Abstract
Small nucleolar RNAs (snoRNAs) constitute a family of non-coding RNAs that are classically known as guide RNAs for processing and modification of ribosomal RNAs. Recently, it was discovered that snoRNAs can be further processed into sno-derived RNAs (sdRNAs), some of which are known to exhibit microRNA-like properties. SdRNAs have been implicated in human cancer; however, a systems-level sdRNA landscape in human cancers is lacking. Through integrative analysis of ~22 nt size-selected smRNA-seq datasets from 10,262 patient samples across 32 cancer types, we mapped a pan-cancer sdRNAome and interrogated its signatures in multiple clinically relevant features, particularly cancer immunity and clinical outcome. Aggregating sdRNA abundances by parental snoRNAs, these expression signatures alone are sufficient to distinguish patients with distinct cancer types. Interestingly, a large panel of sdRNAs are significantly correlated with features of the tumor-immune microenvironment, such as immunosuppressive markers, CD8+ T cell infiltration, cytolytic T cell activity, and tumor vasculature. A set of individual sdRNAs with tumor-immune signatures can also stratify patient survival. These findings implicate snoRNAs and their derivative sdRNAs as a class of prevalent non-coding molecular markers of human cancer immunity.
Introduction
Over the past decade, tumor immunity has realized its central importance in oncology 1. Checkpoint blockade immunotherapies targeting the Program Cell Death Protein 1 (PD-1) and Cytotoxic T Lymphocyte Associated Protein 4 (CTLA-4) pathways have revolutionized cancer therapeutics with unprecedented durable responses across multiple cancer types 2–4. However, not all patients respond to checkpoint inhibitors 5. Identifying the molecular correspondents underlying these differential responses is essential to expanding the patient population that can benefit from cancer immunotherapy. Emerging biomarkers for anti-PD-1 response include the expression level of its ligand PD-L1 2, mutation burden or mismatch-repair deficiency 6, and tumor-infiltrating lymphocytes 7. However, currently available markers are insufficient for accurate prediction of immunotherapy responses. Thus, development and investigation of further molecular markers for tumor immunity, especially in large cohorts of human patients, might provide novel insights for precision diagnostics and treatments in immunooncology.
Somatic mutations in oncogenes and tumor suppressors represent the most classic biomarkers as they are the main direct drivers of cancer progression 8,9. Small non-coding RNAs have great potential as molecular markers due to their important biological roles and bioavailability in circulation, facilitating implementation in liquid biopsy settings 10. Since the last decade, microRNAs have been documented as biomarkers for classifying human cancers. For example, analysis of the miRnome using Fluidigm expression profiling has revealed its functional and prognostic importance in classifying clinical populations 11. More recently, molecular profiling of >3,000 tumors from 11 human cancer types in TCGA has enabled systematic analysis of microRNAs and their targets 12. Several other types of non-coding RNAs, such as long-noncoding RNAs (lncRNAs) 13,14, enhancer RNAs 15,16 and circular RNAs 17 have also been implicated in various cancer types 18. While these markers have enhanced our knowledge of cancer diagnostics and prognosis, the immense heterogeneity observed within and across cancer patients far exceeds our current understanding. Molecular markers that can further inform distinct signatures of tumor initiation, progression, and anti-cancer immunity will continue to be of high clinical importance.
Small nucleolar RNAs (snoRNAs) comprise a class of highly conserved small non-coding RNAs that are primarily localized in the nucleolus 19. SnoRNAs have essential roles as guide RNAs for ribosomal RNA processing and several types of RNA modifications such as methylation and pseudouridylation 20. In addition to their essential role in ribosome biogenesis, snoRNAs have also been implicated in chromatin structure regulation 21, RNA splicing 22,23, and protein signaling 24. A number of biologically and clinically important snoRNAs have recently been reported in several cancer types 25,26, implying that snoRNAs might have more pervasive roles in human cancer than previously appreciated.
Recent studies have revealed that snoRNAs can be further processed into smaller RNAs, termed sno-derived RNAs (sdRNAs) 27–30. Whereas snoRNAs range widely in size from 60–300 nt, sdRNAs generally vary from 20–30 nt 27–33 and are similar in size to microRNAs (19–25 nt) 34. Interestingly, sdRNAs preferentially arise from the 5’ or 3’ ends of snoRNAs 31,32,35,36, and the classical RNAi processing proteins Argonaute and Dicer are important for catalyzing sdRNA biogenesis 30,37. Crucially, a handful of sdRNAs have also been demonstrated to have microRNA-like gene regulatory activity 27. Given the remarkable similarities between microRNAs and sdRNAs, it has been hypothesized that sdRNAs might also contribute to the pathogenesis of diverse diseases – for instance, cancer.
Here we performed a pan-cancer analysis of ~22 nt size-selected small RNA-seq (smRNA-seq) datasets from TCGA, exploring the expression of small RNAs mapping to annotated human snoRNAs in 10,262 patient samples across 32 cancer types. Due to the size selection strategy used for generating the sequencing libraries, it was anticipated that the resultant mapped reads were from sdRNAs rather than full-length snoRNAs. This hypothesis was borne out by subsequent analysis of the read lengths mapping to snoRNAs and the read distributions along individual snoRNAs, which demonstrated heavily skewed read densities that are characteristic of sdRNA biogenesis. We therefore generated a landscape of the sdRNAome, subsequently linking sdRNAs to clinically significant features including tumor immunity and overall survival. Since few sdRNAs have been characterized and officially named, in the interest of clarity, we refer to sdRNAs using the name of the parental snoRNA (i.e., SNORD116 refers to the sdRNA derived from SNORD116) and aggregate sdRNA abundances to the level of snoRNAs. Nevertheless, we emphasize that the data and analyses presented here pertain to sdRNAs, rather than full-length snoRNAs.
Aggregated by parental snoRNA, we found that sdRNA expression signatures alone can classify patients from distinct cancer types at high resolution. Over 40 single sdRNAs can stratify patient survival in two or more cancer types, alongside with numerous single-cancer-type-specific ones. Furthermore, many sdRNAs significantly correlate with tumor-immune microenvironment features such as PD-L1 levels, T cell infiltration, functional anti-cancer cytotoxic scores and tumor vascularization. A panel of sdRNAs are significant markers for both immune and survival features, with a total of 25 sdRNAs scored in 4 or more cancer types, in addition to their angiogenesis, copy number and metastasis signals. Our analyses demonstrate that sdRNAs are significant and prevalent molecular markers across multiple types of human cancer.
Results
A comprehensive map of the sdRNA transcriptome across multiple human cancer types
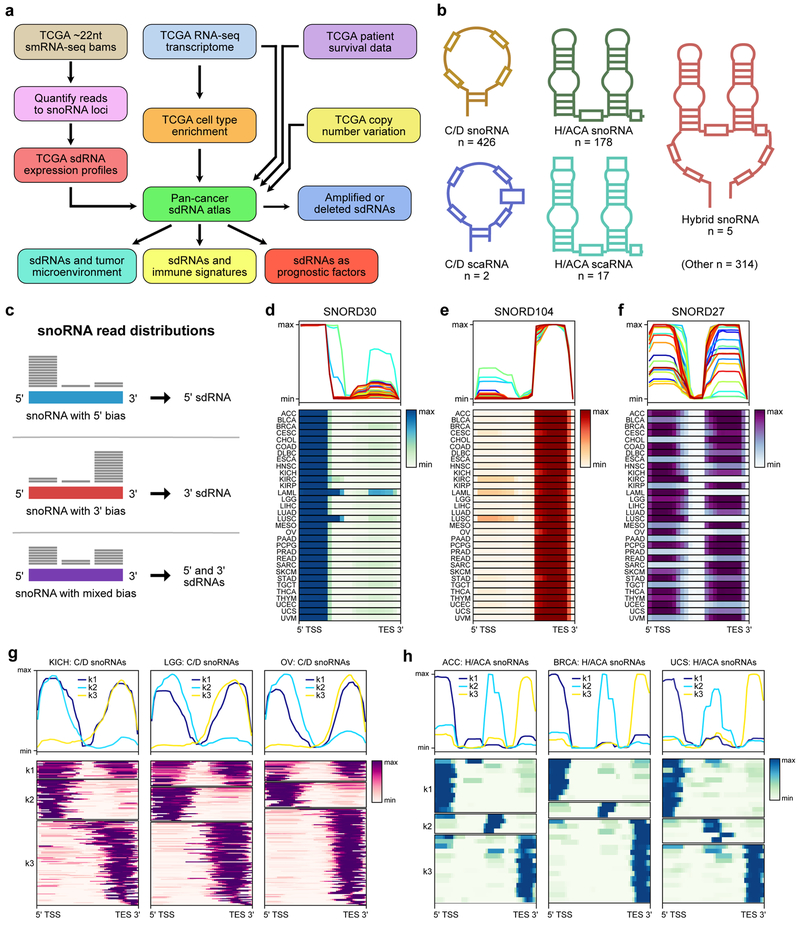
We retrieved all available small RNA-seq (smRNA-seq) reads from TCGA via NCI GDC, which consist of a total of 10,262 patient tumor samples and 675 adjacent normal samples, encompassing 32 cancer types (Methods). The smRNA-seq library construction protocols used by TCGA investigators were designed to enrich for ~22nt sized transcripts, with the primary goal of capturing microRNAs 38. Whereas full-length snoRNAs range in size from 60–300nt and thus would not be expected to be captured, sdRNAs are anticipated to be found within this size range 27–30,30–33,39. We consequently quantified the reads mapping to all annotated snoRNAs (snoRNAome) 40 to construct the sdRNA transcriptome (Figure S1a-b, Figure 1a). This pan-cancer sdRNA transcriptome is derived from several subtypes of snoRNAs with distinct structures and motifs, such as canonical C/D box snoRNAs, H/ACA box snoRNAs, C/D box small Cajal body RNAs (scaRNAs), H/ACA box scaRNAs, hybrid snoRNAs, and several other subtypes (Figure 1b, Table S1, Table S2).
Figure 1: A pan-cancer landscape of the sdRNA transcriptome.
a. Schematic of datasets and analytic flowchart for integrative analysis of the pan-cancer sdRNAome.
b. Schematic depicting the structural features of different snoRNAs. 5 select snoRNA types are illustrated here.
c. Schematic describing the three types of C/D snoRNA read distributions identified in the smRNA-seq datasets. These read distributions correspond to three different types of sdRNAs produced from the parental snoRNAs.
d. Average profile and heat map of SNORD30 read distributions in 32 cancer types. The smRNA-seq reads that mapped to SNORD30 were consistently concentrated on the 5’ end, suggesting that SNORD30 is processed into 5’ sdRNAs.
e. Average profile and heat map of SNORD104 read distributions in 32 cancer types. The smRNA-seq reads that mapped to SNORD104 were consistently concentrated on the 3’ end, suggesting that SNORD30 is processed into 3’ sdRNAs.
f. Average profile and heat map of SNORD27 read distributions in 32 cancer types. The smRNA-seq reads that mapped to SNORD27 were concentrated on either the 5’ or 3’ end, suggesting that SNORD30 is processed into 5’ or 3’ sdRNAs.
g. Average profiles and heat maps of the mapped read distributions from all expressed C/D snoRNAs in kidney chromophobe cancers (KICH, left), low-grade glioma (LGG, center), and ovarian adenocarcinoma (OV, right). The read distributions clustered into three groups by k-means clustering (k1, k2, k3) corresponding to the types depicted in c. Values shown are normalized to maximum read depth for each snoRNA.
h. Average profiles and heat maps of the mapped read distributions from all expressed H/ACA snoRNAs in adrenocortical carcinoma (ACC, left), breast adenocarcinoma (BRCA, center), and uterine carcinosarcoma (UCS, right). The read distributions clustered into three groups by k-means clustering (k1, k2, k3). Values shown are normalized to maximum read depth for each snoRNA.
TSS, transcription start site. TES, transcription end site.
To confirm that the TCGA smRNA-seq datasets are indeed suitable for analysis of sdRNAs, we randomly selected 5 tumors from each cancer type and tabulated the read lengths mapping to the snoRNAome (Figure S2). As anticipated, these analyses revealed that snoRNA-mapping reads mostly ranged in size from 20–30 nt, which is consistent with the size ranges of known sdRNAs. Prior studies have also established that sdRNAs tend to be asymmetrically produced from either the 5’ or 3’ ends of snoRNA transcripts 30. We therefore computed the distribution of the reads mapping to each snoRNA to see if the ~22 nt size-selected transcripts could be consistent with sdRNAs. In the case of C/D snoRNAs, these analyses revealed three classes of read distributions, corresponding to 5’ sdRNAs, 3’ sdRNAs, or mixed sdRNAs (Figure 1c). For instance, reads that mapped to SNORD30 were consistently concentrated on the 5’ end, indicating that SNORD30 is processed into 5’ sdRNAs in a highly conserved manner between different cancer types (Figure 1d). In contrast, reads that mapped to SNORD104 were heavily concentrated on the 3’ end in all cancer types, suggesting that SNORD104 is processed into 3’ sdRNAs (Figure 1e). Unlike with SNORD30 and SNORD104, reads mapping to SNORD27 were instead distributed along both 5’ and 3’ ends (Figure 1f). Of note, different cancer types exhibited distinct balances between the abundance of 5’ and 3’ sdRNAs, suggesting alternate modes of sdRNA biogenesis from SNORD27. Across all expressed C/D snoRNAs, read distributions for each snoRNA were consistently clustered into these three groups regardless of cancer type (Figure 1g, Figure S3). We similarly performed these analyses with H/ACA snoRNAs and found three types of read distributions, corresponding to 5’ sdRNAs, 3’ sdRNAs, and centrally located sdRNAs (Figure 1h, Figure S4). Collectively, these analyses indicate that the TCGA smRNA-seq datasets can be utilized for the study of sdRNAs, and not necessarily for full-length snoRNAs. Though we hereafter refer to the data aggregated by parental snoRNA, we reiterate that the analyses presented here are based on the sdRNA transcriptome, rather than full-length snoRNAs.
To explore the landscape of sdRNA expression across cancers, we calculated the median abundance of each snoRNA (specifically, using aggregated sdRNA levels) within each cancer type (n = 942 snoRNAs) (Figure S1c, Table S3). The sdRNA transcriptome exhibited a wide dynamic range of expression across all cancers (Figure S5a), such that 300.13 ± 4.21 (mean ± s.e.m.) sdRNAs were identified in each cancer type with abundances of median log2 tpm ≥ 1. To test whether certain sdRNAs were more highly expressed in specific cancer types, we assessed the relative expression patterns for each sdRNA across all cancer types (Figure 2a). This revealed that different cancer types are associated with unique sdRNA signatures. Based on median expression within each cancer type, there are three categories of sdRNAs: (1) highly prevalent sdRNAs (expressed in ≥ 10 cancer types, n = 320); (2) subgroup-associated sdRNAs (expressed in 3–9 cancer types, n = 45); and (3) tissue-specific sdRNAs (expressed in only 1 or 2 cancer types, n = 34). For instance, 15 different sdRNAs appeared highly specific for testicular germ cell tumors (TGCT) in terms of median expression (z-score > 5). We next looked to characterize the sdRNA expression landscape in individual tumors, and identified 300 expressed sdRNAs as “high variance” (variance > 0.1, median > 0). By examining co-expression modules, we found that a subset of these 300 high variance sdRNAs clustered into discrete groups (Figure S5b, Table S4). Note that the constituent sdRNAs within a cluster did not necessarily have equivalent median expression levels over the dataset (Figure S5b). This analysis indicates that specific sets of sdRNAs are coordinately expressed across cancers from different tissues of origin, further suggesting that the process of sdRNA biogenesis from snoRNAs is coordinately regulated in cancer.
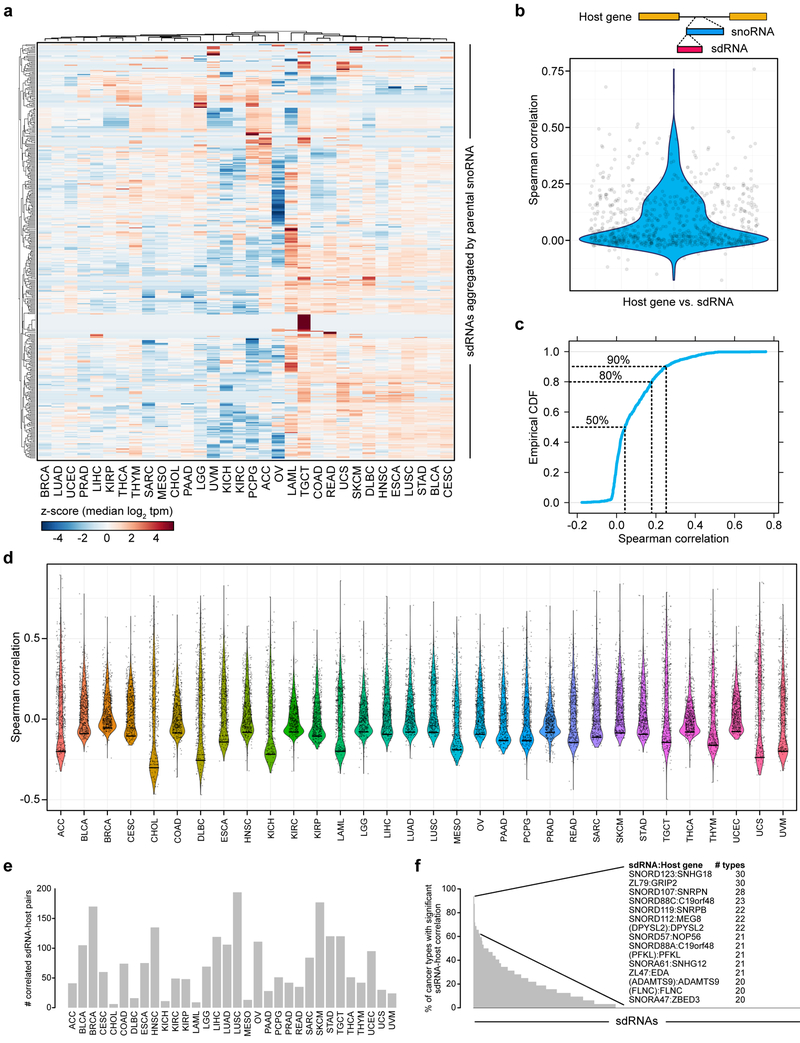
Figure 2: sdRNA expression patterns differ among cancer types and are not adequately explained by host gene transcription.
a. Heat map of relative sdRNA expression aggregated by parental snoRNA across cancer types, filtered for sdRNAs exhibiting non-zero variance in median values. Values shown are z-score transformations of the median log2 tpm, normalized by individual sdRNAs. These data demonstrate that while some sdRNAs are relatively evenly expressed among all cancer types, there are clusters of sdRNAs with highly tissue-specific expression patterns.
b. Violin plot comparing the aggregated abundance of sdRNAs from each snoRNA with its host gene (n = 736 snoRNA – host gene pairs) across all cancers. The top schematic illustrates an intronic snoRNA and the associated sdRNA. The Spearman correlation across all pairs was 0.089 ± 0.004 (mean ± s.e.m.), indicating that sdRNA abundance cannot be adequately explained by host gene transcription.
c. Empirical cumulative density function of the Spearman correlation distribution in b. The dotted lines indicate what cumulative proportion of total snoRNA – host gene pairs (y-axis) have a correlation coefficient up to the indicated point (x-axis).
d. Violin plot comparing the aggregated sdRNA abundance of each snoRNA with its host gene (n = 736 sdRNA – host gene pairs) in each individual cancer type.
e. Bar plot detailing the number of significantly correlated sdRNA-host gene pairs in each cancer type (adjusted p < 0.05).
f. Bar plot detailing the percentage of cancer types in which each sdRNA-host gene pair was found to be significantly correlated (adjusted p < 0.05). Inset, top sdRNA-host gene pairs and the number of cancer types that the pair was found to be significantly correlated.
Of note, previous studies of snoRNAs have occasionally utilized transcription levels of host genes (i.e. the protein-coding genes within which snoRNAs are encoded) as a proxy for snoRNA expression 24. To evaluate this assumption in the context of sdRNAs in cancer, we extracted matching mRNA-seq data from the TCGA database (n = 8,954 cancer samples with corresponding smRNA-seq and mRNA-seq data). Using snoRNA-host gene annotations 40, we calculated the Spearman correlation between each snoRNA (based on sdRNA abundance) and host gene pair (n = 736 annotated pairs with matching data) (Figure 2b, Table S5). Analysis of the distribution of correlation coefficients revealed that the 50% of all pairs had Spearman correlation coefficients less than 0.043 (Figure 2b-c). Additionally, 80% of all pairs had Spearman correlation coefficients less than 0.179, while 90% of all pairs had coefficients less than 0.249 (Figure 2b-c). We further investigated this correlation in a cancer type-specific manner (Figure 2d), again finding that the majority of sdRNA-host gene pairs are not significantly correlated in any single cancer type (Figure 2e-f, Table S6). There were a few notable exceptions, most strikingly SNORD123 and SNHG18, as well as ZL79 and GRIP2. SNORD123 was found to be significantly correlated with SNHG18 in 30 different cancer types, as was ZL79 with GRIP2 (Figure 2f). However, these particular examples were clearly not representative of all sdRNA-host gene pairs. Thus, these analyses suggest that in the TCGA smRNA-seq data, the bulk of sdRNA expression patterns are incompletely captured by the transcription levels of host genes alone, potentially reflecting the importance of regulating sdRNA biogenesis from snoRNAs, which in turn may derive from host gene transcription. Together, this initial analysis generated a pan-cancer dataset of the sdRNA transcriptome (PANCAN32), where the dynamic range and tissue-specific or cancer type-specific patterns enables subsequent analysis of associated quantitative phenotypes and clinical features.
SdRNA transcriptome stratifies distinct groups of patients from various cancer types
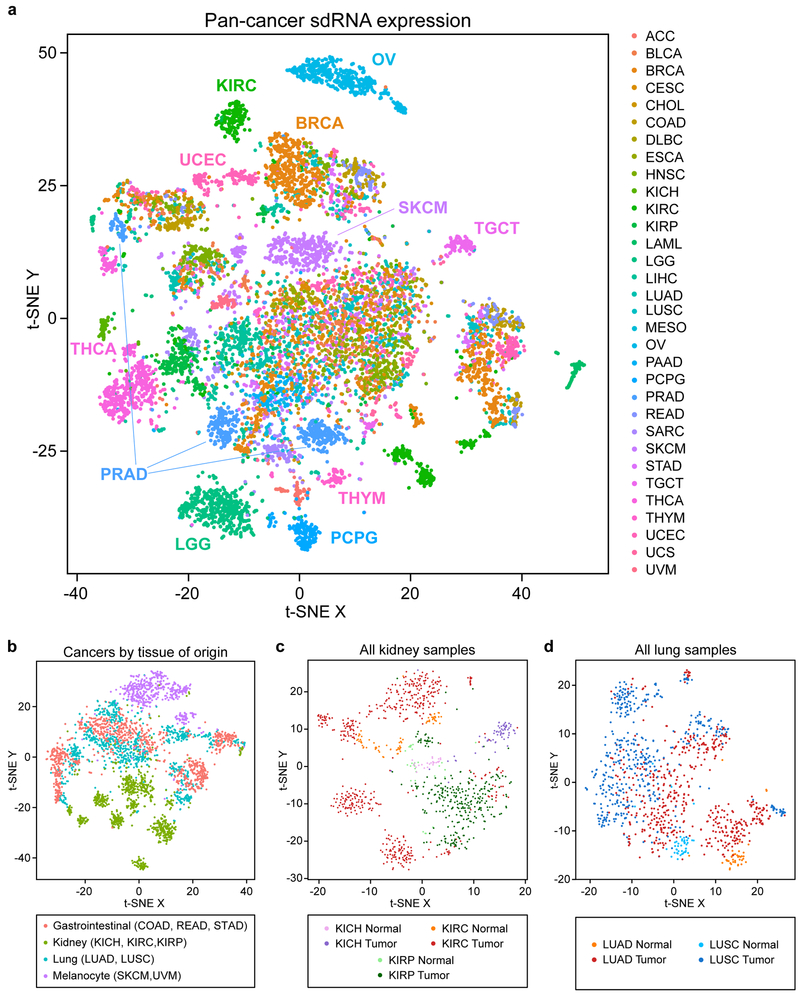
To test whether sdRNA expression can mark molecular signatures or classify distinct cancer types, we first utilized a dimensional reduction approach. We performed t-distributed stochastic neighbor embedding (t-SNE) 41 on the PANCAN32 dataset and visualized the resulting transformations both in individual cancer types (Figure S6) and all cancers as a whole (Figure 3a). The t-SNE visualization revealed a high-level clustering map of all 32 cancer types according to the expression of the sdRNA transcriptome in each patient sample. This map showed that while several cancer types clustered together, there are multiple clusters that were primarily comprised of patient populations from only one cancer type (Figure 3a). These type-specific segregations were particularly apparent for individual cancer types such as thyroid carcinoma (THCA), lower grade gliomas (LGG), pheochromocytomas and paragangliomas (PCPG), skin cutaneous melanoma (SKCM), kidney papillary cell carcinoma (KIRP), uterine corpus endometrial carcinoma (UCEC) and ovarian adenocarcinoma (OV) (Figure 3a, Figure S6). An especially striking example was observed for prostate adenocarcinoma (PRAD), where 3 sub-clusters are readily apparent on t-SNE visualization (Figure 3a, Figure S6). In general, samples from the same cancer type can either cluster tightly as primarily one cluster (e.g. ACC, OV, PAAD, PCPG, SKCM, THCA and UVM), fragment into several discrete sub-clusters (e.g. BRCA, CESC, COAD, KIRC, PRAD), or have a relatively diffuse distribution across the multi-dimensional space (e.g. BLCA, LIHC, LUAD, LUSC, SARC, STAD and UCEC) (Figure S6). Accordingly, several clusters were comprised of patient populations from multiple cancer types, as sub-populations of patients from different types occupied the same multi-dimensional space (Figure 3a). Collectively, these results demonstrate that sdRNA expression signatures alone are sufficient to distinguish certain cancer types, while also uncovering substantial patient heterogeneity within individual cancer types.
Figure 3: High dimensional pan-cancer patient clustering based on sdRNA expression signatures.
a. t-SNE plot of sdRNA expression in tumors from 32 different cancer types (n = 10,262), aggregated by parental snoRNA. Samples are colored by cancer type.
b. t-SNE plot of gastrointestinal, kidney, lung, and melanocyte derived cancers, colored by tissue of origin.
c. t-SNE plot of all normal kidney and kidney tumor samples.
d. t-SNE plot of all normal lung and lung tumor samples.
We wondered if the divergent sdRNA expression signatures were due to cancer-specific molecular changes, or rather simply due to sdRNA expression differences in the normal tissues from which the cancers had risen. Analysis of 675 adjacent normal samples revealed that sdRNA expression patterns were quite distinct across the various normal tissues (Figure S7a), suggesting that the differences in sdRNA expression across different cancer types are at least partly attributable to the tissue of origin. To further investigate this possibility, we selected tissues with multiple cognate cancer types represented: gastrointestinal (COAD, READ, STAD), kidney (KICH, KIRC, KIRP), lung (LUAD, LUSC), and melanocytes (SKCM, UVM). We found that whereas kidney-derived and melanocyte-derived cancers could be readily distinguished, the gastrointestinal-derived and lung-derived cancers were much more heterogeneous (Figure 3b). Even still, the 3 types of kidney cancers and 2 types of melanomas were nevertheless clearly distinguishable from each other (Figure S7b). This finding indicates that despite having arisen from a similar tissue of origin, different cancer types exhibit divergent sdRNA expression patterns. To further investigate this relationship, we next directly compared all normal kidney and kidney tumor samples. These analyses revealed that while the normal kidney samples were grouped together in the center of the multidimensional space, the different kidney cancer subtypes radiated outwards from the center, indicating progressive changes in sdRNA expression signatures from the normal tissue in a cancer type-specific manner (Figure 3c). This finding was similarly corroborated by analysis of all normal lung and lung tumor samples, with normal lung samples grouped together and the different lung cancer types splayed out across the multidimensional space (Figure 3d). Of note, LUSC tumors were more distant from normal samples compared to LUAD samples. This is consistent with our understanding of the different cell types present in LUAD vs LUSC; whereas LUAD is characterized by alveolar-like cells that are present in the normal lung, LUSC is characterized by basal cells that instead mimic the esophageal squamous epithelium 42. In aggregate, these analyses demonstrate that while tissue of origin certainly influences sdRNA expression in cancer, different cancers arising from the same organ can nevertheless be distinguished by their sdRNA signatures.
The PANCAN32 t-SNE visualization coded by tumor immune signature or patient survival presented overviews of the sdRNA transcriptome across all these patients as related to various relevant clinical features (Figure S7c-f), suggesting that sdRNA expression might demarcate these clinical features, which are analyzed in further depth (later in text).
SdRNA transcriptome informs molecular and cellular features of tumor immunity
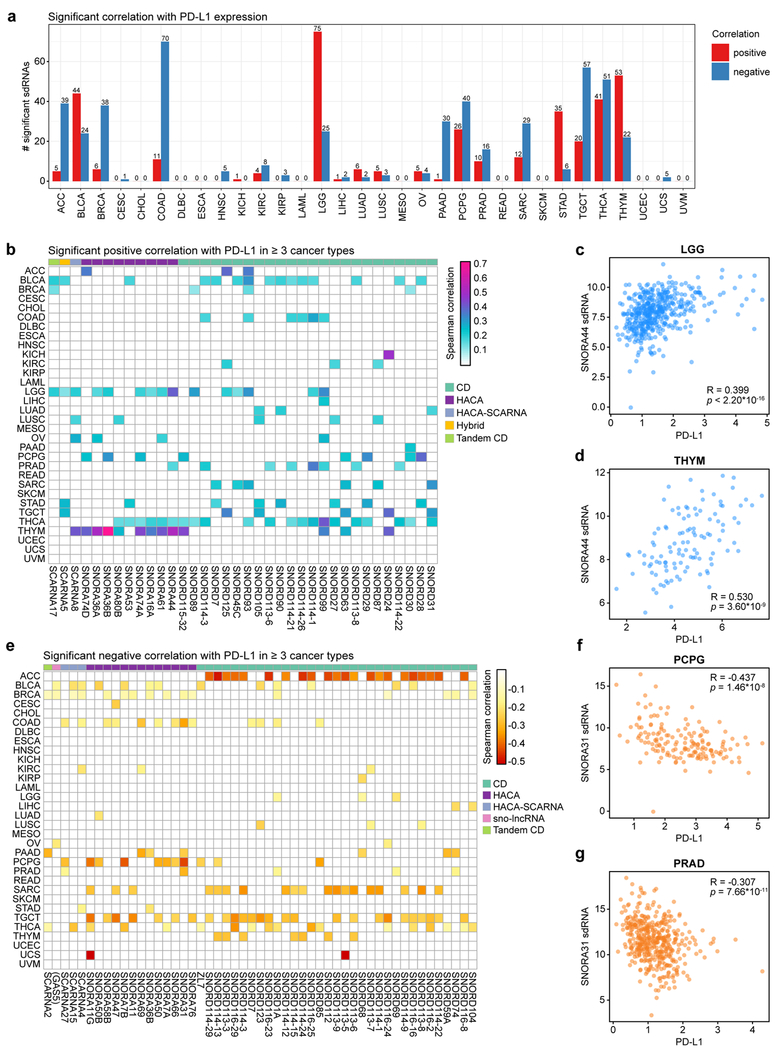
To investigate the potential utility of sdRNAs in understanding tumor immunity, we first systematically assessed the correlation between all individual snoRNAs (through sdRNA abundance levels) and PD-L1 (encoded by the CD274 gene) expression as determined by mRNA-seq. We reiterate that all analyses pertain to sdRNA abundances, but are described in reference to the parental snoRNAs. Remarkably, a total of 350 sdRNAs were found to be significantly correlated with PD-L1 in at least one cancer type (Benjamini-Hochberg adjusted p < 0.05) (Table S8). We analyzed the distribution of the number of predictive sdRNAs for each cancer type, and found that sdRNAs are most predictive (in terms of the number of significant sdRNAs) in cancer types such as adrenocortical carcinoma (ACC), bladder urothelial carcinoma (BLCA), colon adenocarcinoma (COAD), lower grade glioma (LGG), pheochromocytoma and paraganglioma (PCPG), testicular germ cell tumors (TGCT), thyroid carcinoma (THCA), and thymoma (THYM) (Figure 4a). In contrast, several cancer types have few predictive sdRNAs for PD-L1, such as cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), diffuse large B-cell lymphoma (DLBC), esophageal carcinoma (ESCA), kidney chromophobe (KICH), acute myeloid leukemia (LAML), mesothelioma (MESO), rectal adenocarcinoma (READ), skin cutaneous melanoma (SKCM), uterine corpus endometrial carcinoma (UCEC) and uveal melanoma (UVM) (Figure 4a). Among all 32 cancer types, lower grade glioma has the most sdRNAs positively correlated with PD-L1, whereas colon adenocarcinoma has the most sdRNAs negatively correlated with PD-L1 (Figure 4a). Of the significant sdRNAs, 36 were found to be positively correlated in 3 or more cancer types (adjusted p < 0.05) (Figure 4b, Table S8). For instance, sdRNAs derived from SNORA44, an H/ACA-type snoRNA, were significantly correlated with PD-L1 expression in lower grade gliomas (LGG) (Figure 4c), prostate adenocarcinoma (PRAD), thyroid carcinoma (THCA), and thymoma (THYM) (Figure 4d). We further identified sdRNAs that were significantly negatively correlated with PD-L1 expression (adjusted p < 0.05) (Figure 4e, Table S8). A total of 51 sdRNAs were negatively correlated with PD-L1 expression in at least 3 cancer types. For instance, sdRNAs produced from SNORA31, another H/ACA box snoRNA, were inversely correlated with PD-L1 levels in breast adenocarcinoma (BRCA), colon adenocarcinoma (COAD), pancreatic adenocarcinoma (PAAD), pheochromocytomas (PCPG) (Figure 4f) and prostate adenocarcinoma (PRAD) (Figure 4g). Collectively, these findings link sdRNAs to PD-L1 expression in cancers of diverse origins, suggesting that sdRNAs have predictive power in the expression of this immunosuppressive molecule across diverse patient tumors.
Figure 4: sdRNAs are correlated with PD-L1 expression in human cancers.
a. Bar plot depicting the number of significant sdRNAs in each cancer type, aggregated by parental snoRNA, in relation to PD-L1 expression. Red, positive correlation; blue, negative correlation.
b. Heat map of sdRNAs positively correlated with PD-L1 (CD274) expression (adjusted p < 0.05, adjusted within each cancer type). For visibility, only sdRNAs that were positively correlated in three or more cancer types are shown. Boxes are colored according to the Spearman correlation with PD-L1. Parental snoRNAs without annotated names are instead labeled by their host gene in parentheses. SnoRNA classifications are annotated on top based on a color legend on the right panel.
c. Scatter plot depicting the correlation between PD-L1 and SNORA44 sdRNA expression in lower grade gliomas (LGG, n = 453). Spearman correlation R = 0.399, p < 2.20 * 10-16.
d. Scatter plot depicting the correlation between PD-L1 and SNORA44 sdRNA expression in thymomas (THYM, n = 111). R = 0.530, p = 3.60 * 10-9.
e. Heat map of snoRNAs negatively correlated with PD-L1 (CD274) expression (adjusted p < 0.05, adjusted within each cancer type). For visibility, only sdRNAs that were positively correlated in three or more cancer types are shown. Boxes are colored according to the Spearman correlation with PD-L1. Parental snoRNAs without annotated names are instead labeled by their host gene in parentheses. SnoRNA classifications are annotated on top based on a color legend on the right panel.
f. Scatter plot depicting the correlation between PD-L1 and SNORA31 sdRNA expression in pheochromoytomas and paragangliomas (PCPG, n = 157). R = −0.437, p = 1.46 * 10-8.
g. Scatter plot depicting the correlation between PD-L1 and SNORA7B sdRNA expression in prostate adenocarcinoma (PRAD, n = 435). R = −0.307, p = 7.66 * 10-11.
c, d, f, g - Data are shown as tpm, normalized separately for mRNA-seq and smRNA-seq datasets.
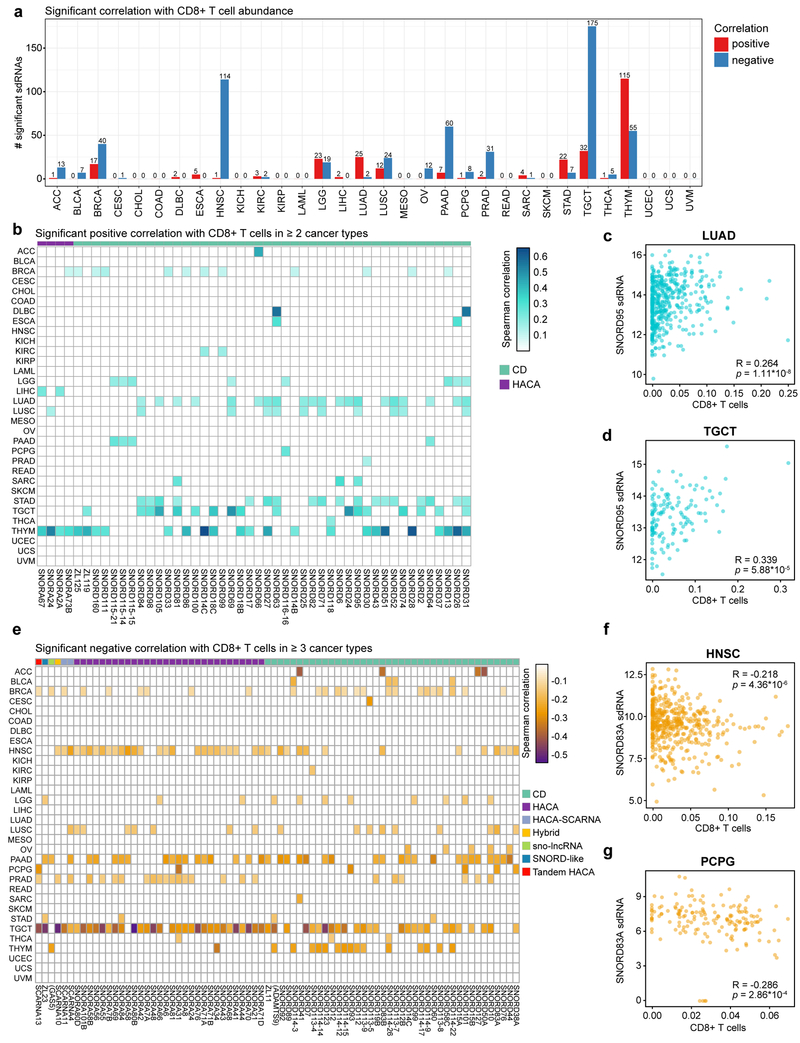
The number of tumor-infiltrating lymphocytes (TILs), especially cytotoxic CD8+ T cells, is a strong positive predictive biomarker of checkpoint blockade immunotherapy efficacy and patient outcome for certain types of solid tumors 2,5. We therefore interrogated the sdRNAome (aggregated by parental snoRNA) in relation to intratumoral CD8+ T cell abundance. We found that a total of 366 sdRNAs were significantly correlated with CD8+ infiltration level in at least one cancer type (adjusted p < 0.05) (Table S9). The number of predictive sdRNAs also varied between cancer types, among which the most predictive ones are breast adenocarcinoma (BRCA), head and neck squamous cell carcinoma (HNSC), lower grade glioma (LGG), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), pancreatic adenocarcinoma (PAAD), prostate adenocarcinoma (PRAD), stomach adenocarcinoma (STAD), testicular germ cell tumors (TGCT) and thymoma (THYM) (Figure 5a). In contrast, cancer types such as CESC, COAD, KIRP, READ, SKCM, UCEC, UCS and UVM have few or no predictive sdRNAs for CD8+ T cell abundance (Figure 5a). Thymoma has the most sdRNAs positively correlated with CD8+ T cell infiltration, whereas testicular germ cell tumor has the most sdRNAs negatively correlated with CD8+ TILs (Figure 5a). 48 sdRNAs were positively correlated with CD8+ T cell abundance in at least 2 cancer types (adjusted p < 0.05) (Figure 5b, Table S9). As an example, sdRNAs from the C/D box snoRNA SNORD95 were significant in lung adenocarcinoma (LUAD) (Figure 5c), lung squamous cell carcinoma (LUSC), sarcoma (SARC), stomach adenocarcinoma (STAD), and testicular germ cell tumors (TGCT) (Figure 5d). On the other hand, 23 sdRNAs were significantly negatively correlated with CD8+ T cell abundance in 4 or more cancer types (adjusted p < 0.05) (Figure 5e, Table S9). These include sdRNAs from SNORD83A, a C/D box snoRNA which was found to be significant in head and neck squamous cell carcinoma (HNSC) (Figure 5f), pheochromocytoma and paraganglioma (PCPG) (Figure 5g), lung squamous cell carcinoma (LUSC), and pancreatic adenocarcinoma (PAAD).
Figure 5: sdRNAs are correlated with CD8+ T cell infiltration in diverse cancers.
a. Bar plot depicting the number of significant sdRNAs in each cancer type, aggregated by parental snoRNA, in relation to CD8+ T cell abundance. Red, positive correlation; blue, negative correlation.
b. Heat map of sdRNAs positively correlated with CD8+ T cell abundance (adjusted p < 0.05, adjusted within each cancer type). For visibility, only sdRNAs that were positively correlated in two or more cancer types are shown. Boxes are colored according to the Spearman correlation with CD8+ T cell abundance. Parental snoRNAs without annotated names are instead labeled by their host gene in parentheses. SnoRNA classifications are annotated on top based on a color legend on the right panel.
c. Scatter plot depicting the correlation between CD8+ T cell abundance and SNORD95 sdRNA expression in lung adenocarcinoma (LUAD, n = 454). R = 0.264, p = 1.11 * 10-8.
d. Scatter plot depicting the correlation between CD8+ T cell abundance and SNORD95 sdRNA expression in testicular germ cell tumors (TGCT, n = 135). R = 0.339, p = 5.88 * 10-5.
e. Heat map of sdRNAs negatively correlated with CD8+ T cell abundance (adjusted p < 0.05, adjusted within each cancer type). For visibility, only sdRNAs that were positively correlated in four or more cancer types are shown. Boxes are colored according to the Spearman correlation with CD8+ T cell abundance. Parental snoRNAs without annotated names are instead labeled by their host gene in parentheses. SnoRNA classifications are annotated on top based on a color legend on the right panel.
f. Scatter plot depicting the correlation between CD8+ T cell abundance and SNORD83A sdRNA expression in head and neck squamous cell carcinoma (HNSC, n = 435). R = −0.218, p = 4.36 * 10-6.
g. Scatter plot depicting the correlation between CD8+ T cell abundance and SNORD83A sdRNA expression in pheochromocytomas and paragangliomas (PCPG, n = 157). R = −0.286, p = 2.86 * 10-4.
c, d, f, g - Data are shown as tpm for smRNA-seq datasets.
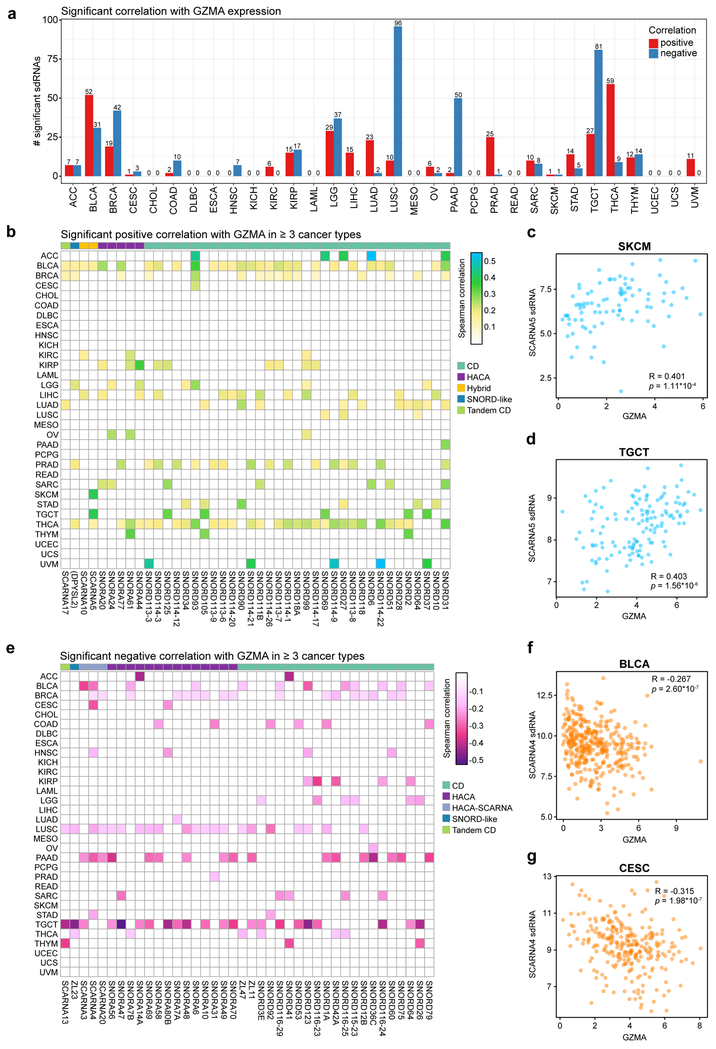
While infiltration of CD8+ T cells is a critical prerequisite for anti-cancer immune responses, intratumoral T cells may be anergic, exhausted or nonfunctional. To assess T cell cytolytic activity, we next investigated the relationship between expression of sdRNAs and GZMA, a gene that encodes the serine protease granzyme A, a key component of cytotoxic T cell granules that has previously been used as a marker of cytolytic activity in human cancer 43. Of note, mRNA levels of Perforin1 (PRF1) across the PANCAN32 dataset strongly correlates with GZMA (correlation = 0.9), and thus the cytotoxic scores using either GZMA, PRF1 or GZMA+PRF1 are highly similar. We pinpointed that a total of 346 sdRNAs were found to be significantly correlated with GZMA level in one or more cancer types (adjusted p < 0.05) (Table S10), where the most predictive cancer types include BLCA, BRCA, LGG, LUAD, LUSC, PAAD, PRAD, TGCT and THCA (Figure 6a). In contrast, cancer types such as ESCA, KICH, PCPG, READ, UCEC and UCS have few predictive sdRNAs for GZMA expression (Figure 6a). Thyroid carcinoma has the most sdRNAs positively correlated with GZMA expression, whereas lung squamous cell carcinoma has most sdRNAs negatively correlated with GZMA (Figure 6a). Among the significant sdRNAs, 42 were positively correlated with GZMA levels in 3 or more cancer types (Figure 6b, Table S10). Of note, sdRNAs from the hybrid snoRNA SCARNA5 were positively correlated in bladder carcinoma (BLCA), melanoma (SKCM) (Figure 6c), testicular germ cell tumors (TGCT) (Figure 6d), and thyroid carcinoma (THCA). On the other hand, 40 sdRNAs were negatively correlated with intratumoral cytolytic activity in at least 3 cancer types (Figure 6e, Table S10). SdRNAs derived from an H/ACA scaRNA SCARNA4 were negatively correlated with GZMA in bladder carcinoma (BLCA) (Figure 6f), breast adenocarcinoma (BRCA) (Figure 6g), cervical squamous cell carcinoma (CESC), head and neck squamous carcinoma (HNSC), lung squamous cell carcinoma (LUSC), pancreatic adenocarcinoma (PAAD), and testicular germ cell tumors (TGCT). These analyses revealed a set of sdRNAs that significantly correlate with intratumoral cytotoxic T cell activity across multiple cancer types.
Figure 6: sdRNAs are correlated with cytolytic T cell activity across multiple cancer types.
a. Bar plot depicting the number of significant sdRNAs in each cancer type, aggregated by parental snoRNA, in relation to GZMA expression. Red, positive correlation; blue, negative correlation.
b. Heat map of sdRNAs positively correlated with GZMA expression (adjusted p < 0.05, adjusted within each cancer type), a marker of cytolytic T cell activity. For visibility, only sdRNAs that were positively correlated in three or more cancer types are shown. Boxes are colored according to the Spearman correlation with GZMA. Parental snoRNAs without annotated names are instead labeled by their host gene in parentheses. SnoRNA classifications are annotated on top based on a color legend on the right panel.
c. Scatter plot depicting the correlation between GZMA and SCARNA5 sdRNA expression in skin cutaneous melanoma (SKCM, n = 89). Spearman correlation R = 0.401, p = 1.11 * 10-4.
d. Scatter plot depicting the correlation between GZMA and SCARNA5 sdRNA expression in testicular germ cell tumors (TGCT, n = 135). R = 0.403, p = 1.56 * 10-6.
e. Heat map of sdRNAs negatively correlated with GZMA expression (adjusted p < 0.05, adjusted within each cancer type). For visibility, only sdRNAs that were positively correlated in three or more cancer types are shown. Boxes are colored according to the Spearman correlation with GZMA. Parental snoRNAs without annotated names are instead labeled by their host gene in parentheses. SnoRNA classifications are annotated on top based on a color legend on the right panel.
f. Scatter plot depicting the correlation between GZMA and SCARNA4 sdRNA expression in bladder carcinomas (BLCA, n = 362). R = −0.267, p = 2.60 * 10-7.
g. Scatter plot depicting the correlation between GZMA and SCARNA4 sdRNA expression in cervical squamous cell carcinoma (CESC, n = 265). R = −0.315, p = 1.98 * 10-7.
c, d, f, g - Data are shown as tpm, normalized separately for mRNA-seq and smRNA-seq datasets.
SdRNA expression associates with angiogenesis in the tumor microenvironment
To further investigate the contribution of sdRNAs to the tumor microenvironment, we explored the connection between sdRNA expression signatures and tumor vascularization. Surprisingly, 449 sdRNAs were significantly correlated with endothelial cell abundance in at least one cancer type (EndothelialScore), among which 61 sdRNAs were positively correlated in 4 or more cancer types (Figure S8a-b, Table S11). Testicular germ cell tumors again have the most sdRNAs positively correlated with endothelial cell abundance, whereas lung squamous cell carcinoma again has the most sdRNAs negatively correlated with endothelial cell abundance (Figure S8a). Strikingly, sdRNAs produced from the C/D snoRNA SNORD114–1 were positively correlated with endothelial cell abundance in 16 different cancer types, including breast adenocarcinoma (BRCA), colon adenocarcinoma (COAD), head and neck squamous cell carcinoma (HNSC), sarcoma (SARC), melanoma (SKCM), stomach adenocarcinoma (STAD), thymoma (THYM), and uterine corpus endometrial carcinoma (UCEC) (Figure S8c). These data suggest that sdRNAs derived from snoRNAs such as SNORD114–1 play highly conserved roles in tumor vascularization across different tissues.
SdRNA expression predicts patient survival across diverse human cancers
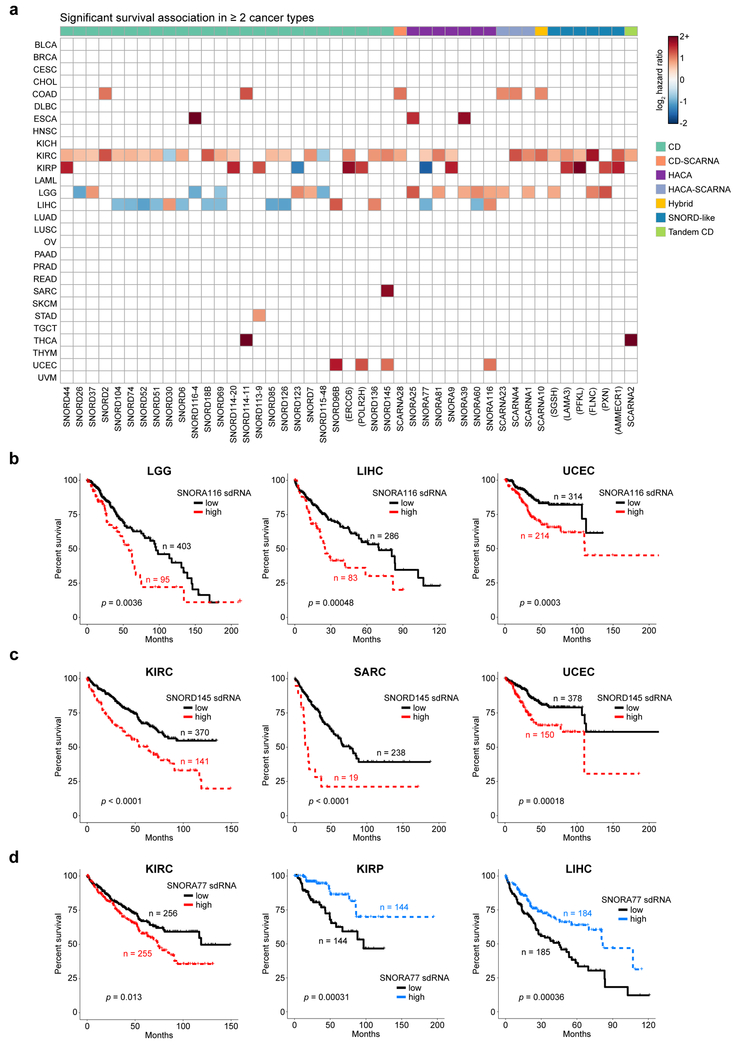
Collectively, our analyses above pointed to a wide-ranging set of sdRNAs as statistically significant molecular markers of important features of cancer immunity. We therefore hypothesized that the expression of sdRNAs might be associated with patient survival, akin to previous studies with snoRNAs 44,45. For each cancer type, we classified patients into either “high” or “low” expression of an sdRNA based on the median expression value (aggregated by parental snoRNA) within the cohort. Using these groups, we performed Kaplan-Meier survival analysis to identify sdRNAs with prognostic significance to overall survival (OS) (adjusted p < 0.05 by log-rank test) (Table S12). In terms of cancer types, KIRC, KIRP, LGG and LIHC have the largest number of OS-predictive sdRNAs (Table S12, Figure 7a). Remarkably, 247 sdRNAs had significant survival associations in one or more cancer type(s), out of which 45 sdRNAs can stratify OS in 2 or more cancer types (Figure 7a). For instance, high expression of sdRNAs derived from SNORA116, an H/ACA snoRNA, was connected to poorer survival in three independent cohorts: lower grade gliomas (LGG), liver hepatocellular carcinoma (LIHC), and uterine corpus endometrial carcinoma (UCEC) (Figure 7b). As another example, high levels of sdRNAs from SNORD145, a CD snoRNA, were associated with shorter survival times in kidney clear cell carcinoma (KIRC), sarcoma (SARC), and uterine corpus endometrial carcinoma (UCEC) (Figure 7c). SdRNAs from SNORA116 and SNORD145 thus appear to be indicators of cancers with a more aggressive course. Interestingly, several sdRNAs are associated with opposite outcomes in different cancer types (Figure 7a). For example, sdRNAs produced from the H/ACA snoRNA SNORA77 were divergently associated with survival. In kidney clear cell carcinoma (KIRC), patients with high SNORA77 had poor survival, whereas the opposite was true in kidney papillary carcinoma (KIRP) and liver hepatocellular carcinoma (LIHC) (Figure 7d). Thus, the same sdRNA can be differentially associated with survival in different cancers, even when these cancers arise from the same organ.
Figure 7: sdRNAs are associated with patient survival across multiple cancer types.
a. Heat map of sdRNAs associated with patient survival, aggregated by parental snoRNA (adjusted p < 0.05 by log-rank test, adjusted within each cancer type). For visibility, only sdRNAs that were significantly associated with survival in two or more cancer types are shown. Boxes are colored according to log2 hazard ratios, where positive values indicate increased mortality risk and negative values denote decreased mortality risk. Parental snoRNAs without annotated names are labeled by their host gene in parentheses. SnoRNA classifications are annotated on top based on a color legend on the right panel.
b. Kaplan-Meier survival curves in lower grade glioma (LGG, n = 498; left), liver hepatocellular carcinoma (LIHC, n = 369; middle), and uterine corpus endometrial carcinoma (UCEC, n = 528; right), stratified by SNORA116 sdRNA expression. High SNORA116 sdRNA expression was concordantly associated with poorer survival in all three cohorts (p = 0.0036, p = 0.0048, p = 0.0003).
c. Kaplan-Meier survival curves in clear cell kidney carcinoma (KIRC, n = 511; left), sarcoma (SARC, n = 257; middle), and uterine corpus endometrial carcinoma (UCEC, n = 528; right), stratified by SNORD145 sdRNA expression. High SNORD145 sdRNA expression was consistently associated with poorer survival in all three cohorts (p < 0.0001, p < 0.0001, p = 0.00018).
d. Kaplan-Meier survival curves in clear cell kidney carcinoma (KIRC, n = 511; left), papillary renal cell carcinoma (KIRP, n = 288; middle), and hepatocellular carcinoma (LIHC, n = 369; right), stratified by SNORA77 sdRNA expression. High SNORA77 sdRNA expression was associated with poorer survival in KIRC, but with better survival in KIRP and LIHC (p = 0.013, p = 0.00031, p = 0.00036).
Integrative analysis of sdRNAome and pan-cancer tumor immunity
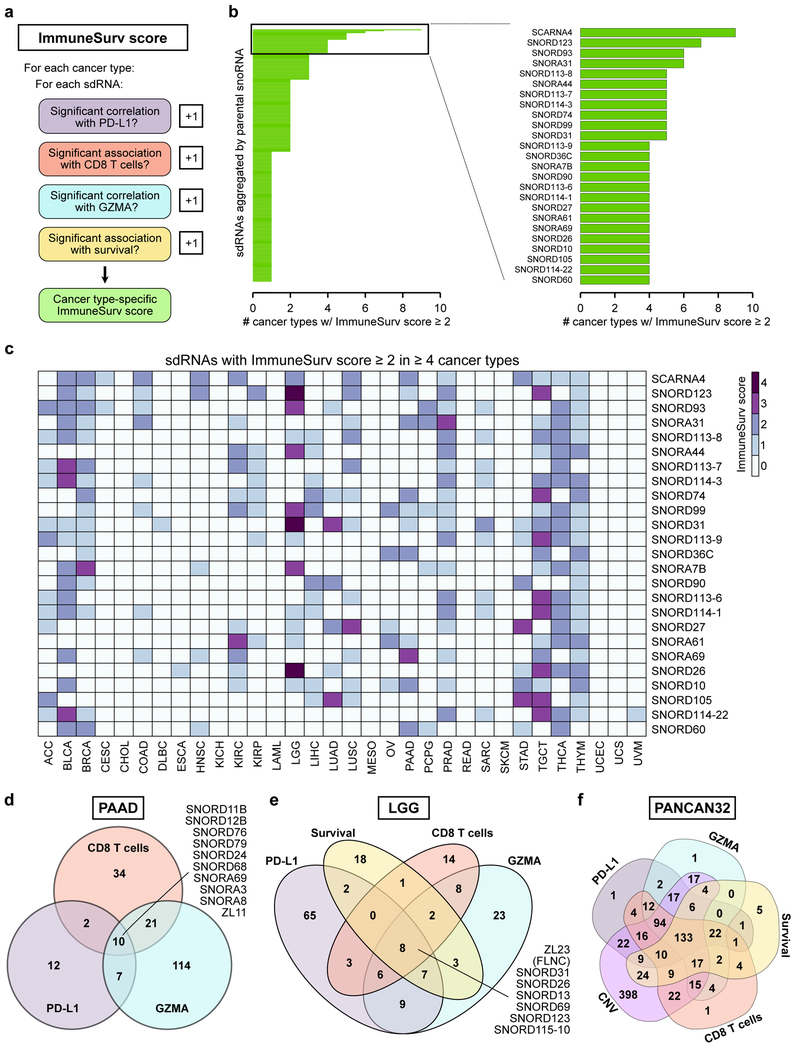
In light of these data, we wondered whether any individual sdRNAs were significantly associated with multiple clinically relevant features. Towards this end, we conceived an ImmuneSurv score to rank sdRNAs (aggregated by snoRNA) based on several dimensions. We calculate the ImmuneSurv score for each sdRNA based on its statistical significance of correlations with PD-L1 expression, CD8+ T cell abundance, GZMA expression, and/or patient survival (when available) in a cancer type-specific manner (Figure 8a, Table S13). As we were particularly interested in sdRNAs with significant associations in multiple aspects of cancer immunity, we focused on sdRNAs with ImmuneSurv scores ≥ 2 (Table S14). A total of 267 sdRNAs were found to meet this criteria in at least one cancer type (Figure 8b, Table S15). Strikingly, 25 sdRNAs met the ImmuneSurv ≥ 2 cutoff in 4 or more cancer types (Figure 8b), suggesting potentially more conserved roles for these short non-coding RNAs. The top sdRNA was that derived from SCARNA4, an H/ACA scaRNA, which had ImmuneSurv scores ≥ 2 in a total of 9 cancer types, including bladder carcinoma (BLCA), breast adenocarcinoma (BRCA), colon adenocarcinoma (COAD), head and neck squamous cell carcinoma (HNSC), kidney clear cell carcinoma (KIRC), lower grade glioma (LGG), lung squamous cell carcinoma (LUSC), pancreatic adenocarcinoma (PAAD), and stomach adenocarcinoma (STAD) (Figure 8c). This surprising finding implicates sdRNAs derived from SCARNA4 as a potential uncharacterized small RNA marker for cancer immunity and clinical outcome across tumors from diverse tissues of origin.
Figure 8: Multidimensional analysis of sdRNAs at the interface of tumor immunity and survival.
a. Schematic of the ImmuneSurv score. In a cancer type-specific manner, each sdRNA is assessed for whether it is significantly associated with PD-L1 expression, CD8+ T cell abundance, GZMA expression, and patient survival. Each significant association adds 1 point to the ImmuneSurv score.
b. Left: bar plot of sdRNAs aggregated by parental snoRNA with ImmuneSurv scores ≥ 2 in at least one cancer type. Right: bar plot of the 25 parental snoRNAs with ImmuneSurv scores ≥ 2 in at least 4 cancer types. Strikingly, SCARNA4 was found to have ImmuneSurv scores ≥ 2 in 9 independent cancer types.
c. Heat map of ImmuneSurv scores for the 25 parental snoRNAs in b. Individual cells are colored based on ImmuneSurv score for the indicated snoRNA in each cancer type.
d. Venn diagram of sdRNAs significantly associated with PD-L1, CD8+ T cells, and/or GZMA in pancreatic adenocarcinoma (PAAD).
e. Venn diagram of sdRNAs significantly associated with PD-L1, CD8+ T cells, GZMA, and/or survival in lower grade glioma (LGG).
f. Venn diagram of sdRNAs significant for PD-L1 correlation, CD8+ T cell abundance, GZMA correlation, patient survival, and/or copy number variation in any cancer type (PANCAN32).
Focusing on individual cancer types, we found that each cancer type has a set of sdRNAs that were associated with more than one ImmuneSurv features (Table S13). For example, 10 sdRNAs were significantly associated with PD-L1 expression, intratumoral CD8+ T cell abundance, and GZMA levels in pancreatic adenocarcinoma (Figure 8d, Table S14). These 10 sdRNAs included those derived from C/D snoRNAs (SNORD76, SNORD79, SNORD24, SNORD12B, SNORD68, SNORD11B, ZL11) and H/ACA snoRNAs (SNORA3, SNORA8, SNORA69, SNORA31). Of note, no individual sdRNAs were significantly associated with survival in the TCGA pancreatic adenocarcinoma cohort, perhaps due to the relatively smaller patient cohort and the highly aggressive course of this disease. Remarkably, among the lower grade glioma patients, 8 sdRNAs had ImmuneSurv scores of 4, meaning that they were significantly associated with all 4 dimensions: PD-L1, CD8+ T cell abundance, GZMA, and patient survival (Figure 8e, Table S14). These 8 parental snoRNAs were SNORD31, SNORD26, SNORD13, SNORD69, SNORD115–10, SNORD123, ZL23, and snoID_0379, an as-of-yet unnamed snoRNA encoded within the host gene FLNC. Because the ImmuneSurv score analyses were conducted in a cancer type-specific manner, we then sought a global assessment of sdRNAs and their relationships to cancer immunity regardless of cancer type (PANCAN32), by compiling all sdRNAs that were found to be significant in any of the 5 categories: PD-L1, CD8+ T cell abundance, GZMA, survival, or copy number variation (supplemental results, Figure S9) in any cancer type. Intersecting these lists, we identified 133 sdRNAs that were found to be significant in all 5 categories (Figure 8f). Together with the metastatic signature (supplemental results, Figure S10), these data collectively suggest a prevalent role of sdRNAs in tumor immunity, thereby influencing significant clinical features of human cancer.
Discussion
SnoRNAs are a class of short RNAs that mainly reside in the nucleolus, though some have also been found in the cytoplasm 46. The known primary functions of snoRNAs are to guide chemical modifications of other types of RNAs, predominantly rRNAs, and small nuclear RNAs (snRNAs). C/D box snoRNAs, together with their protein partners, form small nucleolar ribonucleoprotein (snoRNP) complexes that have catalytic function for RNA methylation 47. Similarly the H/ACA box snoRNPs can catalyze RNA pseudouridylation 48. As mounting evidence points to ribosome biogenesis as a key contributing factor to cancer 49, it is likely that the canonical functions of C/D and H/ACA snoRNAs to guide rRNA processing may play a role in carcinogenesis. ScaRNAs, a subgroup of snoRNAs specifically localized to the Cajal body, a nuclear organelle involved in the biogenesis of (snRNPs), guide the methylation and pseudouridylation of RNA polymerase II (pol II) transcribed spliceosomal RNAs U1, U2, U4, U5 and U12 50. Additionally, scaRNAs have been demonstrated to control the nuclear localization of Cajal bodies, indicating a role in genome organization and thus gene expression 51.
Various cases of snoRNAs have been associated with cancer progression, behaving as oncogenic or tumor suppressive small RNAs. For example, SNORA50A/B have been reported to act as tumor suppressors by opposing the KRAS oncogene 24. On the other hand, SNORD14D and SNORD35A have recently been demonstrated to potentiate the oncogenic effects of the AML1-ETO fusion in leukemia through rRNA methylation 52. A number of other studies have reported prognostically relevant snoRNAs, such as SNORD93 in breast cancer 33, SNORA42 in lung cancer 53, SNORA21 in colon cancer 54, and SNORD47 in glioblastoma 55. Of note, SNORD115 has been demonstrated to act as a regulator of alternative splicing 22,23,56, and its deletion is sufficient to cause Prader-Willi syndrome.
It has been discovered that snoRNAs can be processed into smaller RNAs called sdRNAs, some of which possess microRNA-like functionality 27,57,58. Certain sdRNAs could therefore influence carcinogenesis through gene regulation. As the TCGA smRNA-seq libraries were size-selected for ~22 nt species, these datasets offer the opportunity to investigate the role of sdRNAs in cancer. A recent study of TCGA smRNA-seq datasets looked to explore the associations between snoRNAs and other genomic features, but we note this study did not address the issue of ~22nt size-selection during library preparation nor comprehensively investigate the relationships to tumor immunity, as we have done here 59. For the majority of the several hundred sdRNAs, their roles in cancer have been unexplored until this point. Moreover, the roles of sdRNAs in the tumor microenvironment – for example, whether they are linked to angiogenesis or immunological features – are largely unknown. Here, we successfully generated the expression profiles of the sdRNAome across more than 10,000 patient samples. With this quantitative map of sdRNA abundance across several cancer types, we uncovered a large set of constitutive, cancer type-specific and cancer group-specific sdRNAs. Interestingly, the expression signatures of these sdRNAs alone are sufficient to distinguish samples from differing cancer types, while also revealing sub-clusters within individual cancer types. Though these differences in sdRNA expression are partly driven by the tissue of origin, different cancer types arising from the same organ can nevertheless be distinguished. We found that many sdRNAs (aggregated by parental snoRNA) are significantly correlated with the immunosuppressive biomarker PD-L1, such as sdRNAs derived from SNORA36B in thymoma and SNORA44 in lower grade glioma. Various sdRNAs, such as those derived from SCARNA5, SNORD6 and SNORD114–22, are strongly positively correlated with intratumoral T cell-mediated cytotoxicity by granzyme A. PD-L1 is an indicative biomarker for checkpoint blockade immunotherapy and has been successfully used to guide major clinical trials of immunotherapy. GZMA levels have also been proposed as a predictor of the overall response rate of checkpoint blockade agents. Thus, with further development by the field, the immunological signatures of sdRNAs could potentially be implemented into new paradigms to better identify patients who may potentially benefit from checkpoint blockade. The ImmuneSurv scores presented here, which incorporate both immune and survival signatures, could guide the selection of top candidate sdRNAs for further mechanistic investigation and translational efforts. Though the library design of the TCGA datasets precludes a direct comparison between parental snoRNAs and sdRNAs, it is possible that the differential regulation of sdRNA biogenesis from the parental snoRNAs is a key mechanism through which snoRNAs can affect tumor behavior. Future studies that explicitly compare the relative abundances of snoRNAs and sdRNAs in cancer, as well as the dynamics of snoRNA processing during malignant transformation, may uncover an underexplored mechanism through which ncRNAs can influence tumor biology.
Intratumoral heterogeneity is increasingly recognized as a critical feature of cancers, influencing tumor progression and therapeutic responses 60–62. Given that the TCGA smRNA-seq libraries were prepared from bulk RNA preparations, these datasets cannot be used to explore intratumoral heterogeneity in sdRNA expression. Additional studies using microdissection or single-cell smRNA-seq will be necessary to investigate the potential contribution of intratumoral sdRNA expression heterogeneity towards cancer biology, such as tumor immunity.
In summary, our comprehensive pan-cancer analysis of sdRNAs generated a global view of these transcripts in characterizing different cancer types, leading to the identification of multiple sdRNAs strongly associated with fundamentally important clinical features such as angiogenesis, tumor immunity and overall survival, while simultaneously identifying large sets of novel candidates for further functional studies. Because of their high abundance, short length, tissue-specificity and availability in circulation 63, many novel sdRNAs could be developed as next-generation diagnostic or prognostic biomarkers.
Materials and methods
Data acquisition and pre-processing
TCGA smRNA-seq sequencing bam files were downloaded through the NIH NCI GDC Data Portal (https://portal.gdc.cancer.gov/) in June 2017. Of note, only normal control smRNA-seq data were available for GBM, leaving a total of 32 cancer types for further analysis. To obtain reads specific to sdRNAs, we used featureCounts 64 with settings -Q 20 -largestOverlap -minOverlap 3 -s 1, using the human snoRNAome as the reference region set 40. By this approach, all reads corresponding to sdRNAs were totaled by the parental snoRNA. Since an earlier database was used to map the smRNA-seq files, we removed any snoRNAs that had ≤ 1 read across all the samples. For confirming concordance between the mapped data and our annotations, we visually inspected the bams using IGV 65. We sample-wise normalized sdRNA read counts to transcripts per million (tpm). mRNA-seq gene-level counts were also downloaded from the NIH NCI GDC Data Portal in June 2017, and sample-wise normalized to tpm. SdRNA and RNA tpm values were subsequently log2 transformed. To compare sdRNAs across cancer types, the log2 transformed median expression values were further converted to z-scores normalized within each parental snoRNA. GISTIC 2.0 copy number variation calls were obtained from the GDAC Firehose (http://gdac.broadinstitute.org/) on September 2017. Patient clinical data was obtained through cBioPortal 66,67.
Raw fastq files for independent smRNA-seq datasets (GSE33858, GSE46622, E-MTAB-3494) were accessed by NCBI GEO (https://www.ncbi.nlm.nih.gov/geo/) or EBI (https://www.ebi.ac.uk/). Data were uniformly processed as for the TCGA smRNA-seq datasets, using BWA to first map the reads to the hg38 reference genome prior to quantification with featureCounts. Principal component analysis was performed in R.
Characterization of snoRNA expression profiles
To characterize the sdRNA reads mapping to snoRNAs, we employed a normalized binning approach. We randomly selected 5 samples from each cancer type, and used these to quantify the read density along the length of each snoRNA, with each snoRNA divided into 50 equally sized bins. For visualization, these values were subsequently normalized by maximum intensity, such that the read depths along a given snoRNA were multiplied by a constant scaling factor. Average profiles and hierarchically clustered heat maps were then produced using deepTools v2.5 68. We also tabulated the lengths of reads mapping to snoRNA loci using Samtools 69 and expressed these data as a percentage of total snoRNA-mapped reads.
To explore the overall expression of sdRNAs, we calculated the median expression of each snoRNA within each cancer type. To characterize differences in snoRNA expression across different cancers, we then calculated z-scores for the median snoRNA expression values and visualized them as a heat map using the NMF R library. For unbiased visualization of individual tumors in terms of snoRNA expression, we utilized t-distributed stochastic neighbor embedding (t-SNE) 41. Additional t-SNE plots were generated using alternate coloring schema – CD274 expression, GZMA expression, EndothelialScore (see below), and overall survival. For survival t-SNE visualization, only patients that had died were included to circumvent issues with censored data.
Correlation analysis between sdRNAs and host genes
For correlation analysis between different sdRNAs, we first extracted sdRNAs (aggregated by parental snoRNA) with a variance > 0.1 and median log2 tpm > 0. These high-variance sdRNAs were analyzed by Pearson correlation. For correlation analysis between snoRNAs/sdRNAs and host gene expression, we utilized previously published snoRNA annotations 40 and calculated the Spearman correlation between all defined snoRNA-host gene pairs (n = 736). The empirical cumulative density function of the Spearman correlation distribution was further calculated to illustrate the relative proportion of snoRNA – host gene pairs with Spearman correlations above a specified threshold. Cancer-specific analyses were performed in the same manner. Statistical significance was determined by conversion to a t-statistic, and multiple hypothesis correction was performed by the Benjamini-Hochberg method. The Benjamini-Hochberg procedure is based on controlling the false discovery rate. We set a significance level of adjusted p < 0.05.
Correlation analysis between sdRNAs and other transcriptomic variables
For correlation analysis between sdRNAs and PD-L1 or GZMA expression, we extracted the corresponding mRNA-seq data and computed the Spearman correlation between each sdRNA (aggregated by parental snoRNA) and the gene of interest. Using the correlation coefficient and sample sizes, the correlation coefficients were converted to a t-statistic, from which the associated p-values was calculated. The p-values were then adjusted for multiple comparisons within each cancer type by the Benjamini-Hochberg approach, using an adjusted p-value of 0.05 as the significance threshold. For visualization in the figures, we divided the resulting correlation tables into positive and negative correlations. To improve readability in the figures, we selected sdRNAs that were significantly correlated in multiple cancer types (precise number indicated on figures). SdRNAs that were not significantly correlated in a given cancer type were filtered for visualization purposes (i.e. set to “0”). The complete correlation tables are available in the supplementary tables. To identify sdRNAs associated with CD8+ T cell abundance or tumor vascularization, we utilized the xCell algorithm 70 (http://xcell.ucsf.edu/) and the computed abundances of multiple cell types within TCGA samples. Each sdRNA was then compared to the “CD8+ T-cells” or “Endothelial cells” entry (“EndothelialScore”) in the xCell output matrix. The same statistical procedures were used as above for PD-L1 and GZMA expression.
Survival analysis
For survival analysis, only primary tumor samples were considered such that each patient had exactly one sample for consideration. Within each cancer type, the median expression value for each sdRNA (taking the aggregate abundance of constituent sdRNAs per parental snoRNA) was used as the threshold to define “low” or “high” expression. Any sdRNAs for which either group had less than 4 samples were subsequently excluded. To assess survival differences between patient groups stratified by sdRNA expression, we used the log-rank test. Associated p-values were adjusted for multiple comparisons by the Benjamini-Hochberg approach, adjusting within each cancer type. An adjusted p < 0.05 was considered significant. For the heat map visualization, sdRNAs with significant survival associations were colored by their associated log2 hazard ratio, as determined by a Cox hazards model. Positive log2 hazard ratios indicate increased mortality, while negative log2 hazard ratios indicate decreased mortality.
Copy number variation in snoRNAs
For identification of snoRNAs subject to significant copy number variation, we utilized the GISTIC 2.0 output files (specifically *amp_genes.conf_99.txt and *del_genes.conf_99.txt). As these tables report the precise genomic coordinates in which the amplification or deletion was identified, we utilized a q < 0.05 threshold and subsequently intersected the coordinates with the snoRNA annotations 40. Amplification and deletion calls for individual snoRNAs were then compiled into separate tables. For heat map visualization, the associated GISTIC q-values for a given snoRNA in each cancer type were −log10 transformed.
Differential expression analysis
For differential expression analysis comparing metastases to primary tumors, we utilized limma 71. For multiple hypothesis correction, we used the Benjamini Hochberg method. An adjusted p < 0.05 was used a threshold for significance.
Intersection of significant snoRNA lists
For pan-cancer analysis, we considered all sdRNAs that were found to be significant in at least one cancer type across the following 5 analyses: CD274 correlation, GMZA correlation, CD8+ T cell abundance, copy number variation, and survival. For intersection of these five analyses, the directionality of the association was not considered (i.e. an sdRNA would be included regardless of whether it was positively or negatively correlated with CD274). The pan-cancer 5-way intersection was visualized using the web tool available at http://bioinformatics.psb.ugent.be/webtools/Venn/.
For cancer type-specific analysis, we iterated through each parental snoRNA and tabulated whether it was significant in the 5 aforementioned categories, in addition to tumor vascularization. This information was recorded through a 6 character custom coding scheme (i.e. NNSNSN for snoRNA “X” in cancer type “Y”). “N” denotes “not significant”, “S” denotes “significant”, and “o” signifies that data was not available. Each position corresponds to a specific significant list. Position1: CD274; Position2: GZMA; Position3: Survival; Position4: CD8+ T cells; Position5: CNV; Position 6: EndothelialScore. Thus, for the example above (NNSNSN), sdRNAs from snoRNA X were found to be significantly associated with survival and significant for CNV in cancer type Y. The complete table of codes is available in Table S12. For focused analysis of immune signatures and patient survival, we developed an ImmuneSurv score based on the above coding schema. In this score, the first four letters are considered (corresponding to CD274 correlation, GZMA correlation, survival association, and CD8+ T cell correlation). For a given snoRNA in a specific cancer type, each “S” adds 1 to the ImmuneSurv score, up to a total of 4.
Resource availability
All relevant resources, data and codes will be available to the academic community upon reasonable requests. Please refer to the supplementary tables for the results of the various analyses presented in the study.
Blinding statement and general methods
Investigators were not blinded for sample collection, processing or analysis. No specific methods were used to predetermine sample sizes. No data were excluded.
Regulatory approval
Study involving use of controlled access TCGA patient data has been approved by TCGA DAC and NIH for General Research Use in project titled #15034: “Role of noncoding RNAs in cancer-immune interactions”, with DAR #: 57036–1 at Yale University.
Supplementary Material
Acknowledgments
We thank all members in Chen laboratory for their support. We thank Yale High Performance Computing staff for assistance. S.C. is supported by Yale SBI/Genetics Startup Fund, Damon Runyon (DRG-2117–12; DFS-13–15), Melanoma Research Alliance (412806, 16–003524), St-Baldrick’s Foundation (426685), Breast Cancer Alliance, Cancer Research Institute (CLIP), AACR (499395, 17–20-01-CHEN), The Mary Kay Foundation (017–81), The V Foundation (V2017–022), DoD (W81XWH-17–1-0235) and NIH/NCI (1U54CA209992, 5P50CA196530-A10805, 4P50CA121974-A08306). R.D.C. is supported by the NIH MSTP training grant (T32GM007205).
Footnotes
Conflict of interest
The authors declare no conflicts of interest pertaining to this work.
Contributions: S.C. and R.C. conceived the study. R.D.C. designed the study and performed all data analysis. R.D.C. and S.C. wrote the manuscript. S.C. supervised the work.
References:
- 1.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity 2013; 39: 1–10. [DOI] [PubMed] [Google Scholar]
- 2.Herbst RS, Soria J-C, Kowanetz M, Fine GD, Hamid O, Gordon MS et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014; 515: 563–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ribas A Adaptive Immune Resistance: How Cancer Protects from Immune Attack. Cancer Discov 2015; 5: 915–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF et al. Safety, Activity, and Immune Correlates of Anti–PD-1 Antibody in Cancer. N Engl J Med 2012; 366: 2443–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017; 168: 707–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015; 372: 2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taube JM, Klein A, Brahmer JR, Xu H, Pan X, Kim JH et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res Off J Am Assoc Cancer Res 2014; 20: 5064–5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer Genome Landscapes. Science 2013; 339: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watson IR, Takahashi K, Futreal PA, Chin L. Emerging patterns of somatic mutations in cancer. Nat Rev Genet 2013; 14: nrg3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg 2010; 251: 499–505. [DOI] [PubMed] [Google Scholar]
- 11.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D et al. MicroRNA expression profiles classify human cancers. Nature 2005; 435: nature03702. [DOI] [PubMed] [Google Scholar]
- 12.Jacobsen A, Silber J, Harinath G, Huse JT, Schultz N, Sander C. Analysis of microRNA-target interactions across diverse cancer types. Nat Struct Mol Biol 2013; 20: 1325–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huarte M The emerging role of lncRNAs in cancer. Nat Med 2015; 21: nm.3981. [DOI] [PubMed] [Google Scholar]
- 14.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016; 29: 452–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsieh C-L, Fei T, Chen Y, Li T, Gao Y, Wang X et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc Natl Acad Sci U S A 2014; 111: 7319–7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ørom UA, Shiekhattar R. Long noncoding RNAs usher in a new era in the biology of enhancers. Cell 2013; 154: 1190–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Yang J, Zhou P, Le Y, Zhou C, Wang S et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res 2015; 5: 472–480. [PMC free article] [PubMed] [Google Scholar]
- 18.Esteller M Non-coding RNAs in human disease. Nat Rev Genet 2011; 12: 861–874. [DOI] [PubMed] [Google Scholar]
- 19.Dupuis-Sandoval F, Poirier M, Scott MS. The emerging landscape of small nucleolar RNAs in cell biology. Wiley Interdiscip Rev RNA 2015; 6: 381–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu Rev Biochem 1995; 64: 897–934. [DOI] [PubMed] [Google Scholar]
- 21.Schubert T, Pusch MC, Diermeier S, Benes V, Kremmer E, Imhof A et al. Df31 Protein and snoRNAs Maintain Accessible Higher-Order Structures of Chromatin. Mol Cell 2012; 48: 434–444. [DOI] [PubMed] [Google Scholar]
- 22.Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science 2006; 311: 230–232. [DOI] [PubMed] [Google Scholar]
- 23.Kishore S, Khanna A, Zhang Z, Hui J, Balwierz PJ, Stefan M et al. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum Mol Genet 2010; 19: 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siprashvili Z, Webster DE, Johnston D, Shenoy RM, Ungewickell AJ, Bhaduri A et al. The noncoding RNAs SNORD50A and SNORD50B bind K-Ras and are recurrently deleted in human cancer. Nat Genet 2016; 48: 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mannoor K, Liao J, Jiang F. Small nucleolar RNAs in cancer. Biochim Biophys Acta 2012; 1826: 121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams GT, Farzaneh F. Are snoRNAs and snoRNA host genes new players in cancer? Nat Rev Cancer 2012; 12: 84–88. [DOI] [PubMed] [Google Scholar]
- 27.Ender C, Krek A, Friedländer MR, Beitzinger M, Weinmann L, Chen W et al. A human snoRNA with microRNA-like functions. Mol Cell 2008; 32: 519–528. [DOI] [PubMed] [Google Scholar]
- 28.Martens-Uzunova ES, Olvedy M, Jenster G. Beyond microRNA – Novel RNAs derived from small non-coding RNA and their implication in cancer. Cancer Lett 2013; 340: 201–211. [DOI] [PubMed] [Google Scholar]
- 29.Brameier M, Herwig A, Reinhardt R, Walter L, Gruber J. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res 2011; 39: 675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA 2009; 15: 1233–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pundhir S, Gorodkin J. Differential and coherent processing patterns from small RNAs. Sci Rep 2015; 5: 12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott MS, Ono M, Yamada K, Endo A, Barton GJ, Lamond AI. Human box C/D snoRNA processing conservation across multiple cell types. Nucleic Acids Res 2012; 40: 3676–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patterson DG, Roberts JT, King VM, Houserova D, Barnhill EC, Crucello A et al. Human snoRNA-93 is processed into a microRNA-like RNA that promotes breast cancer cell invasion. Npj Breast Cancer 2017; 3: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Starega-Roslan J, Krol J, Koscianska E, Kozlowski P, Szlachcic WJ, Sobczak K et al. Structural basis of microRNA length variety. Nucleic Acids Res 2011; 39: 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z, Ender C, Meister G, Moore PS, Chang Y, John B. Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res 2012; 40: 6787–6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Falaleeva M, Stamm S. Processing of snoRNAs as a new source of regulatory non-coding RNAs: snoRNA fragments form a new class of functional RNAs. BioEssays News Rev Mol Cell Dev Biol 2013; 35: 46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 2008; 22: 2773–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chu A, Robertson G, Brooks D, Mungall AJ, Birol I, Coope R et al. Large-scale profiling of microRNAs for The Cancer Genome Atlas. Nucleic Acids Res 2016; 44: e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackowiak P, Hojka-Osinska A, Philips A, Zmienko A, Budzko L, Maillard P et al. Small RNA fragments derived from multiple RNA classes - the missing element of multi-omics characteristics of the hepatitis C virus cell culture model. BMC Genomics 2017; 18: 502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jorjani H, Kehr S, Jedlinski DJ, Gumienny R, Hertel J, Stadler PF et al. An updated human snoRNAome. Nucleic Acids Res 2016; 44: 5068–5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Maaten L, Hinton G Visualizing Data using t-SNE. J Mach Learn Res 2008; 9: 2579–2605. [Google Scholar]
- 42.Tata PR, Chow RD, Saladi SV, Tata A, Konkimalla A, Bara A et al. Developmental History Provides a Roadmap for the Emergence of Tumor Plasticity. Dev Cell 2018; 44: 679–693.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and Genetic Properties of Tumors Associated with Local Immune Cytolytic Activity. Cell 2015; 160: 48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao J, Yu L, Mei Y, Guarnera M, Shen J, Li R et al. Small nucleolar RNA signatures as biomarkers for non-small-cell lung cancer. Mol Cancer 2010; 9: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krishnan P, Ghosh S, Wang B, Heyns M, Graham K, Mackey JR et al. Profiling of Small Nucleolar RNAs by Next Generation Sequencing: Potential New Players for Breast Cancer Prognosis. PLOS ONE 2016; 11: e0162622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Michel CI, Holley CL, Scruggs BS, Sidhu R, Brookheart RT, Listenberger LL et al. Small nucleolar RNAs U32a, U33, and U35a are critical mediators of metabolic stress. Cell Metab 2011; 14: 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiss T Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 2002; 109: 145–148. [DOI] [PubMed] [Google Scholar]
- 48.Kiss T, Fayet-Lebaron E, Jády BE. Box H/ACA small ribonucleoproteins. Mol Cell 2010; 37: 597–606. [DOI] [PubMed] [Google Scholar]
- 49.Montanaro L, Treré D, Derenzini M. Nucleolus, ribosomes, and cancer. Am J Pathol 2008; 173: 301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morris GE. The Cajal body. Biochim Biophys Acta BBA - Mol Cell Res 2008; 1783: 2108–2115. [DOI] [PubMed] [Google Scholar]
- 51.Wang Q, Sawyer IA, Sung M-H, Sturgill D, Shevtsov SP, Pegoraro G et al. Cajal bodies are linked to genome conformation. Nat Commun 2016; 7: ncomms10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou F, Liu Y, Rohde C, Pauli C, Gerloff D, Köhn M et al. AML1-ETO requires enhanced C/D box snoRNA/RNP formation to induce self-renewal and leukaemia. Nat Cell Biol 2017; 19: 844–855. [DOI] [PubMed] [Google Scholar]
- 53.Mei Y-P, Liao J-P, Shen J, Yu L, Liu B-L, Liu L et al. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene 2012; 31: 2794–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshida K, Toden S, Weng W, Shigeyasu K, Miyoshi J, Turner J et al. SNORA21 - An Oncogenic Small Nucleolar RNA, with a Prognostic Biomarker Potential in Human Colorectal Cancer. EBioMedicine 2017; 22: 68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu B, Ye M-H, Lv S-G, Wang Q-X, Wu M-J, Xiao B et al. SNORD47, a box C/D snoRNA, suppresses tumorigenesis in glioblastoma. Oncotarget 2017; 8: 43953–43966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sahoo T, del Gaudio D, German JR, Shinawi M, Peters SU, Person RE et al. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat Genet 2008; 40: 719–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ono M, Scott MS, Yamada K, Avolio F, Barton GJ, Lamond AI. Identification of human miRNA precursors that resemble box C/D snoRNAs. Nucleic Acids Res 2011; 39: 3879–3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scott MS, Avolio F, Ono M, Lamond AI, Barton GJ. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput Biol 2009; 5: e1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gong J, Li Y, Liu C, Xiang Y, Li C, Ye Y et al. A Pan-cancer Analysis of the Expression and Clinical Relevance of Small Nucleolar RNAs in Human Cancer. Cell Rep 2017; 21: 1968–1981. [DOI] [PubMed] [Google Scholar]
- 60.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. N Engl J Med 2012; 366: 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McGranahan N, Swanton C. Biological and Therapeutic Impact of Intratumor Heterogeneity in Cancer Evolution. Cancer Cell 2015; 27: 15–26. [DOI] [PubMed] [Google Scholar]
- 62.McGranahan N, Swanton C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017; 168: 613–628. [DOI] [PubMed] [Google Scholar]
- 63.Freedman JE, Gerstein M, Mick E, Rozowsky J, Levy D, Kitchen R et al. Diverse human extracellular RNAs are widely detected in human plasma. Nat Commun 2016; 7: 11106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinforma Oxf Engl 2014; 30: 923–930. [DOI] [PubMed] [Google Scholar]
- 65.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G et al. Integrative Genomics Viewer. Nat Biotechnol 2011; 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov 2012; 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 2014; 42: W187–W191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. The Sequence Alignment/Map format and SAMtools. Bioinforma Oxf Engl 2009; 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 2017; 18 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015; 43: e47–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.