Abstract
The cell division cycle consists of a series of temporally ordered events. Cell cycle kinases and phosphatases provide key regulatory input, but how the correct substrate phosphorylation and dephosphorylation timing is achieved is incompletely understood. Here we identify a PxL substrate recognition motif that instructs dephosphorylation by the budding yeast Cdc14 phosphatase during mitotic exit. The PxL motif was prevalent in Cdc14-binding peptides enriched in a phage display screen of native disordered protein regions. PxL motif removal from the Cdc14 substrate Cbk1 delays its dephosphorylation, whereas addition of the motif advances dephosphorylation of otherwise late Cdc14 substrates. Crystal structures of Cdc14 bound to three PxL motif substrate peptides provide a molecular explanation for PxL motif recognition on the phosphatase surface. Our results illustrate the sophistication of phosphatase-substrate interactions and identify them as an important determinant of ordered cell cycle progression.
Introduction
The division of cells into two daughters is a process fundamental to all forms of life. In order to accomplish this task, eukaryotic cells undergo an archetypical series of events, called the cell division cycle1. The aim is to ensure error-free duplication of the genetic material and all other cellular content, followed by their accurate division to produce two viable progeny cells. The cell division cycle involves numerous cell biological processes, entailing regulated changes to chromatin, cytoskeleton, membranes and metabolism, to name but a few. This enormous regulatory complexity has been entrusted to a ‘master regulator’, the cyclin-dependent kinases (Cdk) and their counteracting phosphatases. Their concerted action steers the cell division cycle through biochemical switches in the form of reversible phosphorylation of serine and threonine residues. Mitosis, in particular, is a period of dynamic changes in the phosphorylation states of hundreds of proteins that direct them to a multitude of molecular fates. These include changes to their localization, interactions and activities.
In the budding yeast Saccharomyces cerevisiae, progression from G1 phase to mitosis is characterized by a quantitative increase in Cdk activity, coupled with changes in the substrate specificities of the sequentially expressed cyclin activators2–4. Modulated by opposing phosphatase activities5, the Cdk activity increase leads to spatiotemporal ordering of protein phosphorylation and cell cycle phases. At its peak, Cdk activates the anaphase promoting complex/cyclosome (APC/C), a large ubiquitin ligase complex that triggers the metaphase-to-anaphase transition and leads to Cdk downregulation by targeting cyclins for destruction. Following anaphase onset, cells enter a stage called mitotic exit which culminates in the physical separation of the two daughter cells. This stage requires exquisite temporal control of cell cycle events. First, the elongating anaphase spindle is stabilized to segregate chromosomes to opposite poles. At the same time, chromosomes reach their greatest compaction to facilitate segregation. This is followed by disassembly of the anaphase spindle and chromosome decondensation, before cell division finally concludes by cytokinesis. Central to these successive processes is protein dephosphorylation by the conserved dual specificity phosphatase, Cdc146–14, which is released from association with its stoichiometric inhibitor Net1 during mitotic exit15,16. Increasing Cdc14 phosphatase activity, in the backdrop of declining Cdk activity, leads to sequential substrate targeting, with each substrate being dephosphorylated at its unique phosphatase-to-kinase threshold17. How Cdc14 differentially recognizes its substrates to set these dephosphorylation thresholds is not known.
Recent systematic and proteomic studies have identified numerous Cdc14 substrates12,18,19. Furthermore, in vitro, Cdc14 strongly favors dephosphorylation of phosphoserines followed by a proline (SP), with an additional positively charged residue downstream (SPxK/R) representing the best Cdc14 substrates. Coincidentally, this corresponds to the preferred Cdk consensus phosphorylation motif. Phosphothreonines in turn are poor Cdc14 substrates, due to a steric clash with the phosphatase active site20–23. However, SP and SPxK/R phosphosite motifs are shared by many substrates, suggesting that additional determinants contribute to Cdc14 substrate discrimination. Here, we use phage display screening of native disordered protein regions24 to enrich Cdc14 binding peptides. Amongst these, we identify a prevalent PxL motif and use a combination of biochemical, structural and molecular genetic approaches to show that it forms a Cdc14 substrate docking motif. The PxL motif enhances phosphatase-substrate interaction and promotes dephosphorylation, forming part of the mechanisms that govern timely substrate dephosphorylation.
Results
Identification of a PxL Cdc14 docking motif
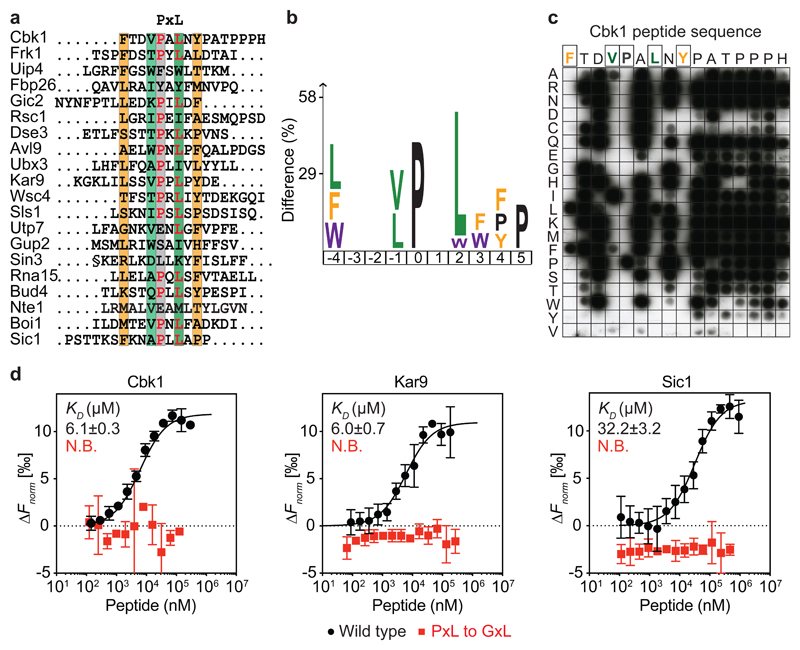
Many protein-protein interactions are mediated by short linear interaction motifs, that are ten or fewer residues in length and are typically found in intrinsically unstructured protein regions25. We therefore screened a 16-mer peptide library representing the predicted disordered regions of the budding yeast proteome (the ‘disorderome’)24 using purified Cdc14 as a bait (Supplementary Fig. 1a). Deep sequencing of the phage inserts after three rounds of selection identified candidate Cdc14 interacting peptides (Fig. 1a and Supplementary Table 1). Visual inspection of these peptides uncovered the prevalence of the amino acid motif ‘PxL’. Enrichment analysis26 revealed an additional aliphatic amino acid at the -1 position, relative to the proline. Large hydrophobic, often aromatic, amino acids were prevalent at -4 and +4 (Fig. 1b). For ease of reference, we refer to this extended hydrophobic ΦxxφPxLxΦ peptide signature (where Φ indicates an aromatic or large hydrophobic and φ a small hydrophobic side chain) as the ‘PxL’ motif. Recovered PxL motif-containing peptides were enriched in known Cdc14 substrates and interactors identified by proteomic profiling (Supplementary Fig. 1b), consistent with a role for a PxL motif in Cdc14 substrate recognition. These included the NDR/LATS kinase Cbk1, spindle positioning protein Kar9 and mitotic exit regulator Sic16,13,18,27.
Fig. 1. Identification of the PxL Cdc14 docking motif.
a, Alignment of the top 20 phage display hits. Conserved positions are highlighted. b, Sequence logo representing enriched residues within the PxL motifs from all peptides with greater than five sequencing counts, generated using a detection threshold of p=0.01 in iceLogo26. c, Mutational Cbk1 peptide array to probe the contribution of individual amino acid positions to Cdc14 binding. d, Microscale thermophoresis profiles of wild type (black) and mutant (red) PxL motif peptides binding to GFP-Cdc14. Shown are the means and standard deviations from three independent experiments.
In order to explore how the PxL motif interacts with Cdc14, we employed a peptide array-based binding experiment. The 16 positions in the top-ranked, Cbk1-derived peptide were permuted to all 20 possible amino acids. These peptides were synthesized next to each other on a membrane that we probed with purified Cdc14. Cdc14 binding to the Cbk1 peptide critically depended on the central P and L residues (Fig. 1c). Hydrophobic amino acids at the -4 and -1 positions were also required. The contribution of the +4 tyrosine remained ambiguous, but could be confirmed in a parallel experiment employing a different peptide solvation approach (Supplementary Fig. 1c). Outside these PxL motif positions, no determinative amino acid preferences were observed.
The peptide array does not report the Cdc14 affinities for PxL motif containing peptides. We therefore used microscale thermophoresis28 to measure affinities between synthetic peptides derived from Cbk1, Kar9 and Sic1 and purified recombinant GFP-Cdc14 (Supplementary Fig. 1a). The Cbk1 and Kar9 peptides bound Cdc14 with a comparable Kd of 6.1 and 6.0 μM, respectively (Fig. 1d), which falls in the range of typical short linear motif interactions25,29. The Sic1 peptide bound the phosphatase with a lower affinity (Kd = 32.2 μM). In all cases, single point mutations in the PxL motif abolished detectable interactions (Fig. 1d and Supplementary Fig. 1d). Although the Cbk1 peptide array revealed little contribution of the final seven residues following the PxL motif, a synthetic Cbk1 peptide lacking these amino acids bound Cdc14 with reduced affinity. Amino acids beyond the PxL consensus motif might make an additional, weaker contribution to Cdc14 binding (Supplementary Fig. 1d). These results biochemically describe a Cdc14-directed substrate interaction motif.
The PxL motif promotes Cbk1 dephosphorylation
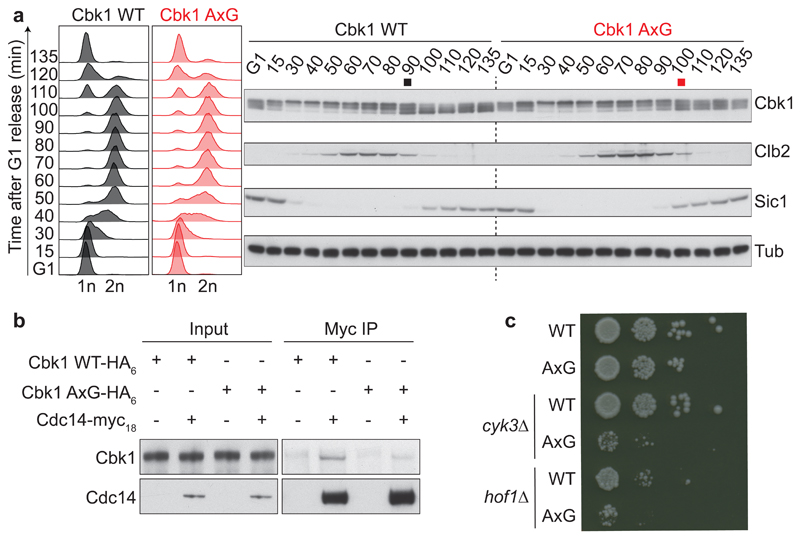
To investigate whether the PxL motif contributes to Cdc14-mediated Cbk1 dephosphorylation in vivo, we created a budding yeast strain in which the central proline and leucine within the PxL motif were mutated to alanine and glycine (AxG). We arrested cells in G1 using the mating pheromone α-factor and released them to progress synchronously through the cell cycle. Cbk1 electrophoretic mobility was monitored as readout for its phosphorylation27. Both Cbk1 and Cbk1 AxG were phosphorylated as cells entered S phase (Fig. 2a). Wild type Cbk1 was dephosphorylated during mitotic exit concomitantly with mitotic cyclin Clb2 degradation. In contrast, Cbk1 AxG dephosphorylation was delayed and never reached completion. This demonstrates that the PxL motif conveys timely Cbk1 dephosphorylation. Cbk1 AxG displayed reduced binding to Cdc14 during co-immunoprecipitation from cell extracts (Fig. 2b), consistent with the idea that the PxL motif enhances Cdc14 substrate recognition.
Fig. 2. The PxL motif promotes Cbk1 dephosphorylation.
a, Strains expressing wild type Cbk1 or Cbk1 AxG progressed synchronously through the cell cycle following α-factor arrest and release. FACS analysis of DNA content confirmed cell cycle synchrony. Western blotting was used to analyze the Cbk1 phosphorylation status, as well as Clb2 and Sic1 levels which served as additional cell cycle markers. The black and red squares mark the approximate midpoints of Cbk1 dephosphorylation. Tubulin served as a loading control. Uncropped images of all the blots are found in Supplementary Data Set 1. b, The Cdc14-Cbk1 interaction depends on the PxL motif. Cdc14-myc18 was immunoprecipitated from asynchronously growing control or Cbk1 AxG cells and Cbk1 co-purification analyzed by western blotting. c, Synthetic growth defect of Cbk1 AxG in the absence of cytokinesis factors. Ten-fold serial dilutions of the indicated strains were spotted onto YPD plates and grown at 25 °C for two days.
Cbk1 dephosphorylation engages a transcriptional program that promotes cytokinesis27,30. To gauge the functional consequences of PxL motif-mediated Cbk1 dephosphorylation, we deleted the genes encoding either Hof1 or Cyk3, actors in two parallel cytokinetic pathways31,32. While these deletions are tolerated in a wild type strain background, they led to severely reduced growth of Cbk1 AxG cells (Fig. 2c). Thus, PxL motif-mediated, timely Cbk1 dephosphorylation is important to robustly link cell cycle progression to cytokinesis.
The PxL peptide inserts into deep surface pockets on the dimeric Cdc14 phosphatase
We next sought structural insight into how Cdc14 recognizes PxL motifs. We expressed and purified the evolutionarily conserved part of budding yeast Cdc14 (amino acids 1-374), consisting of an N-terminal domain A and the catalytic domain B, but lacking the predicted unstructured and ill-conserved domain C21,33 (Supplementary Fig. 2a). Addition of the Cbk1 peptide increased the melting point of Cdc14, suggesting that substrate binding stabilizes the phosphatase (Supplementary Fig. 2b). We obtained crystals of Cdc14 with the Cbk1 peptide in both orthorhombic and monoclinic space groups (Supplementary Fig 2c). These allowed structure determination at resolutions of 1.5 and 2.5 Å (Table 1), respectively, using molecular replacement with human Cdc14B as the template21. We also solved a structure of Cdc14 bound to the Sic1-derived PxL peptide at 1.8 Å resolution (Table 1).
Table 1. Crystallographic data collection and refinement statistics.
| Cdc14–Cbk1 (PDB 6G84) | Cdc14–Cbk1 (PDB 6G85) | Cdc14–Sic1 (PDB 6G86) | Cdc14–Sic1 (Zn peak) | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | C 2 | P 212121 | P 212121 | P 212121 |
| Cell dimensions | ||||
| a, b, c (Å) | 174.5, 73.9, 83.5 | 76.1, 97.4, 129.1 | 75.0, 94.3, 127.6 | 75.1, 94.2, 128.3 |
| α, β, γ (°) | 90, 114.2, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 55.29 - 2.47 | 48.70 - 1.52 | 63.83 - 1.74 | 58.78 - 2.52 |
| (2.55 - 2.47)a | (1.58 - 1.52)a | (1.80 - 1.74)a | (2.56 - 2.52)a | |
| Rmerge | 0.13 (1.47)a | 0.05 (0.85)a | 0.06 (0.95)a | 0.16 (2.5)a |
| I / σI | 8.1 (1.1)a | 10.6 (1.2)a | 11.8 (1.6)a | 17.5 (2.0)a |
| Completeness (%) | 99.7 (99.6)a | 97.3 (92.9)a | 99.5 (98.5)a | 98.2 (96.6)a |
| Redundancy | 6.5 (6.3)a | 5.2 (4.6)a | 5.1 (4.7)a | 26.1 (28.4)a |
| Refinement | ||||
| Resolution (Å) | 55.29 - 2.47 | 48.7 - 1.52 | 63.83 - 1.74 | 58.78 - 2.52 |
| No. reflections | 34,883 | 141,666 | 93,147 | 30,705 |
| Rwork / Rfree | 0.226/0.270 | 0.182/0.208 | 0.164/0.198 | 0.18/0.22 |
| No. atoms | ||||
| Protein | 5,915 | 6,215 | 6,096 | 5,811 |
| Ligand/ion | 7 | 54 | 109 | 74 |
| Water | 97 | 798 | 708 | 44 |
| B-factors | ||||
| Protein | 66.6 | 30.6 | 28.2 | 61.5 |
| Ligand/ion | 88.2 | 49.8 | 54.7 | 97.5 |
| Water | 57.8 | 43.5 | 40 | 57.7 |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.002 | 0.02 | 0.012 | 0.002 |
| Bond angles (°) | 0.43 | 1.31 | 1.11 | 0.55 |
One crystal was used for each structure
Values in parentheses are for highest-resolution shell
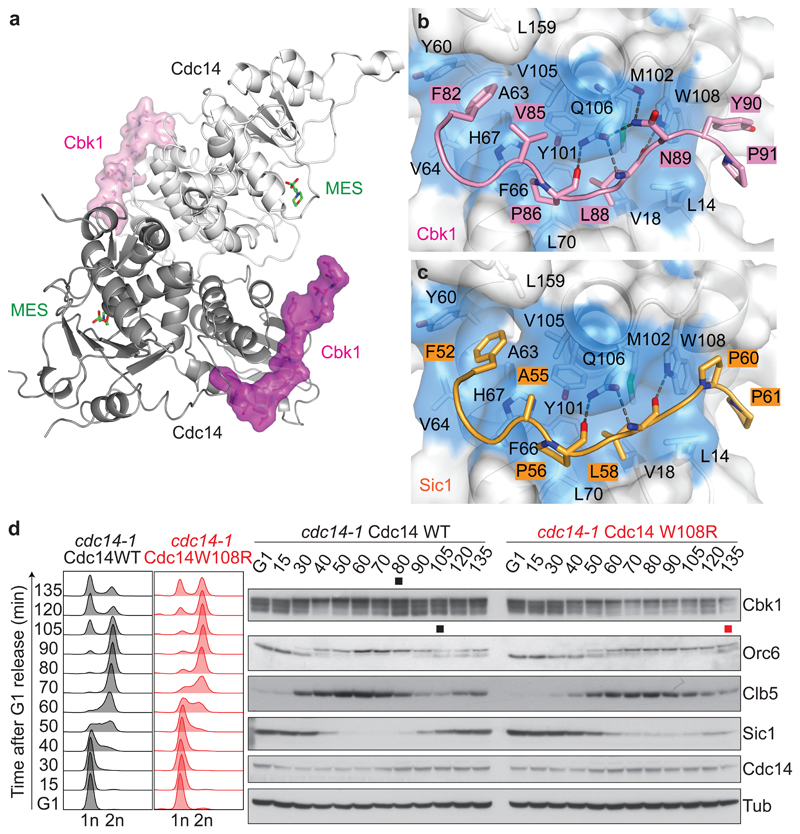
Budding yeast Cdc14 crystalized as a dimer (Fig. 3a), consistent with biochemical data and a recent structural report33,34. The overall fold of each protomer is reminiscent of monomeric human Cdc14B (Supplementary Fig. 3a)21. A difference lies in a zinc binding site that stabilizes a surface loop on the catalytic domain of budding yeast Cdc14, that is absent from human Cdc14B (Supplementary Fig. 3b, c). Both Cdc14 protomers contained PxL peptides bound within a surface-exposed hydrophobic pocket on the non-catalytic domain A. The pocket lies on the opposite face to the phosphatase active site of the same protomer, but adjacent to the active site of the second protomer. The high-resolution structures showed unambiguous densities of the PxL peptides and revealed details of phosphatase binding (Supplementary Fig. 4).
Fig. 3. The PxL motif inserts into deep surface pockets on the dimeric Cdc14 phosphatase.
a, Structure of the Cdc14–Cbk1 peptide complex. Each Cdc14 protomer (shades of grey, cartoon representation) binds a Cbk1 peptide (shades of pink, surface representation). The enzyme active site is filled by 2-(N-morpholino) ethanesulfonic acid (MES) from the crystallization buffer. b, c, Expanded views of the interaction networks between Cbk1 (b) and Sic1-derived PxL motif peptides (c), and a hydrophobic pocket in Cdc14 lined by L14, V18, Y60, A63, V64, F66, H67, L70, Y101, M102, V105, Q106, W108, L159. d, Mitotic exit and Cbk1 dephosphorylation delays caused by a PxL binding pocket mutation. Following α-factor arrest and release, cdc14-1 cells complemented by Cdc14 or Cdc14 W108R progressed synchronously through the cell cycle at 35.5 °C. FACS analysis of DNA content was used to analyze cell cycle progression and western blotting to detect the Cbk1 and Orc6 phosphorylation status and Clb5 levels. The black and red squares mark the approximate midpoints of Orc6 and Cbk1 dephosphorylation in wild type and Cdc14 W108R cells. A Cdc14 antibody45 visualized ectopic Cdc14. Tubulin served as a loading control.
Both Cbk1 and Sic1 peptides adopt a similar conformation and exhibit a comparable array of interactions with the hydrophobic pocket of Cdc14 (Fig. 3b, c and Supplementary Fig. 5). The central proline and leucine side chains of the PxL motif point into the core of the hydrophobic pocket. Three hydrogen bonds between the peptide main chain and the Q106 and W108 side chains of Cdc14 further stabilize these core motif interactions. N-terminal to the PxL, the aliphatic residues at the -1 position (V85 in Cbk1 and A55 in Sic1) engage in hydrophobic interactions with the Cdc14 pocket, whilst the -4 phenylalanines (F82 and F52 in Cbk1 and Sic1, respectively), stack against Cdc14 Y60. Two residues towards the C-termini of the peptides further bolster the peptide-phosphatase interactions. At the +4 position, Y90 in Cbk1 and P60 in Sic1 engage with Cdc14 W108, while the prolines at +5 position are in contact with Cdc14 L14. In the case of Cbk1, N89 forms additional hydrogen bonds with Cdc14, that are absent from Sic1. The deeper hydrophobic interaction of the Cbk1 valine at -1, compared to the Sic1 alanine, and the stacking interaction of the Cbk1 +4 tyrosine compared to the hydrophobic Sic1 proline, could contribute to the greater affinity of the Cbk1 compared to the Sic1 peptide. We have seen above that the final seven residues of the Cbk1 peptide, absent from the Sic1 peptide, also impart a higher affinity. Residues at non-conserved peptide positions were generally more flexible and solvent directed (Supplementary Fig. 4). These observations provide a strong rationale for PxL motif recognition by the Cdc14 phosphatase.
To gauge the importance of the PxL binding pocket for Cdc14 function, we mutated residues that are important for peptide recognition, Q106L or W108R. Neither point mutation affected Cdc14’s enzymatic ability to hydrolyze a small molecule substrate, para-nitrophenyl phosphate (p-NPP). However, these mutations markedly reduced Cbk1 peptide binding (Supplementary Fig. 6a, b). We next expressed GFP-tagged Cdc14 or Cdc14 W108R in cells in which endogenous Cdc14 function depended on the temperature sensitive cdc14-1 allele35. Cells were synchronized in G1 and released to progress through the cell cycle at a restrictive temperature. Cdc14 W108R expressing cells entered S phase approximately 10 minutes later than control cells, maybe because Cdc14 W108R shows reduced inhibition by Net1 (see below). Mitotic exit was markedly delayed and cytokinesis was deferred by at least 30 minutes (Fig. 3d). Little Cbk1 was dephosphorylated, while dephosphorylation of another Cdc14 substrate, Orc636, was also delayed. Thus, a functional PxL motif receptor is required for efficient Cdc14 function. Cdc14 W108R is expected to have formed dimers with Cdc14 encoded by the cdc14-1 allele and these heterodimers might have preserved some Cdc14 functionality. The observed phenotypic defect is therefore a lower estimate for the impact of the Cdc14 W108R mutation.
Sequence alignments of a diverse set of Cdc14 proteins revealed that residues within the hydrophobic pocket critical for PxL motif binding are conserved not only within Saccharomycetes, but also the evolutionarily distant Schizosaccharomyces pombe (Supplementary Fig. 7). Higher eukaryotes also retain the hydrophobicity, but not all residues are strictly conserved.
The Cdc14 inhibitor Net1 uses the PxL motif receptor pocket
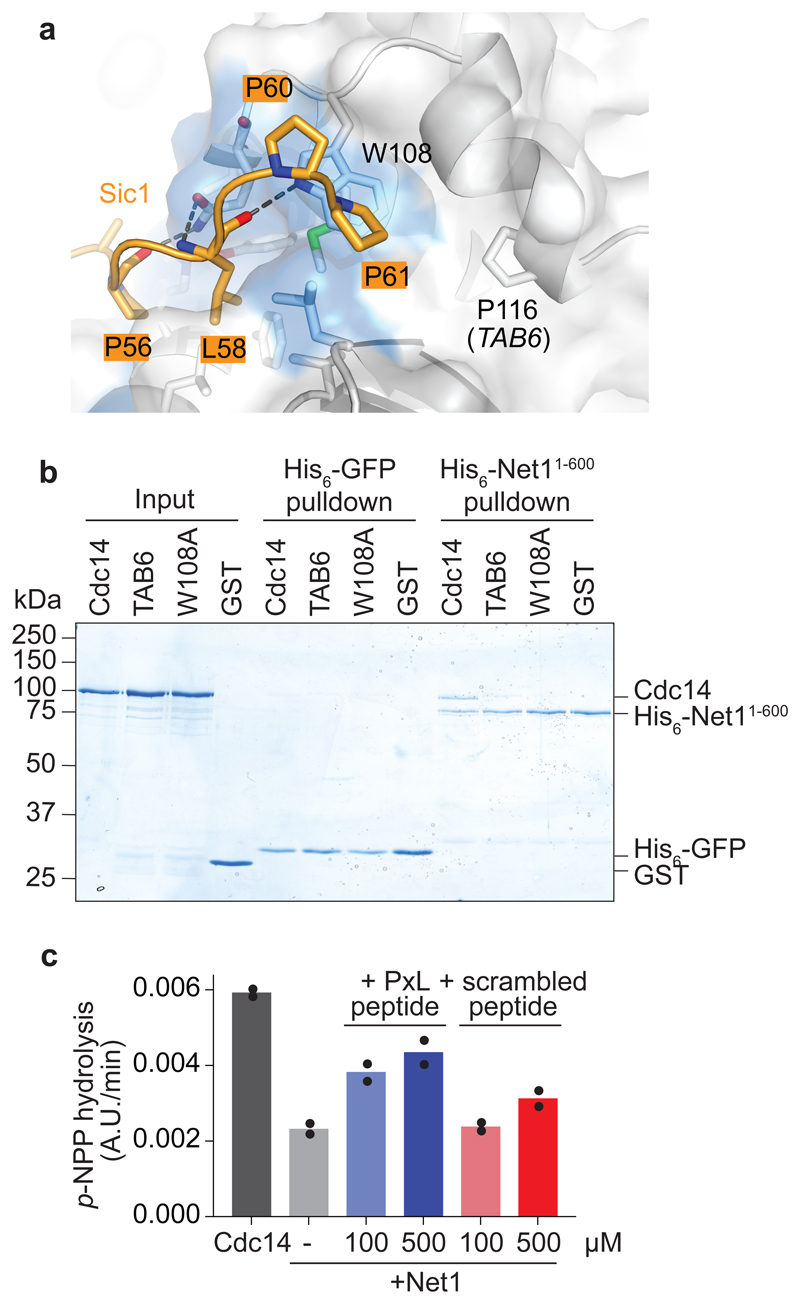
For much of the cell cycle, from G1 until metaphase, Cdc14 is inhibited by sequestration to its stoichiometric inhibitor Net1 in the nucleolus15,16. Previous studies have reported that Net1 both occludes the Cdc14 active site, as well as contacts the phosphatase at its non-catalytic domain A37. We noticed that the location of the previously described dominant active CDC14 TAB6 mutation (Cdc14 P116L), that weakens binding to and inhibition by Net138, lies on the Cdc14 surface adjacent to the PxL receptor pocket (Fig. 4a). We therefore investigated whether Net1 inhibitor binding to Cdc14 makes use of this same docking site.
Fig. 4. The Cdc14 inhibitor Net1 uses the PxL binding pocket.
a, Side view of the Cdc14 PxL binding pocket, bound by the Sic1 peptide. The Cdc14 W108 residue is highlighted, as well as P116, the site of the CDC14 TAB6 mutation. b, His6-pulldown assay to analyze the Net11-600-Cdc14 interaction. The input GST-Cdc14 variants and GST as a control, as well as pulldowns with His6-Net11-600, or with His6-GFP as a control, were analyzed by SDS-PAGE followed by Coomassie Blue staining. c, p-NPP hydrolysis by Cdc14 was analyzed as described in the Methods, with the addition of 200 nM Net11-600 and the indicated concentrations of the Cbk1-derived PxL motif peptide or scrambled control peptide. The results and means from two independent experiments are shown.
We performed pulldown assays using purified Net11-600, a truncate of the protein that is sufficient for Cdc14 regulation39. While wild type Cdc14 was efficiently pulled down by Net11-600, both Cdc14 TAB6 and PxL receptor-mutant Cdc14 W108A exhibited greatly weakened association with the inhibitor (Fig. 4b). This suggests that the Net1 inhibitory binding site on Cdc14 overlaps with the PxL motif substrate binding site. To test whether Net1 competes with PxL motif substrates for binding to Cdc14, we performed an experiment in which we measured the ability of Net1 to inhibit p-NPP hydrolysis by Cdc14. Addition of the Cbk1-derived PxL motif peptide weakened the inhibition of Cdc14 by Net1, an effect that was observed to a much lesser extent with a scrambled peptide comprised of the same amino acids (Fig. 4c). The PxL motif peptide did not alter the rate at which Cdc14 hydrolyzes p-NPP in the absence of Net1 (see below). These results suggest that Net1 shares its binding site on Cdc14 with PxL motif substrates and therefore has characteristics of a pseudosubstrate inhibitor of the Cdc14 phosphatase.
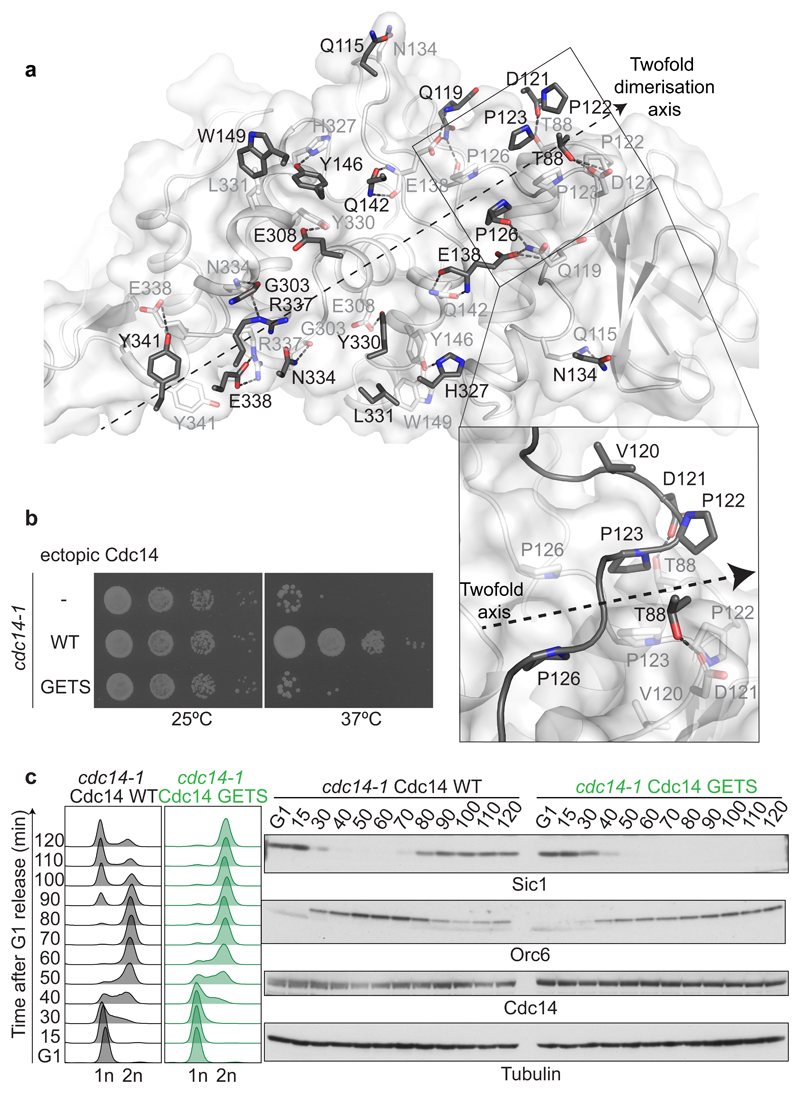
Dimerization is essential for Cdc14 function
S. cerevisiae Cdc14 is a dimer, with an interface40 that spans both domains A and B and buries 1,665 Å2 of surface area. An extensive network of interactions, including salt bridges and hydrophobic contacts, characterizes the interface (Fig. 5a and Supplementary Fig. 8a). To assess the importance of phosphatase dimerization, we introduced four mutations, V120G, D121E, P122T and P123S to disrupt a focal point of protomer interaction (Fig. 5a). The resultant Cdc14 GETS protein exhibits a lower Stokes radius as judged by size exclusion chromatography, consistent with dimer disruption (Supplementary Fig. 8b, c). Cdc14 GETS showed somewhat reduced enzymatic activity against p-NPP, probably because dimer disruption compromises the overall protein fold (Supplementary Fig. 8d). Unlike wild type Cdc14, Cdc14 GETS failed to complement temperature sensitive growth of cdc14-1 cells (Fig. 5b). To analyze the reasons for this lethality we synchronized cdc14-1 cells, complemented by either wild type Cdc14 or Cdc14 GETS, in G1 using α-factor treatment at permissive temperature. Following release at restrictive temperature, although Cdc14 GETS expressing cells traversed through the cell cycle with kinetics similar to the wild type control, they arrested in late mitosis and displayed greatly compromised Cdc14 substrate dephosphorylation (Figure 5b, c). Together with results from other Cdc14 interface mutations34, this suggests that dimerization is crucial for Cdc14 phosphatase function. A requirement to pair a PxL receptor pocket and an active site between two protomers could be a reason for this. Characteristic dimer interface residues are conserved in Cdc14 orthologs in other species, including fission yeast Clp1 and human Cdc14A (Supplementary Fig. 7). In contrast, human Cdc14B lacks dimer interface residues and crystalized as a monomer21, suggesting that dimerization does not extend to all Cdc14 family members.
Fig. 5. Dimerization is essential for Cdc14 function.
a, A two-fold symmetry axis along an extended dimerization interface between two Cdc14 protomers. The interacting surface of one of the protomers is shown in white with interacting residues in stick representation. The pairing residues emanating from the second protomer are shown in dark grey, its surface hidden for clarity. An enlarged view of a cluster of prolines engaged in symmetric hydrophobic interactions is shown, that is critical for dimer formation and was targeted in the dimer disrupting Cdc14 GETS mutant. b, Cdc14 GETS fails to support cell viability. Ten-fold serial dilutions of temperature sensitive cdc14-1 cells, and cdc14-1 cells complemented with wild type Cdc14 or Cdc14 GETS were spotted onto YPD plates and incubated at a permissive temperature of 25 °C (left), or at a restrictive temperature of 37 °C (right). c, Cdc14 GETS blocks mitotic exit. Following α-factor arrest, cdc14-1 cells complemented by wild type Cdc14 or Cdc14 GETS were released to progress through the cell cycle at 35.5 °C. FACS analysis of DNA content was used to analyze cell cycle progression and western blotting detected the Orc6 phosphorylation status and Sic1 levels. A Cdc14 antibody visualized ectopic Cdc14. Tubulin served as a loading control.
A PxL motif enhances peptide substrate dephosphorylation
We next probed the molecular mechanism by which a PxL motif facilitates Cdc14 substrate dephosphorylation. Peptide docking could introduce allosteric changes that enhance Cdc14 phosphatase activity. However, as mentioned above, addition of the Cbk1-derived PxL peptide did not increase Cdc14 activity towards p-NPP (Supplementary Fig. 9a,b), arguing against allosteric phosphatase activation.
To further investigate the impact of a PxL motif on substrate dephosphorylation, we set up an in vitro peptide dephosphorylation assay. We prepared synthetic peptides as substrates, comprised of an N-terminal Cbk1-derived PxL motif (or an AxG mutant variant), followed by an intrinsically unstructured peptide sequence derived from Sic13. Into the latter, we inserted an optimal phospho-SPxKK Cdk consensus site. The distance from the central proline of the PxL motif to the phosphoserine was 17 amino acids, enough to span the distance from the PxL receptor pocket to the Cdc14 active site on the adjacent protomer, but not to reach the active site of the same protomer (Supplementary Fig. 9a). Both peptides were dephosphoryated by Cdc14. Kinetic analysis revealed that KM was less than half for the PxL motif-containing peptide (Supplementary Fig. 9c), as expected for a substrate docking motif. In turn, the maximum dephosphorylation velocity vmax was higher for the peptide lacking the PxL motif. This indicates that, in the context of a small peptide substrate, a PxL motif enhances the affinity but slows down substrate turnover.
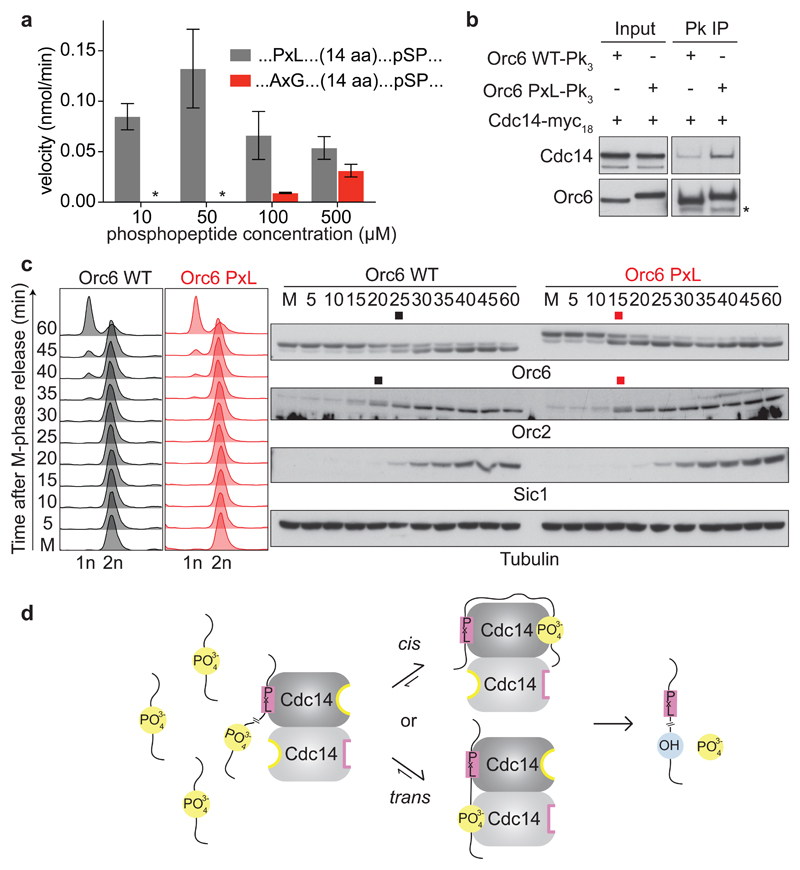
We next analyzed the impact of a PxL motif on dephosphorylation of a suboptimal phospho-SPxAA Cdk consensus site. A peptide with a mutant AxG recognition motif, 17 amino acids upstream of this phosphosite, was poorly dephosphorylated, even at higher concentrations. Strikingly, an intact PxL motif bestowed efficient dephosphorylation to this peptide at concentrations of 10 to 50 μM (Fig. 6a). At higher concentrations, apparent substrate inhibition of Cdc14 was observed, not unexpected from a substrate that interacts with the enzyme at more than one place. This demonstrates that a PxL motif substantially enhances Cdc14 substrate affinity, thereby enhancing dephosphorylation. The result also supports the idea that dephosphorylation can occur in trans, linking PxL motif recognition and substrate dephosphorylation at a short distance between the two Cdc14 protomers.
Fig. 6. A PxL motif advances ORC dephosphorylation.
a, A PxL motif facilitates peptide dephosphorylation. Dephosphorylation velocities were determined using 100 nM Cdc14 and the indicated phosphopeptide concentrations, containing a pSPxAA site preceded by a functional or mutant PxL motif. The means and standard deviations from three independent experiments are shown. b, A PxL motif augments Orc6 interaction with Cdc14. Orc6-Pk3 or Orc6 PxL-Pk3 were immunoprecipitated from asynchronously growing cells and Cdc14-myc18 co-purification analyzed by western blotting. c, The PxL motif advances ORC dephosphorylation. Orc6 and Orc6 PxL expressing cells were arrested in metaphase by Cdc20 depletion and released to exit synchronously from mitosis by Cdc20 reinduction. FACS analysis of DNA content monitored cell cycle progression. Orc6 and Orc2 dephosphorylation was detected by western blotting. The black and red squares mark the approximate midpoints of dephosphorylation. Sic1 served as a cell cycle marker and tubulin as a loading control. d, Model for PxL motif-mediated Cdc14 substrate dephosphorylation. The PxL motif facilitates dephosphorylation in cis or in trans by increasing the local substrate concentration at both Cdc14 active sites.
Cdc14 has been described as a serine-directed phosphatase in vitro22. However, numerous phosphothreonines must also be dephosphorylated during mitotic exit14. To test whether a PxL motif facilitates dephosphorylation of threonine substrates by Cdc14, we employed peptides similar to those above, but carrying a phospho-TP motif. As expected, phospho-TP dephosphorylation remained almost undetectable in case of the AxG…pTP control peptide, even at higher concentrations. Dephosphorylation of the PxL…pTP peptide was substantially enhanced, even though the velocity of dephosphorylation remained well below that observed for pSP substrates (Supplementary Fig. 9d). Whether a PxL motif allows phosphothreonine dephosphorylation by Cdc14 in vivo remains to be tested. These results demonstrate that a PxL motif enhances peptide substrate dephosphorylation in vitro, especially of otherwise suboptimal target sites.
A PxL motif advances ORC dephosphorylation
Lastly, we addressed how a PxL motif enhances substrate dephosphorylation in vivo. To test whether a PxL motif aids dephosphorylation by enhancing substrate interaction, we appended a Cbk1-derived PxL motif peptide to the Orc6 C-terminus (Orc6 PxL). Immunoprecipitation of wild type Orc6 from cell extracts coprecipitated only a small amount of Cdc14, which substantially increased in the case of Orc6 PxL (Fig. 6b). To test whether the tighter Cdc14 substrate interaction accelerates Orc6 dephosphorylation, we synchronized wild type and Orc6 PxL cells in metaphase by transcriptional repression of the APC coactivator Cdc20. Following release into anaphase by Cdc20 reinduction, Orc6 PxL dephosphorylation was advanced by more than 10 minutes compared to wild type Orc6 (Fig. 6c). Orc6 is part of the origin recognition complex (ORC). Dephosphorylation of a second ORC subunit, Orc2, was also advanced in Orc6 PxL cells. These results indicate that increased substrate affinity, bestowed by a PxL motif, has a profound effect on dephosphorylation timing. Furthermore, substrate docking has the potential to aid synchronous dephosphorylation of partnering subunits in a protein complex.
The ability the C-terminal Orc6 PxL fusion to advance Orc6 and Orc2 dephosphorylation prompted us to analyze the relationship between PxL motif occurrence and positioning of Cdk phosphorylation sites. We took the first 10 PxL motif-containing proteins retrieved in the original phage display screen, for which Cdk phosphorylation has been documented, and graphically presented the positions of PxL motifs and known Cdk phosphorylation sites (Supplementary Fig. 10). Visual inspection revealed that PxL motifs are often embedded within clusters of Cdk phosphorylation sites, though sometimes phosphorylation sites are found at a considerable distance. The steric requirements for PxL motif-mediated substrate targeting by Cdc14 will be a fertile subject for further investigation.
Discussion
Eukaryotic cells contain far fewer phosphatases than kinases, which has led to the perception that phosphatases have broad and indistinct substrate specificities. More recently, docking motif-mediated substrate targeting has become apparent for phosphatases, including PP1 and PP2A41,42, revealing a widespread principle governing phosphatase-substrate interactions. Here, we have uncovered a mechanism by which the Cdc14 phosphatase engages with substrates in the form of a PxL substrate docking motif. This motif enhances Cdc14 phosphatase interaction and thereby facilitates substrate dephosphorylation.
Addition or removal of a PxL motif has a profound impact on dephosphorylation timing during mitotic exit. It is therefore tempting to speculate that early dephosphorylated Cdc14 targets display a strong PxL motif, while later dephosphorylated substrates show less optimal or no matches. To analyze whether this is the case, we attempted to use dephosphorylation timing data from a phospho-proteome analysis wherein substrate dephosphorylation was induced by Cdc14 overexpression12, showing correlation to known physiological Cdc14-dependent dephosphorylation timings. We created a position weight matrix based on the known PxL-motif containing peptides and searched the yeast proteome to identify potential Cdc14 binders, contingent on predicted disorder of the PxL motif residues. Despite these efforts, predicted binders were equally frequent amongst proteins dephosphorylated or unresponsive to Cdc1412. This suggests that true motifs are difficult to discern from chance occurrences of similar peptide sequences. Moreover, true interaction motifs identified in our phage display screen show considerable plasticity of conserved residues. This is not uncommon amongst short linear interaction motifs, where compensatory interactions can be provided by non-consensus positions25. This emphasizes the known difficulty to predict functional small linear interaction motifs.
Cdc14 affinity, bestowed by a PxL motif, is one of several factors that impact on a substrate’s dephosphorylation timing. The phosphorylation status of any cellular regulator is the readout of the equilibrium between kinase and phosphatase activities5,17, so the activity and affinity of the corresponding kinase will equally control the dephosphorylation timing. Subcellular localization of both enzymes and substrate add to the collective control of catalysis. Our study revealed that, within the changing milieu of the mitotic exit cell, a PxL motif provides a powerful means to skew the phosphorylation equilibrium towards dephosphorylation, thus enacting accurate dephosphorylation timing.
The PxL binding site on the phosphatase consists of a hydrophobic pocket that lies surface exposed on the conserved, N-terminal domain A. We realized that the Cdc14 inhibitor Net1 also uses this hydrophobic pocket for binding, indicating that the inhibitor and substrates might have molecular features in common. Net1 contains several candidate PxL motif sequences. Whether these indeed engage with Cdc14 to act as pseudosubstrate inhibitors remains to be explored. Equally, the mechanism by which Net1 not only competes with substrate binding, but in addition occludes the Cdc14 active site37 requires further investigation.
Our structural studies revealed a curious position of the PxL receptor pockets relative to the phosphatase active sites on the Cdc14 dimer. Substrate docking could promote dephosphorylation by the same protomer (in cis) or by the adjacent protomers (in trans). While the two possibilities are not mutually exclusive, trans dephosphorylation might be favored due to the spatial proximity of PxL motif receptor and active site between partnering protomers. The impact of disrupting Cdc14 dimerization on phosphatase function is consistent with an important role for trans dephosphorylation. On the other hand, a PxL docking motif also facilitates catalysis of sites at a distance, as illustrated by dephosphorylation of a neighboring subunit of the ORC complex. This opens the possibility that both active sites jointly act on a substrate following the docking interaction. An overview of PxL motif locations relative to Cdk phosphorylation sites showed that the PxL motif often lies embedded in clusters of phosphorylation sites. It is plausible that Cdc14, by the virtue of being a dimer, can direct efficient catalysis both upstream and downstream of the PxL motif. As most proteins contain multiple phosphorylation sites, this might also aid the processive dephosphorylation of target proteins.
The identification of the PxL motif and its receptor pocket on the Cdc14 phosphatase carries potential for additional levels of regulation. For example, the PxL motif identified in the cell fate regulator Dse3 harbors a threonine adjacent to the central proline, which has been detected in a phosphorylated form43. The negative charges that phosphorylation introduces into the otherwise hydrophobic peptide character could well serve to modulate Cdc14 phosphatase binding. In an analogous manner, Cdc14 threonine 109 lies at the edge of the PxL receptor pocket and has been seen phosphorylated in some cellular contexts44. This could equally be envisaged to regulate substrate docking.
Three Cdc14 orthologues CDC14A, B and C are encoded in the human genome. A hydrophobic pocket, where the budding yeast PxL motif receptor lies, is apparent in all of these. This opens the possibility that similar hydrophobic substrate docking is used by human Cdc14 phosphatases. While signs of the dimerization interface are discernible in CDC14A, most interface residues are absent from CDC14B. That the latter phosphatase crystalized as a monomer further lends credence to this isoform being predominantly monomeric21. Deciphering the consequences of these distinct subunit arrangements on Cdc14 catalysis and the cellular roles of the three human CDC14 paralogs remains an important ongoing task.
Online Methods
Peptide phage display
The peptide phage library was designed and constructed as described46 but using the S. cerevisiae proteome. Oligonucleotides for library construction with a diversity of 9 x 104 were purchased from CustomArray Inc. Phage display screening was performed by coating 4 wells of a 96-well plate (Maxisorp, ThermoFisher) with 100 μl of 5 μg/ml Cdc14 overnight, then blocked with 200 μl of blocking buffer (0.2% BSA in PBS) for 1 hour at 4 °C with gentle shaking. 100 μl of the peptide phage library was added to the coated wells and incubated for 1 hour at room temperature with gentle shaking. The plate was washed four times with wash buffer (PBS, 0.5% Tween-20) and bound phages were eluted by adding 100 μl of 100 mM HCl to each well. The eluate was neutralized with 10 μl 1.0 M Tris/HCl pH 11.0. E. coli Omnimax (ThermoFisher) were grown to log phase (A600= ~0.8) and 2 ml culture was infected with 200 μl of the eluted phages for 30 minutes at 37 °C with vigorous shaking at 200 rpm. 1010 phages/ml of M13KO7 helper phage (ThermoFisher) were added, followed by further incubation for 45 minutes at 37 °C with shaking. Cells were then transferred to 50 ml of 2-YT medium including 25 μg/ml kanamycin and 100 μg/ml carbenicillin and incubated overnight at 37 °C with shaking at 200 rpm. Amplified phages were precipitated with 20% PEG-8000 and 2.5 M NaCl to be used for the next round of selection.
Deep sequencing
The enriched phage pool after three rounds was barcoded for Illumina sequencing as described47. Briefly, 5 μl of the phage pool was used as a template for 24 cycles of a 50 μl PCR reaction using 0.5 mM each forward and reverse barcoded primers and KAPA HiFi HotStart Ready Mix (Kapa Biosystems). The PCR products were purified using the QIAquick gel extraction kit (Qiagen), and the concentration of the eluate was determined using the Quant-iT PicoGreen assay (Invitrogen). Samples were sequenced using the MiSeq platform (Illumina) with 250 bp paired-end reads. Reads were demultiplexed by their barcodes and filtered for average Phred quality score of at least 30. Finally, reads mapping to the disordered library were selected and frequencies calculated. Reads not matching the selection criteria were discarded.
Protein purification
Cdc14 and Net11-600 were expressed in E. coli BL21 (DE3) pLysS and purified using Ni-NTA Superflow (Qiagen) or Glutathione Sepharose 4B (GE Healthcare) affinity resins according to manufacturer’s instructions. Following affinity purification, proteins were loaded onto a Superdex 200 Increase 10/300 GL column (GE Healthcare), equilibrated in 50 mM Tris/HCl pH 7.5, 500 mM NaCl, 10% glycerol, 0.5 mM TCEP. Peak fractions were pooled and flash frozen in liquid nitrogen. A list of plasmids used for expression can be found in Supplementary Table 2.
Peptide array binding
The 16-mer Cbk1 peptide and its permuted variants were synthesized on a cellulose membrane using a Multipep peptide synthesizer (Intavis Bioanalytical Instruments)48. The peptide array was solvated in 50% methanol, 10% acetic acid or in 50% propan-2-ol, 10% acetic acid, followed by extensive equilibration in PAB (50 mM Tris/HCl pH 7.5, 150 mM NaCl, 0.1% Tween-20). The array was then blocked in PAB + 2.5% dried skimmed milk (Marvel). For the antibody only control, the array was incubated in PAB + 2.5% skimmed milk, containing α-HA antibody. Following three washes with PAB, the array was incubated with α-mouse horseradish peroxidase-conjugated secondary antibody in PAB + 2.5% milk. The membrane was again washed three times in PAB and developed using ECL reagents (GE Healthcare). For the Cdc14 binding experiment, solvation and blocking steps were as described above, followed by incubated with 100 nM HA-Cdc14 in PAB + 2.5% skimmed milk overnight at 4 °C. Detection with the α-HA antibody was as above.
Microscale thermophoresis
Purified GFP-Cdc14, at a final concentration of 50 nM, was mixed with indicated concentrations of synthetic peptides in 50 mM Tris/HCl pH 7.5, 500 mM NaCl, 0.5 mM TCEP. After incubation in the dark for 30 minutes, each mixture was loaded into glass capillaries (Monolith NT.115 Premium Coated) by capillary action. All measurements were performed at 30% thermophoresis power, 20% blue LED power on the Monolith NT.115 instrument (NanoTemper). Dissociation constants were determined by fitting thermophoresis data (thermophoresis with T-jump) into an equation derived from the law of mass action using the NanoTemper Affinity Analysis software. A list of peptides used can be found in Supplementary Table 3.
Differential Scanning Fluorimetry
For thermal denaturation experiments, 1.5 μg of purified Cdc141-374 was added to a final volume of 25 μl in a buffer containing 50 mM Tris/HCl pH 7.5, 500 mM NaCl, 0.5 mM TCEP and 4.5 μl Sypro Orange fluorescent dye (Invitrogen). A temperature gradient of 0.5 °C/minute was applied from 25 °C to 95 °C in a thermal cycler (Stratagene Mx3005P, Agilent Technologies). All experiments were performed in triplicate. The changes in fluorescence as a function of temperature were fitted to the Boltzmann sigmoidal function in Prism in order to obtain Tm.
Crystallization
For crystallization trials, affinity purification of His-Cdc141-374 was followed by cleavage of the hexahistidine tag by overnight incubation with PreScission Protease (GE Healthcare). The protease was then removed by passing the eluate over Glutathione Sepharose 4B beads before loading onto a HiLoad Superdex 200 26/600 pg column (GE Healthcare) equilibrated in 10 mM Tris/HCl pH 8.5, 750 mM NaCl, 10% glycerol, 0.5 mM TCEP. Peak fractions were pooled and concentrated to 20 mg/ml, as determined by absorbance at 280 nm, by centrifugal filtration.
Cdc141-374 was mixed with either the Cbk1 or Sic1-derived PxL motif peptides (Supplementary Table 3) in a 1:3 molar ratio. The complexes were crystalized at 20 °C using the sitting drop vapor diffusion method. Cdc14:Cbk1 crystalized in two different forms. The first crystal form, referred to hereafter as Cdc14:Cbk1-P212121, grew in a 1 μl drop consisting of a 1:1 (v/v) mixture of protein and a well solution containing 1.5% PEG 6000 and 0.1 M MES pH 6.0. Crystals appeared after 24 hours and reached their maximum size after 4 days (0.7 mm x 0.3 mm x 0.3 mm). The second crystal form, referred to hereafter as Cdc14:Cbk1-C2, grew in a 1 μl drop consisting of a 1:1 (v/v) mixture of protein and a well solution containing 11% PEG 3350 and 0.2 M Calcium acetate. Crystals appeared after 72 hours and reached their maximum size after 10 days (0.5 mm x 0.3 mm x 0.02 mm). Cdc14:Sic1 complex crystals grew in a 1 μl drop consisting of a 1:1 (v/v) mixture of protein and a well solution containing 18% PEG 200, 5% PEG 3000 and 0.1 M MES pH 6. Crystals appeared after 48 hours and reached their maximum size after 7 days (0.08 mm x 0.08 mm x 0.3 mm).
Data collection and refinement of the Cdc14:Cbk1/Sic1 complexes
Crystals were cryoprotected in mother liquor supplemented with 20% glycerol, then flash-frozen in liquid nitrogen. X-ray data sets were collected at 100 K at the IO4 (mx13775-35), IO3 (mx13775-37) and IO4-1 (mx13775-45) beamlines of the Diamond Light Source Synchrotron (Oxford, UK). Data collection and refinement statistics are summarized in Table 1. The data sets were indexed, scaled and merged with xia249. Molecular replacement was achieved by using the atomic coordinates of human Cdc14B from PDB 1OHC (Ref. 21) in PHASER50. Refinement was carried out using PHENIX51. Model building was carried out in Coot52, model validation used PROCHECK53 and figures were prepared using the PyMOL Molecular Graphics System, Version 2.0 (Schrödinger, LLC). The asymmetric unit contains two copies of Cdc14 forming a stable dimer. The difference electron density map covering Cdc14 shows unambiguous density for Cbk1 and Sic1 peptides in the three structures.
Determination of the bound metal using its anomalous signal
To confirm the identity of the metal present in our structure as zinc, we collected a dataset on a Cdc14:Sic1 crystal at the zinc absorption peak (1.281 Å) at 100 K at the IO4 (mx13775-45) beamline of the Diamond Light Source Synchrotron (Oxford, UK). We calculated the anomalous difference density map using the refined molecular model for phase information. The density map showed a clear density peak with a height larger than 8 σ in the metal binding site (Supplementary Fig. 3c). This experiment shows that zinc is coordinated by the four side chains of Cdc14 residues E193, H195, H206 and H238.
In vitro enzymatic phosphatase assay
In the experiment shown in Supplementary Figure 9a, 100 nM His6-Cdc14 and the indicated concentrations of Cbk1 (FTDVPALNYPATPPPH) or scrambled (YVPAPAPTTPLNFDHP) peptide were mixed with 20 μM p-nitrophenyl phosphate (Calbiochem) in the same buffer as that used for thermophoresis experiments (50 mM Tris/HCl pH 7.5, 500 mM NaCl, 0.5 mM TCEP), and incubated at 30 °C. Absorbance at 410 nm over time was recorded using the SpectraMax Plus 384 microplate reader (Molecular Devices). Initial speed determination was performed by fitting the linear increase in product accumulation over time using Prism. For the experiment shown in Supplementary Figure 6a the procedure outlined above was followed, except that incubation was in 50 mM Tris/HCl pH 7.5, 100 mM NaCl, 0.5 mM TCEP. For the experiment shown in Supplementary Figure 8c incubation was in 50 mM Tris/HCl pH 8.5, 750 mM NaCl, 10% glycerol, 0.5 mM TCEP.
In vitro peptide dephosphorylation assay
The following synthetic phosphopeptides were used as Cdc14 substrates in dephosphorylation assays. FTDVPALNYPATPPPH-TSPLGpSPLAAFNGLTSPQRSPFPKSSVK, FTDVAAGNYPATPPPHTSPLGpSPLAAF-NGLTSPQRSPFPKSSVK, FTDVPALNYPATPPPHTSPLGpTPLAAFNGLTSPQRSPFP-KSSVK and FTDVAAGNYPATPPPH-TSPLGpTPLAAFNGLTSPQRSPFPKSSVK. The velocities of phosphate hydrolysis were determined using the BIOMOL Green assay (Enzo Life Sciences) in 50 mM Tris/HCl pH 7.5, 150 mM NaCl, 5 mM 2-mercaptoethanol and the indicated concentrations of Cdc14 (100 nM or 1 μM) and substrate peptides, following the manufacturer’s instructions. At least two time points were taken to ensure linearity of reactions.
Cdc14-Net1 interaction assay
10 μl of Ni-NTA Superflow beads (Qiagen) were equilibrated in 20 mM Tris/HCl pH 7.5, 200 mM NaCl, 0.2% NP-40, 20 mM imidazole, following which they were chelated to 30 pmol His6-Net11-600 or His6-GFP for one hour at 4 °C. 30 pmol GST-Cdc14, GST-Cdc14 TAB6 or GST-Cdc14 W108A were added to the beads for 15 minutes at 30 °C at a final concentration of 0.5 μM. Following washes in the same buffer, the beads were boiled in SDS-PAGE loading buffer and eluates analyzed by SDS-PAGE followed by Coomassie Blue staining.
Analytical gel filtration
100 μl of 100 ng/μl purified Cdc14 or Cdc14 GETS were loaded onto a Superdex 200 Increase 10/300 GL column (GE Healthcare), equilibrated with 50 mM Tris/HCl pH 8.5, 750 mM NaCl, 10% glycerol, 0.5 mM TCEP. The column was calibrated in the same buffer with globular protein markers of known molecular weights. Elution was monitored by measuring protein absorbance at 280 nm.
Yeast strains and techniques
All budding yeast strains used in this study were of the W303 background and are listed in Supplementary Table 4. Cells were grown in YP medium supplemented with 2% glucose, or with a combination of 2% raffinose and 2% galactose as carbon sources. Epitope tagging of endogenous genes, gene deletions and mutagenesis were performed using polymerase chain reaction (PCR)-based gene targeting54. Ectopic Cdc14 expression in the cdc14-1 temperature sensitive background was achieved by integrating a vector carrying wild type or variant CDC14 genes under control of the CDC14 promoter at the TRP1 locus. When fusing the Cbk1-derived PxL motif peptide to Orc6, the non-conserved threonine in the downstream TP feature was substituted with alanine to preempt possible cyclin-dependent kinase phosphorylation. Cell synchronization in G1 phase was performed by addition of 3.25 µg/ml of α-factor to the cultures every hour for ~1.5 population doubling times55. Re-arrest after synchronous passage through one cell cycle was achieved by addition of 7 µg/ml of α-factor. M-phase arrest was obtained by shifting cultures of cells expressing Cdc20 under GAL1 promoter control, growing in medium containing raffinose and galactose, to medium containing 2% raffinose only for ~1.5 population doubling times. Synchronous release from the metaphase arrest was obtained by addition of 2% galactose to re-induce Cdc20 expression56. FACS analysis of DNA content was performed on propidium iodide-stained cells, 10,000 cells were scored at each time point.
Western blotting
Protein extracts for western blotting were prepared following fixation using trichloroacetic acid57. Extracts were separated by SDS-PAGE and processed by western transfer. The following antibodies were used for detection: α-Cdc14 (Ref. 45), α-Clb2 (Santa Cruz, sc-9071), α-Clb5 (Santa Cruz, sc20170), α-Sic1 (Santa Cruz, sc-50441), α-Orc6 (clone SB49), α-Orc2 (a gift from John Diffley)58, α-myc (clone 9E10), α-HA (clone 12CA5), α-Pk (Bio-Rad, clone SV5-Pk1), α-tubulin (Abcam, clone YOL 1/34).
Cdc14-substrate co-immunoprecipitation
To analyze co-immunoprecipitation of Cdc14 with its substrates, 100 ml asynchronously growing mid-log phase cultures were harvested by centrifugation. Protein extracts were prepared in 50 mM Tris/HCl pH 7.5, 150 mM NaCl, 10% glycerol, 1 mM EDTA, 0.1% NP-40, 0.5 mM PMSF, 0.5 mM TCEP, 10 mM β-glycerophosphate, 10 mM sodium fluoride, 0.1 mM sodium vanadate, protease inhibitor cocktail (Roche), benzonase (Merck Biosciences) using glass bead breakage in a Multi Bead Shocker (Yasui Kikai). Extracts were precleared, incubated with the antibody as indicated and immunocomplexes adsorbed to Protein A Dynabeads (Thermo Fisher Scientific). Beads were extensively washed in lysis buffer and elution was carried out by boiling in SDS-PAGE loading buffer.
Supplementary Figures
Acknowledgements
We would like to thank N. Davey and D. Morgan for bringing together the Kim and Uhlmann laboratories, the Diamond Light Source for access to beamlines IO3, IO4, IO4-1 (MX13775), S. Federico, D. Joshi and N. O’Reilly for peptide synthesis, J. Diffley for the Orc2 antibody, V. Christodoulou, M. Hall, S. Kjaer, L. Masino, A. Purkiss and S. Touati for help and advice and M. Godfrey, E. Weiss and members of our laboratory for discussions and critical reading of the manuscript. This work was supported by The Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001198), the UK Medical Research Council (FC001198), and the Wellcome Trust (FC001198).
Footnotes
Data availability
Atomic coordinates and structure factors have been deposited in the Protein Data Bank, with accession numbers PDB 6G84 (Cdc14–Cbk1, C2), PDB 6G85 (Cdc14–Cbk1, P 212121) and PDB 6G86 (Cdc14–Sic1). All additional data that support the findings of this study are available from the corresponding author upon request.
Author Contributions
M.K. and F.U. conceived the study; M.K. performed the experiments; S.M. and M.K. solved the crystal structures; M.-H.S., C.C.-V. and P.M.K. conducted the phage display screen, M.K. and F.U. wrote the manuscript with input from all authors.
Competing interests
The authors declare no competing interests.
References
- 1.Morgan D. The cell cycle: Principles of control. New Science Press; 2007. [Google Scholar]
- 2.Loog M, Morgan DO. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–108. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- 3.Koivomägi M, et al. Dynamics of Cdk1 substrate specificity during the cell cycle. Mol Cell. 2011;42:610–623. doi: 10.1016/j.molcel.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhaduri S, Pryciak PM. Cyclin-specific docking motifs promote phosphorylation of yeast signaling proteins by G1/S Cdk complexes. Curr Biol. 2011;21:1615–1623. doi: 10.1016/j.cub.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Godfrey M, et al. PP2ACdc55 phosphatase imposes ordered cell-cycle phosphorylation by opposing threonine phosphorylation. Mol Cell. 2017;65:393–402. doi: 10.1016/j.molcel.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Visintin R, et al. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- 7.Guacci V, Hogan E, Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol. 1994;125:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Higuchi T, Uhlmann F. Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature. 2005;433:171–176. doi: 10.1038/nature03240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woodbury EL, Morgan DO. Cdk and APC activities limit the spindle-stabilizing function of Fin1 to anaphase. Nat Cell Biol. 2007;9:106–112. doi: 10.1038/ncb1523. [DOI] [PubMed] [Google Scholar]
- 10.Khmelinskii A, Roostalu J, Roque H, Antony C, Schiebel E. Phosphorylation-dependent protein interactions at the spindle midzone mediate cell cycle regulation of spindle elongation. Dev Cell. 2009;17:244–256. doi: 10.1016/j.devcel.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 11.Mirchenko L, Uhlmann F. Sli15INCENP dephosphorylation prevents mitotic checkpoint reengagement due to loss of tension at anaphase onset. Curr Biol. 2010;20:1396–1401. doi: 10.1016/j.cub.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuilman T, et al. Identification of Cdk targets that control cytokinesis. EMBO J. 2014;34:81–96. doi: 10.15252/embj.201488958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bloom J, et al. Global analysis of Cdc14 phosphatase reveals diverse roles in mitotic processes. J Biol Chem. 2011;286:5434–5445. doi: 10.1074/jbc.M110.205054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Touati SA, Kataria M, Jones AW, Snijders AP, Uhlmann F. Phosphoproteome dynamics during mitotic exit in budding yeast. EMBO J. 2018;37:e98745. doi: 10.15252/embj.201798745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Visintin R, Hwang ES, Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- 16.Shou W, et al. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- 17.Bouchoux C, Uhlmann F. A quantitative model for ordered Cdk substrate dephosphorylation during mitotic exit. Cell. 2011;147:803–814. doi: 10.1016/j.cell.2011.09.047. [DOI] [PubMed] [Google Scholar]
- 18.Breitkreutz A, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kao L, et al. Global analysis of Cdc14 dephosphorylation sites reveals essential regulatory role in mitosis and cytokinesis. Mol Cell Proteomics. 2014;13:594–605. doi: 10.1074/mcp.M113.032680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki K, et al. Identification of non-Ser/Thr-Pro consensus motifs for Cdk1 and their roles in mitotic regulation of C2H2 zinc finger proteins and Ect2. Sci Rep. 2015;5:7929. doi: 10.1038/srep07929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gray CH, Good VM, Tonks N K, Barford D. The structure of the cell cycle protein Cdc14 reveals a proline-directed protein phosphatase. EMBO J. 2003;22:3524–3535. doi: 10.1093/emboj/cdg348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bremmer SC, et al. Cdc14 phosphatases preferentially dephosphorylate a subset of cyclin-dependent kinase (Cdk) sites containing phosphoserine. J Biol Chem. 2012;287:1662–1669. doi: 10.1074/jbc.M111.281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eissler CL, et al. The Cdk/Cdc14 module controls activation of the Yen1 Holliday junction resolvase to promote genome stability. Mol Cell. 2014;54:80–93. doi: 10.1016/j.molcel.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seo MH, Nim S, Jeon J, Kim PM. Large-scale interaction profiling of protein domains through proteomic peptide-phage display using custom peptidomes. Methods Mol Biol. 2017;1518:213–226. doi: 10.1007/978-1-4939-6584-7_14. [DOI] [PubMed] [Google Scholar]
- 25.Davey NE, et al. Attributes of short linear motifs. Mol Biosyst. 2012;8:268–281. doi: 10.1039/c1mb05231d. [DOI] [PubMed] [Google Scholar]
- 26.Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K. Improved visualization of protein consensus sequences by iceLogo. Nat Methods. 2009;6:786–787. doi: 10.1038/nmeth1109-786. [DOI] [PubMed] [Google Scholar]
- 27.Brace J, Hsu J, Weiss EL. Mitotic exit control of the Saccharomyces cerevisiae Ndr/LATS kinase Cbk1 regulates daughter cell separation after cytokinesis. Mol Cell Biol. 2011;31:721–735. doi: 10.1128/MCB.00403-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun. 2010;1:100. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- 29.Ivarsson Y, et al. Large-scale interaction profiling of PDZ domains through proteomic peptide-phage display using human and viral phage peptidomes. Proc Natl Acad Sci USA. 2014;111:2542–2547. doi: 10.1073/pnas.1312296111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colman-Lerner A, Chin TE, Brent R. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell. 2001;107:739–750. doi: 10.1016/s0092-8674(01)00596-7. [DOI] [PubMed] [Google Scholar]
- 31.Korinek WS, et al. Cyk3, a novel SH3-domain protein, affects cytokinesis in yeast. Curr Biol. 2000;10:947–950. doi: 10.1016/s0960-9822(00)00626-6. [DOI] [PubMed] [Google Scholar]
- 32.Meitinger F, et al. Targeted localization of Inn1, Cyk3 and Chs2 by the mitotic-exit network regulates cytokinesis in budding yeast. J Cell Sci. 2010;123:1851–1861. doi: 10.1242/jcs.063891. [DOI] [PubMed] [Google Scholar]
- 33.Taylor GS, Liu Y, Baskerville C, Charbonneau H. The activity of Cdc14p, an oligomeric dual specificity protein phosphatase from Saccharomyces cerevisiae, is required for cell cycle progression. J Biol Chem. 1997;272:24054–24063. doi: 10.1074/jbc.272.38.24054. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi J, Matsuura Y. Structure and dimerization of the catalytic domain of the protein phosphatase Cdc14p, a key regulator of mitotic exit in Saccharomyces cerevisiae. Protein Sci. 2017;26:2105–2112. doi: 10.1002/pro.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood JS, Hartwell LH. A dependent pathway of gene functions leading to chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1982;94:718–726. doi: 10.1083/jcb.94.3.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen VQ, Co C, Li JJ. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature. 2001;411:1068–1073. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- 37.Traverso EE, et al. Characterization of the Net1 cell cycle-dependent regulator of the Cdc14 phosphatase from budding yeast. J Biol Chem. 2001;276:21924–21931. doi: 10.1074/jbc.M011689200. [DOI] [PubMed] [Google Scholar]
- 38.Shou W, Deshaies RJ. Multiple telophase arrest bypassed (tab) mutants alleviate the essential requirement for Cdc15 in exit from mitosis in S. cerevisiae. BMC Genet. 2002;3:4. doi: 10.1186/1471-2156-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Azzam R, et al. Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus. Science. 2004;305:516–519. doi: 10.1126/science.1099402. [DOI] [PubMed] [Google Scholar]
- 40.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 41.Bollen M, Peti W, Ragusa MJ, Beullens M. The extended PP1 toolkit: designed to create specificity. Trends Biochem Sci. 2010;35:450–458. doi: 10.1016/j.tibs.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hertz EPT, et al. A conserved motif provides binding specificity for the PP2A-B56 phosphatase. Mol Cell. 2016;63:686–695. doi: 10.1016/j.molcel.2016.06.024. [DOI] [PubMed] [Google Scholar]
- 43.Swanley DL, et al. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat Methods. 2013;10:676–682. doi: 10.1038/nmeth.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holt LJ, et al. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325 doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.López-Avilés S, Kapuy O, Novák B, Uhlmann F. Irreversibility of mitotic exit is the consequence of systems-level feedback. Nature. 2009;459:592–595. doi: 10.1038/nature07984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davey NE, et al. Discovery of short linear motif-mediated interactions through phage display of intrinsically disordered regions of the human proteome. FEBS J. 2017;284:485–498. doi: 10.1111/febs.13995. [DOI] [PubMed] [Google Scholar]
- 47.Sun MG, Seo MH, Nim S, Corbi-Verge C, Kim PM. Protein engineering by highly parallel screening of computationally designed variants. Sci Adv. 2016;2:e1600692. doi: 10.1126/sciadv.1600692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frank R. High-density synthetic peptide microarrays: emerging tools for functional genomics and proteomics. Comb Chem High Throughput Screen. 2002;5:429–440. doi: 10.2174/1386207023330165. [DOI] [PubMed] [Google Scholar]
- 49.Winter G, Lobley CM, Prince SM. Decision making in xia2. Acta Crystallogr. 2013;69:1260–1273. doi: 10.1107/S0907444913015308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vaguine AA, Richelle J, Wodak SJ. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr. 1999;55:191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- 54.Knop M, et al. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 55.O'Reilly N, Charbin A, Lopez-Serra L, Uhlmann F. Facile synthesis of budding yeast a-factor and its use to synchronize cells of a mating type. Yeast. 2012;29:233–240. doi: 10.1002/yea.2906. [DOI] [PubMed] [Google Scholar]
- 56.Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 57.Foiani M, Marini F, Gamba D, Lucchini G, Plevani P. The B subunit of the DNA polymerase a-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol Cell Biol. 1994;14:923–933. doi: 10.1128/mcb.14.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Donovan S, Harwood J, Drury LS, Diffley JF. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chatr-Aryamontri A, et al. The BioGRID interaction database: 2017 update. Nucl Acids Res. 2017;45:D369–D379. doi: 10.1093/nar/gkw1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.