Significance
Transcription in Gram-negative bacteria is greatly influenced by the synergistic interactions of DksA and the nucleotide alarmone guanosine tetraphosphate. Our investigations reveal a unique, previously unknown layer of transcriptional regulation that depends on redox-based protein–protein interactions between DksA and the molecular chaperone DnaJ. Tripartite connections between DksA, DnaJ, and guanosine tetraphosphate afford a dynamic range of transcriptional responses to H2O2 concentrations associated with redox signaling or oxidative stress. The redox-based gene regulation dependent on the combined actions of DksA, DnaJ, and guanosine tetraphosphate confers resistance of Salmonella to the antimicrobial activity of the NADPH phagocyte oxidase.
Keywords: redox signaling, oxidation, stringent response, Salmonella, DnaJ
Abstract
RNA polymerase is the only known protein partner of the transcriptional regulator DksA. Herein, we demonstrate that the chaperone DnaJ establishes direct, redox-based interactions with oxidized DksA. Cysteine residues in the zinc finger of DksA become oxidized in Salmonella exposed to low concentrations of hydrogen peroxide (H2O2). The resulting disulfide bonds unfold the globular domain of DksA, signaling high-affinity interaction of the C-terminal α-helix to DnaJ. Oxidoreductase and chaperone activities of DnaJ reduce the disulfide bonds of its client and promote productive interactions between DksA and RNA polymerase. Simultaneously, guanosine tetraphosphate (ppGpp), which is synthesized by RelA in response to low concentrations of H2O2, binds at site 2 formed at the interface of DksA and RNA polymerase and synergizes with the DksA/DnaJ redox couple, thus activating the transcription of genes involved in amino acid biosynthesis and transport. However, the high concentrations of ppGpp produced by Salmonella experiencing oxidative stress oppose DksA/DnaJ-dependent transcription. Cumulatively, the interplay of DksA, DnaJ, and ppGpp on RNA polymerase protects Salmonella from the antimicrobial activity of the NADPH phagocyte oxidase. Our research has identified redox-based signaling that activates the transcriptional activity of the RNA polymerase regulator DksA.
Starving bacteria down-regulate the transcription of genes encoding translational machinery and up-regulate the expression of loci involved in amino acid biosynthesis and transport. This transcriptional adaptation, which is commonly known as the stringent response (1), is a manifestation of the combined regulatory actions of DksA and the nucleotide alarmones guanosine tetraphosphate and guanosine pentaphosphate [(p)ppGpp] on RNA polymerase. Because the concentration of DksA protein remains constant throughout the bacterial life cycle of Escherichia coli and Salmonella, the stringent response to nutritional starvation is regulated by fluctuating concentrations of (p)ppGpp (2). Responding to shortages in amino acids, iron, fatty acids, or phosphate, RelA and SpoT proteins synthesize (p)ppGpp from GTP, GDP, and ATP. The nucleotide (p)ppGpp binds between the β′ and ω subunits of RNA polymerase (site 1) (3–5), and at the interface of the coiled-coil and C-terminal α-helix of DksA and the β′ helices of RNA polymerase (site 2) (6). Phosphates at 3′ and 5′ positions of (p)ppGpp form salt-bridges with basic residues in the coiled-coil domain and C-terminal α-helix of DksA (7). Binding of (p)ppGpp to site 2 repositions DksA inside RNA polymerase from a stressed to a relaxed conformation, thus guiding Asp71 at the tip of the coiled-coil domain of DksA into the proximity of the bridge-helix loop of RNA polymerase (7, 8). The accommodation of DksA’s coiled-coil tip residues deep into the active site of RNA polymerase destabilizes interactions between the bridge-helix loop and the DNA template, thereby collapsing the open complex. In addition to the aforementioned allosteric regulation, (p)ppGpp increases the affinity of DksA for RNA polymerase (7).
Salmonella DksA consists of a N-terminal coiled-coil domain that is followed by a cysteine-rich globular domain and a C-terminal α-helix (9). As with most homologs, Salmonella DksA harbors four cysteine residues (C4) that coordinate a zinc cation. The existence of zinc-free DksA homologs with one (C1) or two (C2) cysteine residues strongly indicates that the tertiary structure and function of this RNA polymerase regulator can be accomplished without a coordinating metal. Cysteine residues not only play structural roles but also constitute a relay redox system tunable by reactive oxygen and nitrogen species in the bacterial cell (10, 11). The oxidation of cysteine residues in C2 or C4 DksA homologs diminishes the α-helical content of these proteins and decreases their affinity for RNA polymerase (10), offering a rationale for the transcriptional inhibition of amino acid biosynthesis genes in Salmonella experiencing oxidative stress (10, 12, 13). Unexpectedly, the oxidation of DksA also inhibits the transcription of ribosomal proteins (11). As the repression of loci encoding translational components requires the active binding of DksA to RNA polymerase, the inhibition of ribosomal protein gene transcription by oxidized DksA cannot easily be explained by the oxidative destruction of DksA function. To elucidate the mechanisms by which oxidative stress modulates DksA function and transcriptional regulation, we undertook a search for protein partners of DksA. Our investigations herein demonstrate that ppGpp differentially modulates the transcription supported by DksA and the molecular chaperone and oxidoreductase DnaJ in response to divergent concentrations of H2O2 associated with redox signaling and oxidative stress.
Results
DksA Is a Client of the Chaperone DnaJ.
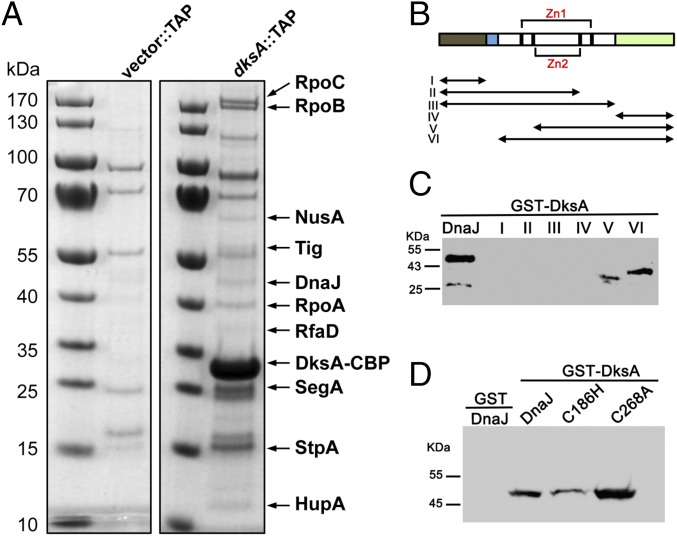
DksA protein levels do not fluctuate during the bacterial life cycle; however, DksA-dependent transcription is influenced by growth phase, nutritional downshifts, and oxidative and nitrosative stress (1, 10). Herein we tested whether, in addition to the allosteric control exerted by (p)ppGpp, DksA is subjected to posttranslational regulation through protein–protein interactions. To identify putative DksA-binding partners, we performed tandem-affinity purification (TAP) of DksA linked at the C terminus with a calmodulin-binding peptide followed by tobacco etch virus (TEV) protease cleavage site and protein A. Mass spectrometric analysis of soluble cytoplasmic proteins isolated from log-phase Salmonella with the DksA-TAP bait identified, among others, RpoA, RpoB, and RpoC proteins (SI Appendix, Table S1), reflecting canonical interactions of DksA with the β′ subunit of RNA polymerase (9, 14). Our screen identified seven additional interacting partners of DksA (Fig. 1A and SI Appendix, Table S1). To validate these putative DksA-binding partners, genes of interest were cloned into a bacterial two-hybrid system that reconstitutes interactions of the T25 and T18 subunits of adenylate cyclase. To identify false positives that may interact with DksA indirectly via RNA polymerase, proteins were also tested for binding to the RpoA α-subunit of RNA polymerase. These screens uncovered trigger factor (encoded by ropA) and DnaJ as putative DksA-specific binding partners (SI Appendix, Fig. S1A). A tertiary screen verified DnaJ as a genuine DksA-binding partner (SI Appendix, Fig. S1 B and C). Interestingly, DksA was discovered as a multicopy suppressor that rescues the temperature sensitivity of a ΔdnaKJ E. coli strain (15).
Fig. 1.
DnaJ is a binding partner of DksA. (A) Putative DksA binding partners were identified by mass spectrometry of cytoplasmic proteins isolated by TAP from ΔdksA Salmonella harboring pWSK29::TAP (Vector::TAP) or pWSK29::dksA::TAP (dksA::TAP) plasmids. TAP-purified proteins separated in SDS/PAGE gels were visualized by Coomassie Brilliant blue staining. The positions of DksA-binding partners are indicated by arrows. (B) Schematic representation of DnaJ domains and DnaJ truncated proteins (I–VI). N-terminal J (gray) and glycine- and phenylalanine-rich GF domains (blue), followed by two zinc fingers (black and red brackets), Zn1 and Zn2, and the C-terminal domain (green). The biochemical pull-down assays using (C and D) GST-DksA as bait and truncated DnaJ variants I–VI (C) or C186H and C268A DnaJ variants (D) as prey were performed in the absence of DTT. The GST protein (D) was used as a negative control. DnaJ proteins isolated in the pull-down assays were detected by Western blotting. Data are representative of three independent experiments.
DnaJ, also known as Hsp40, works as a chaperone and holdase that transfers unfolded, misfolded, or aggregated proteins to DnaK, stimulating the extended ADP-bound DnaK conformation that has high affinity for polypeptides and is endowed with foldase activity (16, 17). Although less efficient than DnaK, DnaJ can act independently as a foldase of denatured polypeptides (18). DnaJ is comprised of J and glycine- and phenylalanine-rich GF N-terminal domains, followed by two zinc fingers, Zn1 and Zn2, and a C-terminal domain (Fig. 1B). To define the DnaJ domains that make contact with DksA, we conducted pull-downs using combinations of a recombinant GST-DksA protein with six DnaJ fragments (SI Appendix, Fig. S1D). These investigations revealed that DksA interacts with C-terminal fragments V and VI containing the C-terminal domain together with Zn2 or both Zn1 and Zn2 sites, respectively (Fig. 1C). Point mutations in Cys186 in the Zn2 site diminished the binding of DnaJ to DksA (Fig. 1D and SI Appendix, Fig. S1E), suggesting that the Zn2 finger is involved in the binding of DnaJ to DksA. These findings are consistent with the importance of the Zn finger region in the recognition of denatured proteins (18, 19). Compared with the wild-type protein, a DnaJ variant bearing a C268A substitution bound with apparently greater strength to DksA (Fig. 1D and SI Appendix, Fig. S1E), suggesting that the Cys268 in the C-terminal domain is not required for binding of DnaJ to DksA. Recombinant DnaJ C186H and C268A variants contained wild-type levels of zinc and the expected reduction in thiol content (SI Appendix, Table S2).
DnaJ Binds to the Unfolded Globular Domain of Oxidized DksA in the Context of the C-Terminal α-Helix.
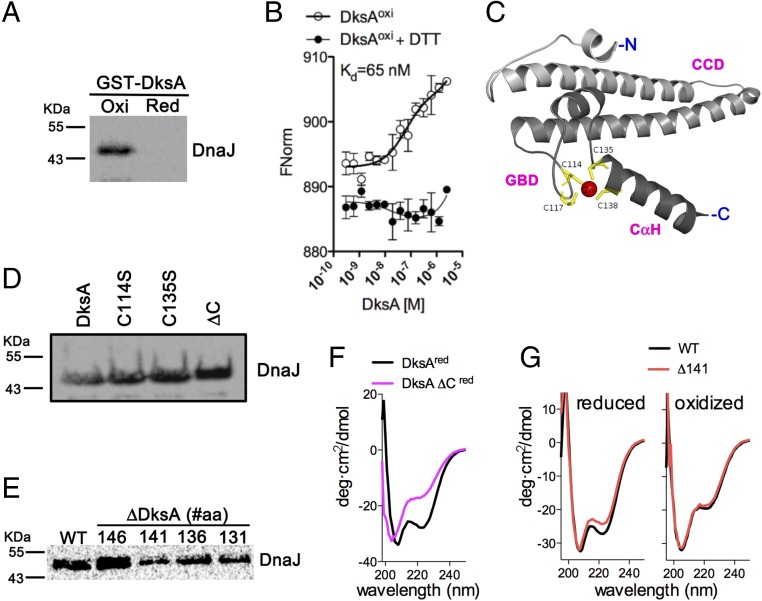
We next tested the effect that the redox state of DksA has on the interaction of this RNA polymerase-binding protein with DnaJ. DnaJ bound to oxidized DksA (Fig. 2A and SI Appendix, Fig. S2A) with an estimated Kd of 65 nM (Fig. 2B). The reduction of oxidized DksA with DTT prevented binding to DnaJ (Fig. 2A), suggesting that oxidation signals the binding of DksA to DnaJ. Unexpectedly, cysteine residues in DksA (Fig. 2C) seem dispensable for the interaction because DksA variants missing Cys114, Cys135, or all four cysteine residues (i.e., DksA ΔC) bound to DnaJ with apparently similar or greater affinity than the wild-type DksA protein (Fig. 2D and SI Appendix, Fig. S2B). Pull-down assays of DnaJ with wild-type and truncated DksA variants showed that DksA Δ141 containing an intact globular domain with normal cysteine and zinc content, but lacking the C-terminal α-helix, bound to DnaJ less efficiently than the wild-type protein (Fig. 2E and SI Appendix, Fig. S2C). Collectively, these findings suggest that DnaJ recognizes the unfolded globular domain of oxidized DksA in the context of the C-terminal α-helix.
Fig. 2.
DksA oxidation triggers high affinity binding to DnaJ. (A) Binding of DksA oxidized with H2O2 to DnaJ was assessed in Western blots using pull-down assays, as described in Fig. 1. (B) Binding affinity of the DksA–DnaJ redox couple was estimated by microscale thermophoresis. The data are the mean ± SD from three independent experiments. (C) Diagram of DksA modeled from the E. coli structure PDB ID code 5W1T (7), showing the coiled-coil domain (CCD), globular domain (GBD) with the four cysteines in the zinc finger, and C-terminal α-helix (CαH). Interactions of DnaJ (prey) with DksA variants (bait) bearing mutations in cysteine residues (D) or truncations in the C-terminal domain (E). DnaJ proteins pulled-down in A, D, and E were detected by Western blotting. A DksA variant missing all four cysteine residues (ΔC) was used for comparison. DksA truncated proteins are identified by the position of the deletion at the C-terminal domain. (F and G) The α-helical secondary structure of DksA proteins was analyzed by CD spectroscopy. Some of the proteins in F and G were reduced with DTT before analysis. The data are representative of two to three independent experiments and each spectrum shows the average of three independent scans.
To get insights into the biophysical determinants underlying the association of oxidized DksA with DnaJ, we examined the properties of various DksA specimens by circular dichroism (CD) spectroscopy. The DksA ΔC variant showed a complete lack of the 223-nm CD minimum compared with the wild-type protein (Fig. 2F), indicating a massive loss of α-helical content. Oxidation of DksA cysteine residues with 1 mM H2O2 (Fig. 2G) or peroxynitrite (ONOO−) (10) also destroyed the 223-nm CD minimum. A DksA Δ141 truncated variant missing all amino acids contributing to the C-terminal α-helix, but expressing a normal zinc finger in the globular domain (SI Appendix, Fig. S2 C and E), manifested a slight reduction in α-helicity in the 223-nm CD region compared with wild-type DksA (Fig. 2G). However, oxidation completely destroyed the 223-nm CD minimum in both wild-type and the Δ141 variant. The 208-nm minimum was neither different between wild-type and the Δ141 variant nor was affected by oxidation. Taken together, these findings suggest that the 223-nm CD minimum is mostly contributed to by the secondary structure of the globular domain. Our research also suggests that oxidation of cysteine residues in the zinc finger destroys the α-helix in the globular domain, not those comprising the coiled-coil region.
DnaJ Reduces Disulfide-Bonded Cysteine Residues in the Globular Domain of Oxidized DksA.
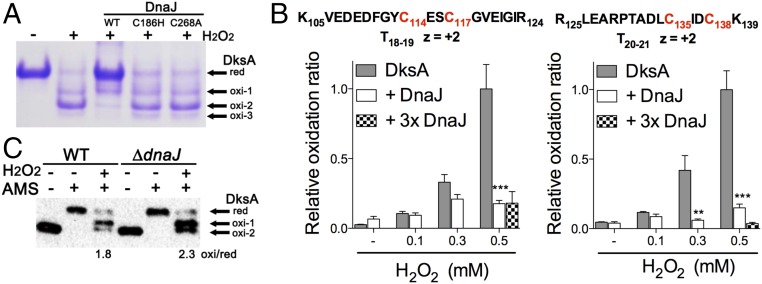
DnaJ catalyzes the formation and reduction of disulfide bonds (20, 21), raising the possibility that DnaJ might not only be a chaperone of unfolded DksA but may also reduce this RNA polymerase regulatory protein. To test this hypothesis, we determined the effect of DnaJ on the redox state of DksA cysteine residues. Nonreducing SDS electrophoresis of DksA derivatized with the thiol trap 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS) showed that H2O2 induces the formation of several DksA redox species (Fig. 3A). This finding is consistent with previous work demonstrating that cysteine residues in the zinc finger of DksA are modifiable by reactive oxygen and nitrogen species (10, 11). In contrast, H2O2 or ONOO− did not oxidize cysteine residues in DnaJ, nor release zinc from the Zn1 or Zn2 sites, as measured by reaction with 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) and 4-(2-pyridylazo) resorcinol (PAR), respectively (SI Appendix, Fig. S3A). The addition of denaturing urea stimulated the reaction of cysteine residues in DnaJ with DTNB and released zinc from DnaJ (SI Appendix, Fig. S3B), suggesting that the native configuration of DnaJ zinc fingers is extraordinarily resilient to thiol modification. These findings are consistent with previous reports that showed diverse reactivity among different zinc fingers according to conformational flexibility and activation entropy (22). Wild-type DnaJ, but not C186H or C268A variants, reduced H2O2-treated DksA (Fig. 3A), indicating that oxidized cysteine residues in DksA can be substrates of the reductase activity of DnaJ. The inability of the DnaJ C186H variant to reduce oxidized DksA could stem from either poor binding to DksA (Fig. 1D) or the destruction of the thiol-disulfide oxidoreductase activity that is associated with the Zn2 zinc finger (23). The lack of oxidoreductase activity of the DnaJ C268A variant cannot be explained by poor binding to DksA (Fig. 1D). Mass spectrometry showed that the oxidoreductase activity of DnaJ targets a pair of intramolecular disulfide bonds formed between Cys114-Cys117 and Cys135-Cys138 in oxidized DksA (Fig. 3B and SI Appendix, Figs. S3C and S4A). DnaJ did not appear to transfer zinc to oxidized, metal-free DksA (SI Appendix, Fig. S3D).
Fig. 3.
The oxidoreductase activity of DnaJ resolves disulfide bonds in oxidized DksA. (A) The redox state of 5 μM DksA was evaluated by nonreducing SDS/PAGE and Commassie Brilliant blue staining after reduced cysteine residues were alkylated with the thiol trapper AMS. Where indicated, the specimens were treated with 1 mM H2O2 in the presence or absence of 5 μM DnaJ. (B) Abundance of intramolecular disulfide bonds between DksA Cys114-Cys117 and Cys135-Cys138 residues was determined by mass spectrometry. DksA proteins were alkylated with NEM after treatment with H2O2 in the presence or absence of DnaJ. Select samples were treated with a threefold molar excess of DnaJ (3× DnaJ). (C) Redox state of DksA in anaerobically grown Salmonella was evaluated by AMS-derivatization and Western blotting. Selected samples were treated with 5 μM H2O2 for 5 min before the thiols were derivatized with AMS. The ratio of the sum of oxidized species to reduced DnaJ within H2O2-treated, AMS-derivatized DksA was calculated by densitometry. Data in A–C are from three independent experiments; **P < 0.01 and ***P < 0.001, according to two-way ANOVA.
We also examined the DksA redox state in vivo. We found that a fraction of the DksA protein is already oxidized in aerobically grown Salmonella (SI Appendix, Fig. S3E), suggesting that DksA cysteine residues are prone to oxidation by oxidants produced during aerobic metabolism. To carefully control levels of oxidants, we followed DksA redox state in Salmonella grown in an anaerobic chamber in the presence or absence of defined concentrations of H2O2. DksA appears to be reduced in Salmonella grown anaerobically (SI Appendix, Fig. S4B). However, a sizable fraction of DksA was oxidized 6 s after the addition of 1 or 10 μM H2O2 to anaerobic Salmonella (SI Appendix, Fig. S4B), persisting for at least 5 min. The fraction of oxidized DksA was greater in ΔdnaJ Salmonella compared with wild-type bacteria (Fig. 3C). Taken together, these findings indicate that DksA cysteine residues are easily oxidized by H2O2 at concentrations that are commonly produced in metabolism or the inflammatory response of phagocytic cells. Our investigations also suggest that the oxidized cysteine residues in DksA are substrates of the oxidoreductase activity of DnaJ in vivo.
DnaJ Contributes to the Antioxidant Defenses of Salmonella.
DksA-dependent transcriptional responses to nutritional limitation and oxidative stress are essential to bacterial physiology and pathogenesis. We therefore examined the phenotypes of ΔdnaJ Salmonella. As expected, strains of Salmonella bearing ΔdnaJ mutations were sensitive to heat shock (SI Appendix, Fig. S5A). In contrast to the ΔdksA strain, ΔdnaJ Salmonella grew in minimal media (SI Appendix, Fig. S5B). This research suggests that the ΔdnaJ mutant, but not ΔdksA Salmonella, can mount an appropriate stringent response to nutritional starvation. The lack of dnaJ did not affect DksA or DnaK protein concentrations in resting or 1 mM H2O2-treated Salmonella (SI Appendix, Fig. S5C). Together, these data indicate that DnaJ does not control the expression or function of DksA during the canonical stringent response to nutritional starvation.
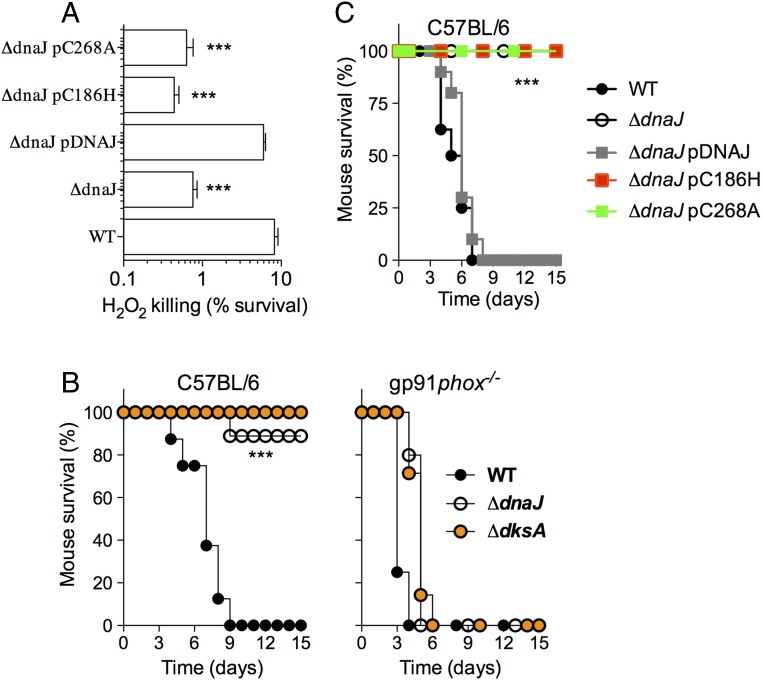
DksA promotes antioxidant defense of Salmonella by activating transcription of various steps in metabolism that supply reducing equivalents to the antioxidant enzyme system (10, 12). Because oxidized DksA is a substrate of DnaJ, we evaluated the contribution of DnaJ to the antioxidant defenses of Salmonella. ΔdnaJ and ΔdksA Salmonella were similarly hypersusceptible to the antimicrobial activity of 200 μM H2O2 (Fig. 4A). Expression of a wild-type dnaJ allele, but not the C186H or C268A variants (SI Appendix, Fig. S5D), reversed the increased susceptibility of ΔdnaJ Salmonella to oxidative stress (Fig. 4A). Salmonella strains deficient in dnaJ or dksA were attenuated in a murine model of acute salmonellosis, but became virulent in gp91phox−/− mice deficient in the catalytic subunit of the NADPH phagocyte oxidase (Fig. 4B). Salmonella strains expressing the dnaJ C186H or dnaJ C268A alleles were also attenuated in a murine model of infection (Fig. 4C). Together, these investigations indicate that DnaJ protects Salmonella against the oxidative stress emanating from the enzymatic activity of the NADPH phagocyte oxidase.
Fig. 4.
DnaJ contributes to antioxidant defense and Salmonella pathogenesis. (A) Susceptibility of wild-type Salmonella and the indicated mutants to 200 μM H2O2; ***P < 0.001 as determined by one-way ANOVA (n = 12). (B and C) C57BL/6 and congenic gp91phox−/− mice were challenged with ∼100 CFU of the indicated Salmonella strains. The fraction of mice that survive the challenge was followed over time. The data are from 9 to 10 mice evaluated in two independent experiments; ***P < 0.001 as determined by log-rank analysis.
DnaJ Facilitates the Binding of Oxidized DksA to RNA Polymerase, Thereby Activating the Transcription of Genes Involved in Amino Acid Biosynthesis.
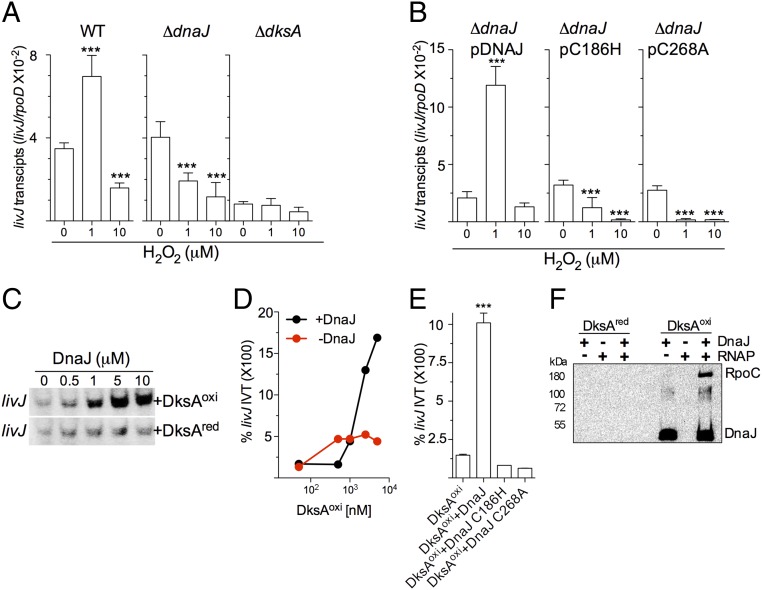
DksA controls the transcription of bacterial genes through its associations with the secondary channel and trigger loop domain in the β′ subunit of RNA polymerase (9, 14). Binding of DksA to RNA polymerase affects open complex stability and transcriptional initiation in response to starvation signals. Binding of DksA protein to RNA polymerase is also regulated in response to oxidation (10). Because oxidized DksA is a substrate of the oxidoreductase activity of DnaJ, we tested whether DnaJ impacts DksA-dependent transcriptional regulation in response to H2O2. As the growth of Salmonella under aerobic conditions oxidizes endogenous DksA, we examined the role of DnaJ in directing DksA-dependent transcriptional changes in Salmonella grown anaerobically, a condition in which cysteine residues in all DksA molecules appear to be reduced (Fig. 3C and SI Appendix, Fig. S4B). Wild-type and ΔdnaJ Salmonella expressed equivalent basal levels of the livJ gene encoding a Leu/Ile/Val/Thr-binding protein, each higher than ΔdksA bacteria (Fig. 5A), indicating that DnaJ does not influence the DksA-dependent activation of the canonical stringent response to nutritional starvation. The addition of 10 μM H2O2 inhibited transcription of livJ in wild-type Salmonella (Fig. 5A), an observation that is consistent with our previous findings in aerobic Salmonella exposed to 1 mM H2O2 (10, 12). To our great surprise, 1 μM H2O2 induced livJ expression (P < 0.001), suggesting that levels of H2O2 associated with redox signaling stimulate amino acid gene transcription. The addition of 1 μM H2O2 to ΔdnaJ Salmonella, however, inhibited livJ transcription. Complementation of ΔdnaJ bacteria with a wild-type dnaJ allele, but not the dnaJ C186H or dnaJ C268A variants, reestablished wild-type levels of livJ transcription (Fig. 5B). Similar phenotypes were seen when hisG, which encodes a gene involved in histidine biosynthesis, was probed (SI Appendix, Fig. S6 A and B). Compared with untreated or controls treated with 1 μM H2O2, 10 μM H2O2 slowed the growth of anaerobic Salmonella (SI Appendix, Fig. S6C). Collectively, these findings suggest that concentrations of H2O2 associated with redox signaling activate livJ and hisG transcription in a DksA and DnaJ codependent manner, whereas levels associated with oxidative stress inhibit transcription.
Fig. 5.
DnaJ facilitates interactions of oxidized DksA with RNA polymerase. Normalized abundance of livJ transcripts in WT, ΔdksA, and ΔdnaJ Salmonella (A) or ΔdnaJ bacteria complemented with the indicated dnaJ alleles (B) during exponential growth in an anaerobic chamber. Indicated cultures were left untreated, or treated with 1 or 10 μM H2O2 in EGCA minimal media. ***P < 0.001 as determined by one-way ANOVA compared with untreated controls (n = 6). The abundance of livJ message was normalized to transcripts of the housekeeping gene rpoD. Data are the mean ± SD from six independent experiments collected on 2 d. (C) Autoradiograms of [32-P]-UTP-labeled livJ in vitro transcripts from reactions containing E. coli RNA polymerase together with reduced or oxidized DksA and increasing concentrations of DnaJ. The products of the reactions were separated by SDS/PAGE. The blot is representative of three independent experiments. (D and E) DnaJ- and DksA-dependent activation of livJ in vitro transcription was evaluated by qRT-PCR. Reactions in D contained increasing concentrations of oxidized DksA in the presence or absence of 5 μM DnaJ. Reactions in E contained 5 μM of DnaJ and 1 μM of the indicated DksA variant. Data are the mean ± SD from three independent experiments. (F) Interactions of reduced and oxidized DksA proteins with S. Typhimurium RNA polymerase in the presence or absence of DnaJ. Oxidized and reduced DksA proteins in F were used as bait. DnaJ and the RpoC subunit of RNA polymerase were detected by immunobloting. The data in F are representative of three independent experiments.
To directly test if DnaJ activates DksA-dependent livJ transcription at low levels of H2O2 associated with redox signaling, we used a reconstituted in vitro transcription system. DnaJ did not stimulate livJ in vitro transcription by reduced DksA (Fig. 5C), but potently activated transcription in combination with oxidized DksA (Fig. 5 C and D). The DnaJ C186H and DnaJ C268A variants did not support livJ expression by oxidized DksA (Fig. 5E), indicating that the potentiation of regulatory activity of oxidized DksA by DnaJ depends on the oxidoreductase enzymatic activity of the latter.
The levels of transcription achieved by oxidized DksA and DnaJ were higher than that of reduced DksA, suggesting that the contribution of DnaJ is not limited to reductase but may also involve chaperone activities. We therefore examined whether DnaJ affects the interactions between DksA and RNA polymerase recombinant proteins from Salmonella (SI Appendix, Fig. S6D). Under the concentrations of proteins used in our investigations, reduced DksA did not appreciably interact with RNA polymerase from Salmonella, and the addition of DnaJ did not increase binding (Fig. 5F). As expected, oxidized DksA interacted directly with DnaJ but not RNA polymerase. However, the presence of DnaJ increased the amount of RNA polymerase that was pulled down by oxidized DksA, suggesting that the tripartite interactions depend on the redox state of DksA.
Guanosine Tetraphosphate Modulates DksA-Dependent Transcription in Response to Increasing Levels of Oxidants.
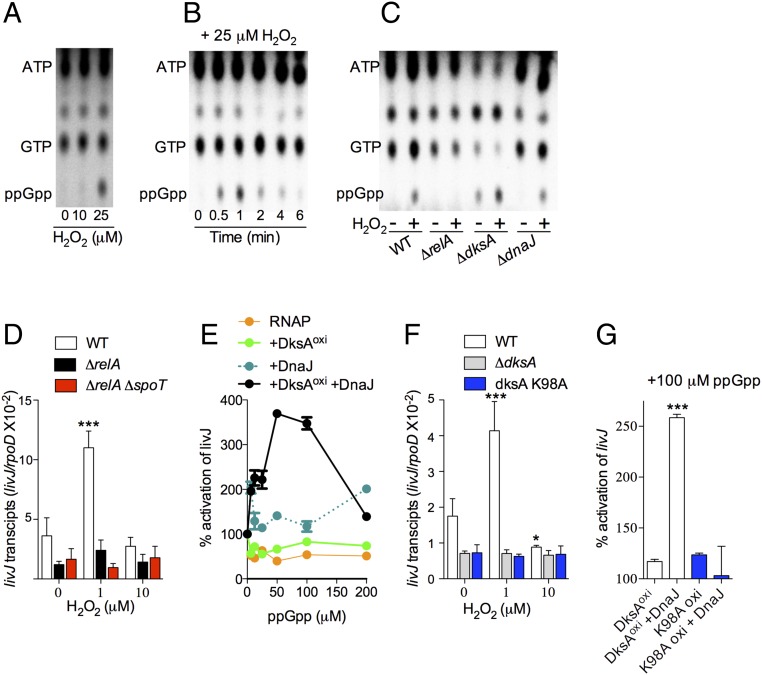
It seems paradoxical that low levels of H2O2 activate DksA- and DnaJ-dependent livJ transcription, whereas high levels inhibit it. Therefore, the outcomes of livJ transcription in response to disparate levels of oxidative stress cannot easily be explained by the actions of DksA and DnaJ alone. Because the regulatory activity of DksA is potentiated by (p)ppGpp (1, 9, 14), we tested whether this nucleotide alarmone contributes to the distinct outcomes in gene transcription observed at different levels of oxidative stress. First, we measured the production of (p)ppGpp in H2O2-treated aerobic Salmonella. H2O2 elicited ppGpp production in a concentration-dependent manner (i.e., 5, 20, and 300 μM ppGpp in untreated and 10 and 25 μM H2O2-treated Salmonella, respectively) (Fig. 6A). As described for NO (24), H2O2 seems to predominantly elicit ppGpp synthesis, not pppGpp. The addition of 25 μM H2O2 to aerobically grown Salmonella elicited a burst of ppGpp 1 min after treatment, followed by rapid disappearance of this nucleotide (Fig. 6B). Synthesis of ppGpp in H2O2-treated Salmonella was catalyzed by the enzymatic activity of the RelA protein and was largely unaffected by the absence of dksA or dnaJ (Fig. 6C). Consistent with the idea that RelA monitoring nonacylated tRNAs in the ribosome is the source of ppGpp seen in H2O2-treated Salmonella, ppGpp synthesis was lost in the presence of the aminoacyl-tRNA attachment inhibitor tetracycline (SI Appendix, Fig. S7A). Collectively, these findings indicate that H2O2 stimulates ppGpp synthesis, likely as a consequence of functional amino acid auxotrophies, as it has recently been demonstrated in Salmonella experiencing nitrosative stress (24).
Fig. 6.
Guanosine tetraphosphate synergizes with the DksA/DnaJ redox couple to regulate transcription. (A–C) TLC autoradiograms of nucleotides labeled with inorganic 32-P. Nucleotides were extracted from aerobic wild-type (A and B) or mutant Salmonella (C) at 1 min (A and C) or at the indicated times (B) after H2O2 exposure. All autoradiograms are representative of three independent experiments. (D and F) Abundance of livJ transcripts in log phase anaerobic Salmonella that were treated with the indicated concentrations of H2O2. The data, which are normalized to internal transcripts of the housekeeping gene rpoD, are the mean ± SD from four independent experiments. (E and G) livJ in vitro transcription reactions containing ppGpp and 5 μM of oxidized DksA and/or DnaJ. The ability of recombinant WT and K98A DksA proteins to activate livJ in vitro transcription is compared in G. Data are the mean ± SD from four independent experiments. *P < 0.05 and ***P < 0.001 compared with untreated (D and F) or DksAoxi (G) controls as determined by two-way (D and F) or one-way (G) ANOVA.
These findings raise the interesting possibility that ppGpp produced at low levels of H2O2 associated with redox signaling could potentiate DnaJ- and DksA-dependent livJ transcription. In support of this model, both ΔrelA and ΔrelA ΔspoT Salmonella strains phenocopied the inability of ΔdksA and ΔdnaJ Salmonella strains to activate livJ transcription in response to 1 μM H2O2 (Fig. 6D). Because production of ppGpp is proportional to the levels of oxidative stress, it is possible that higher concentrations of the nucleotide alarmone inhibit livJ transcription driven by DnaJ and oxidized DksA. To more easily reveal synergistic actions of ppGpp with oxidized DksA and DnaJ, we developed an in vitro transcription system with suboptimal concentrations of RNA polymerase that do not synergize with ppGpp (Fig. 6E). These investigations showed that ppGpp failed to activate livJ transcription in in vitro reactions containing RNA polymerase with either DnaJ or oxidized DksA (Fig. 6E). However, the addition of low levels of ppGpp (50–100 μM) to reactions containing RNA polymerase together with DnaJ and oxidized DksA activated livJ in vitro transcription (Fig. 6E). As predicted by our model, higher concentrations of ppGpp (>100 μM) inhibited livJ transcription. These data suggest that the regulatory activity exerted by DnaJ on oxidized DksA is differentially modulated by varying levels of ppGpp.
The synergism of ppGpp with DksA stems from the binding of this nucleotide to charged residues, such as Lys98,in the coiled-coil domain near the C-terminal α-helix of DksA (6). To determine if the synergistic activation of livJ expression seen in Salmonella experiencing low levels of H2O2 is a direct consequence of the binding of ppGpp at site 2, comprising the interface of DksA and RNA polymerase, we constructed a Salmonella strain expressing the dksA K98A allele. Compared with wild-type controls, dksA K98A Salmonella expressed low levels of livJ (Fig. 6F), indicating that tonal expression of livJ transcription is dependent on both DksA and basal concentrations of (p)ppGpp. Moreover, dksA K98A-expressing Salmonella did not activate livJ transcription in response to 1 μM H2O2, demonstrating that the induction of livJ expression by low levels of H2O2 is dependent on the synergism between DksA and ppGpp. The transcriptional deficiencies of dksA K98A-expresssing Salmonella cannot be explained by differences in DnaK, DnaJ, or DksA proteins (SI Appendix, Fig. S7B). Employing the reconstituted in vitro transcription system, we found that the H2O2-treated DksA K98A protein cannot support livJ expression in response to DnaJ and ppGpp (Fig. 6G and SI Appendix, Fig. S7C). Together, these observations suggest that the stimulation of livJ transcription in Salmonella experiencing low levels of oxidative stress is dependent on the synergistic actions of oxidized DksA, DnaJ, and ppGpp.
The Synergistic Actions of DksA and ppGpp Promote Resistance of Salmonella to the Antimicrobial Activity of the NADPH Phagocyte Oxidase.
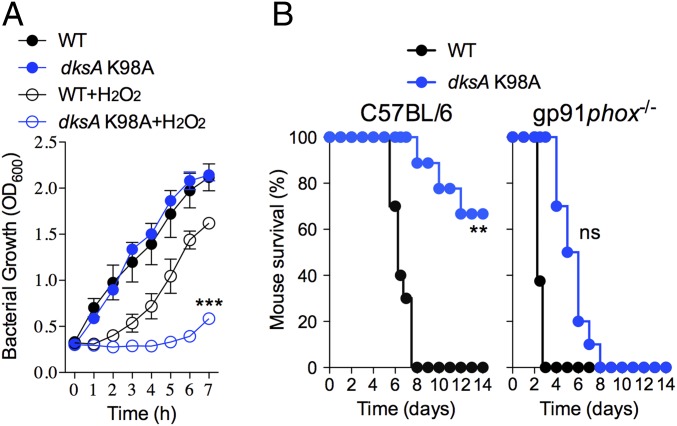
DksA is a critical regulator of the antioxidant defenses of Salmonella (12). Our investigations herein have shown that the regulatory activity of oxidized DksA is dependent on DnaJ and the particular level of ppGpp. Using a dksA K98A-expressing Salmonella strain, we next examined if the antioxidant defenses associated with DksA are dependent on the regulatory activity exerted by ppGpp binding to site 2 at the interface of DksA and RNA polymerase. Salmonella expressing dksA K98A were found to be hypersusceptible to H2O2 (Fig. 7A). Moreover, dksA K98A-expressing Salmonella were attenuated in immunocompetent C57BL/6 mice but virulent in congenic gp91phox−/− mice (Fig. 7B). Cumulatively, these findings demonstrate that together DksA, DnaJ, and ppGpp protect Salmonella against the respiratory burst of the NADPH phagocyte oxidase, an essential aspect of Salmonella pathogenesis.
Fig. 7.
Guanosine tetraphosphate binding to site 2 in the interface formed between DksA and RNA polymerase contributes to the antioxidant defenses of Salmonella. (A) Growth of WT and dksA K98A Salmonella in EGCA minimal medium. Indicated cultures were treated with 500 μM H2O2. ***P < 0.001 compared with WT + H2O2 as determined by two-way ANOVA. (B) C57BL/6 and congenic gp91phox−/− mice were inoculated intraperitoneally with ∼100 CFU of either WT or dksA K98A Salmonella. Mouse survival data are from 9 to 10 mice per bacterial strain and were collected from two independent experiments. **P < 0.01 or nonsignificantly different (ns) as determined by log-rank analysis.
Discussion
Our research has identified H2O2 as a previously unappreciated redox-based signal that activates the DksA-dependent transcription of genes involved in amino acid biosynthesis and transport (Fig. 8). Because redox active groups in enzymes involved in amino acid biosynthesis are prone to inhibition by reactive oxygen and nitrogen species, the redox-based, DksA-dependent activation of transcription of amino acid biosynthesis and transport genes provides a feedback mechanism of establishing homeostasis in cells experiencing low fluxes of oxidants. Guanosine tetraphosphate synthesized in response to low concentrations of H2O2 synergizes with the DksA–DnaJ redox couple to stimulate transcription of amino acid biosynthesis and transport genes. However, Salmonella shuts down transcription of amino acid biosynthesis during the oxidative stress associated with higher concentrations of H2O2. The high micromolar concentrations of ppGpp produced during periods of oxidative stress counteract the positive effects the DnaJ–DksA redox couple has on transcription. Thus, the combined actions of DksA, DnaJ, and ppGpp fine-tune transcriptional regulation in response to divergent levels of reactive oxygen species.
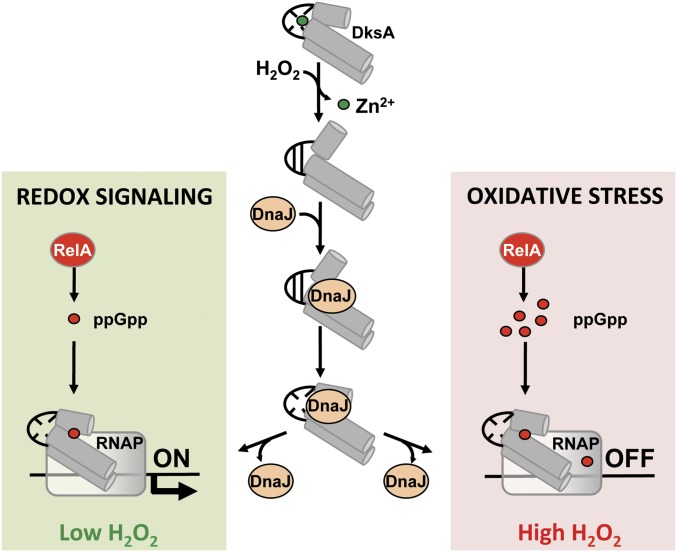
Fig. 8.
Model for the redox-dependent, DksA–DnaJ regulation of transcription. Oxidation of cysteine residues destroys the zinc finger of DksA. The consequent conformational changes in the globular domain and C-terminal α-helix of oxidized DksA trigger high-affinity binding to DnaJ. The oxidoreductase and chaperone activities of DnaJ resolve disulfide bonds and help load DksA into RNA polymerase. Concomitantly, the accumulation of deacylated tRNAs following H2O2-induced amino acid auxotrophies stimulates production of guanosine tetraphosphate (ppGpp) from RelA bound to ribosomes. The low concentrations of ppGpp generated at low concentrations of H2O2 binds at site 2 formed in the interface of DksA and RNA polymerase, activating transcription of amino acid biosynthetic genes as part of the adaptation of Salmonella to redox signaling. However, our biochemical data suggest that the high concentrations of ppGpp produced in response to high levels of H2O2 inhibit transcription in Salmonella undergoing oxidative stress, perhaps by binding to sites 1 and 2 of RNA polymerase.
Our research has also uncovered a previously unknown mechanism for the regulation of DksA function that relies on the redox-based interactions with the molecular chaperone DnaJ triggered in response to H2O2. Oxidation of cysteine residues in the globular domain of DksA triggers binding to DnaJ (Fig. 8). DksA cysteine residues do not seem to be directly involved in this interaction, because a DksA variant lacking all cysteine residues binds with apparently normal affinity to DnaJ. Rather, oxidation of cysteine residues unfolds the globular domain of DksA, and exposes the C-terminal α-helix for binding to DnaJ. It is unlikely that the C-terminal α-helix is unfolded in oxidized DksA as suggested by the structure of a disulfide-bonding C2 DksA paralogue from Pseudomonas (25). Changes in the position of the C-terminal α-helix in oxidized DksA may explain why oxidized DksA binds poorly to RNA polymerase (10) as the C-terminal domain contacts the βlobe/i4 domain that forms the pincer surrounding the DNA-binding channel of RNA polymerase (7). Nonetheless, the changes in the position of the C-terminal α-helix that are triggered in response to the unfolding of the globular domain rationalize the redox-based interactions between DksA and DnaJ, as suggested by the poor binding of the DksA Δ141 variant to DnaJ, despite containing normal thiol and zinc content. The chaperone DnaJ has strong avidity for a hydrophobic core of eight residues enriched for aliphatic and aromatic amino acids, as well as arginine (26). This motif is represented in the C-terminal α-helix of DksA (i.e., LAEIREKQMA). Collectively, our investigations suggest that oxidation of cysteine residues in DksA zinc finger triggers unfolding of the globular domain, signaling binding of DnaJ to hydrophobic and charged residues in the C-terminal α-helix of DksA.
The direct interaction of DnaJ with unfolded DksA is not limited to the holdase function of DnaJ, but also involves the reductase activity of the former (Fig. 8). Exposure of DksA to H2O2 yields Cys114-Cys117 and Cys135-Cys138 disulfide pairs, which are substrates of the reductase activity of DnaJ. A mutation in DnaJ Cys186 prevents the reduction of oxidized DksA, an observation that is consistent with the notion that the Zn2 site of DnaJ harbors the reductase activity (27). However, the DnaJ C186H variant binds with apparently low affinity to DksA, indicating that the Zn2 site may also be important for the recognition of oxidized DksA. The DnaJ C268A variant binds to DksA with apparently better affinity than the wild-type protein, but lacks reductase activity and does not support livJ or hisG gene expression. Together, these findings suggest that the holdase and oxidoreductase activities of DnaJ support transcriptional regulation by oxidized DksA.
It is quite probable that DnaJ acts on oxidized DksA in ways that are not limited to its oxidoreductase and holdase activities, because the DksA–DnaJ redox couple is more effective than reduced DksA at activating transcription. The transcriptional activation of amino acid biosynthesis genes via the DksA/DnaJ redox couple reflects improved binding of DksA to RNA polymerase. In this sense, DnaJ acts as a chaperone that facilitates interactions of its client DksA with RNA polymerase (Fig. 8). DnaJ does not seem to transfer zinc to DksA. Therefore, this chaperone could load apo-DksA onto RNA polymerase, a situation that would be analogous to the binding of zinc-free C1 and C2 DksA variants to RNA polymerase. Guanosine tetraphosphate also facilitates interactions of DksA for RNA polymerase (7). Thus, ppGpp and DnaJ facilitate DksA interaction with RNA polymerase, thereby promoting transcriptional responses to nutritional starvation and redox signaling.
The stimulatory effects of the DnaJ–DksA redox couple cannot be executed in vivo in the absence of ppGpp (Fig. 8). In addition to oxidizing the cysteine residues of DksA, H2O2 also activates ppGpp synthesis by RelA proteins responding to deacylated tRNAs in the ribosomal A-site. The lack of activation of DksA–DnaJ-dependent livJ transcription in ΔrelA or ΔrelA ΔspoT mutants is consistent with this model. Moreover, a strain of Salmonella expressing the DksA K98A variant, unable to establish a salt bridge with the 3′ and 5′ phosphates of ppGpp, does not activate livJ transcription in response to 1 μM H2O2. The proposed model is further supported by the synergistic activation of livJ in vitro transcription by oxidized DksA, DnaJ, and 50–100 μM ppGpp. Low levels of H2O2 associated with redox signaling may oxidize DksA, which—with the help of DnaJ and small amounts of ppGpp—activates transcription of genes involved in amino acid biosynthesis. However, the high concentrations of ppGpp that are produced during oxidative stress inhibit amino acid biosynthesis gene transcription and antagonize the stimulatory effects of the DksA–DnaJ redox couple. At higher concentrations, ppGpp may occupy both sites 1 and 2 in RNA polymerase with detrimental effects on transcription (6). Alternative models are also possible. Our research does not exclude the possibility that, in cells experiencing oxidative stress, oxidized DksA must compete for DnaJ binding with an increasing number of unfolded clients, effectively reducing the amount of DksA that is loaded onto RNA polymerase. It is also possible that oxidative stress oxidizes DnaJ in Salmonella, as has been documented in NaOCl-treated Staphylococcus aureus or Mycobacterium smegmatis (28, 29). Oxidation of DnaJ could prevent interactions with oxidized DksA, providing a mechanism for the failure of anaerobic Salmonella treated with 10 μM H2O2 to activate amino acid biosynthesis gene transcription. The inhibition of amino acid biosynthesis gene transcription may synergize with oxidation-induced amino acid auxotrophies (30) to effectively block translation during periods of oxidative stress. Shutting down translation must be a critical component of the adaptation of bacterial cells to intense oxidative stress. DksA is in competition with GreA and GreB for binding to the secondary channel of RNA polymerase (31). Preferential binding of Gre factors to RNA polymerase in favor of oxidized DksA could improve transcriptional fidelity (32) but could also impede DNA strand-break repair (33).
Zinc-bound C4 DksA proteins, such as that of Salmonella, react with H2O2 with an estimated second-order rate constant of 0.6 M−1 s−1 (11), which contrasts with reactions on the order of 105 M−1 s−1 for dedicated H2O2 sensors, such as OxyR (34). Given these kinetic considerations, we were surprised to find that a significant fraction of DksA is already oxidized in aerobic cultures, and that 6 s of exposure to 1 μM H2O2 effectively oxidizes DksA in anaerobic Salmonella. Because the direct oxidation of DksA cysteine residues by H2O2 occurs at a low kinetic rate, the efficient oxidation of DksA in vivo suggests that DksA is oxidized indirectly. This possibility is consistent with the recent report that thiol peroxidases facilitate the oxidation of the transcription factor Yap1 (35). Future investigations will be needed to establish whether thiol peroxidases or some other partners mediate the oxidation of DksA in Salmonella experiencing levels of H2O2 associated with redox signaling and oxidative stress.
Our investigations indicate that the reductase activity of DnaJ is an integral part of the antioxidant defenses of Salmonella. Given its interactions with DksA, the antioxidant role of DnaJ could be partially mediated through the activation of the stringent response. DnaJ is also likely to play antioxidant roles that are independent of DksA. Oxidative stress denatures a variety of proteins that could become clients of DnaJ. Similarly, some of the functions of DksA seem to be independent of DnaJ, as suggested by the DksA-dependent tonal expression of livJ and hisG genes in ΔdnaJ Salmonella.
In summary, our investigations have revealed an aspect of DksA-dependent transcriptional activation that relies on redox-based interactions with the holdase and reductase DnaJ. Cumulatively, our research provides a deeper molecular understanding for the regulation of RNA polymerase and the adaptive responses undertaken by Salmonella in response to divergent levels of H2O2 associated with signaling or oxidative stress.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions.
The derivatives of Salmonella enterica serovar Typhimurium strain 14028s and E. coli, as well as the plasmids used in this study, are listed in SI Appendix, Tables S3 and S4. Deletion mutants were constructed using the λ-Red homologous recombination system (36). Bacteria were used at log phase after growth at 37 °C in LB broth, EGCA medium [0.2% MgSO4, 2% C6H8O7˙H2O, 10% K2HPO4, 3.5% Na(NH4)HPO4˙4H2O, 0.1% casamino acid, and 4% d-glucose, pH 5.5] (11), or Mops minimal medium [40 mM Mops buffer, pH 7.2, 4 mM Tricine, 2 mM K2HPO4, 10 μM FeSO4 ⋅ 7H2O, 9.5 mM NH4Cl, 276 μM K2SO4, 500 nM CaCl2, 50 mM NaCl, 525 pM MgCl2, 2.9 nM (NH4)6Mo7O24⋅4H2O, 400 nM H3BO3, 30 nM CoCl2, 9.6 nM CuSO4, 80.8 nM MnCl2, 9.74 nM ZnSO4, and 0.4% glucose] (24). Penicillin, chloramphenicol, tetracycline, and kanamycin were added as appropriate at final concentrations of 250, 40, 20, or 50 μg/mL, respectively.
TAP and LC-MS/MS Analysis.
The TAP tag is composed of the IgG-binding site of protein A, a cleavage site for TEV protease, and the calmodulin-binding protein. Log-phase ΔdksA S. Typhimurium strains harboring low-copy pWSK29::TAP or pWSK29::dksA::TAP plasmids were grown at 37 °C in LB broth containing 250 μg/mL penicillin. Bacterial cells were disrupted by ultrasonication in lysis buffer (50 mM Tris⋅HCl, pH 7.4, 50 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 10% glycerol, 0.01% lgepal CA6-30, 0.5 mM β-mercaptoethanol, and 0.5 mM PMSF). Soluble fractions were incubated for 2 h at 4 °C with 500 μL of IgG Sepharose resin (GE Healthcare) while gently mixing. The resins were washed three times with 5 mL of washing buffer (10 mM Tris⋅HCl, pH 7.4, 50 mM NaCl, and 0.01% lgepal CA6-30) and 2 mL of TEV cleavage buffer (10 mM Tris⋅HCl, pH 8.0, 50 mM NaCl, 0.5 mM EDTA, 1 mM DTT, and 0.01% lgepal CA6-30). Columns were equilibrated in 2 mL of TEV cleavage buffer and then incubated at room temperature overnight with 50 U of Halo TEV Protease (Promega) in TEV cleavage buffer. Samples were eluted with TEV cleavage buffer and mixed with calmodulin affinity resin (Agilent Technologies) in calmodulin-binding buffer (10 mM Tris⋅HCl, pH 8.0, 50 mM NaCl, 1 mM magnesium acetate, 1 mM imidazole, 2 mM CaCl2, 0.01% lgepal CA6-30, and 10 mM β-mercaptoethanol) for 2 h at 4 °C with agitation. Proteins eluted with calmodulin elution buffer (10 mM Tris⋅HCl, pH 8.0, 50 mM NaCl, 1 mM magnesium acetate, 1 mM imidazole, 2 mM EGCA, 0.01% lgepal CA6-30, and 10 mM β-mercaptoethanol) were precipitated with 20% trichloroacetic acid (TCA). Precipitates were loaded onto 4–15% NuPAGE gels (Invitrogen) in 2-(N-morpholino)ethanesulfonic acid (Mes) SDS running buffer (Invitrogen) and visualized with Imperial Protein Stain (Invitrogen). DksA interacting partners were identified at the Mass Spectrometry and Proteomics Core of the University of Colorado School of Medicine by LS-MS/MS using an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific).
Bacterial Two-Hybrid System and β-Galactosidase Activity Assay.
A bacterial adenylate cyclase two-hybrid system (Euromedex) was used to validate DksA binding partners. The pKN25-dksA plasmid was cotransformed with the pUT18C vector expressing the bait targets stpA, hupA, tig, and dnaJ. pKT25-rpoA plasmid was used as a negative control. Transformants of E. coli BTH10 were selected at 37 °C on LB agar supplemented with penicillin and kanamycin. β-Galactosidase activity was determined in samples collected at stationary phase using the o-nitrophenyl-β-galactoside substrate. Enzyme activity expressed as Miller units was calculated as A420 × 1,000/[time (min) × A600 × cell volume (mL)].
Overexpression and Purification of Proteins.
Genes encoding wild-type or dksA point mutants were cloned into the GST fusion plasmid pGEX-6P-1 (GE) (10). Full-length or truncated dnaJ, tig, and rpoC genes were directionally cloned as C-terminal 6His fusions into the NdeI and XhoI sites of the pET-22b(+) plasmid (Novagen). All constructs were confirmed by sequence analysis. Plasmids were expressed in E. coli BL21 (DE3) (Invitrogen) or E. coli Origami B (DE3) pLysS (Novagen) (SI Appendix, Table S1). Cells grown in LB broth at 37 °C to an OD600 of 0.5–0.7 were then treated with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After 3 h, the cells were harvested, disrupted by sonication, and centrifuged to obtain cell-free supernatants. GST and 6His-tagged fusion proteins were purified using Glutathione-Sepharose 4B (bioWORLD) and TALON metal-affinity chromatography (Clontech), respectively. To remove the GST tag from recombinant GST-DksA protein, PreScission protease (PSP), prepared in PBS buffer supplemented 10 mM DTT, was added. After an overnight incubation at 4 °C, protein was eluted and further purified by size-exclusion chromatography on Superdex 75 (GE Healthcare Life Sciences). Purified DksA proteins were aliquoted inside a BACTRON anaerobic chamber (Shel Lab). The purity and mass of the recombinant proteins were assessed by SDS/PAGE.
Pull-Down Assays.
Interactions between recombinant proteins were examined using pull-down assays (10, 37). Reduced DksA proteins were prepared by treatment with 2 mM DTT for 1 h at room temperature. Excess DTT was removed by buffer exchange with 50 mM Tris⋅HCl buffer, pH 7.5, using Zeba desalting spin columns (Thermo Fisher Scientific). Reduced DksA proteins were treated with 1 mM H2O2 or 1 mM DTT for 1 h at 37 °C. DTT and H2O2 were eliminated using an Amicon Ultra-10 centrifugal filter [molecular weight cut-off (MWCO) = 10,000]. One micromole of GST-DksA (i.e., bait) was incubated for 2 h with 200 μL Glutathione-Sepharose 4B beads (BioWorld) in 50 mM Tris⋅HCl, pH 7.5, at 4 °C. The columns were washed with 20 bed volumes of 50 mM Tris⋅HCl buffer, pH 7.5, and incubated for 2 h at 4 °C with rotation in the presence of 1 μmole of 6His-tagged DnaJ or RNA polymerase (i.e., prey). The columns were washed with 50 mM Tris⋅HCl buffer, pH 7.5, containing 30 mM NaCl, and the proteins were eluted with 50 mM Tris⋅HCl buffer, pH 7.5, containing 500 mM NaCl. Eluted proteins were precipitated with 10% TCA. 6His-tag fusion proteins were loaded into 12% SDS/PAGE gels and detected by immunoblot analysis using a 1:1,000 dilution of anti-6His antibody (Rockland), followed by a 1:10,000 dilution of goat anti-rabbit IgG (Pierce) conjugated with horseradish peroxidase (HRP). The blots were processed using the ECL prime Western blotting detection reagent (GE). Purified GST protein was used as negative control. To investigate the role that the redox status plays on protein-protein interactions, DksA-GST proteins were treated with 1 mM DTT or 1 mM H2O2 (Sigma) in 50 mM Tris⋅HCl buffer, pH 7.5, at 37 °C for 1 h.
Pull-Down Assays of S. Typhimurium RNA Polymerase.
To isolate the holo RNA polymerase complex from Salmonella, a His6-tag was fused to the 3′ of rpoC in the Salmonella genome. Briefly, an EcoRI/XbaI-cut 539-bp PCR product containing the rpoC-6His construct was directionally cloned together with a chloramphenicol resistance cassette and rpoC flanking region into the EcoRI/HindIII sites of pBluscriptKS(+) (Stratagene) to generate the pSK::rpoC′::6His::Cm plasmid. To abolish endogenous DksA binding to RNA polymerase during purification, this construct was introduced into the rpoC native locus of ΔdksA::FRT Salmonella using the λ-Red recombination system. The cointegrate was verified by sequencing.
To isolate RNA polymerase, 4 L of Salmonella expressing the rpoC-6His construct were growth in LB broth at 37 °C to an OD600 0.5–0.7. Cells were disrupted by sonication and cell-free extracts were collected after centrifugation at 4 °C. The 6His-fused RNA polymerase complex was purified with TALON metal-affinity chromatography (Clontech Laboratories). Eluted samples were exchanged with 50 mM Tris⋅HCl buffer, pH 7.5, containing 50 mM NaCl using an Amicon Ultra-100 centrifugal filter (MWCO = 100,000) (Millipore Sigma). Protein concentration was determined using the bicinchoninic acid assay (BCA), and samples were immediately used for pull-down assays. The purified Salmonella RNA polymerase holoenzyme was assessed by 8% SDS PAGE using E. coli RNA polymerase holoenzyme (New England Biolabs) for comparison. One nanomole of the GST-DksA fusion was immobilized in Glutathione-Sepharose 4B beads in the presence of 1 mM DTT or 1 mM H2O2. Equimolar concentrations of DnaJ-6His and the purified RpoC-6His-tagged RNA polymerase were used as prey. 6His-tagged DnaJ and RNA polymerase that were eluted from the columns were detected by Western blotting.
CD Spectroscopy.
DksA used for CD spectroscopy was oxidized or reduced as described in the pull-down assay section above. CD spectroscopy of 0.2 mg/mL DksA recombinant protein in 200 μL of 50 mM Tris⋅HCl buffer, pH 7.5, was performed at the Biophysics Core of the University of Colorado School of Medicine using a Jasco 815 spectrometer (Jasco) with constant nitrogen flushing at 37 °C. Each spectrum is the average of three independent scans.
Evaluation of DksA Oxidation by Alkylation with AMS.
To determine in vivo thiol-modification of DksA cysteine residues in Salmonella grown anaerobically, culture media, buffers, and reagents were placed in a Bactron anaerobic chamber at least 2 d before use. Wild-type and ΔdnaJ S. Typhimurium expressing the dksA::3xFLAG construct were grown in LB broth at 37 °C overnight in the anaerobic chamber, harvested by centrifugation, and resuspended to an OD600 of 0.3 in PBS buffer. Selected cultures were then treated with 1–10 μM H2O2 at 37 °C for 5 min. Bacteria were treated with 15% TCA and the pellets were resuspended in AMS buffer [1 M Tris⋅HCl, pH 8.0, 1 mM EDTA, 0.1% SDS, and 15 mM 4-acetamido-4′-maleimidyl stilbene-2,2′-disulfonic acid (AMS) (Thermo Fisher Scientific)]. Samples were incubated at 37 °C for 1 h in the dark and loaded onto nonreducing SDS/PAGE gels (Bio-Rad). DksA-3xFLAG proteins were detected by immunoblot analysis. Where indicated, wild-type dksA::3xFLAG was cultured in a shaking incubator under aerobic conditions before thiol groups were alkylated with AMS, as described above. In addition, we examined the redox state of thiol groups in recombinant DksA proteins. Briefly, 5 μM DksA was incubated for 1 h in 100 mM potassium phosphate buffer, pH 7.4. Selected samples were treated with 1 mM H2O2 at 37 °C in the presence or absence of 5 μM DnaJ. Samples were precipitated by 15% TCA, resuspended in AMS buffer, incubated at 37 °C for 1 h in the dark, and loaded onto nonreducing 15% SDS PAGE gels for visualization using Coomassie Brilliant blue staining.
Evaluation of DksA Redox State by Alkylation with N-Ethylmaleimide and HPLC-MS Analysis.
DksA separated in SDS gels was digested with trypsin to detect and quantitate DksA peptides containing cysteine residues in either oxidized (S-S) or reduced state [alkylated, +2 N-ethylmaleimide (NEM)]. Briefly, initial procedure was the same as previous but NEM was used as maleimide instead of AMS. Once SDS/PAGE was performed, bands corresponding to DskA were submitted to in-gel tryptic digestion in ammonium carbonate buffer 0.2 M pH 8, overnight, 37 °C. After digestion, peptides were extracted from gel and analyzed with reverse phase liquid chromatography–MS (HPLC-MS, QTRAP4500; ABSciex). Chromatography was performed in a Vydac 218TP column (C18, 150 × 2.1 mm, 5 μ; GRACE) and an acetonitrile gradient (2–45%). All trypsin digestion peptides were analyzed with the mass spectrometer in search of cysteine-containing ones. Once identified, one peptide for each pair of cysteines was selected for quantitative analysis (T18–19: K105VEDEDFGYC114ESC117GVEIGIR124 and T20–21: R125LEARPTADLC135IDC138K139). After parameters optimization, the mass spectrometer was set in positive mode as an ion trap in Enhanced Resolution mode, in search of ions corresponding to each peptide in different charge (+2, +3) and oxidation states (oxidized, S-S or reduced, +2 NEM). Area corresponding to each ion (m/z ± 0.1) was obtained from HPLC-MS analysis and the oxidation ratio (area oxidized/area +2 NEM) was calculated for each sample. This value allows correction of possible digestion variability between samples, because the ratio is calculated with values from the same analyzed sample. However, this ratio does not strictly represent molar relation between peptides. Thus, a more representative index was calculated for each sample, (i.e., the relative oxidation ratio). This ratio is calculated by normalization of previously described oxidation ratio relative to the maximal oxidation condition (DksA + 0.5 mM H2O2).
Susceptibility to H2O2.
Stationary phase bacteria grown in LB broth at 37 °C were diluted in PBS buffer to a final concentration of 2 × 105 CFU/mL. The specimens were treated with 200 μM of H2O2 at 37 °C for 2 h. The percentage of surviving bacteria was calculated as [(CFU from H2O2 treated sample/CFU from untreated sample) × 100]. The effect of H2O2 on bacterial growth was also examined. Briefly, bacteria were grown overnight in LB broth at 37 °C, subcultured 1:100 in EGCA minimal media, and grown at 37 °C. Selected samples were treated with 500 μM H2O2. Subsequent bacterial growth was measured by monitoring OD600 over time.
Animal Studies.
Six- to 8-wk old C57BL/6 and congenic gp91phox−/− (38) mice were inoculated intraperitoneally with 100–200 CFU of Salmonella grown overnight in LB broth. Mouse survival was monitored over 2 wk. The data are representative of two to three independent experiments. All mice were bred according to protocols approved by the Institutional Animal Care and Use Committee at the University of Colorado School of Medicine.
RNA Isolation and Quantitative RT-PCR.
Salmonella grown overnight in LB broth at 37 °C in an anaerobic chamber were subcultured in EGCA medium, diluted to an OD600 of 0.3, and treated with 1–10 μM H2O2 at 37 °C for 30 min. Total RNA was isolated using the High Pure RNA Isolation Kit (Roche). The synthesis of cDNA was achieved using 1 μg of purified RNA, TaqMan Gene Expression Master Mix (Thermo Fisher Scientific), gene-specific primers, and probes containing 5′ 6-carboxyfluorescein and 3′ black-hole quencher 1 modifications. Reactions were performed using a CFX connect Real-Time System (Bio-Rad). PCR-amplified DNA fragments containing the gene of interest were used to generate standard curves. The abundance of transcripts within each sample was normalized to internal transcripts of the housekeeping gene rpoD.
In Vitro Transcription Reactions.
In vitro transcription reactions were performed as previously described with modifications (39). Briefly, DNA template for livJ in vitro transcription assays was PCR-amplified using the primers listed in SI Appendix, Table S3 and extracted using the GeneJET Gel Extraction Kit (Thermo Fisher Scientific). Reduced DksA proteins were prepared as described above for CD spectroscopy. Oxidized DksA proteins were prepared by treatment with 1 mM H2O2 in 40 mM Hepes, pH 7.4, at 37 °C for 1 h. Excess H2O2 was removed after 30-min treatment on ice with 20 U bovine liver catalase (Sigma). Transcription reactions were performed in 40 mM Hepes, pH 7.4, 2 mM MgCl2, 60 mM potassium glutamate, 0.1% Nonidet P-40, 200 μM of each ATP, GTP, and UTP, 8 U RiboLock RNase inhibitor (Thermo Fisher Scientific), 1 μCi [α-32P]-UTP, 1 nM DNA template, 5 nM E. coli RNA polymerse (holoenzyme; New England Biolabs), and 5 μM of individual DksA proteins. Where indicated, the reactions contained 0.5–10 μM DnaJ. Reactions were incubated at 37 °C for 10 min and then inactivated by heat treatment (70 °C for 10 min). Products were analyzed on 7 M urea-6% polyacrylamide gels and visualized using phosphor screens (Kodak) and the Molecular Imager FX (Bio-Rad).
Where indicated, products of the in vitro transcription reactions were quantified by real-time PCR (qRT-PCR) according to previously published methods (39). In vitro transcription reactions were done as described above in the absence [α-32P]-UTP. Guanosine tetraphosphate (Trilink Biotechnologies) was added to selected reactions. After DNaseI treatment, template DNA was removed from the reactions with DNA-free DNA Removal Kit (Thermo Fisher Scientific), and the resulting material was used as templates to generate cDNA using 100 U MMLV reverse transcriptase (Promega), 0.45 μM N6 random hexamer primers (Thermo Fisher Scientific), and 20 U RNase inhibitor (Promega). The amount of cDNA synthesized following 1 h of incubation at 42 °C was quantified by (qRT-PCR) using gene-specific primers and probes presented in SI Appendix, Table S3. Specific transcripts were normalized to standard curves using known amounts of transcript concentrations.
Protein Binding Affinity.
Binding of DksA to DnaJ was evaluated by MicroScale Thermophoresis by detecting temperature-induced changes in fluorescence of DnaJ in the presence of increasing concentrations of DksA proteins. Briefly, 100 nM DnaJ-6His protein was labeled with NT647 fluorescent dye using the Monolith NT His-Tag Labeling Kit RED-Tris-NTA (Nano Temper Technologies). Oxidized DksA proteins, prepared as described for CD spectroscopy, were serially diluted in PBS buffer with 0.01% Tween 20, mixed with an equal volume of 20-nM labeled protein, and loaded into capillaries. Binding was measured on Monolith NT.115 at LED/excitation and MST power of 40% at Biophysics Core in the University of Colorado School of Medicine. Kd values from three independent experiments were determined from a dose–response curve that was fitted to a one-site binding model.
Measurement of (p)ppGpp.
Measurements of Thiol Content and Zn Release.
Reduced DksA and DnaJ proteins were prepared as described above for CD spectroscopy. Proteins were treated with 50–1,000 μM H2O2 or 1–6 M urea at 37 °C for 1 h. H2O2 remaining in the reactions was eliminated after 30-min treatment with 20 U bovine liver catalase on ice. The specimens were mixed with 2 mM DTNB (Thermo Fisher Scientific), and 0.1 mM diethylenetriaminepentaacetic acid in 100 mM potassium phosphate, pH 7.5. Reactions were incubated on ice for 30 min and the products of the reaction of sulfhydryl groups with DTNB were determined spectrophotometrically at OD412. Thiol content was calculated from the molar extinction coefficient of TNB (14,150 M−1cm−1) as [OD412/(path length × 14,1500)] × dilution factor (41). To determine zinc content, chelates of PAR were quantified spectrometrically at OD500. Zinc concentrations were calculated from a standard curve prepared with ZnCl2 (11).
Statistical Analyses.
Statistical analyses and graphing were performed using GraphPad Prism 4.0 software. Determination of statistical significance between two comparisons was achieved using an unpaired t test. Determination of statistical significance between multiple comparisons was done using a one- and two-way ANOVA followed by Bonferroni multiple-comparison posttest. Statistical significance for mouse survival curves was determined using the log-rank test.
Supplementary Material
Acknowledgments
We thank Dr. Jessica Jones-Carson (University of Colorado) for providing the mice used in this study, and Dr. Valeria Valez (Universidad de la República) for assistance in the artwork. This research was funded by VA Merit Grant BX0002073; NIH Grants AI054959, AI136520, AI118223, and AI052066; and the Burroughs Welcome fund. Additional support was obtained from Comisión Sectorial de Investigación Científica Universidad de la República, Uruguay (CSIC I+D 2016) (to M.T.) and CSIC Groups 2014 (to R.R.).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 12550.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1813572115/-/DCSupplemental.
References
- 1.Potrykus K, Cashel M. (p)ppGpp: Still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 2.Paul BJ, Ross W, Gaal T, Gourse RL. rRNA transcription in Escherichia coli. Annu Rev Genet. 2004;38:749–770. doi: 10.1146/annurev.genet.38.072902.091347. [DOI] [PubMed] [Google Scholar]
- 3.Mechold U, Potrykus K, Murphy H, Murakami KS, Cashel M. Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res. 2013;41:6175–6189. doi: 10.1093/nar/gkt302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ross W, Vrentas CE, Sanchez-Vazquez P, Gaal T, Gourse RL. The magic spot: A ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol Cell. 2013;50:420–429. doi: 10.1016/j.molcel.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuo Y, Wang Y, Steitz TA. The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex. Mol Cell. 2013;50:430–436. doi: 10.1016/j.molcel.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross W, et al. ppGpp binding to a site at the RNAP-DksA interface accounts for its dramatic effects on transcription initiation during the stringent response. Mol Cell. 2016;62:811–823. doi: 10.1016/j.molcel.2016.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molodtsov V, et al. Allosteric effector ppGpp potentiates the inhibition of transcript initiation by DksA. Mol Cell. 2018;69:828–839.e5. doi: 10.1016/j.molcel.2018.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parshin A, et al. DksA regulates RNA polymerase in Escherichia coli through a network of interactions in the secondary channel that includes sequence insertion 1. Proc Natl Acad Sci USA. 2015;112:E6862–E6871. doi: 10.1073/pnas.1521365112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perederina A, et al. Regulation through the secondary channel—Structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 10.Henard CA, et al. The 4-cysteine zinc-finger motif of the RNA polymerase regulator DksA serves as a thiol switch for sensing oxidative and nitrosative stress. Mol Microbiol. 2014;91:790–804. doi: 10.1111/mmi.12498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crawford MA, et al. Redox-active sensing by bacterial DksA transcription factors is determined by cysteine and zinc content. MBio. 2016;7:e02161-15. doi: 10.1128/mBio.02161-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henard CA, Bourret TJ, Song M, Vázquez-Torres A. Control of redox balance by the stringent response regulatory protein promotes antioxidant defenses of Salmonella. J Biol Chem. 2010;285:36785–36793. doi: 10.1074/jbc.M110.160960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crawford MA, et al. DksA-dependent transcriptional regulation in Salmonella experiencing nitrosative stress. Front Microbiol. 2016;7:444. doi: 10.3389/fmicb.2016.00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paul BJ, et al. DksA: A critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 15.Kang PJ, Craig EA. Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J Bacteriol. 1990;172:2055–2064. doi: 10.1128/jb.172.4.2055-2064.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winter J, Jakob U. Beyond transcription—New mechanisms for the regulation of molecular chaperones. Crit Rev Biochem Mol Biol. 2004;39:297–317. doi: 10.1080/10409230490900658. [DOI] [PubMed] [Google Scholar]
- 17.Celaya G, et al. Crowding modulates the conformation, affinity, and activity of the components of the bacterial disaggregase machinery. J Mol Biol. 2016;428:2474–2487. doi: 10.1016/j.jmb.2016.04.027. [DOI] [PubMed] [Google Scholar]
- 18.Szabo A, Korszun R, Hartl FU, Flanagan J. A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. EMBO J. 1996;15:408–417. [PMC free article] [PubMed] [Google Scholar]
- 19.Banecki B, et al. Structure-function analysis of the zinc finger region of the DnaJ molecular chaperone. J Biol Chem. 1996;271:14840–14848. doi: 10.1074/jbc.271.25.14840. [DOI] [PubMed] [Google Scholar]
- 20.de Crouy-Chanel A, Kohiyama M, Richarme G. A novel function of Escherichia coli chaperone DnaJ. Protein-disulfide isomerase. J Biol Chem. 1995;270:22669–22672. doi: 10.1074/jbc.270.39.22669. [DOI] [PubMed] [Google Scholar]
- 21.Tang W, Wang CC. Zinc fingers and thiol-disulfide oxidoreductase activities of chaperone DnaJ. Biochemistry. 2001;40:14985–14994. doi: 10.1021/bi0107593. [DOI] [PubMed] [Google Scholar]
- 22.Isaac M, Latour L, Seneque O. Nucleophilic reactivity of zinc-bound thiolates: Subtle interplay between coordination set and conformational flexibility. Chem Sci. 2012;3:3409–3420. [Google Scholar]
- 23.Linke K, Wolfram T, Bussemer J, Jakob U. The roles of the two zinc binding sites in DnaJ. J Biol Chem. 2003;278:44457–44466. doi: 10.1074/jbc.M307491200. [DOI] [PubMed] [Google Scholar]
- 24.Fitzsimmons LF, Liu L, Kim JS, Jones-Carson J, Vázquez-Torres A. Salmonella reprograms nucleotide metabolism in its adaptation to nitrosative stress. MBio. 2018;9:e00211-18. doi: 10.1128/mBio.00211-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furman R, et al. DksA2, a zinc-independent structural analog of the transcription factor DksA. FEBS Lett. 2013;587:614–619. doi: 10.1016/j.febslet.2013.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rüdiger S, Schneider-Mergener J, Bukau B. Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J. 2001;20:1042–1050. doi: 10.1093/emboj/20.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi YY, Tang W, Hao SF, Wang CC. Contributions of cysteine residues in Zn2 to zinc fingers and thiol-disulfide oxidoreductase activities of chaperone DnaJ. Biochemistry. 2005;44:1683–1689. doi: 10.1021/bi0480943. [DOI] [PubMed] [Google Scholar]
- 28.Imber M, et al. Protein S-bacillithiolation functions in thiol protection and redox regulation of the glyceraldehyde-3-phosphate dehydrogenase gap in Staphylococcus aureus under hypochlorite stress. Antioxid Redox Signal. 2018;28:410–430. doi: 10.1089/ars.2016.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hillion M, et al. Monitoring global protein thiol-oxidation and protein S-mycothiolation in Mycobacterium smegmatis under hypochlorite stress. Sci Rep. 2017;7:1195. doi: 10.1038/s41598-017-01179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carlioz A, Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: Is superoxide dismutase necessary for aerobic life? EMBO J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Potrykus K, et al. Antagonistic regulation of Escherichia coli ribosomal RNA rrnB P1 promoter activity by GreA and DksA. J Biol Chem. 2006;281:15238–15248. doi: 10.1074/jbc.M601531200. [DOI] [PubMed] [Google Scholar]
- 32.Erie DA, Hajiseyedjavadi O, Young MC, von Hippel PH. Multiple RNA polymerase conformations and GreA: Control of the fidelity of transcription. Science. 1993;262:867–873. doi: 10.1126/science.8235608. [DOI] [PubMed] [Google Scholar]
- 33.Sivaramakrishnan P, et al. The transcription fidelity factor GreA impedes DNA break repair. Nature. 2017;550:214–218. doi: 10.1038/nature23907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aslund F, Zheng M, Beckwith J, Storz G. Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc Natl Acad Sci USA. 1999;96:6161–6165. doi: 10.1073/pnas.96.11.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delaunay A, Pflieger D, Barrault MB, Vinh J, Toledano MB. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell. 2002;111:471–481. doi: 10.1016/s0092-8674(02)01048-6. [DOI] [PubMed] [Google Scholar]
- 36.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song M, Kim JS, Liu L, Husain M, Vázquez-Torres A. Antioxidant defense by thioredoxin can occur independently of canonical thiol-disulfide oxidoreductase enzymatic activity. Cell Rep. 2016;14:2901–2911. doi: 10.1016/j.celrep.2016.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pollock JD, et al. Mouse model of X-linked chronic granulomatous disease, an inherited defect in phagocyte superoxide production. Nat Genet. 1995;9:202–209. doi: 10.1038/ng0295-202. [DOI] [PubMed] [Google Scholar]
- 39.Tapscott T, et al. Guanosine tetraphosphate relieves the negative regulation of Salmonella pathogenicity island-2 gene transcription exerted by the AT-rich ssrA discriminator region. Sci Rep. 2018;8:9465. doi: 10.1038/s41598-018-27780-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cashel M. Preparation of guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp) from Escherichia coli ribosomes. Anal Biochem. 1974;57:100–107. doi: 10.1016/0003-2697(74)90056-6. [DOI] [PubMed] [Google Scholar]
- 41.Ellman GL. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.