Abstract
Two DNA methyltransferases, Dam and β-class cell cycle–regulated DNA methyltransferase (CcrM), are key mediators of bacterial epigenetics. CcrM from the bacterium Caulobacter crescentus (CcrM C. crescentus, methylates adenine at 5′-GANTC-3′) displays 105–107-fold sequence discrimination against noncognate sequences. However, the underlying recognition mechanism is unclear. Here, CcrM C. crescentus activity was either improved or mildly attenuated with substrates having one to three mismatched bp within or adjacent to the recognition site, but only if the strand undergoing methylation is left unchanged. By comparison, single-mismatched substrates resulted in up to 106-fold losses of activity with α (Dam) and γ-class (M.HhaI) DNA methyltransferases. We found that CcrM C. crescentus has a greatly expanded DNA-interaction surface, covering six nucleotides on the 5′ side and eight nucleotides on the 3′ side of its recognition site. Such a large interface may contribute to the enzyme's high sequence fidelity. CcrM C. crescentus displayed the same sequence discrimination with single-stranded substrates, and a surprisingly large (>107-fold) discrimination against ssRNA was largely due to the presence of two or more riboses within the cognate (DNA) site but not outside the site. Results from C-terminal truncations and point mutants supported our hypothesis that the recently identified C-terminal, 80-residue segment is essential for dsDNA recognition but is not required for single-stranded substrates. CcrM orthologs from Agrobacterium tumefaciens and Brucella abortus share some of these newly discovered features of the C. crescentus enzyme, suggesting that the recognition mechanism is conserved. In summary, CcrM C. crescentus uses a previously unknown DNA recognition mechanism.
Keywords: DNA methylation, enzyme catalysis, DNA enzyme, DNA-binding protein, DNA damage
Introduction
A key driver of epigenetics in bacteria relies on two “orphan” DNA methyltransferases, Dam (α-class DNA adenine methyltransferase, modifies 5′-GATC-3′ sites) and CcrM (β-class cell cycle regulated methyltransferase, modifies 5′-GANTC-3′ sites) (1). Although Dam is important for DNA replication, mismatch repair, and gene regulation, CcrM2 along with three other proteins (DnaA, GcrA, and CtrA) appears to be exclusively involved in controlling the expression of at least 200 genes (2, 3). CcrM Caulobacter crescentus helps to orchestrate the cell cycle–regulated asymmetric cell division in which a single genome gives rise to two distinct and heritable cell types (flagellated swarmer cell and a stalked cell). At the beginning of the cell cycle in the Gram-negative, aquatic bacterium C. crescentus, the chromosome is fully methylated at CcrM sites. Following replication, the two replication forks proceed bidirectionally, generating hemimethylated DNA. CcrM C. crescentus is only expressed near the end of replication and is present for a short 10–20-min window. It is during this period that the 4,515 GANTC sites are rapidly remethylated (2–4).
Originally discovered in C. crescentus, CcrM orthologs are widespread throughout the α-class of proteobacteria and are important for several organisms, including the human pathogen Brucella abortus (5, 6). Based on the arrangement of conserved motifs, CcrM C. crescentus is a β-class adenine N6-methyltransferase. CcrM C. crescentus (358 amino acids) and some β-class adenine N6-methyltransferases that recognize 5′-GANTC-3′ sites have an unusual C-terminal 80-residue extension of unknown function. We recently reported that CcrM C. crescentus is a functional monomer, with KmDNA (17 nm), kcat (5.2 min−1), kmethylation (0.6–8.0 min−1 depending on the flanking sequence), KdDNA (10–100 nm) values, and the rate-limiting step is either methylation or a step preceding methylation (7). kmethylation is particularly slow when compared with other DNA adenine N6-methyltransferases such as EcoRI (2,460 min−1) and T4 Dam (36 min−1) studied under similar conditions of ionic strength, suggesting that CcrM C. crescentus may have steps missing from the pathways of these other enzymes or are simply slower in CcrM C. crescentus (7). The enzyme shows a 107-fold loss in specificity for an AATTC site versus the cognate GACTC sequence in dsDNA, primarily driven by changes in methylation. This is an unprecedented level of discrimination based on a single-bp change for any DNA methyltransferase by 3–4 orders of magnitude. This may be particularly important for CcrM C. crescentus, because the inappropriate methylation of genomic DNA sequences could disrupt gene regulation (2–4). Our demonstration that CcrM C. crescentus also modifies ssDNA with similar discrimination, efficiency, and high processivity (7) provides an experimental approach to investigate its DNA recognition mechanism. These findings are poorly accommodated by current models for DNA recognition and, in particular, other DNA methyltransferases (8, 9).
DNA recognition is now well-understood, based on many protein–DNA structures (10). There are few examples of protein–ssDNA complexes, particularly those involving high levels of sequence-specific binding. By far the best understood example of a protein that binds single-stranded and dsDNA with some level of sequence discrimination is the Pur protein family (DNA replication, transcription), which is conserved from bacteria to humans (11–14). Pur α binds ssDNA and ssRNA with poor, micromolar affinity. Interestingly, Pur α binds dsDNA with similar affinity and has ATP-independent dsDNA destabilizing activity. By binding the purine-rich strand within promoters, it is proposed to create localized melting, making the pyrimidine-rich strand available for binding by other regulatory proteins, thereby acting as a transcriptional activator. The cocrystal structures of Pur α complexed with ssDNA (5′-GCGGCGG-3′) provides the basis for the model that the protein melts the dsDNA (13, 14). Although DNA methyltransferases, some endonucleases, and repair enzymes are well-known to cause a localized disruption of dsDNA by virtue of “flipping” out the target base (8, 9), this is fundamentally different from the model suggested for Pur α. Based on our findings for CcrM C. crescentus and the recognition model proposed for Pur α, we set out to investigate the underlying recognition mechanism used by CcrM C. crescentus. Our results are inconsistent with CcrM C. crescentus and related enzymes working through the canonical DNA recognition model and support the reliance on a new DNA recognition model.
Results
Mismatched DNA enhances methylation by CcrM C. crescentus and disrupts methylation by other enzymes
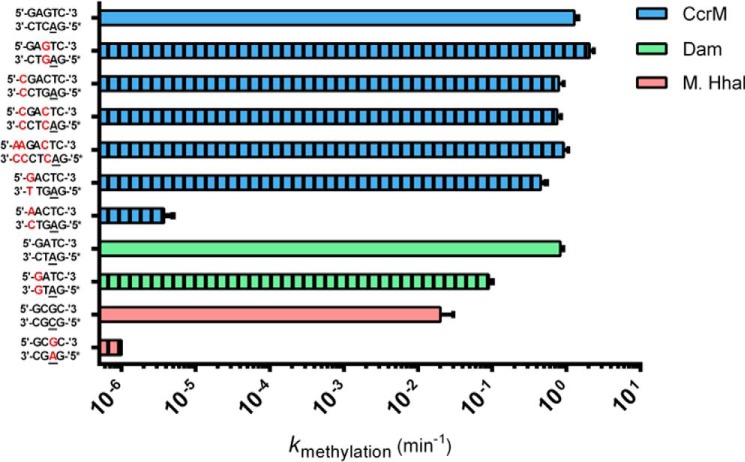
The more than 1,500 structures of protein–DNA complexes now deposited in the Protein Data Bank provide a deep basis for understanding “base readout” and “shape readout” mechanisms, involving diverse protein–DNA interfaces (10). For the vast majority of these complexes, the dsDNA remains largely intact. Interestingly, although DNA methyltransferases rely on a canonical “base flipping” recognition mechanism in which the target base is stabilized in an extrahelical position, they also leave the DNA in its duplex form (8, 9). Thus, placement of a mismatched bp into the recognition site of a sequence specific DNA-binding protein should disrupt stabilizing recognition interactions because at least one base is “incorrect,” and the mismatch significantly disrupts the sugar-phosphate positioning. In all cases we used hemimethylated DNA to force one productive binding orientation; in the case of CcrM C. crescentus, the hemimethylated DNA is what the enzyme is normally presented with within the cell. We tested this first with M.HhaI, a structurally characterized DNA cytosine methyltransferase (methylates the underlined cytosines in 5′-GCGC-3′/5′-GCGC-3′) (15). As is typical for protein–DNA complexes, the M.HhaI-DNA cocrystal structure shows extensive interactions to bases within both strands, as well to the sugar-phosphate backbone of both strands (16). Thus, as anticipated, the G:A mismatch results in at least a 106-fold loss of activity (bold, underlined is the mismatch, italicized cytosine undergoes methylation, 5′ GCGC 3′/5′ GAGC 3′) (Fig. 1). The same experiment with CcrM C. crescentus (Fig. 1, G:G mismatch) results in enhanced activity (kmethylation, 2.05 ± 0.29 min±−1 versus the control 1.28 ± 0.14 min−1), providing good evidence that CcrM C. crescentus does not interact with its substrate like M.HhaI. The results for Dam (another DNA adenine methyltransferase) also show a significant loss of activity (Fig. 1). Placement of multiple mismatches within and just outside of the GANTC recognition sequence has only minor impact on kcat (0.79 ± 0.13, 0.75 ± 0.1, 0.92 ± 0.14 min−1 for substrates containing up to three mismatches (M = mismatch, 5′-MGACTC-3′, 5′-MGAMTC-3′, and 5′-MMGAMTC-3′)).
Figure 1.
Mismatched DNA impact on methylation kinetics of CcrM C. crescentus, M.HhaI, and Dam. The kmethylation constants extracted from single-turnover kinetics are reported above for three enzymes on both their cognate and noncognate mismatched substrates. The data in blue correspond to CcrM C. crescentus kinetics, whereas the data in green correspond to Dam kinetics, and the data in red correspond to M.HhaI kinetics. Striped bars represent data collected on a substrate containing a mismatched bp. Asterisks placed next to a strand of DNA indicate the methylated strand of the hemimethylated duplex, and underlined bases indicate the methylated base. All reactions were conducted with 150 nm enzyme and 100 nm DNA, whereas the CcrM C. crescentus A–C mismatched substrate and the M.HhaI C–G mismatched substrate were characterized at 1.5 μm enzyme and 1 μm DNA. All data were collected in triplicate, and standard errors are shown.
To further probe the interactions between CcrM C. crescentus and dsDNA, we inserted a A:C mismatch into the CcrM C. crescentus recognition site, involving the guanosine immediately adjacent to the target adenine (5′AACTC-3′/5′-GAGTC-3′, A is already methylated, italicized A undergoes methylation, bold and underlined are the mismatched bases), and compared this to the mismatch at the same position, which retains the cognate sequence in the strand that undergoes methylation (involving a G:T mismatch) (Fig. 1). In this case the mismatch derives from the complement (5′-GACTC-3′/5′-GAGTT-3′). Although both substrates contain a mismatch, only the former carries a change in the strand positioned to be methylated. The selective 105-fold decrease observed when the mismatch occurs on the strand undergoing methylation provides compelling evidence not only that CcrM C. crescentus relies on a recognition mechanism distinct from other DNA methyltransferases (8, 9) but that this most likely involves a highly asymmetric reading of the two strands.
The CcrM C. crescentus DNA interface is unusually large as determined by kinetic footprinting
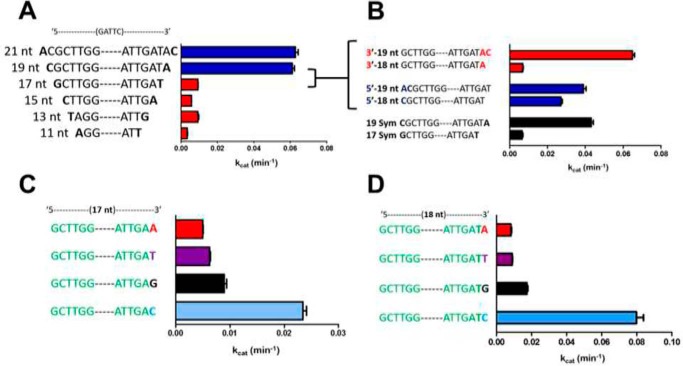
The distinctive response to mismatched DNA shown with CcrM C. crescentus (Fig. 1), compared with other classes of DNA methyltransferases (Dam, M.HhaI), both of which are structurally characterized, suggests a nonstandard protein–DNA interface. We probed this further using a kinetic footprinting strategy with ssDNA, which measures kcat at saturating DNA concentrations; because kcat is determined by kmethylation or a prior step for CcrM C. crescentus (7), it is the appropriate kinetic constant to compare substrates. Based on known protein–DNA structures and particularly those involving DNA methyltransferases (8–10, 16), we anticipated showing the importance of interactions on either side of the recognition GANTC sequence, out to two or perhaps three nucleotides. We systematically changed the overall length from 11 to 21 nucleotides (nt), by keeping the number of 5′ and 3′ nt the same (Fig. 2A). The dramatic drop from 19 to 17 nt was probed further in Fig. 2B, showing that even the removal of a single nt from the 19-nt substrate results in a large decrease in kcat. The blue sequences in Fig. 2B show that it is not simply the length that is important, because the asymmetric 19-nt sequence (addition of nt on the 5′ side at the expense of the 3′ side) shows a loss in kcat.
Figure 2.
CcrM C. crescentus has an unusually large DNA interface. A, a series of ssDNA substrates with equal numbers of nucleotides surrounding the cognate 5′-GANTC-3′ shows that 7 nt on each side (19-nt substrate) are necessary for good activity. B, the asymmetric 19-nt substrate with 6 nt on the 5′ side and 8 nt on the 3′ side is more active than the original, symmetric 19-nt substrate (A). Removal of the 3′ C results in significant loss of activity. The alternative asymmetric 19-nt substrate (8 nt 5′, 6 nt 3′) is nearly as active as the original asymmetric 19-nt substrate. Removal of a single nt from this asymmetric 19-nt substrate results in less of a loss of activity than the original 18-nt substrate. C, a cytosine at the 3′ end of 17-nt substrates (symmetric) shows significantly better activity than the other three bases at this position. D, a cytosine at the 3′ end of 18-nt substrates (asymmetric) shows significantly better activity than the other three bases at this position. Saturating AdoMet (30 μm), CcrM C. crescentus 1 μm, DNA (5 μm), time points at 5 and 10 min, reactions were conducted at room temperature in triplicate, and all data were below 20% product conversion; standard errors are shown.
To determine whether the identity of the 3′ nt contributed to kcat, we replaced this in symmetric 17-nt substrates (Fig. 2C). Surprisingly, the ssDNA with a 3′ cytidine shows 3–5-fold higher kcat values than the guanosine, thymidine, or adenosine. This effect is even more dramatic with the corresponding 18-nt substrates (Fig. 2D), which shows that the cytidine at the 3′ end causes kcat to be 4.5–10-fold larger than with adenosine, thymidine, or guanosine. The identity of the 3′ base is so important that the activity with a 17-nt substrate can be as good as with a 19-nt substrate if the former has an asymmetric positioning of the 3′ cytosine 8 nt from the recognition GANTC site (Fig. S1A). Intriguingly, this effect of a 3′ cytosine is observed at lengths from 11 to 19 nt, although the effect is most dramatic at the 18- and 17-nt lengths (Fig. S1B).
The kinetic footprinting approach used here hides the impact of changes in binding, yet it does reveal functionally important interactions. What remains unclear is how interactions so far removed from the consensus recognition sequence impact methylation. Nevertheless, functionally important interactions covering six nucleotides on the 5′ side and eight nucleotides on the 3′ side of the consensus GANTC sequence is highly unusual. We suggest that this large interface may be an important element of the DNA recognition mechanism used by CcrM C. crescentus.
Ribose replacement within the cognate site is the basis for 106-fold discrimination against RNA
The ability of CcrM C. crescentus to efficiently and sequence-specifically modify ssDNA presents a potential problem because of the high intracellular levels of single-stranded RNA. Our prior demonstration that CcrM C. crescentus is able to discriminate against single-stranded RNA by over a million-fold helps explain this, and we sought to isolate the features of the RNA that contribute to this discrimination. The ability of proteins to discriminate between ssDNA and single-stranded RNA binding has certainly been reported but remains poorly understood (17, 18). The dramatic discrimination displayed by CcrM C. crescentus (7) provides an opportunity to investigate this in some detail. Replacing the five deoxyriboses with riboses within the recognition site (GANTC) dramatically lowers the methylation kinetics (Fig. 3A), whereas similar substitutions to three sugars on either side of the recognition site have minimal impact. These are saturating conditions that remove concerns that the CcrM C. crescentus may not be able to bind some of the substrates (increasing the protein concentration does not change the kinetics). Introduction of riboses at each of the two positions within the site (Fig. 3B, GACTC) results in a 6–41-fold loss of activity, and the doubly substituted sequence is dramatically less active (100-fold decrease). These results suggest that the interactions between CcrM C. crescentus and the sugars at these internal positions within the sequence involving the 2′-OH groups contribute to these dramatic decreases in kinetics rather than any large-scale conformation differences between single-stranded RNA and DNA (Fig. 3A).
Figure 3.
The basis of RNA discrimination by CcrM C. crescentus. Single-turnover experiments with CcrM C. crescentus and selectively modified single-stranded 30-nucleotide substrates were performed. A, oligonucleotide made up exclusively of deoxyriboses displayed with red circles. The blue squares represent the reactivity of the substrate with three-flanking sugars on either side of the GANTC being ribose (5′-rArGrGGACTCrGrCrC-3′). The black triangles have riboses at all five internal positions (5′-AGGrGrArCrTrCGCC-3′). B, the DNA control is given for perspective as a dotted gray line (5′-AGGGACTCGCC-3′), and the all internal ribose is given as a dotted light blue line (5′-AGGrGrArCrTrCGCC-3′). The open upside-down triangles represent a single ribose substitution at the adenine (5′-AGGGrACTCGCC-3′), the right-side-up filled triangles represent a single ribose at the cytosine (5′-AGGGArCTCGCC-3′), and the black diamonds are both positions with a ribose (5′-AGGGrArCTCGCC-3′). Single-turnover assays were conducted in triplicate at room temperature with 1.5 μm CcrM C. crescentus, 1 μm substrate and saturating cofactor AdoMet 15 μm; standard errors are shown.
The conserved C-terminal segment found in CcrM C. crescentus is important for DNA recognition and sequence discrimination
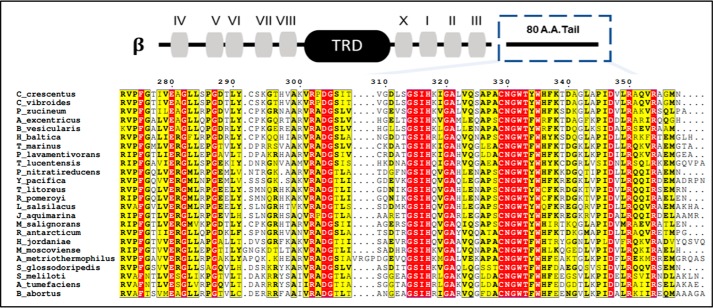
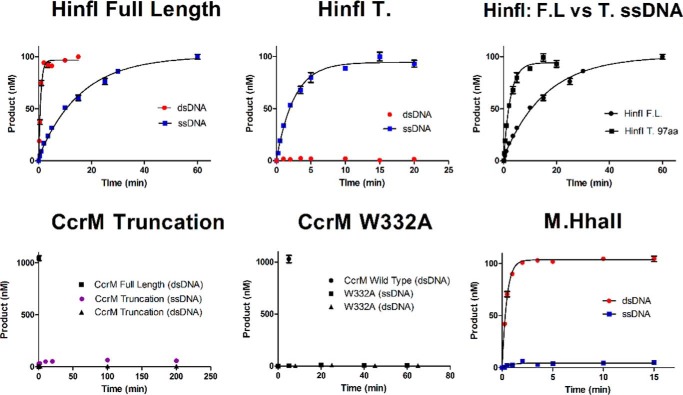
The poorly characterized β-class DNA methyltransferases, which include CcrM C. crescentus and its orthologs, have a distinctive organization of conserved motifs (Fig. 4) in relation to the target recognition domain. We analyzed the protein sequences of functionally characterized β-class enzymes that recognize GANTC sequences. We identified a new motif, located in the C terminus with 13 highly conserved residues; the β-class enzymes are broadly observed throughout proteobacteria. M.HinfI, an archetype β-class methyltransferase, is part of a type II restriction/modification system that was previously shown to have activity with both single- and double-stranded dsDNA (19), although lacking the dramatic sequence discrimination displayed by CcrM. We obtained similar results showing that the activity with dsDNA is 21-fold better than with ssDNA (Table 1). Intriguingly, removal of the C-terminal 97 residues results in a 7-fold enhancement of ssDNA activity, whereas the activity with dsDNA is completely lost. These results strongly implicate this C-terminal region in dsDNA recognition, whereas its contribution toward ssDNA recognition is minimal.
Figure 4.
Sequence alignment of β-Class DNA methyltransferases that recognize GANTC. The motifs of β-class methyltransferases are displayed for the CcrM C. crescentus sequence above the alignment. The C-terminal 80 amino acids (indicated with dashed blue box) contain a newly described motif with a proposed role in dsDNA recognition. The target recognition domain (TRD) corresponds to the target recognition domain. The alignment of sequences was constructed in ESPRIPT 3.0. Organism sequences were collected through a NCBI BlastP search against the CcrM C. crescentus sequence belonging to C. crescentus with an expected value of 10 for a window of 500 sequences. Notable species in relation to pathology or mechanistic insight of methylation that are included are: C. crescentus, A. tumefaciens, B. abortus, and Sinorhizobium meliloti from α-proteobacteria. The remaining organisms belong to α-proteobacteria and contain a putative CcrM as defined by the restrictions on the Blast algorithm. Residues highlighted in red are strictly conserved among every species, whereas residues in yellow signify a column containing residues that are similar or a column with a frequently appearing residue that is not strictly conserved. DG, GSIH, and CNGWT-(F/Y)-W are highly conserved.
Table 1.
Summary of kinetic data and substrate specificity constants, single-turnover assays, and thermodynamic constants from EMSAs
Activity assays included 150 nm enzyme, 100 nm DNA, and 15 μm AdoMet. Reactions were conducted in triplicate and reached completion in 20 min at room temperature. Binding studies included 5 nm DNA and 60 μm sinefungin. Binding experiments were performed at 4 °C for 30 min in methylation reaction buffer. Bold type indicates a lack of the C-terminal segment.
| Enzyme | Double-stranded DNA |
Single-stranded DNA |
||||
|---|---|---|---|---|---|---|
| kmethylation | Kd | kmethylation/Kd | kmethylation | Kd | kmethylation/Kd | |
| min−1 | nm | nm min−1 | min−1 | nm | nm min−1 | |
| CcrM (C. crescentus) | 2.61 ± 0.21 | 70.8 ± 7.2 | 0.037 ± 0.005 | 3.33 ± 0.30 | 39.2 ± 4.5 | 0.085 ± 0.012 |
| CcrM C. crescentus truncation | No activity | No activity | No activity | No activity | No activity | No activity |
| CcrM C. crescentus W332A | No activity | No activity | No activity | No activity | No activity | No activity |
| CcrM A. tumefaciens | 0.50 ± 0.05 | 89.0 ± 3.1 | 0.006 ± 0.001 | 0.73 ± 0.07 | 34.2 ± 5.0 | 0.021 ± 0.004 |
| CcrM B. abortus | 0.42 ± 0.03 | 41.5 ± 7.4 | 0.010 ± 0.002 | 0.46 ± 0.06 | 77.1 ± 14.6 | 0.006 ± 0.001 |
| M.HinfI | 1.27 ± 0.10 | 0.060 ± 0.003 | ||||
| M.HinfI truncation | No activity | >1 μm | No activity | 0.39 ± 0.03 | ||
| M.HhaII | 2.30 ± 0.21 | 31.5 ± 3.0 | 0.073 ± 0.010 | >300 times slower | ||
| DpnA | 0.020 ± 0.001 | 0.051 ± 0.004 | ||||
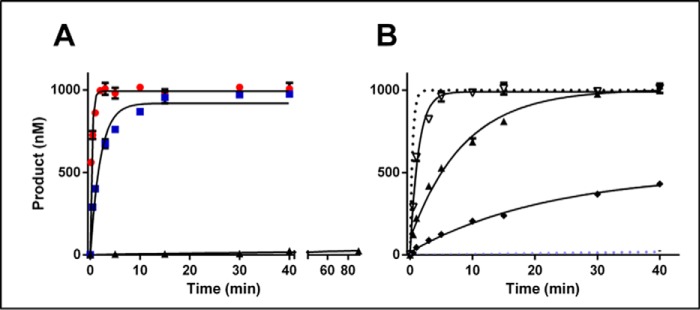
To investigate the importance of the C-terminal region in CcrM C. crescentus, we studied the comparable deletion, removing 80 residues from the C terminus (Fig. 4). This CcrM C. crescentus truncation showed no activity with either single-stranded or dsDNA (Fig. 5) and shows no ability to bind DNA (Fig. S5). Based on our limits of detection, we estimate that the activity of the truncated form is at least 107-fold less than the WT CcrM C. crescentus. The CD spectrum of the truncation is nearly identical to the full-length CcrM C. crescentus, suggesting that little or no overall conformational changes are involved (Fig. S6). Tryptophan 332 is one of the highly conserved residues in this region, and mutation to an alanine results in the same functional consequences as the C-terminal truncation (Table 1). Again, the corresponding CD spectra (Fig. S6) strongly suggests the lack of any large-scale conformational changes resulting from this substitution. These data indicate that the C-terminal segment and its highly conserved residues are likely important contributors both to DNA recognition and to the discrimination of single-stranded and dsDNA. A final challenge of this concept is our demonstration that M.HhaII, a β-class enzyme that lacks the C-terminal region but also methylates GANTC sites, shows 300-fold less activity with ssDNA than dsDNA (Fig. 5).
Figure 5.
The C-terminal domain is important for CcrM C. crescentus function. Single-turnover experiments were performed. Full-length M.HinfI has a preference for hemimethylated 60-bp substrate (red circles) over the single-stranded substrate (60 nt, blue squares). The M.HinfI 97 amino acid C-terminal truncation (HinfI T) loses all dsDNA activity (red circles) while maintaining single-stranded activity (blue squares). A comparison of full-length M.HinfI to the truncated on ssDNA reveals a dramatic enhancement in activity for the truncated M.HinfI (black squares). The CcrM C. crescentus 80-amino acid C-terminal truncation loses all activity on both hemimethylated DNA (60 bp, black triangles) and ssDNA (60 nt, purple circles). A single point mutation in the CcrM C. crescentus C-terminal tail W332A produced an inactive mutant, and the dsDNA time points were offset from the ssDNA to prevent overlap. M.HhaII revealed robust activity on dsDNA hemimethylated DNA (60 bp, red circles) but equally surprisingly no activity on ssDNA (60 nt, blue squares). For the truncated CcrM C. crescentus and mutant a CcrM C. crescentus WT control is given on the same 60-bp hemimethylated dsDNA. The single-turnover assays for M.HinfI and M.HhaII were conducted in triplicate at room temperature with 150 nm enzyme to 100 nm substrate and saturating cofactor AdoMet 15 μm, the CcrM C. crescentus single-turnover conditions used 1.5 μm enzyme to 1 μm substrate. Single-turnover reactions were fits to a one-phase decay. Standard errors are shown.
CcrM orthologs from Agrobacterium tumefaciens and B. abortus and other β-class DNA methyltransferases (Table 1)
The two orthologs of CcrM C. crescentus, CcrM A. tumefaciens and CcrM B. abortus, have 65% sequence identity with the CcrM C. crescentus. Although they do not display the extreme substrate discrimination observed with CcrM C. crescentus (Table 1), this is largely manifested through catalysis, not substrate binding, as is the case for CcrM C. crescentus. Like CcrM C. crescentus, both orthologs have similar single-turnover kinetics on ssDNA substrates when compared with the kinetics they display on dsDNA substrates (Table 1). Furthermore, all three enzymes display slightly faster kinetics on ssDNA than they do on dsDNA (Table 1). In contrast, both M.HinfI and M.HhaII display significantly slower kinetics on ssDNA when compared with dsDNA (Table 1). Thus, the efficient dual activities with single-stranded and dsDNA are not limited to CcrM C. crescentus, suggesting that the underlying recognition mechanism utilized by CcrM B. abortus and CcrM A. tumefaciens is likely to be similar.
Upon comparing the steady-state kinetics of CcrM C. crescentus and its two orthologs, we see that CcrM A. tumefaciens maintains similar kinetic behavior to CcrM C. crescentus, whereas CcrM B. abortus diverges. Both CcrM C. crescentus and CcrM A. tumefaciens lack burst kinetics on both single-stranded and dsDNA substrates, suggesting that the rate-limiting step for both enzymes on either substrate occurs prior to methylation (Fig. S4D). Although CcrM B. abortus also exhibits a lack of burst kinetics on ssDNA substrates, its kinetic profile diverges from that of CcrM C. crescentus and CcrM A. tumefaciens because of the strong substrate inhibition it displays with dsDNA substrates (Fig. S4D). An inhibition assay revealed that CcrM B. abortus displayed ∼50% inhibition at 5:1 substrate to enzyme concentrations and nearly complete inhibition at steady-state conditions (Fig. S4D). Steady-state analysis with dsDNA reveals that M.HhaII displays burst kinetics and that the rate-limiting step is methylation. This further distinguishes M.HhaII mechanistically from CcrM C. crescentus and its orthologs.
Discussion
Investigations of the mechanisms whereby proteins recognize nucleic acids continue to provide surprises. The discovery over 20 years ago that DNA methyltransferases stabilize their target bases in an extrahelical position was unprecedented (20), but this stabilization has since been revealed to occur with many DNA methyltransferases, as well as other classes of enzymes (21). Large numbers of protein–nucleic acid structures provide high resolution insights into the underlying recognition mechanisms (10), whereas dynamic studies have described the contributions of conformational changes in this recognition (22, 23). Proteins that recognize both single- and double-stranded nucleic acids, and particularly those displaying high levels of sequence discrimination, remain a relatively poorly understood subclass (10, 17, 18). Our prior work provided evidence that CcrM C. crescentus is capable of extreme sequence discrimination in both single-stranded and dsDNA, as well as between ssDNA and RNA (7). Here we provide additional support for this interesting combination of activities and suggest that this is not well-accommodated by our current models of nucleic acid recognition.
The canonical recognition mechanism for DNA methyltransferases relies on an extensive molecular interface between the enzyme and major groove moieties with dsDNA (8, 9, 20, 21), as anticipated years ago and confirmed in numerous cocrystal structures for a large variety of DNA-binding proteins (10). Further, this initial recognition complex precedes an extensive rearrangement of the protein and stabilization of the target base (either cytosine or adenine) into an extrahelical position (base flipping) (8, 9, 20, 21). Methyl transfer from the cofactor AdoMet to the target base is followed by repositioning of the methylated base within the duplex DNA, which throughout this process, is left largely unperturbed. Given our prior demonstration that CcrM C. crescentus shows unprecedented discrimination of single-bp substitutions within its recognition site (5′-GANTC-3′) (7), we anticipated that mismatches within this site would result in similar and large losses of activity. This prediction was confirmed with two other DNA methyltransferases (Fig. 1). In contrast, a single-bp mismatch within the CcrM C. crescentus site results in an increase in methylation kinetics, whereas a similar change outside the site results in a minor decrease. Further, substrates with double mismatches, outside and within the recognition site, again have minor consequences on methylation activity. These results, when taken in the context of the predicted and verified dramatic decreases for two other methyltransferases, provide strong support for the hypothesis that CcrM C. crescentus may rely on a novel recognition mechanism.
One model for a protein to recognize double and ssDNA, like CcrM C. crescentus, is based on the Pur α protein; a cocrystal structure of Pur α bound to ssDNA forms the basis of a mechanism in which the protein induces ATP-independent strand separation of duplex DNA, to recognize the single-stranded nucleic acid (11–14). Such a mechanism could rely on the protein “reading” only one of the two strands. We tested this with hemimethylated DNA in which the strand containing the methylated adenine has a single base that is substituted (Fig. 1, M, N6-methyladenine; T, source of mismatch, 5′-GACTC-3′/5′-GMGTT-3′) and compared this with hemimethylated DNA in which the strand that will undergo methylation is substituted at the same position (Fig. 1, M, N6-methyladenine; A, source of mismatch, 5′-AACTC-3′/5′-GMGTC-3′). As predicted, replacing the base on the targeted strand causes a dramatic ∼106-fold loss of activity, whereas replacement of its bp partner results in a minor (less than 3-fold) loss. Thus, not only does CcrM C. crescentus respond to mismatched DNA in a fashion distinct from other DNA tested methyltransferases, the million-fold differential response to which strand has the modified base strongly suggests that the underlying mechanism involves selective strand-specific interrogation.
The level of sequence discrimination shown by CcrM C. crescentus for both single-stranded and dsDNA is unprecedented for a DNA methyltransferase. We suspect that this may stem from its role in controlling the expression of numerous and important C. crescentus genes. Unlike methyltransferases that form part of restriction-modification systems, for which off-target methylation may not be as problematic, the inappropriate methylation of regulatory regions by CcrM C. crescentus may have unacceptable consequences (1–6). Interestingly, the other bacterial orphan DNA adenine methyltransferase, Dam, unlike CcrM C. crescentus, is involved in multiple roles (e.g. mismatch repair, replication, gene regulation) (1) and is at least 1,000-fold less discriminating. Because the discrimination by CcrM C. crescentus occurs with ssDNA, we carried out a functional mapping of the CcrM C. crescentus–ssDNA interface, on the premise that the enhanced discrimination may derive from an unusually large interface (Fig. 2 and Fig. S1). This functional mapping relies on single-turnover measurements under saturating conditions and shows a sharp drop in activity when a sequence with seven nucleotides on either side of the recognition site is shortened to six on either side (Fig. 2A). This initial and somewhat surprising result suggests that CcrM C. crescentus makes contacts over a span of 17–19 nucleotides; although not unprecedented, this is unusual for a relatively small, monomeric enzyme.
Further investigation with this approach revealed that the interface is asymmetric, because changes on the 3′ side of the site extended further (7 nucleotides) than on the 5′ side (5 or 6 nucleotides). Furthermore, the methylation efficiency is quite dependent on the nature of the nucleotide at the 3′ end (Fig. 2, C and D, and Fig. S1). The combined results suggest that not only is the CcrM C. crescentus–DNA interface unusually large for a methyltransferase (8, 9, 20, 21) but that these interactions are important for the correct assembly of the active site. Because CcrM C. crescentus shows the uncharacteristic behavior (for a DNA methyltransferase) that methylation or a prior step defines kcat, the observed changes in methylation most likely do not result from alterations in substrate binding or product release. It is intriguing that genomic methylation analysis shows little evidence for CcrM C. crescentus specificity beyond the recognition hexanucleotide (2, 3). Thus, we suggest that the CcrM C. crescentus–DNA interface provides some other function than in determining specificity. Certainly, a plausible function would be some feature of the recognition mechanism, such as inducing strand separation, as suggested for Pur α (11–14).
The ability to efficiently methylate ssDNA with high fidelity presents an intriguing biological challenge for CcrM C. crescentus, because the predominant cellular single-stranded nucleic acid is RNA. Our previous demonstration that CcrM C. crescentus displays an incredible discrimination against single-stranded RNA provides a plausible solution to this situation (7). However, the basis for protein discrimination of ssDNA over RNA (or its reverse) remains largely obscure (17, 18). By replacing deoxyribose sugars with ribose sugars at various positions, both within and outside the recognition site (Fig. 3), we showed that a small number of riboses within the recognition site make a significant contribution to this discrimination, which derives largely from changes in kmethylation, not binding (Fig. 3 and Table S1). The overall picture that emerges from these DNA and RNA discrimination studies, as well as the sequence discrimination data (Fig. 1 and Table 1), (7) is that discrimination is extreme and overwhelmingly determined at the level of catalysis.
The features of CcrM C. crescentus responsible for high fidelity recognition of single-stranded and dsDNA remain uncertain. The sequence analysis of β-class DNA methyltransferases that recognize GANTC sites, which include CcrM C. crescentus and its orthologs, led to the identification of a highly conserved set of residues in the C terminus (Fig. 4). That this region is involved in recognition was suggested by a truncation study of M.HinfI, a β-class DNA methyltransferase that forms part of a bacterial type II R/M system (19), showing that its dsDNA but not its ssDNA activity is lost when this region is removed. Our results with the WT M.HinfI showed a 21-fold preference for dsDNA, and we obtained similar results with the truncation (Fig. 5) as the prior report. Interestingly, a similar C-terminal truncation, and a single substitution of a conserved tryptophan (Fig. 5 and Fig. S5) with CcrM C. crescentus resulted in complete loss of DNA binding and enzyme activity, despite the retention of the protein's overall conformation, as determined by CD (Fig. S6). Clearly these highly conserved residues and the entire C-terminal segment are critical to CcrM C. crescentus function. M.HhaII, a β-class enzyme that lacks the C-terminal region but also methylates GANTC sites, shows minimal (300-fold lower) activity with ssDNA than dsDNA (Fig. 5), further suggesting the importance of this C-terminal domain in DNA recognition by CcrM C. crescentus.
To determine whether the functional characteristics we report here for CcrM C. crescentus are more broadly observed, we studied two orthologs from A. tumefaciens and B. abortus (Fig. S4) (5, 6). The substrate discrimination for each of these, like the C. crescentus enzyme, is largely manifested at the level of kmethylation, although the discrimination is less extreme (Table 1). Further, both orthologs are fully able to methylate single-stranded and dsDNA. An interesting divergence occurs in that only the CcrM B. abortus shows strong substrate inhibition with dsDNA substrates (Fig. S4D), suggesting that this enzyme may be capable of binding two dsDNA molecules.
Materials and methods
The CcrM C. crescentus, CcrM A. tumefaciens, and CcrM B. abortus genes (UniProt accession numbers B8GZ33, F7U651, and Q2YMK2, respectively) were obtained from Dr. Lucy Shapiro at Stanford. The M.HinfI and M.HhaII genes (UniProt accession numbers P20590 and P00473, respectively) were obtained from Geoff Wolf at New England Biolabs (NEB). The DpnA gene (UniProt accession number P09358) was purchased from Integrated DNA Technologies (IDT) gBLOCK. Unless specifically stated, the following protocols for cloning, protein expression, and purification were used for CcrM C. crescentus, CcrM A. tumefaciens, CcrM B. abortus, M.HinfI, M.HhaII, and DpnA. Using PCR with 1× NEB Taq reaction buffer, 200 μm dNTPs, 0.5 μm forward primer, 0.5 μm reverse primer, <1 μg of template DNA, and 1.25 units Taq polymerase, the respective methyltransferase gene was amplified in preparation for cloning (see Fig. S6 for primer sequences and annealing temperatures). Separately, the pET28a expression vector (purchased from IDT) and resulting amplicons were digested using endonucleases NdeI and EcoRI (purchased from NEB) with the following reaction: 1× NEB CutSmart buffer, 1 μg of DNA, 10 units of restriction enzyme, 1 h of incubation at 37 °C followed by inactivation at 60 °C for 20 min. The digested pET28a expression vectors were subsequently dephosphorylated using phosphatase rSAP (purchased from NEB) with the following reaction: 1× NEB CutSmart buffer, 1 unit of rSAP, 1 pmol of DNA ends (≈1 μg of 3-kb plasmid), with 30 min of incubation at 37 °C followed by inactivation at 65 °C for 5 min. Ligation of the digested amplicons into the processed pET28a vectors was accomplished using T4 DNA ligase (purchased from NEB) and the following reaction: 1× NEB T4 DNA ligase buffer, 1 μm DNA 5′ termini, and 2,000 cohesive end units of T4 DNA ligase at 16 °C for 16 h. Ligation resulted in cloned expression vectors that encoded an N-terminally hexahistidine-tagged methyltransferase. The cloned expression vector was chemically transformed into NiCo21(DE3) Escherichia coli cells (purchased from NEB) via a 20-min incubation with the competent cell solution followed by heat shock at 42 °C for 30 s. Cell growths were carried out in 8 liters of LB medium containing 30 μg/ml kanamycin with vigorous shaking at 37 °C until A600 = 1.0 was achieved. Protein induction for CcrM C. crescentus, CcrM A. tumefaciens, and CcrM B. abortus was initiated by addition of isopropyl β-d-1-thiogalactopyranoside to 1 mm with a subsequent 3 h of incubation at 23 °C with vigorous shaking. Protein induction for M.HinfI, M.HhaII, and DpnA was initiated by the addition of isopropyl β-d-1-thiogalactopyranoside to 1 mm with a subsequent 3 h of incubation at 37 °C with vigorous shaking. The cells were pelleted using a Beckman centrifuge with a JA-10 rotor at 5,000 rpm for 25 min at 4 °C. The resulting cell pellet was collected and resuspended in buffer 1 (50 mm HEPES, 300 mm NaCl, 10% glycerol, and 70 mm imidazole at pH 8.0) at 4 °C. Additionally, phenylmethanesulfonyl fluoride was immediately added to a concentration of 1 mm in the cell suspension. Although the cell suspension was maintained at 4 °C using a water/ice bath, the cells were lysed via sonification with a Branson digital sonifier horn at an amplitude of 75%. The cell lysate was then clarified using a Beckman centrifuge with a JA-20 rotor for 2.5 h at 11,000 rpm at 4 °C. Using the following protocol, the desired methyltransferase was purified from the clarified supernatant with an ÄKTA Start FPLC and a 5-ml HisTrap HP nickel-affinity column (both purchased from General Electric). The clarified supernatant was loaded and subsequently washed with 9.5 CV of buffer 1 at a flow rate of 5 ml/min. A 30-ml isocratic elution was then performed with buffer 2 (50 mm HEPES, 300 mm NaCl, 160 mm imidazole, 10% glycerol, pH 8.0) at a flow rate of 2 ml/min. The eluted fractions were then concentrated using Amicon Ultra 0.5-ml centrifugal filters with a 10-kDa cutoff (purchased from Millipore Sigma) followed by a buffer exchange into the storage buffer (100 mm HEPES, 300 mm NaCl, 50% glycerol, pH 8.0); enzyme aliquots were stored at −80 °C. Protein purity was determined using a 12% acrylamide SDS-PAGE with BSA standards (purchased from Thermo Fisher Scientific) and analyzed by densitometry via ImageJ.
Radiochemical assays for single-turnover and kinetic footprinting
Single-turnover, steady-state, and kinetic footprinting data were collected using the Reaction Buffer (100 mm HEPES, 20 mm NaCl, 1 mm DTT, 1 mm EDTA, 2 mg/ml BSA, pH 8.0) and saturating cofactor AdoMet (15 μm), 1:10 (radiolabeled:unlabeled) in triplicate at 23 °C. AdoMet stocks were made using 32 mm AdoMet supplied by NEB in 10 mm H2SO4, and (3H-CH3 − 1 mCi [82.7 mCi/mmol]) AdoMet supplied by PerkinElmer with a final concentration of 50 μm of 1:10. DNA substrates with and without the N6-methyl adenosine, C5-methyl cytosine were purchased from the Keck Oligo facility at Yale. For single-turnover assays, the reaction conditions were under saturating enzyme (150 nm) to substrate (100 nm). For steady-state assays, the reaction conditions were under saturating substrate (3 μm) with enzyme (100 nm). The reactions were initiated by the addition of substrate into a mix of reaction buffer and enzyme. The reactions were quenched when blotted on to Amersham Biosciences Hybond nucleic acid blotting paper from GE. The papers were then immediately placed in 50 mm KH2PO4 buffer. After completion of all data points, the papers were washed in three rounds of gentle shaking in 500 ml of 50 mm KH2PO4 buffer for 5 min followed by one round of gentle shaking in 500 ml of 80% ethanol solution for 5 min, and one round of gentle shaking in 500 ml of 100% ethanol for 5 min. The papers were then soaked in diethyl ether anhydrous for 5 min followed by 20 min of being air dried. The papers were placed in scintillation vials and submerged in 3 ml of BioSafeII scintillation fluid, and a Beckman Coulter LS-6500 scintillation counter with units of DPM was used to evaluate product formation. Background readings were subtracted from all points, and the data were fit using GraphPad Prism 5 using a one-phase decay for single-turnover reactions. Reactions for collecting kinetic footprinting were in an identical procedure to the single-turnover reactions except with saturating substrate at 5 and 1 μm of CcrM C. crescentus.
EMSA
An EMSA was used to determine the dissociation constants (Kd) for substrates conducted in reaction buffer on ice with FAM-tagged DNA (IDT) as previously described (7).
CD spectroscopy
CD spectroscopy was done on a JASCO J-1500 spectrophotometer using 5 μm protein in 1 mm HEPES, 3 mm NaCl, 1 mm dithiothreitol, pH 8.0. All measurements were at 25 °C, and the buffer contribution was subtracted from all spectra.
Author contributions
N. O. R. conceptualization; N. O. R. formal analysis; N. O. R. funding acquisition; N. O. R. project administration; E. D., M. K., S. P., and C. B. W. investigation.
Supplementary Material
The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Figs. S1–S8.
- CcrM
- cell cycle–regulated DNA methyltransferase of C. crescentus
- M.HinfI
- Haemophilus influenza DNA methyltransferase
- ssDNA
- single-stranded DNA
- M.HhaII
- methyltransferase from Haemophilus haemolyticus
- EMSA
- electrophoretic mobility shift assay
- nt
- nucleotide(s)
- NEB
- New England Biolabs
- IDT
- Integrated DNA Technologies.
References
- 1. Adhikari S., and Curtis P. (2016) DNA methyltransferases and epigenetic regulation in bacteria. FEMS Microbiol. Rev. 40, 575–591 10.1093/femsre/fuw023 [DOI] [PubMed] [Google Scholar]
- 2. Kozdon J. B., Melfi M. D., Luong K., Clark T. A., Boitano M., Wang S., Zhou B., Gonzalez D., Collier J., Turner S. W., Korlach J., Shapiro L., and McAdams H. H. (2013) Global methylation state at base-pair resolution of the Caulobacter genome throughout the cell cycle. Proc. Natl. Acad. Sci. U.S.A. 110, E4658–E4667 10.1073/pnas.1319315110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou B., Schrader J. M., Kalogeraki V. S., Abeliuk E., Dinh C. B., Pham J. Q., Cui Z. Z., Dill D. L., McAdams H. H., and Shapiro L. (2015) The global regulatory architecture of transcription during the Caulobacter cell cycle. PLoS Genet. 11, e10043831 10.1371/journal.pgen.1004831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gonzalez D., Kozdon J. B., McAdams H. H., Shapiro L., and Collier J. (2014) The functions of DNA methylation by CcrM in Caulobacter crescentus: a global approach. Nucleic Acids Res. 42, 3720–3735 10.1093/nar/gkt1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wright R., Stephens C., and Shapiro L. (1997) The CcrM DNA methyltransferase is widespread in the alpha subdivision of proteobacteria, and its essential functions are conserved in Rhizobium meliloti and Caulobacter crescentus. J. Bacteriol. 179, 5869–5877 10.1128/jb.179.18.5869-5877.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Robertson G. T., Reisenauer A., Wright R., Jensen R. B., Jensen A., Shapiro L., and Roop R. M. (2000) The Brucella abortus CcrM DNA methyltransferase is essential for viability, and its overexpression attenuates intracellular replication in murine macrophages. J. Bacteriol. 182, 3842–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Woodcock C. B., Yakubov A. B., and Reich N. O. (2017) Caulobacter crescentus cell cycle–regulated DNA methyltransferase uses a novel mechanism for substrate recognition. Biochemistry 56, 3913–3922 10.1021/acs.biochem.7b00378 [DOI] [PubMed] [Google Scholar]
- 8. Cheng X., and Roberts R. J. (2001) AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 29, 3784–3795 10.1093/nar/29.18.3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng X. (2014) Structural and functional coordination of DNA and histone methylation. Cold Spring Harbor Perspect. Biol. 6, a018747 10.1101/cshperspect.a018747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rohs R., Jin X., West S. M., Joshi R., Honig B., and Mann R. S. (2010) Origins of specificity in protein–DNA recognition. Annu. Rev. Biochem. 79, 233–269 10.1146/annurev-biochem-060408-091030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wortman M. J., Johnson E. M., and Bergemann A. D. (2005) Mechanism of DNA binding and localized strand separation by Purα and comparison with Pur family members. Biochim. Biophys. Acta 1743, 64–78 10.1016/j.bbamcr.2004.08.010 [DOI] [PubMed] [Google Scholar]
- 12. Darbinian N., Gallia G. L., and Khalili K. (2001) Helix-destabilizing properties of the human single stranded DNA- and RNA-binding protein Purα. J. Cell. Biochem. 80, 589–595 10.1002/1097-4644(20010315)80:4%3C589::AID-JCB1013%3E3.0.CO%3B2-0 [DOI] [PubMed] [Google Scholar]
- 13. Graebsch A., Roche S., and Niessing D. (2009) X-ray structure of Purα reveals a Whirly-like fold and an unusual nucleic-acid binding surface. Proc. Natl. Acad. Sci. U.S.A. 106, 18521–18526 10.1073/pnas.0907990106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Weber J., Bao H., Hartlmüller C., Wang Z., Windhager A., Janowski R., Madl T., Jin P., and Niessing D. (2016) Structural basis of nucleic acid-recognition and double-strand unwinding by the essential neuronal protein Purα. eLife 5, e11297 10.7554/eLife.11297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Estabrook R. A., Nguyen T. T., Fera N., and Reich N. O. (2009) Coupling sequence-specific recognition to DNA modification. J. Biol. Chem. 284, 22690–22696 10.1074/jbc.M109.015966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shieh F. K., Youngblood B., and Reich N. O. (2006) The role of Arg165 towards base flipping, base stabilization and catalysis in M.HhaI. J. Mol. Biol. 362, 516–527 10.1016/j.jmb.2006.07.030 [DOI] [PubMed] [Google Scholar]
- 17. Shamoo Y. (2002) Single stranded DNA binding proteins. Encyclopedia of Life Sciences 1–7, John Wiley and sons, New York [Google Scholar]
- 18. Lu D., Searles M. A., and Klug A. (2003) Crystal structure of a zinc-finger RNA complex reveals two modes of molecular recognition. Nature 426, 96–100 10.1038/nature02088 [DOI] [PubMed] [Google Scholar]
- 19. Bassing C. H., Kim Y. G., Li L., and Chandrasegaran S. (1992) Overproduction, purification and characterization of M. HinfI methyltransferase and its deletion mutant. Gene 113, 83–88 10.1016/0378-1119(92)90672-C [DOI] [PubMed] [Google Scholar]
- 20. Klimasauskas S., Kumar S., Roberts R. J., and Cheng X. (1994) HhaI methyltransferase flips its target base out of the DNA helix. Cell 76, 357–369 10.1016/0092-8674(94)90342-5 [DOI] [PubMed] [Google Scholar]
- 21. Roberts R. J., and Cheng X. (1998) Base flipping. Annu. Rev. Biochem. 67, 181–198 10.1146/annurev.biochem.67.1.181 [DOI] [PubMed] [Google Scholar]
- 22. Youngblood B., Buller F., and Reich N. O. (2006) Determinants of sequence-specific DNA methylation: target recognition and catalysis are coupled in M.HhaI. Biochemistry 45, 15563–15572 10.1021/bi061414t [DOI] [PubMed] [Google Scholar]
- 23. Youngblood B., Bonnist E., Dryden D. T., Jones A. C., and Reich N. O. (2008) Differential stabilization of reaction intermediates: specificity checkpoints for M. EcoRI revealed by transient fluorescence and fluorescence lifetime studies. Nucleic Acids Res. 36, 2917–2925 10.1093/nar/gkn131 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.