Nitrate transporter NRT1.1 enhances NH4+ accumulation, disturbs NH4+ metabolism, and aggravates NH4+ toxicity in Arabidopsis grown in a high concentration of NH4+ as the sole N source.
Abstract
A high concentration of ammonium (NH4+) as the sole source of nitrogen in the growth medium often is toxic to plants. The nitrate transporter NRT1.1 is involved in mediating the effects of NH4+ toxicity; however, the mechanism remains undefined. In this study, wild-type Arabidopsis (Arabidopsis thaliana Columbia-0 [Col-0]) and NRT1.1 mutants (chl1-1 and chl1-5) were grown hydroponically in NH4NO3 and (NH4)2SO4 media to assess the function of NRT1.1 in NH4+ stress responses. All the plants grew normally in medium containing mixed nitrogen sources, but Col-0 displayed more chlorosis and lower biomass and photosynthesis than the NRT1.1 mutants in (NH4)2SO4 medium. Grafting experiments between Col-0 and chl1-5 further confirmed that NH4+ toxicity is influenced by NRT1.1. In (NH4)2SO4 medium, NRT1.1 induced the expression of NH4+ transporters, increasing NH4+ uptake. Additionally, the activities of glutamine synthetase and glutamate synthetase in roots of Col-0 plants decreased and soluble sugar accumulated significantly, whereas pyruvate kinase-mediated glycolysis was not affected, all of which contributed to NH4+ accumulation. By contrast, the NRT1.1 mutants showed reduced NH4+ accumulation and enhanced NH4+ assimilation through glutamine synthetase, glutamate synthetase, and glutamate dehydrogenase. Moreover, the up-regulation of genes involved in ethylene synthesis and senescence in Col-0 plants treated with (NH4)2SO4 suggests that ethylene is involved in NH4+ toxicity responses. This study showed that NH4+ toxicity is related to a nitrate-independent signaling function of NRT1.1 in Arabidopsis, characterized by enhanced NH4+ accumulation and altered NH4+ metabolism, which stimulates ethylene synthesis, leading to plant senescence.
Ammonium (NH4+) is a paradoxical inorganic nitrogen (N) source in the soil that is available to plants during growth and development. It is the preferred N source for some species such as rice (Oryza sativa) and tea (Camellia sinensis; Gao et al., 2010; Ruan et al., 2016). However, NH4+ toxicity occasionally occurs in terrestrial plants due to excessive N fertilizer application (Chen et al., 2013; Li et al., 2013). NH4+ toxicity typically takes place when plants are exposed to a high concentration, or alternatively, when plant cells themselves overproduce NH4+ under environmental stresses resulting from alterations in NH4+ metabolism (Barker and Corey, 1991; Feng and Barker, 1992; Bittsánszky et al., 2015). NH4+ toxicity often is accompanied by a depletion of organic acids and inorganic cations and by an accumulation of NH4+ (Hachiya et al., 2012). Several hypotheses have been proposed to explain the toxic effect of NH4+ on plants (Britto et al., 2001; Bittsánszky et al., 2015); however, the primary physiological mechanisms underlying NH4+ toxicity in plants remain elusive (Li et al., 2013).
NH4+ accumulation is widely observed when plants grow under high NH4+ concentration conditions. In rice, the application of high levels of NH4+ results in NH4+ accumulation and futile transmembrane NH4+ cycling (Chen et al., 2013). NH4+ accumulation occurs partially due to over uptake of NH4+. In Arabidopsis (Arabidopsis thaliana), four ammonium transporters (AMTs) function in NH4+ root acquisition, three of which, AtAMT1;1, AtAMT1;2, and AtAMT1;3, are up-regulated by NH4+ and collectively are responsible for up to 90% of NH4+ import (Yuan et al., 2007a). Knockout of the AMT1 gene significantly decreases NH4+ uptake, whereas overexpression of AMT1 results in higher NH4+ permeability in roots (Ranathunge et al., 2014; Li et al., 2016). Wang et al. (2013) verified that, under high-NH4+ stress (30 mm NH4+), AtAMT1;3 is endocytosed from the plasma membrane to reduce NH4+ uptake and mitigate NH4+ toxicity.
NH4+ serves as a ubiquitous intermediate metabolite in N metabolism in plants (Joy, 1988; Glass et al., 1997). Its high concentration in plant cells is toxic to plant tissues. Therefore, the capacity of plant cells to detoxify excessive NH4+ is viewed as an important metabolic response to alleviate the consequences of this stress (Hoai et al., 2003). The incorporation of NH4+ into Gln and the synthesis of Glu are critical pathways of NH4+ assimilation and NH4+ detoxification (Li et al., 2014). Glutamine synthase (GS), glutamate synthase (GOGAT), and glutamate dehydrogenase (GDH) are important enzymes in these processes (Miflin and Habash, 2002; Skopelitis et al., 2006). NH4+-tolerant plants have higher GS activity and less NH4+ accumulation in tissues (Cruz et al., 2006; Omari et al., 2010). Konishi et al. (2017) reported that GLN1 plays a dominant role in regulating NH4+ uptake in Arabidopsis. Indeed, pharmacological studies suggest that Gln, rather than NH4+, could be the real signaling molecule that regulates the expression of NH4+ transport and assimilation genes (Tabuchi et al., 2007). It is likely that NH4+ uptake and assimilation are integrated. Nevertheless, the coordination between NH4+ uptake and assimilation during NH4+ toxicity remains unclear.
NRT1.1 is the primary member of the nitrate transporter (NRT) gene family in higher plants (Tsay et al., 1993). It functions in multiple physiological processes in plants, such as NO3− and auxin transport, NO3− signal sensing, and stomatal movement (Tsay et al., 1993; Guo et al., 2003; Krouk et al., 2010; Bouguyon et al., 2015). Several studies have revealed that NRT1.1 is essential for plants to adapt to unfavorable environments, including cadmium stress, salt stress, and proton toxicity (Mao et al., 2014; Fang et al., 2016; Álvarez-Aragón and Rodríguez-Navarro, 2017). Interestingly, the Arabidopsis NRT1.1 mutants chl1-1 and chl1-5 display higher resistance to a high concentration (10 mm) of NH4+ as the sole source of N than wild-type plants (Hachiya et al., 2011; Hachiya and Noguchi, 2011). However, the mechanisms by which NRT1.1 mediates NH4+ uptake and the subsequent symptoms of NH4+ toxicity have to be elucidated at the physiological and molecular levels.
Here, we analyzed gene expression, metabolites, and the physiological processes and chemical activities in roots and shoots of wild-type Arabidopsis plants and NRT1.1 mutants grown in nutrient growth medium containing high concentrations of NH4+ as the sole N source. We aimed to clarify (1) whether NH4+ toxicity symptoms are NRT1.1 related and (2) the physiological roles of NH4+ uptake and assimilation during NH4+ toxicity. Our results provide further insights into NH4+ toxicity in plants.
RESULTS
NH4+ Toxicity Is Related to NRT1.1 Signaling Function
A previous study showed that 5 mm (NH4)2SO4 (10 mm NH4+) as the sole source of N causes chlorosis in the leaves of Columbia-0 (Col-0), but not in the leaves of chl1-1 and chl1-5 mutant lines, in 11-d-old seedlings (Hachiya and Noguchi, 2011). In this study, with the equivalent stoichiometric sole NH4+ source supplied in different forms [(NH4)2C4H6O4 (ammonium succinate), (NH4)2SO4, or NH4Cl] for 10 d to 4-week-old seedlings, we obtained the same results and observed severe chlorosis in plants treated with (NH4)2SO4 (Fig. 1A). Accordingly, we used (NH4)2SO4 treatment for all the subsequent experiments in this study.
Figure 1.
Effects of high concentration of a sole NH4+ source on Arabidopsis growth and photosynthesis. A, Plants were treated with 5 mm NH4NO3 (Ctl) and with different sources of NH4+ (10 mm) applied to rooting medium for 10 d. B to E, Relative fresh weight (B), relative biomass (C), photosynthetic rate (D), and chlorophyll concentration (E) of Col-0, chl1-1, and chl1-5 grown in 5 mm NH4NO3 (Ctl) and (NH4)2SO4 for 10 d. Fresh weight (Fw) and dry weight were obtained from pooled samples of seven plants from each replicate, and data were calculated relative to the average value of Col-0 under control conditions. F, Grafted plants grown in 5 mm NH4NO3 (Ctl) and (NH4)2SO4 for 10 d. Plant material names on the left indicate the genotypes of the shoot/root combination. G, Chlorophyll concentration of the grafted plants grown in 5 mm NH4NO3 (Ctl) and (NH4)2SO4 for 10 d. H and I, Phenotypes and chlorophyll concentrations of Col-0 and chl1-9 grown in 5 mm NH4NO3 (Ctl) and (NH4)2SO4 for 10 d. Data represent means ± se (n = 4). Bars with the same letter indicate no significant difference at P < 0.05 using the lsd method. Bars = 1 cm in A, F, and H.
High-concentration (NH4)2SO4 treatment resulted in high amounts of sulfur (S) in the growth medium. Considering that S is an essential element for the synthesis of some amino acids, three different conditions were set to assess whether S was involved in (NH4)2SO4-induced plant chlorosis: 5 mm (NH4)2SO4 treatment as described above, 5 mm K2SO4 was added to control medium, or 5 mm NH4NO3 treatment was replaced with 10 mm KNO3. We found that only the Col-0 plants treated with (NH4)2SO4 displayed chlorosis (Supplemental Fig. S1). Therefore, the effect of S on (NH4)2SO4-induced chlorosis in Col-0 plants was dismissed.
The application of sole NH4+ sources significantly reduced the plant size (Fig. 1A). The fresh weight of chl1-1 and chl1-5 was reduced by 44% and 39%, and the biomass was lowered by 10% and 16%, respectively, compared with the 58% and 36% reduction in fresh weight and biomass, respectively, observed in Col-0 (Fig. 1, B and C). There was no difference in photosynthetic rates or chlorophyll concentration between Col-0 and NRT1.1 mutants under NH4NO3 treatment, whereas (NH4)2SO4 reduced chlorophyll and photosynthetic rates in all genotypes, with greater reduction in Col-0 than in chl1-1 and chl1-5 (Fig. 1, D and E). Nevertheless, there was no significant difference in stomatal conductance between these genotypes with (NH4)2SO4 treatment (Supplemental Fig. S2). These results indicate that NRT1.1 is involved in NH4+ toxicity in Arabidopsis.
To further confirm that NH4+ toxicity is NRT1.1 related, we developed grafted Col-0 and chl1-5 plants and subjected them to different NH4+ conditions. As shown in Figure 1, F and G, the plants with Col-0 shoots and/or roots developed chlorosis under (NH4)2SO4 conditions, and the grafted plants with the shoot of Col-0 displayed more serious chlorosis than those with the roots of Col-0, although chlorosis was much milder than that of Col-0. This implies that NRT1.1-related NH4+ toxicity is independent of the nitrate transporter function of NRT1.1. The chl1-9 mutant is defective in nitrate uptake but shows a normal primary nitrate response (Ho et al., 2009). In contrast to chl1-1 and chl1-5, the phenotype of chl1-9 was consistent with that of Col-0 (Fig. 1, H and I) when grown in (NH4)2SO4 medium. These results indicate that the signaling function of NRT1.1 is involved in NH4+ toxicity in Arabidopsis.
NRT1.1 Increases NH4+ Uptake and Accumulation
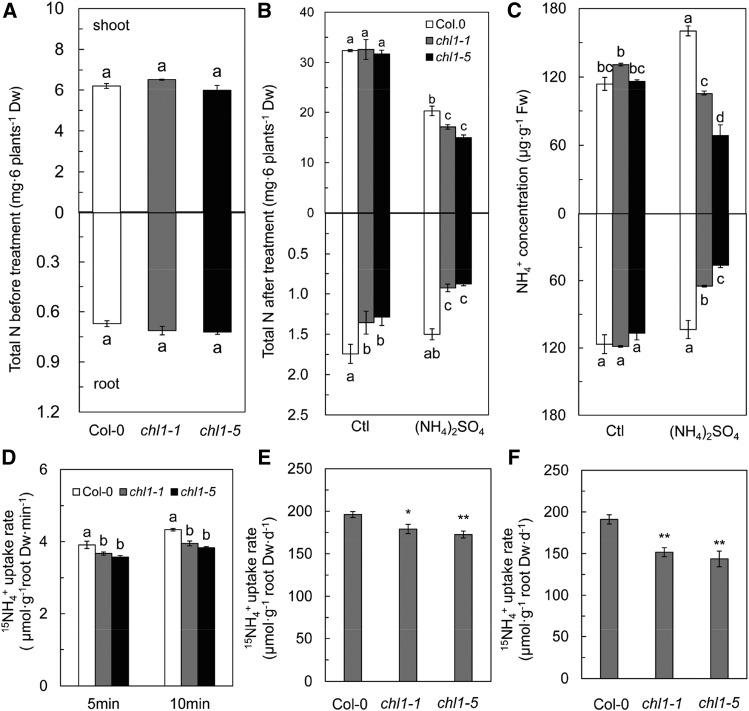
The total N content, NH4+ concentration, and expression of AMT1 genes were assayed to identify the difference in NH4+ uptake between Col-0 and NRT1.1 mutants in response to (NH4)2SO4. There was no difference among Col-0, chl1-1, and chl1-5 plants in total N content before (NH4)2SO4 treatment (Fig. 2A). However, treatment with (NH4)2SO4 for 10 d decreased total N content significantly in the three genotypes, but it remained significantly higher in Col-0 than in chl1-1 and chl1-5 (Fig. 2B). NH4+ concentration in the three genotypes showed no significant difference under control treatment (Fig. 2C); however, (NH4)2SO4 treatment significantly reduced NH4+ concentrations in chl1-1 and chl1-5 roots and shoots, while the NH4+ concentration in Col-0 was stable in the root and increased significantly in the shoot (Fig. 2C). We found that the 15NH4+ uptake rate in Col-0 plants was significantly higher than that of chl1-1 and chl1-5 plants during both the first and second stages of 15NH4+ labeling (Fig. 2, E and F). This result was consistent with those of experiments in which 15NH4+ was not used and further verified that NRT1.1 enhanced NH4+ uptake under (NH4)2SO4 treatment. Short-term 15NH4+ labeling for 5 and 10 min showed that the NH4+ uptake rate of Col-0 was significantly higher than that of chl1-1 and chl1-5 plants (Fig. 2D). The NH4+ uptake rate measured by short-term labeling was higher than that measured by long-term labeling. The decreasing concentration of NH4+ in growth medium over time could be an explanation for the lower NH4+ uptake rate observed by long-term labeling compared with short-term labeling. The circadian rhythm of nutrient uptake in plants also makes the average NH4+ uptake rate of long-term labeling lower than that of short-term labeling. NH4+ fluxes measured with noninvasive microtest technology (NMT) also showed that NH4+ influxed predominantly in Col-0 plants but effluxed in chl1-1 and chl1-5 (Supplemental Fig. S3, A and B).
Figure 2.
Effects of high concentration of a sole NH4+ source on N accumulation and NH4+ uptake in Arabidopsis. A, Total N content of Col-0, chl1-1, and chl1-5 plants grown in 1.125 mm NH4NO3 medium before (NH4)2SO4 treatment. B and C, Total N content (B) and NH4+ concentration (C) of Col-0, chl1-1, and chl1-5 plants grown in 5 mm NH4NO3 (Ctl) and (NH4)2SO4 for 10 d. D, 15NH4+ uptake rates in Col-0, chl1-1, and chl1-5 plants labeled for 5 and 10 min. E and F, 15NH4+ uptake rates in Col-0, chl1-1, and chl1-5 plants during the first (E) and second (F) stages of (15NH4)2SO4 treatment. Total N amount was obtained from pooled samples of six plants from each replicate. Data represent means ± se (n = 4). Bars with the same letter are not significantly different at P < 0.05 using the lsd method. Bars with two and three asterisks indicate significant differences from the control at P < 0.01 and P < 0.001, respectively, using a two-tailed Student’s t test. Dw, Dry weight; Fw, fresh weight.
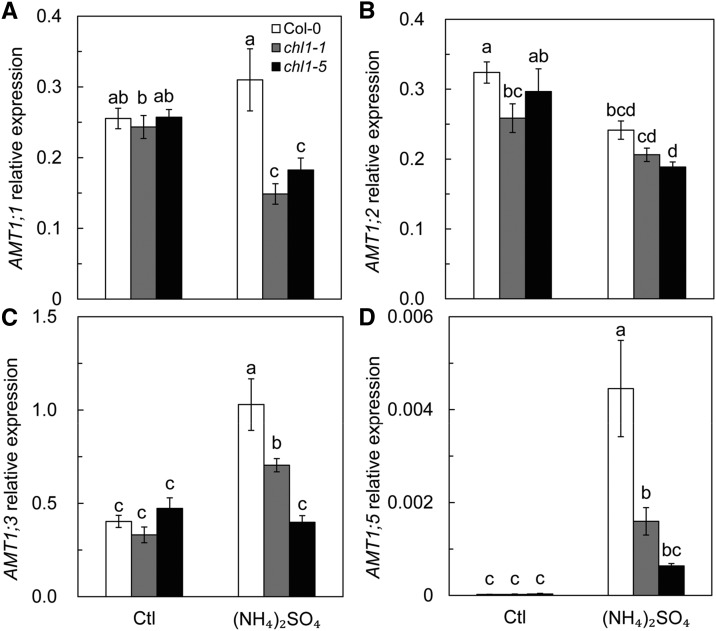
The modifications in NH4+ uptake could be the result of the expression of AMT1 genes. In Arabidopsis, AMT1;1, AMT1;2, AMT1;3, and AMT1;5, which encode AMT1 family transporters, are responsible for the majority of NH4+ acquisition in roots (Tegeder and Masclaux-Daubresse, 2018). The expression of these genes did not differ significantly between Col-0 and nrt1.1 lines under control conditions. Generally, (NH4)2SO4 resulted in the up-regulation of AMT1;1, AMT1;3, and AMT1;5 in Col-0, particularly for AMT1;5, where the expression was 200-fold higher than that in the control (Fig. 3, C and D). However, the expression of AMT1 genes in chl1-1 and chl1-5 decreased significantly (AMT1;1) or was unchanged (AMT1;2 and AMT1;3) under (NH4)2SO4 compared with that of the control and was significantly lower than that in Col-0 (Fig. 3). NRT1.1 facilitates the expression of AMT1s in Arabidopsis in response to (NH4)2SO4, which could lead to increased NH4+ uptake. The deficiency of AMT1;1 causes a 30% decrease in NH4+ uptake by roots (Yuan et al., 2007a). The results in Supplemental Figure S4 show that AMT1;1 deletion lowered the sensitivity of plants to high concentrations of NH4+ as the sole N source. This suggests that NH4+ uptake was responsible for NH4+ toxicity.
Figure 3.
AMT1 gene expression in roots of Col-0, chl1-1, and chl1-5 plants grown in 5 mm NH4NO3 (Ctl) and (NH4)2SO4 for 3 d. Roots were sampled for the expression of AMT1;1 (A), AMT1;2 (B), AMT1;3 (C), and AMT1;5 (D). Data represent means ± se (n = 4). Bars with the same letter are not significantly different at P < 0.05 using lsd.
NRT1.1 Inhibits the Activity of NH4+ Metabolism Enzymes under High Concentration of NH4+ as the Sole N Source
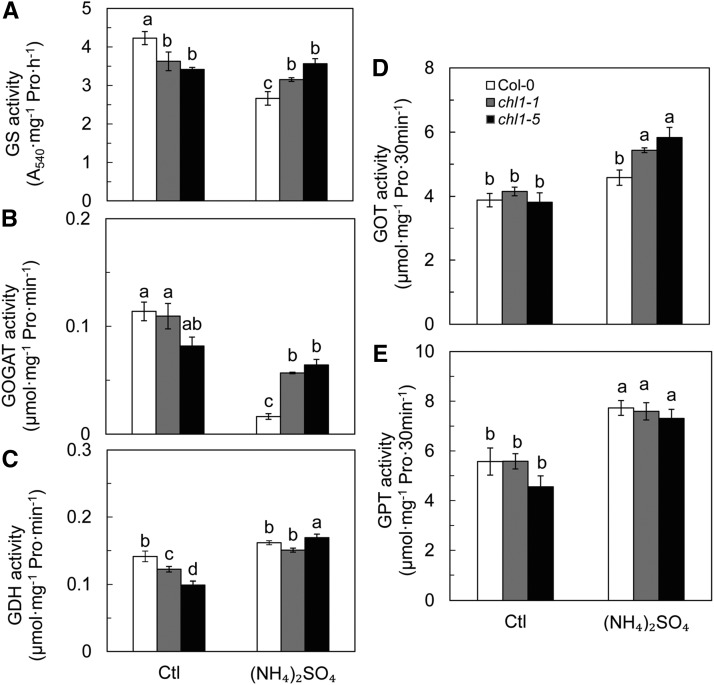
Under control conditions, GS activity was significantly lower in chl1-1 and chl1-5 than in Col-0 in roots (Fig. 4A). The GS activity decreased in Col-0 plants treated with (NH4)2SO4 but was not affected in chl1-1 and chl1-5 under the same growth conditions (Fig. 4A). In shoots, (NH4)2SO4 increased GS activity significantly in Col-0 and chl1-1 but not in chl1-5 (Supplemental Fig. S5A).
Figure 4.
Activities of NH4+ assimilation enzymes and transaminases in roots of Col-0, chl1-1, and chl1-5 plants. GS (A), GOGAT (B), GDH (C), GOT (D), and GPT (E) activities in roots of Col-0, chl1-1, and chl1-5 plants grown in 5 mm NH4NO3 (Ctl) and (NH4)2SO4 for 10 d are shown. Data represent means ± se (n = 4). Bars with the same letter are not significantly different at P < 0.05 using lsd.
The GOGAT activity in the roots did not differ between genotypes under control conditions (Fig. 4B). (NH4)2SO4 reduced GOGAT activity by 85.6% in Col-0 but to lesser extents in chl1-1 and chl1-5, where it decreased by 48.1% and 21.5%, respectively (Fig. 4B). In shoots, (NH4)2SO4 increased GOGAT activity significantly in chl1-1 and chl1-5 but not in Col-0, and no difference was observed among genotypes under control conditions (Supplemental Fig. S5B).
The activity of GDH was significantly higher in Col-0 than in chl1-1 and chl1-5 in roots under control conditions (Fig. 4C), and (NH4)2SO4 treatment had no effect on GDH activity in Col-0 but increased GDH activity significantly in chl1-1 and chl1-5 by 23.4% and 70.6%, respectively, compared with the control (Fig. 4C). In shoots, (NH4)2SO4 increased GDH activity in all genotypes, with no differences among genotypes under control or (NH4)2SO4 treatments (Supplemental Fig. S5C).
No differences in glutamic-oxaloacetic transaminase (GOT) or glutamic-pyruvic transaminase (GPT) activities were observed in roots of the three genotypes under control conditions (Fig. 4, D and E); however, (NH4)2SO4 increased GOT activity in chl1-1 and chl1-5 but not in Col-0 (Fig. 4D). GPT activity in the three genotypes was higher under (NH4)2SO4 treatment than in the control, but with no significant difference between genotypes (Fig. 4E). In shoots, GOT and GPT activities were not affected by (NH4)2SO4 treatment, and no difference in their activities could be measured between the wild-type and mutant genotypes (Supplemental Fig. S5, D and E). Taken together, these data suggest that NRT1.1 had a negative effect on the activities of NH4+ assimilation enzymes and transaminases in response to a high concentration of the sole NH4+ source.
NRT1.1 Disturbs the Balance between Carbon and NH4+ Metabolism under NH4+ as the Sole N Source
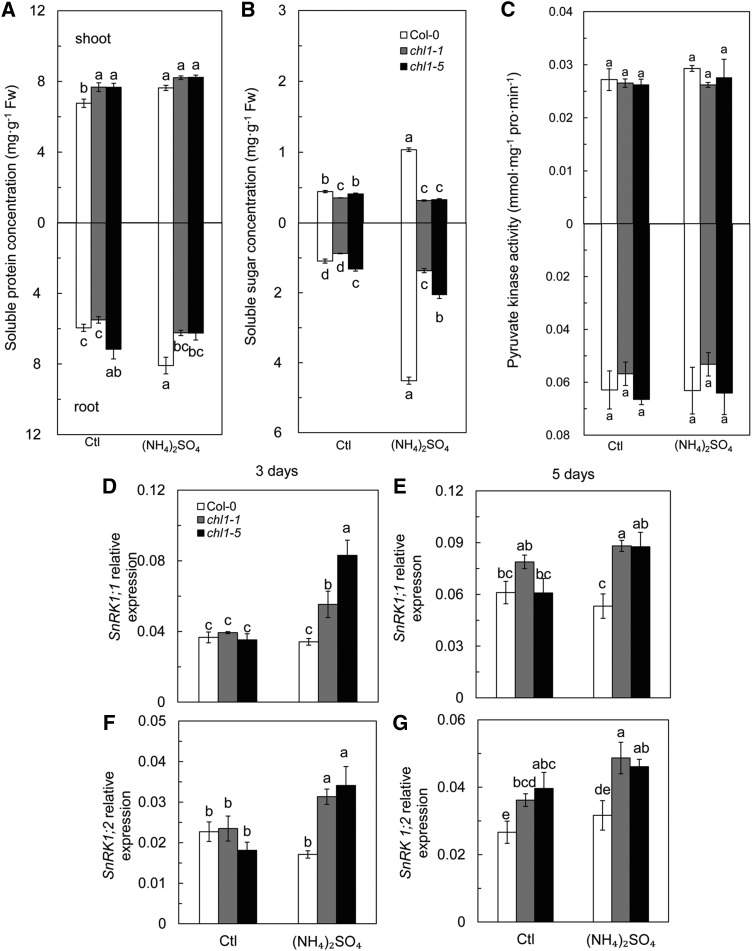
The soluble protein concentration in shoots of chl1-1 and chl1-5 was significantly higher than that in Col-0 under the control condition; however, chl1-5 roots showed higher soluble protein concentrations than those of Col-0 (Fig. 5A). Irrespective of shoot or root, (NH4)2SO4 increased soluble protein concentration in Col-0 but not in chl1-1 and chl1-5. The mutants showed lower concentrations in roots than did Col-0 (Fig. 5A).
Figure 5.
Effects of high concentration of NH4+ as the sole source of N on C and N metabolism. Soluble protein (A), soluble sugar content (B), activity of pyruvate kinase (C), and the expression of SnRK1 genes (D–G) in Col-0, chl1-1, and chl1-5 plants are shown. D and F represent SnRK1;1 and SnRK1;2 expression after 3 d, and E and G represent SnRK1;1 and SnRK1;2 expression after 5 d of (NH4)2SO4 treatment. Plants measured in A to C were grown in 5 mm NH4NO3 (Ctl) and (NH4)2SO4 media for 10 d. Data represent means ± se (n = 4). Bars with the same letter are not significantly different at P < 0.05 using lsd. Fw, Fresh weight.
Soluble sugar concentrations in roots and shoots varied slightly between the wild type and mutants in the control treatment (Fig. 5B) and were 4.14 and 2.33 times higher in roots and shoots of Col-0, respectively, under (NH4)2SO4 conditions than under control conditions (Fig. 5B). However, soluble sugar concentrations in chl1-1 and chl1-5 were not affected by (NH4)2SO4 and were significantly lower than that in Col-0 (Fig. 5B). Pyruvate kinase (PK) activity was consistent among genotypes regardless of (NH4)2SO4 treatment (Fig. 5C).
SNF1-RELATED PROTEIN KINASE1 (SnRK1) regulates the coordination between carbon (C) and N metabolism (Wang et al., 2012). (NH4)2SO4 treatment led to inconsistent responses of SnRK1 genes. As shown in Figure 5, D to G, the expression of SnRK1;1 and SnRK1;2 was unchanged in Col-0 with (NH4)2SO4 treatment but was up-regulated in chl1-1 and chl1-5 when the plants were treated with (NH4)2SO4 for 3 d (Fig. 5, D and F). Their expression on day 5 increased even further (Fig. 5, E and G).
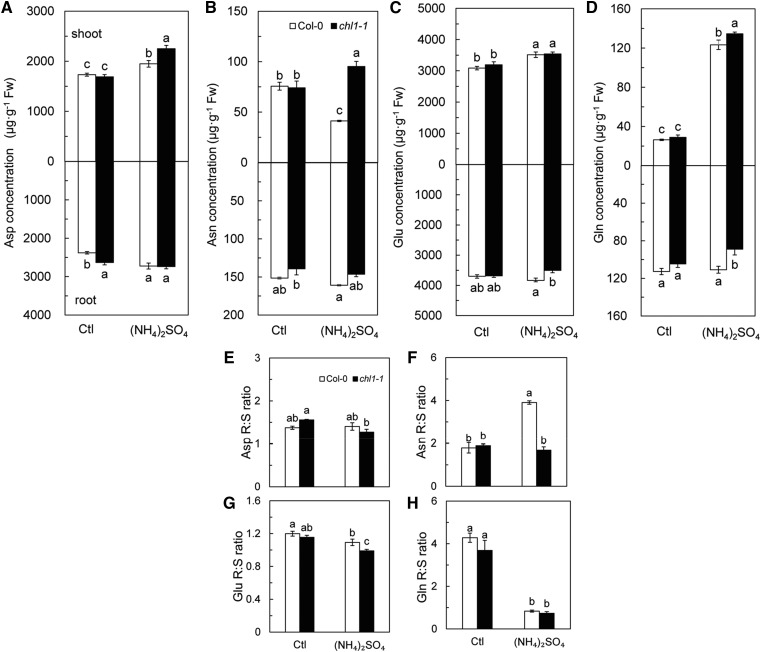
To further investigate the effect of (NH4)2SO4 on N metabolism, we compared the amino acid composition of Col-0 and chl1-1 grown under different N sources. As shown in Figure 6, A to D, the major amino acids such as Asp and Asn were less affected by (NH4)2SO4 in roots than in shoots of the wild-type and mutant lines (Fig. 6, A and B). Glu and Gln in the roots of chl1-1 were significantly lower than in roots of Col-0 under (NH4)2SO4 treatment (Fig. 6, C and D). In shoots, there were no significant differences in amino acid content between Col-0 and chl1-1 under control conditions (Fig. 6, A–D); however, (NH4)2SO4 treatment increased the concentration of all measured amino acids, with the exception of Asn in Col-0, which was lower than that in the control (Fig. 6, A–D). In shoots, Asp, Asn, and Gln, but not Glu, were significantly higher in chl1-1 than in Col-0 under (NH4)2SO4 treatment (Fig. 6, A–D).
Figure 6.
Effects of high concentration of NH4+ as the sole source of N on free amino acid concentration and their R:S ratios in Col-0 and chl1-1. Plants were grown in 5 mm NH4NO3 (Ctl) and (NH4)2SO4 media for 10 d. Asp (A) Asn (B), Glu (C), and Gln (D) were measured in the shoots and roots, and then the R:S ratios of Asp (E), Asn (F), Glu (G), and Gln (H) were calculated. Data represent means ± se (n = 3). Bars with the same letter are not significantly different at P < 0.05 using lsd. Fw, Fresh weight.
Under control conditions, there was no noticeable change in the root-to-shoot (R:S) ratios of amino acids between the wild type and mutants (Fig. 6, E–H). Although (NH4)2SO4 promoted a higher accumulation of Asn in Col-0 roots than in shoots, the R:S ratio of Asn in chl1-1 was not affected significantly (Fig. 6F). The R:S ratio of Gln similarly declined under (NH4)2SO4 treatment in both Col-0 and chl1-1 (Fig. 6H).
(NH4)2SO4 treatment increased Thr, Ala, Ile, and Arg (Supplemental Fig. S6, A–C) in the shoots of chl1-1 but not in those of Col-0. While Lys, Phe, and Ala were reduced or unchanged in chl1-1, their concentrations were lower than in Col-0 under (NH4)2SO4 treatment (Supplemental Fig. S6, D–F).
NRT1.1-Related NH4+ Toxicity Is Associated with Ethylene and Plant Senescence
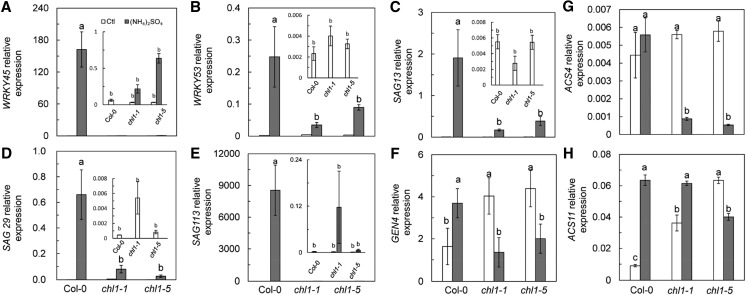
The expression of the senescence-related genes WRKY45, WRKY53, SAG13, SAG29, SAG113, and GEN4 were assayed after treating Col-0, chl1-1, and chl1-5 with (NH4)2SO4. All genes were significantly up-regulated by (NH4)2SO4 treatment in Col-0; but, together with GEN4, these genes were either slightly up-regulated or even down-regulated in chl1-1 and chl1-5 (Fig. 7, A–F). (NH4)2SO4 treatment up-regulated the ethylene synthesis gene ACS4 in Col-0 by 25.6% but inhibited ACS4 expression by 84.3% and 90.7% in chl1-1 and chl1-5, respectively. (NH4)2SO4 treatment up-regulated ACS11 expression by 606.3%, while ACS11 expression in chl1-1 was up-regulated by only 69.9% but inhibited in chl1-5 by 36.8% (Fig. 7, G and H). Thus, it is apparent that ethylene synthesis and plant senescence are associated with NRT1.1-related NH4+ toxicity.
Figure 7.
Gene expression related to senescence and 1-aminocyclopropane carboxylic acid (ACC) synthesis in leaves of Col-0, chl1-1, and chl1-5. Expression of WRKY45 (A), WRKY53 (B), SAG13 (C), SAG29 (D), SAG113 (E), GEN4 (F), ACS4 (G), and ACS11 (H) in Col-0, chl1-1, and chl1-5 grown in 5 mm NH4NO3 (Ctl) and (NH4)2SO4 media is shown. A to F show genes related to plant senescence and were measured 10 d after treatment, and G and H show genes related to ACC synthesis and were measured 3 d after treatment. Data represent means ± se (n = 4). Bars with the same letter are not significantly different at P < 0.05 using lsd.
By using the ethylene signal mutant ein2, along with foliar application of the ethylene precursor ACC and the ethylene antagonist aminoethoxyvinylglycine (AVG) to the wild type and NRT1.1 mutant lines, we found that the disruption of ethylene signaling did not induce plant chlorosis, although the NH4+ concentration in ein2 was as high as that in Col-0 (Supplemental Fig. S7, A–C) and the inhibition of ethylene synthesis suppressed the effect of high NH4+ (Supplemental Fig. S7, D and E).
DISCUSSION
Nitrate-Independent Function of NRT1.1 in NH4+ Toxicity
Chlorosis and decline in biomass are the typical phenotypic markers of NH4+ toxicity in plants grown under high concentrations of sole NH4+ sources (Chen et al., 2013; Bittsánszky et al., 2015). These phenotypes are consistent with the reduced photosynthetic rate and chlorophyll concentration that were observed in Arabidopsis wild-type plants in this study (Fig. 1, A–E). Our results agree with those reported by Hachiya and coauthors (Hachiya and Noguchi, 2011; Hachiya et al., 2011) that the NRT1.1 knockout mutants chl1-1 and chl1-5 exhibited stronger resistance to NH4+ toxicity relative to Col-0, showed weaker chlorosis, and experienced less reduction in biomass, photosynthetic rate, and chlorophyll concentration (Fig. 1, A–E).
These results suggest that NRT1.1 is responsible for NH4+ toxicity in Arabidopsis grown with a high concentration of NH4+ as the sole source of N. Our grafting experiment showed that the chlorosis induced by NH4+ occurred only in the presence of normal NRT1.1. This result further demonstrated that NH4+ toxicity in Arabidopsis is NRT1.1 related regardless of its expression in roots or shoots (Fig. 1, F and G).
NRT1.1 is a multifunctional gene that participates in plant development and in the interaction between plants and their environment (Bouguyon et al., 2015). In addition to being an NO3− transporter, NRT1.1 serves as an NO3− sensor in roots and also is a stomatal regulator in guard cells (Guo et al., 2003). NRT1.1 expression in guard cells mediates the accumulation of NO3− in guard cells during stomatal opening (Guo et al., 2003). Stomatal closure increases the resistance of CO2 diffusion from the ambient air into the intercellular space of leaves and, consequently, limits the rate of photosynthesis (Lawlor and Tezara, 2009). In this study, stomatal conductance in Col-0, chl1-1, and chl1-5 showed no significant response to (NH4)2SO4 treatment (Supplemental Fig. S2), suggesting that the NO3− transport and stomatal regulation functions of NRT1.1 are independent from NH4+ toxicity in Arabidopsis.
NRT1.1 can function in the absence of nitrate. The existence of a discrete nitrate-independent signaling pathway from NRT1.1 has been suggested by Hachiya et al. (2011). Since the chl1-9 mutant shows normal function in primary nitrate signaling, even though it is defective in nitrate transport (Ho et al., 2009), the similar symptoms of chl1-9 and Col-0 in response to (NH4)2SO4 (Fig. 1, H and I) suggest that a specific deficiency in NO3− transport capacity is not sufficient to induce NH4+ toxicity in Arabidopsis in the absence of NO3−; the signaling function of NRT1.1 could play an important role in mediating the effects of NH4+.
This notion was verified indirectly by the finding that the grafted plants with a Col-0 shoot showed more serious chlorosis than those with Col-0 roots when grown under (NH4)2SO4. As the phosphorylation state and NRT1.1 level are similar in both Col-0 and chl1-9 (Hachiya and Noguchi, 2011), we speculate that the altered phosphorylation state of NRT1.1 and the interaction between NRT1.1 and other proteins in metabolic pathways could be related to the occurrence of NH4+ toxicity. The difference in the degree of chlorosis in grafted plants with the shoot or roots of Col-0 also suggests that there could be different mechanisms in the shoot and roots through which NRT1.1 regulates the occurrence of NH4+ toxicity. Further studies are needed to elucidate the detailed mechanisms underlying the nitrate-independent signaling pathway of NRT1.1 in the regulation of NH4+ toxicity.
NRT1.1 Induces NH4+ Accumulation in Tissues under High NH4+ as the Sole Source of N
Excessive accumulation of NH4+ in plant tissues is believed to be one of the major causes of injury in plants under unfavorable conditions (Barker, 1999; Hoai et al., 2003; Nguyen et al., 2005). The accumulation of NH4+ is the first factor that is associated with NH4+ toxicity (Hachiya et al., 2012). In this study, NH4+ accumulated significantly in Col-0 but not in chl1-1 and chl1-5 under high (NH4)2SO4 conditions (Fig. 2C). NH4+ equilibrium in plant cells is determined by NH4+ uptake, transport, and assimilation (Nguyen et al., 2005). Hachiya et al. (2011) proposed that increased NH4+ assimilation due to improved growth, rather than a decrease in NH4+ uptake, lowered the accumulation of NH4+ in NRT1.1 mutants. In contrast, we found that the biomass and plant size of the wide type and NRT1.1 mutant lines decreased under (NH4)2SO4 treatment and that the increased accumulation of NH4+ in Col-0 (or the reduced accumulation of NH4+ in NRT1.1 mutants) was caused largely by increased (or reduced) NH4+ uptake, according to 15NH4+ tracer and NH4+ flux experiments (Fig. 2, D–F; Supplemental Fig. S3). A rice variety sensitive to NH4+ toxicity displayed higher NH4+ accumulation and futile NH4+ cycling across the root-cell plasma membrane (Chen et al., 2013). However, in Arabidopsis, the efflux of NH4+ in chl1-1 and chl1-5 under high-NH4+ conditions could be beneficial by reducing NH4+ accumulation and alleviating its toxicity (Supplemental Fig. S3). Furthermore, the up-regulation of AMT1 genes, which mediate the majority of NH4+ uptake in Arabidopsis roots (Yuan et al., 2007a), is associated with the increased NH4+ accumulation in Col-0 (Fig. 3, A–D).
NRT1.1 can generate an unknown signal to regulate NRT2.1 expression, hence, ensuring significant NO3− uptake in the presence of NH4+ in the external environment (Muños et al., 2004). Here, the distinct expression of AMT1s between Col-0 and NRT1.1 mutants under NH4+ stress suggests that NRT1.1 could be related to the response of AMT1 genes to external NH4+ modification (Fig. 3, A, C, and D).
Proton secretion is accompanied by NH4+ uptake, leading to acidification of the growth medium. The acidification of medium has severe negative effects on Arabidopsis growth (Fang et al., 2016). Chaillou et al. (1991) suggested that NH4+ uptake induces a reduction in pH in the rhizosphere, which is one of the major factors of NH4+ toxicity. Our data excluded the effect of medium acidification on NH4+ toxicity, because the pH of the medium was similar among genotypes (Supplemental Fig. S8). Furthermore, although the chl1 mutant has a pH-dependent phenotype (Fang et al., 2016), it was unharmed under a high concentration of NH4+. Contrarily, exogenously applied NH4+ acidifies the apoplast in plant tissues. However, NH4+ is not always related to cytosolic acidification in plant shoots (Husted and Schjoerring, 1995; Hachiya et al., 2012).
The capacity to ameliorate NH4+ toxicity has been viewed as an important factor in alleviating the consequences of this stress (Hoai et al., 2003; Nguyen et al., 2005). The GS-GOGAT cycle continuously provides the substrate Glu for the incorporation of NH4+ into Gln, a process that removes excess NH4+, thereby reducing toxicity symptoms (Bittsánszky et al., 2015). The reduced activities of GS and GOGAT in Col-0 under high-(NH4)2SO4 conditions observed in this study indicated that attenuated NH4+ assimilation capacity contributed to the accumulation of NH4+ (Figs. 2C and 4, A and B). NADH-GDH plays a unique role independent of GS-GOGAT in N recycling (Masclaux-Daubresse et al., 2006). It has been widely reported that GDH responds positively to abiotic stress and is important for the detoxification of NH4+ under stress conditions (Boussama et al., 1999; Bittsánszky et al., 2015; Zhong et al., 2017). Compared with Col-0, the significantly higher activities of GDH, GS, and GOGAT in chl1-1 and chl1-5 under high-(NH4)2SO4 conditions could decrease the accumulation of NH4+ (Fig. 4, A–C). Our results reveal that the higher N assimilation capacity of chl1-1 and chl1-5 mutants plays a vital role in mitigating NH4+ toxicity.
Taken together, this study revealed that enhanced NH4+ uptake accompanied by reduced NH4+ assimilation causes higher NH4+ accumulation in Col-0 than in the NRT1.1 mutants when grown with high concentrations of (NH4)2SO4. NH4+ uptake and assimilation are tightly linked. Gln is the actual signal regulating NH4+ uptake and assimilation (Tabuchi et al., 2007). The expression of the cytosolic isoform of GS, AtGLN1;2, is induced strongly in the roots by NH4+, contributing to NH4+ assimilation in the root (Konishi et al., 2017). Furthermore, differences in NH4+ uptake and assimilation between Col-0 and NRT1.1 mutants suggest that NRT1.1 plays a significant role in regulating these processes. Bouguyon et al. (2015) hypothesized that NRT1.1 affects AMT1s by regulating N assimilation enzymes, as NRT1.1 regulates the expression level of several genes involved in the N assimilation pathway. The substantial reductions in the activities of GS and GOGAT in Col-0 plants grown in (NH4)2SO4 medium (Fig. 4, A and B) suggest that the pathway for N assimilation regulated by NRT1.1 is perturbed by high concentrations of NH4+.
Numerous studies have demonstrated the combined effects of ethylene production and NH4+ accumulation in the development of NH4+ toxicity symptoms in plants (You and Barker, 2005; Li et al., 2011, 2013). Ethylene production could be the link between NH4+ accumulation and the symptoms of NH4+ toxicity, as the inhibition of ethylene production ameliorated the symptoms of NH4+ toxicity even when NH4+ concentration in leaves was high (Li et al., 2013). In our study, the accumulation of NH4+ was consistent with the up-regulation of ACC synthesis genes and senescence-related genes (Fig. 7). In addition, the ethylene mutant ein2 grown in (NH4)2SO4 medium was not affected, although the concentration of NH4+ was comparable to that in Col-0. NH4+ toxicity symptoms in Col-0 were alleviated by the application of AVG, whereas NH4+ toxicity symptoms in chl1-1 and chl1-5 were accelerated by the application of ACC (Supplemental Fig. S8, A–E). These results suggest that NH4+ accumulation-induced ethylene synthesis and plant senescence are the two major reasons for the development of NH4+ toxicity symptoms.
NRT1.1-Related Perturbation of C and N Metabolism under High Concentration of NH4+ as the Sole Source of N
The interaction between C and N metabolism is of paramount importance to improve plant stress tolerance (Reguera et al., 2013). The assimilation of NH4+ requires carbohydrates to provide the C skeletons of 2-oxoglutarate (2-OG) generated from the tricarboxylic acid cycle (Yuan et al., 2007b). Nguyen et al. (2005) proposed that a reduction in photosynthetic capacity and a decrease in 2-OG production might be responsible for the excessive accumulation of NH4+. The availability of sugar in plant tissues may regulate N (including NH4+) uptake and assimilation but depends on a series of C metabolic steps downstream of hexose phosphorylation (Lejay et al., 2003). PK is at a crucial position to control glycolysis and the assimilation of C to provide energy and C skeletons (2-OG) for N metabolism (Stitt, 1999). In this study, the activity of PK was statistically similar in Col-0 and NRT1.1 mutants and between experimental treatments, suggesting that the availability of C skeletons is not a limiting factor in N assimilation (Fig. 5C). The significant increase in the accumulation of sugars in Col-0 plants under NH4+ stress suggested that C and N metabolism in Col-0 were uncoupled when plants were exposed to high concentrations of NH4+ in the medium (Fig. 5B).
Complex regulatory mechanisms are involved in the coordination between N and C metabolism, including sensing of sugars, nitrate, and amino acids, as well as the effects of phytohormones (Nunes-Nesi et al., 2010). SnRK1 plays a vital role in plant energy homeostasis, growth and development, and stress tolerance (Baena-González et al., 2007; Jossier et al., 2009) by regulating the key enzymes involved in C and N metabolism. The up-regulated expression of SnRK1;1 and SnRK1;2 in NRT1.1 mutants was consistent with the increased N metabolism and resistance to NH4+ toxicity, suggesting that SnRK1 plays an essential role in the homeostasis of C and N metabolism under NH4+ stress (Fig. 5, D–G).
The accumulation of NH4+ often is accompanied by the accumulation of free amino acids (Hoai et al., 2003), probably due to the diminished demand for NH4+ during amino acid biosynthesis. In this study, however, chl1-1 presented a higher amino acid content and a lower NH4+ content under (NH4)2SO4 conditions; this could have resulted from greater NH4+ assimilation capacity and higher amino acid biosynthesis. Gln and Asn are important amides for organic N storage and transport in plant tissues. They function as important signals that reflect the N status in plants and mediate NH4+ uptake and assimilation (Tabuchi et al., 2007; Konishi et al., 2017). The accumulation of Gln and Asn was less in the roots of chl1-1 than in the roots of Col-0 during (NH4)2SO4 treatment (Fig. 6, E and F), which potentially benefits chl1-1 plants. This is because less root Gln and Asn accumulation might increase the requirement for NH4+ incorporation into de novo amino acids and, thus, mitigate NH4+ accumulation. Additionally, Asn plays an important role in NH4+ detoxification (Masclaux-Daubresse et al., 2006). The significantly higher Asn concentration in (NH4)2SO4-treated chl1-1 plants compared with that in Col-0 indicates that Asn could have contributed to the lower accumulation of NH4+ in chl1-1 (Fig. 6B).
N in rooting medium affects the accumulation of amino acids in plants (Noguchi et al., 2015). Increased amino acid synthesis can be advantageous for the adaptation to stress and for the accumulation of precursors for biosynthetic pathways (Joshi et al., 2010). Glu can be converted into Asp and Ala by GOT and GPT, respectively (Brock et al., 1970) and used subsequently for the synthesis of branched-chain amino acids. Branched-chain amino acids such as Met, Thr, and Ile are derived from the Asp pathway and act as precursors for many plant secondary metabolites such as ethylene, which are involved in the responses of plants to stresses (Joshi et al., 2010). In this study, chl1-1 and chl1-5 showed enhanced GOT activity in the roots under (NH4)2SO4 treatment (Fig. 4D), which was accompanied by significant increases in the intracellular pools of Ile and Thr (Supplemental Fig. S6). Thus, our results suggest that the Asp-derived pathway for amino acid synthesis is of great significance to improve the tolerance of plants to NH4+ toxicity.
CONCLUSION
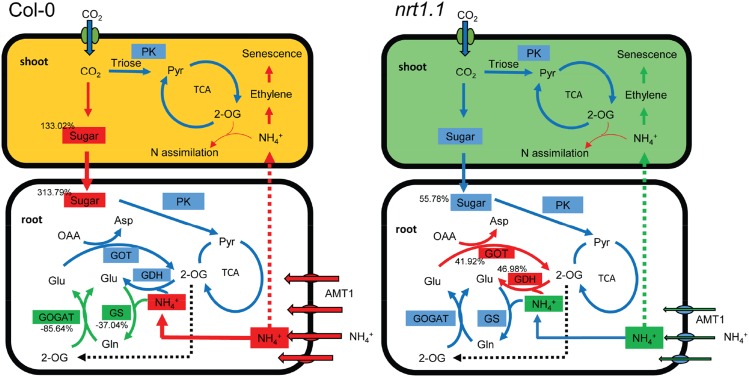
Our results suggest that NRT1.1 plays a signaling role in mediating NH4+ uptake and downstream NH4+ assimilation into amino acid metabolism, thus playing a role in NH4+ toxicity. In wild-type Arabidopsis, NRT1.1 increased the expression of AMT1s and NH4+ uptake when plants were grown in the presence of high concentrations of a sole NH4+ source. However, the ability to assimilate NH4+ decreased when plants were grown under such conditions. Furthermore, with a sole NH4+ source, the metabolism of C and N was uncoupled, causing an accumulation of soluble sugars and reduced amino acid metabolism. As a result, NH4+ accumulated, increasing the production of ethylene and the expression of genes related to plant senescence and, finally, NH4+ toxicity (Fig. 8A).
Figure 8.
Proposed model for NH4+ metabolism in Col-0 and NRT1.1 mutants grown under high concentration of NH4+ (10 mm) as the sole source of N. Effects in Col-0 cells (A) and nrt1.1 mutants cells (B) are shown. The colored rectangles are as follows: red, molecules or physiological processes that increased; blue, molecules or physiological processes that did not change significantly; and green, molecules or physiological processes that decreased. Red or green dotted arrows represent NH4+ flow from roots to shoots speculated as being higher or lower. Red or green solid arrows represent the physiological processes getting stronger or weaker; blue solid arrows represent physiological processes that were not changed significantly; and numbers nearest to a physiological process indicate the change in percentage of that process. Compared with plants grown under control conditions, NRT1.1 up-regulates the expression of AMT1 genes in roots and facilitates NH4+ uptake. Simultaneously, GS and GOGAT activities are inhibited by NH4+ as the sole source of N. Furthermore, sugars accumulate and are stored in roots, disturbing C and NH4+ metabolism. As a result, free NH4+ increases and can be transported to the shoot. High NH4+ concentrations in the shoot induce ethylene synthesis, which causes plant senescence. In NRT1.1 mutants, NH4+ uptake, the GS-GOGAT cycle, and C and NH4+ metabolism are not affected by NH4+ as the sole source of N, and the activities of NADH-dependent GDH and GOT are enhanced under NH4+, which diminish NH4+ accumulation and improve amino acid metabolism. Thus, NRT1.1 mutant plants can better survive environments with high concentrations of NH4+ as the main source of N than the wild type. TCA, Tricarboxylic acid; Pyr, pyruvate; OAA, oxaloacetic acid.
In the NRT1.1 mutants chl1-1 and chl1-5, the expression of AMT1s was lower than that in Col-0. In addition, the metabolism of C and N in the mutants was less affected by the sole NH4+ source, and NH4+ assimilation by the GS-GOGAT and GDH pathways and amino acid synthesis by GOT led to reductions in the accumulation of NH4+. Ultimately, chlorosis did not occur in NRT1.1 mutants (Fig. 8B). Clearly, the interaction between nitrate and AMTs is complex and is an attractive research area. To further explore the regulatory pathways that involve NRT1.1 during NH4+ uptake, transport, and assimilation is of great importance to further elucidate the mechanisms of NH4+ toxicity and to develop effective approaches for NH4+ nutrient management.
MATERIALS AND METHODS
Experimental Materials and Growth Conditions
Seeds of wild-type Arabidopsis (Arabidopsis thaliana Col-0), NRT1.1 knockout mutants (chl1-1 and chl1-5), an NRT1.1 point mutation (chl1-9) line, the amt1;1 mutant, and an ethylene signal mutant (ein2) were sown in nutrient-supplemented soil medium in a growth chamber (300 μmol photons m−2 s−1, 16-h photoperiod, and 22°C) and allowed to germinate and grow for 10 d. Subsequently, the seedlings with one pair of true leaves were transplanted in 600-mL pots and grown hydroponically using nutrient medium containing 1.125 mm NH4NO3, 1.25 mm KCl, 0.625 mm KH2PO4, 0.5 mm MgSO4, 0.5 mm CaCl2, 1.25 μm Fe-EDTA, 17.5 μm H3BO3, 3.5 μm MnCl2, 0.25 μm ZnSO4, 0.05 μm NaMoO4, and 0.125 μm CuSO4. The pH of the nutrient medium was adjusted to 6, and MES (2.5 mm) was added to the nutrient medium to buffer changes in pH. The medium was refreshed every 4 d. Nine plants were grown in each pot, and the growth conditions were the same as described above. After 4 weeks, half of the plants were transferred to 5 mm (NH4)2SO4, 5 mm (NH4)2C4H6O4, or 10 mm NH4Cl medium for an additional 10 d, with the control pots supplied with 5 mm NH4NO3. To assess the effect of SO42− on NH4+ toxicity, 4-week-old seedlings were treated with 5 mm (NH4)2SO4 medium or 5 mm NH4NO3 medium containing 5 mm K2SO4 for 10 d, with the control supplied with 10 mm KNO3. The experiment was arranged in a completely randomized design. The positions of the pots were interchanged when refreshing the medium to eliminate edge effects.
15N Pulse Assay
To investigate the uptake of NH4+ during (NH4)2SO4 treatment, Col-0, chl1-1, and chl1-5 seedlings were hydroponically grown for 4 weeks, then divided into two groups for 15N labeling with 5% atom abundance of (15NH4)2SO4, with one group at the start of (NH4)2SO4 treatment and the other at 5 d after the start of the treatment. To perform a short-term NH4+ uptake analysis, 99.9% atom abundance of (15NH4)2SO4 was used for labeling for 5 and 10 min at 10 am. Growth conditions were as described above. Before 15N labeling, the roots were washed with 0.1 mm CaSO4 for 1 min and then placed in growth medium containing 5 mm (15NH4)2SO4 for 5 d. While sampling, the roots were washed again with 0.1 mm CaSO4 for 1 min and then with deionized water. The root and shoot tissues were harvested separately and oven dried at 70°C for 48 h. The samples were pulverized with TissueLyser-48 (Jingxin), and 15N abundance in the samples was determined using a continuous-flow isotope ratio mass spectrometer coupled with a C-N elemental analyzer (ANCA-MS; PDZ Europa).
Grafting Experiment
A grafting experiment was performed as described by Rus et al. (2006) using an SZX-ILLB200 microscope (Olympus Optical). Col-0 and chl1-5 seeds were sterilized with 75% ethanol and germinated on one-half-strength Murashige and Skoog plates containing 3 mg L−1 benomyl, 0.04 mg L−1 benzyladenine, 0.02 mg L−1 indole-3-acetic acid, and 17 g L−1 agar. All the plates with stratified seeds were placed vertically in a growth chamber as described above. Five-day-old seedlings were grafted on the plate with a MANI Ophthalmic Knife (Straight 15; SWISSMED). The grafted seedlings were placed on plates for another 5 d until the graft union formed. The grafted seedlings without adventitious roots were grown hydroponically and treated with (NH4)2SO4 as described.
Determination of NH4+ Flux at the Root Surface with NMT
NMT is a method that measures the gradient of specific molecules or ions by means of a selective flux microsensor vibrating repeatedly between two predefined points. The molecular/ionic fluxes are calculated based on Fick’s law of diffusion: J = −D0·(dc/dx), where J is the ion flux (pmol cm−2 s−1), dc is the concentration gradient, dx is the distance between the two points, and D0 is the diffusion constant. The direction of flux is derived from Fick’s law of diffusion.
The experiment was carried out at the YoungerUSA-Xuyue BioFunction Institute using NMT Physiolyzer (YoungerUSA, Xuyue) and imFluxes V2.0 (YoungerUSA) software. Measurements were performed at room temperature (24°C–26°C). Before determination, the roots were equilibrated immediately in measuring solution [5 mm (NH4)2SO4, 0.1 mm CaCl2, and 0.3 mm MES, pH 6] for 10 min, then transferred to the measuring chamber, a small dish containing measuring solution. The ion flux microsensor was calibrated using different NH4+ concentrations [10 mm (NH4)2SO4 and 1 mm (NH4)2SO4, with other conditions consistent with the test solution] before NH4+ flux measurement. An NH4+-selective microsensor used for the net NH4+ flux measurement was vibrated in two measuring solutions between two positions, 1 and 30 μm at the meristem. Only the flux microsensor with a Nernstian slope > 50 mV decade−1 was used in this study. The same flux microsensor was calibrated again according to the same procedure and standards after each test. The μV differences were exported as raw data before they were imported and converted into net NH4+ fluxes using the JCal V3.3 software (a free MS Excel spreadsheet; YoungerUSA, Xuyue). Four plants were used for measurements in each treatment with each plant measured once.
Ethylene Regulation Experiment
Four-week-old Col-0, chl1-1, and chl1-5 seedlings were divided into three groups for (NH4)2SO4 treatment. The leaves of the first and second groups were sprayed with 5 μm AVG and 50 μm ACC, respectively, at 8 am every day throughout the (NH4)2SO4 treatment period. The third group was sprayed with deionized water and served as the control. Each plant was sprayed until the leaves were thoroughly wetted. After 10 d of treatment, chlorophyll concentration was determined and photographs were taken to determine the phenotypes.
Photosynthesis Measurements
Photosynthesis of the youngest fully expanded leaves in rosettes was measured 10 d after (NH4)2SO4 treatment using an LI-6400XT portable photosynthesis system (Li-Cor). Air temperature in the cuvette during measurement was maintained at 22°C, and the photosynthetic photon flux intensity was 200 μmol m−2 s−1. The CO2 concentration in the cuvette was adjusted to 500 μmol mol−1 with a CO2 mixer, and the vapor pressure deficit (VPD) was at 1 to 1.5 kPa. Data were recorded after equilibration to a steady state. After photosynthetic determination, the leaf area was measured with a leaf area meter (CI-202 Portable Laser Area Meter; CID Bio Science). The data of gas-exchange parameters were adjusted to the actual leaf areas.
Measurement of Chlorophyll Concentration
Chlorophyll in fresh rosette leaves of NH4NO3 (control)- and (NH4)2SO4-treated plants was extracted according to the method of Wellburn and Lichtenthaler (1984) with 80% (v/v) acetone. The absorbance of the extract was measured at 663 and 645 nm using a UV-VIS spectrophotometer (UV-2600; Shimadzu) to estimate total chlorophyll concentrations.
Measurements of Biomass, Total Nitrogen, and NH4+ and Soluble Sugar Concentrations
Control and (NH4)2SO4-treated plants were harvested, and roots and aerial parts were separated, oven dried at 70°C to a constant weight, and then ground to a fine powder. About 100 mg of each sample was digested with H2SO4-H2O2 at 350°C for N quantification using a high-resolution digital colorimeter (Autoanalyzer 3; SEAL).
NH4+ and soluble sugar were extracted from each sample with deionized water. The NH4+ concentration was measured colorimetrically using the method of Indophenol Blue colorimetry at 630 nm (Zanini, 2001), with (NH4)2SO4 used as a standard. Soluble sugar concentration was determined by the anthrone-sulfuric acid method (Wang et al., 2002).
Amino Acid Quantification
Amino acids in roots and shoots were quantified using HPLC (del Campo et al., 2009). Samples were harvested and immediately frozen in liquid N2, then stored at −80°C until analyses. Frozen leaf samples (200 mg) were pulverized with liquid N2 and homogenized in 1.5 mL of 0.1% phenol and 6 m HCl. The homogenates were hydrolyzed for 22 h at 100°C and cooled, and 1 mL of hydrolysate was dried with organomation (NDK200-2; Hangzhou Mi’ou Instrument) and redissolved in 1 mL of 0.1 m HCl. To determine the amino acids, 200 μL of redissolved hydrolysate was mixed with 20 μL of nor-Leu internal standard solution, 200 μL of triethylamine acetonitrile (pH > 7), and 100 μL of isothiocyanate acetonitrile, then incubated at 25°C for 1 h. Then, 400 μL of hexane was added and the samples were incubated for 10 min with shaking. The underlayer solution was filtered with a 0.45-μm syringe filter. The analyses were performed on a Rigol L3000 device (Beijing Rigol Technology). Chromatographic separation was accomplished using an RP-HPLC ACE column (5C18-HL) with particle size of 5 μm (250 mm × 4.6 mm) at room temperature through a binary gradient. Mobile phase A was 25 mm acetate buffer (pH 6.5) and 70 mL acetonitrile. Mobile phase B was 80% acetonitrile aqueous solution. The flow rate was 1 mL min−1 and the column temperature was 40°C.
Enzyme Activity Assays
Frozen samples (100 mg) were powdered with liquid N and homogenized with 3 mL of 50 mm Tris-HCl buffer (pH 8) containing 2 mm Mg2+, 2 mm DTT, and 0.4 m Suc. The homogenate was centrifuged at 10,000g and 4°C for 10 min. The supernatant was used to determine the activities of the N metabolic enzymes GS, NADH-GOGAT, NADH-GDH, GOT, and GPT.
The activities of GS and GOGAT were measured according to the method described by Zhang et al. (1997) and Singh and Srivastava (1986), respectively. The activity of NADH-GDH was determined according to the method of Loulakakis and Roubelakis-Angelakis (1990). GOT and GPT activities were assayed using the method described by Wu et al. (1998).
Frozen samples were pulverized in liquid N, and about 100 mg of each sample was used to measure PK activity according to Lepper et al. (2010) with some modifications. The samples were extracted with 3 mL of 100 mm Tris-HCl (pH 7.5) containing 10 mm β-mercaptoethanol, 12.5% (v/v) glycerine, l mm EDTA-Na2, 10 mm MgCl2, and 1% (m/v) polyvinylpyrrolidone-40. These solutions were centrifuged at 10,000g and 4°C for 10 min. The reaction mixture contained 100 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 0.16 mm NADH, 75 mm KCl, 5 mm ADP, 7 units of l-lactate dehydrogenase, and 1 mm phosphoenolpyruvate. The reaction was started by adding 0.1 mL of enzyme extract to 0.9 mL of the reaction mixture and was carried out at 25°C to monitor the absorbance changes at 340 nm for 180 s.
Enzyme activities were expressed as mol of metabolite generated/consumed per mg of protein per unit of time. Soluble protein content was measured by the Bradford method with BSA as the standard (Bradford, 1976).
RNA Extraction and Reverse Transcription Quantitative PCR Assay
The seedlings were treated with 5 mm NH4NO3 and (NH4)2SO4 for 3 d before roots and shoots were harvested separately for RNA analysis. Total RNA was extracted with TRIzol (Invitrogen), precipitated with an equal volume of isopropanol, washed with 75% ethanol, and dissolved in RNase-free water. cDNAs were synthesized using the PrimeScript RT Kit with gDNA Eraser (Perfect Real Time; Takara) following the protocol of the manufacturer. The relative expression of genes in roots and shoots was determined by reverse transcription quantitative PCR performed on an Applied Biosystems StepOne Real-Time PCR System with SYBR Premix Ex-Taq (Takara) according to the protocol of the manufacturer. Primers used in the assays are listed in Supplemental Table S1, and the expression data were normalized to ACTIN2 or SAND.
Statistical Analysis
All experiments were conducted using a completely randomized design. Four samples used as replicates and two technical replicates were used for each treatment. Two-way ANOVA was conducted to analyze the effects of N source and plant material. Multiple comparisons were performed using the lsd multiple range test. The differences between control and (NH4)2SO4 treatments in the same plant material were evaluated with Student’s t test. Differences were considered statistically significant at P < 0.05.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AT3G18780 (ACTIN2), AT3G01970 (WRKY4), AT4G23810 (WRKY53), AT2G29350 (SAG13), AT5G13170 (SAG13), AT5G59220 (SAG113), AT4G30270 (SEN4), AT4G13510 (AMT1;1), AT1G64780 (AMT1;2), AT3G24300 (AMT1;3), AT3G24290 (AMT1;5), AT3G01090 (SnRK1.1), AT3G29160 (SnRK1.1), AT2G22810 (ACS4), AT4G08040 (ACS11), and AT2G28390 (SAND).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phenotypes and chlorophyll concentrations of Col-0, chl1-1, and chl1-5 plants treated with (NH4)2SO4, K2SO4, and KNO3.
Supplemental Figure S2. Effects of higher (NH4)2SO4 on the stomatal conductance of Col-0, chl1-1, and chl1-5 plants.
Supplemental Figure S3. Effects of high concentrations of NH4+ as the sole source of N on NH4+ uptake using NMT.
Supplemental Figure S4. Phenotypes of Col-0 and amt1;1 plants treated with 5 mm (NH4)2SO4 for 10 d.
Supplemental Figure S5. Effects of (NH4)2SO4 on the activity of NH4+ assimilation enzymes and transaminase in the shoots of Col-0, chl1-1, and chl1-5 plants.
Supplemental Figure S6. Effects of (NH4)2SO4 on free amino acid content in Col-0 and the chl1-1 mutant.
Supplemental Figure S7. Ethylene is involved in NRT1.1-related NH4+ toxicity.
Supplemental Figure S8. Comparison of pH values of growth media.
Supplemental Table S1. Primer sequences for gene expression used in this study.
Acknowledgments
We thank Li-Xing Yuan (China Agricultural University) for kindly providing the seeds of amt1;1 and Dr. Shuan Meng (Hunan Agricultural University) for technical support and helpful discussions.
Footnotes
This study was supported in part by the National Key R&D Program of China (2017YFD0200103 and 2017YFD0200100), the National Natural Science Foundation of China (31101596 and 31372130), the Hunan Provincial Recruitment Program of Foreign Experts, the National Oilseed Rape Production Technology System of China, the 2011 Plan supported by the Chinese Ministry of Education, the Research and Innovation Project of Postgraduates in Hunan Province (CX2015B242), and the Double First-Class Construction Project of Hunan Agricultural University (kxk201801005).
Articles can be viewed without a subscription.
Senior author.
References
- Álvarez-Aragón R, Rodríguez-Navarro A (2017) Nitrate-dependent shoot sodium accumulation and osmotic functions of sodium in Arabidopsis under saline conditions. Plant J 91: 208–219 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Barker AV. (1999) Foliar ammonium accumulation as an index of stress in plants. Commun Soil Sci Plant Anal 30: 167–174 [Google Scholar]
- Barker AV, Corey KA (1991) Interrelations of ammonium toxicity and ethylene action in tomato. HortScience 26: 177–180 [Google Scholar]
- Bittsánszky A, Pilinszky K, Gyulai G, Komives T (2015) Overcoming ammonium toxicity. Plant Sci 231: 184–190 [DOI] [PubMed] [Google Scholar]
- Bouguyon E, Brun F, Meynard D, Kubeš M, Pervent M, Leran S, Lacombe B, Krouk G, Guiderdoni E, Zažímalová E, et al. (2015) Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nat Plants 1: 15015. [DOI] [PubMed] [Google Scholar]
- Boussama N, Ouariti O, Suzuki A, Ghorbel MH (1999) Cd-stress on nitrogen assimilation. J Plant Physiol 155: 310–317 [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Britto DT, Siddiqi MY, Glass ADM, Kronzucker HJ (2001) Futile transmembrane NH4+ cycling: A cellular hypothesis to explain ammonium toxicity in plants. Proc Natl Acad Sci USA 98: 4255–4258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock BLW, Wilkinson DA, King J (1970) Glyoxylate aminotransferases from oat leaves. Can J Biochem 48: 486–492 [DOI] [PubMed] [Google Scholar]
- Chaillou S, Vessey JK, Morot-Gaudry JF, Raper CD Jr, Henry LT, Boutin JP (1991) Expression of characteristics of ammonium nutrition as affected by pH of the root medium. J Exp Bot 42: 189–196 [DOI] [PubMed] [Google Scholar]
- Chen G, Guo SW, Kronzucker HJ, Shi WM (2013) Nitrogen use efficiency (NUE) in rice links to NH4+ toxicity and futile NH4+ cycling in roots. Plant Soil 369: 351–363 [Google Scholar]
- Cruz C, Bio AFM, Domínguez-Valdivia MD, Aparicio-Tejo PM, Lamsfus C, Martins-Loução MA (2006) How does glutamine synthetase activity determine plant tolerance to ammonium? Planta 223: 1068–1080 [DOI] [PubMed] [Google Scholar]
- del Campo CP, Garde-Cerdán T, Sánchez AM, Maggi L, Carmona M, Alonso GL (2009) Determination of free amino acids and ammonium ion in saffron (Crocus sativus L.) from different geographical origins. Food Chem 114: 1542–1548 [Google Scholar]
- Fang XZ, Tian WH, Liu XX, Lin XY, Jin CW, Zheng SJ (2016) Alleviation of proton toxicity by nitrate uptake specifically depends on nitrate transporter 1.1 in Arabidopsis. New Phytol 211: 149–158 [DOI] [PubMed] [Google Scholar]
- Feng J, Barker AV (1992) Ethylene evolution and ammonium accumulation by tomato plants under water and salinity stresses. Part II. J Plant Nutr 15: 2471–2490 [Google Scholar]
- Gao YX, Li Y, Yang XX, Li HJ, Shen QR, Guo SW (2010) Ammonium nutrition increases water absorption in rice seedlings (Oryza sativa L.) under water stress. Plant Soil 331: 193–201 [Google Scholar]
- Glass ADM, Erner Y, Kronzucker HJ, Schjoerring JK, Siddiqi MY, Wang MY (1997) Ammonium fluxes into plant roots: Energetics, kinetics, and regulation. Zeitschrift Fur Pflanzenernahrung and Bodenkunde 160: 261–268 [Google Scholar]
- Guo FQ, Young J, Crawford NM (2003) The nitrate transporter AtNRT1.1 (CHL1) functions in stomatal opening and contributes to drought susceptibility in Arabidopsis. Plant Cell 15: 107–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachiya T, Noguchi K (2011) Mutation of NRT1.1 enhances ammonium/low pH-tolerance in Arabidopsis thaliana. Plant Signal Behav 6: 706–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachiya T, Mizokami Y, Miyata K, Tholen D, Watanabe CK, Noguchi K (2011) Evidence for a nitrate-independent function of the nitrate sensor NRT1.1 in Arabidopsis thaliana. J Plant Res 124: 425–430 [DOI] [PubMed] [Google Scholar]
- Hachiya T, Watanabe CK, Fujimoto M, Ishikawa T, Takahara K, Kawai-Yamada M, Uchimiya H, Uesono Y, Terashima I, Noguchi K (2012) Nitrate addition alleviates ammonium toxicity without lessening ammonium accumulation, organic acid depletion and inorganic cation depletion in Arabidopsis thaliana shoots. Plant Cell Physiol 53: 577–591 [DOI] [PubMed] [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF (2009) CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194 [DOI] [PubMed] [Google Scholar]
- Hoai NTT, Shim IS, Kobayashi K, Kenji U (2003) Accumulation of some nitrogen compounds in response to salt stress and their relationships with salt tolerance in rice (Oryza sativa L.) seedlings. Plant Growth Regul 41: 159–164 [Google Scholar]
- Husted S, Schjoerring JK (1995) Apoplastic pH and ammonium concentration in leaves of Brassica napus L. Plant Physiol 109: 1453–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi V, Joung JG, Fei Z, Jander G (2010) Interdependence of threonine, methionine and isoleucine metabolism in plants: Accumulation and transcriptional regulation under abiotic stress. Amino Acids 39: 933–947 [DOI] [PubMed] [Google Scholar]
- Jossier M, Bouly JP, Meimoun P, Arjmand A, Lessard P, Hawley S, Grahame Hardie D, Thomas M (2009) SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signalling in Arabidopsis thaliana. Plant J 59: 316–328 [DOI] [PubMed] [Google Scholar]
- Joy KW. (1988) Ammonia, glutamine, and asparagine: A carbon-nitrogen interface. Can J Bot 66: 2103–2109 [Google Scholar]
- Konishi N, Ishiyama K, Beier MP, Inoue E, Kanno K, Yamaya T, Takahashi H, Kojima S (2017) Contributions of two cytosolic glutamine synthetase isozymes to ammonium assimilation in Arabidopsis roots. J Exp Bot 68: 613–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K, et al. (2010) Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell 18: 927–937 [DOI] [PubMed] [Google Scholar]
- Lawlor DW, Tezara W (2009) Causes of decreased photosynthetic rate and metabolic capacity in water-deficient leaf cells: A critical evaluation of mechanisms and integration of processes. Ann Bot 103: 561–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejay L, Gansel X, Cerezo M, Tillard P, Müller C, Krapp A, von Wirén N, Daniel-Vedele F, Gojon A (2003) Regulation of root ion transporters by photosynthesis: Functional importance and relation with hexokinase. Plant Cell 15: 2218–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper TW, Oliveira E, Koch GD, Berlese DB, Feksa LR (2010) Lead inhibits in vitro creatine kinase and pyruvate kinase activity in brain cortex of rats. Toxicol In Vitro 24: 1045–1051 [DOI] [PubMed] [Google Scholar]
- Li B, Li Q, Su Y, Chen H, Xiong L, Mi G, Kronzucker HJ, Shi W (2011) Shoot-supplied ammonium targets the root auxin influx carrier AUX1 and inhibits lateral root emergence in Arabidopsis. Plant Cell Environ 34: 933–946 [DOI] [PubMed] [Google Scholar]
- Li B, Li G, Kronzucker HJ, Baluška F, Shi W (2014) Ammonium stress in Arabidopsis: signaling, genetic loci, and physiological targets. Trends Plant Sci 19: 107–114 [DOI] [PubMed] [Google Scholar]
- Li C, Tang Z, Wei J, Qu H, Xie Y, Xu G (2016) The OsAMT1.1 gene functions in ammonium uptake and ammonium-potassium homeostasis over low and high ammonium concentration ranges. J Genet Genomics 43: 639–649 [DOI] [PubMed] [Google Scholar]
- Li G, Li B, Dong G, Feng X, Kronzucker HJ, Shi W (2013) Ammonium-induced shoot ethylene production is associated with the inhibition of lateral root formation in Arabidopsis. J Exp Bot 64: 1413–1425 [DOI] [PubMed] [Google Scholar]
- Loulakakis CA, Roubelakis-Angelakis KA (1990) Intracellular localization and properties of NADH-glutamate dehydrogenase form Vitis vinifera L.: Purification and characterization of the major leaf isoenzyme. J Exp Bot 41: 1223–1230 [Google Scholar]
- Mao QQ, Guan MY, Lu KX, Du ST, Fan SK, Ye YQ, Lin XY, Jin CW (2014) Inhibition of nitrate transporter 1.1-controlled nitrate uptake reduces cadmium uptake in Arabidopsis. Plant Physiol 166: 934–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Reisdorf-Cren M, Pageau K, Lelandais M, Grandjean O, Kronenberger J, Valadier MH, Feraud M, Jouglet T, Suzuki A (2006) Glutamine synthetase-glutamate synthase pathway and glutamate dehydrogenase play distinct roles in the sink-source nitrogen cycle in tobacco. Plant Physiol 140: 444–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miflin BJ, Habash DZ (2002) The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J Exp Bot 53: 979–987 [DOI] [PubMed] [Google Scholar]
- Muños S, Cazettes C, Fizames C, Gaymard F, Tillard P, Lepetit M, Lejay L, Gojon A (2004) Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1.1 in the regulation of another nitrate transporter, NRT2.1. Plant Cell 16: 2433–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HTT, Shim IS, Kobayashi K, Usui K (2005) Regulation of ammonium accumulation during salt stress in rice (Oryza sativa L.) seedlings. Plant Prod Sci 8: 397–404 [Google Scholar]
- Noguchi K, Watanabe CK, Terashima I (2015) Effects of elevated atmospheric CO2 on primary metabolite levels in Arabidopsis thaliana Col-0 leaves: an examination of metabolome data. Plant Cell Physiol 56: 2069–2078 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Fernie AR, Stitt M (2010) Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol Plant 3: 973–996 [DOI] [PubMed] [Google Scholar]
- Omari RE, Rueda-López M, Avila C, Crespillo R, Nhiri M, Cánovas FM (2010) Ammonium tolerance and the regulation of two cytosolic glutamine synthetases in the roots of sorghum. Funct Plant Biol 37: 55–63 [Google Scholar]
- Ranathunge K, El-Kereamy A, Gidda S, Bi YM, Rothstein SJ (2014) AMT1;1 transgenic rice plants with enhanced NH4+ permeability show superior growth and higher yield under optimal and suboptimal NH4+ conditions. J Exp Bot 65: 965–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera M, Peleg Z, Abdel-Tawab YM, Tumimbang EB, Delatorre CA, Blumwald E (2013) Stress-induced cytokinin synthesis increases drought tolerance through the coordinated regulation of carbon and nitrogen assimilation in rice. Plant Physiol 163: 1609–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan L, Wei K, Wang L, Cheng H, Zhang F, Wu L, Bai P, Zhang C (2016) Characteristics of NH4+ and NO3− fluxes in tea (Camellia sinensis) roots measured by scanning ion-selective electrode technique. Sci Rep 6: 38370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus A, Baxter I, Muthukumar B, Gustin J, Lahner B, Yakubova E, Salt DE (2006) Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet 2: e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RP, Srivastava HS (1986) Increase in glutamate synthase (NADH) activity in maize seedlings in response to nitrate and ammonium nitrogen. Physiol Plant 66: 413–416 [Google Scholar]
- Skopelitis DS, Paranychianakis NV, Paschalidis KA, Pliakonis ED, Delis ID, Yakoumakis DI, Kouvarakis A, Papadakis AK, Stephanou EG, Roubelakis-Angelakis KA (2006) Abiotic stress generates ROS that signal expression of anionic glutamate dehydrogenases to form glutamate for proline synthesis in tobacco and grapevine. Plant Cell 18: 2767–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M. (1999) Nitrate regulation of metabolism and growth. Curr Opin Plant Biol 2: 178–186 [DOI] [PubMed] [Google Scholar]
- Tabuchi M, Abiko T, Yamaya T (2007) Assimilation of ammonium ions and reutilization of nitrogen in rice (Oryza sativa L.). J Exp Bot 58: 2319–2327 [DOI] [PubMed] [Google Scholar]
- Tegeder M, Masclaux-Daubresse C (2018) Source and sink mechanisms of nitrogen transport and use. New Phytol 217: 35–53 [DOI] [PubMed] [Google Scholar]
- Tsay YF, Schroeder JI, Feldmann KA, Crawford NM (1993) The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72: 705–713 [DOI] [PubMed] [Google Scholar]
- Wang Q, Zhao Y, Luo W, Li R, He Q, Fang X, Michele RD, Ast C, von Wirén N, Lin J (2013) Single-particle analysis reveals shutoff control of the Arabidopsis ammonium transporter AMT1;3 by clustering and internalization. Proc Natl Acad Sci USA 110: 13204–13209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Peng F, Li M, Yang L, Li G (2012) Expression of a heterologous SnRK1 in tomato increases carbon assimilation, nitrogen uptake and modifies fruit development. J Plant Physiol 169: 1173–1182 [DOI] [PubMed] [Google Scholar]
- Wang YY, Khoo KH, Chen ST, Lin CC, Wong CH, Lin CH (2002) Studies on the immuno-modulating and antitumor activities of Ganoderma lucidum (Reishi) polysaccharides: Functional and proteomic analyses of a fucose-containing glycoprotein fraction responsible for the activities. Bioorg Med Chem 10: 1057–1062 [DOI] [PubMed] [Google Scholar]
- Wellburn AR, Lichtenthaler HK (1984) Formulae and program to determine total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Advances in Photosynthesis Research 2: 9–12 [Google Scholar]
- Wu LH, Jiang SH, Tao QN (1998) The application of colormetric method on the determination of transaminase activity. Tu Jang Tung Pao 3: 41–43 [Google Scholar]
- You W, Barker AV (2005) Ethylene evolution and ammonium accumulation by tomato plants after root-applied glufosinate-ammonium treatment in the presence of ethylene inhibitors. Commun Soil Sci Plant Anal 35: 1957–1965 [Google Scholar]
- Yuan L, Loqué D, Kojima S, Rauch S, Ishiyama K, Inoue E, Takahashi H, von Wirén N (2007a) The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. Plant Cell 19: 2636–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan YZ, Ou JQ, Wang ZQ, Zhang CF, Zhou ZP, Lin QH (2007b) Regulation of carbon and nitrogen metabolisms in rice roots by 2-oxoglutarate at the level of hexokinase. Physiol Plant 129: 296–306 [Google Scholar]
- Zanini E. (2001) Indophenol blue colorimetric method for measuring cation exchange capacity in sandy soils. Commun Soil Sci Plant Anal 32: 2519–2530 [Google Scholar]
- Zhang C, Peng S, Peng X, Chavez AQ, Bennett J (1997) Response of glutamine synthetase isoforms to nitrogen sources in rice (Oryza sativa L.) roots. Plant Sci 125: 163–170 [Google Scholar]
- Zhong C, Cao X, Hu J, Zhu L, Zhang J, Huang J, Jin Q (2017) Nitrogen metabolism in adaptation of photosynthesis to water stress in rice grown under different nitrogen levels. Front Plant Sci 8: 1079. [DOI] [PMC free article] [PubMed] [Google Scholar]