Abstract
Glycosylation is a biologically important protein modification process by which a carbohydrate chain is enzymatically added to a protein at a specific amino acid residue. This process plays roles in many cellular functions, including intracellular trafficking, cell–cell signaling, protein folding and receptor binding. While glycosylation is a common host cell process, it is utilized by many pathogens as well. Protein glycosylation is widely employed by viruses for both host invasion and evasion of host immune responses. Thus better understanding of viral glycosylation functions has potential applications for improved antiviral therapeutic and vaccine development. Here, we summarize our current knowledge on the broad biological functions of glycans for the Mononegavirales, an order of enveloped negative-sense single-stranded RNA viruses of high medical importance that includes Ebola, rabies, measles and Nipah viruses. We discuss glycobiological findings by genera in alphabetical order within each of eight Mononegavirales families, namely, the bornaviruses, filoviruses, mymonaviruses, nyamiviruses, paramyxoviruses, pneumoviruses, rhabdoviruses and sunviruses.
Keywords: fusion, glycoprotein, glycosylation, Mononegavirales, viral entry
Introduction
Glycosylation is a common cellular protein modification process whereby carbohydrates (glycans) are attached to specific amino acid side chains in proteins, resulting in glycoproteins (GPs). Cellular glycan functions include protein trafficking and folding, cellular differentiation, receptor binding, cell–cell interactions and cell signaling. Additionally, glycosylation is utilized by many pathogens in host cell invasion and immune system evasion (Vigerust and Shepherd 2007). Many viruses rely on specific host cell glycosylation patterns for binding to the cell and/or viral entry, as is well documented for both influenza virus and human immunodeficiency virus (HIV) (Nizet and Esko 2009). Viruses, through their reliance on hijacking host cell machinery for their own reproductive needs, also use host cell machinery to glycosylate their own GPs. These virally encoded, cell-executed GPs have the added benefit of being more likely recognized as “self” proteins by the host immune system, thus helping “hide” the virus from host defenses (Scanlan et al. 2007; Helle et al. 2011; Biering et al. 2012). Viral GPs then contribute to multiple viral functions, including viral entry and immune evasion (Merry and Astrautsova 2010). This has been well characterized in viruses such as HIV, where glycosylation protects the virus from antibody neutralization (Ogert et al. 2001; Wei et al. 2003; Scanlan et al. 2007; Li et al. 2008), and for influenza virus, where glycosylation modulates virulence and immune system targeting (Ohuchi et al. 1997; Wagner et al. 2000; Reading et al. 2009; Tate, Brooks et al. 2011; Tate, Job et al. 2011; Tsuchiya, Sugawara, Hongo, Matsuzaki, Muraki et al. 2002; Tsuchiya, Sugawara, Hongo, Matsuzaki, Muraki et al. 2002; Vigerust and Shepherd 2007; Vigerust et al. 2007). Functions of glycosylation in other viral systems, however, remain relatively less characterized.
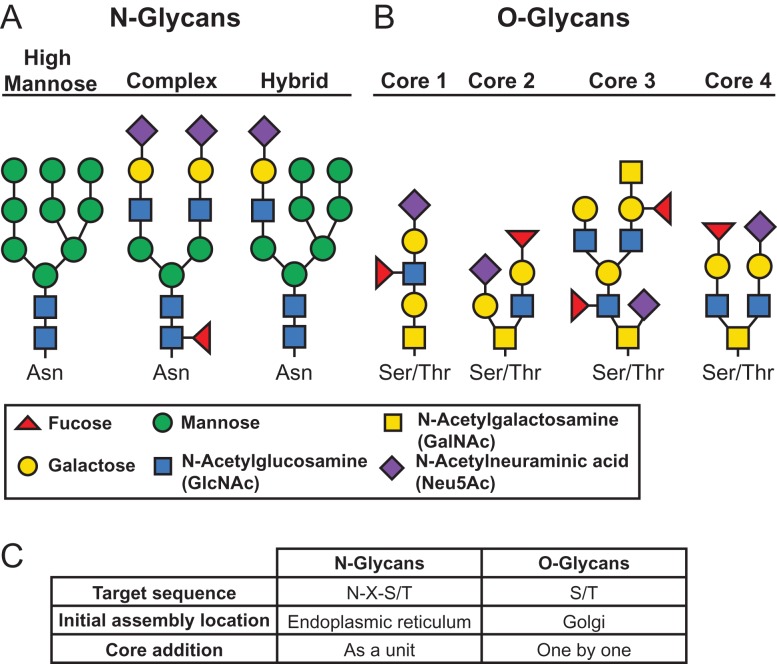
There are two main protein glycan modification types utilized by viruses. The first type, N-linked glycosylation, occurs when an N-acetylglucosamine (GlcNAc) sugar is covalently attached to an asparagine (N) residue within the predicted sequence N–X–S/T, where N is asparagine, X is any amino acid other than proline and S/T is a serine or threonine residue (Stanley et al. 2009). The N–X–S/T motif is necessary but not sufficient for glycosylation, as not all ectodomain N–X–S/T sites are N-glycosylated. Factors such as stable conformation adoption, localization of the protein and N–X–ST site accessibility to solvent likely play roles in determining whether a site is N-glycosylated (Bause 1983; Zielinska et al. 2010). N-glycans are attached to proteins in the endoplasmic reticulum (ER) early in the protein synthesis process when glycosyltransferases transfer a high-mannose carbohydrate core onto the protein. Further modifications occur as the GP progresses through the ER and the Golgi apparatus as additional carbohydrate residues are added and trimmed. N-glycans can be further processed in the Golgi apparatus and identified as high-mannose, hybrid or complex, depending on their carbohydrate content and branching (Nizet and Esko 2009; Stanley et al. 2009; Aebi 2013) (Figure 1A).
Fig. 1.
Glycosylation schematic. (A) Diagram of high-mannose (left), complex (center) and hybrid (right) N-glycans. (B) Diagram of the four main O-glycan core types. (C) Differences between N- and O-glycans.
The second type of glycosylation, O-linked glycosylation, can occur at S or T residues, though most S or T residues are not glycosylated, thus O-glycosylation sites are relatively more difficult to predict than N-glycosylation sites. O-glycans have an N-acetylgalactosamine (GalNAc) sugar linked through an O-linkage (hence the structural term O-glycan) to the free hydroxyl group of the S or T residue (Brockhausen et al. 2009). Additional sugars are then added by numerous glycosyltransferases, resulting in the formation of different core structures, which can then be further modified (Rini et al. 2009). The four most common core structures are shown in Figure 1B. O-glycosylation occurs post-translationally in the Golgi apparatus, and unlike N-glycosylation, sugar residues are added one by one instead of in a block (Figure 1C). As a result, O-glycans tend to be smaller and less branched than N-glycans (Van den Steen et al. 1998; Lodish et al. 2000; Peter-Katalinic 2005).
Each strategy in the study of glycosylation has advantages and disadvantages. For example, mass spectrometry can yield valuable chemical information but often requires quantities of protein that may be impractical for some viral studies (Rawling and Melero 2007). Cells can be treated with enzyme inhibitors, such as tunicamycin, a chemical that inhibits N-glycan synthesis within the ER, through chemical treatment. However, in addition to blocking glycosylation, tunicamycin may have other undesirable side effects within the cell, such as protein misfolding (Esko and Bertozzi 2009). There are also enzymes available for studying glycan composition. Peptide-N-glycosidase F (PNGase F) cleaves all N-glycans from proteins. Endoglycosidase H (endo H) cleaves high-mannose oligosaccharides and hybrid N-glycosylation sites, but not complex N-glycans. Endoglycosidase F (endo F) cleaves complex glycans, leaving high-mannose glycans intact (Maley et al. 1989; Stanley et al. 2009). Additionally, O-glycosidase can remove O-glycans (Brockhausen et al. 2009; Magnelli et al. 2012). Proteins, such as lectins, are additional tools that specifically bind certain types of glycans or antibodies against specific carbohydrates (Rawling and Melero 2007). Other strategies involve mutating the N–X–S/T sequence via site-directed mutagenesis, usually by mutating the N conservatively to a Q (glutamine), though mutations of N to S (serine), N to A (alanine) or S/T to G (glycine) are published as well (Hu et al. 1995; Zimmer et al. 2001; Moll et al. 2004; Schowalter et al. 2006; Aguilar et al. 2016). An additional strategy is the use of glycosylation-defective cell lines to study glycan effects (Esko and Stanley 2009). Viruses or viral GPs lacking some or all glycans can then be used for a variety of studies, and results compared to those obtained using fully glycosylated virus or viral GPs. Noteworthy, different cell types have different sets of glycosylation and de-glycosylation enzymes, thus introducing another level of variability to the study of glycan functions (Stanley and Cummings 2009).
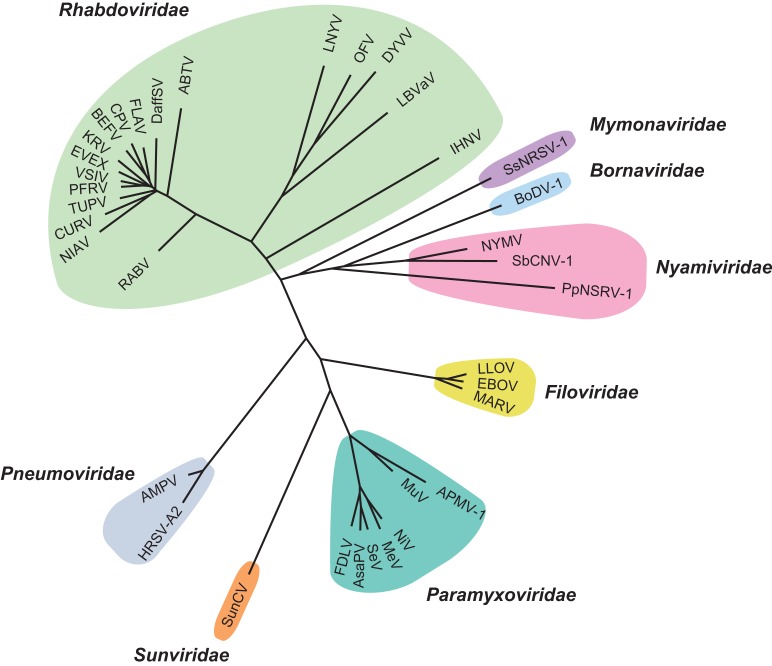
The Mononegavirales order is comprised of negative-sense single-stranded RNA viruses. This order currently contains eight viral families encompassing 36 genera and over a 100 known species. The families are as follows: bornaviridae (e.g. Borna disease virus), filoviridae (e.g. Ebola and Marburg viruses), mymonaviridae (e.g. sclerotimonavirus), nyamiviridae (e.g. nyavirus), paramyxoviridae (e.g. measles, mumps and Nipah viruses), pneumoviridae (e.g. human respiratory syncytial virus and human metapneumovirus), rhabdoviridae (e.g. rabies virus) and sunviridae (e.g. Sunshinevirus) (Amarasinghe et al. 2017) (Figure 2). Though these viruses have vastly differing hosts and tissue tropisms, they all share a similar genomic organization consisting from 3′ to 5′ ends of core protein genes, envelope GP genes and RNA-dependent RNA polymerase gene (Kuhn et al. 2013; Pfaller et al. 2015). Evidently, all Mononegavirales virions are enclosed by host cell-derived membrane envelopes (Kuhn et al. 2013). Here, we summarize the known functions of N- and O-linked glycans for the Mononegavirales and discuss their frequently underestimated importance.
Fig. 2.
Diagram of the Mononegavirales order. The phylogenetic tree was built after obtaining the RNA polymerase/large protein sequences of the viruses from the NCBI Protein Database. The protein sequences were aligned by using the COBALT Multiple alignment tool, by the fast-minimum evolution method and visualized using Figtree. The virus names and GenBank accession numbers are as follows: Bornaviridae—Borna disease virus 1 (BoDV-1; NP_042024), Filoviridae—Marburg virus (MARV; YP_001531159), Lloviu virus (LLOV; YP_004928143.1), Zaire ebola virus virus (EBOV; NP_066251.1), Mymonaviridae—Sclerotinia sclerotiorum negative-stranded RNA virus 1 (SsNSRV-1; 20996185), Nyamiviridae—Nyamanini virus (NYMV; YP_002905337), Pteromalus puparum negative-strand RNA virus 1 (PpNSRV-1; APL97667.1), Soybean cyst nematode virus (SbCNV-1; AEF56729), Paramyxoviridae—Avian paramyxovirus 1 (APMV-1; NP_071471), measles virus (MeV; NP_056924), Mumps virus (MuV; NP_054714), Nipah virus (NiV; AAK50546.1), Sendai virus (SeV; NP_056879), Atlantic salmon paramyxovirus (AsaPV; ABX57743.1), Fer-de-Lance virus (FDLV; AAN18266.1), Pneumoviridae—human respiratory syncytial virus-A2 (HRSV-A2; NP_056866), Avian metapneumovirus (AMPV; YP_443845), Rhabdoviridae—rabies virus (RABV; NP_056797), Tupaia virus (TUPV; YP_238534.1), Drosophila affinis sigmavirus (DAffSV; KR822811.1), pike fry rhabdovirus (PFRV; ACP28002.1), Niakha virus (NIAV; AGO44084.1), vesicular stomatitis Indian virus (VSIV; NP_041716), eel virus European X (EVEX; AHD46104.1), bovine ephemeral fever virus (BEFV; NP_065409), Coastal Plains virus (CPV; ADG86364.1), lettuce necrotic yellow virus (LNYV), orchid fleck virus (OFV; NC_009609.1), Datura yellow vein virus (DYVV; AKH61406.1), lettuce big-vein-associated virus (LBVaV; JN710440.1), Arboretum virus (ABTV; AHU86500.1), Flanders virus (FLAV; AAN73288.1), Kumasi rhabdovirus (KRV; YP_009177014.1), Curionopolis virus (CURV; AIE12119.1), infectious hematopoietic necrosis virus (IHNV; NP_042681), Sunshinevirus—Sunshine Coast virus (SunCV; YP_009094051.1).
Bornaviridae
Within the Bornaviridae family, there is only the genus Bornavirus, which includes the Borna disease virus (BDV). BDV has been shown to infect a wide range of vertebrates, causing encephalitis and behavioral abnormalities (Ludwig and Bode 2000; Richt and Rott 2001; Lipkin et al. 2011). Although its pathogenicity in humans is controversial, possible links to depression, schizophrenia, multiple sclerosis, chronic fatigue syndrome and aggressive brain tumors have been suggested (Ludwig and Bode 2000; Ikuta et al. 2002; Lipkin et al. 2011). Tunicamycin treatment inhibited production of infectious BDV, and glycosidase treatment eliminated virus infectivity, suggesting that glycans play a significant role in BDV pathogenicity (Stoyloff et al. 1994). There are two reported glycosylated BDV proteins. The first is gp18 (or p16), which was initially suggested to be an integral membrane matrix-like GP (Hatalski et al. 1995; Stoyloff et al. 1997). Gp18 was later proposed to be nonglycosylated and to only line the inner leaflet of the lipid bilayer, as do the typical cytoplasmic matrix proteins of the Mononegavirales (Kraus et al. 2001). Gp18 is suggested to be essential for infection and includes epitopes important for virus neutralization (Kliche et al. 1994; Hatalski et al. 1995; Stoyloff et al. 1997). Both lectin-binding and endoglycosidase digestion assays have shown the matrix protein to be N- but not O-glycosylated (Kliche et al. 1994). The specific effects of these N-glycans in BDV infection and potential neutralization, however, are still unknown. The second glycosylated protein in BDV is the G GP (p56), which is thought to have roles in receptor binding and viral entry. BDV G has been shown to contain N-glycans, but no O-glycans, and the specific functions of the N-glycans remain unknown (Gonzalez-Dunia et al. 1997; Schneider et al. 1997).
Filoviridae
The filovirus family comprises the Ebolavirus, Marburgvirus and Cuevavirus genera. Ebolavirus and Marburgvirus infections cause severe hemorrhagic fever with mortality rates of up to 90% (Feldmann and Geisbert 2011; Marcinkiewicz et al. 2014; Zawilinska and Kosz-Vnenchak 2014). These viruses are classified as bio-safety level 4 pathogens due to their high mortality rates and scarcity of approved treatments or cures. The recent 2014 Ebola outbreak in Africa had >28,000 infected individuals and >11,000 deaths, highlighting the need for further study of these deadly viruses (Zeitlin et al. 2016). The filoviruses rely on a single GP for both binding to host cells and viral-cell membrane fusion leading to viral entry (White et al. 2008; Hunt et al. 2012). GP is cleaved by cathepsins B and L into two subunits linked by a disulfide bond: GP1, which binds host cells, and GP2, which executes membrane fusion (Chandran et al. 2005; Schornberg et al. 2006; Kaletsky et al. 2007; White et al. 2008). There is also a soluble small form of the GP, sGP, whose function is still largely unknown but is suspected to act in immune evasion as a decoy for antibody binding (Ritchie et al. 2010; Zawilinska and Kosz-Vnenchak 2014).
Ebolavirus
Ebola virus (EBOV) GP1 has four domains: a base, receptor-binding domain, glycan cap domain consisting of only N-linked glycan sites and a mucin-like domain (MLD), which is heavily O-glycosylated, though it contains N-glycans as well (Feldmann et al. 1991, 1994; Lee et al. 2008; Powlesland et al. 2008; Lennemann et al. 2014; Davey et al. 2017). It has been suggested that the thick glycan layer created around GP by the glycan cap and MLD helps in antibody evasion (Lee et al. 2008). In support of this hypothesis, mutation of two N-glycosylation sites increased immunogenicity, possibly by unmasking epitopes normally covered by glycans (Dowling et al. 2007).
Remarkably, all 15 N-glycosylation sites from GP1 have been successfully mutated concurrently without affecting GP expression (Lennemann et al. 2014). N-glycan removal did, however, increase protease sensitivity, antibody sensitivity and transduction of pseudotyped virions into cells, suggesting that N-glycans protect against proteases and shield against antibody immune responses at the expense of viral entry efficiency (Lennemann et al. 2014). Single N-glycosylation site mutations asparagine to aspartate (N to D), however, were found to have similar processing, expression and transduction levels as wild-type virus (Jeffers et al. 2002). One mutant, N40D, did display less processing and no transduction. Loss of the same N-glycan via a T42D mutation, however, did not result in an altered phenotype, suggesting that the effects observed for the N40D mutant were most likely due to a change in protein conformation (as this site is near a cysteine residue) and not due to loss of the N-glycan itself (Jeffers et al. 2002). Additionally, D is not the most ideal substitution since it introduces a negative charge in addition to the loss of the N-glycan. A separate study, however, found T42A or N40K mutants to be cathepsin B resistant, suggesting that the N40 glycan is important for GP cleavage and processing (Wong et al. 2010). Additionally, the amount of N-glycan processing can also determine which receptors GP will bind. High-mannose N-glycans allow DC-SIGN lectin binding (Lin et al. 2003; Marzi et al. 2006; Powlesland et al. 2008), while the lectin LSECTIN does not recognize high-mannose glycan forms (Gramberg et al. 2005) and has been shown to bind truncated N-glycans (Powlesland et al. 2008). Thus, N-glycans clearly play a role in receptor binding and proper GP cleavage, and these functions may or may not have a direct role in viral entry by modulating receptor-induced membrane fusion.
In comparison to N-glycan removal, deletion of the O-glycan-rich MLD had more prominent effects. Deletion of the entire MLD resulted in increased processing, incorporation and viral entry using a retroviral system expressing EBOV GP. Deletion of this O-glycan-rich region also decreased cellular detachment capabilities, abolished cytopathic effects both in vitro and in vivo and reduced immune responses in mice (Yang et al. 2000; Jeffers et al. 2002; Simmons et al. 2002; Dowling et al. 2007). However, since the MLD region is 188 amino acids in length, it is possible that deletion of this region affected the GP conformation (Tran et al. 2014). In virus-like particles (VLPs) studies, immunization of mice with VLPs expressing full-length GP elicited higher antiviral titers than immunization with VLPs expressing an MLD-deleted GP (Martinez et al. 2011). Additionally, epitopes of protective monoclonal antibodies (MAbs) were mapped to a domain C-terminal of the GP O-glycan-rich region (Wilson et al. 2000). The use of two different models, a retroviral system and VLPs, both highly suggest the MLD is important for an immune response, at least in mice, in addition to playing roles in viral infectivity. It should be noted that many of these studies deleted the entire O-glycan region, and thus, it remains unclear if the loss of O-glycans and/or the loss of the amino acid backbone caused the observed effects. In addition, there are N-glycans within the MLD that may partially contribute to the phenotypes observed.
The GP2 subunit contains two N-glycosylation sites conserved among all five EBOV species (Jeffers et al. 2002; Ritchie et al. 2010). Deletion of one of the two N-glycosylation sites decreased immunogenicity in mice, possibly by disrupting the dimerization of GP1 and GP2 (Dowling et al. 2007). Interestingly, sGP uses five of its six potential N-glycosylation sites and contains no O-glycans (Falzarano et al. 2006; Ritchie et al. 2010). The sGP N-glycans are more processed (containing more complex carbohydrates) than those on GP1, but the reasons for these differences in processing are still undetermined (Lennemann et al. 2014; Ritchie et al. 2010). Since sGP is suspected to act as an immune system decoy, it would be interesting to investigate whether these sGP N-glycans play an immunity role.
Marburgvirus
Marburg virus contains similar numbers of N- and O-glycans as an EBOV (Sanchez et al. 1993). While many of the N-glycans on EBOV are complex, Marburg virus contains mainly high-mannose and hybrid N-glycans that lack terminal sialic acids (Feldmann et al. 1994; Geyer et al. 1992). The reasons or effects of these differences among these filoviruses, however, have yet to be elucidated, as some of these differences may be a consequence of the different cell lines used to study the different viruses.
Cuevavirus
Lloviu virus (LLOV) is the first discovered member of the new Cuevavirus genus. LLOV was isolated in Cueva de Lloviu, Asturias, Spain after a major die-off of Schreiber’s bats (Negredo et al. 2011). Similarly to Ebola and Marburg viruses, Lloviu requires NPC1 (an endosomal membrane protein Niemann–Pick disease type C1, used for transport of cholesterol to cellular membranes) to mediate viral entry (Ng et al. 2014; Wang et al. 2016). Vesicular stomatitis virus (VSV)-pseudotyped virions bearing LLOV GP (VSV–LLOV) yielded a transduction efficiency comparable to Ebola and Marburg viruses (Ng et al. 2014; Hoffmann et al. 2016). Sequence analysis of LLOV GP showed it to contain N-glycans at both the N-terminus of GP1 and C-terminus of GP2. Treatment with PNGase F also suggested that LLOV GP1 contains a high number of N-glycans, similar to Ebola and Marburg viruses. However, the functions of these glycans are unknown (Ng et al. 2014).
Summary for Filoviridae
Both Ebola and Marburg viruses are heavily glycosylated by both N- and O-glycans, suggesting an extreme importance for these secondary modifications. With less research done on Marburg virus and LLOV, it is difficult to determine if the functions of these glycans demonstrated for EBOV are conserved among the Filoviridae. It is interesting to note that the EBOV N-glycans do not appear to play important roles in cell surface expression (CSE), a difference from other mononegaviruses such as many paramyxoviruses. However, filovirus N-glycans do seem to modulate protease sensitivity and thus protein processing. It appears that the O-glycan-rich region is involved more heavily in many of these functions, but since most studies so far deleted this entire region, it remains to be determined whether the O-glycans themselves are responsible for the phenotypes observed.
Mymonaviridae
Sclerotinia sclerotiorum negative-stranded RNA virus 1 (SsNSRV-1) is the only known member of the Sclerotimonavirus genus. SsNSRV-1 was isolated in China from a fungal plant pathogen, Sclerotinia sclerotiorum, and has a filamentous viral particle shape. Importantly, to our knowledge, this is the first fungus-infecting mononegavirus discovered. The open reading frames were analyzed and all six proteins were determined to have multiple potential N-glycosylation sites, but it is unknown if they are used (Liu et al. 2014).
Nyamiviridae
This family has three genera: Nyavirus, Socyvirus and Peropuvirus. To our knowledge, no glycan identification or functional studies have been done for any members of the Nyamiviridae family.
Nyavirus
This genus is currently comprised of Nyamanini virus (NYMV), Midway virus (MIDWV) and Sierra Nevada virus (SNVV) (Pfaller et al. 2015). NYMV and MIDWV viruses are tick-borne and are not known to cause disease in humans or animals, however they can infect birds (Kuhn et al. 2013). SNVV was isolated after an outbreak of epizootic bovine abortions transmitted by Ornithodoros coriaceus ticks in northern California (Rogers et al. 2014). Putative glycosylation sites for SNVV have not been described. Both NYMV and MIDWV are predicted to have multiple glycosylation sites on the ORF V. NYMV has one putative O-linked and eight potential N-linked glycosylation sites in the ORF V (G). The MIDWV ORF V is predicted to have six N-linked glycosylation sites on its ORF V (G) (Mihindukulasuriya et al. 2009).
Socyvirus
The soybean cyst nematode virus-1 (SbCNV) belongs to the Socyvirus genus and was discovered in the plant parasitic nematode. SbCNV has an ORF IV sequence with one potential N-glycosylation site (Afonso et al. 2016; Bekal et al. 2011).
Peropuvirus
This is the newest genus to this family, which consists of the Pteromalus puparum negative-strand RNA virus 1 (PpNSRV-1). PpNSRV-1 was found in tissues of the parasitoid wasp, Pteromalus puparum, and may be transmitted vertically through infected wasps. The ORF IV protein contains seven putative O-linked and four putative N-linked glycosylation sites (Wang et al. 2017).
Paramyxoviridae
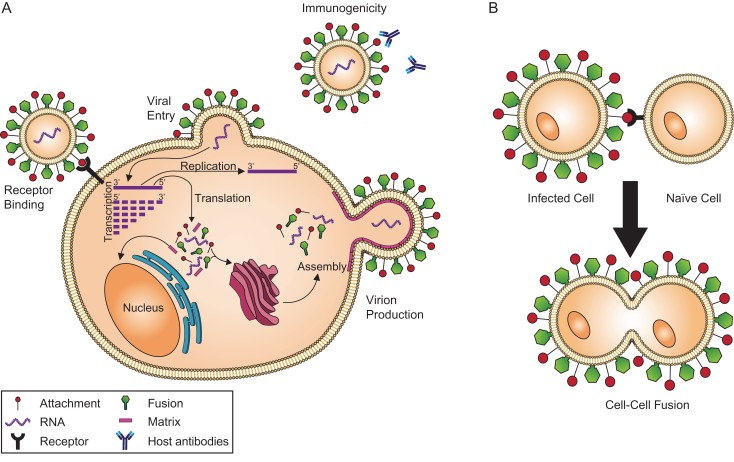
The Paramyxoviridae family contains many veterinary and medically important viruses such as measles virus (MeV), avian paramyxovirus (APMV-1), parainfluenza viruses (PIV), and the deadly Nipah virus (NiV) and Hendra virus (HeV). These viruses rely on the coordinated efforts of two envelope GPs to enter cells via a viral-cell membrane fusion event (Figure 3A). Additionally, paramyxoviral GP-induced membrane fusion does not require a drop in pH, as many other viral families do. The same two GPs execute cell–cell fusion (syncytia) events (Figure 3B), believed to be a pathway for the virus to spread between host cells without being intercepted by the immune system (Chang and Dutch 2012). Binding of the attachment GP (HN, H or G) to a cell surface receptor triggers the fusion GP (F) to insert itself into the host cell membrane, facilitating viral-cell membrane fusion (Iorio et al. 2009; Chang and Dutch 2012). For F to become biologically active, the precursor (F0) is either transported through the trans-Golgi network and cleaved by furin, transported to the cellular surface and subsequently endocytosed and cleaved by cellular proteases, or cleaved by extracellular proteases (Chang and Dutch 2012). This yields the biologically active GP made of F1 and F2 subunits linked by disulfide bonds. F is then transported to the surface of the plasma membrane (Lamb and Parks 2007). Here, we discuss the glycan functions by paramyxoviral genus: Aquaparamyxovirus, Avulavirus, Ferlavirus, Henipavirus, Morbillivirus, Respirovirus and Rubulavirus.
Fig. 3.
Diagram of the viral life cycle. (A) Virions bind to host cell receptors (black), facilitating viral entry into the cell. Once within the cell, transcription and replication occur. This allows the production of viral proteins, which are processed and trafficked through the cell using host cell machinery (endoplasmic reticulum and Golgi apparatus). Viral proteins and RNA are packaged into new virions, which are then released from the cell. These virions can then interact with the host immune system. (B) Infected cells can express viral proteins on their cell membranes, which can react with cell receptors on neighboring naïve cells, causing cell–cell fusion.
Aquaparamyxovirus
The Aquaparamyxovirus genus is currently solely comprised of the Atlantic salmon paramyxovirus (AsaPV). Upon infection, inflammation of the gills and respiratory distress were followed with an accrued mortality of 40% in farmed Atlantic salmon (Kvellestad et al. 2003). The AsaPV F has two putative N-glycosylation sites, one at the F1 subunit one at the F2 subunit, and the HN protein has four potential N-glycosylation sites, all with unknown functions (Falk et al. 2008).
Avulavirus
Newcastle disease virus infects avian species and is endemic in many third-world countries. It is designated as serotype-1 of the APMV-1. APMV-1 causes severe economic losses for many poultry farmers due to its high morbidity, mortality and its infectious nature (Ganar et al. 2014). Studies in Lec1 cells, a cell line lacking the enzymes necessary to convert high-mannose N-glycans to complex or hybrid forms, suggest that APMV-1 fusion and infection are not dependent on hybrid or complex N-glycans. This suggests that high-mannose N-glycans may play a role in cell–cell fusion, however, the folding of these GPs was not studied (Sun et al. 2012).
APMV-1F contains four N-glycosylated sites. Mutation of these sites altered F processing and decreased cell–cell fusion (McGinnes et al. 2001). In contrast, the removal of N-glycans specifically from the heptad repeat regions 1 and 2 of APMV-1F in combination resulted in increased virulence and immunogenicity of virions (Samal et al. 2012), suggesting that the effects of N-glycans on APMV-1F may be location-specific. Interestingly, the latter study also found that the removal of N-glycans did not affect APMV-1F processing or CSE, conflicting with the previously mentioned study. These differences may be due to the use of transfected vs. infected cells or the use of S/T to A mutations in the first study vs. N to Q mutations in the second.
APMV-1 HN utilizes four of its six potential N-glycosylation sites, two of which modulate protein folding and activity (McGinnes and Morrison 1995). N to Q mutations caused altered fusion activity (usually lower, but one double mutant had increased fusogenicity), and all were less virulent (Panda et al. 2004), an interesting result since fusogenicity often directly correlates with virulence. The N481 mutant had lowered CSE, suggesting that the most C-terminal N-glycan is important for processing and/or transport. None of the mutants had altered cell receptor-binding abilities, though two had decreased neuraminidase activities, suggesting that the decreased virulence of these mutants may be due, at least partially, to a decreased cellular detachment ability (Panda et al. 2004). Furthermore, it has been shown that failure to form a disulfide bond results in an additional N-glycosylation, possibly by causing differential folding that exposes an otherwise obscured site to glycosyltransferases (McGinnes and Morrison 1997). However, when a D287N mutation introduced an N-glycan the effect of neutralizing antibodies was reduced. Interestingly, the D287N mutation also reduced fusion from within (fusion induced by infected cells) and increased fusion from without (cell–cell fusion mediated by virus at a high multiplicity of infection). Overall, addition of the N-glycan caused diminished syncytium formation as compared to wild-type HN and enabled the virus to escape from neutralizing MAbs specific for antigenic site 3 (Deng et al. 1994). The effects of this additional glycosylation on virulence and pathogenicity, however, are still somewhat unclear.
Ferlavirus
The Fer-de-Lance virus (FDLV) was isolated after an outbreak in a Swiss serpentarium that caused lethargy, respiratory distress, and death (Clark et al. 1979). FDLV may cause epidemics in captive reptile populations with high mortality rates (Folsch and Leloup 1976). The FDLV HN GP has two putative N-linked glycosylation sites. As for the FDLV F GP, it has three putative N-linked glycosylation sites, two within the F1 subunit and one in F2 (Kurath et al. 2004). No further studies on the role of glycans in the virus life cycle have been conducted.
Henipavirus
The Henipavirus genus contains the deadly emerging zoonotic NiV and HeV, as well as Cedar, Mojiang, Kumasi, and over a dozen new uncharacterized and unnamed henipaviruses recently discovered in bats (Drexler et al. 2012; Marsh and Wang 2012; Wu et al. 2014). All glycobiology studies so far have focused on NiV and HeV. Infection with NiV or HeV results in severe respiratory and neurological symptoms in humans, progressing to fatal encephalitis in 40–100% of cases (Luby et al. 2009). Both viruses are bio-safety level 4 pathogens, with no currently approved treatment or vaccine available for human use.
NiV F is glycosylated at four of its five potential N-glycosylation sites (Moll et al. 2004; Aguilar et al. 2006). Loss of some of the N-glycans by N to Q mutations resulted in increased fusogenicity but also increased susceptibility to antibody neutralization, implying a need for NiV to balance these two important glycan functions, and one N-glycan was relatively more important for protein expression/transport (Aguilar et al. 2006). The same N-glycosylation sites are used in HeV F, and similarly, of these, the equivalent N-glycan as for NiV (N414 in HeV) is important for proper folding and transport, while the removal of the N464 N-glycan yielded an increase in fusion, the other two had no significant effect on fusion (Carter et al. 2005), an interesting difference from NiV. Additionally, Galectin-1, a lectin that binds primarily a specific N-glycan in NiV F, inhibited NiV cell–cell fusion if added post-infection but enhanced viral entry if present at the time of infection by increasing viral attachment to target cells (Garner et al. 2010, 2015; Levroney et al. 2005), demonstrating the importance of the timing of viral exposure to this innate immune factor.
Six of seven potential NiV G N-glycosylation sites are utilized: one in the stalk and five in the globular head domain. The removal of N-glycans from the head region of NiV G generally resulted in increased fusion, with some synergistic effects observed for double or triple mutants, which had higher fusion levels than their individual site counterparts. These mutants were also more susceptible to antibody neutralization, similar to NiV F N-glycans. In contrast, the removal of an N-glycan in the NiV G stalk created a fusion-dead mutant, even in combination with the removal of N-glycans from sites that created hyperfusogenic mutants when removed on their own. NiV G N-glycan removal also affected viral entry and binding avidities with NiV F, though protein conformations and receptor-binding capabilities were not significantly affected (Biering et al. 2012). The removal of N-glycans from HeV G revealed similar trends. However, simultaneous removal of multiple N-glycans from HeV resulted in a much stronger reduction in CSE than from NiV. Additionally, the removal of the N306 and N378 N-glycans from HeV G resulted in higher cell–cell fusion levels than for NiV, while the removal of the N481 and N529 N-glycans of NiV caused higher fusion levels than for HeV (Bradel-Tretheway et al. 2015). These differences suggest conformational or functional differences for the same N-glycans in the two closely related viruses and/or differences in glycan branching/composition. Mass spectrometry analysis of a soluble HeV G showed it contained mainly complex N-glycans (Colgrave et al. 2012), but a similar mass spectrometry analysis has not been performed for NiV G.
Interestingly, the loss of N-glycans for NiV and HeV not only correlated to an enhancement of membrane fusion but also to looser F/G GP–GP interactions. This phenotype illustrates that not only the inherent fusogenic capacities of the GPs, but also their interactions important for their membrane fusion functions can be modulated by N-glycans. These results have also helped to confirm the F/G dissociation model of membrane fusion for this paramyxovirus genus (Aguilar et al. 2006; Aguilar and Iorio 2012; Bradel-Tretheway et al. 2015).
Relatively little is known about the functions of O-glycans for the Mononegavirales, and no function had been described for any paramyxoviral O-glycan until a recent study by Stone et al. (2016). HeV G has been shown to contain O-glycans via mass spectrometry (Colgrave et al. 2012). Loss of specific O-glycosylation sites from the HeV G stalk affected cell–cell fusion, protein conformational changes, binding avidities with HeV F, pseudotyped viral entry, and quite interestingly, HeV F protein processing and incorporation into pseudotyped virions (Stone et al. 2016). Since F and G are thought not to interact until reaching the cell surface, it is quite interesting that loss of an O-glycan on G affects F incorporation into virions, especially since this effect was observed in pseudotyped virions but not in cell lysates. Similar effects were observed for the same O-glycan mutants in NiV G, with the exception that interestingly, NiV F incorporation into pseudotyped virions and pseudotyped viral entry were not altered by the loss of O-glycans on NiV G. Additionally, one mutant in NiV G was unable to express at the cell surface even though its HeV G counterpart expressed at wild-type G levels (Stone et al. 2016). Loss of an O-glycan in either HeV or NiV G had no effect on antibody neutralization susceptibility, demonstrating functional differences between N- and O-glycans for the Henipaviruses. Additionally, O-glycans were shown not to affect G oligomerization. All these results suggest the importance of both N- and O-glycans in the henipavirus life cycle, though there appears to be subtle differences of their effects between NiV and HeV.
Morbillivirus
The morbillivirus genus contains important viruses such as MeV and canine distemper virus (CDV). Measles infections represent one of the largest causes of childhood morbidity and mortality. In addition to acute infection, characterized by the development of a skin rash, there is a risk of developing subacute sclerosing panencephalitis, an often fatal neurological disease resulting from persistent MeV infection (Griffin et al. 2012). CDV causes symptoms such as fever, respiratory distress and neurological disorders and infects dogs and many other carnivore species (Harder and Osterhaus 1997; Martella et al. 2008).
N-glycans on both MeV F and MeV H are important for syncytia formation (Malvoisin and Wild 1994), and the specific processing of these N-glycans is crucial. Treatment with an α-glucosidase inhibitor, castanospermine, revealed that trimming of glucose residues within the ER was required for syncytia formation and for infectivity (Malvoisin and Wild 1994; Bolt et al. 1999). Additionally, tunicamycin treatment to block N-glycan synthesis inhibited virion particle formation (Stallcup and Fields 1981). All three potential MeV F N-glycosylation sites are glycosylated and N-glycan removal from any resulted in loss of protein CSE and/or processing (Alkhatib et al. 1994; von Messling and Cattaneo 2003). Nonconservative mutations (N to S) of N29 and/or N61 resulted in a lack of MeV F processing and transport to the cell surface (Hu et al. 1995). Results were similar for conservative mutations (N to Q), as N-glycan loss resulted in reduced MeV F CSE (Alkhatib et al. 1994). Interestingly, mutant N61Q still had fusion activity, while mutant N29Q did not, a finding differing from the N to S mutations where neither mutant was fusogenic, suggesting that the loss of fusion for mutant N61S was due to the loss of the N residue and not the glycan. Mutation N67Q alone did not affect protein processing or fusogenicity but combined with the loss of either of the other two glycans resulted in impaired cleavage, stability, CSE, processing and fusion (Alkhatib et al. 1994). These results highlight the importance and partial flexibility of N-glycans on MeV F for proper protein processing, expression and function.
MeV H is glycosylated at four out of five potential N-glycosylation sites (Bolt et al. 1999). Two of the four N-glycans affect GP conformation, as measured by conformational MAbs. No individual N-glycan affected folding, oligomerization or transport, but the presence of at least two of the four N-glycans were necessary for these functions, suggesting transport and folding are not dependent on a specific carbohydrate chain but do rely on a collective carbohydrate functions (Hu et al. 1994). Crystallization data suggest that N-glycans may modulate receptor binding by positioning MeV H for receptor binding (Hashiguchi et al. 2007). Supporting this notion, MeV H binding to the CD46 receptor was shown to be dependent on N-glycans, but not O-glycans, on MeV H, suggesting that N-glycans help keep MeV H in a conformation recognized by CD46. Likewise, the CD46 receptor also requires N-glycans to serve as the receptor for MeV H (Maisner et al. 1994, 1996; Maisner and Herrler 1995). However, it has been argued that CD46 is not a relevant in vivo receptor (Noyce and Richardson 2012), thus the biological significance of these findings remains to be determined. The effects of glycosylation on MeV H interactions with signaling lymphocyte activation molecule (SLAM) or Nectin 4, the two other known and biologically relevant receptors of MeV, have not been reported. Regardless, N-glycans on MeV H appear to play a general role in keeping MeV H in a favorable functional conformation.
Similar to MeV F, N to Q mutations demonstrated that CDV F also uses all three potential N-glycosylation sites, and these N-glycans are important for processing, protein stability, CSE and fusion (von Messling and Cattaneo 2003). CDV H contains three N-glycosylated sites conserved among all CDV strains, with up to an additional five sites present in some wild-type strains. Loss of all N-glycosylation sites from CDV H using N to Q mutations resulted in a functional and immunosuppressive CDV H GP, but one that no longer caused disease (Sawatsky and von Messling 2010). It has also been suggested that lack of N-glycans on the attenuated vaccine strain may be responsible for the loss of virulence (Iwatsuki et al. 1997). These results support the importance of N-glycans in GP function, pathogenicity and vaccine development. Notably, N-glycan presence appears to be more conserved between Morbillivirus F GPs (three out of three sites for both MeV and CDV, all in conserved positions) than between the H GPs (four out of five on MeV, and up to eight on CDV), suggesting that H is under higher host immune pressure.
Respirovirus
Human parainfluenza virus (HPIV) types 1–3 are major causes of respiratory tract infections in children, the elderly and immunocompromised individuals (Mishin et al. 2010; Chu et al. 2013; Bailly et al. 2016). Sendai virus (SeV) is a murine virus that has functioned as a model virus in many paramyxoviral studies (Pfaller et al. 2015).
The HN GP of HPIV-3 utilizes three of its four potential N-glycosylation sites. Mutation (N to A) of these sites revealed that N-glycans on HPIV-3 HN are important for receptor binding, F cleavage, fusion, but not in HN avidity for HPIV-3F or neuraminidase activity (Chu et al. 2013). Remarkably, N-glycan loss from HPIV-3 HN affected F cleavage without affecting overall interactions with HPIV-3F, findings that warrant further investigation and suggest the roles of unknown factors involved. Additionally, the N-glycan at HPIV-3 HN N523 was found to cover a second receptor-binding site (Mishin et al. 2010). A similar observation was seen for HPIV-1 HN residue N173 (Alymova et al. 2008), though the implications of these additional binding sites on viral infectivity have yet to be defined.
Similar to HPIV-3, SeV HN utilizes three of its four potential N-glycosylation sites, and 9% of its mass can be attributed to glycan modifications (Kohama et al. 1978; Segawa et al. 2003). These N-glycans play a significant role in intracellular transport and fusion (Segawa et al. 2003). Fifteen percent of the mass of SeV F is attributable to O-glycan modifications (Kohama et al. 1978), and SeV F has three N-glycosylation sites. The removal of the N245 site affected protein folding, trafficking, CSE, and, as a result, fusion (Segawa et al. 2000). High-mannose sugars are thought to be necessary for the immunoreactivity of SeV F, but not SeV HN (Mottet et al. 1986). Additionally, tunicamycin treatment inhibited SeV viral replication (Nakamura et al. 1982). Thus, it is clear that N-glycans play a significant role in protein processing, proper viral function for these viruses, and that targeting these glycans may be a potential therapeutic option.
Rubulavirus
Parainfluenza virus 5 (PIV-5) has a broad host range including cats, dogs, pigs, guinea pigs and hamsters, and PIVs can cause hospitalizations in humans (Chatziandreou et al. 2004). PIV-5 F utilizes all six potential N-glycosylation sites. The removal of the two F2 subunit N-glycans via N to A mutations had no significant effects. However, the removal of any of the four F1 subunit N-glycans resulted in protein degradation, instability and delayed intracellular transport, CSE and fusion (Bagai and Lamb 1995). PIV-5 HN contains four N-glycans that affect protein folding and assembly in a cumulative manner. Additionally, one of these four N-glycans prevents aggregation of oligomers in the ER (Ng et al. 1990). Thus, N-glycans on both PIV-5 F and HN play roles in proper protein folding, thus facilitating proper viral protein function.
Mumps virus (MuV) causes painful swelling of the salivary gland as well as the potential for systemic inflammation in organs including the brain, heart and kidneys. Though rarely fatal, in some cases infection can result in long-term symptoms such as paralysis, seizures and deafness. Before the introduction of a vaccine, mumps was a common childhood infection (Rubin et al. 2015). Tunicamycin treatment inhibits viral release, likely by trapping viral GPs in the cytoplasm of the infected cell. Both the HN and F GPs are N-glycosylated with both complex and high-mannose glycans (HN and F1 subunit) or only high mannose (F2 subunit), as revealed by endoglycosidase treatments (Herrler and Compans 1983). MuV F contains six potential N-glycosylation sites (Alirezaie et al. 2008), while MuV HN contains five to seven potential N-glycosylation sites (Lim et al. 2003; Yates et al. 1996), though site utilization and functions remain unknown. It is thus unknown whether the N-glycan functions among rubulaviruses are conserved.
Summary for Paramyxoviridae
While the paramyxoviruses vary extensively in host range and distribution, they share many similarities within their GPs. Both the F and HN/H/G GPs for each virus within this family contain a number of N-glycans, though the number of N-glycosylation sites utilized varies widely among genera. N-glycans commonly appear to have roles in protein expression, processing, trafficking to the cell surface, cell–cell fusion and production of infectious virions. Additionally, many N-glycans also affect pathogenicity and immunogenicity. Some, such as the glycans on HPIV3 HN, affect receptor binding, while others have no effect on receptor binding at all but affect cellular detachment. Additionally, O-glycans have been detected on some paramyxoviral GPs, and their functions so far have only been determined for NiV and HeV G GPs. Research will determine if these O-glycan functions are conserved among the paramyxoviral genera.
Pneumoviridae
Orthopneumovirus
Human respiratory syncytial virus (HRSV, also referred to as RSV) causes severe bronchiolitis in infants, accounting for over 50% of childhood bronchiolitis cases requiring hospitalization (Rodriguez and Ramilo 2014). It is estimated that most children will have at least one HRSV infection by the age of 2 (Borchers et al. 2013). Interestingly, for HRSV, only the fusion GP is required for cell–cell fusion and viral entry to occur, though the presence of HRSV G enhances fusion (Chang and Dutch 2012). HRSV infectivity is sensitive to N- or O-glycan removal (Lambert 1988). HRSV F utilizes three of its six potential N-glycosylation sites. The removal of these N-glycans through conservative N to Q mutations did not affect HRSV F expression or processing (cleavage activation). However, one mutant, N70Q, had a 40% increase in fusion, while another mutant, N500Q, had a 90% fusion reduction. A mutant lacking all three N-glycosylation sites had almost no fusion (Zimmer et al. 2001). Since conformations of these proteins were not tested, it is possible that the altered fusion phenotypes were due to altered protein conformations. Conformational MAbs may help determine if this is the case. Importantly, it is still unknown whether HRSV F glycans affect the binding of HRSV F to palivizumab, a therapeutic MAb against HRSV and to our knowledge still the only MAb approved for use in humans against any virus.
HRSV G has been shown to be heavily O-glycosylated, with only 3% of its mass from N-glycans but 55% from O-glycans based on gel motility (Lambert 1988). Interestingly, certain MAbs did not bind nonglycosylated G (Palomo et al. 2000), and the recognition of some MAbs depended on cell-specific glycosylation (Garcia-Beato et al. 1996). O-glycans have been shown not to be involved in HRSV G oligomerization (Collins and Mottet 1992). Additionally, modeling studies suggest a positive selection of certain O-glycans in response to immune pressure (Zlateva et al. 2004), suggesting that viral glycosylation may play an important role in the host immune response. Further study of immune responses to glycosylation may lead to more effective vaccines or therapeutic options.
Metapneumovirus
As with HRSV, human metapneumovirus (HMPV) causes severe upper and lower respiratory tract infections and accounts for 5–10% of childhood acute respiratory infection hospitalizations (Panda et al. 2014). Infection generally occurs during childhood, but can be recurrent throughout life. In addition to infecting children, HMPV also poses a threat to immunocompromised individuals, including the elderly (Panda et al. 2014). HMPV F utilizes all three potential N-glycosylation sites: one on the F2 and two on the F1 subunits. N to A mutations resulted in decreased cell–cell fusion, most likely caused by reduced F cleavage, and all mutants expressed well at the cell surface (Schowalter et al. 2006). In another study, mutants lacking the most C-terminal N-glycan, N353, either alone or in combination with the removal of other N-glycosylation sites did not produce virions (Zhang et al. 2011). Although N to Q vs. N to A mutations as well as virus vs. transfected cells were significant differences between these two studies, it appears that N-glycans on HMPV F affect cleavage of F into its F1 and F2 subunits, impacting viral replication and cell–cell fusion (Schowalter et al. 2006; Zhang et al. 2011). Viruses lacking the N172 glycan displayed impaired growth in vitro and in vivo and reduced pathology in both immunocompetent and immunocompromised mice (Zhang et al. 2011; Yu et al. 2012). Administration of a live vaccine attenuated by the loss of the N172 glycan resulted in immune protection in mice (Liu et al. 2013), highlighting the ways in which knowledge of viral glycosylation can aid viral infection prevention strategies.
Like HRSV G, HMPV G is heavily glycosylated, with three to six potential N-glycosylation sites depending on the strain, and an unknown number of O-glycans (Bastien et al. 2004; Galiano et al. 2006; Liu et al. 2007). Sequencing of HMPV G strains, however, has revealed a high serine and threonine content of 30–34%, suggesting that HMPV G may have a high O-glycan content (Liu et al. 2007). Effects of the actual or putative N- or O-glycans are still largely undetermined.
Summary for Pneumoviridae
Members of this family have been observed to undergo attachment protein-independent membrane fusion (Iorio et al. 2009; Smith et al. 2009). There is extensive O-linked as well as N-linked glycosylation of the G GP (Wang et al. 2012). This, along with high proline content, suggests the G GP has a mucin-like structure (Johnson et al. 1987; Langedijk et al. 1996). Though the functions of Pneumoviridae fusion GP N-glycans have been characterized, the purpose of G GP N- and O-glycans remains unknown.
Rhabdoviridae
The widely distributed rhabdoviruses infect both invertebrates and vertebrates, as well as a wide variety of plants. They have been divided into 18 genera. Four of these genera infect mammals: Lyssavirus, containing rabies virus (RABV), Tibrovirus, Vesiculovirus, containing VSV, and Ephemerovirus, containing bovine ephemeral fever virus (BEFV), and the Tupavirus infects birds (Da Poian et al. 2005; Calisher and Ellison 2012). Three genera that infect fish include Novirhabdoviruses, Perhabdoviruses and Spriviviruses. The Cytorhabdoviruses, Nucleorhabdoviruses, Dichorhaviruses and Varicosaviruses infect plants (Falzarano et al. 2006). Members of the Almendravirus, Sigmavirus, Ledantevirus, Peropuvirus and Hapavirus genera circulate within arthropod populations (Contamine 2008; Blasdell et al. 2015; Contreras et al. 2017; Wang et al. 2017). These viruses all have a bullet-shaped morphology and enter cells through receptor-mediated endocytosis followed by membrane fusion induced by low pH. Both host cell receptor binding and membrane fusion are executed by the envelope GP G, which for the two most widely studied viruses from this family, RABV and VSV, contains two N-glycosylation sites (Jayakar et al. 2004; Da Poian et al. 2005).
Almendravirus
Members of the Almendravirus genus were isolated from mosquitoes and found to effectively replicate in mosquito cells (Contreras et al. 2017). The Arboretum virus (ABTV) and Puerto Almendras virus (PTAMV) are suggested to have six putative N-glycosylation sites within its G ORF ectodomain (Vasilakis et al. 2014). The Balsa virus (BALV) and Rio Chico virus (RCHV) have two putative N-glycosylation sites, while Coot Bay virus (CBV) contains five (Contreras et al. 2017). The role of these putative N-glycans have not been determined.
Curiovirus
Members of the curiovirus genus have been reported to be lethal in mice, causing necrosis and pyknosis. The Curionopolis, Ibirius and Itacainus viruses were isolated in Brazil from species of Culicoides insects, causing neuropathogenesis in mice (Gomes-Leal et al. 2006; Diniz et al. 2008). No characterization of the curiovirus GPs has been conducted.
Cytorhabdovirus and Nucleorhabdovirus
The plant rhabdoviruses can be separated into the cytorhabdovirus and nucleorhabdovirus genera depending on their sites of replication and morphogenesis (Jackson et al. 2005). The number of potential N-glycosylation sites for these viruses varies widely. The cytorhabdovirus lettuce necrotic yellow virus (LNYV) G contains three potential N-glycosylation sites, while the Northern cereal mosaic virus (NCMV) contains six (Goldberg et al. 1991; Tanno et al. 2000; Jackson et al. 2005; Dietzgen et al. 2006). The nucleorhabdoviruses rice yellow stunt virus (RYSV) and sonchus yellow net virus (SYNV) contain 10 and 6 potential N-glycosylation sites, respectively (Jackson et al. 2005). Tunicamycin treatment suggests that at least some of these N-glycosylation sites are used (Jackson et al. 2005). Interestingly, there is very little sequence homology between NCMV and SYNV G, yet they are both potentially glycosylated at similar sites, suggesting the importance of glycosylation site conservation (Tanno et al. 2000). The functions of these N-glycans, however, have yet to be determined.
Dichorhavirus
The Dichoraviruses, which include the coffee ringspot virus (CoRSV) and orchid fleck virus (OFV) (synonymous with the citrus leprosis virus nuclear type and citrus necrotic spot virus), cause necrotic symptoms in plant species spread by the Brevipalpus species of mites. The OFV G protein has two potential N-glycosylation sites, and interestingly it has been proposed that the fewer potential N-glycans may contribute to the incomplete budding process of the virions (Kondo et al. 2006, 2009).
Ephemerovirus
BEFV infections occur mainly in cattle and buffalo in Africa, Asia and Australia, causing acute fever and lameness, resulting in high morbidity and economic loss. BEFV can also infect other ruminants without causing disease (Nandi and Negi 1999). BEFV G contains five potential N-glycosylation sites. Additionally, a nonstructural protein with 36% similarity to G, termed Gns, contains eight potential N-glycosylation sites (Johal et al. 2008). Gns is detected within infected cells; however, it is not found in virions and does not appear to induce an immune response. The utilization of the potential N-glycosylation sites and the glycan functions are still unknown (Walker et al. 1992; Johal et al. 2008).
Hapavirus
Several viruses of this new genus can infect mammals, birds, insects or reptiles, though most were isolated from mosquitoes (Causey et al. 1966; Karabatsos 1985; Simo Tchetgna et al. 2017). Ngaingan virus (NGAV) was first isolated from biting midges in Australia and is predicted to have four N-glycosylation sites on its NGAV G (Doherty et al. 1973; Gubala et al. 2010). The Marco virus was isolated from lizards in Brazil and has two putative N-linked glycosylation sites on its transmembrane GP (Causey et al. 1966; Walker et al. 2015). No glycan functions have been studied for any of the 15 Hapavirus species.
Ledantevirus
Within the Ledantevirus genus, the Kolente virus has four putative N-glycosylation sites with unexplored functions within the GP (Ghedin et al. 2013).
Lyssavirus
RABV infects most mammals and is transmitted through bites, scratches and saliva. Infection is characterized by neurological symptoms culminating in severe encephalitis and is fatal in humans if untreated (Calisher and Ellison 2012). RABV G usually contains one (N319) N-glycosylation site highly conserved among different RABV strains (Wunner et al. 1985, 1988; Badrane and Tordo 2001; Yamada et al. 2012). Additionally, an N-glycan at site N247 is found in many lab-adapted strains (Wunner et al. 1985). It has been suggested that the latter N-glycan may play a role in the reduced pathogenicity observed in many of these lab-adapted strains and possibly the increased efficiency for growth in cell culture (Yamada et al. 2012, 2014a). Further, N37 is inefficiently glycosylated and efforts to increase N-glycosylation efficiency reduced pathogenicity of the virus (Wunner et al. 1985; Shakin-Eshleman et al. 1992, 1993; Yamada et al. 2014b). Loss of the N-glycan at N319 resulted in decreased viral production and an inability for RABV G to fold correctly that inhibited membrane fusion (Yamada et al. 2013). Deletion of all three N-glycans completely blocked CSE. A mutant containing only the N37 glycan was still able to express on the cell surface, suggesting that this inefficiently glycosylated site is not important for CSE (Shakin-Eshleman et al. 1992). Addition of an extra N-glycan (N194), created from spontaneous mutation during serial passage, increased virus production (Yamada et al. 2013). Thus, it appears that N-glycans play a crucial role in proper RABV G CSE, though the combined effects of glycans at all the N-glycan sites suggest a crucial balance where too few or too many N-glycans negatively impact viral pathogenicity.
Tunicamycin treatment blocked CSE of RABV G but not intracellular synthesis (Burger et al. 1991; Wojczyk et al. 1995). Additionally, experiments with a panel of various CHO cell lines deficient for different steps in the N-glycan processing pathway revealed that RABV G expression was independent of the type of N-glycan processing, as all cell lines expressed RABV G on the cell surface (Burger et al. 1991). It is still possible that RABV G trafficked at different rates, as the timing of RABV G CSE was not explored. Notably, mutation of site N247 facilitated an additional glycosylation of site N319 (Wojczyk et al. 2005). Recombinant RABV G protein produced in eukaryotic cells (and thus glycosylated) was effective as a vaccine (Kieny et al. 1984; Wiktor et al. 1984), while RABV G protein produced in prokaryotic cells (and thus not glycosylated) was not effective (Yelverton et al. 1983). While these differences could be due to other factors such as protein conformation, it is possible that proper glycosylation of RABV G is necessary for effective immune responses. However, another study indicated that glycans may not be a required target for mouse antibodies (Wunner et al. 1985), suggesting that the immunoreactivity of RABV G is multifactorial. Undoubtedly, N-glycans play a critical role in at least proper CSE of RABV G, though their effects on the host immune response merit further exploration.
Novirhabdovirus
Viruses within the Novirhabdovirus genus infect fish species, causing significant morbidity and mortality. These viruses include infectious hematopoietic necrosis virus (IHNV), hirame rhabdovirus (HIRV) and viral hemorrhagic septicemia virus (VHSV), among others, and cause vast aquaculture and wildlife problems (Purcell et al. 2012). Though the number of potential N-glycans varies by virus, a putative N-glycan at position N438 is present in each virus analyzed (Lorenzen et al. 1993; Bjorklund et al. 1996). IHNV G contains four potential N-glycosylation sites and one potential O-glycosylation site (Ammayappan et al. 2010). VHSV also contains four potential N-glycosylation sites (Chico et al. 2010), and IHNV and VHSV share similar glycosylation patterns: one putative N-glycan at the N-terminus, none in the middle of the protein and three at the C-terminus (Lorenzen et al. 1993). HIRV contains three putative N-glycosylation sites—all at the C-terminus (Bjorklund et al. 1996). Studies on the function of these glycans, however, have yet to be undertaken.
Perhabdovirus
Perhabdoviruses are associated with fish infections. The eel virus European X (EVEX) is a major concern with the farming of freshwater eels, Anguilla species, with increased mortality and symptoms including hemorrhages in the skin and lesions (Caruso et al. 2014). The EVEX G protein has one N-glycosylation site at N185 (Galinier et al. 2012). Functions of glycans in this genus remain unknown.
Sigmavirus
Five members of the Drosophila genus, and one member of the muscidae are natural hosts of sigma viruses that cause paralysis and death upon exposure to carbon dioxide. Sigma viruses have a vertical mode of transmission through the gametes of male or female flies. Drosophila melanogaster sigmavirus (DMelSV) has a surface GP (G) (Longdon et al. 2010, 2011). Not much is known regarding the number or function of glycans in this genus.
Sprivivirus
Viruses within the Sprivivirus genus infect fish species, causing significant morbidity and mortality (Purcell et al. 2012). These viruses include spring viremia of carp virus (SVCV) and pike fry rhabdovirus (PFRV). Not much is known about glycosylation for these viruses, except that the SVCV G GP contains five potential N-glycosylation sites. A variation in the glycosylation site N362 is due to nucleotide insertion differences between SVCV strains (Bjorklund et al. 1996; Zhang et al. 2009). Potential function of these glycans for any of these viruses have yet to be studied.
Sripuvirus
Within the Sripuvirus genus, the Almpiwar virus G GP contains two possible glycosylation sites with unknown biological function (McAllister et al. 2014). The sandfly Niakha virus G GP is reported to possess two potential N-glycosylation sites (Vasilakis et al. 2013).
Tibrovirus
Tibrovirus is transmitted by Culicoides insects and infects members of the bovine species. Tibroviruses have not been observed to induce illness. Both the Tibrogargan virus (TIBV) and the Coastal Plains virus (CPV) G proteins are predicted to contain nine N-glycosylation sites (Cybinski et al. 1980; Gubala et al. 2011). No account has been made on the function of glycosylation sites in this genus.
Tupavirus
The Tupaia virus (TUPV) was first isolated in a liver tumor of a Tupaia tree shrew, Tupaia belangeri (Kurz et al. 1986). The TUPV G protein possesses three potential N-glycosylation sites (Springfeld et al. 2005). The Durham virus (DURV) was first isolated in 2005 in an American coot suffering from neurological symptoms. The DURV G protein was reported to have two possible N-glycosylation sites (Allison et al. 2011). Not much is known regarding the number or function of glycans in this genus.
Varicosavirus
The sole member of the Varicosavirus genus is the lettuce big-vein-associated virus (LBVaV). LBVaV causes pale yellow veins on leaves of lettuce and is characterized by leaf puckering. It is considered to be soil-borne and transmitted by zoospores of the fungus, Olpidium brassicae, and healthy lettuce planted into contaminated soil will develop signs of infection (Tomlinson et al. 1962; Umar et al. 2017). LBVaV has a difficult mechanical transmission which makes studying it problematic (Sasaya et al. 2011). There is no report on the glycobiology of this genus.
Vesiculovirus
VSV infects mainly livestock, causing severe blistering and/or ulceration of the tongue, mouth and feet, often resulting in extensive productivity loss (Letchworth et al. 1999). VSV is also used extensively in the production of pseudotyped viruses due to its ability to incorporate viral GPs from many other viral families (Huang et al. 1974; Witte and Baltimore 1977). VSV GP (G) contains two well-conserved N-glycosylation sites (N181 and N336), which make up approximately 10% of the GP’s mass, and no O-glycosylation sites have been discovered (Etchison and Holland 1974a, 1974b; Robertson et al. 1976; Reading et al. 1978; Farley et al. 2007). Tunicamycin treatment, particularly of the San Juan strain, impaired intracellular transport of VSV G to the plasma membrane (Leavitt et al. 1977a; Morrison et al. 1978; Gibson et al. 1979), inhibited viral replication (Leavitt et al. 1977b) and altered MAb-binding patterns to the VSV G GP (Grigera et al. 1991). However, these effects appeared to be partially strain-specific, as tunicamycin treatment of the New Jersey strain slowed the folding rate of VSV G but did not prevent its eventual expression at the cell surface (Mathieu et al. 1996). Additionally, the Orsay strain showed some resistance to tunicamycin treatment (Gibson et al. 1979).
It appears that these observed effects are reliant on the presence of a glycan itself, but the specific types of carbohydrates added are not important. For example, VSV particles produced in a variety of CHO cells lacking specific steps in the glycosylation pathway were still able to replicate and were as infectious as their fully glycosylated counterparts (Schlesinger et al. 1975; Robertson et al. 1978). It was also shown that either of the two N-glycans was necessary and sufficient for proper G transport, (Machamer et al. 1985). This observation is further supported by a naturally occurring mutant containing only one N-glycan that trafficked properly to the cell surface (Kotwal et al. 1986). It was also observed that the addition of new N-glycosylation sites to unglycosylated VSV G allowed for transport of G to the plasma membrane, though this restored transport ability was only successful for two of the six engineered N-glycosylation sites (Machamer and Rose 1988a).
These findings and experiments with temperature-sensitive mutants have led to the hypothesis that N-glycans on VSV G are important for proper folding and conformation (Chatis and Morrison 1981; Machamer and Rose 1988b), perhaps affecting the stability of disulfide bonds (Grigera et al. 1991), and altered conformations in the absence of N-glycosylation sites results in a lack of transport to the cell surface (Chatis and Morrison 1981; Machamer and Rose 1988b). It has been observed that low amounts of infectious virus are produced even in the presence of tunicamycin and it is possible that some glycan mutant virions with different conformations may thus be able to traffic to the cell surface despite a lack of glycosylation (Gibson et al. 1978). This may also explain why certain strains are more resistant to tunicamycin treatment than others, as different amino acid sequences may result in different conformations that are more or less reliant on N-glycans for their structural integrity (Chatis and Morrison 1981). Thus, it appears that either of the two natural N-glycan is necessary and sufficient for proper VSV G transport and expression, suggesting that proper GP conformation, and not specific N-glycans, is most important for VSV G expression and function.
Summary for Rhabdoviridae
The lyssavirus and vesiculovirus genera have two highly conserved N-glycans on G, which suggests these sites are evolutionarily important. Indeed, sequence comparisons between RABV G and VSV G show approximately 20% overall sequence identity between these two GPs, yet greater than 50% identity around the most C-terminal N-glycosylation site, with exact alignment at the N-glycosylation site itself (Rose et al. 1982). For both genera, N-glycans appear to be necessary for proper transport to the cell surface, with one N-glycan being sufficient for the cell transport function (Machamer et al. 1985; Kotwal et al. 1986; Shakin-Eshleman et al. 1992). Additionally, dependence on N-glycans for proper trafficking is independent on the type of glycan processing, as evidenced by a variety of glycosylation-deficient CHO cells where the G GP was still expressed at the cell surface (Burger et al. 1991; Schlesinger et al. 1975; Robertson et al. 1978). The effects of N-glycans appear to be more strain-specific for VSV. Although RABV strains differ more in the number of N-glycans present, the effects of tunicamycin treatment appear to be more variable among VSV strains than among RABV strains. General comparisons among the other Rhabdoviruses are difficult to make since far fewer studies exist. However, these viruses could potentially contain more N-glycosylation sites than their lyssavirus and vesiculovirus counterparts, though studies determining site usage have yet to be undertaken.
Sunviridae
The Sunviridae family is solely comprised of the Sunshine Coast virus in the Sunshinevirus genus. This virus was initially isolated from Australian python snake populations showing symptoms of neurorespiratory disease and is not known to cause illness in humans (Hyndman et al. 2012). No studies have been performed on the function of glycans for this virus.
Discussion and conclusions
Glycosylation is a common protein modification process utilized by animals, plants, bacteria, fungi, viruses and even some archaea, illustrating its importance (Varki, Freeze et al. 2009). In recent years, many human illnesses, such as autoimmune diseases and cancer, have been linked to altered glycosylation (Varki and Freeze 2009; Varki, Kannagi et al. 2009). Glycosylation is commonly used by pathogens in host invasion and immune evasion. Within the Mononegavirales order there are many conserved functions of glycosylation, such as receptor binding, viral entry, protein processing or trafficking, cell–cell fusion and immunogenicity (Table I, Figure 3), all important roles for the viral life cycle. It should be noted that much more has been discovered for N-glycans than O-glycans, partly due to the difficulty in determining O-glycosylation sites compared to N-glycosylation sites. Few O-glycan functions have been described. Many functions have been attributed to O-glycan-rich regions, such as that of the Ebola GP, yet these functions have yet to be directly attributed to the O-glycans themselves. In a new study for the Henipavirus genus, novel roles were shown for NiV and HeV O-glycans (Stone et al. 2016). It remains to be seen if such roles apply to other paramyxoviruses or mononegaviruses.
Table I.
Many glycan functions are conserved among viral genera. Summary of the different functions documented for each viral genera. N = N-glycans are involved. O = O-glycans are involved. Dashed line = not involved, blan k = not documented, unknown. # marks functions documented for an O-glycan-rich region (but not specifically the O-glycans themselves)
| Family | Genus | Receptor binding | Viral entry | Processing and trafficking | Cell–cell fusion | Infectious virion production | Immunogenicity |
|---|---|---|---|---|---|---|---|
| Bornaviridae | Bornavirus | N | |||||
| Filoviridae | Cuevavirus | N, # | |||||
| Ebolavirus | N | N, # | N, # | # | N, # | ||
| Marburgvirus | |||||||
| Mymonaviridae | Sclerotimonavirus | ||||||
| Nyamiviridae | Nyavirus | ||||||
| Peropuvirus | |||||||
| Socyvirus | |||||||
| Paramyxoviridae | Aquaparamyxovirus | N | N | ||||
| Avulavirus | – | N | N | N | |||
| Ferlavirus | |||||||
| Henipavirus | – | N, O | N, O | N, O | N | ||
| Morbillivirus | N | N | N | N | |||
| Respirovirus | N | N | N | N | N | ||
| Rubulavirus | N | N | N | ||||
| Pneumoviridae | Metapneumovirus | N | N | N | |||
| Orthopneumovirus | N | ||||||
| Rhabdoviridae | Almendavirus | ||||||
| Curiovirus | |||||||
| Cytorhabdovirus | |||||||
| Dichorhavirus | |||||||
| Ephemerovirus | |||||||
| Hapavirus | |||||||
| Ledantevirus | |||||||
| Lyssavirus | N | N | N | ||||
| Novirhabdovirus | |||||||
| Nucleorhabdovirus | |||||||
| Perhabdovirus | |||||||
| Sigmavirus | |||||||
| Sprivivirus | |||||||
| Sripuvirus | |||||||
| Tibrovirus | |||||||
| Tupavirus | |||||||
| Varicosavirus | |||||||
| Vesiculovirus | N | N | N | ||||
| Sunviridae | Sunshinevirus |
A more thorough understanding of viral glycosylation has the potential for therapeutic use. Glycomimetics, compounds that mimic the bioactive properties of carbohydrates, are being developed and tested for a number of different viral infections (Ernst and Magnani 2009; Magnani and Ernst 2009). The glucose-mimetic N-9-methoxynonyl (DNJ), for example, inhibits a part of the glycosylation pathway, thus preventing viral GPs from folding properly (Francois and Balzarini 2012; Dalziel et al. 2014). DNJ has been shown to be effective in combating HIV, hepatitis B and C viruses and has been suggested as a possible therapeutic against the flaviviruses dengue virus and West Nile virus (Merry and Astrautsova 2010). Carbohydrate mimetics are suggested to be good drug candidates because they generally have a low toxicity, are water soluble, can be easily modified by adding different functional groups at multiple locations and have the potential to be used as scaffolds for more complex drugs (Merry and Astrautsova 2010). However, carbohydrates generally have been thought to make poor drug candidates due to their high polarity, preventing passive transport through the intestinal lining (and thus problematic for oral dosing regimens). The use of carbohydrate mimetics enables sufficient manipulation to overcome this barrier (Ernst and Magnani 2009).
The addition of glycans to proteins can also be used as a tool to study protein functions. For example, for the paramyxoviral HN/H/G attachment GPs, N-glycans have been added to study the effects of the protein stalk on membrane fusion and interactions with the fusion protein. This has been done for NDV, PIV-5 and NiV to probe protein function (Melanson and Iorio 2006; Bose et al. 2011; Zhu et al. 2013).
Lectins and other carbohydrate-binding agents are also being investigated as antiviral therapies (Ernst and Magnani 2009). Cyanovirin is a lectin that binds the gp120 GP of HIV and prevents entry and was tested against many other viruses as well (Merry and Astrautsova 2010; Francois and Balzarini 2012). Carbohydrate-binding agents cannot only bind the viral GPs to prevent entry but also can create a long-term selective pressure for viral strains with the protective N-glycan regions removed, thus making the virus more susceptible to immune system neutralization (Francois and Balzarini 2012). Lectins are also found naturally across a wide variety of species (prokaryotes, algae, fungi, plants, invertebrates and vertebrates). Thus, there are many different lectins to explore as potential viral inhibitors (Francois and Balzarini 2012). Antibodies against the carbohydrates of viral proteins, though less common, can also be effective. 2G12 is an antibody that binds an N-glycan region of gp120 on HIV, thus inhibiting viral entry into host cells and limiting HIV infections (Merry and Astrautsova 2010; Francois and Balzarini 2012). Thus, a number of viral glycosylation-targeting therapeutics are in development that have potential to inhibit viral infections. It is also likely that a combination of these strategies will prove best for maximal therapeutic benefit.
While significant advances in understanding and characterizing viral glycosylation have been made, there are still many knowledge gaps. Mass spectrometric data for many viral GPs is still nonexistent, particularly for O-glycans. Some of the methods commonly used to study glycosylation effects, such as tunicamycin treatment, may cause additional cellular effects, thus any effects observed after tunicamycin treatment may be from the treatment itself and not directly from a lack of glycosylation. A significant hurdle to overcome is that there is considerable variation in glycosylation machinery among different cell types; therefore, the effects of glycosylation observed in one study may not hold true in other cell types or in vivo. Nevertheless, the advances made in recent years and the knowledge gained continues to highlight how crucial proper glycosylation of viral GPs is for their function and for viral pathogenicity. Improved understanding of viral glycosylation is essential to pave the way for future therapeutics and vaccines targeting important medical, agricultural and veterinary viruses.
Abbreviations
- ABTV
Arboretum virus
- APMV-1
avian paramyxovirus
- AsaPV
Atlantic salmon paramyxovirus
- BALV
Balsa virus
- BEFV
bovine ephemeral fever virus
- CBV
Coot Bay virus
- CDV
canine distemper virus
- CoRSV
coffee ringspot virus
- CPV
Coastal Plains virus
- CSE
cell surface expression
- DMelSV
Drosophila melanogaster sigmavirus
- DURV
Durham virus
- EBOV
Ebola virus
- ER
endoplasmic reticulum
- EVEX
eel virus European X
- FDLV
Fer-de-Lance virus
- GP
glycoprotein
- HeV
Hendra virus
- HIRV
hirame rhabdovirus
- HIV
human immunodeficiency virus
- HMPV
human metapneumovirus
- HPIV
human parainfluenza virus
- HRSV
human respiratory syncytial virus
- IHNV
infectious hematopoietic necrosis virus
- LBVaV
lettuce big-vein-associated virus
- LLOV
Lloviu virus
- LNYV
lettuce necrotic yellow virus
- Mev
measles virus
- MIDWV
Midway virus
- MLD
mucin-like domain
- MuV
mumps virus
- NCMV
Northern cereal mosaic virus
- NGAV G
Ngaingan virus
- NiV
Nipah virus
- NYMV
Nyamanini virus
- OFV
orchid fleck virus
- PFRV
pike fry rhabdovirus
- PIV
parainfluenza viruses
- PIV-5
parainfluenza virus 5
- PNGase F
peptide-N-glycosidase F
- PpNSRV-1
Pteromalus puparum negative-strand RNA virus 1
- PTAMV
Puerto Almendras virus
- RABV
rabies virus
- RCHV
Rio Chico virus
- RYSV
rice yellow stunt virus
- SbCNV
soybean cyst nematode virus-1
- SeV
Sendai virus
- SLAM
signaling lymphocyte activation molecule
- SNVV
Sierra Nevada virus
- SsNSRV-1
Sclerotinia sclerotiorum negative-stranded RNA virus 1
- SVCV
spring viremia of carp virus
- SYNV
sonchus yellow net virus
- TIBV
Tibrogargan virus
- TUPV
Tupaia virus
- VHSV
viral hemorrhagic septicemia virus
- VLPs
virus-like particles
- VSV
vesicular stomatitis virus.
Funding
This work was supported by the National Institutes of Health (AI109022 to H.A.C., T32GM008336 to J.A.S., E.M.C. and V.O.).
Conflict of interest statement
None declared.
References
- Aebi M. 2013. N-linked protein glycosylation in the ER. Biochim Biophys Acta. 1833:2430–2437. [DOI] [PubMed] [Google Scholar]
- Afonso CL, Amarasinghe GK, Banyai K, Bao YM, Basler CF, Bavari S, Bejerman N, Blasdell KR, Briand FX, Briese T et al. . 2016. Taxonomy of the order Mononegavirales: Update 2016. Arch Virol. 161:2351–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar HC, Henderson BA, Zamora JL, Johnston GP. 2016. Paramyxovirus glycoproteins and the membrane fusion process. Curr Clin Microbiol Rep. 3:142–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar HC, Iorio RM. 2012. Henipavirus membrane fusion and viral entry. Curr Top Microbiol Immunol. 359:79–94. [DOI] [PubMed] [Google Scholar]
- Aguilar HC, Matreyek KA, Filone CM, Hashimi ST, Levroney EL, Negrete OA, Bertolotti-Ciarlet A, Choi DY, McHardy I, Fulcher JA et al. . 2006. N-glycans on Nipah virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J Virol. 80:4878–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alirezaie B, Aghaiypour K, Shafyi A. 2008. Genetic characterization of RS-12 (S-12), an Iranian isolate of mumps virus, by sequence analysis and comparative genomics of F, SH, and HN genes. J Med Virol. 80:702–710. [DOI] [PubMed] [Google Scholar]
- Alkhatib G, Shen SH, Briedis D, Richardson C, Massie B, Weinberg R, Smith D, Taylor J, Paoletti E, Roder J. 1994. Functional analysis of N-linked glycosylation mutants of the measles virus fusion protein synthesized by recombinant vaccinia virus vectors. J Virol. 68:1522–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison AB, Palacios G, da Rosa AT, Popov VL, Lu L, Xiao SY, Detoy K, Briese T, Lipkin WI, Keel MK et al. . 2011. Characterization of Durham virus, a novel rhabdovirus that encodes both a C and SH protein. Virus Res. 155:112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alymova IV, Taylor G, Mishin VP, Watanabe M, Murti KG, Boyd K, Chand P, Babu YS, Portner A. 2008. Loss of the N-linked glycan at residue 173 of human parainfluenza virus type 1 hemagglutinin-neuraminidase exposes a second receptor-binding site. J Virol. 82:8400–8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarasinghe GK, Bao Y, Basler CF, Bavari S, Beer M, Bejerman N, Blasdell KR, Bochnowski A, Briese T, Bukreyev A et al. . 2017. Taxonomy of the order Mononegavirales: Update 2017. Arch Virol. 162:2493–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammayappan A, LaPatra SE, Vakharia VN. 2010. Molecular characterization of the virulent infectious hematopoietic necrosis virus (IHNV) strain 220-90. Virol J. 7:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badrane H, Tordo N. 2001. Host switching in Lyssavirus history from the Chiroptera to the Carnivora orders. J Virol. 75:8096–8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagai S, Lamb RA. 1995. Individual roles of N-linked oligosaccharide chains in intracellular transport of the paramyxovirus SV5 fusion protein. Virology. 209:250–256. [DOI] [PubMed] [Google Scholar]
- Bailly B, Dirr L, El-Deeb IM, Altmeyer R, Guillon P, von Itzstein M. 2016. A dual drug regimen synergistically blocks human parainfluenza virus infection. Sci Rep. 6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastien N, Liu L, Ward D, Taylor T, Li Y. 2004. Genetic variability of the G glycoprotein gene of human metapneumovirus. J Clin Microbiol. 42:3532–3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bause E. 1983. Structural requirements of N-glycosylation of proteins. Studies with proline peptides as conformational probes. Biochem J. 209:331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekal S, Domier LL, Niblack TL, Lambert KN. 2011. Discovery and initial analysis of novel viral genomes in the soybean cyst nematode. J Gen Virol. 92:1870–1879. [DOI] [PubMed] [Google Scholar]
- Biering SB, Huang A, Vu AT, Robinson LR, Bradel-Tretheway B, Choi E, Lee B, Aguilar HC. 2012. N-Glycans on the Nipah virus attachment glycoprotein modulate fusion and viral entry as they protect against antibody neutralization. J Virol. 86:11991–12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorklund HV, Higman KH, Kurath G. 1996. The glycoprotein genes and gene junctions of the fish rhabdoviruses spring viremia of carp virus and hirame rhabdovirus: Analysis of relationships with other rhabdoviruses. Virus Res. 42:65–80. [DOI] [PubMed] [Google Scholar]
- Blasdell KR, Guzman H, Widen SG, Firth C, Wood TG, Holmes EC, Tesh RB, Vasilakis N, Walker PJ. 2015. Ledantevirus: A proposed new genus in the Rhabdoviridae has a strong ecological association with bats. Am J Trop Med Hyg. 92:405–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolt G, Pedersen IR, Blixenkrone-Moller M. 1999. Processing of N-linked oligosaccharides on the measles virus glycoproteins: importance for antigenicity and for production of infectious virus particles. Virus Res. 61:43–51. [DOI] [PubMed] [Google Scholar]
- Borchers AT, Chang C, Gershwin ME, Gershwin LJ. 2013. Respiratory syncytial virus—A comprehensive review. Clin Rev Allergy Immunol. 45:331–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose S, Welch BD, Kors CA, Yuan P, Jardetzky TS, Lamb RA. 2011. Structure and mutagenesis of the parainfluenza virus 5 hemagglutinin-neuraminidase stalk domain reveals a four-helix bundle and the role of the stalk in fusion promotion. J Virol. 85:12855–12866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradel-Tretheway BG, Liu Q, Stone JA, McInally S, Aguilar HC. 2015. Novel functions of hendra virus G N-glycans and comparisons to Nipah virus. Journal of virology. 89:7235–7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhausen I, Schachter H, Stanley P. 2009. O-GalNAc glycans In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of glycobiology. New York: Cold Spring Harbor. p. 115–128. [PubMed] [Google Scholar]
- Burger SR, Remaley AT, Danley JM, Moore J, Muschel RJ, Wunner WH, Spitalnik SL. 1991. Stable expression of rabies virus glycoprotein in Chinese hamster ovary cells. J Gen Virol. 72(Pt 2):359–367. [DOI] [PubMed] [Google Scholar]
- Calisher CH, Ellison JA. 2012. The other rabies viruses: The emergence and importance of lyssaviruses from bats and other vertebrates. Travel Med Infect Dis. 10:69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter JR, Pager CT, Fowler SD, Dutch RE. 2005. Role of N-linked glycosylation of the Hendra virus fusion protein. J Virol. 79:7922–7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso C, Peletto S, Gustinelli A, Arsieni P, Mordenti O, Modesto P, Acutis PL, Masoero L, Fioravanti ML, Prearo M. 2014. Detection of a phylogenetically divergent eel virus European X (EVEX) isolate in European eels (Anguilla anguilla) farmed in experimental tanks in Italy. Aquaculture. 434:115–120. [Google Scholar]
- Causey OR, Shope RE, Bensabath G. 1966. Marco, Timbo, and Chaco, newly recognized arboviruses from lizards of Brazil. Am J Trop Med Hyg. 15:239–243. [DOI] [PubMed] [Google Scholar]
- Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. 2005. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 308:1643–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A, Dutch RE. 2012. Paramyxovirus fusion and entry: Multiple paths to a common end. Viruses. 4:613–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatis PA, Morrison TG. 1981. Mutational changes in the vesicular stomatitis virus glycoprotein affect the requirement of carbohydrate in morphogenesis. J Virol. 37:307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatziandreou N, Stock N, Young D, Andrejeva J, Hagmaier K, McGeoch DJ, Randall RE. 2004. Relationships and host range of human, canine, simian and porcine isolates of simian virus 5 (parainfluenza virus 5). J Gen Virol. 85:3007–3016. [DOI] [PubMed] [Google Scholar]
- Chico V, Martinez-Lopez A, Ortega-Villaizan M, Falco A, Perez L, Coll JM, Estepa A. 2010. Pepscan mapping of viral hemorrhagic septicemia virus glycoprotein G major lineal determinants implicated in triggering host cell antiviral responses mediated by type I interferon. J Virol. 84:7140–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu FL, Wen HL, Hou GH, Lin B, Zhang WQ, Song YY, Ren GJ, Sun CX, Li ZM, Wang Z. 2013. Role of N-linked glycosylation of the human parainfluenza virus type 3 hemagglutinin-neuraminidase protein. Virus Res. 174:137–147. [DOI] [PubMed] [Google Scholar]
- Clark HF, Lief FS, Lunger PD, Waters D, Leloup P, Foelsch DW, Wyler RW. 1979. Fer de Lance virus (FDLV): A probable paramyxovirus isolated from a reptile. J Gen Virol. 44:405–418. [DOI] [PubMed] [Google Scholar]
- Colgrave ML, Snelling HJ, Shiell BJ, Feng YR, Chan YP, Bossart KN, Xu K, Nikolov DB, Broder CC, Michalski WP. 2012. Site occupancy and glycan compositional analysis of two soluble recombinant forms of the attachment glycoprotein of Hendra virus. Glycobiology. 22:572–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins PL, Mottet G. 1992. Oligomerization and post-translational processing of glycoprotein G of human respiratory syncytial virus: Altered O-glycosylation in the presence of brefeldin A. J Gen Virol. 73(Pt 4):849–863. [DOI] [PubMed] [Google Scholar]
- Contamine DaG S. 2008. Sigma rhabdoviruses. Encyclopedia Virol. 5:576–581. [Google Scholar]
- Contreras MA, Eastwood G, Guzman H, Popov V, Savit C, Uribe S, Kramer LD, Wood TG, Widen SG, Fish D et al. . 2017. Almendravirus: A proposed new genus of rhabdoviruses isolated from mosquitoes in tropical regions of the Americas. Am J Trop Med Hyg. 96:100–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybinski DH, Stgeorge TD, Standfast HA, McGregor A. 1980. Isolation of tibrogargan virus, a new Australian rhabdovirus, from Culicoides brevitarsis. Vet Microbiol. 5:301–308. [Google Scholar]
- Da Poian AT, Carneiro FA, Stauffer F. 2005. Viral membrane fusion: Is glycoprotein G of rhabdoviruses a representative of a new class of viral fusion proteins? Braz J Med Biol Res. 38:813–823. [DOI] [PubMed] [Google Scholar]
- Dalziel M, Crispin M, Scanlan CN, Zitzmann N, Dwek RA. 2014. Emerging principles for the therapeutic exploitation of glycosylation. Science. 343:1235681. [DOI] [PubMed] [Google Scholar]
- Davey RA, Shtanko O, Anantpadma M, Sakurai Y, Chandran K, Maury W. 2017. Mechanisms of filovirus entry In: Current topics in microbiology and immunology. Berlin, Heidelberg: Springer Berlin Heidelberg; p. 1–30. [DOI] [PubMed] [Google Scholar]
- Deng R, Wang Z, Glickman RL, Iorio RM. 1994. Glycosylation within an antigenic site on the HN glycoprotein of Newcastle disease virus interferes with its role in the promotion of membrane fusion. Virology. 204:17–26. [DOI] [PubMed] [Google Scholar]
- Dietzgen RG, Callaghan B, Wetzel T, Dale JL. 2006. Completion of the genome sequence of Lettuce necrotic yellows virus, type species of the genus Cytorhabdovirus. Virus Res. 118:16–22. [DOI] [PubMed] [Google Scholar]
- Diniz JAP, dos Santos ZA, Braga MAG, Dias ALB, da Silva DEA, Medeiros DBD, Barros V, Chiang JO, Zoghbi KED, Quaresma JAS et al. . 2008. Early and late pathogenic events of newborn mice encephalitis experimentally induced by Itacaiunas and Curionopolis bracorhabdoviruses infection. PLoS One. 3:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty RL, Carley JG, Standfast HA, Dyce AL, Kay BH, Snowdon WA. 1973. Isolation of arboviruses from mosquitoes, biting midges, sandflies and vertebrates collected in Queensland, 1969 and 1970. Trans R Soc Trop Med Hyg. 67:536–543. [DOI] [PubMed] [Google Scholar]
- Dowling W, Thompson E, Badger C, Mellquist JL, Garrison AR, Smith JM, Paragas J, Hogan RJ, Schmaljohn C. 2007. Influences of glycosylation on antigenicity, immunogenicity, and protective efficacy of ebola virus GP DNA vaccines. J Virol. 81:1821–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler JF, Corman VM, Müller MA, Maganga GD, Vallo P, Binger T, Gloza-Rausch F, Rasche A, Yordanov S, Seebens A et al. . 2012. Bats host major mammalian paramyxoviruses. Nat Commun. 3:796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst B, Magnani JL. 2009. From carbohydrate leads to glycomimetic drugs. Nat Rev Drug Discov. 8:661–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esko J, Bertozzi C. 2009. Chemical tools for inhibiting glycosylation In: Varki A, Cummings R, Esko J et al., editors. Essentials of glycobiology. New York: Cold Spring Harbor. p. 705–718. [Google Scholar]
- Esko JD, Stanley P. 2009. Glycosylation mutants of cultured cells In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of glycobiology. New York: Cold Spring Harbor. p. 649–660. [PubMed] [Google Scholar]
- Etchison JR, Holland JJ. 1974. a. Carbohydrate composition of the membrane glycoprotein of vesicular stomatitis virus. Virology. 60:217–229. [DOI] [PubMed] [Google Scholar]
- Etchison JR, Holland JJ. 1974. b. Carbohydrate composition of the membrane glycoprotein of vesicular stomatitis virus grown in four mammalian cell lines. Proc Natl Acad Sci USA. 71:4011–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk K, Batts WN, Kvellestad A, Kurath G, Wiik-Nielsen J, Winton JR. 2008. Molecular characterisation of Atlantic salmon paramyxovirus (ASPV): A novel paramyxovirus associated with proliferative gill inflammation. Virus Res. 133:218–227. [DOI] [PubMed] [Google Scholar]
- Falzarano D, Krokhin O, Wahl-Jensen V, Seebach J, Wolf K, Schnittler HJ, Feldmann H. 2006. Structure-function analysis of the soluble glycoprotein, sGP, of Ebola virus. Chembiochem. 7:1605–1611. [DOI] [PubMed] [Google Scholar]
- Farley DC, Iqball S, Smith JC, Miskin JE, Kingsman SM, Mitrophanous KA. 2007. Factors that influence VSV-G pseudotyping and transduction efficiency of lentiviral vectors-in vitro and in vivo implications. J Gene Med. 9:345–356. [DOI] [PubMed] [Google Scholar]
- Feldmann H, Geisbert TW. 2011. Ebola haemorrhagic fever. Lancet. 377:849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann H, Nichol ST, Klenk HD, Peters CJ, Sanchez A. 1994. Characterization of filoviruses based on differences in structure and antigenicity of the virion glycoprotein. Virology. 199:469–473. [DOI] [PubMed] [Google Scholar]
- Feldmann H, Will C, Schikore M, Slenczka W, Klenk HD. 1991. Glycosylation and oligomerization of the spike protein of marburg virus. Virology. 182:353–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folsch DW, Leloup P. 1976. Fatal endemic infection in a serpentarium. Diagnosis, treatment and preventive measures. Tierarztl Prax. 4:527–536. [PubMed] [Google Scholar]
- Francois KO, Balzarini J. 2012. Potential of carbohydrate-binding agents as therapeutics against enveloped viruses. Med Res Rev. 32:349–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiano M, Trento A, Ver L, Carballal G, Videla C. 2006. Genetic heterogeneity of G and F protein genes from Argentinean human metapneumovirus strains. J Med Virol. 78:631–637. [DOI] [PubMed] [Google Scholar]
- Galinier R, van Beurden S, Amilhat E, Castric J, Schoehn G, Verneau O, Fazio G, Allienne J-F, Engelsma M, Sasal P et al. . 2012. Complete genomic sequence and taxonomic position of eel virus European X (EVEX), a rhabdovirus of European eel. Virus Res. 166:1–12. [DOI] [PubMed] [Google Scholar]
- Ganar K, Das M, Sinha S, Kumar S. 2014. Newcastle disease virus: Current status and our understanding. Virus Res. 184:71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beato R, Martinez I, Franci C, Real FX, Garcia-Barreno B, Melero JA. 1996. Host cell effect upon glycosylation and antigenicity of human respiratory syncytial virus G glycoprotein. Virology. 221:301–309. [DOI] [PubMed] [Google Scholar]
- Garner OB, Aguilar HC, Fulcher JA, Levroney EL, Harrison R, Wright L, Robinson LR, Aspericueta V, Panico M, Haslam SM et al. . 2010. Endothelial galectin-1 binds to specific glycans on Nipah virus fusion protein and inhibits maturation, mobility, and function to block syncytia formation. PLoS Pathog. 6:e1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner OB, Yun T, Pernet O, Aguilar HC, Park A, Bowden TA, Freiberg AN, Lee B, Baum LG. 2015. Timing of galectin-1 exposure differentially modulates Nipah virus entry and syncytium formation in endothelial cells. J Virol. 89:2520–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer H, Will C, Feldmann H, Klenk HD, Geyer R. 1992. Carbohydrate structure of Marburg virus glycoprotein. Glycobiology. 2:299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghedin E, Rogers MB, Widen SG, Guzman H, da Rosa A, Wood TG, Fitch A, Popov V, Holmes EC, Walker PJ et al. . 2013. Kolente virus, a rhabdovirus species isolated from Communication ticks and bats in the Republic of Guinea. J Gen Virol. 94:2609–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson R, Leavitt R, Kornfeld S, Schlesinger S. 1978. Synthesis and infectivity of vesicular stomatitis virus containing nonglycosylated G protein. Cell. 13:671–679. [DOI] [PubMed] [Google Scholar]
- Gibson R, Schlesinger S, Kornfeld S. 1979. The nonglycosylated glycoprotein of vesicular stomatitis virus is temperature-sensitive and undergoes intracellular aggregation at elevated temperatures. J Biol Chem. 254:3600–3607. [PubMed] [Google Scholar]
- Goldberg KB, Modrell B, Hillman BI, Heaton LA, Choi TJ, Jackson AO. 1991. Structure of the glycoprotein gene of sonchus yellow net virus, a plant rhabdovirus. Virology. 185:32–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes-Leal W, Martins LC, Diniz JAP, Dos Santos ZA, Borges JA, Macedo CAC, Medeiros AC, De Paula LS, Guimaraes JS, Freire MAM et al. . 2006. Neurotropism and neuropathological effects of selected rhabdoviruses on intranasally-infected newborn mice. Acta Trop. 97:126–139. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Dunia D, Cubitt B, Grasser FA, de la Torre JC. 1997. Characterization of Borna disease virus p56 protein, a surface glycoprotein involved in virus entry. J Virol. 71:3208–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramberg T, Hofmann H, Moller P, Lalor PF, Marzi A, Geier M, Krumbiegel M, Winkler T, Kirchhoff F, Adams DH et al. . 2005. LSECtin interacts with filovirus glycoproteins and the spike protein of SARS coronavirus. Virology. 340:224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin DE, Lin WH, Pan CH. 2012. Measles virus, immune control, and persistence. FEMS Microbiol Rev. 36:649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigera PR, Mathieu ME, Wagner RR. 1991. Effect of glycosylation on the conformational epitopes of the glycoprotein of vesicular stomatitis virus (New Jersey serotype). Virology. 180:1–9. [DOI] [PubMed] [Google Scholar]
- Gubala A, Davis S, Weir R, Melville L, Cowled C, Boyle D. 2011. Tibrogargan and Coastal Plains rhabdoviruses: Genomic characterization, evolution of novel genes and seroprevalence in Australian livestock. J Gen Virol. 92:2160–2170. [DOI] [PubMed] [Google Scholar]
- Gubala A, Davis S, Weir R, Melville L, Cowled C, Walker P, Boyle D. 2010. Ngaingan virus, a macropod-associated rhabdovirus, contains a second glycoprotein gene and seven novel open reading frames. Virology. 399:98–108. [DOI] [PubMed] [Google Scholar]
- Harder TC, Osterhaus AD. 1997. Canine distemper virus—A morbillivirus in search of new hosts? Trends Microbiol. 5:120–124. [DOI] [PubMed] [Google Scholar]
- Hashiguchi T, Kajikawa M, Maita N, Takeda M, Kuroki K, Sasaki K, Kohda D, Yanagi Y, Maenaka K. 2007. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc Natl Acad Sci USA. 104:19535–19540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatalski CG, Kliche S, Stitz L, Lipkin WI. 1995. Neutralizing antibodies in Borna disease virus-infected rats. J Virol. 69:741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helle F, Duverlie G, Dubuisson J. 2011. The hepatitis C virus glycan shield and evasion of the humoral immune response. Viruses-Basel. 3:1909–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrler G, Compans RW. 1983. Posttranslational modification and intracellular transport of mumps virus glycoproteins. J Virol. 47:354–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Hernandez MG, Berger E, Marzi A, Pohlmann S. 2016. The glycoproteins of all filovirus species use the same host factors for entry into bat and human cells but entry efficiency is species dependent. PLoS One. 11:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu A, Cathomen T, Cattaneo R, Norrby E. 1995. Influence of N-linked oligosaccharide chains on the processing, cell surface expression and function of the measles virus fusion protein. J Gen Virol. 76(Pt 3):705–710. [DOI] [PubMed] [Google Scholar]
- Hu A, Cattaneo R, Schwartz S, Norrby E. 1994. Role of N-linked oligosaccharide chains in the processing and antigenicity of measles virus haemagglutinin protein. J Gen Virol. 75:1043–1052. [DOI] [PubMed] [Google Scholar]
- Huang AS, Palma EL, Hewlett N, Roizman B. 1974. Pseudotype formation between enveloped RNA and DNA viruses. Nature. 252:743–745. [DOI] [PubMed] [Google Scholar]
- Hunt CL, Lennemann NJ, Maury W. 2012. Filovirus entry: A novelty in the viral fusion world. Viruses. 4:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyndman TH, Shilton CM, Doneley RJT, Nicholls PK. 2012. Sunshine virus in Australian pythons. Vet Microbiol. 161:77–87. [DOI] [PubMed] [Google Scholar]
- Ikuta K, Ibrahim MS, Kobayashi T, Tomonaga K. 2002. Borna disease virus and infection in humans. Front Biosci. 7:470–495. [DOI] [PubMed] [Google Scholar]
- Iorio RM, Melanson VR, Mahon PJ. 2009. Glycoprotein interactions in paramyxovirus fusion. Future Virol. 4:335–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatsuki K, Miyashita N, Yoshida E, Gemma T, Shin YS, Mori T, Hirayama N, Kai C, Mikami T. 1997. Molecular and phylogenetic analyses of the haemagglutinin (H) proteins of field isolates of canine distemper virus from naturally infected dogs. J Gen Virol. 78(Pt 2):373–380. [DOI] [PubMed] [Google Scholar]
- Jackson AO, Dietzgen RG, Goodin MM, Bragg JN, Deng M. 2005. Biology of plant rhabdoviruses. Annu Rev Phytopathol. 43:623–660. [DOI] [PubMed] [Google Scholar]
- Jayakar HR, Jeetendra E, Whitt MA. 2004. Rhabdovirus assembly and budding. Virus Res. 106:117–132. [DOI] [PubMed] [Google Scholar]
- Jeffers SA, Sanders DA, Sanchez A. 2002. Covalent modifications of the Ebola virus glycoprotein. J Virol. 76:12463–12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johal J, Gresty K, Kongsuwan K, Walker PJ. 2008. Antigenic characterization of bovine ephemeral fever rhabdovirus G and GNS glycoproteins expressed from recombinant baculoviruses. Arch Virol. 153:1657–1665. [DOI] [PubMed] [Google Scholar]
- Johnson PR, Spriggs MK, Olmsted RA, Collins PL. 1987. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: Extensive sequence divergence between antigenically related proteins. Proc Natl Acad Sci USA. 84:5625–5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletsky RL, Simmons G, Bates P. 2007. Proteolysis of the Ebola virus glycoproteins enhances virus binding and infectivity. J Virol. 81:13378–13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabatsos N. 1985. International Catalogue of Arboviruses: Including Certain Other Viruses of Vertebrates. San Antonio: American Society for Tropical Medicine and Hygiene. [DOI] [PubMed] [Google Scholar]
- Kieny MP, Lathe R, Drillien R, Spehner D, Skory S, Schmitt D, Wiktor T, Koprowski H, Lecocq JP. 1984. Expression of rabies virus glycoprotein from a recombinant vaccinia virus. Nature. 312:163–166. [DOI] [PubMed] [Google Scholar]
- Kliche S, Briese T, Henschen AH, Stitz L, Lipkin WI. 1994. Characterization of a Borna disease virus glycoprotein, gp18. J Virol. 68:6918–6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohama T, Shimizu K, Ishida N. 1978. Carbohydrate composition of the envelope glycoproteins of Sendai virus. Virology. 90:226–234. [DOI] [PubMed] [Google Scholar]
- Kondo H, Maeda T, Shirako Y, Tamada T. 2006. Orchid fleck virus is a rhabdovirus with an unusual bipartite genome. J Gen Virol. 87:2413–2421. [DOI] [PubMed] [Google Scholar]
- Kondo H, Maeda T, Tamada T. 2009. Identification and characterization of structural proteins of orchid fleck virus. Arch Virol. 154:37–45. [DOI] [PubMed] [Google Scholar]
- Kotwal GJ, Buller ML, Wunner WH, Pringle CR, Ghosh HP. 1986. Role of glycosylation in transport of vesicular stomatitis virus envelope glycoprotein. A new class of mutant defective in glycosylation and transport of G protein. J Biol Chem. 261:8936–8943. [PubMed] [Google Scholar]
- Kraus I, Eickmann M, Kiermayer S, Scheffczik H, Fluess M, Richt JA, Garten W. 2001. Open reading frame III of Borna disease virus encodes a nonglycosylated matrix protein. J Virol. 75:12098–12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JH, Bekal S, Cai Y, Clawson AN, Domier LL, Herrel M, Jahrling PB, Kondo H, Lambert KN, Mihindukulasuriya KA et al. . 2013. Nyamiviridae: Proposal for a new family in the order Mononegavirales. Arch Virol. 158:2209–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurath G, Batts WN, Ahne W, Winton JR. 2004. Complete genome sequence of Fer-de-Lance virus reveals a novel gene in reptilian paramyxoviruses. J Virol. 78:2045–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz W, Gelderblom H, Flugel RM, Darai G. 1986. Isolation and characterization of a tupaia rhabdovirus. Intervirology. 25:88–96. [DOI] [PubMed] [Google Scholar]
- Kvellestad A, Dannevig BH, Falk K. 2003. Isolation and partial characterization of a novel paramyxovirus from the gills of diseased seawater-reared Atlantic salmon (Salmo salar L). J Gen Virol. 84:2179–2189. [DOI] [PubMed] [Google Scholar]
- Lamb R, Parks G. 2007. Paramyxoviridae: The viruses and their replication In: Knipe DM, Howley PM, editors. Fields virology. 2007, 5th ed Wolters Kluwer: Lippincott WIlliams & Wilkins; p. 1449–1496. [Google Scholar]
- Lambert DM. 1988. Role of oligosaccharides in the structure and function of respiratory syncytial virus glycoproteins. Virology. 164:458–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langedijk JP, Schaaper WM, Meloen RH, van Oirschot JT. 1996. Proposed three-dimensional model for the attachment protein G of respiratory syncytial virus. J Gen Virol. 77(Pt 6):1249–1257. [DOI] [PubMed] [Google Scholar]
- Leavitt R, Schlesinger S, Kornfeld S. 1977. a. Impaired intracellular migration and altered solubility of nonglycosylated glycoproteins of vesicular stomatitis virus and Sindbis virus. J Biol Chem. 252:9018–9023. [PubMed] [Google Scholar]
- Leavitt R, Schlesinger S, Kornfeld S. 1977. b. Tunicamycin inhibits glycosylation and multiplication of Sindbis and vesicular stomatitis viruses. J Virol. 21:375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO. 2008. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 454:177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennemann NJ, Rhein BA, Ndungo E, Chandran K, Qiu X, Maury W. 2014. Comprehensive functional analysis of N-linked glycans on Ebola virus GP1. mBio. 5:e00862–00813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letchworth GJ, Rodriguez LL, Del cbarrera J. 1999. Vesicular stomatitis. Vet J. 157:239–260. [DOI] [PubMed] [Google Scholar]
- Levroney EL, Aguilar HC, Fulcher JA, Kohatsu L, Pace KE, Pang M, Gurney KB, Baum LG, Lee B. 2005. Novel innate immune functions for galectin-1: Galectin-1 inhibits cell fusion by Nipah virus envelope glycoproteins and augments dendritic cell secretion of proinflammatory cytokines. J Immunol. 175:413–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Cleveland B, Klots I, Travis B, Richardson BA, Anderson D, Montefiori D, Polacino P, Hu SL. 2008. Removal of a single N-linked glycan in human immunodeficiency virus type 1 gp120 results in an enhanced ability to induce neutralizing antibody responses. J Virol. 82:638–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CS, Chan KP, Goh KT, Chow VT. 2003. Hemagglutinin-neuraminidase sequence and phylogenetic analyses of mumps virus isolates from a vaccinated population in Singapore. J Med Virol. 70:287–292. [DOI] [PubMed] [Google Scholar]
- Lin G, Simmons G, Pohlmann S, Baribaud F, Ni H, Leslie GJ, Haggarty BS, Bates P, Weissman D, Hoxie JA et al. . 2003. Differential N-linked glycosylation of human immunodeficiency virus and Ebola virus envelope glycoproteins modulates interactions with DC-SIGN and DC-SIGNR. J Virol. 77:1337–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipkin WI, Briese T, Hornig M. 2011. Borna disease virus—Fact and fantasy. Virus Res. 162:162–172. [DOI] [PubMed] [Google Scholar]
- Liu L, Bastien N, Li Y. 2007. Intracellular processing, glycosylation, and cell surface expression of human metapneumovirus attachment glycoprotein. J Virol. 81:13435–13443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Shu Z, Qin X, Dou Y, Zhao Y, Zhao X. 2013. A live attenuated human metapneumovirus vaccine strain provides complete protection against homologous viral infection and cross-protection against heterologous viral infection in BALB/c mice. Clin Vaccine Immunol. 20:1246–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Xie J, Cheng J, Fu Y, Li G, Yi X, Jiang D. 2014. Fungal negative-stranded RNA virus that is related to bornaviruses and nyaviruses. Proc Natl Acad Sci USA. 111:12205–12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J. 2000. Protein glycosylation in the ER and Golgi complex In: Molecular cell biology. New York: W.H. Freeman. [Google Scholar]
- Longdon B, Obbard DJ, Jiggins FM. 2010. Sigma viruses from three species of Drosophila form a major new clade in the rhabdovirus phylogeny. Proc R Soc B. 277:35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longdon B, Wilfret L, Arei-Poku J, Cagney H, Obbard DJ, Jiggins FM. 2011. Host switching by a vertically-transmitted rhabdovirus in Drosophila. Biol Lett. 7:747–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzen N, Olesen NJ, Jorgensen PE, Etzerodt M, Holtet TL, Thogersen HC. 1993. Molecular cloning and expression in Escherichia coli of the glycoprotein gene of VHS virus, and immunization of rainbow trout with the recombinant protein. J Gen Virol. 74(Pt 4):623–630. [DOI] [PubMed] [Google Scholar]
- Luby SP, Hossain MJ, Gurley ES, Ahmed BN, Banu S, Khan SU, Homaira N, Rota PA, Rollin PE, Comer JA et al. . 2009. Recurrent zoonotic transmission of Nipah virus into humans, Bangladesh, 2001–2007. Emerg Infect Dis. 15:1229–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig H, Bode L. 2000. Borna disease virus: New aspects on infection, disease, diagnosis and epidemiology. Rev Sci Tech. 19:259–288. [DOI] [PubMed] [Google Scholar]
- Machamer CE, Florkiewicz RZ, Rose JK. 1985. A single N-linked oligosaccharide at either of the two normal sites is sufficient for transport of vesicular stomatitis virus G protein to the cell surface. Mol Cell Biol. 5:3074–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machamer CE, Rose JK. 1988. a. Influence of new glycosylation sites on expression of the vesicular stomatitis virus G protein at the plasma membrane. J Biol Chem. 263:5948–5954. [PubMed] [Google Scholar]
- Machamer CE, Rose JK. 1988. b. Vesicular stomatitis virus G proteins with altered glycosylation sites display temperature-sensitive intracellular transport and are subject to aberrant intermolecular disulfide bonding. J Biol Chem. 263:5955–5960. [PubMed] [Google Scholar]
- Magnani JL, Ernst B. 2009. Glycomimetic drugs—A new source of therapeutic opportunities. Discov Med. 8:247–252. [PubMed] [Google Scholar]
- Magnelli P, Bielik A, Guthrie E. 2012. Identification and characterization of protein glycosylation using specific endo- and exoglycosidases In: Hartley JL, editor. Protein expression in mammalian cells: methods and protocols. Totowa: Humana Press Inc; p. 189–211. [DOI] [PubMed] [Google Scholar]
- Maisner A, Alvarez J, Liszewski MK, Atkinson DJ, Atkinson JP, Herrler G. 1996. The N-glycan of the SCR 2 region is essential for membrane cofactor protein (CD46) to function as a measles virus receptor. J Virol. 70:4973–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisner A, Herrler G. 1995. Membrane cofactor protein with different types of N-glycans can serve as measles virus receptor. Virology. 210:479–481. [DOI] [PubMed] [Google Scholar]
- Maisner A, Schneider-Schaulies J, Liszewski MK, Atkinson JP, Herrler G. 1994. Binding of measles virus to membrane cofactor protein (CD46): Importance of disulfide bonds and N-glycans for the receptor function. J Virol. 68:6299–6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maley F, Trimble RB, Tarentino AL, Plummer TH Jr.. 1989. Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal Biochem. 180:195–204. [DOI] [PubMed] [Google Scholar]
- Malvoisin E, Wild F. 1994. The role of N-glycosylation in cell fusion induced by a vaccinia recombinant virus expressing both measles virus glycoproteins. Virology. 200:11–20. [DOI] [PubMed] [Google Scholar]
- Marcinkiewicz J, Bryniarski K, Nazimek K. 2014. Ebola haemorrhagic fever virus: Pathogenesis, immune responses, potential prevention. Folia Med Cracov. 54:39–48. [PubMed] [Google Scholar]
- Marsh GA, Wang LF. 2012. Hendra and Nipah viruses: Why are they so deadly? Curr Opin virol. 2:242–247. [DOI] [PubMed] [Google Scholar]
- Martella V, Elia G, Buonavoglia C. 2008. Canine distemper virus. The veterinary clinics of North America. Small Anim Pract. 38:787–797, vii–viii. [DOI] [PubMed] [Google Scholar]
- Martinez O, Tantral L, Mulherkar N, Chandran K, Basler CF. 2011. Impact of Ebola mucin-like domain on antiglycoprotein antibody responses induced by Ebola virus-like particles. J Infect Dis. 204(Suppl 3):S825–S832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzi A, Akhavan A, Simmons G, Gramberg T, Hofmann H, Bates P, Lingappa VR, Pohlmann S. 2006. The signal peptide of the ebolavirus glycoprotein influences interaction with the cellular lectins DC-SIGN and DC-SIGNR. J Virol. 80:6305–6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu ME, Grigera PR, Helenius A, Wagner RR. 1996. Folding, unfolding, and refolding of the vesicular stomatitis virus glycoprotein. Biochemistry. 35:4084–4093. [DOI] [PubMed] [Google Scholar]
- McAllister J, Gauci PJ, Mitchell IR, Boyle DB, Bulach DM, Weir RP, Melville LF, Davis SS, Gubala AJ. 2014. Genomic characterisation of Almpiwar virus, Harrison Dam virus and Walkabout Creek virus; three novel rhabdoviruses from northern Australia. Virol Rep. 3:1–17. [Google Scholar]
- McGinnes LW, Morrison TG. 1995. The role of individual oligosaccharide chains in the activities of the HN glycoprotein of Newcastle disease virus. Virology. 212:398–410. [DOI] [PubMed] [Google Scholar]
- McGinnes LW, Morrison TG. 1997. Disulfide bond formation is a determinant of glycosylation site usage in the hemagglutinin-neuraminidase glycoprotein of Newcastle disease virus. J Virol. 71:3083–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnes L, Sergel T, Reitter J, Morrison T. 2001. Carbohydrate modifications of the NDV fusion protein heptad repeat domains influence maturation and fusion activity. Virology. 283:332–342. [DOI] [PubMed] [Google Scholar]
- Melanson VR, Iorio RM. 2006. Addition of N-glycans in the stalk of the Newcastle disease virus HN protein blocks its interaction with the F protein and prevents fusion. J Virol. 80:623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merry T, Astrautsova S. 2010. Alternative approaches to antiviral treatments: Focusing on glycosylation as a target for antiviral therapy. Biotechnol Appl Biochem. 56:103–109. [DOI] [PubMed] [Google Scholar]
- Mihindukulasuriya KA, Nguyen NL, Wu G, Huang HV, Travassos da Rosa APA, Popov VL, Tesh RB, Wang D. 2009. Nyamanini and Midway viruses define a novel taxon of RNA viruses in the order Mononegavirales. J Virol. 83:5109–5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishin VP, Watanabe M, Taylor G, Devincenzo J, Bose M, Portner A, Alymova IV. 2010. N-linked glycan at residue 523 of human parainfluenza virus type 3 hemagglutinin-neuraminidase masks a second receptor-binding site. J Virol. 84:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll M, Kaufmann A, Maisner A. 2004. Influence of N-glycans on processing and biological activity of the Nipah virus fusion protein. J Virol. 78:7274–7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison TG, McQuain CO, Simpson D. 1978. Assembly of viral membranes: Maturation of the vesicular stomatitis virus glycoprotein in the presence of tunicamycin. J Virol. 28:368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottet G, Portner A, Roux L. 1986. Drastic immunoreactivity changes between the immature and mature forms of the Sendai virus HN and F0 glycoproteins. J Virol. 59:132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Homma M, Compans RW. 1982. Effect of tunicamycin on the replication of Sendai virus. Virology. 119:474–487. [DOI] [PubMed] [Google Scholar]
- Nandi S, Negi BS. 1999. Bovine ephemeral fever: A review. Comp Immunol Microbiol Infect Dis. 22:81–91. [DOI] [PubMed] [Google Scholar]
- Negredo A, Palacios G, Vazquez-Moron S, Gonzalez F, Dopazo H, Molero F, Juste J, Quetglas J, Savji N, Martinez MD et al. . 2011. Discovery of an Ebolavirus-like filovirus in Europe. PLoS Pathog. 7:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DT, Hiebert SW, Lamb RA. 1990. Different roles of individual N-linked oligosaccharide chains in folding, assembly, and transport of the simian virus 5 hemagglutinin-neuraminidase. Mol Cell Biol. 10:1989–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M, Ndungo E, Jangra RK, Cai YY, Postnikova E, Radoshitzky SR, Dye JM, de Arellano ER, Negredo A, Palacios G et al. . 2014. Cell entry by a novel European filovirus requires host endosomal cysteine proteases and Niemann–Pick C1. Virology. 468:637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizet V, Esko JD. 2009. Bacterial and viral infections In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of glycobiology. New York: Cold Spring Harbor. p. 537–552. [Google Scholar]
- Noyce RS, Richardson CD. 2012. Nectin 4 is the epithelial cell receptor for measles virus. Trends Microbiol. 20:429–439. [DOI] [PubMed] [Google Scholar]
- Ogert RA, Lee MK, Ross W, Buckler-White A, Martin MA, Cho MW. 2001. N-linked glycosylation sites adjacent to and within the V1/V2 and the V3 loops of dualtropic human immunodeficiency virus type 1 isolate DH12 gp120 affect coreceptor usage and cellular tropism. J Virol. 75:5998–6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchi M, Ohuchi R, Feldmann A, Klenk HD. 1997. Regulation of receptor binding affinity of influenza virus hemagglutinin by its carbohydrate moiety. J Virol. 71:8377–8384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomo C, Cane PA, Melero JA. 2000. Evaluation of the antibody specificities of human convalescent-phase sera against the attachment (G) protein of human respiratory syncytial virus: influence of strain variation and carbohydrate side chains. J Med Virol. 60:468–474. [PubMed] [Google Scholar]
- Panda A, Elankumaran S, Krishnamurthy S, Huang Z, Samal SK. 2004. Loss of N-linked glycosylation from the hemagglutinin-neuraminidase protein alters virulence of Newcastle disease virus. J Virol. 78:4965–4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Mohakud NK, Pena L, Kumar S. 2014. Human metapneumovirus: Review of an important respiratory pathogen. Int J Infect Dis. 25:45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter-Katalinic J. 2005. Methods in enzymology: O-glycosylation of proteins. Methods Enzymol. 405:139–171. [DOI] [PubMed] [Google Scholar]
- Pfaller CK, Cattaneo R, Schnell MJ. 2015. Reverse genetics of Mononegavirales: How they work, new vaccines, and new cancer therapeutics. Virology. 479-480:331–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powlesland AS, Fisch T, Taylor ME, Smith DF, Tissot B, Dell A, Pohlmann S, Drickamer K. 2008. A novel mechanism for LSECtin binding to Ebola virus surface glycoprotein through truncated glycans. J Biol Chem. 283:593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell MK, Laing KJ, Winton JR. 2012. Immunity to fish rhabdoviruses. Viruses. 4:140–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawling J, Melero JA. 2007. The use of monoclonal antibodies and lectins to identify changes in viral glycoproteins that are influenced by glycosylation: The case of human respiratory syncytial virus attachment (G) glycoprotein. Methods Mol Biol. 379:109–125. [DOI] [PubMed] [Google Scholar]
- Reading CL, Penhoet EE, Ballou CE. 1978. Carbohydrate structure of vesicular stomatitis virus glycoprotein. J Biol Chem. 253:5600–5612. [PubMed] [Google Scholar]
- Reading PC, Pickett DL, Tate MD, Whitney PG, Job ER, Brooks AG. 2009. Loss of a single N-linked glycan from the hemagglutinin of influenza virus is associated with resistance to collectins and increased virulence in mice. Respir Res. 10:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richt JA, Rott R. 2001. Borna disease virus: A mystery as an emerging zoonotic pathogen. Vet J. 161:24–40. [DOI] [PubMed] [Google Scholar]
- Rini J, Esko J, Varki A. 2009. Glycosyltransferases and glycan-processing enzymes In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of glycobiology. New York: Cold Spring Harbor. p. 63–74. [PubMed] [Google Scholar]
- Ritchie G, Harvey DJ, Stroeher U, Feldmann F, Feldmann H, Wahl-Jensen V, Royle L, Dwek RA, Rudd PM. 2010. Identification of N-glycans from Ebola virus glycoproteins by matrix-assisted laser desorption/ionisation time-of-flight and negative ion electrospray tandem mass spectrometry. Rapid Commun Mass Spectrom. 24:571–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson MA, Etchison JR, Robertson JS, Summers DF, Stanley P. 1978. Specific changes in the oligosaccharide moieties of VSV grown in different lectin-resistnat CHO cells. Cell. 13:515–526. [DOI] [PubMed] [Google Scholar]
- Robertson JS, Etchison JR, Summers DF. 1976. Glycosylation sites of vesicular stomatitis virus glycoprotein. J Virol. 19:871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R, Ramilo O. 2014. Respiratory syncytial virus: How, why and what to do. J Infect. 68(Suppl 1):S115–S118. [DOI] [PubMed] [Google Scholar]
- Rogers MB, Cui L, Fitch A, Popov V, Travassos da Rosa APA, Vasilakis N, Tesh RB, Ghedin E. 2014. Whole genome analysis of Sierra Nevada virus, a novel mononegavirus in the family Nyamiviridae. Am J Trop Med Hyg. 91:159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JK, Doolittle RF, Anilionis A, Curtis PJ, Wunner WH. 1982. Homology between the glycoproteins of vesicular stomatitis virus and rabies virus. J Virol. 43:361–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin S, Eckhaus M, Rennick LJ, Bamford CG, Duprex WP. 2015. Molecular biology, pathogenesis and pathology of mumps virus. J Pathol. 235:242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samal S, Khattar SK, Kumar S, Collins PL, Samal SK. 2012. Coordinate deletion of N-glycans from the heptad repeats of the fusion F protein of Newcastle disease virus yields a hyperfusogenic virus with increased replication, virulence, and immunogenicity. J Virol. 86:2501–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A, Kiley MP, Holloway BP, Auperin DD. 1993. Sequence analysis of the Ebola virus genome: Organization, genetic elements, and comparison with the genome of Marburg virus. Virus Res. 29:215–240. [DOI] [PubMed] [Google Scholar]
- Sasaya T, Milne RG, Campbell RN, Vetten HJ. 2011. Varicosavirus In: Tidona C, Darai G, editors. The Springer index of viruses. New York: Springer New York; p. 2081–2085. [Google Scholar]
- Sawatsky B, von Messling V. 2010. Canine distemper viruses expressing a hemagglutinin without N-glycans lose virulence but retain immunosuppression. J Virol. 84:2753–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan CN, Offer J, Zitzmann N, Dwek RA. 2007. Exploiting the defensive sugars of HIV-1 for drug and vaccine design. Nature. 446:1038–1045. [DOI] [PubMed] [Google Scholar]
- Schlesinger S, Gottlieb C, Feil P, Gelb N, Kornfeld S. 1975. Growth of enveloped RNA viruses in a line of chinese hamster ovary cells with deficient N-acetylglucosaminyltransferase activity. J Virol. 17:239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider PA, Hatalski CG, Lewis AJ, Lipkin WI. 1997. Biochemical and functional analysis of the Borna disease virus G protein. J Virol. 71:331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornberg K, Matsuyama S, Kabsch K, Delos S, Bouton A, White J. 2006. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J Virol. 80:4174–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schowalter RM, Smith SE, Dutch RE. 2006. Characterization of human metapneumovirus F protein-promoted membrane fusion: Critical roles for proteolytic processing and low pH. J Virol. 80:10931–10941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segawa H, Inakawa A, Yamashita T, Taira H. 2003. Functional analysis of individual oligosaccharide chains of Sendai virus hemagglutinin-neuraminidase protein. Biosci Biotechnol Biochem. 67:592–598. [DOI] [PubMed] [Google Scholar]
- Segawa H, Yamashita T, Kawakita M, Taira H. 2000. Functional analysis of the individual oligosaccharide chains of sendai virus fusion protein. J Biochem. 128:65–72. [DOI] [PubMed] [Google Scholar]
- Shakin-Eshleman SH, Remaley AT, Eshleman JR, Wunner WH, Spitalnik SL. 1992. N-linked glycosylation of rabies virus glycoprotein. Individual sequons differ in their glycosylation efficiencies and influence on cell surface expression. J Biol Chem. 267:10690–10698. [PubMed] [Google Scholar]
- Shakin-Eshleman SH, Wunner WH, Spitalnik SL. 1993. Efficiency of N-linked core glycosylation at asparagine-319 of rabies virus glycoprotein is altered by deletions C-terminal to the glycosylation sequon. Biochemistry. 32:9465–9472. [DOI] [PubMed] [Google Scholar]
- Simmons G, Wool-Lewis RJ, Baribaud F, Netter RC, Bates P. 2002. Ebola virus glycoproteins induce global surface protein down-modulation and loss of cell adherence. J Virol. 76:2518–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simo Tchetgna HD, Nakoune E, Selekon B, Gessain A, Manuguerra JC, Kazanji M, Berthet N. 2017. Molecular Characterization of the Kamese virus, an unassigned rhabdovirus, isolated from Culex pruina in the Central African Republic. Vector Borne Zoonotic Dis. 17:447–451. [DOI] [PubMed] [Google Scholar]
- Smith EC, Popa A, Chang A, Masante C, Dutch RE. 2009. Viral entry mechanisms: The increasing diversity of paramyxovirus entry. FEBS J. 276:7217–7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springfeld C, Darai G, Cattaneo R. 2005. Characterization of the Tupaia rhabdovirus genome reveals a long open reading frame overlapping with P and a novel gene encoding a small hydrophobic protein. J Virol. 79:6781–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallcup KC, Fields BN. 1981. The replication of measles virus in the presence of tunicamycin. Virology. 108:391–404. [DOI] [PubMed] [Google Scholar]
- Stanley P, Cummings RD. 2009. Structures common to different glycans In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of glycobiology. New York: Cold Spring Harbor. p. 175–198. [PubMed] [Google Scholar]
- Stanley P, Schachter H, Taniguchi N. 2009. N-Glycans In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of glycobiology. New York: Cold Spring Harbor. p. 101–114. [PubMed] [Google Scholar]
- Stone JA, Nicola AV, Baum LG, Aguilar HC. 2016. Multiple novel functions of Henipavirus o-glycans: The first o-glycan functions identified in the Paramyxovirus family. PLoS Pathog. 12:e1005445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoyloff R, Briese T, Borchers K, Zimmermann W, Ludwig H. 1994. N-glycosylated protein(s) are important for the infectivity of Borna disease virus (BDV). Arch Virol. 137:405–409. [DOI] [PubMed] [Google Scholar]
- Stoyloff R, Strecker A, Bode L, Franke P, Ludwig H, Hucho F. 1997. The glycosylated matrix protein of Borna disease virus is a tetrameric membrane-bound viral component essential for infection. Eur J Biochem. 246:252–257. [DOI] [PubMed] [Google Scholar]
- Sun Q, Zhao L, Song Q, Wang Z, Qiu X, Zhang W, Zhao M, Zhao G, Liu W, Liu H et al. . 2012. Hybrid- and complex-type N-glycans are not essential for Newcastle disease virus infection and fusion of host cells. Glycobiology. 22:369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanno F, Nakatsu A, Toriyama S, Kojima M. 2000. Complete nucleotide sequence of Northern cereal mosaic virus and its genome organization. Arch Virol. 145:1373–1384. [DOI] [PubMed] [Google Scholar]
- Tate MD, Brooks AG, Reading PC. 2011. Specific sites of N-linked glycosylation on the hemagglutinin of H1N1 subtype influenza A virus determine sensitivity to inhibitors of the innate immune system and virulence in mice. J Immunol. 187:1884–1894. [DOI] [PubMed] [Google Scholar]
- Tate MD, Job ER, Brooks AG, Reading PC. 2011. Glycosylation of the hemagglutinin modulates the sensitivity of H3N2 influenza viruses to innate proteins in airway secretions and virulence in mice. Virology. 413:84–92. [DOI] [PubMed] [Google Scholar]
- Tomlinson JA, Smith BR, Garrett RG. 1962. Graft transmission of lettuce big vein. Nature. 193:599–600. [Google Scholar]
- Tran EEH, Simmons JA, Bartesaghi A, Shoemaker CJ, Nelson E, White JM, Subramaniam S. 2014. Spatial localization of the Ebola virus glycoprotein mucin-like domain determined by cryo-electron tomography. J Virol. 88:10958–10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya E, Sugawara K, Hongo S, Matsuzaki Y, Muraki Y, Li ZN, Nakamura K. 2002. Effect of addition of new oligosaccharide chains to the globular head of influenza A/H2N2 virus haemagglutinin on the intracellular transport and biological activities of the molecule. J Gen Virol. 83:1137–1146. [DOI] [PubMed] [Google Scholar]
- Tsuchiya E, Sugawara K, Hongo S, Matsuzaki Y, Muraki Y, Nakamura K. 2002. Role of overlapping glycosylation sequons in antigenic properties, intracellular transport and biological activities of influenza A/H2N2 virus haemagglutinin. J Gen Virol. 83:3067–3074. [DOI] [PubMed] [Google Scholar]
- Umar M, Amer MA, Al-Saleh MA, Al-Shahwan IM, Shakeel MT, Zakri AM, Katis NI. 2017. Characterization of lettuce big-vein associated virus and Mirafiori lettuce big-vein virus infecting lettuce in Saudi Arabia. Arch Virol. 162:2067–2072. [DOI] [PubMed] [Google Scholar]
- Van den Steen P, Rudd PM, Dwek RA, Opdenakker G. 1998. Concepts and principles of O-linked glycosylation. Crit Rev Biochem Mol Biol. 33:151–208. [DOI] [PubMed] [Google Scholar]
- Varki A, Freeze HH. 2009. Glycans in acquired human diseases In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of glycobiology. New York: Cold Spring Harbor. p. 601–616. [PubMed] [Google Scholar]
- Varki A, Freeze HH, Gagneux P. 2009. Evolution of glycan diversity In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of glycobiology. New York: Cold Spring Harbor. p. 281–292. [PubMed] [Google Scholar]
- Varki A, Kannagi R, Toole BP. 2009. Glycosylation Changes in Cancer In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. New York: Cold Spring Harbor. p. 617–632. [PubMed] [Google Scholar]
- Vasilakis N, Castro-Llanos F, Widen SG, Aguilar PV, Guzman H, Guevara C, Fernandez R, Auguste AJ, Wood TG, Popov V et al. . 2014. Arboretum and Puerto Almendras viruses: Two novel rhabdoviruses isolated from mosquitoes in Peru. J Gen Virol. 95:787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakis N, Widen S, Mayer SV, Seymour R, Wood TG, Popov V, Guzman H, Travassos da Rosa APA, Ghedin E, Holmes EC et al. . 2013. Niakha virus: A novel member of the family Rhabdoviridae isolated from phlebotomine sandflies in Senegal. Virology. 444:80–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigerust DJ, Shepherd VL. 2007. Virus glycosylation: Role in virulence and immune interactions. Trends Microbiol. 15:211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigerust DJ, Ulett KB, Boyd KL, Madsen J, Hawgood S, McCullers JA. 2007. N-linked glycosylation attenuates H3N2 influenza viruses. J Virol. 81:8593–8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Messling V, Cattaneo R. 2003. N-linked glycans with similar location in the fusion protein head modulate paramyxovirus fusion. J Virol. 77:10202–10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R, Wolff T, Herwig A, Pleschka S, Klenk HD. 2000. Interdependence of hemagglutinin glycosylation and neuraminidase as regulators of influenza virus growth: A study by reverse genetics. J Virol. 74:6316–6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker PJ, Byrne KA, Riding GA, Cowley JA, Wang Y, McWilliam S. 1992. The genome of bovine ephemeral fever rhabdovirus contains two related glycoprotein genes. Virology. 191:49–61. [DOI] [PubMed] [Google Scholar]
- Walker PJ, Firth C, Widen SG, Blasdell KR, Guzman H, Wood TG, Paradkar PN, Holmes EC, Tesh RB, Vasilakis N. 2015. Evolution of genome size and complexity in the Rhabdoviridae. PLoS Pathogens. 11:e1004664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LF, Collins PL, Fouchier RAM, Jurath G, Lamb RA, Randall RE, Rima BK. 2012. Subfamily Pneumovirinae In: King AMQ, Adams MJ., Carstens EB, Lefkowitz EJ, editors. Virus taxonomy: Classification and nomenclature of viruses. Ninth Report of the International Committee on Taxonomy of Viruses. London: Academic Press; p. 681–685. [Google Scholar]
- Wang F, Fang Q, Wang B, Yan Z, Hong J, Bao Y, Kuhn JH, Werren JH, Song Q, Ye G. 2017. A novel negative-stranded RNA virus mediates sex ratio in its parasitoid host. PLoS Pathog. 13:e1006201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Shi Y, Song J, Qi J, Lu G, Yan J, Gao GF. 2016. Ebola viral glycoprotein bound to its endosomal receptor Niemann–Pick C1. Cell. 164:258–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS et al. . 2003. Antibody neutralization and escape by HIV-1. Nature. 422:307–312. [DOI] [PubMed] [Google Scholar]
- White JM, Delos SE, Brecher M, Schornberg K. 2008. Structures and mechanisms of viral membrane fusion proteins: Multiple variations on a common theme. Crit Rev Biochem Mol Biol. 43:189–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiktor TJ, Macfarlan RI, Reagan KJ, Dietzschold B, Curtis PJ, Wunner WH, Kieny MP, Lathe R, Lecocq JP, Mackett M et al. . 1984. Protection from rabies by a vaccinia virus recombinant containing the rabies virus glycoprotein gene. Proc Natl Acad Sci U S A. 81:7194–7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JA, Hevey M, Bakken R, Guest S, Bray M, Schmaljohn AL, Hart MK. 2000. Epitopes involved in antibody-mediated protection from Ebola virus. Science. 287:1664–1666. [DOI] [PubMed] [Google Scholar]
- Witte ON, Baltimore D. 1977. Mechanism of formation of pseudotypes between vesicular stomatitis virus and murine leukemia virus. Cell. 11:505–511. [DOI] [PubMed] [Google Scholar]
- Wojczyk B, Shakin-Eshleman SH, Doms RW, Xiang ZQ, Ertl HC, Wunner WH, Spitalnik SL. 1995. Stable secretion of a soluble, oligomeric form of rabies virus glycoprotein: Influence of N-glycan processing on secretion. Biochemistry. 34:2599–2609. [DOI] [PubMed] [Google Scholar]
- Wojczyk BS, Takahashi N, Levy MT, Andrews DW, Abrams WR, Wunner WH, Spitalnik SL. 2005. N-glycosylation at one rabies virus glycoprotein sequon influences N-glycan processing at a distant sequon on the same molecule. Glycobiology. 15:655–666. [DOI] [PubMed] [Google Scholar]
- Wong AC, Sandesara RG, Mulherkar N, Whelan SP, Chandran K. 2010. A forward genetic strategy reveals destabilizing mutations in the Ebolavirus glycoprotein that alter its protease dependence during cell entry. J Virol. 84:163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Yang L, Yang F, Ren X, Jiang J, Dong J, Sun L, Zhu Y, Zhou H, Jin Q. 2014. Novel Henipa-like virus, Mojiang Paramyxovirus, in rats, China, 2012. Emerg Infect Dis. 20:1064–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunner WH, Dietzschold B, Smith CL, Lafon M, Golub E. 1985. Antigenic variants of CVS rabies virus with altered glycosylation sites. Virology. 140:1–12. [DOI] [PubMed] [Google Scholar]
- Wunner WH, Larson JK, Dietzschold B, Smith CL. 1988. The molecular biology of rabies viruses. Rev Infect Dis. 10(Suppl 4):S771–S784. [DOI] [PubMed] [Google Scholar]
- Yamada K, Noguchi K, Nishizono A. 2014. a. Characterization of street rabies virus variants with an additional N-glycan at position 247 in the glycoprotein. Arch Virol. 159:207–216. [DOI] [PubMed] [Google Scholar]
- Yamada K, Noguchi K, Nishizono A. 2014. b. Efficient N-glycosylation at position 37, but not at position 146, in the street rabies virus glycoprotein reduces pathogenicity. Virus Res. 179:169–176. [DOI] [PubMed] [Google Scholar]
- Yamada K, Noguchi K, Nonaka D, Morita M, Yasuda A, Kawazato H, Nishizono A. 2013. Addition of a single N-glycan to street rabies virus glycoprotein enhances virus production. J Gen Virol. 94:270–275. [DOI] [PubMed] [Google Scholar]
- Yamada K, Park CH, Noguchi K, Kojima D, Kubo T, Komiya N, Matsumoto T, Mitui MT, Ahmed K, Morimoto K et al. . 2012. Serial passage of a street rabies virus in mouse neuroblastoma cells resulted in attenuation: potential role of the additional N-glycosylation of a viral glycoprotein in the reduced pathogenicity of street rabies virus. Virus Res. 165:34–45. [DOI] [PubMed] [Google Scholar]
- Yang ZY, Duckers HJ, Sullivan NJ, Sanchez A, Nabel EG, Nabel GJ. 2000. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat Med. 6:886–889. [DOI] [PubMed] [Google Scholar]
- Yates PJ, Afzal MA, Minor PD. 1996. Antigenic and genetic variation of the HN protein of mumps virus strains. J Gen Virol. 77(Pt 10):2491–2497. [DOI] [PubMed] [Google Scholar]
- Yelverton E, Norton S, Obijeski JF, Goeddel DV. 1983. Rabies virus glycoprotein analogs: Biosynthesis in Escherichia coli. Science. 219:614–620. [DOI] [PubMed] [Google Scholar]
- Yu CM, Li RP, Chen X, Liu P, Zhao XD. 2012. Replication and pathogenicity of attenuated human metapneumovirus F mutants in severe combined immunodeficiency mice. Vaccine. 30:231–236. [DOI] [PubMed] [Google Scholar]
- Zawilinska B, Kosz-Vnenchak M. 2014. General introduction into the Ebola virus biology and disease. Folia Med Cracov. 54:57–65. [PubMed] [Google Scholar]
- Zeitlin L, Whaley KJ, Olinger GG, Jacobs M, Gopal R, Qiu X, Kobinger GP. 2016. Antibody therapeutics for Ebola virus disease. Curr Opin Virol. 17:45–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Dou Y, Wu J, She W, Luo L, Zhao Y, Liu P, Zhao X. 2011. Effects of N-linked glycosylation of the fusion protein on replication of human metapneumovirus in vitro and in mouse lungs. J Gen Virol. 92:1666–1675. [DOI] [PubMed] [Google Scholar]
- Zhang NZ, Zhang LF, Jiang YN, Zhang T, Xia C. 2009. Molecular analysis of spring viraemia of carp virus in China: A fatal aquatic viral disease that might spread in East Asian. PLoS One. 4:e6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Biering SB, Mirza AM, Grasseschi BA, Mahon PJ, Lee B, Aguilar HC, Iorio RM. 2013. Individual N-glycans added at intervals along the stalk of the Nipah virus G protein prevent fusion but do not block the interaction with the homologous F protein. J Virol. 87:3119–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinska DF, Gnad F, Wiśniewski JR, Mann M. 2010. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell. 141:897–907. [DOI] [PubMed] [Google Scholar]
- Zimmer G, Trotz I, Herrler G. 2001. N-glycans of F protein differentially affect fusion activity of human respiratory syncytial virus. J Virol. 75:4744–4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlateva KT, Lemey P, Vandamme AM, Van Ranst M. 2004. Molecular evolution and circulation patterns of human respiratory syncytial virus subgroup a: Positively selected sites in the attachment g glycoprotein. J Virol. 78:4675–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]