Significance
The plant phytochrome molecule consists of an N-terminal photosensory domain and a C-terminal dimerization domain, connected by a flexible hinge region. It was previously shown that a serine residue in the hinge region of oat phytochrome A (phyA) could be phosphorylated in vivo and plays an important role in regulating phyA function. However, little is known regarding the function of the hinge region of Arabidopsis phyA. Here, we show that three residues in the hinge region of Arabidopsis phyA are essential in regulating phyA phosphorylation and function. Our study thus provides new insights into the regulatory role of the phytochrome hinge region, suggesting that phosphorylation and function of the hinge region may differ between oat and Arabidopsis phyA photoreceptors.
Keywords: Arabidopsis, far-red light, phyA, hinge region, phosphorylation
Abstract
Phytochrome A (phyA) is the only plant photoreceptor that perceives far-red light and then mediates various responses to this signal. Phosphorylation and dephosphorylation of oat phyA have been extensively studied, and it was shown that phosphorylation of a serine residue in the hinge region of oat phyA could regulate the interaction of phyA with its signal transducers. However, little is known about the role of the hinge region of Arabidopsis phyA. Here, we report that three sites in the hinge region of Arabidopsis phyA (i.e., S590, T593, and S602) are essential in regulating phyA function. Mutating all three of these sites to either alanines or aspartic acids impaired phyA function, changed the interactions of mutant phyA with FHY1 and FHL, and delayed the degradation of mutant phyA upon light exposure. Moreover, the in vivo formation of a phosphorylated phyA form was greatly affected by these mutations, while our data indicated that the abundance of this phosphorylated phyA form correlated well with the extent of phyA function, thus suggesting a pivotal role of the phosphorylated phyA in inducing the far-red light response. Taking these data together, our study reveals the important role of the hinge region of Arabidopsis phyA in regulating phyA phosphorylation and function, thus linking specific residues in the hinge region to the regulatory mechanisms of phyA phosphorylation.
Phytochromes are red (R) and far-red (FR) photoreceptors that play fundamental roles in photoperception of the light environment and triggering the corresponding changes in plant growth and development (1, 2). Phytochromes are generally classified into two types according to their protein stability in light: light-labile type I and light-stable type II. In Arabidopsis thaliana, there are five distinct phytochromes, phyA to phyE; phyA is a type I phytochrome and is the most abundant phytochrome species in etiolated seedlings, whereas phyB to phyE are all type II phytochromes, and phyB is the most abundant phytochrome in light-grown plants (1–3). PhyA is the only photoreceptor in plants responsible for initiating responses in FR light, and thus is unique to higher plants enabling them to de-etiolate in shady conditions characterized by a high FR content (1, 2, 4, 5). Both type I and type II phytochrome molecules can be divided into an N-terminal domain (∼70 kDa) and a C-terminal domain (∼55 kDa), which are connected by a flexible hinge region (2, 6).
Phytochromes are synthesized in the cytosol in their inactive Pr form; upon light illumination, phytochromes are converted to the biologically active Pfr form, and translocate into the nucleus. phyB can enter the nucleus by itself in response to R light, whereas phyA nuclear import depends on two small plant-specific proteins FAR-RED ELONGATED HYPOCOTYL 1 (FHY1) and FHY1-LIKE (FHL) (7–9). Two transposase-derived transcription factors, FHY3 and FAR-RED IMPAIRED RESPONSE1 (FAR1), act together to directly activate the transcription of FHY1 and FHL, thus indirectly regulating phyA nuclear accumulation and phyA responses (10). ELONGATED HYPOCOTYL5 (HY5), a bZIP family transcription factor, has been shown to function as a pivotal positive regulator of photomorphogenic development under a wide spectrum of wavelengths (11–16). CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1), a well-characterized E3 ubiquitin ligase, forms complexes with SUPPRESSOR OF phyA-105 (SPA) proteins (17), and they work in concert to target multiple photomorphogenesis-promoting factors for degradation, such as HY5 (13, 18). It was recently proposed that light-activated phytochromes interact with SPA proteins to disrupt the interaction between COP and SPA proteins and thus inactivate the COP1–SPA complexes, thus allowing rapid accumulation of transcription factors (such as HY5) and initiation of photomorphogenesis (19).
It is well established that phytochromes are phosphoproteins, because they can be readily labeled with 32P in vivo (20–22). An outstanding feature of the N-terminal extension region of all phyA proteins, from both monocots and dicots, is that there is a serine-rich stretch in the first 20 aa, and it has been well-documented that converting these serines to alanines, or deletion of this 20-aa region, results in an increased stability and biological activity of phyA, thus suggesting that these serine residues are involved in desensitization of phyA signaling (23–27). The phosphorylation sites of oat (Avena sativa) phyA have been investigated since the 1980s, and two serine residues in the N-terminal extension region (S8 and S18) and a serine residue in the hinge region (S599) of oat phyA have been well-characterized to be phosphorylation sites (20–22, 28). Interestingly, overexpression of S599A in the Arabidopsis phyA-201 mutant made the transgenic plants hypersensitive to light, and further analyses indicated that phosphorylation of S599 did not affect phytochrome stability, but prevented the interaction of phyA with its signal transducers such as NUCLEOSIDE-DIPHOSPHATE KINASE2 (NDPK2) and PIF3 (29). In contrast to the vast literature on phyA phosphorylation, limited data have been published concerning phyB phosphorylation and its impact on phyB signaling. Very recently the S86 and Y104 residues of Arabidopsis phyB were separately reported to be phosphorylation sites (30, 31). It was shown that phosphorylation of S86 accelerated dark reversion (light-independent reversion of Pfr into Pr) of phyB, whereas light-induced phosphorylation of Y104 inhibited PIF3 binding to the Pfr form of phyB (30, 31). Thus, these reports suggest that phosphorylation of phyB at multiple sites negatively regulates the action of phyB.
The observation that phytochromes are phosphoproteins suggests that there must be protein kinases and phosphatases that phosphorylate and dephosphorylate phytochromes, respectively. Several studies reported the discovery of protein phosphatases that could dephosphorylate phytochromes, such as a phytochrome-associated protein phosphatase 2A (designated FyPP) (32), PHYTOCHROME-ASSOCIATED PROTEIN PHOSPHATASE 5 (PAPP5) (33), and PHYTOCHROME-ASSOCIATED PROTEIN PHOSPHATASE 2C (PAPP2C) (34). However, despite the numerous investigations of phytochrome-interacting proteins, there is no report thus far of a protein kinase that can specifically phosphorylate phytochromes. Instead, accumulating evidence indicates that phytochromes themselves may function as light-regulated serine/threonine kinases (28, 35, 36). It was shown that phytochromes can phosphorylate several substrates, including histone H1, cryptochromes, PHYTOCHROME KINASE SUBSTRATE 1 (PKS1), AUX/IAA proteins, FHY1, PIFs, and themselves in vitro (36–41). Two phosphorylation sites in the N-terminal extension region of oat phyA (i.e., S8 and S18) were shown to be autophosphorylated by phyA itself in vitro (20–22, 27); however, another phosphorylation site of oat phyA, S599, was not autophosphorylated by phyA (27, 29). The protein kinases that phosphorylate this site of oat phyA have not yet been identified.
In this study, we report that three sites in the hinge region of Arabidopsis phyA—S590, T593, and S602—are important for regulating phyA function. We further show that the in vivo formation of a phosphorylated form of Arabidopsis phyA, whose abundance is highly associated with the extent of phyA function, was also influenced by mutations of these sites. Thus, our study demonstrates that the hinge region of Arabidopsis phyA plays an essential role in regulating phosphorylation and function of the phyA photoreceptor.
Results
S590, T593, and S602 in the Hinge Region of Arabidopsis phyA Are Important for phyA Function.
It was previously shown that a phosphorylation site in the hinge region of oat phyA, S599, is important for regulating phyA function (29). However, the amino acid sequences of the hinge region vary widely among monocot and dicot phyA proteins, and S599 is not conserved (SI Appendix, Fig. S1). Notably, serines and threonines are always present in the hinge region of both monocot and dicot phyA proteins (SI Appendix, Fig. S1), implying that this region may contain phosphorylation sites.
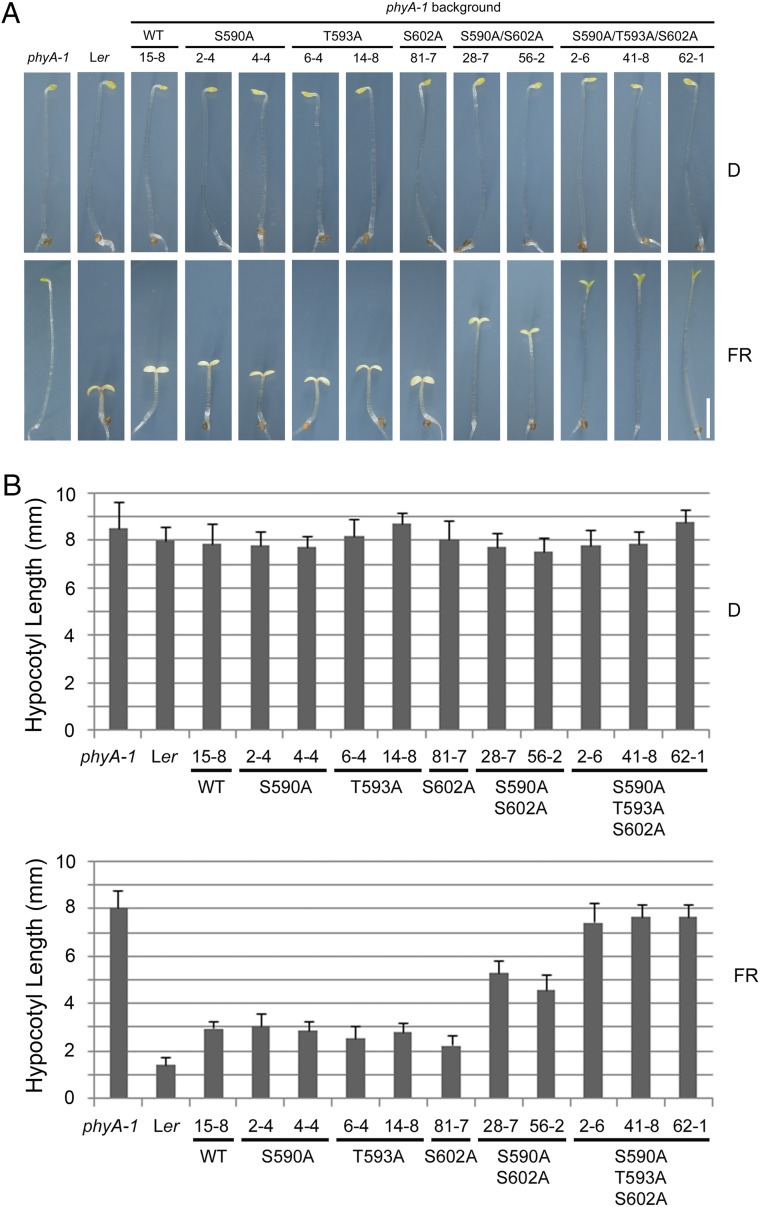
We used the NetPhos 2.0 server (42) (www.cbs.dtu.dk/services/NetPhos/) to predict the possible phosphorylation sites of Arabidopsis phyA, and the output indicated that one residue in the hinge region, T593, was likely to be a phosphorylation site (with NetPhos score 0.910). To test the role of T593 in regulating phyA function, we first generated transgenic Arabidopsis plants to express WT phyA (hereafter abbreviated as phyAWT) or phosphorylation-deficient T593A (by changing T593 to A) phyA, respectively, under the control of the native PHYA promoter in the phyA-1–null mutant background. We produced their T3 homozygous transgenic lines, and the transgene expression levels were determined by immunoblot analysis. Our data showed that WT and T593A phyA levels were largely similar in darkness (SI Appendix, Fig. S2); however, the phenotypes of T593A transgenic lines were indistinguishable from those expressing phyAWT in FR light (Fig. 1), suggesting that mutation of T593 alone to alanine did not affect phyA function.
Fig. 1.
Mutating S590, T593, and S602 to alanines impaired phyA function. (A) Phenotypes of 4-d-old WT (Ler), phyA-1, and homozygous transgenic lines expressing WT or various mutant forms of Arabidopsis phyA (under the control of the native Arabidopsis PHYA promoter in the phyA-1 background) grown in darkness (D) or continuous FR light. (Scale bar: 2 mm.) (B) Quantitative analysis of hypocotyl lengths of the WT (Ler), phyA-1, and various transgenic lines shown in A. Error bars represent SD from 30 seedlings.
There are two serines in the hinge region of Arabidopsis phyA (i.e., S590 and S602); however, their NetPhos scores were low (0.208 and 0.256, respectively, for S590 and S602) (SI Appendix, Fig. S1B). Because overexpression of S599A oat phyA in the Arabidopsis phyA mutant background caused the transgenic plants to be hypersensitive to light (29), we hypothesized that one or more of the serines in the phyA hinge region may be involved in regulating phyA function. We therefore performed site-directed mutagenesis to mutate S590 and S602 to alanine, individually or together, and expressed the mutant phyA proteins in the phyA-1 background under the control of the native PHYA promoter. Our phenotypic analyses showed that whereas mutating either S590 or S602 to alanine separately did not substantially affect phyA function, mutation of S590 and S602 together to alanines resulted in significantly longer hypocotyls of transgenic seedlings grown under FR light (Fig. 1), indicating that phyA function was greatly impaired. Strikingly, when we mutated T593 with S590 and S602 together to alanines, the transgenic seedlings of S590A T593A S602A phyA (hereafter abbreviated as phyAAAA) displayed substantially long hypocotyls in FR light, resembling those of the phyA-1 mutant (Fig. 1). However, phyAAAA transgenic seedlings developed half-open cotyledons (Fig. 1A and SI Appendix, Fig. S3), indicating that phyAAAA still harbored a low level of function.
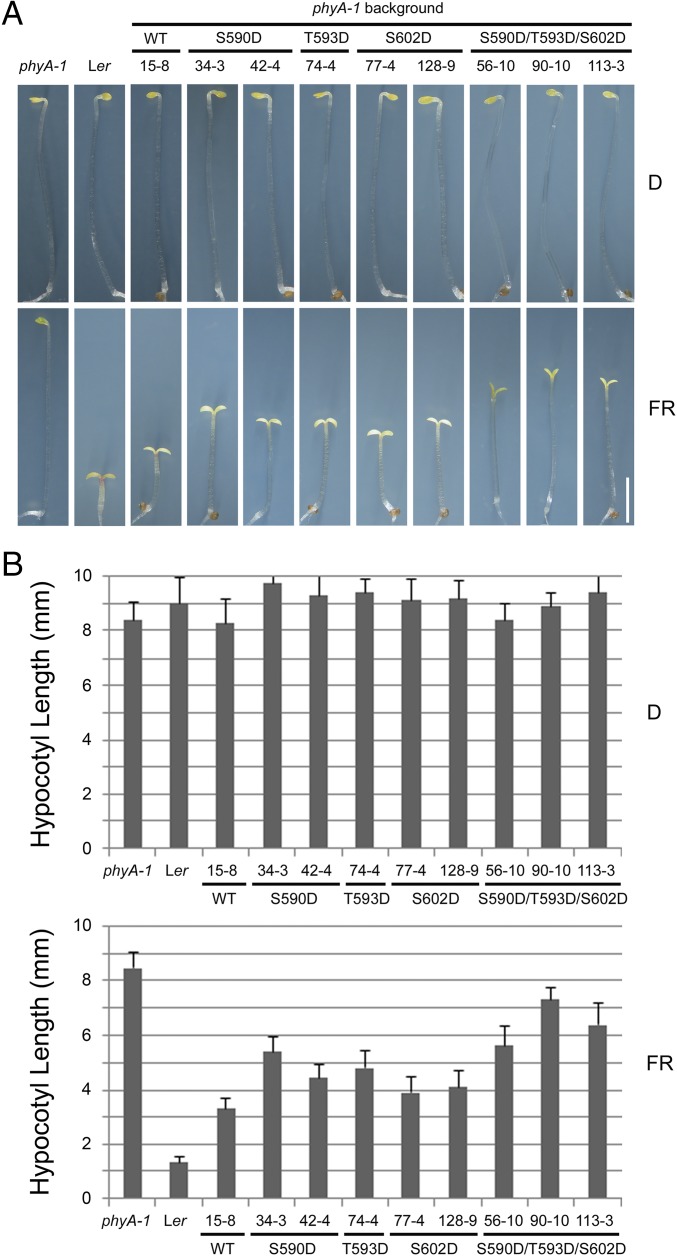
Next, we mutated S590, T593, and S602 of Arabidopsis phyA to Asp (D), individually or together to express phosphorylation-mimic phyA mutants in the phyA-1 background. Surprisingly, we found that mutation of any of these three residues to Asp caused the transgenic seedlings to develop longer hypocotyls than phyAWT when grown in FR light (Fig. 2 and SI Appendix, Fig. S4). Moreover, when all three residues were mutated together to Asp, the S590D T593D S602D phyA (hereafter abbreviated as phyADDD) transgenic seedlings displayed even longer hypocotyls (Fig. 2 and SI Appendix, Fig. S4). Notably, the hypocotyl lengths of phyADDD seedlings were largely shorter than those of phyAAAA (Figs. 1 and 2), indicating that phyADDD lines displayed more photomorphogenic development than phyAAAA seedlings. Collectively, our data indicate that S590, T593, and S602 play important roles in regulating phyA function.
Fig. 2.
Mutating S590, T593, and S602 to aspartic acids also impaired phyA function. (A) Phenotypes of 4-d-old WT (Ler), phyA-1, and homozygous transgenic lines expressing WT or various mutant forms of Arabidopsis phyA (under the control of the native Arabidopsis PHYA promoter in the phyA-1 background) grown in darkness (D) or continuous FR light. (Scale bar: 2 mm.) (B) Quantitative analysis of hypocotyl lengths of the WT (Ler), phyA-1, and various transgenic lines shown in A. Error bars represent SD from 30 seedlings.
Transcriptomic Changes in phyAAAA and phyADDD Seedlings.
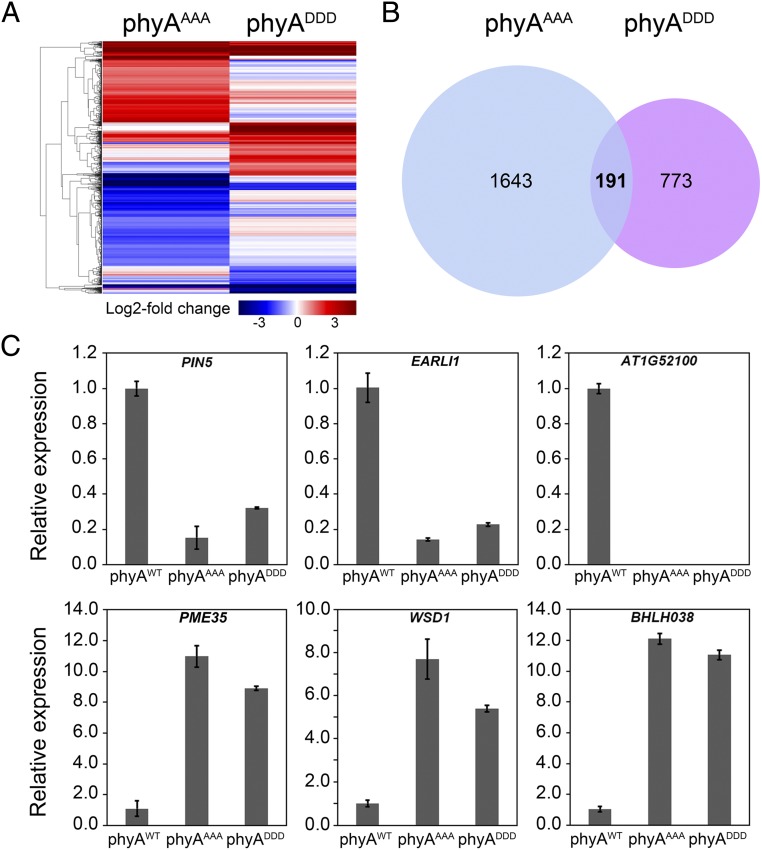
To investigate the global gene-expression changes in phyAAAA and phyADDD seedlings, we examined the transcriptomes of phyAWT, phyAAAA, and phyADDD seedlings by RNA-sequencing analysis. The homozygous transgenic seedlings were grown in FR light for 4 d, and then harvested for RNA extraction and sequencing. RNA-sequencing data were collected from three independent experiments (each sample with 2.0 G clean data) and differential gene-expression analysis was performed using Cufflinks (43) (cufflinks.cbcb.umd.edu). Our results showed that 1,834 and 964 genes displayed statistically significant changes (using Student’s t test with P < 0.05 and fold-change > 2) in phyAAAA and phyADDD seedlings, respectively, compared with those in phyAWT (Fig. 3 A and B). Further comparison of these genes led to the identification of 191 genes whose expression was significantly changed in both phyAAAA and phyADDD seedlings (Fig. 3B), among which 75 and 84 genes were up-regulated and down-regulated, respectively, in both phyAAAA and phyADDD seedlings (Dataset S1).
Fig. 3.
Transcriptomic changes in phyAAAA and phyADDD seedlings. (A) Cluster analysis of genes whose expression was changed in phyAAAA and phyADDD seedlings compared with those in phyAWT. The bar represents the log2 of the ratio. (B) Venn diagram showing the number and overlap of genes whose expression was changed in phyAAAA and phyADDD seedlings. (C) qRT-PCR assays showing the expression of several representative genes whose expression was up-regulated or down-regulated in both phyAAAA and phyADDD seedlings. Error bars represent SD of three technical replicates.
Quantitative RT-PCR (qRT-PCR) assays were performed to validate the expression of several representative genes. The expression of PIN5, encoding a functional auxin transporter involved in regulating intracellular auxin homeostasis and metabolism (44), EARLI1, a member of the proline-rich protein family regulating flowering time and lignin synthesis (45), and AT1G52100, was down-regulated in both phyAAAA and phyADDD seedlings; in contrast, the expression of PME35, encoding a pectin methylesterase regulating the mechanical strength of the Arabidopsis stem (46), WSD1, a member of the bifunctional wax ester synthase/diacylglycerol acyltransferase gene family playing a key role in wax ester synthesis in Arabidopsis (47), and BHLH038, was up-regulated in both phyAAAA and phyADDD seedlings (Fig. 3C). The expression of several key regulatory genes of the phyA signaling pathway, such as FHY1, FHL, FHY3, FAR1, HY5, and HYH, did not display statistically significant changes in RNA-sequencing analysis. qRT-PCR assays were performed to further validate their expression, and the data confirmed that the expression of these genes was not significantly changed in phyAAAA and phyADDD seedlings (SI Appendix, Fig. S5).
Both phyAAAA and phyADDD Interact with FHY1/FHL More Strongly than phyAWT, and Can Enter the Nucleus and Form Nuclear Bodies.
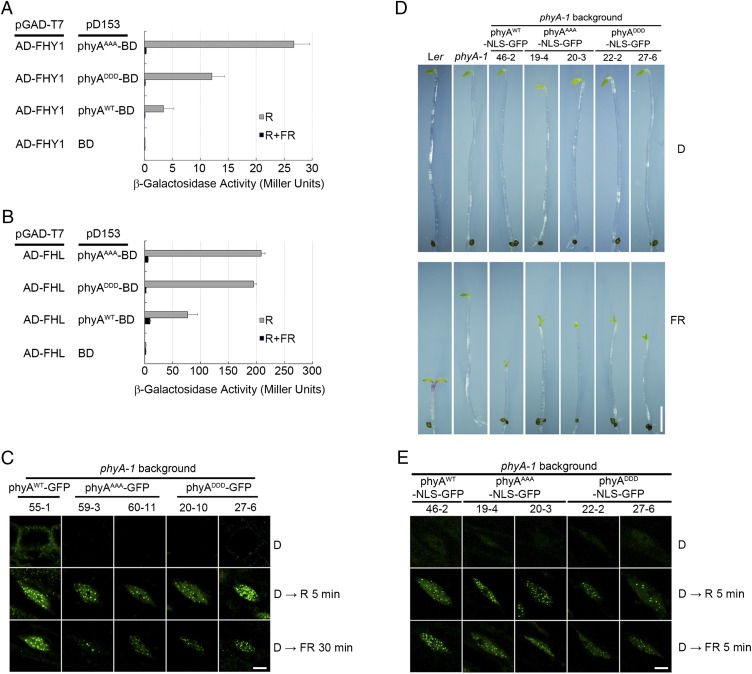
It was previously shown that FHY1/FHL-mediated nuclear translocation of phyA upon light exposure is a prerequisite for phyA signaling (4, 7–10). We therefore asked whether the defect of phyAAAA and phyADDD was due to altered interactions with FHY1/FHL. To address this question, we employed a yeast two-hybrid system (48) to allow phyA to form the Pr and Pfr forms after FR and R light treatments, respectively, by adding phycocyanobilin (PCB) extracted from Spirulina. Consistent with previous reports (4, 7, 8), the Pfr form of phyA specifically interacted with FHY1 and FHL in this system (Fig. 4 A and B). Interestingly, compared with phyAWT, both phyAAAA and phyADDD interacted with FHY1 and FHL more strongly after R light treatment (Fig. 4 A and B).
Fig. 4.
Both phyAAAA and phyADDD interact with FHY1/FHL more strongly and enter the nucleus upon light illumination. (A and B) Both phyAAAA and phyADDD interact with FHY1 (A) and FHL (B) more strongly than phyAWT in yeast cells. Yeast cells transformed with the indicated plasmids were used for ortho-Nitrophenyl-β-galactoside (ONPG) assays. The yeast cultures were irradiated with 5 min of R alone, or with 5 min of R followed by 5 min of FR, and then incubated for another 4 h before measuring the β-galactosidase activity. Error bars represent SD of triplicate experiments. (C) phyAWT-GFP, phyAAAA-GFP and phyADDD-GFP all enter the nucleus after 5 min of R or 30 min of FR irradiation. Homozygous PHYAp:PHYAWT-GFP phyA-1, PHYAp:PHYAAAA-GFP phyA-1, and PHYAp:PHYADDD-GFP phyA-1 seedlings were grown first in darkness (D) for 4 d, and then treated with 5 min of R or 30 min of FR light and examined using fluorescence microscopy. (Scale bar: 10 μm.) (D) Phenotypes of 4-d-old WT (Ler), phyA-1, and homozygous PHYAp:PHYAWT-NLS-GFP phyA-1, PHYAp:PHYAAAA-NLS-GFP phyA-1, and PHYAp:PHYADDD-NLS-GFP phyA-1 seedlings grown in darkness (D) or continuous FR light. (Scale bar: 2 mm.) (E) Nuclear localization of phyAWT-NLS-GFP, phyAAAA-NLS-GFP, and phyADDD-NLS-GFP following exposure to 5 min of R or FR light. Homozygous PHYAp:PHYAWT-NLS-GFP phyA-1, PHYAp:PHYAAAA-NLS-GFP phyA-1, and PHYAp:PHYADDD-NLS-GFP phyA-1 seedlings were grown first in darkness (D) for 4 d, and then irradiated with 5 min of R or FR light and examined using fluorescence microscopy. (Scale bar: 10 μm.)
To investigate whether the mutations affect the nuclear accumulation of phyA, we generated green fluorescent protein (GFP)-tagged phyAWT, phyAAAA, and phyADDD fusions in the phyA-1–null mutant background (SI Appendix, Fig. S6). Our observations made from confocal microscopy indicated that both phyAAAA-GFP and phyADDD-GFP entered the nucleus and formed nuclear bodies as phyAWT-GFP after 5 min of R light or 30 min of FR light treatments (Fig. 4C), indicating that phyAAAA and phyADDD mutations did not obviously affect the nuclear translocation of phyA.
To further investigate whether the phenotypes of phyAAAA and phyADDD mutants were due to impaired nuclear translocation, we generated constitutively nuclear-localized phyAWT-NLS-GFP, phyAAAA-NLS-GFP, and phyADDD-NLS-GFP fusions, respectively, in the phyA-1–null mutant background by inserting a nuclear localization signal (NLS) between phyA and GFP. We selected homozygous lines expressing comparable levels of phyAWT-NLS-GFP, phyAAAA-NLS-GFP, and phyADDD-NLS-GFP, and measured their hypocotyls under FR light. Again, the hypocotyls of phyAAAA-NLS-GFP and phyADDD-NLS-GFP seedlings were still longer than those of phyAWT-NLS-GFP seedlings (Fig. 4D and SI Appendix, Fig. S7). Notably, nuclear bodies appeared extremely rapidly upon light illumination in phyAWT-NLS-GFP, phyAAAA-NLS-GFP, and phyADDD-NLS-GFP plants (Fig. 4E). Taken together, our data indicated that phyAAAA and phyADDD interacted with FHY1/FHL more strongly than phyAWT, but the functional defects of phyAAAA and phyADDD did not seem to be caused by impairment of nuclear translocation.
Both phyAAAA and phyADDD Displayed Normal Activities in Inhibiting COP1–SPA1 Interaction.
It was recently reported that both phyA and phyB could disrupt the interaction of COP1 with SPA proteins, which may serve as a major molecular mechanism for light-activated phytochromes to inactivate the COP1–SPA complexes and to initiate photomorphogenesis (19). Therefore, we next performed yeast three-hybrid assays to investigate whether the phyAAAA and phyADDD mutations affect the inhibitory activity of phyA toward the COP1–SPA1 interaction. The chromophore PCB was added into the yeast system to allow phyA to form Pr and Pfr forms after FR and R light treatments, respectively. Indeed, our data showed that BD–COP1 and AD–SPA1 displayed robust interaction in the yeast cells (SI Appendix, Fig. S8). Unexpectedly, when an NLS was fused with phyA to allow phyA nuclear translocation by itself in yeast cells, both Pr and Pfr forms of the WT phyA displayed strong (with Pfr phyA showing stronger) inhibition of COP1–SPA1 interaction (SI Appendix, Fig. S8). These observations are consistent with those reported by Sheerin et al. (19). Nevertheless, our data showed that the Pfr forms of both phyAAAA and phyADDD displayed normal inhibition of the COP1–SPA1 interaction as phyAWT in yeast cells (SI Appendix, Fig. S8), suggesting that both phyAAAA and phyADDD could form the biologically active Pfr form to disrupt the COP1–SPA complexes.
Both phyAAAA and phyADDD Are Degraded More Slowly.
It was recently suggested by a mathematical modeling approach that light-induced phyA degradation is an important property required for shifting the action peak of phyA from R to FR light (4). Thus, we next examined whether phyAAAA and phyADDD showed different patterns of degradation upon light exposure. Homozygous transgenic plants expressing phyAWT, phyAAAA, and phyADDD proteins were first grown in darkness for 4 d, and then transferred to R light for various time points. Our immunoblot data show that phyAWT, phyAAAA, and phyADDD proteins were all degraded rapidly upon R light exposure, but the degradation of phyAAAA was obviously slower than that of phyAWT (SI Appendix, Fig. S9 A and B). phyADDD proteins were initially degraded similarly as phyAWT, but after 1 h of R light treatment, the degradation of phyADDD slowed relative to that of phyAWT (SI Appendix, Fig. S9 A and B).
We also compared the degradation dynamics of phyAWT, phyAAAA, and phyADDD after FR light treatment. Homozygous transgenic plants expressing phyAWT, phyAAAA, and phyADDD proteins were grown in darkness for 4 d, or in darkness for 1–3 d first and then transferred to FR light for up to 4 d. The seedlings were harvested simultaneously and then subjected to immunoblotting. Although phyA degradation in FR light was much slower than in R, our data showed that both phyAAAA and phyADDD were degraded much slower than phyAWT after FR light treatment (SI Appendix, Fig. S9C) (HRP data). Taken together, our results indicate that both phyAAAA and phyADDD mutants are degraded more slowly upon exposure to R and FR light.
Accumulation of FHY1 and HY5 Proteins in phyAAAA and phyADDD Lines.
Our previous studies showed that phyA mediates the phosphorylation and proteasomal degradation of FHY1 (41, 49). Therefore, we asked whether accumulation of FHY1 protein was affected in phyAAAA and phyADDD lines. Although the expression of FHY1 was largely unchanged in phyAAAA and phyADDD lines (SI Appendix, Fig. S5), our immunoblot data showed that more FHY1 proteins accumulated in both phyAAAA and phyADDD seedlings than in phyAWT (SI Appendix, Fig. S10A), consistent with the conclusion that phyA function is impaired in both phyAAAA and phyADDD lines.
The abundance of HY5 protein was shown to be directly correlated with the extent of photomorphogenesis (13), we next examined HY5 protein levels in phyAAAA and phyADDD lines. Our immunoblot data indicated that HY5 protein levels were much lower in phyAAAA lines (SI Appendix, Fig. S10B), consistent with their phenotypes showing that phyA function was greatly disrupted in phyAAAA lines; in contrast, HY5 protein accumulated in phyADDD lines (SI Appendix, Fig. S10B), which may explain why phyADDD seedlings displayed more photomorphogenic development than phyAAAA seedlings.
A Nuclear Phosphorylated phyA Form Is Less Produced in phyAAAA and phyADDD Seedlings in FR.
A phosphorylated form of Arabidopsis phyA was shown to accumulate in vivo upon irradiation with FR light (50, 51). Our recent report showed that TANDEM ZINC FINGER/PLUS3 (TZP) acts as a key component of phyA signaling required for phosphorylation of phyA, thus suggesting an important role of phosphorylated phyA in inducing the FR light response (51). We then asked whether the production of this phosphorylated phyA form was altered in phyAAAA and phyADDD lines. Interestingly, this phosphorylated phyA form was observed in phyAWT seedlings upon exposure to FR light; however, this form was observed at much lower levels in phyADDD, but was hardly detectable in phyAAAA seedlings (SI Appendix, Fig. S9C) (AP data), indicating that the formation of phosphorylated phyA was influenced by phyAAAA and phyADDD mutations.
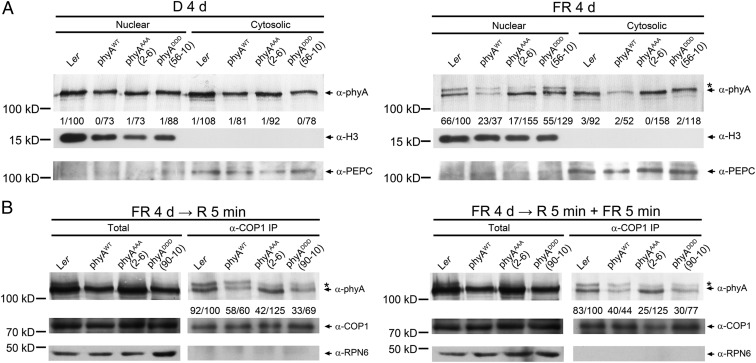
Next, we performed nuclear-cytoplasmic fractionation assays to further localize the changes of phosphorylated phyA production in phyAAAA and phyADDD lines. Consistent with our recent report (51), the phosphorylated phyA form was undetectable in either the nuclear or the cytosolic fractions of phyAWT, phyAAAA, and phyADDD seedlings grown in the dark (Fig. 5A). When the seedlings were grown in FR light, we observed that the phosphorylated form was highly enriched in the nuclear fractions of Ler and phyAWT line (0.66 and 0.62 for the phosphorylated versus unphosphorylated phyA in Ler and phyAWT seedlings, respectively) (Fig. 5A); however, the proportions of the phosphorylated form were dramatically reduced in the nuclear fractions of phyAAAA and phyADDD lines (0.11 and 0.43 for the phosphorylated versus unphosphorylated phyA in phyAAAA and phyADDD lines, respectively) (Fig. 5A). These data indicated that the proportion of the phosphorylated phyA form was decreased in the nucleus in phyADDD lines and further decreased in phyAAAA lines.
Fig. 5.
The abundance of the phosphorylated phyA form is reduced in phyAAAA and phyADDD lines. (A) Immunoblot analyses of nuclear and cytoplasmic fractions from 4-d-old WT (Ler), homozygous PHYAp:PHYAWT phyA-1, PHYAp:PHYAAAA phyA-1, and PHYAp:PHYADDD phyA-1 seedlings grown in darkness (D) or continuous FR light. PEPC and H3 were used as cytosolic and nuclear markers, respectively. (B) Co-IP assays showing that the pool of phyA interacting with COP1 contained less ratio of phosphorylated form in phyAAAA and phyADDD seedlings in FR light. The 4-d-old Ler, phyAWT, phyAAAA, and phyADDD seedlings grown in FR light were subjected to anti-COP1 co-IP assays. Asterisks and arrowheads represent the phosphorylated and unphosphorylated phyA forms, respectively (50). The numbers below each blot indicate relative band intensities for the phosphorylated and unphosphorylated phyA, with the band intensity of unphosphorylated phyA in the first lane set to 100.
Our previous studies showed that COP1 preferentially associates with the phosphorylated phyA form in FR light (50, 51). To investigate whether the pool of phyA interacting with COP1 is altered in phyAAAA and phyADDD seedlings, we performed coimmunoprecipitation (co-IP) assays using anti-COP1 antibodies and 4-d-old Ler, phyAWT, phyAAAA, and phyADDD seedlings grown in FR light. Consistent with our previous report that COP1 is preferentially associated with phosphorylated phyA (50), large ratios of phosphorylated phyA were co-IP by anti-COP1 antibodies in Ler and phyAWT seedlings (0.83 or higher for phosphorylated versus unphosphorylated phyA); in contrast, lower ratios of phosphorylated phyA were associated with COP1 in phyADDD (0.48 and 0.39 for phosphorylated versus unphosphorylated phyA after R and R+FR pulse exposure, respectively) and phyAAAA seedlings (0.34 and 0.2 for phosphorylated versus unphosphorylated phyA after R and R+FR pulse exposure, respectively) (Fig. 5B). Collectively, our data indicated that the pool of phyA interacting with COP1 contained less ratios of the phosphorylated form in phyAAAA and phyADDD seedlings in FR light.
The Phosphorylated phyA Form Is Produced in the Nucleus by the Pfr Form.
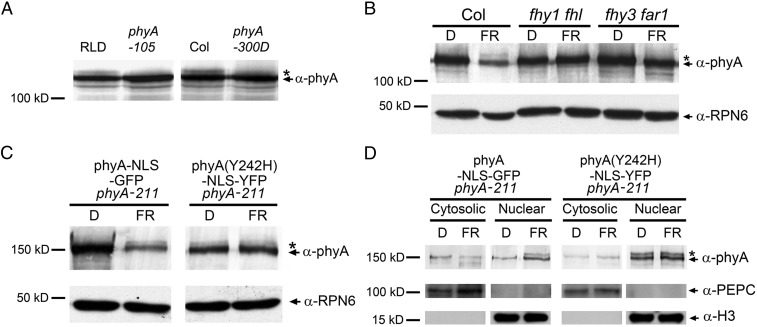
Our recent study showed that the abundance of phosphorylated phyA correlated well with the degree of phyA response (51). To further investigate the importance of the phosphorylated phyA form in FR light response, we examined two phyA point mutants, phyA-105 [A893V (52) and phyA-300D (V631M) (53)], in which point mutations in the C-terminal domain of phyA impaired its function but did not change phyA protein levels in FR light. Our immunoblot data showed that similar levels of mutant phyA proteins accumulated in the phyA-105 and phyA-300D mutants in FR light compared with their corresponding WT (RLD and Col, respectively); however, it was evident that the phosphorylated phyA form was not detected in both phyA-105 and phyA-300D mutants (Fig. 6A). Thus, our data suggest that the formation of the phosphorylated phyA form may be important for phyA to induce the FR light response.
Fig. 6.
The phosphorylated phyA form is produced in the nucleus by the Pfr form. (A) The phosphorylated phyA form was detected in the WT seedlings but not in the phyA-105 and phyA-300D mutants. (B) The phosphorylated phyA form was detected in 4-d-old WT plants but not in fhy1 fhl and fhy3 far1 mutants grown in continuous FR light. (C) The phosphorylated phyA form was produced in phyA-NLS-GFP phyA-211 seedlings only in FR light, but was produced in phyA(Y242H)-NLS-YFP phyA-211 seedlings in both darkness and FR light. (D) Immunoblot analyses of nuclear and cytoplasmic fractions from 4-d-old phyA-NLS-GFP phyA-211 and phyA(Y242H)-NLS-YFP phyA-211 seedlings grown in darkness (D) or continuous FR light. PEPC and H3 were used as cytosolic and nuclear markers, respectively. Asterisks and arrowheads represent the phosphorylated and unphosphorylated phyA forms, respectively (50).
To investigate how phyA phosphorylation is regulated by its subcellular localization, we tested the production of the phosphorylated phyA form in fhy1 fhl and fhy3 far1 mutants, in both of which phyA is localized exclusively in the cytosol (7, 8, 10, 54). Interestingly, we did not detect the phosphorylated phyA form in either mutant in FR light (Fig. 6B), indicating that nuclear localization is required for the formation of this phosphorylated phyA form.
We then asked whether the phosphorylated phyA could be produced by both Pr and Pfr forms when phyA is localized in the nucleus. We grew phyA-NLS-GFP phyA-211 seedlings in dark or FR light for 4 d, and then extracted total proteins and performed immunoblot assays. Our data showed that although phyA-NLS-GFP is constitutively localized in the nucleus (9), the phosphorylated phyA form was only produced in seedlings grown in FR light, but not in darkness (Fig. 6C). The nuclear-cytoplasmic fractionation assay data further support this conclusion (Fig. 6D). These data indicated that the phosphorylated phyA could not be produced by the nuclear-localized Pr form. Furthermore, we examined the constitutively active phyA mutant, phyA(Y242H)-NLS-YFP, which is constitutively locked in the Pfr form and showed strong cop phenotype (4, 55). Strikingly, the phosphorylated phyA form was detected in phyA(Y242H)-NLS-YFP seedlings not only in FR light, but also in darkness (Fig. 6 C and D). Collectively, our data suggested that the phosphorylated phyA form is produced by the nuclear-localized Pfr form, and may be essential for phyA to induce the FR light response.
Discussion
The flexible hinge region of phytochrome connects the N-terminal chromophore-bearing photosensory domain and the C-terminal dimerization domain. It was demonstrated that in the Pr form, the N-terminal domain of Arabidopsis phyB interacts directly with the C-terminal PAS-related domain (PRD), which contains the NLS; thus, the interaction masks the NLS of phyB in the Pr form. Upon photoconversion to the Pfr form, conformational changes allow the exposure of NLS and nuclear translocation of phyB (56). In terms of phyA, although its nuclear translocation has been demonstrated to be dependent on FHY1 and FHL (7–9), it was also shown that the far N-terminal and the distal C-terminal ends of phyA interact in the Pr form, and the interaction seems to shield the hinge region in the Pr form; however, after Pr-to-Pfr photoconversion, the hinge region of phyA becomes exposed in the Pfr form (57). Interestingly, a serine residue in the hinge region of oat phyA (i.e., S599) was shown to be phosphorylated in vivo in a Pfr-dependent manner (21, 22), consistent with the notion that the hinge region is preferentially exposed in the Pfr form. Therefore, it was proposed that the phosphorylation sites in the hinge region may serve as a switch for interdomain interaction between the N- and C-terminal domains (57), and may also affect protein–protein interactions between phytochrome and its signal transducers (29, 33). Our data support this notion because FHY1 and FHL also act as signal transducers of phyA (50, 58, 59). The fact that phyAAAA and phyADDD both showed increased interaction with FHY1 and FHL (Fig. 4 A and B) indicates that these three sites in the hinge region indeed regulate phyA interaction with its signal transducers. Moreover, because FHY1 and FHL interact with both the N-terminal chromophore-binding domain (8) and the C-terminal HKRD domain of phyA (SI Appendix, Fig. S11), FHY1 and FHL may mediate the interdomain cross-talk between the N- and the C-terminal domains of phyA as well. Thus, increased interaction of the Pfr forms of phyAAAA and phyADDD with FHY1 and FHL (Fig. 4 A and B) may reflect altered cross-talk of the N- and C-terminal domains caused by phyAAAA and phyADDD mutations, suggesting that the hinge region of phyA and FHY1/FHL both play roles in regulating interdomain cross-talk of the phyA photoreceptor. Although we tried several times to immunoprecipitate phyA proteins using anti-phyA antibodies from 4-d-old FR-grown WT seedlings and then employed a liquid chromatography-mass spectrometry approach to identify the in vivo phosphorylation sites of Arabidopsis phyA, we were unsuccessful in proving that S590, T593, and S602 are indeed phosphorylated in vivo in FR light, possibly due to the fact that the level of phosphorylated phyA is extremely low in Arabidopsis. Thus, whether S590, T593, and S602 (or some of them) serve as in vivo phosphorylation sites remains to be answered in future studies.
We analyzed the reported phyA mutant alleles caused by amino acid substitutions, and found that all of the reported substitutions occurred at the amino acids conserved among monocot and dicot phyA proteins (SI Appendix, Table S1), whereas S590, T593, and S602 are not conserved residues that only appear in the hinge region of Arabidopsis phyA (SI Appendix, Fig. S1). These observations suggest that the mutations of S590, T593, and S602 may not change the structures of the conserved N- or the C-terminal domain of phyA. The fact that the Pfr forms of both phyAAAA and phyADDD displayed normal activities in inhibiting COP1–SPA1 interaction in yeast cells (SI Appendix, Fig. S8) indicates that the Pfr form formation was not affected by both phyAAAA and phyADDD mutations. In addition, most of the mutations occurred in the regions other than the hinge region, whereas relatively few mutations of both phyA and phyB have been reported to be located in the hinge region. Interestingly, a naturally occurring M548T polymorphism of phyA was identified in the Lm-2 ecotype, and it was shown that this mutation results in the reduced FR sensitivity of Lm-2 (60). Although this residue is also located in the hinge region of phyA, it is within the conserved PHY domain rather than the highly diversified hinge region containing S590, T593, and S602 (SI Appendix, Fig. S1). Moreover, it was also suggested that the Lm-2 phyA variant (M548T) may alter the cross-talk between the light-sensing and output domains of phyA, rather than having a direct structural effect on either domain (60).
Our data indicated that both phyAAAA and phyADDD mutants displayed less (with phyAAAA showing least) activity in vivo in FR light. Although rarely reported, similar observations were also made for Arabidopsis CRY2, for which mutations of 13 serine residues in the CCE domain to either alanines or aspartic acids both caused loss-of-function effect on CRY2 (61). It was reasoned that phosphomimetic mutation does not always mimic or reproduce the changes introduced by protein phosphorylation (62), especially when the phosphorylation of the protein is to establish a charge-dependent structural change, such as blue light-triggered separation of the PHR domain and the CCE domain of CRY2 (61). These notions are consistent with our observations regarding the phyAAAA and phyADDD mutants, especially considering the fact that the hinge region may be involved in regulating the structural changes of photoconversion between the Pr and Pfr forms. Alternatively, dynamic phosphorylation and dephosphorylation of these sites might be important for structural maintenance and function of phyA, which was, however, abolished when these sites were mutated to either alanines or aspartic acids.
An in vivo phosphorylated form of Arabidopsis phyA was first reported in our previous study (50). It was proposed that this phosphorylated phyA form may serve as a preferred substrate for the COP1–SPA complex-mediated degradation (50). Our data of this study, and those shown in our recent report (51), both indicate that the phosphorylated phyA form may play an important role in phyA function based on the following observations. First, the abundance of the phosphorylated phyA form correlated well with the degree of phyA response (51), and it was striking to observe that the phosphorylated phyA form could be produced even in darkness by the constitutively active phyA mutant, phyA(Y242H)-NLS-YFP (Fig. 6 C and D). Second, in phyA-105, phyA-300D, and tzp mutants, all of which were hyposensitive to FR light, unphosphorylated phyA accumulated to normal or even higher levels compared with the WT but the phosphorylated phyA form was not detectable (Fig. 6A) (51), suggesting that the formation of the phosphorylated phyA form may be important for phyA to induce the FR light response. Third, our data indicated that nuclear localization and Pfr form formation are both required for the production of the phosphorylated phyA form, both of which are the well-known prerequisites for phyA to induce the FR light response. Collectively, our data substantiated the tight correlation between the abundance of phosphorylated phyA form and the extent of phyA response, suggesting that the phosphorylated phyA form may represent a more active form of phyA in vivo essential for inducing the FR light response.
Moreover, our data clearly showed that the abundance of phosphorylated phyA form was decreased in both phyAAAA and phyADDD lines; notably, phyAAAA lines, which exhibited greatest defect in phyA function, accumulated the least level of phosphorylated phyA form in vivo. Thus, it seems that the functional defects of phyAAAA and phyADDD could be explained well by the reduced levels of phosphorylated phyA form produced by these mutants in vivo in FR light. However, how many phosphorylation sites are involved in the production of this phosphorylated phyA form in vivo remains currently unknown. Because the phosphorylated phyA form seemed at least 5–10 kDa larger than the unphosphorylated form in immunoblots, it is likely that many phosphorylation sites on phyA contribute to the formation of the phosphorylated phyA form in vivo in FR light. The fact that both phyAAAA and phyADDD mutations caused reduced levels of the phosphorylated phyA form suggests that the hinge region may be involved in regulating the efficiency of phyA phosphorylation at many other sites indirectly.
In this study, our phyAAAA lines showed opposite phenotypes compared with transgenic lines overexpressing S599A oat phyA, which showed hypersensitivity to light (29). These contrasting observations can possibly be explained as follows. First, different experimental conditions were used in these two studies. For example, different promoters (i.e., the Arabidopsis PHYA promoter and the 35S promoter) were employed by this study and the previous study (29), respectively. Second, we hypothesize that phosphorylation and function of the hinge region may differ between oat and Arabidopsis phyA based on the following evidence. (i) It was shown that a single residue in the hinge region of oat phyA, S599, plays an important role in regulating the interaction of phyA with its signal transducers (29); however, S599 of oat phyA is not conserved and the hinge region of Arabidopsis phyA does not have a similarly critical single residue based on our data. (ii) Our previous study showed that the oat phyA (both WT and S599A) did not produce a slower-migrating band (corresponding to the phosphorylated form of Arabidopsis phyA) in Arabidopsis in FR light (figure S7 in ref. 50), indicating that the molecular mechanisms responsible for phosphorylating oat and Arabidopsis phyA may be different.
A recently developed mathematical model suggested that the dissociation rate of the phyA-FHY1/FHL nuclear import complex is a principal determinant for shifting the action peak of phyA from R to FR light (4). Our data indicated that the Pfr forms of both phyAAAA and phyADDD displayed stronger interaction with FHY1 and FHL in yeast cells (Fig. 4 A and B). Notably, phyAAAA, showing the strongest interaction with FHY1, exhibited the greatest defect in phyA function. Thus, increased interaction of phyAAAA and phyADDD with FHY1/FHL may lead to decreased dissociation rate of the phyA-FHY1/FHL complex, which may cause functional defects of phyAAAA and phyADDD in FR light based on the mathematical model. However, it remains obscure how the hinge region of phyA regulates its interaction with FHY1/FHL, and how the increased interaction with FHY1/FHL is finally channeled to the regulation of phyA phosphorylation. Evidently, the elucidation of the precise structural changes of photoconversion between the Pr and Pfr forms of plant phytochromes, and the identification of protein kinases responsible for phosphorylating phyA will help to answer the questions. Taking these data together, our study uncovers an important role of the hinge region of Arabidopsis phyA in regulating phyA phosphorylation and function, thus linking specific residues in the hinge region to the regulatory mechanisms of phyA phosphorylation.
Materials and Methods
The details and procedures of plant materials and growth conditions, plasmid construction, generation of transgenic Arabidopsis plants, yeast assays, real-time qRT-PCR, transcriptome analyses, nuclear-cytoplasmic fractionation, and immunoblotting are provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Peter Quail for the D153 vector, Dr. Christian Fankhauser for the pCF225 vector and the phyA-NLS-GFP phyA-211 seeds, and Dr. Andreas Hiltbrunner for the phyA(Y242H)-NLS-YFP phyA-211 seeds. This work was supported by National Natural Science Foundation of China Grants 31770321 and 31371221, Recruitment Program of Global Youth Experts of China, and US National Institutes of Health Grant GM47850.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1813162115/-/DCSupplemental.
References
- 1.Franklin KA, Quail PH. Phytochrome functions in Arabidopsis development. J Exp Bot. 2010;61:11–24. doi: 10.1093/jxb/erp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li J, Li G, Wang H, Deng XW. Phytochrome signaling mechanisms. Arabidopsis Book. 2011;9:e0148. doi: 10.1199/tab.0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharrock RA, Clack T. Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol. 2002;130:442–456. doi: 10.1104/pp.005389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rausenberger J, et al. Photoconversion and nuclear trafficking cycles determine phytochrome A’s response profile to far-red light. Cell. 2011;146:813–825. doi: 10.1016/j.cell.2011.07.023. [DOI] [PubMed] [Google Scholar]
- 5.Yang C, et al. Phytochrome A negatively regulates the shade avoidance response by increasing auxin/indole acidic acid protein stability. Dev Cell. 2018;44:29–41.e4. doi: 10.1016/j.devcel.2017.11.017. [DOI] [PubMed] [Google Scholar]
- 6.Bae G, Choi G. Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol. 2008;59:281–311. doi: 10.1146/annurev.arplant.59.032607.092859. [DOI] [PubMed] [Google Scholar]
- 7.Hiltbrunner A, et al. Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr Biol. 2005;15:2125–2130. doi: 10.1016/j.cub.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 8.Hiltbrunner A, et al. FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol. 2006;47:1023–1034. doi: 10.1093/pcp/pcj087. [DOI] [PubMed] [Google Scholar]
- 9.Genoud T, et al. FHY1 mediates nuclear import of the light-activated phytochrome A photoreceptor. PLoS Genet. 2008;4:e1000143. doi: 10.1371/journal.pgen.1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin R, et al. Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science. 2007;318:1302–1305. doi: 10.1126/science.1146281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koornneef M, Rolff E, Spruit CJP. Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z Pflanzenphysiol. 1980;100:147–160. [Google Scholar]
- 12.Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- 14.Ulm R, et al. Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA. 2004;101:1397–1402. doi: 10.1073/pnas.0308044100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J, et al. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell. 2007;19:731–749. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, et al. Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell. 2010;22:3634–3649. doi: 10.1105/tpc.110.075788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu D, et al. Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell. 2008;20:2307–2323. doi: 10.1105/tpc.107.056580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saijo Y, et al. The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 2003;17:2642–2647. doi: 10.1101/gad.1122903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheerin DJ, et al. Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell. 2015;27:189–201. doi: 10.1105/tpc.114.134775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McMichael RW, Jr, Lagarias JC. Phosphopeptide mapping of Avena phytochrome phosphorylated by protein kinases in vitro. Biochemistry. 1990;29:3872–3878. doi: 10.1021/bi00468a011. [DOI] [PubMed] [Google Scholar]
- 21.Lapko VN, Jiang XY, Smith DL, Song PS. Posttranslational modification of oat phytochrome A: Phosphorylation of a specific serine in a multiple serine cluster. Biochemistry. 1997;36:10595–10599. doi: 10.1021/bi970708z. [DOI] [PubMed] [Google Scholar]
- 22.Lapko VN, Jiang XY, Smith DL, Song PS. Mass spectrometric characterization of oat phytochrome A: Isoforms and posttranslational modifications. Protein Sci. 1999;8:1032–1044. doi: 10.1110/ps.8.5.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stockhaus J, et al. Serine-to-alanine substitutions at the amino-terminal region of phytochrome A result in an increase in biological activity. Genes Dev. 1992;6:2364–2372. doi: 10.1101/gad.6.12a.2364. [DOI] [PubMed] [Google Scholar]
- 24.Jordan ET, Marita JM, Clough RC, Vierstra RD. Characterization of regions within the N-terminal 6-kilodalton domain of phytochrome A that modulate its biological activity. Plant Physiol. 1997;115:693–704. doi: 10.1104/pp.115.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jordan ET, Cherry JR, Walker JM, Vierstra RD. The amino-terminus of phytochrome A contains two distinct functional domains. Plant J. 1996;9:243–257. doi: 10.1046/j.1365-313x.1996.09020243.x. [DOI] [PubMed] [Google Scholar]
- 26.Casal JJ, et al. The serine-rich N-terminal domain of oat phytochrome A helps regulate light responses and subnuclear localization of the photoreceptor. Plant Physiol. 2002;129:1127–1137. doi: 10.1104/pp.010977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han YJ, et al. Functional characterization of phytochrome autophosphorylation in plant light signaling. Plant Cell Physiol. 2010;51:596–609. doi: 10.1093/pcp/pcq025. [DOI] [PubMed] [Google Scholar]
- 28.Wong YS, Cheng HC, Walsh DA, Lagarias JC. Phosphorylation of Avena phytochrome in vitro as a probe of light-induced conformational changes. J Biol Chem. 1986;261:12089–12097. [PubMed] [Google Scholar]
- 29.Kim JI, et al. Phytochrome phosphorylation modulates light signaling by influencing the protein-protein interaction. Plant Cell. 2004;16:2629–2640. doi: 10.1105/tpc.104.023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Medzihradszky M, et al. Phosphorylation of phytochrome B inhibits light-induced signaling via accelerated dark reversion in Arabidopsis. Plant Cell. 2013;25:535–544. doi: 10.1105/tpc.112.106898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nito K, Wong CC, Yates JR, 3rd, Chory J. Tyrosine phosphorylation regulates the activity of phytochrome photoreceptors. Cell Rep. 2013;3:1970–1979. doi: 10.1016/j.celrep.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim DH, et al. A phytochrome-associated protein phosphatase 2A modulates light signals in flowering time control in Arabidopsis. Plant Cell. 2002;14:3043–3056. doi: 10.1105/tpc.005306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryu JS, et al. Phytochrome-specific type 5 phosphatase controls light signal flux by enhancing phytochrome stability and affinity for a signal transducer. Cell. 2005;120:395–406. doi: 10.1016/j.cell.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 34.Phee BK, et al. A novel protein phosphatase indirectly regulates phytochrome-interacting factor 3 via phytochrome. Biochem J. 2008;415:247–255. doi: 10.1042/BJ20071555. [DOI] [PubMed] [Google Scholar]
- 35.Yeh KC, Lagarias JC. Eukaryotic phytochromes: Light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci USA. 1998;95:13976–13981. doi: 10.1073/pnas.95.23.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin AY, et al. Evidence that phytochrome functions as a protein kinase in plant light signalling. Nat Commun. 2016;7:11545. doi: 10.1038/ncomms11545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong YS, McMichael RW, Lagarias JC. Properties of a polycation-stimulated protein kinase associated with purified Avena phytochrome. Plant Physiol. 1989;91:709–718. doi: 10.1104/pp.91.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahmad M, Jarillo JA, Smirnova O, Cashmore AR. The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol Cell. 1998;1:939–948. doi: 10.1016/s1097-2765(00)80094-5. [DOI] [PubMed] [Google Scholar]
- 39.Fankhauser C, et al. PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science. 1999;284:1539–1541. doi: 10.1126/science.284.5419.1539. [DOI] [PubMed] [Google Scholar]
- 40.Colón-Carmona A, Chen DL, Yeh KC, Abel S. Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol. 2000;124:1728–1738. doi: 10.1104/pp.124.4.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen Y, et al. Phytochrome A mediates rapid red light-induced phosphorylation of Arabidopsis FAR-RED ELONGATED HYPOCOTYL1 in a low fluence response. Plant Cell. 2009;21:494–506. doi: 10.1105/tpc.108.061259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 43.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mravec J, et al. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature. 2009;459:1136–1140. doi: 10.1038/nature08066. [DOI] [PubMed] [Google Scholar]
- 45.Shi Y, et al. Influence of EARLI1-like genes on flowering time and lignin synthesis of Arabidopsis thaliana. Plant Biol (Stuttg) 2011;13:731–739. doi: 10.1111/j.1438-8677.2010.00428.x. [DOI] [PubMed] [Google Scholar]
- 46.Hongo S, Sato K, Yokoyama R, Nishitani K. Demethylesterification of the primary wall by PECTIN METHYLESTERASE35 provides mechanical support to the Arabidopsis stem. Plant Cell. 2012;24:2624–2634. doi: 10.1105/tpc.112.099325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li F, et al. Identification of the wax ester synthase/acyl-coenzyme A: Diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in Arabidopsis. Plant Physiol. 2008;148:97–107. doi: 10.1104/pp.108.123471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. A light-switchable gene promoter system. Nat Biotechnol. 2002;20:1041–1044. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- 49.Shen Y, et al. Arabidopsis FHY1 protein stability is regulated by light via phytochrome A and 26S proteasome. Plant Physiol. 2005;139:1234–1243. doi: 10.1104/pp.105.067645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saijo Y, et al. Arabidopsis COP1/SPA1 complex and FHY1/FHY3 associate with distinct phosphorylated forms of phytochrome A in balancing light signaling. Mol Cell. 2008;31:607–613. doi: 10.1016/j.molcel.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang S, et al. TANDEM ZINC-FINGER/PLUS3 is a key component of phytochrome A signaling. Plant Cell. 2018;30:835–852. doi: 10.1105/tpc.17.00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu Y, Parks BM, Short TW, Quail PH. Missense mutations define a restricted segment in the C-terminal domain of phytochrome A critical to its regulatory activity. Plant Cell. 1995;7:1433–1443. doi: 10.1105/tpc.7.9.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fry RC, Habashi J, Okamoto H, Deng XW. Characterization of a strong dominant phytochrome A mutation unique to phytochrome A signal propagation. Plant Physiol. 2002;130:457–465. doi: 10.1104/pp.005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rösler J, Klein I, Zeidler M. Arabidopsis fhl/fhy1 double mutant reveals a distinct cytoplasmic action of phytochrome A. Proc Natl Acad Sci USA. 2007;104:10737–10742. doi: 10.1073/pnas.0703855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Su YS, Lagarias JC. Light-independent phytochrome signaling mediated by dominant GAF domain tyrosine mutants of Arabidopsis phytochromes in transgenic plants. Plant Cell. 2007;19:2124–2139. doi: 10.1105/tpc.107.051516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen M, Tao Y, Lim J, Shaw A, Chory J. Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr Biol. 2005;15:637–642. doi: 10.1016/j.cub.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 57.Park CM, Bhoo SH, Song PS. Inter-domain crosstalk in the phytochrome molecules. Semin Cell Dev Biol. 2000;11:449–456. doi: 10.1006/scdb.2000.0200. [DOI] [PubMed] [Google Scholar]
- 58.Chen F, et al. Phosphorylation of FAR-RED ELONGATED HYPOCOTYL1 is a key mechanism defining signaling dynamics of phytochrome A under red and far-red light in Arabidopsis. Plant Cell. 2012;24:1907–1920. doi: 10.1105/tpc.112.097733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang SW, Jang IC, Henriques R, Chua NH. FAR-RED ELONGATED HYPOCOTYL1 and FHY1-LIKE associate with the Arabidopsis transcription factors LAF1 and HFR1 to transmit phytochrome A signals for inhibition of hypocotyl elongation. Plant Cell. 2009;21:1341–1359. doi: 10.1105/tpc.109.067215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maloof JN, et al. Natural variation in light sensitivity of Arabidopsis. Nat Genet. 2001;29:441–446. doi: 10.1038/ng777. [DOI] [PubMed] [Google Scholar]
- 61.Wang Q, et al. The blue light-dependent phosphorylation of the CCE domain determines the photosensitivity of Arabidopsis CRY2. Mol Plant. 2015;8:631–643. doi: 10.1016/j.molp.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dephoure N, Gould KL, Gygi SP, Kellogg DR. Mapping and analysis of phosphorylation sites: A quick guide for cell biologists. Mol Biol Cell. 2013;24:535–542. doi: 10.1091/mbc.E12-09-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.