Abstract
Mitochondrial dysfunction is a key pathological feature of many different types of neurodegenerative disease. Sterile alpha and Toll/interleukin receptor motif-containing protein 1 (SARM1) has been attracting much attention as an important molecule for inducing axonal degeneration and neuronal cell death by causing loss of NAD (NADH). However, it has remained unclear what exactly regulates the SARM1 activity. Here, we report that NAD+ cleavage activity of SARM1 is regulated by its own phosphorylation at serine 548. The phosphorylation of SARM1 was mediated by c-jun N-terminal kinase (JNK) under oxidative stress conditions, resulting in inhibition of mitochondrial respiration concomitant with enhanced activity of NAD+ cleavage. Nonphosphorylatable mutation of Ser-548 or treatment with a JNK inhibitor decreased SARM1 activity. Furthermore, neuronal cells derived from a familial Parkinson's disease (PD) patient showed a congenitally increased level of SARM1 phosphorylation compared with that in neuronal cells from a healthy person and were highly sensitive to oxidative stress. These results indicate that JNK-mediated phosphorylation of SARM1 at Ser-548 is a regulator of SARM1 leading to inhibition of mitochondrial respiration. These findings suggest that an abnormal regulation of SARM1 phosphorylation is involved in the pathogenesis of Parkinson's disease and possibly other neurodegenerative diseases.
Keywords: c-Jun N-terminal kinase (JNK), mitochondria, cell death, nicotinamide adenine dinucleotide (NAD), ATP, SARM1, NAD+
Introduction
Parkinson's disease (PD)3 is a neurodegenerative disease that mainly causes degeneration of dopaminergic neurons, which project from the substantia nigra to the striatum. The prevalence is about 100 to 150 per 100,000 people. PD is sporadic in most cases, and its cause of onset is unknown but familial PD accounts for about 10% of PD. Because phenotypes of familial PD are similar to those of sporadic PD, functional analysis of familial PD causative genes is expected to lead to the establishment of effective therapy.
At least 23 loci and 19 disease-causing genes, named PARK1 to PARK23, have been reported to date (1). We have analyzed mainly PARK2 and PARK6, which are involved in juvenile PD of an autosomal recessive form. PARK2 encodes the E3 ubiquitin ligase Parkin, and PARK6 encodes the mitochondrial kinase PTEN-induced putative kinase 1 (PINK1). We reported that PINK1 contributes to the suppression of neuronal cell death through activation of Akt and Bcl-xL, which are involved in cell survival (2, 3). Recently, it has also become clear that Parkin and PINK1 are involved in mitophagy, homeostatic machinery for removal of damaged mitochondria in cells. PINK1 accumulates on the outer mitochondrial membrane when the mitochondrial membrane potential is reduced by either damage or stress to mitochondria (4). The accumulated PINK1 induces phosphorylation of Parkin and ubiquitin (5–7), the activated Parkin ubiquitinates mitochondrial proteins such as mitofusin-1 (MFN1) and voltage-dependent anion-selective channel 1 (VDAC1) (8, 9), and eventually the ubiquitinated mitochondria are removed from cells by the process of autophagy. Hence genetic mutations in PINK1 and Parkin leading to recessive function of them result in a high risk for the appearance of neurodegeneration in PD because defective mitochondria are not properly removed from cells. Accumulating evidence indicates that defective mitochondria readily induce neuronal cell death by causing abundant production of reactive oxygen species (ROS) and significant reduction of ATP synthesis. Besides PINK1 and Parkin, we recently identified another molecule, SARM1, in the PINK1-mediated mitophagy process. We found that SARM1 induces PINK1 accumulation on damaged mitochondria (10).
Osterloh et al. provided the first evidence that SARM1 acts as an inducer of nerve axonal degeneration (11). Axon degeneration is commonly observed in various neurodegenerative diseases such as amyotrophic lateral sclerosis (ALS), multiple sclerosis, Alzheimer's disease, and Parkinson's disease (12–16). It has been reported that axonal degeneration was dramatically reduced in neurons from SARM1-knockout mice (17–19) and that the rate of ischemic neuronal cell death was also significantly decreased in the same engineered mice (20). Interestingly, it has also been reported that axon degeneration and neuronal cell death induced by mitochondrial dysfunction are SARM1-dependent (21). Following those reports, various studies using specific animal models showed one after another that SARM1 deletion improves the pathology of neuronal diseases such as ALS and severe axonopathy (22–25). Human SARM1 is a protein consisting of 724 amino acids and possesses a mitochondrial targeting signal (MTS) (26), armadillo/HEAT motifs (ARM), two sterile alpha motifs (SAM), and Toll/interleukin-1 receptor (TIR) domain (Fig. 1A). Analysis of the biological roles of these domains in SARM1 showed that the ARM domain functions to suppress the activity of SARM1, the SAM domain forms a dimer, and the TIR domain functions as an executioner for axonal degeneration and cell death (17, 18, 27). Surprisingly, Essuman et al. reported that SARM1 has an enzymatic activity for cleavage of NAD+ by utilizing the TIR domain (28). Because NAD+ depletion inhibits ATP production in cells, SARM1-TIR–mediated NAD+ deficiency is thought to be involved in axonal degeneration and cell death. However, little is known about the regulation mechanism(s) of SARM1 activation at molecular levels that leads to NAD+ loss and neuronal cell death. We here report that SARM1 activity is regulated through phosphorylation at Ser-548 by JNK.
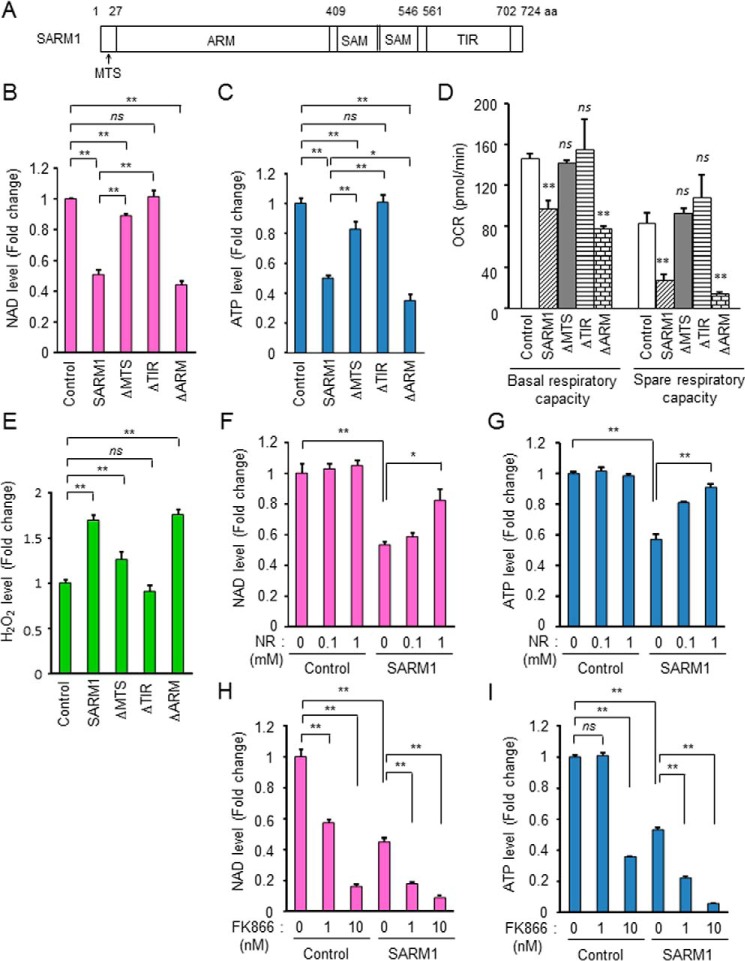
Figure 1.
Inhibition of mitochondrial respiration by SARM1 overexpression. A, schematic representation of human SARM1. MTS, mitochondrial targeting signal; ARM, armadillo/HEAT motifs; SAM, sterile alpha motif; TIR, Toll/interleukin-1 receptor. HEK293T cells were transfected with FLAG-tagged full-length SARM1 or various domain-deletion mutants for 24 h. B, SARM1 overexpression induces reduction of NAD+ in HEK293T cells. NAD+ level was analyzed by the NAD/NADH-Glo assay. C, SARM1 overexpression induces reduction of ATP in HEK293T cells. ATP level was analyzed by the CellTiter-Glo assay. D, measurements of OCR. At 8 h post transfection, the cells were re-seeded in XF96 plates and further incubated for 16 h. OCR measurements were performed using an XF96 extracellular flux analyzer. E, SARM1 overexpression induces ROS production. ROS level was analyzed by ROS-Glo H2O2 assay. F and G, supplementation of NR prevents SARM1-induced NAD+ and ATP reduction. H and I, treatment with the NAMPT inhibitor FK866 enhances SARM1-induced NAD+ and ATP reduction. Data represent the mean ± S.D. of three independent experiments. *, p < 0.05; **, p < 0.01; ns, not significant.
Results
Overexpression of SARM1 inhibits mitochondrial respiration
To confirm the function of SARM1, we overexpressed FLAG-tagged WT SARM1 and various deletion mutants of SARM1 in HEK293T cells. SARM1 has been reported to reduce intracellular NAD+ level (28). WT SARM1 reduced NAD+ level, and this effect was alleviated with MTS deletion (ΔMTS), completely disappeared with TIR deletion (ΔTIR), and almost the same level with that of ARM deletion (ΔARM) (Fig. 1B). The morphology of the cells was changed by overexpression of WT SARM1 and ΔARM (Fig. S1A). Corresponding to the NAD+ level, ATP level was also changed by overexpression of SARM1 constructs (Fig. 1C). It was reported that ΔARM is the constitutively active type (27, 29) and the toxic effect of ΔARM is higher than that of WT SARM1. However, the expression level of ΔARM in HEK293T cells was lower than that of other constructs (Fig. S1B). Therefore, the effect of ΔARM seemed to be almost the same as WT SARM1 in our experimental settings. We next examined mitochondrial respiratory capacity because SARM1 possesses an MTS and the SARM1 (WT)-mediated deleterious effects are very small when the MTS is deleted (ΔMTS). WT SARM1 and ΔARM reduced basal and spare respiratory capacity (SRC), but ΔMTS and ΔTIR had no effect on those respiration states (Fig. 1D and Fig. S1C). Spare respiratory capacity is the extra mitochondrial capacity available in a cell to produce energy under conditions of increased work or stress and is therefore thought to be important for long-term cellular survival (30). WT SARM1 and ΔARM also increased the cellular level of ROS (Fig. 1E and Fig. S1D).
We next investigated the mechanism(s) by which SARM1 alters a mitochondrial function in relation to NAD+ reduction. Di Lisa et al. reported that opening of the mitochondrial permeability transition pore (PTP) causes depletion of mitochondrial and cytosolic NAD+ (31). We tested whether PTP inhibition prevents the SARM1-related NAD+ loss. Cyclosporin A (CsA), a PTP inhibitor, did not affect the SARM1-induced NAD+ and ATP reduction (Fig. S1, E and F), suggesting that PTP is not contributed to the SARM1 activity. We next tested whether increased NAD+ synthesis could counteract the SARM1-induced NAD+ and ATP reduction. Supplementation of NAD+ precursor nicotinamide riboside (NR) suppressed both the SARM1-induced NAD+ and ATP reductions in a dose-dependent manner (Fig. 1, F and G). Nicotinamide phosphoribosyltransferase (NAMPT) is the rate-limiting enzyme in the NAD+ salvage pathway that converts nicotinamide to nicotinamide mononucleotide in mammals, eventually leading to NAD+ biosynthesis. Treatment with the NAMPT inhibitor FK866 enhanced SARM1-induced NAD+ reduction in a dose-dependent manner (Fig. 1H). On the other hand, although FK866 was no effect on the control cells at 1 nm, it markedly induced ATP reduction in the SARM1 overexpressed cells, and the event was further enhanced by the treatment with higher concentration of 10 nm (Fig. 1I). These data indicate that SARM1 plays a suppressive role in mitochondrial function through induction of NAD+ loss.
Previously, we found that SARM1 induces PINK1 accumulation on depolarized mitochondria in a process of mitophagy (10). Overexpression of SARM1 enhanced PINK1 accumulation induced by mitochondrial uncoupler, carbonyl cyanide m-chlorophenyl hydrazone (CCCP), and valinomycin (Fig. S1G). Because SARM1 has mitochondria inhibitory functions such as inhibition of mitochondrial respiration, reduction of ATP synthesis, and production of ROS, PINK1 appears to function to protect cells from SARM1. In addition, being consistent with previous reports (17, 27), we showed that the TIR domain is essential for SARM1-mediated NAD+ and ATP reduction.
SARM1 is phosphorylated at Ser-548
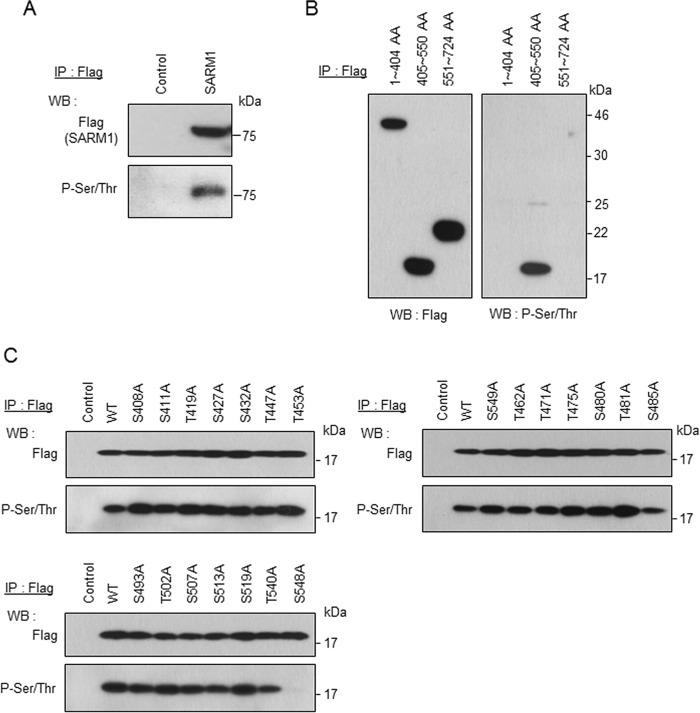
Next, we examined how SARM1 activity is regulated in the process of inhibition of ATP production. We found that the full length of SARM1 is phosphorylated under the condition in which ATP synthesis is inhibited by forced expression of SARM1-FLAG (Fig. 2A). Therefore, we decided to identify the phosphorylation site of SARM1. SARM1 was separated into three regions: 1–404 amino acids (AA) containing the MTS and ARM domain, 405–550 AA containing the SAM domain, and 551–724 AA containing the TIR domain. Although each FLAG-tagged domain was expressed at almost the same level, phosphorylation was only detected in 405–550 AA containing the SAM domain by an anti-phosphoserine/threonine antibody (Fig. 2B). This region (405–550 AA) contains 12 serines and 9 threonines. Each serine and threonine was substituted for nonphosphorylatable alanine to determine the phosphorylation site. As shown in Fig. 2C, we found that the phosphorylation band disappeared only at the replacement site, serine 548. These results indicate that SARM1 is phosphorylated at Ser-548.
Figure 2.
Phosphorylation of SARM1 at Ser-548. A, a control or FLAG-tagged full-length SARM1 vector was transfected in HEK293T cells for 24 h. Phosphorylation of SARM1 was determined by immunoprecipitation (IP) with anti-FLAG agarose, followed by Western blot analysis with anti-phosphorylated serine/threonine antibody. B, the region of 405–550 AA contains a phosphorylation site. FLAG-tagged 1–404 AA, 405–550 AA, or 551–724 AA region was transfected. The experiment was performed under conditions similar to those in (A). C, mutation at Ser-548 abolishes phosphorylation of SARM1. The serine or threonine contained in the region of 405–550 AA was mutated to alanine. Those mutants were transfected in HEK293T cells. The experiment was performed under conditions similar to those in (A). The phosphorylation band disappeared only with the mutation at Ser-548.
JNK phosphorylates SARM1 at Ser-548
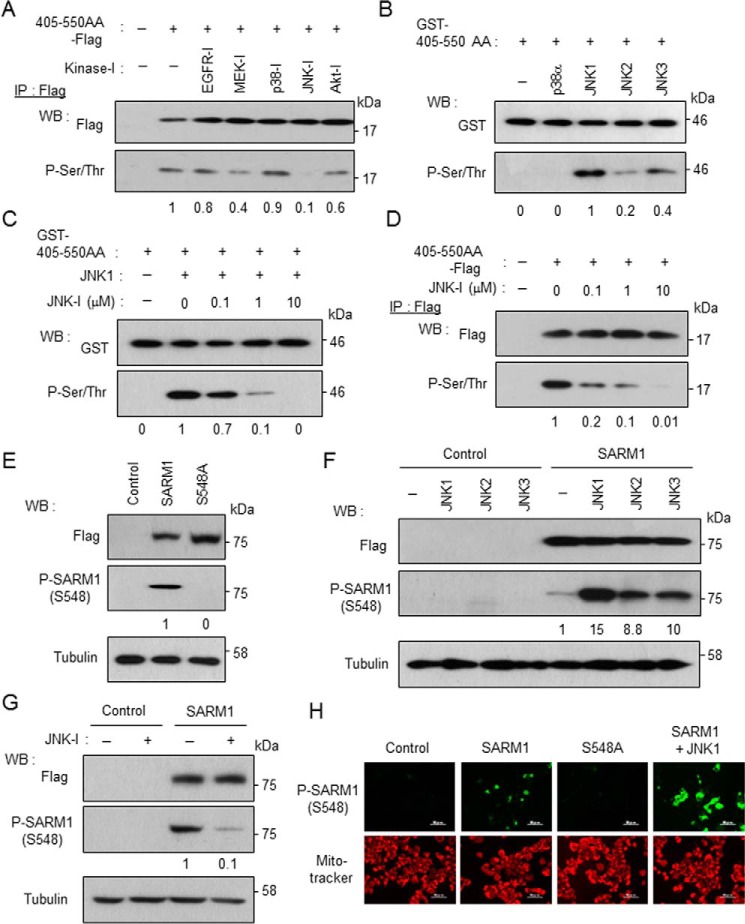
We next investigated which kinase phosphorylates SARM1. The region of 405–550 AA containing Ser-548 was expressed in cells and the cells were treated with some kinase inhibitors. Reliability in selective kinase inhibition of the chemicals used was validated by checking the phosphorylation states of the downstream molecules, which are phosphorylated by the kinases corresponding to a row of the indicated inhibitors (Fig. S2A). In this candidate screening, a JNK inhibitor strongly blocked SARM1 phosphorylation at Ser-548 and a MEK inhibitor and an Akt inhibitor weakly blocked it (Fig. 3A). The chemical-induced inhibitory pattern of the SARM1 phosphorylation was similar to that of the cJun phosphorylation, a representative JNK downstream molecule (Fig. S2A), suggesting that JNK is a potential candidate to recognize SARM1 as a phosphorylation substrate. To confirm whether the phosphorylation of SARM1 is a direct reaction for JNK, the purified GST-tagged 405–550 AA region was incubated with some kinases (p38α, JNK1, JNK2, and JNK3) in vitro. These kinases are all in an active form because they effectively phosphorylated their common substrate ATF2 (Fig. S2B). By this approach, we found that JNK family proteins, but not p38α, phosphorylate the recombinant SARM1 substrate (Fig. 3B). The highest activity was observed when mixed with JNK1. In addition, the JNK1-mediated SARM1 phosphorylation was suppressed in the co-presence of a JNK inhibitor in a dose-dependent manner in vitro (Fig. 3C) and in cells (Fig. 3D). That novel identification spurred us to develop a phospho-SARM1 (Ser-548) antibody to determine the biological significance of SARM1 phosphorylation at Ser-548. The newly developed antibody specifically detected phosphorylation of full-length WT SARM1 at Ser-548 because the antibody was not able to detect phosphorylation of the SARM1 mutant protein with S548A (Fig. 3, E and H). By using the antibody, we confirmed that the phosphorylation of SARM1 is actually enhanced by the forced expression of JNKs (Fig. 3, F and H) and conversely inhibited by a JNK inhibitor or the down-regulation of JNKs (Fig. 3G and Fig. S2C). The antibody also showed that the JNK-mediated SARM1 phosphorylation at Ser-548 is not enhanced by other JNK-relevant MAP kinases (Fig. S2D). Collectively, the results indicate that JNK directly and specifically phosphorylates SARM1 at Ser-548.
Figure 3.
JNK phosphorylates SARM1 at Ser-548. A, screening of SARM1-kinase using kinase inhibitors. The FLAG-tagged 405–550 AA region was transfected in HEK293T cells. At 8 h post transfection, the indicated kinase inhibitors (10 μm EGFR-I, 50 μm MEK-I, 5 μm p38-I, 10 μm JNK-I, and 5 μm Akt-I) were added and further incubated for 16 h. Phosphorylation was determined by immunoprecipitation with anti-FLAG agarose, followed by Western blot analysis with anti-phosphorylated serine/threonine antibody. A JNK inhibitor strongly blocked phosphorylation of FLAG-tagged 405–550 AA. B, JNKs rather than p38 directly phosphorylate Ser-548 of SARM1. The purified GST-tagged 405–550 AA region was incubated with the indicated kinases in kinase reaction buffer. JNK1 strongly phosphorylated GST-tagged 405–550 AA. C, phosphorylation of SARM1–Ser-548 by JNK1 is inhibited by a JNK inhibitor in vitro. The purified GST-tagged 405–550 AA region was incubated with JNK1 and various concentrations of a JNK inhibitor in kinase reaction buffer. D, inhibition of intracellular phosphorylation of FLAG-tagged 405–550 AA by the JNK inhibitor. Various concentrations of the JNK inhibitor were added to HEK293T cells. The experiment was performed under conditions similar to those in (A). E, detection of SARM1–Ser-548 phosphorylation by anti–phospho-SARM1 (Ser-548) antibody. The antibody specifically detects Ser-548 phosphorylation of WT SARM1 but not that of S548A mutant. F, overexpression of JNK family proteins enhances SARM1 phosphorylation. FLAG-tagged SARM1 was co-transfected with HA-tagged JNK1–3 into HEK293T cells. SARM1–Ser-548 phosphorylation was detected by anti-phospho-SARM1 (Ser-548) antibody. G, a JNK inhibitor blocks Ser-548 phosphorylation of full-length SARM1. H, detection of Ser-548 phosphorylation of SARM1 by immunostaining. Mitochondria were stained by MitoTracker Orange. Phosphorylated 405–550 AA or SARM1 was quantified by densitometry (phosphorylated protein/total protein).
JNK-mediated phosphorylation of SARM1 enhances SARM1-NADase activity and SARM1-induced ATP reduction
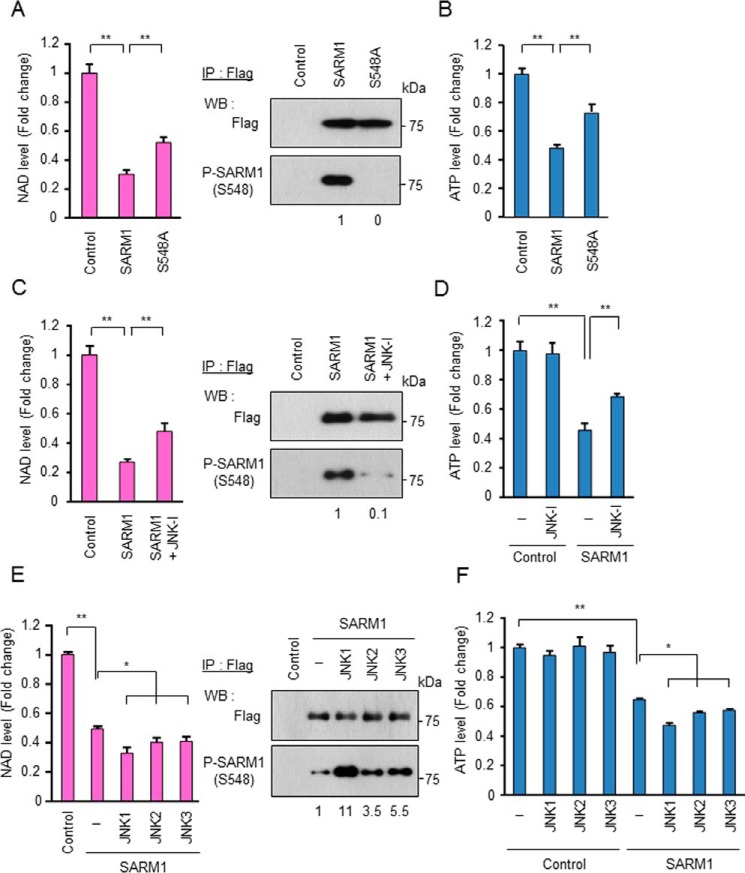
Essuman et al. reported that SARM1 possesses intrinsic NAD+ cleavage activity (28). We therefore investigated the possibility that phosphorylation of SARM1 at Ser-548 enhances NAD+ cleavage activity. To perform the experiment, aberrantly expressed SARM1-FLAG in cells under various conditions was immunopurified by anti-FLAG agarose and incubated with NAD+ and then analyzed for cleavage activities. Cells transfected with an empty vector were used as control cells, and immunoprecipitates from the control cells did not show any reduction of NAD+ level. For comparison, SARM1 wild and S548A mutant types were also immunopurified. The amounts of immunopurified foreign SARM1s were almost the same, but phosphorylation specifically occurred only in WT SARM1 (Fig. 4A, right panel). When we used these preparations for an NAD+ consumption assay, we found that the NAD+ cleavage activity of S548A mutant type was lower than that of the WT (Fig. 4A, left panel). Treatment of the same preparation for WT SARM1 with a JNK inhibitor also reduced both SARM1 phosphorylation and SARM1-NADase activity (Fig. 4C). In contrast, co-overexpression of JNK family proteins with SARM1 WT enhanced SARM1 phosphorylation and SARM1-NADase activity (Fig. 4E). Intracellular ATP and NAD+ levels were changed in correlation with NADase activity of SARM1 (Fig. 4, B, D, and F and Fig. S3, A–C). These results indicate that JNK-mediated phosphorylation of SARM1 at Ser-548 is a regulator of SARM1-NADase activity, which results in reduction of NAD+ and ATP.
Figure 4.
JNK-mediated phosphorylation of SARM1 regulates NAD+ cleavage activity. A, NAD+ cleavage activity of the SARM1-S548A mutant is lower than that of WT. HEK293T cells were transfected with the indicated constructs. FLAG-tagged SARM1 WT or S548A was immunoprecipitated by anti-FLAG agarose. After washing, collected SARM1s were reacted with 1 μm NAD+ for 30 min. NAD+ level was analyzed by the NAD/NADH-Glo assay. B, ATP levels were determined from HEK293T cells expressing the indicated constructs. C and D, a JNK inhibitor blocks SARM1-mediated reduction of NAD+ and ATP. HEK293T cells were transfected with control or SARM1 pDNA. At 8 h post transfection, 10 μm of the JNK inhibitor was added and further incubated for 16 h. E and F, overexpression of JNK1–3 enhances SARM1-mediated reduction of NAD+ and ATP. FLAG-tagged SARM1 was co-transfected with HA-tagged JNK1–3 into HEK293T cells. The experiment was performed under conditions similar to those in (A) and (B). Phosphorylated SARM1 was quantified by densitometry (phosphorylated protein/total protein). Data represent the mean ± S.D. of three independent experiments. *, p < 0.05; **, p < 0.01.
SARM1 is phosphorylated and activated under oxidative stress conditions
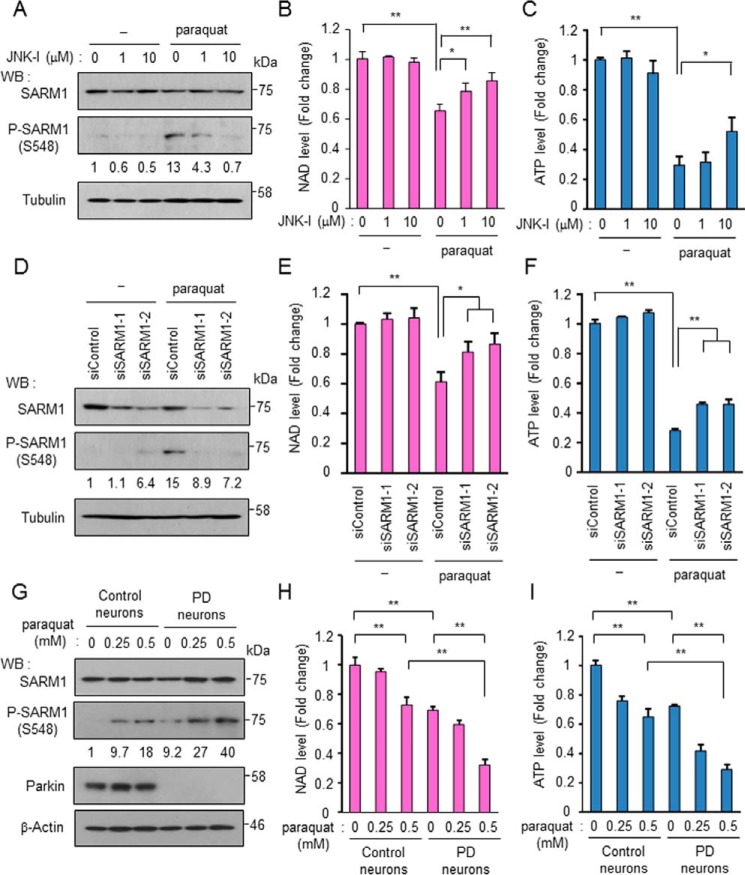
To confirm endogenous SARM1 phosphorylation and activation, neuroblastoma SH-SY5Y cells were treated with various stresses. Oxidative stress treatments such as treatments with 6-hydroxydopamine, paraquat, and rotenone promoted endogenous SARM1 phosphorylation (Fig. S4A). These stresses increased H2O2, one of the ROS, which may be involved in activation of JNK and subsequent SARM1 phosphorylation (Fig. S4B). Inversely, NAD+ and ATP levels were reduced with these stresses (Fig. S4, C and D). We further evaluated paraquat treatment for SARM1 experiments. As expected, the JNK inhibitor blocked paraquat-induced SARM1 phosphorylation (Fig. 5A) and NAD+ and ATP reduction (Fig. 5, B and C) in a dose-dependent manner. At this time, we confirmed proper working of the JNK inhibitor because it blocked cJun phosphorylation but not others, JNK, other MAP kinases, and substrates (Fig. S4E). The JNK inhibitor (JNK inhibitor VIII) was reported to block cJun phosphorylation but not JNK phosphorylation (32). For additional evaluation, SARM1 was down-regulated by siRNAs to determine the significance of SARM1 for paraquat-induced NAD+ and ATP reduction. SARM1 protein level was reduced by siRNA targeting SARM1 (Fig. 5D). The interference procedure did not affect the paraquat-induced phosphorylation of MAP kinases and substrates (Fig. S4F). Notably, down-regulation of SARM1 mitigated the paraquat-induced NAD+ reduction as well as ATP reduction (Fig. 5, E and F). Although the apoptosis rate after paraquat treatment was not so high, it was blocked by treatment with JNK-I or SARM1 siRNA (Fig. S4, G and H). These results suggest that the JNK-SARM1 axis leading to the reduction of NAD+ and ATP is triggered by oxidative stresses.
Figure 5.
SARM1 is phosphorylated and activated under oxidative stress conditions. A, phosphorylation of endogenous SARM1 is induced by paraquat treatment and is blocked by a JNK inhibitor. SH-SY5Y cells were treated with 1 mm paraquat and the indicated concentrations of the JNK inhibitor for 24 h. B and C, NAD+ levels and ATP levels were determined from SH-SY5Y cells treated with paraquat and the JNK inhibitor. D, SARM1 expression was down-regulated by siRNA against SARM1. Control siRNA or SARM1 siRNA was transfected in SH-SY5Y cells. At 48 h post transfection, the cells were treated with 0 or 1 mm paraquat and further incubated for 24 h. E and F, down-regulation of SARM1 inhibits oxidative stress-induced reduction of NAD+ and ATP. NAD+ levels and ATP levels were determined from SH-SY5Y cells treated under conditions similar to those in (D). G, phosphorylation of SARM1 increases in PD patient-derived neurons under oxidative stress conditions. Control iPS cells and PD-derived iPS cells were differentiated into neurons through neural stem cell differentiation. After differentiation induction, the cells were treated with 0, 0.25 or 0.5 mm paraquat for 24 h. H and I, NAD+ levels and ATP levels of control neurons and PD-derived neurons. The treatment of cells was performed under conditions similar to those in (G). Phosphorylated SARM1 was quantified by densitometry (phosphorylated SARM1/total SARM1). *, p < 0.05; **, p < 0.01.
To confirm the clinical importance of SARM1 in PD, we used human induced pluripotent stem cells (iPSCs) from a healthy person (control) and from a familial PD patient with a deletion in the parkin gene (33). The iPSCs were differentiated to mature neurons through neural stem cells (NSC) (Fig. S5A). Although the cellular condition of PD neurons was worse than that of control neurons, the differentiation states of the cells were almost the same. That is, these neurons commonly expressed MAP2 (a neuron marker) and tyrosine hydroxylase (a dopaminergic neuron marker) at similar levels, whereas glial fibrillary acidic protein, an astrocyte marker, was not expressed in any of the cells (Fig. S5B). Control and PD neurons were then treated with paraquat for 24 h, and endogenous SARM1 activation was examined. We found that phosphorylation of SARM1 was increased in control neurons after stimulation with paraquat (Fig. 5G). Surprisingly, phosphorylation levels of SARM1 in PD neurons were already at high levels under the regular culture condition, being comparable with the levels in paraquat-stimulated control neurons. In addition, the basal phosphorylation level in PD neurons was further increased by paraquat treatment (Fig. 5G). Phosphorylation levels of JNK and cJun were correlated with that of SARM1 (Fig. S5C). On the other hand, phosphorylation levels of other JNK-relevant MAP kinases showed different patterns from SARM1, JNK, and cJun phosphorylation except for the ERK phosphorylation between control neurons and PD neurons (Fig. S5C). Rates of reduction in NAD+ and ATP and rates of increase in ROS after paraquat treatment were also markedly higher in PD neurons than in control neurons (Fig. 5, H and I and Fig. S5D). These results indicate that the JNK-SARM1 axis leading to ATP reduction is workable in not only neuronal cell lines but also actual neuronal cells and that the identified pathway is affected by oxidative stress conditions. Considering the sensitivity of SARM1 to activation in PD neurons, SARM1 activation may be involved in the pathogenesis and subsequent progression of PD.
Discussion
In this study, we showed that Ser-548 of SARM1 is phosphorylated by a stress-responsive kinase, JNK, and that the phosphorylation enhances NAD+ cleavage activity of SARM1 and inhibition of mitochondrial respiration. The function of each domain of SARM1 has been uncovered in recent studies. It has been shown that the SAM domain promotes dimer formation of SARM1 and that the TIR domain dimerized via the SAM domain evokes NAD+ loss, eventually leading to axonal degeneration and neuronal cell death (17, 18). It has also been suggested that the ARM domain binds to the TIR domain and inhibits its activity (27, 34). The phosphorylation site, Ser-548, we found is located between the SAM domain and TIR domain. Because mutation or inhibition of Ser-548 phosphorylation reduced the NAD+ cleavage activity of SARM1 (Fig. 4), the phosphorylation may stimulate the structural motion of SARM1 to turn into an active form that probably requires dimerization of the TIR domain.
Yang et al. reported that the MAPK cascade induces axonal degeneration in coordination with SARM1 (35). In their report, they argued that the MAPK cascade is positioned downstream of SARM1 because there is no appreciable difference in the degree of recovery from axonal degeneration in the cases of down-regulation procedures by targeting either or both of MKK4/7 and SARM1 for regular time intervals. On the other hand, Walker et al. suggested the presence of MAPK signaling over the SARM1 position (36). Being consistent with this, we also found that JNK directly phosphorylates SARM1 and regulates its activity, i.e. JNK is located upstream of SARM1. The controversial point may be explained by the unexpected presence of a positive feedback loop between SARM1 and the MAPK cascade. Further studies will be needed to demonstrate it.
Although phosphorylation of SARM1 at Ser-548 is involved in SARM1-mediated NAD+ consumption and ATP reduction, the phospho-modification at Ser-548 is not covered by the entire activity of SARM1. This result suggests the existence of another positive regulatory mechanism such as posttranslational modifications or gene expression that may have an additional role in SARM1 activation coupled with the identified Ser-548 phosphorylation. For SARM1 transcription, it has been reported that activation of the innate immune system by stimulation with lipopolysaccharide, Toll-like receptor (TLR7/9) ligand, or La Crosse virus increases the expression level of SARM1 mRNA and accelerates neuronal cell death (37–39). Thus, the renowned TLR downstream pathways may play a role in positive regulation of SARM1 transcription. Because SARM1 is known as one of the TLR adaptor proteins because of its own TIR domain, the SARM1-TIR domain may have a dual role, NAD+ consumption and TLR adaptor, which may independently or coordinately function in neuronal cells in physiological and pathological settings.
We showed that basal activity of SARM1 is highly increased in PD by using an in vitro model. Endogenous SARM1 phosphorylation was further elevated under oxidative stress conditions, suggesting an unusual role of SARM1 in the pathogenesis of PD. Because SARM1 inhibits mitochondrial respiration, defective mitochondria may accumulate in cells if PINK1 or Parkin do not function by PD genetic mutation. Additional verification that SARM1 contributes to the pathogenesis of PD by using an in vivo model is needed.
In summary, our findings revealed that SARM1 is an inhibitor of mitochondrial respiration through cellular NAD+ and ATP reduction when phosphorylated at Ser-548 by JNK. Because of the congenital up-regulation of the phosphorylation in PD-derived neuronal cells, SARM1 may play a crucial role in the pathogenesis of PD and probably other neurodegeneration-related diseases. Interestingly, several studies have shown that experimental deletion of SARM1 resulted in significant recovery of serious symptoms caused by neurological diseases in animal models (19, 22–25, 40). Therefore, targeting SARM1 phosphorylation may become an effective option among SARM1 targeting strategies for neurodegenerative diseases.
Experimental procedures
Cell culture
HEK293T and SH-SY5Y cells were cultured in DMEM/F12 (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS). SNL76/7 cells were cultured in KnockOut DMEM supplemented with 10% FBS and 2 mm l-glutamine and used as feeder cells after treatment with 12 μg/ml mitomycin C (Sigma-Aldrich) for 2.5 h. To obtain human matured neuronal cells in a culture system, 201B7 iPSCs were purchased from RIKEN BRC (Tsukuba, Japan) as control cells. Familial Parkinson's disease PARK2 patient-derived iPSCs were kindly provided by Dr. Okano (33). Control iPSCs and PD iPSCs were maintained on feeder cells in DMEM/F12 supplemented with 20% KnockOut Serum Replacement (Thermo Fisher Scientific), 1% nonessential amino acid (Sigma-Aldrich), 2 mm l-glutamine, 80 μm β-mercaptoethanol, and 5 ng/ml bFGF (Wako Chemicals). All of the experimental procedures for iPSCs were approved by the Okayama University Ethics Committee (Approval Number: 1609–031). Induction of NSCs from iPSCs was done using PSC neural induction medium (Thermo Fisher Scientific) according to the manufacturer's instructions. After neural induction for 7 days, P0 NSCs were expanded on a Geltrex-coated (Thermo Fisher Scientific) dish with neural expansion medium. For differentiation into neurons, NSCs were cultured on a poly-l-lysine and laminin-coated dish in Neurobasal Plus Medium (Thermo Fisher Scientific) supplemented with 2% B-27 Plus Supplement (Thermo Fisher Scientific), 2 mm GlutaMAX Supplement (Thermo Fisher Scientific), CultureOne Supplement (Thermo Fisher Scientific), 200 μm ascorbic acid (Sigma-Aldrich), 10 ng/ml glial cell-derived neurotrophic factor (Cell Guidance Systems), and 10 ng/ml brain-derived neurotrophic factor (Cell Guidance Systems) for 7 days. Characterization of cells was confirmed with specific cell markers by RT-PCR.
Chemicals and antibodies
Paraquat was purchased from Wako Chemicals. 6-Hydroxydopamine hydrobromide, rotenone, CCCP, valinomycin, cyclosporin A, and an EGFR inhibitor (AG-1478) were purchased from Sigma-Aldrich. A MEK inhibitor (PD98059) and a p38 inhibitor (SB202190) were purchased from Cell Signaling Technologies. A JNK inhibitor (JNK inhibitor VIII), an Akt inhibitor (Akt inhibitor X), FK-866, and NR were purchased from Cayman Chemical. NAD was purchased from SERVA Electrophoresis GmbH. Recombinant p38α, JNK1, JNK2, and JNK3 proteins were purchased from Thermo Fisher Scientific. The antibodies used were as follows: an antibody against SARM1 (13022), PINK1 (6946), Akt (4691), phosoho-Akt (4060), ATF2 (9226), phospho-ATF2 (5112), cJun (9165), phospho-cJun (3270), ERK (4695), phospho-ERK (4370), GSK3β (9315), phospho-GSK3β (9322), HSP27 (2402), phospho-HSP27 (2401), JNK (9252), phospho-JNK (9255), p38 (8690), phospho-p38 (4511), HRP-conjugated anti-mouse, and anti-rabbit secondary antibodies (Cell Signaling Technologies), an antibody against FLAG (DDDDK-tag, PM020), an HRP-conjugated anti-goat secondary antibody (MBL), an antibody against phospho-serine/threonine (612549; BD Transduction Laboratories), an antibody against GST (27457701; GE Healthcare Life Sciences), an antibody against Parkin (mouse monoclonal, sc-32282; Santa Cruz Biotechnology), an antibody against tubulin (mouse monoclonal, T5168; Sigma-Aldrich), and an antibody against β-actin (mouse monoclonal, A2228; Sigma-Aldrich). A mouse mAb against phospho-SARM1 (Ser-548) was generated by ITM Co. (Matsumoto, Japan). The antibody was produced by immunizing animals with a synthetic phospho-peptide corresponding to residues surrounding Ser-548 of human SARM1 (AAREMLHpSPLPCTGG).
Plasmid constructs
Conventional molecular biological techniques were used to generate the following expression constructs: C-terminal FLAG-tagged human WT SARM1, SARM1 mutants with domain deletions (ΔMTS, deletion of 1–27 amino acids (AA); ΔARM, deletion of 28–404 AA; ΔTIR, deletion of 551–724 AA), and SARM1 domain (1–404 AA containing MTS and ARM domain, 405–550 AA containing SAM domain, and 551–724 AA containing TIR domain), and N-terminal HA-tagged JNK1, JNK2, and JNK3. A point mutation of SARM1 or SAM domain was generated by the inverse PCR method using a KOD-Plus-Mutagenesis Kit (Toyobo). These constructs were ligated with the C-TSC vector (41). All expression constructs were sequenced to ensure that the fusion was in the correct reading frame and there were no additional mutations.
Cell transfection
For plasmid transfection, cells were transfected with the indicated plasmids using FuGENE-HD (Promega Biosciences) according to the manufacturer's instructions. Transfection was carried out at 40% cell density. For 12-well plates in transfection experiments, 2 μg of pDNA and 4 μl of FuGENE-HD were used. For double transfection of genes such as SARM1 and JNK1, each 1.5 μg of pDNA and 6 μl of FuGENE-HD were used.
For RNAi, StealthRNAi siRNA targeting SARM1 (HSS177170 and HSS177172; Thermo Fisher Scientific), siGENOME SMARTpool siRNA targeting JNK1 (5599; Dharmacon), JNK2 (5601; Dharmacon), or JNK3 (5602; Dharmacon) were transfected into cells at 20 nm using Lipofectamine RNAiMAX (Invitrogen). Stealth RNAi siRNA negative control (Med GC; Thermo Fisher Scientific) or siGENOME Non-Targeting siRNA pool (Dharmacon) was used as a negative control. The transfection was also performed at 40% cell density. For 12-well plates in transfection experiments, 2 μl of siRNA (final concentration of 20 nm) and 4 μl of Lipofectamine RNAiMAX were used.
Purification of recombinant protein
The DNA region of 405–550 amino acids of SARM1 was inserted into the pGEX-6P1 vector (GE Healthcare). Recombinant protein was produced in Escherichia coli BL21(DE3)-RP (Agilent). Transformed E. coli cells were cultured in Luria-Bertani medium containing 100 μg/ml of ampicillin at 37 °C. When the optical density of the medium at 600 nm had reached ∼0.8, 1 mm isopropyl 1-thio-β-d-galactopyranoside was added and the cells were cultured for another 16 h at 25 °C. Harvested E. coli cells were then lysed by performing a freeze/thaw/sonication cycle twice. Resulting lysates were centrifuged to remove insoluble debris, and recombinant proteins were purified from supernatants by a GSH Sepharose 4B column (GE Healthcare). Briefly, the cell lysate was applied to the column, nonspecifically bound proteins were washed out with PBS containing 0.1% Tween 20, and then specifically bound GST-fused proteins were eluted with 50 mm Tris-HCl (pH 8.0) containing 10 mm of the reduced form of GSH. Recovered GST-fused protein was dialyzed against PBS to remove GSH.
In vitro kinase assay
Purified GST-fused 405–550 amino acids containing the SAM domain or recombinant ATF2 (Abnova) were used as a substrate. Two μg of protein was incubated with 0.1 μg of kinase (p38α, JNK1, JNK2, or JNK3; Thermo Fisher Scientific) in a kinase buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 10 mm MgCl2, 10 mm MnCl2, 1.8 mm ATP 1× PhosphoSTOP (Roche Applied Science), and 1 mm DTT) for 1 h. The reaction was stopped by adding 3× SDS sample buffer. For experiments on inhibition of phosphorylation, a JNK inhibitor (JNK inhibitor VIII) was added to the reaction buffer.
Immunoprecipitation and Western blot analysis
For immunoprecipitation, cells (1,000,000 cells/well in 6-well plates) were lysed in an ice-cold lysis buffer (40 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 1% Triton X-100) with PhosphoSTOP. Supernatants were incubated with 20 μl of a 50% slurry of monoclonal anti-FLAG-M2 agarose (Sigma-Aldrich) for 90 min. Immunoprecipitates were washed three times with the lysis buffer, eluted with 0.1 m Gly-HCl (pH 2.5) solution, and immediately neutralized with 1 m Tris-HCl, pH 9.0. Western blot analysis was performed under conventional conditions after lysing cells using SDS sample buffer with PhosphoSTOP. Ten μg of protein extracts was separated by SDS-PAGE and electrotransferred onto an Immobilon membrane (Millipore). To detect immunoreactive proteins, we used HRP-conjugated anti-mouse, anti-rabbit or anti-goat secondary antibodies and Pierce Western Blotting Substrate Plus (Thermo Fisher Scientific).
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde and permeabilized with 1% Triton X-100 in PBS for 30 min at room temperature. After incubation with the respective first antibodies, the samples were incubated with Alexa Fluor 488 goat anti-mouse IgG antibody (Invitrogen). Mitochondria were stained with MitoTracker Orange CMTMRos (Invitrogen). The specimens were observed using a fluorescence microscope (model BZ-X700; KEYENCE).
ATP assay, ROS assay, and apoptosis assay
CellTiter-Glo Assay (Promega Biosciences) was used to analyze ATP levels. According to the manufacturer's instructions, cells were incubated with CellTiter-Glo detection reagent for 10 min. Luminescence was observed using Fluoroskan Ascent FL (Thermo Fisher Scientific).
ROS-Glo H2O2 assay (Promega Biosciences) was used for an ROS assay. The cells were incubated with test compounds and H2O2 substrate solution for 6 h and then ROS-Glo detection solution was added. Luminescence was observed using Fluoroskan Ascent FL. CM-H2DCFDA staining was also used for an ROS assay. The cells were incubated with 1 μm CM-H2DCFDA for 30 min. After washing, ROS-induced green fluorescence was observed by a fluorescent microscope. Apoptotic cells were identified after staining with Hoechst 33342 (Thermo Fisher Scientific) for 30 min.
NAD assay
To analyze cellular NAD levels, cells were transfected with the indicated plasmids for 8 h and then the cells were re-seeded at 30,000/well in 96-well plates and cultured for 16 h. The NAD/NADH-Glo assay (Promega Biosciences) was used to analyze NAD levels. According to the manufacturer's instructions, cells were incubated with NAD/NADH-Glo reagent for 30 min. Luminescence was observed using Fluoroskan Ascent FL. To analyze NAD cleavage activity of SARM1, a total of 20 μl of anti-FLAG-M2 agarose was reacted with the cell lysates (see the method for immunoprecipitation). After washing, the beads were incubated with 1 μm NAD in PBS for 30 min. NAD level was measured by the NAD/NADH-Glo assay.
Measurement of oxygen consumption rate (OCR)
OCR measurements were performed using the XF96 Extracellular Flux analyzer (Seahorse Bioscience). Transfected cells with indicated plasmids were seeded at 20,000/well in XF96 cell culture plates and incubated for 16 h. Prior to performing an assay, the growth medium in the wells was exchanged with XF base medium supplemented with 17.5 mm glucose, 0.5 mm sodium pyruvate, and 2.5 mm GlutaMAX. To analyze OCR, 1.5 μm oligomycin (Sigma-Aldrich), 0.25 μm FCCP (Cayman Chemical), and 0.5 μm rotenone/antimycin A (Sigma-Aldrich) were used.
Reverse-transcriptional PCR
Total RNA was prepared using an SV Total RNA Isolation System (Promega Biosciences). First-strand cDNA synthesis was performed with total RNA using a SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen). Synthesized cDNA was used for PCR analysis using Taq polymerase (New England Biolabs) with primers targeting Oct3/4 (forward, CTTGCTGCAGAAGTGGGTGGAGGAA; reverse, CTGCAGTGTGGGTTTCGGGCA, 171 bp), MAP2 (forward, CCACCTGAGATTAAGGATCA; reverse, GGCTTACTTTGCTTCTCTGA, 482 bp), tyrosine hydroxylase (forward, TCATCACCTGGTCACCAAGTT; reverse, GGTCGCCGTGCCTGTACT, 126 bp), glial fibrillary acidic protein (forward, GTACCAGGACCTGCTCAAT; reverse, CAACTATCCTGCTTCTGCTC, 321 bp), and GAPDH (forward, ATTCCATGGCACCGTCAAGGCT; reverse, TCAGGTCCACCACTGACACGTT, 571 bp).
Statistical analysis
Prior to statistical analysis, each experiment was repeated three times. The results are expressed as mean ± S.D. For comparison, analysis of variance (ANOVA) was used. If the ANOVA showed a significant difference, the Bonferroni procedure was used as a post hoc test. p values of less than 0.05 were considered statistically significant.
Author contributions
H. M. and M. S. conceptualization; H. M., A. N., and K.-I. Y. data curation; H. M. and A. N. formal analysis; H. M., R. K., and M. S. supervision; H. M. and M. S. funding acquisition; H. M., C. C. K., K.-I. Y., R. K., and M. S. investigation; H. M. and C. C. K. methodology; H. M. writing-original draft; H. M., K.-I. Y., R. K., and M. S. project administration; M. S. writing-review and editing.
Supplementary Material
This work was supported by Japan Science and Technology Agency Grant 16K19037 and Japan Agency for Medical Research and Development Grant JP17km0908001 (to H.M.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5.
- PD
- Parkinson's disease
- AA
- amino acids
- ARM
- armadillo/HEAT motif
- CCCP
- carbonyl cyanide m-chlorophenyl hydrazine
- ERK
- extracellular signal-regulated kinase
- iPSCs
- induced pluripotent stem cells
- JNK
- c-jun N-terminal kinase
- MAP
- mitogen-activated protein
- MAPK
- mitogen-activated protein kinase
- MTS
- mitochondrial targeting signal
- NAMPT
- nicotinamide phosphoribosyltransferase
- NR
- nicotinamide riboside
- NSCs
- neural stem cells
- OCR
- oxygen consumption rate
- PTP
- permeability transition pore
- ROS
- reactive oxygen species
- SAM
- sterile alpha motif
- TIR
- Toll/interleukin-1 receptor
- TLR
- Toll-like receptor.
References
- 1. Deng H., Wang P., and Jankovic J. (2018) The genetics of Parkinson disease. Ageing Res. Rev. 42, 72–85 10.1016/j.arr.2017.12.007 [DOI] [PubMed] [Google Scholar]
- 2. Murata H., Sakaguchi M., Jin Y., Sakaguchi Y., Futami J., Yamada H., Kataoka K., and Huh N. H. (2011) A new cytosolic pathway from a Parkinson disease-associated kinase, BRPK/PINK1: Activation of AKT via mTORC2. J. Biol. Chem. 286, 7182–7189 10.1074/jbc.M110.179390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jin Y., Murata H., Sakaguchi M., Kataoka K., Watanabe M., Nasu Y., Kumon H., and Huh N. H. (2012) Partial sensitization of human bladder cancer cells to a gene-therapeutic adenovirus carrying REIC/Dkk-3 by downregulation of BRPK/PINK1. Oncol. Rep. 27, 695–699 10.3892/or.2011.1543 [DOI] [PubMed] [Google Scholar]
- 4. Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C. A., Sou Y. S., Saiki S., Kawajiri S., Sato F., Kimura M., Komatsu M., Hattori N., and Tanaka K. (2010) PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189, 211–221 10.1083/jcb.200910140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shiba-Fukushima K., Imai Y., Yoshida S., Ishihama Y., Kanao T., Sato S., and Hattori N. (2012) PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci. Rep. 2, 1002 10.1038/srep01002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kane L. A., Lazarou M., Fogel A. I., Li Y., Yamano K., Sarraf S. A., Banerjee S., and Youle R. J. (2014) PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 205, 143–153 10.1083/jcb.201402104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Koyano F., Okatsu K., Kosako H., Tamura Y., Go E., Kimura M., Kimura Y., Tsuchiya H., Yoshihara H., Hirokawa T., Endo T., Fon E. A., Trempe J. F., Saeki Y., Tanaka K., and Matsuda N. (2014) Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510, 162–166 10.1038/nature13392 [DOI] [PubMed] [Google Scholar]
- 8. Gegg M. E., Cooper J. M., Chau K. Y., Rojo M., Schapira A. H., and Taanman J. W. (2010) Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum. Mol. Genet. 19, 4861–4870 10.1093/hmg/ddq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Geisler S., Holmström K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., and Springer W. (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12, 119–131 10.1038/ncb2012 [DOI] [PubMed] [Google Scholar]
- 10. Murata H., Sakaguchi M., Kataoka K., and Huh N. H. (2013) SARM1 and TRAF6 bind to and stabilize PINK1 on depolarized mitochondria. Mol. Biol. Cell 24, 2772–2784 10.1091/mbc.e13-01-0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Osterloh J. M., Yang J., Rooney T. M., Fox A. N., Adalbert R., Powell E. H., Sheehan A. E., Avery M. A., Hackett R., Logan M. A., MacDonald J. M., Ziegenfuss J. S., Milde S., Hou Y. J., Nathan C., et al. (2012) dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science 337, 481–484 10.1126/science.1223899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Conforti L., Gilley J., and Coleman M. P. (2014) Wallerian degeneration: An emerging axon death pathway linking injury and disease. Nat. Rev. Neurosci. 15, 394–409 10.1038/nrn3680 [DOI] [PubMed] [Google Scholar]
- 13. Ito Y., Ofengeim D., Najafov A., Das S., Saberi S., Li Y., Hitomi J., Zhu H., Chen H., Mayo L., Geng J., Amin P., DeWitt J. P., Mookhtiar A. K., Florez M., et al. (2016) RIPK1 mediates axonal degeneration by promoting inflammation and necroptosis in ALS. Science 353, 603–608 10.1126/science.aaf6803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Su K. G., Banker G., Bourdette D., and Forte M. (2009) Axonal degeneration in multiple sclerosis: The mitochondrial hypothesis. Curr. Neurol. Neurosci. Rep. 9, 411–417 10.1007/s11910-009-0060-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kanaan N. M., Pigino G. F., Brady S. T., Lazarov O., Binder L. I., and Morfini G. A. (2013) Axonal degeneration in Alzheimer's disease: When signaling abnormalities meet the axonal transport system. Exp. Neurol. 246, 44–53 10.1016/j.expneurol.2012.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Burke R. E., and O'Malley K. (2013) Axon degeneration in Parkinson's disease. Exp. Neurol. 246, 72–83 10.1016/j.expneurol.2012.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gerdts J., Summers D. W., Sasaki Y., DiAntonio A., and Milbrandt J. (2013) Sarm1-mediated axon degeneration requires both SAM and TIR interactions. J. Neurosci. 33, 13569–13580 10.1523/JNEUROSCI.1197-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gerdts J., Brace E. J., Sasaki Y., DiAntonio A., and Milbrandt J. (2015) SARM1 activation triggers axon degeneration locally via NAD+ destruction. Science 348, 453–457 10.1126/science.1258366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gilley J., Orsomando G., Nascimento-Ferreira I., and Coleman M. P. (2015) Absence of SARM1 rescues development and survival of NMNAT2-deficient axons. Cell Rep. 10, 1974–1981 10.1016/j.celrep.2015.02.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim Y., Zhou P., Qian L., Chuang J. Z., Lee J., Li C., Iadecola C., Nathan C., and Ding A. (2007) MyD88–5 links mitochondria, microtubules, and JNK3 in neurons and regulates neuronal survival. J. Exp. Med. 204, 2063–2074 10.1084/jem.20070868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Summers D. W., DiAntonio A., and Milbrandt J. (2014) Mitochondrial dysfunction induces Sarm1-dependent cell death in sensory neurons. J. Neurosci. 34, 9338–9350 10.1523/JNEUROSCI.0877-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Geisler S., Doan R. A., Strickland A., Huang X., Milbrandt J., and DiAntonio A. (2016) Prevention of vincristine-induced peripheral neuropathy by genetic deletion of SARM1 in mice. Brain 139, 3092–3108 10.1093/brain/aww251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gilley J., Ribchester R. R., and Coleman M. P. (2017) Sarm1 deletion, but not Wld(S), confers lifelong rescue in a mouse model of severe axonopathy. Cell Rep. 21, 10–16 10.1016/j.celrep.2017.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Henninger N., Bouley J., Sikoglu E. M., An J., Moore C. M., King J. A., Bowser R., Freeman M. R., and Brown R. H. Jr. (2016) Attenuated traumatic axonal injury and improved functional outcome after traumatic brain injury in mice lacking Sarm1. Brain 139, 1094–1105 10.1093/brain/aww001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vérièpe J., Fossouo L., and Parker J. A. (2015) Neurodegeneration in C. elegans models of ALS requires TIR-1/Sarm1 immune pathway activation in neurons. Nat. Commun. 6, 7319 10.1038/ncomms8319 [DOI] [PubMed] [Google Scholar]
- 26. Panneerselvam P., Singh L. P., Ho B., Chen J., and Ding J. L. (2012) Targeting of pro-apoptotic TLR adaptor SARM to mitochondria: definition of the critical region and residues in the signal sequence. Biochem. J. 442, 263–271 10.1042/BJ20111653 [DOI] [PubMed] [Google Scholar]
- 27. Summers D. W., Gibson D. A., DiAntonio A., and Milbrandt J. (2016) SARM1-specific motifs in the TIR domain enable NAD+ loss and regulate injury-induced SARM1 activation. Proc. Natl. Acad. Sci. U.S.A. 113, E6271–E6280 10.1073/pnas.1601506113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Essuman K., Summers D. W., Sasaki Y., Mao X., DiAntonio A., and Milbrandt J. (2017) The SARM1 Toll/interleukin-1 receptor domain possesses intrinsic NAD(+) cleavage activity that promotes pathological axonal degeneration. Neuron 93, 1334–1343.e1335 10.1016/j.neuron.2017.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Panneerselvam P., Singh L. P., Selvarajan V., Chng W. J., Ng S. B., Tan N. S., Ho B., Chen J., and Ding J. L. (2013) T-cell death following immune activation is mediated by mitochondria-localized SARM. Cell Death Differ. 20, 478–489 10.1038/cdd.2012.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Choi S. W., Gerencser A. A., and Nicholls D. G. (2009) Bioenergetic analysis of isolated cerebrocortical nerve terminals on a microgram scale: Spare respiratory capacity and stochastic mitochondrial failure. J. Neurochem. 109, 1179–1191 10.1111/j.1471-4159.2009.06055.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Di Lisa F., Menabò R., Canton M., Barile M., and Bernardi P. (2001) Opening of the mitochondrial permeability transition pore causes depletion of mitochondrial and cytosolic NAD+ and is a causative event in the death of myocytes in postischemic reperfusion of the heart. J. Biol. Chem. 276, 2571–2575 10.1074/jbc.M006825200 [DOI] [PubMed] [Google Scholar]
- 32. Zhang T., Inesta-Vaquera F., Niepel M., Zhang J., Ficarro S. B., Machleidt T., Xie T., Marto J. A., Kim N., Sim T., Laughlin J. D., Park H., LoGrasso P. V., Patricelli M., Nomanbhoy T. K., Sorger P. K., Alessi D. R., and Gray N. S. (2012) Discovery of potent and selective covalent inhibitors of JNK. Chem. Biol. 19, 140–154 10.1016/j.chembiol.2011.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Imaizumi Y., Okada Y., Akamatsu W., Koike M., Kuzumaki N., Hayakawa H., Nihira T., Kobayashi T., Ohyama M., Sato S., Takanashi M., Funayama M., Hirayama A., Soga T., Hishiki T., et al. (2012) Mitochondrial dysfunction associated with increased oxidative stress and alpha-synuclein accumulation in PARK2 iPSC-derived neurons and postmortem brain tissue. Mol. Brain 5, 35 10.1186/1756-6606-5-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gerdts J., Summers D. W., Milbrandt J., and DiAntonio A. (2016) Axon self-destruction: New links among SARM1, MAPKs, and NAD+ metabolism. Neuron 89, 449–460 10.1016/j.neuron.2015.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang J., Wu Z., Renier N., Simon D. J., Uryu K., Park D. S., Greer P. A., Tournier C., Davis R. J., and Tessier-Lavigne M. (2015) Pathological axonal death through a MAPK cascade that triggers a local energy deficit. Cell 160, 161–176 10.1016/j.cell.2014.11.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Walker L. J., Summers D. W., Sasaki Y., Brace E. J., Milbrandt J., and DiAntonio A. (2017) MAPK signaling promotes axonal degeneration by speeding the turnover of the axonal maintenance factor NMNAT2. eLife 6, e22540 10.7554/eLife.22540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peng J., Yuan Q., Lin B., Panneerselvam P., Wang X., Luan X. L., Lim S. K., Leung B. P., Ho B., and Ding J. L. (2010) SARM inhibits both TRIF- and MyD88-mediated AP-1 activation. Eur. J. Immunol. 40, 1738–1747 10.1002/eji.200940034 [DOI] [PubMed] [Google Scholar]
- 38. Mukherjee P., Winkler C. W., Taylor K. G., Woods T. A., Nair V., Khan B. A., and Peterson K. E. (2015) SARM1, not MyD88, mediates TLR7/TLR9-induced apoptosis in neurons. J. Immunol. 195, 4913–4921 10.4049/jimmunol.1500953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mukherjee P., Woods T. A., Moore R. A., and Peterson K. E. (2013) Activation of the innate signaling molecule MAVS by bunyavirus infection up-regulates the adaptor protein SARM1, leading to neuronal death. Immunity 38, 705–716 10.1016/j.immuni.2013.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Turkiew E., Falconer D., Reed N., and Höke A. (2017) Deletion of Sarm1 gene is neuroprotective in two models of peripheral neuropathy. J. Peripher. Nerv. Syst. 22, 162–171 10.1111/jns.12219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sakaguchi M., Watanabe M., Kinoshita R., Kaku H., Ueki H., Futami J., Murata H., Inoue Y., Li S. A., Huang P., Putranto E. W., Ruma I. M., Nasu Y., Kumon H., and Huh N. H. (2014) Dramatic increase in expression of a transgene by insertion of promoters downstream of the cargo gene. Mol. Biotechnol. 56, 621–630 10.1007/s12033-014-9738-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.