Abstract
Plants are faced with a barrage of stresses in their environment and must constantly balance their growth and survival. As such, plants have evolved complex control systems that perceive and respond to external and internal stimuli in order to optimize these responses, many of which are mediated by signaling molecules such as phytohormones. One such class of molecules called Brassinosteroids (BRs) are an important group of plant steroid hormones involved in numerous aspects of plant life including growth, development and response to various stresses. The molecular determinants of the BR signaling pathway have been extensively defined, starting with the membrane-localized receptor BRI1 and co-receptor BAK1 and ultimately culminating in the activation of BES1/BZR1 family transcription factors, which direct a transcriptional network controlling the expression of thousands of genes enabling BRs to influence growth and stress programs. Here, we highlight recent progress in understanding the relationship between the BR pathway and plant stress responses and provide an integrated view of the mechanisms mediating cross-talk between BR and stress signaling.
Introduction
As sessile organisms, plants have the exquisite challenge to perceive and withstand the environmental conditions they encounter while optimizing their growth. Stress conditions arise as the consequence of environmental perturbations such as changes in temperature, light, water availability and solute concentrations as well as through interactions of plants with pathogens. Plants employ an array of signaling molecules including phytohormones and peptide regulators to co-ordinate their development and responses to the environment. Among growth-promoting hormones, Brassinosteroids (BRs) are a group of polyhydroxylated plant steroid hormones that have emerged as key agonists and antagonists of pathways controlling growth and stress responses. Mutants deficient in BR signaling or biosynthesis exhibit marked defects in plant growth and reproduction and have altered stress responses [1–4]. Research over the last several decades has defined the BR biosynthesis and signaling pathways, which have been extensively reviewed elsewhere [5–7]. Subsequently, emerging research is characterizing the role of BRs in a tissue- and context-specific manner [8–10], including the way by which BR signaling is co-ordinated with stress and defense responses [11]. In this review, we focus our discussion on two important stresses that BRs have been implicated in: drought and response to pathogens.
Overview of BR signaling
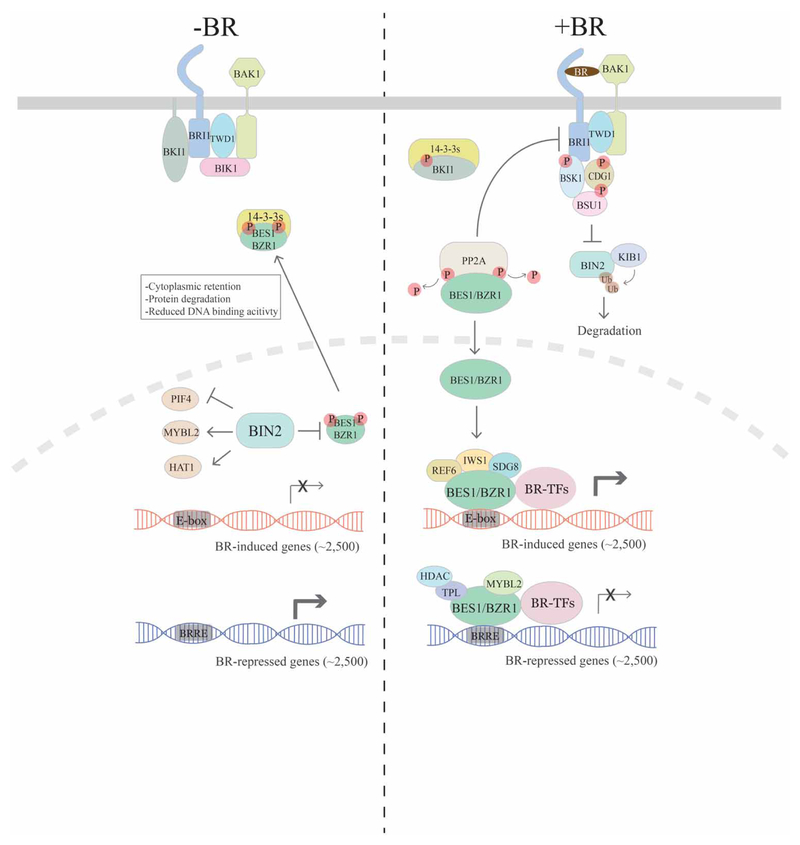
BRs are perceived by BRI1 (BR-INSENSITIVE 1) and its homologs BRL1 (BRI1-LIKE 1) and BRL3 (BRI1-LIKE 3), which are a family of plasma membrane-localized leucine-rich repeat receptor kinases [4,12,13] along with the co-receptor BAK1 (BRI1-ASSOCIATED KINASE 1) and related SERKs (SOMATIC EMBRYOGENESIS RECEPTOR KINASES) [14–16]. When BR levels are low, BR signaling is restrained by multiple mechanisms (Figure 1, left). Firstly, BKI1 (BR KINASE INHIBITOR 1) associates with BRI1 and prevents BRI1–BAK1 interactions [17]. Secondly, BIN2 (BR-INSENSITIVE2), a GSK3-like kinase, phosphorylates a collection of substrates [18] including BES1/BZR1 family transcription factors that function as master regulators of the BR pathway. Phosphorylation of BES1 and BZR1 leads to their inactivation through mechanisms that include cytoplasmic retention via interaction with 14–3-3 proteins [19,20], reduced DNA binding [21] and protein degradation [22,23].
Figure 1. Overview of the BR signaling pathway.
In the absence of BR, several negative regulators (BKI1 and BIK1) act to inhibit BR signaling at BRI1/BAK1 receptors, and BIN2 phosphorylates BES1/BZR1 family transcription factors to inhibit their function through several mechanisms. BIN2 also phosphorylates other transcription factors such as PIF4, MYBL2 and HAT1 to regulate their activities. Without BR signaling, expression of BR-induced genes is relatively low, whereas BR-repressed genes are more highly expressed, leading to suppressed BR responses. When present, BRs bind to receptor BRI1 and co-receptor BAK1, which leads to the disassociation of BKI1 and BIK1 as well as phosphorylation and activation of BRI1/BAK1, which activates BSK1, CDG1 and BSU1. BSU1 then functions to inhibit BIN2 kinase function while KIB1 ubiquitinates BIN2. PP2A activates BES1/BZR1 by dephosphorylation and cytoplasmic BKI1 sequesters 14–3-3s that otherwise sequester BES1/BZR1 in the cytoplasm. These events lead to accumulation of dephosphorylated BES1/BZR1 in the nucleus. BES1/BZR1 binds to E-box elements and interacts with cofactors (such as histone-modifying enzymes REF6 and SDG8 and transcription elongation factor IWS1) and BR-related transcription factors (BR-TFs, such as PIF4 and BIM1) to activate BR-induced gene expression. On the other hand, BES1/BZR1 binds to BRRE sites and interacts with co-repressors (TPL and MYBL2), histone deacetylase (HDAC) and likely other BR-TFs to inhibit BR-repressed genes. The large number of BR-regulated genes (~5000) enables cell elongation and other BR-regulated processes.
When BRs are present, they bind to BRI1 and co-receptor BAK1 to initiate a series of signaling events that ultimately activate BES1/BZR1 family transcription factors (Figure 1, right). Binding of BL to the BRI1–BAK1 complex causes BRI1 to rapidly phosphorylate BKI1 [17], leading to BKI1 dissociation from BRI1 and sequestration of BKI1 by 14–3-3 proteins [24,25]. BRI1 and BAK1 then sequentially phosphorylate and activate one another [26–28], which at least partially requires TWISTED DWARF 1 (TWD1/FKBP42), an immunophilin-like protein, that constitutively interacts with BRI1 but is required for BR-induced association and phosphorylation of the BRI1–BAK1 complex [29,30]. Activated BRI1 phosphorylates receptor-like cytoplasmic kinases BR SIGNALING KINASES (BSKs) and CONSTITUTIVE GROWTH (CDG1) that activate the phosphatase BRI1-SUPPRESSOR 1 (BSU1) [31–34]. BSU1 is proposed to dephosphorylate BIN2 on Y200, leading to inactivation of BIN2 kinase activity [31]. Several additional mechanisms that also regulate BIN2 have been reported recently, which include targeted protein degradation in the presence of BRs by F-box E3 ubiquitin ligase KINK SUPPRESSED IN BZR1–1D (KIB1) [35,36], cell-type specific sequestration of BIN2 at the plasma membrane by OCTOPUS (OPS) in the phloem [37] and inhibition of BIN2 by regulators such as HISTONE DEACTYLASE 6 (HDA6) under energy-limiting conditions [38] and CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1)/SUPPRESSOR of phyA-105 (SPA) in darkness [39]. The inactivation of BIN2 and the action of PROTEIN PHOSPHATASE 2A (PP2A) lead to the desphosphorylation of BES1/BZR1 family transcription factors [40]. Dephosphorylated BES1/BZR1 translocates from the cytoplasm to the nucleus where they function along with a suite of transcription factors and cofactors to regulate the expression of thousands of BR-regulated genes [22,23,41–44].
BES1 and BZR1 function with other transcriptional regulators to promote growth
Many studies have analyzed the BR-responsive transcriptome and have begun to reveal the extent to which BES1 and BZR1 modulate the BR-regulated transcriptional network [6]. Genome-wide ChIP-chip studies identified several thousand genes directly bound by BES1 and/or BZR1, including many BR-regulated genes and many transcription factors [44,45], leading to the idea that BES1 and BZR1 may propagate BR signals by regulating transcription factors in a series of transcriptional waves. Indeed, numerous studies have identified downstream transcriptional regulators in the BR pathway. Many of these transcription factors are themselves regulated by BES1 and/or BZR1 and also physically interact with BES1/BZR1 to carry out BR-regulated gene expression [6,46].
Early studies characterizing the function of BES1 as a transcription factor identified BES1-INTERACTING MYC-LIKE1 (BIM1) as a BES1-interacting transcription factor via yeast two-hybrid screening [43]. BES1 and BIM1 interact and bind synergistically to E-Box (CANNTG) sequences in BR target gene promoters, providing an early clue as to how BES1 co-operates with other transcription factors to regulate BR-responsive gene expression. Further examples of these interactions provide mechanisms connecting the growth-promoting hormones auxin and gibberellins (GAs) with BRs as well as light signaling. PHYTOCHROME-INTERACTING FACTORS (PIFs) are light-regulated transcription factors that function as important regulators of growth and responses to the environment [47]. PIF4 physically interacts with BES1 and BZR1, and these transcription factors share over 2000 common target genes as determined by genome-wide ChIP analysis, which led to a model in which BZR1 and PIFs interact to form heterodimers which bind to G-box (CACGTG) elements in BR target gene promoters [48]. Similar to PIFs, the auxin responsive transcription factors ARF6 and ARF8 interact with BZR1 [49]. ARF6 can also interact with PIF4, and over 40% of ARF6 target genes are shared with both BZR1 and PIF4. While BZR1 and PIFs co-operatively bind to G-box motifs, ARF6 binds to ARF-binding motifs (TGTCTC). BR treatment or increased BZR1/PIF4 in bzr1-d and PIF-OX enhanced ARF binding, suggesting that the three transcription factors bind to target genes co-operatively [49]. Taken together, BZR1, PIFs and ARFs form the so-called BZR1–ARF–PIF module [50] whose targets include the downstream tri-antagonistic bHLH system regulating both growth and defense gene expression. This system includes ATBS1/PRE (ACTIVATION-TAGGED bri1 SUPPRESSOR1/PACLOBUTRAZOL-RESISTANCE) family proteins that antagonize AIFs/IBH1 (ATBS1-INTERACTING FACTORS/INCREASED LAMINA INCLINATION INTERACTING bHLH1) bHLH transcription factors, which in turn inhibit ACEs/HBI1 (ACTIVATORS FOR CELL ELONGATION/HOMOLOG OF BEE2 INTERACTING WITH IBH 1) bHLH proteins that promote cell elongation [51–54].
BRs also function co-operatively with GA, which is at least partially mediated by the DELLA family of repressors inactivating BZR1, PIFs and ARFs to inhibit their function under low GA conditions [55–57]. BRs are also established to regulate GA metabolism and levels in both rice and Arabidopsis [58,59]. Another element of BR–GA cross-talk is manifested through complex interactions revolving around the NAC (NO APICAL MERISTEM, ARABIDOPSIS TRANSCRIPTION ACTIVATION FACTOR AND CUP-SHAPED COTYLEDON) transcription factor JUNGBRUNNEN1 (JUB1). JUB1 also interacts with DELLA proteins, which allows JUB1 to repress transcription of BR and GA biosynthesis genes including DWARF4 (DWF4) and GA3ox1 [60]. Moreover, it was found that BZR1 and PIF4 bind to a G-box in the JUB1 promoter region to repress JUB1 expression, revealing an important regulatory circuit between this NAC transcription factor and BR/GA signaling [60].
In addition to co-operatively binding to the same promoter element, BES1 also regulates target genes via interaction of transcription factors that bind different sites (Figure 1). Some examples include MYB30, and Arabidopsis thaliana HOMEODOMAIN-LEUCINE ZIPPER PROTEIN 1 (HAT1) and ARF6/8. Both MYB30 and HAT1 are direct targets of BES1 and also physically interact with BES1. MYB30 functions with BES1 to control BR-induced genes via binding of BES1 and MYB30 to E-box and MYB sites, respectively [61], whereas BES1 and HAT1 control BR-repressed genes by binding BRRE- and homeodomain-binding sites. Therefore, there are numerous examples of how BES1 and BZR1 co-operate with other transcription factors to mediate various aspects of BR-responsive gene expression. BES1 also co-occupies promoters with transcription factors that exert opposite effects on gene expression. This is the case in the root stem cell niche where BR activation of cell division in the quiescent center (QC) is prevented via antagonism of BES1 with the R2R3–MYB transcription factor, BRAVO (BRs AT VASCULAR AND ORGANIZING CENTER) [9]. BES1 and BRAVO physically interact and together bind the BRAVO promoter via E-box and MYB sites, respectively. BES1 represses BRAVO expression via recruitment of co-repressor TOPLESS (TPL) [62], whereas BRAVO promotes its own expression. Taken together, BES1 and BRAVO create a switch that allows BR regulation of QC cell division to be tightly controlled. Given this paradigm for BES1–BRAVO interactions, it will be interesting to define the genome-wide roles of BES1 and BRAVO and the mechanisms of their interaction.
Several studies also indicated that BES1 regulates gene expression by recruiting histone-modifying enzymes and a transcription elongation factor to differentially control BR-regulated gene expression at both transcription initiation and elongation steps [63–65]. Thus, there is accumulating evidence that BES1 interfaces with many other transcription factors and cofactors to regulate target gene expression and promote BR-mediated growth responses (Figure 1). In some cases, BES1/BZR1 interacts with transcription factors to co-operatively or synergistically regulate target gene expression, whereas in other cases BES1/BZR1 interacts with other transcription factors in an antagonistic manner. Taken together, these interactions provide insights into how BES1/BZR1 regulates a large number of BR-responsive genes; however, the detailed mechanisms that allow BES1 and BRs to regulate specific subsets of BR-regulated genes under different conditions and developmental stages are still under investigation. Future studies are needed to examine the full complement of transcription factors involved in the BR pathway and to define how BES1/BZR1 and other transcription factors interact to regulate various BR-induced and BR-repressed genes, which should yield significant insights into the structure and function of BR-controlled gene regulatory networks.
Cross-talk between BR and drought
In addition to regulating growth, BR signaling also interfaces with various stress outputs. Drought is a major stress that causes dramatic losses of crop yield, and thus, a great deal of effort has been placed on studying drought stress responses [66,67]. Recent progress in understanding the relationship between BR and drought has revealed several mechanisms by which BRs are inhibited during drought stress. Many of these operate to control BR–ABA antagonism at multiple levels of regulation ranging from the receptors complexes to downstream transcription factors. Central to this cross-talk are the BR signaling components BIN2 and BES1/BZR1, which have emerged as key factors promoting and antagonizing drought responses, respectively. Here, we provide an update on the mechanisms controlling BR and drought cross-talk and highlight recent work defining the genetic interactions between these pathways.
Antagonism between BR and ABA pathways
ABA is an important hormone that regulates responses to abiotic stress including drought [67,68]. ABA is synthesized from chloroplast derived carotenoid precursors, and during water deprivation, the rate-limiting enzyme in ABA biosynthesis, nine-cis-epoxycarotenoid dioxygenase (NCED), is rapidly up-regulated [69,70], leading to ABA accumulation that exerts a protective function through mechanisms including stomata closure, growth inhibition and synthesis of osmocompatible solutes [71]. In recent years, a core ABA signaling network has been pieced together. The predominant mechanism for sensing ABA is carried out by a large family of PYR/PLY/RCAR receptors that form a ternary complex with PP2C phosphatases, alleviating PP2C inhibition of SnRK2 kinase [72–75]. SnRK2 can then promote the function of ABA-responsive SnRK2 targets, including AREB/ABF transcription factors and ion channels (Figure 2, left) [76].
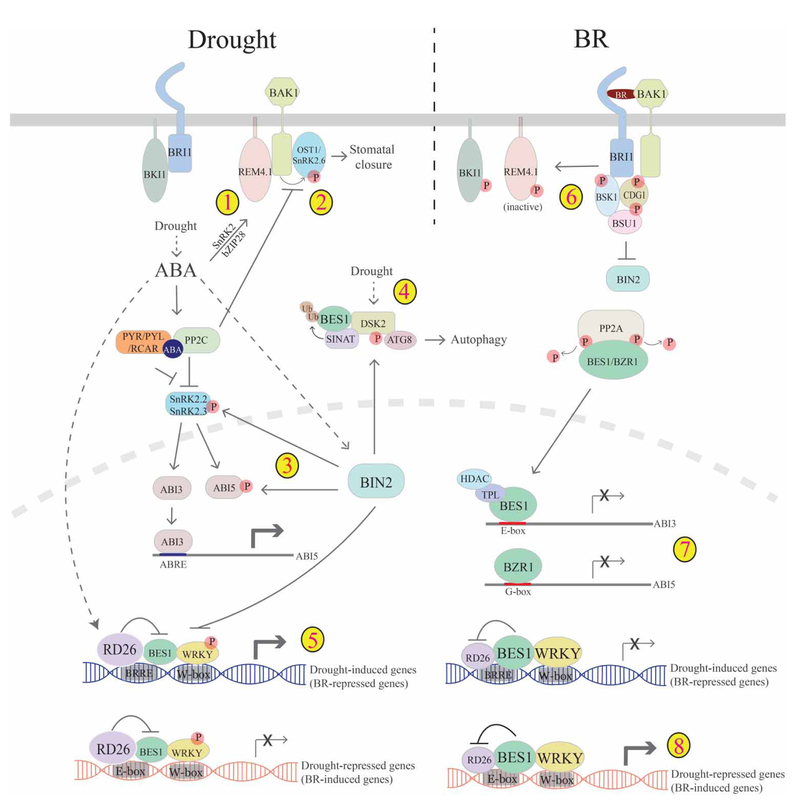
Figure 2. Cross-talk between BR and drought pathways.
Drought induces ABA accumulation, which promotes drought responses. ABA acts through receptors (PYR/PYL/RCAR) to inhibit PP2C repression of SnRKs, allowing SnRKs to phosphorylate downstream transcription factors (ABI3/ABI5 and others), which regulate genes for drought responses. ABA-activated OST1/SnRK2.6 also functions to regulate stomatal closure. There are several mechanisms of cross-talk between drought/ABA and BR pathways that involve the negative regulator of the BR pathway BIN2 and converge on BES1/BZR1. 1. ABA induces the expression of REM4.1 through SnRK2 and bZIP28; and REM4.1 acts to inhibit BRI1/BAK1 and thus BR signaling. 2. BAK1 and ABI1/PP2C oppositely regulate OST1/SnRK2.6 to modulate stomatal closure, providing another layer to BR–ABA cross-talk. 3. Under drought conditions, BIN2 is active, which phosphorylates and activates SnRK2.2/2.3 and ABI5. 4. During stress conditions, BES1 is ubiquitinated by SINAT E3 ubiquitin ligases and targeted for degradation through selective autophagy via phospho-regulated autophagy receptor DSK2. Phosphorylation of DSK2 by BIN2 enhances DSK2–ATG8 interactions, therefore promoting BES1 degradation. 5. Drought and ABA activate NAC family transcription factor RD26 that inhibits BES1 activity to promote drought-induced (BR-repressed) genes and inhibit drought-repressed (BR-induced) genes. 6. In contrast, under BR-promoted growth conditions, BRI1 phosphorylates and inactivates REM4.1. 7. BR-activated BES1/BZR1 inhibits ABI5 expression to inhibit ABA responses either through ABI3 (top) or by directly binding ABI5 promoter (bottom). 8. On the other hand, BES1 also inhibits RD26 transcriptional activity to promote BR-induced (drought-repressed) genes and inhibit drought-induced (BR-repressed) genes, thereby promoting BR-regulated growth.
From early in the BR literature, it has been noted that BR and ABA exhibit an antagonistic relationship. For example, it was shown that roots of BR mutants are hypersensitive to ABA treatment [4,77] and seeds with reduced BR biosynthesis or signaling are more sensitive to ABA [78]. Upon examination of hormone-responsive gene expression at the genome-wide level, it also became apparent that BR and ABA regulate common sets of genes [79]. ABA was shown to modulate BR response outputs including BES1 phosphorylation status and BR marker genes [80]. Since these effects were still observed in BR receptor mutants, but not when the more downstream kinase BIN2 was inhibited, it was postulated that cross-talk between ABA and BR pathways occurs downstream from BR perception, but at or upstream of BIN2 kinase in the BR signaling pathway. More recently, several studies have begun to define these and other molecular mechanisms of BR–ABA cross-talk, showing that BR and ABA pathways interface at multiple points ranging from inhibition of receptor complexes to downstream transcriptional regulation.
BR–ABA cross-talk at BR receptor complexes
Recent evidence in rice suggests that BR–ABA cross-talk may occur as early as the formation of BRI1–BAK1 receptor complexes [81,82]. This occurs via the plant-specific family of membrane-anchored remorin proteins. Specifically, OsREM4.1, which is transcriptionally induced by ABA via the SnRK2-regulated transcription factor OsbZIP28 [81,83], binds to the activation loop of OsSERK1 (homolog of BAK1). This interaction inhibits BRI1–BAK1 complex formation and thus BR signaling, which is reminiscent of the negative regulation provided by BKI1 [17,24]. Similar to the inactivation of BKI1 by BRs, OsREM4.1 can be directly phosphorylated and inactivated by OsBRI1 in the presence of BRs [81]. Thus, OsREM4.1 is activated by ABA to shut down BR responses, whereas BRs inactivate OsREM4.1, providing one mechanism to tradeoff growth and stress response programs depending on the relative amounts of BR and ABA present. It remains to be determined if a similar mechanism operates in Arabidopsis and if so, its contribution to overall ABA–BR cross-talk.
BAK1 is also involved in BR–ABA cross-talk to regulate stomata opening. A recent study found that bak1 mutants lose water more quickly than wild-type plants and are insensitive to ABA in terms of stomata closure [84]. This can be explained by the interaction of BAK1 with OST1/SnRK2.6, which is known to be a major contributor to ABA-induced stomatal closure [85,86]. This study also found that BAK1 can interact with the PP2C family protein ABI1. BAK1 and ABI1 oppositely regulate OST phosphorylation (at least in vitro) and ABI1 interaction with BAK1 inhibits BAK1–OST1 complex formation. Therefore, the BAK1–OST1 complex is promoted by ABA, which leads to stomatal closure, whereas this process is inhibited by BL treatment, providing another layer of BR–ABA cross-talk [84]. Further studies are needed to determine whether BAK1–OST1 interactions affect BR signaling outputs in response to ABA. While these studies seem to support antagonism of BR and ABA in terms of stomata closure, this effect has been somewhat controversial across studies and species and may depend on the relative concentrations of BRs used and whether BRs were exogenously applied or BR mutants were used [87–90]. For example, BR mutants showed an increased response to ABA with enhanced stomatal closure [90–92]; however, BR application could cause stomata opening at low concentrations [89], but promoted stomatal closure at higher concentrations [87,89,90].
BIN2 kinase regulates multiple aspects of BR–ABA cross-talk
Further cross-talk among BR and ABA pathways occurs at the level of BIN2 kinase (Figure 2, left). BIN2 is a GSK3 kinase that functions as a negative regulator in the BR pathway by phosphorylating and inactivating BES1 and BZR1. In addition to BES1 and BZR1, BIN2 has a diverse array of substrates that allow it to regulate numerous processes involved in growth, development and stress responses [93]. Consistent with the reported ABA hypersensitivity of bin2–1D gain-of-function mutants [77], bin2–1D plants were found to be hypersensitive to ABA in root growth inhibition assays and ABA-responsive gene expression and displayed increased phosphorylation of the SnRK2 substrate ABF2. Conversely, BIN2 loss-of-function bin2–3 bil1 bil2 mutants showed compromised ABA responses [94]. A search for BIN2-interacting proteins using immunoprecipitated BIN2-FLAG and liquid chromatography tandem mass spectrometry (LC–MS/MS) identified SnRK2s as BIN2 interactors. BIN2 was shown to specifically interact with SnRK2.2, SnRK2.3 and SnRK2.6 and phosphorylate SnRK2.2 and SnRK2.3 but not SnRK2.6. Mass spectrometry and follow-up analysis identified T180 as a BIN2 phosphorylation site on SnRK2.3 (T181 of SnRK2.2), and a mutant SnRK2.3T180A displayed decreased auto-and trans-phosphorylation activity, indicating that BIN2 phosphorylation is crucial for SnRK2 activity [94]. Therefore, BIN2 phosphorylation and activation of SnRK2 represents one mechanism by which BIN2 promotes ABA responses.
Another point of BR–ABA cross-talk mediated by BIN2 comes in the form of BIN2 interaction with a downstream transcription factor in the ABA pathway, ABI5 (Figure 2, left). ABI5 is a well-known target of SnRK2 kinases and is critical for ABA inhibition of seed germination [95,96]. ABI5 was identified in a yeast two-hybrid screen using BIN2 as bait [97]. Subsequent analysis indicated that BIN2 phosphorylates ABI5 in an ABA dependent manner, likely on distinct residues from those phosphorylated by SnRK2. Genetic evidence indicated that BIN2 phosphorylation stabilizes ABI5, as ABI5 levels were increased in BIN2 gain-of-function mutants, but decreased in the absence of BIN2 or when plants were treated with BRs, which inactivates BIN2 [97]. Together with evidence that BIN2 and its homologs may be induced by stresses [80,93,98,99], it is likely that BIN2 is activated during abiotic stress conditions and positively modulates ABA signaling through both phosphorylation and activation of SnRK2.2 and SnRK2.3 as well as by increasing ABI5 protein stability. Given that several other ABA-regulated ABF transcription factors (ABF1 and ABF3) were found to interact with BIN2 [97], it will be interesting for future studies to determine the extent to which the repertoire of BIN2 substrates extends into the ABA pathway.
Inhibition of ABA signaling by BES1 and BZR1
A more downstream aspect of BR–ABA interactions occurs between ABA- and BR-regulated transcription factors (Figure 2, right). Firstly, BES1 can antagonize ABA signaling via repression of ABI3 expression by the BES1–TPL–HDAC19 complex [100]. The expression of ABA-regulated transcription factors ABI3 and ABI5 was shown to be decreased in bes1-D but up-regulated in a bes1 knockout mutant, which corresponded to the altered ABA sensitivities of these mutants. The mechanistic basis as to how BES1 can repress target gene expression can be explained by interactions of BES1 with TPL and TOPLESS-RELATED (TPR) repressors via an EAR motif that is conserved among BES1/BZR1 family transcription factors. Consistently, expression of a BES1-D construct containing EAR mutations was unable to produce BR- or ABA-related phenotypes normally associated with BES1-D overexpression, reinforcing the notion that the repression function of BES1 is crucial for BR signaling as well as BR–ABA cross-talk. Moreover, BES1 was found to bind to several E-box motifs in the ABI3 promoter, repressing its expression by reducing histone acetylation that is normally associated with gene activation [100]. Since ABI3 functions to promote the expression of ABI5, repression of ABI3 by BES1 could down-regulate both of these transcription factors and thus ABA responses.
Subsequently, another related study also found that BZR1 can directly bind to the promoter of ABI5 [101]. In the present study, it was found that mutant bzr1-D plants are also resistant to ABA treatments, which is at odds with previous reports that bes1-D, but not bzr1-D, displayed ABA insensitivity [100]. Interestingly, bzr1-d/bin2–1D double mutants showed decreased sensitivity to ABA compared with bin2–1D mutants, and a subset of ABA-responsive genes, including ABI5, were differentially expressed in bzr1-D/bin2–1D compared with bin2–1D as monitored by RNA-seq experiments. Follow-up experiments showed that BZR1 could directly bind to G-box elements within the ABI5 promoter. These G-box elements were necessary for BZR1 repression of ABI5 and overexpression of ABI5 in bzr1-d partially rescued the ABA-insensitive phenotype of bzr1-d [101]. These studies suggest that BES1 and BZR1 can transcriptionally modulate ABI5 levels to control ABA signaling either through control of ABI3 and/or via direct binding to the ABI5 promoter. It remains to be determined if the differences between these two studies reflect the specificities of BES1 versus BZR1 or if they can be explained by other experimental differences.
MYB30, a target and interactor of BES1 [61], also functions in regulating ABA responses [102,103]. Similar to BES1 and BZR1, MYB30 appears to negatively regulate ABA responses, which is consistent with BES1 and MYB30 co-operatively regulating BR gene expression. myb30 mutants were more sensitive to ABA treatment, whereas overexpression of MYB30 led to ABA insensitivity [103]. Interestingly, MYB30 is sumoylated by the small ubiquitin-like modifier (SUMO) E3 ligase SIZ1, which leads to stabilization of MYB30 [103]. Conversely, the E3 ubiquitin ligase MIEL1 (MYB30-interacting E3 ligase 1) targets both MYB30 [104] and another ABA-related transcription factor, MYB96, for degradation [102], but the ABA-hypersensitive phenotype of miel1 mutants in seed germination was found to be largely due to accumulation of MYB96 [102], which functions as a positive regulator in the ABA pathway [105]. In contrast, MIEL1 appears to regulate both MYB30 and MYB96 stability in leaf tissue [102], which could have important implications in ABA and pathogen crosstalk. Thus, MYB30 is a negative regulator of ABA responses and is controlled by several post-translational modifications to fine-tune its activity in ABA and related pathways.
Interactions between the BR pathway and drought stress
While there is accumulating evidence that BR and ABA pathways function antagonistically, the relationship between BRs and drought is somewhat more complex. Several studies reported that application of exogenous BRs could actually promote tolerance to drought stress, which is seemingly contradictory to the BR–ABA antagonism [106–110]. However, the effects of BR on drought outcomes seem to depend on the concentrations of BRs used as well as the environment. When high concentrations of BRs are used, they may lead to feedback inhibition or other secondary effects as was noted in tomato, where exogenous BR application leads to elevated ABA levels due to H2O2 production [111].
While the effects of exogenous BRs may be complex, several recent studies have used a genetic approach to address the relationship of the BR pathway with drought responses, showing that the BR pathway inhibits drought response [112–114]. Knockdown of a BRI1 homolog in Brachypodium distachyon, BdBRI1, led to increased tolerance to drought and altered drought-responsive gene expression [115]. Furthermore, Northey et al. [114] connected BR to both ABA and drought by establishing the relationship between the farnesyl transferase ERA1 (ENHANCED RESPONSE TO ABSCISIC ACID 1) and CYP85A2, the cytochrome P450 enzyme that converts castasterone into brassinoloide in the last step of BR biosynthesis. A mutant in CYP85A2 (cyp85a2–2) was identified from a screen targeting candidate ERA1 substrates that contained a motif typically targeted by farnesyl transferases. The cyp85a2–2 mutant showed a phenotype similar to era1–2 including an increased response to ABA and drought tolerance. The known function of CYP85A2 in the BR pathway suggested that the phenotypes observed in era1–2 might be due to decreased BR biosynthesis. Indeed, detailed analysis showed that era1–2 mutants have reduced BR biosynthesis and ERA1-mediated farnesylation of CYP85A2 is required for proper localization and function of CYP85A2 [114]. This study supports the idea that reduced BR biosynthesis can lead to drought tolerance, however, the mechanisms that allow plants to balance BR-regulated growth when drought stress is encountered remained unclear until recently.
Reciprocal inhibition between BR and drought pathways mediated by BES1 and RD26
When plants encounter drought, it is important that growth is quickly inhibited so that resources can be devoted to stress response. Similarly, under optimal growth conditions, resources need not be wasted by unnecessarily activating drought stress responses [116]. Recent studies have shown that several mechanisms converge on BES1 to restrain growth when stress is encountered. One of these mechanisms was revealed through characterization of a BES1 target transcription factor, RESPONSIVE TO DESICCATION 26 (RD26), which allows plants to balance these constrains through modulation of the transcriptional activity of BES1 (Figure 2) [112]. RD26 is a member of the NAC family of transcription factors and is induced under abiotic stress conditions including drought to promote drought tolerance [117–119].
BRs inhibit the expression of RD26 and several of its homologs, which is mediated by BES1 binding to a region of RD26 promoter containing the BES1 BRRE-binding site. When overexpressed (RD26 OX), RD26 caused stunted growth, reduced BR response and suppressed the BR gain-of-function mutant bes1-D. These observations along with the fact that RD26 is induced under abiotic stress [117–119] suggest that RD26 functions to inhibit BR-regulated growth when stress is encountered.
Global gene expression studies were instrumental in deciphering the mechanism of interactions between BES1 and RD26. BRs regulated ~5000 genes, ~35% of which were regulated in an opposite fashion in RD26 OX plants. Investigation of these genes revealed that BR-induced and RD26 OX-repressed genes were enriched for E-box promoter elements, while BR-repressed and RD26 OX-induced genes were enriched for BRRE sites. Since these are BES1-binding sites and closely resembled those previously reported for RD26 and other NACs [117,120], it was postulated that BES1 and RD26 might bind to a common site in these BR- and RD26-regulated genes [112]. Indeed, BES1 and RD26 were found to physically interact and simultaneously bind to the same promoter element where they neutralized each other’s activity on BES1 target genes. For example, BES1 promotes BR-induced genes, whereas RD26 represses these genes, and together the combination of BES1 and RD26 has intermediate activity. The opposite was true on BR-repressed genes, where BES1 repressed their expression and RD26 induced their expression.
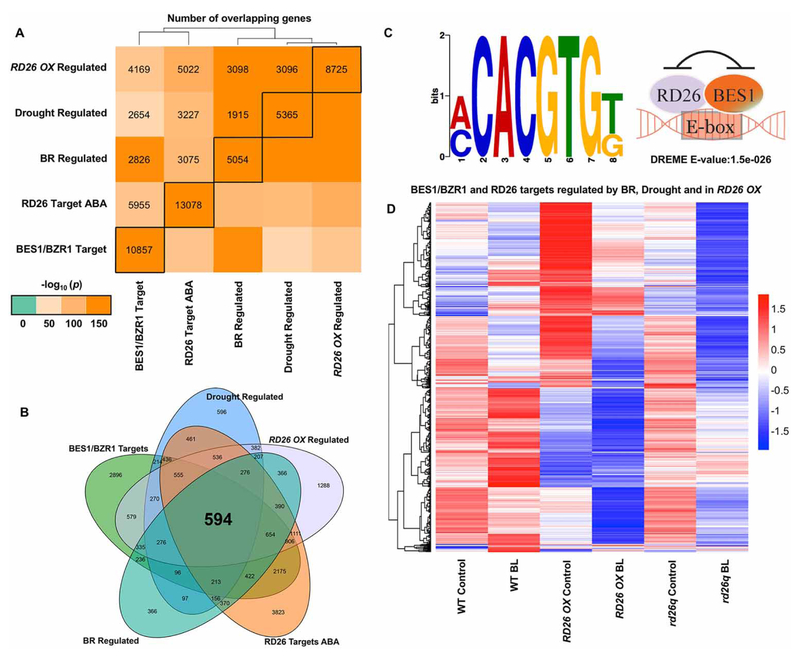
The idea that BES1 and RD26 bind to a common target site is supported by an elegant study by Song et al. [121] that provided the largest set of ABA-responsive ChIP-seq data to date. This study confirmed that the motif enriched in RD26 target genes matched known BES1-binding sites and provided a rich dataset allowing comparisons of RD26 target genes to various BES1 and BR datasets. We combined and analyzed these published datasets to provide a more comprehensive overview of the relationship between BES1 and RD26 target and regulated genes. As expected, RD26 targets from ABA-treated plants [121] show extensive overlaps with RD26 OX-regulated genes (Figure 3A) [112]. Similarly, RD26 target genes have a high degree of overlap with BES1/BZR1 target [44,45,49] and BR-regulated genes (Figure 3A) [63], which confirms the interactions between BES1 and RD26 reported by Ye et al. [112] at the genome-wide level. Consistent with the role of RD26 in drought response, a significant amount of overlap is also observed with drought-regulated genes [122] and RD26 OX/RD26 targets. Comparison of BES1/BZR1 and RD26 target genes with genes regulated by BRs, drought and in RD26 OX revealed a core set of 594 genes (Figure 3B), which are highly enriched for the BES1 and RD26 G-box-binding site (CACGTG; a specific E-box, Figure 3C). We also performed clustering analysis of these 594 genes using published RNA-seq data from plants treated with or without BRs [112], which revealed that many BR-repressed genes are induced in RD26 OX and repressed in rd26q mutants (especially after BR treatment, Figure 3D). Conversely, the subset of BR-induced genes in this core set appear to be highly repressed in RD26 OX, but no longer induced by BRs in rd26q, raising the possibility that RD26 could actually be required for BR-responsive induction of these genes. In any case, perturbation of RD26 leads to dramatic changes in expression of BR and drought-regulated genes that are targets of both BES1/BZR1 and RD26.
Figure 3. RD26 and BES1 regulate a common set of BR and drought genes.
(A) Comparisons of RD26 and BES1/BZR1 target genes with those regulated by RD26 OX, BRs or drought. Gene lists were obtained from previously published datasets [7,44,45,49,112,121] and statistical significance of their intersection was assessed using Fisher’s exact test. Color legend indicates −log10 transformed P-values for the intersection between the given pair of genes; black boxes indicate the total number of genes in each list. (B) Venn diagram showing a core set of 594 genes that are both BES1/BZR1 and RD26 targets and regulated in RD26OX as well as by BRs and drought. (C) The G-box motif is enriched in the core set of genes shown in (B) in DREME promoter motif analysis [171] (left), which supports a model in which BES1 and RD26 bind to a common promoter element to inhibit each other’s function (right). (D) Clustering analysis of BR-responsive gene expression for 594 core genes from (B) using published RD26 OX and rd26q RNA-seq data [112], showing that these genes are strongly influenced by RD26.
The genetic relationship between BR and drought was also confirmed and explained by cross-talk between BES1 and RD26. BR loss-of-function bri1–5 mutants showed increased expression of drought-induced genes and tolerance to drought, while the BR gain-of-function mutant bes1-D suppressed drought gene expression and was more sensitive to drought [112]. A double mutant of bes1-D RD26 OX largely rescued the drought-sensitive phenotype of bes1-D, suggesting that BR repression of RD26 through BES1 plays a major role in controlling drought response. Taken together, these observations suggest a model in which BRs restrain drought responses under normal conditions by repressing the expression of RD26 and other NACs. When drought is encountered, RD26 is quickly induced [119] and interacts with BES1 to inhibit the function of BES1 on target gene promoters. Thus, BR and drought pathways converge by the interaction of BES1 and RD26 on a common promoter element, leading to inactivation of BES1, which ensures a proper growth-stress balance. While BES1 and RD26 oppose each other’s function on many genes, a subset of genes affected by BRs and, in RD26 OX, are regulated in the same direction [112], suggesting that BES1 and RD26 may function co-operatively in certain contexts. Thus, it will be of great interest to determine the mechanisms that lead to antagonism or co-operation between BES1 and RD26. These sets of genes may explain why BRs can sometimes promote resistance to drought [123] and could provide an opportunity to engineer crops for optimal growth and stress responses.
Degradation of BES1 by selective autophagy during drought stress
In addition to inhibition of the activity of BES1 on target gene promoters, more recent evidence suggests that BES1 can also be inhibited during stress by targeted degradation. One pathway that is involved in protein degradation, especially during stress, is autophagy [124,125]. Autophagy can be highly selectively by employing receptor proteins that bind to both cargos destined for degradation and also to the autophagy protein ATG8 (AUTOPHAGY-RELATED 8) [126–128]. Using autophagy inhibitors and autophagy-deficient mutants, Nolan et al. [113] showed that BES1 accumulates when autophagy is blocked, especially during drought and starvation stresses, which is in line with studies showing that TOR, a central regulator of growth and inhibitor of autophagy, promotes accumulation of BZR1 [129].
A mechanistic basis for how BES1 is degraded by autophagy was revealed through yeast two-hybrid screening for BES1-interacting proteins, which led to the discovery that DSK2 (DOMINANT SUPPRESSOR OF KAR2) functions as the autophagy receptor targeting BES1 for degradation [113] and also defined SINAT (SINA of Arabidopsis thaliana) E3 ubiquitin ligases for their role in targeting BES1 for degradation during stress [113] and in response to changing light conditions [130]. DSK2 interacts with ubiquitinated BES1 and is required for recruitment of BES1 to ATG8-labeled autophagosomes, but does not affect bulk autophagy, suggesting that DSK2 is involved in selective autophagy of BES1. Consistently, DSK2 interacts with ATG8 through two regions containing ATG8-interacting motifs (AIMs). DSK2 is phosphorylated by BIN2 kinase around its AIMs, which enhances the interaction between DSK2 and ATG8, thus promoting BES1 degradation [113].
Impairment of BES1 degradation in autophagy mutants or loss-of-function dsk2 RNAi plants led to increased growth in the presence of BR inhibitors compared with wild-type plants, whereas survival during drought stress was compromised in these mutants. Survival of dsk2 RNAi plants could be restored by reduction in BES1 in a dsk2 RNAi bes1 RNAi double mutant, suggesting that BES1 degradation during drought provides a key mechanism to shut down growth in favor of drought responses. This idea is supported by global gene expression studies, which showed that thousands of drought-related genes were misregulated in dsk2 RNAi plants during stress, many of which are BES1 targets [113]. A similar trend was observed during fixed-carbon starvation, indicating that BES1 degradation through autophagy is also critical to balance growth during starvation conditions. Additionally, the SINAT E3 ubiquitin ligase was shown to be induced during starvation stress and control BES1 degradation through autophagy during fixed-carbon starvation [113], however, whether SINAT and/or additional E3 ubiquitin ligases function during drought to degrade BES1 remains to be determined.
Degradation of BES1 during drought conditions was recently confirmed and extended to another family of transcription factors by Chen et al. [131] who characterized the role of WRKY46, WRKY54 and WRKY70 (WRKY46/54/70) in both BR-regulated growth and drought responses. WRKY transcription factors have been extensively studied for their roles in diverse stress responses [132–136], but a role in BR-regulated growth had not been shown previously. Expression of WRKY46/56/70 was induced by BRs in and bes1-D plants, and the dwarf phenotype of a wrky54wrky46wrky70 (wrky54t) mutant indicated that WRKYs are also required for BR-regulated growth. WRKY46/54/70 modulates both BR biosynthesis and also BR signaling, where they interact with BES1 to co-operatively regulate the expression of thousands of genes [131]. A large number of drought-responsive genes are affected in wrky54t mutants, and changes in drought-responsive gene expression corresponded with increased drought resistance of wrky54t mutants. These studies indicate that similar to BES1, WRKY46/54/70 are negative regulators of drought response. WRKY46/54/70 are substrates of BIN2 kinase, and BIN2 phosphorylation of these WRKYs led to their destabilization. During drought conditions, both BES1 and WRKY54 protein levels dramatically decreased [131]. These observations suggest that degradation of growth-promoting transcription factors during drought stress is likely an important mechanism to shut down BR responses, and the transcription factors affected by this process might extend beyond BES1, but the detailed pathways and components that control WRKY46/54/70 stability remain to be investigated.
Taken together, several studies have established BES1 as a key component for BR–drought cross-talk, and BES1 is inhibited through multiple mechanisms when drought stress is encountered. These include inhibition of BES1 transcriptional activity on target gene promoters through interactions with RD26 [112], degradation of BES1 through DSK2-mediated selective autophagy [113] and destabilization of transcription factors that co-operate with BES1 such as WRKY46/54/70 during drought [131]. Additionally, BIN2 kinase has been implicated in several regulatory events that comprise BR–drought cross-talk. BIN2 promotes ABA signaling components such as ABI5 and SnRK2 kinases and also inhibits a BES1 both directly and through modulating BES1 degradation via modulation of DSK2–ATG8 interactions that promote autophagy-mediated degradation of BES1. BIN2 also leads to the destabilization of several other positive regulators of the BR pathway. Thus, it appears that BIN2 is a critical component involved in promoting stress response while inhibiting growth and future studies should reveal the exact mechanisms that control BIN2 activation during stress.
In summary, BR and drought pathways interact at multiple levels (Figure 2). This multi-layer cross-talk happens in both directions (i.e. drought/ABA pathway can inhibit BR signaling and likewise, BR can inhibit drought/ABA signaling), which likely operate to slow down plant growth under drought conditions and also prevent unnecessary activation of drought response during active plant growth. It is likely that the multi-layer cross-talk is needed both to provide genetic redundancy and to fine-tune the growth and stress responses depending on the nature and severity of the imposed stresses.
Cross-talk between BR and plant immunity
In addition to drought, plants are faced with an array of other interactions with their environment, including those with pathogens. Response to pathogen attack must be swift to ensure survival of the plant, but also needs to be carefully controlled to optimize allocation of resources. The BR pathway is extensively intertwined with immunity, and cross-talk starts at receptor complexes that share many components and extends to downstream transcriptional regulators. In this section, we provide an update on BR and immune cross-talk with a focus on the molecular mechanisms controlling interactions between these two pathways.
Interactions between BR signaling and immunity at receptor complexes and the receptor kinase substrates
One major aspect of immune signaling that the BR pathway is involved in is pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) [11]. PAMPs are recognized by pattern-recognition receptors, which result in the activation of PTI responses. For instance, FLAGELLIN SENSING2 (FLS2), a well-studied receptor kinase, recognizes flagellin from bacterial flagella [137]. Recent studies revealed that several signaling components are involved in BR and PTI cross-talk, including receptor-like kinases (RLKs) BAK1 and BIR1 (BAK1-INTERACTING RECEPTOR-LIKE KINASE1), receptor-like cytoplasmic kinases (RLCKs) BSK1 and BIK1, and transcription factors BES1/BZR1 (Figure 4).
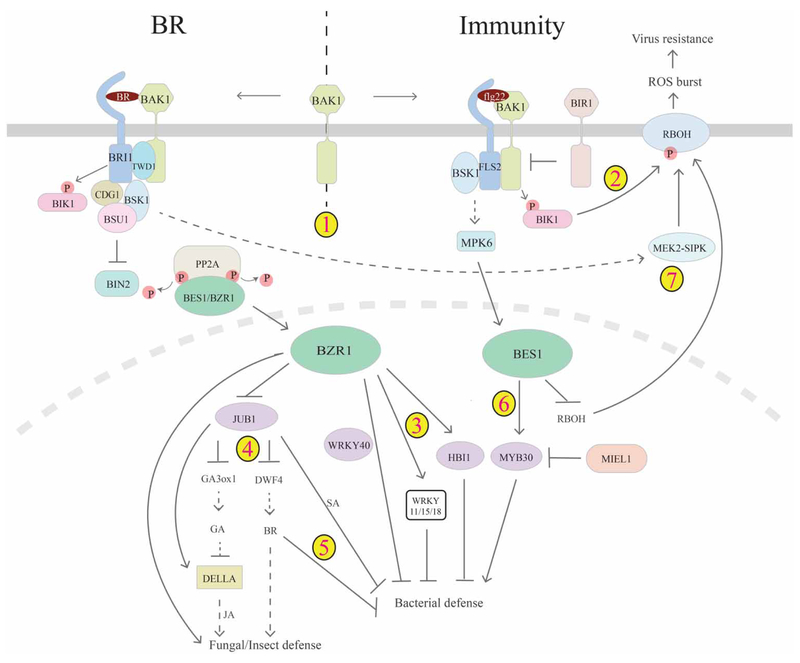
Figure 4. Cross-talk between BR and immunity.
There is multi-layer cross-talk between BR and plant immunity pathways. 1. Genetic studies indicated that BR and PAMP receptors may compete for common co-receptor BAK1, thus enabling BR repression of PTI. 2. Bacterial infection leads to phosphorylation and activation of BIK1, which activates RBOH and ROS burst to confer hypersensitive response. 3. In the nucleus, BR-activated BZR1 can inhibit PTI-mediated defense gene expression through WRKY11/15/18, HBI1 or in collaboration with WRKY40. 4. BZR1 can also function through JA and GA pathways (directly or through NAC transcription factor JUB1) to regulate fungal defense. 5. JUB1 can modulate BR and SA pathways to inhibit bacterial defense. 6. In contrast, BES1 is activated by the MAPK pathway and plays a positive role in bacterial defense, which involves MYB30, its E3 ligase MIEL1 and likely other regulators. 7. Recent studies also suggest that BR signals to activate MEK2–SIPK through BRI1 and BSK1, which in turn triggers RBOH to generate ROS to confer virus resistance. On the other branch, BES1 inhibits the expression of RBOH and thus virus resistance, mediating tradeoffs between BES1-promoted growth and virus resistance.
BAK1 has been considered as a possible candidate to mediate tradeoffs between BR-regulated plant growth and immunity due to its function as a co-receptor in both the BR and PTI pathways. In the BR pathway, BAK1 promotes growth by interacting with BRI1 to initiate BR signal transduction at the plasma membrane [14,15], and knockout of BAK1 and its homologs led to bri1-like BR-insensitive dwarf phenotypes [16]. BAK1 also functions as a co-receptor for several LRR–RLKs (FLS2, EFR and PEPR1) to perceive various PAMP signals (flg22, elf26 and AtPep1) [138–141] and promote PTI responses. In line with this idea, bak1 mutants showed reduced response to PAMPs, suggesting that BAK1 positively regulates PTI in Arabidopsis [138]. Additionally, bak1 mutants suppress the autoimmune phenotype of bir1, an RLK that functions as a negative regulator of plant immunity [142]. Thus, one possibility is that BR signaling and the immunity antagonize each other through competition for BAK1 in their receptor complexes.
Two independent groups reported that the BR pathway inhibits PTI (Belkhadir et al. [144] and Albrecht et al. [145]). However, these two studies had opposite conclusions regarding the role of BAK1 in this process. Belkhadir et al. reported that BAK1 was required for the antagonistic effect of BRs on PTI based on the following observations [143,144]. First, plants overexpressing the BR receptor BRI1 displayed compromised oxidative burst in response to flg22, elf19 and PGN treatment, but not to chitin, a component of the fungal cell wall that activates BAK1-independent defense response. The BRI1 suppression of PTI requires BAK1. Consistent with these observations, overexpression of BR biosynthetic gene DWF4 also showed compromised response to flg22 [143]. Moreover, expression of a hyperactive BRI allele, BRI1sud1, led to enhanced PTI response, likely due to the fact that activated BAK1 (from constitutive active BRI1sud1) could also activate PTI response.
Consistent with a negative role of BRs in PTI response, Albrecht et al. [145] reported that treating Arabidopsis leaves with BR inhibited FLS2-mediated disease resistance to Pseudomonas synringae pv. tomato DC3000 (Pst DC3000), with compromised flg22 or elf18-triggered ROS burst and PTI marker gene expression. BR signaling outputs including BES1 phosphorylation and BR marker gene expression were unaltered by flg22 treatment, suggesting the regulation between BR and PTI is unidirectional [145]. Therefore, both of these studies support negative regulation of PTI by the BR pathway. However, the results of Albrecht et al. suggest that BAK1 is not a rate-limiting factor that causes competition between BR and immune signaling. Co-treatment with BR and flg22 did not reduce the amount of FLS2 that associated with BAK1 [145], which is consistent with a model in which competition for BAK1 by BRI1 and FLS2 does not play a major role balancing growth and immunity. One possibility accounting for the opposite conclusions on the requirement of BAK1 in BR/PTI interaction might be the different approaches, treatments and mutants used in these studies.
Similar to the results regarding the role of BAK1 in immunity, application of BRs versus the use of BR mutants can lead to opposite conclusions about the effect of the BR pathway on plant immune responses. In Hordeum vulgare, BR treatment enhanced the plant tolerance to Fusarium Head Blight (FHB) disease caused by fungi Fusarium culmorum [146]. In Brassica napus, overexpression of AtDWF4 displayed increased tolerance to several fungal pathogens, confirming the results from the BR application experiments [147]. However, semi-dwarf ‘uzu’ barley mutant, which has a mutation (H857A) in the kinase domain of BRI1 and compromised BR signaling, displayed enhanced resistance to a broad range of viral and fungal pathogens, including F. culmorum [148]. The genetic studies indicate a negative role of BR signaling in fungal defense. Similarly, disruption of BRI1 in B. distachyon led to increased tolerance to necrotrophic and hemibiotrophic pathogens but not to biotrophic pathogens [149]. Taken together, these physiological and genetic studies suggest that BRs can play either a negative or positive role in biotic stress responses. The opposite results derived from plants exposed to the exogenous BR and from BR mutants suggest that many factors such as plant age, environment, BR concentrations applied (i.e. signaling strengths) and activation of additional pathways may determine the different outcomes.
Several RLCKs are involved in both BR signaling and immunity response (Figure 4). One RLCK member, BOTRYTIS-INDUCED KINASE 1 (BIK1), is phosphorylated by BAK1 upon flagellin perception and transphosphorylates FLS2/BAK1 via direct interaction to transduce the flagellin signal [150]. bik1 mutants displayed compromised resistance to Pst DC300 infection, indicating that BIK1 also positively regulates flg22-induced immunity [150]. BSK1, another RLCK member, is associated with BR receptor BRI1 upon BR activation and is phosphorylated by BRI1 to transduce the signal to downstream targets [33]. bsk1 knockout mutants display increased susceptibility to pathogens including Pst DC3000 with reduced levels of salicylic acid (SA) [151]. Furthermore, BSK1 directly interacts with FLS2 and is required for ROS burst, indicating a positive role of BSK1 in flg22-induced PTI [151,152]. In contrast with BSK1, BIK1 plays a negative role in BR signaling since bik1 mutants are hypersensitive to BRs, accumulate dephosphorylated-BES1 and have decreased expression of BR biosynthesis genes BR6OX, CPD and DWF4 [153]. BIK1 associates with BRI1 and is directly phosphorylated by BRI1, which is enhanced upon BL treatment [153]. In both BR signaling and FLS2 signaling, BIK1 dissociates from BRI1 and FLS2 receptors upon ligand perception. BAK1 is required for the dissociation of BIK1 with FLS2 in flg22-induced immunity but not in BR signaling [153]. It was further shown that BIK1 regulates flg22-triggered immunity via phosphorylation of the NADPH oxidase RBOHD, which activates ROS burst and controls stomatal movement [154,155].
Both BAK1 and BIK1 were reported to be negatively regulated by phosphatases, which could be alleviated by PAMP treatment [156,157]. PP2A associates with BAK1, negatively regulating BAK1’s activity [157], whereas PP2C38 negatively regulates the activity of BIK1 in immunity [156]. PP2C38 associates with BIK1 and directly dephosphorylates BIK1, leading to a reduction in PAMP-induced ROS production and stomatal immunity. Upon PAMP perception, PP2C38 is phosphorylated, likely by BIK1, which leads to dissociation of PP2C38 from BIK1 thus enabling BIK1 to activate ROS burst [156].
In summary, several points of cross-talk occur between PTI response and BR signaling at the receptor complexes and downstream RLCKs. Given the number of shared signaling components between BR and immunity, understanding how specificity is achieved between the two pathways is an active area of research. Indeed, recent work has suggested that BRI1 and FLS2 receptor complexes are spatially separated at the plasma membrane [158]. Along these lines, it would be interesting to determine why BSK1 and BIK1 both play positive roles in PTI but have positive and negative functions in BR signaling, respectively.
Interactions between BR signaling and immunity at transcription levels
It has been reported that the cross-talk between BR and PTI occurs downstream from BIN2, a central negative regulator in the BR signaling pathway (Figure 4). Flg22-triggered ROS burst was inhibited by BIN2 kinase inhibitors LiCl/bikinin treatment or in loss-of-function of BIN2 mutants [159]. One part of the pathways that this cross-talk occurs at is the downstream transcriptional regulators. The BR-regulated transcription factor BZR1 appears to suppress bacterial defense through several mechanisms. First, BZR1 activates the expression of several WRKY transcription factors, WRKY11, WRKY15 and WRKY18, which negatively control the immunity response [159]. Second, BZR1 interacts with WRKY40 to directly suppress genes required for PTI responses [159]. Finally, the bHLH transcription factor HBI1, which is required for BZR1–PIF4-mediated cell elongation and the activation of BR biosynthetic genes CPD, DWF4 and BR6OX1, inhibits the expression of PTI marker genes and is therefore proposed to mediate the tradeoff between plant growth and bacterial defense [160].
BZR1 is also implicated in fungal and insect defense, which likely involves the plant hormones jasmonic acid (JA) and GA (Figure 4). Gain-of-function bzr1-D mutants exhibited enhanced resistance against thrip feeding with elevated expression of JA-inducible VSP genes [161], indicating that BZR1 positively regulates insect defense, likely by activating JA signaling. In addition, as discussed above, BZR1 acts through NAC transcription factor JUB1 to increase the biosynthesis of GA/BR as well as the expression of DELLA genes, which probably act together to promote fungal defense [60]. Finally, overexpression of JUB1 led to enhanced susceptibility to Pst DC3000 [162]. The contribution of JUB1 to BR regulation of PTI remains to be defined and may represent an indirect mechanism for BZR1 to positively regulate PTI via JUB1. In rice, BRs can antagonize GA-mediated fungal defenses by stabilizing SLR1, an ortholog of Arabidopsis DELLA protein [163,164]; BRs can also suppress SA response to root oomycete Pythium graminicola inoculation [164]. It is proposed that P. graminicola uses BRs as a decoy to suppress SA signaling, operating downstream from SA biosynthesis but upstream of OsNPR1 and OsWRKY45, to achieve pathogenesis [164].
In contrast with BZR1, BES1 was reported to play a positive role in bacterial immunity and a negative role in fungal defense. Loss of function of bes1 mutants showed decreased resistance to Pst DC3000 and BES1 was identified as a direct substrate of MPK6 [165]. Mutation of the MPK6 phosphorylation sites in BES1 (BES1SSAA) led to impaired disease resistance, suggesting a positive role of BES1 in plant immunity downstream from the MAPK pathway [165]. BES1 has been found to negatively regulate the defense response to fungal pathogens as bes1-D gain-of-function mutants showed enhanced susceptibility to a necrotrophic fungus Alternaria brassicicola [166]. The BES1 target transcription factor MYB30 positively regulates the hypersensitive cell death program in plants in response to bacterial and fungal pathogens [61,167], likely mediating some of the function of BES1 in bacterial defense. MIEL1 interacts with and ubiquitinates MYB30, leading to MYB30 degradation, thus weakening MYB30-mediated hypersensitive cell death response [104]. Taken together, there is significant evidence that transcription factors involved in the BR pathway mediate cross-talk with immune responses. Given that BES1 and BZR1 function similarly in controlling BR-regulated growth, it will be interesting to further explore their seemingly contradictory functions in immune responses, which might lead to insights into the complex relationship between BR and immunity.
BR signaling and virus immunity
Early studies in Nicotiana benthamiana indicated that exogenous application of BR enhanced disease resistance to a broad range of pathogens, including virus (TMV), bacteria (Pst DC3000) and fungus (Oidium sp.) [168]. A recent study using the virus-induced gene silencing (VIGS) system revealed potential mechanisms of cross-talk between BR and virus resistance [169]. It was shown that foliar application of BL increased the tolerance of tobacco plants to TMV with accumulation of BR-induced MAPK and RBOHB (NADPH oxidase B) gene expression, which is accompanied by ROS burst and defense-related gene expression. The BR-induced virus tolerance was compromised in BRI1 and BSK1-silenced plants [169]. These results indicated that BRs function through BRI1 and BSK1 to activate MAPK cascade and ROS production to confer TMV tolerance (Figure 4). On the other hand, BR-activated BES1/BZR1 was shown to inhibit RBOHB gene expression, thereby reducing virus resistance and promoting plant growth [169]. The elevated expression of several defense-responsive genes was also observed in the overexpression transgenic line of wheat TaBRI1 in Arabidopsis, confirming the results from VIGS studies [169,170]. BRs thus have dual roles in virus defense and the final outcome is probably determined by the relative signaling strengths of the two branches as well as plant growth and environmental conditions (Figure 4).
Taken together, research into BR and immune cross-talk has shown that BRs and plant defense signaling pathways cross-talk at multiple levels in a complex network at the receptors/coreceptors, their immediate signaling intermediates as well as downstream transcription factors. The outcome of the cross-talk in terms of plant growth and immune response is probably determined by the sum of several interactions. One common feature is that different family members may have different functions (such as BSK1/BIK1 and BES1/BZR1) likely based on their substrates and/or targets.
Summary and future directions
In summary, there is accumulating evidence of cross-talk of BR with both drought and pathogen defense at multiple tiers of these complex signaling pathways. It seems that the role of BRs in drought stress depends on the environment, as well as on whether the BR pathway is manipulated via genetic means or by exogenous application. Similarly, the relationship between BRs and plant immunity may depend on the different pathogens, hosts and the systems used for studies (i.e. exogenously supplied hormone or mutants). The mechanisms controlling the regulation of plant immunity and drought stress by BRs likely operate through complex regulatory networks, including cross-talk with other hormonal pathways. Understanding how these networks function represents a significant challenge for the BR field that should be the focus of future research. Given that many factors involved in BR signaling have already been identified, further studies with system-level approaches are needed to define how the large number of BR signaling components function together and how they are modulated by other pathways, environments and in different developmental contexts. Establishing a global and integrated view of the BR pathway may help clarify the functions and mechanisms of BRs in regulating both the plant immunity and drought pathways, allowing for optimization of BR-regulated growth without compromising tolerance to these important stresses.
Acknowledgements
We apologize to colleagues for not being able to cite all related papers in the review due to space limitations.
Funding
The work in the Yin Laboratory is supported by grants from National Institute of Health [1R01GM120316–01A1], National Science Foundation [IOS-1257631] and the Plant Sciences Institute at Iowa State University.
Footnotes
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Li J, Nagpal P, Vitart V, McMorris TC and Chory J (1996) A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272, 398–401 doi:10.1126/science.272.5260.398 [DOI] [PubMed] [Google Scholar]
- 2.Szekeres M, Németh K, Koncz-Kálmán Z, Mathur J, Kauschmann A, Altmann T et al. (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85, 171–182 doi:10.1016/S0092-8674(00)81094-6 [DOI] [PubMed] [Google Scholar]
- 3.Noguchi T, Fujioka S, Takatsuto S, Sakurai A, Yoshida S, Li JM et al. (1999) Arabidopsis det2 is defective in the conversion of (24R)-24-methylcholest-4-En-3-One to (24R)-24-methyl-5α-cholestan-3-one in brassinosteroid biosynthesis. Plant Physiol. 120, 833–839 doi:10.1104/pp.120.3.833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clouse SD, Langford M and McMorris TC (1996) A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111, 671–678 doi:10.1104/pp.111.3.671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belkhadir Y and Jaillais Y (2015) The molecular circuitry of brassinosteroid signaling. New Phytol. 206, 522–540 doi:10.1111/nph.13269 [DOI] [PubMed] [Google Scholar]
- 6.Guo H, Li L, Aluru M, Aluru S and Yin Y (2013) Mechanisms and networks for brassinosteroid regulated gene expression. Curr. Opin. Plant Biol 16, 545–553 doi:10.1016/j.pbi.2013.08.002 [DOI] [PubMed] [Google Scholar]
- 7.Wang W, Bai M-Y and Wang Z-Y (2014) The brassinosteroid signaling network—a paradigm of signal integration. Curr. Opin. Plant Biol 21, 147–153 doi:10.1016/j.pbi.2014.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaillais Y and Vert G (2016) Brassinosteroid signaling and BRI1 dynamics went underground. Curr. Opin. Plant Biol 33, 92–100 doi:10.1016/j.pbi.2016.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vilarrasa-Blasi J, González-García M-P, Frigola D, Fábregas N, Alexiou KG, López-Bigas N et al. (2014) Regulation of plant stem cell quiescence by a brassinosteroid signaling module. Dev. Cell 30, 36–47 doi:10.1016/j.devcel.2014.05.020 [DOI] [PubMed] [Google Scholar]
- 10.Vragović K, Sela A, Friedlander-Shani L, Fridman Y, Hacham Y, Holland N et al. (2015) Translatome analyses capture of opposing tissue-specific brassinosteroid signals orchestrating root meristem differentiation. Proc. Natl Acad. Sci. U.S.A 112, 923–928 doi:10.1073/pnas.1417947112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lozano-Durán R and Zipfel C (2015) Trade-off between growth and immunity: role of brassinosteroids. Trends Plant Sci. 20, 12–19 doi:10.1016/j.tplants.2014.09.003 [DOI] [PubMed] [Google Scholar]
- 12.Caño-Delgado A, Yin Y, Yu C, Vafeados D, Mora-García S, Cheng J-C et al. (2004) BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 131, 5341–5351 doi:10.1242/dev.01403 [DOI] [PubMed] [Google Scholar]
- 13.Li J and Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938 doi:10.1016/S0092-8674(00)80357-8 [DOI] [PubMed] [Google Scholar]
- 14.Nam KH and Li J (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212 doi:10.1016/S0092-8674(02)00814-0 [DOI] [PubMed] [Google Scholar]
- 15.Li J, Wen J, Lease KA, Doke JT, Tax FE and Walker JC (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222 doi:10.1016/S0092-8674(02)00812-7 [DOI] [PubMed] [Google Scholar]
- 16.Gou X, Yin H, He K, Du J, Yi J, Xu S et al. (2012) Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet. 8, e1002452 doi:10.1371/journal.pgen.1002452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X and Chory J (2006) Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313, 1118–1122 doi:10.1126/science.1127593 [DOI] [PubMed] [Google Scholar]
- 18.Youn J-H and Kim T-W (2015) Functional insights of plant GSK3-like kinases: multi-taskers in diverse cellular signal transduction pathways. Mol. Plant 8, 552–565 doi:10.1016/j.molp.2014.12.006 [DOI] [PubMed] [Google Scholar]
- 19.Gampala SS, Kim T-W, He J-X, Tang W, Deng Z, Bai M-Y et al. (2007) An essential role for 14–3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 13, 177–189 doi:10.1016/j.devcel.2007.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryu H, Kim K, Cho H, Park J, Choe S and Hwang I (2007) Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19, 2749–2762 doi:10.1105/tpc.107.053728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vert G and Chory J (2006) Downstream nuclear events in brassinosteroid signalling. Nature 441, 96–100 doi:10.1038/nature04681 [DOI] [PubMed] [Google Scholar]
- 22.He J-X, Gendron JM, Yang Y, Li J and Wang Z-Y (2002) The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl Acad. Sci. U.S.A 99, 10185–10190 doi:10.1073/pnas.152342599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin Y, Wang Z-Y, Mora-Garcia S, Li J, Yoshida S, Asami T et al. (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109, 181–191 doi:10.1016/S0092-8674(02)00721-3 [DOI] [PubMed] [Google Scholar]
- 24.Wang H, Yang C, Zhang C, Wang N, Lu D, Wang J et al. (2011) Dual role of BKI1 and 14–3-3 s in brassinosteroid signaling to link receptor with transcription factors. Dev. Cell 21, 825–834 doi:10.1016/j.devcel.2011.08.018 [DOI] [PubMed] [Google Scholar]
- 25.Jaillais Y, Hothorn M, Belkhadir Y, Dabi T, Nimchuk ZL, Meyerowitz EM et al. (2011) Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev. 25, 232–237 doi:10.1101/gad.2001911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Goshe MB, Soderblom EJ, Phinney BS, Kuchar JA, Li J et al. (2005) Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17, 1685–1703 doi:10.1105/tpc.105.031393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Kota U, He K, Blackburn K, Li J, Goshe MB et al. (2008) Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev. Cell 15, 220–235 doi:10.1016/j.devcel.2008.06.011 [DOI] [PubMed] [Google Scholar]
- 28.Oh M-H, Wang X, Kota U, Goshe MB, Clouse SD and Huber SC (2009) Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. Proc. Natl Acad. Sci. U.S.A 106, 658–663 doi:10.1073/pnas.0810249106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao B, Lv M, Feng Z, Campbell T, Liscum E and Li J (2016) TWISTED DWARF 1 associates with BRASSINOSTEROID-INSENSITIVE 1 to regulate early events of the brassinosteroid signaling pathway. Mol. Plant 9, 582–592 doi:10.1016/j.molp.2016.01.007 [DOI] [PubMed] [Google Scholar]
- 30.Chaiwanon J, Garcia VJ, Cartwright H, Sun Y and Wang Z-Y (2016) Immunophilin-like FKBP42/TWISTED DWARF1 interacts with the receptor kinase BRI1 to regulate brassinosteroid signaling in Arabidopsis. Mol. Plant 9, 593–600 doi:10.1016/j.molp.2016.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim T-W, Guan S, Sun Y, Deng Z, Tang W, Shang J-X et al. (2009) Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol 11, 1254–1233 doi:10.1038/ncb1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim T-W, Guan S, Burlingame AL and Wang Z-Y (2011) The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol. Cell 43, 561–571 doi:10.1016/j.molcel.2011.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang W, Kim T-W, Oses-Prieto JA, Sun Y, Deng Z, Zhu S et al. (2008) BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321, 557–560 doi:10.1126/science.1156973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sreeramulu S, Mostizky Y, Sunitha S, Shani E, Nahum H, Salomon D et al. (2013) BSKs are partially redundant positive regulators of brassinosteroid signaling in Arabidopsis. Plant J. 74, 905–919 doi:10.1111/tpj.12175 [DOI] [PubMed] [Google Scholar]
- 35.Peng P, Yan Z, Zhu Y and Li J (2008) Regulation of the Arabidopsis GSK3-like kinase brassinosteroid-insensitive 2 through proteasome-mediated protein degradation. Mol. Plant 1, 338–346 doi:10.1093/mp/ssn001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu J-Y, Li Y, Cao D-M, Yang H, Oh E, Bi Y et al. (2017) The F-box protein KIB1 mediates brassinosteroid-induced inactivation and degradation of GSK3-like kinases in Arabidopsis. Mol. Cell 66, 648–657.e4 doi:10.1016/j.molcel.2017.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anne P, Azzopardi M, Gissot L, Beaubiat S, Hématy K and Palauqui J-C (2015) OCTOPUS negatively regulates BIN2 to control phloem differentiation in Arabidopsis thaliana. Curr. Biol 25, 2584–2590 doi:10.1016/j.cub.2015.08.033 [DOI] [PubMed] [Google Scholar]
- 38.Hao Y, Wang H, Qiao S, Leng L and Wang X (2016) Histone deacetylase HDA6 enhances brassinosteroid signaling by inhibiting the BIN2 kinase. Proc. Natl Acad. Sci. U.S.A 113, 10418–10423 doi:10.1073/pnas.1521363113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ling J-J, Li J, Zhu D and Deng XW (2017) Noncanonical role of Arabidopsis COP1/SPA complex in repressing BIN2-mediated PIF3 phosphorylation and degradation in darkness. Proc. Natl Acad. Sci. U.S.A 114, 3539–3544 doi:10.1073/pnas.1700850114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang W, Yuan M, Wang R, Yang Y, Wang C, Oses-Prieto JA et al. (2011) PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol 13, 124–131 doi:10.1038/ncb2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z-Y, Nakano T, Gendron J, He J, Chen M, Vafeados D et al. (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2, 505–513 doi:10.1016/S1534-5807(02)00153-3 [DOI] [PubMed] [Google Scholar]
- 42.Zhao J, Peng P, Schmitz RJ, Decker AD, Tax FE and Li JM (2002) Two putative BIN2 substrates are nuclear components of brassinosteroid signaling. Plant Physiol. 130, 1221–1229 doi:10.1104/pp.102.010918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T and Chory J (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120, 249–259 doi:10.1016/j.cell.2004.11.044 [DOI] [PubMed] [Google Scholar]
- 44.Yu X, Li L, Zola J, Aluru M, Ye H, Foudree A et al. (2011) A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 65, 634–646 doi:10.1111/j.1365-313X.2010.04449.x [DOI] [PubMed] [Google Scholar]
- 45.Sun Y, Fan X-Y, Cao D-M, Tang W, He K, Zhu J-Y et al. (2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19, 765–777 doi:10.1016/j.devcel.2010.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J (2010) Regulation of the nuclear activities of brassinosteroid signaling. Curr. Opin. Plant Biol 13, 540–547 doi:10.1016/j.pbi.2010.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Lucas M and Prat S (2014) PIFs get BRright: PHYTOCHROME INTERACTING FACTORs as integrators of light and hormonal signals. New Phytol. 202, 1126–1141 doi:10.1111/nph.12725 [DOI] [PubMed] [Google Scholar]
- 48.Oh E, Zhu J-Y and Wang Z-Y (2012) Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol 14, 802–809 doi:10.1038/ncb2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oh E, Zhu J-Y, Bai M-Y, Arenhart RA, Sun Y and Wang Z-Y (2014) Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 3, e03031 doi:10.7554/eLife.03031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Z-Y, Bai M-Y, Oh E and Zhu J-Y (2012) Brassinosteroid signaling network and regulation of photomorphogenesis. Ann. Rev. Genet 46, 701–724 doi:10.1146/annurev-genet-102209-163450 [DOI] [PubMed] [Google Scholar]
- 51.Zhang L-Y, Bai M-Y, Wu J, Zhu J-Y, Wang H, Zhang Z et al. (2009) Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21, 3767–3780 doi:10.1105/tpc.109.070441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang H, Zhu Y, Fujioka S, Asami T, Li J and Li J (2009) Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell 21, 3781–3791 doi:10.1105/tpc.109.072504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bai M-Y, Fan M, Oh E and Wang Z-Y (2012) A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 24, 4917–4929 doi:10.1105/tpc.112.105163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ikeda M, Fujiwara S, Mitsuda N and Ohme-Takagi M (2012) A triantagonistic basic helix-loop-helix system regulates cell elongation in Arabidopsis. Plant Cell 24, 4483–4497 doi:10.1105/tpc.112.105023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bai M-Y, Shang J-X, Oh E, Fan M, Bai Y, Zentella R et al. (2012) Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14, 810–817 doi:10.1038/ncb2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gallego-Bartolome J, Minguet EG, Grau-Enguix F, Abbas M, Locascio A, Thomas SG et al. (2012) Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl Acad. Sci. U.S.A 109, 13446–13451 doi:10.1073/pnas.1119992109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Q-F, Wang C, Jiang L, Li S, Sun SSM and He J-X (2012) An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci. Signal 5, ra72 doi:10.1126/scisignal.2002908 [DOI] [PubMed] [Google Scholar]
- 58.Unterholzner SJ, Rozhon W, Papacek M, Ciomas J, Lange T, Kugler KG et al. (2015) Brassinosteroids are master regulators of gibberellin biosynthesis in Arabidopsis. Plant Cell 27, 2261–2272 doi:10.1105/tpc.15.00433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tong H, Xiao Y, Liu D, Gao S, Liu L, Yin Y et al. (2014) Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell 26, 4376–4393 doi:10.1105/tpc.114.132092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shahnejat-Bushehri S, Tarkowska D, Sakuraba Y and Balazadeh S (2016) Arabidopsis NAC transcription factor JUB1 regulates GA/BR metabolism and signalling. Nat. Plants 2, 16013 doi:10.1038/nplants.2016.13 [DOI] [PubMed] [Google Scholar]
- 61.Li L, Yu X, Thompson A, Guo M, Yoshida S, Asami T et al. (2009) Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 58, 275–286 doi:10.1111/j.1365-313X.2008.03778.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Espinosa-Ruiz A, Martínez C, de Lucas M, Fàbregas N, Bosch N, Caño-Delgado AI et al. (2017) TOPLESS mediates brassinosteroid control of shoot boundaries and root meristem development in Arabidopsis thaliana. Development 144, 1619–1628 doi:10.1242/dev.143214 [DOI] [PubMed] [Google Scholar]
- 63.Wang X, Chen J, Xie Z, Liu S, Nolan T, Ye H et al. (2014) Histone lysine methyltransferase SDG8 is involved in brassinosteroid-regulated gene expression in Arabidopsis thaliana. Mol. Plant 7, 1303–1315 doi:10.1093/mp/ssu056 [DOI] [PubMed] [Google Scholar]
- 64.Li L, Ye H, Guo H and Yin Y (2010) Arabidopsis IWS1 interacts with transcription factor BES1 and is involved in plant steroid hormone brassinosteroid regulated gene expression. Proc. Natl Acad. Sci. U.S.A 107, 3918–3923 doi:10.1073/pnas.0909198107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu X, Li L, Li L, Guo M, Chory J and Yin Y (2008) Modulation of brassinosteroid-regulated gene expression by jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc. Natl Acad. Sci. U.S.A 105, 7618–7623 doi:10.1073/pnas.0802254105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nakashima K, Yamaguchi-Shinozaki K and Shinozaki K (2014) The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci 5, 170 doi:10.3389/fpls.2014.00170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoshida T, Mogami J and Yamaguchi-Shinozaki K (2014) ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol 21, 133–139 doi:10.1016/j.pbi.2014.07.009 [DOI] [PubMed] [Google Scholar]
- 68.Raghavendra AS, Gonugunta VK, Christmann A and Grill E (2010) ABA perception and signalling. Trends Plant Sci. 15, 395–401 doi:10.1016/j.tplants.2010.04.006 [DOI] [PubMed] [Google Scholar]
- 69.Espasandin FD, Maiale SJ, Calzadilla P, Ruiz OA and Sansberro PA (2014) Transcriptional regulation of 9-cis-epoxycarotenoid dioxygenase (NCED) gene by putrescine accumulation positively modulates ABA synthesis and drought tolerance in Lotus tenuis plants. Plant Physiol. Biochem 76, 29–35 doi:10.1016/j.plaphy.2013.12.018 [DOI] [PubMed] [Google Scholar]
- 70.Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T et al. (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 27, 325–333 doi:10.1046/j.1365-313x.2001.01096.x [DOI] [PubMed] [Google Scholar]
- 71.Cutler SR, Rodriguez PL, Finkelstein RR and Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Ann. Rev. Plant Biol 61, 651–679 doi:10.1146/annurev-arplant-042809-112122 [DOI] [PubMed] [Google Scholar]
- 72.Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A et al. (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068 doi:10.1126/science.1172408 [DOI] [PubMed] [Google Scholar]
- 73.Park S-Y, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071 doi:10.1126/science.1173041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nishimura N, Sarkeshik A, Nito K, Park S-Y, Wang A, Carvalho PC et al. (2010) PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 61, 290–299 doi:10.1111/j.1365-313X.2009.04054.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F et al. (2009) Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 60, 575–588 doi:10.1111/j.1365-313X.2009.03981.x [DOI] [PubMed] [Google Scholar]
- 76.Yoshida T, Mogami J and Yamaguchi-Shinozaki K (2015) Omics approaches toward defining the comprehensive abscisic acid signaling network in plants. Plant Cell Physiol. 56, 1043–1052 doi:10.1093/pcp/pcv060 [DOI] [PubMed] [Google Scholar]
- 77.Li J, Nam KH, Vafeados D and Chory J (2001) BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 127, 14–22 doi:10.1104/pp.127.1.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Steber CM and McCourt P (2001) A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 125, 763–769 doi:10.1104/pp.125.2.763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nemhauser JL, Hong F and Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126, 467–475 doi:10.1016/j.cell.2006.05.050 [DOI] [PubMed] [Google Scholar]
- 80.Zhang S, Cai Z and Wang X (2009) The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proc. Natl Acad. Sci. U.S.A 106, 4543–4548 doi:10.1073/pnas.0900349106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gui J, Zheng S, Liu C, Shen J, Li J and Li L (2016) OsREM4.1 interacts with OsSERK1 to coordinate the interlinking between abscisic acid and brassinosteroid signaling in rice. Dev. Cell 38, 201–213 doi:10.1016/j.devcel.2016.06.011 [DOI] [PubMed] [Google Scholar]
- 82.Clouse SD (2016) Brassinosteroid/abscisic acid antagonism in balancing growth and stress. Dev. Cell 38, 118–120 doi:10.1016/j.devcel.2016.07.005 [DOI] [PubMed] [Google Scholar]
- 83.Zong W, Tang N, Yang J, Peng L, Ma S, Xu Y et al. (2016) Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes. Plant Physiol. 171, 2810–2825 doi:10.1104/pp.16.00469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shang Y, Dai C, Lee MM, Kwak JM and Nam KH (2016) BRI1-associated receptor kinase 1 regulates guard cell ABA signaling mediated by open stomata 1 in Arabidopsis. Mol. Plant 9, 447–460 doi:10.1016/j.molp.2015.12.014 [DOI] [PubMed] [Google Scholar]
- 85.Acharya BR, Jeon BW, Zhang W and Assmann SM (2013) Open stomata 1 (OST1) is limiting in abscisic acid responses of Arabidopsis guard cells. New Phytol. 200, 1049–1063 doi:10.1111/nph.12469 [DOI] [PubMed] [Google Scholar]
- 86.Mustilli A-C, Merlot S, Vavasseur A, Fenzi F and Giraudat J (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14, 3089–3099 doi:10.1105/tpc.007906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shi C, Qi C, Ren H, Huang A, Hei S and She X (2015) Ethylene mediates brassinosteroid-induced stomatal closure via Gα protein-activated hydrogen peroxide and nitric oxide production in Arabidopsis. Plant J. 82, 280–301 doi:10.1111/tpj.12815 [DOI] [PubMed] [Google Scholar]
- 88.Haubrick LL, Torsethaugen G and Assmann SM (2006) Effect of brassinolide, alone and in concert with abscisic acid, on control of stomatal aperture and potassium currents of Vicia faba guard cell protoplasts. Physiol. Plantarum 128, 134–143 doi:10.1111/j.1399-3054.2006.00708.x [Google Scholar]
- 89.Xia X-J, Gao C-J, Song L-X, Zhou Y-H, Shi K and Yu J-Q (2014) Role of H2O2 dynamics in brassinosteroid-induced stomatal closure and opening in Solanum lycopersicum. Plant Cell Environ. 37, 2036–2050 doi:10.1111/pce.12275 [DOI] [PubMed] [Google Scholar]
- 90.Ha Y, Shang Y and Nam KH (2016) Brassinosteroids modulate ABA-induced stomatal closure in Arabidopsis. J. Exp. Bot 67, 6297–6308 doi:10.1093/jxb/erw385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ephritikhine G, Pagant S, Fujioka S, Takatsuto S, Lapous D, Caboche M et al. (1999) The sax1 mutation defines a new locus involved in the brassinosteroid biosynthesis pathway in Arabidopsis thaliana. Plant J. 18, 315–320 doi:10.1046/j.1365-313X.1999.00455.x [DOI] [PubMed] [Google Scholar]
- 92.Xue LW, Du JB, Yang H, Xu F, Yuan S and Lin HH (2009) Brassinosteroids counteract abscisic acid in germination and growth of Arabidopsis.Z. Naturforsch C 64, 225–230 PMID: [DOI] [PubMed] [Google Scholar]
- 93.Youn J-H and Kim T-W (2015) Functional insights of plant GSK3-like kinases: multi-taskers in diverse cellular signal transduction pathways. Mol. Plant 8, 552–565 doi:10.1016/j.molp.2014.12.006 [DOI] [PubMed] [Google Scholar]
- 94.Cai Z, Liu J, Wang H, Yang C, Chen Y, Li Y et al. (2014) GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc. Natl Acad. Sci. U.S.A 111, 9651–9656 doi:10.1073/pnas.1316717111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Finkelstein RR and Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–609 doi:10.1105/tpc.12.4.599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Skubacz A, Daszkowska-Golec A and Szarejko L (2016) The role and regulation of ABI5 (ABA-Insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci 7, 1884 doi:10.3389/fpls.2016.01884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hu Y and Yu D (2014) BRASSINOSTEROID INSENSITIVE 2 interacts with ABSCISIC ACID INSENSITIVE5 to mediate the antagonism of brassinosteroids to abscisic acid during seed germination in Arabidopsis. Plant Cell 26, 4394–4408 doi:10.1105/tpc.114.130849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Charrier B, Champion A, Henry Y and Kreis M (2002) Expression profiling of the whole Arabidopsis shaggy-like kinase multigene family by real-time reverse transcriptase-polymerase chain reaction. Plant Physiol. 130, 577–590 doi:10.1104/pp.009175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dal Santo S, Stampfl H, Krasensky J, Kempa S, Gibon Y, Petutschnig E et al. (2012) Stress-induced GSK3 regulates the redox stress response by phosphorylating glucose-6-phosphate dehydrogenase in Arabidopsis. Plant Cell 24, 3380–3392 doi:10.1105/tpc.112.101279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ryu H, Cho H, Bae W and Hwang I (2014) Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nat. Commun 5, 4138 doi:10.1038/ncomms5138 [DOI] [PubMed] [Google Scholar]
- 101.Yang X, Bai Y, Shang J, Xin R and Tang W (2016) The antagonistic regulation of abscisic acid-inhibited root growth by brassinosteroids is partially mediated via direct suppression of ABSCISIC ACID INSENSITIVE 5 expression by BRASSINAZOLE RESISTANT 1. Plant Cell Environ. 39, 1994–2003 doi:10.1111/pce.12763 [DOI] [PubMed] [Google Scholar]
- 102.Lee HG and Seo PJ (2016) The Arabidopsis MIEL1 E3 ligase negatively regulates ABA signalling by promoting protein turnover of MYB96. Nat. Commun 7, 12525 doi:10.1038/ncomms12525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zheng Y, Schumaker KS and Guo Y (2012) Sumoylation of transcription factor MYB30 by the small ubiquitin-like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana. Proc. Natl Acad. Sci. U.S.A 109, 12822–12827 doi:10.1073/pnas.1202630109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Marino D, Froidure S, Canonne J, Ben Khaled S, Khafif M, Pouzet C et al. (2013) Arabidopsis ubiquitin ligase MIEL1 mediates degradation of the transcription factor MYB30 weakening plant defence. Nat. Commun 4, 1476 doi:10.1038/ncomms2479 [DOI] [PubMed] [Google Scholar]
- 105.Seo PJ, Xiang F, Qiao M, Park J-Y, Lee YN, Kim S-G et al. (2009) The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 151, 275–289 doi:10.1104/pp.109.144220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Divi UK, Rahman T and Krishna P (2016) Gene expression and functional analyses in brassinosteroid-mediated stress tolerance. Plant Biotechnol. J 14, 419–432 doi:10.1111/pbi.12396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kagale S, Divi UK, Krochko JE, Keller WA and Krishna P (2007) Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta 225, 353–364 doi:10.1007/s00425-006-0361-6 [DOI] [PubMed] [Google Scholar]
- 108.Bajguz A and Hayat S (2009) Effects of brassinosteroids on the plant responses to environmental stresses. Plant Physiol. Biochem 47, 1–8 doi:10.1016/j.plaphy.2008.10.002 [DOI] [PubMed] [Google Scholar]
- 109.Yuan G-F, Jia C-G, Li Z, Sun B, Zhang L-P, Liu N et al. (2010) Effect of brassinosteroids on drought resistance and abscisic acid concentration in tomato under water stress. Sci. Hortic 126, 103–108 doi:10.1016/j.scienta.2010.06.014 [Google Scholar]
- 110.Anjum SA, Wang LC, Farooq M, Hussain M, Xue LL and Zou CM (2011) Brassinolide application improves the drought tolerance in maize through modulation of enzymatic antioxidants and leaf gas exchange. J. Agron. Crop Sci 197, 177–185 doi:10.1111/j.1439-037X.2010.00459.x [Google Scholar]
- 111.Zhou J, Wang J, Li X, Xia X-J, Zhou Y-H, Shi K et al. (2014) H2O2 mediates the crosstalk of brassinosteroid and abscisic acid in tomato responses to heat and oxidative stresses. J. Exp. Bot 65, 4371–4383 doi:10.1093/jxb/eru217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ye H, Liu S, Tang B, Chen J, Xie Z, Nolan TM et al. (2017) RD26 mediates crosstalk between drought and brassinosteroid signalling pathways. Nat. Commun 8, 14573 doi:10.1038/ncomms14573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nolan TM, Brennan B, Yang M, Chen J, Zhang M, Li Z et al. (2017) Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev. Cell 41, 33–46.e7 doi:10.1016/j.devcel.2017.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Northey JGB, Liang S, Jamshed M, Deb S, Foo E, Reid JB et al. (2016) Farnesylation mediates brassinosteroid biosynthesis to regulate abscisic acid responses. Nat. Plants 2, 16114 doi:10.1038/nplants.2016.114 [DOI] [PubMed] [Google Scholar]
- 115.Feng Y, Yin Y and Fei S (2015) Down-regulation of BdBRI1, a putative brassinosteroid receptor gene produces a dwarf phenotype with enhanced drought tolerance in Brachypodium distachyon. Plant Sci. 234, 163–173 doi:10.1016/j.plantsci.2015.02.015 [DOI] [PubMed] [Google Scholar]
- 116.Claeys H and Inze D (2013) The agony of choice: how plants balance growth and survival under water-limiting conditions. Plant Physiol. 162, 1768–1779 doi:10.1104/pp.113.220921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tran L-SP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K et al. (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498 doi:10.1105/tpc.104.022699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chung Y, Kwon SI and Choe S (2014) Antagonistic regulation of Arabidopsis growth by brassinosteroids and abiotic stresses. Mol. Cells 37, 795–803 doi:10.14348/molcells.2014.0127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M et al. (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39, 863–876 doi:10.1111/j.1365-313X.2004.02171.x [DOI] [PubMed] [Google Scholar]
- 120.Guan Q, Yue X, Zeng H and Zhu J (2014) The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress-responsive gene regulation and thermotolerance in Arabidopsis. Plant Cell 26, 438–453 doi:10.1105/tpc.113.118927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Song L, Huang S.-s.-C., Wise A, Castanon R, Nery JR, Chen H et al. (2016) A transcription factor hierarchy defines an environmental stress response network. Science 354, aag1550 doi:10.1126/science.aag1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Maruyama K, Takeda M, Kidokoro S, Yamada K, Sakuma Y, Urano K et al. (2009) Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol. 150, 1972–1980 doi:10.1104/pp.109.135327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Divi UK, Rahman T and Krishna P (2010) Brassinosteroid-mediated stress tolerance in Arabidopsis shows interactions with abscisic acid, ethylene and salicylic acid pathways. BMC Plant Biol. 10, 151 doi:10.1186/1471-2229-10-151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Michaeli S, Galili G, Genschik P, Fernie AR and Avin-Wittenberg T (2016) Autophagy in plants—what’s new on the menu? Trends Plant Sci. 21, 134–144 doi:10.1016/j.tplants.2015.10.008 [DOI] [PubMed] [Google Scholar]
- 125.Floyd BE, Morriss SC, MacIntosh GC and Bassham DC (2012) What to eat: evidence for selective autophagy in plants. J. Integr. Plant Biol 54, 907–920 doi:10.1111/j.1744-7909.2012.01178.x [DOI] [PubMed] [Google Scholar]
- 126.Kraft C, Peter M and Hofmann K (2010) Selective autophagy: ubiquitin-mediated recognition and beyond. Nat. Cell Biol 12, 836–841 doi:10.1038/ncb0910-836 [DOI] [PubMed] [Google Scholar]
- 127.Liu X-D, Yao J, Tripathi DN, Ding Z, Xu Y, Sun M et al. (2015) Autophagy mediates HIF2α degradation and suppresses renal tumorigenesis. Oncogene 34, 2450–2460 doi:10.1038/onc.2014.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gao C, Cao W, Bao L, Zuo W, Xie G, Cai T et al. (2010) Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat. Cell Biol 12, 781–790 doi:10.1038/ncb2082 [DOI] [PubMed] [Google Scholar]
- 129.Zhang Z, Zhu J-Y, Roh J, Marchive C, Kim S-K, Meyer C et al. (2016) TOR signaling promotes accumulation of BZR1 to balance growth with carbon availability in Arabidopsis. Curr. Biol 26, 1854–1860 doi:10.1016/j.cub.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang M, Li C, Cai Z, Hu Y, Nolan T, Yu F et al. (2017) SINAT e3 ligases control the light-mediated stability of the brassinosteroid-activated transcription factor BES1 in Arabidopsis. Dev. Cell 41, 47–58.e4 doi:10.1016/j.devcel.2017.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chen J, Nolan T, Ye H, Zhang M, Tong H, Xin P et al. (2017) Arabidopsis WRKY46, WRKY54 and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought response. Plant Cell 29, tpc.00364.02017 doi:10.1105/tpc.17.00364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li J, Brader G and Palva ET (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16, 319–331 doi:10.1105/tpc.016980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li J, Brader G, Kariola T and Palva ET (2006) WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 46, 477–491 doi:10.1111/j.1365-313X.2006.02712.x [DOI] [PubMed] [Google Scholar]
- 134.Hu Y, Dong Q and Yu D (2012) Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci. 185–186, 288–297 doi:10.1016/j.plantsci.2011.12.003 [DOI] [PubMed] [Google Scholar]
- 135.Ding ZJ, Yan JY, Xu XY, Yu DQ, Li GX, Zhang SQ et al. (2014) Transcription factor WRKY46 regulates osmotic stress responses and stomatal movement independently in Arabidopsis. Plant J. 79, 13–27 doi:10.1111/tpj.12538 [DOI] [PubMed] [Google Scholar]
- 136.Li J, Besseau S, Törönen P, Sipari N, Kollist H, Holm L et al. (2013) Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol. 200, 457–472 doi:10.1111/nph.12378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gómez-Gómez L and Boller T (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011 doi:10.1016/S1097-2765(00)80265-8 [DOI] [PubMed] [Google Scholar]
- 138.Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JDG et al. (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500 doi:10.1038/nature05999 [DOI] [PubMed] [Google Scholar]
- 139.Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JDG, Boller T et al. (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125, 749–760 doi:10.1016/j.cell.2006.03.037 [DOI] [PubMed] [Google Scholar]
- 140.Yamaguchi Y, Pearce G and Ryan CA (2006) The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc. Natl Acad. Sci. U.S.A 103, 10104–10109 doi:10.1073/pnas.0603729103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Schulze B, Mentzel T, Jehle AK, Mueller K, Beeler S, Boller T et al. (2010) Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem 285, 9444–9451 doi:10.1074/jbc.M109.096842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liu Y, Huang X, Li M, He P and Zhang Y (2016) Loss-of-function of Arabidopsis receptor-like kinase BIR1 activates cell death and defense responses mediated by BAK1 and SOBIR1. New Phytol. 212, 637–645 doi:10.1111/nph.14072 [DOI] [PubMed] [Google Scholar]
- 143.Belkhadir Y, Jaillais Y, Epple P, Balsemao-Pires E, Dangl JL and Chory J (2012) Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc. Natl Acad. Sci. U.S.A 109, 297–302 doi:10.1073/pnas.1112840108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Belkhadir Y, Yang L, Hetzel J, Dangl JL and Chory J (2014) The growth-defense pivot: crisis management in plants mediated by LRR-RK surface receptors. Trends Biochem. Sci 39, 447–456 doi:10.1016/j.tibs.2014.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Albrecht C, Boutrot F, Segonzac C, Schwessinger B, Gimenez-Ibanez S, Chinchilla D et al. (2012) Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc. Natl Acad. Sci. U.S.A 109, 303–308 doi:10.1073/pnas.1109921108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ali SS, Kumar GBS, Khan M and Doohan FM (2013) Brassinosteroid enhances resistance to fusarium diseases of barley. Phytopathology 103, 1260–1267 doi:10.1094/PHYTO-05-13-0111-R [DOI] [PubMed] [Google Scholar]
- 147.Sahni S, Prasad BD, Liu Q, Grbic V, Sharpe A, Singh SP et al. (2016) Overexpression of the brassinosteroid biosynthetic gene DWF4 in Brassica napus simultaneously increases seed yield and stress tolerance. Sci. Rep 6, 28298 doi:10.1038/srep28298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ali SS, Gunupuru LR, Kumar GBS, Khan M, Scofield S, Nicholson P et al. (2014) Plant disease resistance is augmented in uzu barley lines modified in the brassinosteroid receptor BRI1. BMC Plant Biol. 14, 227 doi:10.1186/s12870-014-0227-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Goddard R, Peraldi A, Ridout C and Nicholson P (2014) Enhanced disease resistance caused by BRI1 mutation is conserved between Brachypodium distachyon and barley (Hordeum vulgare). Mol. Plant Microbe Interact 27, 1095–1106 doi:10.1094/MPMI-03-14-0069-R [DOI] [PubMed] [Google Scholar]
- 150.Lu D, Wu S, Gao X, Zhang Y, Shan L and He P (2010) A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl Acad. Sci. U.S.A 107, 496–501 doi:10.1073/pnas.0909705107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shi H, Shen Q, Qi Y, Yan H, Nie H, Chen Y et al. (2013) BR-SIGNALING KINASE 1 physically associates with FLAGELLIN SENSING 2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25, 1143–1157 doi:10.1105/tpc.112.107904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Shi H, Yan H, Li J and Tang D (2013) BSK1, a receptor-like cytoplasmic kinase, involved in both BR signaling and innate immunity in Arabidopsis. Plant Signal. Behav 8, e24996 doi:10.4161/psb.24996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lin W, Lu D, Gao X, Jiang S, Ma X, Wang Z et al. (2013) Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor-like cytoplasmic kinase BIK1. Proc. Natl Acad. Sci. U.S.A 110, 12114–12119 doi:10.1073/pnas.1302154110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Li L, Li M, Yu L, Zhou Z, Liang X, Liu Z et al. (2014) The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15, 329–338 doi:10.1016/j.chom.2014.02.009 [DOI] [PubMed] [Google Scholar]
- 155.Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V et al. (2014) Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 54, 43–55 doi:10.1016/j.molcel.2014.02.021 [DOI] [PubMed] [Google Scholar]
- 156.Couto D, Niebergall R, Liang X, Bücherl CA, Sklenar J, Macho AP et al. (2016) The Arabidopsis protein phosphatase PP2C38 negatively regulates the central immune kinase BIK1. PLoS Pathog. 12, e1005811 doi:10.1371/journal.ppat.1005811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Segonzac C, Macho AP, Sanmartin M, Ntoukakis V, Sanchez-Serrano JJ and Zipfel C (2014) Negative control of BAK1 by protein phosphatase 2A during plant innate immunity. EMBO J. 33, 2069–2079 doi:10.15252/embj.201488698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bücherl CA, Jarsch IK, Schudoma C, Segonzac C, Mbengue M, Robatzek S et al. (2017) Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. eLife 6, e25114 doi:10.7554/eLife.25114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lozano-Durán R, Macho AP, Boutrot F, Segonzac C, Somssich IE and Zipfel C (2013) The transcriptional regulator BZR1 mediates trade-off between plant innate immunity and growth. eLife 2, e00983 doi:10.7554/eLife.00983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fan M, Bai M-Y, Kim J-G, Wang T, Oh E, Chen L et al. (2014) The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern-triggered immunity in Arabidopsis. Plant Cell 26, 828–841 doi:10.1105/tpc.113.121111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Miyaji T, Yamagami A, Kume N, Sakuta M, Osada H, Asami T et al. (2014) Brassinosteroid-related transcription factor BIL1/BZR1 increases plant resistance to insect feeding. Biosci. Biotechnol. Biochem 78, 960–968 doi:10.1080/09168451.2014.910093 [DOI] [PubMed] [Google Scholar]
- 162.Shahnejat-Bushehri S, Nobmann B, Devi Allu A and Balazadeh S (2016) JUB1 suppresses Pseudomonas syringae-induced defense responses through accumulation of DELLA proteins. Plant Signal. Behav 11, e1181245 doi:10.1080/15592324.2016.1181245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Navarro L, Bari R, Achard P, Lisón P, Nemri A, Harberd NP et al. (2008) DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr. Biol 18, 650–655 doi:10.1016/j.cub.2008.03.060 [DOI] [PubMed] [Google Scholar]
- 164.De Vleesschauwer D, Van Buyten E, Satoh K, Balidion J, Mauleon R, Choi I-R et al. (2012) Brassinosteroids antagonize gibberellin- and salicylate-mediated root immunity in rice. Plant Physiol. 158, 1833–1846 doi:10.1104/pp.112.193672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kang S, Yang F, Li L, Chen H, Chen S and Zhang J (2015) The Arabidopsis transcription factor BRASSINOSTEROID INSENSITIVE 1-ETHYLMETHANESULFONATE-SUPPRESSOR 1 is a direct substrate of MITOGEN-ACTIVATED PROTEIN KINASE 6 and regulates immunity. Plant Physiol. 167, 1076–1086 doi:10.1104/pp.114.250985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Shin SY, Chung H, Kim SY and Nam KH (2016) BRI1-EMS-suppressor 1 gain-of-function mutant shows higher susceptibility to necrotrophic fungal infection. Biochem. Biophys. Res. Commun 470, 864–869 doi:10.1016/j.bbrc.2016.01.128 [DOI] [PubMed] [Google Scholar]
- 167.Vailleau F, Daniel X, Tronchet M, Montillet J-L, Triantaphylidès C and Roby D (2002) A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc. Natl Acad. Sci. U.S.A 99, 10179–10184 doi:10.1073/pnas.152047199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Nakashita H, Nitta T, Asami T, Fujioka S, Arai Y, Sekimata K et al. (2003) Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J. 33, 887–898 doi:10.1046/j.1365-313X.2003.01675.x [DOI] [PubMed] [Google Scholar]
- 169.Deng X-G, Zhu T, Peng X-J, Xi D-H, Guo H, Yin Y et al. (2016) Role of brassinosteroid signaling in modulating Tobacco mosaic virus resistance in Nicotiana benthamiana. Sci. Rep 6, 20579 doi:10.1038/srep20579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Deng X-G, Zhu T, Zou L-J, Han X-Y, Zhou X, Xi D-H et al. (2016) Orchestration of hydrogen peroxide and nitric oxide in brassinosteroid-mediated systemic virus resistance in Nicotiana benthamiana. Plant J. 85, 478–493 doi:10.1111/tpj.13120 [DOI] [PubMed] [Google Scholar]
- 171.Bailey TL (2011) DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27, 1653–1659 doi:10.1093/bioinformatics/btr261 [DOI] [PMC free article] [PubMed] [Google Scholar]