Significance
While our knowledge of modern plague reservoirs and their hosts is extensive, we have little to no knowledge about the origin of the Medieval plague pandemics or the routes of transmission involved in their spread. Prior genomic data provide a patchy low-resolution picture of the transmission dynamics involved during the Second Plague Pandemic, with only five distinct genomes. We have reevaluated all Medieval strains under the light of archaeological and historical evidence to carefully discuss the involvement of different transmission routes during the Second Plague Pandemic. Our interpretation showcases the importance of trade routes and human movements and further supports the identification of Yersinia pestis as the pathogenic agent of the so-called pestis secunda (1357–1366).
Keywords: plague, Medieval, ancient DNA, Yersinia pestis, Second Pandemic
Abstract
Over the last few years, genomic studies on Yersinia pestis, the causative agent of all known plague epidemics, have considerably increased in numbers, spanning a period of about 5,000 y. Nonetheless, questions concerning historical reservoirs and routes of transmission remain open. Here, we present and describe five genomes from the second half of the 14th century and reconstruct the evolutionary history of Y. pestis by reanalyzing previously published genomes and by building a comprehensive phylogeny focused on strains attributed to the Second Plague Pandemic (14th to 18th century). Corroborated by historical and ecological evidence, the presented phylogeny, which includes our Y. pestis genomes, could support the hypothesis of an entry of plague into Western European ports through distinct waves of introduction during the Medieval Period, possibly by means of fur trade routes, as well as the recirculation of plague within the human population via trade routes and human movement.
The Second Plague Pandemic started in the mid-14th century and lasted until the 19th century (1, 2). Its beginning in Europe is marked by a major epidemic event commonly referred to as the Black Death (1346–1353), which ultimately led to the death of at least 30% of the European population (3). The Black Death reached Western Europe in October 1347 through infected Genoese galleys arriving from Caffa (4) and spread over multiple routes and directions, after having reached the thriving trade center of Constantinople (5). Two other pandemics are historically attested, the first, starting in 541–542 CE with the Justinian Plague and lasting in Europe until the mid of the eighth century, and the third, which originated in 1772 in the Yunnan Province, southwest China, and is still ongoing (2, 6).
Plague is a zoonosis, which occasionally spills over to human populations. However, it is primarily a disease of wildlife and is maintained in reservoirs, which nowadays are present on all continents, with the exception of Australia and Western Europe (2, 6). While our knowledge of modern plague reservoirs and their hosts is extensive, it remains unclear which plague reservoir(s) was the origin of the epidemics recorded in Europe throughout its history. Consequently, we are also lacking knowledge of the main routes and mechanisms of transmission during the historical pandemics. Two scenarios have been suggested in previous studies: (i) After a first introduction during the Black Death, plague periodically spilled over from one or more reservoirs located in Western Europe, from where it was later (re)introduced to China and gave rise to the Third Pandemic (7–9); (ii) plague was repeatedly introduced to Western Europe from a reservoir located in Eastern Europe/Central Asia (1, 2, 10) and spread via commercial trade routes and human movement (11, 12). These two scenarios are mutually exclusive regarding the establishment of local European reservoir of plague during the Second Pandemic.
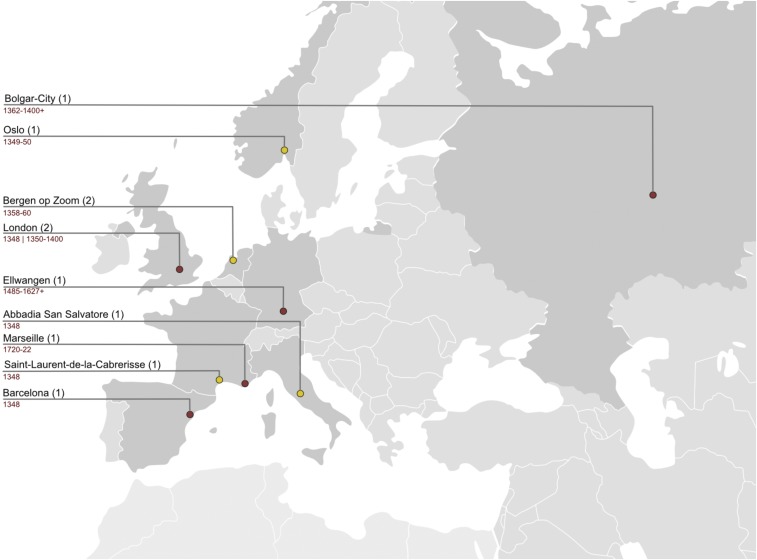
In this study, we provide five sequenced ancient genomes recovered from four archeological sites: Abbadia San Salvatore (BSS) (Italy), Saint-Laurent-de-la-Cabrerisse (SLC) (France), Bergen op Zoom (BER) (The Netherlands), and Oslo (OSL) (Norway) (Fig. 1). The presented genomes all date to the 14th century and represent the focus of our analysis. By supplying additional genomic data of ancient strains recovered from victims of historical epidemics, our study can improve the resolution of the Yersinia pestis phylogeny and can contribute to the discussion on transmission routes and reservoir establishments during historical pandemics. Using state-of-the-art bioinformatics methods, we evaluated the evolutionary history of Y. pestis by incorporating the data reported in this study into a revised phylogeny. We evaluated our results in light of historical, epidemiological, and ecological studies to improve our understanding of the fully documented spectrum of the dynamics at work during the early decades of the Second Plague Pandemic.
Fig. 1.
Geographic locations of previously and presently described ancient genomes. Map of previously and presently described ancient genomes. The red circles represent the locations of previously described ancient genomes. Yellow circles represent ancient genomes described in this study. For the newly described ancient genomes, the indicated years are discussed in Results and Discussion. Numbers in parentheses indicate number of ancient genomes included from each site.
Results
Screening and Authentication of Y. pestis DNA in Human Skeletons.
The presence of Y. pestis in the samples from Saint-Laurent-de-la-Cabrerisse (SLC) and Bergen op Zoom (BER) has been confirmed by PCR in a previous study by Haensch et al. (13). A similar approach was used to screen 36 individuals from Abbadia San Salvatore (BSS) and nine individuals from Oslo (OSL) (see Methods for more details on the experimental work). Positive samples from Abbadia San Salvatore and Oslo were enriched without initial metagenomic screening.
Genomic Data Analyses.
We systematically reanalyzed all available raw sequencing data from ancient Y. pestis metagenomic datasets from the Second Plague Pandemic. Additionally, we reanalyzed the raw data from two Bronze Age samples and two Justinian Plague samples from the First Plague Pandemic. Most sequencing data stemming from modern samples were included as well. By systematically reanalyzing the available raw data, we were able to avoid any incongruity caused by differences in data filtering and SNP calling. Our custom data workflow (SI Appendix, Fig. S5) also accounted for deamination damage patterns typical for ancient DNA (aDNA). For all generated ancient genomes, reads mapping to the genome of Y. pestis, strain CO92, showed a mean read length between 50 and 60 bp, which is consistent with aDNA. The aligned reads also showed a clear deamination profile typical of aDNA misincorporations (SI Appendix, Fig. S6). Moreover, by aligning all generated reads from each sample to Yersinia pseudotuberculosis, strain IP-32953, the edit distance analysis clearly showed that the reads belong to Y. pestis rather than to Y. pseudotuberculosis (SI Appendix, Fig. S7). In fact, for all aDNA reported in this study, the percentage of reads with an edit distance of 0 is higher when aligned against Y. pestis vs. Y. pseudotuberculosis.
After read quality rescaling, the transitions to transversion ratios of all aDNA assemblies were comparable with those of modern genomes (SI Appendix, Fig. S8), which emphasizes the consistency of our methods and data. In addition, to investigate possible cross-contamination or multiple infection, for each sample, the heterozygous profile analysis (SI Appendix, Fig. S9 and Table S4) showed that, for almost all ancient samples, the heterozygous ratio is low (<0.3), as for modern data. Nevertheless, this ratio is high (>0.4) in some modern data.
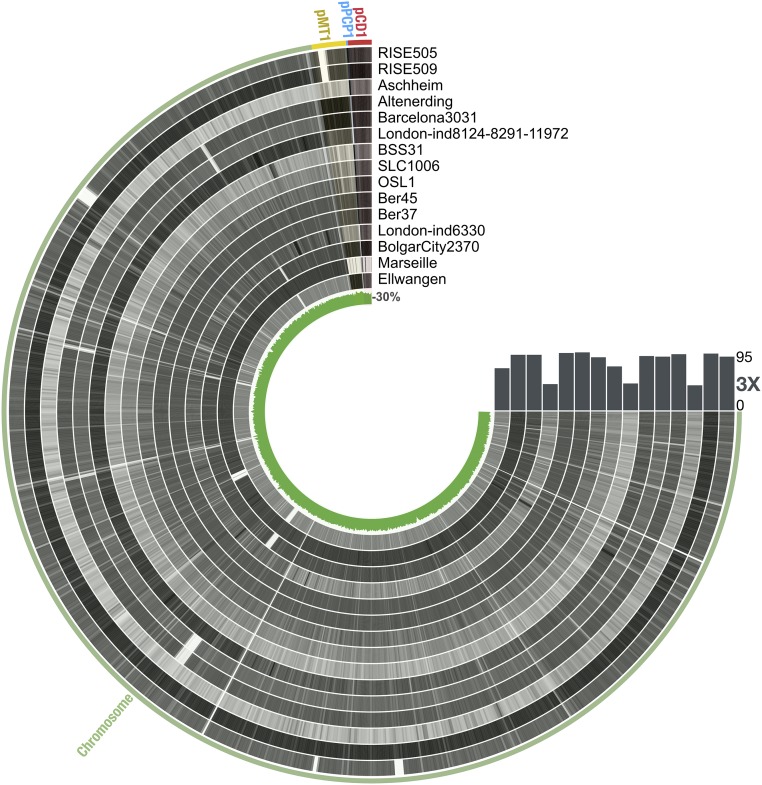
The samples from Bergen op Zoom, Ber37 and Ber45, and the sample from Oslo, OSL1, covered more than 87% of the reference genome, with a minimum of 3× depth of coverage (Fig. 2 and SI Appendix, Table S3). For the samples SLC1006 (Saint-Laurent-de-la-Cabrerisse) and BSS31 (Abbadia San Salvatore), we were able to recover 73% and 44% of the genome at 3× depth of coverage, respectively (Fig. 2 and SI Appendix, Table S3).
Fig. 2.
Overview of the coverage of ancient Y. pestis genomes. Each ring corresponds to one ancient genome. The color intensity is proportional to the coverage across the chromosomal genome and plasmids. The coverage rate, as well as the GC content, was measured throughout the genome using a window size of 200 bp. The bars indicate the fraction of the genome with a depth of coverage of 3×. This figure was generated using anvio (69).
Overall, after having considered all possible patterns of deamination derived from diagenetic processes, along with sequencing inconsistencies, and having rescaled the data accordingly, the comparison of 126 modern Y. pestis genomes with 15 ancient genomes from the Bronze Age and the First and the Second Pandemic (SI Appendix, Table S1) yielded 2,826 polymorphic sites (SI Appendix, Table S5). These SNPs were grouped as follows: 1,456 nonsynonymous SNPs, 625 synonymous SNPs, and 745 intergenic SNPs. Among the 2,826 identified polymorphic sites, 112 were homoplastic (3.9%) (SI Appendix, Table S6).
Dating of the Y. pestis Strain from Abbadia San Salvatore.
Attributed to a major epidemic of the second half of the 14th century by archaeological and stratigraphic evidence, the age of the mass grave found at Abbadia San Salvatore (SI Appendix), as well as the Y. pestis genome retrieved, needed to be refined to help with the interpretation of the phylogenetic tree. The consultation of notorial records from the region, with an impressive increase of death-bed testaments and a decrease of nontestamentary property contracts, revealed the presence of a major dramatic event from late June to early September 1348 (SI Appendix, Fig. S3 and Table S2). The analysis of the data confidently demonstrated that such a catastrophic event, which has no equivalent in the course of the century, was responsible for the death of the large number of victims retrieved in Abbadia San Salvatore.
Evolutionary History Reconstruction.
We built our phylogenetic tree using maximum likelihood, as implemented in IQ-TREE (version 1.5.5) (14). Yersinia pseudotuberculosis, strain IP32953, was used as an outgroup. The adequate substitution model was identified and applied using ModelFinder (15) before building the phylogeny. After testing 484 models, the best-fitting model, according to the Bayesian information criteria (BIC), was the general time reversible (GTR) model (16), with unequal rates and base frequencies with ascertainment (ASC) bias correction and free rate heterogeneity across sites with four rate categories.
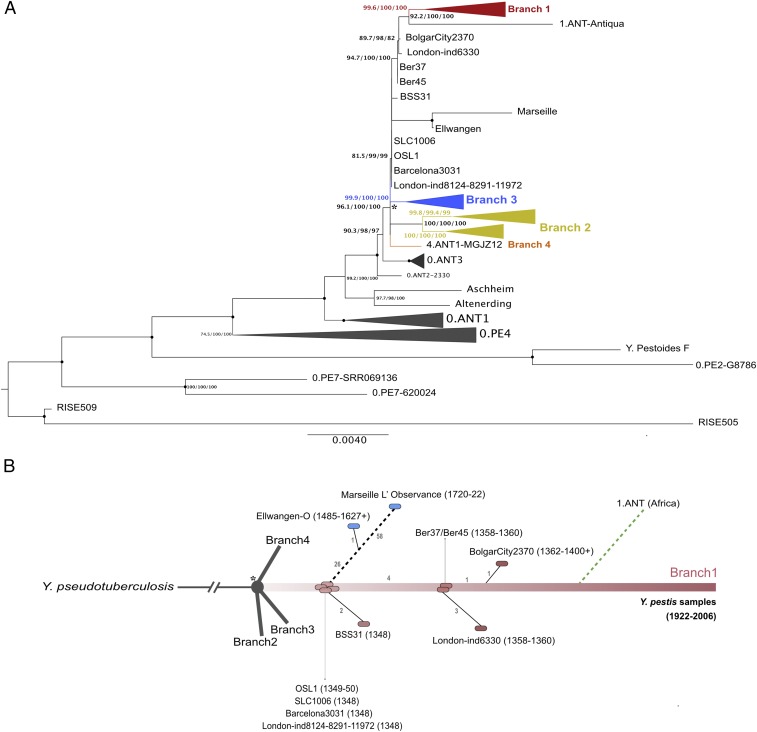
The topology of the generated tree (Fig. 3 and SI Appendix, Fig. S10) was supported by three different tests (bootstrap, SH-alrt, and local bootstrap probability test) and showed a star-like radiation at the end of branch 0, giving rise to branches 1, 2, 3, and 4. As previously reported, two Bronze Age samples from Bateni and Kytmanovo (Russia) (17), as well as the Justinian samples (9, 18), can be found on branch 0 (Fig. 3A). As expected, the five described ancient genomes cluster with all other known Second Plague Pandemic genomes (Fig. 3).
Fig. 3.
Reconstruction of the phylogenetic tree and evolutionary history of Y. pestis. (A) Maximum likelihood (ML) phylogenetic tree for Y. pestis. Consisting of 126 modern Y. pestis strains and 15 ancient genomes, the tree was built using IQ-Tree (version 1.5.5) (14), which implements ModelFinder (15) to find the right substitution model before ML tree search. A total number of 2,826 polymorphic sites were used to construct the ML tree, which was visualized and edited using FigTree (version 1.2.1). Strains belonging to branch 1, branch 2, and branch 3 were collapsed for clarity. The numbers at each node indicate the bootstrap values at 1,000 replicates, SH-alrt support values at 1,000 replicates, and local bootstrap support values, respectively. The nodes indicated by black circles correspond to 100/100/100 of the support values in the same order as previously described. The asterisk marks the polytomy known as the “Big Bang.” (B) Schematic representation of the evolutionary history of Y. pestis genomes dated to the Second Plague Pandemic. Each circle represents one sample. The numbers on each branch correspond to the number of sequentially accumulated SNPs (SI Appendix).
Divergence Dates and Tip Dating.
The reconstructed maximum likelihood tree shows a high correlation between the root-to-tip distance and the sampling time (correlation coefficient r2 = 0.74). The date-randomization test (DRT) showed a different result. In fact, the outcome of the DRT showed that the 95% highest posterior density (HPD) of the substitution rate estimated for both the original and 20 date-randomized datasets is overlapping (SI Appendix, Fig. S11).
Discussion
As mentioned in the Introduction, two scenarios have been proposed in past studies for the persistence of plague during the Second Plague Pandemic. However, the few currently available ancient Y. pestis genomes have proven insufficient to explain the mechanisms underlying the recurrent plague outbreaks in Europe during the Second Plague Pandemic. To provide further insights into the events driving the Second Plague Pandemic and to properly address which of the two proposed scenario has the highest likelihood, we have evaluated the available data along with five sequenced Y. pestis strains stemming from four European archaeological sites of the 14th century, using an integrative approach. By combining historical, archaeological, and genomic data, we attempted to reconstruct the spread of plague in a multifaceted context and to define the routes of transmission following the initial introduction of plague during the Black Death, accordingly. The need of an integrative approach is also supported by the lack of temporal signal in the data, which could lead to inaccurate estimates of substitution rates and timescale, as shown by the date-randomization test, which, so far, represents one of the most robust methods to estimate temporal signal.
Two of our Y. pestis strains, SLC1006 and OSL1, had genomes identical to the clones of London1348 and Barcelona3031 (Fig. 3B), both attributed to the Black Death (7, 19). The strains were extracted from skeletons excavated in Saint-Laurent-de-la-Cabrerisse (France) and Oslo (Norway). These results allowed us to place the outbreak in Saint-Laurent-de-la-Cabrerisse into the Black Death period, as had been proposed by Haensch et al. (13). While no direct historical description of the arrival of plague in Saint-Laurent-de-la-Cabrerisse is available, reports of plague epidemics in the surrounding region (Southern France) in the second half of the 14th century exist (13, 20). Historically, the regions surrounding the site of Saint-Laurent-de-la-Cabrerisse (Aude, Occitanie) are known to have been ravaged by plague from November 1347 onwards; specifically, plague was present in Narbonne and Carcassonne, cities close to Saint-Laurent-de-la-Cabrerisse, from March to April 1348. Interestingly, studies have proposed that plague reached Barcelona from the very same region by April–May 1348 (3, 21). The genomic similarity between the strains SLC1006 and Barcelona3031 seems to corroborate the hypothesis of a common origin. The strain from the St Nicolay’s Church’s cemetery in Oslo (Norway) also clusters with Black Death strains. The time of insurgence of the Medieval plague epidemic in Oslo is debated. However, radiocarbon dating and archaeological data (SI Appendix) place the strain OSL1 at around 1350, which is in line with time lines proposed by some historical studies (22, 23), but in disagreement with others, which had given the Black Death in Oslo an earlier origin (1348) (24, 25). It is important to note that, although the samples identified in Saint-Laurent-de-la-Cabrerisse, Barcelona, London, and Oslo seem to be identical, this similarity is based solely on the regions of the chromosome covered across all of these samples. Regions not covered, as well as structural and plasmid variation, which could account for additional differences among the strains, have not been evaluated or considered in this study or any other previous whole genome aDNA plague phylogeny.
In contrast to the above-mentioned strains, the clone recovered from Abbadia San Salvatore (Siena, Italy) accumulated two additional, specific SNPs (T3529404C and A3897987T), compared with the other Black Death samples (Fig. 3B), and might therefore be interpreted as a more recent derivate. While no direct historical sources are available to confirm the presence of plague in Abbadia San Salvatore in the 14th century, historical data collected and evaluated for this study have revealed a dramatic increase in death-bed testaments in the region from June to September 1348. These data support an account by Agnolo di Tura (26), who reported the desolation of the city of Siena and its surrounding countryside by the plague in 1348 (27). Starting from the port city of Pisa in mid-January 1348 (4), plague made its way through Tuscany and was subsequently reported in Florence (March–April 1348) before reaching Siena and its countryside in May 1348, likely over the Via Francigena, a well-frequented route by pilgrims (4). Considering this chain of events, it is reasonable to assume that the Abbadia San Salvatore strain reached Pisa over sea and continued to spread over land by infected individuals or goods. Given the number of subsequent outbreaks recorded over this short period of time, it is plausible that the additional two SNPs were acquired through the large transmission chain in Italy, rather than having been gained before the strain’s arrival in Pisa or within a newly established local wildlife reservoir.
While the Black Death branches exhibit low variability, the subbranch departing from the Black Death clones, which gave rise to the outbreak in Ellwangen (1485–1627 by radiocarbon dating) (7) and culminated in the outbreak of Marseille (1720–1721), sees a considerable increase in variability (Fig. 3B). Previous literature (7) has attributed this branch to a novel stable reservoir established at the time of the Black Death, in which favorable environmental conditions have enabled a permanent presence of Y. pestis in a host population of wild rodents. The study argued that this ancient reservoir was probably situated in the Alpine region (28). However, ecological and historical–epidemiological studies have concluded that the presence of a plague reservoir in Western Europe during the Second Plague Pandemic was highly improbable (10, 12). Taking a closer look at historical sources, it becomes clear that at least the Great Plague of Marseille (1720–1722) (29, 30) had an extra-Western European source. In fact, historical records clearly associate the start of the outbreak with the successive arrival of several ships from Sidon (Lebanon) (31). While no additional information regarding the geographic location of the wildlife reservoir feeding plague to Europe during the 17th and 18th century is available, studies have argued that, before being introduced to the Mediterranean basin in 1347, plague spread to the territories around the Black Sea and to the Middle East (32, 33) and thus had an initial opportunity to be disseminated in this region.
On the Y. pestis phylogenetic tree, specifically on branch 1, we found two additional genomes isolated from skeletons recovered in Bergen op Zoom. The two identical isolates show four additional SNPs, compared with the Black Death strains [T699494C, G2218046T (homoplastic), T2262577G, and A2894703G] (Fig. 3B) and are basal to the clone found in the St. Mary Grace Cemetery in London (London-ind6330) and in Bolgar (BolgarCity2370). Although no direct historical evidence exists (13), the large number of victims retrieved in Bergen op Zoom likely died during documented plague outbreaks, which occurred in the Belgian Hainault region and the region of Flanders and Antwerp between 1358 and 1363 (34). Based on the phylogeny, the strain London-ind6330 (with three specific SNPs G1130135T, C1159914A, and G4301295T) could be attributed to the wave of plague outbreaks that hit Medieval Europe in 1361, known as “pestis secunda,” and might have been imported directly from Bergen op Zoom, which supports the fact that St. Mary Grace is in general considered a post-Black Death cemetery. The strain isolated from Bolgar (7) was placed one SNP (T3806677C) further on branch 1 from the clones of Bergen op Zoom and is characterized by one additional specific SNP (G3643387T). The Y. pestis clone found in Bolgar, which had previously been attributed to 1362+ (7), could now be more precisely attributed to the outbreaks, which started in 1364 in Nizhnii Novgorod (Fig. 4) and swept throughout all towns in Russia until 1366, killing up to 100 people per day (35). During an excavation of the marketplace of Bolgar, which was active from the 1340s/1350s and was destroyed by a fire in the 1360s/1370s, archaeologists recovered a range of trade goods, in particular artifacts originating from Flemish towns (Tournai, Ypres, etc.) (36, 37), which constitutes direct evidence for trading contacts between the Low Countries and the Volga region. Therefore, it cannot be excluded, that, as reported in previous studies (7), plague was imported to Bolgar from Western Europe, where it had established in a reservoir from the Black Death. Nevertheless, the possibility that the three places (London, Bergen op Zoom, and Bolgar) were infected independently from extra-Western European reservoirs in the 1360s should not be ignored.
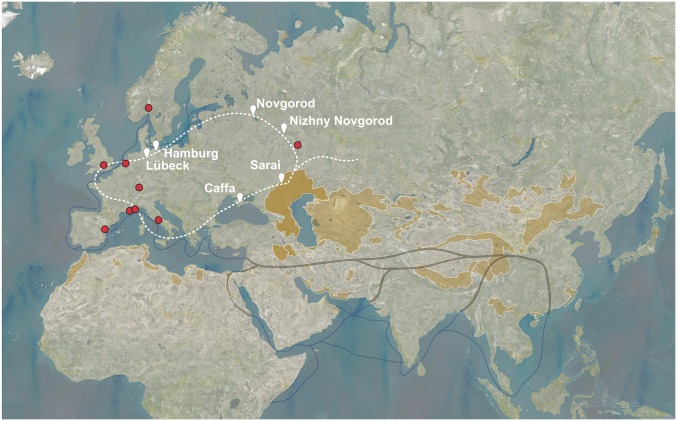
Fig. 4.
Schematic representation of the connection between the fur trade routes and the spread of plague during the early stage of the Second Plague Pandemic (14th century). This simplified map shows cities strategically situated along the fur trade routes (indicated as a white line). The city of Novgorod (Russia) has played a central role in fur export to cities like Hamburg and Lübeck. The regions highlighted in orange represent known modern plague reservoirs. The darker orange delimits the region in which we believe secondary plague reservoirs were established before the Black Death. The red dots represent the locations of all known ancient Y. pestis genomes. Black lines, Silk Road; dark-blue lines, maritime trade routes.
Here, we considered historical evidence and propose that the phylogeny of the plague strains of the 14th century (branch 1) may be also explained by independent introductions on the fur trade routes, a complex network of interconnected maritime, riverine, and overland routes into Western Europe. During the second half of the 14th century, Bolgar and Novgorod (Russia) (Fig. 4) had established an economic dominance as major fur trade centers. Fur was historically an important trade good, which was widely commercialized on established itineraries all over Eurasia. Starting with the second half of the 13th century, Novgorod had gained access to the Western European markets through the Hanseatic League, which it eventually became part of (35). The flourishing of the League in the 14th century permitted Novgorod to export big consignments of fur to London via Lübeck and Hamburg (35, 38). Thus, the three specific SNPs found in London-ind6330 might have been acquired over the course of a more complex transmission chain, similar to Abbadia San Salvatore (Fig. 3B), since the territories of the Hanseatic League were also affected by plague during the pestis secunda (39). However, as plague also struck many Mediterranean ports between 1357 and 1361 (e.g., Alexandria in 1357–1358 and 1361, Venice in 1359–1361, Genoa in 1360–1361, and Ragusa/Dubrovnik in 1361 and Constantinople) (40), it becomes difficult to determine precisely the point of entry and the routes of transmission of the pestis secunda (40). However, a reintroduction from outside seems most likely both from historical (5) and climatic (10) evidence.
On the other hand, the Black Death in Western Europe may also be due to importations on the fur trade network. During the 14th century, Russia was under the domination of the Golden Horde, which had imposed new regulations on the fur trade, and merchants from Bolgar had started importing their fur from the “Land of Darkness,” a region speculated to be situated on the Kama river (41). After exporting it to Sarai (Southern Volga region), which is placed in the easternmost part of Europe, fur was distributed to all parts of Eurasia along with other goods. Remarkably, in the 1340s, there was a significant change that impacted the routes of the fur trade. In fact, a new mainland route connecting Sarai, Tana, and Caffa had been established with the support of the Golden Horde. Traded goods were then further exported from Caffa on the Black Sea trading network by the Genoese (41). Historians have remarked that a considerable variety of fur had started to appear in the ports of the Black Sea by the 1340s (41). These events seem to coincide chronologically with the onset of the Black Death, whose origins remain unclear before its arrival from Caffa. Suggestively, Ibn al-Wardī, an Arab historian, who died of plague in Aleppo (Syria) in 1349, described how returning merchants from Crimea, attributed the beginning of the “contagion” to the “Land of Darkness,” in which the pestilence had been present for 15 years before reaching the West (3). Without having visited the West-Ural region from which fur was imported to the Volga centers, the Arab merchants likely just meant that the source of the Black Death was associated with the fur routes, placed somewhere before Sarai (seen from Caffa), where plague was present before 1347 (3), and hence probably a region next to the Caspian Sea—an area with well-established plague reservoirs today (Fig. 4). From this region, plague may have been transported on the fur trade network to the whole of Europe, not only during the Black Death, but also later, with (possibly) independent goods deliveries to Bergen op Zoom, London, and Bolgar. Extra-SNPs, which developed on subbranches lateral to branch 1, might also have been accumulated during chains of epidemics on the long journey to Western Europe.
Overall, evidence from different disciplines increasingly suggests successive and independent introductions of plague to Western Europe via the transport of infected individuals and goods on trade routes during the Second Plague Pandemic. By analyzing five ancient genomes from the Second Plague Pandemic, adding them to the established Y. pestis phylogeny, and evaluating them with a historical background, our study aimed to answer some of the most debated questions on plague in Western Europe during the Second Plague Pandemic. All things considered, the hypothesis that Y. pestis reached Europe through multiple introductions during the Middle Ages through different routes, including the fur trade, appears very plausible, at least during the Second Plague Pandemic.
Methods
Samples.
The Y. pestis strains presented in this study stem from four European sites dating to the second half of the 14th century (Fig. 1): Saint-Laurent-de-la-Cabrerisse (France) (13), Bergen op Zoom (The Netherlands) (13), Oslo (Norway) (42), and Abbadia San Salvatore (Italy).
For this study, a total of 126 Y. pestis strains (SI Appendix, Table S1) and 15 published ancient DNA samples were used, including 2 Bronze Age samples (17), 2 Justinian plague samples (9, 43), and 10 samples from the Second Plague Pandemic (7, 19).
Dating the Strain of Abbadia San Salvatore with Historical Data.
We collected data of testaments and contracts from 1340 to 1381 (SI Appendix, Fig. S3 and Table S2), a period followed by a gap in documentation until 1440, as documented in inventories (available for consultation at the Archivio di Stato, Florence, Italy), which summarize parchments from Monte Amiata, the region where Abbadia San Salvatore is placed.
Sample Preparation.
We screened a total of 6 teeth from the site of Saint-Laurent-de-la-Cabrerisse, 41 teeth from the site of Abbadia San Salvatore, 3 teeth from the site of Bergen op Zoom, and 18 teeth from the site of Oslo. Laboratory work was performed at the Paleogenetics Laboratories at the University of Mainz, (Germany) and at the Ancient DNA Laboratory at the University of Oslo (Norway). Both laboratories are solely dedicated to the analysis of ancient samples and are subjected to strict anticontamination protocols, including full overnight UV irradiation. Target enrichment was performed at the post-PCR capture laboratory of the Centre for Ecological and Evolutionary Synthesis (CEES), University of Oslo, Oslo.
Extraction and PCR Screening.
The teeth were decontaminated, sandblasted, and milled to fine powder, as previously described (13). aDNA was extracted using either a previously published phenol chloroform protocol (13) or modified versions of silica-based protocols based on Brotherton et al. (44) or Dabney et al. (45). We used 0.2 to 0.5 g of tooth powder for phenol chloroform extractions and 0.1 to 0.26 g for the silica-based extraction methods. Two samples from Bergen op Zoom (Ber37 and Ber45), 6 samples from Saint-Laurent-de-la-Cabrerisse, and 39 samples from Abbadia San Salvatore were extracted by phenol chloroform extraction (protocol A) (SI Appendix). Three teeth from Saint-Laurent-de-la-Cabrerisse, 2 teeth from Abbadia San Salvatore, and 18 teeth from Oslo were extracted via silica extraction based on Brotherton et al. (44) (protocol B) (SI Appendix). Lastly, one tooth from Abbadia San Salvatore was extracted using a protocol based on Dabney et al. (45) (protocol C) (SI Appendix). All extractions included negative milling and extraction controls.
All extracts, which had not previously been screened for Y. pestis in Haensch et al. (13), were screened for human and Y. pestis DNA using previously published primers: pla YP11D/YP10R as published in Raoult et al. (46), caf1 caf1U2/L2 as published in Haensch et al. (13), and human mitochondrial HVR1 primers L16209 (47) and H16348 (48). PCR conditions were as described in Haensch et al. (13). Positive samples were shotgun sequenced on an Illumina HiSeq2500 system [125 bp paired end (PE)] at the Norwegian Sequencing Centre (NSC) at the University of Oslo.
Library Preparation and Target Enrichment.
Library preparation was done following a modified Meyer and Kircher (49) protocol (for more details, see SI Appendix). Amplified libraries were purified using commercial kits (Stratec PCRapace or Qiagen MinElute PCR purification kits, followed by an AMPureXP beads purification) and subsequently quantified on a Bioanalyzer 2100 expert dsDNA High Sensitivity Chip and using a Qubit HS assay kit. When necessary, reamplifications were performed with IS5 and IS6 primers following the original protocol by Meyer and Kircher (49). Positive samples, screened via standard PCR and/or shotgun metagenomics, were enriched for Y. pestis DNA. Over the course of this study, we used two different custom bait kits from different manufacturers for in-solution target enrichment (for more details, see SI Appendix).
Sequencing.
High throughput sequencing (125 bp PE) of the captured libraries was performed on an Illumina HiSeq2500 system at the NSC (University of Oslo). Capture products from Ber45 were pooled on one lane, SLC1006 products were split over two lanes (single capture and double capture products were sequenced separately), and Bss 31d and Ber37c products were each sequenced and pooled with other samples on different lanes and flow cells. Libraries for OSL1A were sequenced and multiplexed with other libraries over two lanes.
Shotgun Metagenomics Data Analysis.
The presence of Y. pestis in shotgun metagenomic datasets was investigated using two methods. The first method is based on read abundance of specific genes and was implemented in Metaphlan (50). The second method is based on the exact alignments of k-mers and was implemented in Kraken (51). This procedure was only applied when the first screening using qPCR was ambiguous.
Mapping-Based SNP Analysis.
We designed a custom pipeline (SI Appendix, Fig. S5) to analyze raw sequencing data from modern and ancient genomes using snakemake as the workflow manager system (52). The genome of the Y. pestis strain CO92 was used as reference. Paired-end reads in fastq format were trimmed and merged using ClipAndmerge (https://github.com/apeltzer/ClipAndMerge). Thereafter, we used bwa-aln (53) to align the merged reads against Y. pestis strain CO92 by disabling the seed option (-l) and setting the option -n to 0.1 (for modern data, the -n option was set to 0.01). Duplicated reads were identified and marked using picard-tools (broadinstitute.github.io/picard/). Local alignment of reads was performed around gapped regions before SNP calling using the GATK IndelRealigner module (https://software.broadinstitute.org/gatk/). DeDup (https://github.com/apeltzer/DeDup) was also used to remove duplicated sequences as it has been specifically designed for merged reads. Aligned Binary Alignment Map (BAM) files of ancient DNA samples were analyzed using MapDamage2 (54) to assess and recalibrate aDNA damage patterns in the form of by C-to-T or G-to-A conversions.
Edit Distance.
To verify that the generated reads belong to Y. pestis rather than Y. pseudotuberculosis, all reads from each sample were mapped to the genome of Y. pseudotuberculosis, strain IP-32953, as described above. The edit distance, which defines the minimal number of substitutions, insertions, and deletions to transform the read sequence to its homologous sequence in the reference genome, was calculated using BAMStats (bamstats.sourceforge.net/).
SNPs Call.
SNP calling was performed using samtools (55, 56) and bcftools (https://samtools.github.io/bcftools/). SNPs located within a frame of 10 bp from indels were excluded with samtools. For each sample, all identified SNPs were filtered and annotated using a freely available tool, snpToolkit (https://bitbucket.org/Amine-Namouchi/snptoolkit); snpToolkit allows one to filter and annotate SNPs from vcf files by providing the genbank file of the reference sequence used during reads alignment. SNPs were filtered according to three criteria: quality score (≥30), depth of coverage (≥3), and allele frequency (90%). In addition, SNPs that are close to each other by less than 10 bp were excluded during the annotation process using snpToolkit with option -f. All generated annotation output files where thereafter compared against each other using the snpToolkit option combine. For each polymorphic site, snpToolkit checks the corresponding alignment region in the bam file to accurately determine the distribution of each polymorphic site by considering the same region in all other aDNA strains of the Second Plague Pandemic. The efficiency of the method implemented in snpToolkit was assessed after manual inspection of these polymorphic sites using Integrative Genomics Viewer (IGV) (57, 58). The option combine produces two output files. The first one represents the distribution of all identified polymorphic sites of all analyzed samples. The second file is a concatenation of all polymorphic sites in fasta format.
Phylogenetic Analysis.
Phylogenies were inferred using IQ-TREE (14). IQ-TREE was run using ModelFinder (15), with the option -m MFP to infer the best substitution model for building the maximum likelihood phylogenetic tree. A total number of 484 models were tested. One thousand fast bootstrap replicates were performed to assess statistical support (59). In addition, branch supports were also assessed through the single branch test SH-aLRT (60) with 1,000 replicated as well as local bootstrap probability tests (61). As the concatenated SNPs include missing information indicated by an exclamation mark in the fasta file generated using snpToolkit, we also used the ASC option to account for ascertainment bias correction. The generated tree was visualized using FigTree (version 1.2.1, tree.bio.ed.ac.uk/software/figtree/), and the substitutions leading to each SNP were mapped in the phylogenetic tree using Fastml v3.1 (62).
Divergence Time and Tip Dating Analysis.
Temporal signal detection in the sequence data was performed using TempEst (63). As previously described (17, 64, 65), the lognormal relaxed clock model and constant population size models implemented in BEAST (66) were applied to the alignment sequence and phylogenetic tree to determine the divergence dates at each node. To evaluate the reliability of the Bayesian inference, we applied the date-randomization test (DRT) as implemented in the R package tipdatingbeast (67) to generate 20 BEAST input files in xml format with randomized dates. As the out dataset included a large number of modern sequences, the randomization process was only applied to ancient sequences, as previously recommended (68). BEAST was run on each file for 50 million iterations.
Data Availability.
Raw sequencing reads produced for this study have been deposited in the European Nucleotide Archive (ENA) under accession number ENA: PRJEB24499.
Supplementary Material
Acknowledgments
We thank Lars Walløe for useful comments and discussion; and Joachim Burger for providing access to the aDNA facilities at the University of Mainz, Germany. This project was funded by the European Research Council (ERC) under the FP7-IDEAS-ERC Program (Grant 324249). Part of the data analysis was performed on the Abel Cluster, owned by the University of Oslo and the Norwegian metacenter for High Performance Computing (NOTUR) and operated by the Department for Research Computing at the University Center for Information Technology Services (USIT), University of Oslo. C.O. is funded by Satsningsmiljø Grant Comparative Infection Biology (COMPI), Department of Biosciences, University of Oslo.
Footnotes
The authors declare no conflict of interest.
Data deposition: Raw sequencing reads have been deposited in the European Nucleotide Archive (ENA) (accession no. PRJEB24499).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812865115/-/DCSupplemental.
References
- 1.Stenseth NC, et al. Plague: Past, present, and future. PLoS Med. 2008;5:e13. doi: 10.1371/journal.pmed.0050003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bramanti B, Stenseth NC, Walløe L, Lei X. Plague: A disease which changed the path of human civilization. Adv Exp Med Biol. 2016;918:1–26. doi: 10.1007/978-94-024-0890-4_1. [DOI] [PubMed] [Google Scholar]
- 3.Benedictow OJ. The Black Death 1346-1353: The Complete History. Boydell Press; Woodbridge, UK: 2004. [Google Scholar]
- 4.Michele da Piazza . In: Cronaca. Giuffrida A, editor. ILA Palma; Palermo, Italy: 1980. [Google Scholar]
- 5.Campbell BMS. The Great Transition: Climate, Disease and Society in the Late-Medieval World. Cambridge Univ Press; Cambridge, UK: 2016. [Google Scholar]
- 6.Vogler AJ, et al. A decade of plague in Mahajanga, Madagascar: Insights into the global maritime spread of pandemic plague. MBio. 2013;4:e00623-12. doi: 10.1128/mBio.00623-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spyrou MA, et al. Historical Y. pestis genomes reveal the European Black Death as the source of ancient and modern plague pandemics. Cell Host Microbe. 2016;19:874–881. doi: 10.1016/j.chom.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 8.Bos KI, et al. A draft genome of Yersinia pestis from victims of the Black Death. Nature. 2011;478:506–510. doi: 10.1038/nature10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagner DM, et al. Yersinia pestis and the plague of Justinian 541-543 AD: A genomic analysis. Lancet Infect Dis. 2014;14:319–326. doi: 10.1016/S1473-3099(13)70323-2. [DOI] [PubMed] [Google Scholar]
- 10.Schmid BV, et al. Climate-driven introduction of the Black Death and successive plague reintroductions into Europe. Proc Natl Acad Sci USA. 2015;112:3020–3025. doi: 10.1073/pnas.1412887112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yue RPH, Lee HF, Wu CYH. Navigable rivers facilitated the spread and recurrence of plague in pre-industrial Europe. Sci Rep. 2016;6:34867. doi: 10.1038/srep34867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yue RPH, Lee HF, Wu CYH. Trade routes and plague transmission in pre-industrial Europe. Sci Rep. 2017;7:12973. doi: 10.1038/s41598-017-13481-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haensch S, et al. Distinct clones of Yersinia pestis caused the Black Death. PLoS Pathog. 2010;6:e1001134. doi: 10.1371/journal.ppat.1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tavaré S. American Mathematical Society: Lectures on Mathematics in the Life Sciences. Vol 17. American Mathematical Society; Fort Collins, CO: 1986. Some probabilistic and statistical problems in the analysis of DNA sequences; pp. 57–86. [Google Scholar]
- 17.Rasmussen S, et al. Early divergent strains of Yersinia pestis in Eurasia 5,000 years ago. Cell. 2015;163:571–582. doi: 10.1016/j.cell.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harbeck M, et al. Yersinia pestis DNA from skeletal remains from the 6(th) century AD reveals insights into Justinianic plague. PLoS Pathog. 2013;9:e1003349. doi: 10.1371/journal.ppat.1003349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bos KI, et al. Eighteenth century Yersinia pestis genomes reveal the long-term persistence of an historical plague focus. eLife. 2016;5:e12994. doi: 10.7554/eLife.12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kacki S, Rahalison L, Rajerison M, Ferroglio E, Bianucci R. Black Death in the rural cemetery of Saint-Laurent-de-la-Cabrerisse Aude-Languedoc, southern France, 14th century: Immunological evidence. J Archaeol Sci. 2011;38:581–587. [Google Scholar]
- 21.Hirsch A. Handbuch der Historisch-Geographischen Pathologie. Stuttgart Enke; Stuttgart, Germany: 1881. [Google Scholar]
- 22.Walløe L. 1982. Pest og folketall 1350–1750 [Plague and Population: Norway 1350–1750]. Historisk Tidsskrift (Norge) 61:1–45. Norwegian.
- 23.Oeding P. [The Black Death in Norway] Tidsskr Nor Laegeforen. 1990;110:2204–2208. Norwegian. [PubMed] [Google Scholar]
- 24.Brothen JA. Population decline and plague in late medieval Norway. Ann Demogr Hist (Paris) 1996:137–149. doi: 10.3406/adh.1996.1915. [DOI] [PubMed] [Google Scholar]
- 25.Benedictow OJ. The Black Death and Later Plague Epidemics in the Scandinavian Countries: Perspectives and Controversies. Sciendo; Warsaw: 2016. [Google Scholar]
- 26.Cohn SK., Jr . Epidemics: Hate and Compassion from the Plague of Athens to AIDS. Oxford Univ Press; Oxford: 2018. [Google Scholar]
- 27.Bowsky WM. The impact of the Black Death upon Sienese government and society. Speculum. 1964;39:1–34. doi: 10.2307/2850126. [DOI] [PubMed] [Google Scholar]
- 28.Carmichael AG. 2014. Plague Persistence in Western Europe: A Hypothesis. The Medieval Globe: Vol 1, No 1, Article 8.
- 29.Russell P. A Treatise of the Plague: Containing an Historical Journal, and Medical Account, of the Plague, at Aleppo, in the Years 1760, 1761, and 1762. G.G.J. and J. Robinson; London: 1791. [Google Scholar]
- 30.Devaux CA. Small oversights that led to the Great Plague of Marseille (1720-1723): Lessons from the past. Infect Genet Evol. 2013;14:169–185. doi: 10.1016/j.meegid.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 31.Simpson WJ. A Treatise on Plague: Dealing with the Historical, Epidemiological, Clinical, Therapeutic and Preventive Aspects of the Disease. Cambridge Univ Press; Cambridge, UK: 2010. [Google Scholar]
- 32.Wheelis M. Biological warfare at the 1346 siege of Caffa. Emerg Infect Dis. 2002;8:971–975. doi: 10.3201/eid0809.010536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ditrich H. The transmission of the Black Death to western Europe: A critical review of the existing evidence. Mediterr Hist Rev. 2017;32:25–39. [Google Scholar]
- 34.Curtis DR, Roosen J. The sex-selective impact of the Black Death and recurring plagues in the Southern Netherlands, 1349-1450. Am J Phys Anthropol. 2017;164:246–259. doi: 10.1002/ajpa.23266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Donald Ostrowski MTP. Portraits of Old Russia: Imagined Lives of Ordinary People, 1300-1745. Routledge; London: 2011. [Google Scholar]
- 36.Коваль ВЮ. 2013 Торговый инвентарь из раскопок базара середины XIV века в Болгаре. Поволжская Археология. Available at archaeologie.pro/ru/archive/6/97/. Accessed June 11, 2018.
- 37.Yu KV. Trade stock from excavation the market of the middle of the XIV century at Bolgar. Volga River Reg Archaeol. 2013;4:9–33. [Google Scholar]
- 38.Veale EM. 2003 The English Fur Trade in the Later Middle Ages (London Record Society, London), 2nd Ed, Vol 38. Available at https://www.british-history.ac.uk/london-record-soc/vol38. Accessed July 3, 2018.
- 39.Suhm PF, Nyerup R. 2012 Historie AF Danmark: Fra AAR 1340 Til 1375 (Nabu Press, Charleston, SC). Available at https://www.betterworldbooks.com/product/detail/historie-af-danmark-fra-aar-1340-til-1375-volume-13-1274946565. Accessed July 3, 2018.
- 40.Varlik N. Plague and Empire in the Early Modern Mediterranean World: The Ottoman Experience, 1347–1600. Cambridge Univ Press; Cambridge, UK: 2015. [Google Scholar]
- 41.Martin J. The Land of Darkness and the Golden Horde. The fur trade under the Mongols XIII–XIVth centuries. Cah Monde Russe Sov. 1978;19:401–421. [Google Scholar]
- 42.Guellil M, et al. Genomic blueprint of a relapsing fever pathogen in 15th century Scandinavia. Proc Natl Acad Sci USA. 2018;115:10422–10427. doi: 10.1073/pnas.1807266115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feldman M, et al. A high-coverage Yersinia pestis genome from a sixth-century Justinianic plague victim. Mol Biol Evol. 2016;33:2911–2923. doi: 10.1093/molbev/msw170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brotherton P, et al. Genographic Consortium Neolithic mitochondrial haplogroup H genomes and the genetic origins of Europeans. Nat Commun. 2013;4:1764. doi: 10.1038/ncomms2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dabney J, et al. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc Natl Acad Sci USA. 2013;110:15758–15763. doi: 10.1073/pnas.1314445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raoult D, et al. Molecular identification by “suicide PCR” of Yersinia pestis as the agent of medieval Black Death. Proc Natl Acad Sci USA. 2000;97:12800–12803. doi: 10.1073/pnas.220225197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Handt O, Krings M, Ward RH, Pääbo S. The retrieval of ancient human DNA sequences. Am J Hum Genet. 1996;59:368–376. [PMC free article] [PubMed] [Google Scholar]
- 48.Haak W, et al. Ancient DNA from the first European farmers in 7500-year-old Neolithic sites. Science. 2005;310:1016–1018. doi: 10.1126/science.1118725. [DOI] [PubMed] [Google Scholar]
- 49.Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010;2010:db.prot5448. doi: 10.1101/pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 50.Truong DT, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 51.Wood DE, Salzberg SL. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Köster J, Rahmann S. Snakemake–A scalable bioinformatics workflow engine. Bioinformatics. 2012;28:2520–2522. doi: 10.1093/bioinformatics/bts480. [DOI] [PubMed] [Google Scholar]
- 53.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jónsson H, Ginolhac A, Schubert M, Johnson PLF, Orlando L. mapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics. 2013;29:1682–1684. doi: 10.1093/bioinformatics/btt193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, et al. 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Minh BQ, Nguyen MAT, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 2013;30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 61.Adachi J, Hasegawa M. 1996 MOLPHY version 2.3: Programs for molecular phylogenetics based on maximum likelihood. Available at citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.476.8552. Accessed September 5, 2018.
- 62.Ashkenazy H, et al. FastML: A web server for probabilistic reconstruction of ancestral sequences. Nucleic Acids Res. 2012;40:W580–W584. doi: 10.1093/nar/gks498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rambaut A, Lam TT, Max Carvalho L, Pybus OG. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen) Virus Evol. 2016;2:vew007. doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cui Y, Song Y. Genome and evolution of Yersinia pestis. In: Yang R, Anisimov A, editors. Yersinia pestis: Retrospective and Perspective. Springer; Dordrecht, The Netherlands: 2016. pp. 171–192. [Google Scholar]
- 65.Andrades Valtueña A, et al. The stone age plague and its persistence in Eurasia. Curr Biol. 2017;27:3683–3691.e8. doi: 10.1016/j.cub.2017.10.025. [DOI] [PubMed] [Google Scholar]
- 66.Drummond AJ, Bouckaert RR. Bayesian Evolutionary Analysis with BEAST 2. Cambridge Univ Press; Cambridge, UK: 2015. [Google Scholar]
- 67.Rieux A, Khatchikian CE. tipdatingbeast: An R package to assist the implementation of phylogenetic tip-dating tests using beast. Mol Ecol Resour. 2017;17:608–613. doi: 10.1111/1755-0998.12603. [DOI] [PubMed] [Google Scholar]
- 68.Ho SYW, et al. Bayesian estimation of substitution rates from ancient DNA sequences with low information content. Syst Biol. 2011;60:366–375. doi: 10.1093/sysbio/syq099. [DOI] [PubMed] [Google Scholar]
- 69.Eren AM, et al. Anvi’o: An advanced analysis and visualization platform for ‘omics data. PeerJ. 2015;3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing reads produced for this study have been deposited in the European Nucleotide Archive (ENA) under accession number ENA: PRJEB24499.