Significance
Endogenous retrovirus (ERV) sequences make up a large fraction of our genome, yet little is understood about their function and biological relevance. Deep-sequencing data contain valuable information on a genome-wide scale. Yet, due to their highly repetitive nature, analysis of ERVs has been computationally challenging. We describe a bioinformatics tool called ERVmap to analyze transcription of unique sets of human ERVs in a range of cell types in health and disease settings. Our open-source code and accompanied web tool should facilitate researchers in all fields to study the expression patterns of ERVs in sequencing data and should lead to significant advancement in understanding the biological relevance of ERVs in health and disease.
Keywords: endogenous retroviruses, retroelements, RNA sequencing, lupus, cancer
Abstract
Endogenous retroviruses (ERVs) are integrated retroviral elements that make up 8% of the human genome. However, the impact of ERVs on human health and disease is not well understood. While select ERVs have been implicated in diseases, including autoimmune disease and cancer, the lack of tools to analyze genome-wide, locus-specific expression of proviral autonomous ERVs has hampered the progress in the field. Here we describe a method called ERVmap, consisting of an annotated database of 3,220 human proviral ERVs and a pipeline that allows for locus-specific genome-wide identification of proviral ERVs that are transcribed based on RNA-sequencing data, and provide examples of the utility of this tool. Using ERVmap, we revealed cell-type–specific ERV expression patterns in commonly used cell lines as well as in primary cells. We identified 124 unique ERV loci that are significantly elevated in the peripheral blood mononuclear cells of patients with systemic lupus erythematosus that represent an IFN-independent signature. Finally, we identified additional tumor-associated ERVs that correlate with cytolytic activity represented by granzyme and perforin expression in breast cancer tissue samples. The open-source code of ERVmap and the accompanied web tool are made publicly available to quantify proviral ERVs in RNA-sequencing data with ease. Use of ERVmap across a range of diseases and experimental conditions has the potential to uncover novel disease-associated antigens and effectors involved in human health that is currently missed by focusing on protein-coding sequences.
The virome is a collection of viruses that are part of our metagenome (1). Many members of the virome are maintained for the life of the host, and thus can have a significant impact on human health. Herpesviruses, such as herpes simplex virus and cytomegalovirus are well-known examples of the virome that are prevalent in greater than 50% and up to 90% of the human population, respectively. While these viruses can become pathogenic during states of immune suppression (2, 3), they may provide beneficial immunologic stimuli to the host at steady state (4, 5). Endogenous retroviruses (ERVs) are integrated retroviral elements that comprise 8% of the human genome compared with 2% that code for proteins (6). Unlike other virome members that require acquisition, ERVs are present in the genome of all humans and constitute one of the largest and most stable members of the human virome (1). However, the contribution of ERVs to human health is less understood and understudied.
Many ERVs are expressed during embryogenesis and are subsequently epigenetically silenced (7). However, certain ERV sequences are actively transcribed and are elevated in diseases, including various cancers, multiple sclerosis, amyotrophic lateral sclerosis, and HIV-1 infection (8–15). Most ERV sequences have acquired numerous mutations over time and therefore do not have protein-coding potential or the potential to generate infectious viral particles. However, such ERVs can function as genomic regulators of transcription. For example, the MER41B family of ERV sequences contains a STAT1 binding site and regulates expression of IFN-γ–responsive genes, such as AIM2, APOL1, IFI6, and SECTM1 (16). ERV elements can drive transcription of genes, generate chimeric transcripts with protein-coding genes in cancer, serve as splice donors or acceptors for neighboring genes, and be targets of recombination and increase genomic diversity (17, 18). ERVs that are elevated in breast cancer tissues correlate with the expression of granzyme and perforin levels, implying a possible role of ERVs in immune surveillance of tumors (19). A small number of ERVs have protein-coding potential. An ERV envelope protein, Syncytin-1, plays a critical role in placental development (20, 21), but when expressed in the wrong context, it can be inflammatory in astrocytes and microglial cells (22, 23). ERV-K envelope protein stimulates the adaptive immune response in breast cancer and during HIV-1 infection (24, 25), and it is also reportedly involved in the activation of the ERK pathway and causes neurotoxicity (26, 27). A recent study identified an ERV protein HEMO, which is secreted in the blood of pregnant women that is also expressed in stem cells and in tumor cells (28). These studies are beginning to shed light on the importance of ERVs in biology. However, many of these studies are confined to a few ERV loci out of the thousands of copies of ERVs present in the genome. Thus, a greater accuracy with which to study global ERV transcription is necessary to reveal the full extent to which ERVs contribute to human health.
Most of the ERV sequences in our genome are non-autonomous long terminal repeat (LTR) elements that are either solo-LTRs or LTRs flanking a small segment of internal ERV sequences and are short in length. These sequences arose through recombination of LTRs and deletion of protein-coding sequences through the course of evolution. Autonomous LTRs are composed of LTRs that flank potential protein-coding sequences, are much longer in length, and are near full-length proviral sequences. Non-autonomous LTR elements are likely to serve as genomic regulators and affect transcription or splicing of nearby genes in cis. Autonomous LTR elements could also serve as genomic regulators, but these elements are more likely to also encode functional proteins, disease-associated antigens, or functional RNAs that regulate gene expression in trans, and are important to study in the context of human health.
There are many challenges in quantifying ERV expression on a genome-wide scale using sequencing data due to their repetitive nature (29). Default pipelines used for gene-expression analysis discard most reads derived from repetitive elements because they do not map to a unique locus. Studies that have quantified ERV expression using RNA sequencing data have all used different methods for analysis, many without disclosing the computational scripts, making it difficult for the field to standardize the methodology (19, 30, 31). It is also challenging to determine the specific chromosomal location of expressed ERVs, due in part to mapping methods and in part to incomplete annotation of ERVs in the human genome. This information is critical for downstream mechanistic studies. ERV annotations in databases, such as Repbase, Repeatmasker, DFAM, HERVgdB4, HERVd, and PostParser HERV Browser, contain a mixture of autonomous LTR elements and noncontiguous non-autonomous LTR elements, which for studies of ERVs as genomic regulators may be adequate, but not sufficient for identification of ERVs that potentially code for disease-associated antigens involved in disease (32–37). These databases have not yet incorporated autonomous LTR elements that are near full-length proviral ERVs that have been specifically identified in disease contexts using conventional amplification methods and in silico analysis. Therefore, we sought to create an open-source pipeline for quantification of locus-specific ERV expression that combines a stringent filtering criteria for RNA sequencing reads that map to ERV loci, and a reference ERV annotation database that focuses on a large number of previously unannotated autonomous ERVs that closely resemble a full-length proviral sequence.
Here, we describe a pipeline called ERVmap to analyze the expression of human ERVs in RNA sequencing data based on a newly annotated database of 3,220 autonomous ERVs that mirror a full-length provirus. To facilitate usage by researchers, our code is available on GitHub and also via a web-based tool on https://www.ervmap.com. To illustrate the utility of ERVmap, we identified ERVs that are expressed in various cell types, in systemic lupus erythematosus (SLE) disease, and in breast cancer tissues. ERVmap can be used with any RNA sequencing data to potentially reveal novel ERV antigens and proteins involved in diseases, such as cancer, neurodegenerative diseases, infectious diseases, and autoimmunity.
Results
ERVmap: A Bioinformatic Tool to Map RNA Sequencing Reads to Human Proviral ERVs.
To obtain a complete high-resolution genome-wide human ERV compendium, or ERVome, we compiled a curated list of 3,220 ERV proviral loci (Dataset S1). These ERVs were either transcribed in various disease contexts or identified as ERVs based on sequence analysis in silico (14, 38–46). We included ERV loci with unique chromosomal locations that had been described as autonomous/proviral ERVs and did not intentionally exclude any loci. For loci that overlapped between studies, we selected the one with longer sequence coverage. Unlike the RepeatMasker annotation in which the ERVs are mainly noncontiguous non-autonomous LTR elements with an average length of 368 bp, the ERVmap database contains ERVs that are mostly autonomous LTR elements with an average length of 7.5 kb (37). RepeatMasker annotation has 885 loci above 5 kb, whereas ERVmap has 2,722 loci above 5 kb. The average length of the rest of the ERVs below 5 kb for ERVmap is 3.6 kb, whereas for RepeatMasker it is 360 bp. ERVmap captures all known proviral ERV sequences to date and is ideal for analysis of specific autonomous ERV genomic loci throughout the host genome.
Using this database, we aligned processed RNA sequencing reads to the human genome (hg38) using Burrows-Wheeler Aligner (BWA), used scripts specifically designed for ERVmap to filter the mapped reads according to our stringent criteria, quantified filtered reads that mapped to the ERV coordinates from our database, and normalized the counts to size factors obtained through standard cellular gene-expression analysis (SI Appendix, Fig. S1). The pipeline yields normalized values based on filtered read counts mapped per locus. To obtain high-fidelity mapping of sequence reads to these repetitive ERV loci, we employed a very stringent filtering criteria to the mapped reads, such that each mapped read: (i) could only have one best match, (ii) the second best match must have at least one more mismatch, and (iii) excluded if it has more than three mismatches (14). This criterion is for 150-bp pair-end reads and is proportionally adjusted according to read length of the sequencing data. In this algorithm, which we call ERVmap, we intentionally excluded reads that mapped to conserved regions in the proviral sequence and reads that mapped to polymorphic loci, both of which would fall under the third criteria of having more than three mismatches per sequenced read to favor locus specificity over overall abundance. Our code is available through GitHub (https://mtokuyama.github.io/ERVmap/) (SI Appendix, Fig. S2). Finally, we developed a web-based tool that is available for users to obtain the human ERVome in any RNA sequencing data by simply uploading raw RNA sequencing files (www.ervmap.com).
ERV Expression Patterns in Human Cell Lines.
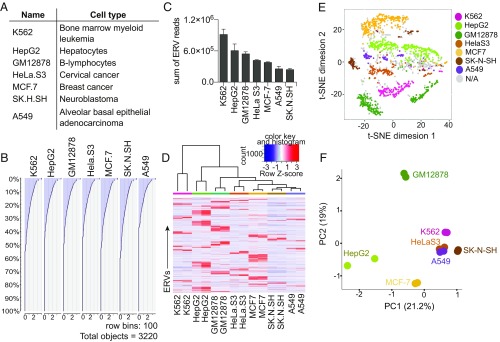
We obtained RNA sequencing data from ENCODE for several common cell lines (Fig. 1A) to analyze the ERVome in these cells. We selected cell lines that have accompanying ChIP-sequencing data. In all of the analyzed cells, we observed ∼40% of ERVs at detectable levels (Fig. 1B). K562 cells expressed the highest level of ERVs, not because they expressed more ERV loci but because the expressed ERVs are transcribed at higher levels (Fig. 1 B and C). In contrast, A549 cells expressed the lowest level of ERVs, roughly one-third of the amount expressed by K562 cells. Comparison of the ERVs between cell lines revealed clusters of ERVs that are uniquely expressed in each cell line (Fig. 1D). These ERVs clustered distinctly using a t-distributed stochastic neighbor-embedding (t-SNE) algorithm, implying that unique sets of ERVs are expressed in each cell line (Fig. 1E). Additionally, ERV expression alone was sufficient to segregate cell types based on principle component analysis (PCA), suggesting that ERV expression is unique enough to allow discrimination between cell types (Fig. 1F). We confirmed expression of a set of ERVs using qRT-PCR (SI Appendix, Fig. S3). Finally, we analyzed ChIP-sequencing data available for these cell lines and observed H3K4me3 and H3K27Ac histone marks at actively transcribed ERV loci, and also observed a positive correlation between active histone marks and higher ERV expression (SI Appendix, Fig. S4 A and B). In contrast, very few repressive histone marks H3K9me3 and H3K27me3 were present at the transcribed ERV loci, suggesting that the lack of silencing histone modifications and the presence of active histone modifications accompany ERV expression in these cells (SI Appendix, Fig. S4 C and D).
Fig. 1.
Cell-type–specific ERV expression in cell lines. (A) Description of cell lines used in the ERV analysis. (B) Histogram of the amount of reads attributed to each of the 3,220 ERV loci sorted in order of highest to lowest expressed ERVs for each cell line. (C) Sum of all ERV reads per cell line compared across cell types. (D) Heatmap of ERVs that are expressed across indicated cell types. ERVs with zero reads across all cell lines were excluded. A total of 1,704 ERVs are displayed. Two-dimensional t-SNE analysis (E) and PCA (F) of ERVs expressed by indicated cell types using the same set of 1,704 ERVs as in D. t-SNE analysis was performed using a perplexity of 30 and maximum iteration of 1,000. N/A, cell assignment not possible due to multiple cell lines expressing the same exact amount of the particular ERV.
In contrast to ERVmap, less resolution was observed when RepeatMasker annotation was used to analyze ERV expression using a published method called RepEnrich (47). RepEnrich quantifies LTR elements at the level of subfamilies, each of which contains hundreds of copies in the genome (SI Appendix, Table S1) and does not yield quantification of reads at specific ERV loci. RepEnrich analysis did not reveal clusters of cell-type–specific ERV elements (SI Appendix, Fig. S5A), but there were enough differences in the expression of ERV families between cell types to segregate cells based on hierarchical clustering and PCA analysis (SI Appendix, Fig. S5B). Thus, ERVmap provides locus-specific profiling of the ERVome using RNA-sequencing (RNA-seq) datasets that should facilitate downstream mechanistic studies.
Differential ERV Expression in Primary Cell Types.
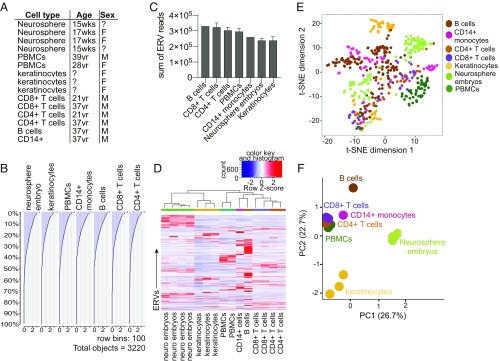
We next used RNA-seq data from primary cells in ENCODE to analyze the ERVome in seven different cell types, both immune and nonimmune cells, to obtain ERV expression in a range of cell types (Fig. 2A). Similar to cell lines, roughly 50% of the ERV loci were expressed by any given cell type (Fig. 2B). All cells expressed similar total levels of ERV transcripts, but distinct sets of ERVs were transcribed in a given cell type (Fig. 2 C and D). Neurosphere embryos and B cells in particular expressed clusters of highly cell-type–specific ERVs. Using the t-SNE algorithm, we observed unique clusters of ERVs expressed in each cell type; however, we observed similar ERV clusters between CD4+ and CD8+ T cells, suggesting that ERVs expressed by these cell types are similar relative to other cell types (Fig. 2E). This likely reflects the biological similarity within the two T cell populations. Cell types segregated based solely on ERV expression profiles and revealed that the ERVome is largely distinct between lymphocytes, keratinocytes, and neurosphere embryos (Fig. 2F). Finally, in comparison with cell lines, primary cells expressed lower levels of ERVs overall, suggesting that the process of transformation or cell culture might lead to elevated ERV expression (SI Appendix, Fig. S6).
Fig. 2.
Cell-type–specific ERV expression in primary cells. (A) Cell types and associated information for each sample used in the ERV analysis. (B) Histogram of the amount of reads attributed to each of the 3,220 ERV loci sorted in order of highest to lowest expressed ERVs for each cell type. For cell types with multiple samples, the average number of reads per locus was plotted. (C) Sum of all ERV reads per sample compared across cell types. For cell types with multiple datasets, the average and SEM are graphed. (D) Heatmap of ERVs that are expressed across indicated cell types. The 500 most varying ERVs were used for the analysis to reduce noise. Two-dimensional t-SNE analysis (E) and PCA (F) of ERVs expressed by indicated cell types using the same set of 500 ERVs as in D. t-SNE analysis was performed using a perplexity of 30 and maximum iteration of 1,000.
ERVome Is Elevated in SLE Patients.
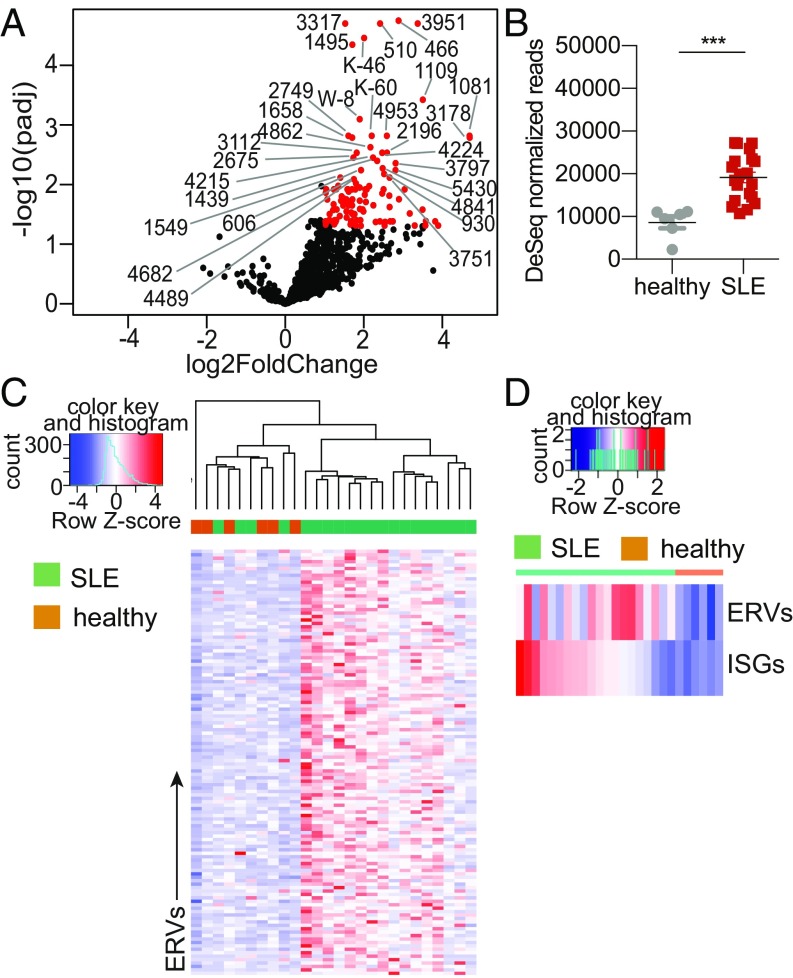
ERVs have been implicated in various diseases, including cancer and autoimmunity. SLE is a multigenic autoimmune disease with diverse clinical manifestations and still lacks a cure. Many drugs that target various immune effectors have been tested, but they have had varying levels of success (48). One of the biggest hurdles in designing effective drugs is the poor understanding of the underlying cause for the diverse array of symptoms associated with the disease. While studies have observed elevated expression of ERV sequences in SLE patients (49–53), these studies have focused on one or two ERVs and the field could benefit from a genome-wide analysis of the ERVome in SLE patients to reveal relevance of ERVs in disease. Thus, we obtained peripheral blood mononuclear cells (PBMCs) from female SLE patients and healthy females (SI Appendix, Table S2), because SLE is a female-dominant disease, and performed RNA sequencing followed by ERVmap analysis. In this cohort, we identified 124 ERVs that were significantly elevated in SLE patients’ PBMCs compared with healthy controls, but none that were repressed (Fig. 3A). SLE patients expressed significantly higher levels of ERV transcripts as a whole as well as at the individual locus, and ERV expression largely segregated SLE patients from healthy controls (Fig. 3 B and C). Finally, we observed that ERV expression is not a direct correlate of the interferon (IFN) signature for many patients, as illustrated by comparison between total ERV expression and the total IFN-stimulated gene (ISG) expression per patient (Fig. 3D). The ISG expression was calculated using a previously published list of ISG signature observed in SLE patients (54). Together, ERVmap revealed a global elevation of the ERVome and identified specific ERV loci that are elevated in SLE patients that together may reflect an ISG-independent signature of SLE.
Fig. 3.
Patients with SLE have elevated ERV expression. (A) A volcano plot depicting differential expression of all 3,220 ERVs. Red ERVs are significantly elevated in SLE patients compared with healthy controls (padj < 0.05, log2 fold-change > 1.0). The top 30 significantly elevated ERVs are indicated by their names. (B) Comparison of the sum of all significantly different ERV reads between healthy and SLE donors (SLE, n = 20; healthy, n = 6). Error bars represent SEM and nonparametric Mann–Whitney U test was performed to calculate significance. ***P < 0.001. (C) Heatmap of the 124 significantly elevated ERVs in SLE patients compared with healthy controls as determined by using a cut-off of padj < 0.05. (D) Heatmap of the sum of reads for significantly elevated ERVs and the sum of reads for ISGs per patient sample.
Identification of Additional ERVs That Are Elevated and Correlate with Cytolytic Activity in Breast Cancer Tissues.
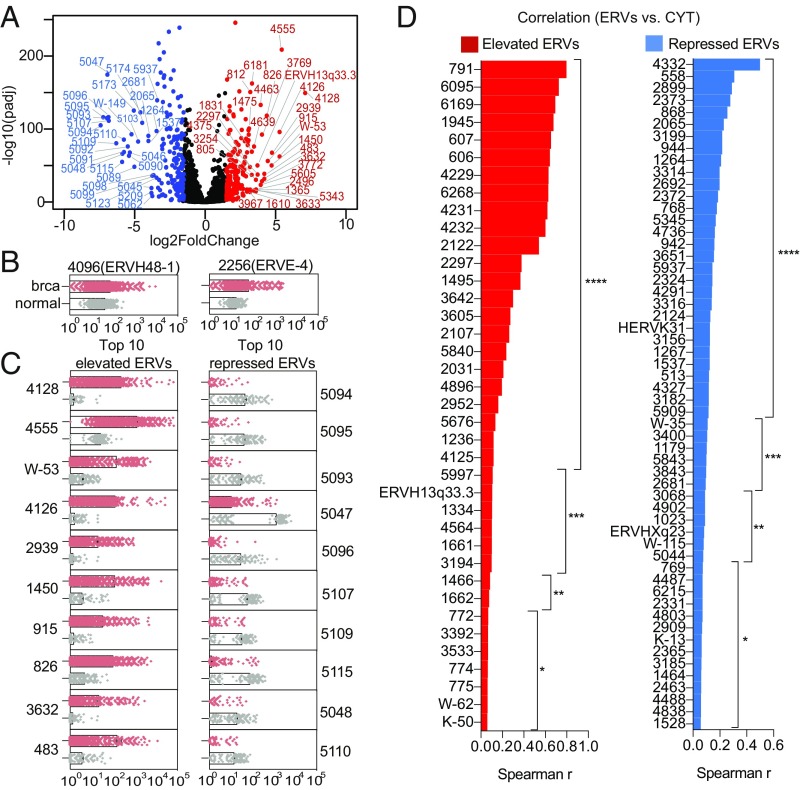
Cytotoxic T cells and natural killer cells are important effectors of tumor surveillance. They are armed with granzyme and perforin to directly kill tumor cells. A recent study using a set of 66 ERVs as a reference showed that 8 of the 66 ERVs positively correlated with the expression of granzyme and perforin in breast cancer tissues, implying a potential role of ERVs in immune surveillance (19). We applied ERVmap to the same breast cancer tissue dataset generated from The Cancer Genome Atlas (TCGA) Research Network to determine whether we are able to identify additional ERVs that are elevated in breast cancer tissues and associate with the granzyme and perforin cytolytic activity measure (CYT), as reported previously. We observed a large number of significantly elevated, as well as repressed, ERVs in breast cancer tissues compared with normal breast tissues (Fig. 4A). We confirmed elevated expression of two of the three tumor-specific ERVs (TSERVs) identified by the Hacohen group (19), ERVH48-1 and ERVE-4 (Fig. 4B). We also identified an additional 203 ERVs that were significantly elevated in breast cancer tissues, as well as 195 repressed ERVs (Fig. 4 A and C). Five of the eight ERVs that positively correlated with CYT in the Hacohen and colleagues (19) paper also showed a positive correlation using ERVmap, but none of these ERVs were significantly elevated in breast cancer tissues. Instead, we identified 38 ERVs that were both significantly elevated and showed positive correlation with CYT (Fig. 4D). We also identified 56 ERVs that were significantly repressed but showed a positive correlation with CYT (Fig. 4D). Together the data illustrated that ERVmap can reveal tumor-associated ERVs, which may play a role in tumor surveillance.
Fig. 4.
ERVmap reveals additional breast cancer-associated ERVs that correlate with cytolytic activity. (A) A volcano plot depicting all 3,220 ERVs. Blue ERVs are significantly repressed and red ERVs are significantly elevated in breast cancer tissues compared with healthy controls (padj < 0.05, log2 fold-change < −1.5 or > 1.5). The top 30 significantly elevated and repressed ERVs are indicated by their names. (B) Normalized DESeq ERV read counts for previously reported TSERVs or the top 10 significantly elevated or significantly repressed ERVs identified in this report (C) are plotted as dot plots for the indicated ERVs for each sample (brca, breast cancer n = 1,246, red; normal n = 221, gray). (D) Spearman’s r correlation was calculated between the significantly elevated or repressed ERVs (padj < 0.05, log2 fold-change > 1.5 or < −1.5) and the average expression level of granzyme and perforin (CYT) for all breast cancer tissue samples. (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
Discussion
ERVs make up a large fraction of our genome, far greater than protein-coding sequences, yet the relevance of ERVs in biology is only beginning to be uncovered. In mice, ERVs retain the capacity to generate infectious virions, whereas in humans, ERV sequences have acquired numerous mutations and large deletions over the course of evolution and lack the ability to produce infectious viral particles. However, ERV sequences are actively transcribed and can function as genomic regulators and functional or inflammatory proteins. Understanding the relevance of each ERV in various contexts—whether as functional proteins, as disease-associated antigens, or as genomic regulators—is necessary to reveal the full impact of ERVs in human health and disease. Here we describe a tool to analyze locus-specific expression of human ERVs in deep-sequencing data. We illustrate the utility of this tool by revealing cell-type–specific expression of distinct ERV networks in both cell lines and primary cells. We also observed elevated expression of ERVs in SLE patients, which was largely independent of the IFN signature. Finally, we observed a large number of differentially expressed ERVs in breast cancer tissues and identified a number of elevated ERVs that significantly correlate with cytolytic activity. These data begin to shed light on the biological relevance of ERVs and illustrate the need to further investigate various conditions in which ERVs are expressed and the subsequent function of ERV expression.
There are several databases and tools to analyze ERVs. Repbase database has the largest collection of consensus repetitive sequences and is most commonly used with the RepeatMasker program to annotate repetitive elements in the genome, including ERV sequences (32, 37). DFAM and HERVd are additional repetitive element and ERV databases that use Repbase and RepeatMasker annotations to compile ERV sequences (33, 35). All of these databases are largely composed of non-autonomous LTR elements (SI Appendix, Table S3), reflecting the distribution of LTR elements in nature, but do not include many of the autonomous LTR elements that have been published in various disease contexts or identified through in silico analysis. The Reference Viral Database (RVDB) contains a significant number of LTR elements but the proportion of human LTR elements is small (55). HERVgDB4 also contains a significant number of LTR elements, but these sequences are used for detection of ERV transcripts in a hybridization assay, which is less sensitive and does not give as high of a resolution as deep sequencing (34). Beyond efforts to create large ERV databases, using the annotation to analyze locus-specific ERV expression in deep-sequencing data requires algorithms to specifically handle the challenges of aligning relatively short sequencing reads to repetitive sequences in the genome. All of these considerations were taken to develop ERVmap, which focuses on autonomous LTR elements that mirror full-length proviral sequences with the potential to code for proteins or antigens and uses an algorithm with stringent filtering criteria to increase confidence in assigning reads to specific ERV loci.
The expression of ERVs is highly dynamic in pluripotent embryonic stem cells. There is evidence that ERVs are important for embryonic stem cells to maintain a pluripotent state, suggesting a role for ERVs in regulating cell differentiation (56, 57). Using ERVmap, we revealed cell-type–specific expression of ERVs in both commonly used cell lines and in primary cells. The data showed that somatic cells at steady state are also capable of expressing ERVs, and transformed cells express even higher levels of ERVs. There is evidence that the envelope protein of ERV-K drives the process of transformation (26, 58), suggesting possible roles of the highly expressed ERVs in transformation of various cell types. For both cell lines and primary cells, the pattern of ERV expression alone was sufficient to discriminate between cell types, and different cell types expressed distinct clusters of ERVs. These data together might suggest a larger role for ERVs in determining cell fate and differentiation. Furthermore, perhaps, clusters of ERVs are more or less fixed as ERV networks and are coordinately regulated through signaling pathways or transcriptional regulatory mechanisms that are cell type-dependent. With ERVmap, future studies will be possible to determine the contributions of specific ERV loci in regulating the process of transformation and cell differentiation.
Epigenetic silencing of many ERVs occurs through the recruitment of TRIM28 to ERV loci via Kruppel-associated box zinc-finger proteins (KRAB-ZNFs) that bind to ERVs in a sequence-specific manner. Subsequently, recruitment of the NURD/HDAC complex along with histone methyltransferase catalyzes the addition of H3K9me3 silencing marks, and DNA methyltransferases methylates the DNA (56, 59). Our data showing that highly transcribed ERV loci are marked with active histone modifications and lack silencing histone modifications suggest that ERVs that were likely silenced postembryogenesis have been epigenetically reversed to allow for expression in somatic cells. It will be important to determine the exact mechanism that allows for ERV transcription in these settings. Future studies may also provide insights into the molecular mechanisms that discriminate between ERVs that are highly expressed versus those are that repressed in each cell type.
Previous studies have shown elevated expression of two distinct ERVs in SLE patients: HERV-K clone 4-1 in PBMCs and HRES1/p28 in B cells (49, 52). Using ERVmap, we identified an additional 124 unique ERV loci that are significantly elevated in PBMCs from SLE patients compared with healthy controls. The gag protein derived from HERV-K clone 4-1 is antigenic in SLE patients and expression of HERV-K clone 4-1 positively correlates with plasma levels of anti-U1RNP, anti–Scl-70, and anti-Sm antibodies (53, 60), implicating a role for ERVs in this disease. Based on this, the SLE-associated ERVs we identified in this study may potentially reflect a large pool of previously unknown autoantigens that contribute to disease. A recent study demonstrated that a number of nonproviral ERV elements are elevated in SLE blood (30). Due to the non-autonomous nature of these LTR sequences, these ERVs likely do not contribute to the antigenic pool, but rather function as genomic regulators. Our set of ERVs potentially functions both as genomic regulators and autoantigens and complements this former study in furthering our understanding of ERVs in SLE. Based on findings that DNA hypomethylation is widely observed in a range of cell types in SLE and that hypomethylation of HRES1/p28 and a few ERV LTR elements positively correlates with disease, SLE-associated ERVs identified in this study may potentially be transcribed due to hypomethylation at these loci (52, 61). Future studies characterizing the function of these ERVs and its involvement in various aspects of the disease should shed light on their exact roles in disease.
A number of studies have observed elevated expression of ERV transcripts in cancer tissues, ranging from breast cancer, ovarian cancer, osteosarcoma, and glioblastoma to melanoma (8–11, 13). In some cases, antibody or cellular reactivity are observed against ERV proteins, but in most cases, these studies have only shown correlation between ERV expression and disease states, and the exact role of ERVs in disease pathogenesis have yet to be revealed. A recent analysis (19) of breast cancer tissues showed a positive correlation between elevated ERV expression and expression of granzyme and perforin, two effector molecules for cytotoxic T cell and NK cells, and was the first to illustrate a potential role of ERVs in tumor immune surveillance. This study (19) used 66 ERVs as a reference. Using ERVmap that is based on 3,220 ERVs, we were able expand the number of ERVs that positively correlate with the expression of granzyme and perforin from 8 to 38 ERVs, highlighting the power of ERVmap. There were some differences between the studies. The potential differences may result from the dataset; since 2015, the number of breast cancer tissue samples in the TCGA dataset has increased from 920 to 1,246 and normal breast samples in GTEX from 133 to 221. Second, the previous study mapped the reads solely to ERVs (19), whereas ERVmap maps reads to the whole genome. Both of these factors may account for the differences and highlights the need for the field to standardize ERV analysis. Importantly, the use of our pipeline and expanded ERV database to analyze ERVs in various cancers may offer promising insights into the biology of ERVs and tumor immune surveillance.
The nomenclature for ERVs is an evolving field of its own (38, 62). Despite attempts to standardize the nomenclature, investigators have employed various means to name newly identified ERVs. There is a useful proposal to unify and standardize ERV names; however, this is only at the proposal stage and is too early to implement (62). Therefore, our database has maintained the published names whenever possible, and for ERVs that only had chromosomal locations in the original studies, we assigned a numerical ID following the family name of ERVs (e.g., K-58). We have also indicated alternative names given to the specific ERV loci by other studies (Dataset S1). These names can be updated in the future when the new ERV nomenclature system is established. The most critical identifier is the chromosomal location of each locus, and we have maintained this information from published studies and unified all of the loci based on the GRCh38 genome assembly.
In conclusion, ERVmap is a powerful tool that can be used to identify specific ERV expression patterns in RNA sequencing data and is highly versatile for use in a wide variety of studies. As deep-sequencing and bioinformatic capabilities continue to improve, we plan to update our database to reflect the most up to date annotations of autonomous LTRs. The use of ERVmap to analyze a range of datasets should make significant strides toward uncovering the biological relevance of each ERV locus in a range of biological processes that are important for human health.
Materials and Methods
ERVmap Database.
The exact locus information for each ERV was extracted from previously published studies (14, 38–46). These were all lifted over to GRCh38/hg38 genome assembly using the LiftOver tool (University of California, Santa Cruz Genome Browser). The ERV loci were then checked for overlapping intervals using the Intersect function on Galaxy (https://usegalaxy.org/). Overlapping ERVs were filtered based on length, and ERVs that were longer in sequence length were kept in the database. Shorter ERVs that overlap with ERVs in the database but were not used in the algorithm are provided as alternative names in Dataset S1. Finally, all of the published nomenclature for ERVs were kept, except in cases where there was no name associated with the locus. For these ERVs, we assigned a numerical value in ascending order. Edit distance (Levenshtein distance) was calculated for all pairs of 3,220 ERVs in the ERVmap database in both directions and the minimum distance of the strands were normalized to the length of each locus and reported as a heatmap (SI Appendix, Fig. S7).
RNA Sequencing Datasets.
We obtained the following RNA sequencing datasets through ENCODE: A549 (SRR4235534 and SRR4235535), K562 (SRR4235541 and SRR4235542), GM12878 (SRR4235527 and SRR4235528), HepG2 (SRR5048081 and SRR50480820), HeLa-S3 (SRR4235529 and SRR4235530), MCF-7 (SRR5048099 and SRR5048100), SK-N-SH (SRR5048153 and SRR5048154), and keratinocytes (SRR3192487, SRR3192488, and SRR3192489). We obtained the following RNA sequencing datasets through Roadmap: neurosphere embryo (SRR2173245, SRR2173235, SRR2173237, and SRR2173254), PBMCs (SRR2173284 and SRR2173278), CD8+ T cells (SRR644512 and SRR644514), CD4+ T cells (SRR643766 and SRR644513), B cells (SRR980471), and CD14+ monocytes (SRR980470). Raw RNA-Seq fastq and BAM files for breast cancer tissue analysis were accessed from the TCGA (phs000178.v9.p8) GDC Data Portal and from the GTEx web portal (phs000424.v7.p2, GTEx Consortium, 2013) using gdc-client_v1.3.0 and sratoolkit.2.8.2–1. All data were downloaded into secure, password-protected directories of the Yale High Performance Computing Cluster.
RNA-seq Analysis.
ERVmap: The Illumina reads were first trimmed by Btrim (63) to remove sequencing adaptors and low-quality regions. The trimmed reads were mapped to the human genome (GRCh38) using BWA mem with default parameters (64). The unmapped reads were filtered out using SAMtools and the mapped reads in SAM format were further processed as the following (65). The CIGAR field in the SAM file is used to check the number of hard or soft clipping. If the ratio of sum of hard and soft clipping to the length of the read (base pair) is greater than or equal to 0.02 (equivalent to three mismatches per 150-bp read length), then the read will be discarded. The remaining reads are checked for the field of edit distance compared with the locus reference (NM field). If the ratio of the edit distance to the sequence read length (base pair) is greater or equal to 0.02, the read is discarded (equivalent to three mismatches per 150-bp read length). Finally, the difference between the alignment score from BWA (field AS) and the suboptimal alignment score from BWA (field XS) is compared. If the difference is less than 5, the read is discarded (equivalent to second best match has one or more mismatches than the best match). The SAM file containing the mapped reads that pass the filtering steps described above is converted to a BAM file using SAMtools. This BAM file, together with a BED file containing ERV coordinates in the human genome (GRCh38) in bed format, is used as input for bedtools to count the read mapping at each ERV locus (66). The read counts are normalized by the size factors obtained from the cellular genes of the same sample, calculated using the DESeq2 normalization method (67). Briefly, the standard cellular gene-expression analysis was carried out by trimming off sequencing adaptors and low-quality regions by Btrim. The trimmed reads were mapped to human genome (GRCh38) by TopHat2 (68). The counts of reads for each gene were based on Ensembl annotation. After the counts are collected, DEseq2 was used to calculate size factor for each sample. The core scripts are provided in SI Appendix, Fig. S2.
For the TCGA samples, the BAM files were converted to fastq files using SAMtools1.5 and BEDTools2.27.1, and fastq files were aligned using STAR aligner 2.5.3a and subsequent counts were made using the coveragebed function in BEDtools.
The accompanied ERVmap web tool allows users to upload FASTQ files and obtain results of ERVmap analysis as an excel spreadsheet. The web tool can be accessed through https://www.ervmap.com.
The repetitive elements analysis was done using RepEnrich (47). The bed file of ERVs was obtained from the Repeatmasker track of University of California, Santa Cruz genome table browser for hg38.
SLE Patient Samples.
Patients with SLE were recruited from the rheumatology clinic of the Yale School of Medicine and Yale New Haven hospital in accordance with a protocol approved by the Institutional Review Committee of Yale University (#0303025105). The diagnosis of SLE was established according to the 1997 update of the 1982 revised American College of Rheumatology criteria (69). After obtaining informed consent, peripheral blood was collected in heparin tubes from human subjects. Demographic and clinical characteristics of patients with SLE were collected from patients and by reviewing medical records.
RNA Sequencing.
Whole blood collected in heparin tubes were centrifuged to obtained plasma, and the rest of the blood was used to isolate PBMCs using Ficoll-Paque density centrifugation separation. PBMCs were stored in Buffer RLT (Qiagen), and RNA was isolated according to manufacturer’s protocol (RNeasy kit; Qiagen). High-throughput sequencing was performed on the RNA samples using HiSeq and NextSeq Illumina sequencing machines. TruSeq DNA LT Sample Prep Kits were used for library preparation according to manufacturer’s instructions (Illumina). Roughly 100 million reads were obtained for each sample using 150-bp pair-end reads.
Supplementary Material
Acknowledgments
We thank Laurie Kramer (Yale University) and Barbara Siconolfi (Yale University) for technical support; and Albert Shaw (Yale University) and Ruth Montgomery (Yale University) for help with sample acquisition. This work was supported in part by grants provided by AbbVie (to A.I.); a Translation Accelerator grant from the Brigham and Women’s Hospital (to A.I.); Roslyn and Jerome Meyer Pilot Awards for Research in Melanoma and Immuno-Oncology (to A.I.); and NIH Awards T32 AI89704-3 (to M.T.) and 2R56AG028069-06A1 (to I.K.). A.I. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/ (accession no. GSE122459).
See Profile on page 12544.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814589115/-/DCSupplemental.
References
- 1.Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009;138:30–50. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 2.Xu F, et al. Trends in herpes simplex virus type 1 and type 2 seroprevalence in the United States. JAMA. 2006;296:964–973. doi: 10.1001/jama.296.8.964. [DOI] [PubMed] [Google Scholar]
- 3.Staras SAS, et al. Seroprevalence of cytomegalovirus infection in the United States, 1988-1994. Clin Infect Dis. 2006;43:1143–1151. doi: 10.1086/508173. [DOI] [PubMed] [Google Scholar]
- 4.Barton ES, et al. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447:326–329. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- 5.Furman D, et al. Cytomegalovirus infection enhances the immune response to influenza. Sci Transl Med. 2015;7:281ra43. doi: 10.1126/scitranslmed.aaa2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lander ES, et al. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921, and correction (2001) 412:565. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 7.Grow EJ, et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522:221–225. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang-Johanning F, et al. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int J Cancer. 2007;120:81–90. doi: 10.1002/ijc.22256. [DOI] [PubMed] [Google Scholar]
- 9.Ho XD, et al. Analysis of the expression of repetitive DNA elements in osteosarcoma. Front Genet. 2017;8:193. doi: 10.3389/fgene.2017.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Díaz-Carballo D, et al. Cytotoxic stress induces transfer of mitochondria-associated human endogenous retroviral RNA and proteins between cancer cells. Oncotarget. 2017;8:95945–95964. doi: 10.18632/oncotarget.21606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Büscher K, et al. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res. 2005;65:4172–4180. doi: 10.1158/0008-5472.CAN-04-2983. [DOI] [PubMed] [Google Scholar]
- 12.Douville R, Liu J, Rothstein J, Nath A. Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann Neurol. 2011;69:141–151. doi: 10.1002/ana.22149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang-Johanning F, et al. Expression of human endogenous retrovirus K envelope transcripts in human breast cancer. Clin Cancer Res. 2001;7:1553–1560. [PubMed] [Google Scholar]
- 14.Schmitt K, et al. Comprehensive analysis of human endogenous retrovirus group HERV-W locus transcription in multiple sclerosis brain lesions by high-throughput amplicon sequencing. J Virol. 2013;87:13837–13852. doi: 10.1128/JVI.02388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Contreras-Galindo R, et al. Characterization of human endogenous retroviral elements in the blood of HIV-1-infected individuals. J Virol. 2012;86:262–276. doi: 10.1128/JVI.00602-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chuong EB, Elde NC, Feschotte C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science. 2016;351:1083–1087. doi: 10.1126/science.aad5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Babaian A, Mager DL. Endogenous retroviral promoter exaptation in human cancer. Mob DNA. 2016;7:24. doi: 10.1186/s13100-016-0080-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jern P, Coffin JM. Effects of retroviruses on host genome function. Annu Rev Genet. 2008;42:709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- 19.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mi S, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 21.Frendo JL, et al. Direct involvement of HERV-W Env glycoprotein in human trophoblast cell fusion and differentiation. Mol Cell Biol. 2003;23:3566–3574. doi: 10.1128/MCB.23.10.3566-3574.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antony JM, et al. Human endogenous retrovirus glycoprotein-mediated induction of redox reactants causes oligodendrocyte death and demyelination. Nat Neurosci. 2004;7:1088–1095. doi: 10.1038/nn1319. [DOI] [PubMed] [Google Scholar]
- 23.Xiao R, et al. Human endogenous retrovirus W env increases nitric oxide production and enhances the migration ability of microglia by regulating the expression of inducible nitric oxide synthase. Virol Sin. 2017;32:216–225. doi: 10.1007/s12250-017-3997-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang-Johanning F, et al. Human endogenous retrovirus K triggers an antigen-specific immune response in breast cancer patients. Cancer Res. 2008;68:5869–5877. doi: 10.1158/0008-5472.CAN-07-6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michaud H-A, et al. Trans-activation, post-transcriptional maturation, and induction of antibodies to HERV-K (HML-2) envelope transmembrane protein in HIV-1 infection. Retrovirology. 2014;11:10. doi: 10.1186/1742-4690-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemaître C, Tsang J, Bireau C, Heidmann T, Dewannieux M. A human endogenous retrovirus-derived gene that can contribute to oncogenesis by activating the ERK pathway and inducing migration and invasion. PLoS Pathog. 2017;13:e1006451. doi: 10.1371/journal.ppat.1006451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W, et al. Human endogenous retrovirus-K contributes to motor neuron disease. Sci Transl Med. 2015;7:307ra153. doi: 10.1126/scitranslmed.aac8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heidmann O, et al. HEMO, an ancestral endogenous retroviral envelope protein shed in the blood of pregnant women and expressed in pluripotent stem cells and tumors. Proc Natl Acad Sci USA. 2017;114:E6642–E6651. doi: 10.1073/pnas.1702204114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: Computational challenges and solutions. Nat Rev Genet. 2011;13:36–46, and correction (2012) 13:146. doi: 10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Attig J, Young GR, Stoye JP, Kassiotis G. Physiological and pathological transcriptional activation of endogenous retroelements assessed by RNA-sequencing of B lymphocytes. Front Microbiol. 2017;8:2489. doi: 10.3389/fmicb.2017.02489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi L, et al. The SLE transcriptome exhibits evidence of chronic endotoxin exposure and has widespread dysregulation of non-coding and coding RNAs. PLoS One. 2014;9:e93846. doi: 10.1371/journal.pone.0093846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bao W, Kojima KK, Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hubley R, et al. The Dfam database of repetitive DNA families. Nucleic Acids Res. 2016;44:D81–D89. doi: 10.1093/nar/gkv1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Becker J, et al. A comprehensive hybridization model allows whole HERV transcriptome profiling using high density microarray. BMC Genomics. 2017;18:286. doi: 10.1186/s12864-017-3669-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paces J, Pavlícek A, Paces V. HERVd: Database of human endogenous retroviruses. Nucleic Acids Res. 2002;30:205–206. doi: 10.1093/nar/30.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garazha A, et al. New bioinformatic tool for quick identification of functionally relevant endogenous retroviral inserts in human genome. Cell Cycle. 2015;14:1476–1484. doi: 10.1080/15384101.2015.1022696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smit AFA, Hubley R, Green P. 2013 RepeatMasker Open-4.0. Available at www.repeatmasker.org. Accessed November 13, 2018.
- 38.Mayer J, Blomberg J, Seal RL. A revised nomenclature for transcribed human endogenous retroviral loci. Mob DNA. 2011;2:7. doi: 10.1186/1759-8753-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang Q, Xu Z, Xu R, Wu L, Zheng S. Expression patterns of non-coding spliced transcripts from human endogenous retrovirus HERV-H elements in colon cancer. PLoS One. 2012;7:e29950. doi: 10.1371/journal.pone.0029950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderssen S, Sjøttem E, Svineng G, Johansen T. Comparative analyses of LTRs of the ERV-H family of primate-specific retrovirus-like elements isolated from marmoset, African green monkey, and man. Virology. 1997;234:14–30. doi: 10.1006/viro.1997.8590. [DOI] [PubMed] [Google Scholar]
- 41.Tristem M. Identification and characterization of novel human endogenous retrovirus families by phylogenetic screening of the human genome mapping project database. J Virol. 2000;74:3715–3730. doi: 10.1128/jvi.74.8.3715-3730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitt K, Reichrath J, Roesch A, Meese E, Mayer J. Transcriptional profiling of human endogenous retrovirus group HERV-K(HML-2) loci in melanoma. Genome Biol Evol. 2013;5:307–328. doi: 10.1093/gbe/evt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vargiu L, et al. Classification and characterization of human endogenous retroviruses; mosaic forms are common. Retrovirology. 2016;13:7. doi: 10.1186/s12977-015-0232-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subramanian RP, Wildschutte JH, Russo C, Coffin JM. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology. 2011;8:90. doi: 10.1186/1742-4690-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Parseval N, Lazar V, Casella JF, Benit L, Heidmann T. Survey of human genes of retroviral origin: Identification and transcriptome of the genes with coding capacity for complete envelope proteins. J Virol. 2003;77:10414–10422. doi: 10.1128/JVI.77.19.10414-10422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mayer J, et al. Human endogenous retrovirus HERV-K(HML-2) proviruses with Rec protein coding capacity and transcriptional activity. Virology. 2004;322:190–198. doi: 10.1016/j.virol.2004.01.023. [DOI] [PubMed] [Google Scholar]
- 47.Criscione SW, Zhang Y, Thompson W, Sedivy JM, Neretti N. Transcriptional landscape of repetitive elements in normal and cancer human cells. BMC Genomics. 2014;15:583. doi: 10.1186/1471-2164-15-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramanujam M, Davidson A. Targeting of the immune system in systemic lupus erythematosus. Expert Rev Mol Med. 2008;10:e2. doi: 10.1017/S1462399408000562. [DOI] [PubMed] [Google Scholar]
- 49.Ogasawara H, et al. Sequence analysis of human endogenous retrovirus clone 4-1 in systemic lupus erythematosus. Autoimmunity. 2000;33:15–21. doi: 10.3109/08916930108994105. [DOI] [PubMed] [Google Scholar]
- 50.Mellors RC, Mellors JW. Antigen related to mammalian type-C RNA viral p30 proteins is located in renal glomeruli in human systemic lupus erythematosus. Proc Natl Acad Sci USA. 1976;73:233–237. doi: 10.1073/pnas.73.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strand M, August JT. Type-C RNA virus gene expression in human tissue. J Virol. 1974;14:1584–1596. doi: 10.1128/jvi.14.6.1584-1596.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fali T, et al. DNA methylation modulates HRES1/p28 expression in B cells from patients with Lupus. Autoimmunity. 2014;47:265–271. doi: 10.3109/08916934.2013.826207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Piotrowski PC, Duriagin S, Jagodzinski PP. Expression of human endogenous retrovirus clone 4-1 may correlate with blood plasma concentration of anti-U1 RNP and anti-Sm nuclear antibodies. Clin Rheumatol. 2005;24:620–624. doi: 10.1007/s10067-005-1123-8. [DOI] [PubMed] [Google Scholar]
- 54.Kennedy WP, et al. Association of the interferon signature metric with serological disease manifestations but not global activity scores in multiple cohorts of patients with SLE. Lupus Sci Med. 2015;2:e000080. doi: 10.1136/lupus-2014-000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goodacre N, Aljanahi A, Nandakumar S, Mikailov M, Khan AS. A reference viral database (RVDB) to enhance bioinformatics analysis of high-throughput sequencing for novel virus detection. mSphere. 2018;3:e00069-18. doi: 10.1128/mSphereDirect.00069-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schlesinger S, Goff SP. Retroviral transcriptional regulation and embryonic stem cells: War and peace. Mol Cell Biol. 2015;35:770–777. doi: 10.1128/MCB.01293-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu X, et al. Brief communications. Nat Publish Group. 2014;21:423–425. [Google Scholar]
- 58.Zhou F, et al. Activation of HERV-K Env protein is essential for tumorigenesis and metastasis of breast cancer cells. Oncotarget. 2016;7:84093–84117. doi: 10.18632/oncotarget.11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolf G, Greenberg D, Macfarlan TS. Spotting the enemy within: Targeted silencing of foreign DNA in mammalian genomes by the Krüppel-associated box zinc finger protein family. Mob DNA. 2015 doi: 10.1186/s13100-015-0050-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hishikawa T, et al. Detection of antibodies to a recombinant gag protein derived from human endogenous retrovirus clone 4-1 in autoimmune diseases. Viral Immunol. 1997;10:137–147. doi: 10.1089/vim.1997.10.137. [DOI] [PubMed] [Google Scholar]
- 61.Nakkuntod J, Sukkapan P, Avihingsanon Y, Mutirangura A, Hirankarn N. DNA methylation of human endogenous retrovirus in systemic lupus erythematosus. J Hum Genet. 2013;58:241–249. doi: 10.1038/jhg.2013.6. [DOI] [PubMed] [Google Scholar]
- 62.Gifford RJ, et al. Nomenclature for endogenous retrovirus (ERV) loci. Retrovirology. 2018;15:59. doi: 10.1186/s12977-018-0442-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kong Y. Btrim: A fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics. 2011;98:152–153. doi: 10.1016/j.ygeno.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 64.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim D, et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.