Abstract
Alcohol use disorders (AUD) are complex traits, meaning that variations in many genes contribute to the risk, as does the environment. Although the total genetic contribution to risk is substantial, most individual variations make only very small contributions. By far the strongest contributors are functional variations in two genes involved in alcohol (ethanol) metabolism. A functional variant in alcohol dehydrogenase 1B (ADH1B) is protective in people of European and Asian descent, and a different functional variant in the same gene is protective in those of African descent. A strongly protective variant in aldehyde dehydrogenase 2 (ALDH2) is essentially only found in Asians. This highlights the need to study a wide range of populations. The likely mechanism of protection against heavy drinking and AUD in both cases is alteration in the rate of metabolism of ethanol that at least transiently elevates acetaldehyde. Other ADH and ALDH variants, including functional variations in ADH1C, have also been implicated in affecting drinking behavior and risk for alcoholism. The pattern of linkage disequilibrium in the ADH region, and the differences among populations, complicate analyses, particularly of regulatory variants. This critical review focuses upon the ADH and ALDH genes as they affect AUDs.
Introduction
Alcohol use disorders (AUD) are common, complex disorders, the risk for which is contributed by genetic differences, environmental differences, and their interactions. AUDs lack an objective test. The current clinical definition (The Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition; DSM-5) requires meeting at least 2 out of 11 criteria that reflect problems caused by consuming alcohol (American Psychiatric Association, 2013). The checklist definition means that theoretically one can meet DSM-5 criteria for AUD in 2036 different ways. Many studies have used DSM-IV definitions of alcohol dependence (AD; 3 or more of 7 criteria), which is more severe than a minimal DSM-5 definition, but still heterogeneous (99 possible combinations). This heterogeneity has obvious implications for the study of AUD. The requirement for alcohol consumption adds additional complexity, because there are large environmental differences in access to and acceptance of alcohol in different social groups and across time and location, and these can vary even within an individual’s life. Average drinks per week is widely studied, but is highly skewed, with most people consuming less than 2 drinks per week and with a small fraction consuming very large quantities. It does not capture the pattern of drinking (e.g., bingeing). There is only a modest genetic correlation between average drinks per week and AUD (from 0.37 – 0.70) (Walters et al., 2018).
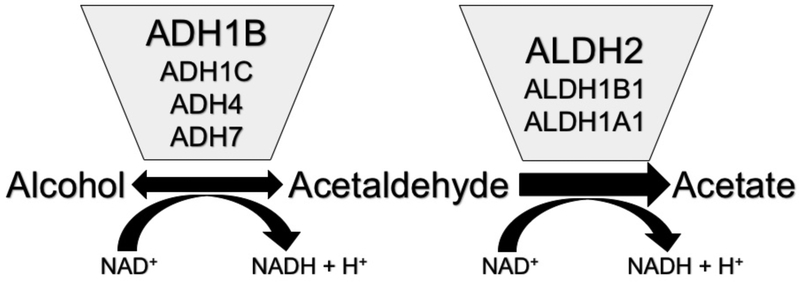
Ethanol is absorbed from the gastrointestinal tract, primarily in the small intestine, then travels to the liver, and from there is distributed throughout the body water (Hurley et al., 2002). The first step in the major pathway of its metabolism is oxidation to acetaldehyde by alcohol dehydrogenases (ADHs) (Figure 1). Metabolism by cytochrome P450s and catalase make only minor contributions (Hurley et al., 2002). Acetaldehyde binds readily to proteins, RNA and DNA, and can be aversive and toxic (Zakhari, 2006). Acetaldehyde is rapidly oxidized to acetate by aldehyde dehydrogenases (ALDHs). First pass metabolism (metabolism before the ethanol reaches the general circulation) occurs in the digestive tract and on its first pass from there through the liver. From then on, most metabolism occurs in the liver, catalyzed by ADH and ALDH enzymes1. Levels of ethanol can get high: the blood alcohol concentration that is defined as legal intoxication in the US (0.08%) corresponds to 17 mM ethanol. The oxidation of acetaldehyde is extremely efficient, such that circulating levels of acetaldehyde are usually more than 1000-fold less; they are generally barely detectable, ≤ 3 μM (Mizoi et al., 1994, Peng et al., 2014a, Harada et al., 1983, Nuutinen et al., 1984), although they are higher in liver (~15 μM after ingestion of 0.8 g/kg ethanol) (Nuutinen et al., 1984).
Figure 1. Primary pathway of alcohol metabolism.
The oxidation of alcohol to acetaldehyde is reversible in vitro, but in vivo the overall reaction goes strongly toward acetate due to the activity of ALDH2. The ADH and ALDH enzymes that carry out most of the metabolism are shown.
The contribution of genetic variants to risk for AUD is spread across a large number of genes, probably at least hundreds, that act through many pathways and interact with the environment (for recent reviews see (Edenberg and Foroud, 2013, Rietschel and Treutlein, 2013, Hart and Kranzler, 2015)). Most variants have very small effects on risk. This critical review will focus on the set of genes with the strongest effect on risk for AUD, the alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH) genes. There is very strong evidence that variations in ADH and ALDH genes affect alcohol consumption and the risk for AUD.
Alcohol dehydrogenases
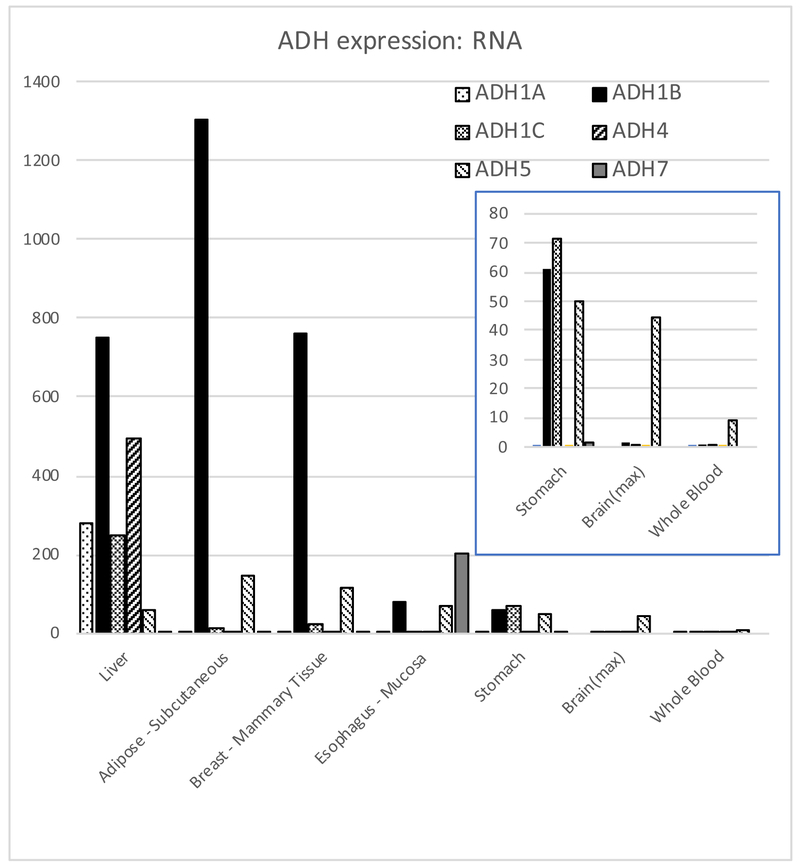
There are 6 closely related ADHs whose structure and enzymology have been studied; a seventh (ADH6) has not been found as a protein in vivo (Table 1) (Bosron et al., 1993, Hurley et al., 2002, Edenberg and Bosron, 2018). Their pattern of expression in tissues differs (Figure 2). ADH1A, ADH1B, and ADH1C are called class I ADHs; they are more than 90% identical in amino acid sequence, and can hetero-dimerize with each other. These three ADHs have Km for ethanol in the range of 0.013 to 27 mM (Chi et al., 2018, Hurley et al., 2002, Hurley and Edenberg, 2012) (Table 1), and carry out most of the ethanol oxidation in liver. The other ADH enzymes function as homodimers. When ethanol levels are high (e.g., intoxicating), ADH4 could contribute substantially, perhaps 1/3 of the overall metabolism (Lee et al., 2004), although a recent model shows a smaller contribution (Chi et al., 2018). ADH7 is the only ADH enzyme not expressed in liver; it contributes to ethanol oxidation and local generation of acetaldehyde primarily in the stomach and esophagus. ADH5 is ubiquitously expressed; although it doesn’t make a major contribution to ethanol oxidation in liver, it can contribute to metabolism in other tissues, including the GI tract and brain, and thereby generate acetaldehyde locally. ADH6 has never been isolated from human tissue, although its RNA is present; computational modeling suggests it is likely to be both highly unstable and inactive (Ostberg et al., 2016) and therefore not likely to impact alcohol metabolism.
Table 1.
ADH genes and enzyme kinetics
| Approved Gene Symbola |
Approved Gene Namea | Synonymsb | RNA: RefSeq Accession ID |
Subunit encodedc |
KM, ethanol (mM) |
Activity Vmax (min)-1 |
Activity at 22 mM ethanol |
RefSeq position |
|---|---|---|---|---|---|---|---|---|
| ADH1A | alcohol dehydrogenase 1A (class I), alpha polypeptide | ADH1 | NM_000667 | α-ADH, ADH1 | 4 | 30 | 25 | 4:99,276,366-99,291,028 |
| ADH1B | alcohol dehydrogenase 1B (class I), beta polypeptide | ADH2 | NM_000668 | β-ADH, ADH2 | 4:99,306,387-99,321,442 | |||
| ADH1B*1; ADH1B[Arg48/Arg370] | ADH2*1 | β1-ADH, ADH2*1 | 0.013 | 5.2 | 5.2 | |||
| ADH1B*2; ADH1B[His48/Arg370] | ADH2*2 | β2-ADH, ADH2*2 | 1.8 | 190 | 176 | |||
| ADH1B*3; ADH1B[Arg48/Cys370] | ADH2*3 | β3-ADH, ADH2*3 | 61 | 140 | 37 | |||
| ADH1C | alcohol dehydrogenase 1C (class I), gamma polypeptide | ADH3 | NM_000669 | γ-ADH, ADH3 | 4:99,336,492-99,353,045 | |||
| ADH1C*1; ADH1C[Arg272/Ile350] | ADH1C*1 | γ1-ADH, ADH1C*1 | 0.1 | 32 | 32 | |||
| ADH1C*2; ADH1C[Gln272/Val350] | ADH1C*2 | γ2-ADH, ADH1C*2 | 0.14 | 20 | 20 | |||
| ADH4 | alcohol dehydrogenase 4 (class II), pi polypeptide | class II ADH, ADH2 | NM_000670 | π-ADH, ADH4 | 11 | 9 | 6 | 4:99,123,667-99,144,298 |
| ADH5 | alcohol dehydrogenase 5 (class III), chi polypeptide | class III ADH, ADH3 | NM_000671 | χ-ADH, ADH5 | >1,000 | 100 | <2 | 4:99,070,978-99,088,788 |
| ADH6 | alcohol dehydrogenase 6 (class V) | class V ADH, ADH5 | NM_000672 | - | - | - | - | 4:99,202,638-99,219,246 |
| ADH7 | alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide | class IV ADH, ADH4 | NM_000673 | μ-ADH, σ-ADH, ADH7 |
30 | 1800 | 760 | 4:99,412,261-99,435,510 |
Data on kinetics are from studies at 0.1 M sodium phosphate, pH 7.5, 25C (Chi et al., 2018). RefSeq positions are from the Human GRCh38/hg38 genome assembly.
HUGO Gene Nomenclature Committee.
Synonyms based on class designations (Duester et al., 1999) create much confusion in the literature, because one must determine what is meant by, for example, “ADH4”: class II (officially ADH4) or class IV (officially ADH7). We use the approved symbols throughout this article.
Protein subunits have traditionally been named with Greek symbols, but can also be named based upon the gene encoding them; genes are in italics, proteins in roman font.
RNA detected, protein not detected.
Figure 2. Expression of ADH mRNA in selected tissues.
Data are in median transcripts per million transcripts (tpm), from GTEx version 7 (gtexportal.org, exported 15 April 2018) (GTEx Consortium, 2013). ADH genes are shown in numerical order, left to right, within each tissue. Inset shows enlarged image of stomach, brain (maximum tpm across all brain tissues) and whole blood.
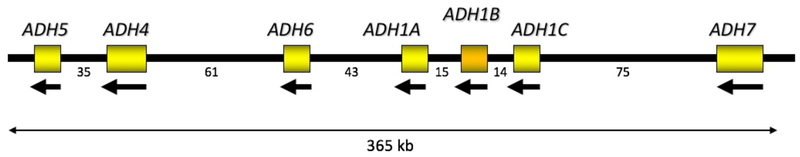
The ADH region of the genome (Figure 3) arose from repeated gene duplication, and many genetic variations in this region are in high linkage disequilibrium (LD), i.e. are inherited together. There are many and often large ethnic differences in allele frequencies and LD patterns. For example, out of 110 SNPs analyzed in a set of European-American and African-American families, 88 had minor allele frequencies (MAF) that differed between the two groups by more than 0.05 (Edenberg et al., 2006) (Table 2). These factors complicate interpretation of the genetic association data and emphasize that it is important to separately analyze different populations and combine data only at the meta-analysis stage.
Figure 3. ADH region of chromosome 4.
ADH genes are arranged head-to-tail along chromosome 4, and transcribed in the opposite direction. Numbers below the line are distances between genes, in kb.
Table 2.
ADH and ALDH2 allele frequencies
| ADH1B*2 | ADH1B*3 | ADH1C*1 | ALDH2*2 | |||
|---|---|---|---|---|---|---|
| rs1229984 | rs2066702 | rs1693482** | rs671 | |||
| Position | 4:99,318,162 | 4:100,229,017 | 4:99,304,835 | 12:111,803,962 | ||
| Genome Allele | T | A | C | A | ||
| RNA Allele | A | T | G | A | ||
| Amino acid | His48 | Cys370 | Arg272 | Lys504 | ||
| Group | Code | Population | ||||
| AFR | ACB | African Carribbeans in Barbados | 0.010 | 0.193 | 0.891 | 0.005 |
| AFR | ASW | Americans of African Ancestry in SW USA | 0.205 | 0.861 | ||
| AFR | ESN | Esan in Nigera | 0.273 | 0.929 | ||
| AFR | GWD | Gambian in Western Divisons in The Gambia | 0.142 | 0.912 | 0.004 | |
| AFR | LWK | Luhya in Webuye, Kenya | 0.141 | 0.859 | ||
| AFR | MSK | Mende in Sierra Leone | 0.088 | 0.906 | ||
| AFR | YRI | Yoruba in Ibadan, Nigera | 0.282 | 0.926 | ||
| AMR | CLM | Colombians from Medellin, Colombia | 0.074 | 0.755 | ||
| AMR | MXL | Mexican Ancestry from Los Angeles USA | 0.086 | 0.719 | 0.008 | |
| AMR | PEL | Peruvians from Lima, Peru | 0.012 | 0.824 | 0.006 | |
| AMR | PUR | Puerto Ricans from Puerto Rico | 0.063 | 0.635 | ||
| ASN | CDX | Chinese Dai in Xishuangbanna, China | 0.634 | 0.887 | 0.043 | |
| ASN | CHB | Han Chinese in Bejing, China | 0.709 | 0.951 | 0.160 | |
| ASN | CHS | Southern Han Chinese | 0.757 | 0.929 | 0.271 | |
| ASN | JPT | Japanese in Tokyo, Japan | 0.731 | 0.928 | 0.240 | |
| ASN | KHV | Kinh in Ho Chi Minh City, Vietnam | 0.646 | 0.919 | 0.136 | |
| EUR | CEU | Utah Residents (CEPH) with Northern and Western European ancestry | 0.015 | 0.525 | ||
| EUR | FIN | Finnish in Finland | 0.490 | |||
| EUR | GBR | British in England and Scotland | 0.005 | 0.560 | ||
| EUR | IBS | Iberian population in Spain | 0.065 | 0.692 | ||
| EUR | TSI | Toscani in Italia | 0.051 | 0.692 | ||
| SAN | BEB | Bengali from Bangladesh | 0.017 | 0.820 | ||
| SAN | GIH | Gujarati Indian from Houston, Texas | 0.024 | 0.718 | ||
| SAN | ITU | Indian Telugu from the UK | 0.015 | 0.765 | ||
| SAN | PJL | Punjabi from Lahore, Pakistan | 0.042 | 0.672 | ||
| SAN | STU | Sri Lankan Tamil from the UK | 0.005 | 0.662 |
Allele corresponding to the variant noted is shown on the genomic reference strand and in the RNA (note that the ADH transcripts run in the opposite direction). Positions are on the GRCh38/hg38 human genome assembly. Data are from the 1000 genomes project, Phase 3 (The 1000 Genomes Project Consortium et al., 2015). Blanks are <0.001. Groups: AFR = African, AMR=American, ASN=East Asian, EUR=European, SAN=South Asian. Code: 3 letter identifier for population.
Frequencies are identical to the more frequently studied SNP rs698 (genome C, RNA G, Val350), except that for rs698, YRI=0.931, ITU= 0.760. For data on a wider range of populations and SNPs, see the alfred database https://alfred.med.yale.edu (Rajeevan et al., 2012).
ADH1B
The kinetic properties of ADH1B and its high levels of expression in liver suggest that it has the largest impact on alcohol consumption and the risk for alcohol dependence; several aspects were reviewed recently (Polimanti and Gelernter, 2018, Edenberg and Bosron, 2018). It is among the top 100 genes expressed in liver, adipose and mammary tissues. It is expressed at lower levels in many other tissues, but at barely detectable levels in brain and whole blood (Figure 2). There are many single nucleotide polymorphisms (SNPs) that affect its expression in one or more tissues (eQTLs), with many concentrated in the region between ADH1C and ADH7 and others between ADH4 and ADH6 (Supplementary Figure 1; all eQTL data are from gtexportal.org (GTEx Consortium, 2013)); many of these SNPs are in strong LD.
There are 3 isoforms of ADH1B that are relatively common in at least some populations. The ADH1B enzyme with arginine at both positions 48 and 3702 is commonly known as ADH1B*1 (in earlier literature it is called β1-ADH or ADHB*1; Table 1). ADH1B*1 metabolizes ethanol at the slowest rate among the 3 isoforms. It is the most common isoform globally except in much of East Asia, and is the form to which others are compared. The isoform with histidine at position 48 is called ADH1B*2 (β2-ADH or ADHB*2), and differs only due to rs1229984. In vitro, ADH1B*2 oxidizes ethanol much faster than ADH1B*1 (Table 1). Computer modeling suggests that at 17 mM ethanol, ADH1B*2 homodimers could oxidize ethanol at about 11 times the rate of ADH1B*1 homodimers (interpolated from (Chi et al., 2018)); heterodimers behave as equal mixtures of the homodimers (Edenberg and Bosron, 2018). The difference in metabolic rate is much smaller in vivo, due to contributions of the other ADH enzymes and limitations by cofactor levels. Neumark et al. (Neumark et al., 2004) found a small (~14%) but significant difference in alcohol elimination rate between subjects of European descent with at least one ADH1B*2 allele compared to those homozygous for ADH1B*1; there was an apparently linear relationship with the number of ADH1B*2 alleles, but the number of ADH1B*2 homozygotes tested was too small for that difference to reach significance. ADH1B*2 increased the frequency of facial flushing in Asians, although the intensity of the flushing was not nearly as great as caused by ALDH2*2 alleles (Takeshita et al., 1996).
The isoform with cysteine at position 370, called ADH1B*3 (β3-ADH or ADHB*3), differs from ADH1B*1 due to rs2066702. The turnover number for ADH1B*3 is more than 60-fold that of ADH1B*1 in vitro (Table 1); at 17 mM ethanol, ADH1B*3/*3 could oxidize ethanol at about 3 times the rate of ADH1B*1/*1 (interpolated from (Chi et al., 2018)). There are only 2 other coding variants with frequencies over 1%, and these have not, in general, been studied for any ADH (Supplementary Information).
ADH1B*2
The kinetic properties of ADH1B*2 and its high frequency in China and Japan (~0.70, Table 2) prompted candidate gene studies founded upon the hypothesis that a variant that affects alcohol metabolism would affect drinking behavior and thereby the risk for AD. Thomasson et al. (Thomasson et al., 1991) found the protective effect of ADH1B*2 was strong (allelic odds ratio (OR) = 0.33) in male Chinese, and independent of that of ALDH2*2 (the inactive aldehyde dehydrogenase; see below). This was followed by many candidate gene studies and meta-analyses in Asian populations. Wherever the frequency of ADH1B*2 was high enough, the same result was obtained: presence of a single ADH1B*2 allele strongly reduced the risk for alcoholism, and in those homozygous for ADH1B*2, the risk was even further reduced (Chen et al., 1999b, Luczak et al., 2006, Whitfield, 2002, Li et al., 2011, Zintzaras et al., 2006, Park et al., 2013)3.
There is heterogeneity among Asian populations in the allele frequency and in the strength of the protection. Han Chinese and Japanese men show the strongest protection (the OR for heterozygotes = 0.18–0.26) (Whitfield, 2002, Chen et al., 1999b, Luczak et al., 2006, Park et al., 2013). Logistic regression of combined ADH2 and ALDH2 genotypes in Han Chinese found that in the presence of active ALDH2 (ALDH2*1 homozygosity), a single ADH1B*2 allele gave an odds ratio (OR) of 0.22, and two ADH1B*2 alleles gave OR = 0.14, both with p<10−6 (Chen et al., 1999b). Minority populations in Asia show less protection (Shen et al., 1997, Thomasson et al., 1994). Meta-analyses that lump all Asian groups show less protection (OR ~ 0.4; p = 10−33 to 7×10−42) (Zintzaras et al., 2006, Li et al., 2011, Luczak et al., 2006, Whitfield, 1997). Because drinking was not as common among Asian women, their overall risk was less and therefore the protective effect were also less (Luczak et al., 2006, Zintzaras et al., 2006).
In a small genome-wide association study (GWAS) plus follow-up in Koreans, rs1229984 (ADH1B*2) gave by far the strongest association with AD (OR = 0.42; p = 2.6×10−21), and once conditioned on rs1229984, no other associations in the region remained significant (Park et al., 2013). In a GWAS among methamphetamine dependent subjects and users in Thailand, rs1229984 was associated with the count of DSM-IV AD symptoms (p = 2.7×10−5) (Gelernter et al., 2018). Rs1229984 was associated with drinking vs. non-drinking in Japan (OR = 1.20, P < 3.6 × 10–4) (Takeuchi et al., 2011). Surprisingly, the East Asians in a US study did not show a significant effect of ADH1B*2 on alcohol consumption (Jorgenson et al., 2017), perhaps due to low overall consumption.
The frequency of ADH1B*2 is very low in most European populations and near zero in African populations (Table 2), making studies of ADH1B*2 outside Asia difficult. An exception is among individuals of Middle Eastern descent (Li et al., 2007), and small studies have shown that the presence of ADH1B*2 in individuals of Jewish descent (MAF ~ 0.2) was associated with reduced consumption, binge drinking, risk, and severity of alcoholism (Hasin et al., 2002, Meyers et al., 2015, Carr et al., 2002, Neumark et al., 1998). Early meta-analysis of small European studies showed that ADH1B*2 was protective, with an OR of 0.28 in men and 0.41 in women (p = 0.0016) (Borras et al., 2000) or 0.47 (Whitfield, 2002). It was also protective in Mexican Americans (OR = 0.28) (Ehlers et al., 2012).
Stronger evidence for association of rs1229984 with alcohol-related phenotypes in individuals of European descent began to accumulate from larger studies. In Denmark, ADH1B*2 was associated with hospitalization for AD (OR = 0.26 in men, 0.37 in women) and with fewer drinks/week and less heavy drinking in both men and women (Tolstrup et al., 2008, Linneberg et al., 2010). Germans with an ADH1B*2 allele drank less per day than those without (Drogan et al., 2012), and ADH1B*2 was strongly associated with AD (p=1.8×10−9) (Treutlein et al., 2014). A US study showed the protective effect of ADH1B*2 on risk for AD was close to that seen in East Asians (OR = 0.34, p= 6.6×10−10) and reduced the maximum drinks in a 24 h period (p=3×10−13) (Bierut et al., 2012). A study in Great Britain showed a similar effect, OR = 0.26 vs. all controls, 0.19 vs. screened controls (p=2.7×10−8) (Way et al., 2015).
In European Americans, rs1229984 was associated with Maxdrinks (p = 6×10−15)(Hart et al., 2016) (Xu et al., 2015) and with the number of DSM-IV and DSM5 criteria (p = 1.4×10−13, 5.3×10−14 respectively), among which withdrawal was the strongest (Hart et al., 2016). In Australian twins, 97% of European descent, those carrying an ADH1B*2 allele reported more flushing after consuming small amounts of alcohol (p = 8.2×10−7), a lower number of Maxdrinks (p = 2.7×10−6), lower total alcohol consumption (p = 8.9×10−8), and fewer DSM-IIIR symptoms of dependence (p = 0.0016) (Macgregor et al., 2009). Jorgenson et al. found rs1229984 was associated with drinker vs. nondrinker status in Americans of both European (p=2.5×10−20) and Hispanic (p=4.4×10−7) descent, and with average drinks per week (p=1.9×10−35 in EA and 2.6×10−6 in Hispanics) (Jorgenson et al., 2017). In a Spanish cohort selected for heavy alcohol consumption and matched controls, ADH1B*2 was associated with protection from heavy drinking in both men (OR = 0.19, p = 4.8×10−10) and women (OR = 0.48 p = 0.0067); other ADH SNPs were not significant when conditioned upon rs1229984 (Munoz et al., 2012). Interestingly, rs12299842 was recently associated with attendance at a pub or social club in Great Britain (p = 4.2 × 10−25) (Day et al., 2018).
A meta-analysis provided strong evidence for association of ADH1B*2 with AD (p=1.2×10−31) and symptom count (p=1.9×10−23) (Gelernter et al., 2014). The latest and largest meta-analysis to date also provides strong evidence for the association of ADH1B*2 with AD in individuals of European ancestry, p= 9.8×10−13 (Walters et al., 2018).
Data on rs1229984 are not available in many GWAS, because it was not included in many genotyping arrays, is not well imputed, its MAF in Europeans falls below the usual cutoff (0.05), and it may fail QC due to differences in MAF among subgroups that lead to apparent violation of Hardy-Weinberg equilibrium (e.g. (Clarke et al., 2017)). In the PGC-SUD meta-analysis, there are data on rs1229984 in only 40% of the subjects (Walters et al., 2018). Thus, in some studies the strongest association of AD is with other SNPs that are in LD with rs1229984.
An initial study from the UK Biobank found 4 SNPs across the ADH region were associated with alcohol consumption, rs145452708, rs29001570, rs35081954, and rs193099203; rs1229984 was not tested because it deviated from Hardy Weinberg equilibrium (Clarke et al., 2017). Their findings at least in part reflect the effects of ADH1B*2, since the associated SNPs are in LD with rs1229984 (D’ = 1, 0.74, 0.91, 0.56, respectively, based on 5 EUR populations, Table 2). In a later UK Biobank GWAS of a partially overlapping sample, ADH1B*2 was very strongly associated with total AUDIT score (p = 5.8×10−72), AUDIT-C (items 1–3, consumption; p = 2.6×10−56), and AUDIT-P (items 7–10, problems; p = 9.9×10−46) (Sanchez-Roige et al., 2018). Conditioning the analysis on rs1229984 rendered other nearby SNPs (except rs13107325) no longer significant, demonstrating that the signal derived from ADH1B*2 (Sanchez-Roige et al., 2018). Meta-analysis of AUDIT scores in the UK biobank and 23andme participants of European ancestry (rs1229984 was not available in 23andme (Sanchez-Roige et al., 2017)) showed rs138495951, in ADH1B, was strongly associated with total AUDIT score (p = 10.7×10−36) (Sanchez-Roige et al., 2018); that SNP (and other associated SNPs in the region) is in LD with rs1229984 (D’ = 1; r2 = 0.54) (Supplementary Figure 2)
The effects of an allele even as strong as ADH1B*2 can be modulated by the environment: the delayed age of first intoxication and first DSM5 symptom in adolescents was reduced if most of their friends drink (Olfson et al., 2014). ADH1B*2 has a stronger effect on alcohol consumption and risk for AUD among those who experience childhood adversity (Meyers et al., 2015).
ADH1B*3
ADH1B*3 is found almost exclusively in individuals of African origin (Table 2). Individuals with an ADH1B*3 allele (ADH1B*369Cys; rs2066702) metabolize ethanol somewhat faster than those with only ADH1B*1 alleles (Thomasson et al., 1995). Within Africa and in African Americans, allele frequencies for ADH1B*3 range from 0.09 to 0.28; in other populations it is essentially absent (Table 2). There are many fewer studies of African populations, an omission that needs to be corrected.
ADH1B*3 has a significant protective effect on risk for alcoholism in African Americans (Edenberg et al., 2006, Gelernter et al., 2014, Walters et al., 2018), and Afro-Trinidadians (Ehlers et al., 2007), and with AD and withdrawal symptoms in Native Americans in southwest California (Wall et al., 2003, Gizer et al., 2011). It appears to be protective against fetal alcohol syndrome, likely by reducing consumption (Warren and Li, 2005, Scott and Taylor, 2007). In a GWAS of African-Americans, rs2066702 was associated with the number of DSM-IV and DSM5 criteria (p = 1.9×10−9, 1.4×10−9, respectively), among which tolerance was the strongest, and with maxdrinks (p = 6.4×10−8) (Hart et al., 2016). A meta-analysis of that sample plus samples from SAGE (Bierut et al., 2010) found strong association with alcohol dependence (OR ~ 0.7; p = 3.7×10−13), DSM-IV symptom counts (p = 6.3×10−17) (Gelernter et al., 2014), and Maxdrinks (p = 2.5×10−10) (Xu et al., 2015). The most recent meta-analysis of African Americans (n = 6280) showed association of ADH1B*3 with AD (p = 2.2×10−9) (Walters et al., 2018). Many SNPs extending across most of the ADH region, from ADH1C to past ADH5, are in LD with rs2066702, and provided supporting evidence (Supplementary Figure 2).
ADH1C
ADH1C is expressed at modest levels in liver (1/3 that of ADH1B), and to a smaller extent in stomach, with little expression in other tissues (Figure 2). There are two major isoforms of ADH1C, and they differ at 2 sites simultaneously: ADH1C*1 (γ1 ADH, ADH1C[Arg272; Ile350]) and ADH1C*2 (γ2 ADH, ADH1C[Gln272;Val350]). The Arg/Gln at position 272 is encoded by rs1693482 and the Ile/Val at 350 by rs698. In vitro kinetic assays show ADH1C*1 is about 1.5 to 2-fold more active than ADH1C*2 (Hurley et al., 2002, Chi et al., 2018) (Table 1). These kinetic differences are almost certainly due to the difference at amino acid 272 (Arg/Gln; rs1693482). Most genetic literature focuses on the other SNP, rs698, for historic and technical reasons (Xu et al., 1988). This does not affect conclusions, because Arg272 is virtually always found together with Ile350, and Gln272 with Val350: the correlation between these SNPs is complete (r2 = 1.0) in 24 of the 26 populations in the 1000 genomes database, and nearly so in the other 2 (r2 = 0.97 in ITU, 0.93 in YRI). Thus measuring either SNP gives essentially the same information. Many other SNPs are highly correlated with rs698/rs1693482. In both Asians (e.g. CHB) and European-Americans (e.g. CEU) more than 100 SNPs with r2>0.90 span a 38 kb region that also covers much of ADH1B.
The association of ADH1C with alcohol dependence is less robust than that of ADH1B. ADH1C*2 is associated with AD and consumption in East Asians (e.g. (Thomasson et al., 1991, Thomasson et al., 1994, Matsuo et al., 2007)), where ADH1C*1 (the higher activity, protective form) is at high frequency (Table 2) and tends to travel with ADH1B*2 (higher activity, protective); D’ = 0.78 in CHB+JPT. The LD pattern led to suggestions that the evidence for an effect of ADH1C*1 independent of ADH1B*2 was weak (Osier et al., 1999, Chen et al., 1999b, Choi et al., 2005). A meta-analysis suggested that ADH1C*1 was protective (OR = 0.52) (Zintzaras et al., 2006). A later meta-analysis found stronger evidence that ADH1C*1 was protective against AD in Asians (OR = 0.47, p = 4×10−33) but was not significant in Europeans (Li et al., 2012a). Neither meta-analysis explicitly examined whether the effect was independent of ADH1B genotype.
In people of European origin, where ADH1B*2 is at very low frequency, there is less confounding. Several studies have shown no (Neumark et al., 2004, Luo et al., 2006b, Borras et al., 2000) or only nominal (Edenberg et al., 2006, Agrawal et al., 2011, Kuo et al., 2008, Li et al., 2012a) allelic association between AD and rs698, rs1693482 or rs1789891 (D’ = 1) in Europeans. In a GWAS of a factor score derived from symptoms of alcohol dependence among controls from a study of schizophrenia, no SNP reached significance, but the strongest result from a candidate-gene-based analysis was ADH1C (p = 0.003 in European-Americans)(Kendler et al., 2011); this was not a finding for a single SNP, but rather a group of SNPs in the region.
Several more recent studies have provided evidence for an independent effect of ADH1C*1 on alcohol dependence, but extensive LD in the ADH region has led to associations of different SNPs. A GWAS and follow-up of key SNPs in German males with early onset alcohol dependence provided evidence of association of rs1614972 (in LD with rs698 and rs1693482, D’=1, r2 = 0.31) with AD (p = 1.4×10−4) but it did not withstand correction for multiple testing (Treutlein et al., 2009). Enlarging that sample provided genome-wide significant evidence for association of a different SNP, rs1789891 (D’ with rs1693482 =1, r2 = 0.22) with alcohol dependence (1.3×10−8, OR4 = 0.68) (Frank et al., 2012). A follow up of SNPs from the Treutlein study and provided limited statistical support (p=0.0017) for association of rs1614972 with AD in a different population (OR = 0.8) (Biernacka et al., 2013). Rs1789891 was associated with alcohol dependence in a study of British and Irish (p = 7.2×10−5; OR =0.71), and the association remained significant when conditioned on rs1229984 (ADH1B*2; p = 1.7×10−4) (Way et al., 2015). In the PGC-SUD trans-ancestral meta-analysis, rs1789912 was associated with alcohol dependence (p = 1.47×10−9) (Walters et al., 2018); it is in complete LD with rs698/rs1693482 (r2 = 1). In analyses of Europeans, conditional on ADH1B*2, the 2 SNPs that define ADH1C*1 and 2 others (rs1789912, rs1154445; pconditional = 7.7×10−4, 1.7×10−4) that were in complete LD (r2 = 1) with them retained some evidence of association (Walters et al., 2018).
A GWAS on AUDIT score in a basically healthy European-American (EA) population provided evidence for association with rs141973904, an uncommon allele (MAF = 0.016) in ADH1C (p = 4.4×10−7) (Sanchez-Roige et al., 2017). The minor allele of rs141973904 is found with the allele of rs1693482 that encodes ADH1C*1 (D’=1) but because of the large difference in allele frequencies their correlation is very low (r2 = 0.01). Rs141973904 is also in high LD with rs1229984 (ADH1B*2; D’ = 1, r2 = 0.54), which was not available for testing but might well have been the functional allele responsible for the finding.
Association with alcohol consumption among Europeans has given mixed results. A study of alcohol elimination in Australian twins did not find evidence for an effect of either rs698 or rs1693482 (Birley et al., 2009), but a later study showed an effect of rs1693482 on maxdrinks that was still nominally significant after controlling for rs1229984 (p=7×10−4, with 50 SNPs tested) (Macgregor et al., 2009). Several studies reported no independent effect of ADH1C*1 on average drinking (Latella et al., 2009, Drogan et al., 2012) or upon likelihood of very heavy drinking (Munoz et al., 2012). In a meta-analysis of average drinking (g/kg/day) among Europeans, rs1789891 was nominally significant (p = 1.2×10−3) if controls were restricted to drinkers (Schumann et al., 2011). A larger follow-up showed suggestive evidence for association of SNPs in the ADH region (1.4×10−6 to 8.5×10−5), most in ADH1C and ADH7 (Schumann et al., 2016); many of those SNPs were in complete LD with rs698 (r2>0.99, D’=1) and also in LD with rs1229984 (D’>0.9). A large study in Denmark found an association of rs698 with heavy drinking in both men and women (OR for ADH1C*1 ~ 0.75), excessive drinking in men (OR = 0.63) and hospitalization for AD in women (OR = 0.71 – 0.45 for heterozygotes and homozygotes) (Tolstrup et al., 2008); secondary analysis excluding individuals carrying ADH1B*2 gave similar results.
Overall, there is evidence that ADH1C*1 is protective against alcohol dependence, but the LD in the region, particularly across ADH1B and ADH1C, makes interpretation of many of the studies difficult. In particular, the high LD with ADH1B*2 (D’ = 0.91 in Europeans, although r2 is low) is generally not acknowledged. Given the strong effect of ADH1B*2 on these phenotypes conditional analyses are important. Another way to disambiguate the situation would be to separately analyze the data in the large group without any ADH1B*2 alleles, as was done in the Danish study (Tolstrup et al., 2008).
ADH4
ADH4 (π-ADH) has Km for ethanol of 34 mM (Table 1). Its expression is relatively high in liver and extremely low elsewhere. A paradox is that there are over 5500 eQTL for ADH4, but all are in tissues in which expression is extremely low; there are no significant eQTLs affecting the expression of ADH4 in liver. There are few coding variants in ADH4, one of which (Ile309Val, rs1126671) affects the stability of the enzyme and its binding of ethanol (Stromberg et al., 2002); it is relatively common in Europeans (MAF = 0.30) and Africans (MAF=0.14) but rare in East Asians (MAF ~ 0.001).
In a family-based study that used the pedigree disequilibrium test, 12 SNPs in and near ADH4 were associated with DSM-IV-defined alcohol dependence in European American families; the top SNP was rs4148886 (Edenberg et al., 2006). Eleven of the SNPs are in LD and mark a region from intron 1 past the 3’ untranslated region that contains many additional SNPs (Edenberg et al., 2006). Neither of 2 non-synonymous SNPs, rs1126671 and rs1126673 nor a functional promoter SNP, rs1800759 (Edenberg et al., 1999) were significant, although rs1800759 had been in an earlier study in Brazil (Guindalini et al., 2005). None of a set of 7 SNPs (in nearly complete LD) showed significant association, but deviation from Hardy-Weinberg equilibrium in European Americans suggested a recessive effect; there was no evidence for association in African Americans (Luo et al., 2006a). In the Irish, there was no association of ADH4 with AD (Kuo et al., 2008). A rare variant downstream of ADH4 (rs187709743) was associated with symptom count in American Indians (Peng et al., 2017). An Australian study found suggestive evidence for association of rs1800759 with lifetime maxdrinks (p = 0.0075), frequency of drinking (p = 0.0055), and total consumption (p = 0.0023), and of rs3762894 with maxdrinks during the past year (p = 0.00048) and usual number of drinks (p = 0.00078) (Macgregor et al., 2009). The evidence dropped substantially after conditioning on ADH1B*2, but some evidence for association of rs3762894 with maxdrinks remained (p = 0.004) (Macgregor et al., 2009). In Koreans, several ADH4 SNPs were significant, the best being rs3805322 (p = 2.0×10−13); however conditioning the analysis on ADH1B*2 genotype reduced all of the SNPs to not significant (p>0.23) (Park et al., 2013).
ADH5
ADH5 encodes χ-ADH (Table 1), which is also a glutathione-dependent formaldehyde dehydrogenase. ADH5 is the most widely expressed of the ADHs, present in essentially all tissues (Figure 2). It has very low affinity for ethanol, but mouse studies suggest that its role might be more significant than originally thought when alcohol levels are high (Haseba and Ohno, 2010). In a small study of first pass metabolism, ADH5 made a contribution when the concentration of alcohol ingested was high (40%) (Dohmen et al., 1996).There are 2667 eQTLs affecting expression of ADH5 in various tissues, 221 in liver and 228 in cerebellum.
A number of studies have provided modest evidence for association of SNPs in the ADH4-ADH5 region with AD (Edenberg et al., 2006) (Kuo et al., 2008) (Kendler et al., 2011) (Luo et al., 2006b). A key issue to keep in mind is that there is a very strong LD block that extends across ADH4 and ADH5, so findings in ADH5 might relate to effects in ADH4, to regulatory effects on other genes, or to LD with rs1229984 in ADH1B. In a Korean GWAS, the initial evidence for association of 2 SNPs in ADH5 with AD disappeared when conditioned on ADH1B*2 (Park et al., 2013).
ADH7
ADH7 (σ-ADH or μ-ADH; Table 1), is the only member of the ADH family that is not expressed in liver (Figure 2). It has a high turnover number, and its high Km suggests it will be most active when ethanol concentrations are high, as they are during ethanol consumption in the esophagus (where its level of expression is affected by 62 eQTLs) and stomach, precisely its locations. A small study showed a significant contribution of ADH7 to first pass metabolism, particularly after low concentrations of oral ethanol (Dohmen et al., 1996).
A single SNP in ADH7 (rs284786) was nominally associated with a DSM-IIIR-based definition of alcohol dependence (Edenberg et al., 2006), one downstream of ADH7 was suggestively associated with AD in Mexican-Americans (Norden-Krichmar et al., 2014), and several in that region were associated with maxdrinks in Native Americans (Peng et al., 2014b). Analysis of alcohol levels after an oral alcohol challenge with 103 SNPs across the ADH region showed early effects from SNPs in and near the 5’ region of ADH7 through intron 6, with only nominally significant effects of SNPs across the region between ADH7 and ADH1A (Birley et al., 2009). In a meta-analysis of average drinking (g/kg/day) among Europeans the most significant SNP in the ADH region was rs2584448 in ADH7 (p=3.9×10−4); when the analysis restricted the controls to drinkers, the top SNP was rs2165672, also in ADH7 (Schumann et al., 2011), neither was genome-wide significant.
ADH1A:
ADH1A is expressed at lower levels in liver than ADH1B or ADH1C, and is barely expressed in other adult tissues (Figure 2). ADH1A is expressed early in fetal development, and may play a role there (Smith et al., 1971). Coding variations are essentially non-existent, with none having an allele frequency of 1% or above in any population studied (Lek et al., 2016). The lack of coding variants and low level of expression in adults suggests that variations in ADH1A are not likely to play major roles in affecting risk for alcoholism. There is nominal evidence that several SNPs are associated with AD (Edenberg et al., 2006, Kuo et al., 2008) but that is likely due to LD with SNPs in ADH1B.
ADH Regulatory variants
The strong effect of ADH1B and ADH1C coding variants may obscure more modest effects of regulatory variants. Coding SNPs that lead to more active ADH enzymes are protective, so it is logical to anticipate that regulatory variants that increase expression of those enzymes have a similar, if more modest, effect. Individual SNPs and haplotypes have been shown to affect expression of ADH genes, including ADH1B (Pochareddy and Edenberg, 2011), ADH1C (Chen et al., 2005), ADH4 (Edenberg et al., 1999, Pochareddy and Edenberg, 2010), and ADH7 (Jairam and Edenberg, 2014a, Jairam and Edenberg, 2014b). Some mapped regulatory elements that affect ADH1B expression in liver-derived cells lie in the region between ADH1B and ADH7 (Chen et al., 2005, Jairam and Edenberg, 2014a, Jairam and Edenberg, 2014b). There are many eQTLs, extending broadly across the region, that affect expression of one or more ADH genes. These differ in different tissues; for example, in subcutaneous adipose there is a dense cluster between ADH7 and ADH1C and a small cluster over 700 kb away, whereas in visceral adipose there is a cluster between ADH4 and ADH6, extending beyond ADH5 (Supplementary Figure 1).
The large trans-ethnic meta-analysis of subjects of European and African descent carried out by the Psychiatric Genomics Consortium Substance Use Disorders working group (PGC-SUD) found that rs10516440 (associated with AD at p = 9.9×10−8; p conditioned on rs1229984 =7.4×10−5) was a significant eQTL for ADH1B in a trans-tissue analysis (p = 1.4×10−76, gtexportal.org), although only nominally significant in liver (Walters et al., 2018). The major allele of rs10516440 (A) was associated with increased ADH1B expression and reduced AD risk, concordant with the expected direction.
ADH results to date
There is very strong evidence, both biochemical and genetic, that two coding variants in ADH1B that affect its kinetic properties (rs1229984 and rs2066702; ADH1B*2 and ADH1B*3 respectively) affect alcohol consumption and risk for alcohol dependence. Their effect on risk for AD is among the strongest of any variant. There is also good evidence for an independent effect of a coding variant in ADH1C (rs698 and rs1693482), although with less effect. There is weaker evidence that other ADH genes affect risk and consumption. Supplementary Table 1 shows ADH SNPs reported at p values < 10−6. The extensive LD in the region, however, makes association of specific SNPs other than the coding variants in ADH1B and ADH1C with AD difficult. Supplementary Figure 2 shows the strong LD among all of the SNPs in the ADH region that are listed in Supplementary Table 1. SNPs that lie within or near the other ADH genes, as well as some outside the area, are in strong LD with those coding variants, and might also act by altering expression of one of the ADH genes.
Aldehyde dehydrogenase enzymes
The second step in the metabolism of ethanol, the oxidation of acetaldehyde to acetate, is important for eliminating the potentially toxic acetaldehyde (Zakhari, 2006). Unlike the oxidation of ethanol to acetaldehyde, this step is essentially irreversible (Hurley et al., 2002). There are 19 human aldehyde dehydrogenases, but three closely related ones (68% amino acid sequence identity) are most relevant to the metabolism of acetaldehyde: ALDH1A1, ALDH1B1 and ALDH2 (Jackson et al., 2011, Vasiliou et al., 2004). All three act as homotetramers, and have broad substrate specificities.
ALDH2
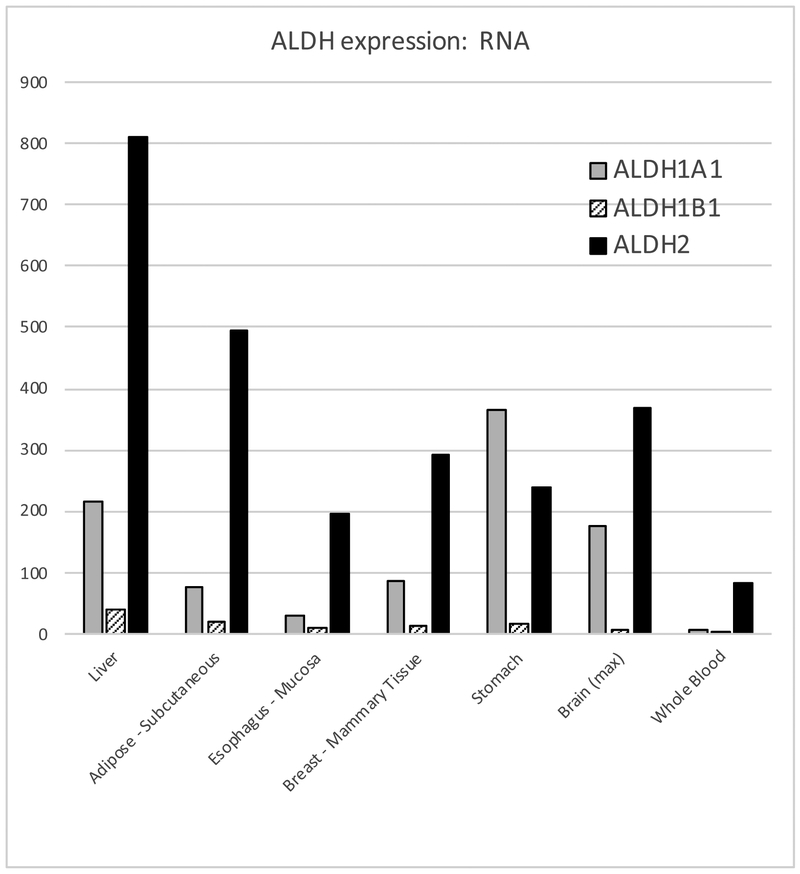
ALDH2, the mitochondrial ALDH, has a very high affinity for acetaldehyde (KM = 0.2 μM) and a high reaction velocity (Vmax = 280/min) (Hurley et al., 2002, Klyosov, 1996) (Table 3). ALDH2 rapidly eliminates most of the acetaldehyde, unless it is inhibited by disulfiram or by an inactivating mutation (see below). In individuals with active ALDH2 enzyme, acetaldehyde in the bloodstream ranges from undetectable to about 3 μM, roughly 1000-fold less than the levels of ethanol (Mizoi et al., 1994, Peng et al., 2014a, Harada et al., 1983, Nuutinen et al., 1984). ALDH2 is expressed ubiquitously, with highest levels in liver and adipose (Figure 4); it is among the top 100 genes expressed in liver. No eQTLs affect its expression in liver.
Table 3.
ALDH genes and enzyme kinetics
| Approve d Gene Symbola |
Approved Gene Namea |
Synonyms | RNA: RefSeq Accession ID |
Subunit encoded |
KM, acetaldehyde (mM) |
Activity Kcat (min-1) |
RefSeq position |
|---|---|---|---|---|---|---|---|
| ALDH2 | aldehyde dehydrogenase 2 | ALDHI; ALDH-E2; ALDM | NM_000690 | - | - | 12:111766887-111809985 | |
| ALDH2*1 | ALDH2*Glu504 | ALDH2*1 ALDH2[Glu504]b | 0.2 | 280 | |||
| ALDH2*2 | ALDH2*Lys504 | ALDH2*2 ALDH2[Lys504]b | c | ||||
| ALDH1B1 | aldehyde dehydrogenase 1 family member B1 | ALDH5; ALDHX | NM_000692 | 55 | 655 | 9:38392664-38398665 | |
| ALDH1A1 | aldehyde dehydrogenase 1 family member A1 | ALDH1; ALDH-E1; ALDH11; RALDH1; ALDC | NM_000689 | 180 | 380 | 9:72900662-72953317 |
RefSeq positions are from the Human GRCh38/hg38 genome assembly. Data on kinetics are from (Klyosov, 1996) (ALDH2, ALDH1A2) and (Stagos et al., 2010) (ALDH1B1).
HUGO Gene Nomenclature Committee.
Position in precursor protein; aa487 in the mature protein.
ALDH2 is essentially inactive under physiological conditions.
Figure 4. Expression of key ALDH RNAs in selected tissues.
Data are median tpm, from GTEx version 7 (exported 15 April 2018) (GTEx Consortium, 2013).
There are 2 main isoforms of the ALDH2 enzyme. The one common in most of the world, ALDH2*1, has glutamate at amino acid 487 of the mature protein (504 of the precursor). A variant, ALDH2*2, has a lysine there instead, encoded by rs671. Allele frequencies for ALDH2*2 are highest in Han Chinese and Japanese, with lower frequencies elsewhere in Asia; it is rarely found outside Asia (Table 2). Even a single ALDH2*2 subunit renders the tetrameric enzyme nearly inactive under physiological conditions (Crabb et al., 1989, Zhou and Weiner, 2000, Hurley et al., 2002) and it is also more rapidly degraded (Xiao et al., 1996).
Presence of a single ALDH2*2 allele is protective against heavy drinking and alcohol dependence in a semi-dominant manner (Crabb et al., 1989). People carrying even one ALDH2*2 allele can have blood acetaldehyde levels of 30 – 75 μM or higher, more than 10 times the normal level (Peng et al., 2014a, Harada et al., 1983, Adachi et al., 1989). This causes a severe form of flushing that includes increased skin temperature, nausea, vomiting, headaches, and increased pulse rate (Goedde et al., 1979, Goedde et al., 1983, Harada et al., 1981, Mizoi et al., 1983, Shibuya et al., 1989). The effects are similar to those of disulfiram (Antabuse®), a drug approved for treatment of AUD. This aversive reaction reduces the propensity to drink, the amount consumed per occasion, and thereby the risk for alcoholism (Bosron and Li, 1981, Harada et al., 1982, Thomasson et al., 1991, Hurley et al., 2002, Edenberg, 2007, Crabb et al., 1989, Chen et al., 2009, Luczak et al., 2006, Goedde et al., 1992, Edenberg, 2012, Chen et al., 1999b, Whitfield, 2002, Hurley and Edenberg, 2012). In Han Chinese, the presence of a single ALDH2*2 allele in a background of homozygous ADH1B*1 gave an OR of 0.40; when combined with a single ADH1B*2 allele the OR dropped to 0.06 (Chen et al., 1999b). Meta-analysis of 22 datasets showed an OR of 0.22 for ALDH2*2 heterozygotes (Luczak et al., 2006). A later meta-analysis showed similarly strong protection against AD: OR = 0.22 (p = 1 × 10−44) under a dominant model that is probably close to the physiological effects of the variant (Li et al., 2012b). The protective alleles at ADH1B and ALDH2 act synergistically to give a relative risk of alcoholism in Asians of 1–10% (Chen et al., 1999a, Luczak et al., 2006).
Heterozygotes have a small fraction of ALDH2*1 homotetramers that provide some residual activity. Homozygotes for ALDH2*2 have no detectable ALDH2 activity and are essentially completely protected against alcohol dependence because they cannot tolerate even one standard drink of alcohol (Higuchi et al., 1994).
In Chinese subjects from rural Northern Hunan Province ALDH2*2 was associated with flushing (p=4.8 ×10−26), reduced the number of maxdrinks (p = 1.5 ×10−16), and was protective against alcohol dependence (p=4.7 ×10−8) (Quillen et al., 2014). SNPs in nearby genes also appeared to be associated (Supplementary Table 2), due to the extensive LD in the region (D’ between rs671 and many SNPs across 1 Mb is over 0.6; Supplementary Figure 3): conditioning on rs671 did not leave any others significant, including a previously reported association in CCDC63 (Quillen et al., 2014). ALDH2*2 explained a substantial fraction of the total phenotypic variance, 7.9% for AD, 22.9% for maxdrinks and 29.3% for flushing (Quillen et al., 2014). Women in that study had very low levels of alcohol consumption, so analyses of women had little or no power. A GWAS on a small number of Korean men found rs671 was associated with alcohol dependence (p = 8.4 ×10−8; OR = 0.22) (Park et al., 2013). A recent GWAS in Thai subjects (ascertained for methamphetamine dependence or use) gave similar results: significant association of rs671 with flushing (5.2×10−14), maxdrinks (1.3×10−10) and DSM-IV criterion count (4.5 ×10−9) (Gelernter et al., 2018).
Ten SNPs on chromosome 12 were significantly associated with the log of the average drinks/day in Korea (Baik et al., 2011). Surprisingly, they did not test rs671; all 10 SNPs are in LD with rs671 (D’ = 0.54 to 0.85), which was almost certainly driving the associations. In Japan, rs671 was very strongly associated with drinkers vs. non-drinkers (OR = 0.16, p = 3.6×10–211); the significance of other SNPs within 0.7 Mb disappeared when adjusted for rs671 (Takeuchi et al., 2011).
Among young adult students of Asian background in the US, those with ALDH2*1/*2 drank less frequently and lower quantities of alcohol, and had fewer heavy drinking episodes and lower maxdrinks (Otto et al., 2013). In a GWAS of Americans of East Asian background, rs671 was very strongly associated with drinking status (drinking at least once per week, OR = 0.40, p = 2.3 ×10−72), but had a weaker effect on typical number of drinks per week among drinkers (p = 5.4×10−4) (Jorgenson et al., 2017); conditional analysis showed no other significant signals in the ALDH2 region.
Regulation of the amount of ALDH2 enzyme produced would also be expected to alter the reaction to alcohol, but studies of the promoter variant rs886205 (Chou et al., 1999) have not shown an independent effect on AD (Harada et al., 1999, Kimura et al., 2006) or risky drinking (Haschemi Nassab et al., 2015).
In Japan, the protection against AD from a single ALDH2*2 allele dropped sharply from 1979 to 1992, as the pressure to drink socially and as part of business culture increased (Higuchi et al., 1994). This is a striking example of gene x environment interaction.
ALDH1B1
ALDH1B1 is 75% identical to ALDH2, and is also located in mitochondria (Stagos et al., 2010, Jackson et al., 2011, Vasiliou et al., 2013, Stewart et al., 1995). It is expressed at much lower levels than ALDH2 (Figure 4). Both because of its lower expression and its much lower affinity for acetaldehyde (Table 3) ALDH1B1 does not normally play a large role in acetaldehyde oxidation. However, knocking out Aldh1b1 in mice led to a significant increase in blood acetaldehyde after ethanol consumption (Singh et al., 2015).
Two missense variants in ALDH1B1 are predicted to be damaging (Way et al., 2017). The Ala86Val variant (ALDH1B1*2; rs2228093) was inactive when expressed in vitro (Jackson et al., 2015). In a Danish allergy cohort, rs2228093 was correlated with fewer drinks/week and alcohol-induced hypersensitivity (rash, itch), although rs2073478, in LD with it, was not (Husemoen et al., 2008, Linneberg et al., 2010). Rs2073478 (Arg107Leu) was associated with heavy drinking in Inuit in Greenland (Bjerregaard et al., 2014). However, neither rs2228093 nor rs2073478 was associated with alcohol dependence in a larger study of British individuals (Way et al., 2017).
ALDH1A1
ALDH1A1 is a cytosolic enzyme that has a low affinity for acetaldehyde (Table 3). It is expressed at lower levels than ALDH2 in most tissues except stomach (Figure 4). As with ALDH1B1, it probably plays only a small role in acetaldehyde elimination, predominantly when ALDH2 is not active and thus acetaldehyde levels are high. Low activity of this enzyme (measured in erythrocytes) correlated with a mild flushing reaction in Europeans that did not affect alcohol consumption (Ward et al., 1994, Yoshida et al., 1989).
There are several low frequency variants of ALDH1A1 that have been nominally associated with alcoholism-related phenotypes, including ALDH1A*2, a 17 bp promoter deletion, and ALDH1A*3, a 3 bp promoter insertion5 that showed a weak trend toward association with alcoholism in African Americans (Spence et al., 2003). ALDH1A1*2 variant was reported to be associated with higher consumption and increased risk for AD among Trinidadians of Indian descent (Moore et al., 2007), but in Mission Indians it showed the opposite direction (Ehlers et al., 2004), and there was no association with drinking in young adult students of Asian background in the US (Otto et al., 2013). An uncommon intronic variant, rs8187974 was nominally associated with both DSM-IV AD and maxdrinks in European Americans (Sherva et al., 2009). Three SNPs were nominally associated with an alcohol consumption score factor in European American women (Agrawal et al., 2011), and several with problem drinking and AD in European populations (Lind et al., 2008). None have shown up in GWAS. Taken together, the evidence that variants affect AD or drinking behavior is weak.
ALDH results to date
There is overwhelming evidence, both biochemical and genetic, that ALDH2*2 reduces alcohol consumption, particularly heavy drinking, and greatly reduces the risk for AD, through its triggering of a strong flushing reaction. Reports of association of other genes on chromosome 12 within 1 – 2 Mb of ALDH2 in populations in which ALDH2*2 is present are nearly certain to be due to strong LD with this functional variant (Supplementary Figure 3), and the evidence for effects of the other variants disappears when conditioned on rs671. Evidence for association of ALDH1A1 and ALDH1B1 is very weak.
Conclusions
The coding variants ADH1B*2, ADH1B*3, ADH1C*1 and ALDH2*2 all provide some protection against excessive alcohol consumption and thereby against alcohol dependence. The effect sizes for ADH1B*2 (rs1229984) and ALDH2*2 (rs671) are high for a complex disease. Presence of even a single ALDH2*2 allele leads to high levels of acetaldehyde in blood and a very strong flushing reaction. Although the ADH1B*2, ADH1B*3 and ADH1C*1 variants do not by themselves lead to high levels of acetaldehyde because an active ALDH2 enzyme so efficiently oxidizes it to acetate, they also provide significant protection. Allele frequencies of these coding SNPs differ widely among populations, as do the patterns of LD, and the impact of a variant can be modified by different environments, so it is important to broaden studies to a wider range of populations.
The evidence for effects of other ADH and ALDH genes is much weaker. Regulatory variants and other coding variants in and around the ADH region and the key ALDHs are also likely to affect risk for AUDs and alcohol consumption, but because they have much smaller effects and because analyses are complicated by the LD in the region, larger and more diverse datasets are needed to reliably determine their independent effects.
Beyond the genes encoding these metabolic enzymes, there are probably at least hundreds to thousands of additional genes, interacting with the environment, that affect the risk for AUDs and excessive alcohol consumption. With the exception of the protection offered by homozygosity for ALDH2*2, no one gene or combination of genes is determinative. Understanding which other genes affect risk, and the mechanisms by which they do, should enable progress in prevention and treatment. Much larger, well-characterized samples are needed to identify these variants of small effect, and thereby to better understand AUDs and the other effects of alcohol. Even variants that individually make only a very small contribution to risk can reveal key pathways and mechanisms of risk, which can then be targeted for treatment and prevention.
Supplementary Material
Acknowledgments:
Related work in the authors’ laboratory is supported by NIH Grants U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA), and U01MH109532 from the National Institute on Mental Health (NIMH) and the National Institute on Drug Abuse (NIDA).
Footnotes
Genes are in italics, proteins in roman font.
Current nomenclature counts from the initiating methionine of the initially synthesized peptide. Older literature and the protein database count from the first amino acid of the mature protein, and therefore calls these 47 and 369
References to many earlier studies are in the reviews cited.
Some report the OR for ADH1C*2 (risk allele), but to be consistent with how we discuss ADH1B, these have been converted to show the OR for the protective allele.
These sequences were not found in dbSNP, but there are two 17 bp deletions at approximately the site of ALDH1A1*2, rs81887866 and rs6151031.
References
- Adachi J, Mizoi Y, Fukunaga T, Ogawa Y, Imamichi H (1989) Comparative study on ethanol elimination and blood acetaldehyde between alcoholics and control subjects. Alcohol Clin Exp Res 13:601–604. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Lynskey MT, Todorov AA, Schrage AJ, Littlefield AK, Grant JD, Zhu Q, Nelson EC, Madden PA, Bucholz KK, Sher KJ, Heath AC (2011) A candidate gene association study of alcohol consumption in young women. Alcohol Clin Exp Res 35:550–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association (2013) Diagnostic and Statistical Manual of Mental Disorders: DSM-5, AMERICAN PSYCHIATRIC PUBLISHING. [Google Scholar]
- Baik I, Cho NH, Kim SH, Han BG, Shin C (2011) Genome-wide association studies identify genetic loci related to alcohol consumption in Korean men. Am J Clin Nutr 93:809–816. [DOI] [PubMed] [Google Scholar]
- Biernacka JM, Geske JR, Schneekloth TD, Frye MA, Cunningham JM, Choi DS, Tapp CL, Lewis BR, Drews MS, T LP, Colby CL, Hall-Flavin DK, Loukianova LL, Heit JA, Mrazek DA, Karpyak VM (2013) Replication of genome wide association studies of alcohol dependence: support for association with variation in ADH1C. PLoS One 8:e58798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, Hinrichs AL, Almasy L, Breslau N, Culverhouse RC, Dick DM, Edenberg HJ, Foroud T, Grucza RA, Hatsukami D, Hesselbrock V, Johnson EO, Kramer J, Krueger RF, Kuperman S, Lynskey M, Mann K, Neuman RJ, Nothen MM, Nurnberger JI Jr., Porjesz B, Ridinger M, Saccone NL, Saccone SF, Schuckit MA, Tischfield JA, Wang JC, Rietschel M, Goate AM, Rice JP, Gene EASC (2010) A genome-wide association study of alcohol dependence. Proc Natl Acad Sci U S A 107:5082–5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Goate AM, Breslau N, Johnson EO, Bertelsen S, Fox L, Agrawal A, Bucholz KK, Grucza R, Hesselbrock V, Kramer J, Kuperman S, Nurnberger J, Porjesz B, Saccone NL, Schuckit M, Tischfield J, Wang JC, Foroud T, Rice JP, Edenberg HJ (2012) ADH1B is associated with alcohol dependence and alcohol consumption in populations of European and African ancestry. Mol Psychiatry 17:445–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birley AJ, James MR, Dickson PA, Montgomery GW, Heath AC, Martin NG, Whitfield JB (2009) ADH single nucleotide polymorphism associations with alcohol metabolism in vivo. Hum Mol Genet 18:1533–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerregaard P, Mikkelsen SS, Becker U, Hansen T, Tolstrup JS (2014) Genetic variation in alcohol metabolizing enzymes among Inuit and its relation to drinking patterns. Drug Alcohol Depend 144:239–244. [DOI] [PubMed] [Google Scholar]
- Borras E, Coutelle C, Rosell A, Fernandez-Muixi F, Broch M, Crosas B, Hjelmqvist L, Lorenzo A, Gutierrez C, Santos M, Szczepanek M, Heilig M, Quattrocchi P, Farres J, Vidal F, Richart C, Mach T, Bogdal J, Jornvall H, Seitz HK, Couzigou P, Pares X (2000) Genetic polymorphism of alcohol dehydrogenase in Europeans: the ADH2*2 allele decreases the risk for alcoholism and is associated with ADH3*1. Hepatol. 31:984–989. [DOI] [PubMed] [Google Scholar]
- Bosron WF, Ehrig T, Li T-K (1993) Genetic factors in alcohol metabolism and alcoholism. Semin.Liver.Dis. 13:126–135. [DOI] [PubMed] [Google Scholar]
- Bosron WF, Li T-K (1981) Genetic determinants of alcohol and aldehyde dehydrogenases and alcohol metabolism. Semin.Liver.Dis. 1:179–188. [DOI] [PubMed] [Google Scholar]
- Carr LG, Foroud T, Stewart T, Castelluccio P, Edenberg HJ, Li TK (2002) Influence of ADH1B polymorphism on alcohol use and its subjective effects in a Jewish population. Am J Med Genet 112:138–143. [DOI] [PubMed] [Google Scholar]
- Chen CC, Lu RB, Chen YC, Wang MF, Chang YC, Li TK, Yin SJ (1999a) Interaction between the functional polymorphisms of the alcohol-metabolism genes in protection against alcoholism. American journal of human genetics 65:795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HJ, Tian H, Edenberg HJ (2005) Natural haplotypes in the regulatory sequences affect human alcohol dehydrogenase 1C (ADH1C) gene expression. Hum Mutat 25:150–155. [DOI] [PubMed] [Google Scholar]
- Chen YC, Lu RB, Peng GS, Wang MF, Wang HK, Ko HC, Chang YC, Lu JJ, Li TK, Yin SJ (1999b) Alcohol metabolism and cardiovascular response in an alcoholic patient homozygous for the ALDH2*2 variant gene allele. Alcoholism, clinical and experimental research 23:1853–1860. [PubMed] [Google Scholar]
- Chen YC, Peng GS, Tsao TP, Wang MF, Lu RB, Yin SJ (2009) Pharmacokinetic and pharmacodynamic basis for overcoming acetaldehyde-induced adverse reaction in Asian alcoholics, heterozygous for the variant ALDH2*2 gene allele. Pharmacogenetics and genomics 19:588–599. [DOI] [PubMed] [Google Scholar]
- Chi Y-C, Lee S-L, Lee Y-P, Lai C-L, Yin S-J (2018) Modeling of Human Hepatic and Gastrointestinal Ethanol Metabolism With Kinetic Mechanism-Based Full Rate Equations of the Component Alcohol Dehydrogenase Isozymes and Allozymes. Chemical Research in Toxicology. [DOI] [PubMed] [Google Scholar]
- Choi IG, Son HG, Yang BH, Kim SH, Lee JS, Chai YG, Son BK, Kee BS, Park BL, Kim LH, Choi YH, Shin HD (2005) Scanning of genetic effects of alcohol metabolism gene (ADH1B and ADH1C) polymorphisms on the risk of alcoholism. Hum Mutat 26:224–234. [DOI] [PubMed] [Google Scholar]
- Chou W-Y, Stewart MJ, Carr LG, Zheng D, Stewart TR, Williams A, Pinaire J, Crabb DW (1999) An A/G polymorphism in the promoter of mitochondrial aldehyde dehydrogenase (ALDH2): effects of the sequence variant on transcription factor binding and promoter strength. Alcohol.Clin.Exp.Res. 26:963–968. [PubMed] [Google Scholar]
- Clarke TK, Adams MJ, Davies G, Howard DM, Hall LS, Padmanabhan S, Murray AD, Smith BH, Campbell A, Hayward C, Porteous DJ, Deary IJ, McIntosh AM (2017) Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N=112 117). Mol Psychiatry 22:1376–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb DW, Edenberg HJ, Bosron WF, Li TK (1989) Genotypes for aldehyde dehydrogenase deficiency and alcohol sensitivity. The inactive ALDH2(2) allele is dominant. J Clin Invest 83:314–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day FR, Ong KK, Perry JRB (2018) Elucidating the genetic basis of social interaction and isolation. Nature communications 9:2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen K, Baraona E, Ishibashi H, Pozzato G, Moretti M, Matsunaga C, Fujimoto K, Lieber CS (1996) Ethnic differences in gastric s-alcohol dehydrogenase activity and ethanol first-pass metabolism. Alcohol.Clin.Exp.Res. 20:1569–1576. [DOI] [PubMed] [Google Scholar]
- Drogan D, Sheldrick AJ, Schutze M, Knuppel S, Andersohn F, di Giuseppe R, Herrmann B, Willich SN, Garbe E, Bergmann MM, Boeing H, Weikert C (2012) Alcohol consumption, genetic variants in alcohol deydrogenases, and risk of cardiovascular diseases: a prospective study and meta-analysis. PLoS One 7:e32176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duester G, Farres J, Felder MR, Holmes RS, Hoog JO, Pares X, Plapp BV, Yin SJ, Jornvall H (1999) Recommended nomenclature for the vertebrate alcohol dehydrogenase gene family. Biochem Pharmacol 58:389–395. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ (2007) The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health 30:5–13. [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ (2012) Genes contributing to the development of alcoholism: an overview. Alcohol research : current reviews 34:336–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Bosron WF (2018) Alcohol Dehydrogenases, in Comprehensive Toxicology, Third Edition, Vol. 10, Comprehensive Toxicology, Third Edition. (MCQUEEN CA: ed, pp 126–145, Oxford:Elsevier Ltd. [Google Scholar]
- Edenberg HJ, Foroud T (2013) Genetics and alcoholism. Nature reviews. Gastroenterology & hepatology 10:487–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Jerome RE, Li M (1999) Polymorphism of the human alcohol dehydrogenase 4 (ADH4) promoter affects gene expression. Pharmacogenetics 9:25–30. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Xuei X, Chen HJ, Tian H, Wetherill LF, Dick DM, Almasy L, Bierut L, Bucholz KK, Goate A, Hesselbrock V, Kuperman S, Nurnberger J, Porjesz B, Rice J, Schuckit M, Tischfield J, Begleiter H, Foroud T (2006) Association of alcohol dehydrogenase genes with alcohol dependence: a comprehensive analysis. Hum Mol Genet 15:1539–1549. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Liang T, Gizer IR (2012) ADH and ALDH polymorphisms and alcohol dependence in Mexican and Native Americans. Am J Drug Alcohol Abuse 38:389–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Montane-Jaime K, Moore S, Shafe S, Joseph R, Carr LG (2007) Association of the ADHIB*3 allele with alcohol-related phenotypes in Trinidad. Alcohol Clin Exp Res 31:216–220. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Spence JP, Wall TL, Gilder DA, Carr LG (2004) Association of ALDH1 promoter polymorphisms with alcohol-related phenotypes in southwest California Indians. Alcohol Clin Exp Res 28:1481–1486. [DOI] [PubMed] [Google Scholar]
- Frank J, Cichon S, Treutlein J, Ridinger M, Mattheisen M, Hoffmann P, Herms S, Wodarz N, Soyka M, Zill P, Maier W, Mossner R, Gaebel W, Dahmen N, Scherbaum N, Schmal C, Steffens M, Lucae S, Ising M, Muller-Myhsok B, Nothen MM, Mann K, Kiefer F, Rietschel M (2012) Genome-wide significant association between alcohol dependence and a variant in the ADH gene cluster. Addict Biol 17:171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, Anton R, Preuss UW, Ridinger M, Rujescu D, Wodarz N, Zill P, Zhao H, Farrer LA (2014) Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Mol Psychiatry 19:41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Zhou H, Nunez YZ, Mutirangura A, Malison RT, Kalayasiri R (2018) Genomewide Association Study of Alcohol Dependence and Related Traits in a Thai Population. Alcohol Clin Exp Res 42:861–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gizer IR, Edenberg HJ, Gilder DA, Wilhelmsen KC, Ehlers CL (2011) Association of alcohol dehydrogenase genes with alcohol-related phenotypes in a Native American community sample. Alcohol Clin Exp Res 35:2008–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedde HW, Agarwal DP, Fritze G, Meier-Tackmann D, Singh S, Beckmann G, Bhatia K, Chen LZ, Fang B, Lisker R, et al. (1992) Distribution of ADH2 and ALDH2 genotypes in different populations. Hum Genet 88:344–346. [DOI] [PubMed] [Google Scholar]
- Goedde HW, Agarwal DP, Harada S (1983) The role of alcohol dehydrogenase and aldehyde dehydrogenase isozymes in alcohol metabolism, alcohol sensitivity, and alcoholism. Isozymes Curr Top Biol Med Res 8:175–193. [PubMed] [Google Scholar]
- Goedde HW, Harada S, Agarwal DP (1979) Racial differences in alcohol sensitivity: a new hypothesis. Hum Genet 51:331–334. [DOI] [PubMed] [Google Scholar]
- Consortium GTEx (2013) The Genotype-Tissue Expression (GTEx) project. Nat Genet 45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindalini C, Scivoletto S, Ferreira RG, Breen G, Zilberman M, Peluso MA, Zatz M (2005) Association of genetic variants in alcohol dehydrogenase 4 with alcohol dependence in Brazilian patients. Am J Psychiatry 162:1005–1007. [DOI] [PubMed] [Google Scholar]
- Harada S, Agarwal DP, Goedde HW (1981) Aldehyde dehydrogenase deficiency as cause of facial flushing reaction to alcohol in Japanese. Lancet 2:982. [DOI] [PubMed] [Google Scholar]
- Harada S, Agarwal DP, Goedde HW, Tagaki S, Ishikawa B (1982) Possible protective role against alcoholism for aldehyde dehydrogenase isozyme deficiency in Japan [letter]. Lancet 2:827. [DOI] [PubMed] [Google Scholar]
- Harada S, Agarwal DP, Goedde HW, Takagi S (1983) Blood ethanol and acetaldehyde levels in Japanese alcoholics and controls. Pharmacol Biochem Behav 18 Suppl 1:139–140. [DOI] [PubMed] [Google Scholar]
- Harada S, Okubo T, Nakamura T, Fujii C, Nomura F, Higuchi S, Tsutsumi M (1999) A novel polymorphism (−357G/A) of the ALDH2 gene: linkage disequilibrium and an association with alcoholism. Alcohol.Clin.Exp.Res. 23:958–962. [PubMed] [Google Scholar]
- Hart AB, Kranzler HR (2015) Alcohol Dependence Genetics: Lessons Learned From Genome-Wide Association Studies (GWAS) and Post-GWAS Analyses. Alcohol Clin Exp Res 39:1312–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AB, Lynch KG, Farrer L, Gelernter J, Kranzler HR (2016) Which alcohol use disorder criteria contribute to the association of ADH1B with alcohol dependence? Addict Biol 21:924–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haschemi Nassab M, Rhein M, Heese P, Glahn A, Frieling H, Linnebank M, Bleich S, Kornhuber J, Heberlein A, Grallert H, Peters A, Rawal R, Strauch K, Hillemacher T (2015) No association between the ALDH2 promoter polymorphism rs886205, alcohol dependence, and risky alcohol consumption in a German population. Psychiatr Genet 25:41–42. [DOI] [PubMed] [Google Scholar]
- Haseba T, Ohno Y (2010) A new view of alcohol metabolism and alcoholism--role of the high-Km Class III alcohol dehydrogenase (ADH3). Int J Environ Res Public Health 7:1076–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin D, Aharonovich E, Liu X, Mamman Z, Matseoane K, Carr And LG, Li TK (2002) Alcohol dependence symptoms and alcohol dehydrogenase 2 polymorphism: Israeli Ashkenazis, Sephardics, and recent Russian immigrants. Alcohol Clin Exp Res 26:1315–1321. [DOI] [PubMed] [Google Scholar]
- Higuchi S, Matsushita S, Imazeki H, Kinoshita T, Takagi S, Kono H (1994) Aldehyde dehydrogenase genotypes in Japanese alcoholics. Lancet 343:741–742. [DOI] [PubMed] [Google Scholar]
- Hurley TD, Edenberg HJ (2012) Genes encoding enzymes involved in ethanol metabolism. Alcohol research : current reviews 34:339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley TD, Edenberg HJ, Li T-K (2002) The Pharmacogenomics of alcoholism, in Pharmacogenomics: The Search for Individualized Therapies, Pharmacogenomics: The Search for Individualized Therapies, pp 417–441, Wiley-VCH. [Google Scholar]
- Husemoen LL, Fenger M, Friedrich N, Tolstrup JS, Beenfeldt Fredriksen S, Linneberg A (2008) The association of ADH and ALDH gene variants with alcohol drinking habits and cardiovascular disease risk factors. Alcoholism, clinical and experimental research 32:1984–1991. [DOI] [PubMed] [Google Scholar]
- Jackson B, Brocker C, Thompson DC, Black W, Vasiliou K, Nebert DW, Vasiliou V (2011) Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Hum Genomics 5:283–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson BC, Reigan P, Miller B, Thompson DC, Vasiliou V (2015) Human ALDH1B1 polymorphisms may affect the metabolism of acetaldehyde and all-trans retinaldehyde--in vitro studies and computational modeling. Pharm Res 32:1648–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jairam S, Edenberg HJ (2014a) An enhancer-blocking element regulates the cell-specific expression of alcohol dehydrogenase 7. Gene 547:239–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jairam S, Edenberg HJ (2014b) Single-nucleotide polymorphisms interact to affect ADH7 transcription. Alcohol Clin Exp Res 38:921–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgenson E, Thai KK, Hoffmann TJ, Sakoda LC, Kvale MN, Banda Y, Schaefer C, Risch N, Mertens J, Weisner C, Choquet H (2017) Genetic contributors to variation in alcohol consumption vary by race/ethnicity in a large multi-ethnic genome-wide association study. Mol Psychiatry 22:1359–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Kalsi G, Holmans PA, Sanders AR, Aggen SH, Dick DM, Aliev F, Shi J, Levinson DF, Gejman PV (2011) Genomewide Association Analysis of Symptoms of Alcohol Dependence in the Molecular Genetics of Schizophrenia (MGS2) Control Sample. Alcoholism, clinical and experimental research 35:963–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Kimura S, Matsushita S, Kashima H, Higuchi S (2006) ALDH2 promoter polymorphism has no effect on the risk for alcoholism. Alcohol Alcohol 41:368–371. [DOI] [PubMed] [Google Scholar]
- Klyosov AA (1996) Kinetics and specificity of human liver aldehyde dehydrogenases toward aliphatic, aromatic, and fused polycyclic aldehydes. Biochemistry 35:4457–4467. [DOI] [PubMed] [Google Scholar]
- Kuo PH, Kalsi G, Prescott CA, Hodgkinson CA, Goldman D, van den Oord EJ, Alexander J, Jiang C, Sullivan PF, Patterson DG, Walsh D, Kendler KS, Riley BP (2008) Association of ADH and ALDH genes with alcohol dependence in the Irish Affected Sib Pair Study of alcohol dependence (IASPSAD) sample. Alcoholism, clinical and experimental research 32:785–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latella MC, Di Castelnuovo A, de Lorgeril M, Arnout J, Cappuccio FP, Krogh V, Siani A, van Dongen M, Donati MB, de Gaetano G, Iacoviello L, European Collaborative Group of the IP (2009) Genetic variation of alcohol dehydrogenase type 1C (ADH1C), alcohol consumption, and metabolic cardiovascular risk factors: results from the IMMIDIET study. Atherosclerosis 207:284–290. [DOI] [PubMed] [Google Scholar]
- Lee SL, Hoog JO, Yin SJ (2004) Functionality of allelic variations in human alcohol dehydrogenase gene family: assessment of a functional window for protection against alcoholism. Pharmacogenetics 14:725–732. [DOI] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG, Exome Aggregation C (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhao H, Gelernter J (2011) Strong association of the alcohol dehydrogenase 1B gene (ADH1B) with alcohol dependence and alcohol-induced medical diseases. Biol Psychiatry 70:504–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhao H, Gelernter J (2012a) Further clarification of the contribution of the ADH1C gene to vulnerability of alcoholism and selected liver diseases. Hum Genet 131:1361–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhao H, Gelernter J (2012b) Strong protective effect of the aldehyde dehydrogenase gene (ALDH2) 504lys (*2) allele against alcoholism and alcohol-induced medical diseases in Asians. Hum Genet 131:725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Mukherjee N, Soundararajan U, Tarnok Z, Barta C, Khaliq S, Mohyuddin A, Kajuna SL, Mehdi SQ, Kidd JR, Kidd KK (2007) Geographically separate increases in the frequency of the derived ADH1B*47His allele in eastern and western Asia. Am J Hum Genet 81:842–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PA, Eriksson CJ, Wilhelmsen KC (2008) The role of aldehyde dehydrogenase-1 (ALDH1A1) polymorphisms in harmful alcohol consumption in a Finnish population. Hum Genomics 3:24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linneberg A, Gonzalez-Quintela A, Vidal C, Jorgensen T, Fenger M, Hansen T, Pedersen O, Husemoen LL (2010) Genetic determinants of both ethanol and acetaldehyde metabolism influence alcohol hypersensitivity and drinking behaviour among Scandinavians. Clin Exp Allergy 40:123–130. [DOI] [PubMed] [Google Scholar]
- Luczak SE, Glatt SJ, Wall TL (2006) Meta-analyses of ALDH2 and ADH1B with alcohol dependence in Asians. Psychol Bull 132:607–621. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Lappalainen J, Yang BZ, Gelernter J (2006a) ADH4 gene variation is associated with alcohol dependence and drug dependence in European Americans: results from HWD tests and case-control association studies. Neuropsychopharmacology 31:1085–1095. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Wang S, Schork NJ, Gelernter J (2006b) Diplotype trend regression analysis of the ADH gene cluster and the ALDH2 gene: multiple significant associations with alcohol dependence. Am J Hum Genet 78:973–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor S, Lind PA, Bucholz KK, Hansell NK, Madden PA, Richter MM, Montgomery GW, Martin NG, Heath AC, Whitfield JB (2009) Associations of ADH and ALDH2 gene variation with self report alcohol reactions, consumption and dependence: an integrated analysis. Human molecular genetics 18:580–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo K, Hiraki A, Hirose K, Ito H, Suzuki T, Wakai K, Tajima K (2007) Impact of the alcohol-dehydrogenase (ADH) 1C and ADH1B polymorphisms on drinking behavior in nonalcoholic Japanese. Hum Mutat 28:506–510. [DOI] [PubMed] [Google Scholar]
- Meyers JL, Shmulewitz D, Wall MM, Keyes KM, Aharonovich E, Spivak B, Weizman A, Frisch A, Edenberg HJ, Gelernter J, Grant BF, Hasin D (2015) Childhood adversity moderates the effect of ADH1B on risk for alcohol-related phenotypes in Jewish Israeli drinkers. Addict Biol 20:205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoi Y, Tatsuno Y, Adachi J, Kogame M, Fukunaga T, Fujiwara S, Hishida S, Ijiri I (1983) Alcohol sensitivity related to polymorphism of alcohol-metabolizing enzymes in Japanese. Pharmacol Biochem Behav 18 Suppl 1:127–133. [DOI] [PubMed] [Google Scholar]
- Mizoi Y, Yamamoto K, Ueno Y, Fukunaga T, Harada S (1994) Involvement of genetic polymorphism of alcohol and aldehyde dehydrogenases in individual variation of alcohol metabolism. Alcohol Alcohol 29:707–710. [PubMed] [Google Scholar]
- Moore S, Montane-Jaime K, Shafe S, Joseph R, Crooks H, Carr LG, Ehlers CL (2007) Association of ALDH1 promoter polymorphisms with alcohol-related phenotypes in Trinidad and Tobago. J Stud Alcohol Drugs 68:192–196. [DOI] [PubMed] [Google Scholar]
- Munoz X, Amiano P, Celorrio D, Dorronsoro M, Sanchez MJ, Huerta JM, Barricarte A, Arriola L, Navarro C, Molina-Montes E, Chirlaque MD, Ardanaz E, Rodriguez L, Duell EJ, Hijona E, Herreros-Villanueva M, Sala N, Bujanda L (2012) Association of alcohol dehydrogenase polymorphisms and life-style factors with excessive alcohol intake within the Spanish population (EPIC-Spain). Addiction 107:2117–2127. [DOI] [PubMed] [Google Scholar]
- Neumark YD, Friedlander Y, Durst R, Leitersdorf E, Jaffe D, Ramchandani VA, O’Connor S, Carr LG, Li TK (2004) Alcohol dehydrogenase polymorphisms influence alcohol-elimination rates in a male Jewish population. Alcohol Clin Exp Res 28:10–14. [DOI] [PubMed] [Google Scholar]
- Neumark YD, Friedlander Y, Thomasson HR, Li TK (1998) Association of the ADH2*2 allele with reduced ethanol consumption in Jewish men in Israel: a pilot study. J Stud Alcohol 59:133–139. [DOI] [PubMed] [Google Scholar]
- Norden-Krichmar TM, Gizer IR, Wilhelmsen KC, Schork NJ, Ehlers CL (2014) Protective variant associated with alcohol dependence in a Mexican American cohort. BMC Med Genet 15:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuutinen HU, Salaspuro MP, Valle M, Lindros KO (1984) Blood acetaldehyde concentration gradient between hepatic and antecubital venous blood in ethanol-intoxicated alcoholics and controls. Eur J Clin Invest 14:306–311. [DOI] [PubMed] [Google Scholar]
- Olfson E, Edenberg HJ, Nurnberger J Jr., Agrawal A, Bucholz KK, Almasy LA, Chorlian D, Dick DM, Hesselbrock VM, Kramer JR, Kuperman S, Porjesz B, Schuckit MA, Tischfield JA, Wang JC, Wetherill L, Foroud TM, Rice J, Goate A, Bierut LJ (2014) An ADH1B variant and peer drinking in progression to adolescent drinking milestones: evidence of a gene-by-environment interaction. Alcohol Clin Exp Res 38:2541–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osier M, Pakstis AJ, Kidd JR, Lee JF, Yin SJ, Ko HC, Edenberg HJ, Lu RB, Kidd KK (1999) Linkage disequilibrium at the ADH2 and ADH3 loci and risk of alcoholism. Am J Hum Genet 64:1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostberg LJ, Persson B, Hoog JO (2016) Computational studies of human class V alcohol dehydrogenase - the odd sibling. BMC Biochem 17:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto JM, Hendershot CS, Collins SE, Liang T, Wall TL (2013) Association of the ALDH1A1*2 promoter polymorphism with alcohol phenotypes in young adults with or without ALDH2*2. Alcohol Clin Exp Res 37:164–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BL, Kim JW, Cheong HS, Kim LH, Lee BC, Seo CH, Kang TC, Nam YW, Kim GB, Shin HD, Choi IG (2013) Extended genetic effects of ADH cluster genes on the risk of alcohol dependence: from GWAS to replication. Hum Genet 132:657–668. [DOI] [PubMed] [Google Scholar]
- Peng GS, Chen YC, Wang MF, Lai CL, Yin SJ (2014a) ALDH2*2 but not ADH1B*2 is a causative variant gene allele for Asian alcohol flushing after a low-dose challenge: correlation of the pharmacokinetic and pharmacodynamic findings. Pharmacogenet Genomics 24:607–617. [DOI] [PubMed] [Google Scholar]
- Peng Q, Gizer IR, Libiger O, Bizon C, Wilhelmsen KC, Schork NJ, Ehlers CL (2014b) Association and ancestry analysis of sequence variants in ADH and ALDH using alcohol-related phenotypes in a Native American community sample. Am J Med Genet B Neuropsychiatr Genet 165B:673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Q, Gizer IR, Wilhelmsen KC, Ehlers CL (2017) Associations Between Genomic Variants in Alcohol Dehydrogenase Genes and Alcohol Symptomatology in American Indians and European Americans: Distinctions and Convergence. Alcohol Clin Exp Res 41:1695–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochareddy S, Edenberg HJ (2010) Identification of a FOXA-dependent enhancer of human alcohol dehydrogenase 4 (ADH4). Gene 460:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochareddy S, Edenberg HJ (2011) Variation in the ADH1B proximal promoter affects expression. Chem Biol Interact 191:38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polimanti R, Gelernter J (2018) ADH1B: From alcoholism, natural selection, and cancer to the human phenome. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics 177:113–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillen EE, Chen XD, Almasy L, Yang F, He H, Li X, Wang XY, Liu TQ, Hao W, Deng HW, Kranzler HR, Gelernter J (2014) ALDH2 is associated to alcohol dependence and is the major genetic determinant of “daily maximum drinks” in a GWAS study of an isolated rural Chinese sample. Am J Med Genet B Neuropsychiatr Genet 165B:103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajeevan H, Soundararajan U, Kidd JR, Pakstis AJ, Kidd KK (2012) ALFRED: an allele frequency resource for research and teaching. Nucleic Acids Research 40:D1010–D1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietschel M, Treutlein J (2013) The genetics of alcohol dependence. Ann N Y Acad Sci 1282:39–70. [DOI] [PubMed] [Google Scholar]
- Sanchez-Roige S, Fontanillas P, Elson SL, andMe Research T, Gray JC, de Wit H, Davis LK, MacKillop J, Palmer AA (2017) Genome-wide association study of alcohol use disorder identification test (AUDIT) scores in 20 328 research participants of European ancestry. Addict Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Roige S, Palmer AA, Fontanillas P, Elson SL, 23andMe Research Team, Substance Use Disorder Working Group of the Psychiatric Genetics Consortium, Adams MJ, Howard DM, Edenberg HJ, Davies G, Crist RC, Deary IJ, McIntosh A, Clarke TK (2018) Genome-wide association study meta-analysis of the Alcohol Use Disorder Identification Test (AUDIT) in two population-based cohorts (N=141,958). bioRxiv 275917. [Google Scholar]
- Schumann G, Coin LJ, Lourdusamy A, Charoen P, Berger KH, Stacey D, Desrivieres S, Aliev FA, Khan AA, Amin N, Aulchenko YS, Bakalkin G, Bakker SJ, Balkau B, Beulens JW, Bilbao A, de Boer RA, Beury D, Bots ML, Breetvelt EJ, Cauchi S, Cavalcanti-Proenca C, Chambers JC, Clarke TK, Dahmen N, de Geus EJ, Dick D, Ducci F, Easton A, Edenberg HJ, Esko T, Fernandez-Medarde A, Foroud T, Freimer NB, Girault JA, Grobbee DE, Guarrera S, Gudbjartsson DF, Hartikainen AL, Heath AC, Hesselbrock V, Hofman A, Hottenga JJ, Isohanni MK, Kaprio J, Khaw KT, Kuehnel B, Laitinen J, Lobbens S, Luan J, Mangino M, Maroteaux M, Matullo G, McCarthy MI, Mueller C, Navis G, Numans ME, Nunez A, Nyholt DR, Onland-Moret CN, Oostra BA, O’Reilly PF, Palkovits M, Penninx BW, Polidoro S, Pouta A, Prokopenko I, Ricceri F, Santos E, Smit JH, Soranzo N, Song K, Sovio U, Stumvoll M, Surakk I, Thorgeirsson TE, Thorsteinsdottir U, Troakes C, Tyrfingsson T, Tonjes A, Uiterwaal CS, Uitterlinden AG, van der Harst P, van der Schouw YT, Staehlin O, Vogelzangs N, Vollenweider P, Waeber G, Wareham NJ, Waterworth DM, Whitfield JB, Wichmann EH, Willemsen G, Witteman JC, Yuan X, Zhai G, Zhao JH, Zhang W, Martin NG, Metspalu A, Doering A, Scott J, Spector TD, Loos RJ, Boomsma DI, Mooser V, Peltonen L, Stefansson K, van Duijn CM, Vineis P, Sommer WH, Kooner JS, Spanagel R, Heberlein UA, Jarvelin MR, Elliott P (2011) Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc Natl Acad Sci U S A 108:7119–7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann G, Liu C, O’Reilly P, Gao H, Song P, Xu B, Ruggeri B, Amin N, Jia T, Preis S, Segura Lepe M, Akira S, Barbieri C, Baumeister S, Cauchi S, Clarke T-K, Enroth S, Fischer K, Hällfors J, Harris SE, Hieber S, Hofer E, Hottenga J-J, Johansson Å, Joshi PK, Kaartinen N, Laitinen J, Lemaitre R, Loukola A, Luan Ja, Lyytikäinen L-P, Mangino M, Manichaikul A, Mbarek H, Milaneschi Y, Moayyeri A, Mukamal K, Nelson C, Nettleton J, Partinen E, Rawal R, Robino A, Rose L, Sala C, Satoh T, Schmidt R, Schraut K, Scott R, Smith AV, Starr JM, Teumer A, Trompet S, Uitterlinden AG, Venturini C, Vergnaud A-C, Verweij N, Vitart V, Vuckovic D, Wedenoja J, Yengo L, Yu B, Zhang W, Zhao JH, Boomsma DI, Chambers J, Chasman DI, Daniela T, de Geus E, Deary I, Eriksson JG, Esko T, Eulenburg V, Franco OH, Froguel P, Gieger C, Grabe HJ, Gudnason V, Gyllensten U, Harris TB, Hartikainen A-L, Heath AC, Hocking L, Hofman A, Huth C, Jarvelin M-R, Jukema JW, Kaprio J, Kooner JS, Kutalik Z, Lahti J, Langenberg C, Lehtimäki T, Liu Y, Madden PAF, Martin N, Morrison A, Penninx B, Pirastu N, Psaty B, Raitakari O, Ridker P, Rose R, Rotter JI, Samani NJ, Schmidt H, Spector TD, Stott D, Strachan D, Tzoulaki I, van der Harst P, van Duijn CM, Marques-Vidal P, Vollenweider P, Wareham NJ, Whitfield JB, Wilson J, Wolffenbuttel B, Bakalkin G, Evangelou E, Liu Y, Rice KM, Desrivières S, Kliewer SA, Mangelsdorf DJ, Müller CP, Levy D, Elliott P (2016) KLB is associated with alcohol drinking, and its gene product β-Klotho is necessary for FGF21 regulation of alcohol preference. Proceedings of the National Academy of Sciences 113:14372–14377 PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DM, Taylor RE (2007) Health-related effects of genetic variations of alcohol-metabolizing enzymes in African Americans. Alcohol Res Health 30:18–21. [PMC free article] [PubMed] [Google Scholar]
- Shen YC, Fan JH, Edenberg HJ, Li TK, Cui YH, Wang YF, Tian CH, Zhou CF, Zhou RL, Wang J, Zhao ZL, Xia GY (1997) Polymorphism of ADH and ALDH genes among four ethnic groups in China and effects upon the risk for alcoholism. Alcohol Clin Exp Res 21:1272–1277. [PubMed] [Google Scholar]
- Sherva R, Rice JP, Neuman RJ, Rochberg N, Saccone NL, Bierut LJ (2009) Associations and interactions between SNPs in the alcohol metabolizing genes and alcoholism phenotypes in European Americans. Alcohol Clin Exp Res 33:848–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya A, Yasunami M, Yoshida A (1989) Genotype of alcohol dehydrogenase and aldehyde dehydrogenase loci in Japanese alcohol flushers and nonflushers. Hum.Genet. 82:14–16. [DOI] [PubMed] [Google Scholar]
- Singh S, Chen Y, Matsumoto A, Orlicky DJ, Dong H, Thompson DC, Vasiliou V (2015) ALDH1B1 links alcohol consumption and diabetes. Biochem Biophys Res Commun 463:768–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M, Hopkinson DA, Harris H (1971) Developmental changes and polymorphism in human alcohol dehydrogenase. Ann Hum Genet 34:251–271. [DOI] [PubMed] [Google Scholar]
- Spence JP, Liang T, Eriksson CJ, Taylor RE, Wall TL, Ehlers CL, Carr LG (2003) Evaluation of aldehyde dehydrogenase 1 promoter polymorphisms identified in human populations. Alcohol Clin Exp Res 27:1389–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagos D, Chen Y, Brocker C, Donald E, Jackson BC, Orlicky DJ, Thompson DC, Vasiliou V (2010) Aldehyde dehydrogenase 1B1: molecular cloning and characterization of a novel mitochondrial acetaldehyde-metabolizing enzyme. Drug Metab Dispos 38:1679–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart MJ, Malek K, Xiao Q, Dipple KM, Crabb DW (1995) The novel aldehyde dehydrogenase gene, ALDH5, encodes an active aldehyde dehydrogenase enzyme. Biochem Biophys Res Commun 211:144–151. [DOI] [PubMed] [Google Scholar]
- Stromberg P, Svensson S, Hedberg JJ, Nordling E, Hoog JO (2002) Identification and characterisation of two allelic forms of human alcohol dehydrogenase 2. Cell Mol Life Sci 59:552–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita T, Mao XQ, Morimoto K (1996) The contribution of polymorphism in the alcohol dehydrogenase beta subunit to alcohol sensitivity in a Japanese population. Hum.Genet. 97:409–413. [DOI] [PubMed] [Google Scholar]
- Takeuchi F, Isono M, Nabika T, Katsuya T, Sugiyama T, Yamaguchi S, Kobayashi S, Ogihara T, Yamori Y, Fujioka A, Kato N (2011) Confirmation of ALDH2 as a Major locus of drinking behavior and of its variants regulating multiple metabolic phenotypes in a Japanese population. Circ J 75:911–918. [DOI] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR (2015) A global reference for human genetic variation. Nature 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomasson HR, Beard JD, Li T-K (1995) ADH2 gene polymorphisms are determinants of alcohol pharmacokinetics. Alcohol.Clin.Exp.Res. 19:1494–1499. [DOI] [PubMed] [Google Scholar]
- Thomasson HR, Crabb DW, Edenberg HJ, Li TK, Hwu HG, Chen CC, Yeh EK, Yin SJ (1994) Low frequency of the ADH2*2 allele among Atayal natives of Taiwan with alcohol use disorders. Alcohol Clin Exp Res 18:640–643. [DOI] [PubMed] [Google Scholar]
- Thomasson HR, Edenberg HJ, Crabb DW, Mai XL, Jerome RE, Li TK, Wang SP, Lin YT, Lu RB, Yin SJ (1991) Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am J Hum Genet 48:677–681. [PMC free article] [PubMed] [Google Scholar]
- Tolstrup JS, Nordestgaard BG, Rasmussen S, Tybjaerg-Hansen A, Gronbaek M (2008) Alcoholism and alcohol drinking habits predicted from alcohol dehydrogenase genes. Pharmacogenomics J 8:220–227. [DOI] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N, Fehr C, Scherbaum N, Steffens M, Ludwig KU, Frank J, Wichmann HE, Schreiber S, Dragano N, Sommer WH, Leonardi-Essmann F, Lourdusamy A, Gebicke-Haerter P, Wienker TF, Sullivan PF, Nothen MM, Kiefer F, Spanagel R, Mann K, Rietschel M (2009) Genome-wide association study of alcohol dependence. Arch Gen Psychiatry 66:773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein J, Frank J, Kiefer F, Rietschel M (2014) ADH1B Arg48His allele frequency map: filling in the gap for Central Europe. Biol Psychiatry 75:e15. [DOI] [PubMed] [Google Scholar]
- Vasiliou V, Pappa A, Estey T (2004) Role of human aldehyde dehydrogenases in endobiotic and xenobiotic metabolism. Drug Metab Rev 36:279–299. [DOI] [PubMed] [Google Scholar]
- Vasiliou V, Thompson DC, Smith C, Fujita M, Chen Y (2013) Aldehyde dehydrogenases: from eye crystallins to metabolic disease and cancer stem cells. Chem Biol Interact 202:2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall TL, Carr LG, Ehlers CL (2003) Protective association of genetic variation in alcohol dehydrogenase with alcohol dependence in Native American Mission Indians. Am J Psychiatry 160:41–46. [DOI] [PubMed] [Google Scholar]
- Walters RK, Adams MJ, Adkins AE, Aliev F, Bacanu S-A, Batzler A, Bertelsen S, Biernacka J, Bigdeli TB, Chen L-S, Clarke T-K, Chou Y-L, Degenhardt F, Docherty AR, Fontanillas P, Foo J, Fox L, Frank J, Giegling I, Gordon S, Hack L, Hartz SM, Heilmann-Heimbach S, Herms S, Hodgkinson C, Hoffmann P, Hottenga J-J, Kennedy MA, Alanne-Kinnunen M, Konte B, Lahti J, Lahti-Pulkkinen M, Ligthart L, Loukola A-M, Maher BS, Mbarek H, McIntosh AM, McQueen MB, Milaneschi Y, Palviainen T, Pearson JF, Peterson RE, Polimanti R, Ripatti S, Ryu E, Saccone NL, Salvatore JE, Sanchez-Roige S, Schwandt M, Sherva R, Streit F, Strohmaier J, Thomas N, Wang J-C, Webb BT, Wedow R, Wetherill L, Wills AG, Boardman JD, Chen D, Choi D-S, Copeland WE, Culverhouse RC, Dahmen N, Degenhardt L, Domingue BW, Elson SL, Frye M, Gäbel W, Ising M, Johnson EC, Keyes M, Kiefer F, Kramer J, Kuperman S, Lucae S, Lynskey MT, Maier W, Mann K, Männistö S, McClintick JN, Meyers JL, Müller-Myhsok B, Nurnberger JI, Palotie A, Preuss U, Räikkönen K, Reynolds MD, Ridinger M, Scherbaum N, Shuckit M, Soyka M, Treutlein J, Witt S, Wodarz N, Zill P, Adkins DE, Boden JM, Boomsma D, Bierut LJ, Brown SA, Bucholz KK, Cichon S, Costello EJ, de Wit H, Diazgranados N, Dick DM, Eriksson JG, Farrer LA, Foroud TM, Gillespie NA, Goate AA, Goldman D, Grucza RA, Hancock DB, Harris KM, Heath AC, Hesselbrock V, Hewitt JK, Hopfer C, Horwood J, Iacono W, Johnson EO, Kaprio JA, Karpyak V, Kendler KS, Kranzler HR, Krauter K, Lichtenstein P, Lind PA, McGue M, MacKillop J, Madden PAF, Maes H, Magnusson P, Martin NG, Medland SE, Montgomery GW, Nelson EC, Nöthen M, Palmer AA, Pedersen NL, Penninx BWJH, Porjesz B, Rice JP, Rietschel M, Riley BP, Rose R, Rujescu D, Shen P-H, Silberg J, Stallings MC, Tarter RE, Vanyukov MM, Vrieze S, Wall TL, Whitfield JB, Zhao H, Neale BM, Gelernter J, Edenberg HJ, Agrawal A(2018) Trans-ancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. bioRxiv 257311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RJ, McPherson AJ, Chow C, Ealing J, Sherman DI, Yoshida A, Peters TJ (1994) Identification and characterisation of alcohol-induced flushing in Caucasian subjects. Alcohol Alcohol 29:433–438. [PubMed] [Google Scholar]
- Warren KR, Li TK (2005) Genetic polymorphisms: impact on the risk of fetal alcohol spectrum disorders. Birth Defects Res A Clin Mol Teratol 73:195–203. [DOI] [PubMed] [Google Scholar]
- Way M, McQuillin A, Saini J, Ruparelia K, Lydall GJ, Guerrini I, Ball D, Smith I, Quadri G, Thomson AD, Kasiakogia-Worlley K, Cherian R, Gunwardena P, Rao H, Kottalgi G, Patel S, Hillman A, Douglas E, Qureshi SY, Reynolds G, Jauhar S, O’Kane A, Dedman A, Sharp S, Kandaswamy R, Dar K, Curtis D, Morgan MY, Gurling HM (2015) Genetic variants in or near ADH1B and ADH1C affect susceptibility to alcohol dependence in a British and Irish population. Addict Biol 20:594–604. [DOI] [PubMed] [Google Scholar]
- Way MJ, Ali MA, McQuillin A, Morgan MY (2017) Genetic variants in ALDH1B1 and alcohol dependence risk in a British and Irish population: A bioinformatic and genetic study. PLoS One 12:e0177009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield JB (1997) Meta-analysis of the effects of alcohol dehydrogenase genotype on alcohol dependence and alcoholic liver disease. Alcohol.Alcohol. 32:613–619. [DOI] [PubMed] [Google Scholar]
- Whitfield JB (2002) Alcohol dehydrogenase and alcohol dependence: variation in genotype-associated risk between populations. Am J Hum Genet 71:1247–1250; author reply 1250–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q, Weiner H, Crabb DW (1996) The mutation in the mitochondrial aldehyde dehydrogenase (ALDH2) gene responsible for alcohol-induced flushing increases turnover of the enzyme tetramers in a dominant fashion. J Clin Invest 98:2027–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Kranzler HR, Sherva R, Sartor CE, Almasy L, Koesterer R, Zhao H, Farrer LA, Gelernter J (2015) Genomewide Association Study for Maximum Number of Alcoholic Drinks in European Americans and African Americans. Alcohol Clin Exp Res 39:1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YL, Carr LG, Bosron WF, Li TK, Edenberg HJ (1988) Genotyping of human alcohol dehydrogenases at the ADH2 and ADH3 loci following DNA sequence amplification. Genomics 2:209–214. [DOI] [PubMed] [Google Scholar]
- Yoshida A, Dave V, Ward RJ, Peters TJ (1989) Cytosolic aldehyde dehydrogenase (ALDH1) variants found in alcohol flushers. Ann.Hum.Genet. 53:1–7. [DOI] [PubMed] [Google Scholar]
- Zakhari S (2006) Overview: how is alcohol metabolized by the body? Alcohol Res Health 29:245–254. [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Weiner H (2000) Basis for half-of-the-site reactivity and the dominance of the K487 oriental subunit over the E487 subunit in heterotetrameric human liver mitochondrial aldehyde dehydrogenase. Biochemistry 39:12019–12024. [DOI] [PubMed] [Google Scholar]
- Zintzaras E, Stefanidis I, Santos M, Vidal F (2006) Do alcohol-metabolizing enzyme gene polymorphisms increase the risk of alcoholism and alcoholic liver disease? Hepatology 43:352–361. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.