Abstract
An imbalance in the colonic microbiota might underlie m any hum an diseases, but the mechanisms maintaining homeostasis remain elusive. Recent insights suggest that colonocyte metabolism functions as a control switch, mediating a shift between homeostatic and dysbiotic communities. During homeostasis, colonocyte metabolism is directed towards oxidative phosphorylation, resulting in high epithelial oxygen consumption. The consequent epithelial hypoxia helps maintain a microbial com m unity dominated by obligate anaerobic bacteria, which provide benefit by converting fiber into fermentation products absorbed by the host. Conditions that alter the m etabolism of the colonic epithelium increase epithelial oxygenation, thereby driving an expansion of facultative anaerobic bacteria, a hallmark of dysbiosis in the colon. Enteric pathogens subvert colonocyte metabolism to escape niche protection conferred by the gut microbiota. The reverse strategy, a m etabolic reprogramming to restore colonocyte hypoxia, represents a promising new therapeutic approach for rebalancing the colonic microbiota in a broad spectrum of hum an diseases.
Single sentence summary:
The energy metabolism of colonic epithelial cells functions as a control switch of the gut microbiota, mediating shifts between homeostatic and dysbiotic communities.
One major task of our immune system is to defend against microbial pathogens, such as bacteria, viruses, parasites or fungi, by recognizing these intruders and removing them from the body. The emerging field of microbiota research has raised awareness that our immune system might also act to balance microbial communities inhabiting our skin and mucosal surfaces (the microbiota). Compared to our detailed understanding of the immune functions that control microbial pathogens, we know little about the host cell types and mechanisms involved in balancing our microbial self. Understanding how our immune system maintains homeostasis is of particular significance in the colon, because this organ harbors the largest microbial community in our body and recent advances in high-throughput sequencing link an imbalance in this microbial community (dysbiosis) to many chronic human illnesses, including colorectal cancer, obesity, diabetes, arthritis, asthma, cardiovascular disease and neurological disorders (reviewed in (1)). Yet, it is a daunting task to define what is a balanced microbial community in the colon, because the resident microbiota is highly diverse (2), differs between individuals (3) and shifts with changes in the diet (4). In turn, not knowing what features characterize a balanced colonic microbiota has hampered progress in specifying immune functions or cell types required for maintaining homeostasis in the colon.
Homeostasis: pulling diffuse concepts into focus
Clues about immune functions important for balancing the microbiota have emerged by viewing coevolution of microbial communities with their hosts from an ecological perspective, which suggests that the immune system maintains homeostasis by shaping the microbiota to be beneficial (5). Applying this concept to the colonic microbiota puts the spotlight on the benefit provided by this microbial community, which is to aid in the digestion of nutrients that cannot be processed by host enzymes. Specifically, complex dietary carbohydrates (fiber) are broken down by the colonic microbiota into fermentation products that are absorbed by the host (6) and contribute to host nutrition (7), immune development (8–11) and niche protection against enteric pathogens (12). Bacterial diversity in the colon is beneficial, because it increases the probability of including species that can break down any complex carbohydrate into fermentation products (13). The community of obligate anaerobic bacteria in the adult colon, which is dominated by members of the classes Clostridia (phylum Firmicutes) and Bacteroidia (phylum Bacteroidetes), provides benefit because they encode a broad spectrum of enzymes for hydrolyzing different complex carbohydrates (14). In contrast, facultative anaerobic bacteria, such as members of the phylum Proteobacteria, do not specialize in consuming fiber and might even interfere with host nutrition by metabolizing fermentation products to carbon dioxide when oxygen is present (15, 16). Thus, viewing host control over microbes from an ecological perspective predicts that our immune system maintains homeostasis by shaping the colonic microbiota to be diverse and dominated by obligate anaerobic bacteria, thereby ensuring this microbial community provides benefit by generating fermentation products from fiber (17).
Studies on host control mechanisms that shape the colonic microbiota to be beneficial suggest that colonic epithelial cells (colonocytes) play a central role in this process (12), which is somewhat surprising because immune functions are typically associated with cells of the hematopoietic lineage. The colonic epithelium is continually renewed by colonic stem cells located at the base of intestinal glands, termed the crypts of Lieberkühn. Asymmetric cell division of colonic stem cells generates transit-amplifying cells, an early intermediate cell type involved in tissue regeneration that divides a finite number of times until terminally differentiating into various epithelial cell types, including colonocytes, enteroendocrine cells and goblet cells (18). The dividing cells located at the base of crypts obtain energy through anaerobic glycolysis, which is characterized by conversion of glucose into lactate even in the presence of oxygen (19), a process known as the Warburg metabolism (20). Epithelial differentiation requires PPARγ (peroxisome proliferator-activated receptor gamma) (21), a nuclear receptor primarily synthesized in differentiated cells of the colonic epithelium of rodents and humans (22). PPARγ activates fatty acid metabolism, resulting in mitochondrial β-oxidation of long-chain and short-chain fatty acids and oxygen consumption through oxidative phosphorylation (23–25). This energy metabolism of mature colonocytes is characterized by high oxygen consumption, resulting in an oxygen partial pressure [pO2] of less than 7.6 mmHg (< 1% oxygen), a condition known as epithelial hypoxia (26). Since oxygen freely diffuses across biological membranes, epithelial hypoxia limits the amount of oxygen emanating from the mucosal surface, which helps maintain anaerobiosis in the intestinal lumen (Fig. 1A) (27). In turn, anaerobiosis ensures the colonic microbiota is dominated by obligate anaerobic bacteria, which provide benefit to the host by converting fiber into fermentation products (17). Through this mechanism, the colonic epithelium shapes the microbiota to be beneficial, thereby maintaining gut homeostasis.
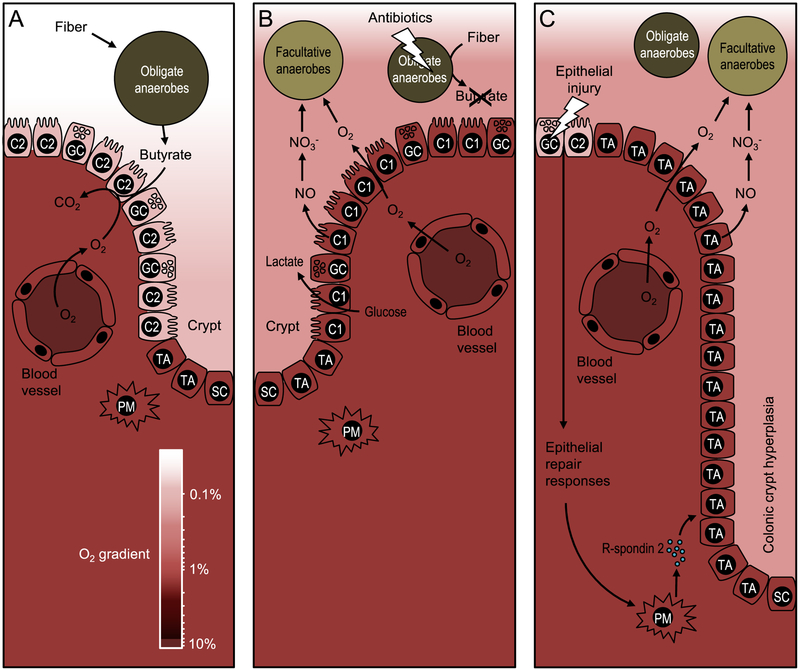
Figure 1: Epithelial metabolism shapes the colonic microbiota.
(A) During gut homeostasis, obligate anaerobic bacteria convert fiber into fermentation products, such as butyrate, to maintain differentiated colonocytes in a C2-skwed metabolic state. The metabolism of C2-colonocytes is characterized by high oxygen consumption, which maintains epithelial hypoxia (<1 % oxygen) to limit the amount of oxygen diffusing into the gut lumen. The color scale on the bottom indicates oxygen (O2) levels, which are between 3 % and 10 % in normoxic tissue (85). (B) Disruption of the gut microbiota by antibiotic treatment depletes microbe-derived fermentation products, causing a metabolic reorientation of terminally differentiated colonocytes towards a C1-skewed metabolism, which is characterized by high lactate release, low oxygen consumption and elevated synthesis of iNOS, an enzyme that generates nitric oxide (NO). Conversion of nitric oxide into nitrate (NO3−) in the gut lumen together with oxygen (O2) emanating from C1-colonocytes provide electron acceptors that drive an expansion of facultative anaerobic bacteria. (C) Epithelial injury activates epithelial repair responses, including a release of R-spondin-2 to stimulate cell division of undifferentiated transit-amplifying cells. Excessive cell division of undifferentiated transit-amplifying cells leads to colonic crypt hyperplasia and an increased epithelial oxygenation. Nitrate and oxygen emanating from the mucosal surface during colonic crypt hyperplasia drive an expansion of facultative anaerobic bacteria. PM, pericryptal myofibroblast; SC, stem cell, TA, undifferentiated transit-amplifying cell; C2, terminally differentiated C2-colonocyte; C1, terminally differentiated C1-colonocyte; GC, goblet cell.
Colonocyte metabolism drives gut dysbiosis
Above considerations suggest that the colonic epithelium might contribute to immune functions that maintain homeostasis by shaping the microbiota to be beneficial (5). Therefore an imbalance in the microbial community could be caused by an underlying defect in epithelial immune functions that maintain homeostasis in the colon (17). Dysbiosis in the colon is commonly associated with an increased abundance of facultative anaerobic bacteria (28, 29), which is observed in individuals undergoing antibiotic therapy (30), consuming a high-fat Western-style diet (31, 32), or suffering from inflammatory bowel disease (33), colorectal cancer (34), irritable bowel syndrome (35, 36), or necrotizing enterocolitis (37). Since only facultative anaerobic bacteria can respire oxygen, it has been proposed that a shift in the microbial community from obligate to facultative anaerobic bacteria might be associated with a disruption in anaerobiosis, a concept known as the “oxygen hypothesis” (38). We now know that the population of facultative anaerobic bacteria expands in the colon during dysbiosis because a disruption of epithelial hypoxia increases the amount of oxygen emanating from the colonic epithelium (39). Thus, the shift in the colonic microbiota composition from obligate to facultative anaerobic bacteria, associated with many chronic human illnesses, might have a common underpinning in colonocyte dysfunction (27).
First insights into mechanisms that disrupt gut homeostasis were obtained by studying the effect of antibiotic treatment (40), which alters epithelial metabolism by depleting microbes producing fermentation products, including the short-chain fatty acids butyrate, propionate and acetate (12). Short-chain fatty acids are absorbed in the colon, where they bind to G-protein coupled receptors to maintain the regulatory T-cell pool in the murine mucosa, thereby inhibiting intestinal inflammation (8–11). Importantly, butyrate activates PPARγ signaling in human epithelial cells (41) to drive the metabolism of surface colonocytes towards mitochondrial β-oxidation of fatty acids (23–25), which is important for maintaining epithelial hypoxia (12). Thus, antibiotic-mediated depletion of short-chain fatty acids silences epithelial PPARγ signaling (12) and lowers the number of regulatory T-cells in mouse models (8, 11). As a result, antibiotic treatment increases the inflammatory tone of colonic mucosa (42). The concomitant elevation of inflammatory signals shifts the metabolism of terminally differentiated surface colonocytes towards anaerobic glycolysis, a metabolism characterized by low oxygen consumption, high glucose consumption and high lactate release (12, 43). This metabolic reprogramming results in a loss of epithelial hypoxia (40). In turn, increased epithelial oxygenation elevates the amount of oxygen emanating from the mucosal surface, thereby driving an expansion of facultative anaerobic bacteria by aerobic respiration (Fig. 1B) (12, 44).
Metabolic polarization of colonocytes
The picture emerging from these studies is that in a healthy gut, butyrate-activated PPARγ signaling increases oxygen consumption in terminally differentiated colonocytes by polarizing their intracellular metabolism towards mitochondrial β-oxidation of fatty acids. However, pro-inflammatory signals can change the colonocyte phenotype by reorienting their metabolism towards anaerobic glycolysis, thereby lowering oxygen consumption, which ultimately leads to a shift in the microbial community from obligate to facultative anaerobic bacteria (17). These two opposing colonocyte phenotypes are strikingly similar to alternatively activated (M2) and classically activated (M1) polarization states of macrophages, which also require a reversible metabolic reprogramming. Although the M1–M2 dichotomy is an oversimplification, macrophages in vivo regularly mimic M1 or M2 polarization stages, which continues to make this nomenclature useful (45). During M2 polarization, interleukin (IL)-4 and IL-13 induce STAT6 signaling to activate PPARγ, which drives the energy metabolism of macrophages towards mitochondrial β-oxidation of fatty acids and oxidative phosphorylation (Fig. 2A) (46). In contrast, classically activated (M1) macrophages emerge when pro-inflammatory signals, such as gamma interferon (IFN-γ) or lipopolysaccharide (LPS), polarize the macrophage metabolism towards anaerobic glycolysis, thereby increasing glucose consumption and lactate release (Fig. 2B) (45). One metabolic signature of M1 macrophages is the conversion of L-arginine and oxygen into nitric oxide and L-citrulline by inducible nitric oxide synthase (iNOS). Similarly, metabolic polarization of colonocytes towards anaerobic glycolysis is accompanied by elevated iNOS synthesis and increased nitric oxide production (12). In the intestinal lumen, nitric oxide is converted into nitrate, which can be used by many facultative anaerobic bacteria as an electron acceptor for anaerobic respiration (47). In turn, colonocyte-derived nitrate contributes to a dysbiotic expansion of facultative anaerobic Proteobacteria, such as Escherichia coli, in the colon of antibiotic-treated mice (12, 42, 44). In conclusion, the different metabolic states colonocytes adopt during gut homeostasis or dysbiosis closely resemble the metabolic programming of M2 or M1 macrophages.
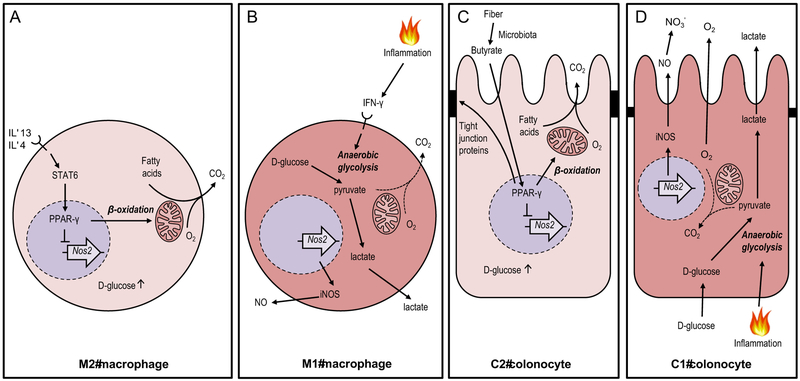
Figure 2: Extending the M1/M2 paradigm to colonocytes.
(A and B) Cytokine signaling can polarize macrophage metabolism and function, a process that is reversible. (A) Interleukin (IL)-4 and IL-13 stimulate polarization into alternatively-activated M2 macrophages by inducing STAT6 signaling to drive a PPARγ-dependent activation of mitochondrial β-oxidation and concomitant repression of the Nos2 gene. (B) Pro-inflammatory signals, such as gamma interferon (IFN-γ), stimulate polarization into classically-activated M1 macrophages by shifting the host cell metabolism towards anaerobic glycolysis. (C) The microbiota converts fiber into fermentation products, such as butyrate, which stimulates a metabolic polarization into homeostatically-activated C2 colonocytes by inducing a PPARγ-dependent activation of mitochondrial β-oxidation, thereby lowering epithelial oxygenation. (D) Pro-inflammatory signals stimulate a metabolic polarization into C1 colonocytes by shifting the host cell metabolism towards anaerobic glycolysis, thereby increasing epithelial oxygenation, which results in oxygen (O2) emanating from the epithelial surface. Lactate produced during anaerobic glycolysis is released into the gut lumen, whereas nitric oxide (NO) produced by iNOS is converted to nitrate (NO3−). CO2, carbon dioxide.
To better conceptualize how the host shapes its colonic microbiota, it seems useful to expand the concept that metabolic features of even non-hematopoietic host cells are deeply associated with immune functions. By analogy with macrophage polarization, microbiota-derived signals, such as short-chain fatty acids, induce a homeostatic (C2) activation state in colonocytes to maintain anaerobiosis (Fig. 2C), which in turn ensures that obligate anaerobic bacteria dominate the microbial community, thereby completing the circuit and guaranteeing microbial conversion of fiber into fermentation products (Fig. 1A). Conversely, dysbiosis ensues when the colonic epithelium loses its homeostatic C2 state. For example, a metabolic reprogramming of terminally differentiated colonocytes towards an inflammation-associated (C1) activation state (Fig. 2D) (12, 43) elevates the luminal availability of host-derived respiratory electron acceptors, thereby fueling a dysbiotic expansion of facultative anaerobic Enterobacteriaceae, a family within the phylum Proteobacteria (12). Thus, maintenance of a microbiota composition characteristic of a healthy gut is intimately linked to a homeostatic C2-skewed metabolism of differentiated surface colonocytes in the colon.
Colonocyte metabolism in ulcerative colitis
A homeostatic C2-skewed colonic epithelial surface can also be lost during excessive epithelial repair, which is observed during ulcerative colitis, an inflammatory bowel disease affecting the colon. To repair epithelial injury, pericryptal myofibroblasts secrete the mitogen R-spondin 2, which triggers division of colonic stem cells and undifferentiated transit-amplifying cells near the base of the crypts (48). Excessive epithelial repair promotes crypt elongation due to an accumulation of dividing transit-amplifying cells, which is known as colonic crypt hyperplasia (48), a common feature of ulcerative colitis. This hyperplasia gives rise to a reduction in the number of terminally differentiated epithelial cells, such as goblet cells, and a concomitant thinning of the mucus layer in patients with ulcerative colitis (49, 50). Since PPARγ is primarily synthesized in terminally differentiated epithelial cells (22), the accumulation of undifferentiated transit-amplifying cells is expected to lower epithelial PPARγ synthesis during colonic crypt hyperplasia, thereby reducing mitochondrial β-oxidation of fatty acids in the colonic epithelium. Consistent with this prediction, colonic epithelial cells from ulcerative colitis patients exhibit lower epithelial PPARγ synthesis (51) and reduced mitochondrial β-oxidation of butyrate to carbon dioxide (52). Low epithelial PPARγ synthesis resulting from the accumulation of transit-amplifying cells might also contribute to the development of a “leaky gut”, as diminished epithelial PPARγ signaling increases colonic permeability in a rat colitis model (53), presumably because PPARγ upregulates tight junction molecules in epithelial cells (Fig. 2C) (54). Importantly, as the metabolism of transit-amplifying cells is characterized by low oxygen consumption (19), an accumulation of these cells increases epithelial oxygenation (55). Another by-product of colitis is the generation of nitrate in the gut lumen (56), which depends on synthesis of the host enzyme iNOS (47). The increased luminal availability of oxygen and nitrate allow populations of facultative anaerobic Enterobactericeae, such as E. coli, to expand in mouse models of ulcerative colitis (Fig. 1C) (47, 57). Thus, the hypothesis that dysanaerobiosis is a driver of dysbiosis during ulcerative colitis (38, 58) can be explained mechanistically in animal models by the loss of a C2-skewed epithelial surface during colonic crypt hyperplasia.
The observation that ulcerative colitis can respond to antibiotic treatment suggests that dysbiosis exacerbates intestinal inflammation (59), although the underlying mechanism is not fully resolved. However, disruption of the gut microbiota with broad-spectrum antibiotics is itself associated with dysbiosis (60), indicating that a rational manipulation of the gut microbiota by targeting only potentially harmful microbes could provide greater benefit. Consistent with this idea, intestinal inflammation can be moderated by selectively inhibiting an expansion of facultative anaerobic Enterobactericeae through precision editing of the gut microbiota in mouse models of ulcerative colitis (61). This approach is based on selective inhibition of molybdenum-cofactor-dependent microbial respiratory pathways that are operational only during episodes of inflammation (61), thus providing a proof of concept that rational chemical modulation of specific enzymatic activities in complex microbial communities could be developed as an intervention strategy (62).
Since the host maintains homeostasis using a C2-skewed colonocyte metabolism, harnessing this epithelial control mechanism for therapeutic purposes could be an alternative strategy for rebalancing the microbiota. Specifically, a PPARγ-mediated differentiation of transit-amplifying cells into colonocytes would be expected to restore a C2-type epithelial surface, thereby remediating dysbiosis and reducing inflammation. Consistent with this idea, treatment with PPARγ agonists reduces crypt hyperplasia by inhibiting excessive division of transit-amplifying cells (63). Furthermore, topical treatment of the epithelial surface with the PPARγ agonist 5-aminosalicylic acid (5-ASA) is a first-line therapy for bringing mild to moderate cases of ulcerative colitis into remission (64–67) and is associated with lowering the abundance of Proteobacteria in the colonic microbiota (68). These data support the hypothesis that rebalancing the gut microbiota by reinstating a homeostatic C2 state of the colonic surface (12) represents a feasible therapeutic approach for restoring homeostasis (68).
Pathogens manipulate colonocyte metabolism for growth
Some intracellular bacterial pathogens can alter macrophage polarization to obtain nutrients from host cells to support microbial growth and long-term persistence in host tissue (69). One example is Brucella abortus, a pathogen that persists in M2-polarized macrophages of the murine liver and spleen. PPARγ-activated β-oxidation of fatty acids leads to a high availability of glucose (Fig. 2A), a carbon source that fuels pathogen growth within alternatively activated macrophages (70). Similar to an exploitation of macrophage polarization, recent findings suggest that bacterial pathogens can also manipulate colonocyte metabolism to favor their luminal growth during competition with the gut microbiota.
An important function of a C2-skewed colonic surface is to limit the luminal availability of host-derived respiratory electron acceptors, which confers niche protection against facultative anaerobic enteric pathogens (60). The enteric pathogen Salmonella enterica (family Enterobacteriaceae) overcomes such niche protection by using its virulence factors to trigger severe acute intestinal inflammation (71). Neutrophils migrating into the intestinal lumen during gut inflammation deplete Clostridia spp. (72, 73). This has the effect of decreasing the concentration of short-chain fatty acids, which steers the metabolism of terminally differentiated colonocytes towards an inflammation-associated C1 activation state and increased epithelial oxygenation (43, 74). The respiratory burst of phagocytes migrating into the intestinal lumen during S. enterica-induced colitis generates additional electron acceptors for anaerobic respiration, including nitrate (75, 76) and tetrathionate (77) (Fig. 3A). S. enterica expands in the lumen of the inflamed gut using a combination of aerobic and anaerobic respiration (74). Respiration provides S. enterica a growth advantage over obligate anaerobic bacteria, because it enables the pathogen to consume microbiota-derived fermentation products, such as succinate (16), butyrate (15), ethanolamine (78) or 1,2-propanediol (79). Interestingly, the high lactate release characteristic of a C1-skewed epithelial surface (Fig. 1B) also provides S. enterica with a host-derived carbon source to support its respiratory growth (43) (Fig. 3A). Thus, S. enterica uses its virulence factors to direct the epithelial surface towards a C1 activation state, which provides the pathogen with both a host-derived electron acceptor (i.e., oxygen) and a host-derived carbon source (i.e., lactate) and thus allows it to outcompete obligate anaerobic bacteria in the gut lumen (43, 74).
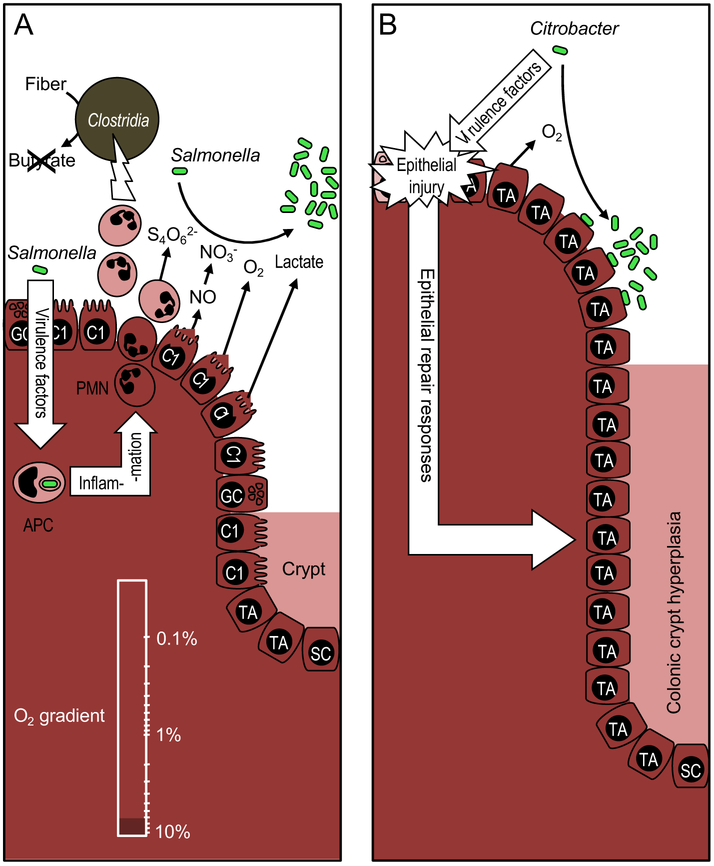
Figure 3: Enteric pathogens overcome niche protection by manipulating colonocyte metabolism.
(A) S. enterica (Salmonella) uses its virulence factors to trigger neutrophil transepithelial migration, which leads to a depletion of Clostridia, thereby lowering the luminal concentration of short-chain fatty acids, such as butyrate. The consequent metabolic reprogramming of the epithelium increases the luminal bioavailability of oxygen (O2) and lactate. The inflammatory response generates additional electron acceptors, including tetrathionate (S4O62-) and nitrate (NO3−). These host-derived resources drive an expansion of the facultative anaerobic pathogen. (B) Virulence factors of C. rodentium (Citrobacter) cause epithelial injury, thereby triggering epithelial repair responses leading to colonic crypt hyperplasia. The resulting increase in epithelial oxygenation drives a C. rodentium expansion by aerobic respiration. APC, antigen presenting cell; PMN, neutrophil; SC, stem cell, TA, undifferentiated transit-amplifying cell; C2, terminally differentiated C2-colonocyte; C1, terminally differentiated C1-colonocyte; GC, goblet cell.
Citrobacter rodentium (family Enterobacteriaceae) is an enteric pathogen of mice that uses virulence mechanisms similar to those employed by human attaching-and-effacing pathogens, such as enteropathogenic E. coli (EPEC) or enterohaemorrhagic E. coli (EHEC) (80). C. rodentium uses its virulence factors to intimately attach to the colonic surface, which enables the pathogen to compete with the gut microbiota (81, 82). Epithelial injury caused by C. rodentium virulence factors triggers excessive epithelial repair, leading to colonic crypt hyperplasia and accumulation of undifferentiated transit-amplifying cells at the mucosal surface (Fig. 1C) (80). The resulting loss of a C2-skewed epithelium increases the amount of oxygen emanating from the mucosal surface and drives growth of C. rodentium through aerobic respiration (55) (Fig. 3B). Respiration supports growth of C. rodentium on microbiota-derived fermentation products, such as formate, which enables the pathogen to compete with the gut microbiota (55).
In conclusion, both S. enterica and C. rodentium use their virulence factors to ablate a homeostatic C2-skewed colonic surface, albeit through different mechanisms. However, in each case the pathogen alters colonocyte metabolism to obtain critical resources from host cells to allow it to compete against the resident gut microbiota. Thus, subversion of colonocyte cell metabolism by enteric pathogens is emerging as a novel strategy to overcome niche protection conferred by the gut microbiota.
Future directions
The finding that colonocytes play a central role in sculpting the gut microbiota points to changes in colonocyte metabolism as a common driver of dysbiosis in the large bowel (17). Here, we make the case that metabolic polarization of colonocytes functions as a control switch for the gut microbiota, mediating the shift between balanced and dysbiotic communities. There is compelling evidence that loss of C2-colonocytes contributes to dysbiosis in antibiotic-treated mice (12), chemically-induced colitis (47, 57) or infection with enteric pathogens (43, 55, 74). Interestingly, decreased colonic PPARγ synthesis is observed in chronically simian immunodeficiency virus (SIV)-infected rhesus macaques (83), suggesting that a change in colonocyte metabolism might also underpin the expansion of Proteobacteria observed in human immunodeficiency virus (HIV) infected subjects (84). However, additional work is needed to investigate whether changes in the microbial community from obligate to facultative anaerobic bacteria observed in patients with colorectal cancer (34), irritable bowel syndrome (35, 36) or individuals consuming a high-fat Western-style diet (31, 32) is caused by an underlying loss of C2-colonocytes. The view that our immune system balances the colonic microbiota by maintaining a C2-skewed epithelial surface suggests that harnessing this host control mechanism for therapeutic means could provide an alternative to targeting the microbes themselves for remediating dysbiosis and could provide new therapeutic strategies for rebalancing the colonic microbiota in a broad spectrum of human diseases.
Acknowledgements
Work in A.J.B.’s laboratory is supported by USDA award 2014-09901 and Public Health Service Grants AI044170, AI096528, AI112445 and AI112949. M.X.B. and A.J.B. filed invention report number 0577501-16-0038 at iEdison.gov for a treatment to prevent post-antibiotic expansion of Enterobacteriaceae.
Literature
- 1.Cani PD, Gut microbiota - at the intersection of everything? Nat Rev Gastroenterol Hepatol, (2017). [DOI] [PubMed] [Google Scholar]
- 2.Eckburg PB et al. , Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tap J et al. , Towards the human intestinal microbiota phylogenetic core. Environ Microbiol 11, 2574–2584 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Turnbaugh PJ et al. , The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 1, 6ra14 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foster KR, Schluter J, Coyte KZ, Rakoff-Nahoum S, The evolution of the host microbiome as an ecosystem on a leash. Nature 548, 43–51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.den Besten G et al. , The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res 54, 2325–2340 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Velazquez OC, Lederer HM, Rombeau JL, Butyrate and the colonocyte. Production, absorption, metabolism, and therapeutic implications. Adv Exp Med Biol 427, 123–134 (1997). [PubMed] [Google Scholar]
- 8.Atarashi K et al. , Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arpaia N et al. , Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furusawa Y et al. , Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Smith PM et al. , The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Byndloss MX et al. , Microbiota-activated PPAR-g signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 357, 570–575 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porter NT, Martens EC, The Critical Roles of Polysaccharides in Gut Microbial Ecology and Physiology. Annu Rev Microbiol 71, 349–369 (2017). [DOI] [PubMed] [Google Scholar]
- 14.El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B, The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol 11, 497–504 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Bronner DN et al. , Genetic Ablation of Butyrate Utilization Attenuates Gastrointestinal Salmonella Disease. Cell Host Microbe 23, 266–273 e264 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spiga L et al. , An Oxidative Central Metabolism Enables Salmonella to Utilize Microbiota-Derived Succinate. Cell Host Microbe 22, 291–301 e296 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Byndloss MX, Baumler AJ, The germ-organ theory of non-communicable diseases. Nat Rev Microbiol 16, 103–110 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Barker N, van de Wetering M, Clevers H, The intestinal stem cell. Genes Dev 22, 1856–1864 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan YY et al. , A bioassay to measure energy metabolism in mouse colonic crypts, organoids, and sorted stem cells. Am J Physiol Gastrointest Liver Physiol 309, G1–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warburg O, Wind F, Negelein E, The Metabolism of Tumors in the Body. J Gen Physiol 8, 519–530 (1927). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tylichova Z et al. , Activation of autophagy and PPARgamma protect colon cancer cells against apoptosis induced by interactive effects of butyrate and DHA in a cell type-dependent manner: The role of cell differentiation. J Nutr Biochem 39, 145–155 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Lefebvre M et al. , Peroxisome proliferator-activated receptor gamma is induced during differentiation of colon epithelium cells. J Endocrinol 162, 331–340 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Duszka K, Oresic M, Le May C, Konig J, Wahli W, PPARgamma Modulates Long Chain Fatty Acid Processing in the Intestinal Epithelium. Int J Mol Sci 18, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donohoe DR et al. , The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab 13, 517–526 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roediger WE, Role of anaerobic bacteria in the metabolic welfare of the colonic mucosa in man. Gut 21, 793–798 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furuta GT et al. , Hypoxia-inducible factor 1-dependent induction of intestinal trefoil factor protects barrier function during hypoxia. J Exp Med 193, 1027–1034 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Litvak Y, Byndloss MX, Tsolis RM, Baumler AJ, Dysbiotic Proteobacteria expansion: a microbial signature of epithelial dysfunction. Curr Opin Microbiol 39, 1–6 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Shin NR, Whon TW, Bae JW, Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol 33, 496–503 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Rizzatti G, Lopetuso LR, Gibiino G, Binda C, Gasbarrini A, Proteobacteria: A Common Factor in Human Diseases. Biomed Res Int 2017, 9351507 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vollaard EJ, Clasener HA, Janssen AJ, Co-trimoxazole impairs colonization resistance in healthy volunteers. J Antimicrob Chemother 30, 685–691 (1992). [DOI] [PubMed] [Google Scholar]
- 31.Devkota S et al. , Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in 1110−/− mice. Nature 487, 104–108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez-Medina M et al. , Western diet induces dysbiosis with increased E coli in CEABAC10 mice, alters host barrier function favouring AIEC colonisation. Gut 63, 116–124 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Morgan XC et al. , Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 13, R79 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arthur JC et al. , Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carroll IM, Ringel-Kulka T, Siddle JP, Ringel Y, Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterology and motility : the official journal of the European Gastrointestinal Motility Society 24, 521–530, e248 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krogius-Kurikka L et al. , Microbial community analysis reveals high level phylogenetic alterations in the overall gastrointestinal microbiota of diarrhoea-predominant irritable bowel syndrome sufferers. BMC gastroenterology 9, 95 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Normann E, Fahlen A, Engstrand L, Lilja HE, Intestinal microbial profiles in extremely preterm infants with and without necrotizing enterocolitis. Acta paediatrica 102, 129–136 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Rigottier-Gois L, Dysbiosis in inflammatory bowel diseases: the oxygen hypothesis. ISME J 7, 1256–1261 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rivera-Chavez F, Lopez CA, Baumler AJ, Oxygen as a driver of gut dysbiosis. Free Radic Biol Med 105, 93–101 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Kelly CJ et al. , Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host Microbe 17, 662–671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alex S et al. , Short-chain fatty acids stimulate angiopoietin-like 4 synthesis in human colon adenocarcinoma cells by activating peroxisome proliferator-activated receptor gamma. Mol Cell Biol 33, 1303–1316 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spees AM et al. , Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. MBio 4, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gillis CC et al. , Dysbiosis-Associated Change in Host Metabolism Generates Lactate to Support Salmonella Growth. Cell Host Microbe 23, 54–64 e56 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reese AT et al. , Antibiotic-induced changes in the microbiota disrupt redox dynamics in the gut. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geeraerts X, Bolli E, Fendt SM, Van Ginderachter JA, Macrophage Metabolism As Therapeutic Target for Cancer, Atherosclerosis, and Obesity. Front Immunol 8, 289 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Charo IF, Macrophage polarization and insulin resistance: PPARgamma in control. Cell Metab 6, 96–98 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Winter SE et al. , Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339, 708–711 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Papapietro O et al. , R-spondin 2 signalling mediates susceptibility to fatal infectious diarrhoea. Nat Commun 4, 1898 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strugala V, Dettmar PW, Pearson JP, Thickness and continuity of the adherent colonic mucus barrier in active and quiescent ulcerative colitis and Crohn’s disease. Int J Clin Pract 62, 762–769 (2008). [DOI] [PubMed] [Google Scholar]
- 50.McCormick DA, Horton LW, Mee AS, Mucin depletion in inflammatory bowel disease. J Clin Pathol 43, 143–146 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dubuquoy L et al. , Impaired expression of peroxisome proliferator-activated receptor gamma in ulcerative colitis. Gastroenterology 124, 1265–1276 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Roediger WE, The colonic epithelium in ulcerative colitis: an energy-deficiency disease? Lancet 2, 712–715 (1980). [DOI] [PubMed] [Google Scholar]
- 53.Ponferrada A et al. , The role of PPARgamma on restoration of colonic homeostasis after experimental stress-induced inflammation and dysfunction. Gastroenterology 132, 1791–1803 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Ogasawara N et al. , PPARgamma agonists upregulate the barrier function of tight junctions via a PKC pathway in human nasal epithelial cells. Pharmacol Res 61, 489–498 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Lopez CA et al. , Virulence factors enhance Citrobacter rodentium expansion through aerobic respiration. Science 353, 1249–1253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dudhgaonkar SP, Tandan SK, Kumar D, Raviprakash V, Kataria M, Influence of simultaneous inhibition of cyclooxygenase-2 and inducible nitric oxide synthase in experimental colitis in rats. Inflammopharmacology 15, 188–195 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Hughes ER et al. , Microbial Respiration and Formate Oxidation as Metabolic Signatures of Inflammation-Associated Dysbiosis. Cell Host Microbe 21, 208–219 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Henson MA, Phalak P, Microbiota dysbiosis in inflammatory bowel diseases: in silico investigation of the oxygen hypothesis. BMC Syst Biol 11, 145 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khan KJ et al. , Antibiotic therapy in inflammatory bowel disease: a systematic review and meta-analysis. Am J Gastroenterol 106, 661–673 (2011). [DOI] [PubMed] [Google Scholar]
- 60.Olsan EE et al. , Colonization resistance: The deconvolution of a complex trait. J Biol Chem 292, 8577–8581 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu W et al. , Precision editing of the gut microbiota ameliorates colitis. Nature 553, 208–211 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rekdal M, Balskus EP, Gut Microbiota: Rational Manipulation of Gut Bacterial Metalloenzymes Provides Insights into Dysbiosis and Inflammation. Biochemistry 57, 2291–2293 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Rousseaux C et al. , The 5-aminosalicylic acid antineoplastic effect in the intestine is mediated by PPARgamma. Carcinogenesis 34, 2580–2586 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brogden RN, Sorkin EM, Mesalazine. A review of its pharmacodynamic and pharmacokinetic properties, and therapeutic potential in chronic inflammatory bowel disease. Drugs 38, 500–523 (1989). [DOI] [PubMed] [Google Scholar]
- 65.Greenfield SM, Punchard NA, Teare JP, Thompson RP, Review article: the mode of action of the aminosalicylates in inflammatory bowel disease. Aliment Pharmacol Ther 7, 369–383 (1993). [DOI] [PubMed] [Google Scholar]
- 66.Zhou SY et al. , Intestinal metabolism and transport of 5-aminosalicylate. Drug Metab Dispos 27, 479–485 (1999). [PubMed] [Google Scholar]
- 67.Rousseaux C et al. , Intestinal antiinflammatory effect of 5-aminosalicylic acid is dependent on peroxisome proliferator-activated receptor-gamma. J Exp Med 201, 1205–1215 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu J et al. , 5-Aminosalicylic Acid Alters the Gut Bacterial Microbiota in Patients With Ulcerative Colitis. Front Microbiol 9, 1274 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Muraille E, Leo O, Moser M, TH1/TH2 paradigm extended: macrophage polarization as an unappreciated pathogen-driven escape mechanism? Front Immunol 5, 603 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xavier MN et al. , PPARgamma-mediated increase in glucose availability sustains chronic Brucella abortus infection in alternatively activated macrophages. Cell Host Microbe 14, 159–170 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stecher B et al. , Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol 5, 2177–2189 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sekirov I et al. , Salmonella SPI-1-mediated neutrophil recruitment during enteric colitis is associated with reduction and alteration in intestinal microbiota. Gut Microbes 1, 30–41 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gill N et al. , Neutrophil elastase alters the murine gut microbiota resulting in enhanced Salmonella colonization. PLoS One 7, e49646 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rivera-Chavez F et al. , Depletion of Butyrate-Producing Clostridia from the Gut Microbiota Drives an Aerobic Luminal Expansion of Salmonella. Cell Host Microbe 19, 443–454 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lopez CA et al. , Phage-mediated acquisition of a type III secreted effector protein boosts growth of salmonella by nitrate respiration. MBio 3, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lopez CA, Rivera-Chavez F, Byndloss MX, Baumler AJ, The Periplasmic Nitrate Reductase NapABC Supports Luminal Growth of Salmonella enterica Serovar Typhimurium during Colitis. Infect Immun 83, 3470–3478 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Winter SE et al. , Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thiennimitr P et al. , Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A 108, 17480–17485 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Faber F et al. , Respiration of Microbiota-Derived 1,2-propanediol Drives Salmonella Expansion during Colitis. PLoS Pathog 13, e1006129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Collins JW et al. , Citrobacter rodentium: infection, inflammation and the microbiota. Nat Rev Microbiol 12, 612–623 (2014). [DOI] [PubMed] [Google Scholar]
- 81.Kamada N et al. , Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 336, 1325–1329 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lupp C et al. , Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2, 204 (2007). [DOI] [PubMed] [Google Scholar]
- 83.Kumar V et al. , miR-130a and miR-212 Disrupt the Intestinal Epithelial Barrier through Modulation of PPARgamma and Occludin Expression in Chronic Simian Immunodeficiency Virus-Infected Rhesus Macaques. J Immunol 200, 2677–2689 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mutlu EA et al. , A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog 10, e1003829 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Carreau A, El Hafny-Rahbi B, Matejuk A, Grillon C, Kieda C, Why is the partial oxygen pressure of human tissues a crucial parameter? Small molecules and hypoxia. J Cell Mol Med 15, 1239–1253 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]