ABSTRACT
Background
It is unknown whether dietary quality modifies genetic association with body mass index (BMI).
Objective
This study examined whether dietary quality modifies genetic association with BMI.
Design
We calculated 3 diet quality scores including the Alternative Healthy Eating Index 2010 (AHEI-2010), the Alternative Mediterranean Diet score (AMED), and the Dietary Approach to Stop Hypertension (DASH) diet score. We examined the interactions of a genetic risk score (GRS) based on 97 BMI-associated variants with the 3 diet quality scores on BMI in 30,904 participants from 3 large cohorts.
Results
We found significant interactions between total GRS and all 3 diet scores on BMI assessed after 2–3 y, with an attenuated genetic effect observed in individuals with healthier diets (AHEI: P-interaction = 0.003; AMED: P = 0.001; DASH: P = 0.004). For example, the difference in BMI (kg/m2) per 10-unit increment of the GRS was smaller among participants in the highest tertile of AHEI score compared with those in the lowest tertile (0.84; 95% CI: 0.72, 0.96 compared with 1.14; 95% CI: 0.99, 1.29). Results were consistent across the 3 cohorts with no significant heterogeneity. The interactions with diet scores on BMI appeared more significant for central nervous system GRSs (P < 0.01 for 3 diet scores) than for non–central nervous system GRSs (P > 0.05 for 3 diet scores).
Conclusions
A higher diet quality attenuated genetic predisposition to obesity. These findings underscore the importance of maintaining a healthful diet for the prevention of obesity, particularly for those individuals with a strong genetic predisposition to obesity. This trial was registered with the Clinical Trial Registry as NCT03577639.
Keywords: body mass index, genetic risk score, diet scores
INTRODUCTION
Genetic factors have been shown to play a role in the development of obesity (1–3). To date, based on genome-wide association studies, 97 loci have been identified as associated with BMI (3). A recent study suggested that the genetic association with BMI could be intensified by an obesogenic environment (4), but specific factors that interact with genetic variants remain largely unknown. Therefore, it is critical to identify those environmental factors, particularly diet, which might modify genetic associations with BMI. Moreover, identification of gene-diet interactions may provide further evidence for benefits of maintaining a healthful diet, especially among those with a strong genetic preposition to obesity.
A few studies have examined interactions of food and nutrient intake with genetic variants in relation to BMI. Studies found that a higher consumption of sugar-sweetened beverages (SSBs) and fried foods was associated with a more pronounced genetic preposition to higher BMI in the Nurses’ Health Study (NHS), the Health Professional Follow-up Study (HPFS), and the Women's Genome Health Study (WGHS) (5, 6). However, nutrients and foods are not consumed in isolation, but are distributed across correlated food networks with complex consumption patterns. It remains unclear whether an overall diet pattern, which represents a broader picture of food and nutrient consumption and captures the complex nature of diet, may modify genetic associations with BMI (7). One previous study found a significant interaction between a diet score and FTO variant on BMI (8), whereas another study did not find any significant interaction between a healthful diet and a genetic risk score (GRS) based on 32 variants on BMI (9). These studies might be limited by cross-sectional study design and measurement error in diet quality assessment resulting from insufficient food components. Furthermore, although the biological functions of most BMI loci are not clearly known, more than half of these loci harbor genes that are highly expressed, or known to function, in the central nervous system (CNS) (3), the key site of central appetite regulation. Given that excessive food and energy intake are the leading risk factors for obesity (10), we classified the SNPs into CNS and non-CNS subgroups based on the potential biological role of nearby genes in the CNS.
Three diet quality scores [the alternative Healthy Eating Index (AHEI) (11), the Alternative Mediterranean Diet score (AMED) (12), and the Dietary Approach to Stop Hypertension (DASH) diet score (13)] have been widely used to measure adherence to a certain dietary pattern. In this study, we examined interactions of AHEI, AMED, and DASH with a GRS based on 97 BMI-associated single-nucleotide polymorphisms (SNPs) (3). We further classified the SNPs into CNS and non-CNS subgroups and examined the effect of their interactions with diet quality scores on BMI.
METHODS
Study population
The NHS began in 1976, and was composed of 121,700 female registered nurses aged 30–55 y. The HPFS was initiated in 1986, and was composed of 51,529 male professionals aged 40–75 y. The loss to follow-up rate of both cohorts was <10%. The current analysis included 5730 women and 3588 men of European ancestry whose genotype data were available. Those participants were sampled as controls in 16 case-control data sets, and a flow diagram of the sample selection process is shown in Supplemental Figure 1A and B.
The WGHS is a prospective cohort study of 28,345 initially healthy US women. Study participants were health professionals aged ≥45 y, and free of cardiovascular disease and cancer at baseline from 1992 to 1995 (14). In our study, we included 21,740 participants of confirmed self-reported European ancestry. A flow diagram of sample selection is shown in Supplemental Figure 1C.
The study protocol was approved by the institutional review boards of Brigham and Women's Hospital and Harvard School of Public Health. This study was registered at clinicaltrials.gov as NCT03577639.
Assessment of diet quality
Dietary data were collected from the NHS in 1984 and from the HPFS in 1986. Since 1986, dietary data were collected every 4 y thereafter until 2008 in both cohorts. Dietary data were collected from the WGHS only at baseline. For the 3 cohorts, we used a 131-item food-frequency questionnaire to obtain information on usual intake of food and beverages. The validity and reproducibility of this questionnaire have been described in detail elsewhere (15–18).
Scoring for the AHEI-2010 was based on intake levels of 11 components, including fruit, vegetables, whole grains, long-chain n–3 fats, nuts and legumes, polyunsaturated fatty acids, sugar-sweetened beverages, alcohol, red and processed meat, trans fat, and sodium. The components were chosen on the basis of their association with chronic disease and mortality risk in observational and interventional studies (11). The total score ranged from 0 (nonadherence) to 110 (perfect adherence). The AMED score was modified and adapted to the Mediterranean diet scale, and included 9 food items, namely fruits, vegetables, whole grains, fish, nuts, legumes, red and processed meat, alcohol consumption, and monounsaturated fat-to-saturated fat ratios (12). The total score ranged from 0 to 9 in the NHS and the HPFS, and from 0 to 55 in the WGHS, with a higher score representing closer adherence to the Mediterranean diet. The DASH score was developed based on foods and nutrients emphasized in the DASH diet and focused on 8 components, namely vegetables, fruits, nuts and legumes, whole grains, low-fat dairy products, SSBs, sodium, and red and processed meat. The total score for the DASH diet ranged from 8 to 40 points (13). For each diet score, a detailed description of component selection and score calculation can be found elsewhere (11–13), and are shown in the Supplemental Methods.
Assessment of BMI
In the NHS and HPFS, height and weight were assessed by questionnaire at baseline, and weight was collected by questionnaire every other year. Self-reported weight was highly correlated with measured weight (r = 0.97 for men and women) (19). In the WGHS, weight and height were assessed at baseline and 36 mo by self-reported questionnaires. BMI was calculated as weight (kg) divided by height squared (m2). We defined obesity as BMI ≥30 kg/m2.
Assessment of covariates
In the questionnaires at baseline in the NHS, the HPFS, and the WGHS, information was collected on age, smoking status, physical activity, and self-reported diagnosis of hypertension and hypercholesterolemia. In the NHS and the HPFS, the information was updated in the biennial follow-up questionnaires. In the WGHS, we used EIGENSTRAT analysis to calculate eigenvectors accounting for population structure (20).
Genotyping and classification of SNPs
We selected 97 SNPs that were known to be associated with BMI. Of the 97 SNPs, 77 SNPs were identified in Caucasians and 20 SNPs were identified in mixed populations including 95% Caucasians (3). SNP genotyping and imputation have been described in detail elsewhere (21). Most of the SNPs were genotyped (sample call rate = 97%) or had a high imputation quality score (r2 ≥ 0.8), as assessed with the use of MACH software. In the WGHS, 1000 genomes version 1 phase 3 was used as reference panel for imputation (sample call rate = 97%). Two SNPs were excluded due to missing (rs12016871) and low imputation quality (rs2245368: r2 = 0.01).
CNS-related SNPs were identified if biological functions of the nearest gene of those BMI-associated loci were in the following categories suggested by Locke et al (3): 1) neuronal developmental process, 2) neurotransmission, 3) hypothalamic expression and regulatory function, and 4) neuronal expression (Supplemental Table 1). In total, 54 out of the 97 SNPs were classified as CNS SNPs.
Genetic risk score
The total, CNS, and non-CNS GRSs were calculated as the weighted average of the 97 SNPs, 54 CNS GRS, and 43 non-CNS GRSs, respectively. Each SNP was weighted proportionally to the magnitude of the per-allele-coefficient extracted from the most recent genome-wide association study on BMI (3). The weights of total, CNS, and non-CNS SNPs were rescaled to sum to 194, 108, and 86, respectively. The total, CNS, and non-CNS scores were in the ranges 0–194, 0–108, and 0–86, with a higher score indicating greater genetic predisposition to obesity.
Statistical analysis
To examine interactions between total GRS and diet scores with BMI, generalized estimation equations were used in the NHS and the HPFS, and a linear model was used in the WGHS as diet was only assessed at baseline. To minimize reverse causation, we used BMI data measured 2–4 y later than assessment of dietary intake in the NHS and the HPFS, and BMI measured at 36 mo after assessment of diet in the WGHS as outcome. We assumed additive interaction between diet quality and GRS on BMI, and interaction was tested using the Wald test by including an interaction term (e.g., AHEI × GRS) into the model, with a P value of the interaction term <0.05 indicating significant interaction. We further conducted stratified analyses to examine associations of total GRS with BMI by tertiles of diet scores. We adjusted for age, level of physical activity, smoking status, total energy intake, and history of hypertension and hypercholesterolemia. We further adjusted for case-control data sets in the NHS and the HPFS, and geographic region and population structure in the WGHS. The findings across cohorts were pooled using fixed-effects models. Statistical heterogeneity across studies was assessed by Cochrane Q test, with P < 0.1 indicating significant between-study heterogeneity. Similar analyses were conducted to examine interactions of CNS GRS and non-CNS GRS with individual food components in relation to BMI in the 3 cohorts.
To obtain predicted BMI from total GRS and tertiles of diet score, we fitted 2 models with and without the interaction term as described above. From each model, we obtained predicted BMI at 10 consecutive points of total GRS between 60 and 110 within each tertile of diet score using median value of covariates among total participants. We pooled predicted BMI across the 3 cohorts using a fixed-effects model. Associations of predicted BMI with total GRS were plotted with the use of the “plot” command in R.
We conducted stratified analyses by age, sex, smoking status, and physical activity levels, as those variables might modify the interaction between genetic score and diet quality on BMI. We further examined interactions between individual SNPs and diet score and food components on BMI, and we used Bonferroni correction to adjust for multiple testing (critical P value after Bonferroni correction = 0.05/[97 × (3 + 14)] = 3.0 × 10−5, where 97 was the number of SNPs, 3 was the number of diet scores, and 14 was the number of food components). All statistical tests were 2-sided with P < 0.05 indicating significance, and were performed with SAS version 9.4 (SAS Institute) and R version 3.2.0 (R Foundation).
RESULTS
Baseline characteristics of the 3 cohorts
Participants with higher diet quality scores had lower BMIs and higher levels of physical activity compared with those who had lower diet scores across the 3 cohorts (Table 1). AHEI, AMED, and DASH scores were correlated with each other. Each diet score was significantly and positively correlated with healthy food components, and negatively correlated with unhealthy components (Supplemental Table 2).
TABLE 1.
Baseline characteristics of participants in the 3 cohorts according to tertiles of AHEI, AMED, and DASH1
| AHEI | AMED | DASH | |||||||
|---|---|---|---|---|---|---|---|---|---|
| T1 | T2 | T3 | T1 | T2 | T3 | T1 | T2 | T3 | |
| NHS | |||||||||
| Diet score | 37.0 ± 4.7 | 47.8 ± 2.7 | 60.9 ± 6.8 | 2.2 ± 0.8 | 4.0 ± 0 | 5.9 ± 0.9 | 18.1 ± 2.5 | 23.5 ± 1.1 | 28.6 ± 2.2 |
| Participants, n | 1832 | 1888 | 2010 | 2243 | 1120 | 2367 | 1970 | 1761 | 1999 |
| Age, y | 50.7 ± 6.7 | 51.9 ± 6.7 | 53.5 ± 6.3 | 51.0 ± 6.7 | 52.0 ± 6.7 | 53.1 ± 6.5 | 50.2 ± 6.6 | 51.9 ± 6.6 | 54.0 ± 6.2 |
| BMI, kg/m2 | 25.1 ± 4.8 | 25.0 ± 4.4 | 24.5 ± 4.1 | 25.0 ± 4.7 | 25.0 ± 4.6 | 24.7 ± 4.1 | 25.0 ± 4.6 | 25.0 ± 4.5 | 24.6 ± 4.2 |
| Physical activity, MET-h/wk | 10.4 ± 12.6 | 14.2 ± 16.7 | 18.2 ± 24.0 | 11.4 ± 14.7 | 14.3 ± 17.7 | 17.2 ± 22.2 | 10.6 ± 13.4 | 14.2 ± 17.0 | 18.3 ± 23.6 |
| Total energy intake, kcal/d | 1900 ± 499 | 1720 ± 506 | 1625 ± 500 | 1593 ± 469 | 1715 ± 509 | 1900 ± 514 | 1738 ± 525 | 1723 ± 526 | 1769 ± 492 |
| Total GRS | 88.1 ± 6.2 | 88.3 ± 6.0 | 88.6 ± 6.4 | 88.2 ± 6.2 | 88.4 ± 6.4 | 88.5 ± 6.2 | 88.1 ± 6.3 | 88.3 ± 6.2 | 88.5 ± 6.2 |
| CNS GRS | 58.2 ± 4.9 | 58.5 ± 4.9 | 58.6 ± 5.1 | 58.2 ± 4.9 | 58.4 ± 5.1 | 58.6 ± 5.0 | 58.2 ± 5.0 | 58.4 ± 4.9 | 58.7 ± 5.1 |
| Non-CNS GRS | 30.1 ± 3.6 | 30.1 ± 3.7 | 30.2 ± 3.5 | 30.2 ± 3.7 | 30.1 ± 3.6 | 30.1 ± 3.5 | 30.2 ± 3.6 | 30.1 ± 3.6 | 30.1 ± 3.6 |
| Never smoker, % | 47.7 | 47.5 | 42.0 | 43.9 | 44.5 | 47.7 | 42.7 | 46.3 | 47.9 |
| Hypertension, % | 6.0 | 8.2 | 7.2 | 6.4 | 7.1 | 7.8 | 5.9 | 6.8 | 8.7 |
| Hypercholesterolemia, % | 2.7 | 3.8 | 5.5 | 3.1 | 3.8 | 5.1 | 2.7 | 4.1 | 5.3 |
| HPFS | |||||||||
| Diet score | 40.2 ± 5.3 | 52.3 ± 3.1 | 66.2 ± 6.6 | 2.2 ± 0.9 | 4.5 ± 0.5 | 6.7 ± 0.8 | 18.1 ± 2.5 | 24.0 ± 1.4 | 30.2 ± 2.7 |
| Participants, n | 1172 | 1164 | 1252 | 1176 | 1295 | 1117 | 1063 | 1236 | 1289 |
| Age, y | 53.3 ± 8.5 | 54.8 ± 8.7 | 56.2 ± 8.3 | 53.4 ± 8.7 | 54.9 ± 8.3 | 56.3 ± 8.6 | 52.9 ± 8.6 | 54.9 ± 8.3 | 56.4 ± 8.6 |
| BMI, kg/m2 | 25.7 ± 3.2 | 25.2 ± 2.8 | 25.0 ± 2.7 | 25.7 ± 3.2 | 25.3 ± 2.8 | 24.8 ± 2.7 | 25.7 ± 3.1 | 25.4 ± 3.0 | 24.8 ± 2.7 |
| Physical activity, MET-h/wk | 17.1 ± 24.9 | 20.4 ± 26.7 | 26.1 ± 27.4 | 17.9 ± 26.0 | 20.4 ± 26.0 | 25.9 ± 27.4 | 16.5 ± 24.4 | 19.8 ± 22.8 | 26.7 ± 30.6 |
| Total energy intake, kcal/d | 2136 ± 603 | 2008 ± 612 | 1938 ± 581 | 1846 ± 546 | 2030 ± 610 | 2208 ± 599 | 1889 ± 577 | 2020 ± 605 | 2143 ± 601 |
| Total GRS | 88.0 ± 6.4 | 88.3 ± 6.3 | 88.1 ± 6.4 | 88.2 ± 6.3 | 88.4 ± 6.4 | 87.8 ± 6.4 | 88.2 ± 6.2 | 88.1 ± 6.5 | 88.2 ± 6.3 |
| CNS GRS | 58.3 ± 5.1 | 58.4 ± 5.0 | 58.3 ± 5.1 | 58.4 ± 5.1 | 58.5 ± 5.1 | 58.1 ± 5.0 | 58.3 ± 5.0 | 58.3 ± 5.1 | 58.4 ± 5.0 |
| Non-CNS GRS | 30.0 ± 3.6 | 30.1 ± 3.6 | 30.0 ± 3.8 | 30.0 ± 3.6 | 30.1 ± 3.6 | 29.9 ± 3.7 | 30.0 ± 3.5 | 30.0 ± 3.7 | 30.0 ± 3.7 |
| Never smoker, % | 48.0 | 46.0 | 48.2 | 45.8 | 46.9 | 49.8 | 42.1 | 46.8 | 52.5 |
| Hypertension, % | 15.0 | 18.0 | 17.7 | 16.8 | 16.8 | 17.0 | 15.2 | 17.0 | 18.1 |
| Hypercholesterolemia, % | 9.6 | 11.2 | 15.2 | 9.3 | 12.0 | 15.1 | 8.9 | 10.9 | 15.8 |
| WGHS | |||||||||
| Diet score | 46.8 ± 5.7 | 58.9 ± 2.8 | 71.2 ± 5.8 | 21.3 ± 2.4 | 26.0 ± 0.8 | 30.3 ± 2.1 | 19.2 ± 2.5 | 24.5 ± 1.1 | 29.2 ± 2.0 |
| Participants, n | 7174 | 7174 | 7392 | 8477 | 5879 | 7384 | 8359 | 6937 | 6444 |
| Age, y | 53.4 ± 6.6 | 54.7 ± 7.0 | 55.8 ± 7.4 | 53.7 ± 6.8 | 54.6 ± 6.7 | 55.6 ± 7.3 | 53.4 ± 6.5 | 54.7 ± 6.9 | 56.2 ± 7.6 |
| BMI, kg/m2 | 27.1 ± 5.5 | 26.4 ± 5.0 | 25.6 ± 4.6 | 27.0 ± 5.5 | 26.3 ± 4.9 | 25.8 ± 4.7 | 26.9 ± 5.4 | 26.4 ± 5.0 | 25.8 ± 4.7 |
| Physical activity, MET-h/wk | 10.0 ± 14.3 | 14.0 ± 17.3 | 20.2 ± 21.3 | 11.2 ± 16.3 | 14.4 ± 17.1 | 19.3 ± 20.5 | 10.7 ± 15.2 | 14.8 ± 18.6 | 20.1 ± 20.5 |
| Total energy intake, kcal/d | 1637 ± 527 | 1694 ± 511 | 1864 ± 508 | 1604 ± 522 | 1714 ± 495 | 1897 ± 505 | 1646 ± 529 | 1736 ± 522 | 1844 ± 498 |
| Total GRS | 87.6 ± 6.1 | 87.7 ± 6.0 | 87.9 ± 6.1 | 87.7 ± 6.2 | 87.7 ± 6.0 | 87.9 ± 6.0 | 87.6 ± 6.1 | 87.8 ± 6.0 | 87.9 ± 6.1 |
| CNS GRS | 58.5 ± 4.9 | 58.5 ± 4.8 | 58.7 ± 4.9 | 58.5 ± 5.0 | 58.5 ± 4.9 | 58.7 ± 4.8 | 58.4 ± 4.9 | 58.6 ± 4.8 | 58.7 ± 4.9 |
| Non-CNS GRS | 29.3 ± 3.5 | 29.3 ± 3.5 | 29.4 ± 3.5 | 29.4 ± 3.5 | 29.3 ± 3.5 | 29.4 ± 3.5 | 29.3 ± 3.5 | 29.4 ± 3.5 | 29.4 ± 3.5 |
| Never smoker, % | 53.2 | 51.6 | 49.4 | 54.2 | 48.6 | 50.3 | 48.0 | 52.9 | 54.0 |
| Hypertension, % | 24.3 | 24.5 | 22.1 | 25.1 | 22.9 | 22.4 | 24.4 | 23.3 | 22.9 |
| Hypercholesterolemia, % | 28.5 | 29.7 | 29.3 | 28.6 | 28.1 | 30.6 | 28.3 | 28.6 | 30.8 |
1Values are means ± SDs for continuous variables. AHEI, Alternative Healthy Eating Index 2010; AMED, Alternative Mediterranean Diet score; CNS, central nervous system; DASH, Dietary Approach to Stop Hypertension diet score; GRS, genetic risk score; MET-h, metabolic equivalent task hour; T, tertile.
Total, CNS, and non-CNS GRSs were normally distributed in the 3 cohorts (Supplemental Figures 2 and 3). As expected, participants with higher GRSs had higher BMIs.
Interaction between diet scores and GRS on BMI
We found significant interactions between total GRS and all 3 diet scores on BMI (AHEI: P-interaction = 0.003; AMED: P = 0.001; DASH: P = 0.004) (Table 2). No significant heterogeneity was observed across the 3 cohorts (P-heterogeneity ≥ 0.50). In the pooled data for the highest and lowest tertile, the difference in BMI (kg/m2) per 10-allele increment (a 10-allele increment was found to be equivalent to a 1.7-SD change in GRS) was 0.84 (95% CI: 0.72, 0.96) and 1.14 (95% CI: 0.99, 1.29) for the AHEI score; 0.83 (95% CI: 0.71, 0.96) and 1.17 (95% CI: 1.03, 1.31) for the AMED score, and 0.78 (95% CI: 0.66, 0.91) and 1.09 (95% CI: 0.95, 1.23) for the DASH score. The CNS GRS significantly interacted with all 3 diet scores on BMI (AHEI: P-interaction = 0.009; AMED and DASH: P < 0.001), but no significant interactions were observed between the non-CNS GRS and diet quality scores (all P-interaction >0.10). We further found inverse associations of BMI with diet quality scores stratified by tertiles of GRSs, and these inverse associations were strongest among participants in the highest tertile of total and CNS GRSs (Supplemental Table 3).
TABLE 2.
Difference in BMI per 10 risk allele increase in total, CNS, and non-CNS GRSs stratified by diet scores in the 3 cohorts1
| Lowest tertile | Second lowest tertile | Highest tertile | P-interaction | |
|---|---|---|---|---|
| AHEI | ||||
| Total GRS | ||||
| NHS | 1.25 (0.92, 1.59) | 1.06 (0.78, 1.35) | 0.76 (0.50, 1.02) | 0.02 |
| HPFS | 0.75 (0.45, 1.04) | 0.53 (0.28, 0.77) | 0.49 (0.26, 0.71) | 0.13 |
| WGHS | 1.28 (1.08, 1.48) | 0.99 (0.80, 1.18) | 1.07 (0.90, 1.24) | 0.12 |
| Pooled | 1.14 (0.99, 1.29) | 0.87 (0.74, 1.00) | 0.84 (0.72, 0.96) | 0.003 |
| CNS GRS | ||||
| NHS | 1.30 (0.88, 1.72) | 1.01 (0.66, 1.36) | 0.66 (0.36, 0.97) | 0.01 |
| HPFS | 0.84 (0.49, 1.19) | 0.39 (0.08, 0.71) | 0.63 (0.35, 0.91) | 0.28 |
| WGHS | 1.31 (1.06, 1.57) | 1.02 (0.79, 1.26) | 1.12 (0.90, 1.37) | 0.22 |
| Pooled | 1.18 (1.00, 1.36) | 0.85 (0.68, 1.01) | 0.87 (0.73, 1.02) | 0.009 |
| Non-CNS GRS | ||||
| NHS | 1.17 (0.60, 1.73) | 1.15 (0.67, 1.63) | 0.94 (0.48, 1.40) | 0.53 |
| HPFS | 0.56 (0.04, 1.07) | 0.76 (0.30, 1.22) | 0.23 (-0.15, 0.60) | 0.21 |
| WGHS | 1.27 (0.92, 1.62) | 0.99 (0.66, 1.32) | 1.04 (0.75, 1.34) | 0.32 |
| Pooled | 1.07 (0.81, 1.33) | 0.97 (0.74, 1.20) | 0.78 (0.57, 0.98) | 0.10 |
| AMED | ||||
| Total GRS | ||||
| NHS | 1.32 (1.01, 1.63) | 1.02 (0.73, 1.31) | 0.74 (0.48, 1.01) | 0.01 |
| HPFS | 0.78 (0.46, 1.10) | 0.50 (0.29, 0.70) | 0.50 (0.26, 0.75) | 0.14 |
| WGHS | 1.23 (1.05, 1.42) | 1.01 (0.81, 1.21) | 1.04 (0.86, 1.21) | 0.11 |
| Pooled | 1.17 (1.03, 1.31) | 0.81 (0.68, 0.94) | 0.83 (0.71, 0.96) | 0.001 |
| CNS GRS | ||||
| NHS | 1.30 (0.92, 1.69) | 1.01 (0.64, 1.38) | 0.64 (0.32, 0.96) | 0.01 |
| HPFS | 0.90 (0.54, 1.27) | 0.52 (0.25, 0.79) | 0.48 (0.16, 0.81) | 0.07 |
| WGHS | 1.35 (1.12, 1.58) | 1.04 (0.79, 1.29) | 1.02 (0.80, 1.23) | 0.03 |
| Pooled | 1.24 (1.07, 1.41) | 0.84 (0.68, 1.01) | 0.80 (0.64, 0.96) | <0.001 |
| Non-CNS GRS | ||||
| NHS | 1.33 (0.82, 1.84) | 1.03 (0.51, 1.56) | 0.93 (0.47, 1.39) | 0.33 |
| HPFS | 0.54 (-0.01, 1.09) | 0.45 (0.07, 0.83) | 0.53 (0.11, 0.95) | 0.81 |
| WGHS | 1.12 (0.80, 1.45) | 0.99 (0.64, 1.35) | 1.11 (0.81, 1.41) | 0.96 |
| Pooled | 1.07 (0.82, 1.32) | 0.81 (0.57, 1.04) | 0.93 (0.71, 1.14) | 0.50 |
| DASH | ||||
| Total GRS | ||||
| NHS | 1.39 (1.07, 1.71) | 1.00 (0.69, 1.30) | 0.71 (0.47, 0.96) | 0.001 |
| HPFS | 0.49 (0.20, 0.78) | 0.67 (0.41, 0.94) | 0.58 (0.35, 0.81) | 0.70 |
| WGHS | 1.23 (1.04, 1.41) | 1.11 (0.92, 1.30) | 0.96 (0.78, 1.15) | 0.05 |
| Pooled | 1.09 (0.95, 1.23) | 0.98 (0.84, 1.12) | 0.78 (0.66, 0.91) | 0.004 |
| CNS GRS | ||||
| NHS | 1.48 (1.08, 1.87) | 0.98 (0.60, 1.36) | 0.54 (0.24, 0.84) | 0.0002 |
| HPFS | 0.51 (0.16, 0.86) | 0.72 (0.40, 1.03) | 0.66 (0.37, 0.95) | 0.65 |
| WGHS | 1.30 (1.07, 1.53) | 1.23 (0.99, 1.47) | 0.87 (0.64, 1.10) | 0.01 |
| Pooled | 1.15 (0.98, 1.33) | 1.03 (0.86, 1.20) | 0.72 (0.56, 0.87) | < 0.001 |
| Non-CNS GRS | ||||
| NHS | 1.23 (0.69, 1.77) | 1.02 (0.52, 1.53) | 1.04 (0.58, 1.50) | 0.56 |
| HPFS | 0.46 (-0.06, 0.97) | 0.59 (0.13, 1.05) | 0.44 (0.06, 0.82) | 0.93 |
| WGHS | 1.18 (0.86, 1.50) | 0.95 (0.62, 1.28) | 1.16 (0.84, 1.49) | 0.87 |
| Pooled | 1.03 (0.79, 1.27) | 0.89 (0.65, 1.13) | 0.90 (0.69, 1.12) | 0.68 |
1Values are BMI change (kg/m2) with 95% CIs in parentheses. For the interaction term, diet scores of 0, 1, and 2 were assigned to the lowest, second lowest, and highest tertiles. Tertiles of diet score and GRS were modeled as continuous variables. Models were adjusted for age (quintiles), physical activity (quintiles), smoking status (never smoker, past smoker with 1–15 cigarettes/d, past smoker with >15 cigarettes/d, current smoker with 1–15 cigarettes/d, or current smoker with >15 cigarettes/d), total energy intake (quintiles), hypertension (yes or no), hypercholesterolemia (yes or no), case-control data sets (9 categories in the NHS and 7 categories in the HPFS), geographic region (categories, WGHS only), and population structure (eigenvectors, WGHS only). AHEI, Alternative Healthy Eating Index 2010; AMED, Alternative Mediterranean Diet score; CNS, central nervous system; DASH, Dietary Approach to Stop Hypertension diet score; GRS, genetic risk score; HPFS, Health Professional Follow-up Study; NHS, Nurses’ Health Study; WGHS, Women's Genome Health Study.
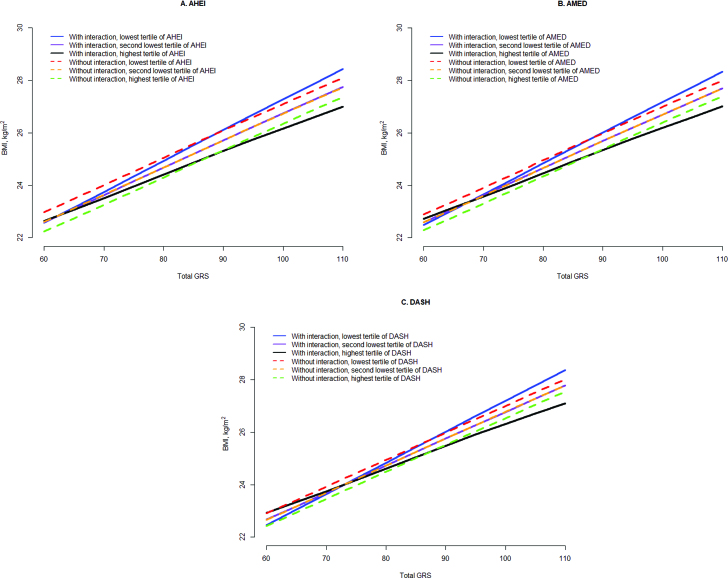
To further illustrate the results in Table 2, we presented predicted BMI with total GRS stratified by tertiles of diet quality scores estimated from multivariate models with the interaction effect in Figure 1 and Supplemental Figure 4, and then, as a comparison, we further obtained predicted BMI assuming no interaction effect. Our figures showed that due to interaction, the effect of GRS on BMI was attenuated with a high-quality diet, and was intensified by a low-quality diet.
FIGURE 1.
Predicted BMI from total GRS and tertiles of diet scores using models with and without interaction terms by pooling the 3 cohorts. Models adjusted for age (quintiles), physical activity (quintiles), smoking status (never smoker, past smoker with 1–15 cigarettes/d, past smoker with >15 cigarettes/d, current smoker with 1–15 cigarettes/d, or current smoker with >15 cigarettes/d), total energy intake (quintiles), hypertension (yes or no), hypercholesterolemia (yes or no), case-control data sets (9 categories in the NHS and 7 categories in the HPFS), geographic region (categories, WGHS only), and population structure (eigenvectors, WGHS only). AHEI, Alternate Healthy Eating Index 2010; AMED, Alternative Mediaterranean Diet score; DASH, Dietary Approach to Stop Hypertension diet score; GRS, genetic risk score; HPFS, Health Professional Follow-up Study; NHS, Nurses’ Health Study; WGHS, Women's Genome Health Study.
Interactions of food components with GRSs on BMI
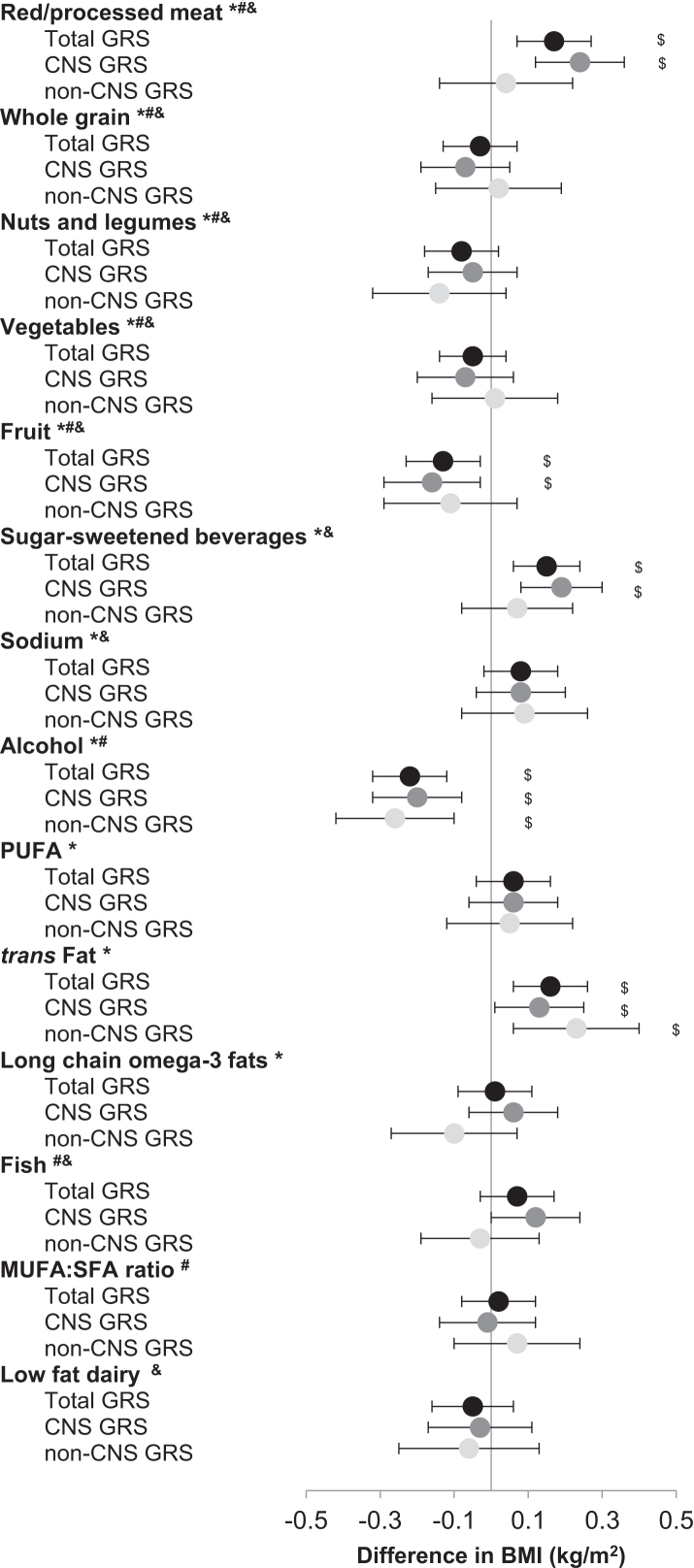
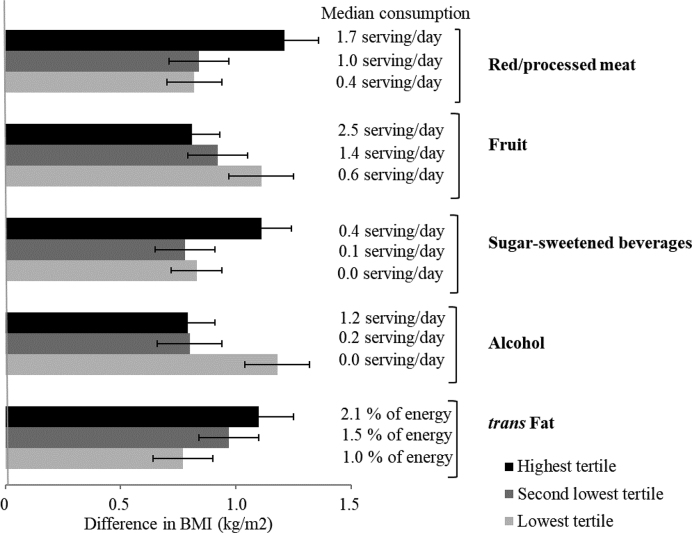
Interactions between food components and GRSs in relation with BMI were examined. Positive interactions of total GRS on BMI were found for consumption of red/processed meat, SSBs, and trans fat (P = 0.001), and negative interactions were found for consumption of fruit and moderate alcohol (P < 0.01) (Figure 2, Supplemental Table 4). For participants in the highest tertile of intake of red/processed meat, SSBs, trans fat, fruit, and alcohol, the difference in BMI was 1.21, 1.11, 1.10, 0.81, and 0.79, respectively, per 10-allele increment in total GRS; for those in the lowest tertile, the difference was 0.82, 0.83, 0.77, 1.11, and 1.18, respectively (Figure 3).
FIGURE 2.
Difference in BMI (kg/m2) per 10 risk allele increase in BMI GRS and per unit increase in diet score components by pooling the 3 cohorts. For the interaction term, food components were assigned 0, 1, and 2 to the lowest, second lowest, and highest tertiles. Tertiles of food components and GRS were modeled as continuous variables. Model adjusted for age (quintiles), physical activity (quintiles), smoking status (never smoker, past smoker with 1–15 cigarettes/d, past smoker with >15 cigarettes/d, current smoker with 1–15 cigarettes/d, or current smoker with >15 cigarettes/d), total energy intake (quintiles), hypertension (yes or no), hypercholesterolemia (yes or no), case-control data sets (9 categories in the NHS and 7 categories in the HPFS), geographic region (categories, WGHS only), and population structure (eigenvectors, WGHS only). $P-interaction < 0.05; *components in AHEI score; #components in AMED scores; &components in DASH scores. GRS, genetic risk score; HPFS, Health Professional Follow-up Study; NHS, Nurses’ Health Study; WGHS, Women's Genome Health Study.
FIGURE 3.
Difference in BMI (kg/m2) per 10 risk allele increase in total BMI GRS stratified by consumptions of red/processed meat, fruit, sugar-sweetened beverages, alcohol, and trans fat by pooling the 3 cohorts. For the interaction term, food components were assigned 0, 1, and 2 to the lowest, second lowest, and highest tertiles. Tertiles of food components and GRS were modeled as continuous variables. Model adjusted for age (quintiles), physical activity (quintiles), smoking status (never smoker, past smoker with 1–15 cigarettes/d, past smoker with >15 cigarettes/d, current smoker with 1–15 cigarettes/d, or current smoker with >15 cigarettes/d), total energy intake (quintiles), hypertension (yes or no), hypercholesterolemia (yes or no), case-control data sets (9 categories in the NHS and 7 categories in the HPFS), geographic region (categories, WGHS only), and population structure (eigenvectors, WGHS only). GRS, genetic risk score; HPFS, Health Professional Follow-up Study; NHS, Nurses’ Health Study; WGHS, Women's Genome Health Study.
Secondary and sensitivity analyses
In a stratified analysis, there were no significant differences in interactions between diet scores and GRSs on BMI when stratified by age, smoking status, physical activity, and sex (Supplemental Table 5). In the NHS and the HPFS, we used BMI assessed 4 y after dietary measurement as the outcome, and the results remained similar to those from the primary analysis (Supplemental Table 6). We further used simply updated diet scores up until 2008 as the exposure, and the results were consistent with those from the primary analysis (Supplemental Table 7).
DISCUSSION
For 3 observational cohorts of US women and men in which BMI was ascertained after assessment of dietary preferences, we found that higher diet quality attenuated the genetic associations with BMI, adjusting for covariates including total energy intake and physical activity. When we considered food components, we found that higher intakes of red/processed meat, SSBs, and trans fat accentuated genetic associations with BMI, whereas higher intakes of fruit and moderate intakes of alcohol attenuated the genetic effects on BMI.
Several previous studies have reported that certain dietary nutrients and foods/beverages may interact with BMI-associated GRSs. For example, by using the same population including the NHS, HPFS, and WGHS, previous studies found that the genetic association with BMI was stronger among participants with higher intakes of SSBs and fried foods than among those with lower intakes (5, 6). However, nutrients and foods are not consumed in isolation, but are distributed across correlated food networks with complex consumption patterns. In the past 2 decades, the dietary pattern approach has achieved considerable success in understanding the dietary determinants of health outcomes and translating public health messages (7). Thus, in the current study, we examined the interactions for diet quality scores that combine various nutrients and foods into “dietary patterns.” Our study demonstrated that higher diet quality could attenuate the genetic association with BMI, and the results were consistent across 3 types of diet scores. Consistent with our study, one recent study in the NHS and HPFS showed that improving adherence to healthy dietary patterns could attenuate the genetic association with weight gain (22). Among individual food or nutrient components, besides the interaction of SSBs with BMI that was found in a previous study (5), our study added new information that higher intakes of red/processed meat and trans fat accentuated the genetic association with BMI, whereas higher intakes of fruits attenuated the association. Moreover, our findings are consistent with previous studies in which higher intakes of red/processed meat, SSBs, and trans fat were found to be associated with a higher BMI, whereas intakes of fruits and moderate alcohol were associated with a lower BMI (10).
Previous studies have examined interactions of individual SNPs with dietary factors on BMI, but results were inconsistent. Some studies reported that associations of the fat-mass and obesity gene (FTO), APOA2, and PPAR-γ with BMI were stronger among participants with higher intakes of saturated fat than among those with lower intakes (23–25), whereas others did not find such interactions (26). These inconsistent observations might be due to insufficient statistical power of individual studies to detect significant interactions between individual SNPs and dietary factors on BMI, as most single variants have very modest effects on BMI (3). This might also be the reason why few SNPs significantly interacted with genetic score on BMI after Bonferroni correction in our study. Therefore, we used the GRS approach, which has been shown to be more powerful in gene-environment interaction analysis than using single SNPs.
Although the biological functions of most BMI loci remain largely unclear, there is increasing evidence supporting the role of some obesity genes in the regulation of food intake and energy balance. For example, the FTO obesity-predisposing allele was associated with higher energy intakes in children (27, 28), and higher intakes of fat and protein in adults independent of BMI (26, 29, 30). Risk variants within brain-derived neurotrophic factor (BDNF), Src homology 2-B1 (SH2B1), troponin I-3 interacting kinase (TNNI3K), potassium channel tetramerization domain containing 15 (KCTD15), mitochondrial carrier 2 (MTCH2), and neuronal growth regulator 1 (NEGR1) were also associated with certain food and nutrient intakes (31–33). Furthermore, risk variants in MTCH2, TNNI3K, zinc finger CCCH-type containing 4 (ZC3H4), and FTO were found to be associated with unhealthy eating behaviors independent of BMI (34). Interestingly, MC4R and BDNF, common monogenic causes of severe obesity, are highly expressed in the brain and regulate appetite and energy control (35, 36). FTO, SH2B1, NEGR1, KCTD15, glucosamine-6-phosphate deaminase 2 (GNPDA2), and transmembrane protein 18 (TMEM18) are also believed to be primarily expressed and functional in the CNS (37). This might help explain the observed significant interactions between GRS and diet scores on BMI, suggesting that appetite-regulating genes may be involved. Moreover, we constructed 2 GRSs based on BMI loci according to potential expression in the CNS, the key site of central appetite regulation, and found that the interactions with diet scores appeared more evident for CNS GRSs than for non-CNS GRSs. This finding may help better understand the mechanisms underlying the observations of gene-diet interactions, although the significant interaction found for CNS GRSs might be because the association of CNS GRSs with BMI was stronger than that of non-CNS GRSs.
Our study has several strengths. First, our study included 3 cohorts with 30,904 participants, which provided us with ample power to examine interactions of total, CNS, and non-CNS GRSs with diet scores separately. Second, BMI was measured after dietary intake with a 2- to 3-y lag in the 3 cohorts to minimize reverse causation. Third, dietary intake was measured repeatedly in the NHS and the HPFS, which minimized measurement errors. Moreover, we analyzed diet quality scores instead of individual nutrients or foods in our primary analyses, which took into account the complex and multidimensional nature of the diet. Fourth, we measured diet quality using 3 diet scores, and found significant interactions of GRS on BMI for all types of diet scores. Our results provide individuals with multiple options of a healthy diet.
Our study also has several limitations. First, interactions between diet quality and GRS might be confounded by other lifestyle factors. Previous studies have showed that physical activity, television watching, and sleep modified genetic associations with BMI (8, 38). Although we have carefully adjusted for other lifestyle factors, confounding by other unmeasured or unknown factors might exist. Second, measurement errors of dietary intake are inevitable. However, measurement error in environmental exposure typically biases the interaction effect toward the null and also substantially decreases the power to detect subtle interaction effects. Third, because biological functions of included SNPs are not known precisely, the classification of CNS and non-CNS SNPs might be arbitrary; however, we still observed stronger gene-diet interactions for CNS GRSs than for non-CNS GRSs.
In conclusion, our study showed that a higher diet quality attenuated genetic association with BMI. These findings provide important insights into the complex interplays between diet and the genetic predisposition to obesity, and underscore the importance of maintaining a healthful diet.
Supplementary Material
ACKNOWLEDGEMENTS
The authors' responsibilities were as follows—MKJ, GCC, LRP, JHK, JLW, DJH, WCW, EBR, PK, DIC, LQ, and FBH: designed the study and collected data; MD and CE: mainly conducted the analyses; TH: contributed to the analyses; MD: wrote the manuscript; QQ: supervised the data analysis and reviewed/edited the manuscript; and all authors: contributed substantially to the interpretation of data and the drafting or critical revision of the manuscript for important intellectual content. None of the authors declared a conflict of interest.
Notes
Supported by grants (UM1 CA167552, R01 HL35464, UM1 CA186107, P01 CA87969, R01 CA49449, R01 HL034594, R01 HL088521, HL60712, P30 DK46200, DK091718, HL071981, HL073168, CA87969, CA49449, CA055075, HL34594, HL088521, U01HG004399, DK080140, 5P30DK46200, U54CA155626, DK58845, U01HG004728-02, EY015473, DK70756, and DK46200) from the National Institutes of Health, with additional support for genotyping from Merck Research Laboratories; an American Heart Association Scientist Development Award (0730094N, to LQ); and a Research to Prevent Blindness award and a Harvard Ophthalmology Scholar Award (both to LRP) from the Harvard Glaucoma Center of Excellence. QQ is supported by a Scientist Development Award (K01HL129892) from the NHLBI. The WGHS is supported by grants (HL043851, HL69757, and CA047988) from the National Institutes of Health, with collaborative scientific support and funding for genotyping provided by Amgen.
Supplemental Figures 1–4, Supplemental Methods, and Supplemental Tables 1–7 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/ajcn/.
Abbreviations used:
- AHEI-2010
Alternative Healthy Eating Index 2010
- AMED
the Alternative Mediterranean Diet score
- CNS
central nervous system
- DASH
the Dietary Approach to Stop Hypertension
- FTO
fat-mass and obesity gene
- GRS
genetic risk score
- HPFS
Health Professional Follow-up Study
- NHS
Nurses’ Health Study
- SNP
single-nucleotide polymorphism
- SSB
sugar-sweetened beverage
- WGHS
Women's Genome Health Study
REFERENCE
- 1. Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, Styrkarsdottir U, Gretarsdottir S, Thorlacius S, Jonsdottir I et al.. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41(1):18–24. [DOI] [PubMed] [Google Scholar]
- 2. Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Lango Allen H, Lindgren CM, Luan J, Mägi R et al.. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(11):937–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J et al.. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Walter S, Mejia-Guevara I, Estrada K, Liu SY, Glymour MM. Association of a genetic risk score with body mass index across different birth cohorts. JAMA. 2016;316(1):63–9. [DOI] [PubMed] [Google Scholar]
- 5. Qi Q, Chu AY, Kang JH, Jensen MK, Curhan GC, Pasquale LR, Ridker PM, Hunter DJ, Willett WC, Rimm EB et al.. Sugar-sweetened beverages and genetic risk of obesity. N Engl J Med. 2012;367(15):1387–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Qi Q, Chu AY, Kang JH, Huang J, Rose LM, Jensen MK, Liang L, Curhan GC, Pasquale LR, Wiggs JL et al.. Fried food consumption, genetic risk, and body mass index: gene-diet interaction analysis in three US cohort studies. BMJ. 2014;348:g1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hu FB. Dietary pattern analysis: a new direction in nutritional epidemiology. Curr Opin Lipidol. 2002;13(1):3–9. [DOI] [PubMed] [Google Scholar]
- 8. Young AI, Wauthier F, Donnelly P. Multiple novel gene-by-environment interactions modify the effect of FTO variants on body mass index. Nat Commun. 2016;7:12724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nettleton JA, Follis JL, Ngwa JS, Smith CE, Ahmad S, Tanaka T, Wojczynski MK, Voortman T, Lemaitre RN, Kristiansson K et al.. Gene x dietary pattern interactions in obesity: analysis of up to 68 317 adults of European ancestry. Hum Mol Genet. 2015;24(16):4728–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Malik VS, Willett WC, Hu FB. Global obesity: trends, risk factors and policy implications. Nat Rev Endocrinol. 2013;9(1):13–27. [DOI] [PubMed] [Google Scholar]
- 11. Chiuve SE, Fung TT, Rimm EB, Hu FB, McCullough ML, Wang M, Stampfer MJ, Willett WC. Alternative dietary indices both strongly predict risk of chronic disease. J Nutr. 2012;142(6):1009–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Trichopoulou A, Costacou T, Bamia C, Trichopoulos D. Adherence to a Mediterranean diet and survival in a Greek population. N Engl J Med. 2003;348(26):2599–608. [DOI] [PubMed] [Google Scholar]
- 13. Fung TT, Chiuve SE, McCullough ML, Rexrode KM, Logroscino G, Hu FB. Adherence to a DASH-style diet and risk of coronary heart disease and stroke in women. Arch Intern Med. 2008;168(7):713–20. [DOI] [PubMed] [Google Scholar]
- 14. Ridker PM, Chasman DI, Zee RY, Parker A, Rose L, Cook NR, Buring JE, Women's Genome Health Study Working Group . Rationale, design, and methodology of the Women's Genome Health Study: a genome-wide association study of more than 25,000 initially healthy american women. Clin Chem. 2008;54(2):249–55. [DOI] [PubMed] [Google Scholar]
- 15. Feskanich D, Rimm EB, Giovannucci EL, Colditz GA, Stampfer MJ, Litin LB, Willett WC. Reproducibility and validity of food intake measurements from a semiquantitative food frequency questionnaire. J Am Diet Assoc. 1993;93(7):790–6. [DOI] [PubMed] [Google Scholar]
- 16. Rimm EB, Giovannucci EL, Stampfer MJ, Colditz GA, Litin LB, Willett WC. Reproducibility and validity of an expanded self-administered semiquantitative food frequency questionnaire among male health professionals. Am J Epidemiol. 1992;135(10):1114–26.; discussion 1127–36. [DOI] [PubMed] [Google Scholar]
- 17. Salvini S, Hunter DJ, Sampson L, Stampfer MJ, Colditz GA, Rosner B, Willett WC. Food-based validation of a dietary questionnaire: the effects of week-to-week variation in food consumption. Int J Epidemiol. 1989;18(4):858–67. [DOI] [PubMed] [Google Scholar]
- 18. Willett WC, Sampson L, Stampfer MJ, Rosner B, Bain C, Witschi J, Hennekens CH, Speizer FE. Reproducibility and validity of a semiquantitative food frequency questionnaire. Am J Epidemiol. 1985;122(1):51–65. [DOI] [PubMed] [Google Scholar]
- 19. Rimm EB, Stampfer MJ, Colditz GA, Chute CG, Litin LB, Willett WC. Validity of self-reported waist and hip circumferences in men and women. Epidemiology. 1990;1(6):466–73. [DOI] [PubMed] [Google Scholar]
- 20. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–9. [DOI] [PubMed] [Google Scholar]
- 21. Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A et al.. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39(7):870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang T, Heianza Y, Sun D, Huang T, Ma W, Rimm EB, Manson JE, Hu FB, Willett WC, Qi L et al.. Improving adherence to healthy dietary patterns, genetic risk, and long term weight gain: gene-diet interaction analysis in two prospective cohort studies. BMJ. 2018;360:j5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Phillips CM, Kesse-Guyot E, McManus R, Hercberg S, Lairon D, Planells R, Roche HM. High dietary saturated fat intake accentuates obesity risk associated with the fat mass and obesity-associated gene in adults. J Nutr. 2012;142(5):824–31. [DOI] [PubMed] [Google Scholar]
- 24. Corella D, Peloso G, Arnett DK, Demissie S, Cupples LA, Tucker K, Lai CQ, Parnell LD, Coltell O, Lee YC et al.. APOA2, dietary fat, and body mass index: replication of a gene-diet interaction in 3 independent populations. Arch Intern Med. 2009;169(20):1897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Memisoglu A, Hu FB, Hankinson SE, Manson JE, De Vivo I, Willett WC, Hunter DJ. Interaction between a peroxisome proliferator-activated receptor gamma gene polymorphism and dietary fat intake in relation to body mass. Hum Mol Genet. 2003;12(22):2923–9. [DOI] [PubMed] [Google Scholar]
- 26. Qi Q, Kilpelainen TO, Downer MK, Tanaka T, Smith CE, Sluijs I, Sonestedt E, Chu AY, Renström F, Lin X et al.. FTO genetic variants, dietary intake and body mass index: insights from 177,330 individuals. Hum Mol Genet. 2014;23(25):6961–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cecil JE, Tavendale R, Watt P, Hetherington MM, Palmer CN. An obesity-associated FTO gene variant and increased energy intake in children. N Engl J Med. 2008;359(24):2558–66. [DOI] [PubMed] [Google Scholar]
- 28. Qi Q, Downer MK, Kilpelainen TO, Taal HR, Barton SJ, Ntalla I, Standl M, Boraska V, Huikari V, Kiefte-de Jong JC et al.. Dietary intake, FTO genetic variants, and adiposity: a combined analysis of over 16,000 children and adolescents. Diabetes. 2015;64(7):2467–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Livingstone KM, Celis-Morales C, Lara J, Ashor AW, Lovegrove JA, Martinez JA, Saris WH, Gibney M, Manios Y, Traczyk I et al.. Associations between FTO genotype and total energy and macronutrient intake in adults: a systematic review and meta-analysis. Obes Rev. 2015;16(8):666–78. [DOI] [PubMed] [Google Scholar]
- 30. Chu AY, Workalemahu T, Paynter NP, Rose LM, Giulianini F, Tanaka T, Ngwa JS, Qi Q, Curhan GCCHARGE Nutrition Working Group, et al., CHARGE Nutrition Working Group . Novel locus including FGF21 is associated with dietary macronutrient intake. Hum Mol Genet. 2013;22(9):1895–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rukh G, Sonestedt E, Melander O, Hedblad B, Wirfalt E, Ericson U, Orho-Melander M. Genetic susceptibility to obesity and diet intakes: association and interaction analyses in the Malmo Diet and Cancer Study. Genes Nutr. 2013;8(6):535–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McCaffery JM, Papandonatos GD, Peter I, Huggins GS, Raynor HA, Delahanty LM, Cheskin LJ, Balasubramanyam A, Wagenknecht LE, Wing RR, Genetic Subgroup of Look AHEAD; Look AHEAD Research Group . Obesity susceptibility loci and dietary intake in the Look AHEAD Trial. Am J Clin Nutr. 2012;95(6):1477–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bauer F, Elbers CC, Adan RA, Loos RJ, Onland-Moret NC, Grobbee DE, van Vliet-Ostaptchouk JV, Wijmenga C, van der Schouw YT. Obesity genes identified in genome-wide association studies are associated with adiposity measures and potentially with nutrient-specific food preference. Am J Clin Nutr. 2009;90(4):951–9. [DOI] [PubMed] [Google Scholar]
- 34. Cornelis MC, Rimm EB, Curhan GC, Kraft P, Hunter DJ, Hu FB, van Dam RM. Obesity susceptibility loci and uncontrolled eating, emotional eating and cognitive restraint behaviors in men and women. Obesity (Silver Spring). 2014;22(5):E135–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Farooqi IS, Keogh JM, Yeo GS, Lank EJ, Cheetham T, O'Rahilly S. Clinical spectrum of obesity and mutations in the melanocortin 4 receptor gene. N Engl J Med. 2003;348(12):1085–95. [DOI] [PubMed] [Google Scholar]
- 36. Han JC, Liu QR, Jones M, Levinn RL, Menzie CM, Jefferson-George KS et al.. Brain-derived neurotrophic factor and obesity in the WAGR syndrome. N Engl J Med. 2008;359(9):918–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yeo GS, Heisler LK. Unraveling the brain regulation of appetite: lessons from genetics. Nat Neurosci. 2012;15(10):1343–9. [DOI] [PubMed] [Google Scholar]
- 38. Qi Q, Li Y, Chomistek AK, Kang JH, Curhan GC, Pasquale LR, Willett WC, Rimm EB, Hu FB, Qi L. Television watching, leisure time physical activity, and the genetic predisposition in relation to body mass index in women and men. Circulation. 2012;126(15):1821–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.