Abstract
This paper is in recognition of the 100th birthday of Dr. Herbert Tabor, a true pioneer in the polyamine field for over 70 years, who served as the editor-in-chief of the Journal of Biological Chemistry from 1971 to 2010. We review current knowledge of MYC proteins (c-MYC, MYCN, and MYCL) and focus on ornithine decarboxylase 1 (ODC1), an important bona fide gene target of MYC, which encodes the sentinel, rate-limiting enzyme in polyamine biosynthesis. Although notable advances have been made in designing inhibitors against the “undruggable” MYCs, their downstream targets and pathways are currently the main avenue for therapeutic anticancer interventions. To this end, the MYC–ODC axis presents an attractive target for managing cancers such as neuroblastoma, a pediatric malignancy in which MYCN gene amplification correlates with poor prognosis and high-risk disease. ODC and polyamine levels are often up-regulated and contribute to tumor hyperproliferation, especially of MYC-driven cancers. We therefore had proposed to repurpose α-difluoromethylornithine (DFMO), an FDA-approved, orally available ODC inhibitor, for management of neuroblastoma, and this intervention is now being pursued in several clinical trials. We discuss the regulation of ODC and polyamines, which besides their well-known interactions with DNA and tRNA/rRNA, are involved in regulating RNA transcription and translation, ribosome function, proteasomal degradation, the circadian clock, and immunity, events that are also controlled by MYC proteins.
Keywords: polyamine, Myc (c-Myc), neuroblastoma, cancer, proteasome, translation, translation initiation factor, circadian clock, glycolysis, DFMO, MYC, Neuroblastoma clinical trials, ODC, polyamines
Introduction
This article is dedicated to the 100th birthday of Dr. Herbert Tabor, a scientist with unparalleled knowledge and enthusiasm for the scientific exploration of polyamines. Although Dr. André Bachmann only had the privilege of meeting Dr. Tabor once in his career, at the 1996 Tokyo International Symposium on Polyamines at Shonan Village in Japan, it was an encounter that remains vivid in his memory to this day. More than a decade later, while chatting with one of Dr. Tabor's postdoctoral fellows on his way to the Gordon Research Conference on Polyamines, Bachmann learned that Dr. Tabor – then in his 90s – still arrives early every morning at his office and works in the lab until later in the afternoon, when he transitions to reviewing manuscripts for JBC. His relentless dedication to science and his untethered vision to unravel the mysteries around polyamines have greatly influenced all of us and inspired Bachmann's career. The polyamine group (also fervently referred to as the “Polyamigos”) would have never evolved to where it stands today had it not been for Dr. Tabor's hard work and dedication and for his countless contributions to the scientific literature. We are blessed and grateful to know Dr. Tabor, an extraordinary scientist and human being. Happy 100th birthday!
The Bachmann laboratory has a longstanding interest in polyamines and, in particular, in the MYC–ODC axis, which forms the center point of this Minireview. Bachmann's work on polyamines began in 1992 when he studied the role of polyamines during foliar senescence of plant leaves (1). Because of the enthusiasm and encouragements by Dr. Alan Slusarenko and the late Dr. Philippe Matile (University of Zürich, Switzerland), he further delved into the world of polyamines in a quest to identify novel plant ornithine decarboxylase (ODC)2 inhibitors, using α-difluoromethylornithine (DFMO) as a positive control (2–4). DFMO, also known as eflornithine and Ornidyl, is as a catalytic irreversible (suicide) inhibitor of ODC synthesized in 1978 by researchers at the Merrell-Dow Research Institute (5). DFMO reached FDA approval for the treatment of West African sleeping sickness (trypanosomiasis; intravenous formulation) (6) and for the treatment of excessive facial hair growth (hirsutism; topical formulation). More recently, DFMO, in an oral formulation (powder or tablets), has been under investigation in multiple clinical trials, for example, the chemoprevention of colorectal cancer and pediatric neuroblastoma (7–9).
In the late 1990s, Bachmann slowly drifted away from plant research toward the ODC/cancer field, and in 2002 began investigations toward repurposing DFMO for pediatric neuroblastoma (10, 11). Although the worlds of plant and cancer research are seemingly unrelated, polyamines exist in nearly all living cells. It was those early experiences in plant science that gave him the scientific knowledge on polyamines, which ultimately led to the idea of repurposing DFMO for the treatment of children with (MYCN-amplified) neuroblastoma.
Bachmann met the co-author of this Minireview, Dr. Dirk Geerts, in 2002 at the “Advances in Neuroblastoma Research” conference in Paris, France. It was at this conference that Bachmann “connected the dots” and concluded that DFMO should be beneficial to neuroblastoma patients, due to fact that MYCN activates the bona fide gene ODC1 (10, 11). Although ODC as a drug target had been well-established by that time (12–14), the specific use of DFMO for the treatment of neuroblastoma–in the clinical setting-had not been seriously considered in the literature (10, 11). Over the following years, the authors continued their collaboration to investigate MYCN-driven ODC expression and polyamine regulation in neuroblastoma (8, 10, 11, 15–25). These preclinical efforts ultimately led to the first phase I neuroblastoma clinical trial with DFMO in 2010 (8). Today, multiple independent phase I and II DFMO neuroblastoma clinical trials are ongoing across the United States of America and in Australia (9).
MYC oncogenes
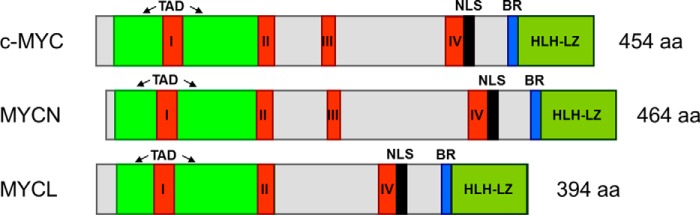
The MYC family of transcription factors is one of the most central–and most studied–gene groups in development and cancer. Three different MYC genes have been described: c-MYC (MYCC), MYCN, and MYCL (when all three genes are described in this Minireview, they will be named “MYC”). Initially discovered in the late 1970s (26), these three genes are aberrant in the majority of cancer types, performing oncogenic functions that correlate with aggressive tumor growth and poor patient prognosis. The human MYC proteins are around 400–450 amino acids in length (see Fig. 1) and are very homologous. This homology is highest in the four short “MYC homology boxes” I–IV that are important for MYC protein activity and oncogenic function. Two other longer and conserved regions exist in MYC: the N-terminal transactivation domain that can transfer transcription activity to the DNA-binding domain of another protein, and the C-terminal basic region (BR), necessary for binding to MYC target sites (the -CACGTG- “E-boxes”) on downstream target genes. The BR is coupled to the helix-loop-helix–leucine zipper (HLH–LZ) domain that allows MYC to bind partner proteins, such as MAX, that are needed for efficient target gene activation.
Figure 1.
Human MYC proteins. Shown is a schematic alignment of human c-MYC, MYCN, and MYCL. I–IV, MYC homology boxes I–IV; TAD, transactivation domain; NLS, nuclear localization signal; BR, basic region; HLH–LZ, helix-loop-helix–leucine zipper domain; aa, amino acid. Depicted are the longest RefSeq isoforms at NCBI_gene (https://www.ncbi.nlm.nih.gov/gene): NP_002458.2 (c-MYC IF1), NP_005369.2 (MYCN IF1), and NP_001028254.2 (MYCL IF3). Domains are assigned based on Ref. 32 for c-MYC, and combined NCBI_gene annotation and BlastP alignment (https://blast.ncbi.nlm.nih.gov/Blast.cgi) for the other MYC proteins.
The three MYC genes were all discovered in relation to cancer: c-MYC as a eukaryotic homolog of the v-myc avian virus oncogene; MYCN in neuroblastoma; and MYCL in lung cancer (26). The MYC genes are located on different chromosomes but share a simple gene structure that suggests they derive from an insertion of a v-myc–like, viral oncogene and later gene multiplication. Because MYC studies have primarily focused on c-MYC, most data exist on c-MYC functions, but MYCN and MYCL are equally powerful oncogenes. Considering the extensive homology between the MYC genes, their functional differences are in part a consequence of their differential mRNA expression during development and among tissue types. c-MYC is expressed throughout development and has ubiquitous expression in most–especially proliferative–tissues. c-MYC is the highest expressed MYC gene. By comparison, MYCN shows the highest expression during development, especially in the nervous system, which then declines considerably, but remains detectable in brain, genital tract, kidney, and stomach. MYCL shows restricted expression, with levels in between c-MYC and MYCN, mainly in the bladder, colon, esophagus, pancreas, and skin (for an overview, see https://www.ncbi.nlm.nih.gov/gene).
The cancer field received major new insights by the publication of the Hanahan and Weinberg reviews in 2000 and 2011 (27, 28). The authors present a list of cellular functions that a cancer cell needs to control by changing genome and gene expression. These functions are called “cancer hallmarks.” MYC genes are unique among oncogenes in that they can achieve most, if not all, of these hallmarks. One reason for this is that MYC genes, as “super transcription factors,” can regulate the activity of ∼15% of all human genes (29). Another reason is that MYC proteins act as obligate partners of other BR–HLH–LZ transcription factors, in the MAX–MLX network (30). It has long been known that MYC genes boost RNA production, ribosome biogenesis, and mRNA translation. MYC genes thereby support the classic hallmarks of sustained proliferation and replication, evasion of growth suppression and cell death, and activation of adhesion/migration (27, 31, 32). More recently, MYC genes were also shown to regulate the new hallmarks of genome integrity, metabolism, immune evasion, and inflammation (28, 32–34). Importantly, MYC genes can activate ornithine decarboxylase 1 (ODC1), a sentinel gene in polyamine synthesis (12–14) suggesting that MYC genes are central regulators of polyamine metabolism, as further discussed in this Minireview (see under “Polyamine synthesis and regulation”).
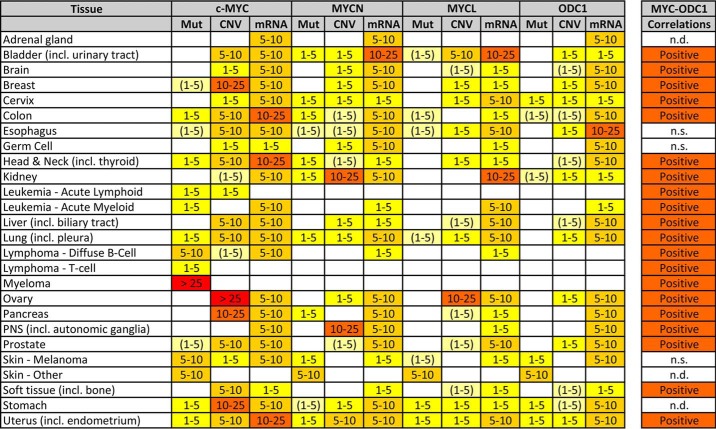
MYC genes can fulfill these oncogenic functions by escaping their normal, strict regulation. MYC genes are among the most frequently deregulated oncogenes in up to 25% of tumors and in many different cancer types (Table 1) (35, 36). The MYC genes only rarely accumulate coding sequence–altering mutations, with a notable exception for MYC gene fusions in lymphomas and myelomas (37). MYC gene amplifications have long been considered the most common MYC deregulation events (35, 36) and are often accompanied by “enhancer hijacking” to up-regulate MYC expression even further (38, 39). Tumors can contain multiple copies of one, two, or three different MYC genes. Occasionally, a specific MYC gene can govern a specific cancer subtype, for example, in brain or breast. For an overview of cancer types and MYC gene amplifications, see Table 1 and Refs. 26, 35, 36, 40. More recently, MYC gene DNA methylation and mRNA expression have also received attention as more dynamic strategies for MYC dysregulation (see also Table 1). As central transcription factors, MYC genes are prime candidates for establishing “tipping points” in cell fate (30), so that even small differences in expression could result in oncogenesis.
Table 1.
MYC and ODC1 aberrations in human cancer
Public human cancer data were queried for coding mutations (Mut), copy number variations (CNV), and mRNA dysregulation (mRNA) of the (three) MYC and ODC1 genes. Numbers represent % of samples with an aberration: white fields represent <1% aberrations; colored fields represent 1–5, 5–10, 10–25, and >25%. (1–5) means that 1–5% aberrations were found in specific tumor subtypes only. All datasets containing human cancer samples available on the public websites COSMIC (https://cancer.sanger.ac.uk/cosmic), cBioPortal (http://www.cbioportal.org/) (167, 168), and OncoMine (https://www.oncomine.org/) (169) were analyzed for a total of >1000 sets. Tumor types are represented by a minimum of three datasets. (Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.) Aberrations from identical samples in different datasets were not over-counted. Single mutations in a dataset or datasets containing <100 samples, were not used for calculations. Presented are only the significant changes in mutant reads, CNV, or mRNA expression, according to the website default analysis parameters. Correlations between (any) MYC and ODC1 mRNA expressions were calculated on the R2 website (http://r2.amc.nl/), using a 2log Pearson test on all tumor datasets on Affymetrix arrays (n = 242). Analysis was as in Ref. 17. n.d. = no data, n.s. = not significant.
MYC, the “undruggable target”
The central oncogenic role of the MYC proteins in many cancer types has evoked significant effort in their targeting for novel, specific cancer therapy; however, MYC proteins have proven to be difficult to drug (hence often referred to as the “undruggable target”). The three reasons for this are as follows: 1) because the MYC proteins act as transcription factors, they lack specific active sites with a defined 3D structure, but instead they function by using larger, flexible protein domains such as the leucine zipper; 2) MYC proteins are so-called “intrinsically disordered proteins” (41), making the design of tight-binding moieties even more difficult; and 3) MYC proteins are active in the nucleus, which precludes targeting with larger molecules (e.g. antibodies) (42). MYC targeting has recently been excellently reviewed (31, 42, 43), and we present only a brief summary. Targeting efforts have focused on four biological processes.
MYC transcription
MYC genes contain a 5′ G-quadruplex, a G-rich folded strand DNA structure upstream of the promoter that has to be resolved for transcription to occur. Compounds like enniantin-B, TH3, and APTO-253 that stabilize this structure decrease MYC transcription (44, 45). Furthermore, efficient MYC gene transcription needs docking of BRD proteins and other co-activating molecules, including IKZF, NME2, and CDK9 at the promoter. Especially, the inhibition of BRD4 with compounds such as JQ1 (46), OTX015 (47), and CPI-0610 (48) appears very promising for indirect blocking of MYC protein function. Drugs that target the other co-activators are also being investigated (49–51).
MYC translation
There has also been some progress in targeting MYC translation, in particular via specific translation inhibition by MYC siRNA (52) or targeting of ribosomal function using CX-5461 (53) and Inauhzin (54). Significant contributions have been made by the Ruggero group in understanding MYC-controlled protein synthesis and deregulation of translational control in cancer (55–59). MYC regulates multiple drivers in the translational machinery, including ribosomal proteins and eukaryotic initiation factors of translation (eIFs), leading to an increase in protein synthesis that is required for cell growth, cell cycle progression, and genome instability as a mechanism for cancer initiation (56).
MYC protein stability
MYC proteins are normally unstable and show cell cycle–regulated expression. Only present at low concentrations in quiescent cells, MYC protein expression is rapidly induced as cells enter the G1 phase of the cell cycle in response to serum or mitogens. In noncancerous cells, MYC levels then decrease to a low steady-state concentration as long as the cells proliferate, as a result of timed MYC removal by the proteasome. This degradation depends upon phosphorylation of residues in MYC homology box I and subsequent ubiquitination by FBXW7, for example (60). In cancer cells, MYC degradation can be prevented by high expression of deubiquitinases (61, 62) or kinases that interfere with FBXW7, notably Aurora-A, PLK1, and proteins in the PI3K/AKT/mTOR route (63, 64). These proteins enable MYC-dependent, sustained cell cycle progression by permitting continuous high-MYC protein expression. Therefore, specific inhibition of these molecules has received much attention. In particular, Aurora-A has been targeted with inhibitors, including alisertib (65, 66), MLN8054 (67), MLN8237 (68), and CD532 (69). Also, PLK1 can be inhibited with volasertib (70) and BI2536 (71). Finally, the P22077 compound inhibits the USP7 deubiquitinase necessary for high MYCN expression in neuroblastoma (62). It is important to mention the many efforts in MYC metabolic targeting aimed at the PI3K/AKT/mTOR axis. In particular, mTOR was targeted with multiple compounds, for instance temsirolimus, everolimus, dactolisib, and INK128 (reviewed in Refs. 43, 72).
MYC DNA binding
The transcription factor function of MYC proteins depends on dimerization with proteins like MAX. The leucine zipper domain involved in this process has been targeted with several compounds, such as 10058-F4 (73), 10074-G5 (74), and Mycro3 (75). Very interesting is the recent development of compounds that can bind to multiple conformations of the leucine zipper (41). Screening of large compound libraries resulted in the characterization of additional molecules that inhibit the MYC–MAX protein interface (76–78) and in a mimic of the MYC–MAX complex E-box–binding domain (76–78) that represents interesting candidates for further development.
Although direct targeting of the MYC genes and proteins is the subject of intense study, it is still a relatively new field. Over the years, the indirect targeting of MYC by inhibition of one or more MYC downstream processes, including cell cycle, apoptosis evasion, and recently also tumor metabolism and immune function, has been more successful. This is due to MYC's “Achilles heel”: MYC genes obtain their powerful oncogenic functions in part by changes in the cancer cell that, paradoxically, make the tumor more vulnerable to specific insults. Decreased MYC function, in combination with inhibition of a downstream process that is not normally toxic to the cell, can then lead to catastrophic cell death. This phenomenon called synthetic lethality (79–81) has been extensively investigated in MYC-addicted cancers (reviewed in Refs. 82–84). For instance, tumors with high MYC activity are especially sensitive to inhibition of the CHK1 cell cycle checkpoint with SB21807 or TCS2312 (85), the BCL2/BCLxl apoptotic axis using ABT-199 (86), and spliceosome activity using SD6 (87). A very promising synthetic lethal association is between MYC overexpression and RAS mutation, making tumors very sensitive to otherwise innocuous CDK2 inhibition (88–90).
Finally, it should be noted that we found DFMO treatment in neuroblastoma leads to significant down-regulation of MYCN protein, an indirect effect of DFMO that was further enhanced in combination with SAM486A, an AMD1 (also known as SAMDC or AdoMetDC) inhibitor (10, 11). Our observation suggests a polyamine-dependent, negative feedback loop that regulates MYC expression. Indeed, Tabib and Bachrach (91) demonstrated in Kirsten sarcoma virus–infected rat kidney cells that putrescine triggers the transcription of c-MYC mRNA and that inhibition of ODC activity by DFMO, which depletes putrescine, prevented the c-MYC transcription. The involvement of putrescine in the transcription of c-MYC mRNA was further confirmed by adding putrescine to cells, which resulted in the formation of c-MYC transcripts. These findings support the notion that putrescine (and possibly spermidine and spermine) is involved in the transcription of c-MYC and may in part explain our own findings that DFMO and DFMO/SAM486A treatments lead to reduced levels of MYCN protein in neuroblastoma cells (11).
Polyamine synthesis and regulation
The polyamine spermine has been first described in 1677 (92) by Antonie van Leeuwenhoek (also spelled Anthonii Lewenhoeck), preceding the discovery of DNA in 1868 by ∼200 years, when the Swiss physician Friedrich Miescher isolated “nuclein” from the nuclei of white blood cells (93). Polyamines are aliphatic cations that interact with negatively charged molecules, including nucleic acids (94, 95). In 1957, Hershey (97) showed that polyamines can bind to phage DNA, and this was confirmed by Ames et al. in 1958 (96). The secondary structures of polyamines in association with DNA were revealed using X-ray diffraction technology (98, 99). Polyamines were also found in association with tRNA and rRNA (100, 101) and contribute to chromatin modification and histone acetylation (102, 103). Over the years, it was discovered that polyamines are implicated in many cellular processes, including the regulation of ion channels (104) and their participation in transcriptional, translational, and post-translational events (105).
Ornithine decarboxylase (EC 4.1.1.17) is a sentinel enzyme that is mandatory to polyamine synthesis. ODC catalyzes the conversion of ornithine to putrescine (a diamine) through the release of carbon dioxide (CO2). Putrescine is the precursor in the synthesis of spermidine (a triamine) and spermine (a tetraamine) which are formed by the action of spermidine synthase and spermine synthase, respectively (Fig. 2). Collectively, putrescine, spermidine, and spermine are referred to as polyamines. ODC has also transforming and oncogenic abilities independent of MYC (106, 107). ODC and AMD1 are rate-limiting enzymes in polyamine synthesis (108–110) and are frequently dysregulated in cancer, with a critical role in cell proliferation (108, 111–114).
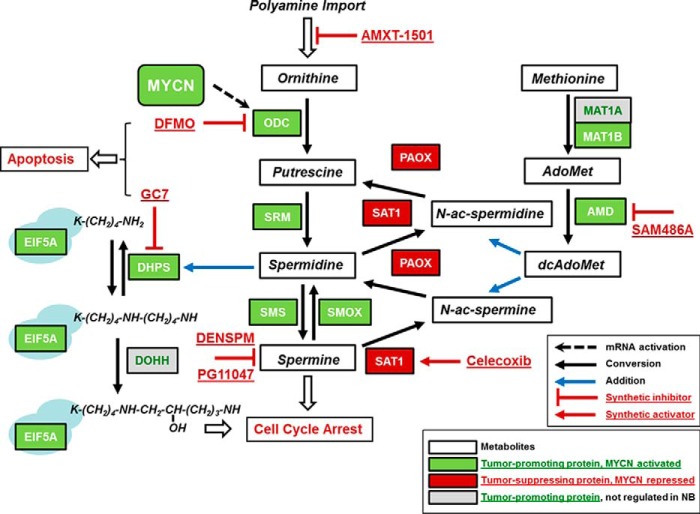
Figure 2.
Hypusine–polyamine pathway in human neuroblastoma. Shown is a graphical overview of the genes involved in eIF5A hypusination and polyamine metabolism with their protein activities and metabolites. Spermidine is the sole substrate and is mandatory for the synthesis of hypusine in eIF5A. Also shown are the action sites of the inhibitors used in neuroblastoma (NB) studies. References for additional inhibitor studies in neuroblastoma not named in the main text are as follows: DENSPM (164) and PG11047 (165). Genes are highlighted in green or red according to their prognostic value in Zhang-498, the largest publicly available RNA-Seq dataset for human neuroblastoma (166). Green or red indicates that high mRNA expression is significantly prognostic for poor or good patient survival in Kaplan-Meier analysis, respectively. In addition, gene mRNA expression was analyzed for correlation with MYCN gene amplification and MYCN mRNA expression using the R2 website (http://r2.amc.nl). (Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.) With the exception of DOHH and MAT1B, all other pathway genes that are prognostic for neuroblastoma also appear to be regulated by MYCN in this tumor. For more details on the analysis see Refs. 17, 18, 24. Data from Zhang-498 are available at the Gene Expression Omnibus database under accession number GSE62564. Reproduced with permission from Ref. 24. This research was originally published in Biochemical Journal. C. R. Schultz, D. Geerts, M. Mooney, R. El-Khawaja, J. Koster, and A. S. Bachmann. Synergistic drug combination GC7/DFMO suppresses hypusine/spermidine-dependent eIF5A activation and induces apoptotic cell death in neuroblastoma.. Biochem. J.2018; 475, 531–545. © Portland Press Ltd.
It has been well documented that ODC antizyme 1–3 (OAZ1–3) proteins are negative regulators of cellular polyamine content, and OAZ expression is controlled via a unique feedback mechanism that involves a +1 frameshift during translation, induced by high cellular polyamine levels (115). ODC is functional as a homodimer, which creates two active sites at the dimer interface that contain residues contributed by each subunit. ODC monomer interactions are relatively weak and the protein cycles between monomeric and dimeric forms. Therefore, a catalytically dead ODC protein can exhibit dominant-negative properties (116). In addition, OAZ binding to ODC monomer leads to its inactivation and ubiquitin-independent proteasomal degradation (117–120). The predominant OAZ family member is antizyme 1 (OAZ1); OAZ2 is usually expressed at significantly lower concentrations, and OAZ3 is restricted to expression in testis (120, 121). Two ODC antizyme inhibitors, AZIN1 and AZIN2, further contribute to the regulation of ODC enzyme activity, a testament to the level of complexity that governs cellular ODC expression in maintaining polyamine homeostasis. Both AZINs are strikingly similar to ODC but lack any decarboxylase activity due to critical amino acid substitutions (122). Notably, AZINs bind to OAZ with greater affinity than ODC, thus leading to a natural competition and liberation of ODC from the inactive ODC–AZ heterodimer complex (123).
OAZs are the only well-established proteins that physically interact with ODC, thereby regulating its activity and degradation. We recently proposed that the ODC–OAZ–AZIN regulatory mechanism is further controlled by a new player, sepiapterin reductase (SPR), an enzyme that forms tetrahydrobiopterin (BH4), a cofactor of nitric-oxide synthase, and together form a quartet that co-regulates both polyamine and NO biosyntheses (22). Because the predicted SPR interaction sites are close to those of ODC, there might be a competitive mechanism in which ODC and AZINs compete for binding to SPR. Sulfasalazine (SSZ) is a salicylate-based anti-inflammatory and immune-modulatory drug approved by the FDA for the treatment of ulcerative colitis and rheumatoid arthritis. SSZ is an SPR inhibitor (124), and we confirmed this finding with purified and active SPR enzyme.3 We found that SPR knockdown by RNAi reduces endogenous ODC enzyme activity and leads to neuroblastoma tumor cell growth inhibition (22). In addition, high SPR mRNA expression in human neuroblastoma tumors samples correlated significantly with poor survival prognosis. We further showed that pharmacological interference with SSZ inhibits neuroblastoma tumor cell growth, with significant synergisms if combined with DFMO (25).
MYC–ODC axis
A number of reports in the mid-1980s hinted at a potential connection between MYC and ODC, for example through observations that included the concomitant overexpression or co-amplification of MYC and ODC1 (125–128). However, a study led by George et al. (129) at the UK Children's Cancer Study Group reported that co-amplification of MYCN and ODC1 was not detected in seven cell lines and 87 primary tumors. In contrast, more recent reports by the Hogarty group proposed that 13–20% of MYCN-amplified neuroblastoma tumors have co-amplification of ODC1 (130, 131). This MYCN and ODC1 co-amplification demonstrates the first example of targeted deregulation of an oncogenic transcription factor (MYCN) and its oncogenic target gene (ODC1) (130).
Although MYC transcription factors activate a large number of genes (25), ODC1 is one of the first and best characterized bona fide targets. In 1992, the Cleveland group first reported that the ODC1 gene is a direct transcriptional target of c-MYC, resulting in a growth factor-independent expression of ODC1 (12, 13). The ODC1 gene harbors canonical MYC-binding sites in its promoter that contain the conserved E-box motif. The Cleveland group later demonstrated in vivo in Eμ-Myc transgenic mice that ODC1 is a critical MYC transcription target and that targeting ODC with DFMO prevents tumor formation in MYC-induced lymphogenesis (132). In this regard, it is noteworthy that Zell et al. (133) described a single nucleotide polymorphism (SNP) in the region of the ODC1 gene E-boxes that affects MYC and MAD binding to ODC1, and this SNP is linked to colon cancer recurrence.
In 2004, we proposed the repurposing of DFMO for neuroblastoma in the clinic (10). This was based on the principal idea that MYCN amplification is a key prognostic feature in high-risk patients that confers poor prognosis (134), thus making the MYCN–ODC axis an attractive drug target. At that time, DFMO was already approved by the FDA for (intravenous) treatment of West African sleeping sickness (trypanosomiasis). DFMO was also available in oral form and had an excellent safety record, thus making it a prime candidate for pediatric cancer clinical studies. We performed a number of preclinical studies over the years to investigate polyamine pathway–associated enzymes and the impact of their inhibitors, including ODC/DFMO, SAMDC/SAM486A, DHPS/GC7, polyamine uptake receptor/AMXT-1501, SPR/sulfasalazine (8, 10, 11, 15–25), and combinations thereof in neuroblastoma. More recently, we studied DFMO in osteosarcoma (135) and endometrial cancer (136). Notably, by early 2009, two excellent papers by the Hogarty and Cleveland groups had independently confirmed that DFMO inhibits tumor growth in vivo using the transgenic TH-MYCN neuroblastoma mouse model (131, 137). A landmark study with DFMO as a chemopreventive agent in colorectal cancer, in combination with sulindac, was published in 2008 by Meyskens et al. (7). A number of recent contributions by the Hogarty group further expanded our current knowledge on the functional relevance of ODC and polyamines in neuroblastoma (59, 130, 138–140).
Although polyamine synthesis has received widespread attention as a target for cancer therapy for close to 30 years, the focus has primarily been on epithelial tumors (carcinomas) (141). Clinical trials have predominantly been performed on breast, cervix, colon, lung, and prostate carcinomas, with a few additional studies in glioma and lymphoma (16, 141–143). Most trials with DFMO monotherapy had no or moderate success, except when used in a chemoprevention setting. However, combination therapies with NSAIDs or conventional chemotherapy agents, for example, appeared to be much more effective (7, 141, 142). Indeed, these cancer types show frequent aberration of one or more MYC genes, together with overexpression of ODC1 mRNA that appears regulated by MYC activation (see Table 1). Clearly, several additional carcinomas, including head and neck, liver, ovary, pancreas, and uterus, show this MYC–ODC1 activation pattern as well as some blastomas (for example, glioblastoma, the peripheral nervous system (PNS) tumor neuroblastoma, and the kidney tumor nephroblastoma) and sarcomas. Most of these cancer types could be selected for polyamine synthesis intervention studies, and several successful animal models have been available for a long time (144).
Neuroblastoma
Neuroblastoma appears amenable to polyamine inhibition as cancer therapy. High-stage tumors have almost 50% MYCN gene amplification and concomitant MYCN mRNA and protein overexpression (134), together with amplification and up-regulation of the sentinel ODC1 polyamine synthesis gene (12, 13, 130, 131). MYCN up-regulation of AMD1, another sentinel polyamine synthesis gene and bona fide target of MYC, was also shown in neuroblastoma (145, 146). Later studies in the TH-MYCN neuroblastoma mouse model (131, 137), and bio-informatic analysis of large sets of human neuroblastoma samples (see Fig. 2) (17, 18, 24), have now shown that genes connected to polyamine biosynthesis (AMD1, AZIN1, DHPS, EIF5A, MAT1B, ODC1, SMS, and SRM) are all up-regulated in neuroblastomas with MYCN amplification/up-regulation. In contrast, the OAZ2, PAOX, and SAT1 genes, involved in polyamine catabolism, are all down-regulated in these tumors. Strikingly, all these genes are also significantly prognostic for patient survival (18, 24, 130, 131).
The MYC–hypusine–polyamine network (147) appears to be central to the progression of MYC(N)-driven cancers, such as neuroblastoma, and is therefore eminently suited for novel therapeutic intervention. Hypusine-dependent activation of the eukaryotic translation initiation factor eIF5A is a unique post-translational process that exquisitely depends on the availability of spermidine, thus directly linking MYC(N), ODC, and the polyamines (spermidine) to eIF5A-controlled translational mechanisms that drive the increased biomass production necessary during tumorigenesis. In addition, eIF5A has been connected to the LIN28/let-7 pathway, which is active in neuroblastoma (148–150).
In support of this concept, the groups of Bachmann, Cleveland, and Hogarty (11, 18–25, 130, 131, 137, 147) have performed a number of preclinical studies with specific targeting of the neuroblastoma MYC–hypusine–polyamine network (see Fig. 2 for the inhibitors used. The legend lists the studies.). DFMO was a potent inhibitor of the neuroblastoma cell cycle, proliferation, and invasion (11, 20, 21). Other drugs that also target the MYC–hypusine–polyamine network, GC7, SAM486A, and AMXT-1501, were additive or synergistic with DFMO, resulting in cell cycle arrest and/or apoptosis (11, 17, 19, 21, 23, 24). In studies with the TH-MYCN mouse model, the Hogarty group showed that SAM486A and the NSAID celecoxib, which can activate SAT1, also potentiate DFMO activity (130). Finally, DFMO enhances the activity of standard chemotherapeutic agents such as cyclophosphamide and cisplatin in their studies (131).
Together, these preclinical studies strongly supported the use of DFMO for neuroblastoma therapy in the clinical setting. The first phase I trial with DFMO plus etoposide was launched in 2010 in neuroblastoma patients with relapsed or refractory disease, at Helen DeVos Children's Hospital (Grand Rapids, MI) and multiple consortium hospitals (coordinated through the Beat Childhood Cancer Consortium, formerly known as NMTRC). DFMO was well tolerated, and the therapy resulted in longer progression-free survival (8). Currently, several DFMO clinical trials are in progress (see Table 2) and include a recently completed DFMO phase II maintenance trial with neuroblastoma patients that are in remission but at high risk for relapse (9). Table 2 also shows that DFMO is being tested in addition to standard chemotherapy in newly diagnosed patients with high-risk disease. The results are eagerly awaited.
Table 2.
Polyamine metabolism interventions in neuroblastoma: Clinical trials
All clinical trials on neuroblastoma intervention are based on polyamine level inhibition, as listed in the National Institutes of Health clinical trial database (https://clinicaltrials.gov/; query date Aug. 8, 2018).
| NCT no. | Status | Phase | Patient status | Interventions | Drugs |
|---|---|---|---|---|---|
| NCT01059071 | Completea | 1 | Refractory/relapsed | DFMO to prevent recurrence | Plus etoposide |
| NCT01586260 | Active, not recruitinga | 2 | Remission | DFMO to prevent recurrence | Monotherapy |
| NCT02030964 | Active, not recruiting | 1 | Refractory/relapsed | DFMO to prevent recurrence | Plus three agentsb |
| NCT02139397 | Recruiting | 1/2 | Refractory/relapsed | DFMO to prevent recurrence | Plus bortezomib |
| NCT02395666 | Active, not recruitinga | 2 | Remission | DFMO to prevent recurrence | Monotherapy |
| NCT02559778 | Recruiting | 2 | New diagnosis | DFMO in high-risk therapy | Plus six agentsc |
| NCT02679144 | Recruiting | 2 | Remission | DFMO to prevent recurrence | Monotherapy |
Considering the central role of polyamine metabolism in many cancer hallmarks, ample possibilities remain for further neuroblastoma combination therapies. One obvious strategy is to combine the inhibition of polyamine biosynthesis with targeting polyamine import, which was successful in neuroblastoma cell culture studies (23) and in a mouse melanoma xenograft model (151). Interestingly, this combined polyamine blocking treatment in vivo resulted in increased immune response (151). Also, MYC genes are involved in tumor immune response: they induce immune checkpoints like CD47 and PD-L1, regulate immune molecules like cytokines, and thereby allow tumor cells to escape immune surveillance (152). In exchange, this MYC dependence might make tumors vulnerable to immunotherapy. An interesting link between MYC and polyamines is arginase; it converts arginine to ornithine and thus directly feeds into polyamine biosynthesis. High arginase expression in neuroblastoma is prognostic for poor patient outcome and significantly is positively correlated to MYCN amplification and mRNA expression (results not shown and see Refs. 153,154). Indeed, arginase activity in the TH-MYCN mouse model was linked to decreased immune surveillance and impaired immune therapy response through a yet uncharacterized mechanism (153, 154). It is not unthinkable that combination therapies targeting MYC and polyamine metabolism will evoke a synthetic lethal response, especially in high-risk neuroblastomas.
Novel roles for polyamines
It has been well established that ODC is one of few proteins rapidly regulated by a ubiquitin-independent proteasomal degradation mechanism that requires interaction with ODC antizyme (117–120). The C-terminal destabilization region (37 amino acids) of ODC is critical for ODC degradation in the proteasome. Interestingly, it was shown that MYC interacts with OAZ2 in the nucleus and nucleolus and can also accelerate MYC degradation by the proteasome (155). Nucleolar MYC plays a key role in positively regulating ribosomal RNA (rRNA) synthesis, and therefore, OAZ2 contributes to pre-rRNA synthesis through control of nucleolar MYC levels. This suggests that OAZ2 is regulating ribosome biogenesis through MYC degradation, but the detailed mechanisms remain unclear (155). One explanation might be that OAZ2 contributes to ribosome biogenesis by controlling the proteasomal degradation of both MYC and ODC, thereby regulating available spermidine pools needed for the following: 1) spermidine-dependent eIF5A hypusination (activation) and/or 2) spermidine-dependent, mTORC-mediated eIF4E activation, both proteins that play a role in the initiation, elongation, and termination of mRNA translation and have been connected to tumor formation (55–59). Pharmacological strategies to specifically block spermidine-dependent hypusination of eIF5A by inhibition of DHPS with GC7 alone or in combination with DFMO have been explored for neuroblastoma in our lab (17, 18, 24). Inactivation of eIF5A (and/or possibly eIF5A2) is expected to suppress mRNA translation and biomass production, a prerequisite for the survival of hyper-proliferating cells and tumorigenesis (147).
Polyamine metabolism has been linked to glycolysis in neuroblastoma cells (156). Impairment of glycolysis is able to trigger signaling events that lead to the reduction of MYCN protein levels and subsequent decrease of both ODC expression and polyamine levels, accompanied by cell cycle blockade preceding cell death (156). Moreover, c-MYC regulates a transcriptional program that stimulates mitochondrial glutaminolysis, which leads to glutamine addiction (157). Notably, c-MYC controls metabolic reprogramming upon T lymphocyte activation and links glutaminolysis to the biosynthesis of polyamines (33, 158). These findings suggest that polyamines contribute critically to the uptake and metabolism of nutrients and the addiction of cancer cells to glucose as initially described by Warburg and co-workers (159, 160).
An exciting new discovery was recently made by Zwighaft and co-workers unraveling a novel mechanism through which polyamines regulate circadian rhythms (161, 162). The authors showed that polyamine levels oscillate in a daily manner. Both clock- and feeding-dependent mechanisms regulate key enzymes of polyamine biosynthesis through engagement of BMAL1:CLOCK and core clock repressors PER2 and CRY1. Notably, BMAL1:CLOCK is a heterodimeric master circadian transcription factor that has E-box–binding sites and therefore binds c-MYC and MYCN (163). Moreover, MYC alters the oscillation of glucose metabolism and perturbs glutaminolysis, thus suggesting a new link between oncogenic transformation and circadian and metabolic dysrhythmia that involves polyamines and contributes to oncogenicity (163).
Finally, our group recently discovered a novel de novo pathogenic variant in the ODC1 gene in a 32-month-old girl with developmental delay, alopecia, and dysmorphic features. The mutation leads to the deletion of 14 amino acids at the ODC C terminus which causes ODC protein and putrescine accumulation in red blood cells. This is the first human case reported that presents with this new syndrome (also referred to as Bachmann–Bupp syndrome) and shows the importance of ODC in human development (170). Treatment with DFMO to counteract the increased ODC levels might be a therapeutic option for this and future patients.
Conclusions
MYC genes are master regulators and importantly regulate the development of many cancers. ODC1 encodes the sentinel, rate-limiting enzyme ODC, which contributes to the biosynthesis of polyamines and represents an important bona fide gene target of MYC genes. Polyamines are well-studied polycationic molecules that orchestrate complex processes in both normal cell growth and cancer development. Because direct MYC inhibition continues to be a challenge, the MYC–ODC-linked polyamine synthesis pathway presents an attractive downstream target for therapy and chemoprevention. DFMO is a well-tolerated ODC inhibitor that has entered multiple clinical trials and appears to be most effective against MYC-driven cancers that display a polyamine addiction phenotype, such as neuroblastoma. The polyamine spermidine is the only available substrate to activate eIF5A via hypusination, a highly specific, post-translational modification that is instrumental in coordinating eIF5A-dependent events during protein synthesis and the translational control of cancer. Novel roles for polyamines continue to be discovered and include proteasomal degradation, the circadian clock, and immunity, all events that are controlled by MYC proteins.
Acknowledgments
We acknowledge the many excellent scientists that have made invaluable contributions to the MYC and ODC/polyamine field. We have made every attempt in this Minireview to cite the large number of important contributions of our esteemed colleagues, and we sincerely apologize for any oversight and omissions in the referenced material.
This article is part of a series on “Polyamines,” written in honor of Dr. Herbert Tabor's 100th birthday. A. S. B. is the sole inventor of U. S. patent 9,072,778 issued on July 7, 2015, entitled “Treatment Regimen for N-Myc, C-Myc, and L-Myc amplified and overexpressed tumors”. A. S. B. is the co-founder, board member, and President of Hibiskus Biopharma, Inc.
A. S. Bachmann and D. Geerts, unpublished data.
- ODC
- ornithine decarboxylase
- DFMO
- α-difluoromethylornithine
- SPR
- sepiapterin reductase
- FDA
- Food and Drug Administration
- SSZ
- sulfasalazine
- SNP
- single nucleotide polymorphism
- BR
- basic region
- HLH–LZ
- helix-loop-helix–leucine zipper
- eIF
- eukaryotic initiation factor
- PI3K
- phosphatidylinositol 3-kinase
- mTOR
- mechanistic target of rapamycin
- DHPS
- deoxyhypusine synthase
- NSAID
- nonsteroidal anti-inflammatory drug.
References
- 1. Bachmann A. S., Fernandez-Lopez J., Ginsburg S., Thomas H., Bouwkamp J. C., Solomos T., and Matile P. (1994) Stay-green genotypes of Phaseolus vulgaris L.: chloroplast proteins and chlorophyll catabolites during foliar senescence. New Phytol. 126, 593–600 10.1111/j.1469-8137.1994.tb02953.x [DOI] [Google Scholar]
- 2. Bachmann A. S., Matile P., and Slusarenko A. J. (1998) Inhibition of ornithine decarboxylase activity by phaseolotoxin: implications for symptom production in halo blight of French bean. Physiol. Mol. Plant Pathol. 53, 287–299 10.1006/pmpp.1998.0183 [DOI] [Google Scholar]
- 3. Bachmann A. S., and Patil S. S. (2003) Characterization of ornithine decarboxylase from Pseudomonas syringae pv. phaseolicola and its inhibition by phaseolotoxin. Physiol. Mol. Plant Pathol. 63, 57–63 10.1016/j.pmpp.2003.09.005 [DOI] [Google Scholar]
- 4. Bachmann A. S., Xu R., Ratnapala L., and Patil S. S. (2004) Inhibitory effects of phaseolotoxin on proliferation of leukemia cells HL-60, K-562 and L1210 and pancreatic cells RIN-m5F. Leuk. Res. 28, 301–306 10.1016/j.leukres.2003.07.002 [DOI] [PubMed] [Google Scholar]
- 5. Metcalf B. W., Bey P., Danzin C., Jung M. J., Casara P., and Vevert J. P. (1978) Catalytic irreversible inhibition of mammalian ornithine decarboxylase (E.C. 4.1.1.17) by substrate and product analogs. J. Am. Chem. Soc. 100, 2551–2553 10.1021/ja00476a050 [DOI] [Google Scholar]
- 6. Priotto G., Kasparian S., Mutombo W., Ngouama D., Ghorashian S., Arnold U., Ghabri S., Baudin E., Buard V., Kazadi-Kyanza S., Ilunga M., Mutangala W., Pohlig G., Schmid C., Karunakara U., et al. (2009) Nifurtimox-eflornithine combination therapy for second-stage African Trypanosoma brucei gambiense trypanosomiasis: a multicentre, randomised, phase III, non-inferiority trial. Lancet 374, 56–64 10.1016/S0140-6736(09)61117-X [DOI] [PubMed] [Google Scholar]
- 7. Meyskens F. L. Jr., McLaren C. E., Pelot D., Fujikawa-Brooks S., Carpenter P. M., Hawk E., Kelloff G., Lawson M. J., Kidao J., McCracken J., Albers C. G., Ahnen D. J., Turgeon D. K., Goldschmid S., Lance P., et al. (2008) Difluoromethylornithine plus sulindac for the prevention of sporadic colorectal adenomas: a randomized placebo-controlled, double-blind trial. Cancer Prev. Res. 1, 32–38 10.1158/1940-6207.CAPR-08-0042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Saulnier Sholler G. L., Gerner E. W., Bergendahl G., MacArthur R. B., VanderWerff A., Ashikaga T., Bond J. P., Ferguson W., Roberts W., Wada R. K., Eslin D., Kraveka J. M., Kaplan J., Mitchell D., Parikh N. S., et al. (2015) A Phase I trial of DFMO targeting polyamine addiction in patients with relapsed/refractory neuroblastoma. PLoS ONE 10, e0127246 10.1371/journal.pone.0127246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sholler G. L. S., Ferguson W., Bergendahl G., Bond J. P., Neville K., Eslin D., Brown V., Roberts W., Wada R. K., Oesterheld J., Mitchell D., Foley J., Parikh N. S., Eshun F., Zage P., et al. (2018) Maintenance DFMO increases survival in high risk neuroblastoma. Sci. Rep. 8, 14445 10.1038/s41598-018-32659-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bachmann A. S. (2004) The role of polyamines in human cancer: prospects for drug combination therapies. Hawaii Med. J. 63, 371–374 [PubMed] [Google Scholar]
- 11. Wallick C. J., Gamper I., Thorne M., Feith D. J., Takasaki K. Y., Wilson S. M., Seki J. A., Pegg A. E., Byus C. V., and Bachmann A. S. (2005) Key role for p27Kip1, retinoblastoma protein Rb, and MYCN in polyamine inhibitor-induced G1 cell cycle arrest in MYCN-amplified human neuroblastoma cells. Oncogene 24, 5606–5618 10.1038/sj.onc.1208808 [DOI] [PubMed] [Google Scholar]
- 12. Bello-Fernandez C., and Cleveland J. L. (1992) c-myc transactivates the ornithine decarboxylase gene. Curr. Top. Microbiol. Immunol. 182, 445–452 [DOI] [PubMed] [Google Scholar]
- 13. Bello-Fernandez C., Packham G., and Cleveland J. L. (1993) The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc. Natl. Acad. Sci. U.S.A. 90, 7804–7808 10.1073/pnas.90.16.7804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ben-Yosef T., Yanuka O., Halle D., and Benvenisty N. (1998) Involvement of Myc targets in c-myc and N-myc induced human tumors. Oncogene 17, 165–171 10.1038/sj.onc.1201939 [DOI] [PubMed] [Google Scholar]
- 15. Bachmann A. S., Geerts D., and Sholler G. (2012) in Pediatric Cancer, Neuroblastoma: Diagnosis, Therapy, and Prognosis (Hayat M. A., ed) pp. 91–103, Springer, Dordrecht, Netherlands [Google Scholar]
- 16. Bachmann A. S., and Levin V. A. (2012) in Polyamine Drug Discovery (Woster P. M., and Casero R. A. Jr., eds) pp. 257–276, Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 17. Bandino A., Geerts D., Koster J., and Bachmann A. S. (2014) Deoxyhypusine synthase (DHPS) inhibitor GC7 induces p21/Rb-mediated inhibition of tumor cell growth and DHPS expression correlates with poor prognosis in neuroblastoma patients. Cell Oncol. 37, 387–398 10.1007/s13402-014-0201-9 [DOI] [PubMed] [Google Scholar]
- 18. Geerts D., Koster J., Albert D., Koomoa D. L., Feith D. J., Pegg A. E., Volckmann R., Caron H., Versteeg R., and Bachmann A. S. (2010) The polyamine metabolism genes ornithine decarboxylase and antizyme 2 predict aggressive behavior in neuroblastomas with and without MYCN amplification. Int. J. Cancer 126, 2012–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koomoa D. L., Borsics T., Feith D. J., Coleman C. C., Wallick C. J., Gamper I., Pegg A. E., and Bachmann A. S. (2009) Inhibition of S-adenosylmethionine decarboxylase by inhibitor SAM486A connects polyamine metabolism with p53-Mdm2-Akt/protein kinase B regulation and apoptosis in neuroblastoma. Mol. Cancer Ther. 8, 2067–2075 10.1158/1535-7163.MCT-08-1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Koomoa D. L., Geerts D., Lange I., Koster J., Pegg A. E., Feith D. J., and Bachmann A. S. (2013) DFMO/eflornithine inhibits migration and invasion downstream of MYCN and involves p27Kip1 activity in neuroblastoma. Int. J. Oncol. 42, 1219–1228 10.3892/ijo.2013.1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koomoa D. L., Yco L. P., Borsics T., Wallick C. J., and Bachmann A. S. (2008) Ornithine decarboxylase inhibition by α-difluoromethylornithine activates opposing signaling pathways via phosphorylation of both Akt/protein kinase B and p27Kip1 in neuroblastoma. Cancer Res. 68, 9825–9831 10.1158/0008-5472.CAN-08-1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lange I., Geerts D., Feith D. J., Mocz G., Koster J., and Bachmann A. S. (2014) Novel interaction of ornithine decarboxylase with sepiapterin reductase regulates neuroblastoma cell proliferation. J. Mol. Biol. 426, 332–346 10.1016/j.jmb.2013.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Samal K., Zhao P., Kendzicky A., Yco L. P., McClung H., Gerner E., Burns M., Bachmann A. S., and Sholler G. (2013) AMXT-1501, a novel polyamine transport inhibitor, synergizes with DFMO in inhibiting neuroblastoma cell proliferation by targeting both ornithine decarboxylase and polyamine transport. Int. J. Cancer 133, 1323–1333 10.1002/ijc.28139 [DOI] [PubMed] [Google Scholar]
- 24. Schultz C. R., Geerts D., Mooney M., El-Khawaja R., Koster J., and Bachmann A. S. (2018) Synergistic drug combination GC7/DFMO suppresses hypusine/spermidine-dependent eIF5A activation and induces apoptotic cell death in neuroblastoma. Biochem. J. 475, 531–545 10.1042/BCJ20170597 [DOI] [PubMed] [Google Scholar]
- 25. Yco L. P., Geerts D., Mocz G., Koster J., and Bachmann A. S. (2015) Effect of sulfasalazine on human neuroblastoma: analysis of sepiapterin reductase (SPR) as a new therapeutic target. BMC Cancer 15, 477 10.1186/s12885-015-1447-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rickman D. S., Schulte J. H., and Eilers M. (2018) The expanding world of N-MYC-driven tumors. Cancer Discov. 8, 150–163 10.1158/2159-8290.CD-17-0273 [DOI] [PubMed] [Google Scholar]
- 27. Hanahan D., and Weinberg R. A. (2000) The hallmarks of cancer. Cell 100, 57–70 10.1016/S0092-8674(00)81683-9 [DOI] [PubMed] [Google Scholar]
- 28. Hanahan D., and Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 29. Dang C. V., O'Donnell K. A., Zeller K. I., Nguyen T., Osthus R. C., and Li F. (2006) The c-Myc target gene network. Semin. Cancer Biol. 16, 253–264 10.1016/j.semcancer.2006.07.014 [DOI] [PubMed] [Google Scholar]
- 30. Carroll P. A., Freie B. W., Mathsyaraja H., and Eisenman R. N. (2018) The MYC transcription factor network: balancing metabolism, proliferation and oncogenesis. Front. Med. 12, 412–425 10.1007/s11684-018-0650-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Koh C. M., Sabò A., and Guccione E. (2016) Targeting MYC in cancer therapy: RNA processing offers new opportunities. Bioessays 38, 266–275 10.1002/bies.201500134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Meyer N., and Penn L. Z. (2008) Reflecting on 25 years with MYC. Nat. Rev. Cancer 8, 976–990 10.1038/nrc2231 [DOI] [PubMed] [Google Scholar]
- 33. Dejure F. R., and Eilers M. (2017) MYC and tumor metabolism: chicken and egg. EMBO J. 36, 3409–3420 10.15252/embj.201796438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pistoia V., Morandi F., Pezzolo A., Raffaghello L., and Prigione I. (2012) MYCN: from oncoprotein to tumor-associated antigen. Front. Oncol. 2, 174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kalkat M., De Melo J., Hickman K. A., Lourenco C., Redel C., Resetca D., Tamachi A., Tu W. B., and Penn L. Z. (2017) MYC deregulation in primary human cancers. Genes (Basel) 8, pii: E151 10.3390/genes8060151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schaub F. X., Dhankani V., Berger A. C., Trivedi M., Richardson A. B., Shaw R., Zhao W., Zhang X., Ventura A., Liu Y., Ayer D. E., Hurlin P. J., Cherniack A. D., Eisenman R. N., Bernard B., et al. (2018) Pan-cancer alterations of the MYC oncogene and its proximal network across the Cancer Genome Atlas. Cell Syst. 6, 282–300.e2 10.1016/j.cels.2018.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sewastianik T., Prochorec-Sobieszek M., Chapuy B., and Juszczyński P. (2014) MYC deregulation in lymphoid tumors: molecular mechanisms, clinical consequences and therapeutic implications. Biochim. Biophys. Acta 1846, 457–467 [DOI] [PubMed] [Google Scholar]
- 38. Chipumuro E., Marco E., Christensen C. L., Kwiatkowski N., Zhang T., Hatheway C. M., Abraham B. J., Sharma B., Yeung C., Altabef A., Perez-Atayde A., Wong K. K., Yuan G. C., Gray N. S., Young R. A., and George R. E. (2014) CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell 159, 1126–1139 10.1016/j.cell.2014.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Herranz D., Ambesi-Impiombato A., Palomero T., Schnell S. A., Belver L., Wendorff A. A., Xu L., Castillo-Martin M., Llobet-Navás D., Cordon-Cardo C., Clappier E., Soulier J., and Ferrando A. A. (2014) A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nat. Med. 20, 1130–1137 10.1038/nm.3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ruiz-Pérez M. V., Henley A. B., and Arsenian-Henriksson M. (2017) The MYCN protein in health and disease Genes (Basel) 8, E113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu C., Niu X., Jin F., Liu Z., Jin C., and Lai L. (2016) Structure-based inhibitor design for the intrinsically disordered protein c-Myc. Sci. Rep. 6, 22298 10.1038/srep22298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen H., Liu H., and Qing G. (2018) Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct. Target Ther. 3, 5 10.1038/s41392-018-0008-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sabnis H. S., Somasagara R. R., and Bunting K. D. (2017) Targeting MYC dependence by metabolic inhibitors in cancer. Genes (Basel) 8, E114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dutta D., Debnath M., Müller D., Paul R., Das T., Bessi I., Schwalbe H., and Dash J. (2018) Cell penetrating thiazole peptides inhibit c-MYC expression via site-specific targeting of c-MYC G-quadruplex. Nucleic Acids Res. 46, 5355–5365 10.1093/nar/gky385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Local A., Zhang H., Benbatoul K. D., Folger P., Sheng X., Tsai C. Y., Howell S. B., and Rice W. G. (2018) APTO-253 stabilizes G-quadruplex DNA, inhibits MYC expression, and induces DNA damage in acute myeloid leukemia cells. Mol. Cancer Ther. 17, 1177–1186 10.1158/1535-7163.MCT-17-1209 [DOI] [PubMed] [Google Scholar]
- 46. Puissant A., Frumm S. M., Alexe G., Bassil C. F., Qi J., Chanthery Y. H., Nekritz E. A., Zeid R., Gustafson W. C., Greninger P., Garnett M. J., McDermott U., Benes C. H., Kung A. L., Weiss W. A., et al. (2013) Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 3, 308–323 10.1158/2159-8290.CD-12-0418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Coudé M. M., Braun T., Berrou J., Dupont M., Bertrand S., Masse A., Raffoux E., Itzykson R., Delord M., Riveiro M. E., Herait P., Baruchel A., Dombret H., and Gardin C. (2015) BET inhibitor OTX015 targets BRD2 and BRD4 and decreases c-MYC in acute leukemia cells. Oncotarget 6, 17698–17712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Siu K. T., Ramachandran J., Yee A. J., Eda H., Santo L., Panaroni C., Mertz J. A., Sims Iii R. J., Cooper M. R., and Raje N. (2017) Preclinical activity of CPI-0610, a novel small-molecule bromodomain and extra-terminal protein inhibitor in the therapy of multiple myeloma. Leukemia 31, 1760–1769 10.1038/leu.2016.355 [DOI] [PubMed] [Google Scholar]
- 49. Bouvard C., Lim S. M., Ludka J., Yazdani N., Woods A. K., Chatterjee A. K., Schultz P. G., and Zhu S. (2017) Small molecule selectively suppresses MYC transcription in cancer cells. Proc. Natl. Acad. Sci. U.S.A. 114, 3497–3502 10.1073/pnas.1702663114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen Z., Wang Z., Pang J. C., Yu Y., Bieerkehazhi S., Lu J., Hu T., Zhao Y., Xu X., Zhang H., Yi J. S., Liu S., and Yang J. (2016) Multiple CDK inhibitor dinaciclib suppresses neuroblastoma growth via inhibiting CDK2 and CDK9 activity. Sci. Rep. 6, 29090 10.1038/srep29090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shan C., Lin J., Hou J. Q., Liu H. Y., Chen S. B., Chen A. C., Ou T. M., Tan J. H., Li D., Gu L. Q., and Huang Z. S. (2015) Chemical intervention of the NM23-H2 transcriptional programme on c-MYC via a novel small molecule. Nucleic Acids Res. 43, 6677–6691 10.1093/nar/gkv641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Reyes-González J. M., Armaiz-Peña G. N., Mangala L. S., Valiyeva F., Ivan C., Pradeep S., Echevarría-Vargas I. M., Rivera-Reyes A., Sood A. K., and Vivas-Mejía P. E. (2015) Targeting c-MYC in platinum-resistant ovarian cancer. Mol. Cancer Ther. 14, 2260–2269 10.1158/1535-7163.MCT-14-0801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rosenwald I. B., Rhoads D. B., Callanan L. D., Isselbacher K. J., and Schmidt E. V. (1993) Increased expression of eukaryotic translation initiation factors eIF-4E and eIF-2α in response to growth induction by c-myc. Proc. Natl. Acad. Sci. U.S.A. 90, 6175–6178 10.1073/pnas.90.13.6175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jung J. H., Liao J. M., Zhang Q., Zeng S., Nguyen D., Hao Q., Zhou X., Cao B., Kim S. H., and Lu H. (2015) Inauhzin(c) inactivates c-Myc independently of p53. Cancer Biol. Ther. 16, 412–419 10.1080/15384047.2014.1002698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pourdehnad M., Truitt M. L., Siddiqi I. N., Ducker G. S., Shokat K. M., and Ruggero D. (2013) Myc and mTOR converge on a common node in protein synthesis control that confers synthetic lethality in Myc-driven cancers. Proc. Natl. Acad. Sci. U.S.A. 110, 11988–11993 10.1073/pnas.1310230110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ruggero D. (2009) The role of Myc-induced protein synthesis in cancer. Cancer Res. 69, 8839–8843 10.1158/0008-5472.CAN-09-1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ruggero D., Montanaro L., Ma L., Xu W., Londei P., Cordon-Cardo C., and Pandolfi P. P. (2004) The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat. Med. 10, 484–486 10.1038/nm1042 [DOI] [PubMed] [Google Scholar]
- 58. Truitt M. L., and Ruggero D. (2016) New frontiers in translational control of the cancer genome. Nat. Rev. Cancer 16, 288–304 10.1038/nrc.2016.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Flynn A. T., and Hogarty M. D. (2018) Myc, oncogenic protein translation, and the role of polyamines. Med. Sci. 6, E41 10.3390/medsci6020041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Welcker M., Orian A., Jin J., Grim J. A., Harper J. W., Eisenman R. N., and Clurman B. E. (2004) The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. U.S.A. 101, 9085–9090 10.1073/pnas.0402770101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Popov N., Wanzel M., Madiredjo M., Zhang D., Beijersbergen R., Bernards R., Moll R., Elledge S. J., and Eilers M. (2007) The ubiquitin-specific protease USP28 is required for MYC stability. Nat. Cell Biol. 9, 765–774 10.1038/ncb1601 [DOI] [PubMed] [Google Scholar]
- 62. Tavana O., Li D., Dai C., Lopez G., Banerjee D., Kon N., Chen C., Califano A., Yamashiro D. J., Sun H., and Gu W. (2016) HAUSP deubiquitinates and stabilizes N-Myc in neuroblastoma. Nat. Med. 22, 1180–1186 10.1038/nm.4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Richards M. W., Burgess S. G., Poon E., Carstensen A., Eilers M., Chesler L., and Bayliss R. (2016) Structural basis of N-Myc binding by Aurora-A and its destabilization by kinase inhibitors. Proc. Natl. Acad. Sci. U.S.A. 113, 13726–13731 10.1073/pnas.1610626113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xiao D., Yue M., Su H., Ren P., Jiang J., Li F., Hu Y., Du H., Liu H., and Qing G. (2016) Polo-like kinase-1 regulates Myc stabilization and activates a feedforward circuit promoting tumor cell survival. Mol. Cell 64, 493–506 10.1016/j.molcel.2016.09.016 [DOI] [PubMed] [Google Scholar]
- 65. Brockmann M., Poon E., Berry T., Carstensen A., Deubzer H. E., Rycak L., Jamin Y., Thway K., Robinson S. P., Roels F., Witt O., Fischer M., Chesler L., and Eilers M. (2013) Small molecule inhibitors of aurora-A induce proteasomal degradation of N-myc in childhood neuroblastoma. Cancer Cell 24, 75–89 10.1016/j.ccr.2013.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mollaoglu G., Guthrie M. R., Böhm S., Brägelmann J., Can I., Ballieu P. M., Marx A., George J., Heinen C., Chalishazar M. D., Cheng H., Ireland A. S., Denning K. E., Mukhopadhyay A., Vahrenkamp J. M., et al. (2017) MYC drives progression of small cell lung cancer to a variant neuroendocrine subtype with vulnerability to Aurora kinase inhibition. Cancer Cell 31, 270–285 10.1016/j.ccell.2016.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Brockmann M., Poon E., Berry T., Carstensen A., Deubzer H. E., Rycak L., Jamin Y., Thway K., Robinson S. P., Roels F., Witt O., Fischer M., Chesler L., and Eilers M. (2016) Small molecule inhibitors of Aurora-A induce proteasomal degradation of N-Myc in childhood neuroblastoma. Cancer Cell 30, 357–358 10.1016/j.ccell.2016.07.002 [DOI] [PubMed] [Google Scholar]
- 68. Dauch D., Rudalska R., Cossa G., Nault J. C., Kang T. W., Wuestefeld T., Hohmeyer A., Imbeaud S., Yevsa T., Hoenicke L., Pantsar T., Bozko P., Malek N. P., Longerich T., Laufer S., et al. (2016) A MYC-Aurora kinase A protein complex represents an actionable drug target in p53-altered liver cancer. Nat. Med. 22, 744–753 10.1038/nm.4107 [DOI] [PubMed] [Google Scholar]
- 69. Gustafson W. C., Meyerowitz J. G., Nekritz E. A., Chen J., Benes C., Charron E., Simonds E. F., Seeger R., Matthay K. K., Hertz N. T., Eilers M., Shokat K. M., and Weiss W. A. (2014) Drugging MYCN through an allosteric transition in Aurora kinase A. Cancer Cell 26, 414–427 10.1016/j.ccr.2014.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gjertsen B. T., and Schöffski P. (2015) Discovery and development of the Polo-like kinase inhibitor volasertib in cancer therapy. Leukemia 29, 11–19 10.1038/leu.2014.222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sebastian M., Reck M., Waller C. F., Kortsik C., Frickhofen N., Schuler M., Fritsch H., Gaschler-Markefski B., Hanft G., Munzert G., and von Pawel J. (2010) The efficacy and safety of BI 2536, a novel Plk-1 inhibitor, in patients with stage IIIB/IV non-small cell lung cancer who had relapsed after, or failed, chemotherapy: results from an open-label, randomized phase II clinical trial. J. Thorac. Oncol. 5, 1060–1067 10.1097/JTO.0b013e3181d95dd4 [DOI] [PubMed] [Google Scholar]
- 72. Li B., and Simon M. C. (2013) Molecular pathways: targeting MYC-induced metabolic reprogramming and oncogenic stress in cancer. Clin. Cancer Res. 19, 5835–5841 10.1158/1078-0432.CCR-12-3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zirath H., Frenzel A., Oliynyk G., Segerström L., Westermark U. K., Larsson K., Munksgaard Persson M., Hultenby K., Lehtiö J., Einvik C., Påhlman S., Kogner P., Jakobsson P. J., and Henriksson M. A. (2013) MYC inhibition induces metabolic changes leading to accumulation of lipid droplets in tumor cells. Proc. Natl. Acad. Sci. U.S.A. 110, 10258–10263 10.1073/pnas.1222404110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang H., Teriete P., Hu A., Raveendra-Panickar D., Pendelton K., Lazo J. S., Eiseman J., Holien T., Misund K., Oliynyk G., Arsenian-Henriksson M., Cosford N. D., Sundan A., and Prochownik E. V. (2015) Direct inhibition of c-MYC–MAX heterodimers by celastrol and celastrol-inspired triterpenoids. Oncotarget 6, 32380–32395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Stellas D., Szabolcs M., Koul S., Li Z., Polyzos A., Anagnostopoulos C., Cournia Z., Tamvakopoulos C., Klinakis A., and Efstratiadis A. (2014) Therapeutic effects of an anti-Myc drug on mouse pancreatic cancer. J. Natl. Cancer Inst. 106, dju320 [DOI] [PubMed] [Google Scholar]
- 76. Jacob N. T., Miranda P. O., Shirey R. J., Gautam R., Zhou B., de Orbe Izquierdo M. E., Hixon M. S., Hart J. R., Ueno L., Vogt P. K., and Janda K. D. (2018) Synthetic molecules for disruption of the MYC protein-protein interface. Bioorg. Med. Chem. 26, 4234–4239 10.1016/j.bmc.2018.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jeong K. C., Ahn K. O., and Yang C. H. (2010) Small-molecule inhibitors of c-Myc transcriptional factor suppress proliferation and induce apoptosis of promyelocytic leukemia cell via cell cycle arrest. Mol. Biosyst. 6, 1503–1509 10.1039/c002534h [DOI] [PubMed] [Google Scholar]
- 78. Ruiz Garcia Y., Pabon-Martinez Y. V., Smith C. I. E., and Madder A. (2017) Specific dsDNA recognition by a mimic of the DNA binding domain of the c-Myc/Max transcription factor. Chem. Commun. 53, 6653–6656 10.1039/C7CC01705G [DOI] [PubMed] [Google Scholar]
- 79. Chan D. A., and Giaccia A. J. (2011) Harnessing synthetic lethal interactions in anticancer drug discovery. Nat. Rev. Drug Discov. 10, 351–364 10.1038/nrd3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kaelin W. G., Jr. (2005) The concept of synthetic lethality in the context of anticancer therapy. Nat. Rev. Cancer 5, 689–698 10.1038/nrc1691 [DOI] [PubMed] [Google Scholar]
- 81. O'Neil N. J., Bailey M. L., and Hieter P. (2017) Synthetic lethality and cancer. Nat. Rev. Genet. 18, 613–623 10.1038/nrg.2017.47 [DOI] [PubMed] [Google Scholar]
- 82. Cermelli S., Jang I. S., Bernard B., and Grandori C. (2014) Synthetic lethal screens as a means to understand and treat MYC-driven cancers. Cold Spring Harb. Perspect. Med. 4, a014209 10.1101/cshperspect.a014209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Li X., Zhang X. A., Li X., Xie W., and Huang S. (2015) MYC-mediated synthetic lethality for treating tumors. Curr. Cancer Drug Targets 15, 99–115 10.2174/1568009615666150121162921 [DOI] [PubMed] [Google Scholar]
- 84. Li X., Zhang X. A., Xie W., Li X., and Huang S. (2015) MYC-mediated synthetic lethality for treatment of hematological malignancies. Curr. Cancer Drug Targets 15, 53–70 10.2174/1568009615666150105120055 [DOI] [PubMed] [Google Scholar]
- 85. Cole K. A., Huggins J., Laquaglia M., Hulderman C. E., Russell M. R., Bosse K., Diskin S. J., Attiyeh E. F., Sennett R., Norris G., Laudenslager M., Wood A. C., Mayes P. A., Jagannathan J., Winter C., et al. (2011) RNAi screen of the protein kinome identifies checkpoint kinase 1 (CHK1) as a therapeutic target in neuroblastoma. Proc. Natl. Acad. Sci. U.S.A. 108, 3336–3341 10.1073/pnas.1012351108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ham J., Costa C., Sano R., Lochmann T. L., Sennott E. M., Patel N. U., Dastur A., Gomez-Caraballo M., Krytska K., Hata A. N., Floros K. V., Hughes M. T., Jakubik C. T., Heisey D. A., Ferrell J. T., et al. (2016) Exploitation of the apoptosis-primed state of MYCN-amplified neuroblastoma to develop a potent and specific targeted therapy combination. Cancer Cell 29, 159–172 10.1016/j.ccell.2016.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hsu T. Y., Simon L. M., Neill N. J., Marcotte R., Sayad A., Bland C. S., Echeverria G. V., Sun T., Kurley S. J., Tyagi S., Karlin K. L., Dominguez-Vidaña R., Hartman J. D., Renwick A., Scorsone K., et al. (2015) The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature 525, 384–388 10.1038/nature14985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Campaner S., Doni M., Hydbring P., Verrecchia A., Bianchi L., Sardella D., Schleker T., Perna D., Tronnersjö S., Murga M., Fernandez-Capetillo O., Barbacid M., Larsson L. G., and Amati B. (2010) Cdk2 suppresses cellular senescence induced by the c-myc oncogene. Nat. Cell Biol. 12, 54–59; sup pp 1–14 10.1038/ncb2004 [DOI] [PubMed] [Google Scholar]
- 89. Hydbring P., Bahram F., Su Y., Tronnersjö S., Högstrand K., von der Lehr N., Sharifi H. R., Lilischkis R., Hein N., Wu S., Vervoorts J., Henriksson M., Grandien A., Lüscher B., and Larsson L. G. (2010) Phosphorylation by Cdk2 is required for Myc to repress Ras-induced senescence in cotransformation. Proc. Natl. Acad. Sci. U.S.A. 107, 58–63 10.1073/pnas.0900121106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Molenaar J. J., Ebus M. E., Geerts D., Koster J., Lamers F., Valentijn L. J., Westerhout E. M., Versteeg R., and Caron H. N. (2009) Inactivation of CDK2 is synthetically lethal to MYCN over-expressing cancer cells. Proc. Natl. Acad. Sci. U.S.A. 106, 12968–12973 10.1073/pnas.0901418106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Tabib A., and Bachrach U. (1994) Activation of the proto-oncogene c-myc and c-fos by c-ras: involvement of polyamines. Biochem. Biophys. Res. Commun. 202, 720–727 10.1006/bbrc.1994.1990 [DOI] [PubMed] [Google Scholar]
- 92. Lewenhoeck D. A. (1677) Observationes D. Anthonii Lewenhoeck, de natis e semine genitali aminalculis. Phil. Trans. 12, 1040–1046 [Google Scholar]
- 93. Bachrach U. (2010) The early history of polyamine research. Plant Physiol. Biochem. 48, 490–495 10.1016/j.plaphy.2010.02.003 [DOI] [PubMed] [Google Scholar]
- 94. Cohen S. S. (1998) A Guide to the Polyamines, pp. 512–543, Oxford University Press, New York [Google Scholar]
- 95. Tabor C. W., and Tabor H. (1984) Polyamines. Annu. Rev. Biochem. 53, 749–790 10.1146/annurev.bi.53.070184.003533 [DOI] [PubMed] [Google Scholar]
- 96. Ames B. N., Dubin D. T., and Rosenthal S. M. (1958) Presence of polyamines in certain bacterial viruses. Science 127, 814–815 10.1126/science.127.3302.814-a [DOI] [PubMed] [Google Scholar]
- 97. Hershey A. D. (1957) Some minor components of bacteriophage T2 particles. Virology 4, 237–264 10.1016/0042-6822(57)90061-2 [DOI] [PubMed] [Google Scholar]
- 98. Liquori A. M., Costantino L., Crescenzi V., Elia V., Giglio E., Puliti R., De Santis Savino M., and Vitagliano V. (1967) Complexes between DNA and polyamines: a molecular model. J. Mol. Biol. 24, 113–122 10.1016/0022-2836(67)90094-0 [DOI] [Google Scholar]
- 99. Suwalsky M., Traub W., Shmueli U., and Subirana J. A. (1969) An X-ray study of the interaction of DNA with spermine. J. Mol. Biol. 42, 363–373 10.1016/0022-2836(69)90049-7 [DOI] [PubMed] [Google Scholar]
- 100. Cohen S. S., and Lichtenstein J. (1960) Polyamines and ribosome structure. J. Biol. Chem. 235, 2112–2116 [PubMed] [Google Scholar]
- 101. Cohen S. S., Morgan S., and Streibel E. (1969) The polyamine content of the tRNA of E. coli. Proc. Natl. Acad. Sci. U.S.A. 64, 669–676 10.1073/pnas.64.2.669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Hobbs C. A., and Gilmour S. K. (2006) in Polyamine Cell Signaling (Wang J.-Y., and Casero R. A. Jr., eds) pp. 75–89, Humana Press, Totowa, NJ [Google Scholar]
- 103. Matthews H. R. (1993) Polyamines, chromatin structure and transcription. Bioessays 15, 561–566 10.1002/bies.950150811 [DOI] [PubMed] [Google Scholar]
- 104. Williams K. (1997) Interactions of polyamines with ion channels. Biochem. J. 325, 289–297 10.1042/bj3250289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Childs A. C., Mehta D. J., and Gerner E. W. (2003) Polyamine-dependent gene expression. Cell. Mol. Life Sci. 60, 1394–1406 10.1007/s00018-003-2332-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Auvinen M., Järvinen K., Hotti A., Okkeri J., Laitinen J., Jänne O. A., Coffino P., Bergman M., Andersson L. C., Alitalo K., and Hölttä E. (2003) Transcriptional regulation of the ornithine decarboxylase gene by c-Myc/Max/Mad network and retinoblastoma protein interacting with c-Myc. Int. J. Biochem. Cell Biol. 35, 496–521 10.1016/S1357-2725(02)00305-9 [DOI] [PubMed] [Google Scholar]
- 107. Auvinen M., Paasinen A., Andersson L. C., and Hölttä E. (1992) Ornithine decarboxylase activity is critical for cell transformation. Nature 360, 355–358 10.1038/360355a0 [DOI] [PubMed] [Google Scholar]
- 108. Pegg A. E. (2016) Functions of polyamines in mammals. J. Biol. Chem. 291, 14904–14912 10.1074/jbc.R116.731661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Shantz L. M., and Pegg A. E. (1999) Translational regulation of ornithine decarboxylase and other enzymes of the polyamine pathway. Int. J. Biochem. Cell Biol. 31, 107–122 10.1016/S1357-2725(98)00135-6 [DOI] [PubMed] [Google Scholar]
- 110. Madeo F., Eisenberg T., Pietrocola F., and Kroemer G. (2018) Spermidine in health and disease. Science 359, eaan2788 10.1126/science.aan2788 [DOI] [PubMed] [Google Scholar]
- 111. Casero R. A. Jr., and Marton L. J. (2007) Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat. Rev. Drug Discov. 6, 373–390 10.1038/nrd2243 [DOI] [PubMed] [Google Scholar]
- 112. Murray-Stewart T. R., Woster P. M., and Casero R. A. Jr. (2016) Targeting polyamine metabolism for cancer therapy and prevention. Biochem. J. 473, 2937–2953 10.1042/BCJ20160383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wallace H. M., Fraser A. V., and Hughes A. (2003) A perspective of polyamine metabolism. Biochem. J. 376, 1–14 10.1042/bj20031327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Casero R. A. Jr, Murray Stewart T., and Pegg A. E. (2018) Polyamine metabolism and cancer: treatments, challenges and opportunities. Nat. Rev. Cancer 18, 681–695 10.1038/s41568-018-0050-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Matsufuji S., Matsufuji T., Miyazaki Y., Murakami Y., Atkins J. F., Gesteland R. F., and Hayashi S. (1995) Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell 80, 51–60 10.1016/0092-8674(95)90450-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Pegg A. E. (2006) Regulation of ornithine decarboxylase. J. Biol. Chem. 281, 14529–14532 10.1074/jbc.R500031200 [DOI] [PubMed] [Google Scholar]
- 117. Li X., and Coffino P. (1993) Degradation of ornithine decarboxylase: exposure of the C-terminal target by a polyamine-inducible inhibitory protein. Mol. Cell. Biol. 13, 2377–2383 10.1128/MCB.13.4.2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Zhang M., Pickart C. M., and Coffino P. (2003) Determinants of proteasome recognition of ornithine decarboxylase, a ubiquitin-independent substrate. EMBO J. 22, 1488–1496 10.1093/emboj/cdg158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Coffino P. (2001) Antizyme, a mediator of ubiquitin-independent proteasomal degradation. Biochimie 83, 319–323 10.1016/S0300-9084(01)01252-4 [DOI] [PubMed] [Google Scholar]
- 120. Coffino P. (2001) Regulation of cellular polyamines by antizyme. Nat. Rev. Mol. Cell Biol. 2, 188–194 10.1038/35056508 [DOI] [PubMed] [Google Scholar]
- 121. Mangold U. (2005) The antizyme family: polyamines and beyond. IUBMB Life 57, 671–676 10.1080/15216540500307031 [DOI] [PubMed] [Google Scholar]
- 122. Albeck S., Dym O., Unger T., Snapir Z., Bercovich Z., and Kahana C. (2008) Crystallographic and biochemical studies revealing the structural basis for antizyme inhibitor function. Protein Sci. 17, 793–802 10.1110/ps.073427208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Mangold U. (2006) Antizyme inhibitor: mysterious modulator of cell proliferation. Cell. Mol. Life Sci. 63, 2095–2101 10.1007/s00018-005-5583-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Chidley C., Haruki H., Pedersen M. G., Muller E., and Johnsson K. (2011) A yeast-based screen reveals that sulfasalazine inhibits tetrahydrobiopterin biosynthesis. Nat. Chem. Biol. 7, 375–383 10.1038/nchembio.557 [DOI] [PubMed] [Google Scholar]
- 125. Guillem J. G., Levy M. F., Hsieh L. L., Johnson M. D., LoGerfo P., Forde K. A., and Weinstein I. B. (1990) Increased levels of phorbin, c-myc, and ornithine decarboxylase RNAs in human colon cancer. Mol. Carcinog. 3, 68–74 10.1002/mc.2940030204 [DOI] [PubMed] [Google Scholar]
- 126. Morris D. R., Allen M. L., Rabinovitch P. S., Kuepfer C. A., and White M. W. (1988) Mitogenic signaling pathways regulating expression of c-myc and ornithine decarboxylase genes in bovine T-lymphocytes. Biochemistry 27, 8689–8693 10.1021/bi00423a027 [DOI] [PubMed] [Google Scholar]
- 127. Stimac E., and Morris D. R. (1987) Messenger RNAs coding for enzymes of polyamine biosynthesis are induced during the G0-G1 transition but not during traverse of the normal G1 phase. J. Cell Physiol. 133, 590–594 10.1002/jcp.1041330323 [DOI] [PubMed] [Google Scholar]
- 128. Tonin P. N., Yeger H., Stallings R. L., Srinivasan P. R., and Lewis W. H. (1989) Amplification of N-myc and ornithine decarboxylase genes in human neuroblastoma and hydroxyurea-resistant hamster cell lines. Oncogene 4, 1117–1121 [PubMed] [Google Scholar]
- 129. George R. E., Kenyon R. M., McGuckin A. G., Malcolm A. J., Pearson A. D., and Lunec J. (1996) Investigation of co-amplification of the candidate genes ornithine decarboxylase, ribonucleotide reductase, syndecan-1 and a DEAD box gene, DDX1, with N-myc in neuroblastoma. United Kingdom Children's Cancer Study Group. Oncogene 12, 1583–1587 [PubMed] [Google Scholar]
- 130. Evageliou N. F., Haber M., Vu A., Laetsch T. W., Murray J., Gamble L. D., Cheng N. C., Liu K., Reese M., Corrigan K. A., Ziegler D. S., Webber H., Hayes C. S., Pawel B., Marshall G. M., et al. (2016) Polyamine antagonist therapies inhibit neuroblastoma initiation and progression. Clin. Cancer Res. 22, 4391–4404 10.1158/1078-0432.CCR-15-2539 [DOI] [PubMed] [Google Scholar]
- 131. Hogarty M. D., Norris M. D., Davis K., Liu X., Evageliou N. F., Hayes C. S., Pawel B., Guo R., Zhao H., Sekyere E., Keating J., Thomas W., Cheng N. C., Murray J., Smith J., et al. (2008) ODC1 is a critical determinant of MYCN oncogenesis and a therapeutic target in neuroblastoma. Cancer Res. 68, 9735–9745 10.1158/0008-5472.CAN-07-6866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Nilsson J. A., Keller U. B., Baudino T. A., Yang C., Norton S., Old J. A., Nilsson L. M., Neale G., Kramer D. L., Porter C. W., and Cleveland J. L. (2005) Targeting ornithine decarboxylase in Myc-induced lymphomagenesis prevents tumor formation. Cancer Cell 7, 433–444 10.1016/j.ccr.2005.03.036 [DOI] [PubMed] [Google Scholar]
- 133. Zell J. A., Ziogas A., Ignatenko N., Honda J., Qu N., Bobbs A. S., Neuhausen S. L., Gerner E. W., and Anton-Culver H. (2009) Associations of a polymorphism in the ornithine decarboxylase gene with colorectal cancer survival. Clin. Cancer Res. 15, 6208–6216 10.1158/1078-0432.CCR-09-0592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Brodeur G. M. (2003) Neuroblastoma: biological insights into a clinical enigma. Nat. Rev. Cancer 3, 203–216 10.1038/nrc1014 [DOI] [PubMed] [Google Scholar]
- 135. Weicht R. R., Schultz C. R., Geerts D., Uhl K. L., and Bachmann A. S. (2018) Polyamine biosynthetic pathway as a drug target for osteosarcoma therapy. Med. Sci. 6, E65 10.3390/medsci6030065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Kim H. I., Schultz C. R., Buras A. L., Friedman E., Fedorko A., Seamon L., Chandramouli G. V. R., Maxwell G. L., Bachmann A. S., and Risinger J. I. (2017) Ornithine decarboxylase as a therapeutic target for endometrial cancer. PLoS ONE 12, e0189044 10.1371/journal.pone.0189044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Rounbehler R. J., Li W., Hall M. A., Yang C., Fallahi M., and Cleveland J. L. (2009) Targeting ornithine decarboxylase impairs development of MYCN-amplified neuroblastoma. Cancer Res. 69, 547–553 10.1158/0008-5472.CAN-08-2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Bassiri H., Benavides A., Haber M., Gilmour S. K., Norris M. D., and Hogarty M. D. (2015) Translational development of difluoromethylornithine (DFMO) for the treatment of neuroblastoma. Transl. Pediatr. 4, 226–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Evageliou N. F., and Hogarty M. D. (2009) Disrupting polyamine homeostasis as a therapeutic strategy for neuroblastoma. Clin. Cancer Res. 15, 5956–5961 10.1158/1078-0432.CCR-08-3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Gamble L. D., Hogarty M. D., Liu X., Ziegler D. S., Marshall G., Norris M. D., and Haber M. (2012) Polyamine pathway inhibition as a novel therapeutic approach to treating neuroblastoma. Front. Oncol. 2, 162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Gerner E. W., and Meyskens F. L. Jr. (2004) Polyamines and cancer: old molecules, new understanding. Nat. Rev. Cancer 4, 781–792 10.1038/nrc1454 [DOI] [PubMed] [Google Scholar]
- 142. Arruabarrena-Aristorena A., Zabala-Letona A., and Carracedo A. (2018) Oil for the cancer engine: the cross-talk between oncogenic signaling and polyamine metabolism. Sci. Adv. 4, eaar2606 10.1126/sciadv.aar2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Wallace H. M. (2007) Targeting polyamine metabolism: a viable therapeutic/preventative solution for cancer? Expert Opin. Pharmacother. 8, 2109–2116 10.1517/14656566.8.13.2109 [DOI] [PubMed] [Google Scholar]
- 144. Raul F. (2007) Revival of 2-(difluoromethyl)ornithine (DFMO), an inhibitor of polyamine biosynthesis, as a cancer chemopreventive agent. Biochem. Soc. Trans. 35, 353–355 10.1042/BST0350353 [DOI] [PubMed] [Google Scholar]
- 145. Dwivedi R. S., Wang L. J., and Mirkin B. L. (1999) S-Adenosylmethionine synthetase is overexpressed in murine neuroblastoma cells resistant to nucleoside analogue inhibitors of S-adenosylhomocysteine hydrolase: a novel mechanism of drug resistance. Cancer Res. 59, 1852–1856 [PubMed] [Google Scholar]
- 146. Fernandez P. C., Frank S. R., Wang L., Schroeder M., Liu S., Greene J., Cocito A., and Amati B. (2003) Genomic targets of the human c-Myc protein. Genes Dev. 17, 1115–1129 10.1101/gad.1067003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Nakanishi S., and Cleveland J. L. (2016) Targeting the polyamine-hypusine circuit for the prevention and treatment of cancer. Amino Acids 48, 2353–2362 10.1007/s00726-016-2275-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Paz E. A., LaFleur B., and Gerner E. W. (2014) Polyamines are oncometabolites that regulate the LIN28/let-7 pathway in colorectal cancer cells. Mol. Carcinog. 53, Suppl. 1, E96–E106 10.1002/mc.22051 [DOI] [PubMed] [Google Scholar]
- 149. Lozier A. M., Rich M. E., Grawe A. P., Peck A. S., Zhao P., Chang A. T., Bond J. P., and Sholler G. S. (2015) Targeting ornithine decarboxylase reverses the LIN28/Let-7 axis and inhibits glycolytic metabolism in neuroblastoma. Oncotarget 6, 196–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Molenaar J. J., Domingo-Fernández R., Ebus M. E., Lindner S., Koster J., Drabek K., Mestdagh P., van Sluis P., Valentijn L. J., van Nes J., Broekmans M., Haneveld F., Volckmann R., Bray I., Heukamp L., et al. (2012) LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat. Genet. 44, 1199–1206 10.1038/ng.2436 [DOI] [PubMed] [Google Scholar]
- 151. Alexander E. T., Minton A., Peters M. C., Phanstiel O. 4th., and Gilmour S. K. (2017) A novel polyamine blockade therapy activates an anti-tumor immune response. Oncotarget 8, 84140–84152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Casey S. C., Baylot V., and Felsher D. W. (2017) MYC: master regulator of immune privilege. Trends Immunol. 38, 298–305 10.1016/j.it.2017.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Hackett C. S., Quigley D. A., Wong R. A., Chen J., Cheng C., Song Y. K., Wei J. S., Pawlikowska L., Bao Y., Goldenberg D. D., Nguyen K., Gustafson W. C., Rallapalli S. K., Cho Y. J., Cook J. M., et al. (2014) Expression quantitative trait loci and receptor pharmacology implicate Arg1 and the GABA-A receptor as therapeutic targets in neuroblastoma. Cell Rep. 9, 1034–1046 10.1016/j.celrep.2014.09.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Mussai F., Egan S., Hunter S., Webber H., Fisher J., Wheat R., McConville C., Sbirkov Y., Wheeler K., Bendle G., Petrie K., Anderson J., Chesler L., and De Santo C. (2015) Neuroblastoma arginase activity creates an immunosuppressive microenvironment that impairs autologous and engineered immunity. Cancer Res. 75, 3043–3053 10.1158/0008-5472.CAN-14-3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Murai N., Murakami Y., Tajima A., and Matsufuji S. (2018) Novel ubiquitin-independent nucleolar c-Myc degradation pathway mediated by antizyme 2. Sci. Rep. 8, 3005 10.1038/s41598-018-21189-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Ruiz-Pérez M. V., Medina M. Á., Urdiales J. L., Keinänen T. A., and Sánchez-Jiménez F. (2015) Polyamine metabolism is sensitive to glycolysis inhibition in human neuroblastoma cells. J. Biol. Chem. 290, 6106–6119 10.1074/jbc.M114.619197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Wise D. R., DeBerardinis R. J., Mancuso A., Sayed N., Zhang X. Y., Pfeiffer H. K., Nissim I., Daikhin E., Yudkoff M., McMahon S. B., and Thompson C. B. (2008) Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. U.S.A. 105, 18782–18787 10.1073/pnas.0810199105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Wang R., Dillon C. P., Shi L. Z., Milasta S., Carter R., Finkelstein D., McCormick L. L., Fitzgerald P., Chi H., Munger J., and Green D. R. (2011) The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882 10.1016/j.immuni.2011.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Warburg O. (1956) On the origin of cancer cells. Science 123, 309–314 10.1126/science.123.3191.309 [DOI] [PubMed] [Google Scholar]
- 160. Warburg O., Posener K., and Negelein E. (1924) Ueber den Stoffwechsel der Tumoren. Biochem. Z. 152, 319–344 [Google Scholar]
- 161. Galluzzi L., Pietrocola F., and Kroemer G. (2015) Molecular regulation of circadian rhythms by polyamines. Cell Metab. 22, 757–758 10.1016/j.cmet.2015.10.007 [DOI] [PubMed] [Google Scholar]
- 162. Zwighaft Z., Aviram R., Shalev M., Rousso-Noori L., Kraut-Cohen J., Golik M., Brandis A., Reinke H., Aharoni A., Kahana C., and Asher G. (2015) Circadian clock control by polyamine levels through a mechanism that declines with age. Cell Metab. 22, 874–885 10.1016/j.cmet.2015.09.011 [DOI] [PubMed] [Google Scholar]
- 163. Altman B. J., Hsieh A. L., Sengupta A., Krishnanaiah S. Y., Stine Z. E., Walton Z. E., Gouw A. M., Venkataraman A., Li B., Goraksha-Hicks P., Diskin S. J., Bellovin D. I., Simon M. C., Rathmell J. C., Lazar M. A., et al. (2015) MYC disrupts the circadian clock and metabolism in cancer cells. Cell Metab. 22, 1009–1019 10.1016/j.cmet.2015.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Söderstjerna E., Holst C. M., Alm K., and Oredsson S. M. (2010) Apoptosis induced by the potential chemotherapeutic drug N1,N11-diethylnorspermine in a neuroblastoma cell line. Anticancer Drugs 21, 917–926 10.1097/CAD.0b013e32833d1cae [DOI] [PubMed] [Google Scholar]
- 165. Smith M. A., Maris J. M., Lock R., Kolb E. A., Gorlick R., Keir S. T., Carol H., Morton C. L., Reynolds C. P., Kang M. H., and Houghton P. J. (2011) Initial testing (stage 1) of the polyamine analog PG11047 by the pediatric preclinical testing program. Pediatr. Blood Cancer 57, 268–274 10.1002/pbc.22797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Zhang W., Yu Y., Hertwig F., Thierry-Mieg J., Zhang W., Thierry-Mieg D., Wang J., Furlanello C., Devanarayan V., Cheng J., Deng Y., Hero B., Hong H., Jia M., Li L., et al. (2015) Comparison of RNA-seq and microarray-based models for clinical endpoint prediction. Genome Biol. 16, 133 10.1186/s13059-015-0694-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., Jacobsen A., Byrne C. J., Heuer M. L., Larsson E., Antipin Y., Reva B., Goldberg A. P., Sander C., and Schultz N. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Gao J., Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., Sumer S. O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., and Schultz N. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Rhodes D. R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., and Chinnaiyan A. M. (2004) ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Bupp C. P., Schultz C. R., Uhl K. L., Rajasekaran S., and Bachmann A. S. (2018) Novel de novo pathogenic variant in the ODC1 gene in a girl with developmental delay, alopecia, and dysmorphic features. Am. J. Med. Genet. A 10.1002/ajmg.a.40523 10.1002/ajmg.a.40523 [DOI] [PubMed] [Google Scholar]