Abstract
Background
Plasmodium falciparum and Plasmodium vivax are two major parasites responsible for malaria which remains a threat to almost 50% of world's population despite decade-long eradication program. One possible reason behind this conundrum is that the bases of clinical variability in malaria caused by either species are complex and poorly understood.
Methods
Whole-genome transcriptome was analyzed to identify the active and predominant pathways in the PBMC of P. falciparum and P. vivax infected malaria patients. Deregulated genes were identified and annotated using R Bioconductor and DAVID/KEGG respectively. Genetic and functional regulation of CD14, a prioritized candidate, were established by quantitative RT-PCR, genotyping using RFLP and resequencing, mapping of transcription factor binding using CONSITE and TFBIND, dual luciferase assay, western blot analysis, RNAi- mediated gene knockdown and chromatin-immunoprecipation.
Findings
The study highlighted that deregulation of host immune and inflammatory genes particularly CD14 as a key event in P. falciparum malaria. An abundance of allele-C of rs5744454, located in CD14 promoter, in severe malaria motivated us to establish an allele-specific regulation of CD14 by SP1. An enhancement of SP1 and CD14 expression was observed in artemisinin treated human monocyte cell line.
Interpretation
Our data not only reinstates that CD14 of TLR pathway plays a predominant role in P. falciparum malaria, it establishes a functional basis for genetic association of rs5744454 with P. falciparum severe malaria by demonstrating a cis-regulatory role of this promoter polymorphism. Moreover, the study points towards a novel pharmacogenetic aspect of artemisinin-based anti-malarial therapy.
Fund
DST-SERB, Govt. of India, SR/SO/HS-0056/2013.
Keywords: Malaria, Plasmodium falciparum, Transcriptome, CD14, SP1, Host Polymorphism
Abbreviations: AF, After fever (convalescence sample); ART, Artemisinin; CD14, Cluster of Differentiation 14; ChIP, Chromatin Immuno Precipitation assay; DC, Dendritic Cell; F, P. falciparum infection; GO, Gene Ontology; HWE, Hardy Weinberg Equilibrium; KEGG, Kyoto Encyclopedia of Genes and Genomes; LPS, Lipopolysaccharide; N, Infection naïve US individuals (GSE45919); PBMC, Peripheral Blood Mononuclear Cell; PCR, Polymerase Chain Reaction; qPCR, Quantitative real time PCR; RFLP, Restriction Fragment Length Polymorphism; RBC, Red Blood Cell; SNP, Single Nucleotide Polymorphism; SP1, Specificity Protein 1; TLR, Toll Like Receptor; V, P. vivax infection
Research in context section.
Despite commendable progress in reduction of mortality, malaria caused by different Plasmodia continues to threaten an estimated 216 million people worldwide. India which is co-endemic for P. falciparum and P. vivax accounts for 80% of the reported cases in South-East Asia. Since, host genetic factors are implicated to account for one quarter of the total variability in malaria infection and severity, our laboratory has been engaged in finding out how host genetic variation may account for the inter-individual differences in disease susceptibility and severity. In the present study, a whole genome approach was adopted to identify specific perturbation of gene expression in the peripheral blood mononuclear cells of P. falciparum infected malaria patients during the acute phase of infection using P. vivax infection as an important comparison group. The microarray data was subsequently subjected to appropriate statistical analysis and functional annotation to prioritize the most likely participating factor (CD14) marking the difference of PBMC gene expression between P. falciparum and P. vivax infections. A battery of experiments was conducted to detect the possible molecular cause of the observed transcriptional deregulation and its possible genetic connection. In the post-genome era, application of genome wide association studies has revolutionized identification of genetic basic of complex traits. Establishment of a functional connection of the variants with disease biology, nevertheless, often remains incomplete as majority of the GWAS associated-SNPs are located in the non-coding region of the genome. To circumvent this challenge, a combination of genomic, molecular, genetic and bioinformatic tools and approaches were used in logical progression to identify and characterize one of the key participating principles of host immune regulation in P. falciparum malaria, which often produces severe clinical outcome. In addition, this study also provides a preliminary indication regarding how host genetic constitution may modulate efficacy of artemisinin, a first-line drug in uncomplicated malaria. Although further investigation is necessary to elaborate to what extent CD14 promoter genotype may modulate clinical prognosis of malaria patients treated with artemisinin, the observation made in the present study is clearly noteworthy given the fact that failure to artemisinin treatment has been reported from Southeast Asian countries.
Alt-text: Unlabelled Box
1. Introduction
Despite commendable progress in reduction of mortality due to malaria in the last decade, the disease continues to threaten an estimated 216 million people worldwide causing 4,45,000 deaths in 2016 [1]. Among the different parasites of Plasmodium genus capable of infecting humans, Plasmodium falciparum, the most virulent species which often develops serious neurological and hematological sequeals in infected patients, is particularly predominant in Sub-Saharan Africa. P. vivax, on the other hand, has a wider geographical distribution and contributes to a significant share of disease burden affecting an estimated 8.55 million people in 2016 worldwide [1,2]. India which is co-endemic for P. falciparum and P. vivax accounts for 6% of the global disease burden and 80% of the reported cases in South-East Asia [1]. According to a report by National Vector Borne Disease Control Board, Government of India, P. falciparum accounts for 65% of malaria cases in India although the relative distribution of two species differs across the country due to variable eco-epidemiological factors and transmission intensities [3,4].
Pathogenesis of malaria is complex and influenced by host age, immunology and genetic background, type of infecting species, its genetics and parasitemia [[5], [6], [7]]. In addition to erythropoietic responses, one of the key events affecting the clinical spectra of malaria includes a balance between parasite growth and its clearance by host's adaptive immunity [8]. P. vivax adopts an additional strategy to persist in humans via the reservoir of hypnozoites in the liver [9,10].
Despite intense research, it is still not clear to what extent different parasite species influence clinical presentation and anti-parasite immunity of individuals inhabiting in regions co-endemic for P. falciparum and P. vivax. In areas of low endemicity during uncomplicated disease, both species have been shown to elicit similar cytokine and antibody responses [11,12]. On contrary, species specific induction of different T cell subsets has also been reported [13]. A study even indicates the possibility of P. vivax suppressing P. falciparum parasites in mixed infection through the induction of CD3 + δ2 + T cells and anti-P. falciparum antibodies, while maturation of DC is shown to be skewed in a substantial proportion of P. vivax infection as part of parasite's immune escape strategy [14,15]. Given this controversy and gap in knowledge and anticipating that different parasite species would furnish different tropism for host cells, we investigated the molecular basis of immune responses induced by P. falciparum which is responsible for more severe and debilitating forms of malaria. The study, thus, focuses on identifying major transcriptional changes in peripheral blood mononuclear cells of patients, from a co-endemic region, with acute malaria by P. falciparum infection with respect to that by P. vivax as an important comparison group. The agenda in the study also includes (i) identification of key participating genes associated with P. falciparum infection and (ii) unraveling the root of transcriptional deregulation of a crucial gene, CD14. Since host genetic factors are estimated to account for one quarter of the total variability in malaria infection and severity [16], the possibility that a genetic variant acting as a cis-regulatory factor of CD14 expression is also explored. Taken together the present study reconstructs the functional basis of an important immunological event during P. falciparum induced pathogenesis and generates a baseline knowledge with regard to species specific host gene expression.
2. Materials and method
2.1. Enrollment of patients
A total number of 181 malaria patients infected with either P. falciparum (n = 139) or P. vivax (n = 42) were collected from Surguja during the period of July 2013–December 2015. Surguja is located in the forested state of Chhattisgarh which is incidentally a tribal dominated region. Chhattisgarh is the second highly endemic state contributing to 14% of malaria in India [3]. Although there are government hospitals and clinics in and around, with facilities for treating malaria in the study area, the people prefer indigenous measures of treatment as an initial choice. Therefore, the majority of patients except those employed in gene expression assays, had already received traditional medicines at the time of enrollment. Confirmation of the disease and detection of parasites were carried out using rapid diagnostic tests based on dual-Antigen and/or thick and thin blood film using microscopy by expert pathologists. Children, pregnant women and patients with mixed infections by P. falciparum and P. vivax were excluded. Detailed information on clinical attributes including patient history, physical symptoms, biochemical findings and demography (age and sex) of the study participants were collected (Table 1).
Table 1.
Clinical and demographic characteristics of malaria patients.
| Plasmodium species |
P. falciparum |
P.vivax |
||
|---|---|---|---|---|
| Total samples | Severe | Uncomplicated | Uncomplicated | |
| n = 181 | n = 73 | n = 66 | n = 42 | |
| Clinical characteristics (n) | ||||
| Characteristics | Anemia, splenomegaly and respiratory distress | Cerebral and neurological complications | Mild febrile illness | |
| Anemia (Hb < 7 g/dl) | 5 | 4 | 2 | – |
| Episodic fever | 24 | 29 | 65 | 42 |
| Chills | 27 | 21 | 46 | 24 |
| Enlarged spleen | 20 | 7 | – | – |
| Respiratory distress | 26 | 16 | – | – |
| Mild jaundice | 7 | – | – | – |
| Vomiting | 15 | 9 | 42 | – |
| Convulsions | – | 17 | – | – |
| Neurological deficits | – | 14 | – | – |
| Headache | 6 | 26 | 46 | 16 |
| Demographic characteristics | ||||
| Gendera (n): M & F | 24 & 18 | 15 & 13 | 34 & 32 | 26 & 16 |
| Age range in years | 24–45 | 29–48 | 26–48 | 32–41 |
| Samples used in gene expression and genotypying | ||||
| Blood samples drawn before receiving any medications | 9 | – | 19 | 12 |
| Samples employed in gene expression analysis | RNA samples with concentration ≥ 25 ng/μl and RIN value 8 | |||
| 1 | 19 | 12 | ||
| Samples used genotyping | Severe malaria (n = 64) | Uncomplicated n = 52 | 30 | |
| Samples with all three loci genotypes (n = 141) | 33 | 26 | 52 | 30 |
M and F represent male and female respectively.
Venous blood (2–3 ml) was collected from each patient after obtaining informed consent. Blood samples from a subset of malaria patients with P. falciparum (n = 28) or P. vivax (n = 12) infection who had not received any anti-malarial medications at the time of study enrollment, were used to isolate peripheral blood mononuclear cells (PBMC) for RNA extraction. Following initiation of treatment, a subset of P. falciparum malaria patients was followed up for 4 weeks. Physical examination and assessment of blood smears were performed to ensure that the subjects were disease and parasite free. Approximately 3 ml of blood samples were collected from them (n = 4) to separate PBMC. Blood samples from remaining 141 patients were used to isolate genomic DNA. Among them, 111 were infected with P. falciparum with mild (n = 52) and severe (n = 59) symptoms as per WHO guidelines [17]. The study was approved by the Research Ethics Committee of Pt. Ravishankar Shukla University, Raipur (IEC Ref No.023/IEC/PRSU/2013 dated 15.4.2013) and Calcutta University, Kolkata (CU/BIOETHICS/HUMAN/1311 dated 17.6.2014).
2.2. Extraction of RNA and genomic DNA
Immediately after drawing blood, PBMC were separated from venous blood of 44 patients (P. falciparum infected = 28; P. vivax infected = 12 and P. falciparum convalescence = 4) using Histopaque 1077 (Sigma Aldrich, St. Louis, MO) and density gradient centrifugation according to manufacturer's protocol. Cells were preserved in RNAlater, transported to University of Calcutta and stored in −80 °C freezer. Cells were thawed gently, washed twice in PBS. Total RNA was isolated following manufacturer's protocol using RNeasy® Plus Minikit (QIAGEN) and quantified using nanodrop 2000C spectrophotometer (Thermo Scientific). Integrity values were checked using Agilent 2100 Bioanalyzer instrument. Samples (n = 35) with total RNA concentration ≥ 25 ng/μl and RIN value 8 were selected for Illumina whole genome microarray and quantitative PCR analyses. Genomic DNA was extracted from peripheral blood leukocytes (n = 141) using a QIAamp DNA Blood Kit (Qiagen, Hilden, Germany), quantified spectrophotometrically and stored in −80 °C until further use.
2.3. Microarray gene expression analysis
Microarray experiment was conducted using 23 RNA samples derived from P. falciparum (n = 13), P. vivax (n = 6) infected patients and P. falciparum patients (n = 4) in convalescent stage of the disease following standard protocol [18]. One P. falciparum sample was used as a technical replicate. Briefly, RNA was reverse transcribed to synthesize cDNA followed by in vitro synthesis of biotin labeled cRNA using Illumina® TotalPrep™ RNA Amplification Kit (life technologies). cRNA samples (750 ng) were then hybridized onto Illumina HumanHT-12 v4 Expression BeadChip targeting >47,000 probes. Staining was carried out using Streptavidin conjugated Cy3 Dye (GE Healthcare Life Sciences) and after washing chips were scanned in Illumina iScan (Illumina, San Diego, CA, USA). Illumina GenomeStudio was used to generate signal intensity estimates from the scans and Bioconductor package R 3.2.2 Limma was used for downstream analysis.
2.4. Quantitative RT-PCR
cDNA was synthesized from 1 μg of total RNA (with RIN value 8) isolated from P. falciparum and P. vivax infected individuals using MultiScribe Reverse Transcriptase (Applied Biosystems). Quantitative PCR was performed following standard protocol using SYBR Green I (06402712001, Roche) [19]. Real time PCR reaction was prepared in a total volume of 10 μl with 5 pmol of each primer and 1 μl of 10× diluted cDNA (Supplementary Material, Table S1). The amplifications were performed in triplicate on a LightCycler 96 real time system (Roche) following thermal cycling conditions 95 °C for 5 mins, 40 denaturation cycles at 95 °C for 30 s, annealing at 60 °C for 30 s and extension at 72 °C for 30 s. ACTB gene was co-amplified and served as an endogenous control. Specificity of PCR amplification for each primer pair was confirmed by melting curve analysis.
2.5. Cell culture, LPS, artemisinin and small interfering RNA treatments
U937 and PC3 cell lines were cultured in RPMI-1640 medium (AL162A, HIMEDIA) and HepG2 cell line was cultured in DMEM medium (D5523, Sigma Aldrich) supplemented with 10% heat inactivated foetal bovine serum (FBS) (RM1112, HIMEDIA) and 1% antibiotic antimycotic solution 100× [10,000 units Penicillin, 10 mg Streptomycin, 25 μg Amphotericin B/ml in 0.9% normal saline (A002A, HIMEDIA)] at 37 °C, 5% CO2.
For treatment 2 × 106 U937 cells were initially exposed to 0.1 μmolar 1,25α dihydroxy vitamin D3 (D1530, Sigma-Aldrich) for 24 h followed by treatment with E. coli (strain:0111:B4) lipopolysaccharide (LPS) (L4391, Sigma-Aldrich). Optimum induction of CD14 was observed at an LPS concentration of 500 ng/ml upon treatment of 6 h. Cell viability was measured by MTT assay. U937 cells were seeded on 96- well plate at a density of 2000 cells/well, cultured and treated with LPS. A total of 10 μl of 3-(4, 5-dimethylthiazol-2yl)-2, 5-diphenyl tetrazolium bromide (MTT) was added at a concentration of 0.5 mg/ml. The plates were incubated at 37 °C for 4 h to dissolve the formazan formed. Cell supernatant was removed from each well and 100 μl of DMSO was added and incubated for 10 min. Reduced MTT was measured on an ELISA reader (Bio-Rad, Hercules, CA, USA) at a wavelength of 570 nm. Values were expressed as percentage of cell viability. RNA and protein were extracted from U937 for analysis of gene expression from treated and control cells. U937 cells induced with 1,25α dihydroxy vitamin D3 were treated with 25 μM artemisinin (ART) drug (361,593, Sigma Aldrich) for 48 h. Cell viability was checked using MTT assay.
U937 and HepG2 cells were transfected at 60% confluence with small interfering RNA (siRNA) specific for SP1 (Qiagen ID numbers SI00150990) or AllStars Negative Control siRNA (Qiagen) using Lipofectamine® 3000 (Thermo Fisher Scientific) and HiPerFect transfection reagent (Qiagen) respectively. Effects of siRNA mediated SP1 gene knockdown was analyzed at the levels of mRNA, protein and reporter gene expression. U937 cells were induced with 0.1μmolar 1,25α dihydroxy vitamin D3 (Sigma) for 24 h prior to siRNA treatment.
2.6. Western blot analysis
Western blot was performed following standard protocol [20]. Protein was extracted from differentially treated U937 cells (approximately 4 × 106) with RIPA buffer and Protease inhibitor cocktail (P8340, Sigma Aldrich) followed by measurement of concentration by Bradford assay. Protein extracts (30–50 μg/lane) were resolved in SDS-PAGE using Prism Ultra Protein Ladder (ab116028, Abcam). Separated proteins transferred to polyvinylidene fluoride membrane (Pall Corporation). The primary antibodies used included rabbit monoclonal anti-CD14 (ab133335), rabbit polyclonal anti-SP1 (ab13370) (Abcam) and mouse monoclonal anti-β-actin (anti-ACTB) (C4: sc-47778) (Santa Cruz Biotechnology). Band intensities were quantified using ImageJ [21]. Average optical density of bands for CD14 and SP1 for each dataset was normalized with respect to that of β-actin.
2.7. Promoter analysis
CD14 promoter fragments spanning a region from −441 to +106 and encompassing rs5744454 were PCR amplified using specific primers containing recognition sites for XhoI and KpnI using genomic DNAs from two individuals harboring either AA or CC genotype (Supplementary Material, Table S1). PCR products were cloned into pTZ57R/T vector (InsTAclone PCR Cloning Kit, Fermentas) followed by sub-cloning in pGL3 basic vector. Plasmid isolation was carried out using QIAprep Spin Miniprep Kit (Qiagen). Integrity of clones and presence of A and C alleles were confirmed by restriction digestion and sequencing.
2.8. Genotyping
CD14 promoter polymorphism rs2569190 was genotyped with PCR coupled RFLP technique using 10U of HaeIII restriction endonuclease (Fermentas) (Supplementary Material, Table S1). To confirm the genotypes ascribed by RFLP, PCR products from 10% of the samples were randomly selected and subjected to sequencing using Big- Dye Terminator v3.1 and ABI Prism-3500 Genetic Analyzer (Applied Biosystems). Loci rs2569191 and rs5744454 were genotyped by resequencing of PCR products.
2.9. Promoter assay
CONSITE and TFBIND were used to map a 1 kb upstream promoter region of CD14 gene to locate putative binding sites for transcription factors. Approximately 5 × 104 cells were plated in each of the 24-well plate in triplicate using serum and antibiotic free DMEM/RPMI medium. Untreated/SP1 siRNA/AllStars negative control siRNA treated U937 and HepG2 cells were transfected with 0.25 μg each of the CD14 promoter constructs containing either A or C allele at position −281 or empty pGL3-Basic vector along with 0.02 μg of pRL-Renilla Luciferase control reporter vector (Promega, Madison,WI). To determine the allele specific (A or C) reporter gene expression, cells were incubated for 48 h and lysed. Firefly and Renilla luciferase activities were evaluated using dual luciferase reporter assay (Promega, Madison, WI). Luminescence was measured as relative light units (RLU) in GloMax 20/20 Luminometer (Promega, Madison, USA) using 7 ml of cell supernatant. Firefly luciferase activity was normalized with respect to Renilla luciferase and total protein produced was estimated using Bradford method. All transfection assays were carried in triplicates. The change in normalized luciferase expression was denoted in fold changes of RLU relative to appropriate control (empty pGL3 basic/negative control siRNA treatment).
2.10. Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed following standard protocol [22]. Approximately 3 × 106 cells were cross linked with 1% formaldehyde solution (F8775, Sigma) for 10 min following which the reaction was stopped with 0.125 M Glycine (Sigma Aldrich). Cells were harvested followed by treatment with cell lysis buffer containing 5 mM PIPES (pH 8.0), 85 mMKCl, 0.5% NP40 (with fresh protease inhibitor cocktail PIC). The cells were next treated with nuclear lysis buffer containing 50 mM Tris-HCl (pH 8.0), 10 mM EDTA, 1% SDS (with fresh PIC) followed by sonication at standard frequency (60 Hz). Pre-clearing of chromatin was carried out using normal sheep serum (NSS) (I5131, Sigma Aldrich). The chromatin was henceforth set for immunoprecipitation assay using anti-SP1 antibody-ChIP Grade (ab13370, Abcam) overnight at 4 °C. Pre-blocked DYNA beads with 0.3 mg/ml salmon sperm DNA (15,632,011, Invitrogen) were used for binding to pulled chromatin complex. DYNA beads were consecutively washed with RIPA buffer, high salt buffer, LiCl buffer, and TE buffer. Following treatment with RNaseA and Proteinase K, the beads were kept for de-cross-linking at 65 °C. The chromatin was extracted using conventional phenol-chloroform method followed by ethanol precipitation. Pellet was dissolved in nuclease free water and binding efficiency of SP1 to CD14 locus of interest was estimated by quantitative PCR using specific primers of interest (Supplementary Material, Table S1). Ct values for both IgG and ChIP samples were normalized to 10% input control and fold change with respect to IgG values was calculated from data of three replicated experiments.
2.11. Statistical analysis
Raw data from GenomeStudio were processed using the R Language (R) 3.2.2 in association with Bioconductor [23]. Limma package for R was used to perform quality control, log2 transformation and quantile normalization [24]. Dataset was submitted to GEO database and accession number obtained was GSE116149. Data was filtered to remove unexpressed genes based on detection call P values <0.05 for at least 3 arrays computed for each probeset out of >47,000 probes present on the Illumina HumanHT-12 v4 array and 24,000 probes were retained for further analysis. Correlation values between the technical replicates were analyzed in Illumina GenomeStudio to look for the reliability of the experiment. Chhattisgarh is a high transmission area for malaria. Therefore the possibility that a self-reporting healthy adult volunteer might already encounter a past Plasmodium infection could not be ruled out. Thus, PBMC transcriptome data of 5 healthy volunteers from University of Minnesota from GEO database (accession number: GSE45919) was additionally used as a surrogate for infection-naive control. The patterns of differential gene expression between the study groups were evaluated by generating linear regression models and moderated t-statistic with the package Limma for R [24]. P values were adjusted with Benjamini-Hochberg false discovery rate (FDR) correction. Differentially expressed probes were identified by a FDR <0.05 and mean fold-difference > +2 and < −2 between pairwise combinations of the study groups. Differentially expressed gene lists were incorporated to DAVID online gene annotation tool to identify genes that overlap with biological processes and pathways at a higher frequency than would normally be expected to occur for a randomly selected set of genes [25]. An FDR <0.05 was used as a threshold to determine whether a particular pathway was statistically enriched by differentially expressed genes. GO categories derived from DAVID were reclassified based on analysis of gene content and similarity of functions of pathways using GeneCards and PubMed [26,27].
Differential expression of candidate genes was quantified in terms of RQ (Relative Quantification = 2−ΔΔCq) value where ∆Cq expression of a gene in P. falciparum infection was normalized to that in P. vivax infection used as control. Percentage reduction in mRNA level calculated as (1 − ∆∆Cq)×100 was represented by bar diagram and statistical significance of a comparison was analyzed by Student-t-test in GraphPad Prism [28]. Allele frequencies of polymorphic loci were computed by gene counting. Any departure from Hardy–Weinberg equilibrium (HWE) for a locus was examined using HaploView [29]. Comparisons of genotype and allele counts were conducted using 3-way and 2-way contingency tables respectively. The fold difference of binding enrichment for ChIP assay were calculated using formula 2−[{Ct Target−(CtInput−6·644)}−{CtIgG−(CtInput−6·644)}] for each primer set, where 6·644 was a dilution factor used for input DNA.
3. Results
3.1. Capturing genes deregulated in P. falciparum infection
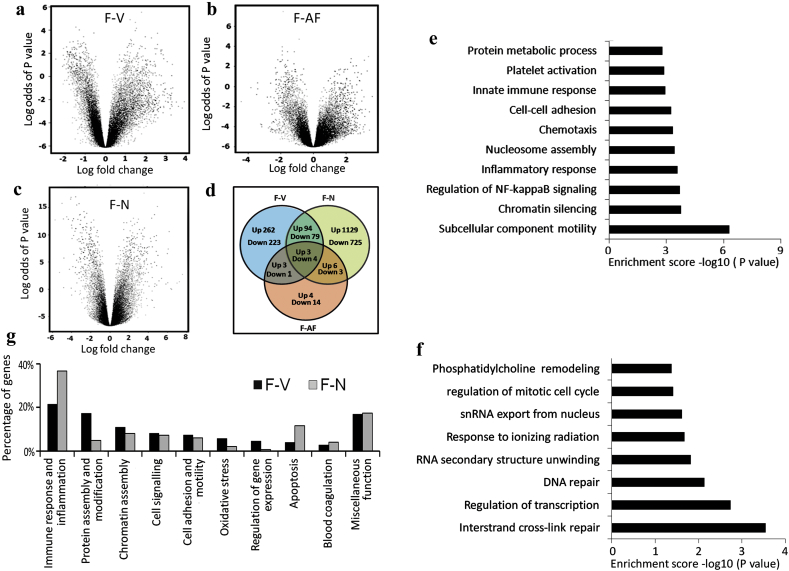
A whole genome microarray analysis was conducted using RNA isolated from PBMC of P. falciparum (F) infected malaria patients (n = 13) in the acute phase of disease using Illumina HumanHT-12 v4 Expression BeadChip. RNA samples from PBMC of P. vivax (V) infected symptomatic patients (n = 6) and a subset of P. falciparum malaria patients (n = 4) during their convalescent phase (AF) were also analyzed. Datasets from V and AF groups were used as baseline controls to identify genes differentially expressed in the P. falciparum malaria (Table 1). Consistency of microarray experiments was demonstrated by a significantly high correlation of average gene expression signals between technical replicates (r2 = 99.4%) and pattern of hybridization of control probes (Supplementary Material, Fig. S1).
A set of 669 genes was differentially expressed in F compared to V using Benjamini Hochberg FDR corrected P < 0.05 and fold change >2.0 for upregulation and fold change <−2.0 for downregulation. Among these, expression of 362 genes was increased in P. falciparum infected patients (Fig. 1a and d, Supplementary Material, Table S2a). Thirty eight genes were alternately regulated between F compared to that in AF, of which expression of 16 genes was significantly higher in acute phase infection (Fig. 1b, Supplementary Material, Table S2b). A subset of 11 genes including C4orf18, CD36, F13A1, MS4A7, RAB4A, VCL, AGAP8, D2HGDH, EYA3, METT11D1 and SLC7A5 was common between F-V and F-AF comparisons (Fig. 1d). Expression of 1232 genes was elevated in F compared to infection naïve US adults (N) from GSE45919 data out of a total number of 2043 differentially regulated genes (Fig. 1c and d, Supplementary Material, Table S2c). Approximately 48% (172 of 362) genes upregulated in F with respect to V were classified into 45 GO enrichment terms using DAVID bioinformatics tool (Fig. 1e, Supplementary Material, Fig. S2). In contrast only 20.5% (63 of 307) genes downregulated in F as compared to V belonged to 8 significantly enriched GO terms (Fig. 1f). Approximately 10% of the genes from 45 GO terms belonged to multiple enrichment categories with similar and overlapping functions. The gene set (n = 172) was manually curated into 10 non-redundant functional categories including immune response and inflammation (n = 37), protein assembly and modification (n = 30), chromatin assembly (n = 19), cell signaling (n = 14), cell adhesion and motility (n = 13), oxidative stress (n = 10), regulation of gene expression (n = 8), apoptosis (n = 7), blood coagulation (n = 5) and miscellaneous function (n = 29) (Fig. 1g, Supplementary Material, Table S3). Similarly, 356 out of 1232 genes from F-N comparison were reorganized into 10 enrichment categories such as immune response and inflammation (n = 131), apoptosis (n = 42), chromatin assembly (n = 29), cell signaling (n = 26), cell adhesion and motility (n = 22), protein assembly and modification (n = 18), blood coagulation (n = 15), oxidative stress (n = 8), regulation of gene expression (n = 3), miscellaneous category (n = 62) (Fig. 1g, Supplementary Material, Fig. S3, Table S4). In summary, the GO enrichment class composed of immune and inflammatory genes emerged as the most deregulated event in P. falciparum infection compared to that in P. vivax or malaria naïve individuals from the US, but not with respect to the convalescent samples (AF).
Fig. 1.
Description of microarray based global gene expression profiles. Volcano plots representing pairwise comparison of data pertaining to patients with (a) P. falciparum and P. vivax infections (b) acute phase of P. falciparum infection and at their convalescence stage (c) P. falciparum infection and GSE 45919 dataset. Coordinates of Volcano plot represent log odds of P value from a modified t-test vs. log ratio of fold change. (d) Venn diagram showing the number of genes differentially expressed in datasets namely F, V, AF and N. (e) Bar chart showing significantly enriched top ten functional categories of genes up regulated in P. falciparum infection compared to P. vivax infection. (f) Bar chart showing all eight significant GO categories for genes down regulated in P. falciparum infection compared to that with P. vivax infection. Absolute values of the –log10 (P value) of the enrichment analysis were used for plotting. Only categories with P value <0.05 threshold were shown. (g) Bar diagram depicting comparison of F-V and F-N datasets with respect to major GO enriched categories composed of genes up regulated in acute phase of P. falciparum infection. F, V, AF and N indicate P. falciparum infection, P. vivax infection, P. falciparum convalescent and malaria naïve US adults from GSE 45919 dataset respectively.
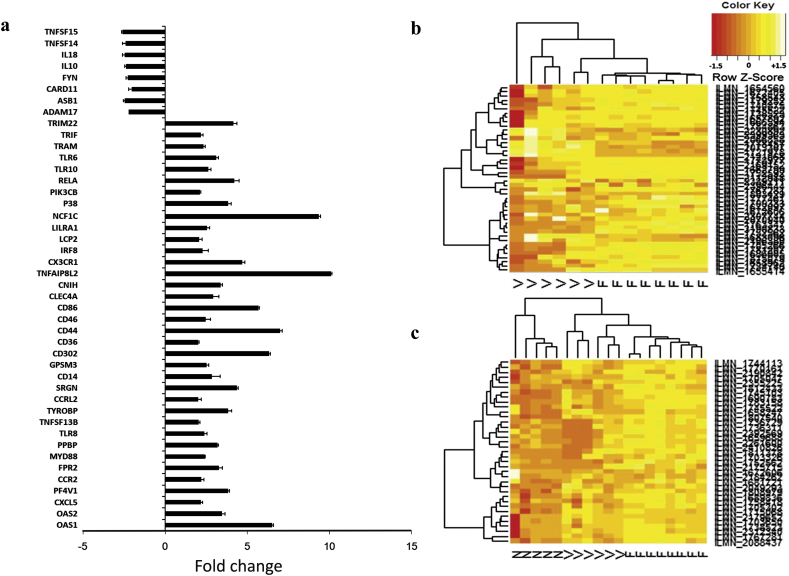
3.2. Differentially regulated immune response genes
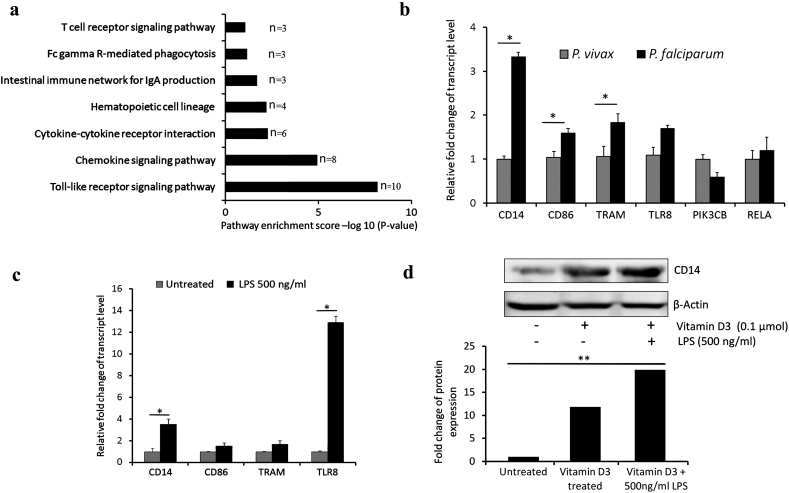
We focused our attention to the immune and inflammatory responsive genes (n = 45) differentially regulated in F (nupregulated = 37 and ndownregulated = 8) using V as a reference group (Fig. 2a). A heatmap constructed using normalized Z-Score of immune and inflammatory responsive genes from microarray data showed differential clustering of 6 V and 8 F samples (Fig. 2b). Similar analysis classified F, V and N into 3 distinct clusters (Fig. 2c). These genes were annotated using Kyoto Encyclopedia of Genes and Genomes of DAVID bioinformatics tool into 7 different immune pathways, namely Toll like receptor signaling pathway (n = 10), Chemokine signaling pathway (n = 8), Cytokine-cytokine receptor interaction (n = 6), Hematopoietic cell lineage (n = 4), Intestinal immune network for IgA production (n = 3), Fc gamma R mediated phagocytosis (n = 3) and T cell receptor signaling pathway (n = 3) (Fig. 3a).
Fig. 2.
Differentially regulated immune and inflammatory responsive genes associated with P. falciparum. (a) List of genes differentially regulated in F compared to V and their respective fold changes. Hierarchical clustering using 45 differentially regulated immune genes classifying samples pertaining to (b) F and V and (c) F, V and N datasets. Heat map was constructed based on Z-Score transformed expression values. A color code of orange from darker shade to light represents negative to positive distribution of Z score.
Fig. 3.
Immune and inflammatory pathways with special reference to TLR signaling in P. falciparum malaria. (a) KEGG pathways pertaining to differentially regulated immune response genes in F compared to V. X axis denotes the enrichment score -log10(P value). n denotes number of genes. (b) Bar plot showing relative mRNA expression levels of candidate genes from TLR pathway as part of validation of microarray results. (c) Bar diagram representing relative mRNA expression of candidate genes in U937 cell line stimulated with 1,25α dihydroxy vitamin D3 and treated with LPS. (d) Representative result of Western blot for CD14 in U937 cell line treated with LPS. β-actin was used as an endogenous control. Histogram depicting relative expression of CD14 in treated and control U937 cells. * denotes P value <0.05 which was considered significant.
Quantitative RT-PCR was carried out to validate the components of Toll-Like Receptor (TLR) pathway (P value <0.05) which was enriched with 10 upregulated genes (CD14, TLR6, TLR8, MYD88, TRAM, TRIF, RELA, PIK3CB, P38, CD86) (Supplementary Material, Fig. S4). Relative mRNA expression of CD14 (3.3 fold), CD86 (1.6 fold) and TRAM (1.8 fold) was significantly elevated in PBMC of P. falciparum (n = 6) compared with that in P. vivax (n = 6) infection. A 1.7 fold increased expression of TLR8 gene was also observed in P. falciparum infection (Fig. 3b).
To examine the suitability of a cell line model which was subsequently used to decipher regulation of CD14 gene in P. falciparum malaria, U937 cell line was treated with 1,25α dihydroxy vitamin D3 and 500 ng/ml of LPS. Relative transcript levels of CD14 (3.5 fold, P < 0.05), CD86 (1.7 fold), TRAM (1.8 fold) and TLR8 (12.9 fold, P < 0.05) were increased following LPS stimulation compared to those in untreated control (Fig. 3c). Western blot analysis of total protein extracted from LPS treated U937 cells showed 1.7 fold (P < 0.01) increase in CD14 expression (Fig. 3d). CD14 which displayed a significantly higher fold difference in P. falciparum infection in both microarray and qPCR was prioritized for further analysis.
3.3. Analysis of CD14 promoter
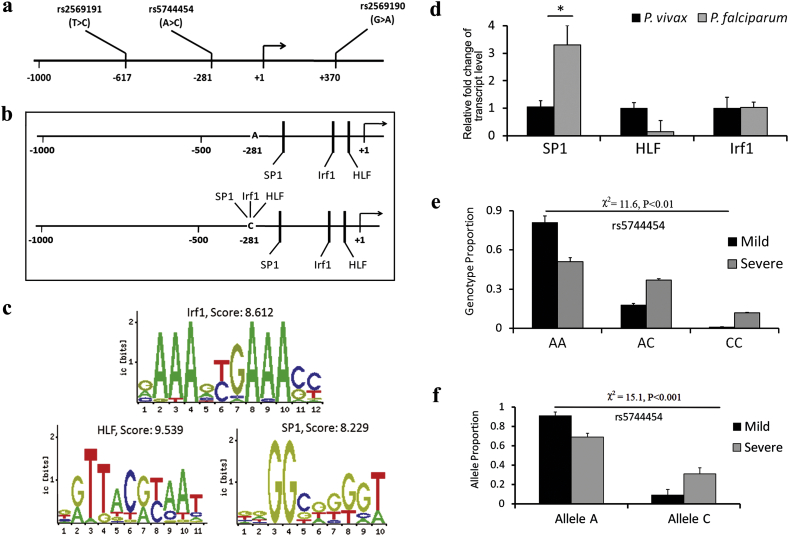
To investigate if CD14 gene was regulated by any host genetic polymorphisms, 1 kb upstream region of the CD14 promoter was scanned using dbSNP to locate rs2569190, rs5744454 and rs2569191 at positions +370, −281 and − 617 respectively with reference to CD14 transcription start site (Fig. 4a) [30]. A bioinformatic search using CONSITE and TFBIND was also conducted to identify whether the allelic variations of above SNPs resulted in differential binding of any transcription factors. Presence of C allele of rs5744454 (−281 A > C) was found to generate binding sites of HLF, IRF1 and SP1 with scores ranging from 6.523–9.785 (Fig. 4b). Each of these transcription factors also displayed an invariant binding site spanning −80 to −70 (HLF), −94 to −83 (IRF1) and − 248 to −239 (SP1) in the CD14 promoter (Fig. 4c). To narrow down the search, relative levels of HLF, IRF1 and SP1 transcripts were assayed in RNA isolated from PBMC of P. falciparum (n = 6) and P. vivax (n = 6) infected patients. A 3.3 fold higher expression (P < 0.05) of SP1 was observed in P. falciparum patients (Fig. 4d).
Fig. 4.
Analysis of CD14 promoter polymorphisms. (a) Diagram of 1.0 kb promoter sequence showing positions of rs2569190, rs2569191 and rs5744454 with respect to CD14 transcriptional start site. (b) Schematic representation of altered transcription factor binding due to allelic (A or C) variation of rs5744454. (c) Consensus binding sequence and scores of Irf1, HLF and SP1 for the invariant locations. (d) Quantitative PCR showing significant up regulation of SP1 in P. falciparum infected samples compared to P. vivax. Graphical representation of (e) genotype and (f) allele proportions of rs5744454 in P. falciparum infected uncomplicated and severe malaria patients. Frequency differences were tested using χ2 statistics. P value <0.05 was considered significant.
Genotypes of rs2569190, rs2569191 and rs5744454 were determined for P. faciparum (n = 111) and P vivax (n = 30) infected patients. Frequencies were compared between P. falciparum and P. vivax infected patients and between P. falciparum infected patients with uncomplicated (n = 52) and severe (n = 59) forms of the disease (Supplementary Material, Table S5). rs2569191 was excluded from the analysis since its frequencies did not conform to Hardy-Weinberg equilibrium. A statistically significant difference of genotype (χ2 = 11.6 and P < 0.01) and allele (χ2 = 15.1 and P < 0.001) frequencies was noted between uncomplicated and severe P. falciparum infected groups for rs5744454 (Fig. 4e and f). Together, these observations motivated us to explore the possibility of any functional interaction between rs5744454 located at −281 of CD14 promoter and SP1.
3.4. Dependence of SP1 binding on CD14 promoter polymorphism
Western blot analysis was performed to examine if CD14 expression was dependent on SP1. Protein levels of SP1 and CD14 were diminished by 99% and 70% respectively in SP1 siRNA treated U937 cell line compared to cells treated with AllStars Negative Control siRNA (Fig. 5a). On the contrary, treatment of U937 cells with 25 μM of artemisinin for 48 h elevated the expression of both SP1 and CD14 (Fig. 5b).
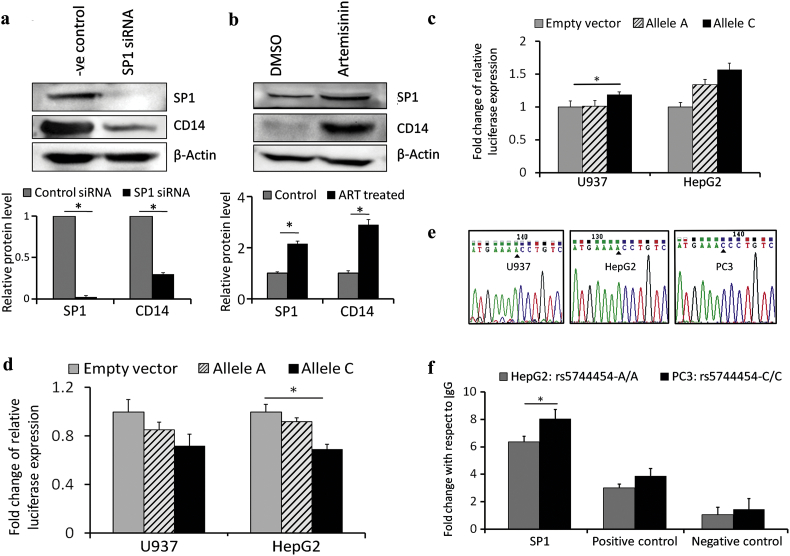
Fig. 5.
SP1 dependent expression and promoter activity of CD14. (a) Western blot in U937 cell line showing siRNA mediated knock down of SP1 and corresponding down regulation of CD14 and (b) Western blot showing artemisinin mediated increase of SP1 and CD14 protein levels in U937 cell line. Bar diagrams representing normalized relative light units obtained from U937 and HepG2 cell lines following (c) transfection with empty vector or promoter construct containing rs5744454: A/C allele and (d) SP1 specific siRNA treatment and transfection with empty vector or rs5744454: A/C allele. (e) Electropherograms showing AA, AA and CC genotypes at −281 position in CD14 in U937, HepG2 and PC3 cell lines respectively. (f) ChIP assay conducted to compare SP1 binding efficiencies in HepG2 and PC3 cell lines on CD14 promoter using quantitative PCR. A higher binding efficiency of SP1 in PC3 compared to HepG2 cells with respect to IgG control was shown. Binding efficiencies of SP1 for a positive control locus in CD14 gene and a negative control locus in DHFR gene were demonstrated. * denotes P value <0.05 which was considered significant.
To examine if CD14 promoter was controlled by SP1, CD14 promoter construct (spanning a region from −441 to +106) containing either A or C allele at position −281 was generated in pGL3 basic vector. Construct containing A allele harbored one SP1 binding site while that with C allele harbored two SP1 sites. Dual luciferase assays showed an increase in promoter activity for allele C compared to that of empty vector in U937 (19%, P < 0.05) and HepG2 (57%) cell lines. The promoter construct harboring A allele did not show any difference of relative firefly luciferase activity in U937 (2%), while in HepG2 the activity of the promoter construct was increased by 34% with respect to that of empty vector (Fig. 5c). The results indicated a higher activity of CD14 promoter associated with C allele. Binding of SP1 with CD14 promoter was further studied by assaying luciferase activity in U937 and HepG2 cells treated with SP1 specific siRNA. SP1 gene knockdown showed a decrease of firefly luciferase activity of A and C alleles by 15% and 28.4% respectively in U937 cell line with respect to empty vector. Similar reduction of promoter activity of A and C alleles by 8% and 31% (P < 0.05) respectively were recorded in HepG2 cell line (Fig. 5d).
Finally, allele specific interaction of SP1 with the CD14 promoter was investigated by using ChIP assay. Primers were designed to quantify the binding enrichment of SP1 through amplification of a region (−372 to −214) on CD14 gene that included the site (−281) at which rs5744454 was located. HepG2 and PC3 were considered for this experiment since these cell lines harbored AA and CC genotypes respectively at −281 in the CD14 promoter (Fig. 5e). U937 cell line with AA genotype at rs5744454 was also used (Supplementary Material, Fig. S5). Quantitative PCR of a 159 bp fragment from CD14 promoter using the immunoprecipitated DNA with SP1 antibody showed higher (8.02 ± 1.247) fold enrichment of SP1 binding in PC3 compared to that in HepG2 (6.36 ± 0.61) with respect to IgG control. Binding enrichments of a positive control (spanning 333 to 487 region) from CD14 gene (PC3 3.86 ± 0.569 and HepG2 3.01 ± 0.28) and a negative control (spanning −526 to −380 region) from DHFR gene (PC3 1.43 ± 1.61 and HepG2 1.05 ± 1.54) justified the validity of the experiment (Fig. 5f). ChIP experiments confirmed a higher affinity of SP1 association to CD14 promoter due to creation of an additional SP1 binding element for C allele of rs5744454.
4. Discussion
Development of clinical outcome in malaria, pathogenic or protective, is mediated through a complex interaction among host, parasite and eco-epidemiological factors [31]. Age, ability to acquire natural immunity and genetic makeup are recognized as important host-related factors. Studies have suggested that induction of a strong inflammatory response occupies centre stage in the beginning of acute phase of malaria followed by suppression of the host immune system in a subset of patients developing severe complications [32]. It has also been deciphered that initial proinflammatory processes in human malaria are triggered in dendritic cells and neutrophils present in the peripheral blood, although the underlying derangements of gene expression in this repertoire of cells in response to different malaria parasites are not fully elucidated [33,34]. Although malaria literature is replete with epidemiologic studies establishing association between multitude of immune and inflammatory response host genes with disease susceptibility and severity, there is still paucity of studies establishing a mechanistic link between host genetics and disease propensity [35].
In an attempt to decipher host transcriptome in acute phase of malaria, the present study captures major biological pathways perturbed in the PBMC of P. falciparum infected patients as compared to that of P. vivax. So far only a limited number of studies had addressed similar topic [[36], [37], [38], [39], [40]]. Since P. falciparum malaria is on the rise in India, to identify the active and predominant pathways associated with P. falciparum infection, we have adopted an improved approach by rigorously comparing PBMC transcriptome of patients suffering from acute phase of P. falciparum malaria with that of P. vivax and convalescence stage of the disease as references [27,41]. The analysis revealed that 669 and 38 genes were deregulated in F-V and F-AF comparisons respectively. The finding that only a limited number of genes were differentially regulated between F-AF may partially be attributed to an inadequate sensitivity of light microscopy based parasite detection method. Immune response and inflammation, protein modification, chromatin assembly, cell signaling, cell adhesion and motility, oxidative stress, regulation of gene expression are major GO categories enriched in P. falciparum malaria. As per our microarray results, the GO category of immune and inflammation composed of 45 differentially expressed genes which together can discriminate the molecular responses of P. falciparum infection from those of P. vivax infection as decorated in the heatmaps. Of the 7 KEGG pathways pertaining to immune and inflammatory response, Toll like receptor signaling pathway is prioritized for further investigation. Activation of TLR ligands and signaling components is a well known event in P. falciparum mediated pro-inflammatory Th1 responses by macrophages and DC as they confront parasite factors released by infected RBCs [42,43]. We focus our attention on CD14 as it emerges to be the most consistent participating principle in P. falciparum infection from microarray and subsequent validation assays conducted in patient samples and U937 cell line. CD14, a GPI-anchored protein and a major inflammatory surface marker of most TLR4 expressing cells is known to play a predominant role regulating parasite density in murine model of severe malaria and CD14+ CD16+ monocytes have been shown to mediate effector function in P. vivax mediated malaria as well [44,45].
A large volume of literature is available on genetic association of CD14 polymorphisms with susceptibility to a variety of diseases including asthma [46], allergic rhinitis [47], inflammatory bowel disease [48], respiratory infection [49] and even P. falciparum mediated malaria [50]. Among these, rs2569190, an e-QTL, was significantly correlated with majority of the above diseases [51]. Genotype frequencies of rs2569190 did not vary significantly between any of the malaria groups under this study. In an effort to establish a functional correlation between the disease, gene and polymorphism, we demonstrate here that CD14 expression in P. falciparum malaria is modulated by SP1 through differential binding of the transcription factor to CD14 promoter due to allelic variation of rs5744454, which display a higher prevalence of allele C in severe malaria.
One limitation of the present study is the small sample size of the study groups which may weaken the strength of the statistical association. However, this statistical shortcoming is overcome by establishing functional significance of the allelic variation at rs5744454 using a battery of experiments including promoter assays, RNAi mediated gene knockdown and ChIP conducted in U937, HepG2 and PC3 cell lines. Earlier research has shown that SP1 is a critical transcription factor for CD14 such that elevated level of phosphorylated SP1 during myeloid differentiation leads to a stronger binding of SP1 to its target sequence in CD14 promoter resulting in monocyte specific CD14 expression [52,53]. However, sequence dependent binding of SP1 to CD14 promoter has never been highlighted before. This, in addition to the observed association of rs5744454 with severe P. falciparum malaria adds value to our current understanding of host factors in malaria pathogenesis.
Finally we focus our attention to one important corollary emerging from the present investigation. As recommended by WHO, artemisinin is the first-line anti-malarial drug since 2001, as it reduces mortality in children with uncomplicated malaria by >95% [54]. The mechanism by which artemisinins exert their antimalarial action is controversial. Majority of studies concur that the activities of most artemisinin derived compounds arise from reductive cleavage of peroxide bridge of parasite proteins by reduced haem iron produced on haemoglobin digestion in the acidic digestive vacuoles [55]. It has been demonstrated in this study that treatment of artemisinin results in concomitant increase of SP1 and CD14 levels in U937 monocyte cell line which carries AA genotype for rs5744454. Since CD14 expression is associated with myeloid differentiation and DC maturation, artemisinin induced CD14 upregulation is expected to trigger host immune and inflammatory responses leading to parasite clearance and fever resolution. Dependence of CD14 expression on SP1 and CD14 promoter genotype, as shown in this study, indicates that host genetic constitution may modulate efficacy of artemisinin treatment in addition to parasite factors [56]. However, further investigation is necessary to clarify to what extent CD14 promoter genotype may modulate clinical prognosis of malaria patients treated with artemisinin. It may be useful to note in this context that artemisinin resistance has been reported from Southeast Asian countries where frequency of allele (C) of rs5744454 is 30% [30].
In summary, the current report reinstates that immune and inflammatory response genes and TLR pathway play a predominant role in P. falciparum malaria pathogenesis. Our data not only indicates a genetic association of CD14 promoter polymorphism, rs5744454, with P. falciparum mediated malaria severity, it establishes that CD14 expression is modulated by SP1 in an allele specific manner implicating rs5744454 to be a cis-regulatory polymorphism. Finally, the study underscores a pharmacogenetic angle of artemisinin therapy that needs to be explored further in view of the global malaria eradication initiative.
Acknowledgments
Acknowledgement
We thank CAS (UGC) and DST-FIST for providing some of the instrument facilities at the Department of Biochemistry, University of Calcutta and School of Studies in Anthropology, Pt. Ravishankar Shukla University, Raipur (SR/FIST/LSI-588/2014 dt. 21.11.2014) respectively. We are grateful to Dr. Arindam Maitra and Dr. Neeta S. Roy of National Institute of Biomedical Genomics, Kalyani for providing with the microarray and Illumina iScan facility. We also thank Dr. Indranil Mukhopadhyay of Human genetics unit, Indian Statistical Institute, Kolkata for his support in statistical analysis.
Funding
This work was supported by Science & Engineering Research Board, Department of Science and Technology (DST-SERB) grants SR/SO/HS-0056/2013.
Conflict of interest statement
The author declares that there is no conflict of interest.
Authors contribution
SS conceived and designed the study. BC performed the experiments and statistical and bioinformatic analysis. PM and CD helped in conducting ChIP assay. PG and MM participated by collecting samples. SS and BC wrote the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.09.049.
Supplementary data
Supplementary material
References
- 1.World Health Organization . 2017. World malaria report. [Google Scholar]
- 2.Battle K.E., Karhunen M.S., Bhatt S., Gething P.W., Howes R.E., Golding N. Geographical variation in Plasmodium vivax relapse. Malar J. 2014;13:144. doi: 10.1186/1475-2875-13-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.http://nvbdcp.gov.in/malaria3.html
- 4.Kumar A., Valecha N., Jain T., Dash A.P. Burden of malaria in India: retrospective and prospective view. Am J Trop Med Hyg. 2007;77:69–78. [PubMed] [Google Scholar]
- 5.Manjurano A., Sepúlveda N., Nadjm B., Mtove G., Wangai H., Maxwell C. USP38, FREM3, SDC1, DDC, and LOC727982 Gene Polymorphisms and Differential Susceptibility to Severe Malaria in Tanzania. J Infect Dis. 2015;212:1129–1139. doi: 10.1093/infdis/jiv192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anvikar A.R., Shah N., Dhariwal A.C., Sonal G.S., Pradhan M.M., Ghosh S.K. Epidemiology of Plasmodium vivax Malaria in India. Am J Trop Med Hyg. 2016;95:108–120. doi: 10.4269/ajtmh.16-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonçalves B.P., Huang C.Y., Morrison R., Holte S., Kabyemela E., Prevots D.R. Parasite burden and severity of malaria in Tanzanian children. N Engl J Med. 2014;370:1799–1808. doi: 10.1056/NEJMoa1303944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mcqueen P.G., McKenzie F.E. Host control of malaria infections: constraints on immune and erythropoeitic response kinetics. PLoS Comput Biol. 2008;4 doi: 10.1371/journal.pcbi.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White N.J. Determinants of relapse periodicity in Plasmodium vivax malaria. Malar J. 2011;10:297. doi: 10.1186/1475-2875-10-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White M.T., Karl S., Koepfli C., Longley R.J., Hofmann N.E., Wampfler R. Plasmodium vivax and Plasmodium falciparum infection dynamics: re-infections, recrudescences and relapses. Malar J. 2018;17(170) doi: 10.1186/s12936-018-2318-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonçalves R.M., Salmazi K.C., Santos B.A., Bastos M.S., Rocha S.C., Boscardin S.B. CD4+ CD25+ Foxp3+ regulatory T cells, dendritic cells, and circulating cytokines in uncomplicated malaria: do different parasite species elicit similar host responses? Infect Immun. 2010;78:4763–4772. doi: 10.1128/IAI.00578-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mclean A.R., Boel M.E., Mcgready R., Ataide R., Drew D., Tsuboi T. Antibody responses to Plasmodium falciparum and Plasmodium vivax blood-stage and sporozoite antigens in the postpartum period. Sci Rep. 2016;6 doi: 10.1038/srep32159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burel J.G., Apte S.H., McCarthy J.S., Doolan D.L. Plasmodium vivax but not Plasmodium falciparum blood-stage infection in humans is associated with the expansion of a CD8+ T cell population with cytotoxic potential. PLoS Negl Trop Dis. 2016;10 doi: 10.1371/journal.pntd.0005031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chuangchaiya S., Jangpatarapongsa K., Chootong P., Sirichaisinthop J., Sattabongkot J., Pattanapanyasat K. Immune response to Plasmodium vivax has a potential to reduce malaria severity. Clin Exp Immunol. 2010;160:233–239. doi: 10.1111/j.1365-2249.2009.04075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woodberry T., Loughland J.R., Minigo G., Burel J.G., Amante F.H., Piera K.A. Early immune regulatory changes in a primary controlled human Plasmodium vivax infection: CD1c myeloid dendritic cell maturation arrest, induction of the Kynurenine pathway, and regulatory T cell activation. Infect Immun. 2017;85(6) doi: 10.1128/IAI.00986-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mackinnon M.J., Mwangi T.W., Snow R.W., Marsh K., Williams T.N. Heritability of malaria in Africa. PLoS Med. 2005;2 doi: 10.1371/journal.pmed.0020340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.http://apps.who.int/iris/bitstream/10665/44294/1/9789241599221_eng.pdf
- 18.Bondar G., Cadeiras M., Wisniewski N., Maque J., Chittoor J., Chang E. Comparison of whole blood and peripheral blood mononuclear cell gene expression for evaluation of the perioperative inflammatory response in patients with advanced heart failure. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0115097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basu S., Majumder S., Bhowal A., Ghosh A., Naskar S., Nandy S. A study of molecular signals deregulating mismatch repair genes in prostate cancer compared to benign prostatic hyperplasia. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0125560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majumder S., Bhowal A., Basu S., Mukherjee P., Chatterji U., Sengupta S. Deregulated E2F5/p38/SMAD3 circuitry reinforces the pro-tumorigenic switch of TGFβ signaling in prostate cancer. J Cell Physiol. 2016;231:2482–2492. doi: 10.1002/jcp.25361. [DOI] [PubMed] [Google Scholar]
- 21.http:/imajej.nih.gov/ij
- 22.Adhikary S., Sanyal S., Basu M., Sengupta I., Sen S., Srivastava D.K. Selective recognition of H3.1K36 dimethylation/H4K16 acetylation facilitates the regulation of all-trans-retinoic acid (ATRA)-responsive genes by putative chromatin reader ZMYND8. J. Biol. Chem. 2016;291:2664–2681. doi: 10.1074/jbc.M115.679985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huber W., Carey V.J., Gentleman R., Anders S., Carlson M., Carvalho B.S. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.https://david.ncifcrf.gov/
- 26.http://www.genecards.org/
- 27.https://www.ncbi.nlm.nih.gov/pubmed/
- 28.http://www.graphpad.com
- 29.http://www.broadinstitute.org/scientific-community/science/programs/medical-and populationgenetics/haploview/haploview
- 30.https://www.ncbi.nlm.nih.gov/projects/SNP/
- 31.Recker M., Bull P.C., Buckee C.O. Recent advances in the molecular epidemiology of clinical malaria. F1000Res. 2018;7 doi: 10.12688/f1000research.14991.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olivier M., Van den Ham K., Shio M.T., Kassa F.A., Fougeray S. Malarial pigment hemozoin and the innate inflammatory response. Front Immunol. 2014;5(25) doi: 10.3389/fimmu.2014.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amorim K.N., Chagas D.C., Sulczewski F.B., Boscardin S.B. Dendritic cells and their multiple roles during malaria infection. J Immunol Res. 2016:2926436. doi: 10.1155/2016/2926436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolf A.S., Sherratt S., Riley E.M. NK cells: uncertain allies against malaria. Front Immunol. 2017;8:212. doi: 10.3389/fimmu.2017.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Driss A., Hibbert J.M., Wilson N.O., Iqbal S.A., Adamkiewicz T.V., Stiles J.K. Genetic polymorphisms linked to susceptibility to malaria. Malar J. 2011;10:271. doi: 10.1186/1475-2875-10-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu W.C. Microarray analysis of PBMC after plasmodium falciparum infection: molecular insights into disease pathogenesis. Asian Pac J Trop Med. 2016;9:313–323. doi: 10.1016/j.apjtm.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 37.Dunachie S., Berthoud T., Hill A.V., Fletcher H.A. Transcriptional changes induced by candidate malaria vaccines and correlation with protection against malaria in a human challenge model. Vaccine. 2015;33:5321–5331. doi: 10.1016/j.vaccine.2015.07.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colborn J.M., Ylöstalo J.H., Koita O.A., Cissé O.H., Krogsta D.J. Human gene expression in uncomplicated plasmodium falciparum malaria. J Immunol Res. 2015;2015:162639. doi: 10.1155/2015/162639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCall M.B., Netea M.G., Hermsen C.C., Jansen T., Jacobs L., Golenbock D. Plasmodium falciparum infection causes proinflammatory priming of human TLR responses. J Immunol. 2007;179:162–171. doi: 10.4049/jimmunol.179.1.162. [DOI] [PubMed] [Google Scholar]
- 40.Franklin B.S., Parroche P., Ataíde M.A., Lauw F., Ropert C., de Oliveira R.B. Malaria primes the innate immune response due to interferon-gamma induced enhancement of toll-like receptor expression and function. Proc Natl Acad Sci U S A. 2009;106:5789–5794. doi: 10.1073/pnas.0809742106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dash A.P., Valecha N., Anvikar A.R., Kumar A. Malaria in India: challenges and opportunities. J Biosci. 2008;33:583–592. doi: 10.1007/s12038-008-0076-x. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y., Zhu X., Feng Y., Pang W., Qi Z., Cui L. TLR4 and TLR9 signals stimulate protective immunity against blood-stage Plasmodium yoelii infection in mice. Exp Parasitol. 2016;170:73–81. doi: 10.1016/j.exppara.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 43.Ing R., Stevenson M.M. Dendritic cell and NK cell reciprocal cross talk promotes gamma interferon-dependent immunity to blood-stage Plasmodium chabaudi AS infection in mice. Infect Immun. 2009;77:770–782. doi: 10.1128/IAI.00994-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Antonelli L.R., Leoratti F.M., Costa P.A., Rocha B.C., Diniz S.Q., Tada M.S. The CD14+CD16+ inflammatory monocyte subset displays increased mitochondrial activity and effector function during acute Plasmodium vivax malaria. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oakley M.S., Majam V., Mahajan B., Gerald N., Anantharaman V., Ward J.M. Pathogenic roles of CD14, galectin-3, and OX40 during experimental cerebral malaria in mice. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0006793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nabih E.S., Kamel H.F., Kamel T.B. Association between CD14 polymorphism (−1145G/a) and childhood bronchial asthma. Biochem Genet. 2016;54:50–60. doi: 10.1007/s10528-015-9699-4. [DOI] [PubMed] [Google Scholar]
- 47.Kang H.J., Choi Y.M., Chae S.W., Woo J.S., Hwang S.J., Lee H.M. Polymorphism of the CD14 gene in perennial allergic rhinitis. Int J Pediatr Otorhinolaryngol. 2006;70:2081–2085. doi: 10.1016/j.ijporl.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 48.Kim E.J., Chung W.C., Lee K.M., Paik C.N., Jung S.H., Lee B.I. Association between toll-like receptors/CD14 gene polymorphisms and inflammatory bowel disease in Korean population. J Korean Med Sci. 2012;27:72–77. doi: 10.3346/jkms.2012.27.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hong S.A., Lee E., Kwon S.O., Kim K.W., Shin Y.H., Ahn K.M. Effect of prenatal antioxidant intake on infants' respiratory infection is modified by a CD14 polymorphism. World J Pediatr. 2017;13:173–182. doi: 10.1007/s12519-016-0054-6. [DOI] [PubMed] [Google Scholar]
- 50.Ojurongbe O., Funwei R.I., Snyder T.J., Aziz N., Li Y., Falade C.O. Genetic diversity of CD14 promoter gene polymorphism (rs2569190) is associated with regulation of malaria parasitemia and susceptibility to infection. Infect Dis (Auckl) 2017;10 doi: 10.1177/1178633617726781. 1178633617726781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.https://www.gtexportal.org/home/eqtls/bySnp?snpId=rs2569190&tissueName=All/
- 52.Zhang D.E., Hetherington C.J., Tan S., Dziennis S.E., Gonzalez D.A., Chen H.M. Sp1 is a critical factor for the monocytic specific expression of human CD14. J Biol Chem. 1994;269:11425–11434. [PubMed] [Google Scholar]
- 53.O'Connor L., Gilmour J., Bonifer C. The role of the ubiquitously expressed transcription factor Sp1 in tissue-specific transcriptional regulation and in disease. Yale J Biol Med. 2016;89:513–525. [PMC free article] [PubMed] [Google Scholar]
- 54.Djuika C.F., Staudacher V., Sanchez C.P., Lanzer M., Deponte M. Knockout of the peroxiredoxin 5 homologue PFAOP does not affect the artemisinin susceptibility of Plasmodium falciparum. Sci Rep. 2017;7:4410. doi: 10.1038/s41598-017-04277-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eastman R.T., Fidock D.A. Artemisinin-based combination therapies: a vital tool in efforts to eliminate malaria. Nat Rev Microbiol. 2009;7:864–874. doi: 10.1038/nrmicro2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Phompradit P., Chaijaroenkul W., Na-Bangchang K. Cellular mechanisms of action and resistance of Plasmodium falciparum to artemisinin. Parasitol Res. 2017;116:3331–3339. doi: 10.1007/s00436-017-5647-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material