Abstract
Background:
Gliomas, a genetically heterogeneous group of primary central nervous system tumors, continue to pose a significant clinical challenge. Discovery of chromosomal rearrangements involving kinase genes has enabled precision therapy, and improved outcomes in several malignancies.
Experimental Design:
Positing that similar benefit could be accomplished for brain cancer patients, we evaluated The Cancer Genome Atlas (TCGA) glioblastoma dataset. Functional validation of the oncogenic potential and inhibitory sensitivity of discovered ROS1 fusions was performed using three independent cell-based model systems, and an in vivo murine xenograft study.
Results:
In silico analysis revealed previously unreported intrachromosomal 6q22 microdeletions that generate ROS1-fusions from TCGA glioblastoma dataset. ROS1 fusions in primary glioma and ependymoma were independently corroborated from MSK-IMPACT and Foundation Medicine clinical datasets. GOPC-ROS1 is a recurrent ROS1 fusion in primary CNS tumors. CEP85L-ROS1 and GOPC-ROS1 are transforming oncogenes in cells of astrocytic lineage, and amenable to pharmacological inhibition with several ROS1 inhibitors even when occurring concurrently with other cancer hotspot aberrations frequently associated with glioblastoma. Oral monotherapy with a potent brain-permeable ROS1 inhibitor, lorlatinib, significantly prolonged survival in an intracranially xenografted tumor model generated from a ROS1 fusion-positive GBM cell line.
Conclusions:
Our findings highlight that CNS tumors should be specifically interrogated for these rare intrachromosomal 6q22 microdeletions that generate actionable ROS1 fusions. ROS1 fusions in primary brain cancer may be amenable for clinical intervention with kinase inhibitors, and this holds the potential of novel treatment paradigms in these treatment-refractory cancer types, particularly in glioblastoma.
Keywords: Glioblastoma, ROS1, inhibitor, oncogene
Introduction
Gliomas are the most common central nervous system (CNS) tumors affecting 6.6 per 100,000 patients in the US (1) and include a heterogeneous group of neoplasms, including diffuse or anaplastic astrocytoma, oligodendroglioma, pilocytic astrocytoma, ependymoma, among several subtypes (2,3). Approximately half of the newly diagnosed gliomas are glioblastoma (GBM), a highly aggressive and infiltrative brain malignancy that is refractory to most standard of care treatment options. Despite multimodal therapeutic interventions, including combinations of surgery, chemotherapy and radiation, relative five year survival for GBM patients is about 5.5% (1). Pediatric (ages 0–19 years) and young adult (ages 20–44 years) patients fare slightly better, with 5-year survival rates of 16.8 and 19.1%, respectively (1). Ten-year survival outlook of patients harboring other types of gliomas varies depending on age at presentation, diffuse or anaplastic tumor characteristics, and genomic profile, with malignant CNS tumor continuing to have some of the worst survival statistics when compared to most other cancer types. Thus, there is a pressing, unmet need for effective, personalized treatments that will improve patient outcomes in malignant CNS tumor patients.
Surveys of clinically relevant oncogenic drivers have the potential to inform novel molecularly-targeted treatment strategies (4). Over the last decade, GBM tumors have been extensively profiled at the genomic and molecular level resulting from efforts initiated by investigators of The Cancer Genome Atlas (TCGA) project (5,6). The TCGA studies systematically catalogued genome alterations from a combined cohort of 506 glioblastoma samples and identified four distinct molecular subgroups: classical, mesenchymal, neural and pro-neural. Tumors from these molecular subgroups harbor unique patterns of somatic mutations and DNA copy number, including EGFR, NF1 and PDGFRA/IDH1 aberrations in classical, mesenchymal and proneural subtypes, respectively (6), and provide an informative framework for ongoing therapeutics development and risk stratification in GBM.
Chromosomal rearrangements that generate oncogenic kinase-fusion(s) are promising drug targets and selectively inhibiting them has led to unprecedented tumor responses in several malignancies (7–9). Rearrangements involving ROS1, an orphan receptor tyrosine kinase gene, was first described in a GBM cell line (U118MG) in 1987 (10), followed by further biochemical validation of this ROS1 fusion protein as a bonafide oncoprotein (11–13). More recently, various ROS1-fusions were identified in subsets of diverse pediatric and adult malignancies, including infantile fibrosarcoma, spitzoid melanoma, non-small cell lung cancer (NSCLC) and cholangiocarcinoma (14). We and others have shown dramatic clinical efficacy of ROS1 tyrosine kinase inhibitors (ROS1i) in ROS1-fusion expressing lung cancer patients (15–17), but their efficacy in ROS1-fusion positive brain tumors has not been examined to date due in part to lack of studies demonstrating the importance of ROS1 fusions in brain tumors and limitations in TKIs that can cross the blood-brain barrier (BBB).
While discovery of single nucleotide variants, insertions, deletions, and copy numbers variants can be reliably identified from massively parallel next-generation sequencing (NGS), discovery of gene fusions is more complicated and has only recently become efficient with the development of powerful in silico fusion-mining tools (7). Recently, the use of an algorithm called TX-Fuse, assisted in the discovery of recurrent FGFR1-TACC1 fusions in the TCGA glioblastoma patient cohort; the FGFR1-TACC1 fusions resulting from chromosomal microdeletions are particularly challenging to identify from large NGS data, and were not reported in the original TCGA publications (18). This suggests that a re-interrogation of the original TCGA datasets may still yield novel information about actionable drivers in gliomas.
Here, we reevaluated TCGA glioma (low-grade glioma and glioblastoma) genomic sequencing datasets with the hypothesis that a subset of patients may harbor chromosomal rearrangements of the ROS1 gene, and that identification and validation of ROS1 fusions may facilitate clinical implementation of effective, brain-permeable ROS1 kinase inhibitors in patients with these lethal tumors.
Methods
Isolation and quantitative reverse transcriptase PCR (qRT-PCR) of GOPC-ROS1 and CEP85L-ROS1 from primary GBM tumors.
Frozen brain tumor samples were obtained from Henry Ford Hospital (Detroit, MI) with written consent from patients under an approved Institutional Review Board protocol. Total RNA was isolated from two to five milligrams of frozen tumors using the RNeasy (Qiagen) system. Two micrograms of total RNA were used to generate complementary DNA (cDNA) using SuperScript™ VILO™ cDNA synthesis kit (ThermoFisher). Standard PCR was performed using AccuPrime™ Taq DNA polymerase with the following primers: CEP85L_ATG-Fwd-M13F (5’ atgtgggggcgcttcctg 3’) and ROS1_TERM-Rev (5’ ttaatcagacccatctccatatc 3’) for amplification of CEP85L-ROS1 fusion from TCGA-06–5418, and FIG_ATG-Fwd (5’ atgtcggcgggcggtccat 3’) with ROS1_TERM-Rev (5’ ttaatcagacccatctccatatc 3’) for amplification of GOPC-ROS1 from TCGA-12–5301 and TCGA-76–6192. Sanger sequencing of the PCR products was performed by using a series of internal sequencing primers. GOPC-ROS and CEP85L-ROS1 cDNAs were subcloned into pMSCV-IRES-GFP retroviral vectors in order to generate stable cell lines. To ascertain relative expression of ROS1-fusions we performed qRT-PCR for ROS1 kinase domain using the following primers: ROS1-Kin-qPCR-F1 (5’AAGAAGGGTTCCACAGACCAGG 3’) and ROS1-Kin-qPCR-R1 (5’ GCAGACAAACTCCAAGCTGCTT 3’). For qRT-PCR we used the PowerUp™ SYBR® Green Master Mix and the StepOnePlus™ Real-Time PCR System, both from Applied Biosystems and followed manufacturers protocol.
Cell culture, cell line generation and transformation assays
U118MG, Ba/F3, and NIH3T3 cells were purchased from American Type Culture Collection, ATCC, and cultured as recommended. SF-268 cells were procured from the National Cancer Institute (NCI-60 collection) via a Material Transfer Agreement. Briefly, for U118MG and Ba/F3 cells, complete medium (R10) contained RPMI medium 1640 with 10% (vol/vol) Fetal Bovine Serum, L-glutamine, penicillin/streptomycin), with additional supplementation with 2 ng/ml of recombinant murine interleukin-3 (Peprotech) in the case of Ba/F3 cells. Human astrocytes stably expressing human telomerase (hTERT), and the E6/E7 viral antigens (HA TERT/E6/E7) were a gift from Dr. Russel O’Pieper’s laboratory (UCSF) as reported in their previous publication (19). These modified human astrocytes were cultured in DMEM with 10% FBS, and sub-cultured using routine practice when 70% confluent. Replication incompetent, infectious ecotropic and amphotropic retroviral particles were generated using Platinum-E and Platinum-A cells (Cell Biolabs, Inc.), respectively. To generate stable Ba/F3 GOPC-ROS1 and CEP85L-ROS1 cell lines, Ba/F3 cells were maintained at a density between 0.5 × 106 to 1 × 106/mL, transduced with ecotropic retrovirus, selected and validated as previously described(20,21). The ecotropic retrovirus was also used to infect NIH3T3 cells, to generate the CEP85L-ROS1 cell line. Amphotropic retrovirus was used to generate stable, transduced HA TERT/E6/E7 GOPC-ROS1 or CEP85L-ROS1 astrocyte cell lines.
Oncogenic potential of the ROS1-fusions was assessed with a IL-3 independent growth assay (IL-3 withdrawal). Briefly, stable cell lines were washed with R10 medium to remove IL-3. Cells were counted every 2–4 days, and those that exhibited sustained outgrowth were considered transformed. Transformed Ba/F3 cells were expanded and used for inhibitor testing via dose-response proliferation assays. Anchorage-independent soft-agar assays using HA TERT/E6/E7 GOPC-ROS1 and CEP85L-ROS1 cell lines were done as described in Sonoda et al(19). Briefly, 10,000 cells in DMEM + 20% calf serum were mixed with 1.4% low melting temperate agarose to yield a final concentration of 0.7% of low melting. The agar layer with cells was laid on top of a pre-set 1.4 % low melting temperate agar layer that had been prepared in advance. Per cell line, a minimum of 6 wells were seeded. Colony growth for monitored over 4 weeks. At the end of the incubation period, number of colonies were imaging 6–8 random fields per well, and using Image J to quantify colony number.
Immunoblotting
Where indicated, cells were treated with tyrosine kinase inhibitor for 2 to 4 hours. Lysates were prepared from cells using a standard cell lysis buffer as described before (21). Protein quantitation of cleared cell lysates was performed using Pierce™ BCA Protein Assay Kit (ThermoFisher Scientific), and 25 μg of total protein was loaded on pre-cast 4–12% Criterion™ XT Bis-Tris Protein Gels (Bio-Rad, # 3450125). Proteins were transferred to nitrocellulose membranes, and probed with phospho-ROS1 [#3078, 1:1,000; Cell Signaling Technology (CST)], total ROS1 (#3266, 1:1,000; CST), phospho-SHP2 (#3751, 1:1000, CST), total SHP2 (#3397, 1:1000; CST), phospho-ERK1/2 (#9101, 1:1,000; CST), total ERK2 (sc-1647, 1:2,000; Santa Cruz, phospho-Akt (#4060, 1:1,000; CST), AKT (#610860, 1:1,000; BD Transduction Laboratories). Blots were imaged using either a LI-COR Odyssey imaging system or the Bio-Rad ChemiDoc imaging station according to the manufacturer’s protocol for immunoblot detection with use of infrared dye or horseradish peroxidase-conjugated secondary antibodies, respectively. Phospho-ROS1 detection required the SuperSignal™ West Femto Maximum Sensitivity Substrate (ThermoFisher Scientific).
U118MG spheroids generation and inhibitor testing
3D U118MG spheroids were generated using the hanging drop method. Specifically, 1000 U118MG cells in 35 μL volume per well were carefully seeded in the top chamber of a HDP1096 Perfecta3D® 96-Well plate (Sigma) to promote spheroid formation. For inhibitor treatment, cells were directly resuspended in R10 medium containing 50 nM of indicated inhibitor or in 0.05% DMSO (vehicle) and 35 μL of this cell suspension was seeded as above. The resulting spheroids were “dropped” into the recipient chamber after 54 hours of incubation by centrifugation, and calcein AM plus ethidium homodimer (using the LIVE/DEAD™ Viability/Cytotoxicity Kit, ThermoFisher Scientific, #L3224) were added to stain as well as quantify live (calcein am, green) and dead (ethidium homodimer, red). Stained spheroids were live-imaged using a Nikon/Yokogawa CSU-W1 spinning disk confocal microscope. Z-sections were reconstructed and green versus red events were quantified using Bitplate Imaris software. Images were compiled and exported using Image J and/or Zen software (Carl Zeiss, Inc.).
Intracranial U118MG xenograft and oral inhibitor administration.
In vivo efficacy studies were performed in accordance with federal standards and protocols approved by the Institutional Animal Care and Use Committee at Oregon Health and Science University. U118MG cells were transduced with a puromycin-resistant luciferase lentivirus (Cellomics, #PLV-10003–50). Transduced cells were selected with puromycin and expanded for implantation. In log-growth phase, 1 × 106 cells were injected into 8-week-old Nu/Nu male mice (Jackson Laboratories, athymic nude: Foxnlnu; Whn-; hairless). Briefly, mice were deeply anesthetized and a small cranial-caudal incision made just left of midline on the scalp. Using a 0.9 mm burr, the intracranial space was carefully accessed just anterior to bregma. A 10 μL pipet tip was inserted to a depth of 2 mm, and cells were slowly injected over 2 minutes. The hole was sealed with absorbable hemostatic cellulose and skin re-approximated with surgical glue. To confirm engraftment, mice were administered D-luciferin (75mg/kg; Promega) via intraperitoneal injection and imaged on a Xenogen IVIS2000 (Perkin-Elmer) 14 days after injection. Mice were then randomized to receive either lorlatinib (n=5, 30mg/kg in solution of Ethanol/PEG200/Water (10/40/50)) or vehicle alone (n=5, solution of Ethanol/PEG200/Water (10/40/50)) daily via oral gavage until death or euthanasia was required. Tumor growth was monitored with IVIS imaging weekly.
Results
ROS1 fusions are present in primary GBM tumor samples
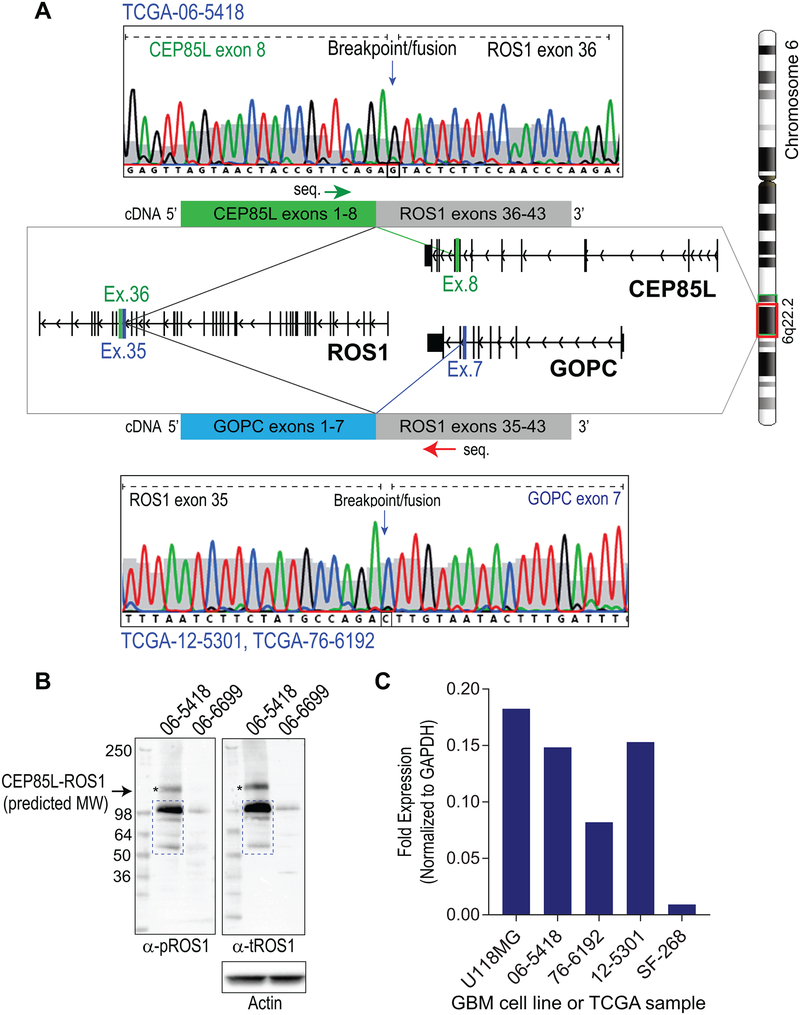
Shah et al. recently developed a bioinformatic algorithm that enables fusion finding from RNA-sequencing data by examining exons exhibiting outlier expression, and assessing if there is a 5’ – 3’ imbalance in expression (22), a potential signature for fusion transcripts. Using this concept, we examined TCGA glioblastoma databases from previously reported studies (5,24). Our analysis of copy number of ROS1 genomic regions concords with the in silico prediction that TCGA-06–5418 GBM patient sample harbors the CEP85L-ROS1 fusion (Supplementary Fig. S1). Interrogating TCGA data using Integrative Genomics Viewer (IGV) (25,26) revealed a characteristic pattern of loss of a chromosomal segment between CEP85L gene exon 8 and ROS1 gene exon 36 (Supplementary Fig. S2). We used this pattern as a hallmark for identifying samples harboring potential ROS1 fusions resulting from chromosomal microdeletion surrounding the ROS1 gene locus, and found two patient samples that we hypothesized harbor the GOPC-ROS1 fusion Supplementary Figs. S3 & S4). mRNA expression of predicted CEP85L-ROS1 (TGCA-06–5418) and GOPC-ROS1 (in TCGA-12–5301 & TCGA-76–6192) fusions was validated by reverse-transcriptase PCR (RT-PCR) reactions where the cDNA templates were synthesized from total RNA that had been extracted from fresh frozen tumor samples from these patients. Sanger sequencing of the RT-PCR reaction products confirmed the presence of CEP85L-ROS1 fusion in TCGA-06–5418 (CEP85L exons 1–8 fused in frame with ROS1 exons 36–43), and GOPC-ROS1 in TCGA-12–5301 and TCGA-76–6192 samples (GOPC exons 1–7 arranged in frame with ROS1 exons 35–43) (Fig. 1A). The cDNA sequence, and the predicted protein sequences for CEP85L-ROS1 (TCGA-06–5418) and GOPC-ROS1 (TCGA-12–5301 & TCGA-76–6192) are reported in supplemental data (Supplementary Figs. S6–9). CEP85L-ROS1 and GOPC-ROS1 result from small intrachromosomal deletion in 6q22.1 (illustrated in Supplementary Fig. S11B). The sample TCGA-06–6699 was also evaluated as it exhibited potential anomalies in the neighborhood of the ROS1 locus, but we were unable to ascertain the identity of the hypothesized ROS1 fusion (Supplementary Figs. S5). Similar interrogation of the low-grade glioma datasets revealed no aberrations in Ch. 6q22.2 region that would be indicative of ROS1 fusion genes.
Figure 1. Validation of GOPC-ROS1 and CEP85L-ROS1 mRNA and protein expression in GBM samples.
A. Sanger sequencing (chromatographs) of the CEP85L-ROS1 and GOPC-ROS1 cDNAs in glioblastoma tumor samples from the TCGA cohort. Green and red arrows indicated above fusion cDNA diagram indicate primer binding location and direction used for sequencing CEP85L-ROS1 and GOPC-ROS1, respectively. Both fusions are generated from intrachromosomal deletion resulting in fusion of exons as depicted in the illustration. B. Immunoblotting lysates generated from frozen TCGA-06–5418 and TCGA-06–6699 tumors shows expression of phosphorylated ROS1 protein. C. qRT-PCR analysis of TCGA samples as compared to established glioblastoma cell lines. Fold expression data for ROS1 are normalized to GAPDH.
Fluorescence in situ hybridization (FISH), a cytogenetic technique routinely used in diagnostic molecular pathology for detection of chromosomal abnormalities, was used to assess whether this standard pathology test could be used to reliably detect ROS1 rearrangement resulting from intrachromosomal deletions (27). To this end, we employed clinical diagnostic ROS1-breakapart FISH probes (Abbott Laboratories) on TCGA-12–5301, TCGA-06–5418, TCGA-06–6699 and TCGA-14–0789. The FISH assay showed that TCGA-06–5418 and TCGA-06–6699 samples harbor a chromosomal rearrangement consistent with an intrachromosomal deletion and translocation, respectively. The TCGA-14–0789 tissue section had extensive necrosis and quality of FISH data were poor, thus precluding confident interpretation by the cytogeneticist. The TCGA-12–5301 and TCGA-76–6192 samples were definitively negative for ROS1-rearrangement by FISH (Supplementary Fig. S11A, TCGA-12–5301 shown) despite being confirmed to express GOPC-ROS1 by Sanger sequencing (Fig. 1A). For TCGA-06–6699, we unsuccessfully attempted rapid amplification of 5’ complementary DNA ends (5’ RACE) assay as well as multiplex PCR using primers designed for known 5’ fusion gene partners; thus it is unclear if TCGA-06–6699 expresses a novel or known ROS1 bonafide fusion gene.
Sufficient fresh frozen tumor was available to biochemically assess protein expression of ROS1 fusions in TCGA-06–5418 and TCGA-06–6699. Immunoblotting with phospho-specific as well as total ROS1 antibodies shows immunoreactive protein migrating at the predicted molecular weight of CEP85L-ROS1 (110.3 kDa) in the TCGA-06–5418 sample, however there are also several lower molecular weight species detected that may be reflecting protein degradation in these archival samples (Fig. 1B, left lane). We were unable to readily identify the ROS1 fusion partner in the TCGA-06–6699 sample but immunoblotting reveals a band with apparent molecular weight that is consistent with several previously reported ROS1 fusion proteins (Fig. 1B, right lane). To assess the relative expression of ROS1 fusions in tumor samples, we performed qRT-PCR comparing the TCGA GBM samples to two established human GBM cell lines: U118MG that was previously shown to express GOPC-ROS1 (10,12,28) as a positive control, and SF-268, an EGFR A289V mutation-driven GBM cell line as a negative control (29). Fold expression data show that both TCGA-06–5418 and TCGA-12–5301 have ROS1 mRNA expression that is at 83% and 85% of that of U118MG, respectively (Fig. 1C). In comparison, TCGA-76–6192 expresses a lower amount of ROS1 mRNA suggesting a smaller subclonal population or other technical factors such as degraded RNA. ROS1 did not amplify from the negative control SF-268 cells, consistent with previous report by Jun et al. (30). Taken together these data suggest that ROS1 chromosomal rearrangements in glioblastoma tumors produce ROS1 fusion-proteins that are expressed at detectable levels.
ROS1 fusion expressing glioblastoma patients had an average overall survival of 83 days as compared to a 357 day median overall survival for all patients in the TCGA cohort (Supplementary Fig. S12). However, we are unable to use statistical methods to ascertain if ROS1-fusion expression affects the survival outcome due to the small sample size.
Validation of the occurrence and frequency ROS1-fusions in independent patient cohorts
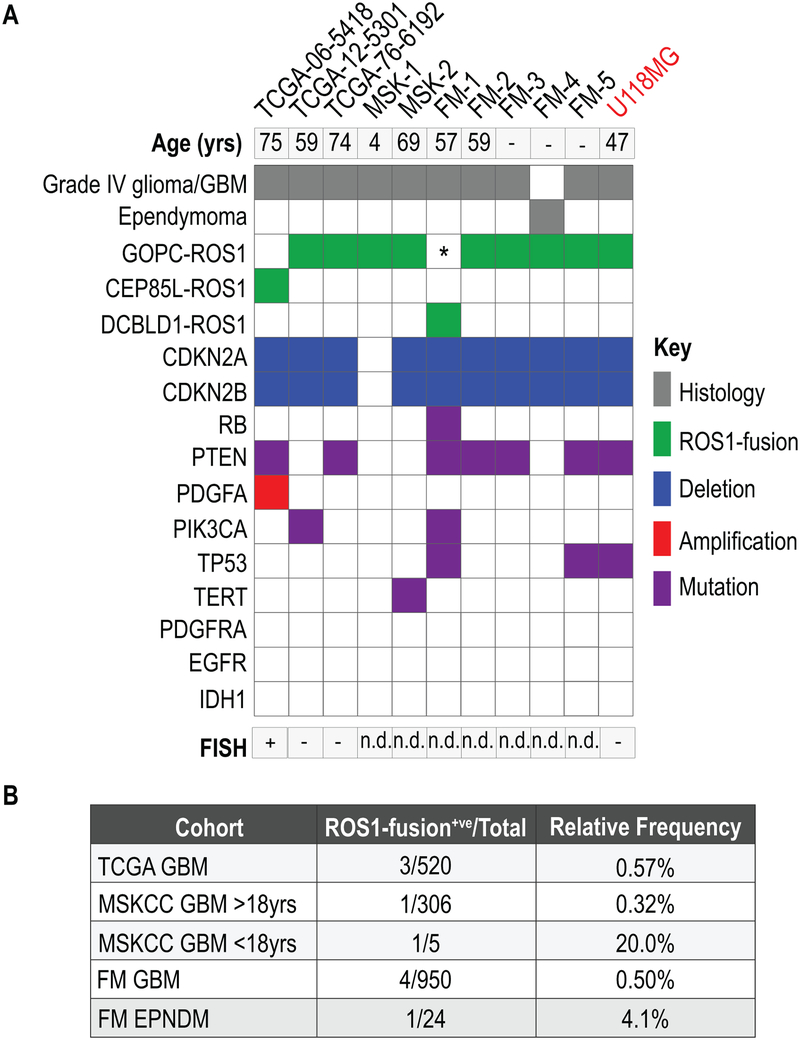
To validate these findings in independent GBM patient cohorts, and to better define the frequency of ROS1 fusions in primary brain tumors, we examined additional cancer genomic data. Specifically, we referred to the NGS data generated by the Memorial Sloan Kettering Cancer Center (MSKCC) MSK-IMPACT (31,32), and the Foundation Medicine™ genomic sequencing panels (33,34) that conducted massively parallel DNA sequencing of 341 and 287 cancer-related genes, respectively. Notably, both these panels achieve sequencing reads depths of about 500–1000X, and analyze exons as well as some introns that are hotspots for mutations or chromosomal rearrangements. In addition, MSKCC also analyzed selected GBM cases using an anchored multiplex PCR targeted RNAseq panel assay (35). These data evidenced additional 7 cases, 6 GBM (5 adult, 1 pediatric) and 1 ependymoma, corroborating that rare but recurrent ROS1 fusions are present in glial tumors as shown in the oncoprint (Fig. 2A, B, Supplementary Fig. S10). The relative frequency of ROS1-fusions in these sample sets is shown in Fig. 2B. The predominant fusion was GOPC-ROS1 (81%), and one case each of CEP85L- and DCBLD1-ROS1 fusions were also identified. Based on a previous case of DCBLD1-ROS1 that was first discovered by DNA capture sequencing (MSK-IMPACT) but turned out to produce a GOPC-ROS1 transcript by RNA-sequencing, here we conclude that DCBLD1-ROS1 discovered in the FM1 sample similarly will produce a GOPC-ROS1 transcript (personal communication, Dr. Marc Ladanyi, MSKCC). As we were unable to validate the identity and sequence of the putative ROS1-fusion transcript in TCGA-06–6699, this sample is excluded from the oncoprint and frequency analysis. The oncoprint (Fig. 2A) also shows the concurrent genomic aberrations in common cancer hot-spot genes. These include (i) homozygous deletion of cell cycle checkpoint proteins, CDKN2A and B (this is uniform in all adult patient, but absent in the only pediatric patient sample), (ii) mutations in PTEN and, (iii) mutations in TERT or TP53. The FM1 GBM sample had an activating mutation in PI3KCA, and a concurrent RB mutation in addition to CDKN2A/B loss (Fig. 2A). The TCGA-06–5418 sample also has concurrent chromosomal amplification of PDFGFA. The pediatric patient sample (MSK-1, 4 years old) had no concurrent aberrations in exons and selected introns of the 341 cancer-related genes on the MSK-IMPACT panel. Notably, ROS1 fusion expression was mutually exclusive of EGFR, PDGFRA and IDH1 aberrations that are recurrent in gliomas. The U118MG cell line (Fig. 2A) has a very similar genomic profile as the primary GBM tumors (36) that harbor GOPC-ROS1, including homozygous deletion of CDKN2A/B, and mutations in PTEN as well as TP53. Given the absence of any new ROS1-rearranged, patient-derived glioma cell lines or xenografts, U118MG that is a genomically comparable cell-based model of the GBM was used as a surrogate for conducting functional studies.
Figure 2. Validation of ROS1 fusion prevalence in GBM cases from independent genomic datasets.
A. Oncoprint plot illustrates ROS1 fusion in primary GBM and ependymoma samples from TCGA, MSK-IMPACT (MSK) and Foundation Medicine (FM) genomic sequencing datasets. Aberrations in cancer associated gene concurrent with ROS1-fusions are shown in various colors as indicated. Sample FM1 is indicated as having both DBCLD1-ROS1 and GOPC-ROS1 as we believe that this rearrangement generates a GOPC-ROS1 transcript due to genomic structure (see Results). The last lane shows the genomic profile of the established human GBM cell line, U118MG (indicated in red). B. Table shows relative frequency of ROS1 fusions in the indicated datasets. Clinical information pertaining to age is absent from most FM data. For the MSKCC MSK-IMPACT data, the adult and pediatric patients are shown as separate rows in the table.
GOPC-ROS1 and CEP85L-ROS1 are dominant, targetable oncogenic drivers
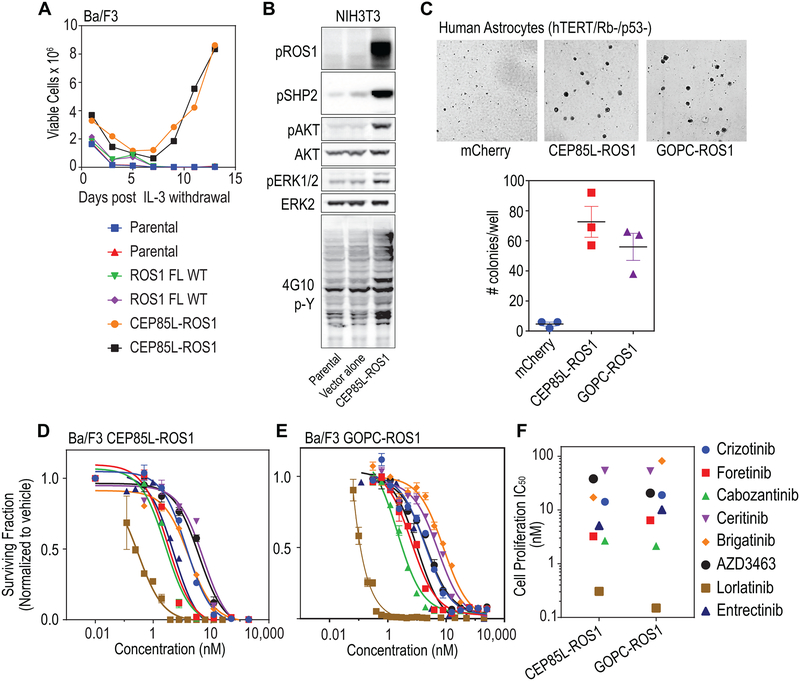
To test the oncogenic potential of the ROS1 fusions, we first used the Ba/F3 cytokine-independent transformation assay system (37). Ba/F3, murine pro-B lineage cells, undergo apoptotic cell death in the absence of interleukin-3. However, ectopic expression of oncogenic tyrosine kinases confers IL-3 independence and neoplastic transformation. CEP85L-ROS1 and GOPC-ROS1 expressing Ba/F3 cells proliferated in the absence of IL-3, whereas ectopic expression of wildtype ROS1 was insufficient to transform Ba/F3 cells (Fig. 3A). Further, expression of CEP85L-ROS1 in NIH3T3 murine fibroblasts robustly upregulates effector phosphorylation (phospho-SHP2 (pSHP2), phospho-AKT (pAKT), and phospho-ERK (pERK1/2)) downstream of ROS1 (Fig. 3B).
Figure 3. CEP95L-ROS1 and GOPC-ROS1 transform human astrocytes and respond to ROS1 kinase inhibitors.
A. Graph depicts data from an interleukin-3 (IL-3) withdrawal assay showing that ectopic expression of CEP85L-ROS1 & GOPC-ROS1 but not native full length ROS1 permits sustained outgrowth of Ba/F3 cells in the absence of IL-3. Parental indicates untransduced Ba/F3 cells (negative control). B. Immunoblot analysis shows ectopic expression of CEP85L-ROS1 upregulates phospho-tyrosine signaling (4G10, generic p-Tyr antibody), as well as canonical ROS1- effector pathway signaling (phosphorylation of SHP2 (pSHP2), AKT (pAKT), ERK1/2 (pERK1/2) in NIH3T3 murine fibroblasts. C. Soft-agar colony forming assay data shows that expression of CEP85L-ROS1 and GOPC-ROS1 confers neoplastic properties to human astrocytes with deficiency in TP53 and RB. Upper panel: representative images; lower panel: quantification of number of colonies. D. Dose response proliferation assay of Ba/F3 CEP85L-ROS1 and, (E) GOPC-ROS1 cells after 72 hour exposure to crizotinib, foretinib, cabozantinib, ceritinib, brigatinib, AZD3463, lorlatinib and entrectinib. Data are normalized to vehicle-treated control, and values shown are the mean ± SEM. (F) Scatter plot of cell proliferation IC50 values for each TKI against Ba/F3 cells. Colored symbols represent different inhibitors as shown on the right.
Next, we tested whether the fusions could transform immortalized human astrocytes (HA TERT) that carry concurrent TP53 and RB functional deficiency (19). We demonstrate that introduction of GOPC-ROS1 and CEP85L-ROS1 expression in cells of human astrocytic lineage enhances the ability of these cells to form anchorage-independent colonies, a hallmark of neoplastic transformation (Fig. 3C). To determine relative efficacy of tyrosine kinase inhibitors (TKIs) with previously reported activity against ROS1 kinase, we performed dose-response cell viability assays using oncogene-addicted, transformed Ba/F3 GOPC-ROS1 and CEP85L-ROS1 cell lines. Cell-based 50% inhibitor concentration (IC50) (scatter plot, Fig. 3F and Supplementary Fig. S13) were derived from dose-response curves (Fig. 3D & E) for CEP85L-ROS1 and GOPC-ROS1. These data demonstrate that multiple tyrosine kinase inhibitors are effective against CEP85L-ROS1 and GOPC-ROS1 (12,13,20,28,38).
Pharmacological inhibition with tyrosine kinase inhibitors blocks autophosphorylation of ROS1, downstream effector protein activation and 3D spheroid formation in the U118MG-GBM model
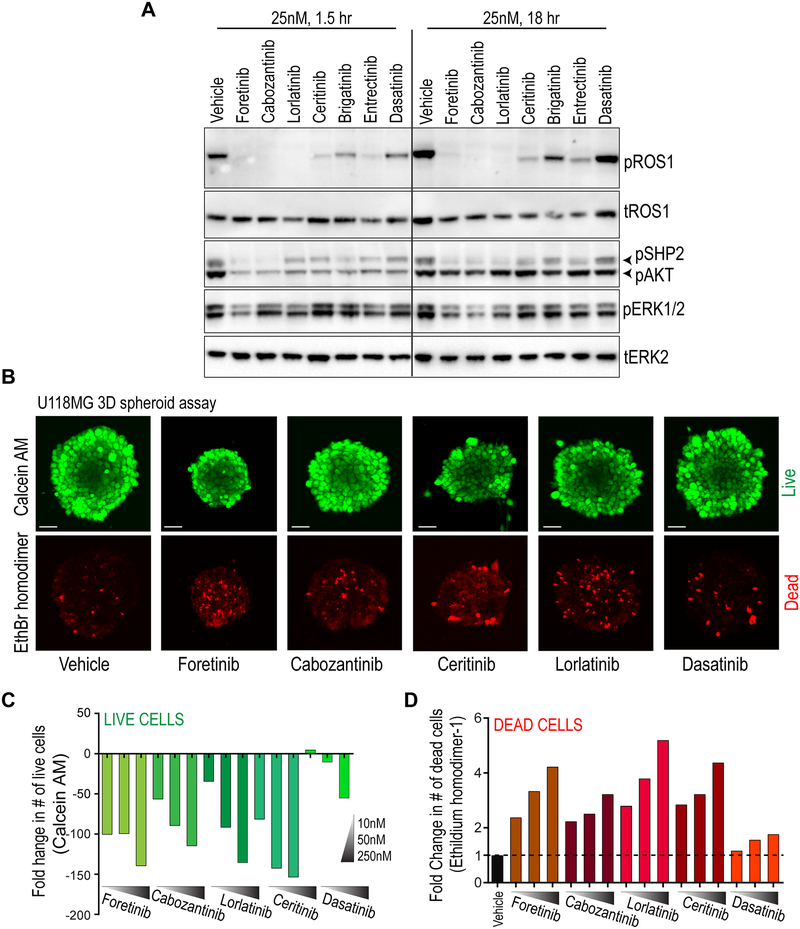
Using the U118MG cell line as a surrogate for human ROS1-fusion positive GBM, we tested the on-target inhibition of GOPC-ROS1 after treatment with six potentially brain-permeable ROS1 kinase inhibitors for 1 and 18 hours. The small molecules exhibited varying degrees of inhibition, with near complete inhibition of ROS1 autophosphorylation accomplished by cabozantinib (XL-184), foretinib (XL-880), lorlatinib (PF-06463922), and entrectinib (RXDX-101) (Fig. 4A). The extent of phospho-inhibition of signaling effectors that are downstream of ROS1, including SHP2, AKT and ERK1/2 kinase phosphorylation corresponded to level of ROS1 catalytic inhibition in these cells. The multi-kinase inhibitor dasatinib was used as a selectivity control, and did not exhibit substantial activity against ROS1.
Figure 4. ROS1 kinase inhibitors suppress catalytic activity, effector phosphorylation, and cell viability in GOPC-ROS1 harboring human glioblastoma cells.
A. Immunoblot analysis of phospho-ROS1 (pROS1), total ROS1 (tROS1), phospho-SHP2 (pSHP2), phospho-AKT (pAKT), phospho-ERK1/2 (pERK1/2) and total ERK2 (tERK2) from U118MG cell lysates generated after treatment with inhibitors (indicated, 25 nM) for 1.5 or 18 hours. B. Suppression of U118MG spheroid growth and increase in cell death after treatment with foretinib, cabozantinib, ceritinib, lorlatinib and dasatinib for 48 hours, as indicated. Top: Calcein-AM staining shows viable cells within hanging drop spheroids (pseudocolored green), and ethidium bromide (EthBr homodimer) staining shows dead cells (pseudocolored red). C. Quantification of live cell numbers (green bars) from spheroids after Z-stack confocal microscopy after treatment with 10, 50 and 250 nM for indicated kinase inhibitor for 48 hours. Data are depicted as fold-change relative to vehicle (DMSO) treated cells. D. Quantification of dead cell numbers (red bars) from spheroids after Z-stack confocal microscopy after treatment with 10, 50 and 250 nM for indicated kinase inhibitor for 48 hours. Data are depicted as fold-change relative to vehicle (DMSO) treated cells.
Given demonstrable potency of these small molecules to inhibit ROS1 autophosphorylation, we tested their efficacy to block U118MG growth in dose-response cell viability assays. Given the robust biochemical inhibition of ROS1 phosphorylation (Fig. 4A), the U118MG cells were unexpectedly resistant to ROS1 inhibitors (Supplementary Fig. S14). We considered these possibilities: (a) U118MG cells are not dependent on ROS1 signaling for cell growth, or (b) when investigated in two-dimensional (2D) culture, ROS1 fusion positive cells experience substantial compensatory signaling from cell adhesion to treated plastic matrix thus enabling bypass survival signaling. Initial dose response experiments were done in 2D culture using standard, tissue-culture treated polystyrene microplates. Tumor cells depend more extensively on cell-cell contact and the native microenvironment for survival and growth in situ; both of these aspects are lost in standard 2D culture. We experimentally tested the latter hypothesis by comparing ROS1i efficacy in dose-response assay with U118MG cells either grown in 2D on standard, cell culture grade plastic dishes or grown as suspended clusters in ultra-low attachment dishes. U118MG cells cultured in low attachment dishes grew as suspended cell clusters, and this culture setting did not intrinsically inhibit their viability or proliferation. Notably, under these conditions the cells responded in a dose-dependent manner to lorlatinib, cabozantinib, foretinib, and entrectinib (Supplementary Fig. S15). In contrast, cells grown at the same time on 2D were nearly completely resistant to these same inhibitors.
To further validate this hypothesis, we tested the efficacy of ROS1i to block U118MG cell growth using a three-dimensional spheroid assay. Specifically, the cells were grown in hanging drop culture plates that promoted the formation of three-dimensional cell spheroids which to large degree recapitulates the cyto-architecture and cell-cell interactions as they occur in situ. The spheroids were smaller in diameter, and contained a larger proportion of dead cells when treated with ROS1i (foretinib, cabozantinib, ceritinib and lorlatinib as compared to dasatinib or vehicle treatment (Fig. 4B, C & D). Compilation of Z-stack images collected using confocal microscope reveal dose-dependent increase in number of dead cells (pseudocolored red) in lorlatinib-treated U118MG GBM spheroids (Movies S1–4).
Monotherapy with brain permeable ROS1i, lorlatinib reduces tumor burden and prolongs survival in the U118MG GOPC-ROS1 driven orthotopic xenograft GBM model
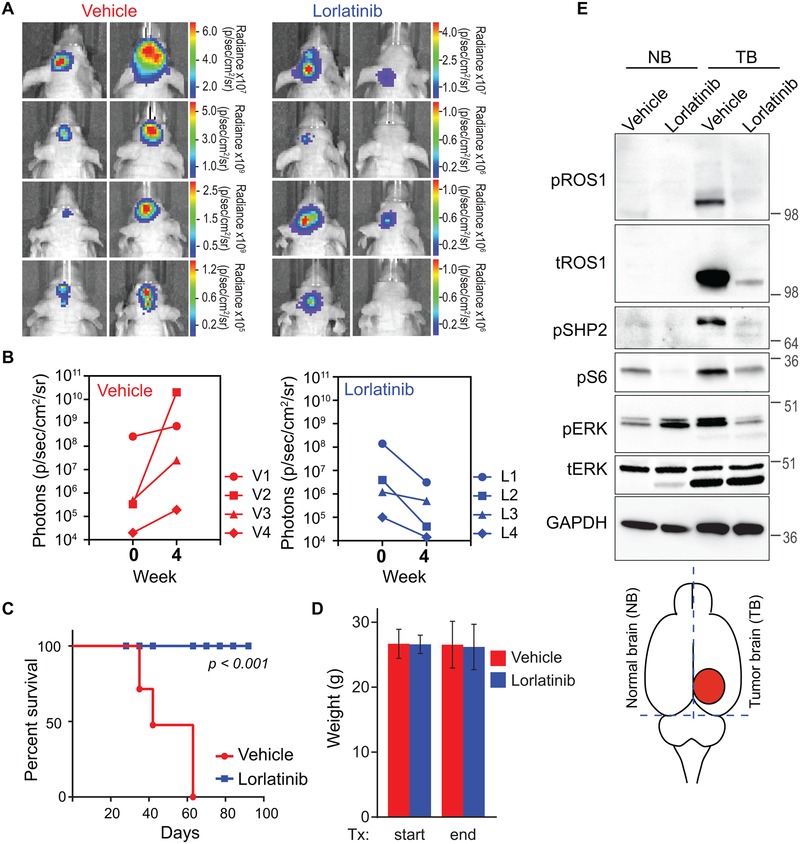
Entrectinib, cabozantinib, foretinib and lorlatinib are all attractive candidates for inhibiting ROS1-fusion driven GBM. Here, lorlatinib was selected for in vivo studies as it exhibited highest level of potency in Ba/F3 cells, and in the U118MG spheroid model. Additionally, having been specifically optimized for improved CNS availability, it has been shown to have favorable plasma:brain ratio (38,39). It should be noted that Zou et al. demonstrated efficacy of lorlatinib to reduce tumor burden in ROS1-fusion driven glioma using a genetically engineered mouse model of the disease (38). However, the survival benefit conferred by ROS1 inhibition in human cell origin GBM has not been previously reported. ROS1-positive adult GBM samples have several concurrent genomic aberrations (Fig. 2A), including mutations in TP53, TERT, PI3KCA, PTEN, RB, amplifications of other RTKs, and/or homozygous loss of CDKN2A/B that may contribute to primary resistance in vivo. To evaluate if there is survival benefit of ROS1-TKI in human ROS1 fusion-expressing GBM, we investigated the efficacy of oral monotherapy with lorlatinib in an orthotopic intracranial xenograft model of the disease. U118MG cells were engineered to express luciferase for bioluminescent monitoring of tumor engraftment, growth and response to inhibitor. Intracranial U118MG implantation resulted in tumor formation in all mice that were injected. These tumors were not infiltrative as native human glioblastoma tumors are, but similar to xenograft tumors formed by other glioblastoma cell lines such as U87 (Supplementary Fig. S17). After engraftment, the mice were divided into two cohorts with equivalent representation of starting tumor volume. Lorlatinib (30 mg/kg) was administered by oral gavage, once daily for 4 weeks. Since lorlatinib has been recently interrogated in vivo for ALK and ROS1-driven tumors, we based the dose of lorlatinib for oral gavage on these studies (38,40). Regardless of the starting tumor size, all lorlatinib-treated mice exhibited decrease in luciferase signal (Fig. 5A, B), and importantly, survived for a significantly longer period (p < 0.001) (Fig. 5C). Even after lorlatinib was halted on day 28 of treatment, the mice exhibited no signs of disease or weight loss for another 36 days after we stopped the treatment (Fig. 5D). We terminated the experiment after 92 days.
Figure 5. Oral monotherapy with lorlatinib decreases tumor burden and prolongs survival in an intracranial U118MG GBM xenograft model.
A. Bioluminescence imaging of the U118MG-xenografted tumors pre- and post-four weeks of Vehicle or lorlatinib treatment (30 mg/kg by oral gavage, once daily). Firefly luciferase-labeled U118MG GBM cells had been implanted into forebrain of NOD-scid mice four weeks prior to starting treatment. B. Photon emission as surrogate readout for tumor volume at start of treatment (week 0) and end of treatment (week 4) in Vehicle treated (left graph) and lorlatinib-treated (right graph) mice. C. Vehicle or lorlatinib-treated mouse weights at start and end of treatment period. D. Kaplan Meier survival curve shows statistically significant difference in survivability of vehicle versus lorlatinib-treated U118MG tumor bearing mice. p<0.001 by Anova. E. Immunoblots from lysate prepared from mice treated as indicated. The illustration at the bottom indicates rough dissection lines used to harvest tissue to create normal brain (NB), and tumor brain (TB) as denoted in image. Lysates were interrogated with antibodies are shown in western blot panels, and dilutions described in Methods.
To confirm on-target activity of lorlatinib in U118MG xenografted tumors, we treated mice with either vehicle or lorlatinib (30 mg/kg) by oral gavage, and harvested tissue after four hours. The cerebral hemisphere contralateral to the one bearing the tumor was used as an internal normal brain control. Immunoblotting with indicated antibodies was performed using protein lysates prepared from treated tissue. These data (Fig. 5E) show near complete inhibition of intratumoral ROS1 catalytic activity after oral lorlatinib treatment, as well as concomitant inhibition of downstream effector pathways, including suppression of phospho-SHP2, phospho-ERK1/2 and phospho-S6. We also observe degradation of total GOPC-ROS1; these data are consistent with a recent report (40). The mechanism of this inhibitor-induced downregulation of total ROS1 is currently unknown and will be clarified in future studies. In summary, these data confirm that oral lorlatinib treatment accomplishes intratumoral inhibition of GOPC-ROS1 as well as its downstream effector pathways in intracranial tumors.
Discussion
ROS1 mRNA expression in tumors that originate in the central nervous system (e.g., astrocytomas, meningiomas, and glioblastoma), has been reported by multiple groups over the last twenty-five years (10,11,41,42). In 2003, Charest et al. discovered the native GOPC-ROS1 fusion gene in U118MG GBM cells, and went on to show that in cooperation with loss of p16Ink4a and p19Arf (murine orthologues of CDKN2A &B), this ROS1-fusion drives formation of glioblastoma in murine models (12,13,28). Since then, there have been multiple inconsistent reports regarding the presence or absence of ROS1-fusions in GBM. These disparate results confound the utility of screening for ROS1 fusions in GBM or other primary CNS cancer patients. These inconsistencies are a result of poorly interpreted experimental results, challenges and limitations associated with mining for fusion genes in NGS data or due to the specific design of the break-apart FISH probes used. First, Das et al. (43) report that 78% of glioblastoma patients they interrogated (15 of 19) expressed the GOPC-ROS1, and these tumors co-expressed phosphorylated ALK and MET. This conclusion was based solely on an immuno-blot that shows a ROS1 antibody-reactive protein band of about 110kDa protein in 15 out of the 19 samples tested. They do not show any additional data that validates this using independent techniques (e.g., Sanger-sequencing, NGS, immunohistochemistry), thereby weakening the conclusions. Second, Lim et al.(44) reported that ROS1 gene rearrangements are absent in GBM. In this study, 109 GBM patient samples were interrogated with FISH break-apart probes, and IHC with a commercial ROS1 antibody. Due to the rarity of ROS1 fusions in GBM (0.5–1% in adults), it is feasible that a sample size of 109 was not large enough for discovery. However, a technical caveat pertaining to the ROS1 FISH break-apart probes may also have resulted in negative findings. GOPC-ROS1 is generated from an intrachromosomal microdeletion of ~250 kbps between the GOPC and ROS1 genes on 6q22. Many commercially available ROS1-breakapart probes do not detect the GOPC-ROS1 fusion because the 5’ ROS1 probe overlaps with the GOPC gene that is 134kbp upstream of ROS1. Indeed, in our study, the GBM TCGA-12–5301 patient sample that definitively expresses GOPC-ROS1 fusion, as validated by Sanger sequencing, was FISH-negative for ROS1 fusion presence when analyzed by the clinical cytogenetics lab using two different commercial FISH probes. This has been previously observed and reported by Suehera et al. (45) in lung cancer.
Since ROS1 fusions in primary CNS tumors are generated from intrachromosomal deletion, careful selection of FISH probes or alternate sequencing methods are required to ensure their detection. Notably, even with NGS, not all computational algorithms or bioinformatics tools make high confidence calls for all fusion genes. For example, GOPC-ROS1 and CEP85L-ROS1 fusions that we discovered by re-examining the TCGA GBM sequencing data have not been reported in landmark TCGA papers (5,24). Further, the Iavarone group reanalyzed TCGA GBM data to discover FGFR receptor fusions that were also missed (18). Given these previous contradictory data pertaining to ROS1-fusions in GBM, our findings now firmly establish the occurrence and the frequency of ROS1-fusions in GBM. We examined independent patient cohorts, and by combining the TCGA, MSKCC and Foundation Medicine™ CNS tumor datasets, propose that the frequency of ROS1 fusions in adult GBM patients is likely to be between 0.5–1%. We also show that pediatric GBM (1 of 5; MSK-IMPACT) and ependymoma patients (1 of 24; Foundation Medicine) also harbor ROS1-fusions, however our sample size for this age group (<18 years) and histology (ependymoma) were too small to definitively report frequency.
To date, twenty-six different 5’ ROS1-fusion partners resulting from various rearrangement mechanisms have been described in multiple malignancies, including NSCLC, cholangiocarcinoma, inflammatory myofibroblastic tumors, spitzoid melanoma, and others (7). Given the relative promiscuity of the ROS1 gene rearrangement that results in these diverse fusions, it is striking that all ROS1-fusions identified in primary CNS tumor samples result from intrachromosomal 6q22 microdeletions. These data are strongly suggestive of susceptibility of that locus or a selection pressure for this type of rearrangement in cells of astroglial or CNS origin. The molecular mechanisms governing this remain unknown. An exception to this is ZCCHC8-ROS1, a novel fusion gene recently identified in a case of congenital GBM, a rare malignancy that occurs in <5% of all tumors in the pediatric population (46). This ROS1-fusion arose from a reciprocal t(6;12)(q21;q24.3) chromosomal translocation. A recent publication showed that GOPC-ROS1 and CEP85L-ROS1 fusion proteins are also present in pediatric low-grade glioma and diffuse astrocytoma (47), confirming our finding of GOPC-ROS1 in a pediatric GBM patient. Taken together, these data suggest that ROS1 fusions may be playing an oncogenic role in subsets of adult glioblastoma, and a more diverse set of pediatric low- and high-grade CNS tumors of glial or ependymal origin.
The activity and oncogenic potential of the CEP85L-ROS1 has not been functionally evaluated previously. We showed that both GOPC-ROS1 and CEP85L-ROS1 are oncogenic kinases in independent model systems examined, including in immortalized human astrocytes. Notably, multiple ROS1 inhibitors potently block catalytic activity, effector phosphorylation, and cell viability in ROS1-fusion driven model systems. In our experiments, we find that established ROS1-fusion expressing cancer cell lines (GBM: U118MG & NSCLC: HCC78 (data not shown), are resistant to ROS1-TKI when assayed in traditional 2D culture systems (treated plastic). Performing dose-response studies of these cells cultured in 3D, or in vivo, using murine tumor models concurs with immunoblotting data, and shows robust inhibitory efficacy of several ROS1 inhibitors (Fig. 4 & 5). Future experiments are warranted to elucidate the mechanisms underlying this intriguing phenotypic discrepancy between cells cultured on plastic versus in 3D. The tyrosine phosphatase, SHP2, is an important effector of ROS1, and its activity is tightly coupled to that of ROS1 kinase function in onco-addicted cells (7,14,20). However, SHP2 is also a negative regulator of focal adhesion kinase (FAK) that is activated predominantly by cell adhesion (48). Therefore, a possible hypothesis to explain this phenotype is that downregulation of SHP2 activity via inhibition of ROS1 may augment the FAK-SRC signaling axis, and promote ROS1i-resistance.
Chromosomal rearrangements that generate ROS1 kinase-fusions are now established as dominant oncogenes in lung adenocarcinoma (14). Clinical trial data show that oral monotherapy with ROS1i in ROS1-fusion driven NSCLC offers significant benefit (3,16), and US Food and Drug Administration (FDA) approved crizotinib for front-line treatment in ROS1-fusion positive, metastatic NSCLC patients. Broadly, these data suggest that detection and pharmacological targeting of ROS1-fusions driven tumors may improve outcomes in ROS1-positive patients bearing cancers of other histologies as well. The expanding pharmacopeia of small molecule ROS1 inhibitors now includes multiple potent, brain-permeable inhibitors, including those advancing through clinical trials (e.g., lorlatinib, entrectinib, and ceritinib). Shaw et al. reported Phase I dose-escalation data showing that oral lorlatinib treatment achieves both systemic and intracranial activity in ALK- and ROS1-positive NSCLC patients, 72% of whom had CNS metastasis (49). Similarly, entrectinib treatment has led to complete CNS response in a patient with metastatic lung cancer (3). Thus, reaching effective CNS concentrations due to blood brain barrier permeability issues is no longer a clinical constraint that would minimize the translational relevance of our findings. Consequently, we propose that the detection of ROS1-fusions in CNS cancer patients may facilitate clinical investigation of ROS1-TKI as a realistic therapeutic modality for improving outcomes.
GBM continues to pose a significant clinical challenge, and while one-year relative survival rates have improved from 4.4% (1999–2000) to 37% in 2016, the five- and ten-year statistics remain dismal, with relative survival estimated to be 4.3% and 2.0%, respectively (50). Innovations in clinical trial design, and the emphasis on biomarker-based selection has paved the way for therapeutic innovations in rare diseases, and in cancers that were once thought to be untenable due to small patient numbers. Multi-armed, targeted trials such as the NCI- Molecular Analysis for Therapy Choice (MATCH), and NCI-COG-MATCH (pediatric), rely on identifying recurring functionally relevant genetic events that can be targeted with small molecules. Our findings suggest that the identification ROS1-fusions will facilitate precision oncology strategies that may demonstrably improve outcomes in this subset of adult and pediatric GBM patients. Future clinical exploration of this hypothesis is warranted.
Supplementary Material
Statement of Translational Relevance.
Actionable ROS1-fusions generated from intrachromosomal 6q22 microdeletions are rare but recurrent in a subset of glioma, and may go undetected using certain types of sequencing or FISH analyses; immediate implementation of specific clinical screening for them may have a measurable impact on disease management.
Acknowledgments:
We would like to thank Rebecca L. Smith (Knight Cancer Institute) for her help with molecular cloning of ROS1-fusions, Dr. Nicolle E. Hofmann for her assistance with U118MG 3D culture, and Madison Wise Nakamoto in the Dr. Jim Olson’s laboratory at the Fred Hutchinson Cancer Research Center for teaching us efficient intracranial orthotopic xenograft techniques.
Funding: This study was supported in part by funding from the Howard Hughes Medical Institute (Brian J. Druker) and Hyundai Hope on Wheels Hope grant (Monika A. Davare)
Footnotes
Conflict(s) of interest: None
Competing interests: None.
Data and materials availability: The TCGA GBM genomic data are in the public domain, and can be accessed via multiple portals, including the NCI Genomic Data Commons, and the cBIO Cancer Genomics Portal (51,52). MSK-IMPACT data are also publicly available at http://cbioportal.org/msk-impact. Foundation Medicine genomic data are available in Hartmaier et al.(34) All cell lines reported here will be shared by OHSU via MTA.
References
- 1.Ostrom QT, Gittleman H, Xu J, Kromer C, Wolinsky Y, Kruchko C, et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2009–2013. Neuro-Oncology 2016;18:v1–v75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoang-Xuan K, Idbaih A. [Advances in molecular genetics and treatment of gliomas]. Bull Acad Natl Med 2011;195:11–20; discussion −1 [PubMed] [Google Scholar]
- 3.Drilon A, Siena S, Ou SI, Patel M, Ahn MJ, Lee J, et al. Safety and Antitumor Activity of the Multitargeted Pan-TRK, ROS1, and ALK Inhibitor Entrectinib: Combined Results from Two Phase I Trials (ALKA-372–001 and STARTRK-1). Cancer Discov 2017;7:400–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pagliarini R, Shao W, Sellers WR. Oncogene addiction: pathways of therapeutic response, resistance, and road maps toward a cure. EMBO Rep 2015;16:280–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455:1061–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010;17:98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davare MA, Tognon CE. Detecting and targetting oncogenic fusion proteins in the genomic era. Biol Cell 2015;107:111–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Druker BJ. Translation of the Philadelphia chromosome into therapy for CML. Blood 2008;112:4808–17 [DOI] [PubMed] [Google Scholar]
- 9.Shaw AT, Hsu PP, Awad MM, Engelman JA. Tyrosine kinase gene rearrangements in epithelial malignancies. Nat Rev Cancer 2013;13:772–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birchmeier C, Sharma S, Wigler M. Expression and rearrangement of the ROS1 gene in human glioblastoma cells. Proc Natl Acad Sci U S A 1987;84:9270–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birchmeier C, O’Neill K, Riggs M, Wigler M. Characterization of ROS1 cDNA from a human glioblastoma cell line. Proc Natl Acad Sci U S A 1990;87:4799–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charest A, Lane K, McMahon K, Park J, Preisinger E, Conroy H, et al. Fusion of FIG to the receptor tyrosine kinase ROS in a glioblastoma with an interstitial del(6)(q21q21). Genes Chromosomes Cancer 2003;37:58–71 [DOI] [PubMed] [Google Scholar]
- 13.Charest A, Wilker EW, McLaughlin ME, Lane K, Gowda R, Coven S, et al. ROS fusion tyrosine kinase activates a SH2 domain-containing phosphatase-2/phosphatidylinositol 3-kinase/mammalian target of rapamycin signaling axis to form glioblastoma in mice. Cancer Res 2006;66:7473–81 [DOI] [PubMed] [Google Scholar]
- 14.Davies KD, Doebele RC. Molecular pathways: ROS1 fusion proteins in cancer. Clin Cancer Res 2013;19:4040–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drilon A, Somwar R, Wagner JP, Vellore NA, Eide CA, Zabriskie MS, et al. A Novel Crizotinib-Resistant Solvent-Front Mutation Responsive to Cabozantinib Therapy in a Patient with ROS1-Rearranged Lung Cancer. Clin Cancer Res 2016;22:2351–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaw AT, Ou SH, Bang YJ, Camidge DR, Solomon BJ, Salgia R, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med 2014;371:1963–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaw AT, Solomon BJ. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med 2015;372:683–4 [DOI] [PubMed] [Google Scholar]
- 18.Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 2012;337:1231–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sonoda Y, Ozawa T, Hirose Y, Aldape KD, McMahon M, Berger MS, et al. Formation of intracranial tumors by genetically modified human astrocytes defines four pathways critical in the development of human anaplastic astrocytoma. Cancer Res 2001;61:4956–60 [PubMed] [Google Scholar]
- 20.Davare MA, Saborowski A, Eide CA, Tognon C, Smith RL, Elferich J, et al. Foretinib is a potent inhibitor of oncogenic ROS1 fusion proteins. Proc Natl Acad Sci U S A 2013;110:19519–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davare MA, Vellore NA, Wagner JP, Eide CA, Goodman JR, Drilon A, et al. Structural insight into selectivity and resistance profiles of ROS1 tyrosine kinase inhibitors. Proc Natl Acad Sci U S A 2015;112:E5381–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shah N, Lankerovich M, Lee H, Yoon JG, Schroeder B, Foltz G. Exploration of the gene fusion landscape of glioblastoma using transcriptome sequencing and copy number data. BMC Genomics 2013;14:818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell 2013;155:462–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 2013;14:178–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol 2011;29:24–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Speicher MR, Carter NP. The new cytogenetics: blurring the boundaries with molecular biology. Nat Rev Genet 2005;6:782–92 [DOI] [PubMed] [Google Scholar]
- 28.Charest A, Kheifets V, Park J, Lane K, McMahon K, Nutt CL, et al. Oncogenic targeting of an activated tyrosine kinase to the Golgi apparatus in a glioblastoma. Proc Natl Acad Sci U S A 2003;100:916–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vivanco I, Robins HI, Rohle D, Campos C, Grommes C, Nghiemphu PL, et al. Differential sensitivity of glioma- versus lung cancer-specific EGFR mutations to EGFR kinase inhibitors. Cancer Discov 2012;2:458–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jun HJ, Woolfenden S, Coven S, Lane K, Bronson R, Housman D, et al. Epigenetic regulation of c-ROS receptor tyrosine kinase expression in malignant gliomas. Cancer Res 2009;69:2180–4 [DOI] [PubMed] [Google Scholar]
- 31.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23:703–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 2015;17:251–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013;31:1023–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartmaier RJ, Albacker LA, Chmielecki J, Bailey M, He J, Goldberg ME, et al. High-Throughput Genomic Profiling of Adult Solid Tumors Reveals Novel Insights into Cancer Pathogenesis. Cancer Res 2017;77:2464–75 [DOI] [PubMed] [Google Scholar]
- 35.Wang L, Busam KJ, Benayed R, Cimera R, Wang J, Denley R, et al. Identification of NTRK3 Fusions in Childhood Melanocytic Neoplasms. J Mol Diagn 2017;19:387–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishii N, Maier D, Merlo A, Tada M, Sawamura Y, Diserens AC, et al. Frequent co-alterations of TP53, p16/CDKN2A, p14ARF, PTEN tumor suppressor genes in human glioma cell lines. Brain Pathol 1999;9:469–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warmuth M, Kim S, Gu XJ, Xia G, Adrian F. Ba/F3 cells and their use in kinase drug discovery. Curr Opin Oncol 2007;19:55–60 [DOI] [PubMed] [Google Scholar]
- 38.Zou HY, Li Q, Engstrom LD, West M, Appleman V, Wong KA, et al. PF-06463922 is a potent and selective next-generation ROS1/ALK inhibitor capable of blocking crizotinib-resistant ROS1 mutations. Proc Natl Acad Sci U S A 2015;112:3493–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson TW, Richardson PF, Bailey S, Brooun A, Burke BJ, Collins MR, et al. Discovery of (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(m etheno)pyrazolo[4,3-h][2,5,11]-benzoxadiazacyclotetradecine-3-carbonitrile (PF-06463922), a macrocyclic inhibitor of anaplastic lymphoma kinase (ALK) and c-ros oncogene 1 (ROS1) with preclinical brain exposure and broad-spectrum potency against ALK-resistant mutations. J Med Chem 2014;57:4720–44 [DOI] [PubMed] [Google Scholar]
- 40.Zou HY, Friboulet L, Kodack DP, Engstrom LD, Li Q, West M, et al. PF-06463922, an ALK/ROS1 Inhibitor, Overcomes Resistance to First and Second Generation ALK Inhibitors in Preclinical Models. Cancer Cell 2015;28:70–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma S, Birchmeier C, Nikawa J, O’Neill K, Rodgers L, Wigler M. Characterization of the ros1-gene products expressed in human glioblastoma cell lines. Oncogene Res 1989;5:91–100 [PubMed] [Google Scholar]
- 42.Wu JK, Chikaraishi DM. Differential expression of ros oncogene in primary human astrocytomas and astrocytoma cell lines. Cancer Res 1990;50:3032–5 [PubMed] [Google Scholar]
- 43.Das A, Cheng RR, Hilbert ML, Dixon-Moh YN, Decandio M, Vandergrift WA 3rd, et al. Synergistic Effects of Crizotinib and Temozolomide in Experimental FIG-ROS1 Fusion-Positive Glioblastoma. Cancer Growth Metastasis 2015;8:51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lim SM, Choi J, Chang JH, Sohn J, Jacobson K, Policht F, et al. Lack of ROS1 Gene Rearrangement in Glioblastoma Multiforme. PLoS One 2015;10:e0137678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suehara Y, Arcila M, Wang L, Hasanovic A, Ang D, Ito T, et al. Identification of KIF5B-RET and GOPC-ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA-based screen for tyrosine kinase fusions. Clin Cancer Res 2012;18:6599–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cocce MC, Mardin BR, Bens S, Stutz AM, Lubieniecki F, Vater I, et al. Identification of ZCCHC8 as fusion partner of ROS1 in a case of congenital glioblastoma multiforme with a t(6;12)(q21;q24.3). Genes Chromosomes Cancer 2016;55:677–87 [DOI] [PubMed] [Google Scholar]
- 47.Johnson A, Severson E, Gay L, Vergilio JA, Elvin J, Suh J, et al. Comprehensive Genomic Profiling of 282 Pediatric Low- and High-Grade Gliomas Reveals Genomic Drivers, Tumor Mutational Burden, and Hypermutation Signatures. Oncologist 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mitra SK, Schlaepfer DD. Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr Opin Cell Biol 2006;18:516–23 [DOI] [PubMed] [Google Scholar]
- 49.Shaw AT, Felip E, Bauer TM, Besse B, Navarro A, Postel-Vinay S, et al. Lorlatinib in non-small-cell lung cancer with ALK or ROS1 rearrangement: an international, multicentre, open-label, single-arm first-in-man phase 1 trial. Lancet Oncol 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ostrom QT, Gittleman H, Xu J, Kromer C, Wolinsky Y, Kruchko C, et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2009–2013. Neuro Oncol 2016;18:v1–v75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.