Abstract
The progressive loss of midbrain (MB) dopaminergic (DA) neurons defines the motor features of Parkinson disease (PD), and modulation of risk by common variants in PD has been well established through genome-wide association studies (GWASs). We acquired open chromatin signatures of purified embryonic mouse MB DA neurons because we anticipated that a fraction of PD-associated genetic variation might mediate the variants’ effects within this neuronal population. Correlation with >2,300 putative enhancers assayed in mice revealed enrichment for MB cis-regulatory elements (CREs), and these data were reinforced by transgenic analyses of six additional sequences in zebrafish and mice. One CRE, within intron 4 of the familial PD gene SNCA, directed reporter expression in catecholaminergic neurons from transgenic mice and zebrafish. Sequencing of this CRE in 986 individuals with PD and 992 controls revealed two common variants associated with elevated PD risk. To assess potential mechanisms of action, we screened >16,000 proteins for DNA binding capacity and identified a subset whose binding is impacted by these enhancer variants. Additional genotyping across the SNCA locus identified a single PD-associated haplotype, containing the minor alleles of both of the aforementioned PD-risk variants. Our work posits a model for how common variation at SNCA might modulate PD risk and highlights the value of cell-context-dependent guided searches for functional non-coding variation.
Keywords: Parkinson disease, enhancer, regulatory variation, ATAC-seq, dopaminergic neurons, chromatin accessibility, alpha-synuclein (SNCA)
Introduction
Parkinson disease (PD) is a common progressive neurodegenerative disorder characterized by preferential and extensive degeneration of dopaminergic (DA) neurons in the substantia nigra.1, 2 This loss of midbrain (MB) DA neurons disrupts the nigrostriatal pathway and results in the movement phenotypes observed in PD. Although this disorder affects approximately 1% of people over 70 years old worldwide,3 the mechanisms underlying genetic risk of sporadic PD in the population remain largely unknown. Familial cases of PD with known pathogenic mutations are better understood but account for ≤10% of PD cases.4
The α-synuclein gene (SNCA) is commonly disrupted in familial PD through missense mutations predicted to promote misfolding5, 6, 7 or genomic multiplications, resulting in an overexpression paradigm.8 The SNCA locus has also been shown by genome-wide association studies (GWASs) to harbor common variants, modulating risk of sporadic PD.9 In the same way, common variants at more than 40 additional loci have been implicated in PD,10 but the causal variants which are responsible for elevating risk, and the genes they modulate in doing so, remain largely undetermined.
That most GWAS-implicated variants are non-coding11 is a major source of this uncertainty, obstructing the identification of: (1) the causative variant at a locus; (2) the context in which a variant is acting; and (3) the mechanism by which a variant asserts its effect on disease risk.
GWASs are inherently biologically agnostic, and their exploitation of linkage disequilibrium (LD) structure frequently results in the implication of many variants at a particular locus, but no one variant is prioritized over those in LD. One method to prioritize non-coding variants is to examine the chromatin status at that locus.11, 12, 13 Accessible chromatin is more likely to be functional, and variants therein might impact that activity more so than those variants residing in inaccessible chromatin. Recent studies have prioritized neuropsychiatric variants through examination of the chromatin status of iPSC-derived neurons or post-mortem whole brain tissues.14, 15 However, chromatin accessibility is dynamic and often varies across cell types and developmental time; therefore, understanding and isolating the in vivo cellular context in which variation acts is critical to increasing our ability to prioritize variants and query their methods of action.11, 16, 17, 18
By exploiting the preferential vulnerability of MB DA neurons in PD, we have prioritized DA neurons as the biological context in which a fraction of PD-associated variants are likely to act. DA neurons in other brain regions, such as the forebrain (FB), provide a related substrate that is less vulnerable to loss in PD. We sought to use chromatin data from ex vivo populations of DA neurons to investigate the contributions of non-coding variation to PD risk. To maximize the specificity of the biological context, we generated chromatin signatures for purified mouse MB and FB DA neurons. We examined the resulting regulatory regions for their ability to direct in vivo reporter expression and developed a regulatory sequence vocabulary specific to DA neurons. In doing so, we identified a novel MB DA regulatory element that falls within intron 4 of SNCA and demonstrated its ability to direct reporter expression in catecholaminergic neurons from transgenic mice and zebrafish, confirming it to be an enhancer. Furthermore, this enhancer harbors two common variants, falling within a haplotype that we determine to be associated with PD risk. We demonstrate that these enhancer variants impact protein binding, and we propose a model for how the variants and the haplotype at large contribute to SNCA regulatory control. This work illustrates the power of cell-context-dependent guided searches for the identification of disease-associated and functional non-coding variation.
Material and Methods
Animal Husbandry
Tg(Th-EGFP)DJ76Gsat mice (Th-EGFP) were generated by the GENSAT project19 and purchased through the Mutant Mouse Resource and Research Centers Repository. Colony-maintenance matings were between hemizygous male Th-EGFP mice and female Swiss Webster (SW) mice, obtained from Charles River Laboratories. This same mating scheme was used for establishing timed matings and thus generating litters for assay; the day on which the vaginal plug was observed was E0.5. Adult AB zebrafish lines were maintained in system water according to standard methods.20 All work involving mice and zebrafish (husbandry, colony maintenance, procedures, and euthanasia) were reviewed and pre-approved by the institutional care and use committee.
Neural Dissociation and Fluorescence-Activated Cell Sorting
Pregnant SW mice were euthanized at E15.5, and the embryos were removed and immediately placed in chilled Eagle’s Minimum Essential Medium (EMEM) on ice. The embryos were decapitated, and the brains were removed into Hank’s Balanced Salt Solution without Mg2+ and Ca2+ (HBSS w/o) on ice. Under a fluorescent microscope, EGFP+ brains were identified and microdissected to yield the desired MB and FB regions. Microdissected regions were placed in fresh HBSS w/o on ice and pooled per litter for dissociation.
Pooled brain regions were dissociated via the Papain Dissociation System (Worthington Biochemical Corporation). The tissue was dissociated in the papain solution for 30 min at 37°C, and gentle trituration was performed every 10 min with a sterile Pasteur pipette. After dissociation, cells were passed through a 40 μm cell strainer into a 50 mL conical tube, centrifuged for 5 min at 300 × g, resuspended in albumin-inhibitor solution containing DNase, applied to a discontinuous density gradient, and centrifuged for 6 min at 70 × g. The resulting cell pellet was resuspended in HBSS with Mg2+ and Ca2+ and submitted to fluorescence-activated cell sorting (FACS). Aliquots of 50,000 EGFP+ cells were sorted directly into 300 μL HBSS with Mg2+ and Ca2+ and 10% FBS for ATAC-seq. Aliquots containing ≥50,000 EGFP+ cells were sorted into kit-provided lysis buffer for RNA-seq. This procedure was repeated such that a single aliquot of cells from each region per litter was submitted to either ATAC-seq or bulk RNA-seq, repeated three (ATAC-seq) or four (RNA-seq) times for each region.
ATAC-seq Library Preparation and Quantification
ATAC-seq library preparation generally followed the steps as set out in the original ATAC-seq paper,21 with minor modifications. Aliquots of 50,000 EGFP+ cells were centrifuged for 5 min at 4°C and 500 × g, washed with 50 μL of chilled PBS, and centrifuged again for 5 mins at 4°C and 500 × g. Next, the cell pellet was resuspended in lysis buffer, as set out in the protocol, and cells were left to lyse for 5 min at 4°C before being centrifuged for 10 min at 4°C at 500 × g. The resulting nuclei pellet was tagmented, as written, using the transposase from the Nextera DNA Library Preparation Kit. After transposition, DNA was purified with the MinElute Reaction Clean-up Kit (QIAGEN) and eluted in 10 μL elution buffer.
The libraries were amplified according to the original ATAC-seq protocol.21 The qPCR surveillance steps were modified such that the additional number of cycles of amplification were calculated as ¼ maximum intensity, so as to limit PCR duplication rates in the final libraries. The amplified libraries were purified with Ampure XP beads (Beckman Coulter) according to the Nextera DNA Library Prep Protocol Guide. The libraries were quantified with the Qubit dsDNA High Sensitivity Assay (Invitrogen) in combination with the High Sensitivity DNA Assay (Agilent) on the Agilent 2100 Bioanalyzer.
ATAC-seq Sequencing, Alignment, and Peak Calling
Individual ATAC-seq libraries were sequenced on the Illumina MiSeq to a minimum depth of 15 million, 2 × 75 bp reads per library. A single MB ATAC-seq library was sequenced on the Illumina HiSeq in rapid run mode with 2 × 100 bp reads to a depth of ≥350 million paired-end reads.
The quality of sequencing was evaluated with FastQC (v0.11.2). Reads were aligned to mm9 with Bowtie222 (v2.2.5) under --local mode. Reads aligning to the mitochondrial genome; unknown and random chromosomes; and PCR duplicates were removed prior to peak calling (SAMtools23). Peaks were called on individual libraries and on a concatenated file combining all MB or all FB (“joint”) libraries via MACS224 (v2.1.1.20160309) “callpeak” with the following options: –nomodel –nolambda -B -f BAMPE –gsize mm –keep-dup all. Peaks overlapping blacklisted regions that were called by ENCODE and that were in the original ATAC-seq paper were removed.21, 25 Peaks were examined for their genomic distribution via CEAS in the Cistrome pipeline.26, 27 The fragment lengths were extracted from the SAM files and plotted with a custom script. Mouse (mm9) transcriptional start site (TSS) coordinates were extracted from the UCSC Genome Browser,28 and deepTools29 was used for quantifying the pileup of reads over TSSs.
RNA-seq Library Preparation and Quantification
Total RNA was extracted with the Purelink RNA Micro Kit (Invitrogen). After FACS isolation into kit-provided lysis buffer, samples were homogenized, and RNA extraction proceeded according to the manufacturer’s recommendations. Total RNA integrity was determined with the RNA Pico Kit (Agilent) on the Agilent 2100 Bioanalyzer. RNA samples were sent to the Sidney Kimmel Comprehensive Cancer Center Next Generation Sequencing Core at Johns Hopkins for library preparation with the Ovation RNA-Seq System V2 (Nugen) and for sequencing.
RNA-seq Sequencing, Alignment, and Transcript Quantification
The libraries were pooled and sequenced on Illumina’s HiSeq 2500 in rapid-run mode with 2 × 100 bp reads to an average depth of >90 million reads per library. The quality of sequencing was evaluated via FastQC. FASTQ files were aligned to mm9 with HISAT230 (v2.0.1-beta) with --dta specified.
Aligned reads from individual samples were quantified against a reference transcriptome with the Rsubread package31, 32, 33 (v1.22.3) function “featureCounts” with the following options: isPairedEnd = TRUE, requireBothEndsMapped = TRUE, isGTFAnnotationFile = TRUE, and useMetaFeature = TRUE. The GENCODE vM9 GTF was downloaded34 (date: March 30, 2016) and lifted over from the mm10 genome to the mm9 genome with CrossMap (v0.2.2) under default parameters.35 This was used for quantification, in which gene-level raw counts were converted to RPKM (reads per kilobase of transcript per million) values and means for each region were calculated.
Relationship between RNA-seq and ATAC-seq
The 1,000 most highly expressed genes and the 1,000 least highly expressed genes (RPKM ≥ 1) in both the MB and FB were identified, and their transcriptional start sites (Ensembl) were extracted from the UCSC Table Browser.28 Intervals of 1, 10, and 100 kb surrounding these TSSs were intersected with the ATAC-seq libraries, and the overlap quantified36 and plotted. These same TSSs were provided to deepTools,29 and the ATAC-seq signal over these most highly- and least highly-expressed genes was quantified and plotted. Additionally, the 1,000 highest and lowest ATAC-seq peaks (by q value) were extracted, and the expression of the nearest gene was quantified and plotted as a final metric to relate the RNA-seq and ATAC-seq datasets.
cDNA Synthesis and RT-qPCR for DA Neuron Markers
RNA was extracted with the RNeasy Mini Kit (QIAGEN) after 50,000 cells were sorted directly into Buffer RLT. Aliquots of 50,000 non-fluorescing cells were also collected and processed in parallel. 100 ng of each RNA sample was submitted to first-strand cDNA synthesis with the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) according to the oligo(dT) method.
Primers (Table S1) were designed with Primer-BLAST37 under default parameters, except the requirement for exon-exon junction spanning was specified. qPCR was performed with Power SYBR Green Master Mix (Applied Biosystems). Reactions were run in triplicate under default SYBR Green Standard cycle specifications on the Viia7 Real-Time PCR System (Applied Biosystems). Relative quantification followed the 2-ΔΔCT method, and results were normalized to Actb in the EGFP− aliquot of cells for each region.
Correlation Analysis between Regions and within Replicates
Peaks from all six ATAC-seq libraries and the two “joint” ATAC-seq libraries were concatenated together, sorted on the basis of chromosomal location, merged into a unified peak set,36 and converted to Simplified Annotation Format. Reads from each BAM file overlapping this unified peak set were quantified with the Rsubread package’s “featureCounts” command, with the following options: isPairedEnd = TRUE and requireBothEndsMapped = FALSE. Read counts were normalized for each library via conditional quantile normalization;38 library size, peak length, and peak GC content were accounted for. Pearson correlation coefficients were calculated from this normalized count matrix and visualized with corrplot, RColorBrewer, and LSD (see Web Resources).
Sequence-Constraint Analysis
Average phastCons39 were calculated for the “joint” peak file for both the MB and FB libraries with Cistrome.27 Beforehand, peaks with overlap of exons or promoters (defined here as ± 2,000 bp from the transcriptional start site) were removed. The exon and promoter BED files were downloaded from the UCSC Table Browser28 (Mouse genome; mm9 assembly; Genes and Gene Predictions; RefSeq Genes track using the table refGene).
Gene Ontology of Nearest Expressed Gene
The Genomic Regions Enrichment of Annotations Tool40 (GREAT; v3.0.0) predicted the gene ontology (GO) term enrichment in the catalogs. Beforehand, peaks were processed so that (1) peaks overlapping commonly open regions would be removed; (2) the top 20,000 peaks would be selected; (3) peaks would overlap the nearest expressed gene’s transcriptional start site (TSS).
First, regions that are commonly open were defined as those regions of the genome that are open in >30% of ENCODE DNase hypersensitivity site (DHS) assays in mouse tissues. These ubiquitously open regions were removed from the peak files. Next, so that the binomial distribution for calculating enrichment was still valid, we limited the number of regions considered by GREAT by only submitting the top 20,000 peaks on the basis of q value.
Finally, in order to limit ourselves to the nearest expressed gene, we supplied a list of the TSSs of the nearest expressed genes, considering only those genes that are used by GREAT.
Only genes that are in this list and for which RPKM >1 were considered to be expressed. The expressed gene nearest to each of the top 20,000 peaks was identified. Each peak is associated with its nearest expressed gene, and to ensure that GREAT only considered these nearest genes for analysis, we submitted these nearest expressed genes’ TSSs as a proxy for each peak. We submitted these proxy peaks to GREAT by using the NCBI build 37 (mm9) assembly under the “whole-genome background regions” setting; we used the single nearest gene as the association rule and included curated regulatory domains.
Quantification of Overlap between CRE Catalogs and the VISTA Enhancer Browser
All elements tested in vivo were downloaded from the VISTA Enhancer Browser on September 4, 2016. These regions were stratified into those annotated as positive and those annotated as negative. BED coordinates of these regions were extracted and intersected with the ATAC-seq catalogs. Positive regions were further stratified into those with annotations for only forebrain, only midbrain, or only hindbrain; combinations of regions (“multiple regions”); or all three regions (“whole brain”). These categories comprised the “neuronal” category. Other regions that were annotated as positive but not driving expression in any of those three regions were placed in the “non-neuronal” category.
Testing Five Putative CREs for In Vivo Reporter Activity
Prioritized regions were amplified (Table S1) from human genomic DNA via PCR and cloned into either pENTR for mouse lacZ assays (Invitrogen) or pDONR221 for zebrafish assays (Invitrogen). Sequences were validated, and regions were cloned via LR cloning (Invitrogen) into either an hsp68-lacZ vector or pXIG vector, with a TdTomato cassette in place of GFP.
Generation of transgenic mice and E11.5 embryo-staining were performed as previously described41, 42, 43 with FVB strain mice. Embryos expressing the lacZ reporter gene were scored and annotated for their expression patterns by multiple curators. For a construct to be considered positive, a minimum of three embryos per construct were required to demonstrate reporter activity in the same tissue. Mouse transient transgenic assays were approved by the Lawrence Berkeley National Laboratory Animal Welfare and Research Committee.
Generation of transgenic zebrafish was performed as previously described44 in AB zebrafish. Reporter expression patterns were evaluated at both 3 and 5 days post-fertilization (dpf). For a construct to be considered as positive, ≥25% of mosaic embryos had to display reporter activity in one or more anatomical structures. Positive zebrafish were quantified for reporter activity in five anatomical regions (forebrain, midbrain, hindbrain, amacrine cells, and the spinal cord).
Development of Regulatory Vocabulary
We applied the machine-learning algorithm gkm-SVM45 to our MB and FB catalogs under default settings. We trained gkm-SVM on the sequences underlying the summits ±250 bp of non-ubiquitously open, top 10,000 peaks by q value versus five negative sets, which were matched for GC content, length, and repeat content. Weights across all five tests were averaged for all 10-mers.
All 10-mers with weight ≥1.50 were clustered on the basis of sequence similarity via Starcode;46 sphere clustering was specified, and the distance was set to 3. clustalOmega47 aligned the sequences within these clusters, and MEME,48 under default parameters except for -dna -maxw 12, generated position weight matrices (PWMs) for these aligned clusters. Tomtom,49 querying the Jolma 2013, JASPAR Core 2014, and Uniprobe mouse databases, identified the top transcription factors corresponding to these PWMs, under default parameters except for -no-ssc -min-overlap 5 -evalue -thresh 10.0.
We used the same procedure to identify transcription factors specifically conveying regulatory potential in the MB library relative to the FB library; except during gkm-SVM training, the positive set was specified as the top 10,000 non-ubiquitously open MB summits, and the negative set was specified to be the top 10,000 non-ubiquitously open FB summits, both ±250 bp.
Transcription-Factor Footprinting
CENTIPEDE50 was used for footprint identification. Sequences underlying the deeply sequenced MB library peaks, less those that were ubiquitously open, were extracted. FIMO,51 with options –text –parse-genomic-coord, identified all locations underlying ATAC-seq peaks of the motifs identified above. Additionally, conservation data from 30-way vertebrate phastCons were considered in the CENTIPEDE calculations; for each PWM site, those with a mean conservation score greater than 0.9 were considered. Finally, the BAM file read-end coordinates were adjusted in response to the shift resulting from the transposase insertion.52 As such, reads were adjusted +4 bp on the positive strand and −5 bp on the negative strand as performed in the original ATAC-seq method.21
Genome-wide Read Pileup over Predicted Motif Sites
FIMO, as above, was used for genome-wide identification of all coordinates of the identified motifs. The deepTools'29 “bamCoverage” tool was run, under default conditions, for conversion of the deeply sequenced MB library BAM to bigwig format. After this, a matrix file was generated with “computeMatrix;” specified options were –referencePoint center -b 1000 -a 1000 -bs 50. Finally, “plotHeatmap” was used for generating plots indicating ATAC-seq read pileup over predicted motif sites.
Intersection of CRE Catalogs and PD-Associated GWAS Variants
Lead SNPs from two of the most recent meta-analyses9, 10 were submitted to rAggr (see Web Resources), and SNPs in LD were identified (1000 Genomes, phase 3, EUR populations; minimum minor-allele frequency [MAF] = 0.05, r2 ≥ 0.8; maximum distance = 5,000 kb). These variants were intersected36 with the CRE catalogs after they were lifted over to hg19 coordinates, and the overlap was extracted and quantified.
In Vivo Validation of the MB-Specific Enhancer
The MB-specific peak was amplified (Table S1) from human genomic DNA via PCR and cloned into pCR8 via TA cloning (Invitrogen). Sequences were validated, and regions were cloned into either an hsp68-lacZ vector or a modified pXIG vector, with a TdTomato cassette in place of GFP, via LR cloning (Invitrogen).
For zebrafish transgenesis, the modified pXIG vector was injected into 1- to 2-cell-stage embryos as previously described44 in AB zebrafish. TdTomato reporter expression was assayed at 72 hr post-fertilization (hpf) and 5 dpf; mosaic embryos positive for TdTomato expression were selected and raised to adulthood, and founders were identified. The progeny of founders were screened at 72 hpf for reporter activity. For mouse transgenesis, the generated hsp68-lacZ vector was purified in a double CsCl gradient (Lofstrand Labs), and stable mouse transgenesis was performed in C57BL/6 mice by Cyagen Biosciences. Multiple founder lines were generated. For lacZ staining, embryos were collected at E12.5, and mouse brains were isolated at E15.5, P7, P30, and P574. Brains were roughly sectioned in 1 mm sections at P7 and P30. The animals were perfused at P574, and fixed brains were sectioned (200 μm) with a vibratome. Specimens were subsequently fixed for 2 hr on ice in 1% formaldehyde, 0.2% glutaraldehyde, and 0.02% Igepal CA-630 in PBS. After fixation, tissues were permeabilized over the course of three 15 min washes in 2 mM MgCl2 and 0.02% Igepal CA-630 in PBS at room temperature. Embryos and/or tissues were incubated overnight at 37°C in a staining solution containing 320 μg/mL X-Gal in N,N-dimethyl formamide, 12 mM K-ferricyanide, 12 mM K-ferrocyanide, 0.002% Igepal CA-630, and 4 mM MgCl2 in PBS. Specimens were washed twice for 30 min each time in 0.2% Igepal CA-630 in PBS and finally stored in 4% formaldehyde, 100 mM sodium phosphate, and 10% methanol.
Individuals with and without PD-Sequencing and Genotyping at SNCA
986 individuals with PD and 992 controls who were all seen at the Mayo Clinic in Jacksonville, FL were sequenced across the putative enhancer and genotyped for 25 variants across the SNCA locus. The variants chosen for genotyping were those identified during the sequencing of the enhancer and a subset of those identified by Guella et al.53 For PD-affected individuals, median age at blood draw was 69 years (range: 28–97 years), median age at PD onset was 67 years (range: 28–97 years), and 631 subjects (64.0%) were male. Median age at blood draw for control individuals was 67 years (range: 18–92 years), and 415 subjects (41.8%) were male. Individuals with PD were diagnosed according to standard clinical criteria.54 All subjects are unrelated, non-Hispanic Caucasians of European descent. The Mayo Clinic Institutional Review Board approved the study, and all subjects provided written informed consent.
Genomic DNA was extracted from whole blood with the Autogen FlexStar. Sanger sequencing of the enhancer region was performed bidirectionally with the ABI 3730xl DNA analyzer (Applied Biosystems) according to standard protocols. Sequence data were analyzed with SeqScape (v2.5; Applied Biosystems). Statistical analyses were performed with both SAS and R (see Web Resources). Of the variants identified within the enhancer, only those with a MAF greater than 5% were evaluated for association with PD in single-variant analysis. Associations between individual variants and PD were evaluated with logistic regression models, adjusted for both sex and age at blood draw, and, when variants were considered, under an additive model (i.e., the effect of each additional minor allele was evaluated). Odds ratios and 95% confidence intervals were estimated. A Bonferroni correction for multiple testing was utilized in single-variant analysis because of the four common variants that were evaluated for association with PD, after which p values ≤0.0125 were considered as statistically significant.
Genotyping of the 25 SNPs across the SNCA locus was performed with the iPLEX Gold protocol on the MassARRAY System and analyzed in TYPER 4.0 software (Agena Bioscience). For the 25 SNPs genotyped across the SNCA locus, all genotype call rates were >95%, and there was no evidence for departure from Hardy-Weinberg equilibrium (all χ2 p values >0.05 after Bonferroni correction). Haplotype frequencies in cases and controls were estimated with the haplo.stats package function “haplo.group” (see Web Resources). The haplo.score function was used to perform score tests for association evaluating the relationships between haplotypes and the risk of PD.55 Tests were adjusted for both sex and age at blood draw, haplotypes occurring in less than 1% of subjects were excluded, and only individuals with no missing genotype calls for any variants were included. A Bonferroni correction for multiple testing was applied as a result of the 12 different common haplotypes that were observed and tested for association with PD risk, after which p values ≤0.0042 were considered as statistically significant.
LD structure and r2 values at the SNCA locus in the 1000 Genomes EUR population were extracted from LDlink56 with the LDmatrix tool and plotted with R. The chromatin structure at SNCA was extracted from the 3D Genome Browser, and POLR2A binding in MCF-7 cells was examined at the SNCA promoter.
Protein Array Testing Differential Binding
HuProt v3.1 human proteome microarrays (Grace Bio-Labs) containing >16,000 unique proteins representing 12,586 genes (CDI laboratories)58 were blocked with 25 mM HEPES (pH 8.0), 50 mM potassium glutamate, 8 mM MgCl2, 3 mM dithiothreitol (DTT), 10% glycerol, 0.1% Triton X-100, and 3% BSA on an orbital shaker at 4°C for ≥3 hr. Allele-specific protein-DNA binding interactions were identified through dye-swap competition of major and minor alleles labeled with either Cy3 or Cy5. DNA fragments for dbSNP: rs2737024 and dbSNP: rs2583959 were synthesized such that each allele was flanked by 15 nucleotides of the upstream and downstream sequence and there was a common priming site at the 3′ end (Table S1).
We created the dsDNA fragments by separately annealing a primer containing a Cy3 or Cy5 label and adding Klenow (New England Biolabs) with dNTP to fill in the complementary strand for each allele.59 Cy3-labeled major allele was mixed with Cy5-labeled minor allele (each at 40 nM) in 1× hybridization buffer (10 mM Tris-Cl [pH 8.0], 50 mM KCl, 1 mM MgCl2, 1 mM DTT, 5% glycerol, 10 μM ZnCl2, and 3 mg/mL BSA) and added to an array; dyes were then swapped for each allele, and the mixture was then added to a second array. DNA was allowed to bind overnight at 4°C on an orbital shaker with protection from light. Chips were washed once with cold 1× Tris-buffered saline-Triton solution (0.1% Triton X-100) for 5 min at 4°C, rinsed, and dried in the centrifuge. Cy5 and Cy3 images were taken separately on a Genepix 4000B scanner and, after alignment to the GAL file, individual spot intensities were extracted with Genepix Pro software.
Allele-specific interactions were identified through dye-swap analysis. The ratio of major to minor allele binding was calculated from the duplicate spot-average, median-foreground signal for each protein according to the following equation:
Mean intensity was calculated from the average foreground signal for the Cy3 and Cy5 channels of the major and minor alleles. MA plots were made for each allele using the calculated mean intensity and the log ratio of the major to the minor allele.
Results
ATAC-seq Identifies Open Chromatin in MB and FB DA Neurons
To identify open chromatin regions (OCRs) in DA neurons, we performed ATAC-seq21 on ∼50,000 FACS-isolated cells (per replicate) from microdissected regions of the MB and FB of embryonic day 15.5 (E15.5) Tg(Th-EGFP)DJ76Gsat BAC transgenic mice60 (Figure 1A). EGFP, expressed under the control of the tyrosine hydroxylase (Th) locus, labels catecholaminergic neurons (i.e.: DA, noradrenergic, and adrenergic neurons) in this mouse line. To confirm capture of the corresponding catecholaminergic neurons, we performed RT-qPCR on the isolated reporter-labeled cells and established them to be enriched for DA neuronal markers in comparison to unlabeled populations from the same dissected tissues (Figure S1).
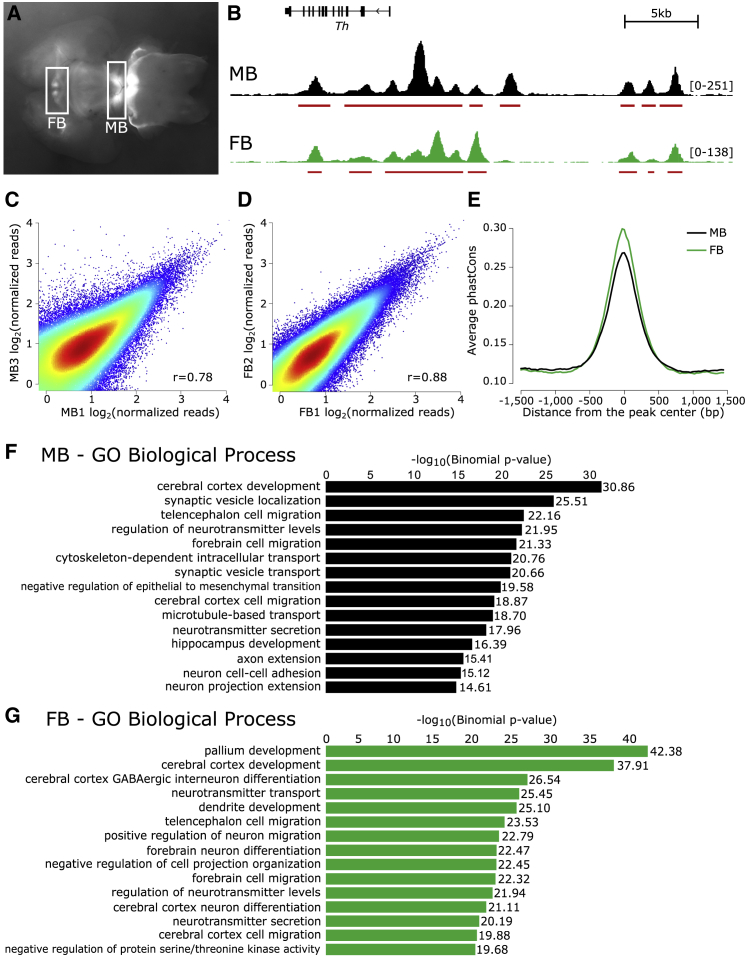
Figure 1.
Preliminary Validation of ATAC-seq Catalogs Generated from Ex Vivo DA Neurons
(A) The midbrain (MB) and forebrain (FB) of E15.5 brains from Tg(Th-EGFP)DJ76Gsat mice are microdissected, dissociated, and isolated by FACS.
(B) Read pileup and called peaks for the MB and FB libraries at the Th locus.
(C and D) Chromatin accessibility, genome-wide, is correlated between replicates.
(E) The sequences underlying MB and FB peaks display a high degree of evolutionary sequence constraint as measured by phastCons scores.
(F and G) For both the MB and FB, gene ontology terms of the expressed genes nearest to each peak reflect the neuronal origin and function of these catalogs.
To evaluate the ATAC-seq libraries, we examined in silico quality-control measures (Figure S2); evaluated the called peaks and read pileups with the Integrative Genomics Viewer (IGV);61, 62 and quantified the correlation between brain regions and within replicates. A representative browser trace at the Th locus in both MB and FB libraries is presented in Figure 1B. Replicates are well correlated: MB library replicates have an average correlation of 0.72 (Figure 1C), and FB replicates are more highly correlated, at r = 0.86 (Figure 1D). Given the robust correlation between replicates, we pooled all reads from the same brain region and called peaks on this unified set to increase our power to detect regions of open chromatin. As a result, we identified 104,217 regions of open chromatin in the MB DA neurons and 87,862 regions in the FB. MB and FB libraries are moderately well correlated (average r = 0.64; Figure S3), and approximately 60% of MB OCRs are also represented in the FB libraries.
To assess these catalogs for characteristics of functionality, we examined the sequence constraint underlying the called regions of open chromatin but excluded peaks that overlap promoters. Promoters are typically accessible,63 and thus we aimed to reduce the inflation that affects sequence conservation as a result of highly conserved promoter-overlapping ATAC-seq peaks. Despite removal of these highly conserved peaks, we found that the degree of sequence constraint underlying open-chromatin peaks was high in comparison to background constraint (Figure 1E). The fact that elements in these libraries of putative cis-regulatory elements (CREs) are constrained highlights their probable functional significance.
To further examine the OCR catalogs for biological relevance, we explored the GO terms of nearby genes. Although CREs are not restricted to acting solely on the nearest gene, this restriction is often used as a proxy in the absence of other information. To bolster our predictions, we have also generated bulk RNA-seq data on these same populations of sorted cells (Figure S4) and used these data to examine the GO terms of the nearest expressed gene (RPKM ≥1). Although still imperfect, implementing this method as a proxy for function results in GO terms that are enriched for neuronal functions in both MB and FB OCR catalogs (Figures 1F and 1G). Thus, we establish that these OCR catalogs are enriched for putative CREs, probably directing the expression of genes with key roles in neuronal biology.
Candidate Regulatory Regions Are Capable of Directing Expression In Vivo
Although sequence conservation and GO suggest that our OCR catalogs are enriched for functional elements, both of these metrics are indirect surrogates for true measures of function. To more directly measure the biological relevance of the catalogs and to identify enhancers, we assessed the capability of the candidate CREs to direct expression in vivo.
We took advantage of the large repository of elements that have already been tested in lacZ reporter assays in vivo and that have been cataloged in the VISTA Enhancer Browser64 (accessed September 4, 2016). Overlap between our catalogs and all 2,387 VISTA Enhancer Browser elements, which were scored for their ability to direct lacZ reporter expression in E11.5 mice, was quantified (Table S2). Of the 1,264 VISTA elements identified as enhancers, 786 were present in the MB catalog, and 719 were present in the FB catalog (Figure 2A). We examined the overlap of the FB and MB catalogs with enhancers that have been demonstrated to direct expression in either non-neuronal or neuronal tissues, and we observed that 42%–47% of enhancers reported to direct expression in non-neuronal tissues are present in the catalogs. In contrast, 71%–76% of enhancers that direct expression in one or more regions of the brain overlapped the FB and MB catalogs (Figure 2B), confirming an abundance of brain enhancers in our catalogs. Stratifying these confirmed neuronal enhancers on the basis of their expression patterns in VISTA, we observed an abundance of MB-specific enhancers in our MB catalog and an abundance of FB-specific enhancers in our FB catalog; 77% of MB- and FB-specific enhancers in VISTA were captured in our MB and FB catalogs, respectively (Figure 2C). Collectively, these data establish that our region-specific OCR catalogs capture region-specific, active CREs with high efficiency.
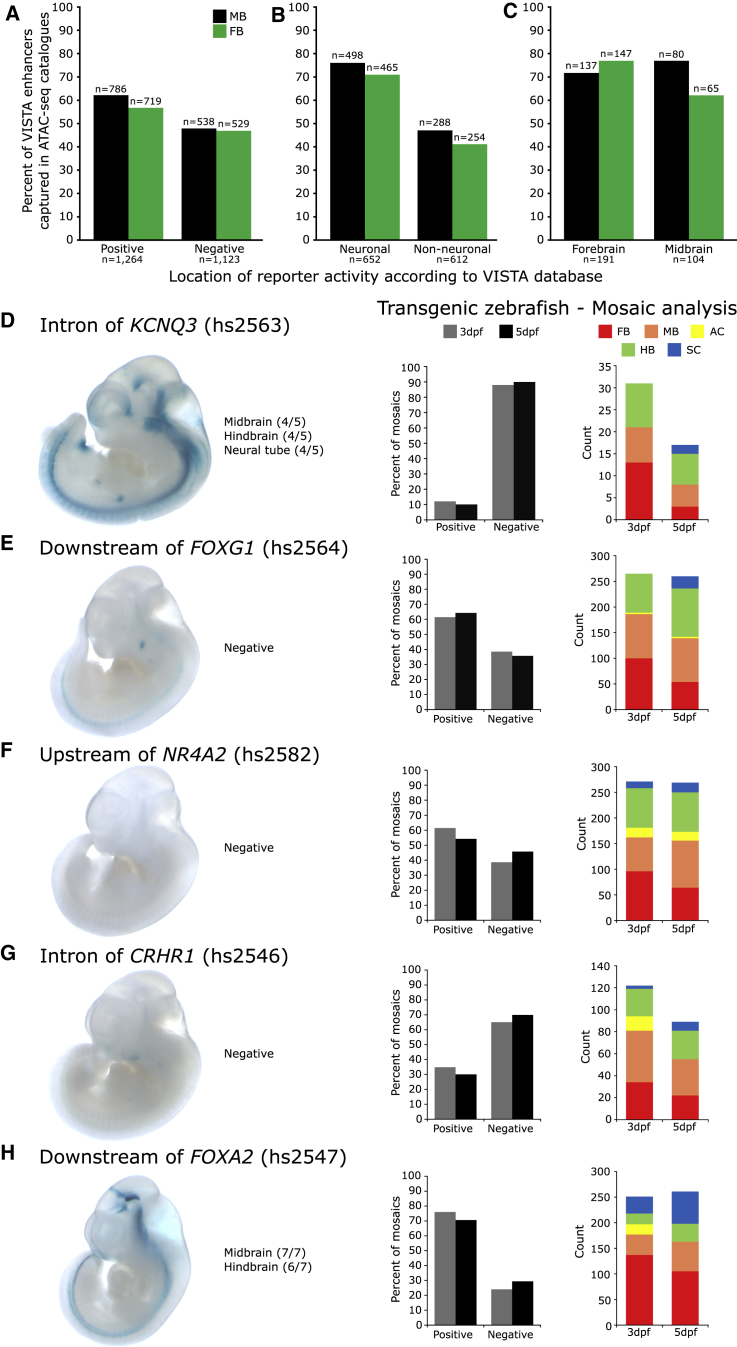
Figure 2.
Validation of the Putative CRE Catalogs In Vivo
(A) Of the elements annotated in VISTA as having enhancer activity, 62% and 56% of these are represented in the MB and FB catalogs, respectively.
(B) An abundance of open-chromatin regions in the MB and FB catalogs overlap confirmed neuronal enhancers (≥70%).
(C) Neuronal enhancers were stratified by the anatomical domains in which they are active; those that are reported active in the MB and FB are enriched in our MB and FB catalogs, respectively.
(D–H) Testing five prioritized putative CREs in vivo identifies five neuronal enhancers.
(D) A putative CRE in intron 1 of KCNQ3 directs expression in the midbrain, hindbrain, and neural tube of E11.5 lacZ reporter mice. It fails to direct expression in a transgenic zebrafish assay at either 3 or 5 days post fertilization (dpf); reporter expression is present in ≤25% of mosaics.
(E, F, and G) Putative CREs downstream of FOXG1, upstream of NR4A2, and in an intron of CRHR1 fail to direct expression in transgenic mice; however, they direct robust neuronal appropriate expression in transgenic zebrafish reporter assays (scored for expression in MB, FB, amacrine cells [ACs], hindbrain [HB], and spinal cord [SC]).
(H) A putative CRE downstream of FOXA2 directs neuronal expression in both transgenic mice and zebrafish assays. n mosaic zebrafish scored: ≥141 for 3 dpf, ≥119 for 5 dpf. All constructs have since been deposited in the VISTA database under the supplied hs numbers.
To extend our assessment of the biological activity of sequences within these OCR catalogs, we focused on an additional five candidate CREs not already tested in the VISTA browser and evaluated their ability to act as enhancers in lacZ reporter mice and in transgenic zebrafish TdTomato reporter assays. All five regions were represented by robust peaks in both the MB and FB catalogs (Figure S5). Two regions, one in the first intron of Kcnq3 and the other downstream of Foxg1, were additionally prioritized via H3K27ac ChIP-seq from a variety of tissues taken from E11.5 and E15.5 embryonic mice because we sought to limit our selection to candidate enhancers predicted to have neuronal-specific activity. The remaining three candidate CREs were selected on the basis of their proximity to genes important in DA neuron biology. We selected sequences at Foxa2 and Nr4a2 because both are key transcription factors (TFs) in the development and maintenance of DA neurons.65, 66, 67, 68 We selected the final region, located in an intron of Crhr1, because this locus has been implicated in PD by GWAS,9 and our group has recently prioritized this gene as a candidate for PD risk.69 All selected sequences were lifted over to hg19, a process which facilitated the identification and assay of their corresponding human sequence intervals.
When tested in transgenic reporter mice at E11.5 (Figure S5), two of the five regions (those near KCNQ3 and FOXA2) were validated as enhancers (Figures 2D and 2H). Recognizing that a disparity exists between the developmental time at which we generated the catalogs (E15.5) and the time at which the mice were assayed (E11.5), and that this disparity might compromise validation rates, we also assayed each sequence across multiple time points in zebrafish. All assayed regions except the region at KCNQ3 directed reporter expression in mosaic transgenic zebrafish (Figures 2E–2H). All five regions displayed enhancer activity in vivo in neuronal tissues in one or both transgenic assays. Our transgenic animal experiments corroborate the results of the retrospective VISTA Enhancer Browser intersection, implying that our OCR catalogs are biologically active and enriched for sequences capable of driving neural expression in vivo.
Candidate CREs Are Enriched for TF Motifs Active in DA Neurons
To identify sequence modules (kmers) predicted to contribute regulatory activity of putative CREs in our catalogs, we applied the machine learning algorithm gkm-SVM.45 The resulting regulatory vocabularies of kmers had high predictive power (auROCMB = 0.915, auROCFB = 0.927). We rank-ordered and collapsed related kmers to reveal motifs enriched in the OCRs and their corresponding TFs (Figures 3A, 3E, 3I, and 3M). In the MB, the four most enriched motifs correspond to RFX1, FOXA2, ASCL2, and NR4A2. Given the degeneracy of binding motifs within TF families, we consulted the bulk RNA-seq data for each of the implicated TF families and examined the relative expression levels to prioritize which TFs are most likely producing the observed motif enrichments (Figures 3B, 3F, 3J, and 3N). For example, the reported DNA binding domain is highly conserved between RFX family members, and as a result, the predicted sequence motif for each is highly similar;70, 71 thus, we must use other means to identify which family member is likely to be acting in these cells. Although no member of the RFX family has been canonically associated with MB DA neurons, we expect Rfx3 and Rfx7, as the two most highly-expressed Rfx genes, to probably be active in MB DA neurons and be driving this motif enrichment (Figure 3B). FOXA1 and, especially, FOXA2 are both known to DA neuron biology65, 72 and both are highly expressed in the MB DA neurons (Figure 3F). Regarding enrichment for the ASCL family, ASCL1 is known to be involved in DA neuron biogenesis73 and is more highly expressed than any other TF in the family (Figure 3J). Finally, NR4A2 is both canonically associated with DA neurons and required for their development;68 we observe it to be highly expressed in MB DA neurons (Figure 3N). Examining the sequences underlying the OCR catalogs, we identified TF families known and unknown to DA neuron biology and further refined the TF associations by using expression data.
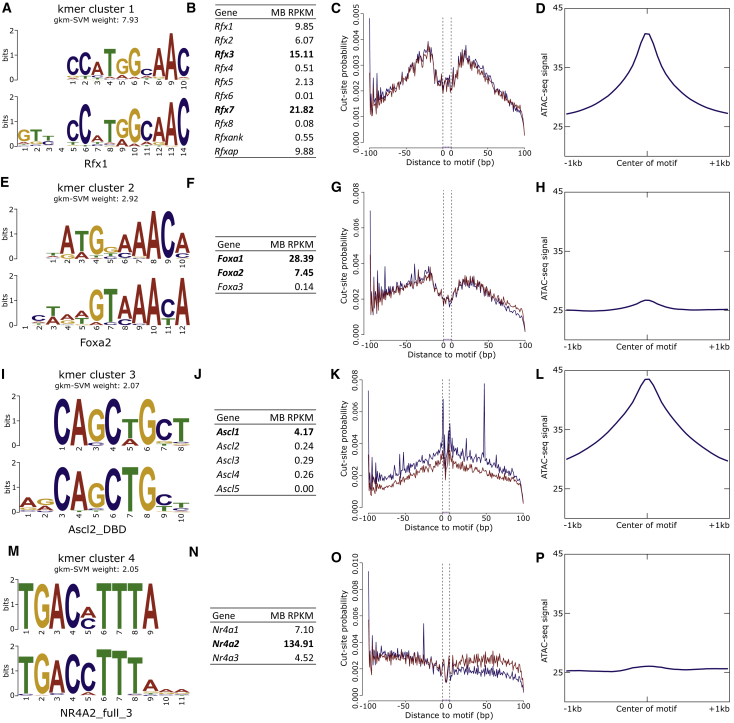
Figure 3.
Identification of Transcription Factors (TFs) Important to DA Neurons
(A) The kmer predicted to have the greatest regulatory potential underlying MB ATAC-seq peaks corresponds to the RFX family of TFs.
(B, C, and D) RNA-seq quantification in these same cells indicates this enrichment is likely due to RFX3 or RFX7 activity. Examining the ATAC-seq signal over predicted binding sites reveals a robust TF footprint (C) and a general enrichment of reads overlapping RFX sites genome-wide (D).
(E–H) Similarly, a kmer corresponding to FOXA1 and/or FOXA2 has similar evidence for the activity of one or both of these TFs.
(I–L) The third-ranked motif most likely corresponds to ASCL1, and although it fails to leave a robust TF footprint (K), there is clear enrichment of ATAC-seq signal overlapping genome-wide predicted ASCL1 binding sites (L).
(M–P) NR4A2, canonically associated with DA neuron biology, is identified as a highly expressed TF that probably contributes to the regulatory potential of the putative CREs; however, it fails to leave a TF footprint in the cut-site patterns around predicted motif sites (O) and is only mildly enriched for ATAC-seq reads over its predicted binding sites (P).
We also examined the qualities that differentiate MB CREs from FB CREs by examining the sequences underlying MB-specific and FB-specific regions. We developed a vocabulary that discriminates MB and FB regions with high predictive power (auROC = 0.926) and identified kmers enriched in MB-specific peaks where the top corresponding TFs are FOXA1 and/or FOXA2 and NR4A2 (Figure S6). We confirmed this MB bias by again considering the bulk RNA-seq for genes encoding these proteins. As expected, these TFs are more highly expressed in the MB where Nr4a2 is present at 12-fold higher levels than in the FB (135 RPKM in the MB versus 11 RPKM in the FB) and Foxa1 and Foxa2 are not expressed in the FB but are present in the MB (Foxa1: 28 RPKM, Foxa2: 7 RPKM). Not only do we identify FOXA1 and/or FOXA2 and NR4A2 as more active in MB DA neurons than in the FB, but we also did so solely by comparing their role in the vocabulary of MB-specific OCRs versus FB-specific OCRs.
In a parallel strategy to identify TFs actively engaging the DNA in MB DA neurons, we performed TF footprinting in a single, deeply sequenced MB ATAC-seq library. In doing so, we confirm that two of the TFs prioritized by gkm-SVM leave robust footprints. The motif corresponding to RFX-binding results in a dearth of cuts directly over predicted binding sites (Figure 3C). The same can be seen to a lesser extent for the motif corresponding to FOXA1 and/or FOXA2 (Figure 3G). By contrast, motifs corresponding to ASCL1 or NR4A2 fail to leave a robust mark on the chromatin availability (Figures 3K and 3O). It has been noted that nuclear receptors, such as NR4A2, only transiently interact with DNA,74 and as a result, it could be that the short DNA residence time fails to result in a robust footprint detectable by transposition. These footprinting data substantiate the claim that the RFX family of TFs and FOXA1 and/or FOXA2 are active in MB DA neuron CREs.
We confirmed that these sequences are indeed enriched in the catalogs by examining the pileup of reads overlapping all genome-wide predicted motif binding sites for each motif identified by gkm-SVM. We see an abundance of reads over predicted binding sites of all four motifs (Figures 3D, 3H, 3L, and 3P); the strongest enrichment overlaps RFX and ASCL1 motif sites (Figures 3D and 3L). Despite the less robust footprint generated at the ASCL1 sites, this TF clearly underlies a larger-than-expected proportion of OCRs in the MB catalog. Integrating a support vector machine-learning algorithm, as applied to the sequences underlying OCRs, with footprinting analysis in the same chromatin substrate both powerfully identifies TFs that are important for DA neuron biology and suggests the RFX family of TFs, FOXA1 and FOXA2, ASCL1, and NR4A2 are actively influencing gene expression in the MB DA neurons.
A Candidate CRE in Intron 4 of SNCA Is Associated with PD Risk
Having established the biological robustness of the OCR catalog, we moved to exploit these data to investigate how non-coding variation therein might be contributing to PD risk. We established two complementary strategies. First, we sought to globally examine the overlap of PD GWAS SNPs9, 10 and those in LD (r2 > 0.8) with our DA OCR catalogs. In doing so, we identified 129 unique PD-associated variants that occur at 20 GWAS-associated loci and that are present in one or both of our OCR catalogs (34 specifically overlap the MB catalog, 14 specifically overlap the FB catalog, and 81 overlap both; Table S3).
Second, we examined the chromatin landscape surrounding familial PD genes by focusing on genes with no obvious overlaps in the first strategy. In doing this, we turned our attention to the SNCA locus. Despite the fact that this locus is the most significant hit in PD GWASs,9, 10 the LD structure surrounding the lead SNP (dbSNP: rs356182) is such that no variants in LD are apparent at our r2 cut-off, and the lead SNP itself is not overlapped by either our MB or FB catalog. Given α-synuclein’s established role in PD pathogenesis and the strength of GWAS signal at SNCA, we prioritized this locus for a closer, more targeted, inspection.
We first noted that Snca expression differs significantly between the MB and FB DA neurons in our bulk RNA-seq analysis (Figure 4B). Examining the chromatin accessibility at the Snca locus, we found that the MB and FB were largely the same with the exception of one robust peak in intron 4 (mm9: chr6: 60,742,503–60,744,726) that is present in the MB but completely absent in the FB (Figure 4A). DNase hypersensitivity site (DHS) linkage57, 63 suggests that this putative CRE interacts with the SNCA promoter. Given the MB specificity of this putative CRE and indications that it interacts with the SNCA promoter, we suspected that this region might be a driving force behind the MB-specific expression of Snca.
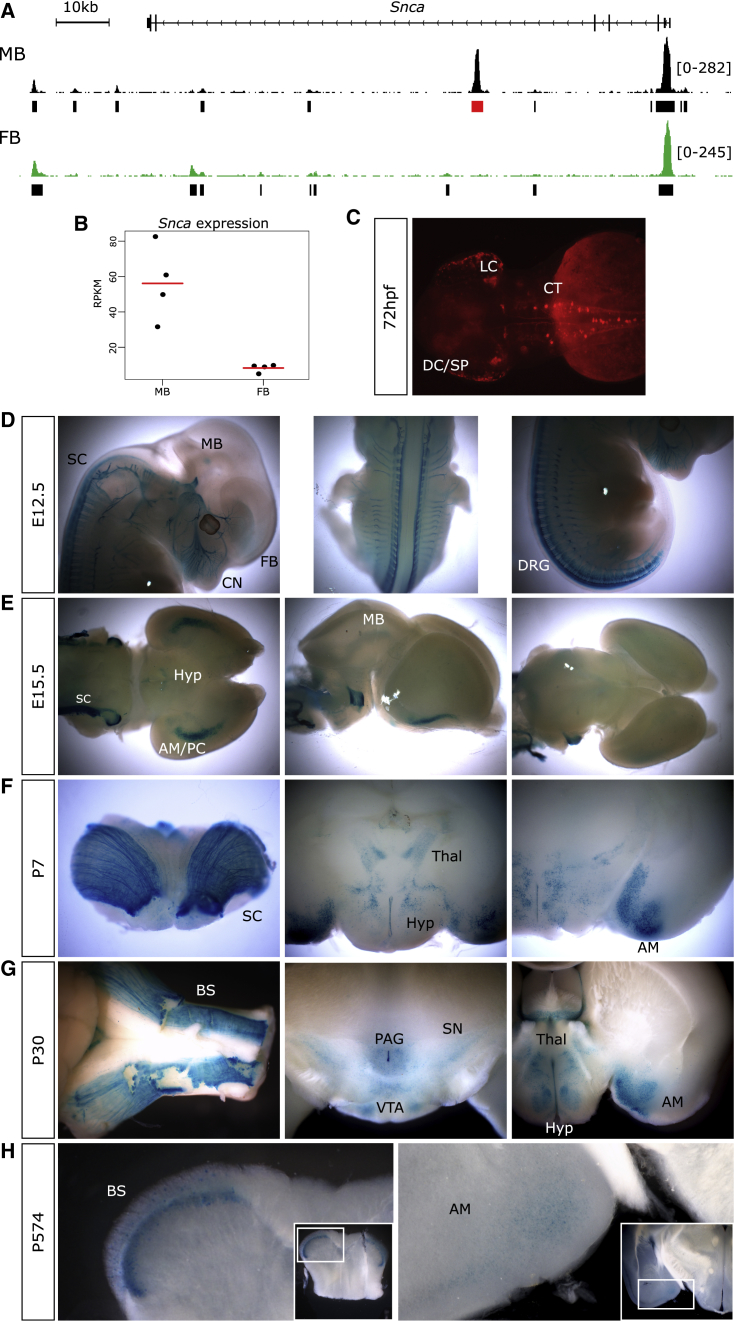
Figure 4.
A MB-Specific Enhancer Directs Expression in Catecholaminergic Populations of Neurons Known to Parkinson Disease Biology
(A) An IGV track indicating the location of the MB-specific region of open chromatin located in intron 4 of Snca.
(B) Snca is differentially expressed between the MB and FB DA neurons. The red bar is the mean expression of the four replicates (black dots).
(C) At 72 hpf, stable transgenic zebrafish reporter assays indicate this putative CRE is capable of directing reporter expression in key catecholaminergic neuronal populations, including the locus coeruleus (LC), the catecholaminergic tract (CT) of the hindbrain, the diencephalic cluster (DC), and the subpallium (SP), into which the DC projects.
(D–H) Further studies in lacZ reporter assays in embryonic (E) and post-natal (P) mice indicate dynamic enhancer usage across developmental time.
(D) This enhancer directs expression throughout the MB, FB, dorsal root ganglia (DRG), sympathetic chain (SC), and cranial nerves (CN) of E12.5 mice.
(E) By E15.5, reporter expression is observed in the amygdala and/or piriform cortex (AM/PC), sympathetic chain, MB, and hypothalamus (Hyp).
(F) Patterns of reporter expression at P7 reflect those seen at E15.5.
(G) Reporter activity is observed at P30 in the amygdala; hypothalamus and thalamus (Thal); brain stem (BS); substantia nigra (SN); ventral tegmental area (VTA); and the periaqueductal gray area (PAG).
(H) In aged mice (P574), reporter expression is detected robustly in the brain stem and faintly in the amygdala.
To test this hypothesis, we assayed whether, when lifted over to hg19, the central portion of this putative CRE (chr4: 90,721,063–90,722,122), is capable of directing appropriate reporter expression in transgenic zebrafish and mouse reporter assays. Stable transgenesis of zebrafish indicates that this CRE directs reporter expression at 72 hpf in the locus coeruleus, a key population of catecholaminergic neurons preferentially degenerated in PD,75 and along the catecholaminergic tract through the hindbrain, which is largely composed of DA neurons76 (Figure 4C). Additionally, we observe reporter expression throughout the diencephalic catecholaminergic cluster with projections to the subpallium, which is analogous to mammalian dopaminergic projections from the ventral midbrain to the striatum.77 Reporter expression in these transgenic zebrafish is largely consistent with an enhancer active in catecholaminergic populations.
To further evaluate this CRE in a mammalian system, we generated lacZ reporter mice and examined reporter activity across developmental time. Whole-mount E12.5 reporter mice indicate this enhancer directs exquisitely restricted expression in Th+ populations, including the dorsal root ganglia, extending into the sympathetic chain and throughout the cranial nerves (particularly the trigeminal). Additional diffuse staining is noted throughout the MB and FB (Figure 4D). Specifically examining the brains of lacZ animals at E15.5, we identified reporter expression in the MB and hypothalamus, as well as strong expression through the amygdala and piriform cortex and along the anterior portion of the sympathetic chain (Figure 4E); we see similar reporter patterns at P7 (Figure 4F). At P30, we detect reporter activity in the amygdala, hypothalamus, thalamus, periaqueductal gray area, brain stem, and importantly, in the substantia nigra and ventral tegmental area (Figure 4G). By contrast, in aged lacZ reporter mice (574 days old [∼19 months]), we detect strong reporter expression only in the brain stem and observe weak reporter expression in the amygdala (Figure 4H). Collectively, the regions in which we detect reporter activity reflect those compromised in PD; Lewy bodies (aggregates of α-synuclein) have been detected in the locus coeruleus, sympathetic chain, amygdala, hypothalamus, ventral tegmental area, and periaqueductal gray area of PD-affected individuals.78, 79, 80, 81, 82 Critically, the preferential degradation of the substantia nigra is the pathological hallmark of PD progression.2 This enhancer directs region-specific appropriate expression throughout development in key locations in concordance with SNCA activity in PD pathogenesis.
After confirming this CRE’s regulatory activity in brain regions associated with PD, we next inspected this sequence for PD-associated variation. We sequenced across this interval in 986 individuals with PD and 992 controls and identified 14 variants (Table S4); four of these variants had an MAF greater than 5% and were common and present in both affected individuals and controls. Of these, two tightly linked variants (r2 = 0.934; Table S5), dbSNP: rs2737024 (OR = 1.25, 95% CI = 1.09–1.44, p value = 0.002) and dbSNP: rs2583959 (OR = 1.22, 95% CI = 1.06–1.40, p value = 0.005), were significantly associated with PD (Table 1). These data support a role for variation within the enhancer in conferring PD risk.
Table 1.
Two Tightly Linked SNPs within the Enhancer are Significantly Associated with PD Risk
| Variant | MA | MAF in PD-Affected Individuals (n = 986) | MAF in Controls (n = 992) |
Association with PD |
|
|---|---|---|---|---|---|
| OR (95% CI) | p Value | ||||
| rs7684892 | A | 0.063 | 0.069 | 0.93 (0.72–1.20) | 0.562 |
| rs17016188 | C | 0.082 | 0.061 | 1.35 (1.04–1.75) | 0.023 |
| rs2583959 | G | 0.317 | 0.271 | 1.22 (1.06–1.40) | 0.005∗ |
| rs2737024 | G | 0.319 | 0.270 | 1.25 (1.09–1.44) | 0.002∗ |
Abbreviations are as follows: MA = minor allele; MAF = minor-allele frequency; OR = odds ratio; and CI = confidence interval.
Only variants with MAF > 0.05 were considered.
ORs, 95% CIs, and p values result from additive logistic regression models adjusted for sex and age at blood draw. p values ≤ 0.0125 were considered as statistically significant after a Bonferroni correction for multiple testing (∗).
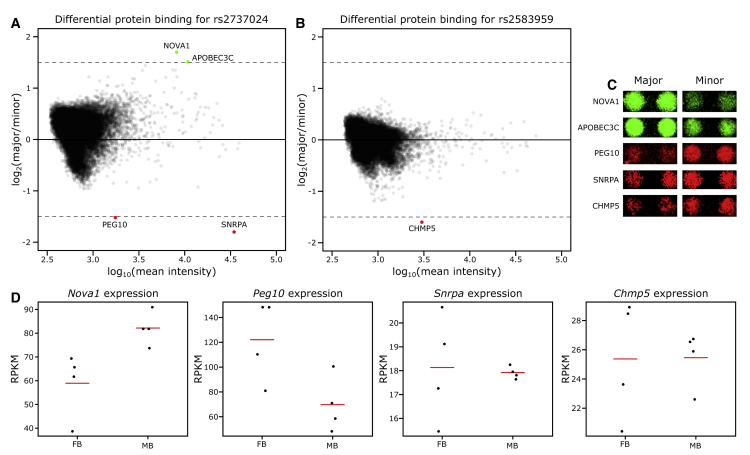
To assess how these variants might impact enhancer function and thus PD risk, we assayed differential protein binding at these variants for >16,000 proteins.58 In doing so, we identified five proteins whose binding is robustly impacted by these implicated variants: NOVA1, APOBEC3C, PEG10, SNRPA, and CHMP5 (Figures 5A–5C). Of these, all except APOBEC3C (RPKM ≤1) are expressed at appreciable levels in both MB and FB DA neurons (Figure 5D). Of the remaining four proteins, three (PEG10, SNRPA, and CHMP5) demonstrate an increased binding affinity for the minor risk allele over the major allele; this direction of effect is consistent with the overexpression paradigm by which SNCA confers PD risk.8 Interestingly, out of those proteins we identified, CHMP5 is the only one whose binding affinity is impacted by variant dbSNP: rs2583959, and our group has recently implicated one of its family members, CHMP7, in conferring PD risk,69 perhaps indicating a role for this family of proteins in PD. Although no single protein stands out, the increased affinity of proteins expressed in DA neurons for the risk alleles of the identified enhancer variants is consistent with a potential mechanistic contribution to SNCA expression and, therefore, to PD risk.
Figure 5.
Identification of Proteins Whose Binding Is Impacted by the Implicated PD-risk SNPs
(A and B) MA plots for both dbSNP: rs2737024 and dbSNP: rs2583959 indicate the magnitude of the effect of the minor and major allele on binding. Cut-off for differential binding: log2(major/minor) ≥ 1.5 or ≤ −1.5.
(A) NOVA1 and APOBEC3C (green circles) bind at dbSNP: rs2737024 with greater affinity for the major allele, but PEG10 and SNRPA (red circles) have a greater affinity for the minor allele.
(B) CHMP5 (red circle) has a greater affinity for the minor allele of dbSNP: rs2583959.
(C) Representative images of the protein binding for each of the differentially bound proteins.
(D) Expression analysis in the MB and FB DA neurons for each of the differentially bound proteins indicates Nova1, Peg10, Snrpa, and Chmp5 to be highly expressed in these populations, yet none of the Apobec family member genes are expressed (RPKMs ≤1, data not shown). The red bar is the mean expression of the four replicates (black dots).
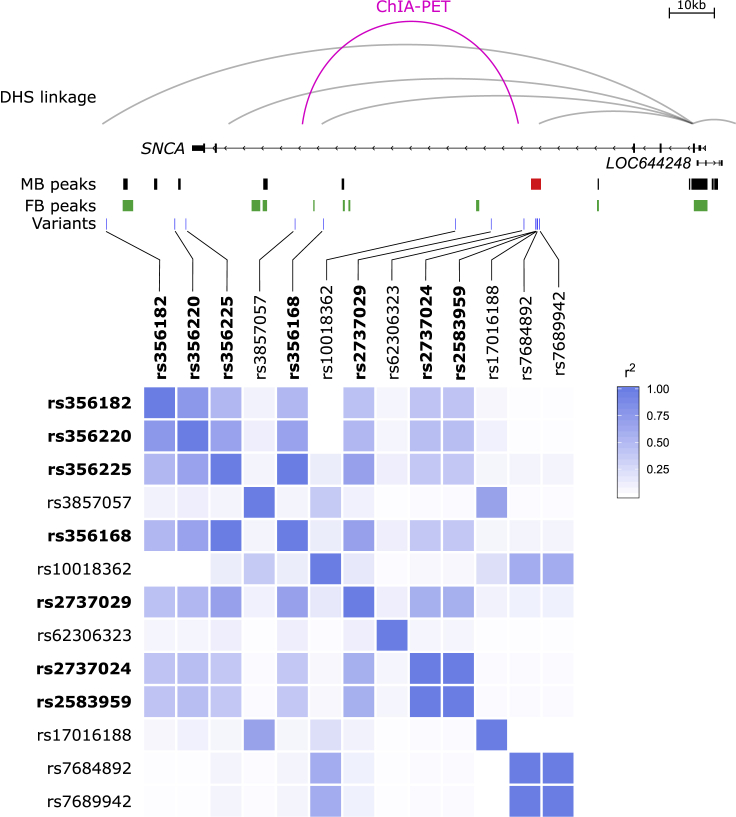
Finally, we set out to refine the haplotype structure and understand how this identified variation might be interacting with other variants at this locus. A panel of common variants had previously been genotyped across SNCA, and PD-associated haplotypes were identified.53 After genotyping our PD-affected individuals and controls for a subset of this panel of variants (in addition to all enhancer-associated variants identified by sequencing [Table S6]), we identified a single haplotype that was significantly associated with PD (p value = 0.003) and that had a higher observed frequency in PD-affected individuals (28.3%) than in controls (23.4%; Table 2). This haplotype implicates some of the same variants as those in Guella et al.53 (dbSNP: rs356220, dbSNP: rs737029) but also implicates dbSNP: rs356225 and dbSNP: rs356168, as well as the two enhancer-associated variants. Additionally, within the 1000 Genomes data, we observe that moderate LD structure exists between the lead GWAS variant (dbSNP: rs356182) and the enhancer variants (r2 = 0.418, D′ = 0.745) in the general European population. Despite the moderate LD, the risk allele of dbSNP: rs356182 falls in our identified PD-associated haplotype 94% of the time. Thus, it is likely that at least part of the risk captured by dbSNP: rs356182 can be attributed to these enhancer variants and the implicated haplotype reported here. Furthermore, this does not preclude additional variants from being present and contributing to the risk captured by the lead SNP because the dbSNP: rs356182 risk allele can occur in the absence of the enhancer-associated variants (i.e.: ∼31% of EUR individuals with the dbSNP: rs356182 risk allele do not carry the risk alleles of the PD-associated enhancer variants). A schematic depiction of the variants, open-chromatin regions, chromatin interactions,57, 63 and LD structure at this locus is presented in Figure 6. Of the variants, including dbSNP: rs356182, whose minor alleles define this PD-associated haplotype, only the two enhancer-associated variants and dbSNP: rs2737029 are identified as eQTLs for SNCA expression in any tissue in the GTEx database (Figure S7). Collectively, these data identify a catecholaminergic enhancer harboring common variation that is part of a larger haplotype associated with PD risk, likely by modulating SNCA activity.
Table 2.
A Single Haplotype, Containing the Minor Alleles of the Implicated SNPs, Is Significantly Associated with PD Risk
 |
Only haplotypes with frequency ≥0.01 were considered. Black boxes indicate the minor allele in Europeans.
p values result from score tests for association that were performed under an additive model and adjusted for sex and age at blood draw.
p values ≤0.0042 were considered as statistically significant after application of a Bonferroni correction for multiple testing (∗).
Figure 6.
A Schematic of the Chromatin Interactions, LD Structure, Variation, and Open Chromatin at the SNCA Locus
Publicly available DNase hypersensitivity site (DHS) linkage analysis suggests that the promoter of SNCA possibly interacts with the identified MB-specific enhancer, the lead GWAS variant (dbSNP: rs356182), and a previously functionally validated variant (dbSNP: rs356168). ChIA-PET data suggest that the MB-specific enhancer might interact with variant dbSNP: rs356168. Open-chromatin data from DA neurons do not overlap with any variants at this locus or haplotype other than at the MB-specific enhancer. LD analysis at this locus indicates that despite the low LD structure between the lead GWAS variant (dbSNP: rs356182) and the enhancer-associated variants (dbSNP: rs2737024 and dbSNP: rs2583959), the variants are in the same haplotype. Therefore, the GWAS signal might, at least in part, be flagging the identified enhancer-associated variants.
Discussion
The identification and prioritization of biologically pertinent non-coding variation associated with disease remains challenging. Recent studies by our and other groups have emphasized the importance of cellular context in the identification of sequences harboring biologically pertinent variation and the genes they regulate. To this end, we used chromatin signatures from ex vivo isolated DA neurons to reveal biologically active sequences that harbor non-coding variation contributing to PD risk. We generated robust OCR catalogs for both MB and FB DA neurons, confirmed their capacity to act as enhancers, identified motifs that confer their regulatory potential, and notably, identified two variants that are located within a MB-specific enhancer and are associated with an increase in PD risk.
In contrast to strategies predicated solely on dissection of post-mortem tissues or on the differentiation of cultured cells, we leveraged the use of transgenic reporter mice to specifically isolate Th-expressing neurons from discrete neuroanatomical (FB and MB) domains. Although our approach assays a more refined population of DA neurons than would be achieved via gross dissection, recent single-cell RNA-seq analyses of these same cells make clear that even within these highly restricted MB and FB populations there exist two primary cellular phenotypes.69 The “homogenous” MB and FB populations each are comprised of an immature neuroblast population and a more mature, domain-specific, post-mitotic population of DA neurons. As a result, our OCR catalogs capture the chromatin accessibility from both of these states. These catalogs are demonstrably biologically relevant for our purposes, but future studies requiring even greater homogeneity might wish to consider single-cell ATAC-seq to refine these domains further.83
Through in silico validation of the catalogs, we established that they are enriched for both sequence constraint and biological relevance in a manner consistent with function and their FB or MB origin. Furthermore, these sequences are frequently domain-appropriate enhancers, and each catalog captures a large fraction (77%) of previously validated MB and FB enhancers. Although more regions are shown to direct neuronal expression than to direct negative or non-neuronal expression, it is interesting to note that almost half of the sequences previously documented as not directing expression in vivo are also represented in one or both of our catalogs.
Given the frequently dynamic nature of CRE activity, this overlap with negative regions most likely results from temporal differences in these assays. Our data indicate that these regions are accessible at E15.5, but the lacZ reporter assays were carried out at E11.5; regions that have been annotated as negative at E11.5 might be active at later time points and, as such, appear in our catalogs. As we moved from these unbiased functional comparisons to more highly selected ones, the potential impact of temporal differences became more pronounced. In mouse transgenic reporter assays, two of five assayed putative CREs directed detectable expression of lacZ in neuronal populations. Consistent with the temporally dynamic nature of CREs, when we tested these same regions in zebrafish across multiple developmental time points, we observed that four of the five sequences acted as neuronal enhancers.
By examining the sequence composition underlying the ATAC-seq peaks, we illuminate powerful vocabularies for both FB and MB DA neuron transcriptional regulatory control. The machine-learning algorithm gkm-SVM prioritized four transcription factor families (RFX, FOXA1/2, NR4A2, ASCL1/2) as conveying significant regulatory potential in the CRE catalogs. Of these, the RFX family had not previously been implicated in DA neuron biology. Although several of the RFX family members have been annotated as having expression in the cerebellum or fetal brain,70 a role specifically in MB DA neurons has not previously been appreciated. By contrast, NR4A2 is canonically associated with MB DA neurons,67, 68 is highly expressed in this population (139 RPKM), and was prioritized as having TF-conferring regulatory potential in these cells; however, TF footprinting fails to provide evidence supporting its activity. We postulate that this lack of a footprint might reflect the transient DNA-binding dynamics of NR4A2. Transcription factors with short DNA residence times often fail to reveal footprints, and nuclear receptors, such as NR4A2, have markedly transient DNA interactions.74
Taken collectively, these data establish a robust biological platform from which PD-associated variation can be evaluated. To this end, an obvious candidate to interrogate was an apparent MB-specific open-chromatin domain within intron 4 of the known PD-associated gene SNCA. We assayed the activity of this putative CRE in zebrafish and across the life course of mice and found it to be active in key catecholaminergic structures (e.g.: the substantia nigra and locus coeruleus) injured in PD, from mid-gestation until at least P30. Thereafter, the utilization of this enhancer in the brain is diminished and by late life appears restricted to the brainstem and amygdala. By the time of clinical presentation, individuals diagnosed with PD have already lost a significant proportion (≥30%) of their nigral DA neurons;2, 84 the observed biology of this CRE is consistent with a progressive pathogenic influence that begins early in life and renders these populations preferentially vulnerable to loss over an extensive period of time.
Sequencing this interval in PD-affected individuals and controls revealed two common variants (dbSNP: rs2737024 and dbSNP: rs2583959), individually associated with an increased risk of PD. After testing these variants for their effect on protein binding, we identified five proteins whose binding is affected, three of which (PEG10, SNRPA, and CHMP5) display greater affinity for the risk allele. Furthermore, we identified a larger haplotype containing these variants; this haplotype is also significantly associated with PD risk. Although none of the other SNPs in this haplotype overlap with CREs identified in the DA neuron catalogs, there is significant functional evidence of the activity and contribution to PD risk by variant dbSNP: rs356168.85 The same DHS correlation analysis57, 63 that suggests an interaction between the SNCA promoter and our identified CRE also suggests an interaction between the SNCA promoter and the dbSNP: rs356168 variant. Additionally, ChIA-PET data25, 57 indicate that sequences encompassing this variant might interact with our enhancer, suggesting a potential cooperative mode of action; Gupta and colleagues86 recently proposed that such a paradigm takes place at the EDN1 locus. We propose that the variants within the enhancer, independently or in concert with other variation within the identified haplotype, might act throughout an individual’s lifespan to render key populations of catecholaminergic neurons vulnerable and thus increase PD risk in individuals harboring this variation.
This work emphasizes the value of biologically informed, cell-context-dependent guided searches for the identification of disease-associated and functional non-coding variation. Given the extent of non-coding GWAS-identified variation, the need for strategies to prioritize variants for functional follow-up is greater than ever. Here, we generate chromatin accessibility data from purified populations of DA neurons to generate catalogs of putative CREs. We have demonstrated how these data can be used to reveal non-coding variation contributing to PD risk; focusing on a single region of open chromatin at the SNCA locus, we uncover PD-associated variation therein and propose a model through which this sequence can contribute to normal DA neuronal biology and PD risk. There remains a plethora of information still to be explored in these catalogs, either through further single-locus investigations or through massively parallel assays. For example, our MB DA-neuron OCR catalog overlaps variants at 20 of 49 (41%) PD-associated loci,9, 10 all of which can be investigated further for the mechanisms by which they impact PD risk. Our work establishes a powerful paradigm, leveraging transgenic model systems to systematically generate cell-type-specific chromatin accessibility data and reveal disease-associated variation, in a manner that can be progressively guided by improved biological understanding.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
This research undertaken at Johns Hopkins University School of Medicine was supported in part by awards from the NIH (NS62972 and MH106522 to A.S.M.; GM111514 to H.Z.; HG007348 to M.A.B.). The Mayo Clinic collection was supported in part by Morris K. Udall Center of Excellence in Parkinson’s Disease Research (P50 NS072187), American Parkinson Disease Association Center, and The Mangurian Foundation for Lewy Body Research. O.A.R. is supported by NS078086 and NS10069 (NIH), W81XWH-17-1-0249 (Department of Defense), The Michael J. Fox Foundation, and The Little Family Foundation. Z.K.W. is supported by the Mayo Clinic Center for Regenerative Medicine; the Mayo Clinic Center for Individualized Medicine; the Mayo Clinic Neuroscience Focused Research Team (Cecilia and Dan Carmichael Family Foundation; and the James C. and Sarah K. Kennedy Fund for Neurodegenerative Disease Research at Mayo Clinic in Florida); a gift from Carl Edward Bolch, Jr., and Susan Bass Bolch; The Sol Goldman Charitable Trust; and Donald G. and Jodi P. Heeringa. Research conducted at the E.O. Lawrence Berkeley National Laboratory was performed under U.S. Department of Energy contract DE-AC02-05CH11231, University of California, and was supported by HG003988 (NIH) to L.A.P.
Published: November 29, 2018
Footnotes
Supplemental Data include seven figures and six tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.10.018.
Accession Numbers
ATAC-sequencing, RNA-sequencing and related data will be available at the Gene Expression Omnibus (GEO) under the accession number GSE122450.
Web Resources
The 3D Genome Browser, http://promoter.bx.psu.edu/hi-c/index.html
Cistrome, http://cistrome.org/ap/
FastQC, https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
The Genomic Regions Enrichment of Annotations Tool (GREAT), http://great.stanford.edu
LDlink, https://ldlink.nci.nih.gov/
The List of Genes and Their TSSs Used by GREAT: http://bejerano.stanford.edu/help/download/attachments/2752609/mm9.great3.0.genes.txt
The UCSC Table Browser, http://genome.ucsc.edu
R: A Language and Environment for Statistical Computing, https://www.R-project.org/
R Corrplot package, https://github.com/taiyun/corrplot
R haplo.stats package, https://CRAN.R-project.org/package=haplo.stats
R LSD package, https://CRAN.R-project.org/package=LSD
R RColorBrewer package, https://CRAN.R-project.org/package=RColorBrewer
rAggr, http://raggr.usc.edu/
VISTA Enhancer Browser, https://enhancer.lbl.gov
Supplemental Data
References
- 1.Ma S.Y., Röyttä M., Rinne J.O., Collan Y., Rinne U.K. Correlation between neuromorphometry in the substantia nigra and clinical features in Parkinson’s disease using disector counts. J. Neurol. Sci. 1997;151:83–87. doi: 10.1016/s0022-510x(97)00100-7. [DOI] [PubMed] [Google Scholar]
- 2.Fearnley J.M., Lees A.J. Ageing and Parkinson’s disease: Substantia nigra regional selectivity. Brain. 1991;114:2283–2301. doi: 10.1093/brain/114.5.2283. [DOI] [PubMed] [Google Scholar]
- 3.Pringsheim T., Jette N., Frolkis A., Steeves T.D.L. The prevalence of Parkinson’s disease: A systematic review and meta-analysis. Mov. Disord. 2014;29:1583–1590. doi: 10.1002/mds.25945. [DOI] [PubMed] [Google Scholar]
- 4.Thomas B., Beal M.F. Parkinson’s disease. Hum. Mol. Genet. 2007;16 Spec No. 2:R183–R194. doi: 10.1093/hmg/ddm159. [DOI] [PubMed] [Google Scholar]
- 5.Zarranz J.J., Alegre J., Gómez-Esteban J.C., Lezcano E., Ros R., Ampuero I., Vidal L., Hoenicka J., Rodriguez O., Atarés B. The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann. Neurol. 2004;55:164–173. doi: 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- 6.Krüger R., Kuhn W., Müller T., Woitalla D., Graeber M., Kösel S., Przuntek H., Epplen J.T., Schöls L., Riess O. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson’s disease. Nat. Genet. 1998;18:106–108. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- 7.Polymeropoulos M.H., Lavedan C., Leroy E., Ide S.E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 8.Singleton A.B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J., Hulihan M., Peuralinna T., Dutra A., Nussbaum R. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 9.Nalls M.A., Pankratz N., Lill C.M., Do C.B., Hernandez D.G., Saad M., DeStefano A.L., Kara E., Bras J., Sharma M., International Parkinson’s Disease Genomics Consortium (IPDGC) Parkinson’s Study Group (PSG) Parkinson’s Research: The Organized GENetics Initiative (PROGENI) 23andMe. GenePD. NeuroGenetics Research Consortium (NGRC) Hussman Institute of Human Genomics (HIHG) Ashkenazi Jewish Dataset Investigator. Cohorts for Health and Aging Research in Genetic Epidemiology (CHARGE) North American Brain Expression Consortium (NABEC) United Kingdom Brain Expression Consortium (UKBEC) Greek Parkinson’s Disease Consortium. Alzheimer Genetic Analysis Group Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 2014;46:989–993. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang D., Nalls M.A., Hallgrímsdóttir I.B., Hunkapiller J., van der Brug M., Cai F., Kerchner G.A., Ayalon G., Bingol B., Sheng M., International Parkinson’s Disease Genomics Consortium. 23andMe Research Team A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat. Genet. 2017;49:1511–1516. doi: 10.1038/ng.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaub M.A., Boyle A.P., Kundaje A., Batzoglou S., Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748–1759. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ernst J., Kheradpour P., Mikkelsen T.S., Shoresh N., Ward L.D., Epstein C.B., Zhang X., Wang L., Issner R., Coyne M. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forrest M.P., Zhang H., Moy W., McGowan H., Leites C., Dionisio L.E., Xu Z., Shi J., Sanders A.R., Greenleaf W.J. Open chromatin profiling in hiPSC-derived neurons prioritizes functional noncoding psychiatric risk variants and highlights neurodevelopmental loci. Cell Stem Cell. 2017;21:305–318.e8. doi: 10.1016/j.stem.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fullard J.F., Giambartolomei C., Hauberg M.E., Xu K., Voloudakis G., Shao Z., Bare C., Dudley J.T., Mattheisen M., Robakis N.K. Open chromatin profiling of human postmortem brain infers functional roles for non-coding schizophrenia loci. Hum. Mol. Genet. 2017;26:1942–1951. doi: 10.1093/hmg/ddx103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyle E.A., Li Y.I., Pritchard J.K. An expanded view of complex traits: From polygenic to omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee D., Gorkin D.U., Baker M., Strober B.J., Asoni A.L., McCallion A.S., Beer M.A. A method to predict the impact of regulatory variants from DNA sequence. Nat. Genet. 2015;47:955–961. doi: 10.1038/ng.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Praetorius C., Grill C., Stacey S.N., Metcalf A.M., Gorkin D.U., Robinson K.C., Van Otterloo E., Kim R.S.Q., Bergsteinsdottir K., Ogmundsdottir M.H. A polymorphism in IRF4 affects human pigmentation through a tyrosinase-dependent MITF/TFAP2A pathway. Cell. 2013;155:1022–1033. doi: 10.1016/j.cell.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gong S., Zheng C., Doughty M.L., Losos K., Didkovsky N., Schambra U.B., Nowak N.J., Joyner A., Leblanc G., Hatten M.E., Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 20.Westerfeld M. Univ. Oregon Press; Eugene: 2007. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) [Google Scholar]
- 21.Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shin H., Liu T., Manrai A.K., Liu X.S. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009;25:2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- 27.Liu T., Ortiz J.A., Taing L., Meyer C.A., Lee B., Zhang Y., Shin H., Wong S.S., Ma J., Lei Y. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 2011;12:R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44(W1) doi: 10.1093/nar/gkw257. W160-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim D., Langmead B., Salzberg S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao Y., Smyth G.K., Shi W. The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huber W., Carey V.J., Gentleman R., Anders S., Carlson M., Carvalho B.S., Bravo H.C., Davis S., Gatto L., Girke T. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mudge J.M., Harrow J. Creating reference gene annotation for the mouse C57BL6/J genome assembly. Mamm. Genome. 2015;26:366–378. doi: 10.1007/s00335-015-9583-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao H., Sun Z., Wang J., Huang H., Kocher J.-P., Wang L. CrossMap: A versatile tool for coordinate conversion between genome assemblies. Bioinformatics. 2014;30:1006–1007. doi: 10.1093/bioinformatics/btt730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hansen K.D., Irizarry R.A., Wu Z. Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics. 2012;13:204–216. doi: 10.1093/biostatistics/kxr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pennacchio L.A., Ahituv N., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 42.Poulin F., Nobrega M.A., Plajzer-Frick I., Holt A., Afzal V., Rubin E.M., Pennacchio L.A. In vivo characterization of a vertebrate ultraconserved enhancer. Genomics. 2005;85:774–781. doi: 10.1016/j.ygeno.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 43.Kothary R., Clapoff S., Darling S., Perry M.D., Moran L.A., Rossant J. Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development. 1989;105:707–714. doi: 10.1242/dev.105.4.707. [DOI] [PubMed] [Google Scholar]
- 44.Fisher S., Grice E.A., Vinton R.M., Bessling S.L., Urasaki A., Kawakami K., McCallion A.S. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat. Protoc. 2006;1:1297–1305. doi: 10.1038/nprot.2006.230. [DOI] [PubMed] [Google Scholar]
- 45.Ghandi M., Lee D., Mohammad-Noori M., Beer M.A. Enhanced regulatory sequence prediction using gapped k-mer features. PLoS Comput. Biol. 2014;10:e1003711. doi: 10.1371/journal.pcbi.1003711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zorita E., Cuscó P., Filion G.J. Starcode: Sequence clustering based on all-pairs search. Bioinformatics. 2015;31:1913–1919. doi: 10.1093/bioinformatics/btv053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bailey T.L., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 49.Gupta S., Stamatoyannopoulos J.A., Bailey T.L., Noble W.S. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pique-Regi R., Degner J.F., Pai A.A., Gaffney D.J., Gilad Y., Pritchard J.K. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011;21:447–455. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grant C.E., Bailey T.L., Noble W.S. FIMO: Scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adey A., Morrison H.G., Asan, Xun X., Kitzman J.O., Turner E.H., Stackhouse B., MacKenzie A.P., Caruccio N.C., Zhang X., Shendure J. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11:R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guella I., Evans D.M., Szu-Tu C., Nosova E., Bortnick S.F., Goldman J.G., Dalrymple-Alford J.C., Geurtsen G.J., Litvan I., Ross O.A., SNCA Cognition Study Group α-synuclein genetic variability: A biomarker for dementia in Parkinson disease. Ann. Neurol. 2016;79:991–999. doi: 10.1002/ana.24664. [DOI] [PubMed] [Google Scholar]
- 54.Hughes A.J., Daniel S.E., Kilford L., Lees A.J. Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: a clinico-pathological study of 100 cases. J. Neurol. Neurosurg. Psychiatry. 1992;55:181–184. doi: 10.1136/jnnp.55.3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schaid D.J., Rowland C.M., Tines D.E., Jacobson R.M., Poland G.A. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am. J. Hum. Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Machiela M.J., Chanock S.J. LDlink: A web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31:3555–3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y., Song F., Zhang B., Zhang L., Xu J., Kuang D., Li D., Choudhary M.N.K., Li Y., Hu M. The 3D Genome Browser: A web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 2018;19:151. doi: 10.1186/s13059-018-1519-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jeong J.S., Jiang L., Albino E., Marrero J., Rho H.S., Hu J., Hu S., Vera C., Bayron-Poueymiroy D., Rivera-Pacheco Z.A. Rapid identification of monospecific monoclonal antibodies using a human proteome microarray. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.O111.016253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu S., Wan J., Su Y., Song Q., Zeng Y., Nguyen H.N., Shin J., Cox E., Rho H.S., Woodard C. DNA methylation presents distinct binding sites for human transcription factors. eLife. 2013;2:e00726. doi: 10.7554/eLife.00726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heintz N. Gene expression nervous system atlas (GENSAT) Nat. Neurosci. 2004;7:483. doi: 10.1038/nn0504-483. [DOI] [PubMed] [Google Scholar]
- 61.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Visel A., Minovitsky S., Dubchak I., Pennacchio L.A. VISTA Enhancer Browser—A database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stott S.R.W., Metzakopian E., Lin W., Kaestner K.H., Hen R., Ang S.-L. Foxa1 and foxa2 are required for the maintenance of dopaminergic properties in ventral midbrain neurons at late embryonic stages. J. Neurosci. 2013;33:8022–8034. doi: 10.1523/JNEUROSCI.4774-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arenas E. Foxa2: The rise and fall of dopamine neurons. Cell Stem Cell. 2008;2:110–112. doi: 10.1016/j.stem.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 67.Prakash N., Wurst W. Development of dopaminergic neurons in the mammalian brain. Cell. Mol. Life Sci. 2006;63:187–206. doi: 10.1007/s00018-005-5387-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smits S.M., Ponnio T., Conneely O.M., Burbach J.P.H., Smidt M.P. Involvement of Nurr1 in specifying the neurotransmitter identity of ventral midbrain dopaminergic neurons. Eur. J. Neurosci. 2003;18:1731–1738. doi: 10.1046/j.1460-9568.2003.02885.x. [DOI] [PubMed] [Google Scholar]
- 69.Hook P.W., McClymont S.A., Cannon G.H., Law W.D., Morton A.J., Goff L.A., McCallion A.S. Single-cell RNA-Seq of mouse dopaminergic neurons informs candidate gene selection for sporadic Parkinson disease. Am. J. Hum. Genet. 2018;102:427–446. doi: 10.1016/j.ajhg.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sugiaman-Trapman D., Vitezic M., Jouhilahti E.-M., Mathelier A., Lauter G., Misra S., Daub C.O., Kere J., Swoboda P. Characterization of the human RFX transcription factor family by regulatory and target gene analysis. BMC Genomics. 2018;19:181. doi: 10.1186/s12864-018-4564-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gajiwala K.S., Chen H., Cornille F., Roques B.P., Reith W., Mach B., Burley S.K. Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature. 2000;403:916–921. doi: 10.1038/35002634. [DOI] [PubMed] [Google Scholar]
- 72.Kittappa R., Chang W.W., Awatramani R.B., McKay R.D.G. The foxa2 gene controls the birth and spontaneous degeneration of dopamine neurons in old age. PLoS Biol. 2007;5:e325. doi: 10.1371/journal.pbio.0050325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Caiazzo M., Dell’Anno M.T., Dvoretskova E., Lazarevic D., Taverna S., Leo D., Sotnikova T.D., Menegon A., Roncaglia P., Colciago G. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- 74.Sung M.-H., Guertin M.J., Baek S., Hager G.L. DNase footprint signatures are dictated by factor dynamics and DNA sequence. Mol. Cell. 2014;56:275–285. doi: 10.1016/j.molcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zarow C., Lyness S.A., Mortimer J.A., Chui H.C. Neuronal loss is greater in the locus coeruleus than nucleus basalis and substantia nigra in Alzheimer and Parkinson diseases. Arch. Neurol. 2003;60:337–341. doi: 10.1001/archneur.60.3.337. [DOI] [PubMed] [Google Scholar]
- 76.Kastenhuber E., Kratochwil C.F., Ryu S., Schweitzer J., Driever W. Genetic dissection of dopaminergic and noradrenergic contributions to catecholaminergic tracts in early larval zebrafish. J. Comp. Neurol. 2010;518:439–458. doi: 10.1002/cne.22214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rink E., Wullimann M.F. The teleostean (zebrafish) dopaminergic system ascending to the subpallium (striatum) is located in the basal diencephalon (posterior tuberculum) Brain Res. 2001;889:316–330. doi: 10.1016/s0006-8993(00)03174-7. [DOI] [PubMed] [Google Scholar]
- 78.Seidel K., Mahlke J., Siswanto S., Krüger R., Heinsen H., Auburger G., Bouzrou M., Grinberg L.T., Wicht H., Korf H.-W. The brainstem pathologies of Parkinson’s disease and dementia with Lewy bodies. Brain Pathol. 2015;25:121–135. doi: 10.1111/bpa.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wakabayashi K., Mori F., Tanji K., Orimo S., Takahashi H. Involvement of the peripheral nervous system in synucleinopathies, tauopathies and other neurodegenerative proteinopathies of the brain. Acta Neuropathol. 2010;120:1–12. doi: 10.1007/s00401-010-0706-x. [DOI] [PubMed] [Google Scholar]
- 80.Wakabayashi K., Takahashi H. Neuropathology of autonomic nervous system in Parkinson’s disease. Eur. Neurol. 1997;38(Suppl 2):2–7. doi: 10.1159/000113469. [DOI] [PubMed] [Google Scholar]
- 81.Braak H., Braak E., Yilmazer D., de Vos R.A., Jansen E.N., Bohl J., Jellinger K. Amygdala pathology in Parkinson’s disease. Acta Neuropathol. 1994;88:493–500. doi: 10.1007/BF00296485. [DOI] [PubMed] [Google Scholar]
- 82.Langston J.W., Forno L.S. The hypothalamus in Parkinson disease. Ann. Neurol. 1978;3:129–133. doi: 10.1002/ana.410030207. [DOI] [PubMed] [Google Scholar]
- 83.Preissl S., Fang R., Huang H., Zhao Y., Raviram R., Gorkin D.U., Zhang Y., Sos B.C., Afzal V., Dickel D.E. Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation. Nat. Neurosci. 2018;21:432–439. doi: 10.1038/s41593-018-0079-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Greffard S., Verny M., Bonnet A.-M., Beinis J.-Y., Gallinari C., Meaume S., Piette F., Hauw J.-J., Duyckaerts C. Motor score of the Unified Parkinson Disease Rating Scale as a good predictor of Lewy body-associated neuronal loss in the substantia nigra. Arch. Neurol. 2006;63:584–588. doi: 10.1001/archneur.63.4.584. [DOI] [PubMed] [Google Scholar]
- 85.Soldner F., Stelzer Y., Shivalila C.S., Abraham B.J., Latourelle J.C., Barrasa M.I., Goldmann J., Myers R.H., Young R.A., Jaenisch R. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature. 2016;533:95–99. doi: 10.1038/nature17939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gupta R.M., Hadaya J., Trehan A., Zekavat S.M., Roselli C., Klarin D., Emdin C.A., Hilvering C.R.E., Bianchi V., Mueller C. A genetic variant associated with five vascular diseases is a distal regulator of endothelin-1 gene expression. Cell. 2017;170:522–533.e15. doi: 10.1016/j.cell.2017.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.