Abstract
Thrips palmi is a widely distributed major agricultural pest in the tropics and subtropics, causing significant losses in cucurbit and solanaceous crops through feeding damage and transmission of tospoviruses. Thrips palmi is a vector of capsicum chlorosis virus (CaCV) in Australia. The present understanding of transmission biology and potential effects of CaCV on T. palmi is limited. To gain insights into molecular responses to CaCV infection, we performed RNA-Seq to identify thrips transcripts that are differentially-abundant during virus infection of adults. De-novo assembly of the transcriptome generated from whole bodies of T. palmi adults generated 166,445 contigs, of which ~24% contained a predicted open reading frame. We identified 1,389 differentially-expressed (DE) transcripts, with comparable numbers up- (708) and down-regulated (681) in virus-exposed thrips compared to non-exposed thrips. Approximately 59% of these DE transcripts had significant matches to NCBI non-redundant proteins (Blastx) and Blast2GO identified provisional functional categories among the up-regulated transcripts in virus-exposed thrips including innate immune response-related genes, salivary gland and/or gut-associated genes and vitellogenin genes. The majority of the immune-related proteins are known to serve functions in lysosome activity and melanisation in insects. Most of the up-regulated oral and extra-oral digestion-associated genes appear to be involved in digestion of proteins, lipids and plant cell wall components which may indirectly enhance the likelihood or frequency of virus transmission or may be involved in the regulation of host defence responses. Most of the down-regulated transcripts fell into the gene ontology functional category of ‘structural constituent of cuticle’. Comparison to DE genes responsive to tomato spotted wilt virus in Frankliniella occidentalis indicates conservation of some thrips molecular responses to infection by different tospoviruses. This study assembled the first transcriptome in the genus Thrips and provides important data to broaden our understanding of networks of molecular interactions between thrips and tospoviruses.
Introduction
Thrips belong to the family Thripidae in the order Thysanoptera which contains nearly 7700 described thrips species [1]. However, less than 1% of them are considered as agricultural pests that cause crop damage directly by feeding and indirectly by transmitting tospoviruses [2]. At present, 15 thrips species have been reported to transmit tospoviruses [3]. Among them, Frankliniella occidentalis is world-wide the most devastating invasive species, with a broad host range, transmitting multiple tospoviruses (genus Orthotospovirus, family Tospoviridae, Order Bunyavirales) including the economically important tomato spotted wilt virus (TSWV) [4]. Melon thrips (Thrips palmi) originated in Southeast Asia [5] and have become a serious invasive pest in tropical and subtropical countries [6]. Several tospoviruses are known to be transmitted by T. palmi including calla lily chlorotic spot virus [7], groundnut bud necrosis virus [8], melon yellow spot virus [9], tomato necrotic ringspot virus [10], watermelon bud necrosis virus [11] and watermelon silver mottle virus [12]. In Australia, capsicum chlorosis virus (CaCV) is transmitted by T. palmi [13].
Thrips transmit tospoviruses in a persistent and propagative mode by which virus circulates and replicates within the thrips body [3]. Thrips acquire virus while feeding on infected plant tissues—most efficiently as first instar larvae–and the virus is retained during larval and pupal molts [14]. While viruliferous late second instar larvae can inoculate plants, adults are vector-competent only if the virus was acquired during the larval stages [15]. After ingestion, virions travel through the esophagus to the midgut—the primary site of virus entry—where they replicate and then disseminate and replicate in the surrounding visceral muscle tissue [16]. Virus also replicates in the primary salivary glands (PSG) of thrips [17]. Virus is then transmitted from salivary glands to plants during thrips feeding. Until recently, there was no evidence to indicate the exact infection route of TSWV from midgut to PSG. However, a recent study revealed progression of TSWV infection in larvae of F. occidentalis spread from midgut to ligaments and tubular salivary glands (TSG), where efferent salivary duct and filament structures connect TSG and PGS [18]. These authors further showed that during thrips development, the primary site of tospovirus replication shifts from midgut and TSG in larvae to PSG in adult thrips.
Tospoviruses have been shown to alter thrips vector performance and behavior both directly and indirectly. Direct negative effects on thrips reproductive potential and developmental time have been reported from TSWV-F. fusca [19, 20] and impatiens necrotic spot virus-F. occidentalis [21] interactions, however experimental evidence indicates no apparent negative effect of TSWV infection on life history traits of F. occidentalis [22, 23] or watermelon silver mottle virus on T. palmi [24]. Effects of TSWV infection on F. occidentalis have been documented, including enhanced reproduction [25], reduced developmental time [26, 27], and altered feeding behaviors [28]. Predictive models developed to study dynamics in virus spread suggest that TSWV infection may change thrips preferential feeding behavior and enhance survival [29]. Indirect effects include plant-mediated effects of virus infection on the performance, development, fecundity, survival and host preference of thrips vectors [3]. In general, the majority of tospovirus-thrips interactions report no apparent negative effects on the fitness of the vector. One hypothesis is that thrips mount molecular defense responses against virus infection that minimize cytopathological effects that could, if unharnessed, negatively impact their development and survival.
Recently, transcriptomes of two Frankliniella species, F. occidentalis and F. fusca, in response to tospovirus infection were reported [30, 31]. Both studies analysed larval, pupal and adult stages for whole-body responses to TSWV infection using high throughput sequencing (RNA-Seq). Gene ontologies that infer processes and functions associated with host defence, insect cuticle structure and development, metabolism and transport were affected by TSWV infection in F. occidentalis [30]. In F. fusca, TSWV-responsive genes were similarly associated with intracellular transport, development and immune responses [31]. Furthermore, the repertoire of responsive genes varied between developmental stages in both systems. In this study, we aimed to investigate a different thrips-tospovirus system involving the genus Thrips for the first time, to broaden our understanding of the molecular responses of thrips vectors exposed to tospovirus infection. We identified transcriptome-wide responses of T. palmi to CaCV infection, some of which were conserved in other thrips species in response to infection with different tospoviruses. This knowledge may be useful in future studies to identify molecular targets to interfere with tospovirus transmission by thrips.
Materials and methods
Maintenance of T. palmi colonies
A T. palmi colony derived from a pure culture maintained at the Vector Laboratory, National Taiwan University, Taipei, Taiwan was reared on bean (Phaseolus coccineus) seedlings following conditions previously established [24]. Oviposition of female thrips was enhanced by allowing them to feed on pollen (Hung Gee, Taiwan) in a sealed Petri plate containing a bean leaf. Cohorts of L1 larvae were transferred into a 2-L beaker enclosed with a fresh bean seedling and reared until adulthood in a growth cabinet at 25°C with 70% relative humidity and 16 h/8 h light/dark photoperiod.
To generate populations of CaCV-exposed and non-exposed adult T. palmi, larvae were given a 24-h acquisition access period (AAP) on CaCV-infected and non-infected Chenopodium quinoa leaves. Briefly, 5–6 weeks old C. quinoa plants were mechanically inoculated with a crude extract of CaCV-infected symptomatic C. quinoa leaves and kept in a growth cabinet at 25°C with 16 h/8 h light/dark photoperiod until symptom development. Cohorts of larvae (<12 h) were obtained from thrips that fed on healthy bean leaves. Batches of 100 larvae were transferred into Petri plates each containing a C. quinoa leaf placed on wet tissue paper. Larvae were given 24 h AAP on CaCV-infected leaves that developed chlorotic lesions seven days after inoculation. As control, larvae were allowed to feed on uninfected leaves. At least 1000 CaCV-exposed and non-exposed larvae were transferred to fresh bean seedlings contained in 2-L beakers and reared until adulthood in separate growth cabinets at 25°C with 70% relative humidity and 16 h/8 h day/night. Infection status of batches of virus-exposed and non-exposed thrips for the presence or absence of CaCV was determined by reverse transcription polymerase chain reaction (RT-PCR) using RNA extracted from sub-samples of each batch of thrips.
Total RNA extraction and library preparation
Virus-exposed and non-exposed adult thrips were collected separately as batches of 100 individuals into 1.5 ml microfuge tubes to obtain three biological replicates for the two treatments. All samples were immediately processed independently. Total RNA was extracted using TRIzol reagent (Life Technologies) following manufacturer’s instructions. RNA extracts were treated with DNase using Turbo DNA-free kit (Ambion, Thermo Fisher Scientific) following manufacturer’s protocol. RNA was quantified using NanoDrop 3000 (Thermo Fisher Scientific). CaCV infection in all RNA samples was assessed using One-step RT-PCR kit (GeneMark) with CaCV-N gene-specific primers [CaCV-N-F1: ATGTCTAACGTCAGGCAACTT and CaCV-N-R1: CACTTCTATAGAAGTACTAGG [32]. Total RNA (2.5–3.0 μg) from three biological replicates of virus-exposed and non-exposed T. palmi was shipped on dry ice from Taiwan to the Australian Genome Research Facility (AGRF, Melbourne) for cDNA library preparation, Illumina sequencing, transcriptome assembly and expression profiling. Rest of the total RNA was stored at -80°C until quantitative PCR (qPCR) analysis.
Illumina sequencing
Illumina cDNA libraries were prepared from total RNA by AGRF following the protocols for TruSeq RNA v2 (2014). Briefly, mRNA in total RNA preparations was enriched by using oligo dT beads prior to library preparation. Purified mRNA was then fragmented with a combination of divalent cations and heat and cDNA was synthesized. First strand cDNA was synthesized by random priming. Six cDNA libraries were prepared from poly(A) mRNA of three replicates each of virus-exposed and non-exposed adult thrips. The six libraries were multiplex-sequenced in one lane of an Illumina HiSeq 2000 sequencer to generate 100-bp paired-end reads using bclsfastq 2.17.1.14 pipeline. Quality control (QC) of resulting sequence reads was done according to AGRF QC standards, Phred 30 across all samples for 100 bp reads [33, 34]. High quality reads were further screened for the presence of any Illumina adapters/overrepresented sequences and CaCV sequences that were then removed.
De novo assembly of T. palmi transcriptome
High quality reads from the six libraries were enriched as described below prior to de novo assembly of T. palmi reference transcriptome. Random errors in Illumina sequencing were corrected by Recorrector software using a k of 31 [35] followed by adapter trimming using Trimmomatic with Phred cut-off ≤ 2 [36]. Following enrichment, reads were de novo assembled using Trinity (v2.2.1), specifying the library type [37]. Quality of the de novo assembly was evaluated using TransRate by mapping all reads to the assembly which gave 0.46 optimal score with 0.38 optimal cut-off [38].
Differential expression analysis
To determine differentially expressed (DE) transcripts in response to exposure to CaCV, reads from 6 Illumina libraries were individually mapped to the de novo assembled T. palmi reference transcriptome using TopHat (v2.0.14) software [39]. Number of Illumina reads that mapped to each contig of the reference transcriptome were estimated and counts were summarized at gene level across the three biological replicates using the featureConts (v1.4.6-p5) [40] utility of the Subread package [41]. Transcripts were assembled with the Stringtie tool v1.1.4 utilizing the reads alignment and in a de novo fashion [42].
DE transcripts between replicates of virus-exposed and non-exposed thrips were determined using Cufflinks tools [39]. Expression values were normalized as read counts per gene per sample with fragments per kilobase of exon per million mapped reads (FPKM). Significantly DE genes were identified using a binary statistical assessment. Briefly, a p-value was calculated for each gene in each sample and each comparison. Then p-values were corrected for multiple tests and comparisons (q-value) using false discovery rate (FDR). In this study, a FDR 0.05 cut off was used to determine p-value threshold. The correlation between virus-exposed and non-exposed transcripts was determined by calculating a Pearson correlation coefficient value [43].
Provisional functional annotation of DE transcripts
Stringent filtering criteria were used to select highly significant DE transcripts for functional annotation. DE transcripts were selected by setting a cut off q-value < 0.01, log2-fold change (FC) > 1 and FPKM > 10, and were classified into functional categories using Blast2GO (B2GO) with default parameters [44]. Initially, transcripts were searched for sequence similarities in the NCBI non-redundant (nr) database using Blastx algorithm. Then transcripts were annotated by retrieving gene ontology (GO) terms associated with BLAST hits using GO databases in NCBI, nr reference protein database including PSD, UniProt, Swiss-Prot, TrEMBL, RefSeq, GenPept and DBXRef. Annotations were further improved by merging InterPro protein signatures and fine-tuned by using Annex-based GO term augmentation followed by removal of First Level GO terms. Enzyme codes were assigned for annotations using B2GO to identify which biological pathways are effected by DE enzymes and these were mapped to the Kyoto Encyclopaedia of Genes and Genomes (KEGG) database [45].
Validation of RNA-Seq expression data with real-time RT-qPCR of selected transcripts
RNA-Seq expression levels of six randomly selected transcripts were validated using RT-qPCR. Actin, β-tubulin and 40S ribosomal protein S14 (RPS 14) were selected as non-DE genes from the dataset as internal references. Primers for target and reference genes were designed using Primer3 [46, 47]. Primer sequences are listed in S1 Table. Complementary DNA was synthesized using oligo dT primers and Superscript III First-strand cDNA synthesis kit (Life Technologies) using the same total RNA preparations (DNase-treated) used for Illumina sequencing. SensiFAST SYBR No-ROX Kit (Bioline) was used in a Rotor-Gene Q real-time PCR cycler (Qiagen) with 20 μl volumes containing 1 μl (10 ng) cDNA, 0.8 μl of each primer (10 μM), 7.4 μl of DNase- and RNase-free water and 10 μl of 2x SYBR No-ROX mix. Reaction conditions were 2 min at 95°C followed by 40 cycles of 95°C for 5s, 60°C for 10s and 72°C for 20s. Experimental design and subsequent data analysis methods were adopted from a previously described protocol [48]. Three biological replicates and two technical replicates were used per sample. Since initial experiments showed stable gene expression for all reference genes across all samples and treatments, actin was selected for future experiments. Real-time PCR amplification efficiencies of target genes and actin were determined by the standard curve method using a ten-fold dilution series of cDNA (from 50 ng to 0.001 ng). PCR efficiencies were calculated from the slopes of standard curves. Threshold cycle (Ct) number was determined from log scale amplification curves. Reaction efficiencies for target and reference genes showed 95–100% efficiency for 1–50 ng of cDNA template input. Hence, we used 10 ng of cDNA template for further experiments. For each reaction, no-template and no-RT control samples were included. Relative expression levels of target genes were calculated as 2−(Ct of target−Ct of reference) [49]. Fold changes in gene expression between treatment and control were calculated using the 2−ΔΔCt method; 2−(ΔCt of treatment−ΔCt of control) [49]. For validation, qPCR derived log2-fold changes were compared with log2-fold values obtained by RNA-Seq analysis. Quantitative PCR results were compared with RNA-Seq data and Pearson correlation coefficient R and associated p values were calculated [43].
Results
De novo transcriptome assembly and differential gene expression
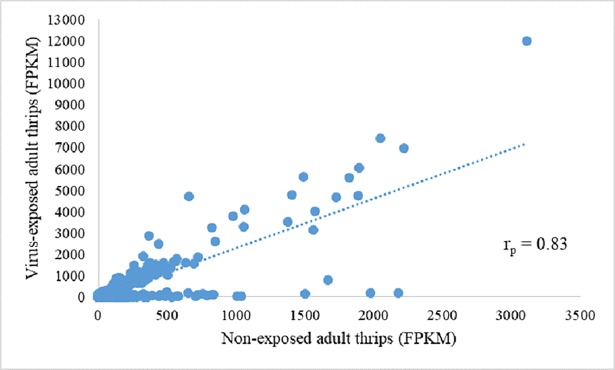
Thrips palmi transcriptome was de novo assembled using 243,546,011 paired-end reads of 100 bp (48.71 Gb of sequence) from three biological replicates of cDNA libraries consisting of virus-exposed and non-exposed thrips. The raw sequence read data generated from the six samples have been deposited in the NCBI Sequence Read Archive as BioProject PRJNA498538 with accession numbers SAMN10316162—SAMN10316167. Per base sequence quality of all six libraries was >93% bases above the Phred quality score of 30. Assembly of high quality reads generated 166,445 contigs with an average length of 918 bp and an N50 of 2114 bp (Table 1). Cleaned reads from individual libraries were aligned to the de novo assembled contigs to estimate read counts that mapped to contigs. All six libraries aligned to the reference transcriptome with at least 75% concordant pair alignment rate. Quality assessment of Cufflinks data showed fairly good quality in all the six libraries. Collectively, there was a positive correlation (Pearson correlation, rp = 0.83, P < 0.0001) between the normalized read counts (FPKM) of the virus-exposed and non-exposed treatments for transcripts that exhibited greater than 2-fold change in expression with a q-value < 0.01, and the virus treatment tended to have a larger range of FPKM values, indicating significant perturbation (Fig 1). For stringency, a transcript was considered DE when the log2-fold change was >1.0, the q-value was < 0.01, and FPKM >10 for at least one of the treatments (CaCV-exposed or non-exposed) in the pairwise comparison. With these criteria, we identified 1,389 DE transcripts, of which 708 were up-regulated and 681 were down-regulated in virus-exposed thrips.
Table 1. Summary statistics for T. palmi de-novo assembled transcriptome.
| Assembly feature | Statistic |
|---|---|
| Total assembled contigs | 166,445 |
| Total assembled bases | 152,899,637 |
| Mean contig length | 919 bp |
| N 50 contig length | 2,114 bp |
| No. contigs with predicted ORFs | 39,449 |
| No. contigs ≥ 400 bp with predicted ORFs | 32,262 |
| Blastx matches (E ≤ 10−5) | 22,582 |
| B2GO annotations | 10,407 |
Fig 1. Comparison of normalized read counts (FPKM) between CaCV-exposed and non-exposed adults of Thrips palmi.
Each point represents one transcript with log2 fold change > 1 in relative abundance between the two treatments and a q-value < 0.01. rp = Pearson’s correlation coefficient.
Annotation of DE transcripts
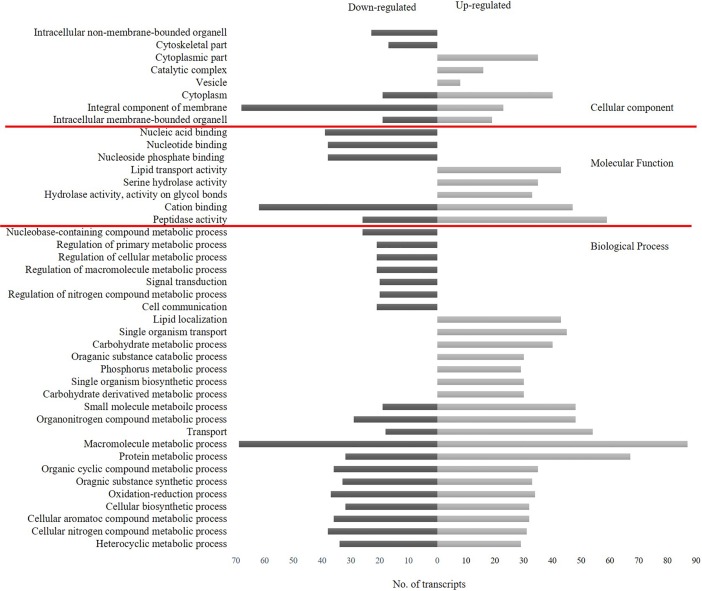
Of the 1,389 identified DE transcripts, 430 (60.7%) up-regulated and 385 (56.5%) down-regulated transcripts matched sequences in the NCBI non-redundant sequence database. Among the DE transcripts that had Blastx hits, 316 up-regulated and 287 down-regulated transcripts were annotated. Forty-five GO terms categorized into biological process (BP), molecular function (MF) and cellular component (CC) were assigned for DE transcripts at level 2. The most dominant GO terms were ‘metabolic process’ in BP domain, ‘peptidase activity’ in MF domain and ‘cytoplasm’ and ‘cytoplasm part’ in CC domain (Fig 2). Within the BP category, GO terms associated with regulation of metabolic processes, signal transduction and cell communication were all down-regulated, whereas lipid localization, carbohydrate/derivative metabolic process, phosphorous metabolic process, single organism transport and biosynthetic process were all up-regulated. Other transcripts that fell into GO terms ‘transport’, ‘metabolic processes’, ‘cellular biosynthetic process’ and ‘oxidation-reduction’ had both up- and down-regulated transcripts. In the MF category, transcripts assigned with GO terms ‘nucleic acid/nucleotide binding’ and ‘nucleoside phosphate binding’ were all down-regulated, whereas ‘lipid transport activity’ and ‘hydrolase activity’ were all up-regulated. In the CC category, GO terms such as ‘integral component of membrane’, ‘cytoskeletal part’ and ‘intracellular non-membrane-bounded organelle’ were among the down-regulated GO terms and cytoplasm and cytoplasmic part were among the up-regulated GO terms. Mapping enzymes into KEGG pathways [45] revealed the most enriched pathways among the up-regulated transcripts were ‘biosynthesis of antibiotics’ and ‘purine metabolism’ with 10 enzymes placed in each. Of the down-regulated transcripts the most enriched pathways were ‘purine metabolism’, ‘tyrosine metabolism’ and ‘isoquinoline alkaloid biosynthesis’ with 3 enzymes placed in each.
Fig 2. Number and functional categories of gene ontology terms assigned for up-regulated and down-regulated transcripts of CaCV-exposed Thips palmi at level 2.
Following GO term assignment, 186 up-regulated DE transcripts could be categorized into three groups that may infer roles in innate immunity, salivary gland processes, and thrips fitness and fecundity based on Blastx descriptions and GO terms (Table 2). The majority of the most highly down-regulated transcripts had Blastx annotations for structural constituents of the cuticle (Table 3). In addition, there were chitinases and nucleic acid binding genes among the down-regulated transcripts.
Table 2. List of significantly up-regulated DE (q < 0.01, log2-FC > 1.0, FPKM >10) transcripts categorized based on Blastx descriptions and GO terms that are predicted to be associated with innate immunity, salivary glands and fitness and fecundity in CaCV-exposed and non-exposed Thrips palmi transcriptome.
| Putative pathway | Blastx description (no. of transcripts) |
FPKM (non-virus-exposed) |
FPKM (virus-exposed) | log2- FC | GO term | Putative function |
|---|---|---|---|---|---|---|
| Cellular immunity | ||||||
| Lysosome | Cathepsin B (10) | 40.9–501.4 | 166–1188.8 | 1.0–2.0 | Viral entry into host cells, Proteolysis, Endopeptidase activity, Lysosome | Innate immunity [50] |
| Lipase 1 (4) | 3.2–33.6 | 20.7–82.5 | 1.3–2.7 | Hydrolase activity, Lipid metabolic process, Membrane | Antiviral [51], innate immunity [52] | |
| Lysosomal aspartic protease (3) | 316.5–360.9 | 934.4–1127.5 | 1.6 | Proteolysis, Lysosome | Innate immunity [53] | |
| Lysozyme C (2) | 37.2–341.7 | 115–784.3 | 1.2 | Lysozyme activity | Innate immunity [54–56] | |
| Acid-phosphatase-1 | 8.4 | 20.8 | 1.3 | Acid phosphatase activity | Antimicrobial [57] | |
| Beta mannosidase | 4.3 | 10.6 | 1.3 | CHO metabolic process | Innate immunity [58] | |
| Melanisation | Phenoloxidase 2 (6) | 2.3–42.5 | 10.0–91.0 | 1.0–2.3 | Melatonin encapsulation of foreign target, L-DOPA monooxygenase activity, Defence, Oxidation-reduction | Innate immunity [59] |
| Phenoloxidase subunit A3 | 16.4 | 51.8 | 1.7 | Oxidation-reduction | Innate immunity [59] | |
| Serine protease ester-like | 14.9 | 47.8 | 1.7 | Proteolysis, Melanisation defence response | Innate immunity [60], Salivary gland [61] | |
| Glucose dehydrogenase [quinone] (9) | 5.2–77.8 | 12.8–193.4 | 1.0–1.6 | Oxidation-reduction process | Innate immunity [62] | |
| Poly(U)-specific endoribonuclease homologue (3) | 93–11.0 | 31.7–38.2 | 1.8 | Hydrolase activity | Innate immunity [63] | |
| Laccase-5 isoform X1 | 19.4 | 61.5 | 1.7 | Oxidation-reduction process | Innate immunity [64, 65] | |
| Troponin isoform 1 like (2) | 13.7–65.8 | 28.4–208.1 | 1.0–1.6 | Protein binding, Calcium binding | Innate immunity [66] | |
| Serine protease 13 | 143.0 | 354.3 | 1.3 | Proteolysis, Serine-type peptidase activity | Innate immunity [67] | |
| Humoral immunity | ||||||
| Complement and coagulation cascades | Carboxypeptidase B like (3) | 4.5–146.3 | 27.8–328.0 | 1.3–3.3 | Proteolysis, Metallo-carboxypeptidase | Innate immunity [68] |
| Coagulation factor ix | 11.6 | 31.0 | 1.4 | Zymogen activation, Extracellular exosome | Innate immunity [69] | |
| Limulus clotting factor C like | 11.1 | 31.6 | 1.5 | Protein binding | Innate immunity [70] | |
| Immune system pathway | ||||||
| Toll | Defensin 2 | 10.4 | 47.2 | 2.2 | Defence response | Innate immunity [65] |
| Gram-negative bacteria-binding 3-like | 132.8 | 469.8 | 1.8 | CHO binding | Innate immunity [71] | |
| Serine protease 44 | 5.9 | 18.9 | 1.7 | Proteolysis, Serine-type peptidase activity | Innate immunity [72, 73] | |
| Antigen processing and presentation | 70 kDa heat shock partial | 20.2 | 54.7 | 1.4 | ATP binding | Antiviral [74] |
| Heat shock 70 a1 partial | 54.7 | 113.0 | 1.0 | ATP binding, Extracellular exosome | Antiviral [74] | |
| Heat shock cognate 71 | 31.7 | 90.6 | 1.1 | ATP binding | Antiviral [55] | |
| Other innate immunity-related genes | ||||||
| Serine proteases | Serine protease (4) | 15.3–95.8 | 31.4–218.7 | 1.0–1.7 | Proteolysis, Serine-type peptidase activity | Innate immunity [75] |
| Serine protease 12 -like (3) | 4.2–37.8 | 13.0–109.9 | 1.6 | Proteolysis, Serine-type peptidase activity | Innate immunity [76] | |
| Serine protease 9-like (5) | 8.2–38.6 | 21.0–92.9 | 1.3–1.5 | Proteolysis, Serine-type peptidase activity | Innate immunity [76] | |
| Trypsins | Trypsin-like serine protease (3) | 2.6–26.2 | 12.8–73.5 | 1.5–2.3 | Proteolysis, Serine-type peptidase activity | Innate immunity [77] |
| Trypsin | 7.6–32.0 | 16.1–85.0 | 1.0–1.5 | Proteolysis, Serine-type peptidase activity | Innate immunity [78] | |
| Apolipo D-like (2) | 12.8–29.1 | 43.6–78.3 | 1.4–1.8 | Pigment binding | Innate immunity [56] | |
| Chitin binding peritrophin-A domain containing (2) | 33.0–48.3 | 110.0–211.8 | 1.7–2.1 | Chitin metabolic process | Innate immunity [79, 80] | |
| Chymotrypsin protease (4) | 8.7–28.5 | 22.1–89.1 | 1.3–1.6 | Proteolysis | Innate immunity [81] | |
| Cytochrome P450 4C1 (2) | 3.5–10.9 | 15.7–31.7 | 1.5–2.1 | Oxidation-reduction process | Immunity [82] | |
| Cytochrome P450 6k1-like | 313.5 | 634.1 | 1.0 | Oxidation-reduction process | Immunity [82] | |
| Extracellular serine threonine kinase FAM20C | 44.2 | 135.8 | 1.6 | Protein phosphorylation | Immunity [83] | |
| Esterase FE4-like | 6.0 | 16.7 | 1.5 | Hydrolase activity | Innate immunity [80] | |
| Facilitated trehalose transporter Tret1-like | 9.7 | 19.6 | 1.0 | Transmembrane transport | Immunity [84] | |
| Elicitin 6 partial | 12.2 | 33.0 | 1.4 | Defence response, Chitin binding | Defence response in plants [85] | |
| Pathogenesis-related protein 5-like | 5.8 | 15.8 | 1.4 | Systemic acquired resistance, Response to virus, apoplastic | Defence response in plants [86] | |
| Salivary gland-associated | ||||||
| Lipase 3 (15) | 2.5–71.8 | 12.3–347.4 | 1.0–2.3 | Lipid metabolic process, Hydrolase activity | General digestion [87, 88] | |
| Seine protease (9) | 5.9–95.8 | 18.8–218.2 | 1.0–1.7 | proteolysis | ||
| Trypsin-like serine protease (3) | 2.6–26.2 | 12.8–73.4 | 1.5–2.3 | Proteolysis, Serine-type peptidase activity | ||
| Carboxypeptidase (3) | 4.5–146.4 | 27.8–328.0 | 1.3–3.3 | Proteolysis | ||
| Pectin lyase (4) | 32.4–216.8 | 68.1–507.0 | 1.0–1.5 | Polysaccharide catabolic process | Digestion of plant cell wall [88] | |
| β-glucosidase | 2.3 | 12.0 | 2.3 | Carbohydrate metabolic process | ||
| Endoglucanase (4) | 66.5–152.1 | 166.7–339.2 | 1.2–1.3 | Fructose, Sucrose metabolic process, Cellulase activity | ||
| α-amylase A-like | 69.0 | 186.8 | 1.4 | Sucrose metabolic process, Extracellular exosome | Sugar metabolism [88] | |
| Angiotensin-converting enzyme-like | 8.4 | 17.0 | 1.0 | Proteolysis | Detoxification and inhibition of plant defence [89] | |
| Phosphoribosyl formylglycinamidine synthase | 24.7 | 56.2 | 1.2 | 'de novo' IMP biosynthetic process | Advantageous for infecting virus [90] | |
| Uridine phosphorylase 1 isoform X1 | 3.1 | 10.9 | 1.8 | Nucleoside metabolic process | Salivary gland [91] | |
| Pyridoxal phosphate phosphatase | 8.3 | 46.7 | 2.5 | Dephosphorylation, Hydrolase activity | Salivary gland expressed gene [92] | |
| Fitness and fecundity-related genes | ||||||
| Vitellogenin (54) | 3.1–2446.1 | 11–7446 | 1.0–2.6 | Lipid transport | Reproduction [93,94], Innate immunity [95] | |
| Other genes | ||||||
| LPXTG-domain-containing cell wall anchor partial | 28.0 | 77.7 | 1.5 | Phosphorylation | Cell surface adhesion [96] | |
FC = fold change of virus-exposed treatment relative to non-virus control treatment.
Table 3. List of significantly down-regulated DE (q < 0.01, log2-FC > 1.0, FPKM >10) transcripts in CaCV-exposed and non-exposed Thrips palmi transcriptome.
| Blastx description (no. of transcripts) |
FPKM (non-virus-exposed) |
FPKM (virus-exposed) | log2- FC | GO term |
|---|---|---|---|---|
| Larval cuticle A2B-like (3) | 117.1–220.8 | 0.3–1.8 | 9.3–8.0 | Structural constituent of cuticle |
| Serine protease inhibitor 3 4 isoform X1 | 44.2 | 0.2 | 7.9 | Extracellular space |
| Pupal cuticle C1B-like (2) | 29.4–100.6 | 0.1–0.6 | 7.4–7.6 | Structural constituent of cuticle |
| Endocuticle structural glyco bd-8-like | 53.6 | 0.4 | 7.1 | Structural constituent of cuticle |
| Alpha-tocopherol transfer -like | 28.2 | 0.2 | 7.0 | Transport |
| Collagen alpha-1(III) chain-like | 47.2 | 0.5 | 6.5 | Chitin binding, Extracellular region |
| Uncharacterized protein LOC106678716 | 29.6 | 0.3 | 6.4 | Integral component of membrane |
| Apolipo D-like | 85.8 | 1.0 | 6.4 | Pigment binding |
| Endocuticle structural glyco bd-4-like (2) | 29.8–203 | 0.4–2.8 | 6.2–6.3 | Structural constituent of cuticle |
| Uncharacterized protein LOC103510819 | 24.2 | 0.3 | 6.1 | Integral component of membrane |
| Trypsin | 24.2 | 0.4 | 6.0 | Proteolysis; serine-type endopeptidase activity |
| Serine ase stubble | 72.8 | 1.1 | 6.0 | GTPase activity, Proteolysis, Serine-type endopeptidase activity |
| Probable chitinase- 3 | 19.4 | 0.3 | 6.0 | Chitinase activity, Extracellular region |
| Fibroin heavy chain | 32.7 | 0.5 | 5.9 | Structural constituent of cuticle |
| Cuticle 7 | 49.6 | 0.8 | 5.9 | Structural constituent of cuticle |
| Glycine-rich cell wall structural -like | 37.4 | 0.7 | 5.8 | Anatomical structure development |
| Cuticular analogous to peritrophins 1-G | 12.5 | 0.2 | 5.7 | Chitin binding, Chitin metabolic process, Extracellular region |
| Cytochrome P450 4g15 | 26.1 | 0.5 | 5.7 | Iron ion binding, Oxidation-reduction process |
| Serine ase stubble isoform X2 | 16.7 | 0.4 | 5.5 | GTP binding, Serine-type endopeptidase activity |
| Location of vulva defective 1 isoform X2 | 12.7 | 0.3 | 5.5 | Membrane, Integral component of membrane |
| COPII coat assembly partial | 87.2 | 1.9 | 5.5 | Neuropeptide signaling pathway |
| Sal 1 | 20.9 | 0.5 | 5.5 | Nucleic acid binding |
| Glucose dehydrogenase [quinone]-like |
14.5 | 0.3 | 5.4 | Oxidation-reduction process |
| Proclotting enzyme-like | 14.8 | 0.3 | 5.4 | Proteolysis, Serine-type endopeptidase activity |
| Larval cuticle A3A-like | 24.2 | 0.7 | 5.2 | Structural constituent of cuticle |
| Glycine-rich cell wall structural -like | 12.9 | 0.3 | 5.2 | Chitin-based cuticle development, Structural constituent of chitin-based cuticle |
| Endocuticle structural glyco bd- partial | 168.6 | 4.8 | 5.1 | Structural constituent of cuticle, Extracellular space |
| Osiris 14 | 15.2 | 0.4 | 5.1 | Integral component of membrane |
| Cuticular precursor | 24.1 | 0.7 | 5.1 | Structural constituent of cuticle, Nucleic acid binding |
| Endocuticle structural glyco bd-2-like | 33.4 | 1.0 | 5.0 | Structural constituent of cuticle, Extracellular space |
FC = fold change of virus-exposed treatment relative to non-virus control treatment.
Real-time quantitative PCR
Fold changes of six randomly selected DE transcripts among the annotated genes were validated by real-time qPCR using the same total RNA preparations used for Illumina library preparation. Expression of target genes was normalized to actin internal reference gene selected from non-DE genes in the dataset. Relative expression levels and fold changes in target gene expression in virus-exposed and non-exposed thrips showed similar trends in qPCR and RNA-Seq for most transcripts tested. The exception was pectin lyase transcripts that showed a reduced ratio between virus-exposed and non-exposed expression levels in qPCR (Fig 3). Pearson correlation coefficient rp was 0.658 (P = 0.003) indicating a significant positive correlation between the RNA-Seq and qPCR data.
Fig 3. Validation of RNA-Seq gene expression by qPCR.
FPKM values obtained by RNA-Seq analysis and relative expression levels obtained by qPCR for six selected genes in CaCV-exposed and non-exposed Thrips palmi are shown. Values were multiplied by a factor of 1000. Error bars represent the standard error for three biological replicates.
Comparison of DE transcripts between T. palmi-CaCV and F. occidentalis-TSWV interactions
A comparison of the adult transcriptomes of T. palmi—CaCV (this study) and F. occidentalis—TSWV [30] using tblastx revealed a small number of transcripts that were differentially up-regulated in both thrips—tospovirus interactions (Table 4). This suggests potentially conserved responses by different thrips species across genera against tospoviruses from different serogroups that may be suitable targets for novel generic pest control.
Table 4. List of conserved up-regulated transcript sequences in two adult-stage thrips—tospovirus interactions.
| Tp transcript annotation | Focc—TSWV transcript code | Tp—CaCV transcript code | Log2 FC Focc | Log2 FC Tp | % sequence identity | Sequence coverage |
|---|---|---|---|---|---|---|
| DNase I | FOCC007280-RA | TP.8153.1 | + 2.62 | + 1.31 | 86 | 1186 nt |
| Uncharacterized protein LOC106129042 | TCONS_00032732 | TP.1784.1 | + 2.06 | + 2.47 | 81 | 194 aa |
| Endoglucanase | FOCC015899-RA | TP.19931.1, TP21769.1, TP.21770.1, TP.21771.1 | + 1.95 | + 1.2 to + 1.3 | 80 | 115 aa |
| Carbonic anhydrase | CUFF.8568.2 | TP.33694.1 | + 2.88 | + 2.08 | 80 | 92 aa |
| Uncharacterized protein LOC105690123 | CUFF.8228.1 CUFF.2322.1 |
>10 Isoforms >10 Isoforms |
+ 2.58 +2.64 |
up up |
62 60 |
< 200 aa < 200 aa |
| Hexamerin | FOCC009367-RA | TP.20198.1# | + 4.20 | + 1.81 | 45 | 33 aa |
| Hemocyanin subunit type 1 precursor | FOCC002013-RA | TP.20202.1# | + 13.31 | + 1.75 | 38 | 50 aa |
| Hexamerin, arylphorin subunit alpha | FOCC012829-RA | TP.25095.1# | + 8.87 | + 1.1 | 38 | 45 aa |
Focc = Frankliniella occidentalis; TSWV = tomato spotted wilt virus; Tp = Thrips palmi; CaCV = capsicum chlorosis virus; nt = nucleotides; aa = amino acids. FC = fold change of virus-exposed treatment in relative to non-virus control treatment.
# Annotated as phenol oxidase 2-like
The up-regulated genes common to both virus-vector interactions are associated with innate immunity and salivary glands. Hexamerins [log2 fold change 1.81 (T. palmi) vs 4.2 (F. occidentalis)] are multi-subunit storage proteins with phenol oxidase activity that are involved in insect innate immunity, humoral immune response to pathogens and disease resistance [59]. Similarly, hemocyanin subunit 1 precursor [log2 fold change 1.75 (T. palmi) vs 13.31 (F. occidentalis)] was shown to have phenol oxidase activity in melanogenesis [97]. Carbonic anhydrase [log2 fold change 2.08 (T. palmi) vs 2.88 (F. occidentalis)] has a physiological role in pH and ion regulation pathways. Endoglucanase [log2 fold change 1.2–1.3 (T. palmi) vs 1.95 (F. occidentalis)] is a salivary gland-associated cell wall digestive enzyme [88]. Four isoforms of this gene were identified in T. palmi transcriptome. Deoxyribonuclease I [log2 fold change 1.31 (T. palmi) vs 2.62 (F. occidentalis)] is an endonuclease that cleaves DNA and has been suggested to play a role in apoptosis [98]. This gene is also listed among the DE contigs with a log2 fold change of 2.82 associated with vector response to TSWV infection in F. fusca adult transcriptome [31], further validating the conserved response of up-regulating this thrips gene.
When down-regulated genes in adult transcriptomes were compared, none of the most highly down-regulated TP transcripts were shared between the two systems. However, genes of endocuticle structural glycoprotein [log2 fold change 5.0–7.1 (T. palmi) vs 4.5 (F. occidentalis)], which is a structural constituent of cuticle and alpha-tocopherol transfer-like [log2 fold change 7.0 (T. palmi) vs 4.9 (F. occidentalis)] which has been shown to be involved in antiviral immunity in Drosophila melanogaster [99] were down-regulated in F. occidentalis larvae [30] and T. palmi adults.
Discussion
Quantifying changes in global gene expression is one means of inferring cellular and physiological responses of insects to infection by insect-pathogenic viruses or insect-vectored plant-viruses [100]. This is especially relevant for viruses like tospoviruses that circulate and propagate inside insect cells and tissue systems, exploiting host cellular machinery to replicate and to complete their life-cycles [101, 102]. This study presents the first transcriptome for an insect in the genus Thrips. We analysed T. palmi transcriptome in response to CaCV infection to identify DE transcripts and to classify these genes by sequence homologies (ontologies) to known proteins to begin to dissect the global response of this thrips species to tospovirus infection. Here, we present hypotheses about the effect of virus infection on three dynamic processes in vector biology: innate immunity, growth and development, and fitness.
Innate immunity-related genes
Based on Blastx descriptions and GO terms for DE transcripts, we identified 86 up-regulated transcripts putatively associated with innate immune response that may have been triggered by CaCV infection (Table 2). The majority of these genes are likely involved in cellular immunity activated in lysosomes such as cathepsin B [50, 103, 104], lysosomal aspartic proteases and lysozyme C. We identified 10 cathepsin B genes that were up-regulated in virus-exposed T. palmi. Cathepsins are known to be involved in various biological processes including protein degradation, apoptosis and signalling activated in late endosome and lysosome and implicated in virus entry and replication [105, 106]. Therefore, it appears that CaCV infection may lead to cytopathological effect in thrips cells through cell damage and apoptosis. Another set of genes that were up-regulated in virus-exposed thrips encode glucose dehydrogenases that are known to participate in the initiation of cellular immune responses by encapsulation of pathogens [62].
Genes associated with melanisation seem to play a significant role in virus-exposed T. palmi. Besides pigmentation, melanin deposits and encapsulates foreign targets such as nematodes, parasitoids and virus-infected tissues and inhibits progression of infection [59, 107]. Enzymes such as phenoloxidases (PO) [59], serine proteases (SP) [60] and laccase [64] are the key players in the melanisation pathway. PO generate highly cytotoxic quinones that can inactivate invading viral pathogens [108]. In T. palmi transcriptome, we identified several PO genes and a CLIP domain-containing SP that were up-regulated in virus-exposed thrips. Previous studies have shown that PO cascade is activated in mosquitoes in response to Semliki Forest virus infection resulting in inhibition of virus spread in cell culture [109, 110]. CLIP domain-containing SP induce melanisation immune response by activating PO [59, 60]. In addition to PO and SP, other genes that participate in the melanisation process were identified, such as poly(U)-specific endoribonuclease homologues and laccase-5 [63–65].
Genes involved in other innate immune system pathways such as Toll, antigen processing and presentation, and complement and coagulation cascades (humoral immunity) were also found up-regulated. Among the up-regulated transcripts in CaCV-exposed thrips, 63 encoded proteolytic enzymes, including 30 serine protease and trypsin genes. According to GO assignment all serine proteases and trypsins in this thrips transcriptome were predicted to exhibit serine-type endopeptidase activity. The role that these proteases may play in virus-exposed thrips is not known. Previously it was shown that insect serine proteases and trypsin-like serine proteases are generally involved in hemolymph coagulation, activation of antimicrobial peptide synthesis, and melanin synthesis [75, 111, 112]. Therefore, these genes may have roles in thrips humoral immunity. Interestingly, among the immune-related genes, two genes showed >67% similarity to plant genes encoding pathogenesis-related (PR) protein 5 and elicitin 6.
Our study of T. palmi identified activation of different immunity-related pathways than the results reported by Zhang and collaborators [52] for F. occidentalis-TSWV, such as Toll, JAK-STAT and RNA interference. Either these pathways have no significant role in adult T. palmi against CaCV or they were active only in larvae at the time of virus acquisition and initial replication and intercellular movement, which was not captured by our adult transcriptome. In another study, Schneweis and colleagues [30] showed that in adult F. occidentalis exposed to TSWV, 75% of innate immune response related transcripts were annotated as insect storage proteins, hexamerins which are also associated with humoral immunity. We did not identify hexamerins among the DE transcripts of T. palmi suggestive of activation of different set of genes in T. palmi’s innate immunity. When compared with F. fusca-TSWV adult DE genes, we identified up-regulation of several similar immune system-related genes such as cathepsin B, aminopeptidase N, serine proteases and heat shock protein 70. Therefore, it is likely that more generally in adult thrips infected with tospoviruses, immune reactions like apoptosis, phagocytosis and proteolysis are activated.
Salivary gland-associated genes
Salivation is an essential process in insect feeding [113, 114] and virus inoculation of hosts [28, 115]. Various components of saliva are involved in extra-oral digestion of plant tissues and suppression or detoxification of host defence responses [116, 117]. In the present study, the majority of up-regulated putative salivary gland genes identified in this study appear to be involved in digestion (Table 2). Of the 20 lipases, up-regulated in virus-exposed thrips, 15 were lipase 3-like, 4 transcripts corresponded to lipase 1, and 1 was a H-B-like lipase. Lipases are also known to be involved in innate immunity [51]. To identify lipases that may be secreted components of saliva, signal peptide sequences were predicted in silico. All lipase 1 sequences lacked a signal peptide, hence were considered unlikely to be components of saliva and more likely involved in innate immunity. Of the 15 lipase 3 genes, nine sequences contained a signal peptide and a predicted cleavage site at their N-terminus, suggestive of secreted proteins. Lipases have previously been identified to be associated with salivary glands of other insects, including the potato leafhopper (Empoasca fabae) [87], whitefly (Bemisia tabaci) [61] and F. occidentalis [88]. Other large groups of enzymes that may be involved in general digestion are proteolytic enzymes, serine proteases and carboxypeptidases. Eleven of 15 serine proteases and trypsin-like serine proteases and carboxypeptidases in virus-exposed T. palmi, contained predicted signal peptides and cleavage sites. Similar proteins were identified in salivary gland transcriptomes of E. fabae and F. occidentalis [87, 88]. One of the serine proteases contained a CLIP domain. Because CLIP domain-containing serine proteases are known to induce melanisation [59] and the presence of a predicted signal peptide cleavage site, this protease may be a component of saliva which provides immunity by activation of Toll signalling pathway or PO cascade leading to melanisation [118, 119]. Up-regulation of several transcripts encoding secreted lipases and proteolytic enzymes implies that they participate in general digestion of lipids and proteins during thrips feeding.
Plant-feeding insects secrete a range of cell wall-degrading enzymes to release cell contents and to facilitate subsequent ingestion [120]. Pectin is one of the major polysaccharides in the plant cell wall and middle lamella [121]. Pectin lyase is a pectin-hydrolysing enzyme expressed in salivary glands of phytophagous insects, including F. occidentalis [88], E. fabae [87] and the plant bug Lygus hesperus [122]. Other cell wall degrading enzymes investigated from insects are laccase, β-glucosidase and endo-β-glucanase [87, 88, 123–125]. Presence of transcripts encoding pectin lyases, β-glucosidases and endoglucanases in the T. palmi transcriptome is consistent with findings in other insects. Based on signal peptide predictions, some of these enzymes may be secreted, a requisite for extra-oral digestion.
A α-amylase that may be involved in sugar metabolism was identified among up-regulated transcripts in virus-exposed thrips. This sequence lacked an obvious signal peptide, but its GO term ‘extracellular exosome’ suggests extracellular localization and a putative digestive role. α-amylases have been identified in the saliva of many insects including honeybee (Apis melifera) [126], mosquito (Ades aegypti) [127], silkworm (Bombyx mori) [128] and red flour beetle (Tribolium castaneum) [129], and also in salivary gland transcriptomes of thrips F. occidentalis [88] and E. fabae, [87]. Genes encoding several other sugar metabolic enzymes such as maltase, sucrase and β-glucosidase were identified in the F. occidentalis salivary gland transcriptome [88] but were not detected among the significantly DE genes in the T. palmi transcriptome.
Among the other salivary gland-associated genes identified in this study were angiotensin-converting enzyme-like (ACE) genes which may have a role in inhibition of plant defence [89, 91]. Wang and colleagues [89] suggest that pea aphid secreted ACE may digest plant peptide hormones in phloem sap and function as signal molecules to mount defence response against herbivorous insects, thereby supressing plant immune responses. The T. palmi ACE may have similar functions.
Fitness and fecundity-associated genes
Among the up-regulated genes in CaCV-exposed thrips, 54 transcripts represented vitellogenin (Vg) homologues (Table 2). Vg contigs were also highly up-regulated (log2 fold change >9) in adult and pupal transcriptome of F. fusca infected with TSWV [31]. The primary role of insect Vg is to transport egg yolk protein components synthesized extra-ovarially in fat bodies into growing oocytes by receptor-mediated endocytosis [130]. In female insects Vg is one of the most abundant proteins [131]. Accumulation of Vg in vesicles in insects is mediated by receptors of the low-density lipoprotein receptor (LDLR) family [111]. Among the up-regulated genes in virus-exposed thrips we identified two Vg receptors and a LDLR; one of the Vg receptor genes was highly abundant with a FPKM value of 1598 and a log2-fold change of 2.0. Abundance of Vg transcripts and their receptors in virus-exposed T. palmi implies transovarial transport of egg yolk protein components to the growing oocytes had been promoted in the presence of CaCV infection. Enhanced vitellin and vitellogenin levels in response to a plant virus infection was first reported from whitefly, MEAM1 (B biotype) B. tabaci that fed on tomato yellow leaf curl China virus-infected plants [93]. These authors speculated that the vector benefits from virus infection due to increased longevity and fecundity. A recent study has shown that TSWV has positive effects on the fecundity of F. occidentalis [132]. In contrast, leafhopper, Recilia dorsalis that fed on rice gall dwarf virus (RGDV)-infected plants showed significantly reduced longevity and fecundity associated with reduced levels of Vg compared to non-viruliferous individuals [94].
There are other reports that have shown involvement of Vg in immune responses of insects including citrus whitefly (Dialeurodes citri) [50], A. aegypti [95] and honeybee (Apis mellifera) [133]. Based on these precedents, enhanced Vg gene expression in virus-exposed T. palmi may play a role in innate immunity to CaCV infection. In support, Vg was found to be 9-fold up-regulated in pupae of F. fusca exposed to TSWV [31]. Another known role that Vg plays in plant virus-vector interactions is facilitation of vertical virus transmission, exemplified by transovarial transmission of rice stripe virus in small brown planthopper (Laodelphax striatellus) [134]. Similar to our findings in virus-exposed T. palmi, Vg was the most abundant transcript in virus-exposed small brown planthopper transcriptome [135]. Previously, it has been shown that TSWV is not vertically transmitted from viruliferous adult females to eggs [22]. Further, there is no evidence for vertical transmission in other tospovirus-thrips pathosystems and it is generally accepted that tospoviruses are not transovarially transmitted. However, existence of transovarial transmission of CaCV in T. palmi cannot be excluded due to a current lack of knowledge in tissue tropism and virus localization in the body of T. palmi. Furthermore, Vg was not among the most abundant unigenes and DE genes in F. occidentalis transcriptome infected with TSWV [52]. Therefore, it is possible that enhanced Vg expression is unique to CaCV-T. palmi interaction, but the exact role that Vg may play needs to be investigated further.
In summary, we have assembled a whole-body transcriptome of adult T. palmi and estimated differential abundance of transcripts when exposed to CaCV infection. The majority of DE transcripts did not contain Blastx annotations or conserved functional domains, but their differential abundance reflects unique roles in T. palmi-virus interaction and T. palmi biology in general. Although we cannot directly compare our data with studies of F. occidentalis [30] and F. fusca [31] exposed to TSWV, during the adult stages of these species, there were a few common GO terms assigned to DE transcripts such as hydrolase activity, nucleotide/nucleic acid binding, carbohydrate metabolic process and peptidase activity. In addition, the majority of annotations assigned to DE transcripts of T. palmi appeared to be species-specific. Aside from the inherent differences across thrips species and tospoviruses species, differences in the experimental methods and computational protocols and parameters used in data analyses could explain the weak overlap in transcripts responsive to virus across the three studies. For example, DE analyses for F. fusca [31] and T. palmi were performed using de novo transcriptome assemblies, whereas Schneweis and colleagues [30] used a draft genome reference for DE analysis for F. occidentalis.
We identified virus-responsive up-regulated DE transcripts that have putative roles in thrips innate immunity, feeding and fecundity. Down-regulated transcripts that are likely associated with structural component of cuticle and integral components of membrane may be responsible for delaying thrips development and increased longevity to ensure virus persistence and transmission. However, functions of those genes need to be experimentally validated in future. For example, transcripts associated with thrips feeding and fecundity could be candidates for potential targets in thrips and tospovirus management. In addition, transcriptomic data generated in this study will enrich genomic information of thrips and will allow functional studies on other economically important thrips and insects more broadly.
Supporting information
(DOCX)
Acknowledgments
We thank Prof Tsung-Chi Chen (Asian University, Taiwan) for providing CaCV isolate and host plants for the experiment, De-Fen Mou (Vector Laboratory, National Taiwan University) for the support extended during maintenance of thrips cultures and Dr Sonika Tyagi (Australian Genome Research Facility Ltd) for her helpful comments.
Data Availability
The raw sequence read data generated in this study have been deposited in the NCBI Sequence Read Archive as BioProject PRJNA498538 with accession numbers SAMN10316162 - 167.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Mound L. Thysanoptera (Thrips) of the World–a checklist 2012. [cited 2016]. Available from: http://anic.ento.csiro.au/thrips/checklist.html. [Google Scholar]
- 2.German TL, Ullman DE, Moyer JW. Tospoviruses: diagnosis, molecular biology, phylogeny, and vector relationships. Annu Rev Phytopathol. 1992;30(1):315–48. [DOI] [PubMed] [Google Scholar]
- 3.Rotenberg D, Jacobson AL, Schneweis DJ, Whitfield AE. Thrips transmission of tospoviruses. Curr Opin Plant Virol. 2015;15:80–9. [DOI] [PubMed] [Google Scholar]
- 4.Riley DG, Joseph SV, Srinivasan R, Diffie S. Thrips vectors of tospoviruses. J Integr Pest Manag. 2011;2(1):1–10. [Google Scholar]
- 5.Mound LA. So many thrips-so few tospoviruses In: Marullo R, Mound L, editors. Thrips and Tospoviruses: Proceedings of the 7th International Symposium on Thysanoptera. Canberra, Australia: Australian National Insect Collection; 2002:15–8. [Google Scholar]
- 6.Rebijith K, Asokan R, Hande HR, Kumar NK. The first report of miRNAs from a thysanopteran insect, Thrips palmi Karny using high-throughput sequencing. PLoS One. 2016; 11(9):e0163635 10.1371/journal.pone.0163635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen C, Chen T, Lin Y, Yeh S, Hsu H. A chlorotic spot disease on calla lilies (Zantedeschia spp.) is caused by a tospovirus serologically but distantly related to watermelon silver mottle virus. Plant Dis. 2005;89(5):440–5. [DOI] [PubMed] [Google Scholar]
- 8.Lakshmi KV, Wightman J, Reddy D, Rao GR, Buiel A, Reddy D. Transmission of peanut bud necrosis virus by Thrips palmi in India In: Parker B, Skinner M, Lewis T, editors. Thrips Biology and Management. NATO ASI Series New York, USA: Springer; 1995:179–84. [Google Scholar]
- 9.Kato K, Handa K, Kameya-Iwaki M. Melon yellow spot virus: a distinct species of the genus Tospovirus isolated from melon. Phytopathology. 2000;90(4):422–6. 10.1094/PHYTO.2000.90.4.422 [DOI] [PubMed] [Google Scholar]
- 10.Seepiban C, Gajanandana O, Attathom T, Attathom S. Tomato necrotic ringspot virus, a new tospovirus isolated in Thailand. Arch Virol. 2011;156(2):263–74. 10.1007/s00705-010-0856-0 [DOI] [PubMed] [Google Scholar]
- 11.Reddy D, Ratna A, Sudarshana M, Poul F, Kumar IK. Serological relationships and purification of bud necrosis virus, a tospovirus occurring in peanut (Arachis hypogaea L.) in India. Ann Appl Biol. 1992;120(2):279–86. [Google Scholar]
- 12.Iwaki M, Honda Y, Hanada K, Tochihara H, Yonaha T, Hokama K, et al. Silver mottle disease of watermelon caused by tomato spotted wilt virus. Plant Dis. 1984;68(11):1006–8. [Google Scholar]
- 13.Persley D, Thomas J, Sharman M. Tospoviruses-an Australian perspective. Australas Plant Pathol. 2006;35(2):161–80. [Google Scholar]
- 14.Kritzman A, Gera A, Raccah B, Van Lent J, Peters D. The route of tomato spotted wilt virus inside the thrips body in relation to transmission efficiency. Arch Virol. 2002;147(11):2143–56. 10.1007/s00705-002-0871-x [DOI] [PubMed] [Google Scholar]
- 15.Nagata T, Inoue-Nagata AK, Smid HM, Goldbach R, Peters D. Tissue tropism related to vector competence of Frankliniella occidentalis for tomato spotted wilt tospovirus. J Gen Virol. 1999;80(2):507–15. [DOI] [PubMed] [Google Scholar]
- 16.Ullman D, German T, Sherwood J, Westcot D, Cantone F. Tospovirus replication in insect vector cells—immunocytochemical evidence that the nonstructural protein encoded by the S-RNA of Tomato spotted wilt tospovirus is present in thrips vector cells. Phytopathology. 1993;83: 456–63. [Google Scholar]
- 17.Whitfield AE, Ullman DE, German TL. Tospovirus-thrips interactions. Annu Rev Phytopathol. 2005;43:459–89. 10.1146/annurev.phyto.43.040204.140017 [DOI] [PubMed] [Google Scholar]
- 18.Montero-Astúa M, Ullman DE, Whitfield AE. Salivary gland morphology, tissue tropism and the progression of tospovirus infection in Frankliniella occidentalis. Virology. 2016;493:39–51. 10.1016/j.virol.2016.03.003 [DOI] [PubMed] [Google Scholar]
- 19.Stumpf CF, Kennedy GG. Effects of tomato spotted wilt virus (TSWV) isolates, host plants, and temperature on survival, size, and development time of Frankliniella fusca. Entomol Exp Appl. 2005;114(3):215–25. [Google Scholar]
- 20.Shrestha A, Srinivasan R, Riley DG, Culbreath AK. Direct and indirect effects of a thrips‐transmitted tospovirus on the preference and fitness of its vector, Frankliniella fusca. Entomol Exp Appl. 2012;145(3):260–71. [Google Scholar]
- 21.De Angelis J, Sether D, Rossignol P. Survival, development, and reproduction in western flower thrips (Thysanoptera: Thripidae) exposed to impatiens necrotic spot virus. Environ Entomol. 1993;22(6):1308–12. [Google Scholar]
- 22.Wijkamp I, Goldbach R, Peters D. Propagation of tomato spotted wilt virus in Frankliniella occidentalis does neither result in pathological effects nor in transovarial passage of the virus. Entomol Exp Appl. 1996;81(3):285–92. [Google Scholar]
- 23.Roca E, Aramburu J, Moriones E. Comparative host reactions and Frankliniella occidentalis transmission of different isolates of tomato spotted wilt tospovirus from Spain. Plant Pathol. 1997;46(3):407–15. [Google Scholar]
- 24.Chen W-T, Tseng C-H, Tsai C-W. Effect of watermelon silver mottle virus on the life history and feeding preference of Thrips palmi. PLoS One. 2014;9(7):e102021 10.1371/journal.pone.0102021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maris P, Joosten N, Goldbach R, Peters D. Tomato spotted wilt virus infection improves host suitability for its vector Frankliniella occidentalis. Phytopathology. 2004;94(7):706–11. 10.1094/PHYTO.2004.94.7.706 [DOI] [PubMed] [Google Scholar]
- 26.Stumpf CF, Kennedy GG. Effects of tomato spotted wilt virus isolates, host plants, and temperature on survival, size, and development time of Frankliniella occidentalis. Entomol Exp Appl. 2007;123(2):139–47. [Google Scholar]
- 27.Belliure B, Janssen A, Maris PC, Peters D, Sabelis MW. Herbivore arthropods benefit from vectoring plant viruses. Ecol Lett. 2005; 8:70–9. [Google Scholar]
- 28.Stafford CA, Walker GP, Ullman DE. Infection with a plant virus modifies vector feeding behavior. Proc Natl Acad Sci USA. 2011;108(23):9350–5. 10.1073/pnas.1100773108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogada PA, Moualeu DP, Poehling H-M. Predictive models for tomato spotted wilt virus spread dynamics, considering Frankliniella occidentalis specific life processes as influenced by the virus. PLoS One. 2016;11(5): e0154533 10.1371/journal.pone.0154533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneweis DJ, Whitfield AE, Rotenberg D. Thrips developmental stage-specific transcriptome response to tomato spotted wilt virus during the virus infection cycle in Frankliniella occidentalis, the primary vector. Virology. 2017;500:226–37. 10.1016/j.virol.2016.10.009 [DOI] [PubMed] [Google Scholar]
- 31.Shrestha A, Champagne DE, Culbreath AK, Rotenberg D, Whitfield AE, Srinivasan R. Transcriptome changes associated with tomato spotted wilt virus infection in various life stages of its thrips vector, Frankliniella fusca (Hinds). J Gen Virol. 2017;98(8):2156–70. 10.1099/jgv.0.000874 [DOI] [PubMed] [Google Scholar]
- 32.Widana Gamage S, Persley DM, Higgins CM, Dietzgen RG. First complete genome sequence of a capsicum chlorosis tospovirus isolate from Australia with an unusually large S RNA intergenic region. Arch Virol. 2015;160(3):869–72. 10.1007/s00705-014-2324-8 [DOI] [PubMed] [Google Scholar]
- 33.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8(3):186–94. [PubMed] [Google Scholar]
- 34.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 1998;8(3):175–85. [DOI] [PubMed] [Google Scholar]
- 35.Song L, Florea L. Rcorrector: efficient and accurate error correction for Illumina RNA-seq reads. GigaScience. 2015;4(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–512. 10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith-Unna R, Boursnell C, Patro R, Hibberd J, Kelly S. TransRate: reference free quality assessment of de novo transcriptome assemblies. Genome Res. 2016;26:1134–44. 10.1101/gr.196469.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–78. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–30. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 41.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41(10):e108 10.1093/nar/gkt214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(3):290–295 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Social Science Statistics [cited 2017]. Available from: www.socscistatistics.com.
- 44.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–6. 10.1093/bioinformatics/bti610 [DOI] [PubMed] [Google Scholar]
- 45.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23(10):1289–91. 10.1093/bioinformatics/btm091 [DOI] [PubMed] [Google Scholar]
- 47.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):e115 10.1093/nar/gks596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3(6):1101–8. [DOI] [PubMed] [Google Scholar]
- 49.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 50.Yu S, Ding L, Luo R, Li X, Yang J, Liu H, et al. Identification of immunity-related genes in Dialeurodes citri against entomopathogenic fungus Lecanicillium attenuatum by RNA-Seq analysis. PLoS One. 2016;11(9):e0162659 10.1371/journal.pone.0162659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ponnuvel KM, Nakazawa H, Furukawa S, Asaoka A, Ishibashi J, Tanaka H, et al. A lipase isolated from the silkworm Bombyx mori shows antiviral activity against nucleopolyhedrovirus. J Virol. 2003;77(19):10725–9. 10.1128/JVI.77.19.10725-10729.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Z, Zhang P, Li W, Zhang J, Huang F, Yang J, et al. De novo transcriptome sequencing in Frankliniella occidentalis to identify genes involved in plant virus transmission and insecticide resistance. Genomics. 2013;101(5):296–305. 10.1016/j.ygeno.2013.02.005 [DOI] [PubMed] [Google Scholar]
- 53.Hamilton C, Lejeune BT, Rosengaus RB. Trophallaxis and prophylaxis: social immunity in the carpenter ant Camponotus pennsylvanicus. Biol Lett. 2011;7(1):89–92. 10.1098/rsbl.2010.0466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanaka H, Yamakawa M. Regulation of the innate immune responses in the silkworm, Bombyx mori. Invert Surv J. 2011;8(1):59–69 [Google Scholar]
- 55.Badillo-Vargas I, Rotenberg D, Schneweis D, Hiromasa Y, Tomich J, Whitfield A. Proteomic analysis of Frankliniella occidentalis and differentially-expressed proteins in response to tomato spotted wilt virus infection. J Virol. 2012;86(16):8793–8809. 10.1128/JVI.00285-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clayton AM, Dong Y, Dimopoulos G. The Anopheles innate immune system in the defense against malaria infection. J Innate Immun. 2014;6(2):169–81. 10.1159/000353602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hernández-Martínez S, Lanz-Mendoza H, Martínez-Barnetche J, Rodríguez MH. Antimicrobial properties of Anopheles albimanus pericardial cells. Cell Tissue Res. 2013;351(1):127–37. 10.1007/s00441-012-1505-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wippler J, Kleiner M, Lott C, Gruhl A, Abraham PE, Giannone RJ, et al. Transcriptomic and proteomic insights into innate immunity and adaptations to a symbiotic lifestyle in the gutless marine worm Olavius algarvensis. BMC genomics. 2016;17(1):942 10.1186/s12864-016-3293-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.González‐Santoyo I, Córdoba‐Aguilar A. Phenoloxidase: a key component of the insect immune system. Entomol Exp Appl. 2012;142(1):1–16. [Google Scholar]
- 60.An C, Zhang M, Chu Y, Zhao Z. Serine protease MP2 activates prophenoloxidase in the melanization immune response of Drosophila melanogaster. PLoS One. 2013;8(11):e79533 10.1371/journal.pone.0079533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Su Y-L, Li J-M, Li M, Luan J-B, Ye X-D, Wang X-W, et al. Transcriptomic analysis of the salivary glands of an invasive whitefly. PLoS One. 2012;7(6):e39303 10.1371/journal.pone.0039303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Antúnez K, Martín‐Hernández R, Prieto L, Meana A, Zunino P, Higes M. Immune suppression in the honey bee (Apis mellifera) following infection by Nosema ceranae (Microsporidia). Environ Microbiol. 2009;11(9):2284–90. 10.1111/j.1462-2920.2009.01953.x [DOI] [PubMed] [Google Scholar]
- 63.Pascale M, Laurino S, Vogel H, Grimaldi A, Monné M, Riviello L, et al. The Lepidopteran endoribonuclease-U domain protein P102 displays dramatically reduced enzymatic activity and forms functional amyloids. Dev Comp Immunol. 2014;47(1):129–39. 10.1016/j.dci.2014.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shao Q, Yang B, Xu Q, Li X, Lu Z, Wang C, et al. Hindgut innate immunity and regulation of fecal microbiota through melanization in insects. J Biol Chem. 2012;287(17):14270–9. 10.1074/jbc.M112.354548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Viljakainen L. Evolutionary genetics of insect innate immunity. Brief Func Genomics. 2015;14(6):407–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ishii K, Hamamoto H, Kamimura M, Nakamura Y, Noda H, Imamura K, et al. The insect cytokine paralytic peptide (PP) induces cellular and humoral immune responses in the silkworm Bombyx mori. J Biol Chem. 2010;285(37):28635–42. 10.1074/jbc.M110.138446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu Y, Shen D, Zhou F, Wang G, An C. Identification of immunity-related genes in Ostrinia furnacalis against entomopathogenic fungi by RNA-seq analysis. PLoS One. 2014;9(1):e86436 10.1371/journal.pone.0086436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lavazec C, Boudin C, Lacroix R, Bonnet S, Diop A, Thiberge S, et al. Carboxypeptidases B of Anopheles gambiae as targets for a Plasmodium falciparum transmission-blocking vaccine. Infect Immun. 2007;75(4):1635–42. 10.1128/IAI.00864-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zou Z, Lopez DL, Kanost MR, Evans JD, Jiang H. Comparative analysis of serine protease‐related genes in the honey bee genome: possible involvement in embryonic development and innate immunity. Insect Mol Biol. 2006;15(5):603–14. 10.1111/j.1365-2583.2006.00684.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang L, Ho B, Ding JL. Transcriptional regulation of limulus Factor C: Repression of an NFKB motif modulates its responsiveness to bacterial lipopolysaccharide. J Biol Chem. 2003: 49428–37. 10.1074/jbc.M306641200 [DOI] [PubMed] [Google Scholar]
- 71.Bulmer MS, Bachelet I, Raman R, Rosengaus RB, Sasisekharan R. Targeting an antimicrobial effector function in insect immunity as a pest control strategy. Proc Natl Acad Sci USA. 2009;106(31):12652–7. 10.1073/pnas.0904063106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ligoxygakis P, Pelte N, Hoffmann JA, Reichhart J-M. Activation of Drosophila Toll during fungal infection by a blood serine protease. Science. 2002;297(5578):114–6. 10.1126/science.1072391 [DOI] [PubMed] [Google Scholar]
- 73.Zou Z, Evans JD, Lu Z, Zhao P, Williams M, Sumathipala N, et al. Comparative genomic analysis of the Tribolium immune system. Genome Biol. 2007;8(8):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Merkling SH, Overheul GJ, Van Mierlo JT, Arends D, Gilissen C, Van Rij RP. The heat shock response restricts virus infection in Drosophila. Sci Rep. 2015;5:12758 10.1038/srep12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gorman MJ, Paskewitz SM. Serine proteases as mediators of mosquito immune responses. Insect Biochem Mol Biol. 2001;31(3):257–62. [DOI] [PubMed] [Google Scholar]
- 76.Ryabov EV, Wood GR, Fannon JM, Moore JD, Bull JC, Chandler D, et al. A virulent strain of deformed wing virus (DWV) of honeybees (Apis mellifera) prevails after Varroa destructor-mediated, or in vitro, transmission. PLoS Pathog. 2014;10(6):e1004230 10.1371/journal.ppat.1004230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gao K, Zhang S. Ovochymase in amphioxus Branchiostoma belcheri is an ovary-specific trypsin-like serine protease with an antibacterial activity. Dev Comp Immunol. 2009;33(12):1219–28. 10.1016/j.dci.2009.07.002 [DOI] [PubMed] [Google Scholar]
- 78.Molina-Cruz A, Gupta L, Richardson J, Bennett K, Black IV W, Barillas-Mury C. Effect of mosquito midgut trypsin activity on dengue-2 virus infection and dissemination in Aedes aegypti. Am J Trop Med Hyg. 2005;72(5):631–7. [PubMed] [Google Scholar]
- 79.Aguilar R, Jedlicka AE, Mintz M, Mahairaki V, Scott AL, Dimopoulos G. Global gene expression analysis of Anopheles gambiae responses to microbial challenge. Insect Biochem Mol Biol. 2005;35(7):709–19. 10.1016/j.ibmb.2005.02.019 [DOI] [PubMed] [Google Scholar]
- 80.Gupta SK, Kupper M, Ratzka C, Feldhaar H, Vilcinskas A, Gross R, et al. Scrutinizing the immune defence inventory of Camponotus floridanus applying total transcriptome sequencing. BMC Genomics. 2015;16(1):540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Robertson AS, Belorgey D, Lilley KS, Lomas DA, Gubb D, Dafforn TR. Characterization of the necrotic protein that regulates the Toll-mediated immune response in Drosophila. J Biol Chem. 2002:278(8): 6175–6180. [DOI] [PubMed] [Google Scholar]
- 82.Wang Y, Xiao D, Wang R, Li F, Zhang F, Wang S. Deep sequencing-based transcriptome analysis reveals the regulatory mechanism of Bemisia tabaci (Hemiptera: Aleyrodidae) nymph parasitized by Encarsia sophia (Hymenoptera: Aphelinidae). PLoS One. 2016;11(6):e0157684 10.1371/journal.pone.0157684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Han B, Fang Y, Feng M, Lu X, Huo X, Meng L, et al. In-depth phosphoproteomic analysis of royal jelly derived from western and eastern honeybee species. J Proteome Res. 2014;13(12):5928–43. 10.1021/pr500843j [DOI] [PubMed] [Google Scholar]
- 84.De Smet L, Ravoet J, Wenseleers T, De Graaf DC. Expression of key components of the RNAi machinery are suppressed in Apis mellifera that suffer a high virus infection. Entomol Sci. 2017;20(1):76–85. [Google Scholar]
- 85.Ponchet M, Panabieres F, Milat M-L, Mikes V, Montillet J-L, Suty L, et al. Are elicitins cryptograms in plant-Oomycete communications? Cell Mol Life Sci. 1999;56(11–12):1020–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van Loon LC, Rep M, Pieterse CM. Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol. 2006;44:135–62. 10.1146/annurev.phyto.44.070505.143425 [DOI] [PubMed] [Google Scholar]
- 87.DeLay B, Mamidala P, Wijeratne A, Wijeratne S, Mittapalli O, Wang J, et al. Transcriptome analysis of the salivary glands of potato leafhopper, Empoasca fabae. J Insect Physiol. 2012;58(12):1626–34. 10.1016/j.jinsphys.2012.10.002 [DOI] [PubMed] [Google Scholar]
- 88.Stafford-Banks CA, Rotenberg D, Johnson BR, Whitfield AE, Ullman DE. Analysis of the salivary gland transcriptome of Frankliniella occidentalis. PLoS One. 2014;9(4):e94447 10.1371/journal.pone.0094447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang W, Luo L, Lu H, Chen S, Kang L, Cui F. Angiotensin-converting enzymes modulate aphid–plant interactions. Sci Rep. 2015; 5:8885 10.1038/srep08885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Girard YA, Mayhew GF, Fuchs JF, Li H, Schneider BS, McGee CE, et al. Transcriptome changes in Culex quinquefasciatus (Diptera: Culicidae) salivary glands during West Nile virus infection. J Med Entomol. 2010;47(3):421–35. [DOI] [PubMed] [Google Scholar]
- 91.Li Z, An X-K, Liu Y-D, Hou M-L. Transcriptomic and expression analysis of the salivary glands in white-backed planthoppers, Sogatella furcifera. PLoS One. 2016;11(7):e0159393 10.1371/journal.pone.0159393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Das S, Radtke A, Choi Y-J, Mendes AM, Valenzuela JG, Dimopoulos G. Transcriptomic and functional analysis of the Anopheles gambiae salivary gland in relation to blood feeding. BMC Genomics. 2010;11(1):566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guo J-Y, Dong S-Z, Yang X-l, Cheng L, Wan F-H, Liu S-S, et al. Enhanced vitellogenesis in a whitefly via feeding on a begomovirus-infected plant. PLoS One. 2012;7(8):e43567 10.1371/journal.pone.0043567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen Y, Lu C, Li M, Wu W, Zhou G, Wei T. Adverse effects of rice gall dwarf virus upon its insect vector Recilia dorsalis (Hemiptera: Cicadellidae). Plant Dis. 2016;100(4):784–90. [DOI] [PubMed] [Google Scholar]
- 95.Raikhel AS, Kokoza VA, Zhu J, Martin D, Wang S-F, Li C, et al. Molecular biology of mosquito vitellogenesis: from basic studies to genetic engineering of antipathogen immunity. Insect Biochem Mol Biol. 2002;32(10):1275–86. [DOI] [PubMed] [Google Scholar]
- 96.Davies JR, Svensäter G, Herzberg MC. Identification of novel LPXTG-linked surface proteins from Streptococcus gordonii. Microbiology. 2009;155(6):1977–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cerenius L, Söderhäll K. The prophenoloxidase‐activating system in invertebrates. Immunol Rev. 2004;198(1):116–26. [DOI] [PubMed] [Google Scholar]
- 98.Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391(6662):43 10.1038/34112 [DOI] [PubMed] [Google Scholar]
- 99.Kemp C, Mueller S, Goto A, Barbier V, Paro S, Bonnay F, et al. Broad RNA interference–mediated antiviral immunity and virus-specific inducible responses in Drosophila. J Immunol. 2013;190(2):650–8. 10.4049/jimmunol.1102486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang L, Tang N, Gao X, Guo D, Chang Z, Fu Y, et al. Understanding the immune system architecture and transcriptome responses to southern rice black-streaked dwarf virus in Sogatella furcifera. Sci Rep. 2016;6:36254 10.1038/srep36254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dietzgen RG, Mann KS, Johnson KN. Plant virus–insect vector interactions: current and potential future research directions. Viruses. 2016;8(11):303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Whitfield AE, Falk BW, Rotenberg D. Insect vector-mediated transmission of plant viruses. Virology. 2015; 479:278–289. 10.1016/j.virol.2015.03.026 [DOI] [PubMed] [Google Scholar]
- 103.Rispe C, Kutsukake M, Doublet V, Hudaverdian S, Legeai F, Simon J-C, et al. Large gene family expansion and variable selective pressures for cathepsin B in aphids. Mol Biol Evol. 2008;25(1):5–17. 10.1093/molbev/msm222 [DOI] [PubMed] [Google Scholar]
- 104.Ford L, Zhang J, Liu J, Hashmi S, Fuhrman JA, Oksov Y, et al. Functional analysis of the cathepsin-like cysteine protease genes in adult Brugia malayi using RNA interference. PLoS Negl Trop Dis. 2009;3(2):e377 10.1371/journal.pntd.0000377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kubo Y, Hayashi H, Matsuyama T, Sato H, Yamamoto N. Retrovirus entry by endocytosis and cathepsin proteases. Adv Virol. 2012: 640894 10.1155/2012/640894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sim S, Ramirez JL, Dimopoulos G. Dengue virus infection of the Aedes aegypti salivary gland and chemosensory apparatus induces genes that modulate infection and blood-feeding behavior. PLoS Pathog. 2012;8(3):e1002631 10.1371/journal.ppat.1002631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rohrmann GF. Host resistance, susceptibility and the effect of viral infection on host molecular biology Baculovirus molecular biology. 3rd ed Bethesda, MD, USA: NCBI; 2013. [Google Scholar]
- 108.Ourth DD, Renis HE. Antiviral melanization reaction of Heliothis virescens hemolymph against DNA and RNA viruses in vitro. Comp Biochem Physiol B Biochem Mol Biol. 1993;105(3):719–23. [DOI] [PubMed] [Google Scholar]
- 109.Rodriguez-Andres J, Rani S, Varjak M, Chase-Topping ME, Beck MH, Ferguson MC, et al. Phenoloxidase activity acts as a mosquito innate immune response against infection with Semliki Forest virus. PLoS Pathog. 2012;8(11):e1002977 10.1371/journal.ppat.1002977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jermy A. Host response: PO-faced response to SFV. Nature Rev Microbiol. 2013;11(1):4–5. [DOI] [PubMed] [Google Scholar]
- 111.Sappington TW, Raikhel AS. Molecular characteristics of insect vitellogenins and vitellogenin receptors. Insect Biochem Molec. 1998;28(5):277–300. [DOI] [PubMed] [Google Scholar]
- 112.Tang H, Kambris Z, Lemaitre B, Hashimoto C. Two proteases defining a melanization cascade in the immune system of Drosophila. J Biol Chem. 2006;281(38):28097–104. 10.1074/jbc.M601642200 [DOI] [PubMed] [Google Scholar]
- 113.Hogenhout SA, Bos JIB. Effector proteins that modulate plant-insect interactions. Curr Opin Plant Biol. 2011; 14:422–8. 10.1016/j.pbi.2011.05.003 [DOI] [PubMed] [Google Scholar]
- 114.Mutti NS, Louis J, Pappan LK, Pappan K, Begum K, Chen MS, Park Y, Dittmer N, Marshall J, Reese JC, Reeck GR. A protein from the salivary glands of the pea aphid, Acyrthosiphon pisum, is essential in feeding on a host plant. Proc Natl Acad Sci USA. 2008; 105:9965–9. 10.1073/pnas.0708958105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Girard YA, Schneider BS, McGee CE, Wen J, Han VC, Popov V, et al. Salivary gland morphology and virus transmission during long-term cytopathologic West Nile virus infection in Culex mosquitoes. Am J Trop Med Hyg. 2007;76(1):118–28. [PubMed] [Google Scholar]
- 116.Kaloshian I, Walling LL. Hemipteran and dipteran pests: Effectors and plant host immune regulators. J Integr Plant Biol. 2016; 58(4):350–61. 10.1111/jipb.12438 [DOI] [PubMed] [Google Scholar]
- 117.Villarroel CA, Jonckheere W, Alba JM, Glas JJ, Dermauw W, Haring MA, Van Leeuwen, T, Schuurink, RC, Kant, MR. Salivary proteins of spider mites suppress defenses in Nicotiana benthamiana and promote mite reproduction. Plant J. 2016; 86(2):119–31. 10.1111/tpj.13152 [DOI] [PubMed] [Google Scholar]
- 118.Kellenberger C, Leone P, Coquet L, Jouenne T, Reichhart J-M, Roussel A. Structure-function analysis of grass clip serine protease involved in Drosophila Toll pathway activation. J Biol Chem. 2011;286(14):12300–7. 10.1074/jbc.M110.182741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kanost MR, Jiang H. Clip-domain serine proteases as immune factors in insect hemolymph. Curr Opin Insect Sci. 2015;11:47–55. 10.1016/j.cois.2015.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhu YC, Yao J, Luttrell R. Identification of genes potentially responsible for extra-oral digestion and overcoming plant defense from salivary glands of the tarnished plant bug (Lygus lineolaris) using cDNA sequencing. J Insect Sci. 2016;16(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Caffall KH, Mohnen D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr Res. 2009;344(14):1879–900. 10.1016/j.carres.2009.05.021 [DOI] [PubMed] [Google Scholar]
- 122.de la Paz Celorio-Mancera M, Greve LC, Teuber LR, Labavitch JM. Activity in Lygus hesperus (knight) salivary glands. Arch Insect Biochem Physiol. 2009;70(2):122–35. 10.1002/arch.20282 [DOI] [PubMed] [Google Scholar]
- 123.Harmel N, Létocart E, Cherqui A, Giordanengo P, Mazzucchelli G, Guillonneau F, et al. Identification of aphid salivary proteins: a proteomic investigation of Myzus persicae. Insect Mol Biol. 2008;17(2):165–74. 10.1111/j.1365-2583.2008.00790.x [DOI] [PubMed] [Google Scholar]
- 124.Hattori M, Tsuchihara K, Noda H, Konishi H, Tamura Y, Shinoda T, et al. Molecular characterization and expression of laccase genes in the salivary glands of the green rice leafhopper, Nephotettix cincticeps (Hemiptera: Cicadellidae). Insect Biochem Molec. 2010;40(4):331–8. [DOI] [PubMed] [Google Scholar]
- 125.Watanabe H, Tokuda G. Cellulolytic systems in insects. Annu Rev Entomol. 2010;55:609–32. 10.1146/annurev-ento-112408-085319 [DOI] [PubMed] [Google Scholar]
- 126.Ohashi K, Natori S, Kubo T. Expression of amylase and glucose oxidase in the hypopharyngeal gland with an age‐dependent role change of the worker honeybee (Apis mellifera L.). Eur J Biochem. 1999;265(1):127–33. [DOI] [PubMed] [Google Scholar]
- 127.Grossman G, James A. The salivary glands of the vector mosquito, Aedes aegypti, express a novel member of the amylase gene family. Insect Mol Biol. 1993;1(4):223–32. [DOI] [PubMed] [Google Scholar]
- 128.Ngernyuang N, Kobayashi I, Promboon A, Ratanapo S, Tamura T, Ngernsiri L. Cloning and expression analysis of the Bombyx mori α-amylase gene (Amy) from the indigenous Thai silkworm strain, Nanglai. J Insect Sci. 2011;11(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Feng GH, Richardson M, Chen MS, Kramer KJ, Morgan TD, Reeck GR. α-Amylase inhibitors from wheat: amino acid sequences and patterns of inhibition of insect and human α-amylases. Insect Biochem Molec. 1996;26(5):419–26. [DOI] [PubMed] [Google Scholar]
- 130.Van Antwerpen R, Pham D, Ziegler R. Accumulation of lipids in insect oocytes. Progress in Vitellogenesis. 2005;9. [Google Scholar]
- 131.Tufail M, Takeda M. Insect vitellogenin/lipophorin receptors: molecular structures, role in oogenesis, and regulatory mechanisms. J Insect Physiol. 2009;55(2):88–104. [DOI] [PubMed] [Google Scholar]
- 132.Shalileh S, Ogada PA, Moualeu DP, Poehling H-M. Manipulation of Frankliniella occidentalis (Thysanoptera: Thripidae) by tomato spotted wilt virus (tospovirus) via the host plant nutrients to enhance its transmission and spread. Environ Entomol. 2016;45(5):1235–42. 10.1093/ee/nvw102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Amdam GV, Simões ZL, Hagen A, Norberg K, Schrøder K, Mikkelsen Ø, et al. Hormonal control of the yolk precursor vitellogenin regulates immune function and longevity in honeybees. Exp Geront. 2004;39(5):767–73. [DOI] [PubMed] [Google Scholar]
- 134.Huo Y, Liu W, Zhang F, Chen X, Li L, Liu Q, et al. Transovarial transmission of a plant virus is mediated by vitellogenin of its insect vector. PLoS Pathog. 2014;10(3):e1003949 10.1371/journal.ppat.1003949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang F, Guo H, Zheng H, Zhou T, Zhou Y, Wang S, et al. Massively parallel pyrosequencing-based transcriptome analyses of small brown planthopper (Laodelphax striatellus), a vector insect transmitting rice stripe virus (RSV). BMC Genomics. 2010;11(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
The raw sequence read data generated in this study have been deposited in the NCBI Sequence Read Archive as BioProject PRJNA498538 with accession numbers SAMN10316162 - 167.