Abstract
Development and implementation of rapid antimicrobial susceptibility testing is critical for guiding patient care and improving clinical outcomes, especially in cases of sepsis. One approach to reduce the time-to-answer for antimicrobial susceptibility is monitoring the inhibition of DNA production, as differences in DNA concentrations are more quickly impacted compared to optical density changes in traditional antimicrobial susceptibility testing. Here, we use real-time PCR to rapidly determine antimicrobial susceptibility after short incubations with antibiotic. Application of this assay to a collection of 144 isolates in mock blood culture, covering medically relevant pathogens displaying high rates of resistance, provided susceptibility data in under 4 hours. This assay provided categorical agreement with a reference method in 96.3% of cases across all species. Sequencing of a subset of PCR amplicons showed accurate genus level identification. Overall, implementation of this method could provide accurate susceptibility results with a reduced time-to-answer for a number of medically relevant bacteria commonly isolated from blood culture.
Introduction
Introduction of antibiotics into patient treatment strategies drastically reduced bacterial infection related morbidity and mortality and opened the door to medical procedures that otherwise carry high risk of infection, including invasive surgeries and medical device implantation. Unfortunately, the spread of antimicrobial resistance and the emergence of multi-drug resistant strains is a global problem limiting treatment options and contributing to prescribing of ineffective antibiotic regimens that fail to resolve infections [1]. Therefore, it is imperative for clinicians to have access to susceptibility data when prescribing antimicrobial agents to avoid the empirical selection of ineffective treatments, especially in life-threatening, time sensitive cases such as sepsis [2, 3]. Traditional, culture-based antimicrobial susceptibility testing (AST) often takes more than 48 h for time-to-answer [4], an inappropriate timeframe for guiding early treatment strategies; thus, development of more rapid AST assays could potentially mitigate these issues [5, 6]. A further benefit of rapid AST is the fostering of the initiative towards antimicrobial stewardship, which aims to reduce unnecessary antibiotic usage to improve patient outcomes and reduce selection-induced antimicrobial resistance [7, 8]. Rapid evolution and propagation of resistance determinants coupled with the challenges of developing novel antimicrobial agents and treatment methods highlights the importance of maintaining the effectiveness of currently available antimicrobial drugs [9].
An ideal AST assay would report phenotypic susceptibility, be applicable across a broad range of human pathogens and antibiotics, function with clinical samples, and deliver a result in the shortest amount of time possible. A variety of novel approaches work towards these goals, including but not limited to: genotyping [10, 11], single cell microscopy [12, 13], Raman spectroscopy [14], mass spectrometry [15–17], and nucleic acid amplification [18–22]. Each of these strategies has strengths and weaknesses, and ongoing work within respective fields continues to push each methodology towards the ideal AST. In particular, strategies focusing on nucleic acid amplification are especially appealing as DNA and RNA cellular synthesis can be very rapid and is proportionally impacted either directly or indirectly by antibiotic inhibition of susceptible cells.
In general, nucleic acid amplification strategies for AST compare relative quantification of DNA or RNA concentrations between antibiotic-treated samples and untreated controls post-antibiotic treatment. Bacterial strains susceptible to the given antibiotic will display growth inhibition resulting in significantly lower concentrations of nucleic acid, measurable by quantitative PCR (qPCR) or other amplification technologies [18]. Previous studies utilizing this approach showed promising results with high levels of categorical agreement with gold standard assays [18–21]. In fact, the method adequately determined susceptibility across multiple classes of antibiotics and was applicable to a variety of bacterial pathogens depending on the primer (and probe) design [19, 21]. More recent studies focused on reducing the overall assay time-to-answer, with specific focus on rapid results obtained through the use of isothermal amplification technologies [20].
The present study aims to assess a previously reported, broad-coverage qPCR primer and probe set, the BactQuant assay, for use in AST assay development [23]. The BactQuant assay targets the V3-V4 region of the 16S gene sequence with specific design features providing improved genus and species coverage over earlier 16S-targeting assays [23]. Here, we modified the method for the RApid Molecular Antibiotic Susceptibility Testing (RAMAST) assay reported by Beuving et al. [19] to reduce the time-to-answer and increase species coverage and evaluated the assay with a large set of important human pathogens. The RAMAST assay was performed on mock blood cultures with three disparate antibiotics per species, demonstrating the applicability of this assay to multiple antibiotic classes.
Materials and methods
Bacterial isolates and culture
Clinical isolates and reference strains were obtained from the American Type Culture Collection (ATCC), the Biodefense and Emerging Infections Research Resources Repository (BEI Resources), the Food and Drug Administration (FDA), the Centers for Disease Control (CDC), and the Unified Culture Collection housed at United States Army Medical Research Institute of Infectious Diseases (USAMRIID). The origin of each strain is listed in Tables A-E in S1 File. Isolates were propagated on blood agar (trypticase soy agar with 5% sheep blood) at 35°C and cultured in tryptic soy broth (TSB) liquid medium at 37°C with shaking unless otherwise noted. Staphylococcus aureus 880 (BR-VRSA) was found to rapidly lose vancomycin resistance in liquid culture and was thus propagated in the presence of 32 μg/mL vancomycin when liquid culture was required; cultures with vancomycin were then diluted at least 1,000 fold before use in any assay (32 ng/mL is well below the vancomycin minimum inhibitory concentration (MIC) of all strains tested). Antibiotics were obtained from Sigma-Aldrich, Thermo Fisher Scientific, or Gold Biotechnology and used at the concentrations given from 100x stock solutions in water.
Gold standard antibiotic susceptibility testing
Reference MICs were determined by the broth microdilution (BMD) method in accordance with Clinical and Laboratory Standards Institute (CLSI) guidelines [24]. Enterococcus faecium NR-32094 did not display visible growth in the standard testing media (cation adjusted Mueller Hinton broth, CAMHB) and was tested in TSB instead. Staphylococcus aureus ATCC 13565 did not display visible growth in the presence of additional 2% NaCl and was tested without the added NaCl for oxacillin. The susceptibility breakpoints for antibiotics were taken from the 2017 CLSI guidelines [25].
Assessment of antibiotic susceptibility using qPCR
Bacterial cultures prepared in TSB media were inoculated from colonies on fresh blood agar plates and were incubated at 37°C with shaking overnight (~16 h). The overnight cultures were used to spike 10 mL of human whole blood (BioIVT, Maryland, USA), and the entire aliquots of blood were injected into BD BACTEC standard/10 aerobic/F bottles (Becton Dickinson, New Jersey, USA). Blood culture (BC) bottles were incubated in a BD BACTEC FX40 instrument until flagged as positive by the instrument software. Within 30 min of positivity, 500 μL of culture was removed from each bottle and diluted into 4.5 mL of room temperature TSB. This diluted culture was then used to make 200 μL aliquots. Either antibiotics at the concentrations to be tested or no-antibiotic controls were added to each aliquot from 100x stocks in water. The aliquots were vortexed briefly and then incubated at 37°C with shaking in a ThermoMixer (Eppendorf, Hamburg, Germany) for 1 or 2 h. After incubation, samples were centrifuged at 16,000 x g for 3 min at room temperature and the supernatant was carefully removed by pipet.
Pellets were resuspended in 200 μL lysis buffer (10 mM Tris-HCl, 1 mM disodium EDTA, pH 8.0, 20 mg/mL lysozyme, 300 U/mL mutanolysin, 1.2% Triton X-100) and incubated at 37°C with shaking at 1,200 rpm in a ThermoMixer for 20 min. Glass beads (0.1 mm PowerBeads; Qiagen, Hilden, Germany) were then added to the samples to an approximate volume of 20 μL and the samples were vortexed on the fastest setting for 5 min. Buffers and spin columns from a QIaAMP DNA Mini Kit (Qiagen) were then used to complete the DNA extraction. To the bead beat samples, 200 μL of buffer AL was added and the samples were mixed by brief vortexing. Then, 200 μL of 100% ethanol was added and the samples were again mixed by brief vortexing. The samples were centrifuged briefly and 450 μL of supernatant was transferred to the kit spin columns. The samples were then washed according to the manufacturer’s instructions and eluted in 200 μL of water.
Eluted DNA was diluted 100 fold in water before use in qPCR. The qPCR was performed as previously described, with slight modifications [23]. Briefly, the PCR reactions were run in 10 μL volumes and were composed of the universal 16S BactQuant primers (5’-CCTACGGGDGGCWGCA-3’ and 5’-GGACTACHVGGGTMTCTAATC-3’) at 1.8 μM, probe (6FAM-5′-CAGCAGCCGCGGTA-3′-MGBNFQ) at 0.2 μM, Platinum qPCR Supermix-UDG (ThermoFisher) at 1x, water, and diluted sample DNA (2.5 μL). The amplification was performed on a LightCycler 480 II instrument (Roche, Basel, Switzerland) with cycling conditions of 50°C for 2 min, 95°C for 2 min, and 40 cycles of 95°C for 15 s and 60°C for 45 s. Quantification cycle (Cq) values were calculated automatically with the second derivative max method. The qPCR for each sample was run as technical triplicates, and the three values were averaged to give the final Cq value used in data analysis.
Data analysis
To determine antibiotic susceptibility, ΔCq values were calculated by subtracting the Cq value obtained from the no-antibiotic control sample from the Cq values for the antibiotic-treated samples. Cutoff ΔCq values were determined for each species (or family for Enterobacteriaceae spp.) from ROC curves derived from the ΔCq values determined for each isolate (Figure A in S1 File). Cutoff values for each species were selected that maximized the likelihood ratio for a correct susceptibility call. ΔCq values used as cutoffs for each species or family were: > 2.25 for Enterobacteriaceae spp.; > 2.00 for Acinetobacter baumannii; > 1.70 for Pseudomonas aeruginosa; > 1.55 for Enterococcus faecium; and > 2.05 for S. aureus. A ΔCq value above the cutoff was used to indicate susceptibility to the antibiotic while a ΔCq value below the cutoff indicated resistance. Errors were defined as minor, major, or very major. Major errors were defined as false-resistant results and the major error rate was calculated as the number of false-resistant results over the total number of susceptible isolates as determined by BMD. Very major errors were defined as false-susceptible results and the very major error rate was calculated as the number of false-susceptible results over the total number of resistant isolates as determined by BMD. Strains with an intermediate resistance result from BMD were treated as resistant for the purposes of calculating categorical agreement, and a susceptible result from the qPCR assay for an intermediate strain was defined as a minor error.
Amplicon sequencing and analysis
Amplicons from the qPCR assay were sequenced with a MiSeq system (Illumina, California, USA). Amplicons were first run through automated PCR purification using an Apollo 324 NGS Library Prep System (Takara Bio, Kusatsu, Japan) running the instrument’s “PCR Cleanup 32” protocol. TruSeq HT adaptors (Illumina) were then ligated using the Apollo 324’s “PrepX ILM 32i DNA” protocol for 520 base pair size, utilizing a SMARTer PrepX Universal DNA Library kit (Takara Bio). PCR enrichment was performed on the library products by adding 25 μL of KAPA HiFi HotStart ReadyMix (Roche), 5 μL of KAPA Library Amplification Primer Mix, and 5 μL of molecular biology grade water to each sample followed by a thermal cycling program of 98°C for 45 s, 10 cycles of [98°C for 15 s, 60°C for 30 s, and 72°C for 30 s], and 72°C for 1 min. PCR purification was performed again as above before DNA was quantified, diluted, and pooled. Pooled DNA was quantified by qPCR using a KAPA Library Quantification Kit (Roche), then further diluted to 2 nM. The 2 nM pooled DNA library was mixed with an equal volume of 0.1 M NaOH and incubated at room temperature for 5 min to denature. A PhiX library was also prepared by mixing equal volumes of 2 nM PhiX Sequencing Control v3 (Illumina) with 0.1 M NaOH and incubating at room temperature for 5 min to denature. Both denatured libraries were diluted to 12 pM with pre-chilled buffer HT1 (from a MiSeq Reagent kit v2 (Illumina)), pooled to give a final ratio of 25% PhiX, and sequenced on a MiSeq with a 600-cycle MiSeq cartridge with paired-end reads.
Analysis was performed using CLC Genomics Workbench (Qiagen). Imported paired end reads were merged and trimmed using a quality score 0.05 and a sequence cutoff length of >100 base pairs. The Ribosomal Database Project (RDP) was used to curate a down selected reference database composed of isolates of type strains from genera of medically relevant pathogens (with the inclusion of all species in those genera) as previously reported [26]. Only entries of greater than 1,200 base pairs with good quality were used in the curated database. A stringent reference-based mapping of sequencing reads to the curated RDP was used with mapping settings as follows: mismatch cost of 10, insertion cost of 3, deletion cost of 3, insertion open cost of 6, insertion extend cost of 1, deletion open cost of 6, deletion extend cost of 1, length fraction of 0.5, and similarity fraction of 1. Non-template controls were used to account for sample bleed and a cutoff was calculated using the mean plus 3 times the standard deviation for each genus. Reads which fell below this cutoff were removed and total reads mapped for each sample were calculated along with the representative percentage of each genus in the sample.
Results and discussion
Assay design and optimization
We chose to perform the RAMAST assay on extracted DNA due to the necessity for determining relative bacterial growth through quantitative DNA concentration measurements. While the addition of a DNA extraction before qPCR increased the turn-around-time of the assay, this step decreased the likelihood of qPCR inhibition by culture components for establishing a proof of concept for this assay. Ensuring the removal of PCR inhibitors is especially important when blood is present in a sample [27], as was the case in this study. Previous work examining AST by qPCR primarily utilized urine samples or pure culture, with one exception which used bacteria from blood culture bottles partially purified with serum separator tubes before the incubation with antibiotics [19]. Because initial testing with the standard lysis procedure stipulated in the DNA extraction kit protocol did not appear to result in complete lysis, especially for dense cultures of Gram-positive strains, our DNA extraction method included a rigorous cell lysis procedure including enzymatic digestion and bead beating to ensure complete lysis (Figure B in S1 File). Moving forward, alternative sample preparation procedures prior to qPCR could be utilized to decrease the assay time while maintaining adequate performance, particularly for Gram-negative species.
Susceptibility testing on positive blood cultures
Empirical assessment is required before any assay can be matured through clinical evaluations. Towards this end, we tested a total of 144 clinical and reference isolates after mock blood culture for antibiotic susceptibility with the RAMAST assay: 50 Enterobacteriaceae spp., 30 Acinetobacter baumannii, 22 Pseudomonas aeruginosa, 22 Enterococcus faecium, and 20 S. aureus (Table 1). Assay optimization for three antibiotics per species (ciprofloxacin, gentamicin, and imipenem for Gram-negatives; ciprofloxacin, vancomycin, and oxacillin for S. aureus; ciprofloxacin, vancomycin, and ampicillin for E. faecium), across multiple concentrations spanning CLSI breakpoints determined the optimal concentration for distinguishing susceptible from resistant strains. Optimal concentrations are found for each species-antibiotic combination in Table 1. Due to the need for different antibiotic concentrations for different species, the assay is best performed after species identification. Temporally, we found an antibiotic incubation time of 2 h to be sufficient across all antibiotics with the exception of ciprofloxacin, which only required a 1 h antibiotic incubation. Shorter incubation times required for ciprofloxacin may be due to the fluoroquinolone antimicrobial inhibitory mode of action, which involves inhibition of DNA gyrase and topoisomerase IV activity through formation of drug-enzyme-DNA complexes with indirect consequences for DNA replication [28]. Inhibition of gyrase activity results in double-strand DNA breaks in metabolically active bacteria, terminating further DNA replication likely with higher rapidity than possible with other antibiotic classes. Other DNA targeting antimicrobials could also afford a shorter required incubation time, as noted in the literature for both nitrofurantoin and ciprofloxacin [20].
Table 1. Number of tested isolates and the antibiotic concentration used in the qPCR assay.
| Enterobacteriaceae spp. (N = 50) | Susceptible* | Intermediate* | Resistant* | Antibiotic conc. tested | |
|---|---|---|---|---|---|
| Ciprofloxacin | 23 | 0 | 27 | 2 μg/mL | |
| Gentamicin | 31 | 2 | 17 | 16 μg/mL | |
| Imipenem | 24 | 1 | 25 | 4 μg/mL | |
| A. baumannii (N = 30) | |||||
| Ciprofloxacin | 5 | 0 | 25 | 2 μg/mL | |
| Gentamicin | 7 | 3 | 20 | 8 μg/mL | |
| Imipenem | 16 | 1 | 13 | 4 μg/mL | |
| P. aeruginosa (N = 22) | |||||
| Ciprofloxacin | 5 | 0 | 17 | 2 μg/mL | |
| Gentamicin | 5 | 0 | 17 | 8 μg/mL | |
| Imipenem | 8 | 1 | 13 | 2 μg/mL | |
| E. faecium (N = 22) | |||||
| Ciprofloxacin | 7 | 3 | 12 | 1 μg/mL | |
| Vancomycin | 14 | 0 | 8 | 1 μg/mL | |
| Ampicillin | 10 | 0 | 12 | 16 μg/mL | |
| S. aureus (N = 20) | |||||
| Ciprofloxacin | 11 | 0 | 9 | 2 μg/mL | |
| Vancomycin | 18 | 0 | 2 | 2 μg/mL | |
| Oxacillin | 8 | 0 | 12 | 0.5 μg/mL | |
* Categorical assignment from reference method (BMD)
Ciprofloxacin is not generally indicated for blood stream infections with E. faecium and S. aureus [24]; however, we included this fluoroquinolone to highlight the shorter incubation time. In addition, this assay could be easily repurposed to different matrices, such as urine, or bacterial species where testing ciprofloxacin would be more appropriate. With the optimized incubation times, the overall assay turn-around-time was approximately 3 h for ciprofloxacin and 4 h for the other antibiotics post blood culture positivity.
With the optimized assay conditions, we tested a total of 432 antibiotic-species combinations. We found an overall categorical agreement of 96.3%, with a major error rate of 3.7% and a very major error rate of 3.1% (Table 2, Fig 1) compared to broth microdilution (BMD). Major and very major error rates were calculated as the numbers of false resistant or false susceptible results divided by the numbers of true susceptible or true resistant isolates, respectively. The numbers of true susceptible and true resistant isolates were used in the denominator rather than the total number of samples (the latter method is occasionally reported in AST assay evaluation but provides misleadingly low error rates). Calculations were performed in this manner to accurately reflect the high error rates resulting from even a single error when a small population of either resistant or susceptible isolates are used for assay evaluation. We treated a susceptible result from the RAMAST assay for an intermediate resistance strain as a minor error, with only 2 minor errors from 12 intermediate strains, as the assay produces a binary result limiting the application for intermediate resistance classification. Retesting major or very major errors from specific antibiotic-strain combinations resulted in 5 of 14 producing correct results upon re-inspection (Table F in S1 File). These conflicting data could be due to operator error, failure in an extraction column, or erroneous qPCR readings. In the future, automation of the sample preparation procedure would reduce the potential for operator error while also minimizing hands-on time and decreasing assay time. A possibility for repeat errors on retesting may be that these strains simply behave differently than other strains of the same species, possibly due to different mechanisms of resistance or genetic variability in other parts of the genome.
Table 2. Categorical agreement and errors for the tested isolates.
| Antibiotic | Isolates resistant by BMD | Isolates susceptible by BMD | Isolates intermediate by BMD | Total replicates | Very major error rate | Major error rate | Number of minor errors | Categorical agreement | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Susceptible by qPCR | Resistant by qPCR | Susceptible by qPCR | Resistant by qPCR | Susceptible by qPCR | Resistant by qPCR | ||||||
| Ciprofloxacin | 1 | 89 | 47 | 3 | 0 | 4 | 144 | 1.1% | 6.0% | 0 | 97.2% |
| Gentamicin | 1 | 52 | 42 | 1 | 1 | 4 | 102 | 3.7% | 2.3% | 1 | 96.1% |
| Imipenem | 1 | 50 | 47 | 1 | 1 | 2 | 102 | 2.0% | 2.1% | 1 | 97.1% |
| Oxacillin | 1 | 11 | 7 | 1 | 0 | 0 | 20 | 8.3% | 12.5% | 0 | 90.0% |
| Ampicillin | 0 | 12 | 10 | 0 | 0 | 0 | 22 | 0.0% | 0.0% | 0 | 100.0% |
| Vancomycin | 2 | 8 | 31 | 1 | 0 | 0 | 42 | 20.0% | 3.1% | 0 | 92.9% |
| Overall | 7 | 222 | 184 | 7 | 2 | 10 | 432 | 3.1% | 3.7% | 2 | 96.3% |
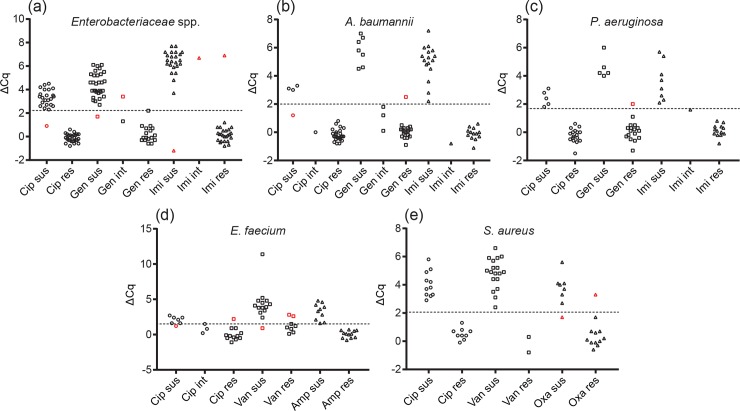
Fig 1. Distribution of ΔCq values for tested isolates.
The ΔCq values for all isolates are shown, separated by antibiotic and susceptibility as determined by the reference method (BMD), for (a) Enterobacteriaceae spp., (b) A. baumannii, (c) P. aeruginosa, (d) E. faecium, and (e) S. aureus. Isolate-antibiotic combinations that resulted in minor, major, or very major errors are colored red. Sus, susceptible; int, intermediate; res, resistant; Cip, ciprofloxacin; Gen, gentamicin; Imi, imipenem; Van, vancomycin; Amp, ampicillin; Oxa, oxacillin.
The RAMAST assay generally performed well when parsing results by species and antibiotic with Enterobacteriaceae spp., A. baumannii, P. aeruginosa, and S. aureus (96–99% categorical agreement), but resulted in a larger number of errors with E. faecium (92.4% categorical agreement, with 2 major errors and 3 very major errors) (Fig 1, Table G in S1 File). The combination of vancomycin with E. faecium proved particularly challenging, with two very major errors and one major error at the optimized antibiotic concentration (25% very major error rate, 7% major error rate). Moreover, repeating the assay on the erroneously called isolates with vancomycin resulted in replication of both very major errors (Table F in S1 File). Attempts to further optimize the assay to reduce the very major error rate through the use of lower vancomycin concentrations resulted in unacceptably large numbers of major errors (Table H in S1 File). We also obtained high very major error rates with ciprofloxacin in E. faecium and oxacillin in S. aureus (both 8.3% very major error rates). The relatively small sample sizes for these antibiotic-species combinations may have contributed to these high very major error rates. A longer antibiotic incubation or other assay optimization is likely necessarily to achieve a more acceptable level of accuracy. Similarly, ciprofloxacin and A. baumannii yielded a major error rate of 25%; but, again, the sample set was small with only 4 susceptible strains.
Several similar studies reported qPCR assays for AST but, to the best of our knowledge, only the study by Beuving et al. used samples from blood culture bottles [19]. As discussed above, the presence of blood cells and other serum components introduces complexities beyond what might be expected for performing qPCR on pure culture or urine samples. Compared to Beuving et al., we reduced the antibiotic incubation time from 6 h to 1 or 2 h, depending on the antibiotic, by using a richer media and minimally diluted positive blood culture soon after positivity. The qPCR cycling conditions used here with the BactQuant primers and probe were also significantly faster, enabling an overall reduction in assay turn-around-time from 9 h to 3 or 4 h (depending on antibiotic).
Bacterial identification through amplicon sequencing
As 16S sequence data is often used for definitive bacterial identification for infrequently isolated pathogens, we explored if the RAMAST assay amplicon could be repurposed for genus identification given the broad coverage of the primer set. As mentioned, the BactQuant primers target variable regions V3 and V4 of the 16S. Stringent analysis of ribosomal sequences from medically relevant pathogens suggest the V3 region is suitable for distinguishing all bacterial regions down to the genus level except for closely related Enterobacteriaceae [29]. Using next-generation sequencing, we batch sequenced a subset of 29 strains used in this study, covering 17 different species, to test whether the RAMAST assay amplicon could be used in this fashion. These experiments resulted in correct genus level concordance for all species (Table 3), with the exception of Klebsiella oxytoca, through read mapping to a medically relevant 16S ribosomal database as described in Stefan et al. [26]. In the exception of Klebsiella, a higher percentage of reads mapped to Enterobacter for K. oxytoca; however, Klebsiella had the second highest percentage of mapped reads and the two genera are both closely related Enterobacteriaceae. Although this method for performing species identification is slower than other available methods (i.e., MALDI-TOF, biochemical), 16S amplicon sequencing is a useful tool for infrequently isolated bacteria that may be misidentified by other methods. Although relatively short, the amplicon from the RAMAST assay provided sufficiently long sequences through variable regions of the 16S gene for differentiation and was adequate for sequencing by Sanger or next generation sequencing techniques.
Table 3. Read mapping of sequenced qPCR amplicons.
| Highest% mapped | 2nd Highest% mapped* | |||
|---|---|---|---|---|
| Input | Genus | % Mapped Reads | Genus | % Mapped Reads |
| E. coli | Escherichia/Shigella | 98.03 | - | - |
| E. coli | Escherichia/Shigella | 92.59 | Vibrio | 3.10 |
| E. coli | Escherichia/Shigella | 94.96 | Vibrio | 1.76 |
| K. pneumoniae | Klebsiella | 77.99 | Enterobacter | 11.92 |
| K. pneumoniae | Klebsiella | 82.78 | Enterobacter | 8.34 |
| K. pneumoniae | Klebsiella | 79.48 | Enterobacter | 10.28 |
| A. baumannii | Acinetobacter | 99.95 | - | - |
| A. baumannii | Acinetobacter | 99.78 | - | - |
| A. baumannii | Acinetobacter | 99.09 | - | - |
| S. aureus | Staphylococcus | 99.70 | - | - |
| S. aureus | Staphylococcus | 99.17 | - | - |
| S. aureus | Staphylococcus | 99.93 | - | - |
| E. faecium | Enterococcus | 99.04 | - | - |
| E. faecium | Enterococcus | 98.32 | - | - |
| E. faecium | Enterococcus | 98.86 | - | - |
| P. aeruginosa | Pseudomonas | 99.89 | - | - |
| P. aeruginosa | Pseudomonas | 99.16 | - | - |
| P. aeruginosa | Pseudomonas | 99.83 | - | - |
| Enterobacter cloacae | Enterobacter | 85.49 | - | - |
| Enterobacter aerogenes | Enterobacter | 66.07 | Raoultella | 10.15 |
| Citrobacter freundii | Citrobacter | 88.76 | Kluyvera | 5.67 |
| Citrobacter koseri | Citrobacter | 75.66 | Salmonella | 15.00 |
| Proteus stuartii | Proteus | 86.64 | Vibrio | 5.74 |
| Serratia marcescens | Serratia | 77.69 | Vibrio | 21.15 |
| Klebsiella oxytoca | Enterobacter | 58.39 | Klebsiella | 25.82 |
| Proteus mirabilis | Proteus | 99.90 | - | - |
| Shigella sonnei | Escherichia/Shigella | 99.25 | - | - |
| Salmonella typhimurium | Salmonella | 98.34 | - | - |
| Salmonella senftenberg | Salmonella | 94.91 | Enterobacter | 3.76 |
* Only samples with less than 95% of reads mapped have a second genus shown
Conclusion
Here, we showed that use of the RAMAST assay for relative DNA quantification after an antibiotic incubation could be used to generate phenotypic antibiotic susceptibility results in as little as 3 to 4 hours from positive blood culture. The use of the universal BactQuant 16S amplification system simplified the assay, requiring only a single set of primers and probes for testing of all bacterial pathogens. Further improvement of this assay could be achieved through the application of recently reported thermo-cold lysis for DNA extraction, potentially decreasing hands-on time and operator error [21]. Overall, our assay showed high categorical agreement at 96.3% while representing a significant decrease in total assay time compared to a similar qPCR AST assay from blood culture.
Supporting information
(PDF)
Acknowledgments
The opinions, interpretations, conclusions, and recommendations contained herein are those of the authors and are not necessarily endorsed by the U.S. Army. The following reagents were obtained through the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH: A. baumannii isolates NR-13380, NR-13383, NR-17780, NR-17781, NR-17782, NR-17784, NR-13379, NR-13378, NR-17786, NR-17778, NR-17787, NR-13375; P. aeruginosa isolates NR-31041, NR-50573, NR-48928; E faecium isolates NR-32065, NR-32094, HM-463, NR-32053, NR-31967, NR-31916, NR-31937, NR-28979, NR-28982, HM-204, NR-31963, NR-31954, NR-28976, NR-31955, NR-31912, NR-31913, NR-32052, NR-32054; S. aureus isolates 880 (BR-VRSA), S. aureus 71080.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by the Defense Threat Reduction Agency (DTRA) project number CB10097. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Laxminarayan R, Duse A, Wattal C, Zaidi AK, Wertheim HF, Sumpradit N, et al. Antibiotic resistance-the need for global solutions. Lancet Infect Dis. 2013;13(12): 1057–98. 10.1016/S1473-3099(13)70318-9 [DOI] [PubMed] [Google Scholar]
- 2.Fraser A, Paul M, Almanasreh N, Tacconelli E, Frank U, Cauda R, et al. Benefit of appropriate empirical antibiotic treatment: thirty-day mortality and duration of hospital stay. Am J Med. 2006;119(11): 970–6. 10.1016/j.amjmed.2006.03.034 [DOI] [PubMed] [Google Scholar]
- 3.Rhodes A, Evans LE, Alhazzani W, Levy MM, Antonelli M, Ferrer R, et al. Surviving Sepsis Campaign: International Guidelines for Management of Sepsis and Septic Shock: 2016. Intensive Care Med. 2017;43(3): 304–77. 10.1007/s00134-017-4683-6 [DOI] [PubMed] [Google Scholar]
- 4.Wiegand I, Hilpert K, Hancock RE. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3(2): 163–75. 10.1038/nprot.2007.521 [DOI] [PubMed] [Google Scholar]
- 5.Barenfanger J, Drake C, Kacich G. Clinical and financial benefits of rapid bacterial identification and antimicrobial susceptibility testing. J Clin Microbiol. 1999;37(5): 1415–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maurer FP, Christner M, Hentschke M, Rohde H. Advances in Rapid Identification and Susceptibility Testing of Bacteria in the Clinical Microbiology Laboratory: Implications for Patient Care and Antimicrobial Stewardship Programs. Infect Dis Rep. 2017;9(1): 6839 10.4081/idr.2017.6839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malani AN, Richards PG, Kapila S, Otto MH, Czerwinski J, Singal B. Clinical and economic outcomes from a community hospital's antimicrobial stewardship program. Am J Infect Control. 2013;41(2): 145–8. 10.1016/j.ajic.2012.02.021 [DOI] [PubMed] [Google Scholar]
- 8.Davey P, Marwick CA, Scott CL, Charani E, McNeil K, Brown E, et al. Interventions to improve antibiotic prescribing practices for hospital inpatients. Cochrane Database Syst Rev. 2017;2: CD003543 10.1002/14651858.CD003543.pub4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. Pharm Ther. 2015;40(4): 277–83. [PMC free article] [PubMed] [Google Scholar]
- 10.Kostic T, Ellis M, Williams MR, Stedtfeld TM, Kaneene JB, Stedtfeld RD, et al. Thirty-minute screening of antibiotic resistance genes in bacterial isolates with minimal sample preparation in static self-dispensing 64 and 384 assay cards. Appl Microbiol Biotechnol. 2015;99(18): 7711–22. 10.1007/s00253-015-6774-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zboromyrska Y, Vergara A, Cosgaya C, Verger G, Mosqueda N, Almela M, et al. Rapid detection of beta-lactamases directly from positive blood cultures using a loop-mediated isothermal amplification (LAMP)-based assay. Int J Antimicrob Agents. 2015;46(3): 355–6. 10.1016/j.ijantimicag.2015.06.002 [DOI] [PubMed] [Google Scholar]
- 12.Fredborg M, Andersen KR, Jorgensen E, Droce A, Olesen T, Jensen BB, et al. Real-time optical antimicrobial susceptibility testing. J Clin Microbiol. 2013;51(7): 2047–53. 10.1128/JCM.00440-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baltekin O, Boucharin A, Tano E, Andersson DI, Elf J. Antibiotic susceptibility testing in less than 30 min using direct single-cell imaging. Proc Natl Acad Sci U S A. 2017;114(34): 9170–5. 10.1073/pnas.1708558114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu CY, Han YY, Shih PH, Lian WN, Wang HH, Lin CH, et al. Rapid bacterial antibiotic susceptibility test based on simple surface-enhanced Raman spectroscopic biomarkers. Sci Rep. 2016;6: 23375 10.1038/srep23375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lange C, Schubert S, Jung J, Kostrzewa M, Sparbier K. Quantitative matrix-assisted laser desorption ionization-time of flight mass spectrometry for rapid resistance detection. J Clin Microbiol. 2014;52(12): 4155–62. 10.1128/JCM.01872-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kostrzewa M, Sparbier K, Maier T, Schubert S. MALDI-TOF MS: an upcoming tool for rapid detection of antibiotic resistance in microorganisms. Proteomics Clin Appl. 2013;7(11–12): 767–78. 10.1002/prca.201300042 [DOI] [PubMed] [Google Scholar]
- 17.Maxson T, Taylor-Howell CL, Minogue TD. Semi-quantitative MALDI-TOF for antimicrobial susceptibility testing in Staphylococcus aureus. PLoS One. 2017;12(8): e0183899 10.1371/journal.pone.0183899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rolain JM, Mallet MN, Fournier PE, Raoult D. Real-time PCR for universal antibiotic susceptibility testing. J Antimicrob Chemother. 2004;54(2): 538–41. 10.1093/jac/dkh324 [DOI] [PubMed] [Google Scholar]
- 19.Beuving J, Verbon A, Gronthoud FA, Stobberingh EE, Wolffs PF. Antibiotic susceptibility testing of grown blood cultures by combining culture and real-time polymerase chain reaction is rapid and effective. PLoS One. 2011;6(12): e27689 10.1371/journal.pone.0027689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schoepp NG, Schlappi TS, Curtis MS, Butkovich SS, Miller S, Humphries RM, et al. R apid pathogen-specific phenotypic antibiotic susceptibility testing using digital LAMP quantification in clinical samples. Sci Transl Med. 2017;9(410): eaal3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo J, Yu J, Yang H, Wei H. Parallel susceptibility testing of bacteria through culture-quantitative PCR in 96-well plates. Int J Infect Dis. 2018;70: 86–92. 10.1016/j.ijid.2018.03.014 [DOI] [PubMed] [Google Scholar]
- 22.Mezger A, Gullberg E, Goransson J, Zorzet A, Herthnek D, Tano E, et al. A general method for rapid determination of antibiotic susceptibility and species in bacterial infections. J Clin Microbiol. 2015;53(2): 425–32. 10.1128/JCM.02434-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu CM, Aziz M, Kachur S, Hsueh PR, Huang YT, Keim P, et al. BactQuant: an enhanced broad-coverage bacterial quantitative real-time PCR assay. BMC Microbiol. 2012;12: 56 10.1186/1471-2180-12-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.CLSI. Performance Standards For Antimicrobial SusceptibilityTesting. 27th Edition CLSI supplement M100. Wayne, PA, USA: Clinical and Laboratory Standards Institute; 2017. [Google Scholar]
- 25.CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard—Tenth Edition. CLSI Document M07-A10. Wayne, PA, USA: Clinical and Laboratory Standards Institute; 2015. [Google Scholar]
- 26.Stefan CP, Hall AT, Minogue TD. Detection of 16S rRNA and KPC Genes from Complex Matrix Utilizing a Molecular Inversion Probe Assay for Next-Generation Sequencing. Sci Rep. 2018;8(1): 2028 10.1038/s41598-018-19501-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schrader C, Schielke A, Ellerbroek L, Johne R. PCR inhibitors—occurrence, properties and removal. J Appl Microbiol. 2012;113(5): 1014–26. 10.1111/j.1365-2672.2012.05384.x [DOI] [PubMed] [Google Scholar]
- 28.Hooper DC. Mechanisms of action of antimicrobials: focus on fluoroquinolones. Clin Infect Dis. 2001;32 Suppl 1: S9–S15. [DOI] [PubMed] [Google Scholar]
- 29.Chakravorty S, Helb D, Burday M, Connell N, Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods. 2007;69(2): 330–9. 10.1016/j.mimet.2007.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.