We engineered a light-responsive, quorum quenching biofilm and demonstrated its application in mitigating membrane biofouling.
Abstract
Quorum quenching (QQ) has been reported to be a promising approach for membrane biofouling control. Entrapment of QQ bacteria in porous matrices is required to retain them in continuously operated membrane processes and to prevent uncontrollable biofilm formation by the QQ bacteria on membrane surfaces. It would be more desirable if the formation and dispersal of biofilms by QQ bacteria could be controlled so that the QQ bacterial cells are self-immobilized, but the QQ biofilm itself still does not compromise membrane performance. In this study, we engineered a QQ bacterial biofilm whose growth and dispersal can be modulated by light through a dichromatic, optogenetic c-di-GMP gene circuit in which the bacterial cells sense near-infrared (NIR) light and blue light to adjust its biofilm formation by regulating the c-di-GMP level. We also demonstrated the potential application of the engineered light-responsive QQ biofilm in mitigating biofouling of water purification forward osmosis membranes. The c-di-GMP–targeted optogenetic approach for controllable biofilm development we have demonstrated here should prove widely applicable for designing other controllable biofilm-enabled applications such as biofilm-based biocatalysis.
INTRODUCTION
As the global demand for fresh water increases, membrane technology has emerged as one of the leading approaches for water treatment from lower-quality water sources (1). In recent years, forward osmosis (FO) technology has been applied to various water treatment processes because of its lower energy consumption and lower fouling propensity than pressure-driven approaches such as nanofiltration and reverse osmosis (2). Although FO processes tend to foul less than pressurized membrane processes, biofouling, i.e., undesirable attachment and growth of microorganisms on membranes, still remains as one of the most important challenges in practical applications of FO (3).
In biofouling, cells form a structured microbial consortium on membrane surfaces known as biofilms, in which cells are embedded within a self-produced extracellular matrix (3). Quorum sensing (QS), i.e., a chemical-based intercellular communication between bacterial cells, has been shown to play an important role in biofilm formation (4). Hence, inhibiting QS system or quorum quenching (QQ) has been reported to be a promising approach for biofouling control (4). Recent studies have shown that N-acyl homoserine lactone (AHL)–based QS was involved in membrane biofouling, and inactivation of QS by enzymes that degrade AHL substantially reduced biofouling (5–7). Purified QQ enzymes are typically characterized by high cost and low stability; in contrast, QQ bacteria that continuously produce AHL-degrading enzymes have attracted great interest because of the cost benefits and their continual production of QQ enzymes (8–10). However, entrapment of QQ bacteria in porous matrices such as hydrogels or hollow fibers (6, 8) is required to retain them in continuously operated membrane processes and to prevent uncontrollable biofilm formation by the QQ bacteria on membrane surfaces. It would be more desirable if the formation of the biofilms by QQ bacteria on membranes could be controlled so that the QQ biofilm itself does not compromise membrane performance. Unfortunately, controlling biofilm development through adjusting physicochemical parameters alone is highly challenging because of the interplay between the dynamic, sophisticated, biofilm signaling networks and heterogeneous microenvironments within biofilms.
Biofilm formation and dispersal in a wide variety of bacteria is controlled by a signal cascade mediated by the intracellular secondary messenger bis-(3′-5′) cyclic dimeric guanosine monophosphate (c-di-GMP). Typically, high c-di-GMP levels promote biofilm formation, while low levels lead to reduced biofilm formation and increased dispersal (11, 12). Hence, modulating the intracellular c-di-GMP level is a promising strategy toward a controllable biofilm development (13). In this study, we engineered a biofilm of Escherichia coli by introducing synthetic gene circuits to control its own biofilm formation by modulating intracellular c-di-GMP concentration and to inhibit biofouling caused by other bacteria via production of QQ enzymes. To control biofilm formation, we introduced near-infrared (NIR; 660 nm) light–responsive diguanylate cyclase (DGC; c-di-GMP synthase) activity (14) and blue (465 nm) light–activated phosphodiesterase (PDE; c-di-GMP hydrolase) activity into E. coli. To enable the production of a QQ enzyme that degrades AHL-based QS signals, we introduced the gene aiiO (10) into the E. coli strain. Following this, an E. coli biofilm with a light-responsive thickness and biovolume was achieved on FO membranes, and this biofilm effectively inhibited biofouling caused by a model organism.
MATERIALS AND METHODS
Bacterial strains and culture conditions
All strains and plasmids used in this study are summarized in Table 1. All strains were routinely maintained in lysogeny broth (LB) at 30°C. Antibiotics were supplemented into the growth media whenever necessary [ampicillin (100 μg/ml), kanamycin (50 μg/ml), and gentamicin (80 μg/ml); Sigma-Aldrich]. Stock cultures were maintained in LB medium with 20% glycerol at −80°C. Seed culture was prepared by transferring 0.5 ml of stock culture to 50 ml of LB medium in a 200-ml Erlenmeyer flask and incubated for 12 hours at 30°C with shaking at 200 rpm.
Table 1. Bacterial strains and plasmids used in this study.
Amp, ampicillin; Km, kanamycin; Gm, gentamicin.
| Bacterial strains |
Relevant characteristics |
Reference or source |
| E. coli | ||
| DH5α | The strain for gene cloning |
Thermo Fisher Scientific |
| BL21(DE) | The strain for protein expression |
Thermo Fisher Scientific |
| Pantoea stewartii | ||
| P. stewartii R067d | AHL producer and model biofouling organism |
(42) |
| P. stewartii YFP | GmR, yfp, AHL producer |
This study |
| Plasmids | ||
| pYYDT | KmR; oriV(pBBR1), Ptac |
(43) |
| pYYDT-L | KmR; oriV(pBBR1), Ptac, bphS, bphO |
(16) |
| pMal-EB1 | AmpR; oriV(pBR322), Ptac, eb1 |
(17) |
| pYYDT-RB | KmR; oriV(pBBR1), Ptac, bphS, bphO, eb1 |
This study |
| pTrcHis2-aiiO | AmpR and KmR; oriV(pBR322), Ptrc, aiiO |
(10) |
| pTrcHis2-maiiO | AmpR and KmR; oriV(pBR322), Ptrc, maiiO |
This study |
| pTrcHis2-aiiO. mCherry |
AmpR and KmR; oriV(pBR322), Ptrc, maiiO; PguaB, mCherry |
This study |
| pTrcHis2-maiiO. mCherry |
AmpR and KmR; oriV(pBR322), Ptrc, maiiO; PguaB, mCherry |
This study |
| pAiiO | AmpR; oriV(pBR322), Ptrc, aiiO; PguaB, mCherry |
This study |
| pmAiiO | AmpR; oriV(pBR322), Ptrc, maiiO; PguaB, mCherry |
This study |
Plasmid construction and transformation
All plasmid constructions were conducted in E. coli DH5α through transformation using the heat shock method. The chemically competent E. coli cells were prepared using the Inoue method (15). To dynamically control biofilm formation and dispersal, a bidirectionally controllable c-di-GMP module, comprising a previously constructed NIR light–responsive c-di-GMP gene module (i.e., bphS and bphO) (14, 16) and a blue light–activated PDE gene eb1 (17), was constructed as follows. The gene eb1 with a Ptac promoter and the Shine-Dalgarno (SD) sequence was amplified by polymerase chain reaction (PCR) from pMal-EB1 using primers Blue-Ptac-F (5′-GGACTAGTTGACAATCATCGGCTCGTAT-3′) and Blue-Ptac-R (5′-GGACTAGTTCATGAGTCCAGACTGATGGTTC-3′) (Spe I enzyme site underlined). The Spe I–digested fragment was ligated with the pYYDT-L fragment, which was cut using Spe I for fragment ligation and dephosphorylated using FastAP thermosensitive alkaline phosphatase, forming the plasmid pYYDT-RB. The constructs were selected by colony PCR using YYD-F (5′-GCCTCAGGCATTTGAGAAGCACA-3′) and YYD-R (5′-AGAGCGTTCACCGACAAACAACAGATAA-3′) and further verified to confirm the orientation of eb1 by DNA sequencing (AITbiotech, Singapore). In pYYDT-RB, bphS and bphO were placed under Ptac as an operon, whereas eb1 with an additional Ptac promoter was inserted downstream of the bphS-bphO operon to achieve coordinated efficient expression of all genes. The gene aiiO encoding a QQ enzyme (10) was then combined with bidirectionally controlled c-di-GMP module in E. coli BL21(DE). A 99–base pair (bp) fragment, flanked by Nru I restriction sites at each end, was amplified by PCR from the multiple cloning sites of pUCP22 (18, 19). The Nru I–digested 99-bp fragment was inserted at an Nru I site, near the C-terminal, of aiiO to generate an aiiO mutant (i.e., maiiO) that does not produce QQ enzyme. The aiiO gene (10) and its mutant maiiO were cloned into pTrcHis2-aiiO and pTrcHis2-maiiO, respectively, with ampicillin- and kanamycin-resistant markers (10). The mCherry gene with a PguaB promoter and the SD sequence, flanked by Pme I restriction sites at each end, was amplified by PCR from pABG5:PguaB-mCherry. The Pme I–digested PguaB.mCherry fragment was inserted at a Pme I site downstream of the kanamycin resistance gene in both pTrcHis2-aiiO and pTrcHis2-maiiO to construct the pTrcHis2-aiiO.mCherry and pTrcHis2-maiiO.mCherry, respectively. To enable the compatibility of pTrcHis2-aiiO.mCherry and pTrcHis2-maiiO.mCherry with pYYDT-RB in a stable double plasmid system, the kanamycin resistance genes flanked by two Hind III restriction sites at each end were removed using Hind III restriction enzyme, and then pTrcHis2-aiiO.mCherry and pTrcHis2-maiiO.mCherry were recircularized by T4 ligase to generate the resulting plasmids pAiiO and pmAiiO. pYYDT-RB and pAiiO (or pmAiiO) were simultaneously transformed into E. coli BL21(DE). The sequence details for the constructed gene circuits are included in table S1.
Congo red assay
To examine the synthesis of curli fimbriae as a proxy for the c-di-GMP level, strains were grown on LB agar media supplemented with Congo red (50 μg/ml) at 25°C for 24 hours and incubated in the dark or irradiated with NIR light (660 nm, 5.3 mW cm−2) or blue light (465 nm, 5.3 mW cm−2; pulses of 15 s light and 60 s dark) (20).
Extraction and quantification of c-di-GMP
The extraction and quantification of c-di-GMP followed established methods (21). The light-responsive strain E. coli/pYYDT-RB was grown at 25°C in modified M9 minimal medium [33.7 mM Na2HPO4, 22.0 mM KH2PO4, 8.55 mM NaCl, and 9.35 mM NH4Cl supplemented with 1/10-strength LB (pH 7.0)] in the dark, NIR light (660 nm, 5.3 mW cm−2), and blue light (465 nm, 5.3 mW cm−2; pulses of 15 s light and 60 s dark). At late exponential growth phase, 15 ml of culture was collected by centrifugation at 10,000g for 5 min at 4°C for c-di-GMP extraction using organic solvents (22). The concentration of c-di-GMP was determined by using a Thermo Accela 1250 series liquid chromatography system with a Thermo Velos Pro Orbitrap mass spectrometer, as described earlier (23). The c-di-GMP concentration was normalized to total protein quantified by Qubit protein assay.
Biofilm formation assay
The biofilm assay was conducted on the basis of a 24-well plate method with slight modifications (24). Diluted overnight cultures of E. coli/pYYDT and E. coli/pYYDT-RB, at a final turbidity of OD600 (optical density at 600 nm) ~ 0.1, were inoculated into 24-well plates with modified M9 minimal media. The cultures were incubated for 24 hours at 25°C without shaking in the dark or irradiated with NIR light or blue light. After the crystal violet was added to each well, the wells were rinsed and dried, and then ethanol was added to dissolve the crystal violet. The total biofilm formation was measured at 550 nm. At least nine independent replicates were used for each strain.
AHL degradation assay
The QQ activity of E. coli/(pYYDT-RB + pAiiO) and E. coli/(pYYDT-RB + pmAiiO) was tested as follows. Two milliliters of bacterial culture in modified M9 minimal medium was collected at 0, 2, 6, and 12 hours. The cell pellet was collected after being centrifuged at 8000g for 10 min, while the cell supernatant was filtered through a 0.2-μm filter (polyethersulfone; Sartorius) and treated as the cell-free supernatant sample. 3-Oxo-hexanoyl-homoserine lactone (3OC6-HSL) was used as a model AHL signal and added to the cell pellet, which has been resuspended in fresh modified M9 medium, and cell-free supernatant, to achieve a final concentration of 1 μM. The mixtures were incubated at 25°C with constant shaking at 100 rpm for 2 hours. After centrifugation and ultraviolet sterilization, residual 3OC6-HSL in each sample was measured using the reporter strain E. coli JB525 (25). The green fluorescence intensity was quantified using a microplate reader (Tecan Infinite M200) at excitation and emission wavelengths of 488 nm and 530 nm, respectively.
Dynamics of the light-responsive biofilm on FO membrane
Biofilm development on water-purifying FO membranes was performed in an in-house–developed, confocal laser scanning microscopy (CLSM)–compatible membrane flow cell as described previously with some modifications (26). The commercial FO membrane, Aquaporin-TFC, was obtained from Sterlitech Corporation. The thickness of the membrane varied from 95 to 125 μm. Three individual membrane coupons with an effective area of 460 mm2 for each were embedded in the different channels of the flow cell. The fully assembled membrane flow cell was sterilized with recirculation of 70% ethanol for 1 hour. The flow cell was further flushed with autoclaved, deionized water to remove the ethanol from the system before commencing the biofouling experiment. The feed solution was composed of modified M9 minimal media, while the draw solution was composed of 1 M NaCl (26). The system was stabilized and the membranes were conditioned for 24 hours to obtain the initial water flux before injecting the bacterial culture into the feed solution. To maintain the osmotic pressure gradient, fresh feed and draw solutions were added to the reservoirs daily at a rate of 50 ml/day to ensure that the exchange of liquid between the two chambers did not interfere with the measurement of permeate water flux and that the system could be operated at a constant osmotic pressure driving force during the experiment. The flow rate of the recirculation on both sides of each membrane was set to 2 ml/hour. Overnight cultures of the light-responsive strain (E. coli/pYYDT-RB) and the control strain (E. coli/pYYDT) were added to the respective feed solution to a final OD600 of ~0.1. Biofilms were initially grown for 48 hours in the dark, and then the 48-hour biofilms on FO membranes were subjected to sequential irradiation by NIR light and blue light. The biofilm was observed every 24 hours using CLSM, and the transmembrane water flux was recorded during the entire biofouling experiment.
Biofouling dynamics of P. stewartii in the presence of the engineered QQ biofilm
The biofilms of E. coli/(pYYDT-RB + pAiiO) and E. coli/(pYYDT-RB + pmAiiO) were grown on the FO membranes for 24 hours in the dark. After 24 hours, fresh feed solution was inoculated with an overnight P. stewartii culture to a final OD600 of ~0.1 and monitored for 96 hours. Biofilm formation by P. stewartii was compared for membranes without the preformed E. coli biofilms. To regulate the QQ biofilm formation, the biofilms on FO membranes were subjected to sequential irradiation by NIR light and blue light, as described above. Biofilms were visualized every 24 hours using CLSM, and the permeate flux was recorded during the entire biofouling experiment.
Confocal microscopy and imaging
Biofilms grown in the membrane flow cell were monitored using a CLSM (Carl Zeiss Microscopy LSM 780). The yellow fluorescent protein (YFP)–tagged P. stewartii was observed with excitation and emission at 514 and 527 nm, respectively. The mCHERRY-tagged E. coli was observed with excitation and emission at 550 and 650 nm, respectively. The CLSM images were analyzed using Imaris (Bitplane, Zurich, Switzerland) to obtain quantitative parameters such as biovolume and thickness (27). To estimate the biofilm biovolume and thickness, five regions on each membrane were randomly picked from the center of the channel for image acquisition, which is a common practice in flow cell–based biofilm studies (28). At least three independent replicates were used for each experiment.
RESULTS AND DISCUSSION
Design of the engineering gene circuits
To form an antifouling biofilm layer on FO membranes, we aimed to develop an engineered E. coli biofilm that itself does not attain a large thickness but is capable of inhibiting biofilm formation of other bacteria. C-di-GMP is an important secondary messenger involved in biofilm regulatory networks that coordinate biofilm formation and detachment in a wide range of bacteria (29). Earlier studies have shown that increased c-di-GMP promotes biofilm formation, whereas a decrease leads to reduced biofilm formation and increased dispersal (11, 30). In general, c-di-GMP levels are controlled by two groups of enzymes: (i) DGCs that synthesize c-di-GMP and (ii) PDEs that hydrolyze c-di-GMP into two GMPs (12, 31). In this study, we engineered a bacterial biofilm whose growth and dispersal can be modulated by light through implementing a dichromatic, optogenetic c-di-GMP gene circuit (17), in which the bacterial cell senses NIR light and blue light to adjust its c-di-GMP level. To do so, we first introduced a synthetic NIR light–responsive c-di-GMP module into E. coli BL21 (14, 16). The module consisted of an engineered protein, BphS, a bacteriophytochrome c-di-GMP synthase, and BphO, a chromophore biliverdin (BV) synthase from Rhodobacter sphaeroides (14). In the presence of NIR light, the BV-bound photoreceptor domain (PAS-GAF-PHY) in BphS undergoes conformational changes and activates the DGC output domain (GGDEF), thereby synthesizing intracellular c-di-GMP to enhance biofilm formation (Fig. 1). In our previous study, the NIR light–responsive activity in Shewanella oneidensis showed an increase in c-di-GMP and enhanced biofilm formation (16). However, a constantly high DGC activity could result in an undesirably thick biofilm, which itself compromises membrane performance by reducing transmembrane water fluxes. To reduce the biofilm thickness when it is overly thick, we introduced a blue light–activated PDE gene into the E. coli strain (17). The blue light–responsive c-di-GMP module consists of the engineered protein EB1, a bacteriophytochrome c-di-GMP hydrolase (17). Following irradiation with blue light, the photoreceptor (BLUF) in EB1 undergoes conformational changes and activates PDE output domain (EAL), thereby degrading intracellular c-di-GMP to disperse the biofilm (Fig. 1).
Fig. 1. Schematic illustration of the gene circuits in engineered E. coli.
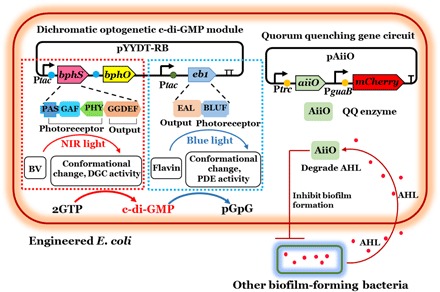
Dichromatic, optogenetic module for regulating intracellular c-di-GMP levels consisting of a NIR light–regulated DGC, BphS, and a blue light–activated PDE, EB1. QQ gene circuit for producing AiiO was used to reduce AHL-regulated biofilm formation by other bacteria. A circle in front of each gene indicates a ribosome-binding site; a T sign at the end of the operon indicates a transcription terminator.
In addition, a gene encoding a QQ enzyme was introduced into the light-responsive E. coli strain. The QQ gene circuit in this study consists of the gene aiiO from Ochrobactrum anthropi (10). The AHL acylase AiiO produced by the light-responsive QQ E. coli strain would then break down AHL produced by other bacteria and thereby inhibit their biofilm formation. A gene encoding a variant of red fluorescent protein, mCHERRY, was constitutively expressed in the QQ gene circuit to track the biofilm formation and dispersal on FO membranes by CLSM imaging (Fig. 1).
Light-controllable development of the engineered E. coli biofilm
The DGC and PDE activities of BphS and EB1 in the light-responsive strain were assayed using c-di-GMP–dependent curli fimbriae formation (32, 33). Colonies of E. coli/pYYDT-RB, expressing the bphS-bphO-eb1, in the dark or with exposure to blue light were nonpigmented, which is indicative of low intracellular c-di-GMP levels (Fig. 2A). However, colonies when grown under NIR light turned red due to the binding of Congo red to curli fimbriae, which is indicative of an elevated c-di-GMP level (Fig. 2A). In contrast, the control strain, i.e., E. coli/pYYDT harboring the empty vector, produced nonpigmented colonies under NIR light (Fig. 2A).
Fig. 2. The performance of the light-responsive gene circuit under NIR light and blue light.
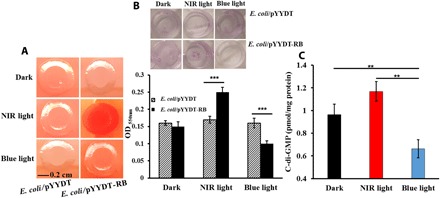
(A) C-di-GMP–dependent curli fimbriae synthesis in E. coli. The control strain E. coli/pYYDT and the light-responsive strain E. coli/pYYDT-RB were grown on Congo red plates for 24 hours at 25°C in the dark or when exposed to NIR light or blue light. (B) Total biofilm biomass (OD550nm) of the control strain E. coli/pYYDT and the light-responsive strain E. coli/pYYDT-RB in each well after incubation for 24 hours at 25°C, without shaking, under dark, NIR light, and blue light. (C) Intracellular c-di-GMP concentration (pmol/mg protein) in the light-responsive strain E. coli/pYYDT-RB after incubation for 24 hours at 25°C in the dark or when exposed to NIR light or blue light. Experiments were conducted in triplicate. Error bar represents SD. **P < 0.005 and ***P < 0.0005, Student’s t test.
The biofilm formation assay for the light-responsive strain and the control strain was conducted in a 24-well microplate. For the light-responsive strain, biofilm biomass under NIR light was ~1.5-fold of the biofilm formed under dark, while the biofilm biomass under blue light was ~60% of the biofilm biomass incubated in the dark (Fig. 2B). In contrast, no significant difference in the biofilm biomass was observed for the control strain under all conditions tested (Fig. 2B). The intracellular c-di-GMP level of the E. coli/pYYDT-RB strain expressing bphS-bphO-eb1 under NIR light was ~1.2-fold of that under dark condition (1.16 versus 0.96 pmol mg−1 total protein) (Fig. 2C). The fold changes for biofilm biomass and c-di-GMP concentration are relatively low. One plausible reason is that, even in the absence of blue light, the PDE EB1 still exhibits a low PDE activity, compromising the effect of the NIR light on the c-di-GMP level (17). A relatively low fold change may be addressed in future studies by enhancing the activity of BphS in the NIR light and/or reducing the activity of EB1 in the absence of blue light.
To examine the effect of NIR light on biofilm formation in biofilm flow cells, we exposed a 24-hour preformed light-responsive biofilm to NIR light and observed a ~20% increase in biofilm biovolume within 24 hours. Following the NIR light irradiation, we exposed the biofilm to blue light and observed a ~60% decrease in biofilm biovolume after 24 hours. In contrast, the preformed control biofilm showed no significant difference in biofilm biovolume over 24 hours under NIR light or blue light (fig. S1).
To further examine the dynamics of biofilm formation on FO membranes under sequential exposure to NIR light or blue light, biofilms of the light-responsive strain and the control strain were grown in an in-housed–developed, CLSM-compatible membrane flow cell (26) with commercially available thin-film composite FO membranes (Fig. 3A). Under dark conditions, no significant difference was observed for the 48-hour biofilms for the light-responsive or control strains in terms of biofilm biovolume (46.9 ± 4.8 μm3 μm−2 versus 39.8 ± 7.0 μm3 μm−2) and biofilm thickness (19.2 ± 1.6 μm versus 16.2 ± 2.3 μm) (Fig. 3, C and D). After exposure to blue light for 24 hours, the biovolume of the light-responsive biofilm decreased to 24.9 ± 8.5 μm3 μm−2, while the control biofilm biovolume increased to 60.2 ± 9.7 μm3 μm−2 at 72 hours (Fig. 3C). Subsequently, when the biofilms were exposed to NIR light (for 24 hours) followed by another exposure to blue light (for 24 hours), biovolume and thickness for the light-responsive biofilm gradually increased and then decreased to 12.9 ± 3.9 μm3 μm−2 and 4.9 ± 1.2 μm, respectively, while the control biofilm did not respond to the light exposure (Fig. 3, C and D).
Fig. 3. Biofouling dynamics of the control, E. coli/pYYDT, and light-responsive, E. coli/pYYDT-RB, strains on FO membrane in membrane flow cell.
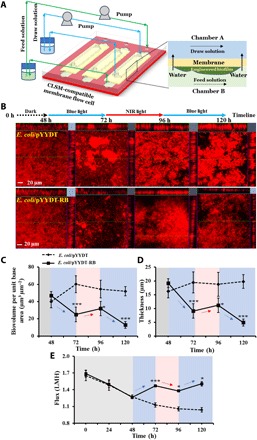
(A) The CLSM-compatible membrane flow cell for nondestructive monitoring of biofouling dynamics on membranes. The device contains three independent channels, with each channel separated into two chambers by a flat sheet membrane. (B) CLSM images of biofilms formed on the FO membrane at 48, 72, 96, and 120 hours. The blue line represents biofilms formed after 24 hours of incubation under blue light, and the red line represents biofilms formed after 24 hours of incubation under NIR light. Each CLSM image contains one top-down view (x-y plane) and two side views (x-z and y-z planes). Scale bar, 20 μm. (C) Biofilm biovolume (expressed as biovolume per unit base area) and (D) biofilm thickness. (E) Comparisons of permeate flux through membranes at different time points. The gray, blue, and red zones represent incubation in the dark and under blue light and NIR light, respectively. Experiments were conducted in triplicate, and for each replicate, five representative images were acquired and quantified. Error bar represents SD. *P < 0.05 and ***P < 0.0005, Student’s t test.
In addition to biofilm dynamics on the FO membranes, we also observed that the transmembrane water flux for the FO membranes with the light-responsive biofilm gradually decreased and then increased under exposure to NIR light and blue light. This correlates with the increase in biofilm observed under NIR light and the reduction in biofilm when exposed to blue light. After the operation of the FO process for 120 hours, the water flux was 1.5 ± 0.04 liters m−2 hour−1 (LMH), which was ~10% lower than the initial water flux (1.7 ± 0.05 LMH). In contrast, the control biofilm resulted in a constant decline of the water flux by ~40% from 1.6 ± 0.09 LMH to 1.0 ± 0.04 LMH (Fig. 3E). Together, our results demonstrate that the biovolume and thickness of the light-responsive E. coli biofilm with the synthetic optogenetic c-di-GMP module could be modulated using light to limit its impact on the transmembrane water flux.
QQ activity of the engineered E. coli biofilm and inhibitory effect on biofilm formation by P. stewartii
The gene encoding AiiO from O. anthropi (aiiO) (10) was introduced to the light-responsive E. coli strain to enable production of the QQ enzyme for mitigating biofouling caused by other bacteria (Fig. 1). This strain was referred to as E. coli/(pYYDT-RB + pAiiO) or the QQ biofilm strain. Another strain, E. coli/(pYYDT-RB + pmAiiO), in which aiiO was mutated so that the strain was not capable of producing the QQ enzyme, was used as a negative control. To examine the QQ activity in the QQ biofilm strain, 3OC6-HSL (final concentration, 1 μM) was added to the cell suspension (OD600 ~ 0.3) and the cell-free conditioned medium (i.e., supernatant from the bacterial culture). Residual 3OC6-HSL was assayed with the E. coli JB525 biosensor (25). There was no obvious degradation of the AHL signal by the negative control strain E. coli/(pYYDT-RB + pmAiiO) (Fig. 4A). In contrast, complete degradation of the added 3OC6-HSL by the E. coli/(pYYDT-RB + pAiiO) cells was observed within 2 hours (Fig. 4A). Inactivation of 3OC6-HSL was also observed for the cell-free conditioned medium, although the activity was much lower than when the cells were present (fig. S2). This result is consistent with previous studies where the QQ enzyme has been shown to be intracellularly localized, and AHL inactivation in most cases was found to occur within bacterial cells (34, 35). We further tested the QQ activity of the QQ biofilm strain under light conditions and found that the exposure to NIR light or blue light did not affect the QQ activity (fig. S3).
Fig. 4. AHL degradation is mediated by QQ enzymatic activities.
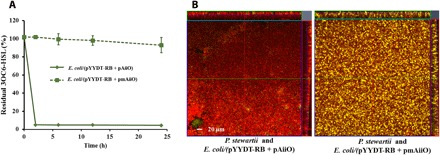
(A) Inactivation of exogenous 3OC6-HSL by the light-responsive QQ strain, E. coli/(pYYDT-RB + pAiiO), was assessed over 24 hours at room temperature. Residual 3OC6-HSL was quantified using the E. coli JB525 bioassay at 0, 2, 6, 12, and 24 hours after incubation. (B) CLSM images of biofilm formation by YFP-tagged P. stewartii on preformed light-responsive QQ biofilm E. coli/(pYYDT-RB + pAiiO) and QQ control biofilm E. coli/(pYYDT-RB + pmAiiO) on biofilm flow cells after 24 hours. Scale bar, 20 μm. Experiments were conducted in triplicate.
To demonstrate that the QQ biofilm strain can inhibit biofilm formation by other bacteria, biofilm formation of P. stewartii on a preformed QQ biofilm was investigated. P. stewartii, an isolate from wastewater, produces a wide range of AHL signals, and its biofilm formation is regulated by AHL-based QS signaling (36, 37). Note that, unlike P. stewartii, the biofilm formation in E. coli was independent from the AHL signals (38), and therefore, the QQ enzyme (AiiO) produced by the engineered E. coli strain has no interference with its own biofilm formation. In addition, the negative control E. coli lacking QQ activity did not inhibit P. stewartii growth in either planktonic (10) or biofilm cocultures (fig. S4). Biofilms of the engineered QQ E. coli strain and the negative control, tagged with mCHERRY, were pregrown on glass slides in biofilm flow cells for 24 hours and then challenged with the YFP-tagged P. stewartii. The preformed biofilm producing the QQ enzyme inhibited the biofilm formation by P. stewartii, while the QQ negative biofilm did not interfere with the biofilm formation by P. stewartii (Fig. 4B). These results show that the engineered light-responsive QQ E. coli strain is capable of effectively reducing biofilm formation by other bacteria.
Light-modulated biofouling dynamics on FO membranes in membrane flow cells
We further tested the activity of the light-responsive QQ biofilm in FO membrane–based water purification processes using an in-housed built membrane flow cell under FO operation mode (Fig. 3A). The membranes were first colonized with the light-responsive QQ strain E. coli/(pYYDT-RB + pAiiO) or the control strain E. coli/(pYYDT-RB + pmAiiO) (the light-responsive strain with no AiiO activity) for 24 hours in FO operation mode. The preformed biofilms on the membranes were challenged with P. stewartii, and then the flow cells were allowed to run for 24 hours in the dark. Biofilm formation by P. stewartii on FO membranes without the preformed E. coli biofilms was also monitored for comparison.
After 24 hours of colonization on the preformed light-responsive QQ biofilm, a small amount of P. stewartii biomass (3.5 ± 1.8 μm3 μm−2) was observed (Fig. 5A). In contrast, the membranes with a preformed control biofilm lacking QQ activity and without preformed biofilms were covered by P. stewartii with a biovolume of 17.4 ± 4.9 μm3 μm−2 and 33 ± 5.6 μm3 μm−2, respectively (Fig. 5A). The results suggest that the light-responsive QQ biofilm was effective in inhibiting FO membrane biofouling caused by other bacteria. However, excessive biofilm formation by the light-responsive QQ E. coli strain could compromise membrane performance (e.g., reduction in transmembrane water flux). Therefore, we tested the use of NIR light and blue light to modulate the engineered light-responsive QQ biofilm growth and the biofouling dynamics on the FO membranes.
Fig. 5. Biofouling dynamics of P. stewartii in the presence of a light-responsive QQ biofilm, E. coli/(pYYDT-RB + pAiiO), and a QQ control biofilm, E. coli/(pYYDT-RB + pmAiiO), in a membrane flow cell.
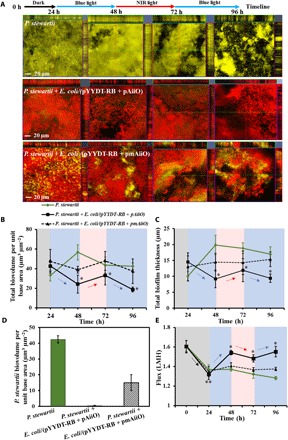
(A) CLSM images of biofilms formed on the FO membrane at 24, 48, 72, and 96 hours. The blue line represents imaging after 24 hours of incubation under blue light, and the red line represents imaging after 24 hours of incubation under NIR light. Each CLSM image contains one top-down view (x-y plane) and two side views (x-z and y-z planes). Scale bar, 20 μm. (B) Total biofilm biovolume of the consortium (expressed as biovolume per unit base area). (C) Total biofilm thickness of the consortium. (D) Biofilm biovolume of P. stewartii in different consortium at 96 hours. (E) Comparisons of permeate flux through membranes at different time points. The gray, blue, and red zones represent incubation in the dark or under blue light and NIR light, respectively. Error bar represents SD. *P < 0.05 and **P < 0.005, Student’s t test.
Preformed (from t = 0 to 24 hours) membrane biofilms of the P. stewartii alone, the P. stewartii + E. coli/(pYYDT-RB + pAiiO) consortium, and the P. stewartii + E. coli/(pYYDT-RB + pmAiiO) consortium were subjected to sequential exposures to different lights: blue light for 24 hours (from t = 24 to 48 hours), then NIR light for 24 hours (t = 48 to 72 hours), and followed by blue light again for 24 hours (t = 72 to 96 hours). Before exposure to light (t = 24 hours), biofilm biovolume and thickness for the three preformed biofilms were comparable (biovolume: 33 ± 5.6 μm3 μm−2 versus 43.8 ± 7.1 μm3 μm−2 versus 47.5 ± 12 μm3 μm−2; thickness: 10 ± 1.9 μm versus 14.5 ± 1.4 μm versus 12.9 ± 4.4 μm). In the presence of the preformed light-responsive QQ biofilm, total biofilm biovolume and thickness decreased to 24.3 ± 7.8 μm3 μm−2 and 9.2 ± 3.0 μm, respectively, after the first exposure to blue light for 24 hours (at t = 48 hours) (Fig. 5, B and C). The amount of biofilms was significantly lower than that formed in the presence of the control biofilm (39 ± 3.4 μm3 μm−2; 14.4 ± 2.7 μm) and the P. stewartii biofilm alone (56.7 ± 9.2 μm3 μm−2; 19.8 ± 2.8 μm) (Fig. 5, B and C). Although the control biofilm formed by E. coli/(pYYDT-RB + pmAiiO) did not produce QQ enzyme, it could respond to the blue light. Hence, after 24 hours of blue light exposure, the total biofilm biovolume formed in the presence of the preformed control biofilm was ~31% lower than the P. stewartii biofilm alone at t = 48 hours (Fig. 5B). Exposure of E. coli/(pYYDT-RB + pAiiO) biofilms to NIR light (for 24 hours from t = 48 to 72 hours) followed by blue light (for 24 hours from t = 72 to 96 hours) resulted in a light-mediated increase and then decrease in total biofilm biomass, reaching a biovolume of 18.7 ± 3.4 μm3 μm−2 at t = 96 hours. In comparison, the total biofilm formed in the presence of the control biofilm that did not produce QQ enzyme exhibited a final biovolume of 37.4 ± 12.5 μm3 μm−2 at t = 96 hours (Fig. 5B). Although P. stewartii lacked the light-responsive module and did not respond to NIR light or blue light, a slightly lower biofilm biovolume at t = 96 hours was observed for P. stewartii biofilm alone. This is likely due to the uncontrolled “dispersal” event that is often observed for a mature biofilm as a result of hydrodynamic sloughing and/or genetically programmed cell detachment triggered by local physicochemical conditions (39, 40). In addition, although the control E. coli lacked QQ activity and a natural inhibition to P. stewartii growth, it seemed that the biofilm development of P. stewartii was inhibited by the presence of a preformed biofilm of the control E. coli (lacking QQ activity) (Fig. 5A). It is important to note that surface properties including physicochemical characteristics such as hydrophobicity, roughness, and surface charges are expected to differ greatly between the FO membrane and the preformed E. coli biofilm. The difference in surface physiochemical properties would affect the biofilm development of P. stewartii (41). Furthermore, the control E. coli (lacking QQ activity) is light responsive, and the blue light–mediated dispersal would result in the displacement of the P. stewartii biofilm colonized on the surface of control E. coli biofilm.
The thickness of the biofilms formed in the presence of preformed light-responsive QQ biofilm exhibited a similar light-mediated increase and decrease trend, which was not observed for the biofilms formed in the presence of preformed control biofilm or the P. stewartii biofilm. The final thickness (at t = 96 hours) of the biofilms formed in the presence of the preformed light-responsive QQ biofilm was 9.4 ± 2.3 μm, which is significantly lower than those for the biofilms formed in the presence of preformed control biofilm (15.3 ± 2.2 μm) and the P. stewartii biofilm alone (16.9 ± 1.3 μm) (Fig. 5C). In addition, at t = 96 hours, about 0.3 ± 0.2 μm3 μm−2 of P. stewartii was found on the membrane surfaces in the presence of the preformed light-responsive QQ biofilm, which is significantly lower than that with the preformed control biofilm (15 ± 5.1 μm3 μm−2) (Fig. 5D). In addition to the AHL-based QS bacteria such as P. stewartii, the presence of extracellular polymeric substances (EPS) and dead bacteria in the filtration system might also pose a biofouling problem, especially considering that they do not respond to QQ activity exhibited by the QQ biofilm. Our results showed that the blue light–mediated QQ biofilm dispersal also resulted in the reduction of residual polysaccharides and a decreased relative amount of dead bacteria in the QQ biofilm (figs. S5 and S6).
Throughout the course of the biofouling experiments, the transmembrane water flux for the membranes with the preformed control biofilm and the P. stewartii biofilm exhibited a decline of ~12% and ~18%, respectively (Fig. 5E). In contrast, in the presence of the light-responsive QQ biofilm, only about 6% reduction in the flux was observed (Fig. 5E). Recent studies have reported that QQ significantly reduced the amount of EPS on membrane surfaces and triggered sloughing off of the biofilms, reducing biofouling (6, 10). The higher flux achieved by the engineered light-responsive QQ biofilm correlated well with the QQ effect reported in other experimental systems (6, 10). The transmembrane water flux for the FO systems in the presence of a preformed light-responsive biofilm also showed light-mediated differences, suggesting that the effect of the preformed light-responsive QQ biofilm on the membrane performance can be effectively controlled by light.
We report the engineering of a light-responsive QQ biofilm by harnessing the power of QS and c-di-GMP regulatory mechanisms in biofilms and demonstrate the potential application of the engineered biofilm in mitigating biofouling of water purification FO membranes. In this study, QQ activity was used to show the effectiveness of the light-responsive biofilm to limit its thickness while exhibiting desirable functions. The c-di-GMP–targeted optogenetic approach for controllable biofilm development we have demonstrated here should prove widely applicable for designing other thickness-controllable biofilm-enabled applications, for example, efficient biofilm-mediated biocatalysis for chemical synthesis. Our study exemplifies the potential for translational research from biofilm biology to biofilm engineering for applied and environmental biotechnology.
Supplementary Material
Acknowledgments
We would like to thank M. Gomelsky from University of Wyoming for providing the plasmid pMal-EB1, and G. Port and M. G. Caparon from Washington University in St. Louis for providing the plasmid pABG5:PguaB-mCherry. Funding: This research was supported by the Ministry of Education (MOE) Academic Research Fund (AcRF) Tier 1 Grant RG50/16 (M4011622.030 to B.C.), AcRF Tier 2 Grant (MOE2017-T2-2-042 to B.C.), and the National Research Foundation and MOE Singapore under its Research Centre of Excellence Programme, Singapore Centre for Environmental Life Sciences Engineering (SCELSE) (M4330005.C70 to B.C.), Nanyang Technological University, Singapore. Author contributions: B.C. conceived the idea. M.M., Y.H., and B.C. designed and performed the experiment and analyzed the data. C.H.T. and S.A.R. contributed to the design of the QQ gene circuit. M.M., Y.H., and B.C. wrote the paper; all authors have contributed to, seen, and approved the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/12/eaau1459/DC1
Table S1. Sequence details for the gene circuits.
Fig. S1. Biofilm formation of the control E. coli/pYYDT and light-responsive E. coli/pYYDT-RB strains in biofilm flow cell.
Fig. S2. Inactivation of exogenous 3OC6-HSL by E. coli/(pYYDT-RB + pAiiO) cells and the cell-free supernatant.
Fig. S3. Inactivation of exogenous 3OC6-HSL by E. coli/(pYYDT-RB + pAiiO) cells incubated in the dark, under NIR light and blue light.
Fig. S4. Coculture biofilm formation by the mCHERRY-tagged control E. coli (QQ negative) and YFP-tagged P. stewartii.
Fig. S5. CLSM images of mCHERRY-tagged light-responsive QQ E. coli biofilm and ConA-stained polysaccharide in the flow cell.
Fig. S6. CLSM images of dead P. stewartii (YFP tagged) on preformed light-responsive QQ E. coli biofilm.
REFERENCES AND NOTES
- 1.Shannon M. A., Bohn P. W., Elimelech M., Georgiadis J. G., Mariñas B. J., Mayes A. M., Science and technology for water purification in the coming decades. Nature 452, 301–310 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Zhao S., Zou L., Tang C. Y., Mulcahy D., Recent developments in forward osmosis: Opportunities and challenges. J. Membr. Sci. 396, 1–21 (2012). [Google Scholar]
- 3.Flemming H.-C., Schaule G., Griebe T., Schmitt J., Tamachkiarowa A., Biofouling—The Achilles heel of membrane processes. Desalination 113, 215–225 (1997). [Google Scholar]
- 4.Davies D. G., Parsek M. R., Pearson J. P., Iglewski B. H., Costerton J. W., Greenberg E. P., The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280, 295–298 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Yeon K.-M., Cheong W.-S., Oh H.-S., Lee W.-N., Hwang B.-K., Lee C.-H., Beyenal H., Lewandowski Z., Quorum sensing: A new biofouling control paradigm in a membrane bioreactor for advanced wastewater treatment. Environ. Sci. Technol. 43, 380–385 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Kim S.-R., Oh H.-S., Jo S.-J., Yeon K.-M., Lee C.-H., Lim D.-J., Lee C.-H., Lee J.-K., Biofouling control with bead-entrapped quorum quenching bacteria in membrane bioreactors: Physical and biological effects. Environ. Sci. Technol. 47, 836–842 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Yeon K.-M., Lee C.-H., Kim J., Magnetic enzyme carrier for effective biofouling control in the membrane bioreactor based on enzymatic quorum quenching. Environ. Sci. Technol. 43, 7403–7409 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Oh H.-S., Yeon K.-M., Yang C.-S., Kim S.-R., Lee C.-H., Park S. Y., Han J. Y., Lee J.-K., Control of membrane biofouling in MBR for wastewater treatment by quorum quenching bacteria encapsulated in microporous membrane. Environ. Sci. Technol. 46, 4877–4884 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Yu H., Liang H., Qu F., He J., Xu G., Hu H., Li G., Biofouling control by biostimulation of quorum-quenching bacteria in a membrane bioreactor for wastewater treatment. Biotechnol. Bioeng. 113, 2624–2632 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Oh H.-S., Tan C. H., Low J. H., Rzechowicz M., Siddiqui M. F., Winters H., Kjelleberg S., Fane A. G., Rice S. A., Quorum quenching bacteria can be used to inhibit the biofouling of reverse osmosis membranes. Water Res. 112, 29–37 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Römling U., Gomelsky M., Galperin M. Y., C-di-GMP: The dawning of a novel bacterial signalling system. Mol. Microbiol. 57, 629–639 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Valentini M., Filloux A., Biofilms and c-di-GMP Signaling: Lessons from Pseudomonas aeruginosa and other bacteria. J. Biol. Chem. 291, 12547–12555 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benedetti I., de Lorenzo V., Nikel P. I., Genetic programming of catalytic Pseudomonas putida biofilms for boosting biodegradation of haloalkanes. Metab. Eng. 33, 109–118 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Ryu M.-H., Gomelsky M., Near-infrared light responsive synthetic c-di-GMP module for optogenetic applications. ACS Synth. Biol. 3, 802–810 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook J., Russell D. W., The Inoue method for preparation and transformation of competent E. coli: “Ultra-competent” cells. CSH Protoc. 1, pdb.prot3944 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Hu Y., Wu Y., Mukherjee M., Cao B., A near-infrared light responsive c-di-GMP module-based AND logic gate in Shewanella oneidensis. Chem Commun. 53, 1646–1648 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Ryu M.-H., Fomicheva A., Moskvin O. V., Gomelsky M., Optogenetic module for dichromatic control of c-di-GMP signaling. J. Bacteriol. 18, e00014-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrero M., de Lorenzo V., Timmis K. N., Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172, 6557–6567 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.West S. E. H., Schweizer H. P., Dall C., Sample A. K., Runyen-Janecky L. J., Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148, 81–86 (1994). [DOI] [PubMed] [Google Scholar]
- 20.Fang X., Gomelsky M., A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol. Microbiol. 76, 1295–1305 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Wu Y., Ding Y., Cohen Y., Cao B., Elevated level of the second messenger c-di-GMP in Comamonas testosteroni enhances biofilm formation and biofilm-based biodegradation of 3-chloroaniline. Appl. Microbiol. Biotechnol. 99, 1967–1976 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Kuchma S. L., Ballok A. E., Merritt J. H., Hammond J. H., Lu W., Rabinowitz J. D., O’Toole G. A., Cyclic-di-GMP-mediated repression of swarming motility by Pseudomonas aeruginosa: The pilY1 gene and its impact on surface-associated behaviors. J. Bacteriol. 192, 2950–2964 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spangler C., Böhm A., Jenal U., Seifert R., Kaever V., A liquid chromatography-coupled tandem mass spectrometry method for quantitation of cyclic di-guanosine monophosphate. J. Microbiol. Methods 81, 226–231 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Ding Y., Peng N., Du Y., Ji L., Cao B., Disruption of putrescine biosynthesis in Shewanella oneidensis enhances biofilm cohesiveness and performance in Cr(VI) immobilization. Appl. Environ. Microbiol. 80, 1498–1506 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andersen J. B., Heydorn A., Hentzer M., Eberl L., Geisenberger O., Christensen B. B., Molin S., Givskov M., gfp-based N-acyl homoserine-lactone sensor systems for detection of bacterial communication. Appl. Environ. Microbiol. 67, 575–585 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mukherjee M., Menon N. V., Liu X., Kang Y., Cao B., Confocal laser scanning microscopy-compatible microfluidic membrane flow cell as a nondestructive tool for studying biofouling dynamics on forward osmosis membranes. Environ. Sci. Technol. Lett. 3, 303–309 (2016). [Google Scholar]
- 27.Mohanty A., Wei L., Lu L., Chen Y., Cao B., Impact of sublethal levels of single-wall carbon nanotubes on pyoverdine production in Pseudomonas aeruginosa and its environmental implications. Environ. Sci. Technol. Lett. 2, 105–111 (2015). [Google Scholar]
- 28.Heydorn A., Ersboll B. K., Hentzer M., Parsek M. R., Givskov M., Molin S., Experimental reproducibility in flow-chamber biofilms. Microbiology 146, 2409–2415 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Hengge R., Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7, 263–273 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Boyd C. D., O’Toole G. A., Second messenger regulation of biofilm formation: Breakthroughs in understanding c-di-GMP effector systems. Annu. Rev. Cell Dev. Biol. 28, 439–462 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borlee B. R., Goldman A. D., Murakami K., Samudrala R., Wozniak D. J., Parsek M. R., Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol. Microbiol. 75, 827–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hammar M., Arnqvist A., Bian Z., Olsén A., Normark S., Expression of two csg operons is required for production of fibronectin and congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 18, 661–670 (1995). [DOI] [PubMed] [Google Scholar]
- 33.Römling U., Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell. Mol. Life Sci. 62, 1234–1246 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Czajkowski R., Krzyżanowska D., Karczewska J., Atkinson S., Przysowa J., Lojkowska E., Williams P., Jafra S., Inactivation of AHLs by Ochrobactrum sp. A44 depends on the activity of a novel class of AHL acylase. Environ. Microbiol. Rep. 3, 59–68 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Jafra S., Przysowa J., Czajkowski R., Michta A., Garbeva P., van der Wolf J. M., Detection and characterization of bacteria from the potato rhizosphere degrading N-acyl-homoserine lactone. Can. J. Microbiol. 52, 1006–1015 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Koutsoudis M. D., Tsaltas D., Minogue T. D., von Bodman S. B., Quorum-sensing regulation governs bacterial adhesion, biofilm development, and host colonization in Pantoea stewartii subspecies stewartii. Proc. Natl. Acad. Sci. U.S.A. 103, 5983–5988 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohanty A., Tan C. H., Cao B., Impacts of nanomaterials on bacterial quorum sensing: Differential effects on different signals. Environ. Sci. Nano 3, 351–356 (2016). [Google Scholar]
- 38.Sharma G., Sharma S., Sharma P., Chandola D., Dang S., Gupta S., Gabrani R., Escherichia coli biofilm: Development and therapeutic strategies. J. Appl. Microbiol. 121, 309–319 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Kaplan J. B., Biofilm dispersal: Mechanisms, clinical implications, and potential therapeutic uses. J. Dent. Res. 89, 205–218 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Toole G., Kaplan H. B., Kolter R., Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 49–79 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Palmer J., Flint S., Brooks J., Bacterial cell attachment, the beginning of a biofilm. J. Ind. Microbiol. Biotechnol. 34, 577–588 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Tan C. H., Koh K. S., Xie C., Zhang J., Tan X. H., Lee G. P., Zhou Y., Ng W. J., Rice S. A., Kjelleberg S., Community quorum sensing signalling and quenching: Microbial granular biofilm assembly. NPJ Biofilms Microbiomes 1, 15006 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y., Ding Y., Hu Y., Cao B., Rice S. A., Kjelleberg S., Song H., Enhancing bidirectional electron transfer of Shewanella oneidensis by a synthetic flavin pathway. ACS Synth. Biol. 4, 815–823 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/12/eaau1459/DC1
Table S1. Sequence details for the gene circuits.
Fig. S1. Biofilm formation of the control E. coli/pYYDT and light-responsive E. coli/pYYDT-RB strains in biofilm flow cell.
Fig. S2. Inactivation of exogenous 3OC6-HSL by E. coli/(pYYDT-RB + pAiiO) cells and the cell-free supernatant.
Fig. S3. Inactivation of exogenous 3OC6-HSL by E. coli/(pYYDT-RB + pAiiO) cells incubated in the dark, under NIR light and blue light.
Fig. S4. Coculture biofilm formation by the mCHERRY-tagged control E. coli (QQ negative) and YFP-tagged P. stewartii.
Fig. S5. CLSM images of mCHERRY-tagged light-responsive QQ E. coli biofilm and ConA-stained polysaccharide in the flow cell.
Fig. S6. CLSM images of dead P. stewartii (YFP tagged) on preformed light-responsive QQ E. coli biofilm.


