Abstract
Background
NSCLC (non-small-cell lung cancer) is the leading cause of cancer-related mortality worldwide. Both epigenetic and genetic changes contribute to the initiation, development and metastasis of NSCLC. Recently, accumulating data have begun to support the notion that long noncoding RNAs (lncRNAs) function as new crucial regulators of diverse biological processes, including proliferation, apoptosis and metastasis, and play crucial roles in tumorigenesis. Nevertheless, further study is warranted to comprehensively determine lncRNAs' functions and potential mechanism.
Methods
In this study, we performed a comprehensive analysis of the lncRNA expression profile of NSCLC using data from TCGA and Gene Expression Omnibus (GEO). PCAT6 expression level in a cohort of 60 pairs of NSCLC tissues using quantitative real-time PCR (qRT-PCR). Additionally, Loss-of-function assays and gain-of-function assays were used to assess the role of PCAT6 in promoting NSCLC progression. Tumor formation assay in a nude mouse model was performed to verity the role of PCAT6 in NSCLC in vivo. Meanwhile, RIP, ChIP, resue experiment and western blot assays were used to highlights the potential molecular mechanism of PCAT6 in NSCLC.
Findings
We identified that an oncogene, PCAT6, was upregulated in NSCLC, and this upregulation was verified in a cohort of 60 pairs of NSCLC tissues. Additionally, the expression level of PCAT6 was correlated with tumor size (P = .036), lymph node metastasis (P = .029) and TNM stage (P = .038). Loss-of-function and gain-of-function assays were used to assess the role of PCAT6 in promoting NSCLC progression. The results revealed that PCAT6 knockdown mitigated NSCLC cell growth by inducing G1-phase cell cycle arrest and apoptosis in vitro and in vivo. Whereas, PCAT6 overexpression could promoted tumor cell growth. Meanwhile, PCAT6 additionally promoted NSCLC cell migration and invasion. Furthermore, mechanistic investigation demonstrated that the oncogenic activity of PCAT6 is partially attributable to its repression of LATS2 via association with the epigenetic repressor EZH2 (Enhancer of zeste homolog 2). Overall, our study highlights the essential role of PCAT6 in NSCLC, suggesting that PCAT6 might be a potent therapeutic target for patients with NSCLC.
Keywords: Long noncoding RNA, PCAT6, Non-Small-Cell Lung Cancer, LATS2, EZH2
Abbreviations: NSCLC, Non-small-cell lung carcinoma; ChIP, Chromatin immunoprecipitation; EZH2, Enhancer of zeste homolog 2; H3K27me3, Histone H3 lysine 27 trimethylation; PRC2, Polycomb repressive complex 2; RIP, RNA immunoprecipitation
Evidence before this study.
Lung cancer is the leading cause of cancer-related mortality worldwide. Recently, accumulating data have begun to support the notion that long noncoding RNAs (lncRNAs) function as new crucial regulators of diverse biological processes, including proliferation, apoptosis and metastasis, and play crucial roles in tumorigenesis. Nevertheless, further study is warranted to comprehensively determine lncRNAs' functions and potential mechanism.
Added value of this study
We identified that an oncogene, PCAT6, was upregulated in NSCLC through a comprehensive analysis of the lncRNA expression profile of NSCLC using data from TCGA and Gene Expression Omnibus (GEO). Clinically, the expression level of PCAT6 was correlated with tumor size, lymph node metastasis and TNM stage. PCAT6 could impact NSCLC cell growth by inducing G1-phase cell cycle arrest and apoptosis in vitro and in vivo. Meanwhile, PCAT6 additionally promoted NSCLC cell migration and invasion. Furthermore, mechanistic investigation demonstrated that the oncogenic activity of PCAT6 is partially attributable to its repression of LATS2 via association with the epigenetic repressor EZH2.
Implication of all the available evidence
Both PCAT6 and LATS2 paly crucial roles in the development of NSCLC. Our study provides new insight into the novel mechanism of PCAT6-mediated NSCLC via epigenetically suppressing LATS2, suggesting that PCAT6 might be a potent therapeutic target for patients with NSCLC.
Alt-text: Unlabelled Box
1. Introduction
Lung cancer remains the leading cause of cancer-related death in men and women, despite improvements in the present standard therapeutics, including cancer surgeries, radiotherapy, and anti-cancer drugs [1]. Lung cancer consists of two principal types: small-cell lung carcinoma and non-small-cell lung carcinoma (NSCLC). NSCLC, accounting for 85% of all cases, is the most prevalent histological type of lung cancer and can be further categorized into two common subtypes, adenocarcinoma (LUAD) and squamous cell carcinoma (LUSC) [2,3]. With of 5-year overall survival as low as 15% [4], lung cancer continues to be a crucial public health problem throughout the world. One reason for this low survival rate is caused by uncontrolled proliferation and metastatic potential of NSCLC cells [5]. Therefore, better understanding of novel mechanisms governing NSCLC cell growth and metastasis is essential for developing early diagnostic strategies, as well as individualized therapy.
In the past several decades, various important oncogenes or tumor suppressors have been identified as involved in the pathogenesis of human tumors. Due to the rapid development of high-throughput sequencing-based gene expression profiling technologies and bioinformatics, researchers focused on the role of long non-coding RNAs (lncRNAs) [6]. Long noncoding RNAs (lncRNAs) are functional noncoding RNA molecules >200 nt in length [7]. LncRNAs are transcribed throughout eukaryotic genomes and can be categorized as genic (exonic, intronic, overlapping) or intergenic lncRNAs based on their location with respect to the nearest protein-coding transcripts [8]. Based on the emerging literature, lncRNAs play important roles in various cellular processes, such as X chromosome imprinting, muscle cell differentiation, cell autophagy, migration and invasion [[9], [10], [11]]. More importantly, large-scale RNA sequencing in cancer or normal tissues demonstrated that several lncRNAs are dysregulated in tumor tissues compared with normal tissues [12], implying that lncRNAs might function as potential onco- or tumor-suppressor RNAs in human cancer progression. Furthermore, subsequent studies suggest that lncRNAs regulated gene expression at both post-transcriptional and transcriptional level [13]. For instance, HOTAIR is significantly overexpressed in several cancers and functions as an oncogene by modifying chromatin structure as a modular scaffold for histone modification complexes in multiple tissues [[14], [15], [16], [17]]. HULC is the first lncRNA with markedly higher expression in HCC and acts as an endogenous ‘sponge’ to promote HCC metastasis [18]. In NSCLC, lncRNAs plays several roles, and their molecular regulatory mechanisms were uncovered in our previous studies. We observed that linc00673 was upregulated in NSCLC and that linc00673 bind with LSD1 resulting in inhibiting NCALD expression and promoting cell proliferation [19]. In addition to linc00673, lncRNA AGAP2-AS1 was significantly overexpression in NSCLC tissues and correlated with poor prognostic outcomes in patients. It might bind to LSD1 and EZH2 resulting in reducing the expression level of KLF2 and LATS2 lead to NSCLC development and progression [20]. In this regard, identifying NSCLC-related lncRNAs the associated molecular mechanisms are necessary for understanding progression and establishing better treatment of NSCLC.
Thus, in the current study, we screened for the novel candidate lncRNAs responsible for the progression of NSCLC. Based on TCGA LUAD data, TCGA LUSC data and two gene profiling datasets from GEO, we determine lncRNAs profiles in NSCLC through genome-wide analyses. We identified that PCAT6 was upregulated in all four datasets and the expression level of PCAT6 was associated with poor prognosis. Moreover, we further focus on the functional roles and potential molecular mechanism of PCAT6 in NSCLC cell lines.
2. Materials and methods
2.1. Gene expression datasets
The RNA sequencing data of paired normal tissue samples and TCGA lung tumor tissues and the corresponding clinical data were downloaded from http://ibl.mdanderson.org/tanric/_design/basic/download.html. Another two public lung RNA sequencing and microarray datasets (GSE19188, GSE18842) were downloaded from Gene Expression Omnibus (GEO). LncRNA expression profiles from GEO microarray datasets was analyzed using GSE18842 (Affymetrix Human Genome U133 Plus 2.0 Array) and GSE19188 (Affymetrix Human Genome U133 Plus 2.0 Array). These microarray data were preprocessed using R software and packages.
2.2. Tissue collection and ethics statement
Sixty adjacent normal lung tissues and paired NSCLC tissues were obtained from patients who received primary surgery at the Department of Thoracic Surgery in the Second Affiliated Hospital, Nanjing Medical University, between February 2016 and July 2017. All collected tissue samples were staged and graded by an experienced pathologist according to the tumor node metastasis (TNM) classification and criteria of the World Health Organization (WHO). All samples were preserved in liquid nitrogen and stored at −80 °C, after immersing in RNA Later stabilization solution (Qiagen, Hilden, Germany). The patients had not received any local or systemic anticancer treatment before surgical resection. Our study protocol was approved by the research ethics committee of the Second Affiliated Hospital, Nanjing Medical University. Written informed consent was signed by all participants.
2.3. Cell culture and transfection
Seven human NSCLC cell lines were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China), including four adenocarcinoma cell lines (A549, SPC-A1, H1299 and H1975) and three squamous carcinoma cell lines (H1703, SK-MES-1 and H520). The cells were cultured in a humidified incubator in a 5% CO2 atmosphere at 37 °C with Dulbecco's modified Eagle's media (DMEM; GIBCO-BRL, Invitrogen, Carlsbad, CA) or RPMI Medium 1640 basic media (GIBCO-BRL, Invitrogen, Carlsbad, CA) containing heat-inactivated 10% fetal bovine serum (FBS) and antibiotics (100 U/ml penicillin and 100 mg/ml streptomycin) (Invitrogen, Carlsbad). The cells were sub-cultured and used for the following experiments. When cells in the logarithmic growth phase reached 80% confluence, transfections were performed using Lipofectamine 2000 (Invitrogen, Carlsbad) according to the manufacturer's instruction. A549 and SK-MES-1 cells were transfected with 20 μM siRNA (Invitrogen, Carlsbad) targeting PCAT6. A549 cells were also transfected with 20 μM siRNA (Invitrogen, Carlsbad) targeting EZH2 and LATS2. The siRNA sequences were as follows: PCTA6, si-1#, UGGCCUAGGAACCCGAACCUGACCC; si-2#, AAACAUUCCAGGGCACCGAGAGAUG. EZH2, si-1#, AAGACTCTGAATGCAGTTGCT; si-2#, CGGCUUCCCAAUAACAGUATT; si-LATS2, CAGGTGGACTCACAATTCCAAATAT; Knockdown of PCAT6, EZH2 and LATS2 was confirmed by qRT-PCR. For the tumor formation assay in a nude mouse model, two PCAT6-specific short hairpin (sh) RNA lentiviruses were produced with the following sequences: sh-PCAT6 1#, 5′-CACCGTGGCCTAGGAACCCGAACCTGACCCCG-AAGGGTCAGGTTCGGGTTCCTAGGCCA-3′; sh-PCAT6 2#, 5′-CA-CCAAACATTCCAGGGCACCGAGAGATGCGAACATCTCTCGGTGCCCTGGATG-3′.
In order to upregulate the expression level of PCAT6, the full-length PCAT6 cDNA sequence was synthesized and cloned into pcDNA3.1 vector (Invitrogen, Shanghai, China). Transfections were performed in SPCA1 using X-tremeGENE HP DNA transfection reagent (Roche, Basel, Switzerland), according to the manufacturer's instructions. QRT-PCR analysis was used to assess for PCAT6 overexpression.
Each experiment was performed in triplicate and repeated at least 3 times.
2.4. RNA isolation and quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR) analyses
Total RNA was extracted from tissue samples or cells by TRIzol (Invitrogen, Carlsbad) method. Then, RNA was reversely transcribed to cDNA by using a reverse transcription kit (TaKaRa, Tokyo, Japan). To evaluate the expression leve of lncRNA or mRNA, qRT-PCR analyses were performed using SYBR Premix Ex Taq II (Perfect Real Time; TaKaRa), according to the manufacturer's instructions. The primers used were as follows: GAPDH forward 5′-AGAAGGCTGGGGCTCATTTG-3′ and reverse 5′-AGGGGCCATCCACAGTCTTC-3′; PCAT6 forward 5′-CCCCTCCTTACTCTTGGACAAC-3′ and reverse 5′-GACCGAATGAGGATGGAGACAC-3′; LATS2 forward 5′-ACCCCAAAGTTCGGACCTTAT-3′ and reverse 5′-CATTTGCCGGTTCACTTCTGC-3′; EZH2 forward 5′-TTGTTGGCGGAAGCGTG-3′ and reverse 5′-TCCCTAGTCCCGCGCAATGTGC-3.
2.5. MTT assay and colony formation assay
For cell viability assay, the Cell Proliferation Reagent Kit I (MTT; Roche Applied Science) was used. 200 μl of the treated or untreated cells suspensions (3000 cells/well) was dispensed in 96-well plates. We estimated the cell viability every 24 h according to the manufacturer's instructions. The spectrophotometric absorbance was measured at 490 nm for each sample. All experiments were performed in quadruplicate and repeated 3 times.
For the colony formation assay, cells were trypsinized and suspended in medium with 10% FBS 24 h after transfection. The cells were re-seeded in 6-well plates at a density of 1000 cells/well in triplicate and cultured in a humidified atmosphere containing 5% CO2 at 37 °C. After 14 days, the cell colonies were washed with PBS and fixed with methanol for 30 min. The colonies were stained with 0.1% crystal purple (Sigma, St. Louis, MO) for 10 min. Colonies containing 450 cells were counted, and the mean colony numbers were calculated. Each treatment group was assessed in triplicate.
2.6. Flow cytometric analysis of apoptosis and cell cycle
The treated or untreated cells were harvested, washed with PBS and fixed overnight in 4% formaldehyde at −20 °C. Then, the DNA content was stained with propidium iodide (PI) using the Cycle TEST TM PLUS DNA reagent kit (BD Biosciences) in accordance with the manufacturer's recommendations. The cells were analyzed by FACScan (BD Biosicences, Franklin Lakes, NJ, USA).
Cell apoptosis was assessed by flow cytometry. According to the manufacturer's recommendations, double staining with fluorescein isothiocyanate (FITC)-Annexin V and propidium iodide was performed using the FITC Annexin V Apoptosis Detection Kit (BD Biosciences). The cells were divided into viable cells, dead cells, early apoptotic cells, and late apoptotic cells. The early or late apoptotic cells were measured.
2.7. Transwell assay
Cell migration and invasion ability was assessed through transwell assays, which were conducted in 24-well plates with 8-μm transwell inserts (Millipore, USA). For the migration assays, the cells were trypsinized, and adjusted to 5 × 104/ml cells. 300 μl of serum-free medium cell supernatant and 700 μl of 10% FBS conditioned medium were then added in the upper and lower chamber, respectively. The cells that remained on the upper side of the inserts were removed, while the cells that invaded the lower chamber were fixed with methanol and stained with 0.5% crystal violet, after culturing for 24 h. The number of invading cells was photographed and calculated by counting five random views. For the invasion assays, before the addition of the cells, upper chamber of Transwell chamber was previously coated with 50 μL of Matrigel (BD Biosciences, San Jose, CA, USA). The cells were trypsinized, and 1 × 105 cells in 300 μl of serum-free medium were plated into the upper chamber. To the lower chamber of the 24-well plates, 10% FBS conditioned medium (700 μl) was added. After culturing for 24 h, the cells that remained on the upper side of the inserts were removed, while the cells invading the lower chamber were fixed with methanol and stained with 0.5% crystal violet. Finally, 5 randomly selected fields were captured for cell count.
2.8. Xenografts
Tumor formation assay were performed as previously described [21]. Four-week-old male BALB/c athymic nude mice were maintained under specific pathogen-free conditions and used for the tumor formation assay. A549 cells (2 × 107/ml) transfected with an empty vector or shRNA PCAT6 were cultured in six-well plates for 48 h. The cells were washed twice with PBS and resuspended in PBS. Then 100 μl of the cell suspension was inoculated into the right and left flank of each mouse. The longest and shortest diameters of tumor masses were measured using calipers every 3 days until 13 days later when the tumors were removed. Tumor volumes were calculated using the formula, volume = (length × width2 × 0.5). At 13 days after injection, the subcutaneous growth of each tumor was examined. All animal allocation, surgery were approved by the Model Animal Research Center of Nanjing Medical University.
2.9. Western blot analysis
After siRNA or vector transfection, the cells were harvested in a lysis buffer containing PMSF (Roche), a protease inhibitor cocktail (Roche, Basel, Switzerland) and a mammalian protein extraction reagent RIPA (Beyotime China). The BCA method was used to determined the total protein concentration. Equal amounts of the total protein were separated by 10% SDS-PAGE and then transferred to 0.22 mm NC membranes (Sigma-Aldrich). After blocking in 5% non-fat milk, the membranes were incubated with specific antibodies including LATS2 (1:1000, Cell Signaling Technology, Inc). Lastly, the immunoreactive protein bands on the membrane were visualized using an ECL Kit (Pierce, Thermo Fisher Scientific, IL, USA).
2.10. RNA-binding protein immunoprecipitation (RIP) assay
In order to assess the interaction between EZH2 and PCAT6, the EZMagna RIP kit (Millipore, Billerica, MA, USA) was used. A549 cells were lysed and the supernatants were incubated with protein A/G Sepharose beads coated with antibodies that recognized EZH2 or with control IgG (EMD Millipore, Darmstadt, Germany). The antibodies were conjugated at 4 °C for 6 h. We used wash buffer to wash the beads and used 0.1% SDS/0.5 mg/ml Proteinase K to digest the proteins (30 min at 55 °C). Then, the purified RNA from the immunoprecipitation materials was used for further assessed by qPCR analysis.
2.11. Chromatin immunoprecipitation (ChIP) assays
Chromatin immunoprecipitation assays were performed using the MagnaChIP Kit (Millipore, Bedford, MA) according to the manufacturer's instructions. Finally, qPCR was used to identify and quantify the precipitated chromatin DNA.
2.12. Statistical analysis
The SPSS 19.0 software (IBM, Chicago, IL, USA) was used for all statistical analyses and P < .05 considered difference was statistically significant. All data were expressed as the mean ± S.D. Student's t-test (two-tailed) and the chi-square test analysis were performed to analyze differences between groups. Graphpad Prism 5 was introduced for image editing.
3. Results
3.1. Identification of lncRNAs alterations in NSCLC tissues
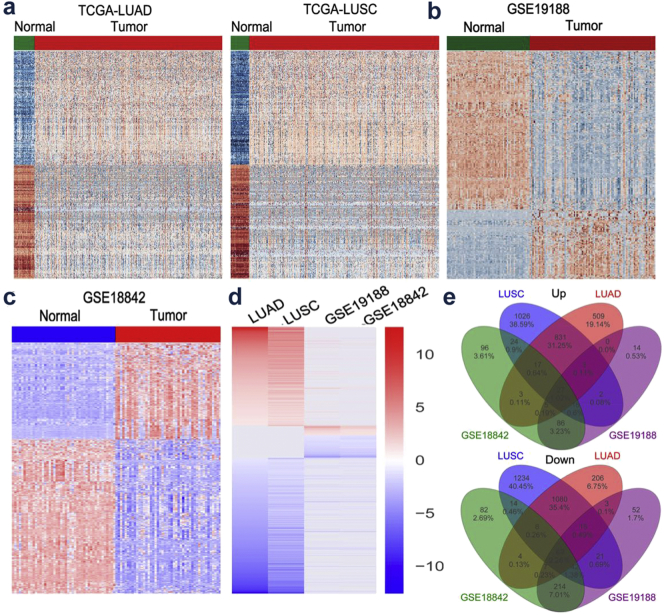
To uncover novel oncogenic lncRNAs essential for NSCLC tumorigenesis, four bioinformatics datasets (TCGA-LUAD, TCGA-LUSC, GSE19188, GSE18842) were utilized to analyze differentially expressed lncRNAs between lung tumors samples and the corresponding non-tumor samples. Analysis of these datasets showed that the expression of 3095 lncRNAs was dysregulated in the TCGA-LUAD dataset (1593 upregulated and 1502 downregulated); the expression of 4429 lncRNAs was dysregulated in the TCGA-LUSC dataset (1946 upregulated and 2483 downregulated); the expression of 576 lncRNAs was dysregulated in the GSE19188 dataset (153 upregulated and 423 downregulated); the expression of 714 lncRNAs was dysregulated in the GSE18842 dataset (274 upregulated and 440 downregulated) (Fig. 1a-d and Supplementary Table 1). Notably, further analyses revealed that 96 lncRNAs were consistently upregulated or downregulated in all datasets (Fig. 1e and Supplementary Table 2). Subsequently, we pay close attention to overexpressed lncRNAs owing to their oncogenic ability.
Fig. 1.
The expression profiles of lncRNAs in NSCLC tissues and normal tissues.
(a) Heatmap of the differentially expressed lncRNAs expression in LUAD or LUSC and normal tissue samples was analyzed using the TCGA datasets. (b and c) Heatmap of the dysregulated lncRNAs in NSCLC was analyzed using the GSE19188, GSE18842 datasets. (d) Heatmap of the altered lncRNAs profiling (consistently altered at least two datasets, fold change) in TCGA-LUAD, TCGA-LUSC, GSE19188, GSE18842 datasets. (e) Venn diagram of altered lncRNAs in TCGA-LUAD, TCGA-LUSC, GSE19188, GSE18842 datasets.
3.2. PCAT6 upregulated in human NSCLC tissues
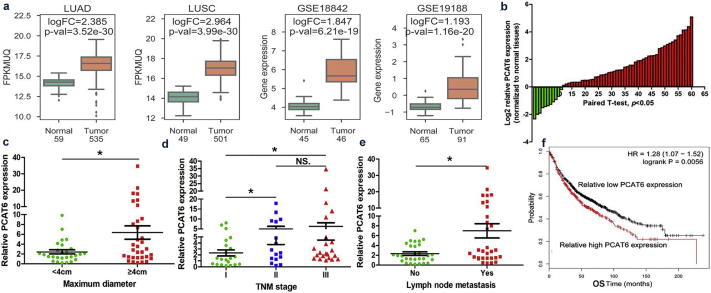
To validate these findings, we selected PCAT6 which was upregulated in four datasets and correlated with a poor OS, for further investigation (Fig. 2a and f). We first analyzed the expression level of PCAT6 in 60 patients with NSCLC tissues and paired normal tissues. The results showed that compared with adjacent normal tissues, PCAT6 was significantly upregulated in clinical NSCLC tissues (Student's t-test, P < .05; Fig. 2b). To further evaluate the relationship between PCAT6 expression and clinicopathological traits (i.e., lymphatic metastasis, maximum diameter or TNM stage), we divided these patients into two groups (low and high) according to the median value. We determined that PCAT6 expression is associated with tumor size (Chi-square test, P = .036), lymph node metastasis (Chi-square test, P = .029) and TNM stage (Chi-square test, P = .038) (Fig. 2c, d, e and Table.1). However, there was no significant correlation between PCAT6 expression and other clinical characteristics, such as age, gender, differentiation and smoking history (Chi-square test, P > .05; Table 1). Taken together, these date indicated that PCAT6 might play a crucial role in NSCLC progression.
Fig. 2.
Higher PCAT6 expression levels in NSCLC and its clinical significance.
(a) The PCAT6 expression levels in NSCLC tissues compared with normal tissues in TCGA-LUAD, TCGA-LUSC, GSE19188, GSE18842 datasets. (b) The PCAT6 expression level in 60 NSCLC tissues and corresponding adjacent non-tumor tissues was quantified by Quantitative real-time PCR analysis and normalized to GAPDH expression. Expression levels were shown as log2-fold change to match non-tumor tissues. Red column represented overexpression and Green column represented down-regulation. (c, d and e) The relationship between PCAT6 expression and clinicopathological parameters (such as maximum diameter, lymphatic metastasis and TNM stage) was shown. (f) Kaplan-Meier survival plots demonstrated that higher PCAT6 abundance correlated with a poor OS, using microarray data from 1926 lung cancer patients. Data are shown as the mean ± SD Based on at least three independent experiments. *P < .05, **P < .01.
Table 1.
Correlation between PCAT6 expression and clinicopathological parameters of NSCLC.
| clinicopathological parameters⁎ | N of cases | Log2 relative PCAT6 expression |
||
|---|---|---|---|---|
| Low | High | P-valuea⁎ | ||
| Age(years) | 0.861 | |||
| <65 | 25 | 12 | 13 | |
| ≥65 | 35 | 16 | 19 | |
| Gender | 0.809 | |||
| male | 32 | 15 | 17 | |
| female | 28 | 14 | 14 | |
| Differentiation | 0.198 | |||
| well, moderate | 17 | 9 | 8 | |
| poor | 43 | 15 | 28 | |
| Tumor size (maximum diameter cm) | 0.036⁎ | |||
| <4 cm | 27 | 23 | 4 | |
| ≥4 cm | 33 | 20 | 13 | |
| Primary location | 0.821 | |||
| left lung | 26 | 13 | 13 | |
| right lung | 34 | 18 | 16 | |
| Histology type | 0.593 | |||
| adenocarcinoma | 35 | 20 | 15 | |
| squamous carcinoma | 25 | 16 | 9 | |
| Smoking history | ||||
| smokers | 23 | 14 | 9 | 0.051 |
| never smokers | 37 | 13 | 24 | |
| Lymph node metastasis | 0.029⁎ | |||
| positive | 31 | 19 | 12 | |
| negative | 29 | 25 | 4 | |
| TMN stage | 0.038⁎ | |||
| I | 21 | 17 | 4 | |
| II/III | 37 | 19 | 18 | |
Chi-square test.
P < .05.
3.3. PCAT6 promotes NSCLC cell proliferation in vitro
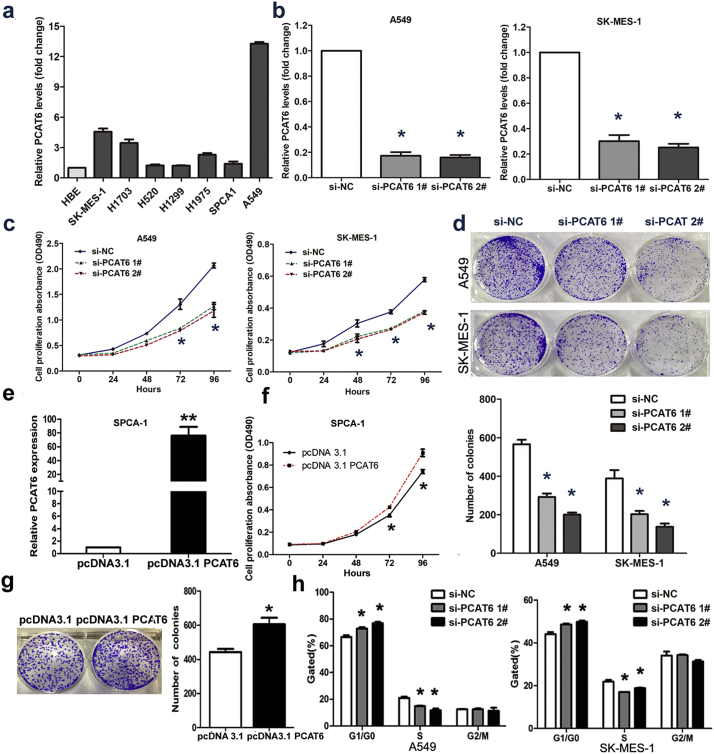
Next, we determined the expression level of PCAT6 in 7 NSCLC cell lines including both squamous carcinoma and adenocarcinoma via qRT-PCR. The result showed that PCAT6 expression was higher in 7 NSCLC cell lines than in HBE (human bronchial epithelial cells) (Fig. 3a). Among them, the expression level of PCAT6 in A549 cell was the highest. With the purpose of manipulating the PCAT6 level in NSCLC cells, we performed loss-of-function study using two discrete chemically synthesized siRNA in A549 (adenocarcinoma cell line) and SK-MES-1 (squamous carcinoma cell line). After 24 h of transfection, compared to the respective control cells, 70% downregulation was confirmed for the siRNAs targeting PCAT6 by qRT-PCR (Fig. 3b). Simultaneously, in order to copy the development process of NSCLC, we performed gain-of-function study in SPCA1 via pcDNA3.1-PCAT6 transfection (Fig. 3e).
Fig. 3.
The effects of PCAT6 on NSCLC cell viability in vitro.
(a) qRT-PCR analysis was performed to identify the expression level of PCAT6 in 7 NSCLC cell lines and human bronchial epithelial cell (HBE). The expression levels are normalized to HBEs. (b) The PCAT6 expression level in A549 and SK-MES-1 transfect with two discrete chemically synthesized siRNAs. (c) MTT assays were used to measure the growth curve of si-PCAT6–transfected A549 and SK-MES-1 cells. Values indicate the mean ± SD from three independent experiments. (d) Colony-forming assays were conducted to determine the proliferation of si-PCAT6-transfected A549 and SK-MES-1 cells. (e) The PCAT6 expression level in SPCA1 transfected with pcDNA3.1 PCAT6 and empty vector. (f) MTT assays were used to measure the growth curve of pcDNA3.1 PCAT6 and empty vector transfected in SPCA1. (g) Colony-forming assays were conducted to determine the proliferation of pcDNA3.1 PCAT6 and empty vector transfected in SPCA1. (h) Flow cytometry assays were performed to analyze the cell cycle progression when NSCLC cells transfected with PCAT6. * P < .05, **P < .01. N.S., not significant.
Because lncRNAs was involved in a spectrum of biological processes, we investigated the impact of PCAT6 in NSCLC cell lines. An MTT assay demonstrated that PCAT6 knockdown inhibits the proliferation rate of A549 and SK-MES-1 (Fig. 3c). Meanwhile, a colony formation assay was performed to evaluate the role of PCAT6 in long-term survival. As shown in Fig. 3d, downregulation of PCAT6 significantly attenuated the colony-forming ability of the population. What's more, overexpression PCAT6 could promote cell growth and protect the colony-forming ability (Fig. 3f and g).
These observations made with siRNA and overexpression plasmid indicated that PCAT6 might be an important factor that contribute to NSCLC progression.
3.4. Knockdown of PCAT6 causes G1 arrest and promotes apoptosis
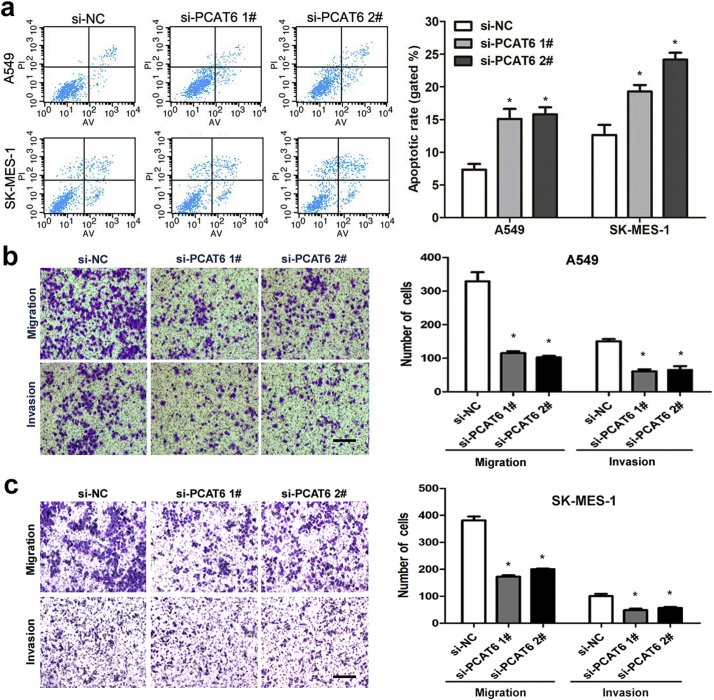
Several lines of evidence show that apoptosis and growth arrest are major mechanism resulted in controlled cell death. To further determine whether the effect of PCAT6 on NSCLC cell proliferation is the result of PCAT6-mediated changes in cell cycle progression, FACS technology was applied. As presented in Fig. 3h, PCAT6 knockdown decreased the percentage of cells in the S phase and increased the percentage of cells in the G0/G1 phase compared with control cells. Additionally, we identify the influence of PCAT6 on apoptosis by performing flow cytometric analysis. The result showed that NSCLC cells treated with siRNA PCAT6–1# or siRNA PCAT6–2# exhibited significant apoptosis compared with control groups in A549 and SK-MES-1(Fig. 4a).
Fig. 4.
The effects of PCAT6 on NSCLC cell apoptosis, cell migration and invasion in vitro.
(a) The apoptosis of A549 and SK-MES-1S were analyzed by flow cytometry. LR, early apoptotic cells. UR, terminal apoptotic cells. (b and c) Transwell assays were used to determine the invasive ability of si-PCAT6 1#, si-PCAT6 2# or si-NC transfected A549 and SK-MES-1 cells. The cells on the lower chamber were stained and presented. Data represent the mean ± S.D. from three independent experiments. Scale bar represents 100 μm. *P < .05, **P < .01.
Accordingly, we proposed that low PCAT6 expression is both important and necessary for apoptosis and normal growth arrest in the NSCLC cell lines.
3.5. PCAT6 knockdown induces suppression of migration and invasion in NSCLC cells
To explore the functional effect of PCAT6 in NSCLC migration and invasion, in vitro transwell assays were performed in A549 and SK-MES-1 cells 24 h after transfection. Silencing of PCAT6 strongly inhibited their capability of migration, compared to that in the control cells (Fig. 4b and c). Simultaneously, a transwell assay with Matrigel revealed that invasion ability was significantly reduced by downregulation of PCAT6 (Fig. 4b and c). This observation supported by the results of the clinicopathological parameter analysis.
3.6. Knockdown of PCAT6 inhibits NSCLC tumorigenesis in vivo
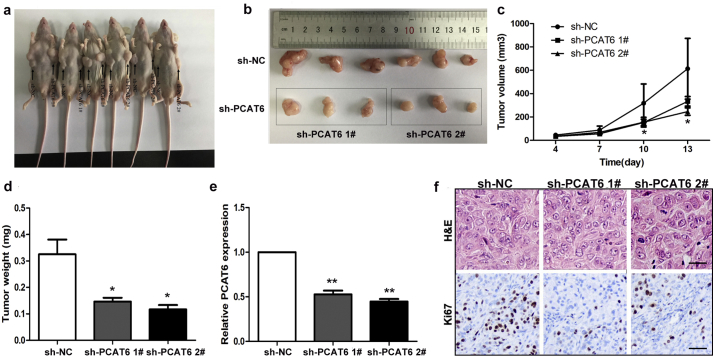
Having examined the biological effect of PCAT6 in vitro, we additionally provide in vivo evidence for the oncogenic role of PCAT6 in NSCLC by injecting control cells and PCAT6 knockdown cells into nude mice. The control, shRNA PCAT6 1# or shRNA PCAT6 2# A549 cells were inoculated subcutaneously into male nude mice. Tumor volumes were measured every three days after injection, and the tumors were removed from all animals 13 days after injection. Compared with the control group, the PCAT6-depleted group showed dramatically decreased tumor growth (Fig. 5a, b and c). Tumor weight of the PCAT6-depleted group was significantly lower than that of the control group (Fig. 5d). Moreover, quantitative RT-PCR analysis demonstrated that the expression level of PCAT6 in the tumors after shRNA PCAT6 transfection were lower than that after shRNA transfection (Fig. 5e). In addition, the HE staining showed typical characteristics of tumor cells, and the Ki-67 staining was performed to verify these results (Fig. 5f). Collectively, these results indicated that downregulation of PCAT6 suppresses tumor progression in vivo.
Fig. 5.
The effects on tumor growth after PCAT6 downregulation in vivo.
(a, b and c) The tumor growth curves were measured 4 days after the injection after injection of A549 cells stably transfected with shRNA PCAT6 1#, shRNA PCAT6 2# or empty vector and tumor volume was calculated once every 3 or 4 days. (d) Tumor weight when the tumors were harvested. (e) qRT-PCR analysis of PCAT6 expression level in tumor tissues formed from shRNA PCAT6 1#, shRNA PCAT6 2# or empty vector transfected A549 cells. (e) Representative images of HE staining and Ki-67 immunohistochemistry of the tumor. Up, H & E staining, Scale bar represents 50 μm; Down, immunostaining, Scale bar represents 20 μm. The data represent the mean ± S.D. from three independent experiments. *P < .05, **P < .01.
3.7. PCAT6 silences LATS2 transcription by binding with H3K27 methyltransferase EZH2
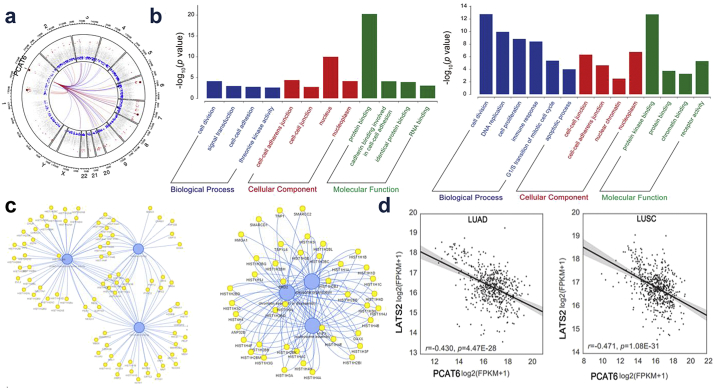
To identify the functional processes that were affected by PCAT6-mediated transcriptional regulation, GO analysis was performed. We determined that cell-cell adhesion was involved in the affected functional processes in LUAD and cell proliferation in LUSC (Fig. 6b), which was consistent with our data. Then, to investigate the mechanism of PCAT6 in NSCLC cell proliferation and metastasis, we analyzed the co-expression patterns of PCAT6 (Fig. 6c). Using qRT-PCR, we confirmed representative genes (Supplementary Fig. 1a) which were identified as tumor suppressor genes in A549 and SK-MES-1. Our results determined that LATS2 was the most upregulation in response to PCAT6 downregulation (Supplementary Fig. 1a), which was consistent with the data of LUAD and LUSC (Fig. 6d). Therefore, we hypothesized that PCAT6 might impact NSCLC cell proliferation and metastasis by regulating LATS2.
Fig. 6.
PCAT6 co-expressed genes and downstream targets in NSCLC.
(a) The co-expression genes of PCAT6. (b) GO pathway analysis for all genes with altered expressions in TCGA-LUAD, TCGA-LUSC datasets. (c) The interaction network of PCAT6. (d) Analysis of the relationship between PCAT6 expression and LATS2 mRNA level in TCGA-LUAD and TCGA-LUSC datasets.
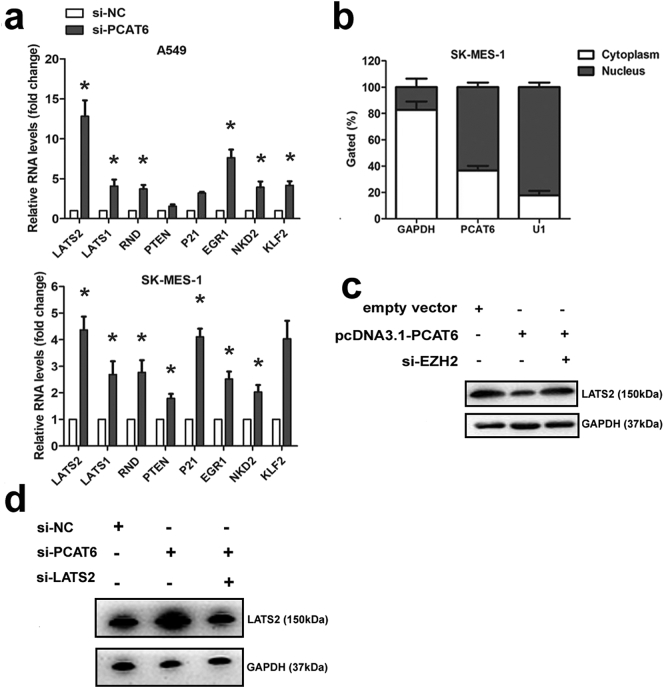
Supplementary Fig. 1.
(a) The expression level of downstream targets of PCAT6. (b) PCAT6 expression levels in different subcellular fractions in SK-MES-1. (c) The the western blot analysis were conducted to detect the expression levels of LATS2 protein in SPCA1 cells transfected with empty vector, pcDNA3.1-PCAT6 and pcDNA3.1-PCAT6 + si-EZH2. (d) The the western blot analysis were conducted to detect the expression levels of LATS2 protein in A549 cells transfected with si-NC, si-PCAT6 and si-PCAT6 + si-LATS2. *P < .05, **P < .01.
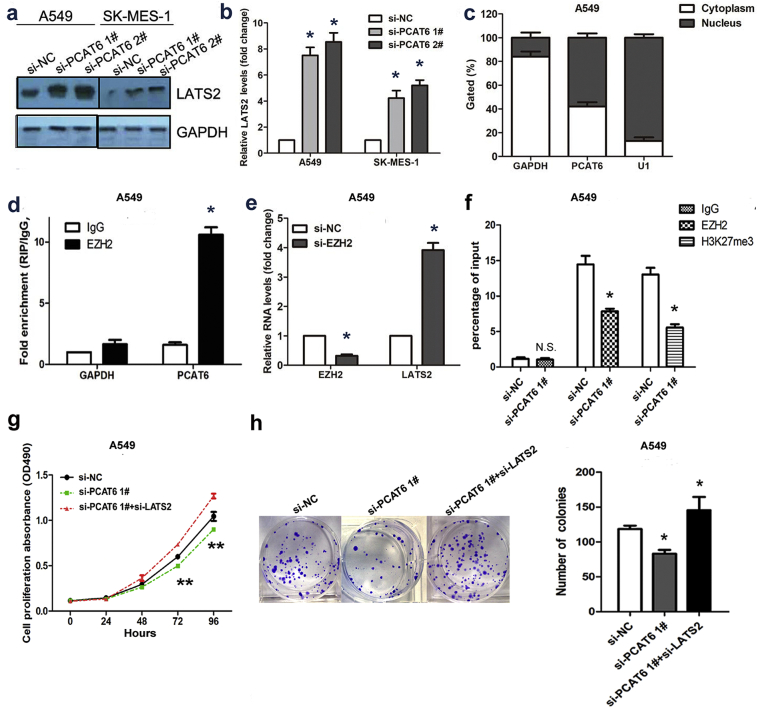
To test this hypothesis, we first investigated whether PCAT6 knockdown affected the expression level of LATS2. As presented in Fig. 7a and b, the expression level of LATS2 protein and mRNA was upregulated after PCAT6 reduction in A549 and SK-MES-1 cells. Furthermore, we measured PCAT6 expression in nuclear and cytosolic fractions from A549 and SK-MES-1 cells. As shown in Fig. 7c and Supplementary Fig. 1b, PCAT6 was mainly localized in the nucleus. Multiple studies have suggested that lncRNAs regulate gene expression at transcriptional level through RNA-binding proteins, such as EZH2, SUZ12, STAU1, and LSD1 [16,22,23]. An RNA-RIP assay was performed to further evaluate whether PCAT6 inhibits LATS2 expression at transcriptional level through a similar mechanism. The result demonstrated that PCAT6 RNA could be pulled down by EZH2 antibody in A549 (Fig. 7d). In our previous study, qRT-PCR results demonstrated that the inhibition of EZH2 expression led to increased LATS2 expression, and chromatin immunoprecipitation analysis indicated that EZH2 could directly bind to the LATS2 promoter region and mediate H3K27 trimethylation modification [20]. Here, the result of qRT-PCR and chromatin immunoprecipitation analysis was corresponded with our previous study (Fig. 7e and f). Furthermore, rescue study was conducted. The western blot indicated that the expression level of LATS2 was downregulated in pcDNA3.1 PCAT6 group while partially upregulated in pcDNA3.1 PCAT6 + si-EZH2 group (Supplementary Fig. 1c). Importantly, The MTT and colony formation as says abilities of cell growth and proliferation were partially recovered by LATS2 knockdown (Fig. 7g and h). Based on these observations, we suggested that PCAT6 overexpression might decrease LATS2 promoter activity via binding to EZH2 resulted in H3K27 trimethylation in the LATS2 promoter region.
Fig. 7.
PCAT6 could recruit EZH2 to LATS2 promoter and represses LATS2 transcription.
(a and b) The western blot analysis and qRT-PCR assay were conducted to detect the expression levels of LATS2 protein and mRNA in A549 and SK-MES-1 cells transfected with si-PCAT6 and si-NC. (c) PCAT6 expression levels in different subcellular fractions in A549 in A549 cell line. (d) RIP with rabbit monoclonal anti-EZH2 and preimmune IgG from A549 cell extracts. RNA levels in immunoprecipitates were detected by qPCR. Expression levels of PCAT6 RNA are presented as fold enrichment in EZH2 relative to IgG immunoprecipitates. (e) The expression level of LATS2 mRNA in A549 cells transfected with si-EZH2 and si-NC. (f) ChIP–qRT-PCR of EZH2 occupancy and H3K27me3 binding in the LATS2 promoter in A549 cells transfected with si-PCAT6 and si-NC. (g) MTT assays were used to measure the growth curve of si-NC, si-PCAT6 and si-PCAT6 + si-LATS2–transfected A549. (h) Colony-forming assays were conducted to determine the proliferation of si-NC, si-PCAT6 and si-PCAT6 + si-LATS2–transfected A549.The data represent the mean ± S.D. from three independent experiments. *P < .05, **P < .01.
4. Discussion
Continuing advances in transcriptomics indicate that the study of epigenetic regulation of lncRNAs in cancer is emerging as a potential research field. In 2010, Tsai MC et al. determined that lincRNA HOTAIR could modify the chromatin structure as a modular scaffold for histone modification complexes and regulate HOXD expression in multiple tissues [16]. Subsequently, multiple lines of evidence demonstrate that lncRNAs lead various chromatin-modifying complexes to specific genomic loci or tumor-cell-specific promoter regions, thereby impacting the cell cycle, differentiation, apoptosis, DNA repair and cell adhesion [24]. For instance, our previous studies showed that lincRNA 00673 interacts with the epigenetic repressor LSD1 and represses NCALD expression in NSCLC [19]. In the present study, we analyzed the expression profile of lncRNA in NSCLC through bioinformatics and observed that lncRNA PCAT6 associated with the prognosis of patients without surgery was upregulation in four bioinformatics datasets (TCGA-LUAD, TCGA-LUSC, GSE19188, GSE18842). We obtained this result by tissue validation. In addition, we explored the biological function of PCAT6 via loss-of-function assays, demonstrating that the knockdown of PCAT6 significantly suppressed the proliferation and metastasis of NSCLC cells in vitro, which was consistent with the result of Li Wan et al. [25]. More recently, Xu et al. reported that PCAT6 was overexpressed in gastric cancer tissues and that PCAT6 impact MKRN3 expression level through endogenously competition with microRNA-30 [26]. Yet, very little is known regarding the molecular mechanisms of PCAT6 in NSCLC carcinogenesis. To this end, we further explored the potential molecular mechanisms involved and determined that LATS2 was remarkably upregulated after PCAT6 knockdown, as analyzed using bioinformatics and qRT-PCR assays. The RIP and ChIP assay revealed that PCAT6 could directly bind with EZH2, and EZH2 could directly bind to the LATS2 promoter region in NSCLC cells. Based on the collective results presented above, we propose that PCAT6 exerts its oncogenic function, at least partly, via binding to EZH2 and inhibiting LATS2 expression in NSCLC tumorigenesis.
Generally, the term “epigenetic regulation” refers to changes that affect gene regulation, including histone modifications and DNA modifications [27]. Multiple evidence suggested that epigenetic regulation plays a key regulatory role in normal development, and epigenetic imbalance is possibly the beginning of tumorigenesis in patients with NSCLC. Histone modifications, as vital factors in transcriptional activation and repression, play important roles in initial tumor formation and progression. It has been revealed that polycomb repressive complex 2 (PRC2) is an essential histone methyltransferase, and the molecular function of PRC2 is responsible for establishing the H3K27me3 mark on specific genes, which promotes transcriptional repression of these genes [[28], [29], [30], [31]]. As a core catalytic subunit of PRC2, Enhancer of zeste homolog 2 (EZH2) is believed to be a key player in tumor progression in multiple cancer types, and EZH2 upregulation correlates with poor prognosis [[32], [33], [34]]. Carmen Behrens et al. determined the clinical relevance of EZH2 protein expression in a large series of surgically resected NSCLCs [32]. They observed that high expression of EZH2 predicts aggressive tumor behavior, and EZH2 serves as a prognostic marker in patients with surgically resected lung adenocarcinomas, when combined with TTF-1 expression. Moreover, Christine M. Fillmore and colleagues demonstrated that EZH2 suppression. May boost the sensitivity of two NSCLC subsets (EGFR mutant or BRG1 mutant) to etoposide [32]. Consequently, accumulated data have begun to advance the idea that EZH2 is a potential novel therapeutic target, and anti-EZH2 therapies are urgent and currently under development.
The LATS gene family, originally isolated from Drosophila, encodes serine/threonine-protein kinases [35,36]. The LATS1 and LATS2 kinases, two of the core members of the LATS gene family, have become the focus of intense research interest in recent years. Multiple lines of evidence demonstrate that they emerged as central regulators of cell fate and play an important role in maintaining cellular homeostasis [37,38]. Importantly, LATS1 and LATS2 are involved in various human malignant tumors by modulating the functions of multiple oncogenic or tumor suppressive effector. For instance, a low expression level of LATS1 and LATS2 is correlated with cell migration via altering p53 function in breast cancer [39]. Moreover, it has been reported that LATS1 could decrease YAP expression and contribute to better prognosis in NSCLC [40]. Given the critical roles of LATS2 in several important life-sustaining processes, we have explored its biological activities in NSCLC cell lines. Consistently, our results revealed that LATS2 overexpression inhibited NSCLC cell growth and induced apoptosis [20]. Therefore, the elucidation of regulatory mechanisms upstream of LATS2 is necessary and essential. Recent studies showed that LATS2 could be suppressed at both the transcriptional and post-transcriptional levels [41,42]. In our study, we observed that LATS2 was negatively regulated by PCAT6. RIP and ChIP assays validated that PCAT6 exerts its oncogenic effect via binding EZH2 and inhibiting tumor suppressor LATS2 expression in NSCLC tumorigenesis.
Taken together, our study establishes an oncogenic role for PCAT6 deregulation in NSCLC. Additionally, we report for the first time that the oncogenic activity of PCAT6 is attributable to its repression of LATS2 through association with the epigenetic repressor EZH2. This study provides a plausible molecular mechanism underlying the deregulation of cancer-associated lncRNA expression in NSCLC. Nonetheless, in this study, we only focus on the key downstream mediators and molecular mechanism of PCAT6. Thus, future upstream mediators and mechanism of PCAT6 are required to uncover. Only by completely elucidating the molecular mechanisms of misregulated PCAT6 in NSCLC can we open avenues for utilizing lncRNAs to identify novel diagnostic or drug targets for NSCLC.
The following are the supplementary data related to this article
The dysregulated lncRNAs in four datasets.
The summary of dysregulated lncRNAs.
Acknowledgments
Acknowledgements
The authors thank Ming Sun, Department of Department of Bioinformatics and computational biology, UT MD Anderson Cancer Center for help with statistical and bioinformatics analysis. We apologize to all researchers whose relevant contributions were not cited due to space limitations.
Conflicts of Interests
The authors declare that they have no competing interests.
Authors' contributions
SXF, NFQ and LZL did the assays in vitro and collected clinical samples and analyzed the data; LZC, FXR, HF and HXX performed experiments in vivo; WB and LKH wrote the manuscript; WB and LKH designed this study. All authors have read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 81602013, No. 81602003, No. 81672949), the medical and health research project of Zhejiang province (No. 2018KY164) and the science and technology development fund projects of Nanjing Medical University (No. 2015NJMU019).
Contributor Information
Bin Wang, Email: 13757295077@139.com.
Kaihua Lu, Email: 13605179453@126.com.
Fengqi Nie, Email: Niefengqi@njmu.edu.cn.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Cooper W.A., Lam D.C., O'Toole S.A., Minna J.D. Molecular biology of lung cancer. J Thorac Dis. 2013;5(Suppl. 5):S479–S490. doi: 10.3978/j.issn.2072-1439.2013.08.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ettinger D.S., Akerley W., Bepler G., Blum M.G., Chang A., Cheney R.T. Non-small cell lung cancer. J Natl Compr Canc Netw. 2010;8(7):740–801. doi: 10.6004/jnccn.2010.0056. [DOI] [PubMed] [Google Scholar]
- 4.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 5.Koudelakova V., Kneblova M., Trojanec R., Drabek J., Hajduch M. Non-small cell lung cancer--genetic predictors. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2013;157(2):125–136. doi: 10.5507/bp.2013.034. [DOI] [PubMed] [Google Scholar]
- 6.Roth A., Diederichs S. vol. 394. 2016. Long Noncoding RNAs in Lung Cancer. Current Topics in Microbiology and Immunology; pp. 57–110. [DOI] [PubMed] [Google Scholar]
- 7.Motzer R.J., Molina A.M. Targeting renal cell carcinoma. J Clin Oncol. 2009;27(20):3274–3276. doi: 10.1200/JCO.2009.21.8461. [DOI] [PubMed] [Google Scholar]
- 8.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang J.L., Zheng L., Hu Y.W., Wang Q. Characteristics of long non-coding RNA and its relation to hepatocellular carcinoma. Carcinogenesis. 2014;35(3):507–514. doi: 10.1093/carcin/bgt405. [DOI] [PubMed] [Google Scholar]
- 10.Fatica A., Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 11.Mercer T.R., Dinger M.E., Sunkin S.M., Mehler M.F., Mattick J.S. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2008;105(2):716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan X., Hu Z., Feng Y., Hu X., Yuan J., Zhao S.D. Comprehensive genomic characterization of long non-coding rnas across human cancers. Cancer Cell. 2015;28(4):529–540. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kornienko A.E., Guenzl P.M., Barlow D.P., Pauler F.M. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013;11:59. doi: 10.1186/1741-7007-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chisholm K.M., Wan Y., Li R., Montgomery K.D., Chang H.Y., West R.B. Detection of long non-coding RNA in archival tissue: correlation with polycomb protein expression in primary and metastatic breast carcinoma. PloS one. 2012;7(10) doi: 10.1371/journal.pone.0047998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li D., Feng J., Wu T., Wang Y., Sun Y., Ren J. Long intergenic noncoding RNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am J Pathol. 2013;182(1):64–70. doi: 10.1016/j.ajpath.2012.08.042. [DOI] [PubMed] [Google Scholar]
- 16.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kogo R., Shimamura T., Mimori K., Kawahara K., Imoto S., Sudo T. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71(20):6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 18.Wang J., Liu X., Wu H., Ni P., Gu Z., Qiao Y. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38(16):5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi X., Ma C., Zhu Q., Yuan D., Sun M., Gu X. Upregulation of long intergenic noncoding RNA 00673 promotes tumor proliferation via LSD1 interaction and repression of NCALD in non-small-cell lung cancer. Oncotarget. 2016;7(18):25558–25575. doi: 10.18632/oncotarget.8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W., Sun M., Zang C., Ma P., He J., Zhang M. Upregulated long non-coding RNA AGAP2-AS1 represses LATS2 and KLF2 expression through interacting with EZH2 and LSD1 in non-small-cell lung cancer cells. Cell Death Dis. 2016;7:e2225. doi: 10.1038/cddis.2016.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi X., Sun M., Liu H., Yao Y., Kong R., Chen F. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Mol Carcinog. 2015;54(Suppl. 1):E1–E12. doi: 10.1002/mc.22120. [DOI] [PubMed] [Google Scholar]
- 22.Nie F.Q., Sun M., Yang J.S., Xie M., Xu T.P., Xia R. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther. 2015;14(1):268–277. doi: 10.1158/1535-7163.MCT-14-0492. [DOI] [PubMed] [Google Scholar]
- 23.Xu T.P., Liu X.X., Xia R., Yin L., Kong R., Chen W.M. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene. 2015;34(45):5648–5661. doi: 10.1038/onc.2015.18. [DOI] [PubMed] [Google Scholar]
- 24.Shi X., Sun M., Liu H., Yao Y., Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 25.Wan L., Zhang L., Fan K., Cheng Z.X., Sun Q.C., Wang J.J. Knockdown of long noncoding RNA PCAT6 Inhibits proliferation and invasion in lung cancer cells. Oncol Res. 2016;24(3):161–170. doi: 10.3727/096504016X14618564639178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y., Sun J.Y., Jin Y.F., Yu H. PCAT6 participates in the development of gastric cancer through endogenously competition with microRNA-30. Eur Rev Med Pharmacol Sci. 2018;22(16):5206–5213. doi: 10.26355/eurrev_201808_15718. [DOI] [PubMed] [Google Scholar]
- 27.Huang Y., Nayak S., Jankowitz R., Davidson N.E., Oesterreich S. Epigenetics in breast cancer: what's new? Breast Cancer Res. 2011;13(6):225. doi: 10.1186/bcr2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441(7091):349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 29.Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298(5595):1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 30.Koyanagi M., Baguet A., Martens J., Margueron R., Jenuwein T., Bix M. EZH2 and histone 3 trimethyl lysine 27 associated with Il4 and Il13 gene silencing in Th1 cells. J Biol Chem. 2005;280(36):31470–31477. doi: 10.1074/jbc.M504766200. [DOI] [PubMed] [Google Scholar]
- 31.Pasini D., Bracken A.P., Jensen M.R., Lazzerini Denchi E., Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23(20):4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fillmore C.M., Xu C., Desai P.T., Berry J.M., Rowbotham S.P., Lin Y.J. EZH2 inhibition sensitizes BRG1 and EGFR mutant lung tumours to TopoII inhibitors. Nature. 2015;520(7546):239–242. doi: 10.1038/nature14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koumangoye R.B., Andl T., Taubenslag K.J., Zilberman S.T., Taylor C.J., Loomans H.A. SOX4 interacts with EZH2 and HDAC3 to suppress microRNA-31 in invasive esophageal cancer cells. Mol Cancer. 2015;14:24. doi: 10.1186/s12943-014-0284-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamaguchi H., Hung M.C. Regulation and role of EZH2 in cancer. Cancer Res Treat. 2014;46(3):209–222. doi: 10.4143/crt.2014.46.3.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Justice R.W., Zilian O., Woods D.F., Noll M., Bryant P.J. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9(5):534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- 36.Xu T., Wang W., Zhang S., Stewart R.A., Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121(4):1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- 37.Visser S., Yang X. LATS tumor suppressor: a new governor of cellular homeostasis. Cell Cycle. 2010;9(19):3892–3903. doi: 10.4161/cc.9.19.13386. [DOI] [PubMed] [Google Scholar]
- 38.Furth N., Aylon Y. The LATS1 and LATS2 tumor suppressors: beyond the Hippo pathway. Cell Death Differ. 2017;24(9):1488–1501. doi: 10.1038/cdd.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Furth N., Bossel Ben-Moshe N., Pozniak Y., Porat Z., Geiger T., Domany E. Down-regulation of LATS kinases alters p53 to promote cell migration. Genes Dev. 2015;29(22):2325–2330. doi: 10.1101/gad.268185.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin X.Y., Zhang X.P., Wu J.H., Qiu X.S., Wang E.H. Expression of LATS1 contributes to good prognosis and can negatively regulate YAP oncoprotein in non-small-cell lung cancer. Tumour Biol. 2014;35(7):6435–6443. doi: 10.1007/s13277-014-1826-z. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi Y., Miyoshi Y., Takahata C., Irahara N., Taguchi T., Tamaki Y. Down-regulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers. Clin Cancer Res. 2005;11(4):1380–1385. doi: 10.1158/1078-0432.CCR-04-1773. [DOI] [PubMed] [Google Scholar]
- 42.Fang L., Du W.W., Yang W., Rutnam Z.J., Peng C., Li H. MiR-93 enhances angiogenesis and metastasis by targeting LATS2. Cell Cycle. 2012;11(23):4352–4365. doi: 10.4161/cc.22670. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The dysregulated lncRNAs in four datasets.
The summary of dysregulated lncRNAs.