Abstract
Spatial and temporal expression of genes is essential for maintaining phenotype integrity. Transcription factors (TFs) modulate expression patterns by binding to specific DNA sequences in the genome. Along with the core binding motif, the flanking sequence context can play a role in DNA–TF recognition. Here, we employ high-throughput in vitro and in silico analyses to understand the influence of sequences flanking the cognate sites in binding of three most prevalent eukaryotic TF families (zinc finger, homeodomain and bZIP). In vitro binding preferences of each TF toward the entire DNA sequence space were correlated with a wide range of DNA structural parameters, including DNA flexibility. Results demonstrate that conformational plasticity of flanking regions modulates binding affinity of certain TF families. DNA duplex stability and minor groove width also play an important role in DNA–TF recognition but differ in how exactly they influence the binding in each specific case. Our analyses further reveal that the structural features of preferred flanking sequences are not universal, as similar DNA-binding folds can employ distinct DNA recognition modes.
INTRODUCTION
Transcription factors (TFs) play a functional role in several vital physiological processes. TFs bind to cis-regulatory elements in DNA to control cellular responses and are generally classified based on the structure of their DNA binding domains (1). DNA–TF interactions are highly sequence-specific and the specificity is dependent on properties of both target DNA and TFs. It is important to bear in mind that the in vivo polymorph of DNA, the B-form, is a dynamically heterogeneous molecule, exploring a large conformational space (2–4). This conformational flexibility depends on sequence-dependent fluctuations in local helical parameters at dinucleotide steps (5–7). While DNA shape is determined by a combination of several structural parameters (4,6,8,9), variations in dinucleotide step parameters can capture variations in DNA shape to a large extent (10). Plasticity in DNA also plays a significant role in DNA–protein recognition, DNA melting, nucleosome assembly and genome integrity. Thus, intrinsic structural properties that define DNA bendability, duplex stability, curvature, groove shape and topography, are more accurate determinants of DNA binding specificities of TFs than the simple nucleotide sequence (10–20).
Recent studies have revealed that presence of an appropriate sequence is not sufficient to explain the high specificity of DNA–TF interaction, considering the large number of putative transcription factor binding sites (TFBSs) that are not bound by respective TFs. The role of the sequence environment of the TFBS is emerging to be an important determinant that confers additional specificity to DNA–TF recognition (21,22). Sequence context effects may vary from the immediate flanking bases of TFBSs to higher-order level (e.g. poly A-tracts in nucleosome positioning) (23). High-throughput DNA–protein binding assays have investigated the role of both proximal and distal flanking sequences of TFBSs in DNA binding events of TFs (21,24–32). In one such study, global analysis of 151 human full-length TFs and 303 DNA binding domains revealed that additional specificity is achieved with A- or T-stretches that flank the core motifs (26). A similar study on core-binding sites of 239 and 56 TFs extracted from in vitro and in vivo datasets, respectively, revealed unique preferences for GC composition and propeller twist of DNA flanks (24). Other studies have concluded that nucleotides directly flanking the cognate sequence significantly affect rate of transcription by inducing structural changes in both DNA and the DNA-binding domain of the associated TF (33,34). Most of these studies have considered flanking sequences in terms of k-mers or GC composition; however, such simple sequence information may not be very informative. Representing DNA sequences in terms of structural features is an alternative approach to elucidate their functional complexities. Compared to the simple nucleotide sequence, structural features have more information content, as similar sequences might possess very different structures while divergent sequences can adopt equivalent local structures (13). With growing recognition of the importance of DNA structure in DNA–protein recognition, it is logical to study flanking sequences in terms of flexibility and other structural features.
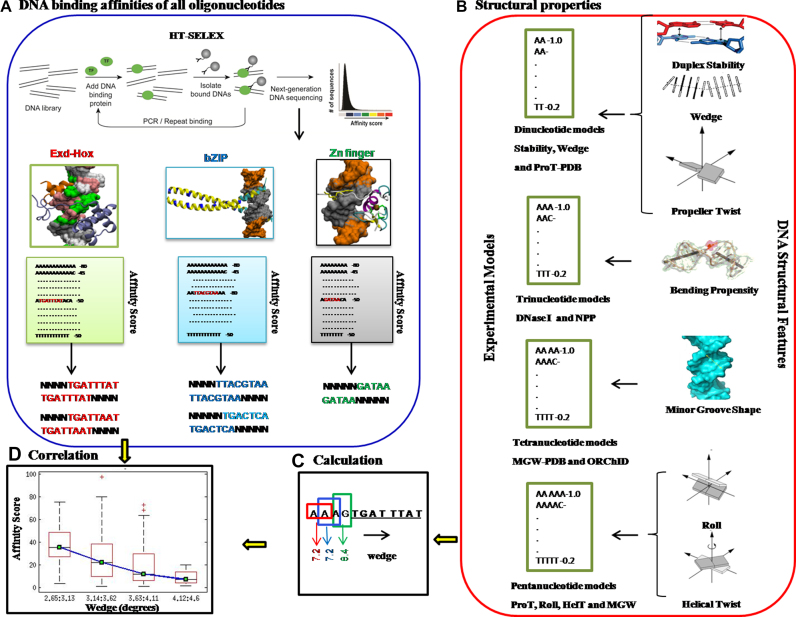
In this study, we present a novel computational approach for sequence-dependent structural analysis of DNA–TF binding specificity. As summarized in Figure 1, our strategy involves correlating DNA structural features of flanking sequences outside the core binding site with comprehensive in vitro DNA-binding preferences of different TFs. Several high-throughput in vivo and in vitro methods have been developed for studying DNA–TF interactions (23). In vivo techniques, like ChIP and DNase I hypersensitivity, measure the occupancy of binding sites along the genome. In vitro techniques, including cognate site identification (CSI), protein binding microarrays (PBMs), high-throughput-systematic evolution of ligands by exponential enrichment (HT-SELEX), and mechanically induced trapping of molecular interactions (MITOMI), quantify the intrinsic binding preferences of TFs based on in vitro affinity measurements (35,36). With recent developments, in vitro methods can provide binding specificity models of given TFs by defining its affinity toward all possible DNA sequences (entire sequence space of typical binding sites up to 20 base pairs). For our study, we compared in vitro DNA binding profiles of seven TFs with physiologically relevant DNA structural features, such as protein induced bendability, stability, wedge, helical twist, propeller twist, roll, and minor groove shape (Figure 1). The seven protein-DNA complexes considered in this study, namely Gata4; Exd-Scr, Exd-Ubx, Exd-AbdA, Exd-AbdB; FOS-JUN and NFIL3, include nine proteins that belong to the three largest classes of eukaryotic DNA binding domain families, namely zinc finger, homeodomain and bZIP, respectively. Gata4 is involved in myocardial development in human and mouse (37). The Hox TFs (Exd-Scr, Exd-Ubx, Exd-AbdA, Exd-AbdB) control proper body pattern formation in organisms as diverse as fruit flies to humans (38–40). FOS-JUN heterodimers, also known as AP1, are involved in a wide variety of cellular responses to extracellular stimuli associated with mitogenesis and differentiation processes (41). Nuclear factor interleukin 3 regulated TF (NFIL3), also known as E4BP4, regulates immune response in humans (42).
Figure 1.
Schematic illustration of analysis pipeline. (A) In vitro DNA-transcription factor (TF) binding preferences were obtained using HT-SELEX method for the entire sequence space of oligomers of length up to 20bps. Subsequently, 10-mer and 12-mer sequences with exact binding sites were considered for structural feature calculation. (B) Seven different physiologically relevant DNA structural features, including stability, wedge, propeller twist, bending propensity, minor groove shape, roll and helix twist were computed using (C) a sliding window, by converting each sequence into overlapping k-nucleotide feature values. (D) Correlation between the structural features of DNA flanks and corresponding binding affinities were illustrated using box plots.
MATERIALS AND METHODS
DNA–protein binding profile analysis
Representatives of three superfamilies of DNA-binding domains were studied to identify their sequence preferences amongst all permutations of a 20bp binding site. His6-tagged Hox proteins Scr, Ubx, AbdA and AbdB and FLAG-tagged Exd were synthesized by wheat-germ cell-free protein expression (CellFree Sciences Co., Ltd., Japan) (43). Protein expression was confirmed by Western blot against the His6 or FLAG epitope tags. HT-SELEX experiments were performed as previously reported (Figure 1) (44). Hox proteins and Exd were equilibrated with a 100 nM DNA library containing central 20 bp randomized region. Binding buffer was prepared as follows: 50 mM HEPES, pH 8, 150 mM potassium glutamate, 2 mM DTT, 40 ng/ul poly(dI:dC), 100 ng/ul BSA, 10% glycerol. Exd-Hox protein complexes were immunoprecipitated with Anti-FLAG M2 Magnetic Beads (Sigma, #M8823). Bound DNA was amplified by PCR with EconoTaq® DNA Polymerase (Lucigen Corporation, Madison, WI, USA), column purified and used for subsequent rounds. After three rounds of binding, Illumina sequencing adapters and a unique 6-bp ‘barcode’ were incorporated by PCR. Samples were pooled and sequenced in a single lane of an Illumina GAIIx sequencer.
Gata4 HT-SELEX data (20-mers) were downloaded from European Nucleotide Archive (ebi.ac.uk/ena; accession numbers ERP001824 and ERP001826) (26). Gata4 PBM data (E-scores for all 8-mers) was downloaded from CIS-BP (http://cisbp.ccbr.utoronto.ca/) (45). Gata4 CSI-array data (Z-score for all 8-mers) was taken from Carlson et al. (21,46). NFIL3 and FOS-JUN HT-SELEX data (20-mers) are from https://ansarilab.biochem.wisc.edu/computation.html (44).
HT-SELEX data (20-mers) was processed by counting the occurrence of every 10- or 12-mer using a sliding window of size 10 or 12. A 5th order Markov model was used to estimate the occurrence of each 10- or 12-mer in the starting DNA library (40). Affinity score for each 10- or 12-mer was calculated by dividing its number of occurrence in the SELEX data by the number of occurrence in the library as estimated from the model.
Affinity scores for all 10- or 12-mer sequences with an exact match with the consensus motif for each TF were considered for structural feature calculations (Table 1). The influence of flanking sequences on DNA-binding affinity was analyzed one flank at a time. Thus, to assess the influence of the 5′-flank, the position of the consensus sequence in the k-mer was fixed and all possible combinations (A, C, G or T) of 5′ flanks were considered. For example, from DNA binding data of FOS-JUN, 12-mer binding sites were considered comprising of 7-mer consensus sequence (TGACTCA) and all possible permutations of the pentameric flanks at the 5′-end (NNNNNTGACTCA) or 3′-end (TGACTCANNNNN), giving rise to a total of ∼1024 (45) sequences in each case (Table 1). Notably, the effect of flanking sequences alone was the focus of the study, hence mismatches to the consensus motif were not considered in the analyses.
Table 1.
Datasets used in the study. DNA binding information for all 12-mers (or 10-mers) were computed for Gata4, Exd-Scr, Exd-Ubx, Exd-AbdA, Exd-AbdB, FOS-JUN and NFIL3 transcription factors, using HT-SELEX method. Sequences that exactly match the consensus binding sites were considered for structural feature calculations. The datasets with 5′- and 3′-flanking tetramer (numbers in black) or pentamer (numbers in blue) sequences are listed in the table. The exact number of sequences in each dataset does not match the expected number (45 = 1024 in case of pentamer flanks or 44 = 256 in case of tetramer flanks) in all cases, since all possible sequences may not be represented in the SELEX data
| Number of sequences analysed | |||
|---|---|---|---|
| Consensus sequence motif | NNNNConsensus or NNNNNConsensus | ConsensusNNNN or ConsensusNNNNN | |
| Zinc Finger | |||
| Gata4 | GATAA | 1024 | 1020 |
| Exd-Hox | |||
| Exd-Scr | TGATTAAT | 256 | 255 |
| Exd-Ubx | TGATTTAT | 254 | 247 |
| Exd-AbdA | TGATTTAT | 256 | 256 |
| Exd-AbdB | TGATTTAT | 251 | 245 |
| bZIP | |||
| FOS-JUN | TGACTCA | 1013 | 1006 |
| NFIL3 | TTACGTAA | 256 | 256 |
DNA structural feature calculations
Seven different non-redundant sequence-dependent DNA structural features (corresponding to 11 different structural scales) were evaluated in this study, including protein induced bendability, stability, wedge, helical twist, propeller twist, roll and minor groove shape (Figure 1B). While almost two dozen properties have been used by studies thus far to describe local structural changes in DNA oligomers (47), variations in the above mentioned features are found to be most commonly associated with events of DNA–TF binding.
Bendability
Protein binding can induce sequence-dependent bending in DNA. DNA bendability is widely measured using two trinucleotide-based models, DNase I sensitivity (DNase I) model and nucleosome positioning preference (NPP) model. The DNase I sensitivity model is based on the increased sensitivity of flexible oligonucleotides to digestion by DNase I (48). DNase I interacts with the minor groove of the target sequences and bends the molecule away from the enzyme towards the major groove. Thus, from experimental DNase I digestion data, the model provides a scale for the propensity of different trinucleotides to bend towards the major groove. The NPP model is based on the finding that the preferential positioning of nucleosome core particles on DNA is determined by the bending ability of the DNA sequence (49). From sequence analysis of 177 different DNA molecules isolated from chicken erythrocyte nucleosome core particles, the NPP model classifies each trinucleotide based on its rotational orientation with respect to the histone core. Thus, the model provides relative values for major groove face preferring or minor groove face preferring trinucleotides, as well as trinucleotides with no rotational position preference, on an absolute scale.
Stability or free energy
DNA duplex stability can be expressed as the sum of free energy of its constituent dinucleotide base pair steps and is dependent on both the sequence as well as the GC/AT content of the DNA. Free energy values of individual dinucleotide steps are taken from unified thermodynamic nearest neighbor parameters obtained from melting studies on 108 oligonucleotides (50).
Wedge
Wedge is a quantitative measure of DNA axis bending caused by subtle variations in roll and tilt angles between adjacent base pairs. According to the wedge model, the global curvature of a DNA duplex is the sum total of local dinucleotide wedge deflections along the molecule. The 16 unique individual dinucleotide wedge angles, used to calculate DNA curvature, are derived from circularization and gel electrophoretic mobility data of 54 synthetic DNA fragments (51).
Minor groove shape
Shape of the DNA minor groove varies along the nucleotide sequence, and is determined as a function of two parameters—groove width and solvent accessible surface area (SASA) of the minor groove. Minor groove width has been calculated using two methods, and the values obtained are referred here as MGW and MGW-PDB, respectively. The MGW values were obtained from a web-based application called DNAshape, wherein a sliding pentamer model is employed to derive the minor groove width of a given DNA sequence (15). The calculations are carried out using the predicted groove width data of all possible 512 pentamers, which were obtained from Monte-Carlo simulations of a large number of distinct oligonucleotides. In the second method, the groove width value (MGW-PDB) of a given DNA sequence is determined by a sliding tetramer model incorporated in our in-house code. The groove width of each unique tetranucleotide is obtained from crystal structures of free and protein-bound DNA complexes (9) available in the Protein Data Bank. Minor groove widths (MGW and MGW-PDB) calculated using the above methods were compared with values calculated for the X-ray crystal structure of the Ultrabithorax-Extradenticle-DNA ternary complex using two different algorithms NUPARM (52) and 3DNA (53), and presented in Figure S1. Both MGW and MGW-PDB values were found to show similarity in trend and reasonable correlation with X-ray structure values.
Solvent accessible surface area (SASA) of the minor groove is directly correlated with the sensitivity of a DNA strand to hydroxyl radical cleavage. Hence, minor groove SASA was calculated using hydroxyl radical cleavage intensity predictions (ORChID) model, which was derived from experimental cleavage patterns for >150 different DNA sequences of 40 bp in length (54).
Propeller twist, helical twist and roll
Several intra- and inter-base pair parameters, especially propeller twist (ProT), helical twist (HelT) and roll, are good measures of the flexibility of DNA and were calculated using the DNAshape analysis described above (15). Propeller twist was also calculated using crystal structure derived dinucleotide values (ProT-PDB) (2) incorporated in our in-house Perl code.
All structural features were calculated using a sliding window, by converting each 12-mer (or 10-mer) sequence into overlapping k-nucleotide feature values (k = 2–5). Duplex stability, ProT-PDB and wedge, were computed using dimer windows. For bending propensity calculations using the trinucleotide-based DNase I and NPP models, k = 3 was used. MGW-PDB and SASA calculations were carried out using sliding tetramer models, while pentamer windows were used to determine MGW, ProT, HelT and roll. GC content of dinucleotides was also calculated to compare with other dinucleotide scales. All experimentally-derived parameters used to compute k-mer structural feature values have been presented in Figure S2. The consensus sequence was identical in all analyzed k-mers for a particular TF class, therefore the variation in structural feature value, although calculated for the entire sequence, essentially represents the differences in properties of the flanks alone. Hence, in subsequent sections, structural features are discussed only in context of the 5′- and 3′-flanks.
RESULTS
To assess the influence of flanking sequences on DNA binding, the average structural feature values of 5′- and 3′-flanking tetramer (for Exd-Hox proteins and NFIL3) or pentamer sequences (for Gata4 and FOS-JUN proteins) were compared with binding scores determined by HT-SELEX (Figure 1A). Solution-based methods have validated that binding intensity values are linearly correlated with association constant or binding affinity (R2 = 0.998) (21,44,46,55).
Correlation between DNA structural features
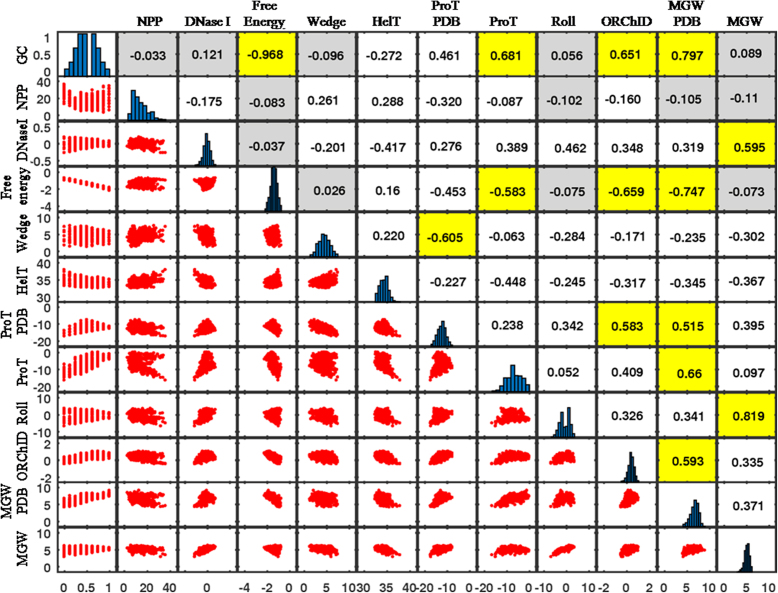
The primary sequence information of DNA can be used to predict several secondary structural features. Various structural models are available for computing DNA structural scales. Eleven different structural scales, namely NPP and DNase I (bendability), free energy, wedge, helical twist (HelT), ProT-PDB and ProT (propeller twist),roll, ORChID (minor groove shape), MGW-PDB and MGW (minor groove width) have been studied in this work (see Materials and Methods for details). Each structural scale was calculated using di, tri, tetra or pentanucleotide models, as described above. In order to compare these structural scales, di, tri and tetranucleotide models were converted to a unanimous pentanucleotide scale by averaging over the overlapping nucleotide steps. Following this, Pearson′s correlation coefficients among all structural scales were calculated from a Student's t-distribution, assuming the analyzed data set follows a normal distribution. While this comparison may be crude, since the exact dependence of few structural features on adjacent bases is not really known, it has been shown to be reliable (56). Figure 2 (upper half triangle) presents the correlation among the eleven different structural scales. Evidently, certain structural features like minor groove width or groove shape (MGW-PDB and ORChID), propeller twist (ProT and ProT-PDB) and free energy are intimately correlated with each other as well as with the GC content. Free energy, being intrinsically dependent on base pairing and stacking interactions of a dinucleotide, is strongly correlated with the GC content (R = −0.968). Similarly, a lower propeller twist and wider minor groove are characteristics of GC-rich sequences, explaining their strong correlation with GC content and free energy of the DNA sequences. Conversely, NPP, which is a measure of trinucleotide flexibility, is not correlated with GC content or other structural features like minor groove width, roll and free energy. Despite the strong correlation among some of the structural features, each of them provides unique insights into the subtle changes occurring in DNA topography during TF binding.
Figure 2.
Correlation between different structural features of DNA. Correlation coefficients between seven structural properties (11 structural scales), namely trinucleotide bendability (DNase I sensitivity [DNase I] and nucleosome positioning preference [NPP]), free energy, wedge, helix twist (HelT), propeller twist (ProT and ProT-pdb), roll and minor groove shape (ORChID, MGW-pdb and MGW) have been depicted. Bar plots on the diagonal represent the distribution of structural features for all possible pentamers. The red scatter diagrams below the corresponding bar plots illustrate the correlation between various scales. The numbers in gray shaded boxes are statistically insignificant at P ≤ 0.0001. The numbers in yellow shaded boxes are strongly correlated (R > 0.5 or R < −0.5) to each other.
DNA binding protein domains with similar structures exhibit distinct binding geometries
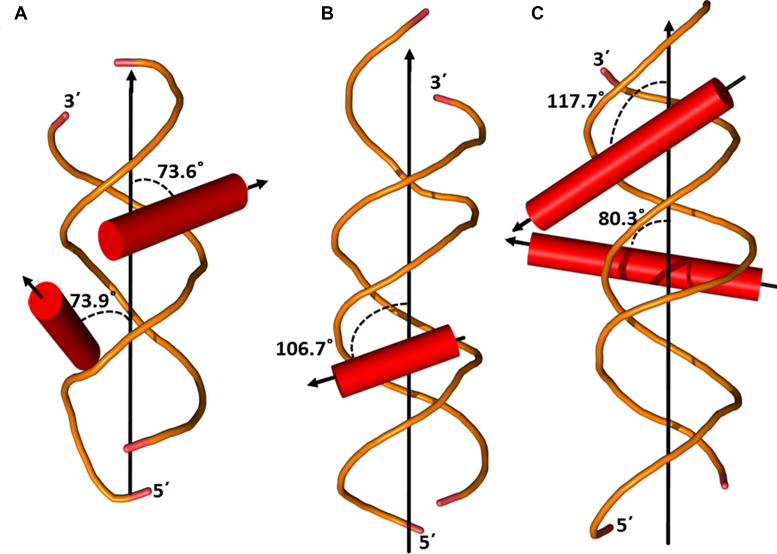
The TFs studied here belong to different structural superfamilies and use distinct folds for DNA-binding, namely the zinc finger domain (Gata4), the helix-turn-helix fold in homeodomain proteins (Exd-Scr, Exd-Ubx, Exd-AbdA and Exd-AbdB), and the basic helix coiled-coil fold in basic leucine zipper TFs (FOS-JUN and NFIL3). Interestingly, while all three DNA-binding domains primarily comprise of α-helix, they differ significantly in their interaction with the cognate DNA. At the site of DNA–protein recognition the convex surface of the α-helix fits into the concave surface of the DNA major groove. The orientation of the recognition helix can be defined by a single angle, denominated here as the orientation angle. The orientation angle can be calculated from the dot product of the direction cosines of the DNA helix and the recognition helix of the binding domain, assuming their rigid body positioning (57). Figure 3 depicts the interaction geometry of cognate DNA binding sites with the recognition helices of the three families of TFs, homeodomain, Zinc finger and bZIP. To evaluate the binding geometries, the coordinates of the crystal structures of the corresponding DNA–protein complexes were retrieved from the Protein Data Bank (58). The axes of the DNA and the recognition helices were determined using in house software packages NUPARM (52) and Helanal-Plus (59), respectively. As evident from the markedly different orientation angles in Figure 3, the recognition helix of each DNA-binding domain is uniquely aligned relative to the axis of its cognate DNA. In order to accommodate the α-helix of a binding protein in its major groove, DNA undergoes several subtle structural changes, including sliding of base pairs, increase in major groove depth, helix unwinding, and change in inclination (5,60). Owing to their distinct binding orientations in the DNA groove, each TF class perturbs the structure of its DNA cognate site in a specific way. Since DNA structure is context dependent, structure of the flanking sequences can significantly modulate DNA–protein recognition and binding. In the subsequent sections, we discuss in detail the effect of different structural features of the flanking sequences on binding affinities of the three TF classes with DNA.
Figure 3.
The three classes of transcription factors, homeodomain, Zinc finger and bZIP, display differential binding geometry. (A) The Exd-Ubx-DNA ternary complex (PDB code: 1B8I) depicts Ubx homeodomain binding to the consensus sequence cooperatively with the cofactor homeodomain protein Exd. The helices (α3, shown in the figure) of Ubx and Exd homeodomains interact with DNA with their helix axis oriented at 73.9° and 73.6°, respectively with respect to the DNA helix axis. (B) The recognition helix of Zinc finger (PDB code: 4HC9) makes an angle of 106.7° and (C) the recognition helices of FOS-JUN (PDB code: 1A02) are oriented at angles of 117.7° and 80.3°, respectively.
Structure of flanking sequence modulates binding affinity of DNA binding domains
The correlation between binding affinities of TFs and the structural features of the DNA sequences flanking the cognate sites was determined for all three TF classes and presented in Table 2.rmation. Pearson's correlation coefficient was calculated with P ≤ 0.0001 being considered as statistically significant. Evidently, some of the structural features of DNA flanks are strongly correlated with the binding affinities of the TFs, indicated by highlighted values in the Table. Interestingly, TFs belonging to different classes are correlated with different DNA structural features. As surmised earlier, this difference arises out of the distinct binding modes of the three TF classes with their cognate binding sites (Figure 3). Correlation between the structural features of DNA flanks and corresponding binding affinities is illustrated using box plots. For each structural feature, all TF-binding sequences were sorted into four bins based on their feature value, with each bin representing one-fourth of the entire range of values observed for the structural feature of interest. Following this, binding affinities of all sequences in a bin, along with their median binding affinity, were calculated and plotted. Since DNA structure is not independent of its sequence, the oligonucleotide composition of the best and weakest binders are presented in Table S1. Taken together these data can be used to comprehend how sequence and structure of DNA flanks are intimately correlated with each other and with binding affinities of TFs.
Table 2.
Correlation between DNA structural features of 5′- and 3′-flanking sequences and the DNA binding affinities of seven TFs studied here. Eleven structural scales listed in the Table have been analyzed to define seven structural properties, namely trinucleotide bendability (DNase I and NPP), free energy, wedge, helical twist (HelT), propeller twist (ProT and ProT-PDB), roll and minor groove shape (ORChID, MGW-PDB and MGW). For comparison, correlation of binding affinities with GC content was also studied, but since GC content is highly correlated with free energy (FE) it is not plotted separately (see Figure 2). Pearson's correlation coefficient was calculated among the structural properties of flanking sequences and binding affinity of seven proteins. The numbers in boldface are statistically significant with P ≤ 0.0001
| GC | NPP | DNase1 | FE | Wedge | HelT | ProT-PDB | ProT | Roll | ORChID | MGW-PDB | MGW | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5′ Flank | ||||||||||||
| Gata4 | 0.312 | 0.061 | 0.168 | 0.305 | 0.172 | 0.075 | 0.189 | 0.152 | 0.063 | 0.168 | 0.305 | 0.081 |
| Exd Scr | 0.145 | 0.050 | 0.022 | 0.181 | 0.236 | 0.005 | 0.176 | 0.060 | 0.074 | 0.251 | 0.004 | 0.091 |
| Exd Ubx | 0.028 | 0.010 | 0.052 | 0.054 | 0.231 | 0.023 | 0.093 | 0.093 | 0.120 | 0.189 | 0.019 | 0.057 |
| Exd AbdA | 0.486 | 0.118 | 0.187 | 0.495 | 0.217 | 0.222 | 0.321 | 0.212 | 0.165 | 0.533 | 0.324 | 0.160 |
| Exd AbdB | 0.236 | 0.207 | 0.190 | 0.200 | 0.079 | 0.212 | 0.167 | 0.302 | 0.157 | 0.198 | 0.381 | 0.326 |
| FOS JUN | 0.031 | 0.065 | 0.237 | 0.054 | 0.310 | 0.172 | 0.003 | 0.083 | 0.182 | 0.095 | 0.045 | 0.174 |
| NFIL3 | 0.203 | 0.107 | 0.238 | 0.253 | 0.440 | 0.348 | 0.037 | 0.022 | 0.347 | 0.011 | 0.177 | 0.347 |
| 3′ Flank | ||||||||||||
| Gata4 | 0.094 | 0.293 | 0.243 | 0.053 | 0.126 | 0.189 | 0.007 | 0.309 | 0.045 | 0.209 | 0.180 | 0.104 |
| Exd Scr | 0.521 | 0.081 | 0.088 | 0.551 | 0.170 | 0.102 | 0.443 | 0.277 | 0.053 | 0.456 | 0.548 | 0.108 |
| Exd Ubx | 0.469 | 0.137 | 0.088 | 0.499 | 0.187 | 0.075 | 0.423 | 0.272 | 0.109 | 0.319 | 0.336 | 0.198 |
| Exd AbdA | 0.620 | 0.115 | 0.002 | 0.671 | 0.043 | 0.082 | 0.415 | 0.315 | 0.036 | 0.456 | 0.414 | 0.063 |
| Exd AbdB | 0.476 | 0.139 | 0.123 | 0.497 | 0.159 | 0.079 | 0.372 | 0.313 | 0.042 | 0.310 | 0.341 | 0.170 |
| FOS JUN | 0.065 | 0.074 | 0.223 | 0.088 | 0.290 | 0.159 | 0.049 | 0.064 | 0.178 | 0.155 | 0.023 | 0.062 |
| NFIL3 | 0.203 | 0.107 | 0.238 | 0.253 | 0.440 | 0.348 | 0.037 | 0.022 | 0.347 | 0.065 | 0.177 | 0.347 |
Correlation between structural features and binding affinities of Gata4 were measured using DNA binding data from three different experimental platforms (Table S2). A good agreement was observed for highly correlated structural properties of DNA flanks between Gata4 binding data obtained from HT-SELEX (human) (26) and CSI-array (mouse) (21). This shows the robustness of our methodology across two experimental platforms as well as the conservation of flanking sequence properties for a TF across two different species. A similar comparison of HT-SELEX with PBM array data yielded a good agreement for structural features of the 5′-flank, but not for the 3′ flank (45). The variation in the results between PBM (mouse) and CSI or HT-SELEX may arise due to the fact that general PBM uses de-Bruijn based arrangement of 8-mer DNA sequences on the array probes (61). Although this method captures the consensus motif, it often fails to extract the relevant flanking sequence effects (36).
Groove width and bendability of DNA flanks attune cognate site for TF binding: The case of Gata4
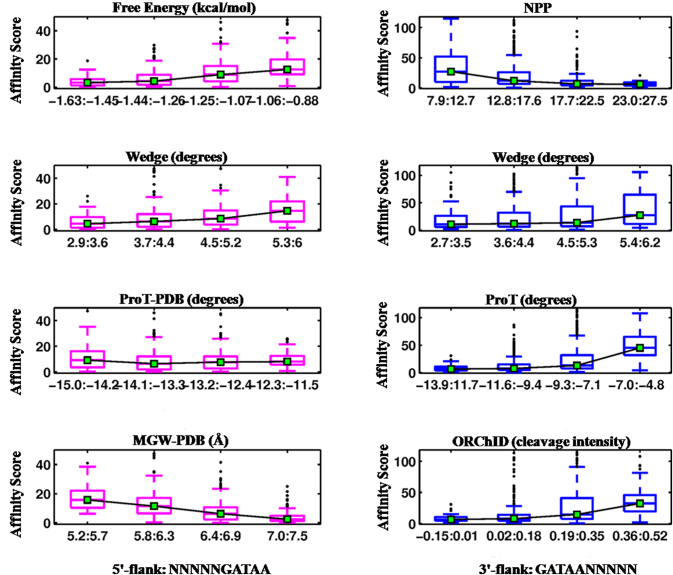
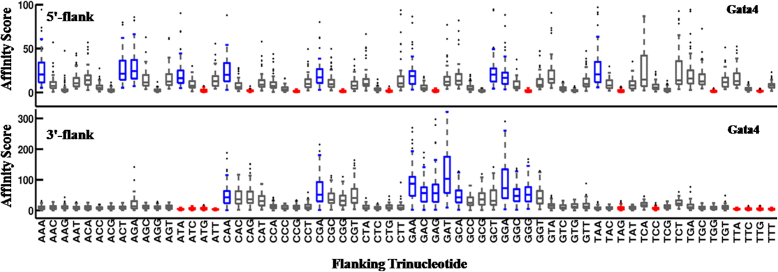
In vitro DNA-binding preferences of the Gata4 binding domain were derived for 10-mers, composed of the 5-bp cognate GATAA site and its 5′- and 3′-flanking pentamers (i.e., 5′-NNNNNGATAA-3′ and 5′-GATAANNNNN-3′). Structural properties that significantly correlate with Gata4 binding (Table 2) are presented as correlation plots in Figure 4. The effect of nucleotide composition of the 5′- or 3′-flanks on Gata4 binding was also determined and is presented in Figure 5. Further a cross platform comparison for binding affinity data obtained from Protein Binding Microarray (8-mer) and HT-SELEX (10-mer) apart from the data from CSI-array (8-mer) was carried out (Table S2). As evident from Figure 4, Gata4 DNA binding correlates negatively with free energy, propeller twist (ProT-PDB) and minor groove width (MGW-PDB) of the 5′-flank. This indicates that lower free energy, a high negative propeller twist and a narrow minor groove at the 5′-flanks are preferred for high-affinity binding of Gata4. These properties corroborate well with AT-rich sequences, as also implied by the higher binding affinities observed for A- and T-containing oligonucleotides at the 5′-end (Figure 5 and Table S1). Conversely, a less negative propeller twist and wider minor groove at the 3′-end, exhibited by G-containing oligonucleotides (Figure 5 and Table S1), are conducive for GATA-binding. Interestingly, flexibility (DNase I or NPP) and related property curvature (represented by wedge) of both flanks were found to affect Gata4 binding at the consensus site. In fact, binding affinity is positively correlated with both DNase I and wedge, indicating that more flexible and curved flanks make the core motif conducive for Gata4 binding.
Figure 4.
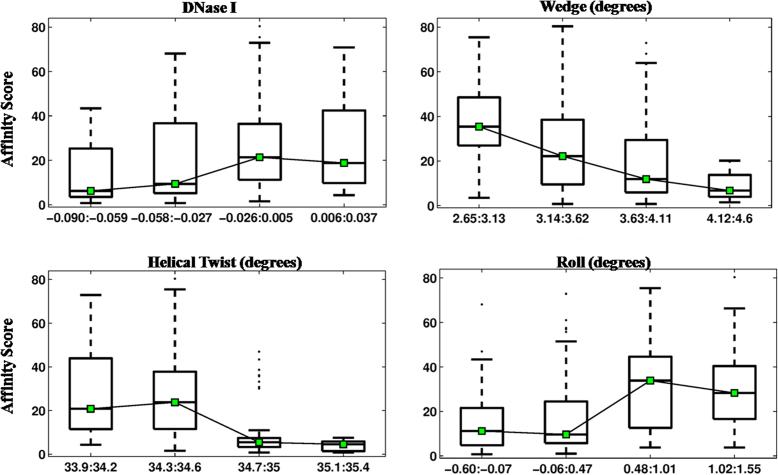
Correlation between binding affinity of Gata4 and the structural features of 5′- (magenta) and 3′-flanking (blue) sequences of cognate DNA is illustrated using box plots. Gata4 binding is highly correlated (P ≤ 0.0001) with four structural parameters of 5′-flanking sequences namely free energy, wedge, propeller twist (ProT-PDB) and minor groove width (MGW-PDB). At the 3′-end, Gata4 binding is modulated by DNA bendability (NPP), wedge, propeller twist (ProT) and minor groove shape (ORChID) of the flanks. DNA bendability (represented by NPP and wedge) of both 5′ and 3′ flanks significantly affect binding affinities of Gata4 TF. The box plots depict the affinity scores corresponding to four ranges of structural feature values. The whiskers indicate values corresponding to ±2.7 s.d. from the mean of the data.
Figure 5.
The effect of oligonucleotide composition of flanking regions on Gata4 binding affinity. The cognate binding motifs with 5′- or 3′-flanking trinucleotides were plotted against corresponding HT-SELEX affinity scores. Highest affinity was shown by A/T-rich flanks at the 5′-end and G-rich flanks at the 3′-end. High affinity binders are indicated by blue boxes and low affinity binders as red boxes. Only the trinucleotide sequence immediately flanking the core motif has been shown.
For a vivid illustration of the effect of the flanking sequences on the binding of GATA proteins to their cognate site, we referred to the X-ray crystal structure of Gata3 (Figure S3). The GATA proteins possess two highly conserved C-X2 -C-X17 -C-X2 -C type zinc fingers at the C- and N-termini. As shown in the figure, the basic region of the C-terminal Zn-finger inserts deep into the major groove of the GATAA site causing a widening of the groove. This in turn leads to a concomitant narrowing of the minor groove at the GATAA site. As a result, narrow minor groove of the 5′-flanks further facilitates high-affinity binding of Gata4 to the cognate binding motif (Figure 4). Moreover, the carboxy terminal tail extending from the Zn-finger domain loops around and inserts into the minor groove towards the 3′-end of the GATAA site. Presumably this causes a slight widening of the minor groove, justifying the need for the presence of G-rich 3′-flanks that possess broad minor grooves (Figure 5 and Table S1). Earlier studies have shown that the negative electrostatic potential of the narrow minor groove at the GATAA site stabilizes binding of C-terminal arginine residues (Figure S3) (9,62). While the role of shape readout within the cognate sites in GATA–DNA binding specificity has been described by several earlier reports, the role of DNA context is only now being considered. Here, we have shown in detail how the compositional and structural properties of sequences flanking the Gata4 cognate site play a significant role in guiding specific DNA–TF binding.
Wider groove at DNA flanks increases TF binding affinity: The case of Hox paralogs
In vivo Hox proteins bind their consensus motif TGATTNAT in association with a co-factor protein, Extradenticle (Exd) (63,64). As illustrated in Figure S4, Exd binds to the 5′-half site TGAT while the Hox partner, Ubx, binds to 3′-half site TTAT. In complex with Exd, all eight Hox paralogs exhibit distinct binding preferences. Based on their binding to specific consensus motifs, the Hox TFs are classified into three groups, Hox-1 (Lab and Pb bind to TGAT), Hox-2 (Dfd and Scr bind to TAAT) and Hox-3 (Ubx, AbdA and AbdB bind to TTAT). Several earlier reports have shown that shape of the consensus site is a significant determinant of Hox specificity (10,40,63,65). Hence it is particularly important to determine if shape of the flanking sequence could also modulate binding affinities of Hox TFs. In our study, we have focused on analyzing the effect of DNA structural features on the binding affinity of the Hox-3 family of TFs.
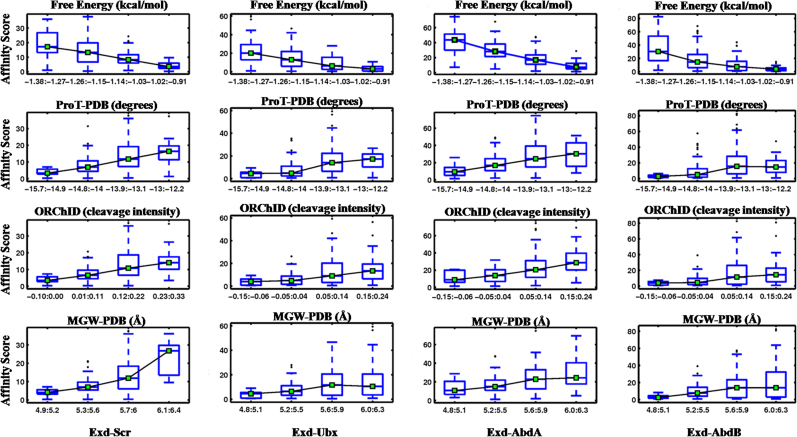
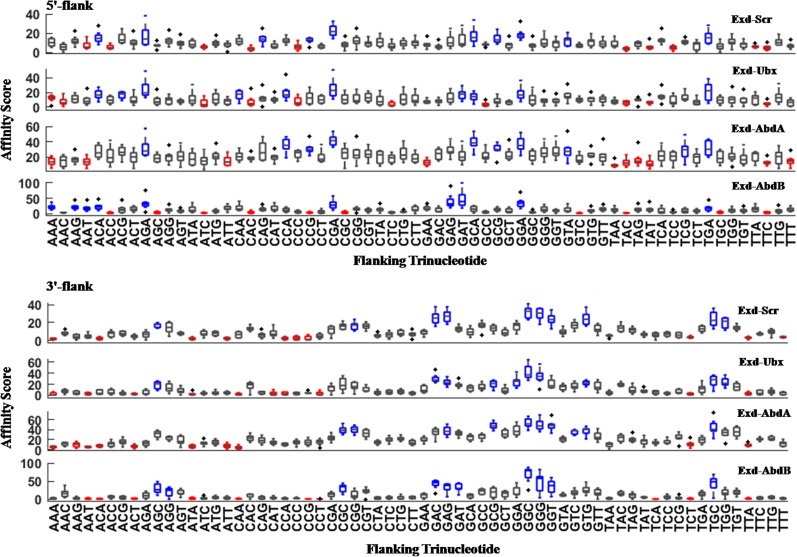
Correlation plots of structural properties that significantly influence binding of all 12-mer Hox-binding sequences (Table 2) are presented in Figure 6. The effects of trinucleotide composition of the 5′- or 3′-flanks on Exd-Hox binding was also determined and are presented in Figure 7 and Table S1. As evident from Table 2, except Exd-AbdA, the other two Hox-3 TFs do not show any significant feature-to-binding correlation at the 5′-flank. Hence, we focused on the 3′-half site and its flank, which are contacted by the Hox counterpart. Figure 6 depicts that binding affinity of Hox-3 TFs shows a negative trend with free energy of the 3′-flank, while it is positively correlated with propeller twist, ORChID, and minor groove width (MGW-PDB). This implies that higher free energy, wider minor groove, and a less negative propeller twist at the 3′-flanks are preferred for binding of Hox-3 TFs. This agrees with our observation that G-rich flanks are preferred at the 3′-end for efficient DNA-Hox binding (Figure 7 and Table S1). The significant role of minor groove width in determining DNA binding specificities of Hox protein families has been reported in earlier studies (40). Minor groove width calculation of the site bound by Exd-Ubx (Figure S1) displays a narrow minor groove at the A3-T4 step and a wider minor groove at T8 of the core binding site TGATTTAT. While a narrow minor groove at the A3-T4 step is conserved across all Hox families, groove width at T8 is variable and is proposed to result in selective binding by different Hox proteins. For example, while the Hox-3 proteins prefer a wider minor groove at T8, Scr and Dfd (Hox-2) bind best to TGATTAAT, which exhibits a narrowing at the A-A step of the Hox half site.
Figure 6.
Correlation between structural features of 3′-flanking sequences of cognate DNA and binding affinities of Hox TFs is illustrated as box-plots. Four Hox proteins Scr, Ubx, AbdA and AbdB, along with cofactor protein Exd, bind to the consensus motif TGATTAAT for Scr or TGATTTAT for other Hox. Binding affinities of Exd-Hox complexes are highly correlated (P ≤ 0.0001) with four properties of 3′-flanks, namely free energy, propeller twist (ProT-PDB), minor groove shape (ORChID) and minor groove width (MGW-PDB). Box plot details are same as in Figure 4.
Figure 7.
The effect of oligonucleotide composition of flanking regions on DNA binding affinities of Hox TFs. All four Exd-Hox complexes show high binding affinity when 3′- flanks are G-rich whereas A/T-rich sequences have an opposite effect. Other details are same as in Figure 5.
To determine if similar distinctions in binding preferences are observed for the flanks as well, we computed the correlation between structural features and binding affinities for the Hox-2 protein Scr. Remarkably, the structural features preferred by Scr at the 3′-flank are identical to the preferences of Hox-3 class of TFs (Table 2, Figures 6 and 7). This implies that despite significant differences in consensus sites, both classes of Hox proteins share similar preferences in the conformation of the flanks. Consistent with this result, the regions of Ubx and Scr DNA recognition helices that interact with the 3′-flank share 100% sequence identity (Figure S5). This possibly explains why both TFs have similar structural preferences for flanking DNA despite having different criteria for selecting the consensus site.
DNA bending at flanking sequence influences TF binding affinity: The case of bZIP TFs
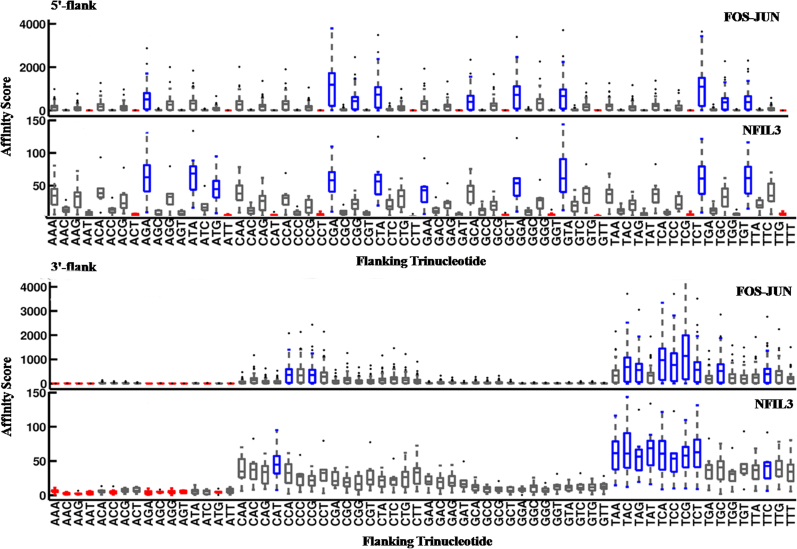
The bZIP proteins, FOS-JUN (AP1) and NFIL3 homodimer, identify their consensus binding site using the classic method of base readout by forming an extensive network of H-bonds with the major groove of the cognate DNA. Hence it is of particular interest to investigate if DNA structural features have any influence on the binding specificities of this protein family. As evident from Table 2, helical twist and wedge are negatively correlated with binding of both FOS-JUN and NFIL3 at the 5′- as well as 3′-flanks. Conversely, minor groove width, roll and bendability of the flanking sequences are positively correlated with binding of the two bZIP proteins to their cognate sites. Notably, for NFIL3 homodimer the correlation coefficients for the structural features of both flanking regions are identical as the binding site is exactly palindromic (see Table 2). The effect of trinucleotide composition of the 5′- or 3′-flanks on CSI intensities was also determined and presented in Figure 8. Interestingly, both proteins were found to have a preference for flanking sequences that resemble the corresponding binding half-sites of the cognate motif (Table S1). This is in interesting agreement with earlier studies suggesting that high affinity binding sites often occur in a homotypic environment (22,24). It has also been reported that FOS-JUN heterodimer is able to bind cognate DNA in two opposite orientations with minimal effect on binding affinity (66). In another study, the same authors have shown that the orientation preference is determined by the flanking sequence composition (67). However, since the entire sequence space has been explored in this work, the effect of sequence composition on differential orientation can be ignored. Thus, we presume that both FOS and JUN monomers contact the 5′- and 3′-binding-half-sites of the consensus motif 5′-TGACTCA-3′ with equal propensity. As shown in Figure 8 (and Table S1), the preferred sequences for high-affinity binding of FOS-JUN are RR and YY steps at the 5′- and 3′-flanks, respectively, owing to their identity with the 5′-GA and TC-3′ steps of the cognate motif. Similarly, for NFIL3, YR steps are preferred at both the 5′- and 3′-flanks. Plots of highly correlated structural properties of the bZIP proteins are presented in Figure 9 using NFIL3 as an example. Binding affinity of NFIL3 shows a negative trend with wedge, helical twist and DNA bendability (DNase I sensitivity) of the flanking sites, and a positive correlation with roll. This indicates that high-affinity binding sites are flanked by sequences that are rigid and possess smaller helical twist and wedge angles. Indeed, the crystal structure of the FOS-JUN-DNA complex reveals an essentially straight DNA with a maximum of 10° bend, possibly because the FOS and JUN monomers are known to bend DNA in opposite directions (Figure S6) (66). This explains the binding preference of the bZIP proteins for cognate DNA flanked by sequences that are less curved, as indicated by the negative correlation with wedge angle and DNase I sensitivity. Notably, such a significant effect of rotational flexibility of flanking regions on modulating binding affinity of TFs has not been reported in earlier studies.
Figure 8.
The effect of oligonucleotide composition of flanking regions on DNA binding affinities of bZIP proteins. Highest affinity was observed when both 5′- and 3′-flanking sequences resembled the corresponding half-binding sites of the consensus sequence for both bZIP proteins, FOS-JUN and NFIL3. Other details are same as in Figure 5.
Figure 9.
Correlation between structural features of DNA flanking the consensus motif and binding affinity of bZIP transcription factor NFIL3 is illustrated as box plots. The palindromic nature of the bZIP binding site results in identical structural feature preferences at both flanks. Binding affinity is primarily modulated by flexibility of the flanking regions represented by bending flexibility (DNase I and wedge) and rotational flexibility (helical twist and roll). Box plot details are same as in Figure 4.
DISCUSSION
The classical approach to understanding the mechanism of DNA–TF interaction is by determining the structure of the DNA–protein complex at atomic resolution and identifying direct contacts between the protein and DNA as well as variations in the local and global DNA helical parameters. However, such an approach cannot be applied to high-throughput data obtained from large scale DNA–TF binding studies and new strategies are required for addressing this problem. For example, DNA structure can be estimated from sequence using information provided by available X-ray or NMR-determined DNA or DNA–protein structures, and other experimental studies or theoretical simulations (12). As the DNA molecule is conformationally variable, several structural parameters have been defined ranging from the global helical axis to groove width and base pair orientation (68).
In this study, we employed structural feature analysis for understanding the influence of conformational plasticity and structure of DNA sequences flanking cognate sites in binding of three most prevalent TF families (Zinc finger, homeodomain, and bZIP) in eukaryotes. While all three TF classes use a common α-helical structural domain for binding, the consensus DNA sequences identified by them have distinct features. Using in vitro data from cognate site identifier and HT-SELEX studies, intrinsic binding preferences or binding specificity model of each TF has been derived by correlating its affinity toward all possible DNA sequences (entire sequence space of a 10- or 12-mer binding site). Furthermore, core motif preferences were correlated with structural features of the flanking DNA, including protein-induced bendability, stability, wedge, helical twist, propeller twist, roll and minor groove shape. Sequence information is generally encoded by various di, tri, tetra- and pentanucleotide structural models. While ∼100 dinucleotide scales are reported in DiProDB database (47), only few of them have been used in this work, since many are redundant. Dinucleotide features, like stability, wedge and propeller twist, are explicitly dependent on identity and orientation of flanking base pairs and are relevant to biomolecular events involving DNA. DNA duplex stability (free energy) is intimately linked to hydrogen bond and stacking interactions. Propeller twist primarily depends on GC content with small variations arising due to the actual dinucleotide composition. DNA flexibility (bendability) has been predicted using two trinucleotide models (DNase I sensitivity and NPP), which are experimentally derived and based on genome sequence context. DNA shape is, on one hand, a function of sequence, but the degeneracy of sequence and shape does not enable a simple mapping of shape to sequence (65). A recent effort has attempted to understand DNA–protein recognition by teasing apart sequence read out and shape read out (65). In another study a mechanism-agnostic model has been presented to quantify binding affinity to consensus sequence alone (10). However, in context of flanking sequences, the contribution of sequence readout is negligible; hence we resorted to relating DNA structural features to both sequence and structural readouts, while examining the 5′- and 3′-flanks separately.
While several studies have investigated the role of motif environment in facilitating the search for consensus binding sites by TFs, our methodology has resulted in some novel findings. Firstly, we have correlated DNA plasticity, in terms of flexure of DNA sequences, with their TF binding affinities. It has been known that certain TF classes prefer bent DNA as a potential interaction site or bend DNA upon binding (69,70). Two out of the three TF classes studied here showed significant correlation of binding affinities with DNA flexibility, both rotational (represented by roll and helical twist) and bending flexibility (represented by wedge, DNase I and NPP models). Notably, currently available DNA-shape based models, while acknowledging the role of flexibility in DNA–TF binding, do not incorporate this key feature in the prediction of potential TFBSs (71), possibly because flexibility of very short oligonucleotides cannot be validated experimentally. Even recent efforts using FRET-based assays have successfully determined bendability of DNA for a length scale of ∼100 bps (72). In this context, DNase I cleavage rates and NPP-based sequence enrichment used in our study can serve as reliable indicators of local flexibility of DNA.
Secondly, the structural features characterizing flanking regions preferred by the three different TF classes showed remarkable agreement with their in vivo binding patterns. For example, while sequences which show high affinity to bind Gata4 were found to possess distinct structural features at both 5′- and 3′-flanks, the palindromic nature of the bZIP binding motifs was reflected in identification of equivalent structural features of both 5′- and 3′-flanks. The homeodomain proteins are distinct from the GATA TFs and bZIP family since they interact not only with the consensus but also with the flanking base pairs. As a result, the flanking sequences play a more direct role in determining binding specificity, which was appropriately identified in our studies. Finally, preferred consensus motifs were found to be flanked by sequences possessing structural features that made the consensus site more conducive for TF binding. For example, Gata4 prefers AT-rich 5′-flanks with narrow minor groove and high propeller twist since this leads to a concomitant widening of the major groove, enabling TF binding.
Earlier investigations on high-throughput data of binding affinities have highlighted certain features of flanking DNA that lead to strong DNA–TF binding, including GC-richness of the flanks and localization of the consensus motif within a homotypic environment (24). Recently, the role of repeat DNA sequences, present in highly extended flanking regions, in controlling DNA–TF binding preferences has been suggested by an in vitro study on human TFs (73). While these features may be useful in identification of a preferred DNA context for binding of an entire TF family, the results may not reflect the complete picture for specific TFs or for the different flanking ends. For example, while flanking sites bearing resemblance with the core motif lead to high affinity binding (Table S1), the general sequence and structure features of preferred flanks are often very different from the features of the core motif (GATA and Hox TFs). This is also evident upon comparing our results with a study on Hox protein family, where the authors identified that cognate sites present in a GC-rich context are preferable (24). Our study on a different and specific set of Hox TFs revealed that while GC-richness is preferred at the 3′-flank, at the 5′-flank low GC is suitable for binding. Hence, we propose that correlating DNA structural features with binding affinities of corresponding TFs might be a more suitable yet less resource consuming protocol for precisely identifying binding preferences of individual TFs.
CONCLUSION
Our in silico study examined the inherent dynamics hidden in the structure of flanking sequences and its influence on the DNA binding affinity of TFs. The results reveal that the structure of immediate flanks may fine-tune the geometrical, rotational and translational settings of TFs in DNA–TF complexes. The set of TFs considered in our analysis belong to three different DNA-binding domain families, viz Zn finger, homeodomain and bZIP. All of these use an α-helix to recognize DNA, yet they display distinct flanking sequence preferences for binding. For example, high affinity binding sites of the Zn-finger TF Gata4 are flanked by flexible DNA sequences, whereas rigid flanks are conducive for binding of bZIP TFs. While homeodomain proteins prefer flanks with wider minor groove for high-affinity binding, Gata4 prefers narrow minor groove at the flanking region. Thus, our results reveal that flanking sequence preferences are not monotonic, as similar DNA-binding folds display distinct modes of DNA engagement. In essence, flexibility, stability and minor groove width of the DNA flanks are found to be important modulators of TF binding to their core motifs. DNA plasticity and mechanistic models employed in this work can provide detailed invaluable mechanistic insights into DNA–protein recognition, which will help refine computational tools for binding site search prediction and modeling of TF binding. The contextual information obtained using this approach can also significantly improve TFBS annotation across genomes. Further, the current study may help in understanding gene regulatory networks based on the integration of DNA structural features, genome sequence, transcription factor binding data, and gene expression data.
Supplementary Material
ACKNOWLEDGEMENTS
M.B. is grateful to Indian National Science Academy and Department of Science and Technology, Government of India, for J.C. Bose fellowship. D.G.D. acknowledges Science Engineering and Research Board, Department of Science and Technology, Government of India, for fellowship.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM120625 to A.Z.A.]. Funding for open access charge: Science Engineering and Research Board, Department of Science and Technology, Government of India [SR/S2/JCB-47/2009 to M.B.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Ptashne M. A Genetic Switch: Gene Control and Phage. Lambda. 1986; Palo Alto: Cell Press and Blackwell Scientific Publications. [Google Scholar]
- 2. Gorin A.A., Zhurkin V.B., Olson W.K.. B-DNA twisting correlates with base-pair morphology. J. Mol. Biol. 1995; 247:34–48. [DOI] [PubMed] [Google Scholar]
- 3. Marathe A., Karandur D., Bansal M.. Small local variations in B-form DNA lead to a large variety of global geometries which can accommodate most DNA-binding protein motifs. BMC Struct. Biol. 2009; 9:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Travers A. The structural basis of DNA flexibility. Philos. Trans. R. Soc. Lond. A. 2004; 362:1423–1438. [DOI] [PubMed] [Google Scholar]
- 5. Bansal M. Structural variations observed in DNA crystal structures and their implications for protein-DNA interaction. Biol. Struct. Dyn. Proc. Ninth Conversation. 1996; 1:121–134. [Google Scholar]
- 6. Harteis S., Schneider S.. Making the bend: DNA tertiary structure and protein-DNA interactions. Int. J. Mol. Sci. 2014; 15:12335–12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Olson W.K., Gorin A.A., Lu X.-J., Hock L.M., Zhurkin V.B.. DNA sequence-dependent deformability deduced from protein–DNA crystal complexes. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:11163–11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kanhere A., Bansal M.. DNA bending and curvature: a ‘turning’point in DNA function. Proc. Indian Natl. Sci. Acad. 2004; B70:239–254. [Google Scholar]
- 9. Rohs R., West S.M., Sosinsky A., Liu P., Mann R.S., Honig B.. The role of DNA shape in protein–DNA recognition. Nature. 2009; 461:1248–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rube H.T., Rastogi C., Kribelbauer J.F., Bussemaker H.J.. A unified approach for quantifying and interpreting DNA shape readout by transcription factors. Mol. Syst. Biol. 2018; 14:e7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bansal M., Kumar A., Yella V.R.. Role of DNA sequence based structural features of promoters in transcription initiation and gene expression. Curr. Opin. Struct. Biol. 2014; 25:77–85. [DOI] [PubMed] [Google Scholar]
- 12. Meysman P., Marchal K., Engelen K.. DNA structural properties in the classification of genomic transcription regulation elements. Bioinform. Biol. Insights. 2012; 6:155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Parker S.C., Hansen L., Abaan H.O., Tullius T.D., Margulies E.H.. Local DNA topography correlates with functional noncoding regions of the human genome. Science. 2009; 324:389–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang L., Zhou T., Dror I., Mathelier A., Wasserman W.W., Gordân R., Rohs R.. TFBSshape: a motif database for DNA shape features of transcription factor binding sites. Nucleic Acids Res. 2013; 42:D148–D155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhou T., Yang L., Lu Y., Dror I., Dantas Machado A.C., Ghane T., Di Felice R., Rohs R.. DNAshape: a method for the high-throughput prediction of DNA structural features on a genomic scale. Nucleic Acids Res. 2013; 41:W56–W62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ponomarenko J.V., Ponomarenko M.P., Frolov A.S., Vorobyev D.G., Overton G.C., Kolchanov N.A.. Conformational and physicochemical DNA features specific for transcription factor binding sites. Bioinformatics. 1999; 15:654–668. [DOI] [PubMed] [Google Scholar]
- 17. Meysman P., Dang T.H., Laukens K., De Smet R., Wu Y., Marchal K., Engelen K.. Use of structural DNA properties for the prediction of transcription-factor binding sites in Escherichia coli. Nucleic Acids Res. 2010; 39:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sarai A., Kono H.. Protein-DNA recognition patterns and predictions. Annu. Rev. Biophys. Biomol. Struct. 2005; 34:379–398. [DOI] [PubMed] [Google Scholar]
- 19. Oshchepkov D.Y., Vityaev E.E., Grigorovich D.A., Ignatieva E.V., Khlebodarova T.M.. SITECON: a tool for detecting conservative conformational and physicochemical properties in transcription factor binding site alignments and for site recognition. Nucleic Acids Res. 2004; 32:W208–W212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Y., Xi Z., Hegde R.S., Shakked Z., Crothers D.M.. Predicting indirect readout effects in protein–DNA interactions. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:8337–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carlson C.D., Warren C.L., Hauschild K.E., Ozers M.S., Qadir N., Bhimsaria D., Lee Y., Cerrina F., Ansari A.Z.. Specificity landscapes of DNA binding molecules elucidate biological function. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:4544–4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dror I., Rohs R., Mandel‐Gutfreund Y.. How motif environment influences transcription factor search dynamics: finding a needle in a haystack. BioEssays. 2016; 38:605–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Levo M., Segal E.. In pursuit of design principles of regulatory sequences. Nat. Rev. Genet. 2014; 15:453–468. [DOI] [PubMed] [Google Scholar]
- 24. Dror I., Golan T., Levy C., Rohs R., Mandel-Gutfreund Y.. A widespread role of the motif environment in transcription factor binding across diverse protein families. Genome Res. 2015; 25:1268–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gordân R., Shen N., Dror I., Zhou T., Horton J., Rohs R., Bulyk M.L.. Genomic regions flanking E-box binding sites influence DNA binding specificity of bHLH transcription factors through DNA shape. Cell Rep. 2013; 3:1093–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jolma A., Yan J., Whitington T., Toivonen J., Nitta K.R., Rastas P., Morgunova E., Enge M., Taipale M., Wei G. et al. DNA-binding specificities of human transcription factors. Cell. 2013; 152:327–339. [DOI] [PubMed] [Google Scholar]
- 27. Levo M., Zalckvar E., Sharon E., Dantas Machado A.C., Kalma Y., Lotam-Pompan M., Weinberger A., Yakhini Z., Rohs R., Segal E.. Unraveling determinants of transcription factor binding outside the core binding site. Genome Res. 2015; 25:1018–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Morin B., Nichols L.A., Holland L.J.. Flanking sequence composition differentially affects the binding and functional characteristics of glucocorticoid receptor homo- and heterodimers. Biochemistry. 2006; 45:7299–7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nagaoka M., Shiraishi Y., Sugiura Y.. Selected base sequence outside the target binding site of zinc finger protein Sp1. Nucleic Acids Res. 2001; 29:4920–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nutiu R., Friedman R.C., Luo S., Khrebtukova I., Silva D., Li R., Zhang L., Schroth G.P., Burge C.B.. Direct measurement of DNA affinity landscapes on a high-throughput sequencing instrument. Nat. Biotechnol. 2011; 29:659–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang L., Orenstein Y., Jolma A., Yin Y., Taipale J., Shamir R., Rohs R.. Transcription factor family‐specific DNA shape readout revealed by quantitative specificity models. Mol. Syst. Biol. 2017; 13:910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Le D.D., Shimko T.C., Aditham A.K., Keys A.M., Longwell S.A., Orenstein Y., Fordyce P.M.. Comprehensive, high-resolution binding energy landscapes reveal context dependencies of transcription factor binding. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E3702–E3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rajkumar A.S., Denervaud N., Maerkl S.J.. Mapping the fine structure of a eukaryotic promoter input-output function. Nat. Genet. 2013; 45:1207–1215. [DOI] [PubMed] [Google Scholar]
- 34. Schone S., Jurk M., Helabad M.B., Dror I., Lebars I., Kieffer B., Imhof P., Rohs R., Vingron M., Thomas-Chollier M. et al. Sequences flanking the core-binding site modulate glucocorticoid receptor structure and activity. Nat. Commun. 2016; 7:12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stormo G.D., Zhao Y.. Determining the specificity of protein–DNA interactions. Nat. Rev. Genet. 2010; 11:751–760. [DOI] [PubMed] [Google Scholar]
- 36. Bhimsaria D., Rodríguez-Martínez J.A., Pan J., Roston D., Korkmaz E.N., Cui Q., Ramanathan P., Ansari A.Z.. Specificity landscapes unmask sub-maximal binding site preferences of transcription factors. Proc. Natl. Acad. Sci. U.S.A. 2018; doi:10.1073/pnas.1811431115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou P., He A., Pu W.T.. Regulation of GATA4 transcriptional activity in cardiovascular development and disease. Curr. Top. Dev. Biol. 2012; 100:143–169. [DOI] [PubMed] [Google Scholar]
- 38. Lemons D., McGinnis W.. Genomic evolution of Hox gene clusters. Science. 2006; 313:1918–1922. [DOI] [PubMed] [Google Scholar]
- 39. Mann R.S., Lelli K.M., Joshi R.. Hox specificity unique roles for cofactors and collaborators. Curr. Top. Dev. Biol. 2009; 88:63–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Slattery M., Riley T., Liu P., Abe N., Gomez-Alcala P., Dror I., Zhou T., Rohs R., Honig B., Bussemaker H.J. et al. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell. 2011; 147:1270–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Curran T. Fos and Jun: oncogenic transcription factors. Tohoku J. Exp. Med. 1992; 168:169–174. [DOI] [PubMed] [Google Scholar]
- 42. Male V., Nisoli I., Gascoyne D.M., Brady H.J.. E4BP4: an unexpected player in the immune response. Trends Immunol. 2012; 33:98–102. [DOI] [PubMed] [Google Scholar]
- 43. Vinarov D.A., Newman C.L.L., Tyler E.M., Markley J.L., Shahan M.N.. Wheat germ Cell-Free expression system for protein production. Curr. Protoc. Protein Sci. 2006; doi:10.1002/0471140864.ps0518s44. [DOI] [PubMed] [Google Scholar]
- 44. Rodríguez-Martínez J.A., Reinke A.W., Bhimsaria D., Keating A.E., Ansari A.Z.. Combinatorial bZIP dimers display complex DNA-binding specificity landscapes. eLife. 2017; 6:e19272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Weirauch M.T., Yang A., Albu M., Cote A.G., Montenegro-Montero A., Drewe P., Najafabadi H.S., Lambert S.A., Mann I., Cook K. et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014; 158:1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Warren C.L., Kratochvil N.C.S., Hauschild K.E., Foister S., Brezinski M.L., Dervan P.B., Phillips G.N., Ansari A.Z.. Defining the sequence-recognition profile of DNA-binding molecules. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:867–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Friedel M., Nikolajewa S., Sühnel J., Wilhelm T.. DiProDB: a database for dinucleotide properties. Nucleic Acids Res. 2008; 37:D37–D40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brukner I., Sanchez R., Suck D., Pongor S.. Sequence‐dependent bending propensity of DNA as revealed by DNase I: parameters for trinucleotides. EMBO J. 1995; 14:1812–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Satchwell S.C., Drew H.R., Travers A.A.. Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol. 1986; 191:659–675. [DOI] [PubMed] [Google Scholar]
- 50. SantaLucia J. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:1460–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bolshoy A., McNamara P., Harrington R.E., Trifonov E.N.. Curved DNA without AA: experimental estimation of all 16 DNA wedge angles. Proc. Natl. Acad. Sci. U.S.A. 1991; 88:2312–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bansal M., Bhattacharyya D., Ravi B.. NUPARM and NUCGEN: software for analysis and generation of sequence dependent nucleic acid structures. Comput. Applic. Biosci.: CABIOS. 1995; 11:281–287. [DOI] [PubMed] [Google Scholar]
- 53. Lu X.J., Olson W.K.. 3DNA: a software package for the analysis, rebuilding and visualization of three‐dimensional nucleic acid structures. Nucleic Acids Res. 2003; 31:5108–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bishop E.P., Rohs R., Parker S.C.J., West S.M., Liu P., Mann R.S., Honig B., Tullius T.D.. A map of minor groove shape and electrostatic potential from hydroxyl radical cleavage patterns of DNA. ACS Chem. Biol. 2011; 6:1314–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tietjen J.R., Donato L.J., Bhimisaria D., Ansari A.Z.. Sequence-specificity and energy landscapes of DNA-binding molecules. Methods Enzymol. 2011; 497:3–30. [DOI] [PubMed] [Google Scholar]
- 56. Baldi P., Chauvin Y., Brunak S., Gorodkin J., Pedersen A.G.. Computational applications of DNA structural scales. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998; 6:35–42. [PubMed] [Google Scholar]
- 57. Suzuki M., Gerstein M.. Binding geometry of α‐helices that recognize DNA. Proteins: Struct. Funct. Bioinformatics. 1995; 23:525–535. [DOI] [PubMed] [Google Scholar]
- 58. Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E.. The Protein Data Bank. Nucleic Acids Res. 2000; 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kumar P., Bansal M.. HELANAL-Plus: a web server for analysis of helix geometry in protein structures. J. Biomol. Struct. Dyn. 2012; 30:773–783. [DOI] [PubMed] [Google Scholar]
- 60. El Hassan M.A., Calladine C.R.. Conformational characteristics of DNA: empirical classifications and a hypothesis for the conformational behaviour of dinucleotide steps. Philos. Trans. R. Soc. Lond. A. 1997; 355:43–100. [Google Scholar]
- 61. Berger M.F., Philippakis A.A., Qureshi A.M., He F.S., Estep P.W III, Bulyk M.L.. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 2006; 24:1429–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chen Y., Bates D.L., Dey R., Chen P.H., Dantas Machado A.C., Laird-Offringa I.A., Rohs R., Chen L.. DNA binding by GATA transcription factor suggests mechanisms of DNA looping and long-range gene regulation. Cell Rep. 2012; 2:1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Joshi R., Passner J.M., Rohs R., Jain R., Sosinsky A., Crickmore M.A., Jacob V., Aggarwal A.K., Honig B., Mann R.S.. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007; 131:530–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Passner J.M., Ryoo H.D., Shen L., Mann R.S., Aggarwal A.K.. Structure of a DNA-bound Ultrabithorax–Extradenticle homeodomain complex. Nature. 1999; 397:714–719. [DOI] [PubMed] [Google Scholar]
- 65. Abe N., Dror I., Yang L., Slattery M., Zhou T., Bussemaker H.J., Rohs R., Mann R.S.. Deconvolving the recognition of DNA shape from sequence. Cell. 2015; 161:307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Leonard D.A., Rajaram N., Kerppola T.K.. Structural basis of DNA bending and oriented heterodimer binding by the basic leucine zipper domains of Fos and Jun. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:4913–4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Leonard D.A., Kerppola T.K.. DNA bending determines Fos–Jun heterodimer orientation. Nat. Struct. Mol. Biol. 1998; 5:877–881. [DOI] [PubMed] [Google Scholar]
- 68. Dickerson R.E., Bansal M., Calladine C., Diekmann S., Hunter W.N., Kennard O., von Kitzing E., Lavery R., Nelson H.C.M., Olson W.K. et al. Definitions and nomenclature of nucleic acid structure parameters. J. Mol. Biol. 1989; 205:787–791. [DOI] [PubMed] [Google Scholar]
- 69. Rozenberg H., Rabinovich D., Frolow F., Hegde R.S., Shakked Z.. Structural code for DNA recognition revealed in crystal structures of papillomavirus E2-DNA targets. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:15194–15199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schultz S.C., Shields G.C., Steitz T.A.. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991; 253:1001–1007. [DOI] [PubMed] [Google Scholar]
- 71. Li J., Sagendorf J.M., Chiu T.-P., Pasi M., Perez A., Rohs R.. Expanding the repertoire of DNA shape features for genome-scale studies of transcription factor binding. Nucleic Acids Res. 2017; 45:12877–12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Vafabakhsh R., Ha T.. Extreme bendability of DNA less than 100 base pairs long revealed by single-molecule cyclization. Science. 2012; 337:1097–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Afek A., Schipper J.L., Horton J., Gordân R., Lukatsky D.B.. Protein− DNA binding in the absence of specific base-pair recognition. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:17140–17145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.