Consequences of transcription out of bounds: a retrograde signal can trigger RNA Polymerase II read-through, upregulating the expression of downstream genes.
Abstract
In plants, the molecular function(s) of the nucleus-localized 5′-3′ EXORIBONUCLEASES (XRNs) are unclear; however, their activity is reported to have a significant effect on gene expression and SAL1-mediated retrograde signaling. Using parallel analysis of RNA ends, we documented a dramatic increase in uncapped RNA substrates of the XRNs in both sal1 and xrn2xrn3 mutants. We found that a major consequence of reducing SAL1 or XRN activity was RNA Polymerase II 3′ read-through. This occurred at 72% of expressed genes, demonstrating a major genome-wide role for the XRN-torpedo model of transcription termination in Arabidopsis (Arabidopsis thaliana). Read-through is speculated to have a negative effect on transcript abundance; however, we did not observe this. Rather, we identified a strong association between read-through and increased transcript abundance of tandemly orientated downstream genes, strongly correlated with the proximity (less than 1,000 bp) and expression of the upstream gene. We observed read-through in the proximity of 903 genes up-regulated in the sal1-8 retrograde signaling mutant; thus, this phenomenon may account directly for up to 23% of genes up-regulated in sal1-8. Using APX2 and AT5G43770 as exemplars, we genetically uncoupled read-through loci from downstream genes to validate the principle of read-through-mediated mRNA regulation, providing one mechanism by which an ostensibly posttranscriptional exoribonuclease that targets uncapped RNAs could modulate gene expression.
Retrograde signaling is the process by which the chloroplasts of plant cells communicate with the nucleus to direct changes in gene expression. This mechanism is required to coordinate chloroplast function during cell differentiation and development (biogenic control) and response to stressful environments (operational control; Pogson et al., 2008; Woodson and Chory, 2012; Chan et al., 2016b). In this way, the chloroplast can act as an environmental stress sensor and utilize retrograde signaling pathways to initiate acclimation responses. Previously, we described the SAL1-PAP-XRN retrograde signaling pathway (Estavillo et al., 2011; Chan et al., 2016a; Pornsiriwong et al., 2017), in which the redox-regulated SAL1 protein controls the levels of the signaling molecule 3′-phosphoadenosine 5′-phosphate (PAP). PAP can move from the chloroplast to the nucleus, where it is it thought to attenuate the activity of the nuclear 5′-3′ exoribonucleases (XRNs), which, in turn, leads to changes in gene expression. However, the mechanism by which this pathway functions in the nucleus is unclear, particularly how an ostensibly posttranscriptional exoribonuclease that targets uncapped RNAs could modulate gene expression.
The XRNs are a family of 5′-3′ exoribonucleases highly conserved and unique to eukaryotes that are responsible for 5′ processing or turnover of a broad range of uncapped RNAs. The XRNs display highly processive 5′-3′ activity toward 5′ monophosphorylated RNA substrates (Zuo and Deutscher, 2001; Nagarajan et al., 2013). In Arabidopsis (Arabidopsis thaliana), there are three XRNs: nucleus-localized XRN2 and XRN3 and cytoplasmic XRN4 (Kastenmayer and Green, 2000). Investigations in Arabidopsis and yeast have established that nuclear 5′ exoribonuclease activity is essential for viability (Gy et al., 2007; Zakrzewska-Placzek et al., 2010). However, beyond knowledge of their 5′ exonuclease activity, the function and substrates of the nuclear XRNs are enigmatic in plants. In a screen to identify repressors of posttranscriptional gene silencing, Gy et al. (2007) discovered that the loss of XRN2 and XRN3 (and SAL1) promotes transgene silencing in a manner similar to that reported previously for XRN4 (Gazzani et al., 2004). In the same study, it was also demonstrated that XRN2 and XRN3 degrade hairpin loops excised during pre-microRNA processing. XRN2 also plays a role in rRNA processing (Zakrzewska-Placzek et al., 2010).
In Arabidopsis, limited accumulation of intergenic transcripts downstream of several thousand genes was observed in sal1 and xrn mutants and was hypothesized to be due to the impairment of a surveillance mechanism for aberrant transcripts (Kurihara et al., 2012). Analogous observations have been reported in yeast for a variety of Xrn1-sensitive unstable transcripts (van Dijk et al., 2011). It is well established that RNA surveillance can take place in the nucleus, where it involves both Rat1/Xrn2 and the nuclear exosome (LaCava et al., 2005; Wyers et al., 2005; Houseley and Tollervey, 2009; Jiao et al., 2010; Jimeno-González et al., 2010; Davidson et al., 2012).
Intriguingly, in addition to the possibility that the XRNs are involved in the turnover of intergenic transcripts, they also may be involved in their production. This hypothesis is based on reported interactions of Xrn2/Rat1 with RNA Polymerase II (Pol II) in yeast, humans, and worms (Kim et al., 2004; West et al., 2004; Miki et al., 2017). In this torpedo model of termination (Proudfoot, 1989), following cleavage and polyadenylation of the mRNA, a 3′ nascent RNA is produced as Pol II continues transcription downstream of the poly(A) site. This RNA is cotranscriptionally decayed by Xrn2/Rat1 faster than Pol II can synthesize it, ultimately dislodging Pol II and removing the nascent noncoding RNA. However, the prevalence of the torpedo mechanism varies between species (Fong et al., 2015; Baejen et al., 2017; Miki et al., 2017), and its functionality in plants is only now being tested. Moreover, the potential consequences of putative transcriptional read-through or proliferation of noncoding RNAs have not been addressed in a systematic manner. These may include the production of small RNAs, which, in turn, could lead to posttranscriptional gene silencing or RNA-directed DNA methylation (Gazzani et al., 2004; Gregory et al., 2008; Matzke and Mosher, 2014). Likewise, read-through may cause generally wasteful or harmful transcription of intergenic space, or the interference of the transcription of downstream genes due to Pol II competition, occlusion, or collision (Shearwin et al., 2005), or may even be linked to the activities of Pol IV and Pol V (Erhard et al., 2015; McKinlay et al., 2018).
Whether the appearance and accumulation of intergenic noncoding RNAs in xrn and sal1 mutants is due to alterations in Pol II termination is unresolved. Here, using a combination of total RNA, mRNA, and degradome short-read sequencing, bioinformatics, and genetic approaches, we elucidate the cause of intergenic noncoding RNA accumulation in sal1 and xrn retrograde signaling mutants and the consequences for the expression of surrounding genes.
RESULTS
XRN-Sensitive Transcripts Identified Using Parallel Analysis of RNA Ends
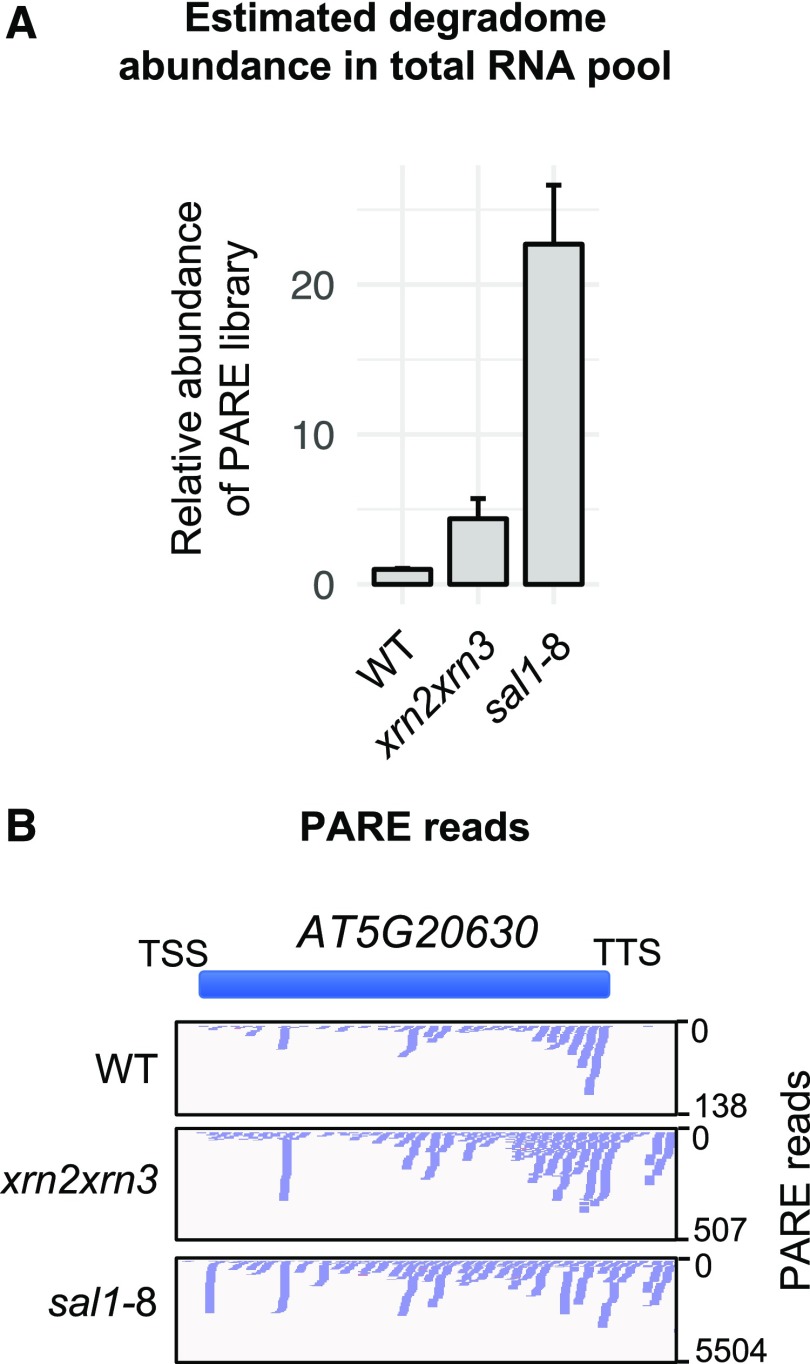
Previously, mRNA profiling and correlation analysis have been used to infer the activities of the XRNs and the role of SAL1 in the accumulation of aberrant genic (Estavillo et al., 2011) and intergenic (Kurihara et al., 2012) transcripts. However, neither prior strategy evaluated whether there was an increase in uncapped genic and intergenic transcripts that can be linked directly to impaired 5′ degradation. To directly identify and characterize the substrates of the nuclear XRNs in Arabidopsis, we profiled mRNA and total RNA using RNA sequencing (RNA-seq) and the 5′ RNA degradome using parallel analysis of RNA ends (PARE) in the xrn2-1xrn3-3 (herein referred to as xrn2xrn3) and sal1-8 (also known as alx8) mutant backgrounds. PARE specifically captures 5′ uncapped, monophosphorylated RNA molecules, which are the canonical substrates of 5′-exoribonucleases (Zuo and Deutscher, 2001; Nagarajan et al., 2013). PARE libraries were constructed by first capturing polyadenylated RNA molecules (Zhai et al., 2014), indicating that both the genic and intergenic RNAs observed in these PARE libraries are polyadenylated. We observed a 1 order of magnitude increase in the abundance of the degradome in sal1-8 and xrn2xrn3 mutants relative to total RNA abundance (Fig. 1; Supplemental Fig. S1A) throughout the length of the chromosomes (Supplemental Fig. S1B), consistent with the accumulation of XRN substrates.
Figure 1.
The degradome of XRN-deficient mutants. A, 5′ Degradome abundance was determined by the quantification of PARE library abundance relative to the wild type (WT) following 10 cycles of PCR and sample purification, expressed as fold change versus the wild type (n = 3; error bars represent sd). B, Example of the distribution of PARE reads over a genomic window in each genotype. The y axis scale varies. TSS, Transcription start site.
Validating and Quantifying Intergenic Transcript Accumulation
PARE reads were observed frequently at the gene terminus and downstream thereof, as shown for AT5G20630 (Fig. 1B). The meta profile of all genes revealed this to be a genome-wide phenomenon in sal1-8 and xrn2xrn3 (Fig. 2A). Read accumulation in the 3′ genic region is consistent with the 5′-3′ direction of RNA turnover and likely represents a snapshot of the cytoplasmic degradome, composed of mRNAs undergoing decay (it should be noted that this also may arise in part due to poly[A] selection). In comparison, the detection of intergenic reads may reflect a nuclear process such as transcriptional termination.
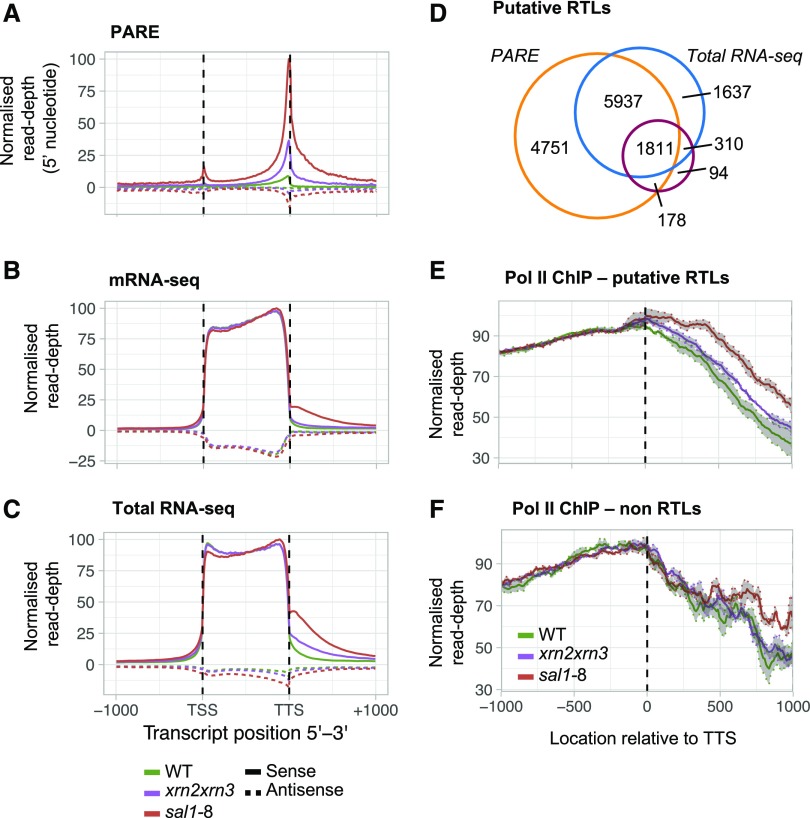
Figure 2.
Identification of read-through loci. A to C, The coding sequences for TAIR10 representative gene models were scaled to uniform length, and read abundance including 1 kb upstream of the TSS and downstream of the transcription termination site (TTS) was plotted for PARE (A), mRNA-seq (B), and total RNA-seq (C). D, Total number and overlap of putative RTLs in the combined sal1-8 and xrn2xrn3 data sets. RTLs were identified by mapping reads to a 500-bp window downstream of the TTS, normalizing to genic coverage, and comparing three biological replicates of each genotype (FDR < 0.05, FC > 2). E, Average Pol II occupancy at the 14,718 putative RTLs in D. Pol II signal was normalized to input and then to genic coverage; shading denotes se (n = 3). F, Average Pol II occupancy at loci with no significant read-through identified in the combined sal1-8 and xrn2xrn3 RNA-seq data sets. WT, Wild type.
Intergenic reads were barely detectable in wild-type plants. An increase in intergenic transcript abundance in the sal1 and xrn mutants was observed in both PARE, poly(A) mRNA-seq profiling (Fig. 2B) and to even greater levels in total RNA-seq (Fig. 2, B and C). This suggests that intergenic transcripts of sal1-8 and xrn2xrn3 are composed of both poly(A) and non-poly(A) RNAs.
Given that intergenic transcripts were most abundant near the TTS and declined to near undetectable levels by 1,000 bp downstream (Fig. 2, A–C), we sought to test the hypothesis that the perturbation to Pol II termination by SAL1-PAP-XRN signaling alters the production of intergenic RNAs. First, to establish a baseline, we surveyed all detectable aberrant transcripts downstream of coding sequences in the sal1-8 and xrn2xrn3 mutants. Using the total RNA-seq data set, for wild-type plants we could detect transcription products (greater than 0.1 cpm) at 912 genes using an overlapping 500-bp window directly downstream of representative TAIR10 gene models. By contrast, a presence-absence test revealed that 10,243 genes in sal1-8 and 3,205 genes in xrn2xrn3 had 3′ intergenic fragments. Sequencing depth could affect the detection of intergenic RNAs; however, each genotype had similar total mapped reads, with 125 million for the wild type, 118 million for xrn2xrn3, and 117 million for sal1-8. Thus, the data sets for each genotype are highly comparable (Supplemental Table S1).
Defining Putative Read-Through Loci
To identify a high-confidence set of putative read-through loci (RTLs), significant changes in intergenic transcript abundance immediately downstream of annotated genes were calculated with reference to both genic abundance (Baejen et al., 2017; McKinlay et al., 2018) and abundance in wild-type plants (false discovery rate [FDR] < 0.05, absolute fold change [FC] over the wild type > 2). Combining this analysis across the total RNA-seq, mRNA-seq, and PARE data sets, we found evidence that impairment of SAL1 or XRN2 and XRN3 led to the accumulation of downstream intergenic RNAs at 14,718 genes, designated the putative RTL set (Fig. 2D; Supplemental Table S2). This strategy captured 84% of the ∼2,000 prior reported loci (Kurihara et al., 2012) and more than 12,000 additional loci. There was strong concordance, with 99% of the 4,324 xrn2xrn3 RTLs overlapping with 14,675 sal1-8 RTLs. We observed that 97% of the mRNA-seq RTLs also were found in the total RNA-seq data, and there were more than 5,000 more RTLs in the total RNA-seq data. Comparatively, only 22% of the total RNA-seq RTLs also were identified by mRNA-seq, initially suggesting that most loci produce predominantly non-poly(A) transcripts. Surprisingly, when we compared the abundance of the 2,107 RTLs that were detected in both the sal1-8 mRNA-seq and total RNA-seq data, we observed slightly higher abundance in the mRNA fraction (Supplemental Fig. S2A). Together, these observations suggest that there are two classes of RTLs: first, poly(A) RTLs that are equally detectable by either total RNA-seq and mRNA-seq; and second, a large class of non-poly(A)-specific loci only reliably detectable through total RNA-seq.
Arabidopsis SAL1, XRN2, and XRN3 Affect Pol II Termination
To assess whether the 14,718 putative RTLs were genuinely associated with Pol II read-through, we analyzed Pol II occupancy using genome-wide Pol II chromatin immunoprecipitation sequencing (ChIP-seq). For RTLs, the meta profile of the region downstream of the TTS revealed a clear and significant increase in Pol II occupancy in sal1-8 and a substantial and consistent increase in xrn2xrn3 compared with the wild type (linear mixed-effects model, ANOVA, P < 0.01). This increase was observed in the combined RTL data set (Fig. 2E) and also when each sequencing method was profiled individually (Supplemental Fig. S2, B–D). In contrast to the intergenic region, Pol II occupancy over the genic region, including the 3′ end, was nearly identical in all genotypes. In comparison with RTLs, at all other genes Pol II levels were far less diverged among the genotypes between 0 and ∼600 bp (Fig. 2F; P > 0.05), although beyond this, Pol II levels in sal1-8 were somewhat elevated. The ChIP-seq protocol was validated independently using quantitative reverse transcription PCR (RT-qPCR; Supplemental Fig. S3, A and B), and we also profiled H3K36me3 as both a positive ChIP control and a mark of active transcription that could potentially mirror any change in Pol II density. For example, mutations affecting human Set2D, an H3K36me3 methyltransferase, also can lead to Pol II read-through (Grosso et al., 2015). In contrast to Pol II ChIP profiles, the distribution of H3K36me3 was similar in all genotypes (Supplemental Fig. S3C). The sal1-8 mutant displayed a higher level of Pol II read-through compared with xrn2xrn3, which is consistent with prior work pointing to the increased severity of the sal1-8 mutation (Estavillo et al., 2011; Pornsiriwong et al., 2017) and the hypomorphic nature of the xrn3-3 allele (Gy et al., 2007; Zakrzewska-Placzek et al., 2010). These results strongly suggest that a major explanation for the increased abundance of intergenic transcripts in xrn2xrn3 and sal1-8 is the deficient Pol II termination and increased Pol II activity in the intergenic regions downstream of genes.
In Saccharomyces cerevisiae, the XRNs are globally required for Pol II termination (Baejen et al., 2017), whereas in Caenorhabditis elegans, the XRNs appear to be required only at a subset of loci (Miki et al., 2017). In total, 14,718 RTLs were identified in the integrated sal1-8 total RNA-seq, mRNA-seq, and PARE data sets, which were correlated with increased Pol II occupancy at the meta level (Fig. 2, D and E). These RTLs occur at 72.4% of all expressed genes (greater than 0.3 cpm, 20,334 genes), suggesting that Pol II read-through is pervasive across the transcriptome. In fact, a feature of the identified RTLs was that they are detected disproportionately at highly expressed genes; for instance, as measured in the total RNA-seq data set (Supplemental Fig. S3D). This suggests that, for genes that are expressed at a low level, read-through may occur below our limit of detection. Indeed, the frequency of genes in the top expression decile (greater than 42 fragments per kilobase of exon per million fragments mapped) that exhibited read-through in the total RNA-seq was greater than 70%. These results are consistent with a global requirement for SAL1 and XRN activities for the efficient Pol II termination of most genes in Arabidopsis.
To further interrogate the characteristics of RTL transcripts and their consequences, we focused on a single sequencing data set. These subsequent analyses required mate-paired reads and correlations with downstream gene expression, ruling out the single-end PARE data. Given that total RNA-seq captures both capped/uncapped and poly(A)/non-poly(A) and also captures 97% of the mRNA-seq RTLs, we focused on this subset comprising 9,965 distinct RTLs identified in sal1-8 or xrn2xrn3 (Supplemental Fig. S2, B and E).
RTL Transcript Structure
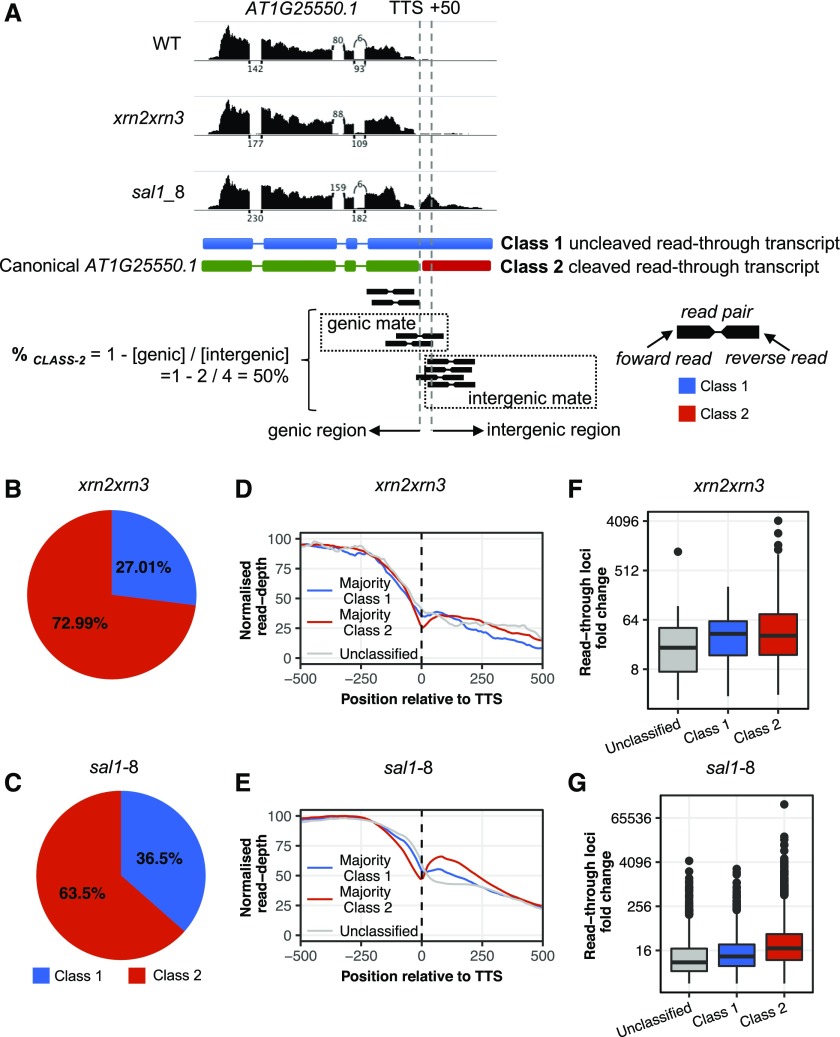
We hypothesized that transcriptional read-through could produce two general transcript classes at the 3′ end of genes. If processing of the primary transcript does not take place, an extended Class 1 transcript comprising the mRNA and the read-through extension will be produced, whereas if processing and cleavage remain functional, a physically separate Class 2 intergenic transcript will result (Fig. 3A). Identifying the prevalence of Class 1 transcripts is important because this may impact the proper functioning of the canonical protein-coding gene by affecting transcription, RNA stability, or translation. In Arabidopsis, it was concluded previously that putative read-through transcripts in the SAL1 mutant fry1-6 were Class 2 based on two genes exhibiting cleaved read-through transcripts (Kurihara et al., 2012). To examine this on a global scale, we estimated the relative abundance of Class 1 and Class 2 putative isoforms using the pattern of mate-paired reads in total RNA-seq data (Fig. 3A). Globally, we estimate that 73% of read-through transcripts are characteristic of Class 2 and 27% are Class 1 for xrn2xrn3 (Fig. 3B); for sal1-8, 63.5% are characteristic of Class 2 and 36.5% are Class 1 (Fig. 3C).
Figure 3.
Assessing the prevalence of continuous read-through products. A, Example RTL AT1G25550.1. Graphs at top display total RNA-seq read coverage over the gene locus and the downstream RTL. Depicted below are the two putative gene/RTL model isoforms, Class 1 and Class 2, and a cartoon depiction of the bioinformatic read-alignment approach to calculating %CLASS 2 (for derivation, see “Materials and Methods”). When one read in a pair overlaps the 50-bp region downstream of the TTS (vertical dashed lines), the mate read then is classified as either genic or intergenic. Using the %CLASS 2 formula, we estimate the fraction of transcripts produced by a locus that fall into Class 2. WT, Wild type. B and C, Estimated cumulative proportion of RTL transcripts that are either Class 1 or Class 2 transcript variants genome wide for xrn2xrn3 (B) and sal1-8 (C). D and E, Each locus was classified as majority Class 1, Class 2, or unclassified, and total RNA-seq read coverage across the TTS ± 500 bp for each RTL category was plotted for xrn2xrn3 (D) and sal1-8 (E). Unclassified loci are represented in gray. F and G, Average abundance of the RTLs in xrn2xrn3 (F) and sal1-8 (G) total RNA-seq compared with that in the wild type (expressed as FC mutant/wild type) for each RTL category. For all plots, blue represents Class 1, red represents Class 2, and gray represents unclassified.
The next question is: for a given gene, are transcripts all Class 1, Class 2, or some proportion of both? Calculating the proportion of Class 2 transcripts on a per-gene basis for xrn2xrn3 (Supplemental Fig. S4A), 79.1% of RTLs produced predominantly Class 2 transcripts (%CLASS 2 > 50), 8.7% produced predominantly Class 1 transcripts (%CLASS 2 < 50), and the remainder were unclassified due to a deficit of intergenic mates indicative of either a misannotated TTS or very short read-through. For sal1-8 (Supplemental Fig. S4B), 63.9% of RTLs produced predominantly Class 2 transcripts (Supplemental Fig. S4C) and 22.4% produced predominantly Class 1 transcripts (Supplemental Fig. S4D). Of note, the two loci examined by Kurihara et al. (2012) were AT1G16410 and AT3G45160, which we found to have %CLASS 2 values in sal1-8 of 61% and 99%, respectively, consistent with the prior report and lending support to our bioinformatic approach. Loci that exhibited a greater proportion of Class 2 cleaved read-through products typically had more abundant read-through relative to both their respective upstream protein-coding gene and wild-type plants, although this trend was clearest for sal1-8 (Fig. 3, D–G). Thus, based on this analysis, the majority of read-through products are cleaved, physically separate intergenic transcripts. However, the prevalence of Class 1 uncleaved continuous read-through products is still substantial across the genome and for a given gene.
Pol II Read-Through Affects Downstream mRNA Expression Proportional to Proximity
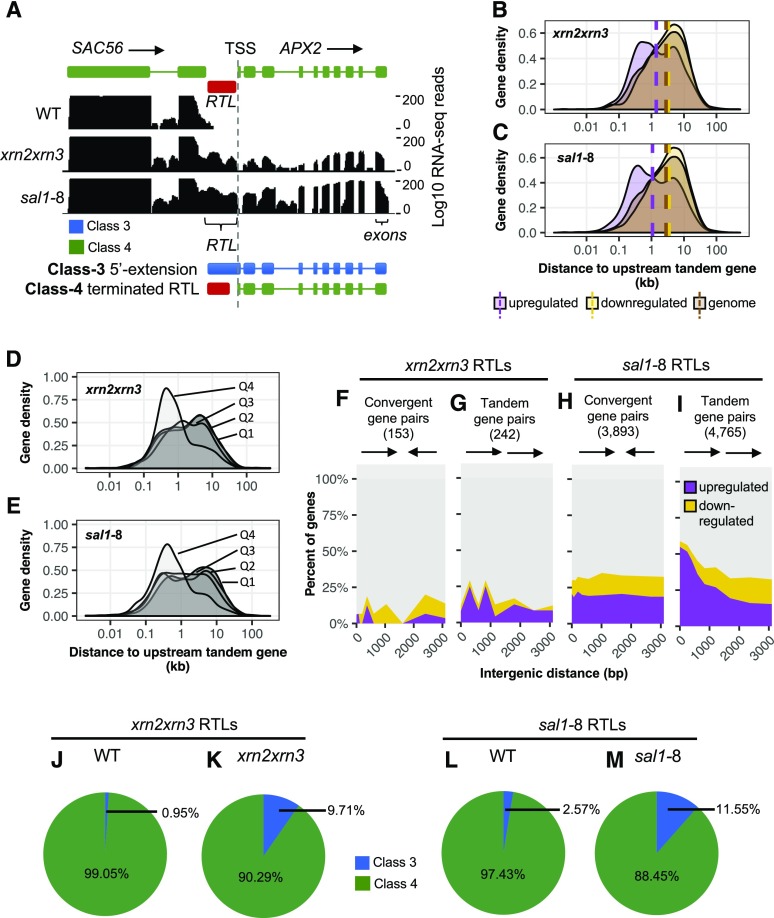
Given that Pol II read-through declines with distance from the TTS, the question arises whether it impacts downstream gene expression either positively by enhancing transcription or negatively by interfering with Pol II, transcription factor binding, or through the generation of antisense transcripts? The sal1-8 mutation was isolated originally in a genetic screen for the up-regulation of APX2. When we examined this locus, we found that APX2 was downstream of a strong RTL, namely SAC56 (Fig. 4A). In the genome, 45% of genes occur in a tandem orientation on the same strand as their nearest upstream gene. All of the 50 most highly up-regulated genes in sal1-8 (50 of 50, 100%) occur in a tandem orientation, compared with only 25% (13 of 50) of the 50 most down-regulated genes. Globally, of the 3,889 up-regulated genes in sal1-8 (Supplemental Tables S3 and S4), 43% were associated with an upstream RTL. In the kernel density plots (Fig. 4, B and C), the median distance for each gene from its upstream neighbor in the genome is 2.8 kb. This reduces to 1.4 kb for xrn2xrn3 up-regulated genes and 1.1 kb for sal1-8 up-regulated genes (purple dashed line), which is significantly and disproportionately closer than the genome average (brown) or the down-regulated set (yellow; P < 0.001 one-sample bootstrap).
Figure 4.
Potential for Pol II run-on to mediate changes in gene expression (global analysis). A, Read-through from SAC56 continues downstream toward the gene APX2. The two potential structures for the downstream APX2 transcript are Class 3 (5′ extension) and Class 4 (canonical start site). Tracks display total RNA-seq read coverage over the gene locus and the downstream RTL. B and C, Gene density plot of up- and down-regulated genes in xrn2xrn3 (B) and sal1-8 (C) against the distance to the nearest upstream tandem gene. Vertical dashed lines represent the median intergenic distance for each subset. D and E, Gene density plots of up-regulated genes in xrn2xrn3 (D) and sal1-8 (E) divided into quartiles (Q1 = first quartile, least up-regulation) against distance to an upstream tandem gene. F to I, Genes downstream of RTLs in a convergent orientation (xrn2xrn3, F; sal1-8, H) or tandem orientation (xrn2xrn3, G; sal1-8, I) were divided into deciles based on intergenic distance and then divided into up-regulated and down-regulated subsets based on mRNA-seq data. The change in the relative proportions of these subsets across deciles was then plotted with each decile set at its median distance in bp. Green represents up-regulated genes, while red represents down-regulated genes. J to M, Classification of genes putatively up-regulated by read-through in xrn2xrn3 (J and K; 20 genes) and sal1-8 (L and M; 903 genes) as Class 3 or Class 4 based on the structure of their TSS using total RNA-seq data. Reads downstream of the TSS (up to 50 bp) were categorized according to their read mate (genic or intergenic) and the ratio of these calculated to represent the relative proportions of Class 3 and Class 4 transcripts. For all plots, blue represents Class 3 transcripts, green represents Class 2 transcripts, purple represents up-regulated mRNAs, and yellow represents down-regulated mRNAs. WT, Wild type.
The normal unimodal distribution for the genome and down-regulated set contrasts with a bimodal distribution for the up-regulated set, with the major peak associated with shorter intergenic distance (Fig. 4, B and C). If read-through were causative of downstream gene activation, then we would anticipate an association between intergenic distance and up-regulation. To test this, we assessed whether the bimodal distribution could be deconvoluted based on expression by dividing the gene sets into four quartiles of expression from low (first quartile) to high (fourth quartile). Indeed, the four quartiles revealed that the distribution of gene expression was associated with the bimodality (Fig. 4, D and E), with the highest expressed quartile being closely associated with the most proximal genes (P < 0.05, Wilcoxon rank-sum test).
Furthermore, we also observed a strong genome-wide correlation between differential expression in xrn2xrn3 and sal1-8 and an interaction of gene orientation and intergenic distance (Fig. 4, F–I). When the gene downstream of an RTL was in a tandem orientation, the frequency of expression up-regulation in sal1-8 increased significantly, from ∼20% at distances greater than 2,000 bp to ∼55% at distances less than 200 bp, with a corresponding decrease in down-regulated and unchanged expression (P < 0.001, Pearson’s χ2 test; Fig. 4I). By contrast, if the downstream gene was in a convergent orientation to the RTL, there were no changes in differential expression as intergenic distance decreased (P > 0.05; Fig. 4H). The smaller number of RTLs in xrn2xrn3 led to more decile-to-decile variability, with proportional change that was not statistically significant (Fig. 4, F and G). However, the expression of tandem downstream genes was more likely to be up-regulated than down-regulated. These results suggest that Pol II read-through can lead to the up-regulation of tandem gene pairs while having no clear association with down-regulation nor a strong effect on genes in colliding orientation.
The association between expression up-regulation in sal1-8 and neighboring gene proximity could potentially be explained by the clustering of active gene expression. Recent reports have identified genes that cluster together in local chromatin domains, such as topologically associating domains (Liu et al., 2017), which may have similar expression levels (Rennie et al., 2017). However, examination of our wild-type RNA-seq data demonstrated that actively expressed genes were not disproportionately close to upstream genes (P > 0.05, pairwise Wilcoxon rank-sum test; Supplemental Fig. S5A).
We subsequently examined genes differentially expressed under drought stress using RNA-seq data (Crisp et al., 2017), which partially overlap with genes differentially expressed in sal1-8, to determine if the up-regulation of gene expression was generally linked to short intergenic distances. To isolate sal1-independent effects, up-regulated genes in this data set were divided into those either up-regulated or unchanged in sal1-8. A clear difference was observed, with the sal1-8 set of up-regulated genes exhibiting a significantly shorter median distance than the sal1-8 set of unchanged genes (Supplemental Fig. S5B; P < 0.05, Wilcoxon rank-sum test). This confirmed that the shift was a specific feature of sal1-8 and not a general characteristic of gene up-regulation.
Collectively, Figures 2, 3, and 4 indicate that a potential threshold for read-through that is able to significantly influence the expression of a downstream gene is on the order of 1,000 bp. In total, 903 up-regulated genes in sal1-8 (23.2% of all up-regulated genes) and 20 up-regulated genes in xrn2xrn3 (1.2% of all up-regulated genes) fall within 1,000 bp of an upstream gene that exhibits read-through.
Defining and Disrupting RTL-Activated Downstream mRNA Expression
An important question is whether the up-regulated transcripts that are downstream of the RTLs have 5′ extensions or whether the 5′ ends map to the canonical TSS. We specifically estimated the proportion of genic transcripts with a 5′ extension spanning the TSS (Class 3) versus canonical transcripts (Class 4) using the mapping location of mate-paired reads. In xrn2xrn3, 9.71% of up-regulated transcripts associated with upstream read-through were characterized as Class 3 (Fig. 4J), meaning that they exhibited 5′ extensions, in contrast to 0.95% of wild-type transcripts at these loci (Fig. 4K). The sal1-8 mutant exhibited the same pattern, with 11.55% of up-regulated transcripts associated with upstream read-through characterized as Class 3 (Fig. 4L), in contrast to 2.57% of wild-type transcripts (Fig. 4M). Thus, read-through does generate some transcripts continuous between the intergenic region and downstream genes; however, by this measure, the large majority of transcripts are canonical and physically separate from the intergenic read-through transcript itself.
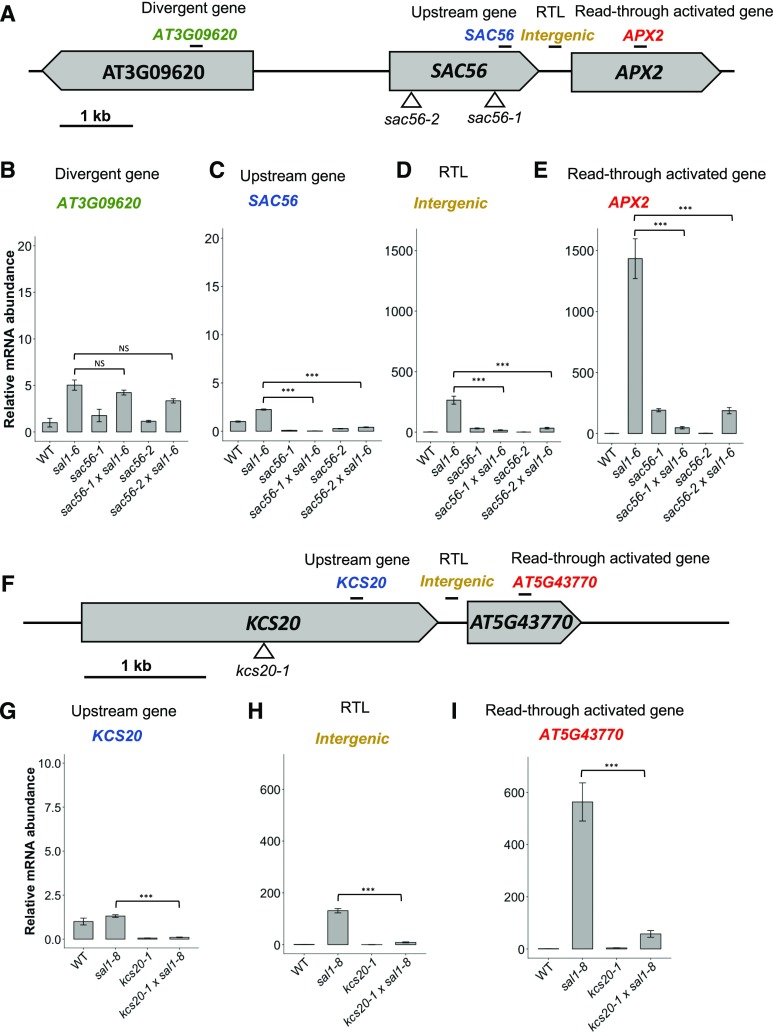
To experimentally verify Pol II read-through as a mechanism capable of promoting downstream gene transcription, we sought to interrupt this process in sal1. T-DNA insertion lines were obtained for genes upstream of two of the most highly up-regulated loci in the set of 903 genes possibly regulated by read-through in sal1-8. These were APX2, which is the most up-regulated gene in sal1-8 and was the basis of the screen that identified a role for SAL1 in retrograde signaling (Wilson et al., 2009), and an uncharacterized Pro-rich gene, AT5G43770, which is induced by abiotic stresses, such as drought and osmotic shock (Kilian et al., 2007), and expressed developmentally to its highest level in dry seed, another tissue high in PAP (Schmid et al., 2005; visualized in the eFP-BAR in Winter et al., 2007). For the gene 432 bp upstream of APX2, namely SAC56, the T-DNA insertion lines sac56-1 and sac56-2 were characterized (Fig. 5A). Similarly, the insertion line kcs20-1 has a T-DNA insertion in the gene KCS20, 192 bp upstream of AT5G43770 (Fig. 5F). The SAC mutations, sac56-1 and sac56-2, and the KCS20 mutation, kcs20-1, were confirmed to substantially and significantly decrease SAC56 and KCS20 mRNA levels, respectively (Fig. 5, C and G; ANOVA, Tukey’s honestly significant difference [HSD] test, P < 0.001), before these lines were crossed with those containing sal1-6 or sal1-8 alleles to obtain homozygous double mutants. Both sal1-6 and sal1-8 lack SAL1 activity and accumulate PAP (Wilson et al., 2009; Estavillo et al., 2011), and sal1-6 has phenocopied sal1-8 in all traits reported to date. We confirmed that read-through levels were highly comparable for the APX2 RTL in lines carrying both sal1 alleles (Supplemental Fig. S6).
Figure 5.
Decoupling of read-through-mediated up-regulation. A, Structure of the APX2 locus and neighboring genes. The APX2 gene is oriented in tandem with the upstream gene SAC56, which is divergent to AT3G09620. B to E, Relative mRNA expression measured by RT-qPCR of AT3G09620 (B), SAC56 (C), the RTL/intergenic region between SAC56 and APX2 (D), and APX2 (E). F, Structure of the AT5G43770 locus and neighboring genes. G to I, Relative mRNA expression of KCS20 (G), the RTL/intergenic region between KCS20 and AT5G43770 (H), and AT5G43770 (I). Samples are normalized within amplicons to that in the wild type (WT) and the reference gene, and error bars indicate se (n = 3). Asterisks indicate significant differences compared with that in the control (ANOVA, Tukey’s HSD test; ***, P < 0.01). NS, Not significant. In A and F, the locations of T-DNA insertions are indicated by triangles and RT-qPCR amplicons are indicated by black bars.
In each double mutant, the insertion of an upstream T-DNA was associated with a significantly attenuated expression of the downstream gene relative to the parental sal1-8 line (Fig. 5, E and I; P < 0.001). We also confirmed that intergenic transcription of the RTL was disrupted significantly (Fig. 5, D and H; P < 0.001). To confirm that these effects were associated with perturbing read-through and not due to a general disruption of the surrounding chromatin, the gene diverging from SAC56 also was assayed. A nonsignificant change in the expression of this gene (AT3G09620) was observed (Fig. 5B). Thus, the major attenuation of intergenic and downstream gene expression can be attributed to the impairment of read-through.
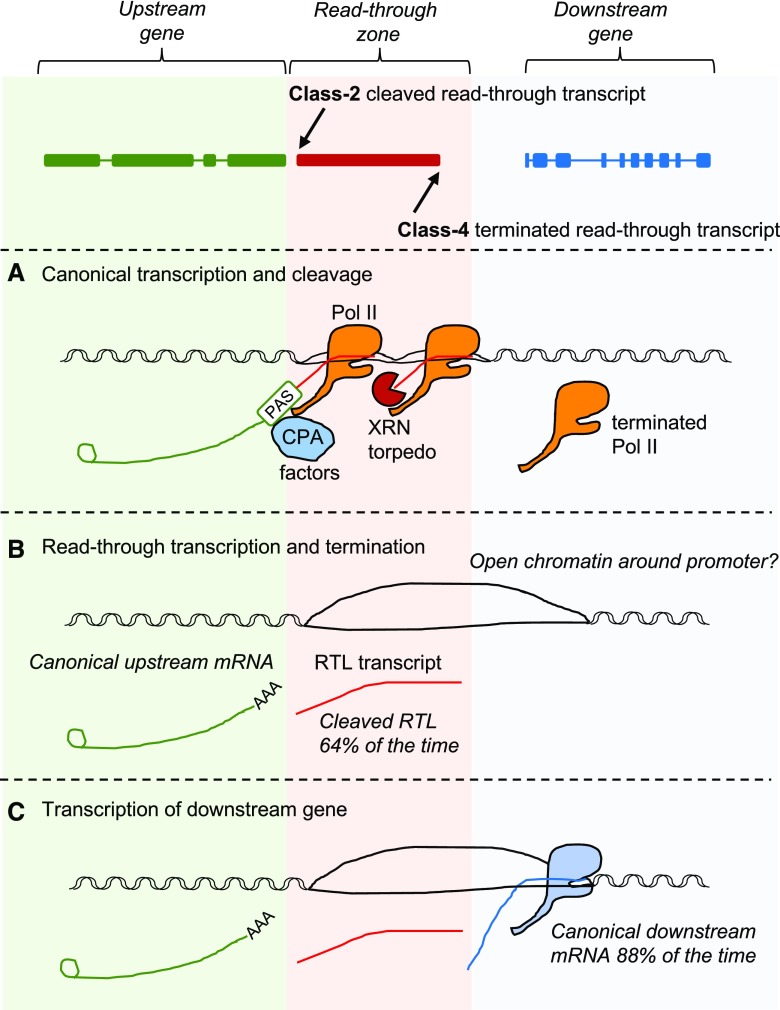
Collectively, the results of our analysis of the structure of RTL transcripts and the expression of downstream genes led us to propose a model whereby read-through can promote the up-regulation of downstream gene expression (Fig. 6).
Figure 6.
Model for read-through-mediated transcript up-regulation. A, When an elongating Pol II transcribes to the end of a protein-coding gene, a concerted process occurs involving coupled 3′ processing of the premRNA and transcription termination. Under a combined allosteric-torpedo model of termination, transcription over the poly(A) site leads to the recruitment of 3′ processing factors and conformational and structural changes that destabilize Pol II. The premRNA is cleaved and polyadenylated to complete the mRNA, and a new 5′ unprotected nascent RNA is generated. Subsequently, an XRN torpedo degrades the nascent RNA from the 5′ end, then catches up with and displaces the Pol II complex from the DNA. CPA, Cleavage and polyadenylation complex; PAS, polyadenylation signal. B, Read-though occurs when Pol II is not effectively terminated and transcribes beyond the end of canonical gene boundaries. In XRN-deficient mutants, we observe this at 14,718 genes. Nevertheless, 64% of the time, cleavage is still functional and the RTL transcript is physically separate from the upstream gene transcript (Class 2). C, Pol II read-through into the intergenic region in the majority of cases is terminated before reaching the TSS of the downstream gene (Class 4). We only found evidence of 3′ extensions 12% of the time; thus, we estimate that 88% of downstream genes are transcribed as canonical mRNAs, with their 5′ ends mapping to the TSS. We speculate that this intergenic Pol II read-through could lead to the up-regulation of the downstream mRNA by altering the chromatin structure or opening the chromatin around promoter regions.
Physiological Significance and Potential Applications for Read-Through
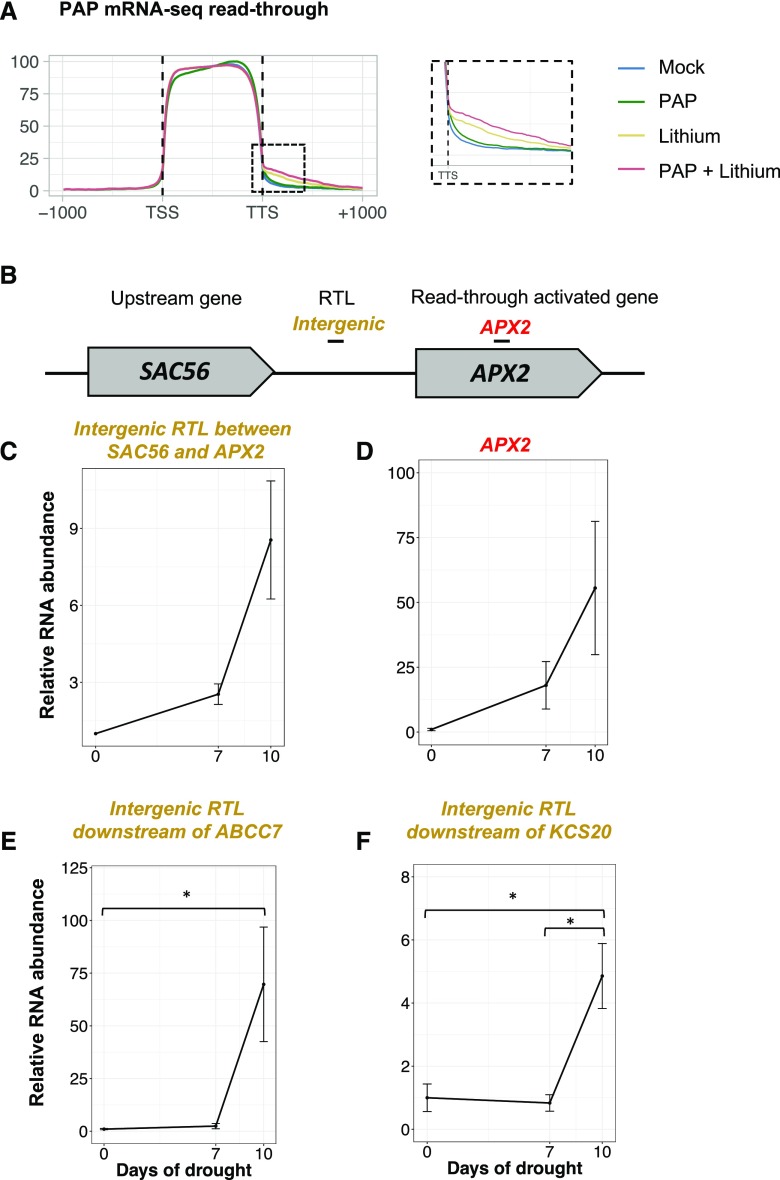
The potential for Pol II read-through to influence the expression of downstream genes in SAL1-PAP mutants raises the possibility that read-through could operate as a novel gene regulatory mechanism under conditions where PAP is elevated. This could include environmental or developmental situations such as during drought and light stress or in dry seed (Estavillo et al., 2011; Pornsiriwong et al., 2017) or could occur as a result of the exogenous application of PAP or PAP analogs. Consistent with this hypothesis, the genes up-regulated in sal1-8 and xrn2xrn3 show a significant overlap (P < 0.001, hypergeometric test) with genes up-regulated under drought (38% and 34%, respectively; Supplemental Fig. S7) and excess light (41% and 53%, respectively). As a second test of this hypothesis, the prevalence of read-through was examined after pharmacological treatments designed to increase cellular PAP levels. The treatment of plant tissues with lithium-containing PAP solutions increases intracellular PAP levels because lithium is a potent inhibitor of PAP catabolism (Murguía et al., 1996; Albert et al., 2000; Estavillo et al., 2011; Pornsiriwong et al., 2017). We reported previously that both PAP+lithium and lithium treatments significantly increase cellular PAP levels (Pornsiriwong et al., 2017). Here, using paired replicate samples from the same experiment, we profiled the transcriptome using RNA-seq. Lithium and PAP+lithium treatments led to the up-regulation of ∼1,500 genes (Table 1; Supplemental Tables S5 and S6), which significantly overlapped with genes up-regulated in sal1-8 (Table 1; P < 0.001, hypergeometric test); however, the down-regulated gene sets did not overlap significantly. No genes were differentially expressed under PAP treatment alone (FC > 1.5, FDR < 0.05). Exogenous PAP application was associated with a global increase in Pol II read-through (Fig. 7A). As expected, treatment with PAP alone did not trigger read-through, likely because of the rapid catabolism of PAP in the absence of lithium. In total, for the tissue samples with increased PAP levels, we identified 62 loci with transcription downstream of the TSS characteristic of RTLs (Table 1; Supplemental Table S7), and in nine cases, the downstream gene was up-regulated. Finally, to examine the possibility of environment-induced read-through, we analyzed the RTL upstream of APX2 (Fig. 7B) in drought-stressed plants using RT-qPCR. After 10 d of soil-drying drought, PAP levels increased moderately from below detectable levels to 15 pmol mg−1 dry weight and APX2 mRNA levels increased 55-fold (Fig. 7D). This was accompanied by the induction of read-through upstream of APX2 at the RTL to 6-fold the level in control plants (Fig. 7C). The transcript abundance of two additional sal1-8 read-through loci was quantified under drought conditions using RT-qPCR, and these loci exhibited ∼5- and ∼70-fold increases, respectively (Fig. 7, E and F).
Table 1. Pharmacological induction of read-through.
Increasing cellular PAP levels are associated with read-through. RTLs were identified in plant samples treated with PAP and lithium solutions, based on mRNA-seq data, using the same method described for sal1 mutants.
| Condition | Differential Expression Analysis | RTL Analysis | |||
|---|---|---|---|---|---|
| Total Up-Regulated Genes | Up-Regulated Genes in Common with sal1-8 | Total RTLs (FC > 2, P < 0.05) | RTLs in Common with sal1-8 | Median RTL FC (Versus Control) | |
| sal1 | 3,454 | N/A | 2,743 | 100 | 160.7 |
| PAP | 0 | N/A | 0 | 0 | N/A |
| Lithium | 1,519 | 23.3% | 29 | 52% (15 of 29) | 20.8 |
| PAP+lithium | 1,326 | 24.9% | 35 | 31% (11 of 35) | 21.7 |
Figure 7.
Read-though under pharmacological treatment and drought stress. A, Increasing cellular PAP levels are associated with increased transcription downstream of genes. PAP levels were increased pharmacologically using petiole feeding to administer mock, PAP, and lithium solutions, then read-through was assessed by summarizing mRNA-seq reads to TAIR10 coding sequence and 1 kb upstream of the TSS and 1 kb downstream of the TTS. B to D, Five-week-old wild-type (Columbia-0) plants were exposed to drought stress by withholding water for 10 D, then the expression of the APX2 RTL was quantified using RT-qPCR. B, Structure of the APX2 locus and the neighboring upstream gene. C, Quantification of the RTL upstream of APX2. D, Quantification of APX2 expression. E and F, The expression of the two additional RTLs, the ABCC7 RTL (E) and the KCS20 RTL (F), under drought conditions was determined in the same manner. Samples are normalized within amplicons to the reference gene, and error bars indicate se (n = 3). Asterisks indicate significant differences compared with that in the control (ANOVA, Tukey’s HSD test; *, P < 0.05).
DISCUSSION
Understanding how the nuclear XRN2 and XRN3 can affect gene expression has important implications generally for gene regulation and specifically for retrograde signaling. In this study, we combined degradome profiling, transcriptome sequencing, Pol II ChIP-seq, and genetic interrogation of Pol II termination in SAL1- and XRN-deficient mutants to elucidate the functions of PAP and the XRNs. In particular, we extended the original applications of PARE (microRNA analysis [Addo-Quaye et al., 2008; German et al., 2008; Gregory et al., 2008] and ribosome footprinting [Hou et al., 2016; Yu et al., 2016; Crisp et al., 2017]) to the identification of XRN substrates. PARE revealed that, in the XRN-deficient sal1-8 and xrn2xrn3 mutants, there was a global increase in the RNA degradome, particularly at the 3′ end of genes and into downstream intergenic regions (Figs. 1 and 2A). This observation is consistent with the known role of XRN2/XRN3 homologs in Pol II termination in yeast, humans, and worms (Kim et al., 2004; West et al., 2004; Miki et al., 2017). Importantly, whereas prior reports have demonstrated correlations between mRNA levels in sal1 and xrn mutants, we show here on a global scale that sal1 mutants accumulate extremely high levels of characteristic XRN substrate RNAs, in particular XRN2 and XRN3 substrates.
Arabidopsis XRN2 and XRN3 Are Required Genome Wide for Pol II Termination
Under the combined allosteric-torpedo model of termination in eukaryotes (Luo and Bentley, 2004; Schreieck et al., 2014; Proudfoot, 2016), the XRNs play a role in terminating Pol II. The torpedo operates globally in S. cerevisiae (Baejen et al., 2017) but is restricted in C. elegans (Miki et al., 2017) and, until recently, was only hypothesized to occur in plants (Kurihara et al., 2012). Genome structure both between and within species can vary a great deal, including gene density and chromatin regulation, from yeast’s compact genome to larger plant genomes with relatively vast intergenic spaces. Here, we find evidence of read-through transcripts for the majority of expressed genes (more than 14,000, or 72%). We demonstrate a global functionality by profiling Pol II occupancy genome wide in xrn2xrn3 and sal1 mutants and report increased levels of Pol II downstream of protein-coding genes. We also extend this to a demonstration that the retrograde regulation of XRNs via SAL1-PAP signaling also can impact Pol II activity. Consistently, we observed higher levels of read-through and related phenotypes in sal1-8 compared with that in xrn2xrn3. This is consistent with prior results suggesting that xrn3-3 is a relatively weak hypomorphic allele (Gy et al., 2007; Zakrzewska-Placzek et al., 2010) and that the sal1-8 mutation likely leads to a greater impairment of XRN activity in comparison with that caused by xrn2xrn3 (Estavillo et al., 2011; Pornsiriwong et al., 2017). In parallel with our observations, a very recent corroborative study in Arabidopsis also concluded that the XRNs are required for Pol II termination and that a more severe xrn3-8 mutant allele resulted in a corresponding increase in Pol II read-through in xrn3-8 compared with the levels we observed in xrn3-3 (Krzyszton et al., 2018). Together, these two parallel investigations independently demonstrate the role of the XRNs in Pol II termination in Arabidopsis.
Pol II Read-Through Up-Regulates the Expression of Downstream Genes
Theoretically, read-through has the potential to negatively affect gene expression, in particular by disrupting the transcription of neighboring genes, thus reducing their mRNA levels (Shearwin et al., 2005). In this context, we were surprised to discover that the effect of read-through was the up-regulation of tandemly oriented downstream genes without any significant or observable changes that could be associated with the down-regulation of adjacent genes. This does not preclude the existence of the latter, but it is clearly a minor effect at a global level (Fig. 4, D and E). We also found that up-regulation correlates with distance from an RTL. As intergenic distance increases, the likelihood of the Pol II complex destabilizing or terminating by alternative mechanisms increases. Read-through into downstream genes is consistent with prior reports in S. cerevisiae, where, in XRN-deficient strains, around 35% of nonterminated Pol II enzymes are estimated to transcribe into downstream genes (Baejen et al., 2017). Likewise, in C. elegans, reporter constructs were employed to demonstrate a capacity for read-through to activate downstream gene expression. In that study, it was also found that promoter elements affect the mode of downstream termination, pointing to mechanisms regulating termination.
Our investigation extends upon prior correlative studies in S. cerevisiae and C. elegans (Baejen et al., 2017; Miki et al., 2017) by decoupling the expression of tandem gene pairs, an important test of causality, by using T-DNA insertional mutants. While the insertion of a large T-DNA could have unanticipated effects on gene expression at an off-target locus, in the case of APX2, only the downstream neighboring gene and not the divergent upstream gene was affected substantially, thus suggesting that the direct impact of the T-DNA was largely localized to the gene it was inserted in. These experiments were performed in sal1 mutant backgrounds because the read-through phenotype was more readily detectable in a sal1 background compared with the hypomorphic xrn2xrn3 and a loss-of-function XRN3 mutant is unavailable because it is embryo lethal (Gy et al., 2007; Zakrzewska-Placzek et al., 2010). Using this approach, we demonstrated that the disruption of upstream genes precluded intergenic read-through and restored the expression of the downstream gene to wild-type levels at the two independent loci examined.
A Functional Role for Read-Through, Implications for Retrograde Signaling and Beyond
While read-through can lead to increased transcript levels of downstream genes, an important consideration is whether these read-through transcripts are cleaved or continuous with the coding sequence. Under the torpedo model of termination, the XRNs theoretically operate after cleavage and polyadenylation of the upstream premRNA, which exposes a 5′ unprotected nascent RNA for degradation from the 5′ end. An earlier study demonstrated cleavage for two genes (Kurihara et al., 2012). Extending this to a global analysis, we estimate that the majority of RTL transcripts are cleaved. Nevertheless, extensions were found at detectable levels at many RTLs, and globally up to 36% of all RTL transcripts could be 3′ extension products (Fig. 3, A and B). RTL transcripts were found to be both poly(A) and non-poly(A). The reason we observe cleaved and continuous read-through products in a poly(A)-derived RNA-seq is not clear. One possibility is that polyadenylation may be part of a mechanism for targeting nuclear RNAs for decay, which has been reported in animals (LaCava et al., 2005; Wyers et al., 2005; Houseley and Tollervey, 2009). Alternatively, the transcribing Pol II may have reached an alternate/cryptic poly(A) signal either in the intergenic region or in a downstream gene, potentially generating chimeric or extended gene read-through products. There are potential limitations to using short-read sequencing to infer transcript structure, and in the future, these conclusions might be tested by full-length cDNA sequencing, for instance using Nanopore or PacBio technology.
Extensions to the 3′ end of mRNAs could affect gene expression; however, extensions are anticipated to have a much greater effect at the 5′ end. For the 903 RTL downstream genes up-regulated in sal1-8, we estimate that 88% of the time they are initiated at the canonical TSS, with 11.6% frequency of 5′ extensions (compared with 2.6% in the wild type). We did not observe evidence of the 5′ extensions leading to gene down-regulation, as for yeast; specifically, yeast Ndc80 is down-regulated by a 5′ extended long undecoded NDC80 transcript isoform (Chia et al., 2017). While evaluation of translation rates for 900 genes is well beyond the scope of this study, for a number of the RTL-activated downstream genes, there is demonstrated physiological and biochemical evidence of increased expression. For example, in the case of the RTL-associated APX2, sal1 mutants have significantly reduced levels of the substrate of APX2, hydrogen peroxide (Rossel et al., 2006; Wilson et al., 2009; Estavillo et al., 2011; Pornsiriwong et al., 2017).
Is read-through restricted to sal1 and xrn mutants? In this context, the sal1-8 and xrn2xrn3 transcriptomes significantly overlap with transcripts up-regulated by drought and excess light, and both SAL1 and XRNs have demonstrated roles in the retrograde signaling of oxidative stress in vivo via reactive oxygen species and redox regulation of chloroplast-localized SAL1 (Wilson et al., 2009; Estavillo et al., 2011; Chan et al., 2016a; Pornsiriwong et al., 2017). In total, 23% of the sal1-8 up-regulated gene set overlaps with the RTL-activated up-regulated set. We also found a strong correlation between the accumulation of PAP during drought and the increased expression of the three RTLs examined, including the RTL upstream of APX2. Several other situations and conditions also are reported to stimulate read-through at specific loci, such as the transcription of Piwi-interacting RNA clusters of Drosophila melanogaster (Mohn et al., 2014), osmotic stress in human cells (Vilborg et al., 2015), cancer (Grosso et al., 2015), and lithium and sodium stress in Arabidopsis (Kurihara et al., 2012). In Arabidopsis, continuous 3′ read-through products from small nuclear RNAs can activate the expression of downstream genes, and this mode of read-through-activated expression was found to be activated by salt stress (Fukudome et al., 2017). Those authors proposed that read-through transcription to produce 3′-extended small nuclear RNA is not a mere transcriptional abnormality but is a part of the transcriptional regulatory strategies in plants. We also found that pharmacological manipulation of PAP levels induced read-through, raising the possibility that exogenous treatments could be applied to plants to manipulate this process. Thus, there is some limited evidence that read-through can occur in response to treatments and drought stress, but whether this is physiologically important or widespread is an open question.
A core goal of this study was to investigate a candidate mechanism by which PAP and the XRNs could modulate gene expression during retrograde signaling. By characterizing sal1 and xrn2xrn3 mutants, we have shown how the SAL1-PAP-XRN pathway could potentially increase transcript abundance via a read-through mechanism (Fig. 6) of up to 903 genes, which is 23% of the sal1-8 up-regulated transcriptome. While we propose that this read-through model is widespread, applying to many genes, it is likely that there are other direct and indirect regulatory mechanisms that affect the expression of many other genes in parallel. We demonstrated that the read-through expression of neighboring genes was affected in a distance-dependent manner up to ∼1,000 bp. Contrary to scenarios where the production of antisense transcripts down-regulates genes, there was no significant change to downstream genes on the complementary strand. Instead, the major effect of read-through in sal1-8 and xrn2xrn3 was to increase mRNAs of genes in a tandem orientation. Significantly, our study moves beyond correlation, demonstrating that 88% of transcripts of the downstream loci are predicted to be transcribed from the canonical TSS and that disruption to upstream gene expression causes the attenuation of intergenic and downstream gene expression.
MATERIALS AND METHODS
Plant Material
For all experiments, Arabidopsis (Arabidopsis thaliana) Columbia-0 plants were cultivated in soil under a 12- or 16-h photoperiod, 100 ± 25 μmol photons m−2 s−1 photosynthetically active radiation, and 23°C/22°C ± 2°C day/night temperatures. The SAL1 mutant alleles sal1-8 (alx8) and sal1-6 (fry1-6) and the xrn2xrn3 double mutant were as described previously (Crisp et al., 2017). The suppressor of acaulis56-1 (sac56-1) mutant (SALK-027492), sac56-2 mutant (SALK-130595), and 3-ketoacyl-CoA synthase20-1 (kcs20-1) mutant (SALK-019727) were recovered in the SALK sequence-indexed T-DNA insertion collections (Sessions et al., 2002; Alonso et al., 2003) obtained from the Arabidopsis Biological Resource Center at Ohio State University. Primer sequences used for genotyping are provided in Supplemental Table S8. Drought treatments were performed as described by Crisp et al. (2017) by withholding water from 5-week-old soil-grown plants for 10 d. PAP and lithium treatments were performed as described by Pornsiriwong et al. (2017) using a petiole feeding technique to administer solutions of 100 mm LiCl, 10 mm ATP, or 1 mm PAP (Sigma-Aldrich) alone or in different combinations, prepared in infiltration buffer (1 mm PIPES-KOH, pH 6, 1 mm sodium citrate, 1 mm KCl, and 15 mm Suc) modified from a cordycepin infiltration buffer (Seeley et al., 1992). Solutions were administered through the petioles of excised leaves. The samples used in this study for transcript profiling were paired replicates of samples from the same experiment described byPornsiriwong et al. (2017) (Fig. 2; Supplemental Fig. S1).
RT-qPCR Analysis
Total RNA was extracted using the Ambion PureLink RNA mini kit with Trizol (Thermo Fisher) and treated on column with DNase (Sigma-Aldrich). Quantification and RNA quality were determined using a LabChIP GXII (PerkinElmer), and 1 μg of RNA was reverse transcribed into cDNA using the Invitrogen SS III First Strand cDNA synthesis kit and oligo(dT18VN) primers. qPCR was performed in a technical duplicate on a LightCycler480 using SybrGreen I (Roche Diagnostics). Data were analyzed using LinReg PCR (Ramakers et al., 2003; Ruijter et al., 2009) and normalized to the reference gene PROTEIN PHOSPHATASE2A SUBUNIT. Primers for RT-qPCR are listed in Supplemental Table S8. Statistical significance was assessed using a one-way ANOVA between genotypes, with posthoc multiple comparison testing using Tukey’s HSD test. Statistically significant differences were defined as pairwise comparisons with an adjusted P value of less than 0.05.
Sequencing Library Preparation and Analysis
A full description of each library preparation, bioinformatic analysis pipeline, and software used is provided in Supplemental Methods, including links for version-controlled online protocols (Supplemental Table S9), and code is available online on GitHub (https://github.com/pedrocrisp/NGS-pipelines). For transcriptome profiling, total RNA was extracted from two whole rosettes (excised from their roots) per sample replicate using an acidic phenol:chloroform method adapted from Box et al. (2011). For mRNA-seq of pharmacological treatments, the Illumina TruSeq mRNA stranded kit was used with minor modifications (TruSeq Stranded mRNA Sample Preparation Guide Revision E), including scaling reactions by one-third. For mRNA-seq of the sal1-8 and xrn2xrn3 mutants, the Illumina TruSeq V2 kit was used (TruSeq User Guide Revision D) with a custom dUTP incorporation step during second strand synthesis to introduce strand specificity (Parkhomchuk et al., 2009). For total RNA-seq, 1 μg of RNA was depleted for rRNA using the Illumina Plant Ribo-Zero kit, following the manufacturer’s protocol (Ribo-Zero Magnetic Kits Guide Revision A and TruSeq Stranded Total RNA Guide Revision E), except that all reactions were scaled by one-half. PARE (degradome) libraries were prepared by capturing a 20- to 21-nucleotide tag of the 5′ end of all uncapped (5′ monophosphate) mRNA molecules. Libraries were prepared from poly(A) RNA captured from 75 μg of total RNA (the same RNA used for RNA-seq), according to Zhai et al. (2014) with minor modifications. Chromatin extraction was adapted from Gendrel et al. (2005) with modifications from Komar et al. (2016). ChIP-seq library preparation was carried out using the ThruPLEX DNA-seq kit (Rubicon Genomics).
Quality control of sequencing data was performed using FastQC. Adapters were trimmed with scythe; read quality was trimmed with sickle. RNA-seq data were mapped to TAIR10 using subjunc (Liao et al., 2013) and summarized using featureCounts (Liao et al., 2014). Differential expression testing was performed following the edgeR-limma-voom approach, with a differential expression threshold of FDR < 0.05 and FC > 1.5. PARE data were mapped using bowtie2 (Langmead and Salzberg, 2012). ChIP data were aligned using subread-align, and quality control was performed using SICER (Xu et al., 2014). Read density gene coverage plots were generated using bedtools (Quinlan and Hall, 2010), removing introns and summarizing and binning the data in R version 3.3.2 (www.r-project.org). Coverage within bins across regions of interest was fitted to a linear mixed-effects model using the R package lmerTest by calculating the mean at each bin across the region of interest followed by an ANOVA to identify a significant genotype effect within the model.
PAP Quantification
Quantification of PAP was performed according to Estavillo et al. (2011) and Pornsiriwong et al. (2017) with minor modifications. Total adenosines were extracted from leaf tissue with 0.1 m HCl, derivatized with chloroacetaldehyde, and quantified fluorometrically after HPLC fractionation. PAP quantification was performed by integrating the HPLC peak area and converting to pmol units using standard curves of 0.2, 10, and 20 pmol of standard (Bürstenbinder et al., 2007). The following modifications were made to the run timetable: equilibration of the column for 0.05 min with 95% (v/v) buffer A (5.7 mm [CH3(CH2)3]4N HSO4 and 30.5 mm KH2PO4, pH 5.8) and 5% (v/v) buffer B (67% [v/v] acetonitrile and 33% buffer A), linear gradient for 13.45 min up to 50% (v/v) buffer B, and reequilibration for 1.8 min with 5% (v/v) buffer B.
Identification of RTLs
All reads were associated with their nearest gene using BEDTools version 2.26.0 genomecov, and coverage depth was summarized in a window up to 500 bp (depending on intergenic distance) on either side of the annotated transcription termination site for genes with cpm > 0.3, excluding intergenic loci of less than 50 bp in length or with a mean coverage of less than 2×. Mean intergenic coverage was normalized to mean genic coverage (McKinlay et al., 2018), and loci with mean intergenic coverage greater than 8 times than of genic coverage were excluded to prevent unannotated loci within the intergenic region from confounding the results. Significant differences between genotypes were identified using limma (FDR < 0.05, FC > 2).
Cleavage and Structural Analysis of RTL Transcripts
Paired-end reads that overlapped with a region composed of the TTS + 50 nucleotides were identified and categorized as either intergenic or genic according to the location of their mate reads. For reads originating from Class 1 transcripts, no bias in either direction would be expected, while for reads originating from Class 2 transcripts, a bias toward intergenic mate reads would be expected. The proportion of Class 2 transcripts was calculated as 100 − genic mates × 100/intergenic mates (for derivation of the formula, see Supplemental Methods). As short Class 2 transcripts (less than the average fragment size of ∼175 nucleotides) are lost during RNA-seq library preparation, this method necessarily underestimates the number of these transcripts.
Accession Numbers
Transcriptomic and ChIP data for sal1 and xrn2xrn3 mutants have been deposited at the National Center for Biotechnology Information Short Read Archive under BioProject PRJNA448270. Transcriptomic data for PAP and lithium treatments have been deposited at the National Center for Biotechnology Information Gene Expression Omnibus data repository (GSE115951).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Sequencing of XRN-sensitive transcripts using PARE.
Supplemental Figure S2. Identification of read-through loci in individual sequencing-type data sets.
Supplemental Figure S3. RTL pipeline quality control.
Supplemental Figure S4. Read-through loci transcript structure at individual loci.
Supplemental Figure S5. Distances between tandem gene pairs in wild-type control and drought-stressed plants.
Supplemental Figure S6. Comparison of read-through between sal1-8 and sal1-6 alleles.
Supplemental Figure S7. Overlap between sal1-8 and stress transcriptomes.
Supplemental Table S1. Summary of transcriptomic data sets.
Supplemental Table S2. RTL expression in xrn2xrn3 and sal1-8.
Supplemental Table S3. Genes differentially expressed in sal1-8 (mRNA-seq).
Supplemental Table S4. Genes differentially expressed in xrn2xrn3 (mRNA-seq).
Supplemental Table S5. Genes differentially expressed in lithium treatment (mRNA-seq).
Supplemental Table S6. Genes differentially expressed in PAP+lithium treatment (mRNA-seq).
Supplemental Table S7. RTL expression in plants treated with lithium or PAP+lithium.
Supplemental Table S8. Primers used in this study.
Supplemental Table S9. Protocol Web links.
Supplemental Methods. Extended technical descriptions of methods.
Acknowledgments
We thank Milos Tanurdizc and Max Nekrasov for advice and assistance with optimizing Pol II ChIP assays and Su Yin Phua and Kai Chan for assistance with pharmacological treatments. We acknowledge the contributions of Terry Neeman toward the statistical analyses performed. We thank Nathan Springer for valuable discussions. We acknowledge the Biomolecular Resource Facility at the Australian National University and the Australian Genome Research Facility for performing Illumina sequencing. This investigation also was supported by the provision of plant growth facilities by the Australian Plant Phenomics Facility and computational infrastructure by the National Computational Infrastructure, both supported under the National Collaborative Research Infrastructure Strategy of the Australian Government.
Footnotes
We received financial assistance from the ARC Centre of Excellence in Plant Energy Biology (CE140100008). P.A.C., A.B.S., and D.R.G. were also supported by Grains Research and Development Council scholarships (GRS184, GRS11010, and GRS10683); S.R.E. by an ARC Discovery Early Career Researcher Award (DE150101206). D.R.G. and A.B.S. were also supported by Australian Research Training Program (RTP) scholarships.
Articles can be viewed without a subscription.
References
- Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ (2008) Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol 18: 758–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert A, Yenush L, Gil-Mascarell MR, Rodriguez PL, Patel S, Martínez-Ripoll M, Blundell TL, Serrano R (2000) X-ray structure of yeast Hal2p, a major target of lithium and sodium toxicity, and identification of framework interactions determining cation sensitivity. J Mol Biol 295: 927–938 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Baejen C, Andreani J, Torkler P, Battaglia S, Schwalb B, Lidschreiber M, Maier KC, Boltendahl A, Rus P, Esslinger S, et al. (2017) Genome-wide analysis of RNA Polymerase II termination at protein-coding genes. Mol Cell 66: 38–49.e6 [DOI] [PubMed] [Google Scholar]
- Box MS, Coustham V, Dean C, Mylne JS (2011) Protocol: a simple phenol-based method for 96-well extraction of high quality RNA from Arabidopsis. Plant Methods 7: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürstenbinder K, Rzewuski G, Wirtz M, Hell R, Sauter M (2007) The role of methionine recycling for ethylene synthesis in Arabidopsis. Plant J 49: 238–249 [DOI] [PubMed] [Google Scholar]
- Chan KX, Mabbitt PD, Phua SY, Mueller JW, Nisar N, Gigolashvili T, Stroeher E, Grassl J, Arlt W, Estavillo GM, et al. (2016a) Sensing and signaling of oxidative stress in chloroplasts by inactivation of the SAL1 phosphoadenosine phosphatase. Proc Natl Acad Sci USA 113: E4567–E4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ (2016b) Learning the languages of the chloroplast: retrograde signaling and beyond. Annu Rev Plant Biol 67: 25–53 [DOI] [PubMed] [Google Scholar]
- Chia M, Tresenrider A, Chen J, Spedale G, Jorgensen V, Ünal E, van Werven FJ (2017) Transcription of a 5′ extended mRNA isoform directs dynamic chromatin changes and interference of a downstream promoter. eLife 6: e27420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp PA, Ganguly D, Smith AB, Murray KD, Estavillo GM, Searle IR, Ford E, Bogdanović O, Lister R, Borevitz JO, et al. (2017) Rapid recovery gene downregulation during excess-light stress and recovery in Arabidopsis. Plant Cell 29: 1836–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson L, Kerr A, West S (2012) Co-transcriptional degradation of aberrant pre-mRNA by Xrn2. EMBO J 31: 2566–2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhard KF Jr, Talbot JERB, Deans NC, McClish AE, Hollick JB (2015) Nascent transcription affected by RNA polymerase IV in Zea mays. Genetics 199: 1107–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estavillo GM, Crisp PA, Pornsiriwong W, Wirtz M, Collinge D, Carrie C, Giraud E, Whelan J, David P, Javot H, et al. (2011) Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23: 3992–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N, Brannan K, Erickson B, Kim H, Cortazar MA, Sheridan RM, Nguyen T, Karp S, Bentley DL (2015) Effects of transcription elongation rate and Xrn2 exonuclease activity on RNA Polymerase II termination suggest widespread kinetic competition. Mol Cell 60: 256–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukudome A, Sun D, Zhang X, Koiwa H (2017) Salt stress and CTD PHOSPHATASE-LIKE4 mediate the switch between production of small nuclear RNAs and mRNAs. Plant Cell 29: 3214–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzani S, Lawrenson T, Woodward C, Headon D, Sablowski R (2004) A link between mRNA turnover and RNA interference in Arabidopsis. Science 306: 1046–1048 [DOI] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Martienssen R, Colot V (2005) Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods 2: 213–218 [DOI] [PubMed] [Google Scholar]
- German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, De et al. (2008) Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol 26: 941–946 [DOI] [PubMed] [Google Scholar]
- Gregory BD, O’Malley RC, Lister R, Urich MA, Tonti-Filippini J, Chen H, Millar AH, Ecker JR (2008) A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell 14: 854–866 [DOI] [PubMed] [Google Scholar]
- Grosso AR, Leite AP, Carvalho S, Matos MR, Martins FB, Vítor AC, Desterro JMP, Carmo-Fonseca M, de Almeida SF (2015) Pervasive transcription read-through promotes aberrant expression of oncogenes and RNA chimeras in renal carcinoma. eLife 4: e09214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gy I, Gasciolli V, Lauressergues D, Morel JB, Gombert J, Proux F, Proux C, Vaucheret H, Mallory AC (2007) Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell 19: 3451–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou CY, Lee WC, Chou HC, Chen AP, Chou SJ, Chen HM (2016) Global analysis of truncated RNA ends reveals new insights into ribosome stalling in plants. Plant Cell 28: 2398–2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Tollervey D (2009) The many pathways of RNA degradation. Cell 136: 763–776 [DOI] [PubMed] [Google Scholar]
- Jiao X, Xiang S, Oh C, Martin CE, Tong L, Kiledjian M (2010) Identification of a quality-control mechanism for mRNA 5′-end capping. Nature 467: 608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno-González S, Haaning LL, Malagon F, Jensen TH (2010) The yeast 5′-3′ exonuclease Rat1p functions during transcription elongation by RNA polymerase II. Mol Cell 37: 580–587 [DOI] [PubMed] [Google Scholar]
- Kastenmayer JP, Green PJ (2000) Novel features of the XRN-family in Arabidopsis: evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc Natl Acad Sci USA 97: 13985–13990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S (2004) The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature 432: 517–522 [DOI] [PubMed] [Google Scholar]
- Komar DN, Mouriz A, Jarillo JA, Piñeiro M (2016) Chromatin Immunoprecipitation Assay for the Identification of Arabidopsis Protein-DNA Interactions In Vivo. J Vis Exp (107): e53422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzyszton M, Zakrzewska-Placzek M, Kwasnik A, Dojer N, Karlowski W, Kufel J (2018) Defective XRN3-mediated transcription termination in Arabidopsis affects the expression of protein-coding genes. Plant J 93: 1017–1031 [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Schmitz RJ, Nery JR, Schultz MD, Okubo-Kurihara E, Morosawa T, Tanaka M, Toyoda T, Seki M, Ecker JR (2012) Surveillance of 3′ noncoding transcripts requires FIERY1 and XRN3 in Arabidopsis. G3 (Bethesda) 2: 487–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D (2005) RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2013) The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41: e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930 [DOI] [PubMed] [Google Scholar]
- Liu C, Cheng YJ, Wang JW, Weigel D (2017) Prominent topologically associated domains differentiate global chromatin packing in rice from Arabidopsis. Nat Plants 3: 742–748 [DOI] [PubMed] [Google Scholar]
- Luo W, Bentley D (2004) A ribonucleolytic rat torpedoes RNA polymerase II. Cell 119: 911–914 [DOI] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA (2014) RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet 15: 394–408 [DOI] [PubMed] [Google Scholar]
- McKinlay A, Podicheti R, Wendte JM, Cocklin R, Rusch DB (2018) RNA polymerases IV and V influence the 3′ boundaries of Polymerase II transcription units in Arabidopsis. RNA Biol 15: 269–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki TS, Carl SH, Großhans H (2017) Two distinct transcription termination modes dictated by promoters. Genes Dev 31: 1870–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Sienski G, Handler D, Brennecke J (2014) The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell 157: 1364–1379 [DOI] [PubMed] [Google Scholar]
- Murguía JR, Bellés JM, Serrano R (1996) The yeast HAL2 nucleotidase is an in vivo target of salt toxicity. J Biol Chem 271: 29029–29033 [DOI] [PubMed] [Google Scholar]
- Nagarajan VK, Jones CI, Newbury SF, Green PJ (2013) XRN 5′ → 3′ exoribonucleases: structure, mechanisms and functions. Biochim Biophys Acta 1829: 590–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, Krobitsch S, Lehrach H, Soldatov A (2009) Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res 37: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson BJ, Woo NS, Förster B, Small ID (2008) Plastid signalling to the nucleus and beyond. Trends Plant Sci 13: 602–609 [DOI] [PubMed] [Google Scholar]
- Pornsiriwong W, Estavillo GM, Chan KX, Tee EE, Ganguly D, Crisp PA, Phua SY, Zhao C, Qiu J, Park J, et al. (2017) A chloroplast retrograde signal, 3′-phosphoadenosine 5′-phosphate, acts as a secondary messenger in abscisic acid signaling in stomatal closure and germination. eLife 6: e23361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ. (1989) How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem Sci 14: 105–110 [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ. (2016) Transcriptional termination in mammals: stopping the RNA polymerase II juggernaut. Science 352: aad9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Rennie S, Dalby M, van Duin L, Andersson R (2017) Transcriptional decomposition reveals active chromatin architectures and cell specific regulatory interactions. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossel JB, Walter PB, Hendrickson L, Chow WS, Poole A, Mullineaux PM, Pogson BJ (2006) A mutation affecting ASCORBATE PEROXIDASE 2 gene expression reveals a link between responses to high light and drought tolerance. Plant Cell Environ 29: 269–281 [DOI] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, van den Hoff MJB, Moorman AFM (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schreieck A, Easter AD, Etzold S, Wiederhold K, Lidschreiber M, Cramer P, Passmore LA (2014) RNA polymerase II termination involves C-terminal-domain tyrosine dephosphorylation by CPF subunit Glc7. Nat Struct Mol Biol 21: 175–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley KA, Byrne DH, Colbert JT (1992) Red light-independent instability of oat phytochrome mRNA in vivo. Plant Cell 4: 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearwin KE, Callen BP, Egan JB (2005) Transcriptional interference: a crash course. Trends Genet 21: 339–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk EL, Chen CL, d’Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix-Né P, Loeillet S, et al. (2011) XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature 475: 114–117 [DOI] [PubMed] [Google Scholar]
- Vilborg A, Passarelli MC, Yario TA, Tycowski KT, Steitz JA (2015) Widespread inducible transcription downstream of human genes. Mol Cell 59: 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Gromak N, Proudfoot NJ (2004) Human 5′ → 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature 432: 522–525 [DOI] [PubMed] [Google Scholar]
- Wilson PB, Estavillo GM, Field KJ, Pornsiriwong W, Carroll AJ, Howell KA, Woo NS, Lake JA, Smith SM, Millar AH, et al. (2009) The nucleotidase/phosphatase SAL1 is a negative regulator of drought tolerance in Arabidopsis. Plant J 58: 299–317 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Chory J (2012) Organelle signaling: how stressed chloroplasts communicate with the nucleus. Curr Biol 22: R690–R692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Régnault B, Devaux F, Namane A, Séraphin B, et al. (2005) Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737 [DOI] [PubMed] [Google Scholar]
- Xu S, Grullon S, Ge K, Peng W (2014) Spatial clustering for identification of ChIP-enriched regions (SICER) to map regions of histone methylation patterns in embryonic stem cells. Methods Mol Biol 1150: 97–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Willmann MR, Anderson SJ, Gregory BD (2016) Genome-wide mapping of uncapped and cleaved transcripts reveals a role for the nuclear mRNA cap-binding complex in cotranslational RNA decay in Arabidopsis. Plant Cell 28: 2385–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrzewska-Placzek M, Souret FF, Sobczyk GJ, Green PJ, Kufel J (2010) Arabidopsis thaliana XRN2 is required for primary cleavage in the pre-ribosomal RNA. Nucleic Acids Res 38: 4487–4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Arikit S, Simon SA, Kingham BF, Meyers BC (2014) Rapid construction of parallel analysis of RNA end (PARE) libraries for Illumina sequencing. Methods 67: 84–90 [DOI] [PubMed] [Google Scholar]
- Zuo Y, Deutscher MP (2001) Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res 29: 1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]