
Quantitative PCR for Bifidobacterium Genus and F. nucleatum
Genomic DNA was extracted from colorectal carcinoma tissue in whole-tissue sections of archival formalin-fixed, paraffin-embedded tissue blocks using the QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany). Custom TaqMan primer-probe sets (Applied Biosystems, Foster City, CA) were used for the 16S ribosomal RNA gene DNA sequence of Bifidobacterium at the genus level and for the reference gene, 16S, as described elsewhere.27, 39, 40 Each reaction contained 80 ng of genomic DNA and was assayed in 20-μL reactions containing 1× final concentration TaqMan Environmental Master Mix 2.0 (Applied Biosystems) and each TaqMan Gene Expression Assay (Applied Biosystems), in a 96-well optical PCR plate. The StepOnePlus Real-Time PCR System (Applied Biosystems) was used for amplification and detection of DNA using the following reaction conditions: 10 minutes at 95°C and 45 cycles of 15 seconds at 95°C and 1 minute at 60°C. The primer and probe sequences for each TaqMan Gene Expression Assay were as follows: Bifidobacterium forward primer, 5′-CGGGTGAGTAATGCGTGACC-3′; Bifidobacterium reverse primer, 5′-TGATAGGACGCGACCCCA-3′; Bifidobacterium FAM probe, 5′-CTCCTGGAAACGGGTG-3′; universal 16S forward primer, 5′-CGGTGAATACGTTCCCGG-3′; universal 16S reverse primer, 5′-TACGGCTACCTTGTTACGACTT-3′; and universal 16S FAM probe, 5′-CTTGTACACACCGCCCGTC-3′. In colorectal carcinoma cases with detectable bifidobacteria, the cycle threshold (CT) values in the quantitative PCR for Bifidobacterium DNA and 16S decreased linearly with the amount of input DNA (in a log scale) from the same specimen (r2 > 0.97) (Supplemental Figure S1). The interassay CV of CT values from the same specimen in five different batches was 1% or less for all targets in this validation study using six colorectal carcinomas (Supplemental Table S1). Each specimen was analyzed in duplicate for each target in a single batch, and the mean of the two CT values for each target was used. The amount of bifidobacteria in each specimen was calculated as a relative unitless value normalized with 16S using the 2−ΔCt method (where ΔCT = the average CT value of Bifidobacterium DNA − the average CT value of 16S), as previously described.41 Cases with detectable bifidobacteria were categorized as low versus high based on the median cut point amount of bifidobacteria, whereas cases without detectable bifidobacteria were categorized as negative.
Quantitative PCR was used for F. nucleatum to measure the amount of tissue F. nucleatum DNA, as previously described.10, 11 We categorized colorectal carcinoma cases with detectable F. nucleatum DNA as low or high in relation to the median cut point amount of F. nucleatum DNA.10, 11
Analysis of MSI, DNA Methylation, and KRAS, BRAF, and PIK3CA Mutations
Genomic DNA was extracted from colorectal carcinoma tissue in whole-tissue sections of archival formalin-fixed, paraffin-embedded tissue blocks. MSI status was determined using PCR of 10 microsatellite markers (D2S123, D5S346, D17S250, BAT25, BAT26, BAT40, D18S55, D18S56, D18S67, and D18S487), and MSI-high was defined as presence of instability in ≥30% of the markers, as previously described.35, 42 Methylation status of eight CpG island methylator phenotype (CIMP)–specific promoters (CACNA1G, CDKN2A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and SOCS1) was determined using MethyLight assay on bisulfite-treated DNA.35 CIMP-high was defined as six or more methylated promoters of eight promoters, and CIMP-low/negative was defined as zero to five methylated promoters.35, 42 Methylation levels at long-interspersed nucleotide element-1 were measured by PCR on bisulfite-treated DNA and pyrosequencing.35 PCR and pyrosequencing were performed for KRAS (codons 12, 13, 61, and 146),43 BRAF (codon 600),42 and PIK3CA (exons 9 and 20).44
Immunohistochemistry
As described previously,45 tissue microarrays that included up to four cores of colorectal cancer tissue from each case and up to two cores of normal adjacent tissue from the same case were constructed. Immunohistochemistry was performed for CD3, CD8, CD45RO (one of PTPRC protein isoforms), and FOXP3, as previously described.30 An automated scanning microscope and the Ariol image analysis system (Genetix, San Jose, CA) were used to measure densities (cells/mm2) of CD3+, CD8+, CD45RO+, and FOXP3+ cells in colorectal cancer tissue. Immunohistochemical analyses for CD274 (PDCD1 ligand 1), PTGS2 (cyclooxygenase-2), and nuclear CTNNB1 (β-catenin) expression were performed using an anti-CD274 antibody (dilution, 1:50; eBioscience, San Diego, CA), an anti-PTGS2 antibody (dilution, 1:300; Cayman Chemical, Ann Arbor, MI), and an anti-CTNNB1 antibody (dilution, 1:400; BD Transduction Laboratories, Franklin Lakes, NJ), respectively, as previously described.16, 46, 47
Statistical Analysis
All statistical analyses were conducted using SAS software version 9.4 (SAS Institute, Cary, NC), and all P values were two sided. The two-sided α level of 0.005 was used.48, 49 The primary hypothesis testing was an assessment of the associations of the amount of Bifidobacterium DNA (negative, low, and high; as an ordinal predictor variable) with the following variables (as an outcome variable): tumor differentiation, extent of signet ring cells, extent of extracellular mucin, the four histopathologic lymphocytic reaction patterns (tumor-infiltrating lymphocytes, intratumoral periglandular reaction, peritumoral lymphocytic reaction, and Crohn-like lymphoid reaction), and the density of T cells in colorectal cancer tissue (CD3+, CD8+, CD45RO+, and FOXP3+ cells).
Multivariable ordinal logistic regression analyses were performed to control for potential confounders. The multivariable ordinal logistic model initially included sex (female versus male), age at diagnosis (continuous), year of diagnosis (continuous), family history of colorectal cancer in any first-degree relative (present versus absent), tumor location (proximal colon versus distal colon versus rectum), MSI status (MSI-high versus non–MSI-high), CIMP status (high versus low/negative), long-interspersed nucleotide element-1 methylation level (continuous), KRAS mutation (mutant versus wild type), BRAF mutation (mutant versus wild type), and PIK3CA mutation (mutant versus wild type). A backward elimination was conducted with a threshold P of 0.05 to select variables for the final models. Cases with missing data were included in the majority category of a given categorical covariate to limit the degrees of freedom: family history of colorectal cancer in a first-degree relative (0.4%), tumor location (0.2%), MSI (4.3%), CIMP (8.2%), KRAS (9.4%), and BRAF (3.5%). For the cases with missing data on long-interspersed nucleotide element-1 methylation (6.5%), a separate indicator variable was assigned. For cases with missing information on PIK3CA mutation (10.3%), a separate missing indicator variable was assigned. It was confirmed that excluding the cases with missing information in any of the covariates did not substantially alter results (data not shown). The proportional odds assumption was assessed in an ordinal logistic regression model, which was generally satisfied (P > 0.90).
To compare clinicopathologic characteristics across ordinal categories of the amount of tissue Bifidobacterium, the χ2 test was used for categorical variables and an analysis of variance assuming equal variances was used for continuous variables. All of those cross-sectional analyses were secondary and exploratory, and hence the two-sided α level of 0.005 was used.48
Kaplan-Meier analysis was conducted to compare survival between patient groups. For analyses of colorectal cancer–specific mortality, deaths as a result of other causes were censored. To control for potential confounders, Cox proportional hazards regression analysis was performed, and hazard ratio was calculated for mortality. The multivariable Cox proportional hazards regression models initially included tumor differentiation (well to moderate versus poor) and disease stage (I/II versus III/IV/missing) in addition to the same set of covariates as the multivariable ordinal logistic regression model. A backward stepwise elimination was performed with a threshold of P = 0.05 to select covariates for the final model. Cases with missing data on tumor differentiation (0.4%) were included in the majority category, and cases with missing data on other covariates were dealt with as in the multivariable ordinal logistic regression model. The proportionality of hazards assumption was assessed by a time-varying covariate, which was an interaction term of survival time and the amount of bifidobacteria (P > 0.1).
In ordinal logistic and Cox regression analyses, the inverse probability weighting (IPW) method was applied to reduce the potential bias attributable to the availability of tumor tissue.50, 51, 52 Using the entire data set of colorectal cancer cases (regardless of available tissue), each patient with complete data was weighted by the inverse probability of the availability of tumor tissue. First, to estimate the probability of study inclusion, the multivariable logistic regression model that initially included sex (female versus male), age at diagnosis (continuous; a linear term and a squared term), year of diagnosis (continuous; a linear term and a squared term), family history of colorectal cancer (absent versus present versus missing), prediagnosis body mass index (<25 versus 25 to 29.9 versus ≥30 kg/m2 versus missing), tumor location (cecum versus ascending colon versus transverse colon versus descending colon versus sigmoid colon versus rectum versus missing), and disease stage (I versus II versus III versus IV versus missing) was constructed. After a selection procedure, the final model included sex, year of diagnosis (a linear term and a squared term), tumor location, and disease stage. Weights >95th percentile were truncated and set to the value of the 95th percentile to reduce outlier effects.52 It was confirmed that results without weight truncation did not change substantially (data not shown). A multivariable IPW-adjusted ordinal logistic regression analysis was performed. Cumulative survival probabilities were estimated using the IPW-adjusted Kaplan-Meier method, and a linear trend in survival probabilities across ordinal categories of the amount of Bifidobacterium DNA was assessed using the weighted log-rank test for trend.53 The logistic and Cox regression analyses without IPW yielded similar results to the IPW-adjusted model (Supplemental Tables S2 and S3).
Results
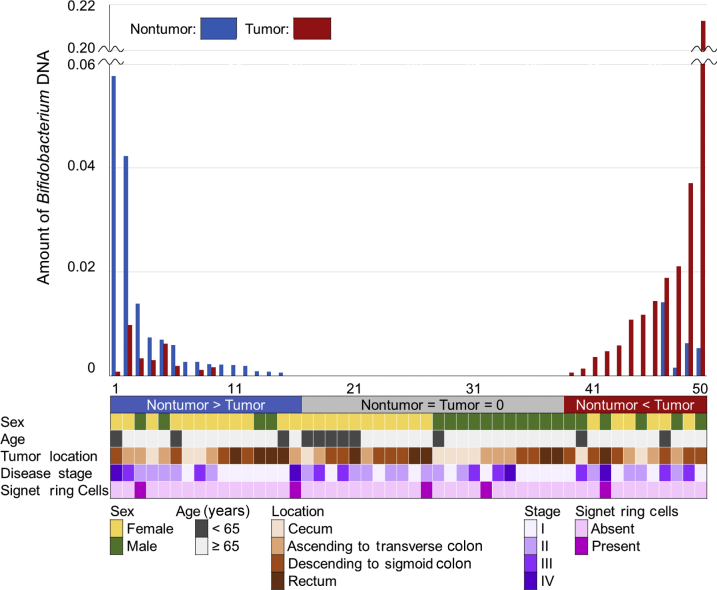
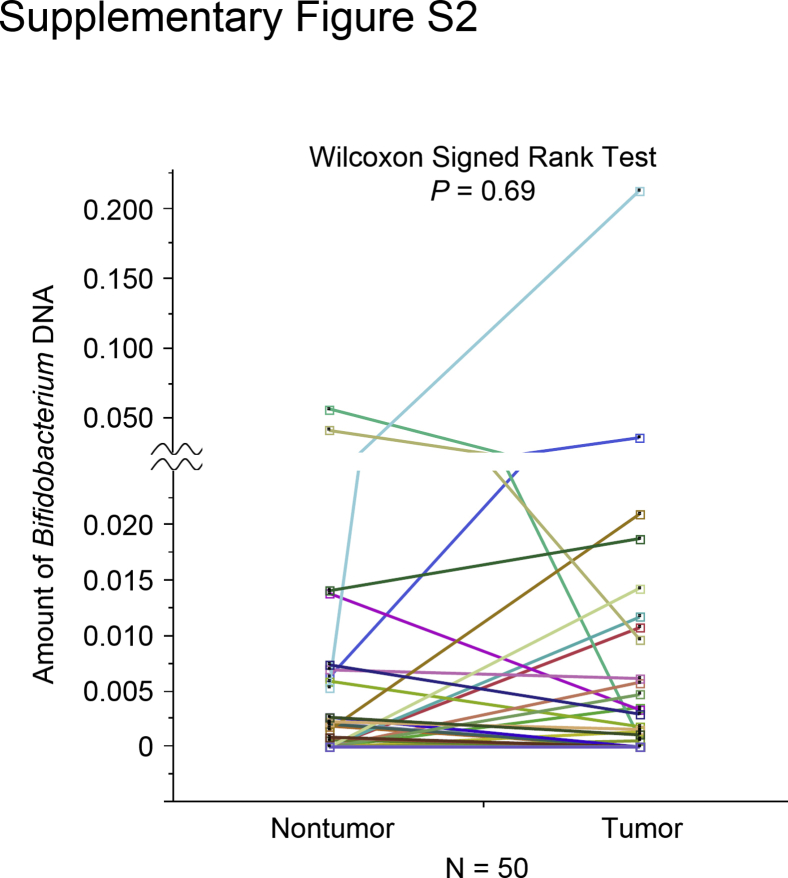
Tumor tissues were analyzed from 1313 incident colorectal carcinoma cases within the Nurses' Health Study and the Health Professionals Follow-Up Study using the quantitative PCR assay, as described elsewhere.27, 39, 40 Members of the Bifidobacterium genus were detected in colorectal carcinoma in 393 cases (30%). Bifidobacterium DNA was measured in tumor and adjacent nontumor tissue pairs from 50 colorectal cancer patients. Bifidobacteria were detected in 20 tumor samples (40%) and 20 nontumor samples (40%). Among the 50 pairs of carcinoma and adjacent nontumor tissue, bifidobacteria were more abundant in carcinomas in 12 pairs, and less abundant in carcinomas in 16 cases. Bifidobacteria were absent in both in the remaining 22 pairs (Figure 2 and Supplemental Figure S2). The difference of bifidobacteria between carcinoma and adjacent nontumor tissue was not associated with sex, age, tumor location, disease stage, or presence of signet ring cells (P > 0.45).
Figure 2.
The amount of Bifidobacterium DNA in 50 pairs of colorectal carcinoma and adjacent nontumor tissue samples. Bars represent the amount of bifidobacteria in carcinoma (red) and paired adjacent nontumor tissue (blue).
Clinical, pathologic, and molecular features are summarized according to the amount of bifidobacteria in colorectal carcinoma tissue overall or by cohort in Table 1 and Supplemental Table S4, respectively. The amount of bifidobacteria was not significantly associated with any of the characteristics examined (P > 0.02; with the α level of 0.005).
Table 1.
Clinical, Pathologic, and Molecular Characteristics of Colorectal Cancer Cases according to the Amount of Bifidobacterium DNA in Colorectal Cancer Tissue
| Characteristic | All cases (n = 1313) | Amount of Bifidobacterium DNA in colorectal cancer tissue |
P value∗ | ||
|---|---|---|---|---|---|
| Negative (n = 920) | Low (n = 197) | High (n = 196) | |||
| Sex | 0.025 | ||||
| Female (NHS) | 744 (57) | 515 (56) | 102 (52) | 127 (65) | |
| Male (HPFS) | 569 (43) | 405 (44) | 95 (48) | 69 (35) | |
| Age in years, means ± SD | 69.3 ± 8.9 | 69.2 ± 8.8 | 68.9 ± 9.8 | 69.9 ± 8.9 | 0.48 |
| Year of diagnosis | 0.089 | ||||
| 1995 or before | 449 (34) | 317 (34) | 74 (38) | 58 (30) | |
| 1996–2000 | 399 (30) | 286 (31) | 61 (31) | 52 (27) | |
| 2001–2008 | 465 (35) | 317 (34) | 62 (31) | 86 (44) | |
| Family history of colorectal cancer in first-degree relative(s) | 0.88 | ||||
| Absent | 1055 (81) | 739 (81) | 160 (82) | 156 (80) | |
| Present | 253 (19) | 177 (19) | 36 (18) | 40 (20) | |
| Tumor location | 0.46 | ||||
| Cecum | 223 (17) | 152 (17) | 41 (21) | 30 (15) | |
| Ascending to transverse colon | 404 (31) | 286 (31) | 54 (27) | 64 (33) | |
| Descending to sigmoid colon | 400 (31) | 290 (32) | 55 (28) | 55 (28) | |
| Rectum | 283 (22) | 189 (21) | 47 (24) | 47 (24) | |
| pT stage (depth of tumor invasion) | 0.39 | ||||
| pT1 (submucosa) | 131 (11) | 90 (11) | 20 (12) | 21 (12) | |
| pT2 (muscularis propria) | 256 (21) | 194 (23) | 28 (16) | 34 (20) | |
| pT3 (subserosa) | 755 (63) | 533 (62) | 112 (65) | 110 (64) | |
| pT4 (serosa or other organs) | 60 (5.0) | 39 (4.6) | 13 (7.5) | 8 (4.6) | |
| pN stage (number of positive lymph nodes) | 0.44 | ||||
| pN0 (0) | 736 (63) | 529 (64) | 109 (63) | 98 (58) | |
| pN1 (1–3) | 267 (23) | 182 (22) | 37 (22) | 48 (29) | |
| pN2 (≥4) | 163 (14) | 115 (14) | 26 (15) | 22 (13) | |
| AJCC disease stage | 0.85 | ||||
| I | 306 (26) | 224 (26) | 41 (24) | 41 (24) | |
| II | 389 (33) | 277 (33) | 59 (35) | 53 (30) | |
| III | 350 (29) | 243 (29) | 52 (31) | 55 (32) | |
| IV | 151 (13) | 108 (13) | 18 (11) | 25 (14) | |
| Tumor size in mm, means ± SD | 43.0 ± 20.9 | 43.0 ± 20.9 | 45.4 ± 22.8 | 40.2 ± 18.7 | 0.076 |
| Tumor necrosis | 0.21 | ||||
| 0 | 880 (70) | 621 (69) | 130 (70) | 129 (71) | |
| 1–10 | 221 (18) | 160 (18) | 25 (13) | 36 (20) | |
| 11–20 | 70 (5.5) | 47 (5.3) | 13 (7.0) | 10 (5.5) | |
| ≥21 | 92 (7.3) | 66 (7.4) | 19 (10) | 7 (3.9) | |
| Tumor growth pattern | 0.21 | ||||
| Expansile | 375 (33) | 283 (35) | 46 (29) | 46 (29) | |
| Intermediate | 599 (53) | 425 (52) | 91 (57) | 83 (53) | |
| Infiltrative | 158 (14) | 105 (13) | 24 (15) | 29 (18) | |
| MSI status | 0.15 | ||||
| Non–MSI-high | 1041 (83) | 744 (84) | 148 (79) | 149 (80) | |
| MSI-high | 216 (17) | 140 (16) | 39 (21) | 37 (20) | |
| CIMP status | 0.45 | ||||
| Low/negative | 979 (81) | 697 (82) | 146 (82) | 136 (78) | |
| High | 227 (19) | 156 (18) | 32 (18) | 39 (22) | |
| Percent LINE-1 methylation level, means ± SD | 63.6 ± 10.2 | 63.2 ± 10.1 | 64.2 ± 10.8 | 64.9 ± 9.5 | 0.11 |
| KRAS mutation | 0.13 | ||||
| Wild type | 690 (58) | 477 (57) | 110 (65) | 103 (58) | |
| Mutant | 500 (42) | 365 (43) | 59 (35) | 76 (42) | |
| BRAF mutation | 0.26 | ||||
| Wild type | 1068 (84) | 756 (85) | 161 (84) | 151 (80) | |
| Mutant | 199 (16) | 132 (15) | 30 (16) | 37 (20) | |
| PIK3CA mutation | 0.19 | ||||
| Wild type | 992 (84) | 683 (83) | 157 (89) | 152 (84) | |
| Mutant | 186 (16) | 138 (17) | 20 (11) | 28 (16) | |
| Fusobacterium nucleatum DNA | 0.038 | ||||
| Negative | 925 (87) | 676 (88) | 113 (80) | 136 (87) | |
| Low | 70 (6.6) | 46 (6.0) | 11 (7.8) | 13 (8.3) | |
| High | 69 (6.5) | 44 (5.7) | 17 (12) | 8 (5.1) | |
| CD274 (PD-L1) expression score | 0.11 | ||||
| 0 | 86 (11) | 54 (9.8) | 12 (12) | 20 (18) | |
| 1 | 216 (28) | 162 (30) | 24 (24) | 30 (26) | |
| 2 | 201 (26) | 140 (26) | 27 (26) | 34 (30) | |
| 3 | 221 (29) | 158 (29) | 35 (34) | 28 (25) | |
| 4 | 41 (5.4) | 35 (6.4) | 4 (3.9) | 2 (1.8) | |
| PTGS2 (cyclooxygenase-2) expression | 0.18 | ||||
| Negative | 440 (39) | 297 (38) | 65 (38) | 78 (45) | |
| Positive | 695 (61) | 495 (62) | 105 (62) | 95 (55) | |
| Nuclear CTNNB1 (β-catenin) expression | 0.36 | ||||
| Negative | 822 (63) | 575 (62) | 117 (59) | 130 (66) | |
| Positive | 491 (37) | 345 (38) | 80 (41) | 66 (34) | |
Data are given as n (%) of each group unless otherwise indicated. Percentage indicates the proportion of patients with a specific clinical, pathologic, or molecular characteristic among all patients or in strata of the amount of Bifidobacterium DNA in colorectal cancer tissue.
AJCC, American Joint Committee on Cancer; CIMP, CpG island methylator phenotype; HPFS, Health Professionals Follow-Up Study; LINE-1, long-interspersed nucleotide element-1; MSI, microsatellite instability; NHS, Nurses' Health Study; PD-L1, PDCD1 ligand 1.
To assess associations between the ordinal categories (negative, low, and high) of the amount of Bifidobacterium DNA in colorectal cancer tissue and categorical data, the χ2 test was performed. To compare age, tumor size, and LINE-1 methylation level, an analysis of variance was performed. Two-sided α level was set at 0.005.
Table 2 shows the distribution of colorectal carcinoma cases according to the amount of bifidobacteria in colorectal carcinoma tissue, and tumor differentiation, extent of signet ring cells, or extent of extracellular mucin. The amount of bifidobacteria correlated with the extent of signet ring cells (P = 0.002; with the α level of 0.005).
Table 2.
Distribution of Colorectal Cancer Cases according to the Amount of Bifidobacterium DNA, and Tumor Differentiation, Extent of Signet Ring Cells, or Extent of Extracellular Mucin
| Characteristic | All cases (n = 1313) | Amount of Bifidobacterium DNA in colorectal cancer tissue |
P value∗ | ||
|---|---|---|---|---|---|
| Negative (n = 920) | Low (n = 197) | High (n = 196) | |||
| Tumor differentiation | 0.25 | ||||
| Well to moderate | 1175 (90) | 832 (91) | 172 (88) | 171 (87) | |
| Poor | 133 (10) | 85 (9.3) | 23 (12) | 25 (13) | |
| Extent of signet ring cells | 0.002 | ||||
| 0 | 1114 (87) | 799 (89) | 164 (87) | 151 (80) | |
| 1–50 | 145 (11) | 94 (10) | 17 (9.0) | 34 (18) | |
| ≥51 | 19 (1.5) | 9 (1.0) | 7 (3.7) | 3 (1.6) | |
| Extent of extracellular mucin (%) | 0.30 | ||||
| 0 | 757 (59) | 535 (59) | 119 (63) | 103 (55) | |
| 1–50 | 369 (29) | 260 (29) | 45 (24) | 64 (34) | |
| ≥51 | 153 (12) | 107 (12) | 25 (13) | 21 (11) | |
Data are given as n (percentage) of each group unless otherwise indicated. Percentage indicates the proportion of patients with a specific clinical, pathologic, or molecular characteristic among all patients or in strata of the amount of Bifidobacterium DNA in colorectal cancer tissue.
To assess associations between the ordinal categories (negative, low, and high) of the amount of Bifidobacterium DNA in colorectal cancer tissue and categorical data, the χ2 test was performed (except for the extent of signet ring cells, for which Fisher's exact test was used). Two-sided α level was set at 0.005.
In the primary hypothesis testing, an ordinal logistic regression analysis was used to assess the association of the amount of bifidobacteria with the extent of signet ring cells (Table 3 and Supplemental Tables S2 and S5). In univariable analysis, compared with Bifidobacterium-negative cases, univariable odds ratios for the extent of signet ring cells were 1.26 (95% CI, 0.75–2.10) for Bifidobacterium-low cases and 1.99 (95% CI, 1.28–3.09) for Bifidobacterium-high cases (Ptrend = 0.003; with the α level of 0.005). In multivariable analysis, compared with Bifidobacterium-negative cases, multivariable odds ratios for the extent of signet ring cells were 1.29 (95% CI, 0.74–2.24) for Bifidobacterium-low cases and 1.87 (95% CI, 1.16–3.02) for Bifidobacterium-high cases (Ptrend = 0.01; with the α level of 0.005).
Table 3.
IPW-Adjusted Ordinal Logistic Regression Analysis to Assess the Association of the Amount of Bifidobacterium DNA (Predictor) with the Extent of Signet Ring Cells (Outcome)
| Variable | Extent of signet ring cells |
|
|---|---|---|
| Univariable OR (95% CI)∗ | Multivariable OR (95% CI)∗† | |
| Amount of Bifidobacterium DNA in colorectal cancer tissue | ||
| Negative | 1 (referent) | 1 (referent) |
| Low | 1.26 (0.75–2.10) | 1.29 (0.74–2.24) |
| High | 1.99 (1.28–3.09) | 1.87 (1.16–3.02) |
| Ptrend‡ | 0.003 | 0.01 |
IPW, inverse probability weighting; OR, odds ratio.
IPW was applied to reduce a bias attributable to the availability of tumor tissue after cancer diagnosis (Statistical Analysis).
The multivariable ordinal logistic regression model initially included age, sex, year of diagnosis, family history of colorectal cancer, tumor location, microsatellite instability, CpG island methylator phenotype, KRAS, BRAF, and PIK3CA mutations, and long-interspersed nucleotide element-1 methylation level. A backward elimination with a threshold of P = 0.05 was used to select variables for the final model. The variables that remained in the final models are shown in Supplemental Table S5.
Ptrend was calculated by the linear trend across the ordinal categories of the amount of Bifidobacterium DNA (negative, low, and high) in the IPW-adjusted logistic regression model for the extent of signet ring cells (absent, low, and high, as an ordinal outcome variable).
Table 4 shows the distribution of colorectal carcinoma cases according to the amount of bifidobacteria in colorectal carcinoma tissue and T-cell densities or histologic lymphocytic reaction patterns. The amount of bifidobacteria was not significantly associated with any of the T-cell densities and histologic lymphocytic reaction patterns (P > 0.08; with the α level of 0.005).
Table 4.
Distribution of Colorectal Cancer Cases according to the Amount of Bifidobacterium DNA and the Densities of T Cells or Histologic Lymphocytic Reaction Patterns
| Characteristic | All cases | Amount of Bifidobacterium DNA in colorectal cancer tissue |
P value∗ | ||
|---|---|---|---|---|---|
| Negative | Low | High | |||
| Tumor-infiltrating lymphocytes (n = 1276) | 0.60 | ||||
| Absent/low | 928 (73) | 652 (72) | 136 (72) | 140 (76) | |
| Intermediate | 212 (17) | 159 (18) | 29 (15) | 24 (13) | |
| High | 136 (11) | 92 (10) | 23 (12) | 21 (11) | |
| Intratumoral periglandular reaction (n = 1278) | 0.98 | ||||
| Absent/low | 147 (12) | 105 (12) | 23 (12) | 19 (10) | |
| Intermediate | 961 (75) | 675 (75) | 146 (77) | 140 (75) | |
| High | 170 (13) | 123 (14) | 20 (11) | 27 (15) | |
| Peritumoral lymphocytic reaction (n = 1275) | 0.98 | ||||
| Absent/low | 164 (13) | 116 (13) | 23 (12) | 25 (13) | |
| Intermediate | 898 (70) | 633 (70) | 138 (73) | 127 (68) | |
| High | 213 (17) | 151 (17) | 28 (15) | 34 (18) | |
| Crohn-like lymphoid reaction (n = 1054) | 0.39 | ||||
| Absent/low | 783 (74) | 553 (73) | 115 (79) | 115 (74) | |
| Intermediate | 190 (18) | 139 (18) | 24 (16) | 27 (17) | |
| High | 81 (7.7) | 61 (8.1) | 7 (4.8) | 13 (8.4) | |
| CD3+ cell density (n = 686) | 0.81 | ||||
| Quartile 1 (lowest) | 172 (25) | 120 (25) | 29 (28) | 23 (24) | |
| Quartile 2 | 171 (25) | 118 (24) | 29 (28) | 24 (25) | |
| Quartile 3 | 172 (25) | 127 (26) | 23 (22) | 22 (23) | |
| Quartile 4 (highest) | 171 (25) | 120 (25) | 24 (23) | 27 (28) | |
| CD8+ cell density (n = 669) | 0.082 | ||||
| Quartile 1 (lowest) | 167 (25) | 113 (24) | 28 (28) | 26 (28) | |
| Quartile 2 | 167 (25) | 115 (24) | 26 (26) | 26 (28) | |
| Quartile 3 | 167 (25) | 119 (25) | 27 (27) | 21 (23) | |
| Quartile 4 (highest) | 168 (25) | 129 (27) | 19 (19) | 20 (22) | |
| CD45RO+ cell density (n = 695) | 0.62 | ||||
| Quartile 1 (lowest) | 174 (25) | 133 (27) | 24 (23) | 17 (18) | |
| Quartile 2 | 173 (25) | 112 (23) | 28 (27) | 33 (35) | |
| Quartile 3 | 175 (25) | 124 (25) | 28 (27) | 23 (24) | |
| Quartile 4 (highest) | 173 (25) | 127 (26) | 24 (23) | 22 (23) | |
| FOXP3+ cell density (n = 659) | 0.79 | ||||
| Quartile 1 (lowest) | 165 (25) | 117 (25) | 31 (33) | 17 (18) | |
| Quartile 2 | 164 (25) | 117 (25) | 24 (25) | 23 (24) | |
| Quartile 3 | 166 (25) | 117 (25) | 21 (22) | 28 (30) | |
| Quartile 4 (highest) | 164 (25) | 119 (25) | 19 (20) | 26 (28) | |
Data are given as n (percentage) of each group unless otherwise indicated. Percentage indicates the proportion of patients with a specific clinical, pathologic, or molecular characteristic in all or in strata of the amount of Bifidobacterium DNA in colorectal cancer tissue.
P value was calculated by Spearman correlation test between the amount of Bifidobacterium DNA in colorectal cancer tissue (negative, low, and high; as an ordinal valuable) and (T) lymphocyte variables, including the densities of T cells (cells/mm2; as continuous variables) and histologic lymphocytic reaction patterns (absent/low, intermediate, and high; as ordinal variables). Because eight primary (T) lymphocyte variables were assessed, the two-sided α level was set at 0.005.
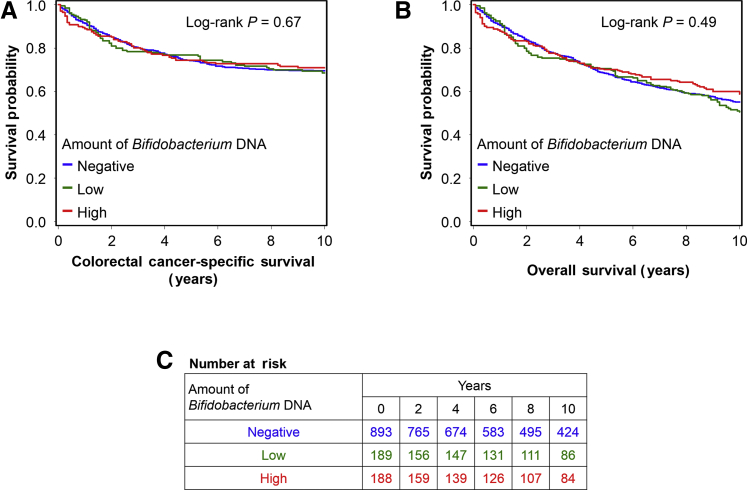
During the median follow-up time of 14.3 years (interquartile range, 10.0 to 18.3 years) for all censored patients, there were 754 all-cause deaths, including 356 colorectal cancer–specific deaths. As exploratory analyses, a Kaplan-Meier analysis and Cox proportional hazards regression analysis were conducted to assess a prognostic role of the amount of bifidobacteria in colorectal cancer tissue. No significant associations of the amount of bifidobacteria with colorectal cancer–specific mortality or overall mortality were observed (Table 5, Supplemental Table S3, and Figure 3).
Table 5.
The Amount of Bifidobacterium DNA in Colorectal Cancer Tissue and Patient Survival with Inverse Probability Weighting
| Variable | Cases, n | Colorectal cancer–specific survival |
Overall survival |
||||
|---|---|---|---|---|---|---|---|
| Events, n | Univariable HR (95% CI)∗ | Multivariable HR (95% CI)∗† | Events, n | Univariable HR (95% CI)∗ | Multivariable HR (95% CI)∗† | ||
| Amount of Bifidobacterium DNA in colorectal cancer tissue | |||||||
| Negative | 893 | 252 | 1 (referent) | 1 (referent) | 531 | 1 (referent) | 1 (referent) |
| Low | 189 | 50 | 1.00 (0.72–1.37) | 0.99 (0.72–1.35) | 116 | 1.04 (0.84–1.30) | 1.01 (0.81–1.26) |
| High | 188 | 54 | 0.93 (0.67–1.29) | 0.93 (0.66–1.32) | 107 | 0.90 (0.70–1.15) | 0.90 (0.71–1.15) |
| Ptrend‡ | 0.70 | 0.71 | 0.51 | 0.48 | |||
HR, hazard ratio.
Inverse probability weighting was applied to reduce a bias attributable to the availability of tumor tissue after cancer diagnosis (Statistical Analysis).
The multivariable Cox regression model initially included sex, age, year of diagnosis, family history of colorectal cancer, tumor location, tumor differentiation, disease stage, microsatellite instability, CpG island methylator phenotype, KRAS, BRAF, and PIK3CA mutations, and long-interspersed nucleotide element-1 methylation level. A backward elimination with a threshold P of 0.05 was used to select variables for the final models.
Ptrend value was calculated across the ordinal categories (negative, low, and high) of the amount of Bifidobacterium DNA in colorectal cancer tissue in the inverse probability weighting–adjusted Cox regression model.
Figure 3.
Inverse probability weighting–adjusted Kaplan-Meier survival analyses of colorectal cancer patients according to the amount of Bifidobacterium DNA in tumor tissue. The P values were calculated using the weighted log-rank test for trend (two sided). A: Colorectal cancer–specific survival. B: Overall survival. C: The number of patients who remained alive and at risk of death at each time point after the diagnosis of colorectal cancer.
Discussion
To test the hypothesis that the amount of Bifidobacterium DNA in colorectal carcinoma tissue might be associated with tumor differentiation, and higher immune response to colorectal cancer, this study was conducted using two US prospective cohort studies. An association between the amount of bifidobacteria and an extent of signet ring cells in colorectal carcinoma tissue was observed. These results suggest a possible role of bifidobacteria in the tumor microenvironment in determining tumor differentiation during colorectal cancer development.
Members of the Bifidobacterium genus are a natural part of the bacterial flora in the human gut lumen, where they are known to produce lactic acid and acetate. In experimental studies, bifidobacteria in the gut appear to inhibit colorectal carcinogenesis through prevention of enteropathogenic infection or acidification, which can reduce secondary bile acid production.22, 54, 55 In the current study, intratumor bifidobacteria were detected in 30% of patients with colorectal cancer, but not in 70% of patients. Further investigations are warranted to examine the association of the amount of Bifidobacterium DNA and colorectal cancer risk. Recent evidence suggests that bifidobacteria promote antitumor immunity and enhance immunotherapeutic efficacy.3, 27 The use of host immunity for regulating cancer progression has attracted much attention, an approach supported by studies illustrating that high-level immune response has been associated with better clinical outcomes of colorectal cancer.13, 14, 29, 30, 31 Collectively, bifidobacteria have the potential to correlate with favorable colorectal cancer survival through increasing the host immune response to cancer cells. By using >1000 human colorectal carcinoma cases, we provide the first study, to our knowledge, examining this relationship. However, no significant associations were observed between the amount of intratumor bifidobacteria and immune response or colorectal cancer survival.
An association was found between the amount of bifidobacteria and the extent of signet ring cells in colorectal carcinoma tissue. A higher proportion of signet ring cells was associated with proximal tumor location, MSI-high status, CIMP-high status, MLH1 promoter hypermethylation, frequent BRAF mutation, higher long-interspersed nucleotide element-1 methylation level, and worse survival.37, 56, 57, 58, 59, 60 Emerging evidence supports that tumorigenesis and oncogenic signaling pathways may be influenced by microbiota in the tumor microenvironment.21, 61 In in vitro and in vivo experiments, bacteria, including bifidobacteria, appear to modulate intestinal epithelial cell differentiation factors.25 Nonetheless, the link between bifidobacteria and signet ring cells remains unexplained. However, these findings present potential avenues for deepening our understanding of cancer, as well as showing once again that further study of the role of the microbiome in tumorigenesis and cancer development may lead to targeted antimicrobial therapies that inhibit cancer initiation or progression.62
Several factors may account for the lack of association between bifidobacteria and immune response or colorectal cancer survival in this cohort. In the tumor microenvironment, extracellular acidification enables cancer cells to progress by promoting proliferation, evasion of apoptosis, metabolic adaptation, migration, and invasion.63 Experimental evidence suggests that high lactic acid production in the tumor microenvironment may promote tumor development through inhibition of tumor immune surveillance and polarization of an M2-like state that is critical for tumor growth.64, 65, 66 Accumulating evidence also supports that acetate produced by microbes can serve as an energy source for carcinoma cells.67, 68, 69 These studies provide support for the assumption that cancer cells can take advantage of the lactic acid and acetate produced by bifidobacteria for progression of the tumor and evasion of the immune response in the tumor microenvironment. Collectively, one possibility is that bifidobacteria behave differently in the tumor microenvironment when compared with their typical functionality in the gut lumen. It is also possible that bifidobacteria contribute to the pathogenesis not individually, but as a member of the overall host-microbial ecosystem. Microbiota other than F. nucleatum, other microbiota that may have an opposing association with bifidobacteria, were not assayed. Given the evidence for the enrichment of bifidobacteria in the hypoxic tumor microenvironment,70, 71, 72 loss of intestinal barrier function because of advanced colorectal cancer might result in entry of bifidobacteria into colorectal carcinoma tissue.73 The slight positive association of bifidobacteria with F. nucleatum might support this scenario. Hence, a better understanding of the roles of Bifidobacterium genus in the tumor microenvironment would have considerable implications in the context of interactions among tumor cells, host cells, and microorganisms.
This study has several limitations. First, the presence of signet ring cells was noted in a minority of colorectal cancer cases, which affected statistical power. The low prevalence of signet ring cells resulted in relatively wide CIs of odds ratio estimates in Table 3. Second, data on cancer recurrence were not available. However, colorectal cancer–specific survival can be considered as a reasonable cancer-specific outcome in a population-based study with long-term follow-up, considering that median survival for colorectal cancer recurrence (metastasis) was approximately 10 to 20 months during the time period of this study.74 Third, data on cancer treatment were also limited. However, decisions on chemotherapy use and regimen are unlikely to differ substantially, according to the amount of bifidobacteria in resected specimens, because these data were not available to treating physicians.
A major strength of this study is the use of a molecular pathologic epidemiology database15, 75 of rectal and colon carcinoma cases in two US-based large prospective cohort studies, which integrates clinicopathologic features, long-term survival data, tumor molecular characteristics, and the amount of bifidobacteria in colorectal carcinoma tissue. In terms of the prevalences of MSI-high and KRAS, BRAF, and PIK3CA mutations, the current study provides data generally consistent with most of previous studies, which indicate reasonable accuracy of these molecular analyses.76, 77, 78 In addition, these findings largely remained similar after applying the IPW method to adjust for the differential availability of tumor tissue samples. This population-based colorectal cancer database enabled us to rigorously examine the interactive prognostic association of the amount of bifidobacteria, controlling for potential confounders. Importantly, our patients with colorectal cancer were derived from a great number of hospitals throughout the United States, which can increase the generalizability of our findings.
In conclusion, we found a strong association between the amount of Bifidobacterium DNA and the extent of signet ring cells in colorectal carcinoma tissue. On validation, these population-based data may inform future research to clarify mechanisms of cancer development influenced by complicated interactions among tumor cells, host cells, and microorganisms.
Acknowledgments
We thank the participants and staff of the Nurses' Health Study and the Health Professionals Follow-Up Study for their valuable contributions and the following state cancer registries: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, and WY. The authors assume full responsibility for analyses and interpretation of these data.
Footnotes
Supported by US NIH grants P01 CA87969 (M.S.), UM1 CA186107, P01 CA55075, UM1 CA167552, U01 CA167552, P50 CA127003 (C.S.F.), R01 CA118553 (C.S.F.), R01 CA169141 (C.S.F.), R01 CA137178 (A.T.C.), K24 DK098311 (A.T.C.), R35 CA197735 (S.O.), R01 CA151993 (S.O.), K07 CA190673 (R.N.), and K07 CA188126 (X.Z.), the Dana-Farber/Harvard Cancer Center Nodal Award (S.O.), Stand Up to Cancer Colorectal Cancer Dream Team Translational Research grant (C.S.F. and M.Gi.), the Project P Fund grant, The Friends of the Dana-Farber Cancer Institute grant, the Bennett Family Fund grant, the Entertainment Industry Foundation through the National Colorectal Cancer Research Alliance grant, Japan Society for the Promotion of Science Overseas Research Fellowship grant JP2017-775 (K.K.), the Mitsukoshi Health and Welfare Foundation (T.H.), the Australia Awards-Endeavour Scholarships and Fellowships Program (J.B.), and China Scholarship Council and a Huazhong University of Science and Technology fellowship grant (L.L.). A.T.C. is a Stuart and Suzanne Steele Massachusetts General Hospital Research Scholar.
K.K., T.H., H.K., and J.B. contributed equally to this work.
A.T.C., J.A.M., C.S.F., and S.O. contributed equally to this work as senior authors.
Disclosures: A.T.C. previously served as a consultant for Bayer Healthcare and Pfizer Inc. This study was not funded by Bayer Healthcare or Pfizer Inc.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Portions of this work were presented at the American Society for Investigative Pathology (ASIP) Annual Meeting at Experimental Biology, held April 21–25, 2018 in San Diego, CA, where S.O. delivered a lecture entitled “Integrative Immunology-MPE (Molecular Pathological Epidemiology): Frontier for Pathobiological Discovery from Big Data”. S.O. is the 2018 recipient of the ASIP Outstanding Investigator Award, which is presented annually to a mid-career investigator with demonstrated excellence in research in experimental pathology.
Supplemental material for this article can be found at https://doi.org/10.1016/j.ajpath.2018.08.015.
Supplemental Data
Supplemental Figure S1.
Quantitative real-time PCR assays for Bifidobacterium DNA and 16S using twofold dilution series (40, 80, 160, and 320 ng) from the same DNA specimen. Symbols indicate mean, and error bars denote SD, of CT values of duplicate runs. The coefficient of determination (r2) in the assays for Bifidobacterium DNA and 16S is shown.
Supplemental Figure S2.
The amount of Bifidobacterium DNA in 50 pairs of colorectal carcinoma and adjacent nontumor tissue samples. Dot plots represent the amount of bifidobacteria in colorectal carcinoma tissue and paired adjacent nontumor tissue. Statistical analyses were performed using a two-sided Wilcoxon signed rank test.
References
- 1.Dzutsev A., Badger J.H., Perez-Chanona E., Roy S., Salcedo R., Smith C.K., Trinchieri G. Microbes and cancer. Annu Rev Immunol. 2017;35:199–228. doi: 10.1146/annurev-immunol-051116-052133. [DOI] [PubMed] [Google Scholar]
- 2.Honda K., Littman D.R. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 3.Snyder A., Pamer E., Wolchok J. IMMUNOTHERAPY: could microbial therapy boost cancer immunotherapy? Science. 2015;350:1031–1032. doi: 10.1126/science.aad7706. [DOI] [PubMed] [Google Scholar]
- 4.Sender R., Fuchs S., Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gopalakrishnan V., Spencer C.N., Nezi L., Reuben A., Andrews M.C., Karpinets T.V. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matson V., Fessler J., Bao R., Chongsuwat T., Zha Y., Alegre M.L., Luke J.J., Gajewski T.F. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Routy B., Le Chatelier E., Derosa L., Duong C.P.M., Alou M.T., Daillere R. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 8.Sears C.L., Pardoll D.M. The intestinal microbiome influences checkpoint blockade. Nat Med. 2018;24:254–255. doi: 10.1038/nm.4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dejea C.M., Fathi P., Craig J.M., Boleij A., Taddese R., Geis A.L., Wu X., DeStefano Shields C.E., Hechenbleikner E.M., Huso D.L., Anders R.A., Giardiello F.M., Wick E.C., Wang H., Wu S., Pardoll D.M., Housseau F., Sears C.L. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 2018;359:592–597. doi: 10.1126/science.aah3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mima K., Nishihara R., Qian Z.R., Cao Y., Sukawa Y., Nowak J.A., Yang J., Dou R., Masugi Y., Song M., Kostic A.D., Giannakis M., Bullman S., Milner D.A., Baba H., Giovannucci E.L., Garraway L.A., Freeman G.J., Dranoff G., Garrett W.S., Huttenhower C., Meyerson M., Meyerhardt J.A., Chan A.T., Fuchs C.S., Ogino S. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65:1973–1980. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mima K., Sukawa Y., Nishihara R., Qian Z.R., Yamauchi M., Inamura K., Kim S.A., Masuda A., Nowak J.A., Nosho K., Kostic A.D., Giannakis M., Watanabe H., Bullman S., Milner D.A., Harris C.C., Giovannucci E., Garraway L.A., Freeman G.J., Dranoff G., Chan A.T., Garrett W.S., Huttenhower C., Fuchs C.S., Ogino S. Fusobacterium nucleatum and T cells in colorectal carcinoma. JAMA Oncol. 2015;1:653–661. doi: 10.1001/jamaoncol.2015.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajpoot M., Sharma A.K., Sharma A., Gupta G.K. Understanding the microbiome: emerging biomarkers for exploiting the microbiota for personalized medicine against cancer. Semin Cancer Biol. 2018;52(Pt 1):1–8. doi: 10.1016/j.semcancer.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Ogino S., Galon J., Fuchs C.S., Dranoff G. Cancer immunology: analysis of host and tumor factors for personalized medicine. Nat Rev Clin Oncol. 2011;8:711–719. doi: 10.1038/nrclinonc.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ogino S., Nowak J.A., Hamada T., Phipps A.I., Peters U., Milner D.A., Jr., Giovannucci E.L., Nishihara R., Giannakis M., Garrett W.S., Song M. Integrative analysis of exogenous, endogenous, tumour and immune factors for precision medicine. Gut. 2018;67:1168–1180. doi: 10.1136/gutjnl-2017-315537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogino S., Nishihara R., VanderWeele T.J., Wang M., Nishi A., Lochhead P., Qian Z.R., Zhang X., Wu K., Nan H., Yoshida K., Milner D.A., Jr., Chan A.T., Field A.E., Camargo C.A., Jr., Williams M.A., Giovannucci E.L. Review article: the role of molecular pathological epidemiology in the study of neoplastic and non-neoplastic diseases in the era of precision medicine. Epidemiology. 2016;27:602–611. doi: 10.1097/EDE.0000000000000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masugi Y., Nishihara R., Yang J., Mima K., da Silva A., Shi Y., Inamura K., Cao Y., Song M., Nowak J.A., Liao X., Nosho K., Chan A.T., Giannakis M., Bass A.J., Hodi F.S., Freeman G.J., Rodig S., Fuchs C.S., Qian Z.R., Ogino S. Tumour CD274 (PD-L1) expression and T cells in colorectal cancer. Gut. 2017;66:1463–1473. doi: 10.1136/gutjnl-2016-311421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bijlsma M.F., Sadanandam A., Tan P., Vermeulen L. Molecular subtypes in cancers of the gastrointestinal tract. Nat Rev Gastroenterol Hepatol. 2017;14:333–342. doi: 10.1038/nrgastro.2017.33. [DOI] [PubMed] [Google Scholar]
- 18.Naxerova K., Reiter J.G., Brachtel E., Lennerz J.K., van de Wetering M., Rowan A., Cai T., Clevers H., Swanton C., Nowak M.A., Elledge S.J., Jain R.K. Origins of lymphatic and distant metastases in human colorectal cancer. Science. 2017;357:55–60. doi: 10.1126/science.aai8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H., Courtois E.T., Sengupta D., Tan Y., Chen K.H., Goh J.J.L., Kong S.L., Chua C., Hon L.K., Tan W.S., Wong M., Choi P.J., Wee L.J.K., Hillmer A.M., Tan I.B., Robson P., Prabhakar S. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat Genet. 2017;49:708–718. doi: 10.1038/ng.3818. [DOI] [PubMed] [Google Scholar]
- 20.Ogino S., Jhun I., Mata D.A., Soong T.R., Hamada T., Liu L., Nishihara R., Giannakis M., Cao Y., Manson J.E., Nowak J.A., Chan A.T. Integration of pharmacology, molecular pathology, and population data science to support precision gastrointestinal oncology. NPJ Precis Oncol. 2017;1:40. doi: 10.1038/s41698-017-0042-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gagliani N., Hu B., Huber S., Elinav E., Flavell R.A. The fire within: microbes inflame tumors. Cell. 2014;157:776–783. doi: 10.1016/j.cell.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 22.Fukuda S., Toh H., Hase K., Oshima K., Nakanishi Y., Yoshimura K., Tobe T., Clarke J.M., Topping D.L., Suzuki T., Taylor T.D., Itoh K., Kikuchi J., Morita H., Hattori M., Ohno H. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 23.Laforest-Lapointe I., Arrieta M.C. Patterns of early-life gut microbial colonization during human immune development: an ecological perspective. Front Immunol. 2017;8:788. doi: 10.3389/fimmu.2017.00788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ubeda C., Djukovic A., Isaac S. Roles of the intestinal microbiota in pathogen protection. Clin Transl Immunology. 2017;6:e128. doi: 10.1038/cti.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becker S., Oelschlaeger T.A., Wullaert A., Vlantis K., Pasparakis M., Wehkamp J., Stange E.F., Gersemann M. Bacteria regulate intestinal epithelial cell differentiation factors both in vitro and in vivo. PLoS One. 2013;8:e55620. doi: 10.1371/journal.pone.0055620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolnerhanssen B.K., Moran A.W., Burdyga G., Meyer-Gerspach A.C., Peterli R., Manz M., Thumshirn M., Daly K., Beglinger C., Shirazi-Beechey S.P. Deregulation of transcription factors controlling intestinal epithelial cell differentiation: a predisposing factor for reduced enteroendocrine cell number in morbidly obese individuals. Sci Rep. 2017;7:8174. doi: 10.1038/s41598-017-08487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sivan A., Corrales L., Hubert N., Williams J.B., Aquino-Michaels K., Earley Z.M., Benyamin F.W., Lei Y.M., Jabri B., Alegre M.L., Chang E.B., Gajewski T.F. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mlecnik B., Bindea G., Angell H.K., Maby P., Angelova M., Tougeron D., Church S.E., Lafontaine L., Fischer M., Fredriksen T., Sasso M., Bilocq A.M., Kirilovsky A., Obenauf A.C., Hamieh M., Berger A., Bruneval P., Tuech J.J., Sabourin J.C., Le Pessot F., Mauillon J., Rafii A., Laurent-Puig P., Speicher M.R., Trajanoski Z., Michel P., Sesboue R., Frebourg T., Pages F., Valge-Archer V., Latouche J.B., Galon J. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity. 2016;44:698–711. doi: 10.1016/j.immuni.2016.02.025. [DOI] [PubMed] [Google Scholar]
- 29.Giannakis M., Mu X.J., Shukla S.A., Qian Z.R., Cohen O., Nishihara R., Bahl S., Cao Y., Amin-Mansour A., Yamauchi M., Sukawa Y., Stewart C., Rosenberg M., Mima K., Inamura K., Nosho K., Nowak J.A., Lawrence M.S., Giovannucci E.L., Chan A.T., Ng K., Meyerhardt J.A., Van Allen E.M., Getz G., Gabriel S.B., Lander E.S., Wu C.J., Fuchs C.S., Ogino S., Garraway L.A. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 2016;15:857–865. doi: 10.1016/j.celrep.2016.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nosho K., Baba Y., Tanaka N., Shima K., Hayashi M., Meyerhardt J.A., Giovannucci E., Dranoff G., Fuchs C.S., Ogino S. Tumour-infiltrating T-cell subsets, molecular changes in colorectal cancer, and prognosis: cohort study and literature review. J Pathol. 2010;222:350–366. doi: 10.1002/path.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogino S., Nosho K., Irahara N., Meyerhardt J.A., Baba Y., Shima K., Glickman J.N., Ferrone C.R., Mino-Kenudson M., Tanaka N., Dranoff G., Giovannucci E.L., Fuchs C.S. Lymphocytic reaction to colorectal cancer is associated with longer survival, independent of lymph node count, microsatellite instability, and CpG island methylator phenotype. Clin Cancer Res. 2009;15:6412–6420. doi: 10.1158/1078-0432.CCR-09-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogino S., Giannakis M. Immunoscore for (colorectal) cancer precision medicine. Lancet. 2018;391:2084–2086. doi: 10.1016/S0140-6736(18)30953-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pages F., Mlecnik B., Marliot F., Bindea G., Ou F.S., Bifulco C. International validation of the consensus immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet. 2018;391:2128–2139. doi: 10.1016/S0140-6736(18)30789-X. [DOI] [PubMed] [Google Scholar]
- 34.Nishihara R., Wu K., Lochhead P., Morikawa T., Liao X., Qian Z.R., Inamura K., Kim S.A., Kuchiba A., Yamauchi M., Imamura Y., Willett W.C., Rosner B.A., Fuchs C.S., Giovannucci E., Ogino S., Chan A.T. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med. 2013;369:1095–1105. doi: 10.1056/NEJMoa1301969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamauchi M., Morikawa T., Kuchiba A., Imamura Y., Qian Z.R., Nishihara R., Liao X., Waldron L., Hoshida Y., Huttenhower C., Chan A.T., Giovannucci E., Fuchs C., Ogino S. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61:847–854. doi: 10.1136/gutjnl-2011-300865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mima K., Cao Y., Chan A.T., Qian Z.R., Nowak J.A., Masugi Y., Shi Y., Song M., da Silva A., Gu M., Li W., Hamada T., Kosumi K., Hanyuda A., Liu L., Kostic A.D., Giannakis M., Bullman S., Brennan C.A., Milner D.A., Baba H., Garraway L.A., Meyerhardt J.A., Garrett W.S., Huttenhower C., Meyerson M., Giovannucci E.L., Fuchs C.S., Nishihara R., Ogino S. Fusobacterium nucleatum in colorectal carcinoma tissue according to tumor location. Clin Transl Gastroenterol. 2016;7:e200. doi: 10.1038/ctg.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inamura K., Yamauchi M., Nishihara R., Kim S.A., Mima K., Sukawa Y., Li T., Yasunari M., Zhang X., Wu K., Meyerhardt J.A., Fuchs C.S., Harris C.C., Qian Z.R., Ogino S. Prognostic significance and molecular features of signet-ring cell and mucinous components in colorectal carcinoma. Ann Surg Oncol. 2015;22:1226–1235. doi: 10.1245/s10434-014-4159-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bosman F.T., World Health Organization. International Agency for Research on Cancer . ed 4. International Agency for Research on Cancer; Lyon: 2010. WHO Classification of Tumours of the Digestive System. [Google Scholar]
- 39.Furet J.P., Firmesse O., Gourmelon M., Bridonneau C., Tap J., Mondot S., Dore J., Corthier G. Comparative assessment of human and farm animal faecal microbiota using real-time quantitative PCR. FEMS Microbiol Ecol. 2009;68:351–362. doi: 10.1111/j.1574-6941.2009.00671.x. [DOI] [PubMed] [Google Scholar]
- 40.Sobhani I., Tap J., Roudot-Thoraval F., Roperch J.P., Letulle S., Langella P., Corthier G., Tran Van Nhieu J., Furet J.P. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One. 2011;6:e16393. doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 42.Ogino S., Nosho K., Kirkner G.J., Kawasaki T., Meyerhardt J.A., Loda M., Giovannucci E.L., Fuchs C.S. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Imamura Y., Lochhead P., Yamauchi M., Kuchiba A., Qian Z.R., Liao X., Nishihara R., Jung S., Wu K., Nosho K., Wang Y.E., Peng S., Bass A.J., Haigis K.M., Meyerhardt J.A., Chan A.T., Fuchs C.S., Ogino S. Analyses of clinicopathological, molecular, and prognostic associations of KRAS codon 61 and codon 146 mutations in colorectal cancer: cohort study and literature review. Mol Cancer. 2014;13:135. doi: 10.1186/1476-4598-13-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao X., Lochhead P., Nishihara R., Morikawa T., Kuchiba A., Yamauchi M., Imamura Y., Qian Z.R., Baba Y., Shima K., Sun R., Nosho K., Meyerhardt J.A., Giovannucci E., Fuchs C.S., Chan A.T., Ogino S. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367:1596–1606. doi: 10.1056/NEJMoa1207756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chan A.T., Ogino S., Fuchs C.S. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. N Engl J Med. 2007;356:2131–2142. doi: 10.1056/NEJMoa067208. [DOI] [PubMed] [Google Scholar]
- 46.Morikawa T., Kuchiba A., Yamauchi M., Meyerhardt J.A., Shima K., Nosho K., Chan A.T., Giovannucci E., Fuchs C.S., Ogino S. Association of CTNNB1 (beta-catenin) alterations, body mass index, and physical activity with survival in patients with colorectal cancer. JAMA. 2011;305:1685–1694. doi: 10.1001/jama.2011.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan A.T., Ogino S., Fuchs C.S. Aspirin use and survival after diagnosis of colorectal cancer. JAMA. 2009;302:649–658. doi: 10.1001/jama.2009.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benjamin D.J., Berger J.O., Johannesson M., Nosek B.A., Wagenmakers E.J., Berk R. Redefine statistical significance. Nat Hum Behav. 2018;2:6–10. doi: 10.1038/s41562-017-0189-z. [DOI] [PubMed] [Google Scholar]
- 49.Ioannidis J.P. How to make more published research true. PLoS Med. 2014;11:e1001747. doi: 10.1371/journal.pmed.1001747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamada T., Cao Y., Qian Z.R., Masugi Y., Nowak J.A., Yang J., Song M., Mima K., Kosumi K., Liu L., Shi Y., da Silva A., Gu M., Li W., Keum N., Zhang X., Wu K., Meyerhardt J.A., Giovannucci E.L., Giannakis M., Rodig S.J., Freeman G.J., Nevo D., Wang M., Chan A.T., Fuchs C.S., Nishihara R., Ogino S. Aspirin use and colorectal cancer survival according to tumor CD274 (programmed cell death 1 ligand 1) expression status. J Clin Oncol. 2017;35:1836–1844. doi: 10.1200/JCO.2016.70.7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu L., Nevo D., Nishihara R., Cao Y., Song M., Twombly T.S., Chan A.T., Giovannucci E.L., VanderWeele T.J., Wang M., Ogino S. Utility of inverse probability weighting in molecular pathological epidemiology. Eur J Epidemiol. 2018;33:381–392. doi: 10.1007/s10654-017-0346-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seaman S.R., White I.R. Review of inverse probability weighting for dealing with missing data. Stat Methods Med Res. 2013;22:278–295. doi: 10.1177/0962280210395740. [DOI] [PubMed] [Google Scholar]
- 53.Xie J., Liu C. Adjusted Kaplan-Meier estimator and log-rank test with inverse probability of treatment weighting for survival data. Stat Med. 2005;24:3089–3110. doi: 10.1002/sim.2174. [DOI] [PubMed] [Google Scholar]
- 54.Hu Y., Le Leu R.K., Christophersen C.T., Somashekar R., Conlon M.A., Meng X.Q., Winter J.M., Woodman R.J., McKinnon R., Young G.P. Manipulation of the gut microbiota using resistant starch is associated with protection against colitis-associated colorectal cancer in rats. Carcinogenesis. 2016;37:366–375. doi: 10.1093/carcin/bgw019. [DOI] [PubMed] [Google Scholar]
- 55.O'Callaghan A., van Sinderen D. Bifidobacteria and their role as members of the human gut microbiota. Front Microbiol. 2016;7:925. doi: 10.3389/fmicb.2016.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alvi M.A., Loughrey M.B., Dunne P., McQuaid S., Turkington R., Fuchs M.A., McGready C., Bingham V., Pang B., Moore W., Maxwell P., Lawler M., James J.A., Murray G.I., Wilson R.H., Salto-Tellez M. Molecular profiling of signet ring cell colorectal cancer provides a strong rationale for genomic targeted and immune checkpoint inhibitor therapies. Br J Cancer. 2017;117:203–209. doi: 10.1038/bjc.2017.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liang Z., Yan D., Li G., Cheng H. Clinical analysis of primary colorectal signet-ring cell carcinoma. Clin Colorectal Cancer. 2018;17:e39–e44. doi: 10.1016/j.clcc.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 58.Razenberg L.G., van Gestel Y.R., Lemmens V.E., de Wilt J.H., Creemers G.J., de Hingh I.H. The prognostic relevance of histological subtype in patients with peritoneal metastases from colorectal cancer: a nationwide population-based study. Clin Colorectal Cancer. 2015;14:e13–e19. doi: 10.1016/j.clcc.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 59.Ogino S., Brahmandam M., Cantor M., Namgyal C., Kawasaki T., Kirkner G., Meyerhardt J.A., Loda M., Fuchs C.S. Distinct molecular features of colorectal carcinoma with signet ring cell component and colorectal carcinoma with mucinous component. Mod Pathol. 2006;19:59–68. doi: 10.1038/modpathol.3800482. [DOI] [PubMed] [Google Scholar]
- 60.Kong X., Zhang X., Huang Y., Tang L., Peng Q., Li J. Characteristics and prognostic factors of colorectal mucinous adenocarcinoma with signet ring cells. Cancer Manag Res. 2017;9:573–580. doi: 10.2147/CMAR.S149582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schneider G., Schmidt-Supprian M., Rad R., Saur D. Tissue-specific tumorigenesis: context matters. Nat Rev Cancer. 2017;17:239–253. doi: 10.1038/nrc.2017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsilimigras M.C., Fodor A., Jobin C. Carcinogenesis and therapeutics: the microbiota perspective. Nat Microbiol. 2017;2:17008. doi: 10.1038/nmicrobiol.2017.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.White K.A., Grillo-Hill B.K., Barber D.L. Cancer cell behaviors mediated by dysregulated pH dynamics at a glance. J Cell Sci. 2017;130:663–669. doi: 10.1242/jcs.195297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brand A., Singer K., Koehl G.E., Kolitzus M., Schoenhammer G., Thiel A. LDHA-associated lactic acid production blunts tumor immunosurveillance by T and NK cells. Cell Metab. 2016;24:657–671. doi: 10.1016/j.cmet.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 65.Dart A. Tumour metabolism: lactic acid: not just a waste product? Nat Rev Cancer. 2016;16:676–677. doi: 10.1038/nrc.2016.109. [DOI] [PubMed] [Google Scholar]
- 66.Colegio O.R., Chu N.Q., Szabo A.L., Chu T., Rhebergen A.M., Jairam V., Cyrus N., Brokowski C.E., Eisenbarth S.C., Phillips G.M., Cline G.W., Phillips A.J., Medzhitov R. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014;513:559–563. doi: 10.1038/nature13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schug Z.T., Vande Voorde J., Gottlieb E. The metabolic fate of acetate in cancer. Nat Rev Cancer. 2016;16:708–717. doi: 10.1038/nrc.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schug Z.T., Peck B., Jones D.T., Zhang Q., Grosskurth S., Alam I.S., Goodwin L.M., Smethurst E., Mason S., Blyth K., McGarry L., James D., Shanks E., Kalna G., Saunders R.E., Jiang M., Howell M., Lassailly F., Thin M.Z., Spencer-Dene B., Stamp G., van den Broek N.J., Mackay G., Bulusu V., Kamphorst J.J., Tardito S., Strachan D., Harris A.L., Aboagye E.O., Critchlow S.E., Wakelam M.J., Schulze A., Gottlieb E. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell. 2015;27:57–71. doi: 10.1016/j.ccell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Routy B., Gopalakrishnan V., Daillere R., Zitvogel L., Wargo J.A., Kroemer G. The gut microbiota influences anticancer immunosurveillance and general health. Nat Rev Clin Oncol. 2018;15:382–396. doi: 10.1038/s41571-018-0006-2. [DOI] [PubMed] [Google Scholar]
- 70.Taniguchi S., Shimatani Y., Fujimori M. Tumor-targeting therapy using gene-engineered anaerobic-nonpathogenic Bifidobacterium longum. Methods Mol Biol. 2016;1409:49–60. doi: 10.1007/978-1-4939-3515-4_5. [DOI] [PubMed] [Google Scholar]
- 71.Lehouritis P., Stanton M., McCarthy F.O., Jeavons M., Tangney M. Activation of multiple chemotherapeutic prodrugs by the natural enzymolome of tumour-localised probiotic bacteria. J Control Release. 2016;222:9–17. doi: 10.1016/j.jconrel.2015.11.030. [DOI] [PubMed] [Google Scholar]
- 72.Kikuchi T., Shimizu H., Akiyama Y., Taniguchi S. In situ delivery and production system of trastuzumab scFv with Bifidobacterium. Biochem Biophys Res Commun. 2017;493:306–312. doi: 10.1016/j.bbrc.2017.09.026. [DOI] [PubMed] [Google Scholar]
- 73.Tjalsma H., Boleij A., Marchesi J.R., Dutilh B.E. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10:575–582. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- 74.Meyerhardt J.A., Mayer R.J. Systemic therapy for colorectal cancer. N Engl J Med. 2005;352:476–487. doi: 10.1056/NEJMra040958. [DOI] [PubMed] [Google Scholar]
- 75.Hamada T., Keum N., Nishihara R., Ogino S. Molecular pathological epidemiology: new developing frontiers of big data science to study etiologies and pathogenesis. J Gastroenterol. 2017;52:265–275. doi: 10.1007/s00535-016-1272-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Loree J.M., Bailey A.M., Johnson A.M., Yu Y., Wu W., Bristow C.A., Davis J.S., Shaw K.R., Broaddus R., Banks K.C., Lanman R.B., Meric-Bernstam F., Overman M.J., Kopetz S., Raghav K. Molecular landscape of ERBB2/ERBB3 mutated colorectal cancer. J Natl Cancer Inst. 2018 doi: 10.1093/jnci/djy067. [Epub ahead of print] doi: 10.1093/jnci/djy067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Loree J.M., Pereira A.A.L., Lam M., Willauer A.N., Raghav K., Dasari A., Morris V.K., Advani S., Menter D.G., Eng C., Shaw K., Broaddus R., Routbort M.J., Liu Y., Morris J.S., Luthra R., Meric-Bernstam F., Overman M.J., Maru D., Kopetz S. Classifying colorectal cancer by tumor location rather than sidedness highlights a continuum in mutation profiles and consensus molecular subtypes. Clin Cancer Res. 2018;24:1062–1072. doi: 10.1158/1078-0432.CCR-17-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Domingo E., Camps C., Kaisaki P.J., Parsons M.J., Mouradov D., Pentony M.M., Makino S., Palmieri M., Ward R.L., Hawkins N.J., Gibbs P., Askautrud H., Oukrif D., Wang H., Wood J., Tomlinson E., Bark Y., Kaur K., Johnstone E.C., Palles C., Church D.N., Novelli M., Danielsen H.E., Sherlock J., Kerr D., Kerr R., Sieber O., Taylor J.C., Tomlinson I. Mutation burden and other molecular markers of prognosis in colorectal cancer treated with curative intent: results from the QUASAR 2 clinical trial and an Australian community-based series. Lancet Gastroenterol Hepatol. 2018;3:635–643. doi: 10.1016/S2468-1253(18)30117-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.