Abstract
The intracellular effects and overall efficacies of anticancer therapies can vary significantly by tumor type. To identify patterns of drug-induced gene modulation that occur in different cancer cell types, we measured gene expression changes across the NCI-60 cell line panel after exposure to 15 anticancer agents. The results were integrated into a combined database and set of interactive analysis tools, designated the NCI Transcriptional Pharmacodynamics Workbench (NCI TPW), that allows exploration of gene expression modulation by molecular pathway, drug target, and association with drug sensitivity. We identified common transcriptional responses across agents and cell types and uncovered gene expression changes associated with drug sensitivity. We also demonstrated the value of this tool for investigating clinically-relevant molecular hypotheses and identifying candidate biomarkers of drug activity. The NCI TPW, publicly available at https://tpwb.nci.nih.gov, provides a comprehensive resource to facilitate understanding of tumor cell characteristics that define sensitivity to commonly used anticancer drugs.
Keywords: transcriptional biomarkers, cancer cell gene expression, therapeutic resistance, chemotherapy, molecularly targeted therapy
Introduction
A better understanding of gene expression changes that occur in response to anticancer agents in genetically heterogeneous tumor cell lines may facilitate prediction of patient response and resistance, and help guide the development of effective clinical combination regimens. We therefore set out to define the drug-induced gene expression profiles of a diverse set of well-characterized cancer cell lines, the NCI-60 panel (1), in response to 15 cytotoxic and targeted anticancer agents (Table 1). In addition to evaluating drug-induced transcriptional changes, we aimed to use computational and visualization tools to make the large datasets generated available to and searchable by the larger research community through the development of the comprehensive NCI Transcriptional Pharmacodynamics Workbench (NCI TPW) interactive web resource (https://tpwb.nci.nih.gov). The NCI TPW integrates cell line sensitivity data for 15 clinically relevant anticancer agents with both basal transcript levels and agent-induced transcriptional changes for 12,704 genes at three time points across the NCI-60 panel. The NCI TPW further relates this information to exon mutation, protein expression, and multidrug resistance data, and enables query of these data in the context of cellular pathways and receptors.
Table 1.
Drugs used in transcriptional response profiling of NCI-60 cell lines for generation of the NCI TPW.
| Drug (NSC) | Mechanism/Target | Cmax (μM)(13) |
NCI-60 mean GI50 (μM)(14) |
Concentration (μM) |
|
|---|---|---|---|---|---|
| Low | High | ||||
| Bortezomib (681239) |
proteasome inhibitor | 0.312 | 0.00051 | 0.01 | 0.1 |
| Sirolimus (226080) |
mTOR inhibitor | 0.016 | 0.050 | 0.01 | 0.1 |
| Geldanamycin† (122750) |
Hsp90 inhibitor | ND | 0.1 | 0.1 | 1 |
| Doxorubicin (123127) |
reactive oxygen species generator, topoisomerase 2 inhibitor | 6.73 | 0.097 | 0.1 | 1 |
| Gemcitabine (613327) |
DNA synthesis inhibitor (chain terminator) | 89.3 | 0.24 | 0.2 | 2 |
| Paclitaxel (125973) |
microtubule stabilizer | 4.27 | 0.025 | 0.01 | 0.1 |
| Dasatinib (732517) |
tyrosine kinase inhibitor | 0.264 | 0.33 | 0.1 | 2 |
| Azacytidine (102816) |
DNA alkyltransferase inhibitor | 3.07 | 0.95 | 1 | 5 |
| Vorinostat (701852) |
HDAC inhibitor | 1.2 | 0.94 | 1 | 5 |
| Sunitinib (750690) |
tyrosine kinase inhibitor | 0.181 | 2.2 | 0.2 | 2 |
| Lapatinib (745750) |
tyrosine kinase inhibitor | 4.18 | 2.9 | 1 | 10 |
| Sorafenib (747971) |
tyrosine kinase inhibitor | 20.1 | 1.9 | 5 | 10 |
| Topotecan (609699) |
topoisomerase 1 inhibitor | 0.15 | 0.031 | 0.01 | 1 |
| Erlotinib (718781) |
kinase inhibitor (EGFR) | 3.15 | 5.5 | 1 | 10 |
| Cisplatin (119875) |
intrastrand DNA crosslinker | 14.4 | 1.4 | 3 | 15 |
ND: not determined
not an FDA-approved agent
The unique NCI TPW dataset complements other publicly available resources, such as the pre-treatment molecular information and pharmacologic response in hundreds of cell lines available from Cancer Cell Line Encyclopedia and the Genomics of Drug Sensitivity in Cancer datasets (2,3), as well as the post-treatment transcriptional response data from the NIH Library of Integrated Network-based Cellular Signatures (LINCS) project. The LINCS project has collected transcriptional data for over 25,000 agents but features separate, independent components that focus on transcriptional response and drug sensitivity, among other areas, with each subproject using a different set of cell lines (4,5); only a small number of core cancer cell lines (< 10) have been tested across all agents, with additional cell lines (many of them non-cancer) used for specific agents. Furthermore, LINCS relies on expression measurements of 978 “landmark” genes, from which levels of all other transcripts have been extrapolated (6), and the bulk of the LINCS transcriptomics dataset is not currently linked to drug sensitivity data (5,7). Compared to these existing resources, the NCI TPW is unique in providing the most detailed systematic compilation of directly measured longitudinal transcriptional responses in a thoroughly characterized panel of cancer cell lines (the NCI-60) to multiple agents at predefined time points.
The range of analysis tools available in the NCI TPW exceeds that for previous genomic data websites, allowing users to analyze transcriptional response data in the context of molecular pathways and mutation spectra and to directly connect this information to cell growth inhibition of clinically important antitumor agents. Workbench users can generate time course plots to visualize expression changes for genes of interest, as well as heatmaps of drug-perturbed genes across the NCI-60 panel based on signaling pathways, interactions with a specific transcription factor, or receptor type. The NCI TPW has recently been applied to the characterization of temporal expression changes in genes involved in DNA methylation (8), glycosylation (9), and endoplasmic reticulum (ER) stress response (Min 2018, in preparation). Here, we describe construction of the database and datasets used to validate it, and then detail use of the NCI TPW to uncover gene expression changes associated with drug sensitivity in 13 curated cell signaling pathways and to gain mechanistic insights that may enable early preclinical evaluation of specific potential combination therapies. For example, analysis of NCI TPW data revealed cell lines in which exposure to the epigenetic agent vorinostat resulted in sustained loss of BRCA1, BRCA2, and RAD51 expression, changes that confer homologous recombination deficiency and susceptibility to PARP inhibition and therefore suggest potential value in vorinostat-PARP inhibitor combination therapies. In contrast, the opposing effects on expression of the early growth response 1 (EGR1) gene induced by the nucleoside analog gemcitabine and the epidermal growth factor receptor (EGFR) inhibitor erlotinib suggested a potential antagonistic relationship between these two agents, which was supported by further cell culture studies and may explain the lack of additional clinical benefit from combinations of gemcitabine and EGFR inhibitors (10-12).
This database and the accompanying set of interactive tools will empower biologists and pharmacologists to generate novel hypotheses and contribute to ongoing wider systems biology efforts evaluating how transcriptional response influences drug sensitivity, ultimately supporting subsequent mechanistic studies.
Materials and Methods
Cell lines, drug treatment, RNA extraction, and sensitivity measurements:
NCI-60 cell lines were obtained from the NCI Developmental Therapeutics Program Tumor Repository (https://dtp.cancer.gov/discovery_development/nci-60/cell_list.htm). Each lot of cells was authenticated through a variety of molecular characterizations, and identity was confirmed from frozen stock of each cell line using Identifiler DNA profiling. Each cell line was tested for mycoplasma when it was accepted into the repository and before being frozen as a working seed stock from which cell lines were limited to 20 passages of growth. Furthermore, randomly selected cultures were tested approximately every 6 months using the MycoAlert Mycoplasma Detection Kit (Lonza, Basel, Switzerland). Testing for drug sensitivity followed NCI-60 drug screening protocols and was completed on the same mixture of cells used for transcriptional profiling. For each cell line, the logGI50 values for bortezomib in the NCI TPW were inferred from a single experiment for each screening concentration of that agent. The logGI50 values for all other agents in NCI-TPW were computed as an average of two logGI50 values derived from two replicate experiments for each cell line.
Authentication results and screening protocols are available at: https://dtp.cancer.gov/discovery_development/nci-60/characterization.htm. For RNA collection, cell lines were inoculated onto 96-well plates at densities of 7,500 – 30,000 cells/well 24 hours prior to addition of drugs. The cells were exposed to 15 anticancer agents (formulated in 0.25% DMSO) at a ‘low’ and ‘high’ concentration, approximating the clinical Cmax (13) and log10(GI50) (14) (Table 1). Total RNA was collected from the untreated control and drug-treated cells at 2, 6, and 24 hours using the Qiagen (Germantown, MD) RNeasy mini kit per manufacturer’s protocol and immediately frozen at −80oC.
Expression profiling on Affymetrix HTA array plates U133A:
RNA quality was determined on a Caliper LabChip GX (Hopkinton, MA) in 96-well format (distinct rRNA band and RQS > 7.5); only high-quality total RNA samples were used. Samples (96-well format, 1 μg each) were labeled using the Affymetrix (Cleveland, OH) HT One-Cycle Eukaryotic Labeling Kit on GeneChip Array Station following the manufacturer’s suggested protocols. Biotin-labeled antisense cRNA (4 μg per sample) was hybridized to Affymetrix GeneChip HT Human Genome U133A 96-Array plates overnight. Plates were washed and stained on a GeneChip Array Station, then scanned on an Affymetrix HT array plate scanner. Data were collected using the Affymetrix GCOS software. Initial data quality control was performed using percent present calls generated by Affymetrix Quality Reporter software. Gene expression data were deposited in the Gene Expression Omnibus (GEO) under accession number GSE116436.
The NCI Transcriptional Pharmacodynamics Workbench:
Tools in NCI TPW include time course graph representation of gene expression, heat map analysis of candidate gene expression, analysis of manually curated genes from selected cancer pathways and receptor lists, and correlation of the gene expression changes with drug sensitivity (log10[GI50]), cell line multidrug resistance status, cell doubling time, and the presence of certain pathogenic mutations. The NCI TPW defines gene expression fold change as the difference in log2 expression between the drug-exposed and corresponding zero concentration within a cell line at each time point. Raw CEL files downloaded from caARRAY were background-subtracted and normalized using the Robust Multi-array Average (RMA) algorithm (15) for all cell lines treated by the same drug. The data containing 22,227 probe sets on Affymetrix U133A were then summarized into 12,704 genes by taking the average of log2 measurements of probe sets for each gene. The data matrices for NCI-60 lines treated with the 15 drugs at different time points and different concentrations, including baseline experiments (zero concentration), were stored in a backend SQLite database. The NCI TPW system operates under the CentOS (version 6.5). The web client interface pages are written in html, css, and JavaScript. Data analysis and graphic display are performed using R packages (version 2.15.0). CGIwithR, which permits the straightforward use of R as a CGI scripting language, is used to facilitate processing of information from web-based forms and reporting of results in the html through the CGI.
High throughput qRT-PCR:
Fluidigm BioMark™ System was used according to the manufacturer’s instructions. High-quality total RNA (500 ng) (RIN > 7 on the Agilent Bioanalyzer) was reverse-transcribed using Invitrogen’s High Capacity cDNA Reverse Transcription Kits. The cDNAs were pre-amplified with a pool of 32 pairs of gene-specific primers for 14 cycles, followed by 40 cycles of real-time qPCR quantification in triplicates on 96.96 dynamic arrays using the Fluidigm gene expression protocol. Real-time PCR and data collection were done using the BioMark system, and data were analyzed with Fluidigm’s Real-Time PCR Analysis software. Affymetrix U133A array results were validated with measurements of the expression of 28 selected genes after treatment with 6 drugs for 24 hours in 24–60 cell lines (Supplementary Table S1).
Confirmation of Genes Using TaqMan qRT-PCR:
Quantitative real-time reverse transcriptase-PCR reactions were monitored using the ABI StepOne Plus and TaqMan Chemistries (Applied Biosystems, Foster City, CA [now Thermo Fisher Scientific, Waltham, MA]). One microgram of total RNA was reverse transcribed in a 20 μL reaction using the ABI High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems); resulting cDNA was stored at −80oC until required. PCR reactions consisted of 5 ng of cDNA, forward and reverse primers for the genes of interest and/or the endogenous control GAPDH (QIAGEN predesigned assays), and TaqMan SYBR Green PCR Master Mix (Applied Biosystems) in 20 μL reactions. Triplicate wells for each sample were analyzed using the comparative Ct method (ABI user bulletin #2), and measurements were expressed as an increase or decrease in relative expression (log2) compared with the untreated control. Commercial primers/probes were purchased from Applied Biosystems.
Identification of genes with consistent expression changes among antitumor agents:
Consistent transcriptional changes of the 12,704 genes were defined as those in which expression of the majority of NCI-60 lines changed in the same direction for that gene, and ≤ 15 cell lines (i.e., ≤ 25% of cell lines) had a change in the opposite direction. Previously, we and others have found that gene signatures identified using this criterion of expression were in good concordance with drug response data that used alternative methods of gene ranking based on the strength of expression response (8,16,17). After identifying genes with concerted expression changes across the lines in individual experiments, we investigated which genes had a transcriptional response to all 15 drugs. Separate analyses were performed at 2, 6, and 24 hours after treatment.
Analyses of most sensitive and least sensitive cell line cohorts:
Genes included in the 13 curated cell signaling pathways are listed in Supplementary Table S2. For heatmaps, average fold change values were calculated for each gene in each pathway across the 10 most sensitive lines and the 10 least sensitive lines, respectively, in response to each of the 15 antitumor agents (high concentration). Mean fold change for all genes in each specific pathway represents the overall expression of that pathway. At each time point, analysis was performed separately for most sensitive and least sensitive cohorts. Heatmaps were generated using the “heatmap.2” package in R. Drugs and genes were aligned in all heatmaps. For correlation analyses of log(IC50) versus log2(fold change) for genes potentially mediating drug insensitivity, p-values were adjusted for multiple testing using the Benjamini and Hochberg method (18).
Analysis of EGR1-mediated antagonism between gemcitabine and erlotinib:
EGR1 protein levels following treatment with gemcitabine or erlotinib were assessed by Western blot using a Novex minigel system with precast 4–20% gels, anti-EGR1 monoclonal antibody (clone 15F7, Cell Signaling Technology), and chemiluminescent HRP visualization. For additivity analyses, cells were incubated with various concentrations of gemcitabine (0.6–2 mM) and erlotinib (0.02–1.25 mM) for 72 hours. Cells were then treated with 50% trichloroacetic acid and stained with Sulforhodamine B, and optical densities were measured at 515 nm. Antagonism was determined by the MacSynergy II model based on Bliss additivity (19,20); 3D response surface plot values, indicating the percent inhibition above or below expected at each set of concentrations, were calculated at the 95% confidence interval.
Assessment of vorinostat-induced HR deficiency and talazoparib sensitization:
T-47D, NCI-H460, UO31, UACC257, and SK-OV-3 cells were inoculated into 96-well plates in 100 μL media and incubated at 37ºC and 5% CO2 for 24 hours prior to addition of vorinostat. Drugs were added to produce a final DMSO concentration of 0.25% in 200 μl. For analysis of BRCA1, BRCA2, and RAD1 expression, cells were exposed to vorinostat (5 μM) for 24 hours, then the media was replaced with vehicle, and cells were cultured for an additional 72 hours. BRCA1 and RAD51 proteins were measured by Western analysis using a Novex minigel system with precast 4–20% gels and chemiluminescent HRP visualization. Anti-RAD51 and anti-BRCA1 antibodies were purchased from Cell Signaling Technologies (Danvers, MA; catalog numbers 8875 and 9010, respectively). For additivity analysis, cells were incubated with vorinostat alone (0.3125–10 μM) for 24 hours, after which the culture medium was replaced with fresh medium containing talazoparib alone (3.125–50 nM), and cells were cultured for an additional 96 hours. Cells were fixed with 50% trichloroacetic acid and stained with Sulforhodamine B, and optical densities were measured at 515 nm. Dose responses and log10(GI50) were calculated, and synergism was determined by the MacSynergy II model based on Bliss additivity (19,20) as described above.
Results
Generation of the NCI TPW web tool
NCI-60 cell lines were exposed to 15 anticancer agents for 2, 6, and 24 hours at two different concentrations that, where possible, spanned the clinical Cmax and mean 50% inhibitory concentration (GI50) for the cells (Table 1). Sensitivity data (log10[GI50]) for each cell line and each drug, measured concurrently with the gene expression changes, can be found at https://tpwb.nci.nih.gov/GeneExpressionNCI60/GI50.html. Changes in gene expression were measured using Affymetrix GeneChip HT Human Genome U133A microarrays, and drug-induced gene expression was compared to basal expression (measured in untreated control cultures) at the same time points to generate positive and negative log2 fold expression change values for each gene in the context of each cell line, drug, and time point; these data form the basis of the NCI TPW.
The NCI TPW online resource (https://tpwb.nci.nih.gov) contains extensive search and display capabilities for this database, allowing users to view the data based on a specific tumor histology, agent of interest, signaling pathway, or user-specified gene list. Options for visualization include: (1) single-gene response to low or high concentration of each agent at 2, 6, and 24 hours, with the ability to stratify data by cancer type; (2) correlation analyses, including gene expression changes most correlated with drug sensitivity, baseline expression of a gene of interest, or the presence of a specific mutation identified by exome sequencing; (3) heatmap or time course graph of the genes most highly modulated by each treatment; and (4) drug-induced effects on expression of a user-defined list of genes. Additionally, users can create a heatmap of drug-perturbed genes based on any one of 65 different signaling pathways, interaction with 121 specific transcription factors, or 55 receptor types; these gene lists were validated using a query signature in the Broad Connectivity Map (17) and the LINCS database (16). Example outputs are displayed in Figure 1A-C.
Figure 1.
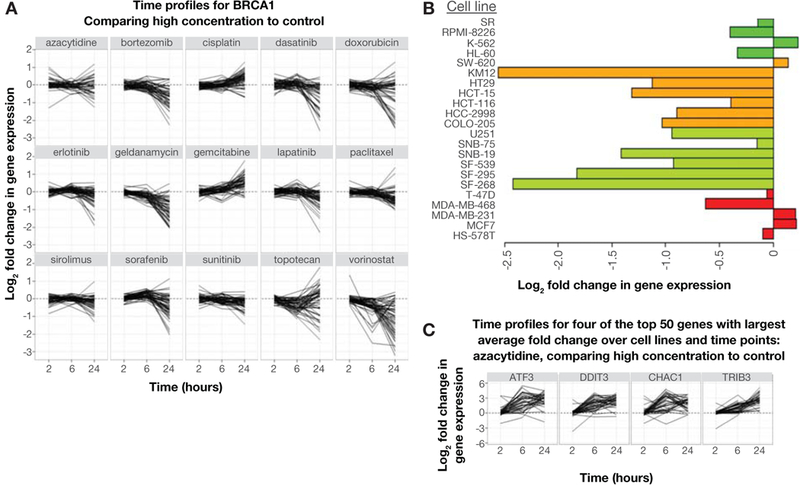
NCI TPW users can download graphs and heatmaps demonstrating changes in the expression levels of 12,704 genes in the NCI-60 panel of cell lines exposed to two different concentrations of 15 anticancer agents. A: single-gene response to all 15 agents at two different concentrations over three time points (shown, BRCA1 against all drugs tested at the high concentration); B: single-gene response (log2 fold expression change) following treatment with selected agent, by cell line (shown, LIF expression following 6 hours of exposure to high-concentration geldanamycin in breast [red], CNS [light green], colon [orange], and leukemia [dark green] cancer cell lines); C: genes most highly modulated by selected treatment parameter (shown, high-concentration azacytidine).
Confirmation and validation of gene expression changes
To confirm the array-based results, independent experiments were performed using a Fluidigm BioMark™ qRT-PCR System to measure expression of 28 selected genes after 24 hours of exposure to 6 agents in subsets of cell lines that were chosen to span a broad range of sensitivities to the agents tested (Supplementary Table S1). The Pearson correlation coefficient for the association between drug-induced gene expression changes in the array experiments and Fluidigm BioMark™ qRT-PCR experiments (n > 11,000), was 0.782, indicating highly similar results from the two types of studies (Figure 2A-D).
Figure 2.
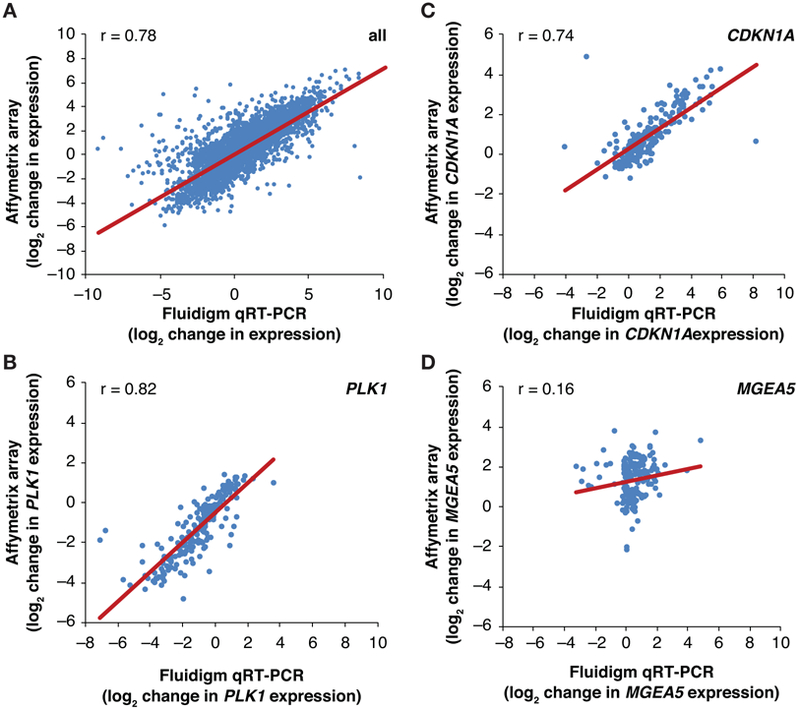
Validation of NCI TPW Affymetrix array gene expression measurements using Fluidigm qRT-PCR. A: Pearson correlation analysis of drug-induced gene expression perturbation measured by Affymetrix array or by HT Fluidigm (qRT-PCR) in an independent experiment. This experiment measured 32 genes in 20–60 cell lines treated with 6 drugs for 24 hours (see Supplementary Table S1); data are expressed as log2 of drug-induced change in expression. B-D: Pearson correlation analyses for expression of select genes (PLK1, CDKN1A, and MGEA5, respectively) measured in the experiment described above (see Supplementary Table S1).
To confirm that NCI TPW data accurately recapitulate the biology and mechanistic effects of targeted therapeutics, we used erlotinib, an FDA-approved inhibitor of EGFR that inhibits mitogen-activated protein kinase (MAPK) signaling (21), to examine downregulation of MAPK pathway transcriptional target genes DUSP6, DUSP4, and SPRY2 (22-24). Consistent with the mechanism of action of erlotinib, we observed an association between cell line sensitivity (log10[GI50]) and erlotinib-induced downregulation of MAPK pathway gene expression at 6 hours, with correlations of 0.48 (DUSP4), 0.56 (SPRY2) and 0.70 (DUSP6) (Supplementary Figure S1).
Comparative analyses of gene expression changes in cell lines of different sensitivity
Using the NCI TPW to compare the average fold change in gene expression across all cell lines and drug treatments, none of the 12,704 genes measured showed a directionally concerted expression change (i.e., up- or down-regulation by all drugs tested and across the majority of cell lines) (8) from exposure to all 15 drugs at either 2 or 6 hours after treatment; however, 8 genes had a common directionally concerted response to all 15 agents across ≥ 75% of NCI-60 cell lines after 24 hours (Supplementary Table S3). Three of these genes were downregulated: histone gene HIST1H4C, involved in chromatin structure; SLC19A1, encoding a folate and drug transporter; and XRCC5, involved in DNA repair. Five of these genes were upregulated: immune-related genes CD55, HLA-E, and HLA-G, proapoptotic gene BTG1, and microtubule-associated gene MAP1LC3B. Furthermore, the magnitudes of these gene expression changes were associated with cellular sensitivity (logGI50) to several agents; the strongest associations with chemosensitivity were observed for dasatinib, lapatinib, erlotinib, and cisplatin (data available on the NCI TPW, https://tpwb.nci.nih.gov).
To uncover signaling pathway–specific gene expression changes associated with drug sensitivity, we generated a list of transcriptionally regulated genes in 13 cell signaling pathways based on published data (see Supplementary Table S2 for details and references). For example, XRCC6, PRKDC, DCLRE1C, NHEJ1, XRCC4, LIG4, BRCA1, RAD52, BRCA2, RAD54L, ATM, ATR, and PARP1 were selected to investigate the modulation of DNA damage and repair mechanisms. We calculated average fold change values for each of these gene subsets in the 10 most sensitive and least sensitive cell lines at 3 time points for the highest concentration of each drug (Figure 3). Few pathway changes were evident at 2 hours, while increasingly substantial changes were observed at 6 and 24 hours. The genes that most prominently responded to all agents were those involved in ER stress activation and apoptosis; these responses were observed in both the most sensitive and least sensitive cohorts, though, in many cases, the least sensitive cell lines exhibited less substantial changes. Treatment with all tyrosine kinase inhibitors except sunitinib was associated with MAPK pathway downregulation of cell cycle checkpoint pathway function at 24 hours among the most sensitive cell lines, as would be expected based on the role of MAPK signaling in promoting cell proliferation, whereas the DNA damaging/cytotoxic agents had negligible effects on these genes.
Figure 3.
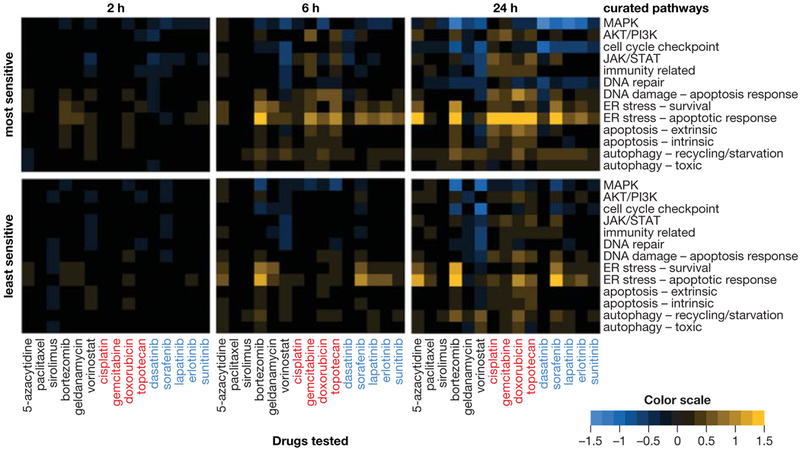
Cell signaling pathway–specific gene expression responses associated with drug sensitivity and insensitivity across the NCI-60 cell line panel. Average responses of the 10 most sensitive (top) or 10 least sensitive (bottom) cell lines after 2, 6, or 24 hours of treatment with the highest concentration of each drug, arranged by drug mechanism of action: kinase inhibitors (labeled in blue), genotoxic agents (red), or agents of other mechanistic classes (black). Pathway upregulation and downregulation are indicated by yellow and blue, respectively. Transcriptionally regulated, pathway-specific genes used to identify pathway modulation are provided in Supplementary Table S2.
We next sought to define potential mechanisms that control responsiveness to each of the 15 agents. We examined fold expression changes in the 15 most sensitive and 15 least sensitive cell lines for each agent, and performed GI50 correlation analyses using the entire set of NCI-60 cell lines, to identify genes for which drug-induced expression changes in the most sensitive cell lines differed from those in the least sensitive cell lines (Supplementary Table S4). Notably, several of the 15 agents did not yield any genes exhibiting large expression differences between the most and least sensitive cohorts for the respective agents and therefore do not appear in this table. Several genes that exhibited lower expression levels in the least sensitive versus most sensitive cell lines are known tumor suppressor genes, while some genes with higher expression levels in the least sensitive cohort, including APOBEC3B, have been implicated as prognostic factors (25,26).
Opposing changes in EGR1 expression underlie antagonism between EGFR inhibitors and DNA damaging agents
To demonstrate the utility of the NCI TPW dataset in enabling early preclinical identification of potential antagonistic interactions that might limit the clinical utility of a combination, we explored agent class–specific modulation of the master transcriptional regulator EGR1. EGR1 promotes cell survival and proliferation, and we found that expression of the EGR1 gene was upregulated or downregulated upon exposure to anticancer agents depending on whether the agent mechanism of action entails a requirement for or suppression of cell proliferation; the EGFR tyrosine kinase inhibitors erlotinib and lapatinib, which exert antitumor activity via suppression of growth factor signaling, generally decreased EGR1 expression, while the DNA damage-inducing agents gemcitabine, topotecan, doxorubicin, and cisplatin generally promoted upregulation of EGR1 expression across the NCI-60 panel (Supplementary Figure S2A-B). Downregulation of EGR1 expression following exposure to high-concentration EGFR tyrosine kinase inhibitors was associated with sensitivity to these agents (e.g., Pearson r = 0.634 or 0.482 after 6 hours of treatment with erlotinib or lapatinib, respectively; Supplementary Figure S2A). In contrast, upregulation of EGR1 expression was associated with sensitivity to high-concentration gemcitabine, topotecan, cisplatin, and doxorubicin (Pearson r = -0.591 or -0.610 after 6 hours of treatment with gemcitabine or topotecan, respectively, and r = -0.434 or -0.347 after 24 hours of treatment with cisplatin or doxorubicin, respectively; P < 0.05 for all; Supplementary Figure S2B).
Given the lack of additional clinical benefit observed for combinations of EGFR inhibitors and gemcitabine relative to respective single-agent therapies (10-12), we postulated that the opposing changes in EGR1 gene expression induced by these two different agent classes might form a basis for antagonism. Focusing on erlotinib and gemcitabine, six cell lines from the NCI-60 panel were selected based on sensitivity to these two agents and the magnitude and direction of agent-induced changes in EGR1 gene expression; NCI-H322M and SK-OV-3 were sensitive to erlotinib but only moderately sensitive to gemcitabine and exhibited large decreases in EGR1 expression following erlotinib treatment; NCI-H460 and U251 were sensitive to gemcitabine but not erlotinib and exhibited large increases in EGR1 expression in response to gemcitabine; PC3 and HCT116 were not sensitive to erlotinib, with only HCT-116 but not PC-3 sensitive to gemcitabine, and did not exhibit appreciably altered EGR1 expression in response to either drug (Figure 4A). Western blot analysis confirmed that gemcitabine- or erlotinib-induced EGR1 gene expression changes were associated with similar changes in EGR1 protein expression (Figure 4B).
Figure 4.
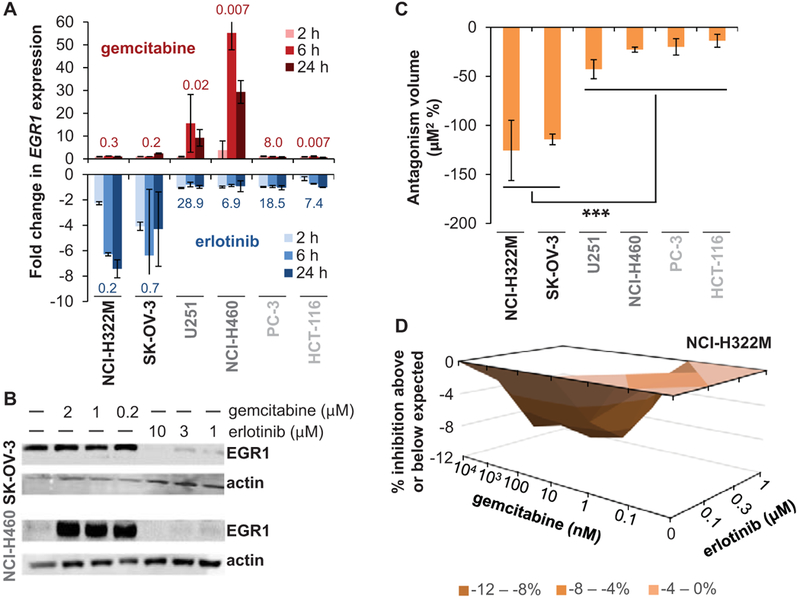
Modulation of EGR1 expression underlies antagonistic interaction between gemcitabine and erlotinib. A: Data from the NCI TPW showing changes in EGR1 gene expression in six selected cell lines after exposure to gemcitabine (top, red) or erlotinib (blue, bottom) for 2, 6, or 24 hours. Each bar represents the average fold change of the two concentrations tested (error bars indicate standard deviation). LogGI50 values (μM) for each cell line and agent are shown above (gemcitabine) or below (erlotinib) each set of bars. Cell lines are grouped and shaded according to shared sensitivity and EGR1 response characteristics (black: sensitive to erlotinib but not gemcitabine, drug-induced downregulation of EGR1; dark gray: sensitive to gemcitabine but not erlotinib, drug-induced upregulation of EGR1; light gray: not sensitive to erlotinib or gemcitabine [other than HCT-116 + gemcitabine], no appreciable drug-induced alteration of EGR1 expression). B: EGR1 protein expression recapitulates EGR1 gene expression changes in SK-OV-3 and NCI-H460 cells following exposure to gemcitabine or erlotinib. Representative Western blots show erlotinib-induced downregulation of EGR1 in SK-OV-3 cells and gemcitabine-induced upregulation of EGR1 in NCI-H460 cells. C: Quantitation of antagonism between gemcitabine and erlotinib. Cell survival was measured after simultaneous exposure to gemcitabine (0.1–104 nM) and erlotinib (0.1–1 μM) for 72 hours in the indicated cell lines, and antagonism was determined by 3D response surface analysis based on the MacSynergy II model (19,20), where a volume of < −100 μM2 % represents significant antagonism. Each column indicates the average of 2–3 separate experiments (error bars indicate standard deviation). The statistically significant difference in mean volume for the two cell lines exhibiting substantial erlotinib-induced EGR1 downregulation (NCI-H322M and SK-OV-3) versus the 4 cell lines for which no such EGR1 downregulation was observed is indicated by asterisks (***P = 0.0007 according to an unpaired, 2-tailed t-test). D: Example 3D plot showing response surface analysis for one experiment with the gemcitabine-erlotinib combination in the NCI-H322M cell line; values shown in (C) represent the average volumes for 3D plots from 2–3 replicate experiments in each cell line.
We next sought to identify any antagonistic interactions over a range of erlotinib and gemcitabine concentrations by performing 3D response surface analysis based on the MacSynergy II model (19,20). This program calculates theoretical additive interactions using the response data for each of the single agents and then subtracts the theoretical from the observed values to generate a 3D surface response plot showing the percent inhibition above or below the expected values (indicated by positive or negative y-axis values, respectively). Calculation of the total volume (area under the curve × % inhibition) for each 3D plot enables identification of significantly greater-than-additive (synergistic) or less-than-additive (antagonistic) interactions, where calculated volumes of < -100 μM2 % represent significant and substantial antagonism. We found that exposure to the gemcitabine-erlotinib combination for 72 hours resulted in substantial antagonism between the drugs in the NCI-H322M and SK-OV-3 cell lines (antagonism volumes of −126 ± 31 and −114 ± 5 μM2 %, respectively), moderate antagonism in the U251 cell line, and weak antagonism in the NCI-H460, PC-3, and HCT-116 cell lines, with the mean antagonism volume for the former two cell lines significantly larger than that of the 4 cell lines for which no substantial erlotinib-induced EGR1 downregulation was observed (P = 0.0007 according to an unpaired, 2-tailed t-test; Figure 4C-D and Supplementary Figure S3A-E).
Vorinostat impairs the homologous recombination pathway and increases sensitivity to PARP inhibitor treatment
As a starting point for further studies on the expression of genes involved in DNA damage and repair, we examined expression of homologous recombination (HR) pathway genes BRCA1, BRCA2, and RAD51 following drug exposure. The HDAC inhibitor vorinostat was one of several drugs that caused a sizeable decrease in relative expression (log2 fold change > 1) of all three genes (BRCA1 shown in Figure 1A; BRCA2 and RAD51 data available on the NCI TPW, https://tpwb.nci.nih.gov). This observation supports published findings that vorinostat, by reducing BRCA1, BRCA2, and RAD51 expression, may confer HR deficiency and drive susceptibility to agents, such as PARP inhibitors, that impair alternative DNA damage repair processes (27-29). We assessed this hypothesis by first more fully exploring the effects of vorinostat on these HR pathway genes. To evaluate whether expression of the HR genes remained low after vorinostat exposure, we exposed 5 vorinostat-sensitive cell lines to the drug for 24 hours, removed the drug, and then continued to culture the cells over 72 hours. Downregulation was sustained in T-47D and NCI-H460 cells for 72 hours after vorinostat washout, whereas gene expression in UO31 and UACC257 cells returned to baseline or higher within 24 hours of vorinostat washout, and no appreciable downregulation of these genes was observed at any time point in the SK-OV-3 cell line (Figure 5A). In addition to RNA levels, BRCA1 and RAD51 protein levels also decreased over 72 hours of exposure to vorinostat (Figure 5B).
Figure 5.
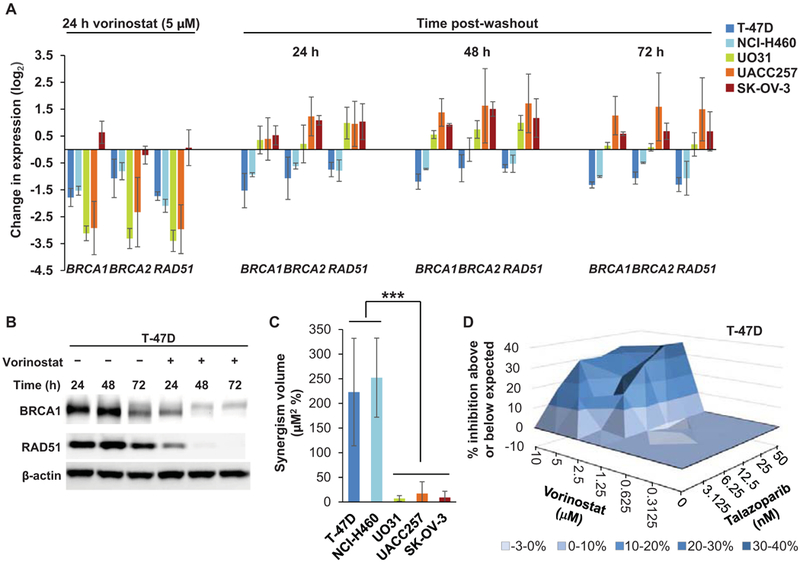
Sustained downregulation of HR genes and sensitivity to the PARP inhibitor talazoparib after vorinostat exposure. A: Sustained downregulation of HR genes BRCA1, BRCA2, and RAD51 after treatment with vorinostat. After 24 hours of exposure to 5 μM vorinostat, vorinostat was removed from the culture, and gene expression changes were determined after the 24-hour vorinostat exposure and after a further 24, 48, and 72 hours of culture in fresh media. Data are average triplicate values from 2–3 separate experiments (error bars indicate standard deviation). B: Decreased BRCA1 and RAD51 protein expression in T-47D cells following continuous exposure to 5 μM vorinostat. Representative Western blot shows sustained decreases in protein expression at the indicated time points throughout 72 hours of vorinostat treatment. C: Vorinostat-mediated sensitization to talazoparib. Cell survival was measured after exposure to vorinostat alone (0.3125–10 μM) for 24 hours, followed by incubation with talazoparib alone (3.125–50 nM) for 96 hours in the indicated cell lines; greater-than-additive inhibition of cell growth was determined by 3D response surface analysis based on the MacSynergy II model (19,20), where a volume of > 100 μM2 % represents significant synergy. Each column indicates the mean volume from 2–3 independent experiments; error bars indicate standard deviation, and asterisks indicate the significant difference in mean volume for the 2 cell lines exhibiting sustained HR gene downregulation (T-47D and NCI-H460) versus the 3 cell lines exhibiting no such sustained downregulation of HR genes, according to an unpaired, 2-tailed t-test (***P = 0.0003). D: Example 3D plot showing the response surface analysis for one experiment in the T-47D cell line; volume measurements shown in (C) represent the average volumes for 3D plots from 2–3 replicate experiments in each cell line.
To further assess the impact of this induced HR deficiency phenotype, we evaluated whether vorinostat-mediated decreases in HR gene expression would enhance sensitivity to PARP inhibition. We assessed greater-than-additive cytotoxicity over a broad range of both vorinostat and talazoparib concentrations (0.3125–10 μM and 3.125–50 nM, respectively) by pre-treating cells with vorinostat for 24 hours, then replacing the culture medium with medium containing only talazoparib and incubating for an additional 96 hours. We again performed 3D response surface analysis based on the MacSynergy II model (19,20), where volumes of > 100 μM2 % represent significant and substantial synergy. This analysis revealed significant synergy between these two agents in the T-47D and NCI-H460 cell lines, with mean synergism volumes of 223 ± 109 and 252 ± 80 μM2 %, respectively, from 3 independent experiments (Figure 5C-D and Supplementary Figure S4A). As expected for UO31 and UACC257 cells (in which HR gene expression was restored within 24 hours after vorinostat exposure) and SK-OV-3 cells (in which HR gene expression was not appreciably downregulated at any time point), increased sensitivity to PARP inhibition was not observed (Figure 5C and Supplementary Figure S4B-D). The mean synergism volume for the cell lines exhibiting sustained suppression of HR gene expression (T-47D and NCI-H460) was significantly greater than that of the 3 cell lines for which no such sustained HR gene downregulation was observed (Figure 5C; P = 0.0003 according to an unpaired, 2-tailed t-test).
Discussion
We measured transcriptional responses to 15 anticancer agents across the NCI-60 cell line panel with the goal of creating a dataset and an accompanying extensive array of interactive, web-based analytical tools to facilitate exploration of gene expression changes induced by various treatment strategies. These tools have been validated to enable the NCI TPW to be used for comprehensive pathway and target effect analyses, complementing hypothesis-driven studies of gene regulation that may be relevant to drug mechanism of action and treatment response.
The examination of genes exhibiting concerted directional responses to all 15 agents may shed light on common mechanisms of drug response or resistance. We found 8 such genes that exhibited concerted responses after 24 hours of drug treatment (Supplementary Table S3): 3 genes were downregulated (HIST1H4C, a replication-dependent histone H4 family gene involved in chromatin structure; SLC19A1, which encodes a folate and drug transporter associated with drug sensitivity (30); and XRCC5, which encodes the protein Ku80, required for non-homologous end joining during DNA double strand break repair); 5 genes were upregulated (the immune-related genes CD55, HLA-E, and HLA-G, the anti-proliferation gene BTG1, and autophagy-associated gene MAP1LC3B). MAP1LC3B encodes the autophagosome component LC3B, which may play a role in chemoresistance and therefore may be a potential target for improving chemotherapy efficacy (31). In contrast, BTG1 promotes stress-induced cell growth arrest and apoptosis (32); BTG1 deletions may be predictive of poor prognosis in some tumor types (33,34), and the concurrence of patient relapse with proliferation of tumor cells containing BTG1 deletions suggests that BTG1 loss may be associated with chemotherapeutic resistance in some instances (35,36). Thus, drug-induced BTG1 expression may be a potential positive biomarker for activity of pharmacological agents.
The increased expression of the immune-related CD55, HLA-E, and HLA-G genes in response to all 15 drugs is consistent with the known roles of the associated proteins in malignant cell survival and may have implications for the development of novel therapies. Expression of the non-classical HLA genes HLA-E and HLA-G promotes tumor evasion, suppression of immune response, and upregulation of matrix metalloproteinases (37); HLA-E and HLA-G expression, particularly in the absence of classical HLA expression, is also associated with poor prognosis in several tumor types (38-40). Similarly, CD55 is a prognostic indicator in multiple cancers and, by enabling cancer cells to evade complement attack, confers resistance to apoptotic stimuli in vitro (41,42). Therefore, drug-induced upregulation of CD55 and these non-classical HLA genes may impede monotherapy treatments, and targeting of these proteins as part of combination therapies may be beneficial. Furthermore, the potential immunosuppressive effects conferred by increased HLA-E, HLA-G, and CD55 expression following treatment with the agents examined in this study suggest the promise of combining these drugs with immune-modulating agents. Indeed, clinical trials investigating several such immunotherapy combinations are currently underway (43).
These NCI TPW data led us to examine other genes relevant to immune function that are affected by the study agents. For example, expression of LIF (leukemia inhibitory factor) decreases following exposure to geldanamycin (Figure 1B). LIF, a multifunctional cytokine, has been shown to promote changes in the tumor microenvironment that result in tumor invasion (44-46). Because LIF promotes tolerance and drives T cells to differentiate into immune-suppressive regulatory T cells, an HSP90 inhibitor such as geldanamycin, which decreases tumor cell LIF production, may be synergistic with agents promoting T cell responses, such as checkpoint inhibitors. However, in vitro studies of immune-related genes must be interpreted with caution, as cell culture does not recapitulate the complex interactions between the immune system and tumor cells.
We identified signaling pathways unique to sensitive or insensitive cells by comparing cohorts of the 10 most sensitive lines and the 10 least sensitive lines for each agent. Upon examining a series of 13 curated pathways, we found that both the most sensitive and least sensitive cell line cohorts had similar pathway signatures, although the magnitude of response was generally smaller in the least sensitive cohort; no pathways were uniquely activated in the least sensitive cohort. Pathways were not appreciably upregulated or downregulated after 2 hours of treatment, indicating that this time point is not optimal for measuring acute transcriptional responses.
Finally, our data demonstrate how the NCI TPW resource can be used to gain mechanistic insights to guide early preclinical evaluation of potential combination therapies. In response to EGFR inhibitors and DNA damaging agents, we observed directionally opposing changes in EGR1 expression that were also differentially associated with sensitivity to these agents; for example, we found that decreased EGR1 expression following erlotinib treatment was associated with sensitivity to erlotinib, consistent with a previous study showing that upregulation of EGR1 expression is associated with in vitro resistance to anti-EGFR antibody treatment and poor clinical outcome for colorectal carcinoma patients receiving cetuximab therapy (47). Our additional experimentation using the combination of erlotinib and gemcitabine indicated antagonism between these agents, which may explain the lack of substantial clinical activity documented for this and other combinations of these two agent classes relative to the respective single agents (10-12); interestingly, recent data suggest that this antagonism may be overcome by changes in the scheduling of drug administration (48-50). Conversely, NCI TPW data revealed that vorinostat treatment induced an HR-deficient phenotype in some cell lines (i.e., loss of BRCA1, BRCA2, and RAD51 expression), which led us to conduct further experiments to demonstrate vorinostat-induced sensitization of these cell lines to a PARP inhibitor, talazoparib. Further investigation of the mechanism by which this response occurs could uncover additional determinants of vorinostat sensitivity.
In conclusion, drug-induced transcriptional profiles were measured in the NCI-60 panel and are publicly available on the NCI TPW (https://tpwb.nci.nih.gov). We utilized these data to identify common transcriptional responses to pharmacological stress across drug mechanisms and multiple cell backgrounds. Evaluating expression differences in response to drugs within the same class or dosing tuned to the sensitivity of each cell line will help identify candidate genes to serve as pharmacodynamic biomarkers or for investigation as potential drug targets. While these data are limited to 24 hours of drug exposure, constraining their relevance to investigations of acquired drug resistance, this resource has already been used to examine expression changes for genes involved in epigenetic regulation and one-carbon metabolism pathways (8), glycosylation (9), and ER stress response (Min 2018, in preparation). The NCI TPW provides a resource to the scientific community to aid in understanding drug mechanisms, finding novel targets for drug combination treatments, and exploring the link between genetic background, transcriptional response, and drug sensitivity.
Supplementary Material
Statement of Significance.
The NCI Transcriptional Pharmacodynamics Workbench represents the most extensive compilation to date of directly-measured longitudinal transcriptional responses to anticancer agents across a thoroughly characterized ensemble of cancer cell lines.
Acknowledgements
The authors wish to thank Dr. Xiaosheng Wang, Mr. Henry Rivera, and Dr. Peter Szabo for their contributions to the development of the TP Workbench program.
Financial support: This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Disclosure of Potential Conflicts of Interest: The authors declare no potential conflicts of interest.
References
- 1.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer 2006;6:813–23. [DOI] [PubMed] [Google Scholar]
- 2.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483:603–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, Schubert M, et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell 2016;166:740–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keenan AB, Jenkins SL, Jagodnik KM, Koplev S, He E, Torre D, et al. The Library of Integrated Network-Based Cellular Signatures NIH Program: System-Level Cataloging of Human Cells Response to Perturbations. Cell Syst 2018;6:13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vidovic D, Koleti A, Schurer SC. Large-scale integration of small molecule-induced genome-wide transcriptional responses, Kinome-wide binding affinities and cell-growth inhibition profiles reveal global trends characterizing systems-level drug action. Front Genet 2014;5:342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017;171:1437–52.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Musa A, Ghoraie LS, Zhang SD, Glazko G, Yli-Harja O, Dehmer M, et al. A review of connectivity map and computational approaches in pharmacogenomics. Brief Bioinform 2018;19:506–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krushkal J, Zhao Y, Hose C, Monks A, Doroshow JH, Simon R. Concerted changes in transcriptional regulation of genes involved in DNA methylation, demethylation, and folate-mediated one-carbon metabolism pathways in the NCI-60 cancer cell line panel in response to cancer drug treatment. Clin Epigenetics 2016;8:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krushkal J, Yingdong Z, Hose C, Monks A, Doroshow JH, Simon R. Longitudinal transcriptional response of glycosylation-related genes, regulators, and targets in cancer cell lines treated with 11 antitumor agents. Cancer Inform 2017;16:1176935117747259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gatzemeier U, Pluzanska A, Szczesna A, Kaukel E, Roubec J, Rosa FD, et al. Phase III Study of Erlotinib in Combination With Cisplatin and Gemcitabine in Advanced Non–Small-Cell Lung Cancer: The Tarceva Lung Cancer Investigation Trial. J Clin Oncol 2007;25:1545–52. [DOI] [PubMed] [Google Scholar]
- 11.Philip PA, Benedetti J, Corless CL, Wong R, O’Reilly EM, Flynn PJ, et al. Phase III Study Comparing Gemcitabine Plus Cetuximab Versus Gemcitabine in Patients With Advanced Pancreatic Adenocarcinoma: Southwest Oncology Group–Directed Intergroup Trial S0205. J Clin Oncol 2010;28:3605–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin S, Park CM, Kwon H, Lee KH. Erlotinib plus gemcitabine versus gemcitabine for pancreatic cancer: real-world analysis of Korean national database. BMC Cancer 2016;16:443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liston DR, Davis M. Clinically relevant concentrations of anticancer drugs: a guide for nonclinical studies. Clin Cancer Res 2017;23:3489–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holbeck SL, Collins JM, Doroshow JH. Analysis of Food and Drug Administration–approved anticancer agents in the NCI60 panel of human tumor cell lines. Mol Cancer Ther 2010;9:1451–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003;4:249–64. [DOI] [PubMed] [Google Scholar]
- 16.Duan Q, Flynn C, Niepel M, Hafner M, Muhlich JL, Fernandez NF, et al. LINCS Canvas Browser: interactive web app to query, browse and interrogate LINCS L1000 gene expression signatures. Nucleic Acids Res 2014;42:W449–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 2006;313:1929–35. [DOI] [PubMed] [Google Scholar]
- 18.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 1995;57:289–300. [Google Scholar]
- 19.Prichard MN, Prichard LE, Shipman C Jr. Strategic design and three-dimensional analysis of antiviral drug combinations. Antimicrob Agents Chemother 1993;37:540–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prichard MN, Shipman C, Jr. A three-dimensional model to analyze drug-drug interactions. Antiviral Res 1990;14:181–205. [DOI] [PubMed] [Google Scholar]
- 21.Miyabayashi K, Ijichi H, Mohri D, Tada M, Yamamoto K, Asaoka Y, et al. Erlotinib prolongs survival in pancreatic cancer by blocking gemcitabine-induced MAPK signals. Cancer Res 2013;73:2221–34. [DOI] [PubMed] [Google Scholar]
- 22.Owens DM, Keyse SM. Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene 2007;26:3203–13. [DOI] [PubMed] [Google Scholar]
- 23.Hanafusa H, Torii S, Yasunaga T, Nishida E. Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat Cell Biol 2002;4:850–8. [DOI] [PubMed] [Google Scholar]
- 24.De Vriendt V, De Roock W, Di Narzo AF, Tian S, Biesmans B, Jacobs B, et al. DUSP 4 expression identifies a subset of colorectal cancer tumors that differ in MAPK activation, regardless of the genotype. Biomarkers 2013;18:516–24. [DOI] [PubMed] [Google Scholar]
- 25.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen T-W, Lee C- C, Liu H, Wu C- S, Pickering CR, Huang P- J, et al. APOBEC3A is an oral cancer prognostic biomarker in Taiwanese carriers of an APOBEC deletion polymorphism. Nat Commun 2017;8:465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ha K, Fiskus W, Choi DS, Bhaskara S, Cerchietti L, Devaraj SG, et al. Histone deacetylase inhibitor treatment induces ‘BRCAness’ and synergistic lethality with PARP inhibitor and cisplatin against human triple negative breast cancer cells. Oncotarget 2014;5:5637–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konstantinopoulos PA, Wilson AJ, Saskowski J, Wass E, Khabele D. Suberoylanilide hydroxamic acid (SAHA) enhances olaparib activity by targeting homologous recombination DNA repair in ovarian cancer. Gynecol Oncol 2014;133:599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yalon M, Tuval-Kochen L, Castel D, Moshe I, Mazal I, Cohen O, et al. Overcoming resistance of cancer cells to PARP-1 inhibitors with three different drug combinations. PLoS One 2016;11:e0155711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Charasson V, Hillaire-Buys D, Solassol I, Laurand-Quancard A, Pinguet F, Le Morvan V, et al. Involvement of gene polymorphisms of the folate pathway enzymes in gene expression and anticancer drug sensitivity using the NCI-60 panel as a model. Eur J Cancer 2009;45:2391–401. [DOI] [PubMed] [Google Scholar]
- 31.Thorburn A, Thamm DH, Gustafson DL. Autophagy and cancer therapy. Mol Pharmacol 2014;85:830–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuniati L, van der Meer LT, Tijchon E, van Ingen Schenau D, van Emst L, Levers M, et al. Tumor suppressor BTG1 promotes PRMT1-mediated ATF4 function in response to cellular stress. Oncotarget 2016;7:3128–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheijen B, Boer JM, Marke R, Tijchon E, van Ingen Schenau D, Waanders E, et al. Tumor suppressors BTG1 and IKZF1 cooperate during mouse leukemia development and increase relapse risk in B-cell precursor acute lymphoblastic leukemia patients. Haematologica 2017;102:541–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao S, Chen SR, Yang XF, Shen DF, Takano Y, Su RJ, et al. BTG1 might be employed as a biomarker for carcinogenesis and a target for gene therapy in colorectal cancers. Oncotarget 2017;8:7502–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waanders E, Scheijen B, van der Meer LT, van Reijmersdal SV, van Emst L, Kroeze Y, et al. The origin and nature of tightly clustered BTG1 deletions in precursor B-cell acute lymphoblastic leukemia support a model of multiclonal evolution. PLoS Genet 2012;8:e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science 2008;322:1377–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin A, Yan WH. HLA-G expression in cancers: roles in immune evasion, metastasis and target for therapy. Mol Med 2015;21:782–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Kruijf EM, Sajet A, van Nes JGH, Natanov R, Putter H, Smit VTHBM, et al. HLA-E and HLA-G expression in classical HLA class I-negative tumors is of prognostic value for clinical outcome of early breast cancer patients. J Immunol 2010;185:7452–9. [DOI] [PubMed] [Google Scholar]
- 39.Zeestraten EC, Reimers MS, Saadatmand S, Goossens-Beumer IJ, Dekker JW, Liefers GJ, et al. Combined analysis of HLA class I, HLA-E and HLA-G predicts prognosis in colon cancer patients. Br J Cancer 2014;110:459–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andersson E, Poschke I, Villabona L, Carlson JW, Lundqvist A, Kiessling R, et al. Non-classical HLA-class I expression in serous ovarian carcinoma: correlation with the HLA-genotype, tumor infiltrating immune cells and prognosis. Oncoimmunology 2016;5:e1052213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kesselring R, Thiel A, Pries R, Fichtner-Feigl S, Brunner S, Seidel P, et al. The complement receptors CD46, CD55 and CD59 are regulated by the tumour microenvironment of head and neck cancer to facilitate escape of complement attack. Eur J Cancer 2014;50:2152–61. [DOI] [PubMed] [Google Scholar]
- 42.Ikeda J, Morii E, Liu Y, Qiu Y, Nakamichi N, Jokoji R, et al. Prognostic significance of CD55 expression in breast cancer. Clin Cancer Res 2008;14:4780–6. [DOI] [PubMed] [Google Scholar]
- 43.Hughes PE, Caenepeel S, Wu LC. Targeted therapy and checkpoint immunotherapy combinations for the treatment of cancer. Trends Immunol 2016;37:462–76. [DOI] [PubMed] [Google Scholar]
- 44.Yue X, Zhao Y, Zhang C, Li Z, Liu J, Hu W. Leukemia inhibitory factor promotes EMT through STAT3-dependent miR-21 induction. Oncotarget 2015;7:3777–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X, Yang Q, Yu H, Wu L, Zhao Y, Zhang C, et al. LIF promotes tumorigenesis and metastasis of breast cancer through the AKT-mTOR pathway. Oncotarget 2014;5:788–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wysoczynski M, Miekus K, Jankowski K, Wanzeck J, Bertolone S, Janowska-Wieczorek A, et al. Leukemia Inhibitory Factor: A newly identified metastatic factor in rhabdomyosarcomas. Cancer Res 2007;67:2131–40. [DOI] [PubMed] [Google Scholar]
- 47.Kumar SS, Tomita Y, Wrin J, Bruhn M, Swalling A, Mohammed M, et al. High early growth response 1 (EGR1) expression correlates with resistance to anti-EGFR treatment in vitro and with poorer outcome in metastatic colorectal cancer patients treated with cetuximab. Clinical and Translational Oncology 2017;19:718–26. [DOI] [PubMed] [Google Scholar]
- 48.Li M, Li H, Cheng X, Wang X, Li L, Zhou T, et al. Preclinical Pharmacokinetic/Pharmacodynamic Models to Predict Schedule-Dependent Interaction Between Erlotinib and Gemcitabine. Pharm Res 2013;30:1400–8. [DOI] [PubMed] [Google Scholar]
- 49.Semrad T, Barzi A, Lenz HJ, Hutchins IM, Kim EJ, Gong IY, et al. Pharmacodynamic separation of gemcitabine and erlotinib in locally advanced or metastatic pancreatic cancer: therapeutic and biomarker results. Int J Clin Oncol 2015;20:518–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ubezio P, Falcetta F, Carrassa L, Lupi M. Integrated experimental and simulation study of the response to sequential treatment with erlotinib and gemcitabine in pancreatic cancer. Oncotarget 2016;7:15492–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


