Abstract
Background
Posttraumatic stress disorder (PTSD) and stress/trauma exposure are cross-sectionally associated with advanced DNA methylation age relative to chronological age. However, longitudinal inquiry and examination of associations between advanced DNA methylation age and a broader range of psychiatric disorders is lacking. The aim of this study was to examine if PTSD, depression, generalized anxiety, and alcohol-use disorders predicted acceleration of DNA methylation age over time (i.e., an increasing pace, or rate of advancement, of the epigenetic clock).
Methods
Genome-wide DNA methylation and a comprehensive set of psychiatric symptoms and diagnoses were assessed in 179 Iraq/Afghanistan war veterans who completed two assessments over the course of approximately two years. Two DNA methylation age indices (Horvath and Hannum), each a weighted index of an array of genome-wide DNA methylation probes, were quantified. The pace of the epigenetic clock was operationalized as change in DNA methylation age as a function of time between assessments.
Results
Analyses revealed that alcohol-use disorders (p = .001) and PTSD avoidance and numbing symptoms (p = .02) at Time 1 were associated with an increasing pace of the epigenetic clock over time, per the Horvath (but not the Hannum) index of cellular aging.
Conclusions
This is the first study to suggest that posttraumatic psychopathology is longitudinally associated with a quickened pace of the epigenetic clock. Results raise the possibility that accelerated cellular aging is a common biological consequence of stress-related psychopathology, which carries implications for identifying mechanisms of stress-related cellular aging and developing interventions to slow its pace.
Keywords: Epigenetic age, DNA methylation age, PTSD, alcohol-use disorders, longitudinal
Mounting evidence suggests that stress-related psychopathology, such as posttraumatic stress disorder (PTSD), is associated with accelerated cellular aging in the epigenome. Specifically, DNA methylation (DNAm) data can be used to estimate cellular age from established epigenetic age algorithms developed by Horvath (2013) and Hannum et al. (2013). When estimated from whole blood, these estimates of epigenetic age (i.e., “DNAm age”) show strong and equivalent patterns of association with chronological age, despite being based on different methods and tissues and despite the two estimates sharing very few DNAm loci. DNAm age estimates that exceed chronological age (i.e., advanced DNAm age) have been associated with a variety of important clinical phenotypes including shortened time until death (Marioni et al., 2015; Chen et al., 2016; Perna et al., 2016), cancer (Zheng et al., 2016), and neurocognitive decline among those with Alzheimer’s disease (Levine et al., 2015) and PTSD (Wolf et al., 2016). Thus advanced DNAm age may be an important part of understanding the link between PTSD and premature health and neurocognitive decline (see Wolf & Morrison, 2017, for a review).
Using these age calculators, a number of studies have suggested that PTSD symptoms are cross-sectionally associated with advanced epigenetic age relative to chronological age, albeit the effect tends to be small in magnitude and not uniform across studies. Specifically, Wolf et al. (2016) showed that an index of PTSD symptom severity across the lifespan was associated with advanced epigenetic age (relative to chronological age) using the Hannum, but not the Horvath, age algorithm in a sample of 281 young veterans returning from the wars in Iraq and Afghanistan (an overlapping cohort is featured in this study). Following this, Wolf et al. (2017) demonstrated that PTSD hyperarousal symptoms were more specifically associated with advanced Hannum epigenetic age in a middle-aged sample of 339 mixed war era veterans. In contrast, Zannas et al. (2015) found no association between PTSD and advanced epigenetic age using the Horvath index (the Hannum index was not evaluated in that study) in a sample of 392 predominately female civilians with substantial exposure to community violence. However, Zannas et al. did report an association between the cumulative burden of daily life stressors and advanced epigenetic age per the Horvath algorithm. Similarly, in a sample of 101 African American children living in an urban environment, Jovanovic et al. (2017) showed that exposure to violence was associated with advanced Horvath DNAm age (the Hannum index was not evaluated). In the only longitudinal PTSD and epigenetic age study to date, Boks et al. (2015) reported that in a sample of 96 Dutch male veterans, increased PTSD symptoms from immediately pre-to-post warzone exposure (i.e., difference scores) were associated with decreased epigenetic age over the same interval while combat exposure was associated with increased epigenetic age per the Horvath algorithm (the Hannum algorithm was not examined).
In an effort to clarify differential patterns of association, Wolf et al. (in press) conducted a cross-sectional meta-analysis of the associations between trauma exposure, PTSD diagnosis and symptom severity, and epigenetic age (per both the Horvath and Hannum algorithms) relative to chronological age using data from 9 cohorts contributing to the Psychiatric Genomics Consortium PTSD Epigenetics Workgroup (Ratanatharathorn et al., 2017) which included over 2000 participants. Results suggested that lifetime PTSD symptom severity was associated with advanced epigenetic age per the Hannum, but not the Horvath, index and that childhood trauma exposure, when assessed with a uniform measure, was also associated with advanced epigenetic age per the Hannum metric. The meta-analytic effect for PTSD was estimated at unstandardized β = .01, implying that the difference between individuals with no PTSD symptoms versus substantial symptoms (i.e., severity scores of 0 versus 100 on the Clinician Administered PTSD Scale) was about 1.1 years, though the cumulative burden of such symptoms over time on cellular age was unknown.
There are two primary limitations of the foregoing studies. The first is that, with few exceptions (reviewed below), the psychopathology/DNAm age literature has largely focused on trauma and PTSD, as opposed to other forms of psychopathology, as predictors of cellular aging. The second limitation is that all but one study employed a cross-sectional design. There is no reason to suspect that accelerated DNAm age is unique to PTSD. Rather, we hypothesize that accelerated epigenetic age may be a common biological correlate of a variety of stress-related psychiatric disorders. Support for this comes from multiple lines of converging research. This includes evidence that shortened telomere length is a common correlate of many forms of stress-related psychopathology (Darrow et al., 2016) and that sleep disturbance, a transdiagnostic symptom across psychological disorders, was associated with advanced age per a Hannum-based metric of epigenetic age that also incorporated age-related components of the immune system (though it was not associated with advanced Horvath age; Carroll et al., 2017). Moreover, PTSD is highly comorbid with a range of other psychiatric conditions including those spanning internalizing (e.g., depression and anxiety) and externalizing (e.g., substance use disorders) psychopathology dimensions (Kessler et al., 1995; Miller et al., 2012) and this raises the possibility that PTSD comorbidity may also contribute to accelerated aging. In addition, accelerated DNAm age has been associated with a variety of other conditions including Alzheimer’s disease (Levine et al., 2015), cancer (Durso et al., 2017), and obesity (Quach et al., 2017), which would argue against the specificity of accelerated DNAm age to trauma and PTSD. That said, it is important to note that the few studies which did examine epigenetic age and other forms of psychopathology found null or negative associations. One post-mortem study found reduced Horvath epigenetic age (relative to chronological age) in neurons in the orbitofrontal cortex among 37 heroin users who overdosed in comparison to both controls and non-overdose suicide cases (Kozlenkov et al., 2017). Two studies reported null effects between schizophrenia and Horvath DNAm age residuals assessed in temporal (McKinney et al., 2017) and frontal (Voisey et al., 2017) post-mortem brain tissue.
The general focus on cross-sectional studies has precluded evaluation of the predictive strength of psychopathology on changes in DNAm age acceleration. This concern is accentuated in light of preliminary evidence from epidemiological cohorts suggesting that accelerated Horvath epigenetic age is largely stable over time (Kananen et al., 2016), raising the possibility that observed relationships with PTSD may reflect co-occurring phenomena, as opposed to a causal association. It is important to test this by examining potential pathogenic environmental factors that might impact the stability of accelerated aging. The primary aim of this study was to examine the predictive strength of posttraumatic psychopathology (PTSD, major depressive disorder [MDD], generalized anxiety disorder [GAD], and alcohol-use disorders) in association with accelerated epigenetic aging over time. A second aim was to test the utility of a novel approach to quantifying accelerated DNAm age over time using a metric that captured changes in the pace of epigenetic aging relative to intervening time. This was accomplished in a longitudinal sample of 179 veterans post-deployment to the wars in Iraq and/or Afghanistan, who had DNAm data available at two time points, spanning, on average, 1.89 years.
Methods
Participants
Participants were U.S. military veterans of the post 9/11 conflicts who were enrolled in the ongoing research protocol, the Translational Research Center for TBI and Stress Disorders (TRACTS), a US Department of Veterans Affairs (VA) Rehabilitation Research and Development Traumatic Brain Injury Center of Excellence at VA Boston Healthcare System. TRACTS (McGlinchey et al., 2017) is a longitudinal investigation of traumatic brain injury (TBI), traumatic stress, health, and neural and cognitive factors among returning veterans. Study exclusion criteria were neurological illness, history of seizures unrelated to head injury, acute psychotic or bipolar disorder, severe or unstable psychological diagnosis that prevented participation, acute homicidal and/or suicidal ideation with intent to act, and cognitive disorder due to general medical condition other than TBI. Analyses for this study were based on a subset of TRACTS participants with Time 1 (T1) and Time 2 (T2) DNAm data at the time of our second data freeze in which DNA was sent for processing. We had previously obtained DNAm data using the Illumina HumanMethylation 450k beadchip on 281 participants at their T1 visit and reported on cross-sectional associations between PTSD and accelerated DNAm age in that initial cohort in Wolf et al. (2016). Of that group, 153 returned for a second visit and an additional 26 new T1 veterans enrolled in the protocol and returned for a T2 visit prior to the data freeze, yielding a total T1/T2 sample size of 179. DNAm values were simultaneously obtained on the Illumina EPIC beadchip for both T1 and T2, including for those with existing T1 DNAm data from the 450k beadchip.
Of the longitudinal sample of n = 179, 88.3% were male veterans and the mean age at T1 was 32.84 years (range: 19 to 65; SD = 9.28; Table 1). The majority of the sample (75.7%) self-reported as white, 12.4% as Hispanic or Latino/a, 9.6% as black, 1.7% as Asian, and 0.6% as American Indian (Table 1).
Table 1.
Demographic and Psychiatric Characteristics of the Longitudinal Sample (n = 179)
| Variable | Mean (SD) | % (n) |
|---|---|---|
| Sex (male) | 88.3 (158) | |
| Race | ||
| White | 75.4 (135) | |
| Black | 9.5 (17) | |
| Latino/a | 11.7 (21) | |
| Asian | 1.1 (2) | |
| Age (T1) | 32.84 (9.28) | |
| Years between T1 and T2 | 1.89 (.65) | |
| Education (# of years of schooling) | 13.97 (1.92) | |
| Current PTSD diagnosis (T1) | 59.2 (106) | |
| Current PTSD symptom severity (T1) | 45.57 (30.02) | |
| Lifetime PTSD diagnosis (T1) | 76.5 (137) | |
| Current MDD diagnosis (T1) | 20.1 (36) | |
| Current GAD diagnosis (T1) | 7.3 (13) | |
| Current alcohol-use disorder (T1) | 16.2 (29) |
Note. T1 = time 1; T2 = time 2; PTSD = posttraumatic stress disorder; MDD = major depressive disorder; GAD = generalized anxiety disorder.
Procedure
After providing written informed consent, participants completed a comprehensive interview- and self-report-based psychological assessment, with diagnostic interviews administered by doctoral-level psychology professionals. Each interview was reviewed by a team of psychologists to inform consensus ratings of presence/absence of psychological diagnoses. Blood was drawn at each time point for DNA extraction and metabolic assays. The two assessment points occurred an average of 1.89 years apart (Table 1). The study was approved by the VA Boston Healthcare System IRB.
Measures
Traumatic Life Events Questionnaire (TLEQ; Kubany et al., 2000), a self-report assessment of exposure to 21 trauma types, was used to quantify the number of different types of traumatic experiences reported at T1. PTSD was assessed with the Clinician Administered PTSD Scale for DSM-IV (CAPS; Blake et al., 1995). This well-validated diagnostic interview was administered to capture current (past month) symptoms, the worst period of symptoms post-military discharge, and any pre-military symptoms. Follow-up analyses focused on PTSD symptom clusters using factor scores representing latent lifespan severity of PTSD symptoms at T1. Other psychiatric disorders (mood, anxiety, and substance use) were measured with the well-established Structured Clinical Interview for DSM-IV Disorders (SCID; First et al., 1997; Spitzer et al., 1998).
DNA Data
Genotypes
DNA was extracted from peripheral blood samples and genotyping obtained via the Illumina HumanOmni2.5-8 beadchip. Genotypes are relevant as they pertain to the development of principal components (PCs) to model ancestry (Supplementary Materials).
Methylation
DNA was hybridized to the Infinium MethylationEPIC beadchip according to the manufacturer’s protocol. To reduce the possibility of systematic bias, T1 and T2 samples were run together on the same chip using a balanced chip design. We followed the processing pipeline established by the PGC PTSD Epigenetics workgroup (Ratanatharathorn et al., 2017) for the Illumina HumanMethylation450 BeadChip, but applied here to the EPIC chip (Supplementary Materials).
Calculation of Epigenetic Age
Horvath DNAm age estimates were computed with an R script based on the 335 probes that were assessed on the EPIC chip and passed quality control (QC). For the Hannum algorithm, data were normalized using the beta mixture quantile dilation (BMIQ), as in prior publications (Wolf et al., 2016; Wolf et al., 2017; Wolf et al., in press; Supplementary Materials). We have previously demonstrated (Logue et al., 2017) that the correlations between EPIC chip-derived Horvath and Hannum age estimates with chronological age (rs = .90 and .87, respectively) were very similar to those reported for Infinium-derived Horvath and Hannum estimates (rs = .88; Wolf et al., 2016).
White Blood Cell Count Estimation
Proportional white blood cell (WBC) counts at T1 and T2 were estimated from the methylation data (Supplementary Materials).
Data Analyses
Operational Definition of Accelerated Aging in DNAm over Time
We examined a new metric of accelerated aging over time focused on the pace of change in the raw DNAm age estimates (i.e., the rate at which the epigenetic clock advances). This was defined as the difference in raw DNAm age estimates (T2 – T1) over time as a function of the time between the two assessments:
A score of 1.0 indicates that for each year between the two assessments, DNAm age also increased by 1 year. Values greater than 1.0 suggest an accelerated pace of DNAm age change relative to the number of intervening years, while values less than 1.0 are indicative of slower pace of the epigenetic clock relative to time between assessments.
Analytic Plan
We first calculated the correlation between the DNAm age estimates and chronological age at each time point. We then extracted DNAm age residuals for T1 and T2 data (defined by the residuals from an equation in which T1/T2 chronological age was regressed out of the T1/T2 Hannum and Horvath DNAm age estimates, respectively) and compared DNAm and DNAm residual correlations across the Horvath and Hannum algorithms. Positive residuals reflect over-predicted DNAm age (i.e., advanced epigenetic age) at a given time point. We examined the stability of DNAm age and DNAm age residuals over time both within and across algorithms via correlation and by examining the descriptive statistics pertaining to the average rate change in raw DNAm age estimates per calendar year per the operational definition defined above. We also examined the stability of the estimated WBCs using correlation.
We conducted two multiple regressions apiece for the two (Horvath/Hannum) DNAm age rate variables: the first simultaneously evaluated the contribution of T1 current diagnoses of PTSD, MDD, GAD, and alcohol-use disorders1 to the pace of epigenetic aging and the second evaluated the contribution of T1 lifespan PTSD symptom cluster severity to this phenomenon. T1 lifespan PTSD symptom cluster severity was represented by factor scores from a confirmatory factor analysis (CFA) of scores on pre-military, post-military, and current (T1) PTSD symptom clusters. Each regression controlled for sex, the top two ancestry PCs, and T1 total trauma exposure. We did not include WBCs as covariates because the dependent variable captured change across T1 and T2 DNAm age and it was not evident which set of WBCs (if any) should be included. Rather, we examined this question in preliminary regressions (detailed in Supplementary Materials) which separately tested T1 and T2 WBCs as predictors of the pace of DNAm change. In follow-up analyses detailed in Supplementary Materials, we evaluated interactions between psychopathology and demographic variables and examined potential confounds (cigarette use, body mass index, traumatic brain injury, psychotropic medication use) of our primary associations of interest. In additional follow-up analyses detailed in Supplementary Materials, we also evaluated if change in psychopathology variables over the two assessment points covaried with the rate of DNAm age change. As each analysis was executed twice (once for Horvath and once for Hannum-based indices), we performed a statistical procedure to correct the p-values using a Monte Carlo null simulation with 1,000 replicates that also took into account the correlational structure of the dependent variables.
Results
Estimated DNAm Age
DNAm age estimates were strongly associated with chronological age. Specifically, correlations between Horvath DNAm age and chronological age were r = .90 (p < .001) and r = .91 (p < .001) at T1 and T2, respectively. Hannum DNAm age correlations with chronological age were r = .88 (p < .001) and r = .85 (p < .001) at T1 and T2, respectively. At T1, the Horvath and Hannum estimates correlated with each other at r = .88 (p < .001) and at T2, at r = .87 (p < .001). At both T1 and T2, the correlation between the Horvath and Hannum age residuals was r = .44 (p < .001).
Stability of DNAm Age Estimates and WBCs over Time
The raw DNAm age estimates were highly correlated over time: rHorvath = .95, p < .001; rHannum = .93, p < .001. The DNAm age residuals were also highly correlated over time (Table 2). Horvath DNAm age estimates increased at a rate of, on average, 0.76 years per calendar year (SD: 1.75, range: −3.62 to 7.11 years) while Hannum DNAm age estimates increased by an average rate of 1.10 years per calendar year (SD: 1.68, range: −5.35 to 5.86). WBC proportional estimates were highly correlated (across the same cell type) over time (range: rs = .67 to .75; Table 3). The rate of change in the raw DNAm age estimates was weakly correlated across the Horvath and Hannum algorithms (r = .26, p < .001). Neither T1 nor T2 WBCs were associated with epigenetic age acceleration and thus WBCs were not included in subsequent models (Supplementary Materials).
Table 2.
Descriptive Statistics (and Correlations) Pertaining to DNAm age Residuals and the Rate of DNAm Age Change over Time
| Horvath |
Hannum |
Correlations (Horvath/Hannum) | |||||
|---|---|---|---|---|---|---|---|
| M (SD) | Range | M (SD) | Range | 1. | 2. | 3. | |
| 1. T1 DNAm age residual | .00 (3.85) |
−9.96 to 10.61 | .00 (3.61) |
−11.91 to 9.51 | – | ||
| 2. T2 DNAm age residual | .00 (3.53) |
−9.11 to 9.43 | .00 (3.86) |
−13.51 to 8.90 | .72/.76 | – | |
| 3. Δ Raw DNAm age/year | .76 (1.75) |
−3.62 to 7.11 | 1.10 (1.68) |
−5.35 to 5.86 | −.49/−.25 | .22/.40 | – |
Note. All correlations were statistically significant at the p < .01 or better level. DNAm = DNA methylation; T1 = time 1; T2 = time 2; diff = difference.
Table 3.
Stability (Pearson Correlations) of White Blood Cell Proportional Estimates over Time
| Time 2 |
|||||
|---|---|---|---|---|---|
| Time 1 | 1. | 2. | 3. | 4. | 5. |
| 1. CD8T | .75** | −.13 | −.06 | .02 | −.07 |
| 2. CD4T | −.15* | .71** | .04 | .14 | −.21** |
| 3. NK | −.07 | −.08 | .72** | .02 | .03 |
| 4. B cells | .04 | .16* | −.01 | .75** | −.10 |
| 5. Monocytes | −.11 | −.07 | −.02 | −.09 | .67** |
Note.
p < .05.
p < .01.
p < .001.
PTSD, MDD, GAD, and Alcohol-use Disorders as Predictors of the Rate of DNAm Age Acceleration
We examined if T1 PTSD, MDD, GAD, or alcohol-use disorders predicted the rate of epigenetic aging.2 Alcohol use diagnoses at T1 were associated with the rate of epigenetic aging estimated via the Horvath algorithm (β = .26, p = .001, corrected p = .002; Table 4). The mean rate of epigenetic aging as a function of T1 alcohol-use diagnosis was 1.58 years, indicating that individuals with a T1 alcohol use disorder evidenced a 1.58 year DNAm age increase per calendar year (compared to 0.60 among those without a T1 alcohol use disorder; p = .006; Figure 1). None of the mental health diagnoses predicted the rate of epigenetic aging estimated with the Hannum method (Table 4). The association between alcohol-use disorders and the rate of change in DNAm age was not moderated by demographic characteristics (Supplementary Materials).
Table 4.
Regression Results Predicting the Rate of Epigenetic Aging Over Time
| Model | Horvath |
Hannum |
||||||
|---|---|---|---|---|---|---|---|---|
| Model | B | SE | β | p | B | SE | β | p |
| 1. Stress-Related Dxs | ||||||||
| PC1 | .1.77 | 1.78 | .08 | .32 | −2.18 | 1.66 | −.10 | .19 |
| PC2 | −1.05 | 1.76 | −.05 | .55 | −.03 | 1.64 | .001 | .99 |
| Sex | −.67 | .43 | −.12 | .12 | −.56 | .40 | −.11 | .16 |
| T1 trauma | −.07 | .08 | −.07 | .39 | .05 | .08 | .05 | .53 |
| T1 current PTSD dx | −.03 | .30 | −.01 | .92 | −.30 | .28 | −.09 | .28 |
| T1 current MDD dx | −.07 | .35 | −.02 | .85 | −.51 | .33 | −.13 | .12 |
| T1 current GAD dx | .48 | .50 | .07 | .34 | −.41 | .47 | −.07 | .38 |
| T1 current Alc. dx | 1.24 | .37 | .26 | .001 | .50 | .35 | .11 | .16 |
| 2. PTSD Sx Clusters | ||||||||
| PC1 | 2.40 | 1.81 | .10 | .19 | −2.03 | 1.68 | −.10 | .23 |
| PC2 | .24 | 1.78 | .01 | .89 | .10 | 1.65 | .01 | .95 |
| Sex | −.65 | .43 | −.12 | .13 | −.58 | .40 | −.12 | .14 |
| T1 trauma | −.04 | .09 | −.04 | .62 | .03 | .08 | .04 | .69 |
| T1 reexp | −.05 | .05 | −.21 | .27 | −.03 | .04 | −.15 | .46 |
| T1 avd & numb | .08 | .04 | .43 | .02 | .03 | .03 | .15 | .42 |
| T1 hyperarousal | −.04 | .04 | −.16 | .34 | −.02 | .04 | −.09 | .59 |
Note. All PTSD severity variables were factor scores derived from a confirmatory factor analysis of pre-military, post-military, and current PTSD symptoms as assessed at T1. DNAm = DNA methylation; PTSD = posttraumatic stress disorder; PC = principal component; T1 = time 1; sx = symptom; dx = diagnosis; reexp = re-experiencing; avd = avoidance; numb = numbing; MDD = major depressive disorder; GAD = generalized anxiety disorder; Alc = alcohol.
Figure 1.
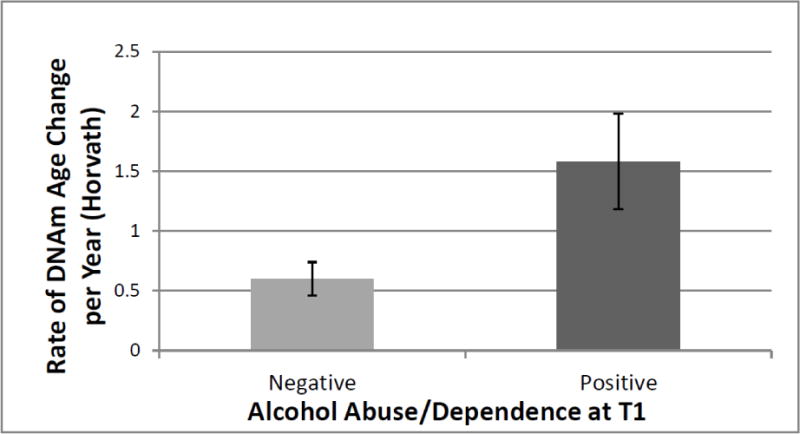
The figure shows the mean differential rate of change in the Horvath DNAm age estimate per year as a function of alcohol-use disorder diagnoses at time 1. Error bars represent standard errors. DNAm = DNA methylation; T1 = time 1.
PTSD Symptom Cluster Severity as a Predictor of the Rate of DNAm Age Acceleration
In the PTSD symptom cluster factor score3 models, T1 avoidance and numbing symptoms were associated with an increased pace of epigenetic aging per the Horvath index (β = .43, p = .016, corrected p = .029; Table 4, Figure 2).4,5 There were no significant predictors of the rate of epigenetic aging as estimated by the Hannum method (Table 4). There was no evidence that demographic variables moderated the associations between PTSD symptom clusters and epigenetic aging and no evidence that change in psychiatric diagnoses or symptoms across the two assessment points was associated with the rate of DNAm age acceleration (Supplementary Materials).
Figure 2.
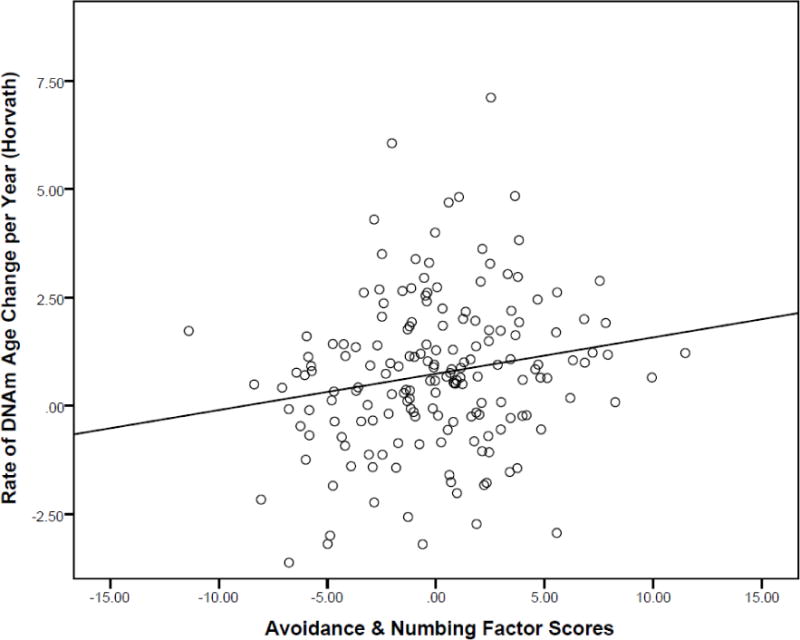
The figure shows the relationship between time 1 avoidance and numbing PTSD latent variable factor scores in association with the rate of change in estimated DNAm age each year per the Horvath algorithm (controlling for all other covariates in the model as per Table 4, Model 2). DNAm = DNA methylation.
Discussion
Longitudinal study is critical for distinguishing the neurobiological correlates of trauma- and stress-related psychopathology from the neurobiological consequences of such symptoms. In this first longitudinal study of the associations between a range of posttraumatic psychiatric disorders and change in epigenetic age, we found that PTSD avoidance and numbing symptoms and alcohol-use disorders predicted increases in the pace of the epigenetic clock. This study represents an important first step in extending results of prior cross-sectional research and advancing our understanding of the neurobiological consequences of psychopathology. A quickened pace of the epigenetic clock could contribute to early onset of age-related health decline, such as cardiometabolic disorders (Perna et al., 2016), cognitive decline (Levine et al., 2015; Marioni et al., 2015), and mortality (Marioni et al., 2015; Chen et al., 2016; Wolf et al., 2017). Results also highlight the utility of a novel metric to represent the changing pace of the epigenetic clock that was directly interpretable. We suggest that prior cross-sectional studies, including our own, should be conceptualized as examining associations between psychiatric phenotypes and advanced, but not necessarily accelerated, DNAm age. In contrast, by modeling the changing speed of the epigenetic clock compared to the passage of time, we were able to more definitively quantify acceleration of cellular age.
Though alcohol-use disorders per se have not previously been investigated in association with a changing pace of the epigenetic clock, our results are broadly consistent with one prior cross-sectional study which suggested that problematic alcohol use was associated with advanced DNAm age. Specifically, in a population study that included DNAm data from 105 individuals, heavy alcohol use was related to advanced epigenetic age using a DNAm age index developed by (Weidner and Wagner, 2014), based on just three loci obtained via pyrosequencing. We did not evaluate that index because it did not evidence as strong an association with chronological age in the validation samples (Weidner and Wagner, 2014) compared to the (Horvath, 2013) and (Hannum et al., 2013) metrics and because we did not have DNAm data on loci that were not on the Illumina chip. In contrast to Weidner and Wagner (2014), a recent cross-sectional study suggested that moderate alcohol use was not related to DNAm age residuals per the Horvath metric, but it was negatively associated with a related index hypothesized to reflect immune system age more generally (Quach et al., 2017). That index was referred to as “extrinsic epigenetic age acceleration” and it was operationalized as the residuals obtained from an equation in which chronological age was regressed from Hannum DNAm age estimates combined with age-weighted WBC estimates (Quach et al., 2017). We did not examine the extrinsic epigenetic age index in this study because: (a) we wanted to extend our prior work on advanced DNAm age specifically; (b) doing so would complicate interpretation of the rate variable as WBCs are not intrinsically linked to the passage of time in the way DNAm age is; and (c) it is not evident how DNAm versus WBCs contribute to effects observed for the extrinsic epigenetic age index. Despite differential quantification of DNAm age indices across our study and the aforementioned ones, results collectively raise the possibility that alcohol-use disorders may accelerate epigenetic aging while moderate alcohol use may be protective; additional research is needed to test this directly.
Prior epigenetic studies suggest that alcohol-related phenotypes are associated with differential DNAm profiles (Zhang and Gelernter, 2017). A number of epigenome-wide association studies (EWAS) have together implicated alcohol-related phenotypes in association with methylation of genes involved in immune response, signal transduction, metabolism, and apoptosis-related gene networks (Zhang et al., 2013; Zhao et al., 2013; Harlaar et al., 2014; Philibert et al., 2014; Liu et al., 2016). There are also numerous candidate gene DNAm studies of alcohol-use, two of which are notable because the genes are relevant to the Horvath algorithm. Specifically, differential methylation of the gene promoter region of ANP (which encodes for the atrial natriuretic peptide, a vasodilator secreted by heart muscle cells) was reported in association with alcohol withdrawal (Glahn et al., 2014). A probe in the gene encoding the corresponding receptor (ANPRC: atrial natriuretic peptide receptor) is part of the Horvath DNAm age algorithm, which could potentially contribute to the sensitivity of DNAm age to alcohol-related phenotypes. ANP has strong anxiolytic effects, and attenuates the stress response by inhibiting the hypothalamic-pituitary-adrenal (HPA) axis, via moderation of adrenocorticotropin (ACTH) and cortisol (Antoni et al., 1992; Wiedemann et al., 2001; Strohle et al., 2006). Likewise, methylation of the 5’ upstream promoter of the hormone precursor polypeptide pro-opiomelanocortin (POMC) gene evidenced marginal associations with alcohol-use disorders and was specifically associated with alcohol cravings among those with alcohol-related disorders (Muschler et al., 2010). POMC is also involved in regulation of the HPA axis and a probe in POMC is included in the Horvath DNAm age algorithm. These findings are broadly consistent with evidence that the probes that comprise the Horvath DNAm age index are responsive to glucocorticoid modulation (Zannas et al., 2015) and that advanced Horvath DNAm age is associated with greater diurnal cortisol (Davis et al., 2017).
In addition to shared sensitivity to glucocorticoids, there is evidence for a common role of inflammation and immune system dysregulation across PTSD, alcohol-use disorders, and cellular aging. Specifically, heavy alcohol-use is associated with increases in memory T-cells and reductions in naïve T-cells (Cook et al., 1994; Cook et al., 1995), which could contribute to the association between alcohol use and infections (Szabo and Mandrekar, 2009). Consistent with this, a gene expression study conducted in a small sample found that alcohol dependence was associated with enrichment in T-cell receptor genetic pathways (Beech et al., 2012). PTSD has been similarly associated with changes in adaptive immunity, including increases in late-stage differentiated T-cells and decreases in naïve T-cell populations (Sommershof et al., 2009; Aiello et al., 2016), as well as gene expression profiles enriched for innate immune system signaling (Breen et al., 2015). Shifts in T-cell populations from naïve to differentiated types is often conceptualized as part of an immunosenescent profile that is itself a biological indicator of aging (Macaulay et al., 2013; Fülöp et al., 2016), and one that can contribute to increased inflammation via pro-inflammatory cytokines (Macaulay et al., 2013). Both alcohol use disorders (Achur et al., 2010; Leclercq et al., 2012) and PTSD (Passos et al., 2015; Miller et al., 2017) are also associated with increased inflammatory parameters including C-reactive protein, IL-6, and TNFα; inflammation may be a common pathway across the two disorders. Finally, accelerated DNAm age has also been associated with both increased inflammation (Quach et al., 2017) and reductions in CD4-T cell proportions (Marioni et al., 2015; Chen et al., 2016; Wolf et al., in press). Collectively, this suggests that one possible mechanism for the common association between PTSD and alcohol-use disorders and an increased pace of the epigenetic clock is alterations in key immune and inflammation pathways.
Differential Findings Relative to Prior Work
We have previously reported (Wolf et al., 2016; Wolf et al., 2017; Wolf et al., in press) cross-sectional associations between PTSD and the Hannum, but not Horvath, index of DNAm age residuals. The only prior study to report an association between PTSD and the Horvath index of epigenetic age suggested that increased PTSD symptoms were associated with decreased epigenetic age, but the relative relationship with chronological age was not modeled (Boks et al., 2015). In contrast, stress (Zannas et al., 2015), trauma (Boks et al., 2015), and violence exposure (Jovanovic et al., 2017) have previously shown positive associations with Horvath advanced age. One possible explanation for these different patterns of association is that the effects of stress, trauma, and psychopathology on Horvath-defined epigenetic aging may be slow to manifest and cumulative, and therefore only evident in longitudinal studies, such as this one, and studies that examine the effects of prior trauma exposure over the course of many years. The Horvath estimate is a reliable index of chronological age across many tissue types (Horvath, 2013) and thus may be fairly robust to the acute or transient effects of biological processes in any one particular tissue type (e.g., metabolic aberrations found in blood). Additional research is needed to examine this possibility and account for differences in the two metrics of accelerated epigenetic age.
Another difference between the results of this study and that of Wolf et al. (2017) is that we previously demonstrated cross-sectional associations between the hyperarousal PTSD symptom cluster and advanced Hannum DNAm age whereas these longitudinal results supported an effect for the avoidance and numbing symptom cluster. This could be related to differences in the two age algorithms or to differences in the study methodology (cross-sectional versus longitudinal, use of different Illumina beadchips) and sample composition (e.g., young versus middle-aged veterans; mixed ancestry compared to a white, non-Hispanic sample) across the two studies. Replication in other studies is needed to determine if a given PTSD symptom cluster is more strongly associated with accelerated cellular aging.
Limitations and Conclusions
Results of this study should be considered in light of a number of limitations. The sample was comprised primarily of white, non-Hispanic men who were military veterans thus the generalizability of the results is limited and additional research in more heterogeneous samples is necessary. This is all the more important given evidence from prior work that age acceleration may differ by sex (Hannum et al., 2013; Marioni et al., 2015; Wolf et al., 2017) and possibly ethnicity (Horvath et al., 2016). As well, as all participants were trauma-exposed, we could not fully evaluate potential effects attributable to trauma exposure (but did include a metric of trauma load in the analyses). The stability of DNAm age estimates over time yields little variability in the rate of change over time and is likely one reason that effect sizes are relatively small in magnitude, consistent with prior work (Wolf et al., 2016; 2017; in press). Thus, caution is needed in interpreting results as the implications on gene expression are currently unknown (Breton et al., 2017). Related to this, although the longitudinal design is a strength of the study, the interval between assessments was fairly small (on average, just under 2 years), and it is possible that greater time between assessments is necessary to fully understand the cumulative effects of psychological stress on accelerated aging. As with any study, we cannot rule out possible confounds that could account for the primary findings of the study and there is much that is not yet known about the mechanisms underlying accelerated epigenetic aging that could be important for understanding and contextualizing these results. All of these limitations highlight the importance of future replication efforts in large, independent cohorts.
In conclusion, this study is the first to suggest that PTSD symptoms and alcohol-use disorders were associated with a quickening pace of the epigenetic clock, per the Horvath-derived estimate of DNAm age. Findings raise the possibility of overlap in the biological consequences of the two disorders. Results also demonstrated the applicability of the epigenetic age algorithms to DNAm data obtained from the latest Illumina methylation beadchip, the Infinium MethylationEPIC array, and highlight a novel approach to quantifying change in age acceleration. The study addresses a number of critical gaps in the literature to date, through use of a longitudinal design and the focus on the predictive strength of an array of posttraumatic psychiatric conditions that may have a shared pathophysiology. Findings from this study highlight the need to take the critical next steps in understanding accelerated epigenetic age and its contribution to health and disease. In particular, it is important to identify the biological mechanisms underlying accelerated aging and determine if intervention, including pharmacological approaches, meaningfully alter or improve the pace of epigenetic aging and thereby reduce the risk of premature health decline.
Supplementary Material
Acknowledgments
Funding
This work was supported by Merit Review Award Number I01 CX-001276-01 from the United States (U.S.) Department of Veterans Affairs Clinical Sciences R&D (CSRD) Service, by NIA grant 3R03AG051877-02S1, and by a Presidential Early Career Award for Scientists and Engineers (PECASE 2013A) to EJW, as administered by U.S. Department of Veterans Affairs Office of Research and Development. Funding for this work is also supported by NIMH grant R21MH102834 to MWM and the Translational Research Center for TBI and Stress Disorders (TRACTS), a VA Rehabilitation Research and Development Traumatic Brain Injury Center of Excellence (B9254-C), and the Cooperative Studies Program, Department of Veterans Affairs. FGM’s contribution to this work was supported by National Institute of Mental Health award # 5T32MH019836-16. This research is the result of work supported with resources and the use of facilities at the Pharmacogenomics Analysis Laboratory, Research and Development Service, Central Arkansas Veterans Healthcare System, Little Rock, Arkansas. The contents of this article do not represent the views of the U.S. Department of Veterans Affairs, the National Institutes of Health, or the United States Government.
Footnotes
We did not have interview-based symptom severity scores for DSM-IV diagnoses other than PTSD, thus the first regression focused on case/control status for each diagnosis.
Cross-sectional associations for all psychopathology models in association with T1 DNAm age residuals are reported in the Supplementary Materials for completeness (but overlap with results reported in Wolf et al., 2016).
The CFA used to develop the lifespan PTSD symptom factor scores at T1 fit the data well: χ2 (15) = 13.91, p = .53, root mean square error of approximation < .001, confirmatory fit index = 1.0, Tucker-Lewis index = 1.0, standardized root mean square residual = .02. All symptom cluster scores from each time period loaded significantly on each T1 lifespan symptom cluster factor.
Results were unchanged when we covaried for childhood trauma exposure (prior to age 18) in place of total trauma exposure.
We also tested a model in which PTSD symptom clusters and alcohol-use disorders were included in the same regression and found that both avoidance and numbing symptoms (β = .38, p = .032) and alcohol-use disorders (β = .24, p = .002) were significantly associated with the pace of Horvath epigenetic age.
Disclosures
All authors report no financial or other conflicts of interest in relationship to the contents of this article.
References
- Achur RN, Freeman WM, Vrana KE. Circulating cytokines as biomarkers of alcohol abuse and alcoholism. Journal of Neuroimmune Pharmacology. 2010;5:83–91. doi: 10.1007/s11481-009-9185-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiello AE, Dowd JB, Jayabalasingham B, Feinstein L, Uddin M, Simanek AM, Cheng CK, Galea S, Wildman DE, Koenen K. PTSD is associated with an increase in aged T cell phenotypes in adults living in Detroit. Psychoneuroendocrinology. 2016;67:133–141. doi: 10.1016/j.psyneuen.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoni FA, Hunter EF, Lowry PJ, Noble JM, Seckl JR. Atriopeptin: an endogenous corticotropin-release inhibiting hormone. Endocrinology. 1992;130:1753–5. doi: 10.1210/endo.130.3.1311248. [DOI] [PubMed] [Google Scholar]
- Beech RD, Qu J, Leffert JJ, Lin A, Hong KA, Hansen J, Umlauf S, Mane S, Zhao H, Sinha R. Altered expression of cytokine signaling pathway genes in peripheral blood cells of alcohol dependent subjects: preliminary findings. Alcoholism: Clinical and Experimental Research. 2012;36:1487–1496. doi: 10.1111/j.1530-0277.2012.01775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake DD, Weathers FW, Nagy LM, Kaloupek DG, Gusman FD, Charney DS, Keane TM. The development of a Clinician-Administered PTSD Scale. Journal of Traumatic Stress. 1995;8:75–90. doi: 10.1007/BF02105408. [DOI] [PubMed] [Google Scholar]
- Boks MP, van Mierlo HC, Rutten BP, Radstake TR, De Witte L, Geuze E, Horvath S, Schalkwyk LC, Vinkers CH, Broen JC, Vermetten E. Longitudinal changes of telomere length and epigenetic age related to traumatic stress and post-traumatic stress disorder. Psychoneuroendocrinology. 2015;51:506–12. doi: 10.1016/j.psyneuen.2014.07.011. [DOI] [PubMed] [Google Scholar]
- Breen MS, Maihofer AX, Glatt SJ, Tylee DS, Chandler SD, Tsuang MT, Risbrough VB, Baker DG, O’Connor DT, Nievergelt CM, Woelk CH. Gene networks specific for innate immunity define post-traumatic stress disorder. Molecular Psychiatry. 2015;20:1538–45. doi: 10.1038/mp.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JE, Irwin MR, Levine M, Seeman TE, Absher D, Assimes T, Horvath S. Epigenetic Aging and Immune Senescence in Women With Insomnia Symptoms: Findings From the Women’s Health Initiative Study. Biological Psychiatry. 2017;81:136–144. doi: 10.1016/j.biopsych.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BH, Marioni RE, Colicino E, Peters MJ, Ward-Caviness CK, Tsai PC, Roetker NS, Just AC, Demerath EW, Guan W, Bressler J, Fornage M, Studenski S, Vandiver AR, Moore AZ, Tanaka T, Kiel DP, Liang L, Vokonas P, Schwartz J, Lunetta KL, Murabito JM, Bandinelli S, Hernandez DG, Melzer D, Nalls M, Pilling LC, Price TR, Singleton AB, Gieger C, Holle R, Kretschmer A, Kronenberg F, Kunze S, Linseisen J, Meisinger C, Rathmann W, Waldenberger M, Visscher PM, Shah S, Wray NR, McRae AF, Franco OH, Hofman A, Uitterlinden AG, Absher D, Assimes T, Levine ME, Lu AT, Tsao PS, Hou L, Manson JE, Carty CL, LaCroix AZ, Reiner AP, Spector TD, Feinberg AP, Levy D, Baccarelli A, van Meurs J, Bell JT, Peters A, Deary IJ, Pankow JS, Ferrucci L, Horvath S. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY) 2016;8:1844–1865. doi: 10.18632/aging.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook RT, Ballas ZK, Waldschmidt TJ, Vandersteen D, LaBrecque DR, Cook BL. Modulation of T‐Cell Adhesion Markers, and the CD45R and CD57 Antigens in Human Alcoholics. Alcoholism: Clinical and Experimental Research. 1995;19:555–563. doi: 10.1111/j.1530-0277.1995.tb01548.x. [DOI] [PubMed] [Google Scholar]
- Cook RT, Waldschmidt TJ, Ballas ZK, Cook BL, Booth BM, Stewart BC, Garvey MJ. Fine T-Cell Subsets in Alcoholics as Determined by the Expression of l-Selectin, Leukocyte Common Antigen, and β-Integrin. Alcoholism: Clinical and Experimental Research. 1994;18:71–80. doi: 10.1111/j.1530-0277.1994.tb00883.x. [DOI] [PubMed] [Google Scholar]
- Darrow SM, Verhoeven JE, Revesz D, Lindqvist D, Penninx BW, Delucchi KL, Wolkowitz OM, Mathews CA. The Association Between Psychiatric Disorders and Telomere Length: A Meta-Analysis Involving 14,827 Persons. Psychosomatic Medicine. 2016;78:776–87. doi: 10.1097/PSY.0000000000000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis EG, Humphreys KL, McEwen LM, Sacchet MD, Camacho MC, MacIsaac JL, Lin DTS, Kobor MS, Gotlib IH. Accelerated DNA methylation age in adolescent girls: associations with elevated diurnal cortisol and reduced hippocampal volume. Translational Psychiatry. 2017;7:e1223. doi: 10.1038/tp.2017.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durso DF, Bacalini MG, Sala C, Pirazzini C, Marasco E, Bonafé M, do Valle ÍF, Gentilini D, Castellani G, Faria AMC. Acceleration of leukocytes’ epigenetic age as an early tumor and sex-specific marker of breast and colorectal cancer. Oncotarget. 2017;8:23237. doi: 10.18632/oncotarget.15573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- First MB, Gibbon M, Spitzer RL, Benjamin LS. User’s guide for the structured clinical interview for DSM-IV axis II personality disorders: SCID-II. American Psychiatric Association Publishing; 1997. [Google Scholar]
- Fülöp T, Dupuis G, Witkowski JM, Larbi A. The role of immunosenescence in the development of age-related diseases. Revista de Investigación Clínica. 2016;68:84–91. [PubMed] [Google Scholar]
- Glahn A, Riera Knorrenschild R, Rhein M, Haschemi Nassab M, Groschl M, Heberlein A, Muschler M, Frieling H, Bleich S, Hillemacher T. Alcohol-induced changes in methylation status of individual CpG sites, and serum levels of vasopressin and atrial natriuretic peptide in alcohol-dependent patients during detoxification treatment. European Addiction Research. 2014;20:143–50. doi: 10.1159/000357473. [DOI] [PubMed] [Google Scholar]
- Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y, Deconde R, Chen M, Rajapakse I, Friend S, Ideker T, Zhang K. Genome-wide methylation profiles reveal quantitative views of human aging rates. Molecular Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlaar N, Bryan AD, Thayer RE, Karoly HC, Oien N, Hutchison KE. Methylation of a CpG site near the ALDH1A2 gene is associated with loss of control over drinking and related phenotypes. Alcoholism: Clinical and Experimental Research. 2014;38:713–721. doi: 10.1111/acer.12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biology. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Gurven M, Levine ME, Trumble BC, Kaplan H, Allayee H, Ritz BR, Chen B, Lu AT, Rickabaugh TM, Jamieson BD, Sun D, Li S, Chen W, Quintana-Murci L, Fagny M, Kobor MS, Tsao PS, Reiner AP, Edlefsen KL, Absher D, Assimes TL. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biology. 2016;17:171. doi: 10.1186/s13059-016-1030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic T, Vance LA, Cross D, Knight AK, Kilaru V, Michopoulos V, Klengel T, Smith AK. Exposure to Violence Accelerates Epigenetic Aging in Children. Scientific Reports. 2017;7:8962. doi: 10.1038/s41598-017-09235-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kananen L, Marttila S, Nevalainen T, Kummola L, Junttila I, Mononen N, Kähönen M, Raitakari O, Hervonen A, Jylhä M. The trajectory of the blood DNA methylome ageing rate is largely set before adulthood: evidence from two longitudinal studies. Age. 2016;38:65. doi: 10.1007/s11357-016-9927-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler RC, Sonnega A, Bromet E, Hughes M, Nelson CB. Posttraumatic stress disorder in the National Comorbidity Survey. Archives of General Psychiatry. 1995;52:1048–60. doi: 10.1001/archpsyc.1995.03950240066012. [DOI] [PubMed] [Google Scholar]
- Kozlenkov A, Jaffe AE, Timashpolsky A, Apontes P, Rudchenko S, Barbu M, Byne W, Hurd YL, Horvath S, Dracheva S. DNA Methylation Profiling of Human Prefrontal Cortex Neurons in Heroin Users Shows Significant Difference between Genomic Contexts of Hyper-and Hypomethylation and a Younger Epigenetic Age. Genes. 2017;8:152. doi: 10.3390/genes8060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubany ES, Haynes SN, Leisen MB, Owens JA, Kaplan AS, Watson SB, Burns K. Development and preliminary validation of a brief broad-spectrum measure of trauma exposure: the Traumatic Life Events Questionnaire. Psychological Assessment. 2000;12:210–24. doi: 10.1037//1040-3590.12.2.210. [DOI] [PubMed] [Google Scholar]
- Leclercq S, Cani PD, Neyrinck AM, Starkel P, Jamar F, Mikolajczak M, Delzenne NM, de Timary P. Role of intestinal permeability and inflammation in the biological and behavioral control of alcohol-dependent subjects. Brain, Behavior, and Immunity. 2012;26:911–8. doi: 10.1016/j.bbi.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Levine ME, Lu AT, Bennett DA, Horvath S. Epigenetic age of the pre-frontal cortex is associated with neuritic plaques, amyloid load, and Alzheimer’s disease related cognitive functioning. Aging (Albany NY) 2015;7:1198–211. doi: 10.18632/aging.100864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Marioni RE, Hedman AK, Pfeiffer L, Tsai PC, Reynolds LM, Just AC, Duan Q, Boer CG, Tanaka T, Elks CE, Aslibekyan S, Brody JA, Kuhnel B, Herder C, Almli LM, Zhi D, Wang Y, Huan T, Yao C, Mendelson MM, Joehanes R, Liang L, Love SA, Guan W, Shah S, McRae AF, Kretschmer A, Prokisch H, Strauch K, Peters A, Visscher PM, Wray NR, Guo X, Wiggins KL, Smith AK, Binder EB, Ressler KJ, Irvin MR, Absher DM, Hernandez D, Ferrucci L, Bandinelli S, Lohman K, Ding J, Trevisi L, Gustafsson S, Sandling JH, Stolk L, Uitterlinden AG, Yet I, Castillo-Fernandez JE, Spector TD, Schwartz JD, Vokonas P, Lind L, Li Y, Fornage M, Arnett DK, Wareham NJ, Sotoodehnia N, Ong KK, van Meurs JB, Conneely KN, Baccarelli AA, Deary IJ, Bell JT, North KE, Liu Y, Waldenberger M, London SJ, Ingelsson E, Levy D. A DNA methylation biomarker of alcohol consumption. Molecular Psychiatry. 2016 doi: 10.1038/mp.2016.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue MW, Smith AK, Wolf EJ, Maniates H, Stone A, Schichman SA, McGlinchey RE, Milberg W, Miller MW. The correlation of methylation levels measured using Illumina 450K and EPIC BeadChips in blood samples. Epigenomics. 2017 doi: 10.2217/epi-2017-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay R, Akbar AN, Henson SM. The role of the T cell in age-related inflammation. Age. 2013;35:563–572. doi: 10.1007/s11357-012-9381-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni RE, Shah S, McRae AF, Chen BH, Colicino E, Harris SE, Gibson J, Henders AK, Redmond P, Cox SR, Pattie A, Corley J, Murphy L, Martin NG, Montgomery GW, Feinberg AP, Fallin MD, Multhaup ML, Jaffe AE, Joehanes R, Schwartz J, Just AC, Lunetta KL, Murabito JM, Starr JM, Horvath S, Baccarelli AA, Levy D, Visscher PM, Wray NR, Deary IJ. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biology. 2015;16:25. doi: 10.1186/s13059-015-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlinchey RE, Milberg WP, Fonda JR, Fortier CB. A methodology for assessing deployment trauma and its consequences in OEF/OIF/OND veterans: The TRACTS longitudinal prospective cohort study. International Journal of Methods in Psychiatric Research. 2017 doi: 10.1002/mpr.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney BC, Lin H, Ding Y, Lewis DA, Sweet RA. DNA methylation evidence against the accelerated aging hypothesis of schizophrenia. npj Schizophrenia. 2017;3:13. doi: 10.1038/s41537-017-0017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MW, Greif JL, Smith AA. Multidimensional Personality Questionnaire profiles of veterans with traumatic combat exposure: externalizing and internalizing subtypes. Psychological Assessment. 2003;15:205–15. doi: 10.1037/1040-3590.15.2.205. [DOI] [PubMed] [Google Scholar]
- Miller MW, Lin AP, Wolf EJ, Miller DR. Oxidative Stress, Inflammation, and Neuroprogression in Chronic PTSD. Harvard Review of Psychiatry. 2017 doi: 10.1097/HRP.0000000000000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MW, Wolf EJ, Reardon A, Greene A, Ofrat S, McInerney S. Personality and the latent structure of PTSD comorbidity. Journal of Anxiety Disorders. 2012;26:599–607. doi: 10.1016/j.janxdis.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschler MA, Hillemacher T, Kraus C, Kornhuber J, Bleich S, Frieling H. DNA methylation of the POMC gene promoter is associated with craving in alcohol dependence. Journal of Neural Transmission (Vienna) 2010;117:513–9. doi: 10.1007/s00702-010-0378-7. [DOI] [PubMed] [Google Scholar]
- Passos IC, Vasconcelos-Moreno MP, Costa LG, Kunz M, Brietzke E, Quevedo J, Salum G, Magalhães PV, Kapczinski F, Kauer-Sant’Anna M. Inflammatory markers in post-traumatic stress disorder: a systematic review, meta-analysis, and meta-regression. The Lancet Psychiatry. 2015;2:1002–1012. doi: 10.1016/S2215-0366(15)00309-0. [DOI] [PubMed] [Google Scholar]
- Perna L, Zhang Y, Mons U, Holleczek B, Saum KU, Brenner H. Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clinical Epigenetics. 2016;8:64. doi: 10.1186/s13148-016-0228-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Penaluna B, White T, Shires S, Gunter T, Liesveld J, Erwin C, Hollenbeck N, Osborn T. A pilot examination of the genome-wide DNA methylation signatures of subjects entering and exiting short-term alcohol dependence treatment programs. Epigenetics. 2014;9:1212–9. doi: 10.4161/epi.32252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach A, Levine ME, Tanaka T, Lu AT, Chen BH, Ferrucci L, Ritz B, Bandinelli S, Neuhouser ML, Beasley JM. Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging (Albany NY) 2017;9:419. doi: 10.18632/aging.101168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratanatharathorn A, Boks MP, Maihofer AX, Aiello AE, Amstadter AB, Ashley-Koch AE, Baker DG, Beckham JC, Bromet E, Dennis M, Garrett ME, Geuze E, Guffanti G, Hauser MA, Kilaru V, Kimbrel NA, Koenen KC, Kuan PF, Logue MW, Luft BJ, Miller MW, Mitchell C, Nugent NR, Ressler KJ, Rutten BPF, Stein MB, Vermetten E, Vinkers CH, Youssef NA, Workgroup VAMAM, Workgroup PPE, Uddin M, Nievergelt CM, Smith AK. Epigenome-wide association of PTSD from heterogeneous cohorts with a common multi-site analysis pipeline. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2017;174:619–630. doi: 10.1002/ajmg.b.32568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommershof A, Aichinger H, Engler H, Adenauer H, Catani C, Boneberg EM, Elbert T, Groettrup M, Kolassa IT. Substantial reduction of naive and regulatory T cells following traumatic stress. Brain, Behavior, and Immunity. 2009;23:1117–1124. doi: 10.1016/j.bbi.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Spitzer R, Gibbon M, Williams J. Structured clinical interview for DSM-IV axis I disorders: Patient Edition (February 1996 Final) Biometrics Research Department, New York State Psychiatric Institute; 1998. [Google Scholar]
- Strohle A, Feller C, Strasburger CJ, Heinz A, Dimeo F. Anxiety modulation by the heart? Aerobic exercise and atrial natriuretic peptide. Psychoneuroendocrinology. 2006;31:1127–30. doi: 10.1016/j.psyneuen.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Szabo G, Mandrekar P. A recent perspective on alcohol, immunity, and host defense. Alcoholism: Clinical and Experimental Research. 2009;33:220–232. doi: 10.1111/j.1530-0277.2008.00842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisey J, Lawford BR, Morris CP, Wockner LF, Noble EP, Young RM, Mehta D. Epigenetic analysis confirms no accelerated brain aging in schizophrenia. npj Schizophrenia. 2017;3 doi: 10.1038/s41537-017-0026-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidner CI, Wagner W. The epigenetic tracks of aging. Biological Chemistry. 2014;395:1307–14. doi: 10.1515/hsz-2014-0180. [DOI] [PubMed] [Google Scholar]
- Wiedemann K, Jahn H, Yassouridis A, Kellner M. Anxiolyticlike effects of atrial natriuretic peptide on cholecystokinin tetrapeptide-induced panic attacks: preliminary findings. Archives of General Psychiatry. 2001;58:371–7. doi: 10.1001/archpsyc.58.4.371. [DOI] [PubMed] [Google Scholar]
- Wolf EJ, Logue MW, Hayes JP, Sadeh N, Schichman SA, Stone A, Salat DH, Milberg W, McGlinchey R, Miller MW. Accelerated DNA methylation age: Associations with PTSD and neural integrity. Psychoneuroendocrinology. 2016;63:155–62. doi: 10.1016/j.psyneuen.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf EJ, Logue MW, Stoop TB, Schichman SA, Stone A, Sadeh N, Hayes JP, Miller MW. Accelerated DNA Methylation Age: Associations with PTSD and Mortality. Psychosomatic Medicine. 2017 doi: 10.1097/PSY.0000000000000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf EJ, Maniates H, Nugent N, Maihofer AX, Armstrong D, Ratanatharathorn A, et al. Traumatic stress and accelerated DNA methylation age: a meta-analysis. Psychoneuroendocrinology. doi: 10.1016/j.psyneuen.2017.12.007. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf EJ, Morrison FM. Traumatic stress and accelerated cellular aging: From epigenetics to cardiometabolic disease. Current Psychiatry Reports. 2017;19:75. doi: 10.1007/s11920-017-0823-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannas AS, Arloth J, Carrillo-Roa T, Iurato S, Roh S, Ressler KJ, Nemeroff CB, Smith AK, Bradley B, Heim C, Menke A, Lange JF, Bruckl T, Ising M, Wray NR, Erhardt A, Binder EB, Mehta D. Lifetime stress accelerates epigenetic aging in an urban, African American cohort: relevance of glucocorticoid signaling. Genome Biology. 2015;16:266. doi: 10.1186/s13059-015-0828-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gelernter J. DNA methylation and alcohol use disorders: Progress and challenges. The American Journal on Addictions. 2017;26:502–515. doi: 10.1111/ajad.12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Miao Q, Wang C, Zhao R, Li W, Haile CN, Hao W, Zhang XY. Genome-wide DNA methylation analysis in alcohol dependence. Addiction Biology. 2013;18:392–403. doi: 10.1111/adb.12037. [DOI] [PubMed] [Google Scholar]
- Zhao R, Zhang R, Li W, Liao Y, Tang J, Miao Q, Hao W. Genome-wide DNA methylation patterns in discordant sib pairs with alcohol dependence. Asia-Pacific Psychiatry. 2013;5:39–50. doi: 10.1111/appy.12010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


