Abstract
Bacteria can use the second messenger c-di-GMP to adjust their motility in response to environmental cues. The protein YcgR, upon binding of c-di-GMP, interacts with the flagellar motor to affect the motor behavior. However, the full feature of the effects of c-di-GMP::YcgR on the flagellar motor remains unclear, and its interacting partners on the motor is still controversial. Here, we characterized the effects of c-di-GMP::YcgR on the torque-speed curve of the flagellar motor, one of the most important properties of the motor, finding that it affects the motor behavior throughout the full range of load conditions from zero to high loads by shifting the motor torque-speed curve downward. We also investigated the interacting partner on the motor through dynamical fluorescent studies, finding that c-di-GMP::YcgR mainly interacts with the motor-switch complex instead of the torque-generating units (stators). To directly test the behavioral consequence of elevated c-di-GMP levels, we measured the distribution of bacteria swimming near a surface, finding that elevated c-di-GMP levels promote bacterial aggregation on surfaces. The effects of c-di-GMP on bacterial motile behavior that we characterized here are consistent with the key role that c-di-GMP plays in the transition between motile and sedentary forms of bacterial life.
Introduction
Bacteria can move in liquid environments to migrate toward favorable conditions by sensing gradients of attractants or repellents via the chemotaxis signaling pathway. The output of the chemotaxis pathway of flagellated bacteria, such as Escherichia coli, is the bacterial flagellar rotary motor, which drives the bacteria to move in a random walk pattern, alternating between runs and tumbles (1). When all of the flagellar motors on a cell rotate in a counterclockwise (CCW) direction, the bacterium runs, whereas the bacterium tumbles when one or more motors rotate in a clockwise (CW) direction (2). The random walk pattern is biased in the presence of a chemical gradient. The response regulator of the chemotaxis signaling pathway, CheY, can be phosphorylated to CheY-P, which binds to the component of flagellar motor-switch complex, FliM, increasing the probability of the motor rotating in a CW direction (raising the CW bias) (3, 4, 5, 6).
The energy source of the bacteria flagellar motor of E. coli is the difference in proton electrochemical potential across the inner cell membrane, called proton motive force (PMF) (7). PMF drives the flow of protons through the transmembrane channels formed inside the stator complex, which is composed of the proteins MotA and MotB (stoichiometry MotA4MotB2) (8). The stators interact with the rotor through electrostatic interactions to generate the motor torque. The stators were shown to be dynamically coming on and off the motor, with a maximal stator number per motor of 11 (9). The rotor consists of ∼26 copies of FliG, 34 copies of FliM, and 136 copies of FliN (9, 10, 11), forming the switching complex at the intracellular base of the motor. The switch complex, upon binding of CheY-P (to FliM), undergoes conformational changes to regulate the rotational direction of the motor. Switching of the motor rotational direction provides the basis for regulating the run-tumble pattern of flagellated bacteria. The rotational speed of the flagellar motor determines bacterial swimming speed and depends strongly on the external load conditions from zero to high loads.
The second messenger c-di-GMP was reported to inhibit the motility of the bacteria (12, 13, 14) and may be coupled to the chemotaxis machinery (15). This molecule, which is synthesized by diguanylate cyclases and degraded by phosphodiesterases in response to environmental cues (16), plays a critical role in the transformation between motile and sedentary forms of the bacteria. The c-di-GMP-binding protein YcgR, upon binding of c-di-GMP, interacts with the flagellar motor to adjust its behavior (17, 18). However, whether the interacting partner comprises the stators or the switch complex remains controversial. One previous study showed that c-di-GMP::YcgR reduces bacterial swimming speed by interacting with the stator protein MotA (12). Another study reported that c-di-GMP::YcgR interacts with the switch-complex proteins FliG and FliM to reduce bacterial swimming and swarming motility and induce CCW motor bias (13). A third study suggested that c-di-GMP::YcgR interacts with FliG (14). To clarify this, we labeled YcgR with eGFP and compared the binding of c-di-GMP::YcgR with individual motors dynamically when the stators were present or absent. Previous studies have only investigated the effect of elevated c-di-GMP levels on the bacterial swimming or swarming speed, which corresponds to one or few load conditions of the flagellar motor. To explore the effects on the motor under the full range of load conditions from zero to high loads, we measured the effect of elevated c-di-GMP levels on the torque-speed curve of the motor. To understand the behavioral consequences of elevated c-di-GMP levels directly, we compared the steady-state distributions of bacteria swimming between two parallel surfaces for wild-type cells and cells with elevated c-di-GMP levels.
Materials and Methods
Strains and plasmids
Strains and plasmids are listed in Table S1. Strains JY27 (ΔfliC cheY), RW1 (ΔfliC cheY yhjH), JY26 (ΔfliC), RW2 (ΔfliC yhjH), and RW3 (ΔfliC cheY yhjH ycgR) are derived from the E. coli K12 strain RP437 (HCB33). The plasmid pKAF131 constitutively expresses FliCst. The plasmid pBAD33fliC expresses wild-type flagellin FliC under the control of an arabinose-inducible promotor. To measure the bacteria flagellar motor rotation speed at zero load, JY27 and RW1 were used. To measure the bacteria flagellar motor rotation speed at low to high loads, JY27 and RW1 transformed with the plasmid pKAF131 were used. To measure the bacteria flagellar motor rotation bias, we used JY26 and RW2 strains transformed with the plasmid pKAF131. To measure the flagellar motor CW rotation, we transformed JY27 and RW1 with pKAF131 and pWB5, which expresses CheY under the control of an IPTG-inducible promoter in the vector pTrc99a. For the fluorescent measurements, we made an eGFP-YcgR fusion using a linker with the sequence “GGAGGCGGAGGCGGA” and inserted it into the vector pTrc99a. For the resurrection experiments, RW1 transformed with the plasmid pHS1, which expresses proteorhodopsin under an arabinose-inducible promoter (19), was used. For the measurement of cell density distributions near surfaces, JY26 and RW2 transformed with pBAD33fliC were used. For the measurement of motor speeds and CW bias with overexpression of MotA and MotB, RW2 transformed with the plasmid pFD313, which constitutively expresses FliCst (20), and the plasmid pJY7, which expresses wild-type MotA and MotB under the control of an arabinose-inducible promoter (21), were used.
Cell culture
Cells were grown at 33°C in T-broth with the appropriate antibiotics (25 μg/mL chloramphenicol and 100 μg/mL ampicillin) and the appropriate inducers (50 μM IPTG for CheY expression, 10 μM IPTG for eGFP-YcgR expression, and 0.01% arabinose for FliC expression). All of them were grown to an OD600 between 0.45 and 0.50 and were harvested by centrifugation at 4000 × g for 1 min (except for the experiments measuring distributions of bacteria swimming near a surface; see below) twice with motility medium (10 mM potassium phosphate, 0.1 mM ethylenediaminetetraacetic acid, 10 mM lactic acid, and 70 mM NaCl (pH 7.0)) and were resuspended in this medium. They were used immediately for experiments or stored at 4°C for up to 3 h.
The bead assay
Cells were sheared to truncate flagella by passing 1 mL of the washed cell suspension 50 times between two syringes equipped with 23-gauge needles and connected by a 7-cm length of polyethylene tubing (0.58 mm inside diameter, No. 427411; Becton Dickinson, Franklin Lakes, NJ). Coverslips were coated by poly-L-lysine, and a chamber was formed with two pieces of double-sided tape spaced between the coverslip and a glass slide. To measure the motor rotating at low to high loads, 40 μL cells were placed on the glass coverslip coated with poly-L-lysine (Sigma, St. Louis, MO) and allowed to stand for 3 min, rinsed with 100 μL motility medium; then, 0.35-μm-diameter polystyrene latex beads (Polysciences, Warrington, PA) were attached to the sheared flagellar stubs, incubated for 3 min, and rinsed with 200 μL motility medium with various Ficoll concentrations (0, 2, 3, 5, 7, 9, 12, or 15% w/v). The chamber was sealed with vacuum grease. The polystyrene beads were observed by phase-contrast microscopy. Other measurements with the bead assay were performed similarly with 1.0- or 1.5-μm-diameter latex beads. Phase-contrast images were recorded at frame rates of 300−2000 fps using a scientific complementary metal-oxide semiconductor (CMOS) camera (C11440; Hamamatsu, Hamamatsu, Japan). To measure the motor rotating at zero load, we attached 100-nm-diameter gold beads to the hook with antibodies, following the procedure, and observed using the laser-darkfield setup described previously (22, 23). Motor resurrection experiments were carried out following the procedure and using the setup described previously (19). Data analysis was carried out using custom scripts in MATLAB (The MathWorks, Natick, MA). The position of the beads were determined by two-dimensional Gaussian fitting. The torque-speed curves were constructed as described previously (24).
Fluorescence measurements
Before use, we thoroughly cleaned the glass coverslips and photobleached them for 1 h with a high-powered mercury lamp. Cells were tethered to a glass coverslip by hooks using an antihook antibody, following a protocol described previously (25). The fluorescence measurements were performed using a total internal reflection fluorescence (TIRF) microscope (Eclipse Ti-E TIRF; Nikon, Tokyo, Japan) with an external objective phase ring that allows simultaneous observation of fluorescence and phase-contrast images. The fluorescence images were recorded with a back-illuminated, cooled (−70°C), electron-multiplying charge-coupled device camera (DU897U-CS0-BV; Andor Technology, Belfast, Northern Ireland) and the phase-contrast images were recorded with a CMOS camera (DCC3240M; Thorlabs, Newton, NJ). TIRF excitation was provided by an optically pumped semiconductor laser (488-nm Sapphire488LP; Coherent, Santa Clara, CA). Details of the optical path for the TIRF setup are shown in Fig. S3. The Andor camera was controlled by Andor Solis software running on a desktop computer. For Fig. 4, image acquisition was conducted under Andor Solis “kinetic” mode, with 500-ms exposure used for each image. For comparison of eGFP-YcgR binding before and after stators fell off the motor, images were taken before and after flowing NaN3 for 30 min, each with an exposure time of 500 ms. The laser illumination was blocked in between exposures. The width of the fluorescent spots was ∼5 pixels (325 nm). The fluorescent intensity centroid for each motor was calculated using a Gaussian mask method, and the motor intensity was calculated following a procedure described previously (25).
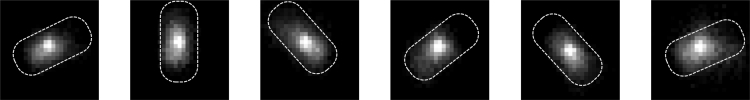
Figure 4.
eGFP-YcgR fluorescence spot detected on an active motor driving the rotation of the tethered cell body. Dashed lines are outlines of the cell body. Images were taken with a TIRF setup continuously every 0.5 s.
Measurements of cell distribution near surfaces
0.5 mL of the cell cultures were pelleted by centrifuging at 1200 × g for 6 min and resuspended gently in 1 mL of 0.005% PVP-40 in motility medium (to prevent adsorption of cells to the glass). 0.4 mL of the washed cell solution was mixed with 0.5 mL Percoll to match the medium and cell buoyant densities. A chamber was constructed with two pieces of double-sided tape spaced between the coverslip and a glass slide, resulting in a sample thickness of ∼150 μm. 100 μL cells were added to the chamber, which was then sealed with vacuum grease. The sample was allowed to stand for 5 min so that the cell distribution would reach a steady state. A phase-contrast Nikon Ti-E inverted microscope with a 40× objective (numerical aperture: 0.60; depth of field: 6.0 μm) and a scientific CMOS camera (C11440) were used to image the population of swimming cells. Images were recorded starting at 3 μm above the lower glass surface and at steps of 9.5 μm until the position within 3 μm of the upper glass surface was reached. At each position, two images separated by 1 s were recorded with an exposure time of 0.1 s, and the nonmotile cells were identified by comparison of the two images and were discarded from the counting of cell numbers. The measurements were repeated with other cell samples 14 times for JY26 and 12 times for RW2 cells transformed with pBAD33fliC.
Results
Elevated c-di-GMP levels shift the torque-speed curve downward
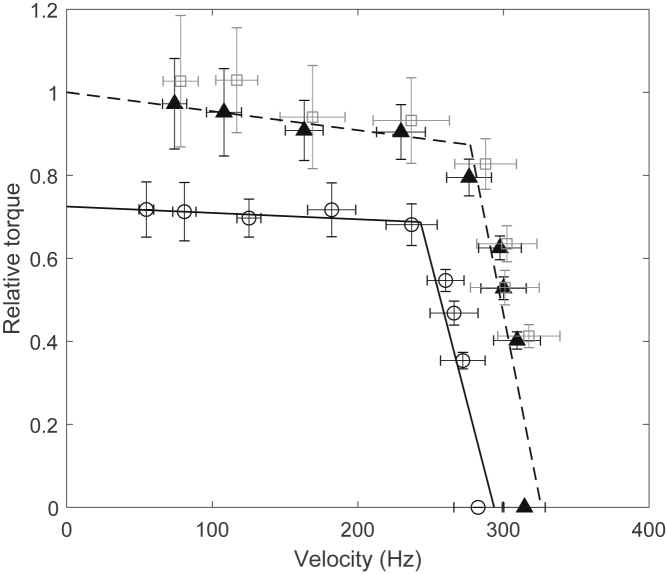
The relationship between torque and speed is one of the most important characteristics of the motor, which was used to distinguish specific mechanisms of motor rotation (22, 24, 26, 27, 28). In E. coli, YhjH is a phosphodiesterase. A wild-type cell with functional YhjH shows a minimal level of c-di-GMP; in contrast, a ΔyhjH mutant has elevated c-di-GMP levels (12). We used the E. coli K12 strain JY27 (ΔcheY ΔfliC), denoted as wild type, to measure the torque-speed curve for a wild-type motor. We further deleted yhjH from JY27, yielding RW1, to study the effect of elevated c-di-GMP levels on the motor torque-speed curve. Both strains were transformed with the plasmid pKAF131, which constitutively expresses the sticky flagellar filament FliCst. To measure the torque-speed curve, we labeled 0.35-μm-diameter latex beads to shortened filament stubs and varied the viscosity of the buffer using different concentrations of Ficoll to cover the load range from low to high loads (24). We used 100-nm-diameter gold spheres on hooks to compare the motor speeds at zero load for both strains. Fig. 1 shows the CCW torque-speed relationship of the wild-type strain JY27 (solid triangles and dashed line) and the mutant strains RW1 (circles and line). The symbols and error bars are means and SDs, respectively, whereas the lines are linear regressions. The symbols from left to right in Fig. 1 correspond to 0.35-μm-diameter latex beads labeled to short filament stubs in motility medium with different concentrations of Ficoll (15, 12, 9, 7, 5, 3, 2, or 0% w/v) and with 100-nm-diameter gold spheres labeled to hooks in motility medium (22). All torques were normalized to JY27 motor stall torque (the torque at zero speed). To calculate the torque precisely, we measured the viscosity of Ficoll solutions at different concentrations using a viscosity meter (data in the Supporting Materials and Methods). From Fig. 1, elevated c-di-GMP levels affect the torque-speed curve for the whole range of load conditions, shifting the curve downward. We further deleted YcgR from RW1, yielding a ΔyhjH ΔycgR double mutant (RW3), and measured its torque-speed relationship (squares in Fig. 1), which is similar to the wild type (JY27). Thus, the effect of elevated c-di-GMP levels in shifting the torque-speed curve is through YcgR.
Figure 1.
Normalized torque versus speed curves for the wild-type strain JY27 (ΔfliC cheY) (solid triangles and dashed line), the ΔyhjH mutant strain RW1 (circles and solid line), and the ΔyhjH ΔycgR double mutant RW3 (gray squares). The symbols and error bars are the means and SDs for the cell population at each load. The lines are linear regressions. The numbers of cells studied for JY27 at each data point were 46, 34, 31, 28, 53, 24, 25, 39, and 24, respectively; the corresponding numbers for RW1 were 13, 15, 25, 14, 28, 29, 19, 34, and 17, respectively; the corresponding numbers for RW3 were 21, 18, 20, 28, 36, 29, 39, and 27, respectively.
Looking at our measurements in another way, elevated c-di-GMP levels reduce the motor speed at each load condition, but the reduction effect is larger at a high load. At the highest load tested (1.5-μm-diameter latex beads on shortened filament stubs), the motor speed was reduced by ∼30%, whereas it was reduced by 10% for motors at zero load. The relative reduction in motor speed with elevated c-di-GMP levels is shown in Fig. S1 for motors labeled with 0.35-μm-diameter latex beads in different concentrations of Ficoll solutions.
Effect of elevated c-di-GMP levels on CCW and CW rotations of the motor
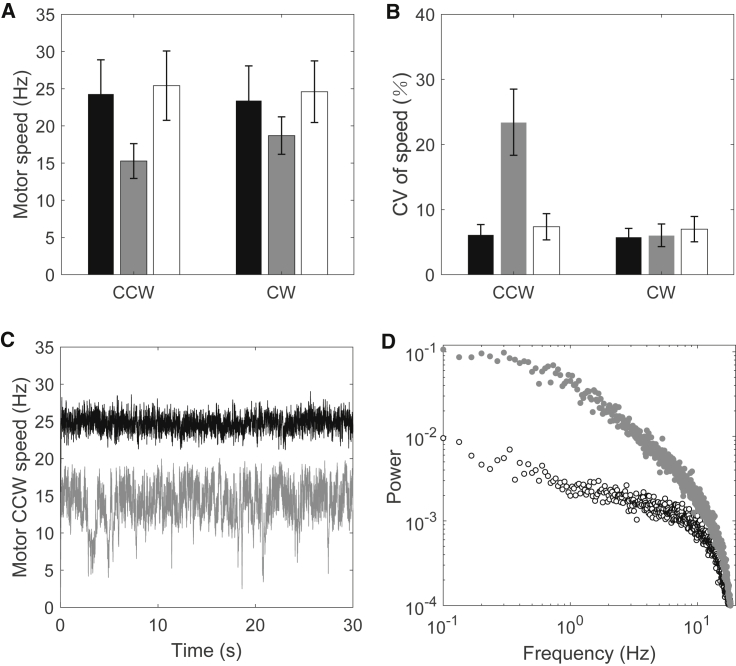
To test whether elevated c-di-GMP levels affect CCW and CW rotations differently, the wild-type strain JY27 and the mutant strain RW1 were both transformed with the plasmid pWB5, which expresses wild-type CheY under the control of an isopropyl-β-D-thiogalactoside (IPTG)-inducible promoter. To make the motor rotate in a CW direction only, high expression of CheY was induced in these strains with 0.05 mM IPTG. We attached 1.5-μm-diameter latex beads on shortened filament stubs of the four types of strains to compare the motor speeds: wild-type with the motor in CCW rotation, wild-type in CW rotation, ΔyhjH strain with the motor in CCW rotation, and ΔyhjH strain with the motor in CW rotation. Fig. 2 A shows the speeds: the average motor speeds of the wild-type strain are 24.33 Hz (CCW) and 23.44 Hz (CW), respectively, and the average motor speeds of the ΔyhjH strain with elevated c-di-GMP levels are 15.28 Hz (CCW) and 18.70 Hz (CW), respectively. Therefore, the influence of elevated c-di-GMP levels on CCW rotation is greater than on CW rotation. The speed of CCW rotation is reduced by 37% with elevated c-di-GMP levels, whereas the CW speed is reduced by 20%. We also measured the CCW and CW motor speeds for a ΔyhjH ΔycgR double-mutant strain and found that both speeds were similar to the corresponding wild-type values (Fig. 2, A and B), demonstrating that the speed-reduction effect of elevated c-di-GMP levels is dependent on YcgR.
Figure 2.
(A) The CCW/CW rotational speeds of the wild-type strain JY27 (ΔfliC cheY) without/with CheY overexpression (black), ΔyhjH mutant strain (gray), and ΔyhjH ΔycgR double-mutant strain (white) at a high load (1.5-μm-diameter latex bead). (B) The coefficient of variation (CV) (SD/mean × 100%) of the CCW/CW rotation of wild-type (black), ΔyhjH mutant (gray), and ΔyhjH ΔycgR double-mutant strains (white). The numbers of motors measured at each point were 48, 43, 27, 46, 57, and 30, respectively. The bars and errors are means and SDs. (C) Two examples of motor CCW speed traces for the wild-type (black) and ΔyhjH (gray) strains, showing the instability of CCW rotation for the ΔyhjH strain with elevated c-di-GMP levels. (D) The power spectra for the motor CCW speed traces for the wild-type (black circles) and ΔyhjH (gray dots) strains.
Furthermore, with elevated c-di-GMP concentration, the CCW rotation became more unstable. The coefficient of variation (CV), defined as the ratio of the motor speed SD to the mean speed for the whole speed trace (∼1 min for each motor), was used to characterize the stability of the rotation. As shown in Fig. 2 B, the CVs were small for motors of the wild-type strain. With elevated c-di-GMP level, the CV was still small for the CW rotation, whereas it increased dramatically for the CCW rotation, demonstrating the instability of CCW rotation with elevated c-di-GMP levels. Two examples of CCW speed traces for the wild-type and ΔyhjH strains are shown in Fig. 2 C, and the power spectra for the CCW speed traces for the wild-type and ΔyhjH strains are shown in Fig. 2 D, demonstrating a much higher low-frequency noise in the CCW rotation of the ΔyhjH strain.
Elevated c-di-GMP levels reduce CW rotational bias of the motor
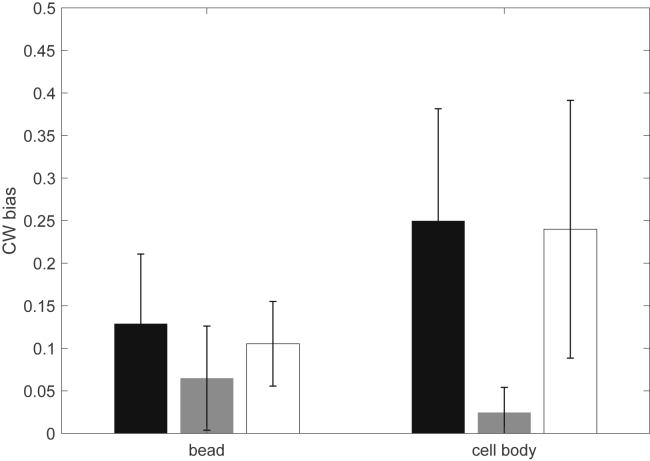
To test the effect of elevated c-di-GMP levels on CW bias of the motor in strains with wild-type levels of CheY-P concentration, we measured the CW biases under two load conditions: with bead assay (1.0-μm-diameter latex bead) and tethered cell rotation experiments. The strains JY26 (ΔfliC) and RW2 (ΔfliC ΔyhjH) were transformed with the plasmid pKAF131 to express FliCst. The CW biases of the motors of JY26 and the ΔyhjH strain RW2 are shown in Fig. 3 under the two load conditions. Clearly, with elevated c-di-GMP levels, the CW bias of the motor reduced, consistent with previous observations (13, 14). In addition, we found that the reduction is more dramatic at a higher load (tethered cell rotation). The effect of elevated c-di-GMP levels on reducing CW bias is dependent on YcgR because a ΔyhjH ΔycgR double mutant showed similar CW bias of the motor as the wild-type (Fig. 3).
Figure 3.
Bead assay (1.0-μm-diameter bead) and tethered cell rotation experiments with wild-type strain JY26 (ΔfliC) (black), ΔyhjH mutant strain (gray), and ΔyhjH ΔycgR double-mutant strain (white). The number of motors were 28, 33, 29, 49, 55, and 33, respectively.
We overexpressed MotA and MotB in strain RW2 (ΔfliC ΔyhjH) using the plasmid pJY7, which expresses MotA and MotB under the control of an arabinose-inducible promoter, and measured the CCW and CW motor speeds and CW bias with the bead assay (1.0-μm-diameter latex bead), finding that they were similar to the values for the wild-type strain JY26: CCW speed of 57.1 ± 6.7 Hz, CW speed of 53.5 ± 6.0 Hz, and CW bias of 12.1 ± 7.8% (mean ± SD), which were obtained from measurements of 24 motors. Therefore, overexpression of the stator proteins compensated the effects of elevated c-di-GMP levels on reducing speed and CW bias, consistent with a previous observation (13).
A dynamical fluorescence experiment determined the binding partner of c-di-GMP::YcgR
There were conflicting previous reports regarding the binding partner of YcgR::c-di-GMP (12, 13, 14) and whether it binds to the stator or the C-ring of the rotor.
To clarify this, we designed a dynamical fluorescent experiment. We further deleted the ycgR gene from RW1, yielding RW3. We constructed an eGFP-YcgR fusion and cloned the fusion gene to a pTrc99a vector, which expressed eGFP-YcgR under the control of an IPTG-inducible promoter. The pTrc99a-eGFP-YcgR plasmid was transformed into RW3.
To verify that the eGFP-YcgR fusion protein can function normally, we tested the motor speed at a high load (with a 1.0-μm-diameter latex bead), as shown in Fig. S2. The motor speed of JY27 (wild-type) is 60 Hz, whereas the motor speed of RW1 (ΔyhjH) is 42 Hz. When the genes yhjH and ycgR in the bacteria were deleted simultaneously (RW3), the motor speed was restored to its wild-type level (60 Hz). When the RW3 mutant strain transformed with the pTrc99a-eGFP-YcgR plasmid was induced by 0.01 mM IPTG, the motor speed was 40 Hz, similar to the motor of RW1 with wild-type YcgR. Therefore, the eGFP-YcgR fusion protein functions normally.
We tried overexpressing the eGFP-YcgR fusion protein in RW3 by induction with 0.1 mM IPTG, finding that the motor speed is similar to when it is induced with 0.01 mM IPTG: 35.6 ± 6.0 Hz (obtained from measurements of nine motors). Thus, overexpressing YcgR did not produce a clearly stronger speed-reduction effect on motor rotation probably because the c-di-GMP concentration is still the limiting factor in the ΔyhjH strain.
We tethered the cells of RW3 (induced with 0.01 mM IPTG) to the glass via the hook using antihook antibodies and used a TIRF setup to monitor the fluorescent intensity of the motor spot. Details of the optical paths for the setup are shown in Fig. S3. Fig. 4 shows the example images of the eGFP-YcgR fluorescent spot on the motor, which drives the rotation of the tethered cell body.
We used 20 mM NaN3 to deplete the cellular PMF. The motors usually stopped rotating within 5 min of flowing NaN3 because of both the depletion of PMF and loss of stators. Previous reports demonstrated that the stators come off the motor when PMF is depleted (19, 29, 30). We compared the fluorescent intensity of the motor spot before flowing NaN3 and after flowing it for 30 min. The ratio was 1.06 ± 0.09 (mean ± SEM) from measurements of nine motors. We also performed control experiments comparing the intensity before and after flowing motility medium for 30 min in which the PMF was kept constant; the ratio was 1.08 ± 0.09 from measurements of 12 motors. Therefore, the fluorescent intensity of eGFP-YcgR spot did not change after the stators came off the motor.
To make sure all or most of the stators came off the motor after flowing 20 mM NaN3 for 30 min, we performed motor resurrection experiments (19). We transformed the plasmid pHS1, which expresses proteorhodopsin under an arabinose-inducible promoter, to RW1. After flowing NaN3 for 30 min, 532 nm light was turned on to restore the PMF immediately (in less than a second), and the motor speed was observed to increase step by step when stators came back on the motor one by one. Sample resurrection traces are shown in Fig. S4. The initial motor speed after restoring PMF was zero or very low (corresponding to zero or one stator). This initial stator number corresponds to the number of stators that remained on the motor after flowing NaN3 for 30 min. Therefore, all or most of the stators came off the motor in our fluorescence measurements.
The intensity of the eGFP-YcgR spot on the motor did not change after the stators came off. Therefore, the binding partner of YcgR::c-di-GMP on the motor is mainly the rotor instead of the stator.
Elevated c-di-GMP levels promote bacterial aggregation on surfaces
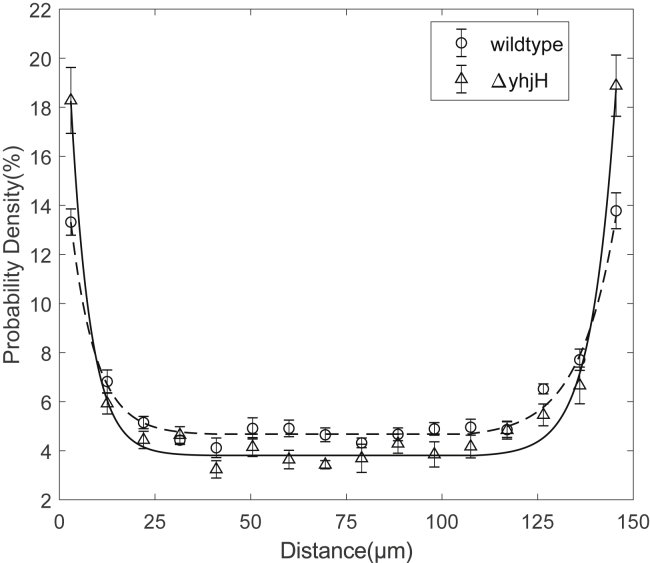
When bacteria swim near a surface, multiple physical effects lead to the accumulation of bacteria on the surface. This phenomenon was clearly demonstrated by measuring the steady-state cell distribution for cells swimming between two parallel surfaces, with cell density strongly increasing near the surface (31, 32). The reduction of motor speed and CW bias by elevated c-di-GMP levels would further help the accumulation near a surface, and here, we found that both effects of reduction are more dramatic for motors at higher loads, which are the load conditions the bacteria experience near surfaces in which the viscous drag is higher. Therefore, we reasoned that a behavioral consequence of the elevated c-di-GMP levels would be the promotion of bacterial aggregation near surfaces. To test this, we measured the steady-state distribution of bacteria swimming between two glass surfaces (separated by 150 μm). We compared the distributions for wild-type cells (JY26) and cells with elevated c-di-GMP levels (RW2), finding that the latter accumulates near surfaces more dramatically (Fig. 5). We fitted the probability density distributions with an exponential decay function. The decay length for the wild-type cells are 8.1 ± 0.5 μm, whereas it is 5.7 ± 0.5 μm for cells with elevated c-di-GMP levels, which is 30% sharper in the density distribution. Therefore, elevated c-di-GMP levels promote bacterial aggregation on surfaces.
Figure 5.
Density distributions of bacteria swimming between two parallel glass surfaces separated by 150 μm for wild-type bacteria (JY26, circles) and bacteria with elevated c-di-GMP levels (RW2, triangles). The lines are fitted to the data with an exponential function near each surface connected by a line of constant.
Discussion
In summary, we measured the motor torque-speed curves for wild-type cells and cells with elevated c-di-GMP levels, finding that c-di-GMP::YcgR binding affects the motor behavior throughout the full load range from zero to high loads. It reduces the motor speed at all load conditions, although the reduction effect is larger at higher loads. We also found that c-di-GMP::YcgR binding affects the CW and CCW rotations differently, exerting larger speed-reduction effects on the CCW rotation and making the CCW rotation more unstable. The c-di-GMP::YcgR binding also makes CCW rotation more probable (that is, it reduces the CW bias), and the reduction is more dramatic at a higher load.
Previous static fluorescent colocalization studies yielded conflicting results regarding where YcgR binds to the flagellar motor. We tested the binding partner of c-di-GMP::YcgR on the motor by dynamical fluorescent measurements. We used a TIRF setup on tethered cells to measure the eGFP-YcgR spot intensity while monitoring cell-body rotation, making sure that the measured eGFP-YcgR binding spots corresponded to functional motors. We found that the spot intensity did not change before or after the stators fell off the motor, thereby determining that c-di-GMP::YcgR binds mainly to the rotor instead of the stators. The fact that c-di-GMP::YcgR binding affects CW and CCW rotations differently and reduces CW bias of the motor also suggests that c-di-GMP::YcgR binds to the motor-switch complex.
A previous study indicated that a ΔmotA mutant reduces YcgR localization to the motor (12). Here, we found that YcgR can maintain on the motor after the stators come off. In light of both findings, MotA might help in recruiting YcgR to the motor but does not play a significant part in maintaining the YcgR binding to the motor. We did not determine whether YcgR binds to FliG or FliM on the motor-switch complex through dynamical fluorescent measurements. A previous study suggested that FliG is the primary interacting partner of YcgR (14). Our results were consistent with this suggestion. C-di-GMP::YcgR probably binds to FliG to affect the stator-rotor electrostatic interaction, thereby reducing the motor torque, and this manifests in shifting the torque-speed curve downward. The reduction of CW bias of the motor by binding of c-di-GMP::YcgR was probably induced by this reduction in motor torque, as a recent study showed that CW bias of the motor depends on the motor torque, decreasing with a decrease in motor torque (19). Our results indicated that overexpressing MotA and MotB compensated the effects of reducing motor speeds and CW bias. This is probably because more stators were incorporated into the motor with overexpression of MotA and MotB, thereby increasing the motor torque (and motor speeds), and CW bias also increased with this increase in motor torque. The previous finding (12) that certain MotA suppressors, which overrode the speed-reduction effect of c-di-GMP::YcgR, involved two residues in the immediate vicinity of MotA-FliG-interacting residues was consistent with the suggestion that these MotA suppressors probably aligned the MotA-FliG electrostatic interface back to normal.
Recent studies identified surface-sensing mechanisms that stimulate the production of c-di-GMP (33, 34). With an elevated c-di-GMP level near the surface after surface sensing, the CW bias of the flagellar motors is reduced. Our study shows that the reduction is more dramatic at a high load, which is the load condition the flagellar motors experience near the surface at which the viscous drag is higher. The reduced CW bias leads to much less tumbling, thereby reducing the probability that the bacterium moves away from the surface. In congruence with the fact that elevated c-di-GMP levels near the surface induce CCW rotation, our study also shows that elevated c-di-GMP levels have a larger speed-reduction effect on CCW rotation than on CW rotation, thus making the bacteria easier to associate with the surface. To directly demonstrate this, we measured distributions of bacteria swimming near a surface for wild-type bacteria and bacteria with elevated c-di-GMP levels, finding that elevated c-di-GMP levels promote bacterial aggregation on surfaces. Therefore, the effects of elevated c-di-GMP levels on the flagellar motor we characterized here are consistent with the critical role that c-di-GMP plays in the transition between free-swimming and motile and surface-attached and sedentary forms of bacterial life.
Author Contributions
J.Y. and R.Z. designed the work. R.W. and F.W. performed the measurements with help on the surface-accumulation experiment from R.H. All authors wrote the article. R.W. and F.W. contributed equally to this work.
Acknowledgments
We thank Howard Berg for the antihook antibody.
This work was supported by National Natural Science Foundation of China (grants 11374282, 21573214, and 11872358), a grant from the Ministry of Science and Technology of China (2016YFA0500700), and Fundamental Research Funds for the Central Universities (WK2030020028). J.Y. and R.Z. are supported by the Chinese Government 1000 Youth Talents Program. R.H. is supported by a National Postdoctoral Program for Innovative Talents (BX201700226).
Editor: Dennis Bray.
Footnotes
Renjie Wang and Fangbin Wang contributed equally to this work.
Supporting Materials and Methods, four figures, and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)31168-8.
Contributor Information
Rongjing Zhang, Email: rjzhang@ustc.edu.cn.
Junhua Yuan, Email: jhyuan@ustc.edu.cn.
Supporting Material
References
- 1.Berg H.C. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 2.Turner L., Ryu W.S., Berg H.C. Real-time imaging of fluorescent flagellar filaments. J. Bacteriol. 2000;182:2793–2801. doi: 10.1128/jb.182.10.2793-2801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Welch M., Oosawa K., Eisenbach M. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc. Natl. Acad. Sci. USA. 1993;90:8787–8791. doi: 10.1073/pnas.90.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuan J., Berg H.C. Ultrasensitivity of an adaptive bacterial motor. J. Mol. Biol. 2013;425:1760–1764. doi: 10.1016/j.jmb.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sourjik V., Berg H.C. Binding of the Escherichia coli response regulator CheY to its target measured in vivo by fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. USA. 2002;99:12669–12674. doi: 10.1073/pnas.192463199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cluzel P., Surette M., Leibler S. An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science. 2000;287:1652–1655. doi: 10.1126/science.287.5458.1652. [DOI] [PubMed] [Google Scholar]
- 7.Gabel C.V., Berg H.C. The speed of the flagellar rotary motor of Escherichia coli varies linearly with protonmotive force. Proc. Natl. Acad. Sci. USA. 2003;100:8748–8751. doi: 10.1073/pnas.1533395100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kojima S., Blair D.F. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry. 2004;43:26–34. doi: 10.1021/bi035405l. [DOI] [PubMed] [Google Scholar]
- 9.Reid S.W., Leake M.C., Berry R.M. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc. Natl. Acad. Sci. USA. 2006;103:8066–8071. doi: 10.1073/pnas.0509932103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukuoka H., Inoue Y., Ishijima A. Exchange of rotor components in functioning bacterial flagellar motor. Biochem. Biophys. Res. Commun. 2010;394:130–135. doi: 10.1016/j.bbrc.2010.02.129. [DOI] [PubMed] [Google Scholar]
- 11.Minamino T., Imada K. The bacterial flagellar motor and its structural diversity. Trends Microbiol. 2015;23:267–274. doi: 10.1016/j.tim.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 12.Boehm A., Kaiser M., Jenal U. Second messenger-mediated adjustment of bacterial swimming velocity. Cell. 2010;141:107–116. doi: 10.1016/j.cell.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 13.Paul K., Nieto V., Harshey R.M. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol. Cell. 2010;38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang X., Gomelsky M. A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol. Microbiol. 2010;76:1295–1305. doi: 10.1111/j.1365-2958.2010.07179.x. [DOI] [PubMed] [Google Scholar]
- 15.Kulasekara B.R., Kamischke C., Miller S.I. c-di-GMP heterogeneity is generated by the chemotaxis machinery to regulate flagellar motility. eLife. 2013;2:e01402. doi: 10.7554/eLife.01402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schirmer T., Jenal U. Structural and mechanistic determinants of c-di-GMP signalling. Nat. Rev. Microbiol. 2009;7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- 17.Ryjenkov D.A., Simm R., Gomelsky M. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 2006;281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- 18.Wolfe A.J., Visick K.L. Get the message out: cyclic-Di-GMP regulates multiple levels of flagellum-based motility. J. Bacteriol. 2008;190:463–475. doi: 10.1128/JB.01418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang F., Shi H., Yuan J. Non-equilibrium effect in the allosteric regulation of the bacterial flagellar switch. Nat. Phys. 2017;13:710–714. [Google Scholar]
- 20.Scharf B.E., Fahrner K.A., Berg H.C. Control of direction of flagellar rotation in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA. 1998;95:201–206. doi: 10.1073/pnas.95.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang F., Yuan J., Berg H.C. Switching dynamics of the bacterial flagellar motor near zero load. Proc. Natl. Acad. Sci. USA. 2014;111:15752–15755. doi: 10.1073/pnas.1418548111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan J., Berg H.C. Resurrection of the flagellar rotary motor near zero load. Proc. Natl. Acad. Sci. USA. 2008;105:1182–1185. doi: 10.1073/pnas.0711539105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang B., Zhang R., Yuan J. Limiting (zero-load) speed of the rotary motor of Escherichia coli is independent of the number of torque-generating units. Proc. Natl. Acad. Sci. USA. 2017;114:12478–12482. doi: 10.1073/pnas.1713655114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan J., Fahrner K.A., Berg H.C. Asymmetry in the clockwise and counterclockwise rotation of the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA. 2010;107:12846–12849. doi: 10.1073/pnas.1007333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan J., Branch R.W., Berg H.C. Adaptation at the output of the chemotaxis signalling pathway. Nature. 2012;484:233–236. doi: 10.1038/nature10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X., Berg H.C. Torque-speed relationship of the flagellar rotary motor of Escherichia coli. Biophys. J. 2000;78:1036–1041. doi: 10.1016/S0006-3495(00)76662-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xing J., Bai F., Oster G. Torque-speed relationship of the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA. 2006;103:1260–1265. doi: 10.1073/pnas.0507959103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meacci G., Tu Y. Dynamics of the bacterial flagellar motor with multiple stators. Proc. Natl. Acad. Sci. USA. 2009;106:3746–3751. doi: 10.1073/pnas.0809929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fung D.C., Berg H.C. Powering the flagellar motor of Escherichia coli with an external voltage source. Nature. 1995;375:809–812. doi: 10.1038/375809a0. [DOI] [PubMed] [Google Scholar]
- 30.Tipping M.J., Steel B.C., Armitage J.P. Quantification of flagellar motor stator dynamics through in vivo proton-motive force control. Mol. Microbiol. 2013;87:338–347. doi: 10.1111/mmi.12098. [DOI] [PubMed] [Google Scholar]
- 31.Berke A.P., Turner L., Lauga E. Hydrodynamic attraction of swimming microorganisms by surfaces. Phys. Rev. Lett. 2008;101:038102. doi: 10.1103/PhysRevLett.101.038102. [DOI] [PubMed] [Google Scholar]
- 32.Li G., Tang J.X. Accumulation of microswimmers near a surface mediated by collision and rotational Brownian motion. Phys. Rev. Lett. 2009;103:078101. doi: 10.1103/PhysRevLett.103.078101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hug I., Deshpande S., Jenal U. Second messenger-mediated tactile response by a bacterial rotary motor. Science. 2017;358:531–534. doi: 10.1126/science.aan5353. [DOI] [PubMed] [Google Scholar]
- 34.Ellison C.K., Kan J., Brun Y.V. Obstruction of pilus retraction stimulates bacterial surface sensing. Science. 2017;358:535–538. doi: 10.1126/science.aan5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.