Abstract
G Protein-Coupled Receptors (GPCRs) represent a large superfamily of membrane proteins that mediate cell signaling and regulate a variety of physiological processes in the human body. Structure-function studies of this superfamily have been enabled a decade ago by multiple breakthroughs in technology that included receptor stabilization, crystallization in a membrane environment, and microcrystallography. The recent emergence of X-ray free electron lasers (XFELs) has further accelerated structural studies of GPCRs and other challenging proteins by overcoming radiation damage and providing access to high-resolution structures and dynamics using micrometer-sized crystals. Here, we summarize key technology advancements and major milestones of GPCR research at XFELs, and provide a brief outlook on future developments in the field.
Keywords: G protein-coupled receptor, Lipidic cubic phase, Serial femtosecond crystallography, Structure-function, X-ray free electron laser
INTRODUCTION
During the last several decades structural biology has considerably advanced our understanding of biological processes at molecular level. Over 130,000 three-dimensional macromolecular structures deposited in the Protein Data Bank (8) provide invaluable templates for elucidating functional mechanisms and assisting in the rational design of new therapeutics. Recently, the field of structural biology has undergone a quantum leap propelled by the resolution revolution in cryo-electron microscopy (Cryo-EM) (4), and by the development of X-ray free electron lasers (XFELs) (90). XFELs generate extremely bright (9–10 orders of magnitude brighter than third-generation synchrotrons) and extremely short in duration (femtoseconds) pulses of coherent X-rays. With such unprecedented properties, XFELs enable high-resolution structure determination of radiation-sensitive (41) and difficult to crystallize macromolecules (66) as well as provide access to dynamics through the analysis of room temperature structures (98) and time-resolved pump-probe experiments (86).
The first hard energy XFEL, the Linac Coherent Light Source (LCLS) at the SLAC National Laboratory in Menlo Park, USA, was commissioned in 2009 (28), followed by the Spring-8 Angstrom Coherent Laser (SACLA) in Harima, Japan in 2011 (78). Since each XFEL pulse can totally destroy the crystal it interacts with, the data are usually collected using a serial femtosecond crystallography (SFX) approach (15), in which crystals are rapidly delivered in the beam in random orientations and diffraction patterns are recorded from tens to hundreds of thousands individual crystals. SFX required the development of new sample preparation protocols and crystal delivery hardware, as well as new data processing software, which have quickly progressed to the stage where the SFX method has started to yield new exciting results for many important biological systems (48).
This review is focused on the applications of SFX to G Protein-Coupled Receptors (GPCRs), one of the most challenging protein families for structural studies.
STRUCTURAL BIOLOGY OF G PROTEIN-COUPLED RECEPTORS
GPCR Structure and Function
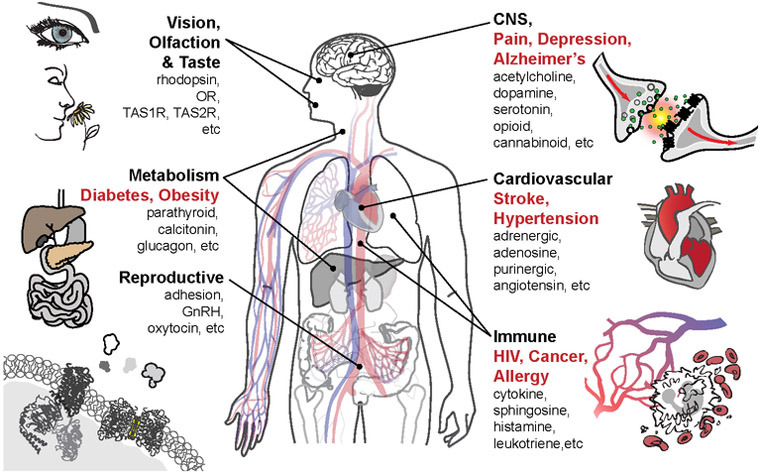
GPCRs constitute the largest membrane protein superfamily in the human genome, with over 800 unique members, typically grouped into 5 classes (A, B, C, Frizzled, and Adhesion) according to receptor topology and sequence homology (2, 44). GPCR-mediated signaling pathways play a key role in all physiological systems (Figure 1) as well as pathophysiological conditions including cancer, immune disorders, cardiovascular diseases, metabolic disorders (e.g. obesity, diabetes), pain and addiction, etc. (38, 99) and are therefore important drug targets; over 30% of all prescription drugs on the market act via these receptors (76, 81). GPCRs have a seven-transmembrane-helix (7TM) topology and contain multiple binding sites for orthosteric ligands and allosteric modulators. They recognize a diverse array of native signaling molecules, including ions, biogenic amines, nucleotides, neurotransmitters, lipids, hormones, peptides, and small proteins (38, 99). Upon ligand binding, a signal is transmitted across the cell membrane to intracellular partner proteins, such as G proteins, β-arrestins, and other effectors (3, 31, 79). Generally, GPCRs exist in the plasma membrane in a dynamic equilibrium between multiple ground and signaling states. Different native and synthetic ligands, depending on their chemical structures, can stabilize different states exhibiting various signaling efficacies (i.e. acting as agonists, biased agonists, antagonists, inverse agonists, or allosteric modulators) (31).
Figure 1. GPCR function and pervasiveness of in physiology and pathology.
Physiological systems (bold black) and pathological conditions (bold red) linked to GPCR families (black) are indicated.
Detailed understanding of the mechanism of GPCR action requires high-resolution structural information for many representative members of the family captured in different conformational states, as well as in complex with different signaling partners. This level of structural detail, in general, can only be achieved via crystallography, which requires obtaining sufficiently large and well-diffracting crystals.
GPCR Structure Determination Pipeline
Owing to the large scale conformational transitions and their dynamic nature, GPCRs are inherently highly flexible and unstable, especially when extracted from their native membrane environment. This property conflicts with their propensity to crystallize, which requires a conformationally stable and pure receptor population that can form well-defined crystal contacts. GPCRs can be stabilized in a number of ways, and in practice, often several of them have to be used in order to encourage crystallization.
Firstly, a high-affinity ligand can be used that keeps the receptor predominantly in a single, active (agonist), or inactive (neutral antagonist, or inverse agonist) conformation (115). The inactive state usually being the ground state of the receptor, crystallization is often more straightforward with antagonists. Indeed, more antagonist-bound than agonist-bound structures have been determined to date (107).
Secondly, the receptor can be engineered so as to be more stable than the wild-type protein. This can be achieved by truncation of N- and C-termini, which for some receptors can be long and (at least in absence of interaction partners) unstructured, point mutations (40), and substitution of loop regions by compact soluble protein domains (fusion partners) (23). Stabilizing point mutations can be identified by brute force, such as alanine scanning (68, 87), or using in vitro evolution approaches (26, 82), designed rationally (97), and sometimes transferred between different receptors.
Thirdly, a specific receptor conformation, in particular, an active or active-like, can be stabilized by receptor binding partners, including G proteins and arrestin (51, 80), or their mimetic, such as engineered mini-G proteins (14), antibodies or nanobodies (24, 32, 47).
Additional to the benefit of stabilizing receptor for crystallization, both ligands and receptor engineering also help to alleviate the second bottleneck of GPCR crystallization, low protein yield. Ligands can often increase protein yield drastically, be it when adding them to the expression medium, or during protein purification. Similarly, it is not uncommon to observe yield increases by several folds when introducing fusion partners, or identifying beneficial point mutations.
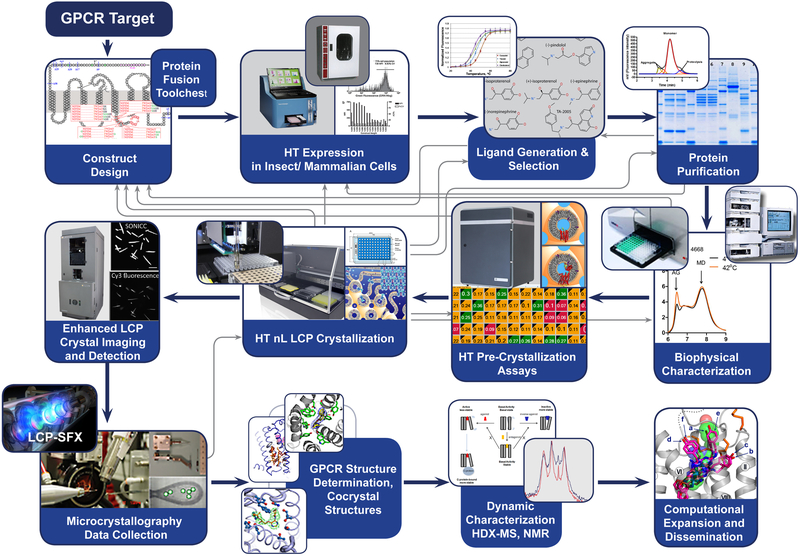
GPCR structure determination is, therefore, commonly achieved using a pipeline (93), which can be used to operate in parallel with several constructs and even several targets (Figure 2). Within this process, attempts are made to achieve several intermediate goals: the large-scale production of stable constructs and their characterization, the crystallization of these constructs, the acquisition of high-quality diffraction data, and finally the determination and refinement of the structure. These goals are commonly achieved through an iterative process using protocols and technologies described in a number of publications (12, 17, 23, 29, 95). The process requires design, production, and testing of a large number of protein-constructs (typically a few hundred) and screening dozens of ligands to identify the construct-ligand combination that can be crystallized. The pipeline relies on a number of metrics measured at specific process steps, which have proven to be an important guide in reducing processing work (and cost) on nonproductive constructs.
Figure 2. GPCR structure determination pipeline.
The pipeline contains several feedback loops and readout options (biochemical assays) which allow monitoring project progress, often bringing the process back to initial construct design.
For each new structure solved, extensive computational modeling is conducted to analyze conformational states and structural features in the context of other receptors, as well as the structural role of SNPs and other disease-related mutations. Molecular dynamics simulations and molecular docking studies are often used to probe the dynamic nature of the receptors and their interaction with ligands. Each structure determination is accompanied by comprehensive functional and mutagenesis studies to decipher the impact of different residues and important structural features on ligand recognition and signal transduction.
Lipidic Cubic Phase Crystallization
One of the important technology advancements that enabled structural studies of GPCRs was the development of crystallization in a membrane mimetic environment, known as the lipidic cubic phase (LCP) (12, 16). LCP represents a liquid-crystalline mesophase that spontaneously forms upon mixing a specific lipid and aqueous phase at a certain ratio. LCP consists of a single lipid bilayer forming a triply periodic structure with zero mean curvature and cubic symmetry (12). Topologically, the single lipid bilayer divides the space into two interpenetrating networks of continuous water channels. Therefore, LCP is often referred to as a bicontinuous lipidic cubic phase, meaning the existence of long-range space continuity in both hydrophilic and hydrophobic milieus. Such bicontinuity is responsible for many unique properties and applications of LCP, including its ability to support nucleation and growth of membrane protein crystals. Since its first introduction in 1996 (60), LCP crystallization has contributed high-resolution structures of over 120 unique membrane proteins from most major families.
Among a large variety of lipids, only two lipid classes, monoacylglycerols (MAGs) (13) and isoprenoid-chain lipids (36, 46, 108), consistently form LCP at or below room temperature, rendering them suitable as host lipids for membrane protein crystallization. The most successful of them are monounsaturated monoacylglycerols with a commonly used N.T MAG notation, in which N represents the number of carbon atoms between the ester group and the double bond and T – the number of carbons between the double bond and the terminal methyl group. Depending on their chemical structure, temperature, hydration and other parameters, N.T MAGs can form a large variety of mesophases, therefore a detailed knowledge of their respective phase diagram is critical for successful application of these lipids for LCP crystallization. While N.T MAGs are not native lipids of biological membranes, they can be doped with native lipids, which, in certain cases, may be essential for crystallization (18). For example, 10% w/w cholesterol/ 90% w/w monoolein (9.9 MAG) mixture was established as a default host lipid for GPCR crystallization.
Macroscopically, LCP is a transparent, optically isotropic and highly viscous gel-like material. Such a gel-like consistency makes handling these materials very challenging, which prompted the development of special tools, protocols, and instruments to miniaturize and automate crystallization tasks and interrogate behavior of proteins and their interaction with lipids in LCP (16).
The success of the LCP crystallization approach can be attributed mainly to two factors. First, the lipid bilayer of LCP provides a more native-like stabilizing environment for integral membrane proteins compared to detergent micelles (16, 63). Second, in contrast to protein-detergent complexes, in which the transmembrane hydrophobic part of the protein is shielded from forming specific protein-protein interactions, membrane proteins embedded in LCP have the ability to interact with each other through their hydrophobic domains. The result is the formation of type I crystal lattice with extensive hydrophilic as well as hydrophobic (often lipid-mediated) contacts between protein molecules, contributing to better ordering of molecules in the crystal lattice and consequently higher resolution diffraction (16).
The intrinsic microstructure of LCP, including the curvature of its lipid bilayer, and the dimensions of its water channels (~50 Å in diameter in their narrowest parts), imposes steric limits on the size of membrane proteins or their oligomeric aggregates which can diffuse within the lipid bilayer of LCP. To overcome this limitation, crystallization of large membrane proteins (>100 kDa) may require swelling LCP by using special additives (19, 21), or even complete transformation into a liquid-like sponge phase. Due to a higher propensity to nucleation and relatively slow diffusion, LCP crystallization typically leads to a large number of small micrometer-sized crystals, optimization of which may be challenging.
Current Status of GPCR structure determination
Multiple technological breakthroughs related to GPCR stabilization, expression, purification, crystallization and crystallographic data collection (32, 107) have enabled, since 2007, high-resolution structure determination of ~45 unique receptors (44), contributing over 200 entries in the Protein Data Bank (8, 44). Most of these receptors were captured in their ground inactive state, while structures of 13 unique receptors are available in an active-like state or in a fully engaged active state (44) in complex with a heterotrimeric Gs protein (80) and arrestin (51).
These structure-function studies have helped to understand ligand selectivity, to establish common and diverse structural elements, to identify activation microswitches and major structural re-arrangements during receptor activation (52, 99, 111). They led to understanding that GPCRs function as intrinsic allosteric machines, which are not only controlled by their native signaling molecules, but also modulated by lipids, such as cholesterol (22, 34), sodium ions (53, 62), and water molecules (1). They helped to identify a variety of allosteric sites in GPCRs for modulation by drug-like molecules (42), including those on the receptor-lipid interface (110) and at the intracellular surface (117). Lastly, they provided plausible explanations for unusual signaling behavior in some receptors (112).
Notwithstanding such formidable progress, we are still far away from complete understanding the whole GPCR superfamily with known structures covering only 5% of the superfamily as a whole, and, considering extension by homology, only ~30% of non-olfactory receptors. Lack of structural coverage for GPCRs is one of the key bottlenecks for rapid expansion of available tool compounds and drug discovery efforts on new GPCR targets. It is becoming obvious that new technologies are needed to accelerate new discoveries, and quite timely the emergence of XFELs and the advancements in Cryo-EM are starting to fulfill this need.
SERIAL FEMTOSECOND CRYSTALLOGRAPHY
SFX vs Traditional Crystallography
Despite all advances in GPCR sample preparation and crystallization, obtaining large, well-diffracting crystals for synchrotron data collection remains a tedious, expensive, and often frustrating process. GPCR crystallization in LCP typically leads to the formation of initially very small micrometer-sized crystals, optimization of which is time-consuming, may introduce growth defects, and sometimes may just fail. The amount of crystallographic information that can be obtained from such well-ordered but small microcrystals using state-of-the-art microfocus beamlines at third-generation synchrotron sources is strongly limited by radiation damage. Current practices include collecting small wedges of data of a few degrees from many frozen crystals and merging them together in a dataset (20). This procedure requires extensive optimization of crystal growth, harvesting and cryo-cooling hundreds of crystals, aligning them by beam rastering, using special algorithms for data processing, all of which involve substantial commitments in time and effort, while at the end still having to tolerate a certain amount of radiation damage.
All these time-consuming steps and, most importantly, effects of radiation damage can be essentially avoided by taking advantage of a new generation XFEL sources. XFELs produce extremely high brilliance X-ray pulses of few femtoseconds duration, allowing one to outrun radiation damage and collect high-resolution data on microcrystals at room temperature employing “diffraction before destruction” principle (10, 75). SFX makes this “one crystal, one shot” approach practicable by constantly replenishing crystals. Rather than traditional oscillation crystallography, where diffraction is collected from multiple exposures of a rotating single (several) crystal(s), SFX using XFELs collects single exposures of tens to hundreds of thousands, randomly oriented micrometer and submicrometer-sized crystals. These small crystals are often found to be better ordered and having fewer growth defects, resulting in similar or even better diffraction at XFELs compared to larger frozen crystals at synchrotron sources, making the SFX method especially enticing for challenging systems such as membrane proteins, and GPCRs in particular.
The “one crystal, one shot” approach also facilitates time-resolved crystallography of irreversible processes and therefore access to dynamics, without limiting achievable time resolution more than the femtosecond time scale of the X-ray pulses. This opens up the possibility to study conformational transitions at time scales from sub-picoseconds to seconds and beyond, and has already found many attractive applications (5, 54, 55, 58, 59, 73, 77, 85, 89, 91, 94).
Practical aspects of GPCR sample preparation for SFX along with necessary developments in instrumentation and data processing will be outlined in the following sections.
LCP-SFX: Sample Preparation and Data Collection
The development of viscous media injectors (103, 104) has allowed combining the advantages of LCP crystallization and SFX data collection by facilitating sample delivery directly in the native crystal growth matrix, thereby circumventing the need to harvest individual crystals. The high viscosity of the crystal delivery matrix allows for a wide range of flow rates, better matching to the XFEL pulse repetition rates and leading to a significantly reduced crystal consumption compared to a popular liquid media Gas Dynamic Virtual Nozzle (GDVN) injector (25). The viscous media injector (104) consists of a reservoir (typically, 20, 40 or 100 μL) for sample loading, a capillary nozzle (20 – 100 μm inner diameter) through which the sample is extruded, and a hydraulic plunger that applies high pressure necessary for extrusion of LCP through the narrow capillary. The plunger is connected through a water line with an HPLC pump, which can be remotely controlled to monitor the pressure and adjust the flow rate. The LCP stream exiting from the injector nozzle is supported by a co-axial sheath flow of gas (typically helium or nitrogen) to prevent it from curling back and sticking to the nozzle. The gas pressure can also be adjusted remotely to ensure a stable flow. The injector can stream samples both inside of a vacuum chamber and in a helium atmosphere. In case of a vacuum chamber, the LCP matrix should be prepared from a short chain MAG, such as 9.7 MAG or 7.9 MAG (70), or doped with one of these lipids just before loading in the injector, in order to prevent its transition into a lamellar crystalline phase upon evaporative cooling (104).
The use of injectors for SFX has imposed new requirements for sample preparation (45, 64). Rather than optimizing crystals to grow sparse and large, such as the ones desired for synchrotron data collection, approximately 30 – 100 μL of LCP densely packed with small and uniform crystals is needed for LCP-SFX. An optimal protocol to achieve these requirements includes initial screening and optimization by high-throughput nanovolume crystallization in 96-well glass sandwich plates, followed up with scaling up the volume by ~1,000 times in gas-tight syringes with the objective of high crystal density and uniform, small crystal size. Achieving a high crystal density is extremely important because crystals grown in LCP can only be further diluted but not concentrated due to the high viscosity of the matrix. Low crystal density leads to low efficiency of data collection. As with any technique, sample quality is paramount for success. Since each sample change and evaluation takes ~30–60 min of valuable XFEL beam time, all samples should be carefully pre-screened to characterize their average crystal size and density before loading them in the injector.
While for synchrotron data collection, several dozens of individual large crystals can be harvested from a few, or even a single nanoliter volume LCP drop in glass sandwich plates, for a complete LCP-SFX dataset often thousands to tens of thousands of microcrystals are required in a volume of a few tens of microliters. These very different sample requirements necessitate divergent optimization objectives after obtaining initial crystal hits.
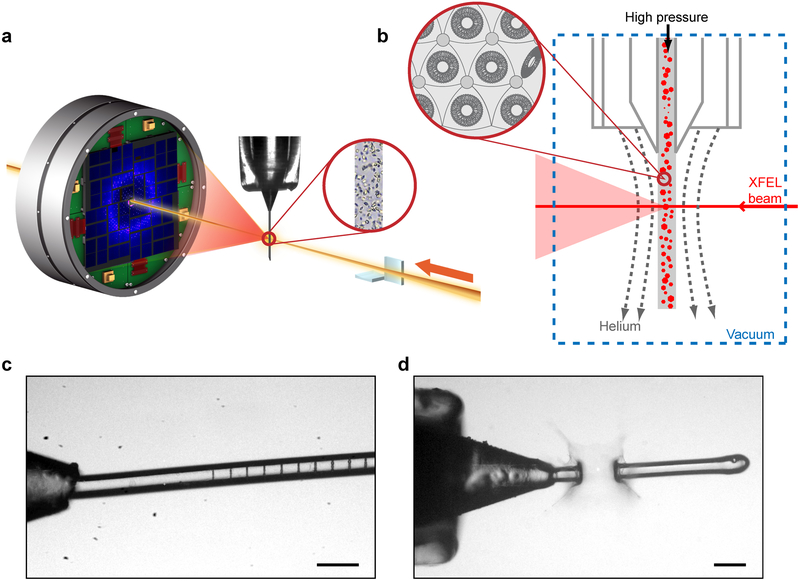
An LCP-SFX data collection experiment requires careful consideration and optimization of several parameters with the ultimate goal of minimizing data collection time while acquiring the most accurate data. Thus, real-time data monitoring and evaluation are essential for ensuring the most efficient use of the scarce XFEL beamtime (69). Microcrystals, randomly dispersed in LCP, are injected into the sample chamber, intersecting with the pulsed XFEL beam with each exposure being recorded by a detector (Figure 3). These detector images are analyzed in real time to identify those that contain at least a certain amount of Bragg diffraction peaks with a specified signal-to-noise threshold. Such patterns are referred to as crystal hits. Crystal hit rates depend on the crystal density and size, the injector nozzle diameter, the XFEL beam size and pulse intensity, but not on the LCP flow rate, because on the time scale of each pulse the crystals appear stationary. The choice of the injector nozzle capillary is dictated by the following considerations. Smaller capillary diameters can minimize the unwanted background scattering, however, they lead to lower crystal hit rates, require higher pressure for LCP extrusion and are prone to clogging. In practice, we established that 50 μm inner diameter capillaries provide a reasonable compromise between the reliability and ease of the injector operation and the obtained data quality. Larger or smaller diameter capillary can be used in some cases depending on the crystal size and density. The optimal crystal size for LCP-SFX is about 2 – 20 μm. Crystals larger than 20 μm can typically be used at synchrotrons. Crystals smaller than 2 μm do not produce sufficiently strong diffraction signal at high-resolution, considering relatively large background scattering from the LCP stream of ~50 μm in diameter. The XFEL beam size ideally should match to the average crystal size, however, it is typically fixed for a given sample chamber by the X-ray optics used. A highly intense XFEL beam passing through an LCP stream leaves a trail of gas bubbles (Figure 3c) (92). The extent of the affected area depends on the X-ray flux density. The LCP flow rate should be adjusted, therefore, to be sufficiently fast to clear out the damaged material and expose intact fresh crystals to the next incoming pulse. Increasing XFEL beam intensity eventually leads to a complete disruption of the LCP flow, recovery from which is not practically possible, thus limiting the maximum intensity for LCP-SFX (Figure 3d). In our experience, this occurs at approximately 10% of the total LCLS intensity operating in the nominal regime and producing 9.5 keV X-rays with the total pulse energy of 4 mJ, when experiments are conducted in the 1 μm sample chamber at the Coherent X-ray Imaging (CXI) beamline (9).
Figure 3. SFX-LCP process.
(a) Schematic of an LCP-SFX data collection setup. XFEL beam is focused to a diameter of ~1 μm by a pair of KB mirrors on a stream of LCP delivering micrometer-sized crystals intersecting the beam in random orientations. Diffraction patterns are collected by a CSPAD detector at 120 Hz. (b) Zoom in on the sample interaction region and LCP microstructure. (c) XFEL beam footprints at ~1% intensity (8·109 photons/pulse). (d) XFEL beam at ~50% intensity (4·1011 photons/pulse) creates an explosion of ~100 μm in size.
Typical LCP-SFX data collection parameters used for GPCR structure determination are listed in Table 1. All experiments were conducted on the CXI beamline at LCLS (9). LCP injectors with 50 μm capillary nozzles were used in all experiments with the LCP flow rates ranging between 0.17 and 0.22 μL/min, corresponding to ~15 μm displacement of the matrix between two consecutive XFEL pulses arriving at 120 Hz (104). The total data collection time and the corresponding total number of images depended primarily on the crystal hit rate, but often were limited by the amount of sample and/or beam time available. While the crystal hit rates and indexing rates are the two parameters typically reported in publications, the definition of a crystal hit is often different between different experiments, thus, the most consistent parameter defining the efficiency of data collection is the percent of indexed images. This parameter was as low as 0.38% for the most challenging rhodopsin-arrestin sample (51) and as high as 7.7% in case of the “easiest” GPCR sample, the adenosine A2A receptor, which is often used as a test sample in various new developments. The total sample volume used for each dataset varied between 30 and 140 μL while the total amount of consumed protein was estimated to be in the range between 100 and 800 μg. Based on our experience, a minimal LCP-SFX dataset should contain at least 10,000 images to sample all possible crystal orientations and to average out fluctuations due to a large pulse-to-pulse variability in the XFEL beam parameters as well as uncertainties in crystal size, quality, and orientation. Therefore, we normalized the total protein consumption in each dataset, which resulted on average in 100 μg protein used (from 20 to 425 μg) per 10,000 indexed images. The accuracy of the structure factor amplitudes, in general, increases with the number of merged diffraction images, however, the improvements above 30,000 – 50,000 images are typically marginal, and in most cases insignificant, especially for structures solved by the molecular replacement method. On the other hand, it has been shown that achieving sufficient accuracy for experimental phasing by sulfur Single-Wavelength Anomalous Diffraction (S-SAD) required about 10 times more data (>500,000 indexed images) with correspondingly increased data collection time (~17 hours) and the amount of consumed protein (~2.7 mg) (7).
Table 1.
LCP-SFX data collection parameters and statistics
| Receptor/ ligand |
5-HT2B/ Erg |
ΔCRD-SMO/ cyclopamine |
δ-OR/ DIPP-NH2 |
AT1R/ ZD7155 |
AT2R/ Cpd 1 |
Rho-Arr | GCGR/ NNC0640- Fab |
SMO/ TC114 |
A2AAR/ ZM24138 5 |
5-HT2B/ Erg-Fab |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| PDB code | 4NC3 | 4O9R | 4RWD | 4YAY | 5UNF | 5UNG | 4ZWJ | 5XEZ | 5V56 | 5K2C* | 5TUD |
| Resolution (Å) | 2.8 | 3.4 / 3.2 / 4.0 | 2.7 | 2.9 | 2.8 | 2.8 | 3.8 / 3.8 / 3.3 | 3.0 | 2.9 | 1.9 | 3.0 |
| Spacegroup | C2221 | P21 | C2 | C2 | P21 | P21221 | P212121 | P21 | P21 | C2221 | P21 |
| Crystal size (μm) | 5×5×5 | 10×2×2 | 5×2×2 | 10×2×2 | 5×2×2 | 10×2×2 | ~5 | 5×5×2 | 5×5×2 | 1-10 | |
| Data acquisition time (min) | 590 | 490 | 275 | 385 | 375 | 700 | 140 | 300 | 135 | 260 | |
| Flow rate (μL/min) | 0.17 | 0.17 | 0.17 | 0.17 | 0.22 | 0.2 | 0.2 | 0.2 | 0.22 | 0.2 | |
| Sample volume used (μl) | 100 | 85 | 50 | 65 | 85 | 140 | 30 | 60 | 30 | 55 | |
| No. of collected images | 4,217,508 | 3,510,525 | 1,967,539 | 2,764,739 | 2,701,530 | ~5,000,000 | ~1,000,000 | 2,102,907 | 948,961 | 1,877,040 | |
| No. of indexed images | 32,819 | 66,165 | 36,083 | 73,130 | 22,774 | 15,804 | 18,864 | 57,573 | 65,560 | 72,735 | 52,291 |
| % indexed images | 0.78 | 1.9 | 1.8 | 2.6 | 0.84 | 0.59 | 0.38 | 5.8 | 3.1 | 7.7 | 2.8 |
| Total amount of protein used (μg) | 300 | 500 | 300 | 292 | 190 | 800 | 115 | 400 | 270 | 420 | |
| Protein consumption per 10,000 indexed images (μg) | 90 | 75 | 85 | 40 | 50 | 425 | 20 | 60 | 37 | 80 | |
| Reference | (65) | (104) | (30) | (114) | (112) | (51) | (113) | (116) | (7) | (47) | |
Detectors for XFEL have a very different profile of requirements than those for synchrotron data collection. While crystals at synchrotrons are exposed to radiation for milliseconds to seconds, XFEL pulses have a much shorter, femtosecond duration, and arrive at a high pulse repetition rate, such as 120 Hz at LCLS operates at a repetition rate of 120 Hz, and up to 4.5 MHz at the newly commissioned European XFEL. All GPCR data to date have been acquired using a CSPAD (Cornell-SLAC Pixel Array Detector) (35) which has a high sensitivity and fast readout, however, suffers from a low dynamic range and prone to damage by high-intensity diffraction, such as from accidental salt crystals or ice. These drawbacks along with even faster readout rates are being addressed in the development of next-generation detectors for XFELs (39, 71).
SFX Data Processing
Along with progress in instrumentation and sample delivery, the advancement of new approaches and protocols for data processing has been critical for the success of SFX. Several specialized software packages have been developed for monitoring data collection in real time, for applying detector corrections and identifying images with Bragg spots, as well as for indexing, integration, scaling and merging individual reflections (6, 27, 33, 37, 49, 50, 56, 83, 84, 96, 106, 109). The common challenges of SFX data processing include location of sharp spots, which often consist of a single or few pixels, background subtraction, which in case of crystal delivery in LCP can be substantial, indexing of single patterns and related to that indexing ambiguity, scaling and merging of individual partial reflections that is typically done using a Monte Carlo approach (56). The algorithms for SFX data processing have constantly been improved with recent notable additions including refinement of the detector geometry (109), estimation of partiality of the reflections (33) and post-refinement (83, 105), as well as methods for resolving indexing ambiguity (11). All these advancements make possible to reduce the amount of data collected while increasing the quality of obtained structures.
APPLICATIONS OF LCP-SFX TO STRUCTURAL STUDIES OF GPCRS
From Initial Validation to Ligand Co-Crystal Structures
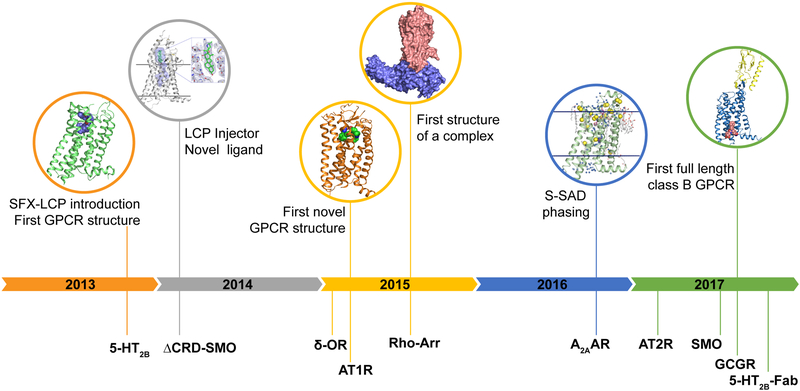
Early applications of LCP-SFX to GPCR structural biology have quickly surpassed the proof-of-principle stage. LCP-SFX was first introduced in 2013 with the determination of the room temperature structure of the serotonin 5-HT2B receptor bound to the migraine drug ergotamine (Figure 4) (65). The new approach was validated by comparing this structure to the corresponding synchrotron cryo structure (100). Despite the substantial differences in crystal size (5×5×5 μm3 vs. 80×20×10 μm3) and data collection temperature (294 K vs. 100 K) both structures showed comparable resolution (2.8 Å vs. 2.7 Å) and final quality of the model. Overall, while the backbones of the structures overlapped closely, the room temperature XFEL structure displayed a unique distribution of thermal motions and conformations of some residues, likely more accurately representing the receptor structure and dynamics in its native environment. Subsequently, the co-crystal structure of the transmembrane part lacking the extracellular Cystein-Rich Domain (CRD) of the smoothened receptor (ΔCRD-Smo), an important anti-tumor target, bound to the teratogen cyclopamine was solved using LCP-SFX (104). While several structures of ΔCRD-Smo bound to different ligands had been solved at a synchrotron source earlier (101, 102), the cyclopamine complex had eluded structure determination due to high crystal mosaicity and inconsistent diffraction. At the same time, smaller, micrometer-sized crystals displayed much lower mosaicity and better diffraction at LCLS, resulting in a 3.2 Å (anisotropic) resolution structure, which revealed the location of cyclopamine inside a long and narrow cavity in the transmembrane part of the receptor. Similarly, for the delta opioid receptor (δ-OR) bound to a bi-functional peptide acting as a potential non-addictive painkiller, LCP-SFX data collection substantially improved the resolution from 3.4 Å achieved at a synchrotron source to 2.7 Å, enabling unambiguous ligand placement in the pocket and uncovering the molecular details of its recognition (30).
Figure 4. Timeline of GPCR structure determination at XFELs.
Important milestones (top) and determined GPCR structures (bottom) are shown on a timeline.
First Novel GPCR Structures Solved by LCP-SFX
The next important milestone of LCP-SFX was the determination of receptor structures that had previously been completely unknown (Figure 4). The structure of the angiotensin II receptor type 1 (AT1R) (114), a major blood pressure regulator and a target for many antihypertensive drugs, was the first novel GPCR structure solved by XFEL. This achievement was followed by the structure determination of the AT2 receptor (112), an angiotensin receptor subtype with unique signaling properties. The structures provided important insights into the distinct functions of the two angiotensin receptors and outlined the structural basis for ligand binding and selectivity.
De Novo Phasing of GPCR LCP-SFX Data
The crystallographic phase problem for new GPCR structures can usually be solved by molecular replacement (MR), thanks to the common 7TM topology of their transmembrane domain. However, with XFELs opening up a novel space of structure determination for difficult-to-crystallize proteins, and LCP-SFX promising to achieve the same for membrane proteins, de novo phasing techniques had to be developed for these most challenging systems that use XFEL data and go beyond the most simple proof-of-principle cases (72, 74).
While some common de novo phasing techniques can utilize heavy atoms incorporated into the crystal either by soaking or mutagenesis, practical implementation of these techniques requires extensive screening and is not always successful. It has been demonstrated that the extremely weak anomalous signal from endogenous sulfur atoms present in most proteins can be sufficient for S-SAD phasing, provided data can be collected with high accuracy (61). Achieving such high accuracy for SFX data collected at XFELs has been challenging.
Recently, it has been demonstrated that SFX-LCP data can be phased using S-SAD to automatically solve a GPCR structure (Figure 4) leading to the bias-free structure of the human adenosine A2A receptor (A2AAR) (7). This success has underlined the impressive progress in data collection and data processing of LCP-SFX. Crucially, XFEL beam energy can be tuned so as to achieve optimal anomalous scattering strength for a given element, while simultaneously maximizing resolution. Anomalous data have been collected at the X-ray energy of 6 keV, as a reasonable compromise between the strength of the signal, effects of X-ray absorption, dependence of resolution on the X-ray wavelength, and the efficiency of the beamline optics and the detector. Notably, the crystallized A2AAR construct only contained 24 sulfur atoms (15 Cys and 9 Met) per its 447 residues, 12 of which were sufficient for phasing; 88% of human proteins contain more than this ratio of sulfur atoms per residue, suggesting that S-SAD should be a generally applicable method for solving the crystallographic phase problem using XFELs (7).
GPCR Complexes
One of the current frontiers of GPCR structural biology is the elucidation of the structure of membrane protein complexes. As previously described, the receptor-ligand interactions allosterically modulate the interactions of the receptor with a multitude of intracellular protein binding partners and effectors that initiate or inhibit different signaling pathways, thereby allowing the cell to respond to environmental cues. Since stable receptor-effector complexes require a particular and well-defined receptor conformation, this adds an extra level of complexity and makes crystallization even more challenging than for the receptor-ligand complex alone, and systems have to be chosen and optimized even more carefully.
Until recently, the only receptor-effector complex for which a structure was known was that of the β2 adrenergic receptor (β2AR) in complex with Gs protein (80). Canonical signaling through G proteins, however, represents only a fraction of physiologically relevant signaling, and so considerable efforts have been made to obtain the complex structure of a GPCR bound to an arrestin. Arrestins bind to activated and phosphorylated receptors, terminating G protein signaling and leading to receptor desensitization and internalization, all the while initiating G protein-independent signaling pathways that lead to disparate cellular responses (67). This dual role in signaling has led to a considerable interest in molecular determinants of this interaction, as specifically modulating those with so-called signaling biased ligands bears the promise to alleviate many problems associated with unwanted side effects such as those observed for opiate painkillers.
Rhodopsin-arresting co-crystals that were obtained could not be optimized beyond 20 micrometers in size and diffracted to only 7–8 Å at synchrotron; however, at LCLS similar crystals diffracted to approximately 3.3 Å (anisotropic) resolution revealing specific interactions between rhodopsin and arrestin (51). Later the resolution was further improved to 3.0 Å (anisotropic) allowing to identify specific phosphorylation codes for arrestin recruitment by GPCRs (118).
Another type of complexes that has garnered considerable interest in recent years is that of receptors bound to monoclonal antibodies. Some of these antibodies can bind extracellulary and stabilize the receptor in distinct activation states, thereby offering an attractive alternative to traditional therapeutic accession points that becomes increasingly important given the traditionally high attrition rate of small molecule drug development (43). Using LCP-SFX, the structure of the complex between the human 5-hydroxytryptamine 2B (5-HT2B) (serotonin) receptor, the target for many anxiety and mood-regulating drugs, and an antibody Fab fragment could be solved (47). This interaction involves a receptor epitope formed by all three extracellular loops, and for the first time illustrates this important modulation by biologics (47).
Multidomain GPCRs
Initial structural studies of GPCRs have been limited to mostly transmembrane class A receptors and to 7TM domains of multidomain class B, C and Frizzled receptors. Most recently with advancements in receptor stabilization technologies, the attention of the community has turned to full-length non-class A GPCRs in attempts to understand how their extracellular domains (ECDs) modulate receptor structure and function. The first full-length structures to be determined were those for the class Frizzled smoothened receptor (116) and the class B glucagon receptor GCGR (113), aided and enabled by LCP-SFX.
The relevance of smoothened in the cancer context has been described above, and while the previously obtained structures of ligands bound to the 7TM part have provided a molecular foundation for ligand modulation (101, 102), this picture had not been complete without an understanding of how the ECD influences ligand recognition and receptor activation through allosteric effects.
Similarly, for GCGR, which is a key player in glucose homeostasis and the pathophysiology of type 2 diabetes, separate structures for ECD (57) and 7TM were known (88), but the full-length structure had eluded structure determination due to difficulties with crystallization. Using LCP-SFX, the full-length structure could be determined to 3.0 Å resolution and made visible the alternate conformation of the N-terminal “stalk” region, linking ECD and 7TM by forming a short beta strand with ECL1 (113).
In both cases, LCP-SFX brought with it a considerable improvement in resolution as compared to synchrotron data collection.
CONCLUSIONS AND FUTURE OUTLOOK
Since its first introduction in 2013, LCP-SFX has demonstrated a tremendous success with ten GPCR structures published within the last 4 years, significantly advancing our understanding of this biomedically important protein superfamily. The most substantial current limitation that prevents even wider spread and impact of this approach is the shortage of XFEL beamtime. Three new XFELs (European XFEL in Hamburg, Germany; PAL-XFEL in Pohang, South Korea and SwissFEL in Villigen, Switzerland) have been recently commissioned and are scheduled to start their user program operations in 2017–2018, to be followed by the LCLS-II upgrade in 2020.
A number of new developments related to structural biology studies of GPCRs at XFELs are anticipated within next few years. They will likely include the establishment of a Structure-Based Drug Development (SBDD) platform which will take advantage of small crystal size and streamlined procedures of co-crystal preparation and SFX data acquisition with many different receptors in complex with a large variety of ligands. Complementing injector-based crystal delivery methods with fixed targets approach may help to decrease relatively strong background from the surrounding crystals LCP matrix, enable the use of the full XFEL power, obtain high-resolution data from even smaller sub-micrometer-sized crystals, and further lower sample consumption. New XFELs (European XFEL, LCLS-II) will enable faster data acquisition with higher pulse repetition rates, along with matching new fast-readout detectors and sample delivery systems. Last but not least, many open questions about the dynamic nature of the GPCR signaling will be addressed by collecting time-resolved molecular movies of conformational changes during GPCR activation triggered by photoswitchable or photocaged ligands. All these advancements should substantially accelerate the pace of structural biology studies of the whole GPCR superfamily to holistically understand the structural diversity of these receptors and their signaling mechanisms.
ACKNOWLEDGEMENTS
Research work in the author’s laboratory covered in this review has been supported in parts by the National Institutes of Health (NIH) grant R01 GM108635 and the National Science Foundation (NSF) grant 1231306. Authors thank S. Boutet and C. Stan for their help with acquiring photographs shown in Fig. 3c,d, and K. Kadyshevskaya for making illustrations.
References
- 1.Angel TE, Chance MR, Palczewski K. 2009. Conserved waters mediate structural and functional activation of family A (rhodopsin-like) G protein-coupled receptors. Proc. Natl. Acad. Sci. U. S. A. 106:8555–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Attwood TK, Findlay JB. 1994. Fingerprinting G-protein-coupled receptors. Protein Eng 7:195–203 [DOI] [PubMed] [Google Scholar]
- 3.Audet M, Bouvier M. 2012. Restructuring G-protein- coupled receptor activation. Cell 151:14–23 [DOI] [PubMed] [Google Scholar]
- 4.Bai X-c, McMullan G, Scheres SHW. 2015. How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci 40:49–57 [DOI] [PubMed] [Google Scholar]
- 5.Barends TR, Foucar L, Ardevol A, Nass K, Aquila A, et al. 2015. Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation. Science 350:445–50 [DOI] [PubMed] [Google Scholar]
- 6.Battye TGG, Kontogiannis L, Johnson O, Powell HR, Leslie AGW. 2011. Acta Crystallogr. D Biol. Crystallogr 67:271–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Batyuk A, Galli L, Ishchenko A, Han GW, Gati C, et al. 2016. Native phasing of x-ray free-electron laser data for a G protein-coupled receptor. Sci. Adv 2:e1600292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, et al. 2000. The Protein Data Bank. Nucleic Acids Res 28:235–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boutet S J Williams G. 2010. The Coherent X-ray Imaging (CXI) instrument at the Linac Coherent Light Source (LCLS). New J. Phys 12:035024 [Google Scholar]
- 10.Boutet S, Lomb L, Williams GJ, Barends TR, Aquila A, et al. 2012. High-Resolution Protein Structure Determination by Serial Femtosecond Crystallography. Science 337:362–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brehm W, Diederichs K. 2014. Breaking the indexing ambiguity in serial crystallography. Acta Crystallogr. D Biol. Crystallogr 70:101–9 [DOI] [PubMed] [Google Scholar]
- 12.Caffrey M, Cherezov V. 2009. Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc 4:706–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caffrey M, Lyons J, Smyth T, Hart DJ. 2009. Chapter 4 Monoacylglycerols: The Workhorse Lipids for Crystallizing Membrane Proteins in Mesophases In Current Topics in Membranes, 63:83–108. Academic Press [Google Scholar]
- 14.Carpenter B, Nehme R, Warne T, Leslie AG, Tate CG. 2016. Structure of the adenosine A(2A) receptor bound to an engineered G protein. Nature 536:104–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chapman HN, Fromme P, Barty A, White TA, Kirian RA, et al. 2011. Femtosecond X-ray protein nanocrystallography. Nature 470:73–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cherezov V 2011. Lipidic cubic phase technologies for membrane protein structural studies. Curr. Opin. Struct. Biol 21:559–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cherezov V, Abola E, Stevens RC. 2010. Recent progress in the structure determination of GPCRs, a membrane protein family with high potential as pharmaceutical targets. Methods Mol. Biol 654:141–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cherezov V, Clogston J, Misquitta Y, Abdel-Gawad W, Caffrey M. 2002. Membrane protein crystallization in meso: Lipid type-tailoring of the cubic phase. Biophys. J. 83:3393–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cherezov V, Clogston J, Papiz MZ, Caffrey M. 2006. Room to move: crystallizing membrane proteins in swollen lipidic mesophases. J. Mol. Biol 357:1605–18 [DOI] [PubMed] [Google Scholar]
- 20.Cherezov V, Hanson MA, Griffith MT, Hilgart MC, Sanishvili R, et al. 2009. Rastering strategy for screening and centring of microcrystal samples of human membrane proteins with a sub-10 µm size X-ray synchrotron beam. J. R. Soc. Interface 6:S587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cherezov V, Liu J, Griffith M, Hanson MA, Stevens RC. 2008. LCP-FRAP assay for pre-screening membrane proteins for in meso crystallization. Cryst. Growth Des 8:4307–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, et al. 2007. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science 318:1258–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chun E, Thompson AA, Liu W, Roth CB, Griffith MT, et al. 2012. Fusion partner toolchest for the stabilization and crystallization of g protein-coupled receptors. Structure 20:967–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Day PW, Rasmussen SG, Parnot C, Fung JJ, Masood A, et al. 2007. A monoclonal antibody for G protein-coupled receptor crystallography. Nat. Methods 4:927–9 [DOI] [PubMed] [Google Scholar]
- 25.DePonte DP, Weierstall U, Schmidt K, Warner J, Starodub D, et al. 2008. Gas dynamic virtual nozzle for generation of microscopic droplet streams. J. Phys. D: Appl. Phys 41:195505 [Google Scholar]
- 26.Dodevski I, Pluckthun A. 2011. Evolution of three human GPCRs for higher expression and stability. J. Mol. Biol 408:599–615 [DOI] [PubMed] [Google Scholar]
- 27.Duisenberg A 1992. Indexing in single-crystal diffractometry with an obstinate list of reflections. J. Appl. Crystallogr 25:92–6 [Google Scholar]
- 28.Emma P, Akre R, Arthur J, Bionta R, Bostedt C, et al. 2010. First lasing and operation of an angstrom-wavelength free-electron laser. Nat. Photon. 4:641–7 [Google Scholar]
- 29.Fenalti G, Abola EE, Wang C, Wu B, Cherezov V. 2015. Fluorescence Recovery After Photobleaching in Lipidic Cubic Phase (LCP-FRAP): A Precrystallization Assay for Membrane Proteins. Methods Enzymol 557:417–37 [DOI] [PubMed] [Google Scholar]
- 30.Fenalti G, Zatsepin NA, Betti C, Giguere P, Han GW, et al. 2015. Structural basis for bifunctional peptide recognition at human delta-opioid receptor. Nat. Struct. Mol. Biol 22:265–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galandrin S, Oligny-Longpré G, Bouvier M. 2007. The evasive nature of drug efficacy: implications for drug discovery. Trends Pharmacol. Sci 28:423–30 [DOI] [PubMed] [Google Scholar]
- 32.Ghosh E, Kumari P, Jaiman D, Shukla AK. 2015. Methodological advances: the unsung heroes of the GPCR structural revolution. Nat. Rev. Mol. Cell. Biol 16:69–81 [DOI] [PubMed] [Google Scholar]
- 33.Ginn HM, Brewster AS, Hattne J, Evans G, Wagner A, et al. 2015. A revised partiality model and post-refinement algorithm for X-ray free-electron laser data. Acta Crystallogr. D Biol. Crystallogr 71:1400–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, et al. 2008. A specific cholesterol binding site is established by the 2.8 A structure of the human beta2-adrenergic receptor. Structure 16:897–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hart P, Boutet S, Carini G, Dragone A, Duda B, et al. 2012. The Cornell-SLAC Pixel Array Detector at LCLS. SLAC-PUB-15284 [Google Scholar]
- 36.Hato M, Yamashita J, Shiono M. 2009. Aqueous Phase Behavior of Lipids with Isoprenoid Type Hydrophobic Chains. J. Phys. Chem. B 113:10196–209 [DOI] [PubMed] [Google Scholar]
- 37.Hattne J, Echols N, Tran R, Kern J, Gildea RJ, et al. 2014. Accurate macromolecular structures using minimal measurements from X-ray free-electron lasers. Nat. Methods 11:545–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heng BC, Aubel D, Fussenegger M. 2013. An overview of the diverse roles of G-protein coupled receptors (GPCRs) in the pathophysiology of various human diseases. Biotechnol. Adv 31:1676–94 [DOI] [PubMed] [Google Scholar]
- 39.Henrich B, Becker J, Dinapoli R, Goettlicher P, Graafsma H, et al. 2011. The adaptive gain integrating pixel detector AGIPD a detector for the European XFEL. Nucl. Inst. Methods Phys. A 633:S11–S4 [Google Scholar]
- 40.Heydenreich FM, Vuckovic Z, Matkovic M, Veprintsev DB. 2015. Stabilization of G protein-coupled receptors by point mutations. Front. Pharmacol 6:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirata K, Shinzawa-Itoh K, Yano N, Takemura S, Kato K, et al. 2014. Determination of damage-free crystal structure of an X-ray-sensitive protein using an XFEL. Nat. Methods 11:734–6 [DOI] [PubMed] [Google Scholar]
- 42.Hollenstein K, Kean J, Bortolato A, Cheng RK, Doré AS, et al. 2013. Structure of class B GPCR corticotropin-releasing factor receptor 1. Nature 499:438–43 [DOI] [PubMed] [Google Scholar]
- 43.Hutchings CJ, Koglin M, Olson WC, Marshall FH. 2017. Opportunities for therapeutic antibodies directed at G-protein-coupled receptors. Nat. Rev. Drug Discov advance online publication:1–24 [Google Scholar]
- 44.Isberg V, Mordalski S, Munk C, Rataj K, Harpsøe K, et al. 2016. GPCRdb: an information system for G protein-coupled receptors. Nucleic Acids Res 44:D356–D64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishchenko A, Cherezov V, Liu W. 2016. Preparation and Delivery of Microcrystals in Lipidic Cubic Phase for Serial Femtosecond Crystallography. J. Vis. Exp 115: e54463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishchenko A, Peng L, Zinovev E, Vlasov A, Lee SC, et al. 2017. Chemically Stable Lipids for Membrane Protein Crystallization. Cryst. Growth Des 17:3502–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishchenko A, Wacker D, Kapoor M, Zhang A, Han GW, et al. 2017. Structural insights into the extracellular recognition of the human serotonin 2B receptor by an antibody. Proc. Natl. Acad. Sci. U. S. A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johansson LC, Stauch B, Ishchenko A, Cherezov V. 2017. A Bright Future for Serial Femtosecond Crystallography with XFELs. Trends Biochem. Sci 42:749–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kabsch W 2010. XDS. Acta Crystallogr. D Biol. Crystallogr 66:125–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kabsch W 2014. Processing of X-ray snapshots from crystals in random orientations. Acta Crystallogr. D Biol. Crystallogr 70:2204–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kang Y, Zhou XE, Gao X, He Y, Liu W, et al. 2015. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature 523:561–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katritch V, Cherezov V, Stevens RC. 2013. Structure-function of the G protein-coupled receptor superfamily. Annu. Rev. Pharmacol. Toxicol 53:531–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Katritch V, Fenalti G, Abola EE, Roth BL, Cherezov V, Stevens RC. 2014. Allosteric sodium in class A GPCR signaling. Trends Biochem. Sci 39:233–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kern J, Alonso-Mori R, Hellmich J, Tran R, Hattne J, et al. 2012. Room temperature femtosecond X-ray diffraction of photosystem II microcrystals. Proc. Natl. Acad. Sci. U. S. A. 109:9721–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kern J, Tran R, Alonso-Mori R, Koroidov S, Echols N, et al. 2014. Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy. Nat. Commun 5:4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kirian RA, Wang X, Weierstall U, Schmidt KE, Spence JC, et al. 2010. Femtosecond protein nanocrystallography-data analysis methods. Opt. Express 18:5713–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koth CM, Murray JM, Mukund S, Madjidi A, Minn A, et al. 2012. Molecular basis for negative regulation of the glucagon receptor. Proc. Natl. Acad. Sci. U. S. A. 109:14393–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kupitz C, Basu S, Grotjohann I, Fromme R, Zatsepin NA, et al. 2014. Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser. Nature 513:261–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kupitz C, Olmos JL, Holl M, Tremblay L, Pande K, et al. 2017. Structural enzymology using X-ray free electron lasers. Struct. Dyn 4:044003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Landau EM, Rosenbusch JP. 1996. Lipidic cubic phases: a novel concept for the crystallization of membrane proteins. Proc. Natl. Acad. Sci. U. S. A. 93:14532–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Q, Dahmane T, Zhang Z, Assur Z, Brasch J, et al. 2012. Structures from anomalous diffraction of native biological macromolecules. Science 336:1033–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu W, Chun E, Thompson AA, Chubukov P, Xu F, et al. 2012. Structural basis for allosteric regulation of GPCRs by sodium ions. Science 337:232–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu W, Hanson MA, Stevens RC, Cherezov V. 2010. LCP-Tm: an assay to measure and understand stability of membrane proteins in a membrane environment. Biophys. J. 98:1539–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu W, Ishchenko A, Cherezov V. 2014. Preparation of microcrystals in lipidic cubic phase for serial femtosecond crystallography. Nat. Protoc 9:2123–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu W, Wacker D, Gati C, Han GW, James D, et al. 2013. Serial femtosecond crystallography of G protein-coupled receptors. Science 342:1521–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu W, Wacker D, Wang C, Abola E, Cherezov V. 2014. Femtosecond crystallography of membrane proteins in the lipidic cubic phase. Philos. Trans. R. Soc. Lond B Biol. Sci 369:20130314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luttrell LM, Lefkowitz RJ. 2002. The role of beta-arrestins in the termination and transduction of G-protein-coupled receptor signals. J. Cell. Sci 115:455–65 [DOI] [PubMed] [Google Scholar]
- 68.Magnani F, Shibata Y, Serrano-Vega MJ, Tate CG. 2008. Co-evolving stability and conformational homogeneity of the human adenosine A2a receptor. Proc. Natl. Acad. Sci. U. S. A. 105:10744–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mariani V, Morgan A, Yoon CH, Lane TJ, White TA, et al. 2016. OnDA: online data analysis and feedback for serial X-ray imaging. J. Appl. Crystallogr 49:1073–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Misquitta Y, Cherezov V, Havas F, Patterson S, Mohan JM, et al. 2004. Rational design of lipid for membrane protein crystallization. J. Struct. Biol 148:169–75 [DOI] [PubMed] [Google Scholar]
- 71.Mozzanica A, Bergamaschi A, Brueckner M, Cartier S, Dinapoli R, et al. 2016. Characterization results of the JUNGFRAU full scale readout ASIC. J. Instrument 11:C02047 [Google Scholar]
- 72.Nakane T, Song C, Suzuki M, Nango E, Kobayashi J, et al. 2015. Native sulfur/chlorine SAD phasing for serial femtosecond crystallography. Acta Crystallogr. D Biol. Crystallogr 71:2519–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nango E, Royant A, Kubo M, Nakane T, Wickstrand C, et al. 2016. A three-dimensional movie of structural changes in bacteriorhodopsin. Science 354:1552–7 [DOI] [PubMed] [Google Scholar]
- 74.Nass K, Meinhart A, Barends TR, Foucar L, Gorel A, et al. 2016. Protein structure determination by single-wavelength anomalous diffraction phasing of X-ray free-electron laser data. IUCrJ 3:180–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Neutze R, Wouts R, van der Spoel D, Weckert E, Hajdu J. 2000. Potential for biomolecular imaging with femtosecond X-ray pulses. Nature 406:752–7 [DOI] [PubMed] [Google Scholar]
- 76.Overington JP, Al-Lazikani B, HA L. 2006. How many drug targets are there? Nat. Rev. Drug Discov 5:993–6 [DOI] [PubMed] [Google Scholar]
- 77.Pande K, Hutchison CD, Groenhof G, Aquila A, Robinson JS, et al. 2016. Femtosecond structural dynamics drives the trans/cis isomerization in photoactive yellow protein. Science 352:725–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pile D 2011. X-rays: First light from SACLA. Nat. Photon. 5:456–7 [Google Scholar]
- 79.Rajagopal S, Rajagopal K, Lefkowitz RJ. 2010. Teaching old receptors new tricks: biasing seven-transmembrane receptors. Nat. Rev. Drug Discov 9:373–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, et al. 2011. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature 477:549–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Santos R, Ursu O, Gaulton A, Bento AP, Donadi RS, et al. 2017. A comprehensive map of molecular drug targets. Nat Rev. Drug Discov 16:19–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sarkar CA, Dodevski I, Kenig M, Dudli S, Mohr A, et al. 2008. Directed evolution of a G protein-coupled receptor for expression, stability, and binding selectivity. Proc. Natl. Acad. Sci. U. S. A. 105:14808–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sauter N 2015. XFEL diffraction: developing processing methods to optimize data quality. J. Synchrotron Radiat 22:239–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sauter NK, Hattne J, Brewster AS, Echols N, Zwart PH, Adams PD. 2014. Improved crystal orientation and physical properties from single-shot XFEL stills. Acta Crystallogr. D 70:3299–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schmidt M 2013. Mix and Inject: Reaction Initiation by Diffusion for Time-Resolved Macromolecular Crystallography. Adv. Condens. Mat. Phys 2013:10 [Google Scholar]
- 86.Schmidt M 2017. Time-Resolved Macromolecular Crystallography at Modern X-Ray Sources In Protein Crystallography: Methods and Protocols, ed. Wlodawer A, Dauter Z, Jaskolski M:273–94. New York, NY: Springer New York. . [DOI] [PubMed] [Google Scholar]
- 87.Serrano-Vega MJ, Magnani F, Shibata Y, Tate CG. 2008. Conformational thermostabilization of the beta1-adrenergic receptor in a detergent-resistant form. Proc. Natl. Acad. Sci. U. S. A. 105:877–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Siu FY, He M, de Graaf C, Han GW, Yang D, et al. 2013. Structure of the human glucagon class B G-protein-coupled receptor. Nature 499:444–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Spence J, Lattman E. 2016. Imaging enzyme kinetics at atomic resolution. IUCrJ 3:228–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Spence JC, Weierstall U, Chapman HN. 2012. X-ray lasers for structural and dynamic biology. Rep. Prog. Phys 75:102601. [DOI] [PubMed] [Google Scholar]
- 91.Stagno JR, Liu Y, Bhandari YR, Conrad CE, Panja S, et al. 2017. Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography. Nature 541:242–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stan CA, Milathianaki D, Laksmono H, Sierra RG, McQueen TA, et al. 2016. Liquid explosions induced by X-ray laser pulses. Nat. Phys 12:966–71 [Google Scholar]
- 93.Stevens RC, Cherezov V, Katritch V, Abagyan R, Kuhn P, et al. 2013. GPCR Network: a large-scale collaboration on GPCR structure and function. Nat. Rev. Drug Discov 12:25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tenboer J, Basu S, Zatsepin N, Pande K, Milathianaki D, et al. 2014. Time-resolved serial crystallography captures high-resolution intermediates of photoactive yellow protein. Science 346:1242–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thompson AA, Liu JJ, Chun E, Wacker D, Wu H, et al. 2011. GPCR stabilization using the bicelle-like architecture of mixed sterol-detergent micelles. Methods 55:310–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Uervirojnangkoorn M, Zeldin OB, Lyubimov AY, Hattne J, Brewster AS, et al. 2015. Enabling X-ray free electron laser crystallography for challenging biological systems from a limited number of crystals. Elife 4:e05421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vaidehi N, Grisshammer R, Tate CG. 2016. How do mutations thermostabilize G protein-coupled receptors? Trends Pharmacol. Sci 37:37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.van den Bedem H, Fraser JS. 2015. Integrative, dynamic structural biology at atomic resolution-it’s about time. Nat. Methods 12:307–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. 2013. Molecular signatures of G-protein-coupled receptors. Nature 494:185–94 [DOI] [PubMed] [Google Scholar]
- 100.Wacker D, Wang C, Katritch V, Han GW, Huang XP, et al. 2013. Structural features for functional selectivity at serotonin receptors. Science 340:615–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang C, Wu H, Evron T, Vardy E, Han GW, et al. 2014. Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs. Nat. Commun 5:4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang C, Wu H, Katritch V, Han GW, Huang XP, et al. 2013. Structure of the human smoothened receptor bound to an antitumour agent. Nature 497:338–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Weierstall U 2014. Liquid sample delivery techniques for serial femtosecond crystallography. Philos. Trans. R. So.c Lond. B Biol. Sci 369:20130337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Weierstall U, James D, Wang C, White TA, Wang D, et al. 2014. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nat. Commun 5:3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.White TA. 2014. Post-refinement method for snapshot serial crystallography. Philos. Trans. R. Soc. Lond. B Biol. Sci 369:20130330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.White TA, Mariani V, Brehm W, Yefanov O, Barty A, et al. 2016. Recent developments in CrystFEL. J. Appl. Crystallogr 49:680–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xiang J, Chun E, Liu C, Jing L, Al-Sahouri Z, et al. 2016. Successful Strategies to Determine High-Resolution Structures of GPCRs. Trends Pharmacol. Sci 37:1055–69 [DOI] [PubMed] [Google Scholar]
- 108.Yamashita J, Shiono M, Hato M. 2008. New Lipid Family That Forms Inverted Cubic Phases in Equilibrium with Excess Water: Molecular Structure−Aqueous Phase Structure Relationship for Lipids with 5,9,13,17-Tetramethyloctadecyl and 5,9,13,17-Tetramethyloctadecanoyl Chains. J. Phys. Chem. B 112:12286–96 [DOI] [PubMed] [Google Scholar]
- 109.Yefanov O, Mariani V, Gati C, White TA, Chapman HN, Barty A. 2015. Accurate determination of segmented X-ray detector geometry. Opt. Express 23:28459–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang D, Gao Z-G, Zhang K, Kiselev E, Crane S, et al. 2015. Two disparate ligand-binding sites in the human P2Y(1) receptor. Nature 520:317–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang D, Zhao Q, Wu B. 2015. Structural Studies of G Protein-Coupled Receptors. Mol. Cells 38:836–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang H, Han GW, Batyuk A, Ishchenko A, White KL, et al. 2017. Structural basis for selectivity and diversity in angiotensin II receptors. Nature 544:327–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang H, Qiao A, Yang D, Yang L, Dai A, et al. 2017. Structure of the full-length glucagon class B G-protein-coupled receptor. Nature 546:259–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang H, Unal H, Gati C, Han GW, Liu W, et al. 2015. Structure of the Angiotensin receptor revealed by serial femtosecond crystallography. Cell 161:833–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang X, Stevens RC, Xu F. 2015. The importance of ligands for G protein-coupled receptor stability. Trends Biochem. Sci 40:79–87 [DOI] [PubMed] [Google Scholar]
- 116.Zhang X, Zhao F, Wu Y, Yang J, Han GW, et al. 2017. Crystal structure of a multi-domain human smoothened receptor in complex with a super stabilizing ligand. Nat. Commun 8:15383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zheng Y, Qin L, Zacarías NV, de Vries H, Han GW, et al. 2016. Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists. Nature 540:458–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhou XE, He Y, de Waal PW, Gao X, Kang Y, et al. 2017. Identification of Phosphorylation Codes for Arrestin Recruitment by G Protein-Coupled Receptors. Cell 170:457–69 [DOI] [PMC free article] [PubMed] [Google Scholar]