Short abstract
Background
Paclitaxel is one of the most commonly used drugs to treat breast cancer. Its major dose-limiting toxicity is paclitaxel-induced peripheral neuropathy (PIPN). PIPN persists into survivorship and has a negative impact on patient’s mood, functional status, and quality of life. No interventions are available to treat PIPN. A critical barrier to the development of efficacious interventions is the lack of understanding of the mechanisms that underlie PIPN. Mitochondrial dysfunction has been evaluated in preclinical studies as a hypothesized mechanism for PIPN, but clinical data to support this hypothesis are limited. The purpose of this pilot study was to evaluate for differential gene expression and perturbed pathways between breast cancer survivors with and without PIPN.
Methods
Gene expression in peripheral blood was assayed using RNA-seq. Differentially expressed genes (DEG) and pathways associated with mitochondrial dysfunction were identified between survivors who received paclitaxel and did (n = 25) and did not (n = 25) develop PIPN.
Results
Breast cancer survivors with PIPN were significantly older; more likely to be unemployed; reported lower alcohol use; had a higher body mass index and poorer functional status; and had a higher number of lower extremity sites with loss of light touch, cold, and pain sensations and higher vibration thresholds. No between-group differences were found in the cumulative dose of paclitaxel received or in the percentage of patients who had a dose reduction or delay due to PIPN. Five DEGs and nine perturbed pathways were associated with mitochondrial dysfunction related to oxidative stress, iron homeostasis, mitochondrial fission, apoptosis, and autophagy.
Conclusions
This study is the first to provide molecular evidence that a number of mitochondrial dysfunction mechanisms identified in preclinical models of various types of neuropathic pain including chemotherapy-induced peripheral neuropathy are found in breast cancer survivors with persistent PIPN and suggest genes for validation and as potential therapeutic targets.
Keywords: Taxanes, mitochondria, neuropathy, breast cancer, survivor, paclitaxel, differential gene expression, pathway analysis
Introduction
Paclitaxel is one of the most commonly used drugs to treat breast, ovarian, and lung cancers.1 Its major dose-limiting toxicity is paclitaxel-induced peripheral neuropathy (PIPN). Prevalence rates for PIPN are extremely high, ranging from 59% to 87%.2,3 PIPN is characterized by paresthesias and dysesthesias that occur in a “stocking-glove” distribution. PIPN persists into the cancer survivorship period and has a negative impact on individuals’ mood, functional status, and quality of life (QOL).4 In particular, the negative impact of PIPN on breast cancer (BC) survivors has been identified as a gap in QOL for BC patients.5,6
Paclitaxel exerts its primary antitumor effects by binding to beta-tubulin in microtubules, forming extremely stable and nonfunctional microtubules that results in apoptosis.7 In terms of its neurotoxic effects, the exact mechanisms that underlie the development of PIPN remain unclear. However, several lines of evidence suggest that interruption of microtubule function in neuron impairs axonal transport and results in a dying back neuropathy. In addition, paclitaxel alters mitochondrial function through the depletion of mRNAs that encode the mitochondrial fission/fusion machinery in distal axons.8 This toxic effect leads to a deficit in axonal energy supply and subsequent axonal degeneration.9,10
Using the National Cancer Institute’s Common Terminology Criteria for Adverse Events, PIPN of grades 2 to 4 occurs at cumulative doses of between 715 and 1500 mg/m2.11 However, not all patients who receive this dose of paclitaxel develop PIPN. The wide range in occurrence rates suggests that genetic factors may play a role in the development of PIPN. While preclinical studies suggest that a number of mechanisms are involved in the development of PIPN,9,10,12 the majority of the genetic association studies of PIPN evaluated for polymorphisms in candidate genes that influence the metabolism and transport of neurotoxic drugs. As noted in a recent systematic review and meta-analysis of the molecular genetics of chemotherapy-induced peripheral neuropathy (CIPN),13 notable methodological issues, including lack of standardization and detail in the phenotype definition and acknowledgment of potential confounding factors, prevented the authors from identifying any candidate genes that were associated with CIPN in general or PIPN specifically.
No studies were identified that evaluated for associations between changes in the expression of genes involved in mitochondrial function and chronic PIPN in cancer survivors. Given the preclinical evidence that paclitaxel alters mitochondrial function in primary afferent neurons,9,10 the purpose of this study was to identify differentially expressed mitochondrial dysfunction (MD)-related genes and perturbed pathways in BC survivors with (n = 25) and without (n = 25) chronic PIPN.
Materials and methods
Survivors and settings
The methods for this study, which is part of a larger study, are described in detail elsewhere.14 In brief, survivors were recruited from throughout the San Francisco Bay area and met prespecified inclusion and exclusion criteria (see Supplemental Material). The National Coalition for Cancer Survivorship’s definition of cancer survivor was used in this study (i.e., a person is a cancer survivor from the moment of diagnosis through the balance of life).15 Of the 1450 survivors who were screened, 754 were enrolled and 623 completed the self-report questionnaires and the study visit. Data from a randomly selected sample of BC survivors with (n = 25) and without (n = 25) chronic PIPN were used in this analysis.
Study procedures
Research nurses screened and consented the survivors over the phone, sent and asked them to complete the self-report questionnaires prior to their study visit, and scheduled the in-person assessment. At this assessment, written informed consent was obtained, questionnaires were reviewed for completeness, and objective measurements were done. Blood samples were drawn, processed, and stored for subsequent molecular analyses in PAXgene® Blood RNA tubes (Qiagen, Inc.). This study was approved by the institutional review board of the University of California, San Francisco.
Study measures
Demographic and clinical characteristics
Survivors provided information on demographic characteristics and completed the Alcohol Use Identification Test (AUDIT),16 Karnofsky Performance Status (KPS) Scale,17–19 and the Self-Administered Comorbidity Questionnaire.20,21
Pain measures
Survivors with PIPN rated the intensity of their pain using a 0 to 10 numeric rating scale and completed the pain interference scale from the Brief Pain Inventory.22 The qualities of PIPN were evaluated using the Pain Quality Assessment Scale.23,24
Objective measures of sensation
Light touch was evaluated using Semmes Weinstein monofilaments.25 Cold sensation was evaluated using the Tiptherm Rod.26,27 Pain sensation was evaluated using the Neurotip.27 Vibration threshold was assessed using a vibrometer.28 For all of the measures of sensation, both the upper and lower extremities on the dominant side were tested.
Balance
Self-report questions from the CIPN Assessment Tool were used to assess the balance.29 The objective measures of balance were the timed get up and go test (TUG)30 and the Fullerton Advanced Balance (FAB) test.31,32
Phenotypic data analysis
Data were analyzed using SPSS version 23.33 Descriptive statistics and frequency distributions were calculated for survivors’ demographic and clinical characteristics. For the four objective measures of sensation (i.e., light touch, cold, pain, and vibration), composite scores, over all of the sites that were tested on the dominant upper and lower extremities, were created. For light touch, cold, and pain, the number of sites with loss of each sensation was summed. For vibration, the mean score across the sites was calculated. Differences between the PIPN groups in phenotypic characteristics and balance were evaluated using independent sample t tests, χ2 analyses, Fisher's Exact test (FET), and Mann–Whitney U tests. A p value of <.05 was considered statistically significant.
RNA sample preparation
Total RNA was isolated using the PAXgene® Blood miRNA Kit (Qiagen, Inc.) using established procedures.34 Total RNA from the 50 survivors was sent to the UC Davis Genomics Core Facility for library preparation and for sequencing. Prior to library preparation, 600 ng of total RNA was treated with the Illumina Globin-Zero Gold rRNA Removal Kit (Illumina Inc., San Diego, CA) to deplete cytoplasmic ribosomal RNA35 and human globin mRNA.36,37 The globin/ribo depleted RNA was cleaned with Agencourt RNAClean XP (Beckman Coulter, Indianapolis, IN), and the sequencing libraries were prepared with KAPA RNA HyperPrep Kit (Roche Diagnostics Corp., Indianapolis, IN) according to the manufacturer’s protocol. Fourteen cycles of polymerase chain reaction (PCR) amplification were used for double six base pair index addition and library fragment enrichment. Prepared libraries were quantified on a Roche LightCycler 480II (Roche Diagnostics Corp.) using KAPA Illumina library quantitative PCR reagents (Roche Diagnostics Corp.).
RNA sequencing (RNA-seq)
Sequencing of the 50 samples was done on an Illumina HiSeq 4000 apparatus (Illumina Inc., San Diego, CA). All 50 samples were multiplexed into a single pool, with each sample labeled with a dual-indexed adapter.38 The sample pool was sequenced on four lanes for 100 cycles of single-end reads with a 1% PhiX v3 control library spike (Illumina Inc., San Diego, CA). Postsequencing basecall files (bclfiles) were demultiplexed and converted into an FASTQ file format using the bcl2fastq v2.17 software (Illumina Inc., San Diego, CA). Data were posted and retrieved from a secured site hosted by the Core Facility.
RNA-seq alignment, data processing, and quality control (QC)
RNA-seq data processing was performed based on best practices39,40 and our previous experience.34,41 Illuimina adapters and leading or trailing low-quality bases were removed, and reads with an average quality per base below 15 in a 4-base sliding window or below a minimum length of 36 bases were removed using trimmomatic.42 Individual samples were inspected with FASTQC43 and in aggregate with MultiQC.44 After initial QC, 10 bases were trimmed from the beginning of all reads and reads were reinspected with FASTQC. The reference genome was prepared using the GRCh38 assembly (gencode.v24.GRCh38.p5.fa).45 Transcriptome annotations were obtained from the Gencode v24 primary assembly (gencode.v24.primary_assembly.annotation.gtf).45 Trimmed reads were aligned to the annotated reference genome using the Spliced Transcripts Alignment to a Reference aligner.46 Output SAM files were validated using ValidateSam. Read groups were added to the SAM file using the Picard tool AddOrReplaceReadGroups. Sorted BAM files were inspected using RNA-SeQC47 and joined for each sample. Abundance of RNA was estimated from the combined aligned reads using featureCounts.48
Replicate count data were processed in edgeR.49 Ensembl50 transcripts were annotated with Entrez gene ID and symbol.51 Lowly expressed tags were filtered out by retaining only those with 3.27 reads per million (10/L where L is the minimum library size in millions) in at least 25 samples (i.e., the smallest group size). Count estimates were normalized with the trimmed means of M values (TMM) method.52 TMM normalization was applied to the data set in edgeR using calcNormFactors. Data were explored using multidimensional scaling (MDS) plots for all samples to identify sample outliers and potential batch effects due to technical artifacts (i.e., RNA integrity number (RIN), date of RNA extraction). The same technician performed all of the RNA extractions in one laboratory. Associations between technical variables and PIPN group were assessed using Fisher’s exact test or a generalized linear model in R. Significance was assessed at a nominal p value of .05. To evaluate the reproducibility of our measurements,53 we utilized the one additional library54 that was generated using an alternative preparation protocol (i.e., without globin/ribo depletion step, but from the same RNA materials) and sequenced in the same lane with the entire sample. We used the weighted kappa statistic to measure the agreement between the count data of this sample produced by the two libraries.55 The count data were summarized as log2 counts per million (after TMM normalization) and grouped into 10 levels with a range of 0 to 20 with a bin size of 2.
The technical variables that demonstrated an association with PIPN group were flagged for subsequent evaluation with surrogate variable analysis (SVA). SVA was used to identify variations that contributed to heterogeneity in the sample (e.g., batch effects) which were not due to the variable of interest (i.e., PIPN group membership) or significant demographic covariates.56 To identify which surrogate variables to include, all surrogate variables were tested for an association with the phenotype and the retained technical variables. Significance was assessed at a nominal p value of .05. Any surrogate variable (SV) that was significantly associated with the phenotype was excluded. Any remaining surrogate variables that were significantly associated with a technical variable were included as covariates in the model for differential expression testing.
Differential expression (DE) analysis
Using edgeR, DE was determined under a variance modeling strategy that addressed the overdispersion observed in gene expression (GE) count data.57 EdgeR is widely used for DE analysis and performs well relative to other strategies.58,59 We followed published best practices60,61 and our previous experience.34,41 The total sample size is 50 survivors. For this analysis, the overall dispersion and the gene-wise and tag-wise dispersion were estimated using general linear models estimated using the Cox–Reid-adjusted likelihood method.62,63 Differences between the PIPN groups were tested using likelihood ratio tests. Demographic and clinical characteristics that differed between the PIPN groups, as well as surrogate variables, were included as covariates in the model. We assessed the significance of the transcriptome-wide analysis to identify diffentially expressed genes (DEGs) using a strict false discovery rate (FDR) of 1% under the Benjamini–Hochberg (BH) procedure and no minimal fold-change as evaluated by the topTags and p.adjust R functions.64,65
Pathway impact analysis
Pathway analysis approaches have been classified into three classes or generations of approaches (reviewed in 66,67): overrepresentation analysis (ORA), functional class scoring (FCS), and pathway topology-based (PTB). ORA evaluates for the enrichment of DEGs in a pathway. Although ORA are widely used, but are limited by the predefined set of input genes (i.e., the DE list and the cutoff used to determine it), the statistic used is independent of the measured changes, no differentiation is made between the genes, and all pathways are considered independent. FCS addresses only the first three limitations. In addition, both ORA and FCS consider pathways as lists of genes (i.e., only the counts) and ignore the additional information available in the pathway representation (i.e., the topology). PTB approaches utilize this additional information to evaluate for biological-relevant interactions between genes in the pathway (reviewed in Nguyen et al.68). Pathway impact analysis (PIA) is a PTB that implements and impact factor analytic approach, which includes potentially important biological factors (e.g., gene–gene interactions, flow signals in a pathway, pathway topologies) as well as the magnitude (i.e., log fold-change) and the significance (i.e., p values) of the biological differences observed in the DE analysis.69 We used Pathway Express70 to perform the PIA. This PTB impact factor analysis approach is widely used and has 1200 citations to date.68 The analysis included all genes (i.e., cutoff free) and the DE analysis results (i.e., p value and log fold-change) to determine the probability of a pathway perturbation (pPERT). By including all genes in the analysis, and using the DE analysis results to represent the biological differences between the groups, we are also able to capture the adjustments made for the demographic, clinical, and technical (i.e., SVs) variation in the sample. A total of 208 signaling pathways were defined using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.71 Sequence loci data were annotated with Entrez gene IDs. The gene names were annotated using the HUGO Gene Nomenclature Committee resource database.72 We assessed for the significance of the pathway analyses using a strict FDR of 1% under the BH procedure.64,65
Results
Differences in demographic and clinical characteristics
As shown in Table 1, survivors with PIPN were significantly older (p = .006) and were more likely to be unemployed (p = .022). In terms of clinical characteristics (see Table 2), survivors with PIPN had a lower AUDIT score (p = .012), a higher body mass index (BMI; p = .017), and a lower KPS score (p < .001). Of note, no between-group differences were found in the total cumulative dose of paclitaxel received or in the percentage of patients who had a dose reduction or delay due to PIPN.
Table 1.
Differences in demographic characteristics between breast cancer survivors with and without paclitaxel-induced neuropathy.
| Characteristic | No neuropathy | Neuropathy | Test, p value |
|---|---|---|---|
|
50.0% (n = 25) |
50.0% (n = 25) |
||
| Mean (SD) | Mean (SD) | ||
| Age (years) | 52.2 (9.5) | 60.0 (9.4) | t = −2.89, p = .006 |
| Education (years) | 16.6 (2.4) | 16.3 (2.9) | t = 0.32, p = .750 |
|
% (n) |
% (n) |
||
| Married/partnered | 80.0 (20) | 76.0 (19) | FE, p = 1.000 |
| Lives alone | 16.0 (4) | 12.0 (3) | FE, p = 1.000 |
| Employed | 72.0 (18) | 36.0 (9) | FE, p = .022 |
| Ethnicity | |||
| White | 80.0 (20) | 76.0 (19) | FE, p = 1.000 |
| Nonwhite | 20.0 (5) | 24.0 (6) | |
| Annual household income | |||
| <$30,000 | 16.7 (4) | 19.0 (4) | |
| $30,000–$69,999 | 16.7 (4) | 19.0 (4) | U, p = .317 |
| $70,000–$99,999 | 16.7 (4) | 33.3 (7) | |
| >$100,000 | 50.0 (12) | 28.6 (6) | |
| Child care responsibilities | 25.0 (6) | 20.0 (5) | FE, p = .742 |
| Adult care responsibilities | 4.3 (1) | 4.3 (1) | FE, p = 1.000 |
FE: Fisher’s exact test; SD: standard deviation; U: Mann–Whitney U test.
Table 2.
Differences in clinical characteristics between breast cancer survivors with and without paclitaxel-induced peripheral neuropathy.
| Characteristic |
No neuropathy50.0% (n=25) |
Neuropathy50.0% (n=25) |
Test, p value |
|---|---|---|---|
| Mean (SD) | Mean (SD) | ||
| Karnofsky Performance Status score | 92.9 (6.2) | 80.0 (11.2) | t = 5.02, p <.001 |
| Body mass index (kg/m2) | 23.5 (3.5) | 27.2 (6.5) | t = −2.50, p = .017 |
| Number of comorbidities | 1.3 (1.2) | 1.5 (1.7) | t = −0.55, p = .589 |
| Self-Administered Comorbidity Questionnaire score | 2.7 (2.7) | 3.6 (4.2) | t = −0.88, p = .384 |
| AUDIT score | 3.0 (1.7) | 1.7 (1.8) | t = 2.63, p = .012 |
| Years since cancer diagnosis | 4.2 (4.7) | 4.6 (3.9) | t = −0.34, p = .739 |
| Number of prior cancer treatments | 3.6 (0.8) | 3.6 (0.8) | t = −0.17, p = .863 |
| Number of current cancer treatments | 0.7 (0.7) | 0.6 (0.7) | t = 0.61, p = .543 |
| Number of metastatic sites (out of seven) | 1.0 (0.8) | 0.6 (0.5) | t = 1.72, p = .092 |
| Number of metastatic sites without lymph node involvement | 0.2 (0.6) | 0.04 (0.2) | t = 0.92, p = .365 |
|
% (n) |
% (n) |
||
| Smoker (ever) | 37.5 (9) | 32.0 (8) | FE, p = .769 |
| Exercise on a regular basis (% yes) | 87.5 (21) | 60.0 (15) | FE, p = .051 |
| Born prematurely (% yes) | 0.0 (0) | 12.5 (3) | FE, p = .110 |
| Comorbid conditions (% yes) | |||
| Osteoarthritis | 8.0 (2) | 28.0 (7) | FE, p = .138 |
| Back pain | 28.0 (7) | 24.0 (6) | FE, p = 1.000 |
| Depression | 28.0 (7) | 16.0 (4) | FE, p = .496 |
| High blood pressure | 12.0 (3) | 28.0 (7) | FE, p = .289 |
| Heart disease | 0.0 (0) | 4.0 (1) | FE, p = 1.000 |
| Diabetes | 0.0 (0) | 8.0 (2) | FE, p = .490 |
| Lung disease | 4.0 (1) | 4.0 (1) | FE, p = 1.000 |
| Anemia or blood disease | 0.0 (0) | 0.0 (0) | Test not run |
| Ulcer or stomach disease | 0.0 (0) | 4.0 (1) | FE, p = 1.000 |
| Kidney disease | 0.0 (0) | 0.0 (0) | Test not run |
| Liver disease | 0.0 (0) | 4.0 (1) | FE, p = 1.000 |
| Rheumatoid arthritis | 0.0 (0) | 4.0 (1) | FE, p = 1.000 |
| Total cumulative dose of taxol (mg/m2)** | 782.8 (228.6) | 814.7 (217.0) | t = 0.14, p = .893 |
| Patients who had a dose reduction or delay due to neuropathy (% (n))** | 0.0 (0) | 12.0 (3) | FE, p = .235 |
Note: AUDIT: Alcohol Use Disorders Identification Test; FE: Fisher’s exact test; kg: kilograms; m2: meters squared; mg: milligrams; SD: standard deviation.
**Doses are reported as milligrams per meter squared.
Pain characteristics
Table 3 summarizes the self-reported pain characteristics of the survivors with PIPN. Worst pain severity was reported as 6.3 (±2.1) of the 10, and the duration of PIPN was 3.8 (±3.9) years.
Table 3.
Pain characteristics of the breast cancer survivors with paclitaxel-induced peripheral neuropathy.
| Characteristic |
Lower extremity(n = 25) |
|---|---|
| Mean (SD) | |
| Pain characteristics | |
| Duration of neuropathy (years) | 3.8 (3.9) |
| Pain now | 3.1 (2.2) |
| Average pain | 3.6 (2.0) |
| Worst pain | 6.3 (2.1) |
| Days per week in pain | 4.0 (3.2) |
| Hours per day in pain | 13.0 (8.8) |
| Pain Interference Scale (0–10) | |
| Walking ability | 4.2 (3.2) |
| Balance | 4.0 (3.1) |
| General activity | 3.0 (2.9) |
| Enjoyment of life | 2.9 (2.8) |
| Sleep | 2.9 (3.0) |
| Normal work | 2.8 (3.0) |
| Mood | 1.9 (2.0) |
| Relations with other people | 1.5 (1.9) |
| Sexual activity | 0.8 (1.9) |
| Mean interference score | 2.7 (2.2) |
| Pain Qualities Assessment Scale scores (0–10) | |
| Numb | 5.8 (3.0) |
| Tingling | 4.8 (3.1) |
| Unpleasant | 4.6 (2.3) |
| Dull | 3.6 (3.3) |
| Intense | 3.5 (2.5) |
| Electrical | 2.9 (3.2) |
| Cramping | 2.8 (3.3) |
| Aching | 2.5 (2.6) |
| Hot | 2.4 (3.1) |
| Radiating | 2.3 (3.0) |
| Shooting | 2.2 (3.1) |
| Sharp | 2.0 (2.5) |
| Heavy | 2.0 (2.7) |
| Tender | 1.7 (2.3) |
| Throbbing | 1.6 (2.6) |
| Sensitive skin | 1.4 (1.8) |
| Itchy | 1.0 (1.6) |
| Cold | 0.9 (1.7) |
| Intense—surface pain | 3.7 (3.0) |
| Intense—deep pain | 3.5 (2.9) |
SD: standard deviation.
Differences in sensation
Survivors with PIPN had a higher number of lower extremity sites with loss of light touch, cold, and pain sensations (all, p < .05). Vibration thresholds were significantly higher in the PIPN group (p = .009, Table 4).
Table 4.
Differences in sensation and balance measures between breast cancer survivors with and without paclitaxel-induced peripheral neuropathy.
| Characteristic | No neuropathy | Neuropathy | Statistic; p value |
|---|---|---|---|
|
50.0% (n = 25) |
50.0% (n = 25) |
||
| Mean (SD) | Mean (SD) | ||
| Sensation measuresa | |||
| Light touch—lower extremity sites (out of nine)b | 0.1 (0.3) | 1.5 (1.5) | t = −4.42, p <.001 |
| Cold—lower extremity sites out of fourc | 1.5 (1.3) | 2.3 (1.0) | t = −2.54, p = .014 |
| Pain—lower extremity sites (out of nine)d | 1.6 (1.6) | 3.0 (2.1) | t = −2.71, p = .009 |
| Vibration—lower extremity sites (volts)e | 16.2 (8.1) | 25.1 (14.0) | t = −2.76, p = .009 |
| Balance measures | |||
| Trouble with balance (% yes (n))f | 13.0 (3) | 76.0 (19) | FE, p <.001 |
| Severity of balance trouble (0–10)g | 1.7 (1.2) | 5.6 (2.9) | t = −4.16, p = .004 |
| Frequency of balance trouble (0–10)h | 2.3 (1.5) | 5.1 (3.2) | t = −2.37, p = .060 |
| Distress from balance trouble (0–10)i | 1.7 (1.2) | 5.8 (3.2) | t = −4.17, p = .003 |
| Timed get up and go test (>13.5 s = higher risk for falls) | 6.1 (1.1) | 8.6 (3.1) | t = −3.76, p = .001 |
| Fullerton Advanced Balance test (≤25 is associated with a higher risk of falls) | 38.0 (3.1) | 33.4 (6.7) | t = 3.14, p = .004 |
Note: FE: Fisher’s exact test; SD: standard deviation.
aChanges in sensation are reported for the dominant extremity.
bLower extremity sites for light touch were as follows: pad of great toe, pad of third toe, pad of fifth toe, base of heel, metocarpophalangeal (MP) joint of great toe, MP joint of third toe, MP joint of fifth toe, midway along tibia, and patella.
cLower extremity sites for cold were as follows: top of great toe at first MP joint, pad of great toe, dorsum of foot midpoint, and medial malleolus.
dLower extremity sites for pain were as follows: pad of great toe, pad of third toe, pad of fifth toe, base of heel, MP joint of great toe, MP joint of third toe, MP joint of fifth toe, midway along tibia, and patella.
eLower extremity sites for vibration were as follows: dorsal IP joint of great toe, medial malleolus, and patella.
fSince your chemotherapy, have you had trouble with your balance?
gAt its worst, how severe is the trouble with your balance (0 = not at all severe to 10 = extremely severe)?.
hHow often do you have trouble with your balance (0 = never to 10 = always)?
iAt its worst, how distressing is the trouble with your balance (0 = not at all distressing to 10 = extremely distressing)?
Differences in balance
Survivors with PIPN were more likely to report trouble with balance (p < .001) as well as higher severity and distress (both p < .05) scores associated with balance problems (Table 4). In addition, these survivors reported worse TUG (p = .001) and worse FAB (p = .004) scores.
GE measurements
Mean RIN was 8.27 (minimum = 7.2), median sequenced library size was 27,458,032, and median aligned library size was 4,412,000 reads. The within-replicate agreement (kappa = 0.82) was consistent with high agreement measures observed in a recent study (i.e., 0.62–0.81) that evaluated the variation of technical replicates within RNA-seq data using non-clinical (i.e., Drosophila) samples.55 After QC filtering, 12,491 transcripts were retained from the total of 60,725 GENCODE transcript targets. No samples were identified as outliers in the MDS plots. No batch effects were identified for sample preparation order, RIN value, or RNA processing date in the first four dimensions of MDS plots. PIPN group was associated with the RNA extraction date (FET p value = .01) but not with RIN score or level. RNA extraction date was strongly associated with one of the six identified surrogate variables. This surrogate variable was included in the model as a covariate to control for potential batch effects. The phenotype characteristics that were associated with PIPN group membership (i.e., KPS, employment, age, BMI, and AUDIT score) were included as covariates in the model. The overall dispersion was observed to be 0.076 (biological coefficient of variation = 0.276), which is lower than values reported in other clinical data sets.62 Using RNA-seq power,73 we estimate that with 25 replicates in each condition (i.e., with and without PIPN), an average depth of coverage of 15 million reads per replicate, and the observed biological coefficient of variation (i.e., 0.276), we are powered to detect 1.5-fold changes for 83% of genes, and 2-fold changes for 99% of genes, at a type I error rate of 0.01.
Differences in whole-transcriptome GE
The DE analysis included 11,487 genes. After correction for multiple hypothesis testing at a conservative FDR of 1% (adjusted p value < .01), 20 genes were identified as differentially expressed between the survivors with and without PIPN. Of these, four genes were related to MD (Table 5).
Table 5.
Differentially expressed mitochondrial dysfunction-related genes between breast cancer survivors with and without paclitaxel-induced peripheral neuropathy (PIPN).
| Ensemble gene ID | Gene symbol | Name | Entrez ID | logFCa | Adjusted p valueb |
|---|---|---|---|---|---|
| ENSG00000126432 | PRDX5 | Peroxiredoxin 5 | 25824 | −0.987 | .0098 |
| ENSG00000182512 | GLRX5 | Glutaredoxin 5 | 51218 | −1.703 | .0062 |
| ENSG00000214253 | FIS1 | Fission, mitochondrial 1 | 51024 | −1.062 | .0062 |
| ENSG00000170315 | UBB | Ubiquitin B | 7314 | −1.374 | .0062 |
alogFC: Fold change of the log2 transformed normalized gene expression counts between the groups. A negative logFC denotes lower expression in survivors with PIPN as compared to those without PIPN.
bp Value adjusted using the Benjamini–Hochberg method.
Successful annotation with ENTREZ IDs was performed for 11,174 unique genes. Fold changes and p values from the DE analysis for these genes were included in the pPERT analysis of the 208 KEGG signaling pathways. PIA identified 53 KEGG signaling pathways that were significantly perturbed between the PIPN groups after correction for multiple hypothesis testing at a conservative FDR of 1% (adjusted perturbation p value <.01). Of these, 10 KEGG signaling pathways were related to MD (Table 6; Figure 1).
Table 6.
Significantly perturbed mitochondrial dysfunction-related Kyoto Encyclopedia of Genes and Genomes pathways between breast cancer survivors with and without paclitaxel-induced peripheral neuropathy.
| Pathway ID | Pathway name | Total perturbation | Adjusted pPerta |
|---|---|---|---|
| hsa04137 | Mitophagy–animal | 11.79 | .004 |
| hsa04150 | mTOR signaling pathway | 9.60 | .004 |
| hsa04152 | AMPK signaling pathway | 8.99 | .004 |
| hsa04151 | PI3K-AKT signaling pathway | 8.04 | .004 |
| hsa04066 | HIF-1 signaling pathway | 7.45 | .004 |
| hsa04216 | Ferroptosis | 6.98 | .004 |
| hsa04115 | p53 signaling pathway | 5.68 | .007 |
| hsa04146 | Peroxisome | 5.32 | .007 |
| hsa04068 | FoxO signaling pathway | 5.32 | .008 |
| hsa04218 | Cellular senescence | 5.36 | .008 |
apPert: Perturbation p value adjusted using the Benjamini–Hochberg method.
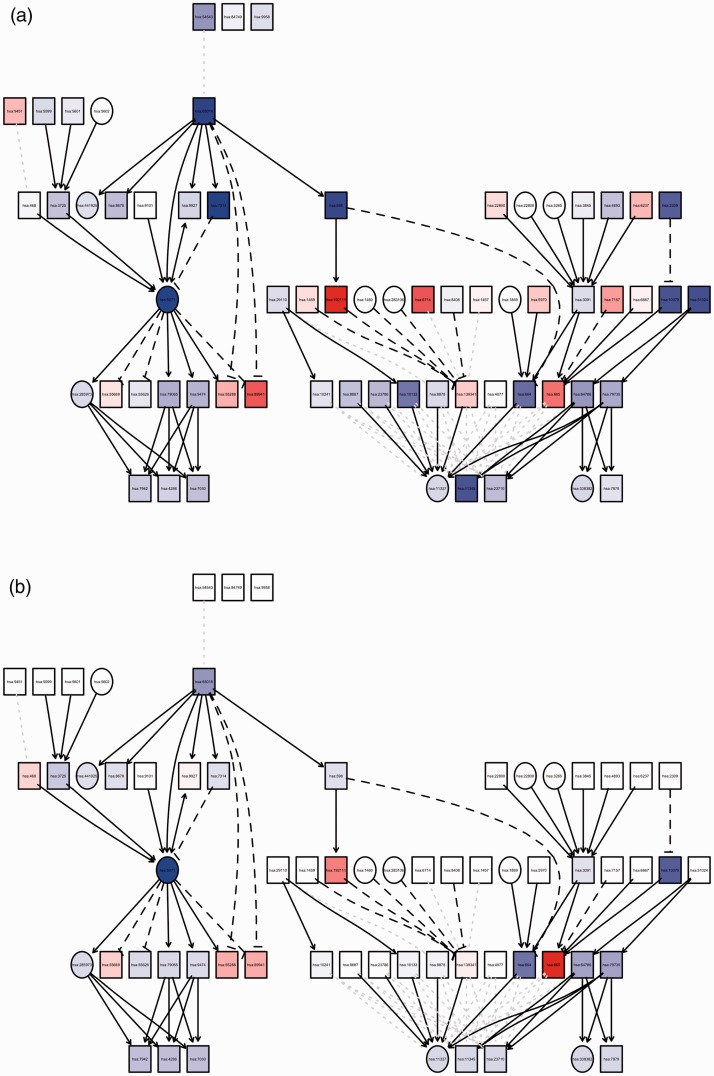
Figure 1.
Graph summary of pathway level statistics. (a) The measured expression change versus (b) perturbation accumulation in the Mitophagy–Animal Kyoto Encyclopedia of Genes and Genomes pathway (hsa04137). The square nodes denote genes with gene expression change, and the circle nodes denote all other nodes. The color of each node represents the perturbation (red = positive, blue = negative), and the shade represents the strength of the perturbation. Note that the square nodes with no parents have no accumulation.
Discussion
This study is the first to provide molecular evidence that some of the mitochondrial mechanisms identified in preclinical models of PIPN12,74,75 are found in cancer survivors. While a number of mechanisms are hypothesized to underlie the development of PIPN,10 the mitotoxicity hypothesis proposes that damage to primary afferent sensory neurons leads to impairments in a variety of mitochondrial functions.76 None of the previously published candidate gene or genome-wide association studies identified any MD-related genes. In this study, we found support for the differential expression of genes and significant perturbations in a number of pathways associated with MD including oxidative stress, iron homeostasis, mitochondrial fission, and apoptosis and autophagy.10
Oxidative stress
Mitochondria play a vital regulatory role in cellular physiology. Within neurons, more than 90% of the mitochondria are localized in axons where they are necessary for energy generation.76,77 Paclitaxel opens the mitochondrial permeability transition pore, which is associated with increased generation of reactive oxygen species (ROS).76 This type of oxidative stress leads to structural and functional damage to mitochondria. In animal studies, the administration of paclitaxel was associated with the appearance of swollen and vacuolated mitochondria in peripheral nerves and dorsal root ganglion (DRG) cells.12,78
In this study, we found differentially expressed genes (i.e., peroxiredoxin 5 (PRDX5), glutaredoxin 5 (GLRX5)) and perturbed pathways (i.e., HIF-1 signaling pathway,79 PI3K-Akt signaling pathway,80 Peroxisome81) that are regulated or mediated by oxidative stress. Several lines of preclinical evidence support our clinical findings.
While not specifically evaluated in PIPN, in murine models of peripheral nerve injury, the generation of ROS is upregulated, and antioxidant pathways are downregulated or functionally impaired.82,83 This redox imbalance is enhanced by mitochondrial damage.84 However, most neurons survive this imbalance, which suggests that endogenous defense systems are activated to restore homeostasis. Peroxiredoxin, glutaredoxins, and thioredoxins are part of this endogenous defense system that remove ROS and mediate the response to the redox stress.85 In one study that evaluated changes in mRNA in DRG cell bodies and axons after unilateral sciatic nerve injury (SNI),86 enriched mRNAs in the axonal compartment on the side of the SNI included mitochondrial genes as well as genes of the peroxidase family including Prdx5 (the rat homolog of human PDRX5, which is downregulated in our survivors with PIPN in this study).
In another study that evaluated the effects of thioredoxin-fold proteins in an SNI model,87 while both glutaredoxin (i.e., Glrx5) and peroxiredotoxins (i.e., Prdx4, Prdx5) were found in the DRG, only Prdx4 and Prdx5 were upregulated following the SNI. Of note, Prdx5 upregulation was associated with an increase in respective mRNA and protein accumulation in peripheral fibers proximal to the injury. This finding is consistent with an accumulation of mitochondria as a result of blocked axonal transport.87 In addition, this upregulation of Prdx5 was dependent on the presence of endogenous HIF-1 (a perturbed pathway in our survivors with PIPN), a global regulator of oxygen homeostasis that facilitates oxygen delivery and adaptation to oxygen deprivation.88,89 The authors hypothesized that failure of Prdx5 upregulation increases the risk for chronic neuropathy.87 Interestingly, differences in PDRX5 GE were observed in sural nerves from patients with progressing compared to nonprogressing diabetic neuropathy.90
Iron homeostasis
Perturbation in the ferroptosis pathway and the decreased expression of GLRX5 was found in our cancer survivors with PIPN. Ferroptosis is a regulated cell death driven by lipid ROS91 and is characterized by abnormal mitochondria morphology.92 Iron is an important cofactor in metabolically active cells like neurons. A carefully controlled supply of iron is needed for mitochondrial function, axonal transport, and myelination.93–95 While glutaredoxins (e.g., GLRX5) are involved in redox reactions,85 they are needed for iron homeostatsis.96,97 A growing body of evidence suggests that alterations in mitochondrial iron metabolism are involved in a number of neurological disorders (e.g., Parkinson’s disease) as well as in motor and cognitive decline associated with aging.98 In addition, the products of iron-regulatory and iron-transport-pathway genes may play a role in the development of diabetic99,100 and HIV neuropathy.101
While no preclinical studies have identified an association between CIPN and ferroptosis, an interesting finding that warrants additional investigation is related to the mitochondrial iron-binding protein called frataxin (FXN). This protein is critical for mitochondrial iron metabolism, overall cellular iron homeostasis, and antioxidant protection.102–104 In an in vitro study of CIPN,105 the administration of alpha-lipoic acid increased the expression of frataxin in vehicle-treated DRG cells as well as in DRG cells treated with paclitaxel or cisplatin. The authors concluded FXN may play a key role in neuroprotective pathways.
Mitochondrial fission
Mitochondrial fission is the process in which mitochondria are divided into smaller units. Mitochondrial fission enhances the number of mitochondria in neural cells and helps to maintain their energy demands.77,106 Fission mitochondrial 1 (FIS1, which was downregulated in survivors with PIPN) is a primary mediator of mitochondrial fission and has been implicated in MD and autophagy.107,108 FIS1 recruits dynamin-related protein 1 (DLRP1/Drp1) from the cytosol, 109 which is the primary executor of fission.106 Defects in mitochondrial fission are implicated in a number of neurodegenerative disorders (e.g., hereditary optic neuropathy, Parkinson’s disease).106,110,111
Evidence from preclinical studies supports the association found in our cancer survivors. In a study that evaluated whether Drp1 catalyzed the process of mitochondrial fission, Sprague Dawley rats were treated with 2′,3′-dideoxycitidine (ddC) or oxaliplatin.112 In this study, intrathecal administration of an oligodeoxynucleotide antisense against Drp1 resulted in a decrease in its expression in peripheral nerves and markedly attenuated neuropathic hyperalgesia associated with both drugs. In a more recent study that evaluated the role of Drp1 in neuropathic pain induced by perineural human immunodeficiency virus gp120,113 intrathecal administration of an oligodeoxynucleotide antisense against Drp1 decreased mechanical allodynia and decreased the spinal expression of increased Drp1 protein induced by gp120.
While not studied in terms of neuropathic pain, the interconnection between mitochondria and the peroxisome (a perturbed pathway in our cancer survivors) is an area of intense investigation.81,114,115 Peroxisomes represent a class of ubiquitous and dynamic single membrane bound organelles in eukaryotic cells.114 Substantial evidence suggests that peroxisomes and mitochondria have a close functional interplay. In particular, peroxisomes and mitochondria contribute to ROS homeostasis and share a redox-sensitive relationship.114 It is plausible that this pathway could be involved in PIPN.
Apoptosis and autophagy
Several signaling pathways involved in mitochondrial apoptosis and autophagy (i.e., PI3K-Akt, mTOR, FoxO, and AMPK) were perturbed in this study. In terms of the PI3K/Akt/mTOR pathways, several lines of evidence suggest that they are involved in the development of neuropathic pain.116 For example, in a rat model of chronic constriction injury (CCI),117 compared to control and sham animals, increased mRNA expression and protein levels were observed for PI3K, Akt, and mTOR in the CCI animals. In a more recent study that evaluated for changes in PI3K/Akt signaling in spinal cord slice preparations following CCI,118 PI3K/Akt signaling increased in the L4-L6 spinal cord sections. The authors concluded that PI3K/Akt signaling is required for central sensitization in the CCI neuropathic pain model.
The FoxO signaling pathway, that is regulated by the PI3K, Akt, and mTOR signaling pathways,119 is a potential target for a number of neurodegenerative disorders 120 including diabetes.121 In a recent study that evaluated for changes in genes and pathways associated with neuropathic pain using a bioinformatics approach,122 upregulated genes in the FoxO signaling pathway were associated with spinal nerve ligation.
Recent evidence suggests that AMPK may be a potential target for the treatment of chronic pain.123–125 AMPK and peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α) signaling axes sense the mitochondrial demands of the cell and regulate mitochondrial function.126 In a recent preclinical study,127 phosphorylation and expression of AMPK/PGC-1α and mitochondrial chain complex proteins were downregulated in the DRG of streptozotocin-diabetic rats. In addition, the administration of resveratrol (i.e., a polyphenol that augments APMK phosphorylation and axonal growth128) was associated with the reversal of thermal hypoalgesia, attenuation of foot skin intradermal fiber loss and a reduction in myelinated fiber mean axonal caliber in streptozotocin-diabetic rats. The authors concluded that the development of neuropathy was linked to nutrient excess and MD through defective signaling of the AMPK/PGC-1α pathway. In terms of CIPN, the coadministration of metformin (i.e., a widely used antidiabetic drug that activates AMPK129) with cisplatin or paclitaxel prevented the occurrence of mechanical allodynia.130 In addition, metformin prevented the cisplatin-induced increase in the latency to detect an adhesive patch (a measure of sensory deficits) and the reduction in the density of intradermal nerve fibers in the paw.
Perturbations in the p53 signaling pathway were identified in our cancer survivors with PIPN. This pathway regulates a complex transcriptional program involved in a variety of biological processes, including cell cycle arrest and apoptosis.131 Mitochondrial translocation of the p53 molecule is associated with MD in a number of neurological disorders.132,133 In terms of CIPN, cisplatin treatment rapidly increased mitochondrial levels of p53 in DRG neurons as well as reduced mitochondrial membrane potential and disrupted mitochondrial integrity and function.134 In addition, the administration of the mitochondrial protectant pifithrin-μ (PFT-μ; which prevents the mitochondrial accumulation of p53) prevented both paclitaxel- and cisplatin-induced mechanical allodynia and sensory loss.135 Equally important, in other studies, the administration of PFT-μ protected against loss of mitochondrial membrane potential, abnormalities in mitochondrial morphology, and functional mitochondrial deficiencies in DRG neurons.12,134,135
In the current study, perturbations in two-related pathways (i.e., mitophagy and HIF-1 signaling) and decreased expression of a related gene (i.e., ubiquitin) were identified in survivors with PIPN. Ubiquitination of mitochondrial proteins, including HIF-1A transcription factor,136 regulates many aspects of mitochondrial homeostasis including mitophagy.137 Mitophagy, a selective form of autophagy, mediates the selective removal of mitochondria138 and plays an essential role in mitochondrial homeostatis.139 While no studies were identified that found associations between CIPN and ubiquitination or mitophagy, alterations in these two processes are associated with neurodegeneration.140
Limitations
Several limitations warrant consideration. While our sample size was relatively small, we had an extremely well-characterized sample of BC survivors with and without PIPN. Of note, no differences were found in the total cumulative dose of paclitaxel that these survivors received. Given that one cannot test for differences in RNA in DRG neurons from living patients, as have others evaluating for GE differences in patients with painful neuropathies,141,142 we have evaluated for differences in RNA expression from peripheral blood. Because our molecular analyses were done using blood samples, we can only infer that the perturbations identified are consistent with changes within the peripheral nervous system. Our findings warrant confirmation in an independent sample. However, to decrease the possibility of spurious associations, we sequenced RNA to a considerable depth, applied best practices for the RNA-seq analyses, followed a strict QC procedure, and evaluated for significance with a severely stringent FDR.
Future research
This study is the first to provide molecular evidence that a number of mitochondrial mechanisms identified in preclinical models of various types of neuropathic pain, including CIPN,12,74,75 are found in cancer survivors with persistent PIPN. The limitations of the analyses of RNA from blood and nonneural tissue notwithstanding, we did observe differential profiles of expression and perturbation in these processes, which suggest persistent damage and/or changes in the regulation of mitochondria. Future studies need to evaluate for differences in epigenetic changes (i.e., methylation, microRNA) between survivors with and without PIPN, which may reflect changes in regulation patterns. In addition, our findings suggest that chronic PIPN involves multiple MD-related mechanisms. Therefore, studies are warranted that evaluate for common and distinct MD-related mechanisms associated with other neurotoxic chemotherapy drugs (e.g., platinum, platinum, and taxane combination). It must be acknowledged that a cross-sectional study cannot demonstrate causality. Longitudinal studies are needed that evaluate for changes over time in the molecular mechanisms associated with CIPN. These longitudinal studies need to include an in-depth characterization of the CIPN phenotype as well as pretreatment and multiple posttreatment assessments of a number of molecular markers (i.e., genes, GE, epigenetics). Translation of these findings, if validated, may be accelerated by investigating drugs that target mitochondrial function to be repurposed to treat CIPN.143 The use of sophisticated bioinformatics approaches and network analyses may allow us to determine the timing of the disruptions in mitochondrial function associated with neurotoxic chemotherapy so that interventions can be initiated to prevent this dose-limiting and devastating adverse effect.
Supplemental Material
Supplemental Material for Expression of mitochondrial dysfunction-related genes and pathways in paclitaxel-induced peripheral neuropathy in breast cancer survivors by Kord M Kober, Adam Olshen, Yvettte P Conley, Mark Schumacher, Kimberly Topp, Betty Smoot, Melissa Mazor, Margaret Chesney, Marilyn Hammer, Steven M Paul, Jon D Levine and Christine Miaskowski in Molecular Pain
Acknowledgments
The contents of this study are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health. Recruitment was facilitated by Dr. Susan Love Research Foundation’s Army of Women® Program. Anatol Sucher provided storage and processing of the biospecimens.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was funded by the National Cancer Institute (NCI; CA151692) and the American Cancer Society (ACS, IRG-97–150-13). Dr. Miaskowski is supported by grants from the ACS and NCI (CA168960). This project was supported by the National Center for Advancing Translational Sciences, National Institutes of Health, through UCSF-CTSI Grant Number UL1 TR000004.
Supplemental Material
Supplemental material is available for this article online.
References
- 1.Kudlowitz D, Muggia F. Defining risks of taxane neuropathy: insights from randomized clinical trials. Clin Cancer Res 2013; 19: 4570–4577. [DOI] [PubMed] [Google Scholar]
- 2.Jones SE, Erban J, Overmoyer B, Budd GT, Hutchins L, Lower E, Laufman L, Sundaram S, Urba WJ, Pritchard KI, Mennel R, Richards D, Olsen S, Meyers ML, Ravdin PM. Randomized phase III study of docetaxel compared with paclitaxel in metastatic breast cancer. J Clin Oncol 2005; 23: 5542–5551. [DOI] [PubMed] [Google Scholar]
- 3.Sarosy G, Kohn E, Stone DA, Rothenberg M, Jacob J, Adamo DO, Ognibene FP, Cunnion RE, Reed E. Phase I study of taxol and granulocyte colony-stimulating factor in patients with refractory ovarian cancer. J Clin Oncol 1992; 10: 1165–1170. [DOI] [PubMed] [Google Scholar]
- 4.Miaskowski C, Mastick J, Paul SM, Abrams G, Cheung S, Sabes JH, Kober KM, Schumacher M, Conley YP, Topp K, Smoot B, Mausisa G, Mazor M, Wallhagen M, Levine JD. Impact of chemotherapy-induced neurotoxicities on adult cancer survivors’ symptom burden and quality of life. J Cancer Surviv 2018; 12: 234–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eccles SA, Aboagye EO, Ali S, Anderson AS, Armes J, Berditchevski F, Blaydes JP, Brennan K, Brown NJ, Bryant HE, Bundred NJ, Burchell JM, Campbell AM, Carroll JS, Clarke RB, Coles CE, Cook GJR, Cox A, Curtin NJ, Dekker LV, Silva IdS, Duffy SW, Easton DF, Eccles DM, Edwards DR, Edwards J, Evans D, Fenlon DF, Flanagan JM, Foster C, Gallagher WM, Garcia-Closas M, Gee JMW, Gescher AJ, Goh V, Groves AM, Harvey AJ, Harvie M, Hennessy BT, Hiscox S, Holen I, Howell SJ, Howell A, Hubbard G, Hulbert-Williams N, Hunter MS, Jasani B, Jones LJ, Key TJ, Kirwan CC, Kong A, Kunkler IH, Langdon SP, Leach MO, Mann DJ, Marshall JF, Martin L, Martin SG, Macdougall JE, Miles DW, Miller WR, Morris JR, Moss SM, Mullan P, Natrajan R, O'Connor JPB, O'Connor R, Palmieri C, Pharoah PDP, Rakha EA, Reed E, Robinson SP, Sahai E, Saxton JM, Schmid P, Smalley MJ, Speirs V, Stein R, Stingl J, Streuli CH, Tutt ANJ, Velikova G, Walker RA, Watson CJ, Williams KJ, Young LS, Thompson AM. Critical research gaps and translational priorities for the successful prevention and treatment of breast cancer. Breast Cancer Res 2013; 15: R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson A, Brennan K, Cox A, Gee J, Harcourt D, Harris A, Harvie M, Holen I, Howell A, Nicholson R, Steel M, Streuli C. Evaluation of the current knowledge limitations in breast cancer research: a gap analysis. Breast Cancer Res 2008; 10: R26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rowinsky EK, Cazenave LA, Donehower RC. Taxol: a novel investigational antimicrotubule agent. J Natl Cancer Inst 1990; 82: 1247–1259. [DOI] [PubMed] [Google Scholar]
- 8.Bobylev I, Joshi AR, Barham M, Ritter C, Neiss WF, Hoke A, Lehmann HC. Paclitaxel inhibits mRNA transport in axons. Neurobiol Dis 2015; 82: 321–331. [DOI] [PubMed] [Google Scholar]
- 9.Fukuda Y, Li Y, Segal RA. A mechanistic understanding of axon degeneration in chemotherapy-induced peripheral neuropathy. Front Neurosci 2017; 11: 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Starobova H, Vetter I. Pathophysiology of chemotherapy-induced peripheral neuropathy. Front Mol Neurosci 2017; 10: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miltenburg NC, Boogerd W. Chemotherapy-induced neuropathy: a comprehensive survey. Cancer Treat Rev 2014; 40: 872–882. [DOI] [PubMed] [Google Scholar]
- 12.Ma J, Kavelaars A, Dougherty PM, Heijnen CJ. Beyond symptomatic relief for chemotherapy-induced peripheral neuropathy: targeting the source. Cancer 2018; 124: 2289–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cliff J, Jorgensen AL, Lord R, Azam F, Cossar L, Carr DF, Pirmohamed M. The molecular genetics of chemotherapy-induced peripheral neuropathy: a systematic review and meta-analysis. Crit Rev Oncol Hematol 2017; 120: 127–140. [DOI] [PubMed] [Google Scholar]
- 14.Miaskowski C, Mastick J, Paul SM, Topp K, Smoot B, Abrams G, Chen LM, Kober KM, Conley YP, Chesney M, Bolla K, Mausisa G, Mazor M, Wong M, Schumacher M, Levine JD. Chemotherapy-induced neuropathy in cancer survivors. J Pain Symptom Manage 2017; 54: 204–218 e202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark EJ. Teamwork—the cancer patient’s guide to talking with your doctor. 5th ed Silver Springs: National Coalition for Cancer Survivorship, 2011. [Google Scholar]
- 16.Bohn MJ, Babor TF, Kranzler HR. The Alcohol Use Disorders Identification Test (AUDIT): validation of a screening instrument for use in medical settings. J Stud Alcohol 1995; 56: 423–432. [DOI] [PubMed] [Google Scholar]
- 17.Karnofsky D. Performance scale. New York: Plenum Press, 1977. [Google Scholar]
- 18.Karnofsky D, Abelmann WH, Craver LV, Burchenal JH. The use of nitrogen mustards in the palliative treatment of carcinoma. Cancer 1948; 1: 634–656. [Google Scholar]
- 19.Schnadig ID, Fromme EK, Loprinzi CL, Sloan JA, Mori M, Li H, Beer TM. Patient-physician disagreement regarding performance status is associated with worse survivorship in patients with advanced cancer. Cancer 2008; 113: 2205–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brunner F, Bachmann LM, Weber U, Kessels AG, Perez RS, Marinus J, Kissling R. Complex regional pain syndrome 1—the Swiss cohort study. BMC Musculoskelet Disord 2008; 9: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cieza A, Geyh S, Chatterji S, Kostanjsek N, Ustun BT, Stucki G. Identification of candidate categories of the International Classification of Functioning Disability and Health (ICF) for a Generic ICF Core Set based on regression modelling. BMC Med Res Methodol 2006; 6: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daut RL, Cleeland CS, Flanery RC. Development of the Wisconsin Brief Pain Questionnaire to assess pain in cancer and other diseases. Pain 1983; 17: 197–210. [DOI] [PubMed] [Google Scholar]
- 23.Jensen MP, Gammaitoni AR, Olaleye DO, Oleka N, Nalamachu SR, Galer BS. The Pain Quality Assessment Scale: assessment of pain quality in carpal tunnel syndrome. J Pain 2006; 7: 823–832. [DOI] [PubMed] [Google Scholar]
- 24.Victor TW, Jensen MP, Gammaitoni AR, Gould EM, White RE, Galer BS. The dimensions of pain quality: factor analysis of the Pain Quality Assessment Scale. Clin J Pain 2008; 24: 550–555. [DOI] [PubMed] [Google Scholar]
- 25.Bell-Krotoski JA. Sensibility testing with Semmes-Weinstein monofilaments In: Hunter JM, Mackin EJ, Callahan ED. (eds) Rehabilitation of the hand and upper extremity. 5th ed St. Louis: Mosby, Inc, 2002. [Google Scholar]
- 26.Viswanathan V, Snehalatha C, Seena R, Ramachandran A. Early recognition of diabetic neuropathy: evaluation of a simple outpatient procedure using thermal perception. Postgrad Med J 2002; 78: 541–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papanas N, Ziegler D. New diagnostic tests for diabetic distal symmetric polyneuropathy. J Diabetes Complications 2011; 25: 44–51. [DOI] [PubMed] [Google Scholar]
- 28.Duke J, McEvoy M, Sibbritt D, Guest M, Smith W, Attia J. Vibrotactile threshold measurement for detecting peripheral neuropathy: defining variability and a normal range for clinical and research use. Diabetologia 2007; 50: 2305–2312. [DOI] [PubMed] [Google Scholar]
- 29.Tofthagen CS, McMillan SC, Kip KE. Development and psychometric evaluation of the chemotherapy-induced peripheral neuropathy assessment tool. Cancer Nurs 2011; 34: E10–E20. [DOI] [PubMed] [Google Scholar]
- 30.Mathias S, Nayak US, Isaacs B. Balance in elderly patients: the “get-up and go” test. Arch Phys Med Rehabil 1986; 67: 387–389. [PubMed] [Google Scholar]
- 31.Hernandez D, Rose DJ. Predicting which older adults will or will not fall using the Fullerton Advanced Balance Scale. Arch Phys Med Rehabil 2008; 89: 2309–2315. [DOI] [PubMed] [Google Scholar]
- 32.Rose DJ, Lucchese N, Wiersma LD. Development of a multidimensional balance scale for use with functionally independent older adults. Arch Phys Med Rehabil 2006; 87: 1478–1485. [DOI] [PubMed] [Google Scholar]
- 33.SPSS. IBM SPSS for windows (version 23 ) Armonk: SPSS, Inc, 2015. [Google Scholar]
- 34.Flentje A, Kober KM, Carrico AW, Neilands TB, Flowers E, Heck NC, Aouizerat BE. Minority stress and leukocyte gene expression in sexual minority men living with treated HIV infection. Brain Behav Immun 2018; 70: 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Neil D, Glowatz H, Schlumpberger M. Ribosomal RNA depletion for efficient use of RNA-seq capacity. Curr Protoc Mol Biol 2013; 103: 4–19. [DOI] [PubMed] [Google Scholar]
- 36.Mastrokolias A, den Dunnen JT, van Ommen GB, AC't Hoen PA, van Roon-Mom WM. Increased sensitivity of next generation sequencing-based expression profiling after globin reduction in human blood RNA. BMC Genomics 2012; 13: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi I, Bao H, Kommadath A, Hosseini A, Sun X, Meng Y, Stothard P, Plastow GS, Tuggle CK, Reecy JM, Fritz-Waters E, Abrams SM, Lunney JK, Guan Le L. Increasing gene discovery and coverage using RNA-seq of globin RNA reduced porcine blood samples. BMC Genomics 2014; 15: 954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sinha R, Stanley G, Gulati GS, Ezran C, Travaglini KJ, Wei E, Chan CKF, Nabhan AN, Su T, Morganti RM, Conley SD, Chaib H, Red-Horse K, Longaker MT, Snyder MP, Krasnow MA, Weissman IL. Index switching causes “spreading-of-signal” among multiplexed samples in Illumina HiSeq 4000 DNA sequencing bioRxiv. Epub ahead of print 9 April 2017. DOI: 10.1101/125724. [Google Scholar]
- 39.Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Szczesniak MW, Gaffney DJ, Elo LL, Zhang X, Mortazavi A. A survey of best practices for RNA-seq data analysis. Genome Biol 2016; 17: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kukurba KR, Montgomery SB. RNA sequencing and analysis. Cold Spring Harb Protoc 2015; 2015: 951–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carrico AW, Flentje A, Kober K, Lee S, Hunt P, Riley ED, Shoptaw S, Flowers RNE, Dilworth SE, Pahwa S, Aouizerat BE. Recent stimulant use and leukocyte gene expression in methamphetamine users with treated HIV infection. Brain Behav Immun 2018; 71: 108–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.FASTQC—A quality control tool for high throughput sequence data, www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2018, accessed 19 November 2018).
- 44.Ewels P, Magnusson M, Lundin S, Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016; 32: 3047–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigo R, Hubbard TJ. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 2012; 22: 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeLuca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire MD, Williams C, Reich M, Winckler W, Getz G. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 2012; 28: 1530–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014; 30: 923–930. [DOI] [PubMed] [Google Scholar]
- 49.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Giron CG, Gil L, Gordon L, Haggerty L, Haskell E, Hourlier T, Izuogu OG, Janacek SH, Juettemann T, To JK, Laird MR, Lavidas I, Liu Z, Loveland JE, Maurel T, McLaren W, Moore B, Mudge J, Murphy DN, Newman V, Nuhn M, Ogeh D, Ong CK, Parker A, Patricio M, Riat HS, Schuilenburg H, Sheppard D, Sparrow H, Taylor K, Thormann A, Vullo A, Walts B, Zadissa A, Frankish A, Hunt SE, Kostadima M, Langridge N, Martin FJ, Muffato M, Perry E, Ruffier M, Staines DM, Trevanion SJ, Aken BL, Cunningham F, Yates A, Flicek P. Ensembl 2018. Nucleic Acids Res 2018; 46: D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res 2011; 39: D52–D57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 2010; 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet 2006; 7: 55–65. [DOI] [PubMed] [Google Scholar]
- 54.Fang Z, Cui X. Design and validation issues in RNA-seq experiments. Brief Bioinform 2011; 12: 280–287. [DOI] [PubMed] [Google Scholar]
- 55.McIntyre LM, Lopiano KK, Morse AM, Amin V, Oberg AL, Young LJ, Nuzhdin SV. RNA-seq: technical variability and sampling. BMC Genomics 2011; 12: 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet 2007; 3: 1724–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Landau WM, Liu P. Dispersion estimation and its effect on test performance in RNA-seq data analysis: a simulation-based comparison of methods. PLoS One 2013; 8: e81415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soneson C, Delorenzi M. A comparison of methods for differential expression analysis of RNA-seq data. BMC Bioinformatics 2013; 14: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rapaport F, Khanin R, Liang Y, Pirun M, Krek A, Zumbo P, Mason CE, Socci ND, Betel D. Comprehensive evaluation of differential gene expression analysis methods for RNA-seq data. Genome Biol 2013; 14: R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, Robinson MD. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc 2013; 8: 1765–1786. [DOI] [PubMed] [Google Scholar]
- 61.Love MI, Anders S, Kim V, Huber W. RNA-seq workflow: gene-level exploratory analysis and differential expression. F1000Res 2015; 4: 1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res 2012; 40: 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cox D, Reid N. Parameter orthogonality and approximate conditional inference. J R Stat Soc Ser B Method 1987; 49: 1–39. [Google Scholar]
- 64.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med 1990; 9: 811–818. [DOI] [PubMed] [Google Scholar]
- 65.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Method 1995; 57: 289–300. [Google Scholar]
- 66.Khatri P, Sirota M, Butte AJ. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol 2012; 8: e1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garcia-Campos MA, Espinal-Enriquez J, Hernandez-Lemus E. Pathway analysis: state of the art. Front Physiol 2015; 6: 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nguyen T, Mitrea C, Draghici S. Network-based approaches for pathway level analysis. Curr Protoc Bioinformatics 2018; 61: 8–25. [DOI] [PubMed] [Google Scholar]
- 69.Mitrea C, Taghavi Z, Bokanizad B, Hanoudi S, Tagett R, Donato M, Voichita C, Draghici S. Methods and approaches in the topology-based analysis of biological pathways. Front Physiol 2013; 4: 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero R. A systems biology approach for pathway level analysis. Genome Res 2007; 17: 1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aoki-Kinoshita KF, Kanehisa M. Gene annotation and pathway mapping in KEGG. Methods Mol Biol 2007; 396: 71–91. [DOI] [PubMed] [Google Scholar]
- 72.Gray KA, Daugherty LC, Gordon SM, Seal RL, Wright MW, Bruford EA. Genenames.org: the HGNC resources in 2013. Nucleic Acids Res 2012; 41: D545–D552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hart SN, Therneau TM, Zhang Y, Poland GA, Kocher JP. Calculating sample size estimates for RNA sequencing data. J Comput Biol 2013; 20: 970–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Waseem M, Kaushik P, Tabassum H, Parvez S. Role of mitochondrial mechanism in chemotherapy-induced peripheral neuropathy. Curr Drug Metab. 2018; 19: 47–54. [DOI] [PubMed] [Google Scholar]
- 75.Flatters SJL, Dougherty PM, Colvin LA. Clinical and preclinical perspectives on chemotherapy-induced peripheral neuropathy (CIPN): a narrative review. Br J Anaesth 2017; 119: 737–749. [DOI] [PubMed] [Google Scholar]
- 76.Canta A, Pozzi E, Carozzi VA. Mitochondrial dysfunction in chemotherapy-induced peripheral neuropathy (CIPN). Toxics 2015; 3: 198–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Areti A, Yerra VG, Komirishetty P, Kumar A. Potential therapeutic benefits of maintaining mitochondrial health in peripheral neuropathies. Curr Neuropharmacol 2016; 14: 593–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Flatters SJ, Bennett GJ. Studies of peripheral sensory nerves in paclitaxel-induced painful peripheral neuropathy: evidence for mitochondrial dysfunction. Pain 2006; 122: 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zepeda AB, Pessoa A, Jr, Castillo RL, Figueroa CA, Pulgar VM, Farias JG. Cellular and molecular mechanisms in the hypoxic tissue: role of HIF-1 and ROS. Cell Biochem Funct 2013; 31: 451–459. [DOI] [PubMed] [Google Scholar]
- 80.Movafagh S, Crook S, Vo K. Regulation of hypoxia-inducible factor-1a by reactive oxygen species: new developments in an old debate. J Cell Biochem 2015; 116: 696–703. [DOI] [PubMed] [Google Scholar]
- 81.Fransen M, Lismont C, Walton P. The peroxisome-mitochondria connection: how and why? Int J Mol Sci 2017; 18: 1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schwartz ES, Lee I, Chung K, Chung JM. Oxidative stress in the spinal cord is an important contributor in capsaicin-induced mechanical secondary hyperalgesia in mice. Pain 2008; 138: 514–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim HK, Park SK, Zhou JL, Taglialatela G, Chung K, Coggeshall RE, Chung JM. Reactive oxygen species (ROS) play an important role in a rat model of neuropathic pain. Pain 2004; 111: 116–124. [DOI] [PubMed] [Google Scholar]
- 84.Chen KH, Lin CR, Cheng JT, Cheng JK, Liao WT, Yang CH. Altered mitochondrial ATP synthase expression in the rat dorsal root ganglion after sciatic nerve injury and analgesic effects of intrathecal ATP. Cell Mol Neurobiol 2014; 34: 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hanschmann EM, Godoy JR, Berndt C, Hudemann C, Lillig CH. Thioredoxins, glutaredoxins, and peroxiredoxins–molecular mechanisms and health significance: from cofactors to antioxidants to redox signaling. Antioxid Redox Signal 2013; 19: 1539–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hirai T, Mulpuri Y, Cheng Y, Xia Z, Li W, Ruangsri S, Spigelman I, Nishimura I. Aberrant plasticity of peripheral sensory axons in a painful neuropathy. Sci Rep 2017; 7: 3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Valek L, Kanngießer M, Häussler A, Agarwal N, Lillig C H, Tegeder I. Redoxins in peripheral neurons after sciatic nerve injury. Free Radic Biol Med 2015; 89: 581–592. [DOI] [PubMed] [Google Scholar]
- 88.Semenza GL. Oxygen homeostasis. Wiley Interdiscip Rev Syst Biol Med 2010; 2: 336–361. [DOI] [PubMed] [Google Scholar]
- 89.Semenza GL. Hypoxia-inducible factor 1: regulator of mitochondrial metabolism and mediator of ischemic preconditioning. Biochim Biophys Acta 2011; 1813: 1263–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hur J, Sullivan KA, Pande M, Hong Y, Sima AA, Jagadish HV, Kretzler M, Feldman EL. The identification of gene expression profiles associated with progression of human diabetic neuropathy. Brain 2011; 134: 3222–3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascon S, Hatzios SK, Kagan VE, Noel K, Jiang X, Linkermann A, Murphy ME, Overholtzer M, Oyagi A, Pagnussat GC, Park J, Ran Q, Rosenfeld CS, Salnikow K, Tang D, Torti FM, Torti SV, Toyokuni S, Woerpel KA, Zhang DD. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell 2017; 171: 273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xie Y, Hou W, Song X, Yu Y, Huang J, Sun X, Kang R, Tang D. Ferroptosis: process and function. Cell Death Differ 2016; 23: 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Galy B, Ferring-Appel D, Sauer S W, Kaden S, Lyoumi S, Puy H, Kölker S, Gröne H-J, Hentze MW. Iron regulatory proteins secure mitochondrial iron sufficiency and function. Cell Metab 2010; 12: 194–201. [DOI] [PubMed] [Google Scholar]
- 94.Millecamps S, Julien JP. Axonal transport deficits and neurodegenerative diseases. Nat Rev Neurosci 2013; 14: 161–176. [DOI] [PubMed] [Google Scholar]
- 95.Sheftel AD, Lill R. The power plant of the cell is also a smithy: the emerging role of mitochondria in cellular iron homeostasis. Ann Med 2009; 41: 82–99. [DOI] [PubMed] [Google Scholar]
- 96.Rouhier N, Couturier J, Johnson MK, Jacquot JP. Glutaredoxins: roles in iron homeostasis. Trends Biochem Sci 2010; 35: 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Berndt C, Lillig CH. Glutathione, Glutaredoxins, and Iron. Antioxid Redox Signal. 2017; 27: 1235–1251. DOI: 10.1089/ars.2017.7132. [DOI] [PubMed] [Google Scholar]
- 98.Isaya G. Mitochondrial iron-sulfur cluster dysfunction in neurodegenerative disease. Front Pharmacol 2014; 5: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Levi S, Taveggia C. Iron homeostasis in peripheral nervous system, still a black box? Antioxid Redox Signal 2014; 21: 634–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhao S, Zhang L, Xu Z, Chen W. Neurotoxic effects of iron overload under high glucose concentration. Neural Regen Res 2013; 8: 3423–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kallianpur AR, Jia P, Ellis RJ, Zhao Z, Bloss C, Wen W, Marra CM, Hulgan T, Simpson DM, Morgello S, McArthur JC, Clifford DB, Collier AC, Gelman BB, McCutchan JA, Franklin D, Samuels DC, Rosario D, Holzinger E, Murdock DG, Letendre S, Grant I, Group CS. Genetic variation in iron metabolism is associated with neuropathic pain and pain severity in HIV-infected patients on antiretroviral therapy. PLoS One 2014; 9: e103123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Braymer JJ, Lill R. Iron-sulfur cluster biogenesis and trafficking in mitochondria. J Biol Chem 2017; 292: 12754–12763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chiang S Kovacevic Z Sahni S Lane DJ Merlot AM Kalinowski DS Huang ML, andRichardson DR.. Frataxin and the molecular mechanism of mitochondrial iron-loading in Friedreich’s ataxia. Clin Sci (Lond) 2016; 130: 853–870. [DOI] [PubMed] [Google Scholar]
- 104.Vanlander AV, Van Coster R. Clinical and genetic aspects of defects in the mitochondrial iron-sulfur cluster synthesis pathway. J Biol Inorg Chem 2018; 23: 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Melli G, Taiana M, Camozzi F, Triolo D, Podini P, Quattrini A, Taroni F, Lauria G. Alpha-lipoic acid prevents mitochondrial damage and neurotoxicity in experimental chemotherapy neuropathy. Exp Neurol 2008; 214: 276–284. [DOI] [PubMed] [Google Scholar]
- 106.Serasinghe MN, Chipuk JE. Mitochondrial fission in human diseases. Handb Exp Pharmacol 2017; 240: 159–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gomes LC, Scorrano L. High levels of Fis1, a pro-fission mitochondrial protein, trigger autophagy. Biochim Biophys Acta 2008; 1777: 860–866. [DOI] [PubMed] [Google Scholar]
- 108.Twig G, Shirihai OS. The interplay between mitochondrial dynamics and mitophagy. Antioxid Redox Signal 2011; 14: 1939–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yoon Y, Krueger EW, Oswald BJ, McNiven MA. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol 2003; 23: 5409–5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Frank S. Dysregulation of mitochondrial fusion and fission: an emerging concept in neurodegeneration. Acta Neuropathol 2006; 111: 93–100. [DOI] [PubMed] [Google Scholar]
- 111.Itoh K, Nakamura K, Iijima M, Sesaki H. Mitochondrial dynamics in neurodegeneration. Trends Cell Biol 2013; 23: 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ferrari LF, Chum A, Bogen O, Reichling DB, Levine JD. Role of Drp1, a key mitochondrial fission protein, in neuropathic pain. J Neurosci 2011; 31: 11404–11410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kanda H, Liu S, Iida T, Yi H, Huang W, Levitt RC, Lubarsky DA, Candiotti KA, Hao S. Inhibition of mitochondrial fission protein reduced mechanical allodynia and suppressed spinal mitochondrial superoxide induced by perineural human immunodeficiency virus gp120 in rats. Anesth Analg 2016; 122: 264–272. [DOI] [PubMed] [Google Scholar]
- 114.Schrader M, Costello J, Godinho LF, Islinger M. Peroxisome-mitochondria interplay and disease. J Inherit Metab Dis 2015; 38: 681–702. [DOI] [PubMed] [Google Scholar]
- 115.Pascual-Ahuir A, Manzanares-Estreder S, Proft M. Pro- and antioxidant functions of the peroxisome-mitochondria connection and its impact on aging and disease. Oxid Med Cell Longev 2017; 2017: 9860841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen SP, Zhou YQ, Liu DQ, Zhang W, Manyande A, Guan XH, Tian YK, Ye DW, Omar DM. PI3K/Akt pathway: a potential therapeutic target for chronic pain. Curr Pharm Des 2017; 23: 1860–1868. [DOI] [PubMed] [Google Scholar]
- 117.Guo JR, Wang H, Jin XJ, Jia DL, Zhou X, Tao Q. Effect and mechanism of inhibition of PI3K/Akt/mTOR signal pathway on chronic neuropathic pain and spinal microglia in a rat model of chronic constriction injury. Oncotarget 2017; 8: 52923–52934. DOI: 10.18632/oncotarget.17629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu W, Lv Y, Ren F. PI3K/Akt pathway is required for spinal central sensitization in neuropathic pain. Cell Mol Neurobiol 2018; 38: 747–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT. The PI3K pathway in human disease. Cell 2017; 170: 605–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Maiese K. FoxO proteins in the nervous system. Anal Cell Pathol (Amst) 2015; 2015: 569392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Maiese K. FoxO transcription factors and regenerative pathways in diabetes mellitus. Curr Neurovasc Res 2015; 12: 404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen CJ, Liu DZ, Yao WF, Gu Y, Huang F, Hei ZQ, Li X. Identification of key genes and pathways associated with neuropathic pain in uninjured dorsal root ganglion by using bioinformatic analysis. J Pain Res 10: 2665–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Asiedu MN, Dussor G, Price TJ. Targeting AMPK for the alleviation of pathological pain. EXS 2016; 107: 257–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Price TJ, Das V, Dussor G. Adenosine monophosphate-activated protein kinase (AMPK) activators for the prevention, treatment and potential reversal of pathological pain. Curr Drug Targets 2016; 17: 908–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Price TJ, Gold MS. . From mechanism to cure: renewing the goal to eliminate the disease of pain. Pain Med 2017; 19: 1525–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhao B, Qiang L, Joseph J, Kalyanaraman B, Viollet B, He YY. Mitochondrial dysfunction activates the AMPK signaling and autophagy to promote cell survival. Genes Dis 2016; 3: 82–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Roy Chowdhury SK, Smith DR, Saleh A, Schapansky J, Marquez A, Gomes S, Akude E, Morrow D, Calcutt NA, Fernyhough P. Impaired adenosine monophosphate-activated protein kinase signalling in dorsal root ganglia neurons is linked to mitochondrial dysfunction and peripheral neuropathy in diabetes. Brain 2012; 135: 1751–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dasgupta B, Milbrandt J. Resveratrol stimulates AMP kinase activity in neurons. Proc Natl Acad Sci U S A 2007; 104: 7217–7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bridgeman SC, Ellison GC, Melton PE, Newsholme P, Mamotte CDS. . Epigenetic effects of metformin: from molecular mechanisms to clinical implications. Diabetes Obes Metab 2018; 20: 1553–1562. [DOI] [PubMed] [Google Scholar]
- 130.Mao-Ying QL, Kavelaars A, Krukowski K, Huo XJ, Zhou W, Price TJ, Cleeland C, Heijnen CJ. The anti-diabetic drug metformin protects against chemotherapy-induced peripheral neuropathy in a mouse model. PLoS One 2014; 9: e100701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kastenhuber ER, Lowe SW. Putting p53 in context. Cell 2017; 170: 1062–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nahalkova J. The protein-interaction network with functional roles in tumorigenesis, neurodegeneration, and aging. Mol Cell Biochem 2016; 423: 187–196. [DOI] [PubMed] [Google Scholar]
- 133.Dai CQ, Luo TT, Luo SC, Wang JQ, Wang SM, Bai YH, Yang YL, Wang YY. p53 and mitochondrial dysfunction: novel insight of neurodegenerative diseases. J Bioenerg Biomembr 2016; 48: 337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Maj MA, Ma J, Krukowski KN, Kavelaars A, Heijnen CJ. Inhibition of mitochondrial p53 accumulation by PFT-μ prevents cisplatin-induced peripheral neuropathy. Front Mol Neurosci 2017; 10: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Krukowski K, Nijboer CH, Huo X, Kavelaars A, Heijnen CJ. Prevention of chemotherapy-induced peripheral neuropathy by the small-molecule inhibitor pifithrin-µ. Pain 2015; 156: 2184–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kallio PJ, Wilson WJ, O’Brien S, Makino Y, Poellinger L. Regulation of the hypoxia-inducible transcription factor 1alpha by the ubiquitin-proteasome pathway. J Biol Chem 1999; 274: 6519–6525. [DOI] [PubMed] [Google Scholar]
- 137.Yamano K, Matsuda N, Tanaka K. The ubiquitin signal and autophagy: an orchestrated dance leading to mitochondrial degradation. EMBO Rep 2016; 17: 300–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol 2011; 12: 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem 2008; 283: 10892–10903. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 140.Covill-Cooke C, Howden JH, Birsa N, Kittler JT. Ubiquitination at the mitochondria in neuronal health and disease. Neurochem Int 2018; 117: 55–64. [DOI] [PubMed] [Google Scholar]
- 141.Langjahr M, Schubert AL, Sommer C, Uceyler N. Increased pro-inflammatory cytokine gene expression in peripheral blood mononuclear cells of patients with polyneuropathies. J Neurol 2018; 265: 618–627. [DOI] [PubMed] [Google Scholar]
- 142.Uceyler N, Rogausch JP, Toyka KV, Sommer C. Differential expression of cytokines in painful and painless neuropathies. Neurology 2007; 69: 42–49. [DOI] [PubMed] [Google Scholar]
- 143.Nosengo N. Can you teach old drugs new tricks? Nature 2016; 534: 314–316. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material for Expression of mitochondrial dysfunction-related genes and pathways in paclitaxel-induced peripheral neuropathy in breast cancer survivors by Kord M Kober, Adam Olshen, Yvettte P Conley, Mark Schumacher, Kimberly Topp, Betty Smoot, Melissa Mazor, Margaret Chesney, Marilyn Hammer, Steven M Paul, Jon D Levine and Christine Miaskowski in Molecular Pain