Abstract
The cell plasma membrane serves as a nexus integrating extra- and intracellular components, which together enable many of the fundamental cellular signaling processes that sustain life. In order to perform this key function, plasma membrane components assemble into well-defined domains exhibiting distinct biochemical and biophysical properties that modulate various signaling events. Dysregulation of these highly dynamic membrane domains can promote oncogenic signaling. Recently, it has been demonstrated that select membrane-targeted dietary bioactives (MTDBs) have the ability to remodel plasma membrane domains and subsequently reduce cancer risk. In this review, we focus on the importance of plasma membrane domain structural and signaling functionalities as well as how loss of membrane homeostasis can drive aberrant signaling. Additionally, we discuss the intricacies associated with the investigation of these membrane domain features and their associations with cancer biology. Lastly, we describe the current literature focusing on MTDBs, including mechanisms of chemoprevention and therapeutics in order to establish a functional link between these membrane-altering biomolecules, tuning of plasma membrane hierarchal organization, and their implications in cancer prevention.
Keywords: Ras, Wnt, n-3 PUFA, Membrane order, Nanoclustering, Membrane therapy, Cancer prevention
1. Introduction
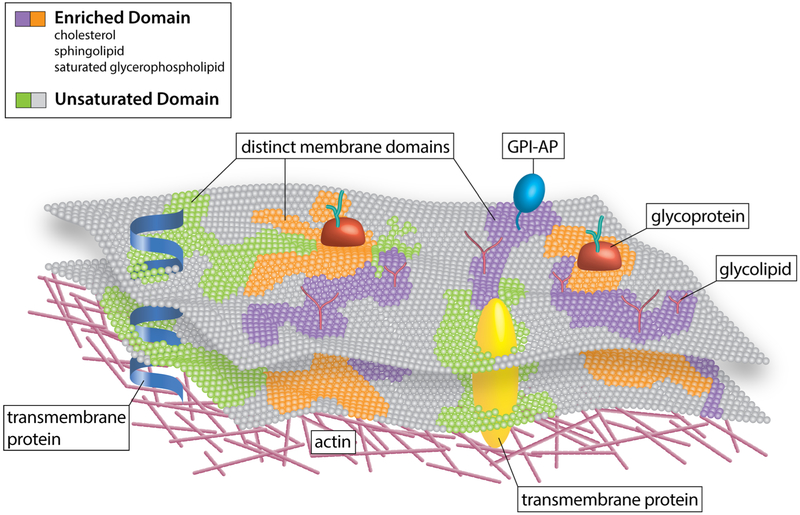
The plasma membrane of cells acts as a barrier that controls the bidirectional movement of a plethora of molecules between the extracellular and cytosolic space of cells. This fundamental property affords protection to cells from their surroundings. In addition, the highly dynamic fluid-mosaic structure of the plasma membrane serves as a point of the first contact between cells and various signaling elements that allow cells to “talk to each other.” Moreover, the plasma membrane is responsible for the compartmentalization of proteins, lipids, and their chemically modified counterparts into distinct assemblies that perform many of the fundamental cellular activities required for life (Fig. 1).
Fig. 1.
The compartmentalization of lipid-protein assemblies into specialized domains in the plasma membrane. Different lipids and proteins organized in an orchestrated manner to form distinct membrane domains, thus creating lateral heterogeneity in the plasma membrane. Moreover, plasma membrane heterogeneity is displayed across the lipid bilayer vertical axis, resulting in highly organized domains both in the plasma membrane outer and inner leaflet. The latter are not disconnected from one another rather they are coupled, bringing about bilayer crosstalk, which is essential for cellular signaling. These domains display distinct biochemical compositions and biophysical features. Some of these domains are enriched in cholesterol, sphingolipids, glycolipids, and saturated glycerophospholipids as well as lipidated and glycosylphosphatidylinositol (GPI)-anchored proteins (GPI-APs), thus generating lipid raft or raft-like domains. In contrast, others domains, termed non-rafts, are enriched in polyunsaturated and branched lipids. Additionally, formation and organization of these membrane domains are believed to involve cortical actin. Finally, these diverse proteolipid domains, exhibiting varying biochemical and biophysical features, exclude one another, yet they can coexist in the plasma membrane, thus creating a multifunctional highly complex and dynamic signaling platform
Highly relevant to the cancer biology field, it is now recognized that the geometry, biochemical composition, and biophysical characteristics of the plasma membrane are tightly associated with its signal processing capability [1–4]. According to this emerging picture, protein and lipid assemblies can be organized to form distinct micro- or nanodomains that facilitate signaling events [5–9]. The formation, organization, and membrane biophysical status, e.g., the degree of membrane rigidity, of these specialized signaling domains are modulated by cortical actin and the presence of specific lipids and proteins (Fig. 1) [10]. Currently, these domains are considered a predominant feature of the plasma membrane and appear to mediate critical signaling processes [11, 12], including the stabilization of β-catenin via stimulation of Wnt pathway-associated receptors [13–15], epidermal growth factor receptor (EGFR) signaling [16–18], and the activation of MAPK/ERK pathway components through membrane-bound Ras proteins [19–21], to name a few examples (Table 1). Interestingly, dysregulation of plasma membrane homeostasis, in part, due to the products of gene mutations as well as changes in protein and lipid localization, alters the degree of clustering and other biochemical and biophysical trademarks, thereby providing a suitable environment for the initiation of cancer-related signaling processes [22–24].
Table 1.
Nanoscale proteolipid clusters relevant to cancer biology
| Main protein | Lipids | Effectors | Size (~) | Technique | Function | References (PMID) |
|---|---|---|---|---|---|---|
| EGFR | PA, PIP2 | Grb2, PI3K, SOS1 | 20–30 nm, 200 NL, 300 LC | EM, dSTORM | Proliferation, migration | 20516214, 25001389 |
| KRas-GTP | PA, PIP3, PS | PI3K, Raf | 6 nm | EM, FLIM-FRET | Proliferation, survival | 25234412, 19115142 |
| HRas-GDP | PIP2, PIP3, PI3P, PI4P, PS, Chol | 11 nm | EM, FLIM-FRET | 25234412, 19115142 | ||
| HRas-GTP | PIP3, PI4P, PS | PI3K, Raf | 6 nm | EM, FLIM-FRET | Proliferation, survival, migration | 25234412, 19115142 |
| Rac1 | PIP2, PIP3 | Tiam1, WAVE | 200 nm | STORM, TIRF SPT-PALM | Cytoskeleton, migration | 29141223 |
| Frizzled | chol | Dvl, LRP5/6 | β-Catenin, stemness | 25024088, 28377511, 15459103, 19619488 | ||
| Lrp5/6 | PIP2 | Axin, Ck1, Frizzled, GSK3 | 100 nm, +WNT 200 nm | STORM | β-Catenin, stemness | 23400998, 19619488 |
| Lypd6 | LRP5/6 | 100 nm | STED | β-Catenin, stemness | 28404749 | |
| Dvl | PA, PIP2, PS, Chol | Axin, Frizzled | > 500 nm | EM, FRAP, FLIP, TIRF | β-Catenin, stemness | 25024088, 11101902, 24606934, 19234454, 17529994, 16263762, 19619488 |
Currently, there is evidence indicating that cell membranes can be “targeted” by pleiotropic dietary bioactives (MTDBs), resulting in the remodeling of plasma membrane domains [25–27]. This suggests that select MTDBs could reduce cancer risk by attenuating oncogenic protein activity by modulating the membrane organization of essential proteins and lipids. Thus, MTDBs could potentially serve as chemoprotective agents for cancer therapy [28–30]. In this review, we describe the importance of the plasma membrane as a central cellular hub that integrates numerous cues to orchestrate the modulation of various signaling networks, which can become hyperactivated, resulting in tumorigenesis. In addition, we discuss currently available tools utilized to investigate these complexes and, sometimes, subtle processes. Lastly, we review the current literature focusing on dietary components that modulate membrane structure, including mechanisms of chemoprevention and therapeutics.
2. Plasma membrane micro- and nanodomain structure and organization
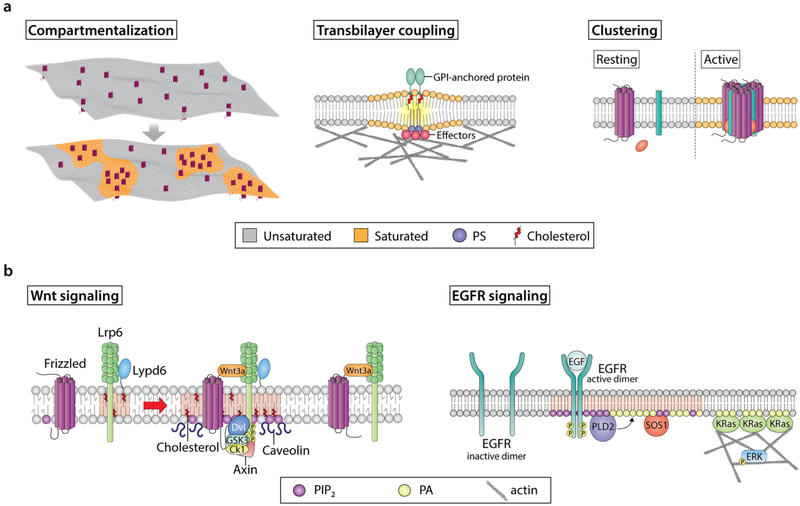
The plasma membrane is composed of a heterogeneous mixture of lipids, proteins, and carbohydrates, whose distinct organization is essential for its function [31, 32]. Lipids self-assemble into lipid bilayers, driven in great part by hydrophobic forces. Plasma membrane lipids can diffuse laterally via different modes and undergo phase separations to form non-homogenous nanoscopic domains (Fig. 2a, compartmentalization). Initial evidence supporting the concept of plasma membrane domain heterogeneity arose from observations that biological membranes can be separated into detergent-resistant and detergent-labile fractions [33, 34]. The presence, composition, and dynamics of these distinct plasma membrane compartments have been studied in great detail ever since. Several findings have pointed to the existence of ordered or rigid (Lo) and disordered or fluid (Ld) phases in the plasma membrane [35–38]. It is widely accepted that the Lo phase, a highly condensed/ordered domain, is enriched in cholesterol and predominantly saturated sphingolipids, while the Ld phase, a relatively disordered domain, is enriched in unsaturated glycerophospholipids [39, 40]. From the existing body of evidence associated with these domains, the plasma membrane lipid raft model emerged [41, 42].
Fig. 2.
Modulation of lipid and protein organization in plasma membrane domains and their functions. a Examples of various membrane domain features. Highly dynamic interactions between lipid and protein molecules shape many of the features display by specialized plasma membrane domains. For example, the preferential interactions between specific proteins and cholesterol, sphingolipids, and, in multiple cases, charged signaling lipids can induce precise spatial compartmentalization of key membrane components, thus creating molecularly well-defined domains. This, in turn, regulates cellular signaling by mediating the recruitment of specific signaling effectors at an exact location and time. Another fundamental feature of membrane domains is proteolipid clustering. In many cases, assembly of clusters requires a stimulus to initiate the movement of cluster forming molecules between different membrane domains, resulting in the activation and oligomerization of these effectors. Plasma membrane clusters contain a variety of protein functionalities that can originate from both the plasma membrane and cytosol. Clustering of multiple functionalities at the membrane modulates high specificity and low membrane molecule diffusion, which in turn enhances signaling robustness. Lastly, proteins and lipids (e.g., cholesterol, GPI-anchored proteins, PS) are organized in plasma membrane domains within a bilayer with distinctive outer exoplasmic and inner cytoplasmic leaflets. These leaflets differ in terms of their lipid and protein compositions. Accordingly, specific membrane domains can induce the formation of proteolipid assemblies in the opposing leaflet. Inner leaflet effector organization is regulated by complex interactions between actin, lipids, and other proteins. These inner leaflet proteolipid assemblies can influence other effectors located in the outer leaflet and engage in transbilayer coupling. This is important, because transbilayer coupling is a mechanism by which membrane domain components are brought together at the two sides of the plasma membrane to efficiently signal. b Examples of the role of membrane domains in signaling events associated with cancer. Wnt signaling receptors, i.e., LRP6 and Fz, localize in both raft and non-raft domains. In the presence of Wnt, cholesterol bilayer asymmetry is triggered, which leads to enrichment of cholesterol in the inner leaflet. Moreover, Wnt-bound Fz-LRP6 complexes preferably localize to cholesterol-enriched membrane domains containing caveolin. This, in turn, leads to the recruitment of the Wnt signaling effector Dvl. The ability of Dvl to oligomerize promotes Fz and LRP6 clustering and recruitment of Axin, leading to LRP6 phosphorylation by GSK3 and CK1 in lipid rafts. Simultaneously, lipid kinases (e.g., PI4KII and PIP5KI, not shown) are recruited to these sites and promote production of PIP2, which in turn promotes LRP6 and Fz clustering and phosphorylation. Importantly, although Wnt-bound LRP6-Fz complexes can localize to non-raft domains, Lypd6, a GPI-anchored protein that localizes specifically in lipid rafts, ensures that LRP6 phosphorylation and thus receptor activation and efficient signaling occur in lipid raft domains. EGFR signaling is initiated from highly organized nanoscale proteolipid domains, driven by the spatiotemporal production of specific lipids. For example, EGFR activation by EGF initiates the formation of lipid domains, which are enriched in PIP2 and PA. The PA generated by PLD2 acts as a beacon to recruit KRas and SOS1, and along with PIP2, acts as a cofactor for SOS1-mediated activation of KRas. Furthermore, this pool of PA stabilizes the actin cytoskeleton, of which KRas nanoclustering is dependent. Together, these steps contribute to efficient ERK activation and downstream signaling
The current consensus describes lipid rafts as heterogeneous, highly dynamic domains that range from 10 to 200 nm in size [43]. These nanodomains are enriched in cholesterol, sphingolipids, glycolipids, and saturated glycerophospholipids as well as lipidated and glycosylphosphatidylinositol (GPI)-anchored proteins and have the ability to form higher order microdomains (> 300 nm) under specific conditions [44, 45]. The process that drives formation of the latter has been postulated to be dependent on selective lipid-lipid and protein-lipid interactions or clustering. Interestingly, although lipid rafts and raft-like domains appear small in size, they are believed to constitute a relatively large fraction of the plasma membrane, if not its majority [34, 44]. Importantly, lipid domain organization is cell type specific and variations between distinct cell types can be observed [44, 46]. The latter is driven by complex interactions between these membrane domains and the cytoskeleton as well as the relative concentrations of bioactive lipids in these domains. Plasma membrane ordered domains appear to have diverse lipid and protein compositions. Various different lipids and proteins have been shown to coalesce into specialized nanoscale proteolipid clusters (Table 1). Consequently, it has been proposed that the plasma membrane is not organized in a binary fashion, but instead contains numerous lipid raft and raft-like domains as well as non-raft domains displaying discrete compositions and features [34]. As an example, caveolin and monosialotetrahexosylganglioside (GM1), both which are considered components of rafts, are not always colocalized [47].
Highly organized domains are found both in the outer and inner leaflets of the plasma membrane (Fig. 1). In that respect, the plasma membrane is also heterogeneous across the lipid bilayer vertical axis. However, lipid raft and raft-like domains are not disconnected from one another but rather it has been postulated that they are coupled in these asymmetric lipid bilayers (Fig. 2a, transbilayer coupling) [48–50]. It has been previously shown that inner leaflet ordered domains can be induced by outer leaflet ordered domains in a reconstituted system. Additionally, experimental and simulation data have provided evidence that clustering of GPI-anchored proteins in the outer leaflet is affected by transbilayer coupling of phosphatidylserine (PS) in the inner leaflet containing a saturated fatty acyl chain [12]. Furthermore, there is evidence indicating that some raft components, e.g., cholesterol, which is found in different concentrations across lipid leaflets, can be modulated in a stimulus-dependent manner and support signal transduction in ordered domains [51].
Due to the great complexity of plasma membrane ordered domains, various lipid models and tools have been employed to study the factors affecting their formation and dynamics. Interestingly, due in great part to the controversies surrounding this field, the toolbox utilized to investigate these membrane domains has been consistently evolving since the initial lipid raft studies in the 1970s. As mentioned above, the first evidence of the existence of distinct domains in the plasma membrane arose from differential solubilization of these biological structures using non-ionic detergents [33]. This allowed scientists to fractionate membranes by density centrifugation, revealing the presence of distinctive domains exhibiting different lipid-protein compositions. However, due to the high variation in the recovery of membrane components, which was partly influenced by differences in detergent types, concentration, and temperature employed during the assay, other more robust and reliable techniques have been the preferred methods of choice recently. For example, researchers studying these membrane domains have recently utilized super-resolution imaging technologies which provide improved size and temporal resolution. Experiments performed utilizing stochastic optical reconstruction microscopy (STORM), photo-activated localization microscopy (PALM), total internal reflection fluorescence microscopy (TIRFM), stimulated emission depletion (STED), immunoelectron microscopy (immuno-EM), and the combination of fluorescence lifetime imaging microscopy and Förster resonance energy transfer (FLIM-FRET) have revealed nanoscale details regarding plasma membrane morphology as well as lipid and protein composition and clustering [52–56]. Additionally, fluorescence recovery after photobleaching (FRAP) has provided evidence of proteins clustered in distinct noncholesterol-dependent signaling microdomains on the inner plasma membrane surface [56, 57]. Other techniques such as single-particle tracking (SPT) and fluorescence correlation spectroscopy (FCS) have evaluated the diffusion of membrane molecules over various length scales as well as other dynamic parameters [58–61]. Recent advances in high-resolution chemical-based imaging approaches such a nanoscale ion mass spectrometry (NanoSIMS) offer lipid resolution at the ~ 100-nm level [62–64]. Furthermore, time of flight secondary ion mass spectrometry (TOF-SIMS), which is based on event by event impacts of single clusters of particles, has achieved a resolution of ~ 10 nm [65–69]. These are very promising techniques that enhance the ability to detect co-localization of raft lipids in preserved human tissues [70]. Finally, electron microscopy (EM), owing to its high resolution, has allowed the study of the arrangement of various domain components in the outer and inner leaflet of the membrane bilayer [71, 72]. Altogether, these techniques have greatly advanced the study of plasma membrane domain organization.
Many imaging techniques rely primarily on membrane probes that can selectively distinguish/reveal distinct plasma membrane domains by exhibiting discrete emission spectra when localized in domains with different physicochemical properties, interacting with specific resident components or localizing within particular domains or by disrupting domain structure and function. Reporters that can selectively locate within rafts or bind to raft components include cholesterol-binding molecules such as filipin and the protein domain 4 of perfringolysin O; prototypical ganglioside-binding molecules such as cholera toxin B; sphingolipid reporters such as ostreolysin A, lysenin, and pleurotolysin; and raft and non-raft markers such as truncated Ras motifs and cyanine dyes (e.g., DiO, DiI, and DiD) [56, 73–79]. Most of these probes are detected utilizing fluorescence techniques. This raises some concerns since these probes are typically labeled with chromophores that are often close to the size of lipid molecules, which themselves could alter the natural state of the lipid environment under investigation. Fluorescent probe-free techniques such as mass spectrometry, Raman spectroscopy, and, as already mentioned, EM can be used to circumvent some of these issues. However, these techniques require cell fixation and longer acquisition times as well as a high degree of expertise and sophisticated equipment [73, 80, 81]. A different set of fluorescent ratiometric probes, which are sensitive to differences in physicochemical membrane properties and can distinguish between a spectrum of tightly packed raft and relatively fluid non-raft domains, include Laurdan and ANEP (e.g., di-4-ANEPPS, di-4-ANEPPDHQ, and di-8-ANEPPS) dyes. These dyes provide a fast, concentration-independent evaluation of the plasma membrane lipid environment [82–85]. On the other hand, these dyes suffer from varying rates of cell internalization accompanied by undesired staining of intracellular membranes [86]. Moreover, due to their relatively low brightness, some of these probes require high concentrations (e.g., 5 μM) for cellular imaging [87]. Finally, raft-targeting drugs have been utilized to study the various roles of membrane domains under a more focused functional context. A few examples include cholesterol-disrupting reagents such as methyl-β-cyclodextrin, mevastatin, and cholesterol oxidase and sphingolipid-disrupting molecules such as fumonisin B1, myoricin, and sphingomyelinase [88–91]. All of these molecules target essential components found in lipid rafts. However, in many cases, these molecules display pleiotropic effects beyond their intended application and require the use of high concentrations [92–96]. Moreover, these pharmacological agents often exhibit low selectivity and consequently suffer from off-target effects. Lastly, pharmacological targeting of cholesterol in the non-raft domain is possible [97]. Overall, the abovementioned reagents provide a valuable membrane-centric toolbox, capable of enhancing our understanding of plasma membrane domain structure, composition, dynamics, and function.
3. Dynamic assembly of plasma membrane signaling “hot spots”
Plasma membrane lipid rafts and raft-like domains are increasingly receiving attention as platforms that directly influence cell signaling pathways. These membrane domains are able to modulate signals received from the extracellular space, amplify these signals, multiplex them with other signals, and mediate dissemination to various cytoplasmic pathways in a two- or three-dimensional fashion. From a mechanistic perspective, an increasing body of evidence suggests that raft-like proteolipid domains coexist as a heterogeneous diversity of protein and lipid compositions in the plasma membrane (Fig. 1) [98, 99]. The organization and function of these domains are postulated to be highly dependent on membrane partitioning into distinct compartments, which in turn is driven by the actin membrane skeleton, as well as the oligomerization of proteins and lipids into large membrane-associated complexes also known as clusters (Fig. 2a, clustering). Currently, some of these domains are described as being highly dynamic, exhibiting short lifetimes (in the 1 ns–1 s range) which are thought to be stabilized and enlarged via protein-lipid clustering following stimulation by a ligand [100–102].
The non-random distribution of discrete organized domains exhibiting diverse biophysical and biochemical characteristics plays an essential role in modulating various signal transduction pathways (Fig. 2b). One relevant example is the canonical Wnt/β-catenin signaling pathway. Canonical Wnt/β-catenin signaling plays a central role in embryonic development as well as adult stem cell homeostasis and tumorigenesis [14, 103, 104]. This signaling pathway is initiated by the interactions between lipid-modified Wnt molecules and the integral plasma membrane receptors, low density lipoprotein receptor-related proteins 5/6 (LRP5/6), and Frizzled (Fz). Activation of these receptors by Wnt, which involves receptor phosphorylation and endocytosis, leads to the stabilization, translocation, and accumulation of β-catenin in the nucleus. Once in the nucleus, the transcriptional co-activator nuclear β-catenin, together with transcription factors of the lymphoid enhancer-binding factor (Lef) and T cell factor (Tcf) family, will switch on various β-catenin related genes.
Recently, it has become apparent that components of the Wnt/β-catenin signaling pathway (Fig. 2b, Wnt signaling) form specialized nanoclusters in order to signal [13, 105–110]. These membrane clusters require the essential Wnt membrane receptors, LRP6 and Fz as well as the cytosolic proteins Dvl and Axin to form active clusters, i.e., Wnt signalosomes. Wnt/β-catenin pathway-related nanoclusters are small in size (< 100 nm) in the absence of Wnt ligands. However, Wnt ligands can induce nanoclustering and a shift in cluster size distribution. For example, in the presence of Wnt, large cluster patches containing LRP6 have been observed (> 200 nm). More recently, evidence from various studies suggests that select proteins, lipids, and even saccharides are involved in the formation of Wnt/β-catenin signaling-associated nanoclusters [14, 111–114]. The Wnt/β-catenin signaling pathway positive feedback regulator Ly6 family protein LY6/PLAUR domain-containing 6 (Lypd6) is required for LRP6 phosphorylation and activation in lipid rafts. This GPI-anchored protein, which preferentially localizes to lipid rafts, has been shown to form ~ 100 nm protein clusters [115, 116]. Interestingly, GPI-anchored proteins have been shown to trigger recruitment of kinases at the inner leaflet of the plasma membrane [117–119]. Additionally, casein kinase 1 gamma (Ck1γ), a kinase that phosphorylates LRP6, has been shown to be primarily present in lipid raft domains [120, 121]. Thus, clustering of Lypd6 might play an important regulatory role in the recruitment of Ck1γ and other Wnt-associated kinases, phosphorylation of Wnt receptors, and stabilization of active Wnt-related clusters in lipid rafts. Similarly, other plasma membrane components such as heparin sulfate (HS), cholesterol, and phosphatidylinositol-4,5-bisphosphate (PIP2) might also play a role in Wnt/β-catenin pathway-related nanoclustering. Two types of HS molecules, “N-sulfo-rich HS” (N-sulfo HS) and “N-acetyl-rich HS” (N-acetyl HS), have been shown to form distinct 200-nm clusters, as measured by STED microscopy [114]. Interestingly, N-sulfo HS, not N-acetyl-HS, preferably colocalize with the Wnt receptors Fz and LRP6 as well as LRP6’s active counterpart, phosphor-LRP6 [114]. N-sulfo HS clusters have also been shown to colocalize with Wnt ligands inside cells and have been postulated to function as pre-existing cores for signalosome formation [122]. Finally, the multifunctional lipid messenger, PIP2, whose synthesis is induced by Wnt stimulation, has been shown to be required for LRP6 aggregation, recruitment of Axin, and other effector proteins to the membrane inner leaflet, resulting in the formation of signalosomes [14, 123]. Thus, it is evident that multiple plasma membrane components such as lipids, proteins, carbohydrates, and the membrane cytoskeleton influence, in a highly dynamic yet precise manner, the plasma membrane microenvironment and organize Wnt signaling events into distinct domain platforms. These highly compartmentalized and diverse membrane domains, in turn, meticulously modulate numerous crucial cellular signaling events.
Ras nanoclustering in the plasma membrane has also been well documented. These nanoclusters are ~ 9 nm in radius, contain ~ 6–7 Ras proteins per cluster, display a rapid turnover and short lifetime (0.1–1 s), and contain ~ 40% of the total Ras proteins [124–126]. Interestingly, nanoclusters of various Ras isoforms contain distinct lipid compositions. For example, studies have shown that HRas-GDP nanoclusters reside in cholesterol-rich domains, while HRas-GTP clusters form in a cholesterol-independent manner [127–129]. In comparison, GDP-bound and GTP-bound KRas both form discrete nanoclusters independently of cholesterol [56, 124, 125]. Similarly, other lipids, such as PIP2, PS, and phosphatidic acid (PA), have been shown to distribute differently in certain types of Ras clusters [124, 130, 131].
With respect to membrane order, Wnt/β-catenin signaling transduction is thought to initiate from raft domains [132] and is modulated by various key lipid raft components (Fig. 2b, Wnt signaling). Early evidence suggested that caveolin, a main component of the raft subdomain known as caveolae, is necessary for the cellular internalization of active Wnt receptors and stabilization of nuclear β-catenin [133]. The localization of Wnt-associated receptors in raft domains is also believed to be essential for signal transduction and activation of downstream effectors. For example, Dickkopf 1 (Dkk1), a Wnt/β-catenin pathway antagonist, has been shown to inhibit Wnt/β-catenin signaling by removing LRP5/6 from lipid rafts, reducing its phosphorylation in these domains and inducing its internalization via clathrin-mediated endocytosis [120, 121]. Interestingly, even though both are necessary for β-catenin activation, receptor phosphorylation and internalization are able to occur independently of each other, suggesting that these two steps are important checkpoints in Wnt/β-catenin signaling modulation. More recently, another essential raft component was found to regulate Wnt/β-catenin signaling activity. There is now cogent evidence indicating that cholesterol selectively activates canonical Wnt signaling over non-canonical Wnt signaling [134]. Specifically, cholesterol promotes interactions of Disheveled (Dvl), a scaffold cytosolic protein associated with the Wnt/β-catenin pathway, with LRP5/6, Axin, and the anionic lipid PIP2 in the plasma membrane, promoting activation of this pathway. Interestingly, binding of canonical Wnt to Fz and LRP5/6 has been shown to induce the local enrichment of cholesterol in the vicinity of activated receptors by bringing Fz closer to LRP6 in a cholesterol-enriched microenvironment, reminiscent of lipid rafts. This is noteworthy, because the concept that raft domains are dramatically different before and after ligand stimulation has been proposed in order to describe, in part, their highly dynamic nature and changes in membrane raft coverage. Other protein effectors known to associate with lipid rafts have been shown to regulate the Wnt/β-catenin pathway. For example, Lypd6 has been shown to physically interact with the LRP5/6, promoting LRP5/6 phosphorylation in lipid rafts and enhancing Wnt/β-catenin signaling. Interestingly, mislocalization of Lypd6 to non-raft membrane domains is sufficient to alter LRP5/6 phosphorylation and inhibit the Wnt/β-catenin pathway [115]. It is also important to note that lipid rafts and their components act as plasma membrane signaling organizers in the Wnt/β-catenin pathway via mediating plasma membrane lipid asymmetry. Interestingly, quantitative live-cell imaging studies have demonstrated a high correlation between the inner leaflet plasma membrane cholesterol levels and cellular Wnt/β-catenin signaling activity [51]. This suggests that stimulus-induced plasma membrane cholesterol redistribution is a key modulator of raft-dependent Wnt/β-catenin signaling.
EGF-mediated activation of ERK serves as yet another excellent paradigm for nanoscale proteolipid-based activation, recruitment, and signaling. In this example, we will describe the binding of EGF to EGFR and the subsequent series of events that lead to the propagation of the MAPK/ERK signal (Fig. 2b, EGFR signaling). Non-active EGFR is believed to reside in a non-raft domain. The binding of EGF to EGFR results in its dimerization, activation (autophosphorylation), and the subsequent coalescence of different lipid raft domains including ganglioside GM1 and glycosylphosphatidylinositol (GPI)-anchored proteins [135, 136]. The formation of these domains may in part be driven by the generation the ionic lipids, including PA by phospholipase D2 (PLD2) [137–139] and PIP2 by PI(4)P5 kinase [140, 141], as these lipids are known to influence the nanoscale organization of EGFR [142, 143]. Furthermore, these ionic lipids serve the role of beacons, which recruit specific effector proteins to the newly forming proteolipid complex. This includes son of sevenless 1 (SOS1) by way of its affinity for PA and PIP2 [144, 145]. This is relevant because SOS1 functions as a guanine nucleotide exchange factor (GEF) responsible for activating Ras [138]. Not surprisingly, the inactive GDP-bound version of H- and KRas has strong affinities for PIP2 and PA, respectively [124, 130], allowing them to preferentially interact with, and therefore be activated by, SOS1 in a predefined time and space. Newly activated GTP-bound HRas then moves into a new nanodomain spatially segregated from its inactive GDP-bound counterpart, while activated KRas remains near its original location [146, 147].
Interestingly, the formation of inactive HRas and active KRas nanoclusters is dependent on the integrity of the actin cytoskeleton, while active HRas shows no such dependency [125]. In many cases, Ras nanocluster formation is tightly correlated with ERK signaling [148], which is in agreement with the observation that actin disruption attenuates oncogenic active GTP-bound KRasG12V signaling, while having no effect on HRasG12V [125]. This is relevant because both PA and PIP2 influence the active remodeling and stabilization of the actin cytoskeleton [149–151]. The net result of these seemingly complex interactions is the formation of a proteolipid cluster, whose heterogeneous composition is exquisitely defined both spatially and temporally, allowing the selective propagation of a signal. Collectively, this evidence establishes how critical plasma membrane domain organization is required for the controlled regulation of cellular processes.
4. Dysregulation of plasma membrane biochemical and biophysical characteristics as a cancer driver
Plasma membrane biochemical and biophysical homeostasis, e.g., precise spatial compartmentalization of signaling components, regulates intracellular signal transduction (Fig. 3a) [152]. As discussed above, multiple signaling pathways, as well as several of their effectors, have been reported to occur in lipid raft domains, including EGFR/Ras, Wnt/β-catenin, insulin receptor, and T cell and B cell receptor signaling [115, 132, 153–158]. Moreover, it has been demonstrated that multiple effectors associated with these pathways require proper membrane organization, i.e., the formation of nanoclusters, in order to signal efficiently (Fig. 3a) [13, 111, 125, 148, 159–161]. Interestingly, all of these signaling pathways and several effectors have been shown to be directly involved in signaling processes associated with cancer. This suggests that alterations in lipid raft domains may mediate tumorigenesis. In this regard, several studies have demonstrated that cancer cells exhibit higher levels of rafts/caveolae compared to normal cells and that disruption of lipid rafts in cancer can lead to increased responsiveness to anti-cancer therapies [122, 162–166]. Additionally, some anti-cancer drugs have beneficial effects through alteration of the protein content of lipid rafts [166]. Thus, a growing number of experimental, epidemiological, and preclinical studies now support the membrane-centric hypothesis that the dysregulation of bio-chemical and biophysical factors associated with the plasma membrane actively mediates cancer risk (Fig. 3b). Consequently, the plasma membrane and these specialized membrane domains have received increased attention, due to their ability to fine-tune, stabilize, and, most importantly, mediate signaling interfaces.
Fig. 3.
Dysregulation of plasma membrane domains and the role of MTDBs as countermeasures for loss of membrane homeostasis. Plasma membrane domain biochemical and biophysical homeostasis, e.g., membrane order, proteolipid clustering, and precise spatial compartmentalization of signaling components, regulates intracellular signaling. a In healthy cells, membrane domains display “normal” levels of order and proteolipid clustering and signaling-associated effectors localize at the right place in the right time. Together, this ensures canonical cell signaling. b During cancer, alterations of membrane domain features are observed such as increased membrane order, clustering, and effector recruitment as well as mislocalization of domain effectors. Loss of membrane domain homeostasis leads to an atypical cellular state, thus driving aberrant signaling. c Interestingly, MTDBs have the ability to target and reshape plasma membrane domains. In some cases, MTDBs have been shown to alter membrane fluidity, proteolipid complexes, and effector localization and suppress oncogenic signaling. In this way, MTDBs are able to restore, to a significant extent, healthy cell signaling
4.1. Cholesterol dysregulation
Unesterified free cholesterol is primarily localized to the plasma membrane, where it constitutes up to 90% of total cell cholesterol and 10–45 mol% of total plasma membrane lipids [167–169]. RNA expression and DNA mutational profiling studies have identified dysregulation of the cholesterol synthetic pathway in several cancers [170–172]. For example, data compiled by The Cancer Genome Atlas (TCGA) reveal the increased activity of genes in the cholesterol biosynthesis pathway in various cancers. Interestingly, a number of oncogenic signals, such as PI3K/AKT/mTOR, EGFR/RAS, and TP53, have been shown to stimulate the activity of the transcription factor SREBP, a major regulator of genes involved in de novo cholesterol biosynthesis [173]. It is also noteworthy that from a translational perspective, EGFR’s association with cholesterol-rich lipid rafts underlies its resistance to EGFR tyrosine kinase inhibitors [174]. In addition, the size and number of EGFR nanoclusters is increased in lung cancer in comparison to normal lung cells [143], and both PIP2 [143] and cholesterol [175] are necessary for the formation of EGFR nanoclusters.
Activating mutations in Wnt/β-catenin pathway effectors have been shown to upregulate genes associated with de novo cholesterol synthesis in various cell lines [176, 177]. Moreover, a number of epidemiologic studies have documented a positive association between elevated serum cholesterol level and risk of certain cancer types [178–182]. For example, a 10 mg/dL increase in cholesterol was associated with a 9% increase in prostate cancer relapse [178]. In this regard, several studies involving the use of HMG-CoA reductase inhibitors, e.g., statins, showed a reduced risk of melanoma, non-Hodgkin lymphoma, endometrial, breast, and colorectal cancers [183–186]. Consistent with these reports, studies have demonstrated that cholesterol levels in tumor cells are higher than those in healthy cells [187–189]. Additionally, cancer tissues display increased upregulation of HMG-CoA reductase, loss of feedback inhibition, decreased expression of the cholesterol exporter ATP binding cassette transporter A1 (ABCA1), and increased extracellular cholesterol uptake via LDL receptor [187, 190–194]. Collectively, these metabolic perturbations contribute to the accumulation of cellular cholesterol. Thus, it now appears that dysregulation of cholesterol homeostasis is a significant contributing factor to cancer risk.
4.2. Dysregulation of other lipids
Dysregulation of other plasma membrane lipids has also been identified in various cancers. Although overlooked in the past, changes in lipid-associated pathways in tumors are now being described more frequently. Thus, plasma membrane lipid reprogramming is an emerging hallmark of cancer. Large-scale microarray profiling studies have revealed alterations in different metabolic pathways associated with lipid second messengers (phosphatidylinositols), lipid mediators (leukotrienes), and structural lipids (glycosphingolipids) [194]. Likewise, a relative increase in lipid levels such as phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), phosphatidic acid (PA), and triglycerides (TG) as well as an increase in saturated fatty acid content has also been observed in a variety of tumor types [195–199]. In addition, increased expression of lipogenic enzymes, such as acetyl-CoA carboxylase (ACC) and fatty acid synthase (FASN), lipid-modifying enzymes such as phosphoinositide 3-kinase (PI3K), phospholipase C (PLC) and phospholipase D (PLD) and ATP citrate lyase (ACLY), which collectively promote cholesterol biosynthesis, represent a phenotypic alteration often observed in different cancer types and frequently predict a poor prognosis in cancer patients [199–204]. Furthermore, state-of-the-art analytical and imaging tools, such as electrospray ionization, matrix-assisted laser desorption/ionization, tandem mass spectrometry (MS/MS), and Raman scattering microscopy, have provided valuable lipidomic data that describe alterations in cellular lipid phenotype and fatty acid composition as well as specific spatial cellular distribution in cancer-related conditions [23, 196, 205–208]. It is apparent that cancer cells display a high demand for various lipids and their excess is considered a trademark of cancer. However, some important questions remain to be addressed. How does loss of membrane lipid homeostasis influence other crucial plasma membrane elements and, thus, modulate cellular signaling? How does deviation from the healthy steady-state promote cancer risk?
4.3. Dysregulation of membrane domain order and clustering and its role in signaling
Different plasma membrane components such as lipids, proteins, and the membrane skeleton influence the plasma membrane microenvironment in a direct and dynamic manner. Alternatively, plasma membrane composition and relative fluidity can regulate the organization, the activity, and, consequently, the function of several membrane effectors (Fig. 3a) [34, 209–211]. Plasma membrane organization, in part, is modulated by the preferential interaction of specific types of lipids. Due to their biochemical and biophysical properties, these lipids can form specialized discrete domains. As discussed above, cholesterol, sphingolipids, and saturated phospholipids allow the formation of relative ordered and condensed domains. In contrast, polyunsaturated and branched lipids form disordered domains [211–213]. These domains, which exhibit different biophysical and biochemical properties, are mutually excluded and coexist in the plasma membrane allowing it to display diverse properties and functionalities. Collectively, these membrane attributes give rise to a distinguishable macroscopic biophysical membrane phenotype, i.e., membrane order.
One of the most popular methods to quantify membrane order involves the use of polarity-sensitive lipophilic dyes or membrane order dyes (e.g., Laurdan and di-4-ANEPPDHQ). The vast majority of membrane order dyes are solvatochromic and display shifts in their emission maxima when found in polar vs. non-polar solvents [214–218]. For example, in Laurdan’s case, the less-polar membrane environment of the ordered phase (often regarded as synonymous with lipid rafts) induces a 50-nm blue shift in the emission maxima. This shift in emission profile between disordered and ordered domains allows the construction of a generalized polarization (GP) image. Hence, the calculation of a ratiometric measurement of the fluorescence intensity recorded in the two “ordered” and “disordered” channels serves as a quantitative assessment of relative plasma membrane order.
Measurements of plasma membrane order have been performed in whole organisms, intact tissue, and live cells as well as biologically derived and synthetic membranes. For example, the formation of selective, ordered lipid domains in membrane vesicles, i.e., giant plasma membrane vesicles (GPMVs), which are derived from live cells, has been documented extensively [87, 219–221]. Most importantly, these findings have served as an integral confirmation of the potential for domain formation in live cells. Not surprisingly, several studies have confirmed the presence of differences in plasma membrane order in domains with higher degrees of complexity such as live cells and tissues. As an example, membrane order experiments performed on human white blood cells showed that higher membrane rigidity resides at the immunological synapse periphery of T cells [222, 223]. Additionally, the first ever report of membrane order in an intact living organism showed high membrane order in the apical surfaces of polarized epithelial cells of zebrafish [219]. Membrane order maps have been acquired from different tissues, including the gut epithelium, muscle, kidney, retina, and neurons [219]. These and multiple other similar studies represent an important step towards understanding the physiological relevance of plasma membrane order and the role of this biophysical parameter in cellular signaling.
Plasma membrane functionality is modulated by heterogeneous lipid and protein domain composition, organization, and component interactions [211, 224, 225]. Thus, membrane order is believed to play a crucial role in plasma membrane homeostasis. Several proteins and lipids have been shown to reside in, and others to be excluded from, domains displaying distinct membrane order. Confocal, single-particle, FLIM-FRET, and FCS measurements have documented the confinement of specific effectors in distinct plasma membrane domains [142, 160, 226–229]. Moreover, membrane order has been implicated in the modulation of spatially localized signaling events. For example, large-scale membrane polarization, such as that which occurs for T cells at the immunological synapse, depends, in part, on membrane order [209, 222]. In addition, it has been established that non-activated HRas partitions into ordered domains, whereas active HRas preferably localizes in non-raft domains, which represent the hot spots of Ras activation and signaling [125, 126, 230, 231]. Lastly, as noted above, it has been recognized that effective canonical Wnt/β-catenin signaling requires the accumulation of its plasma membrane receptors, LRP6 and Fz, in cholesterol-rich domains, which in turn allows receptor phosphorylation, activation, and recruitment of effectors at the inner leaflet to transduce the Wnt-mediated signal intracellularly [51, 134, 232].
In cancer, the uncontrolled alteration of cellular plasma membrane homeostasis can lead to dramatic changes in the biochemical and biophysical characteristics of membrane domains (Fig. 3b). These changes can be triggered by various stimuli, including genetic mutations, abnormal epigenetic modifications, and carcinogenic substances [23, 233–236]. If allowed to operate unchecked, the resulting loss of homeostasis can lead to reprogramming of vital cellular features such as plasma membrane order and nanoclustering (Fig. 3b). Most importantly, this new abnormal cellular state, which is associated with aberrant signaling events, can increase cancer risk. Although a general trend has not been established regarding the direction of membrane order dysregulation, it is clear that alterations in plasma membrane fluidity can profoundly affect several processes such as protein expression, endocytosis, apoptosis, cell proliferation, and metastasis [166]. Thus, it comes as no surprise that membrane order of cancer cells differs from that of normal cells. For example, hepatoma tumor and breast cancer cells as well as hairy cells from hairy cell leukemia have been shown to display higher membrane rigidity compared to normal cells [237, 238]. Additionally, multidrug resistant cancer cell types have been shown to exhibit an increase in membrane rigidity when compared to their non-resistant counterparts and normal cells [239, 240]. In most cases, this increase in rigidity is accompanied by an increase of membrane cholesterol and sphingomyelin. However, changes of plasma membrane order in cancer cells do not always result in higher membrane rigidity [241–243]. Fluidization of membranes in cancer cells appears to be correlated with their malignant potential and competency to metastasize. Interestingly, the ability of a cell to alter its membrane fluidity has been shown to be a key step in modulating anti-cancer drug cell penetration, transport, and responsiveness, thus leading ultimately to drug resistance [166, 238, 239]. This evidence further highlights the importance of plasma membrane order in several steps of cancer. Similar to the case of plasma membrane order, dysregulation of protein, lipid, and saccharide nanoclustering at the plasma membrane can increase cancer risk [24, 244–247]. It has been reported that cancer-associated mutations in HRas, KRas, and NRas oncoproteins increase their activity through augmented nanoclustering of Ras on membranes [22]. Similarly, Ras clustering modulation by the overexpression of various galectins (e.g., Galectin-1 and Galectin-3), which is observed in breast, pancreatic, colorectal, and other cancers, can increase Ras nanoclustering and its oncogenic output [24, 246, 248–251]. Likewise, nanoclustering of other plasma membrane components, i.e., carbohydrates and lipids, plays an important role in cancer. For example, oligomers of galectin, the same proteins shown to increase cancer-associated Ras activity, are known to induce aggregation of carbohydrates on the surface of cells [252–255]. Several studies have revealed that the membrane of cancer cells display distinct distribution patterns of various types of saccharides relative to normal cell membranes [247]. In fact, several types of cancer cells show higher carbohydrate expression levels as well as higher membrane cluster coverage and larger carbohydrate clusters as compared to their normal cell membrane counterparts [247]. These carbohydrate alteration phenotypes are thought to promote tumorigenesis and metastasis. Finally, alterations in lipid membrane organization are also involved in clustering-associated cancer risk. As an example, analysis of large panels of tumor types shows loss of PS asymmetry and increased PS exposure on the surface of malignant cells relative to normal cells [256–259]. Cancer treatments utilizing radiotherapy and chemotherapy can exacerbate this phenotype [260–262]. Interestingly, imaging MRI studies have revealed the presence of cluster structures, stained by a PS-specific MRI-contrast agent, in tumor tissues [263–265]. As noted above, other charged lipids, involved in important cellular signaling processes, such as PIP2 and PA have been shown to cluster in the plasma membrane and modulate cellular signaling [142, 143, 266–270]. Due to their role in several key cancer-associated processes, the modulation of these plasma membrane biochemical and biophysical phenotypes by membrane-targeted countermeasures could serve as a cancer chemoprevention therapeutic strategy.
5. Cancer chemoprevention by membrane-targeted dietary bioactives
“Dietary bioactives” are constituents in foods or dietary supplements, other than those needed to meet basic human nutritional needs, which are responsible for changes in health status. While genetic factors contribute to cancer risk, environmental factors account for the majority of risk [271]. For example, colorectal cancer risk can be significantly reduced by dietary modification, including increased dietary fiber intake and reduced fat intake [272, 273]. Omega 3 (n-3) (e.g., α-linolenic acid, ALA) and omega 6 (n-6) (e.g., linoleic acid, LA) polyunsaturated fatty acids (PUFAs) are essential bioactive nutrients, obtained from the diet, that incorporate into cellular membranes in tissues and influence many physiological processes, including production of eicosanoids [274–277]. Two key n-3 PUFAs found in fish oil are eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) [278]. In humans, production of EPA and DHA from dietary precursors, e.g., ALA, is highly inefficient [279]. Additionally, there is competition with synthesis of long-chain n-6 PUFA, e.g., arachidonic acid (AA), which is produced from LA and is found in much greater abundance in a typical Western diet [280]. In general, n-6 PUFAs are associated with pro-inflammatory responses whereas n-3 PUFAs tend to cause opposing effects and are anti-inflammatory [281, 282]. Over the past few decades, it has been observed that an increase in the intake of n-6 PUFA over n-3 PUFA, e.g., high n-6 PUFA-to-n-3 PUFA ratio diet, which is associated with greater metabolism of n-6 PUFA, coincides with increases in chronic inflammatory diseases such as non-alcoholic fatty liver disease (NAFLD), cardiovascular disease, obesity, inflammatory bowel disease (IBD), rheumatoid arthritis, and Alzheimer’s disease (AD) [281]. Given that inflammation is strongly intertwined with cancer, higher intake of n-3 PUFA could provide a biological chemoprotective effect [274, 283].
With respect to clinical studies, increasing evidence suggests that the consumption of fish oil, containing EPA and DHA, may reduce cancer risk in humans [284–288]. Interestingly, EPA and DHA appear to be ideally suited to work either alone or in combination with other chemoprotective drugs, e.g., NSAIDs [289, 290]. Recently, it was demonstrated that EPA reduced rectal polyp number and size in patients with familial adenomatous polyposis (FAP), a condition associated with colorectal cancer [283, 291]. Most impressive was the fact that fish oil derived n-3 PUFA suppressed FAP to a degree similar to celecoxib, an FDA approved drug for the treatment of FAP. Additionally, in a different clinical study, it was demonstrated that EPA and DHA, contrary to LA and AA, may reduce the risk of total and advanced prostate cancer [292, 293]. Collectively, these data indicate that n-3 PUFAs hold promise as chemoprevention agents for cancer.
The consumption of n-3 PUFA in combination with other agents with complementary anti-tumor action, e.g., curcumin [294] and drugs [295], may improve their overall efficacies as cancer prevention therapies. Polyphenolic and terpenoid phytochemicals have become increasingly popular with consumers in great part because of their putative health benefits. Of these, turmeric (Curcuma longa Linn) extracts including curcumin (diferuloylmethane), a yellow color pigment of turmeric, have shown some promising effects in patients with various pro-inflammatory diseases and cancer [296–303]. Completed and ongoing clinical trials (Clinical Trials.gov) examined the role of curcumin with respect to aberrant crypt foci (NCT00365209), cell proliferation and apoptosis in colonic mucosa (NCT00118989), drug combination therapy (NCT00745134), and treatment of FAP (NCT00927485, NCT00641147). Recent data from one of these phase IIa clinical trials indicates that consumption of curcumin at 4 g per day for 30 days significantly (40%) reduces the number of aberrant crypt foci (ACF) in a CRC model [304] and in men and women [305]. More recently, our laboratory examined the effect of n-3 PUFA and curcumin in a mouse colorectal cancer initiation model and found that these agents when combined synergistically reduced (75%) the number of ACF in a mouse CRC model [28]. Importantly, several studies have confirmed that curcumin is well tolerated and that the effect is potentially mediated by curcumin delivered to the target tissue, i.e., it is directly incorporated into the intestinal mucosa [28, 305, 306]. However, before a dietary combinatorial therapeutic approach can be adopted, it is critical that we fully elucidate the molecular mechanisms associated with n-3 PUFA, curcumin, and other potentially beneficial MTDB chemoprotective action. Thus, establishing a causal role of n-3 PUFA, curcumin, and other MTDBs in cancer prevention would have a major translational impact because these dietary nutrients are safe, well tolerated, relatively inexpensive and provide additional health benefits.
One of the criticisms facing the dietary chemoprevention field is the fact that dietary bioactives affect diverse physiological processes including cell membrane structure/function, eicosanoid signaling, nuclear receptor activation, gene transcription and translation, and inflammatory responses [21, 307–312]. Consequently, a huge challenge is to explain and unify these apparently disconnected signaling nodes. We propose a unifying mechanistic hypothesis to explain the function of these dietary bioactives, with an emphasis in colon cancer. We posit that n-3 PUFA and curcumin/curcuminoids fall into a unique class of MTDBs which, because of their unique amphiphilic properties, are capable of modulating plasma membrane hierarchical organization (Fig. 3c). We also believe that this ability to modulate plasma membrane biophysical properties may underlie the chemoprotective effects of other bioactives, such as cathecins and procyanidins [25, 313].
With respect to the molecular mechanism of action of fish oil, there is a growing body of in vitro and in vivo evidence indicating that n-3 PUFA reshape plasma membrane domains. For example, EPA and DHA are rapidly incorporated into cell primarily into membrane phospholipids at the sn-2 position [314, 315]. This is noteworthy, because the presence of long-chain n-3 PUFA in membrane phospholipids imparts unique biophysical properties which have been linked to alterations in plasma membrane structure and function [315–318]. DHA is known to influence membrane fluidity, ion permeability, fatty acid exchange, and resident protein function [319–321], including the inhibition of EGFR signaling in tumor-bearing mice by disassociating EGFR from lipid rafts [322]. Of particular interest is the ability of DHA to attenuate EGFR signaling. Specifically, immortalized mouse colonic cells incubated with DHA exhibited decreased GTP-bound K, H, and NRas levels following EGF stimulation [322]. Similarly, intact colonic crypts isolated from mice fed n-3 vs. n-6 PUFA (control)-enriched diets had reduced levels of HRas-GTP following EGF stimulation [317]. With respect to a molecular target capable of modulating Ras activation, our lab recently identified a DHA-induced “break point” (following Grb2 recruitment to EGFR) in the Ras signaling pathway [322]. This combined with the fact that DHA can increase EGFR/SOS1 interaction, while reciprocally decreasing SOS1/Ras interaction in three cancer lines [323], suggests that plasma membrane-targeted GEFs may be modulated by DHA.
SOS1 is recruited to the plasma membrane by activated EGFR, which subsequently engages Ras [324]. This process is subject to complex regulation by the temporal and spatial production of membrane phosphoinositides through interactions with SOS1 Dbl homology (DH)/pleckstrin homology (PH) regulatory domains [325]. Two of the lipids involved in SOS1 recruitment are PA [138] and PIP2 [144, 145, 326]. In proof-of-principle experiments, we demonstrated that n-3 PUFAs reduce the levels of PIP2 in CD4+ T cells [327], implying that MTDBs may be capable of modulating Ras activation in vivo. Therefore, we hypothesized that DHA reduces wild-type and oncogenic Ras signaling by disrupting SOS/Ras complex formation through altered temporal spatial production of PA and PIP2. Importantly, the SOS/Ras interaction is a current target of pharmacological agents designed to suppress oncognenic Ras signaling [328–330]. Furthermore, oncogenic KRas-driven cancers are dependent on wild-type H- and NRas signaling, which suggests that Ras may be a target for MTDB therapy [331, 332]. This is highly relevant because approximately 30–50% of colorectal cancers (CRCs) harbor KRas mutations, and KRas-mutant CRCs exhibit resistance to standard therapy [333], thereby reducing survival [334]. Moreover, to date, targeting of mutant RAS proteins in cancers has not been possible [334]. Interestingly, the effects of n-3 PUFA are not limited to wild-type Ras, as DHA also attenuates activation of ERK and reduces the growth of mouse colonocytes expressing oncogenic HRas [322, 335]. This is consistent with work documenting the ability of n-3 PUFA to reduce cell proliferation, signaling, and anchorage-independent growth in different colon cancer cell lines (HCT116, SW480, SW620), harboring unique KRas G12V and G13D mutations [323, 336, 337]. Importantly n-3 PUFAs reduce murine pancreatic cancer development and delay progression of pancreatic ductal adenocarcinoma in mutant KRas-driven mouse models [338, 339].
High fidelity signaling of KRas is dependent on its spatial organization into defined dimers [340, 341] or nanoclusters [125, 148]. Recently, it was demonstrated that select amphiphilic agents, through direct modulation of the biophysical properties of the plasma membrane, compromise oncogenic KRas nanoclustering to modulate signal transduction [20, 124]. These findings suggest that Ras nanoclusters could be a novel therapeutic target [342]. Consistent with this hypothesis, experiments conducted by our lab using immuno-gold electron microscopy of plasma membrane sheets suggest that plasma membrane organization of inner leaflets is fundamentally altered by EPA and DHA [343, 344]. Specifically, n-3 PUFA treatment altered the nanoclustering of truncated forms of wild-type H- and KRas in cervical adenocarcinoma (HeLa) and colorectal carcinoma (HCT-116) cells [343, 344]. Our most recent work targeting the gastrointestinal tract (currently under revision) has demonstrated that dietary n-3 PUFA remodels plasma membrane phospholipids, reducing the lateral segregation of cholesterol-dependent and cholesterol-independent nanoclusters, and suppressing PA-dependent oncogenic KRas effector interactions. This results in the attenuation of oncogenic Ras-driven colonic hyperproliferation in both Drosophila and murine models. These findings demonstrate the unique properties of dietary n-3 PUFA in the shaping of Ras nanoscale proteolipid complexes and support the emerging role of plasma membrane-targeted therapies.
Wnt signaling constitutes the major driving force behind the homeostatic self-renewal of the intestinal crypt [345, 346]. Dysregulation of Wnt signaling has been linked to cancer in multiple tissues [347–352]. It is noteworthy that mediators of Wnt signaling, LRP6 and Fz, are plasma membrane receptors that play a critical role in cell polarity [353, 354], stemness [355, 356], differentiation [356, 357], and neoplastic transformation [345, 355, 356, 358]. From a functional perspective, LRP6 and Fz require lipid raft localization and nanoclustering for efficient signaling and, most notably, stabilization of β-catenin [13, 110, 115, 133]. Extrinsic effectors, e.g., nystatin, which bind cholesterol and disrupt lipid rafts, alter LRP6 and Fz downstream signaling [121]. Similarly, secreted cellular effectors, e.g., Dkk1, bind and displace LRP6 from lipid rafts, leading to a reduction in LRP6 phosphorylated state and levels in lipid rafts [120]. Both distinct mechanisms of action result in the suppression of β-catenin transcriptional activity. Unfortunately, attempts to target aberrant Wnt signaling using drugs still face multiple hurdles due to poor tumor cell targeting, negative side effects associated with required long-term treatments, and a poor understanding of the mechanisms of action [359]. Consequently, there is an urgent need to develop toxicologically innocuous Wnt signaling therapeutic approaches. Nevertheless, previous studies, which have focused on suppressing β-catenin expression [360], preventing transcriptional activation of β-catenin/TCF complexes [361, 362] and inhibiting cyclooxygenase-2 (COX-2) [363, 364], have laid a strong knowledge foundation for the development of potential therapeutic tools for cancer. From a cancer prevention perspective, our laboratory examined the combined effect of n-3 PUFAs and curcumin in a mouse colorectal cancer initiation model and found that these MTDBs synergistically reduce nuclear β-catenin in aberrant crypt foci by threefold [28]. However, further studies are needed to determine precisely how MTDBs function to suppress aberrant Wnt signaling.
6. Summary and future challenges
There is an impending chronic disease crisis in our country. Thus, if the healthcare community hopes to head off the impending cancer storm, we need to get more serious about cancer prevention [365]. Unfortunately, less than 1.5% of total biomedical research is targeted to early detection and prevention of chronic disease [366]. With respect to all human malignancies, 35% are linked directly to diet and an additional 14–20% to obesity [367]. Consistent with these data, cancer risk can be lowered by 36% when humans adhere to healthy dietary principles [368]. In terms of primary cancer prevention, there is a growing body of experimental, epidemiological, and preclinical evidence indicating that diets enriched in MTDBs are protective against various types of cancers. Importantly, these diet-derived agents are toxicologically innocuous and generally free of safety problems intrinsic to drugs administered over long periods of time. However, additional mechanistic insights linking membrane therapy, diet, and cancer risk are required before this application can be used in the advancement of the chemoprevention field. Nonetheless, recent provocative work in this field has afforded us with a new mechanistic perspective in this regard. Thus, we propose that MTDBs, because of their unique properties and pleiotropic effects, are capable of modulating plasma membrane hierarchical organization, thereby suppressing various signaling pathways that are essential for tumorigenesis (Fig. 3c). Elucidation of the role of MTDBs in cancer prevention would have a major translational impact because these dietary bioactives are well tolerated and relatively inexpensive and provide additional health benefits, such as a reduction in mortality.
Acknowledgments
Funding information This work was supported by the National Institutes of Health (NIH) grants R35CA197707 and P30ES023512, the Cancer Prevention Research Institute of Texas (CPRIT), and funds from the Allen Endowed Chair in Nutrition and Chronic Disease Prevention. Natividad Fuentes is a recipient of a Predoctoral Fellowship in Pharmacology/Toxicology from the PhRMA Foundation and the National Science Foundation Texas A&M University System Louis Stokes Alliance for Minority Participation (TAMUS LSAMP) Bridge to the Doctorate Fellowship (HRD-1249272). Alfredo Erazo-Oliveras is a recipient of the 2017 Ford Foundation Postdoctoral Fellowship from The National Academies of Sciences, Engineering and Medicine and the National Institutes of Health-National Cancer Institute (NIH-NCI) Research Supplement to Promote Diversity in Health-Related Research Award (3R35CA197707-02S1).
Abbreviations
- Chol
Cholesterol
- EM
Electron microscopy
- FLIM-FRET
Fluorescence lifetime imaging microscopy combined with fluorescence resonance energy transfer
- LC
Lung cancer
- NL
Normal lung
- PA
Phosphatidic acid
- PALM
Photo-activated localization microscopy
- PIP2
Phosphatidylinositol 4,5-biphosphate
- PIP3
Phosphatidylinositol 3,4,5-trisphosphate
- PI3P
Phosphatidylinositol 3-phosphate
- PI4P
Phosphatidylinositol 4-phosphate
- PS
Phosphatidylserine
- SPT
Single-particle tracking
- STED
Stimulated emission depletion
- STORM
Stochastic optical reconstruction microscopy
- dSTORM
Direct stochastic optical reconstruction microscopy
- TIRF
Total internal reflection fluorescence
Footnotes
Conflict of interest The authors declare that they have no conflict of interest.
References
- 1.Grecco Hernán E., Schmick M, & Bastiaens Philippe I. H. Signaling from the living plasma membrane. Cell, 144(6), 897–909, 10.1016/j.cell.2011.01.029. [DOI] [PubMed] [Google Scholar]
- 2.van Meer G, Voelker DR, & Feigenson GW (2008). Membrane lipids: where they are and how they behave. Nature Reviews. Molecular Cell Biology, 9(2), 112–124. 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chabanon M, Stachowiak JC, & Rangamani P (2017). Systems biology of cellular membranes: a convergence with biophysics. Wiley Interdisciplinary Reviews. Systems Biology and Medicine, 9(5). 10.1002/wsbm.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmick M, & Bastiaens PI (2014). The interdependence of membrane shape and cellular signal processing. Cell, 156(6), 1132–1138. 10.1016/j.cell.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 5.Tocanne JF, Cezanne L, Lopez A, Piknova B, Schram V, Tournier JF, et al. (1994). Lipid domains and lipid/protein interactions in biological membranes. Chemistry and Physics of Lipids, 73(1–2), 139–158. [DOI] [PubMed] [Google Scholar]
- 6.Lamaze C, & Blouin CM (2017). Receptor lipid nanodomain partitioning and signaling. Cell Cycle, 16(3), 237–238. 10.1080/15384101.2016.1245573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foti M, Porcheron G, Fournier M, Maeder C, & Carpentier JL (2007). The neck of caveolae is a distinct plasma membrane subdomain that concentrates insulin receptors in 3T3-L1 adipocytes. Proceedings of the National Academy of Sciences of the United States of America, 104(4), 1242–1247. 10.1073/pnas.0610523104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ziolkowska NE, Christiano R, & Walther TC (2012). Organized living: formation mechanisms and functions of plasma membrane domains in yeast. Trends in Cell Biology, 22(3), 151–158. 10.1016/j.tcb.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida S, Hoppe AD, Araki N, & Swanson JA (2009). Sequential signaling in plasma-membrane domains during macropinosome formation in macrophages. Journal of Cell Science, 122(Pt 18), 3250–3261. 10.1242/jcs.053207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Parajo MF, Cambi A, Torreno-Pina JA, Thompson N, & Jacobson K (2014). Nanoclustering as a dominant feature of plasma membrane organization. Journal of Cell Science, 127(Pt 23), 4995–5005. 10.1242/jcs.146340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ariotti N, Fernandez-Rojo MA, Zhou Y, Hill MM, Rodkey TL, Inder KL, et al. (2014). Caveolae regulate the nanoscale organization of the plasma membrane to remotely control Ras signaling. The Journal of Cell Biology, 204(5), 777–792. 10.1083/jcb.201307055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raghupathy R, Anilkumar AA, Polley A, Singh PP, Yadav M, Johnson C, et al. (2015). Transbilayer lipid interactions mediate nanoclustering of lipid-anchored proteins. Cell, 161(3), 581–594. 10.1016/j.cell.2015.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bilić J, Huang Y-L, Davidson G, Zimmermann T, Cruciat C-M, Bienz M, et al. (2007). Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science, 316(5831), 1619 10.1126/science.1137065. [DOI] [PubMed] [Google Scholar]
- 14.MacDonald BT, Tamai K, & He X (2009). Wnt/beta-catenin signaling: components, mechanisms, and diseases. Developmental Cell, 17(1), 9–26. 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niehrs C (2012). The complex world of WNT receptor signalling. Nature Reviews. Molecular Cell Biology, 13(12), 767–779. 10.1038/nrm3470. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z, Wang L, Du J, Li Y, Yang H, Li C, et al. (2016). Lipid raft localization of epidermal growth factor receptor alters matrix metalloproteinase-1 expression in SiHa cells via the MAPK/ERK signaling pathway. Oncology Letters, 12(6), 4991–4998, 10.3892/ol.2016.5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roepstorff K, Thomsen P, Sandvig K, & van Deurs B (2002). Sequestration of epidermal growth factor receptors in non-caveolar lipid rafts inhibits ligand binding. The Journal of Biological Chemistry, 277(21), 18954–18960. 10.1074/jbc.M201422200. [DOI] [PubMed] [Google Scholar]
- 18.Gueguinou M, Gambade A, Felix R, Chantome A, Fourbon Y, Bougnoux P, et al. (2015). Lipid rafts, KCa/ClCa/Ca2+ channel complexes and EGFR signaling: Novel targets to reduce tumor development by lipids? Biochimica et Biophysica Acta, 1848(10 Pt B), 2603–2620. 10.1016/j.bbamem.2014.10.036. [DOI] [PubMed] [Google Scholar]
- 19.Janosi L, Li Z, Hancock JF, & Gorfe AA (2012). Organization, dynamics, and segregation of Ras nanoclusters in membrane domains. Proceedings of the National Academy of Sciences of the United States of America, 109(21), 8097–8102. 10.1073/pnas.1200773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Y, Cho K-J, Plowman SJ, & Hancock JF (2012). Nonsteroidal anti-inflammatory drugs alter the spatiotemporal organization of Ras proteins on the plasma membrane. Journal of Biological Chemistry, 287(20), 16586–16595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma DW, Seo J, Switzer KC, Fan YY, McMurray DN, Lupton JR, et al. (2004). n-3 PUFA and membrane microdomains: a new frontier in bioactive lipid research. The Journal of Nutritional Biochemistry, 15(11), 700–706. 10.1016/j.jnutbio.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 22.Solman M, Ligabue A, Blazevits O, Jaiswal A, Zhou Y, Liang H, et al. (2015). Specific cancer-associated mutations in the switch III region of Ras increase tumorigenicity by nanocluster augmentation. Elife, 4, e08905 10.7554/eLife.08905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beloribi-Djefaflia S, Vasseur S, & Guillaumond F (2016). Lipid metabolic reprogramming in cancer cells. Oncogenesis, 5, e189 10.1038/oncsis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shalom-Feuerstein R, Plowman SJ, Rotblat B, Ariotti N, Tian T, Hancock JF, et al. (2008). K-ras nanoclustering is subverted by overexpression of the scaffold protein galectin-3. Cancer Research, 68(16), 6608–6616. 10.1158/0008-5472.CAN-08-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuentes NR, Salinas ML, Kim E, & Chapkin RS (2017). Emerging role of chemoprotective agents in the dynamic shaping of plasma membrane organization. Biochimica et Biophysica Acta, 1859(9 Pt B), 1668–1678. 10.1016/j.bbamem.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou TY, McMurray DN, & Chapkin RS (2016). Omega-3 fatty acids, lipid rafts, and T cell signaling. European Journal of Pharmacology, 785, 2–9. 10.1016/j.ejphar.2015.03.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turk HF, & Chapkin RS (2013). Membrane lipid raft organization is uniquely modified by n-3 polyunsaturated fatty acids. Prostaglandins, Leukotrienes, and Essential Fatty Acids, 88(1), 43–47. 10.1016/j.plefa.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim E, Davidson LA, Zoh RS, Hensel ME, Salinas ML, Patil BS, et al. (2016). Rapidly cycling Lgr5+ stem cells are exquisitely sensitive to extrinsic dietary factors that modulate colon cancer risk. Cell Death & Disease, 7(11), e2460 10.1038/cddis.2016.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chapkin RS, DeClercq V, Kim E, Fuentes NR, & Fan Y-Y (2014). Mechanisms by which pleiotropic amphiphilic n–3 PUFA reduce colon cancer risk. Current Colorectal Cancer Reports, 10(4), 442–452. 10.1007/s11888-014-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Triff K, Kim E, & Chapkin RS (2015). Chemoprotective epigenetic mechanisms in a colorectal cancer model: modulation by n-3 PUFA in combination with fermentable fiber. Current Pharmacology Reports, 1(1), 11–20. 10.1007/s40495-014-0005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kraft ML (2013). Plasma membrane organization and function: moving past lipid rafts. Molecular Biology of the Cell, 24(18), 2765–2768. 10.1091/mbc.E13-03-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Curthoys NM, Parent M, Mlodzianoski M, Nelson AJ, Lilieholm J, Butler MB, et al. (2015). Chapter three—dances with membranes: breakthroughs from super-resolution imaging In Kenworthy AK (Ed.), Current topics in membranes (Vol. 75, pp. 59–123): Academic press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu J, Fischman DA, & Steck TL (1973). Selective solubilization of proteins and phospholipids from red blood cell membranes by nonionic detergents. Journal of Supramolecular Structure, 1(3), 233–248. 10.1002/jss.400010308. [DOI] [PubMed] [Google Scholar]
- 34.Sezgin E, Levental I, Mayor S, & Eggeling C (2017). The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nature Reviews. Molecular Cell Biology, 18(6), 361–374. 10.1038/nrm.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ipsen JH, Karlstrom G, Mouritsen OG, Wennerstrom H, & Zuckermann MJ (1987). Phase equilibria in the phosphatidylcholine-cholesterol system. Biochimica et Biophysica Acta, 905(1), 162–172. [DOI] [PubMed] [Google Scholar]
- 36.Lentz BR, Barrow DA, & Hoechli M (1980). Cholesterol-phosphatidylcholine interactions in multilamellar vesicles. Biochemistry, 19(9), 1943–1954. [DOI] [PubMed] [Google Scholar]
- 37.Bacia K, Schwille P, & Kurzchalia T (2005). Sterol structure determines the separation of phases and the curvature of the liquid-ordered phase in model membranes. Proceedings of the National Academy of Sciences of the United States of America, 102(9), 3272–3277. 10.1073/pnas.0408215102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baumgart T, Hess ST, & Webb WW (2003). Imaging coexisting fluid domains in biomembrane models coupling curvature and line tension. Nature, 425(6960), 821–824. 10.1038/nature02013. [DOI] [PubMed] [Google Scholar]
- 39.Oncul S, Klymchenko AS, Kucherak OA, Demchenko AP, Martin S, Dontenwill M, et al. (2010). Liquid ordered phase in cell membranes evidenced by a hydration-sensitive probe: effects of cholesterol depletion and apoptosis. Biochimica et Biophysica Acta, 1798(7), 1436–1443. 10.1016/j.bbamem.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 40.M'Baye G, Mely Y, Duportail G, & Klymchenko AS (2008). Liquid ordered and gel phases of lipid bilayers: fluorescent probes reveal close fluidity but different hydration. Biophysical Journal, 95(3), 1217–1225. 10.1529/biophysj.107.127480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simons K, & Ikonen E (1997). Functional rafts in cell membranes. Nature, 387(6633), 569–572. 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 42.Simons K, & Vaz WL (2004). Model systems, lipid rafts, and cell membranes. Annual Review of Biophysics and Biomolecular Structure, 33, 269–295. 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- 43.Pike LJ (2006). Rafts defined: a report on the keystone symposium on lipid rafts and cell function. Journal of Lipid Research, 47(7), 1597–1598. 10.1194/jlr.E600002-JLR200. [DOI] [PubMed] [Google Scholar]
- 44.Pike LJ (2003). Lipid rafts: bringing order to chaos. Journal of Lipid Research, 44(4), 655–667. 10.1194/jlr.R200021-JLR200. [DOI] [PubMed] [Google Scholar]
- 45.Zuidscherwoude M, de Winde CM, Cambi A, & van Spriel AB (2014). Microdomains in the membrane landscape shape antigen-presenting cell function. Journal of Leukocyte Biology, 95(2), 251–263. 10.1189/jlb.0813440. [DOI] [PubMed] [Google Scholar]
- 46.Liu Y, Yang G, Bu X, Liu G, Ding J, Li P, et al. (2011). Cell-type-specific regulation of raft-associated Akt signaling. Cell Death & Disease, 2, e145 10.1038/cddis.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gorodinsky A, & Harris DA (1995). Glycolipid-anchored proteins in neuroblastoma cells form detergent-resistant complexes without caveolin. The Journal of Cell Biology, 129(3), 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kiessling V, Wan C, & Tamm LK (2009). Domain coupling in asymmetric lipid bilayers. Biochimica et Biophysica Acta, 1788(1), 64–71. 10.1016/j.bbamem.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eisenberg S, Shvartsman DE, Ehrlich M, & Henis YI (2006). Clustering of raft-associated proteins in the external membrane leaflet modulates internal leaflet H-ras diffusion and signaling. Molecular and Cellular Biology, 26(19), 7190–7200. 10.1128/MCB.01059-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Devaux PF, & Morris R (2004). Transmembrane asymmetry and lateral domains in biological membranes. Traffic, 5(4), 241–246. 10.1111/j.1600-0854.2004.0170.x. [DOI] [PubMed] [Google Scholar]
- 51.Liu SL, Sheng R, Jung JH, Wang L, Stec E, O'Connor MJ, et al. (2017). Orthogonal lipid sensors identify transbilayer asymmetry of plasma membrane cholesterol. Nature Chemical Biology, 13(3), 268–274. 10.1038/nchembio.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Truong-Quang BA, & Lenne PF (2014). Membrane microdomains: from seeing to understanding. Frontiers in Plant Science, 5, 18 10.3389/fpls.2014.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shashkova S, & Leake MC (2017). Single-molecule fluorescence microscopy review: shedding new light on old problems. Bioscience Reports, 37(4). 10.1042/BSR20170031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou Y, Wong CO, Cho KJ, van der Hoeven D, Liang H, Thakur DP, et al. (2015). Signal transduction. Membrane potential modulates plasma membrane phospholipid dynamics and K-Ras signaling. Science, 349(6250), 873–876. 10.1126/science.aaa5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mattheyses AL, Simon SM, & Rappoport JZ (2010). Imaging with total internal reflection fluorescence microscopy for the cell biologist. Journal of Cell Science, 123(Pt 21), 3621–3628. 10.1242/jcs.056218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prior IA, Muncke C, Parton RG, & Hancock JF (2003). Direct visualization of Ras proteins in spatially distinct cell surface microdomains. The Journal of Cell Biology, 160(2), 165–170. 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niv H, Gutman O, Kloog Y, & Henis YI (2002). Activated K-Ras and H-Ras display different interactions with saturable nonraft sites at the surface of live cells. The Journal of Cell Biology, 157(5), 865–872. 10.1083/jcb.200202009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manley S, Gillette JM, Patterson GH, Shroff H, Hess HF, Betzig E, et al. (2008). High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nature Methods, 5(2), 155–157. 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 59.Ritchie K, Iino R, Fujiwara T, Murase K, & Kusumi A (2003). The fence and picket structure of the plasma membrane of live cells as revealed by single molecule techniques (review). Molecular Membrane Biology, 20(1), 13–18. [DOI] [PubMed] [Google Scholar]
- 60.Kusumi A, Sako Y, & Yamamoto M (1993). Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophysical Journal, 65(5), 2021–2040. 10.1016/S0006-3495(93)81253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Machan R, & Hof M (2010). Lipid diffusion in planar membranes investigated by fluorescence correlation spectroscopy. Biochimica et Biophysica Acta, 1798(7), 1377–1391. 10.1016/j.bbamem.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 62.Frisz JF, Lou K, Klitzing HA, Hanafin WP, Lizunov V, Wilson RL, et al. (2013). Direct chemical evidence for sphingolipid domains in the plasma membranes of fibroblasts. Proceedings of the National Academy of Sciences of the United States of America, 110(8), E613–E622. 10.1073/pnas.1216585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He C, Hu X, Jung RS, Weston TA, Sandoval NP, Tontonoz P, et al. (2017). High-resolution imaging and quantification of plasma membrane cholesterol by NanoSIMS. Proceedings of the National Academy of Sciences of the United States of America, 114(8), 2000–2005. 10.1073/pnas.1621432114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kraft ML, & Klitzing HA (2014). Imaging lipids with secondary ion mass spectrometry. Biochimica et Biophysica Acta, 1841(8), 1108–1119. 10.1016/j.bbalip.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 65.Chen LJ, Shah SS, Silangcruz J, Eller MJ, Verkhoturov SV, Revzin A, et al. (2011). Characterization and quantification of nanoparticle-antibody conjugates on cells using C(60) ToF SIMS in the event-by-event bombardment/detection mode. International Journal of Mass Spectrometry, 303(2–3), 97–102. 10.1016/j.ijms.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Debord JD, Della-Negra S, Fernandez-Lima FA, Verkhoturov SV, & Schweikert EA (2012). Bi-directional ion emission from massive gold cluster impacts on nanometric carbon foils. The Journal of Physical Chemistry. C, Nanomaterials and Interfaces, 116(14), 8138–8144. 10.1021/jp212126m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eller MJ, Verkhoturov SV, & Schweikert EA (2016). Testing molecular homogeneity at the nanoscale with massive cluster secondary ion mass spectrometry. Analytical Chemistry, 88(15), 7639–7646. 10.1021/acs.analchem.6b01466. [DOI] [PubMed] [Google Scholar]
- 68.Liang CK, Verkhoturov SV, Bisrat Y, Dikler S, Debord JD, Fernandez-Lima FA, et al. (2013). Characterization of individual nano-objects with nanoprojectile-SIMS. Surface and Interface Analysis, 45(1), 329–332. 10.1002/sia.5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Verkhoturov SV, Eller MJ, Rickman RD, Della-Negra S, & Schweikert EA (2010). Single impacts of C60 on solids: emission of electrons, ions and prospects for surface mapping. The Journal of Physical Chemistry C, 114(12), 5637–5644. 10.1021/jp9073703. [DOI] [Google Scholar]
- 70.Fernandez-Lima FA, Post J, DeBord JD, Eller MJ, Verkhoturov SV, Della-Negra S, et al. (2011). Analysis of native biological surfaces using a 100 kV massive gold cluster source. Analytical Chemistry, 83(22), 8448–8453. 10.1021/ac201481r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krager KJ, Sarkar M, Twait EC, Lill NL, & Koland JG (2012). A novel biotinylated lipid raft reporter for electron microscopic imaging of plasma membrane microdomains. Journal of Lipid Research, 53(10), 2214–2225. 10.1194/jlr.D026468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Head BP, Patel HH, Roth DM, Murray F, Swaney JS, Niesman IR, et al. (2006). Microtubules and actin microfilaments regulate lipid raft/caveolae localization of adenylyl cyclase signaling components. The Journal of Biological Chemistry, 281(36), 26391–26399. 10.1074/jbc.M602577200. [DOI] [PubMed] [Google Scholar]
- 73.Klymchenko AS, & Kreder R (2014). Fluorescent probes for lipid rafts: from model membranes to living cells. Chemistry & Biology, 21(1), 97–113. 10.1016/j.chembiol.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 74.Ohno-Iwashita Y, Shimada Y, Waheed AA, Hayashi M, Inomata M, Nakamura M, et al. (2004). Perfringolysin O, a cholesterol-binding cytolysin, as a probe for lipid rafts. Anaerobe, 10(2), 125–134. 10.1016/j.anaerobe.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 75.Skocaj M, Resnik N, Grundner M, Ota K, Rojko N, Hodnik V, et al. (2014). Tracking cholesterol/sphingomyelin-rich membrane domains with the ostreolysin A-mCherry protein. PLoS One, 9(3), e92783 10.1371/journal.pone.0092783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yamaji A, Sekizawa Y, Emoto K, Sakuraba H, Inoue K, Kobayashi H, et al. (1998). Lysenin, a novel sphingomyelin-specific binding protein. The Journal of Biological Chemistry, 273(9), 5300–5306. [DOI] [PubMed] [Google Scholar]
- 77.Bhat HB, Kishimoto T, Abe M, Makino A, Inaba T, Murate M, et al. (2013). Binding of a pleurotolysin ortholog from Pleurotus eryngii to sphingomyelin and cholesterol-rich membrane domains. Journal of Lipid Research, 54(10), 2933–2943. 10.1194/jlr.D041731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baumgart T, Hunt G, Farkas ER, Webb WW, & Feigenson GW (2007). Fluorescence probe partitioning between Lo/Ld phases in lipid membranes. Biochimica et Biophysica Acta, 1768(9), 2182–2194. 10.1016/j.bbamem.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Prior IA, Harding A, Yan J, Sluimer J, Parton RG, & Hancock JF (2001). GTP-dependent segregation of H-ras from lipid rafts is required for biological activity. Nature Cell Biology, 3(4), 368–375. 10.1038/35070050. [DOI] [PubMed] [Google Scholar]
- 80.Mariani MM, Lampen P, Popp J, Wood BR, & Deckert V (2009). Impact of fixation on in vitro cell culture lines monitored with Raman spectroscopy. Analyst, 134(6), 1154–1161. 10.1039/b822408k. [DOI] [PubMed] [Google Scholar]
- 81.Schaepe K, Kokesch-Himmelreich J, Rohnke M, Wagner AS, Schaaf T, Wenisch S, et al. (2015). Assessment of different sample preparation routes for mass spectrometric monitoring and imaging of lipids in bone cells via ToF-SIMS. Biointerphases, 10(1), 019016 10.1116/1.4915263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dietrich C, Bagatolli LA, Volovyk ZN, Thompson NL, Levi M, Jacobson K, et al. (2001). Lipid rafts reconstituted in model membranes. Biophysical Journal, 80(3), 1417–1428. 10.1016/S0006-3495(01)76114-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jin L, Millard AC, Wuskell JP, Dong X, Wu D, Clark HA, et al. (2006). Characterization and application of a new optical probe for membrane lipid domains. Biophysical Journal, 90(7), 2563–2575. 10.1529/biophysj.105.072884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kwiatek JM, Owen DM, Abu-Siniyeh A, Yan P, Loew LM, & Gaus K (2013). Characterization of a new series of fluorescent probes for imaging membrane order. PLoS One, 8(2), e52960 10.1371/journal.pone.0052960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Loew LM, Cohen LB, Dix J, Fluhler EN, Montana V, Salama G, et al. (1992). A naphthyl analog of the aminostyryl pyridinium class of potentiometric membrane dyes shows consistent sensitivity in a variety of tissue, cell, and model membrane preparations. The Journal of Membrane Biology, 130(1), 1–10. [DOI] [PubMed] [Google Scholar]
- 86.Sezgin E, Sadowski T, & Simons K (2014). Measuring lipid packing of model and cellular membranes with environment sensitive probes. Langmuir, 30(27), 8160–8166. 10.1021/la501226v. [DOI] [PubMed] [Google Scholar]
- 87.Owen DM, Rentero C, Magenau A, Abu-Siniyeh A, & Gaus K (2011). Quantitative imaging of membrane lipid order in cells and organisms. Nature Protocols, 7(1), 24–35. 10.1038/nprot.2011.419. [DOI] [PubMed] [Google Scholar]
- 88.Schilling T, & Eder C (2010). Importance of lipid rafts for lysophosphatidylcholine-induced caspase-1 activation and reactive oxygen species generation. Cellular Immunology, 265(2), 87–90. 10.1016/j.cellimm.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 89.Bang B, Gniadecki R, & Gajkowska B (2005). Disruption of lipid rafts causes apoptotic cell death in HaCaT keratinocytes. Experimental Dermatology, 14(4), 266–272. 10.1111/j.0906-6705.2005.00283.x. [DOI] [PubMed] [Google Scholar]
- 90.Saghy E, Szoke E, Payrits M, Helyes Z, Borzsei R, Erostyak J, et al. (2015). Evidence for the role of lipid rafts and sphingomyelin in Ca2+-gating of transient receptor potential channels in trigeminal sensory neurons and peripheral nerve terminals. Pharmacological Research, 100, 101–116. 10.1016/j.phrs.2015.07.028. [DOI] [PubMed] [Google Scholar]
- 91.Schwan C, Nolke T, Kruppke AS, Schubert DM, Lang AE, & Aktories K (2011). Cholesterol- and sphingolipid-rich microdomains are essential for microtubule-based membrane protrusions induced by clostridium difficile transferase (CDT). The Journal of Biological Chemistry, 286(33), 29356–29365. 10.1074/jbc.M111.261925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zidovetzki R, & Levitan I (2007). Use of cyclodextrins to manipulate plasma membrane cholesterol content: evidence, misconceptions and control strategies. Biochimica et Biophysica Acta, 1768(6), 1311–1324. 10.1016/j.bbamem.2007.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Linetti A, Fratangeli A, Taverna E, Valnegri P, Francolini M, Cappello V, et al. (2010). Cholesterol reduction impairs exocytosis of synaptic vesicles. Journal of Cell Science, 123(Pt 4), 595–605. 10.1242/jcs.060681. [DOI] [PubMed] [Google Scholar]
- 94.Voss KA, Liu J, Anderson SP, Dunn C, Miller JD, Owen JR, et al. (2006). Toxic effects of fumonisin in mouse liver are independent of the peroxisome proliferator-activated receptor alpha. Toxicological Sciences, 89(1), 108–119. 10.1093/toxsci/kfj019. [DOI] [PubMed] [Google Scholar]
- 95.Contreras FX, Villar AV, Alonso A, Kolesnick RN, & Goni FM (2003). Sphingomyelinase activity causes transbilayer lipid translocation in model and cell membranes. The Journal of Biological Chemistry, 278(39), 37169–37174. 10.1074/jbc.M303206200. [DOI] [PubMed] [Google Scholar]
- 96.Merrill AH Jr., Sullards MC, Wang E, Voss KA, & Riley RT (2001). Sphingolipid metabolism: roles in signal transduction and disruption by fumonisins. Environmental Health Perspectives, 109(Suppl 2), 283–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Arita Y, Nishimura S, Ishitsuka R, Kishimoto T, Ikenouchi J, Ishii K, et al. (2015). Targeting cholesterol in a liquid-disordered environment by theonellamides modulates cell membrane order and cell shape. Chemistry & Biology, 22(5), 604–610. 10.1016/j.chembiol.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 98.Pike LJ (2004). Lipid rafts: heterogeneity on the high seas. The Biochemical Journal, 378(Pt 2), 281–292. 10.1042/BJ20031672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mishra S, & Joshi PG (2007). Lipid raft heterogeneity: an enigma. Journal of Neurochemistry, 103(Suppl 1), 135–142. 10.1111/j.1471-4159.2007.04720.x. [DOI] [PubMed] [Google Scholar]
- 100.Hancock JF (2006). Lipid rafts: contentious only from simplistic standpoints. Nature Reviews. Molecular Cell Biology, 7(6), 456–462. 10.1038/nrm1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stockl M, Plazzo AP, Korte T, & Herrmann A (2008). Detection of lipid domains in model and cell membranes by fluorescence lifetime imaging microscopy of fluorescent lipid analogues. The Journal of Biological Chemistry, 283(45), 30828–30837. 10.1074/jbc.M801418200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kusumi A, Koyama-Honda I, & Suzuki K (2004). Molecular dynamics and interactions for creation of stimulation-induced stabilized rafts from small unstable steady-state rafts. Traffic, 5(4), 213–230. 10.1111/j.1600-0854.2004.0178.x. [DOI] [PubMed] [Google Scholar]
- 103.Reya T, & Clevers H (2005). Wnt signalling in stem cells and cancer. Nature, 434(7035), 843–850. 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 104.Clevers H (2006). Wnt/beta-catenin signaling in development and disease. Cell, 127(3), 469–480. 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 105.Chen Q, Su Y, Wesslowski J, Hagemann AI, Ramialison M, Wittbrodt J, et al. (2014). Tyrosine phosphorylation of LRP6 by Src and Fer inhibits Wnt/beta-catenin signalling. EMBO Reports, 15(12), 1254–1267. 10.15252/embr.201439644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hagemann AI, Kurz J, Kauffeld S, Chen Q, Reeves PM, Weber S, et al. (2014). In vivo analysis of formation and endocytosis of the Wnt/beta-catenin signaling complex in zebrafish embryos. Journal of Cell Science, 127(Pt 18), 3970–3982. 10.1242/jcs.148767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cervenka I, Wolf J, Masek J, Krejci P, Wilcox WR, Kozubik A, et al. (2011). Mitogen-activated protein kinases promote WNT/beta-catenin signaling via phosphorylation of LRP6. Molecular and Cellular Biology, 31(1), 179–189. 10.1128/MCB.00550-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Henriquez JP, Webb A, Bence M, Bildsoe H, Sahores M, Hughes SM, et al. (2008). Wnt signaling promotes AChR aggregation at the neuromuscular synapse in collaboration with agrin. Proceedings of the National Academy of Sciences of the United States of America, 105(48), 18812–18817. 10.1073/pnas.0806300105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kim I, Pan W, Jones SA, Zhang Y, Zhuang X, & Wu D (2013). Clathrin and AP2 are required for PtdIns(4,5)P2-mediated formation of LRP6 signalosomes. The Journal of Cell Biology, 200(4), 419–428. 10.1083/jcb.201206096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schwarz-Romond T, Fiedler M, Shibata N, Butler PJ, Kikuchi A, Higuchi Y, et al. (2007). The DIX domain of Dishevelled confers Wnt signaling by dynamic polymerization. Nature Structural & Molecular Biology, 14(6), 484–492. 10.1038/nsmb1247. [DOI] [PubMed] [Google Scholar]
- 111.Metcalfe C, Mendoza-Topaz C, Mieszczanek J, & Bienz M (2010). Stability elements in the LRP6 cytoplasmic tail confer efficient signalling upon DIX-dependent polymerization. Journal of Cell Science, 123(Pt 9), 1588–1599. 10.1242/jcs.067546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.MacDonald BT, Yokota C, Tamai K, Zeng X, & He X (2008). Wnt signal amplification via activity, cooperativity, and regulation of multiple intracellular PPPSP motifs in the Wnt coreceptor LRP6. The Journal of Biological Chemistry, 283(23), 16115–16123. 10.1074/jbc.M800327200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.van Amerongen R, & Nusse R (2009). Towards an integrated view of Wnt signaling in development. Development, 136(19), 3205–3214. 10.1242/dev.033910. [DOI] [PubMed] [Google Scholar]
- 114.Mii Y, Yamamoto T, Takada R, Mizumoto S, Matsuyama M, Yamada S, et al. (2017). Roles of two types of heparan sulfate clusters in Wnt distribution and signaling in Xenopus. Nature Communications, 8(1), 1973 10.1038/s41467-017-02076-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Özhan G, Sezgin E, Wehner D, Pfister AS, Kühl SJ, Kagermeier-Schenk B, et al. (2013). Lypd6 enhances Wnt/β-catenin signaling by promoting Lrp6 phosphorylation in raft plasma membrane domains. Developmental Cell, 26(4), 331–345. 10.1016/j.devcel.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 116.Schneider F, Waithe D, Clausen MP, Galiani S, Koller T, Ozhan G, et al. (2017). Diffusion of lipids and GPI-anchored proteins in actin-free plasma membrane vesicles measured by STED-FCS. Molecular Biology of the Cell, 28(11), 1507–1518. 10.1091/mbc.E16-07-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Davy A, Gale NW, Murray EW, Klinghoffer RA, Soriano P, Feuerstein C, et al. (1999). Compartmentalized signaling by GPI-anchored ephrin-A5 requires the Fyn tyrosine kinase to regulate cellular adhesion. Genes & Development, 13(23), 3125–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Saha S, Anilkumar AA, & Mayor S (2016). GPI-anchored protein organization and dynamics at the cell surface. Journal of Lipid Research, 57(2), 159–175. 10.1194/jlr.R062885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Harder T, Scheiffele P, Verkade P, & Simons K (1998). Lipid domain structure of the plasma membrane revealed by patching of membrane components. The Journal of Cell Biology, 141(4), 929–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yamamoto H, Sakane H, Yamamoto H, Michiue T, & Kikuchi A (2008). Wnt3a and Dkk1 regulate distinct internalization pathways of LRP6 to tune the activation of β-catenin signaling. Developmental Cell, 15(1), 37–48. 10.1016/j.devcel.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 121.Sakane H, Yamamoto H, & Kikuchi A (2010). LRP6 is internalized by Dkk1 to suppress its phosphorylation in the lipid raft and is recycled for reuse. Journal of Cell Science, 123(3), 360–368. 10.1242/jcs.058008. [DOI] [PubMed] [Google Scholar]
- 122.Lucken-Ardjomande S, Montessuit S, & Martinou JC (2008). Bax activation and stress-induced apoptosis delayed by the accumulation of cholesterol in mitochondrial membranes. Cell Death and Differentiation, 15(3), 484–493. 10.1038/sj.cdd.4402280. [DOI] [PubMed] [Google Scholar]
- 123.Pan W, Choi SC, Wang H, Qin Y, Volpicelli-Daley L, Swan L, et al. (2008). Wnt3a-mediated formation of phosphatidylinositol 4,5-bisphosphate regulates LRP6 phosphorylation. Science, 321(5894), 1350–1353. 10.1126/science.1160741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhou Y, & Hancock JF (2015). Ras nanoclusters: versatile lipid-based signaling platforms. Biochimica et Biophysica Acta, 1853(4), 841–849. 10.1016/j.bbamcr.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 125.Plowman SJ, Muncke C, Parton RG, & Hancock JF (2005). H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proceedings of the National Academy of Sciences of the United States of America, 102(43), 15500–15505. 10.1073/pnas.0504114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Murakoshi H, Iino R, Kobayashi T, Fujiwara T, Ohshima C, Yoshimura A, et al. (2004). Single-molecule imaging analysis of Ras activation in living cells. Proceedings of the National Academy of Sciences of the United States of America, 101(19), 7317–7322. 10.1073/pnas.0401354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Abankwa D, Gorfe AA, & Hancock JF (2007). Ras nanoclusters: molecular structure and assembly. Seminars in Cell & Developmental Biology, 18(5), 599–607. 10.1016/j.semcdb.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Abankwa D, Gorfe AA, & Hancock JF (2008). Mechanisms of Ras membrane organization and signalling: Ras on a rocker. Cell Cycle, 7(17), 2667–2673. 10.4161/cc.7.17.6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Guzman C, Solman M, Ligabue A, Blazevits O, Andrade DM, Reymond L, et al. (2014). The efficacy of Raf kinase recruitment to the GTPase H-ras depends on H-ras membrane conformer-specific nanoclustering. The Journal of Biological Chemistry, 289(14), 9519–9533. 10.1074/jbc.M113.537001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhou Y, Liang H, Rodkey T, Ariotti N, Parton RG, & Hancock JF (2014). Signal integration by lipid-mediated spatial cross talk between Ras nanoclusters. Molecular and Cellular Biology, 34(5), 862–876. 10.1128/MCB.01227-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Plowman SJ, Ariotti N, Goodall A, Parton RG, & Hancock JF (2008). Electrostatic interactions positively regulate K-Ras nanocluster formation and function. Molecular and Cellular Biology, 28(13), 4377–4385. 10.1128/MCB.00050-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sezgin E, Azbazdar Y, Ng XW, Teh C, Simons K, Weidinger G, et al. (2017). Binding of canonical Wnt ligands to their receptor complexes occurs in ordered plasma membrane environments. The FEBS Journal, 284(15), 2513–2526. 10.1111/febs.14139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yamamoto H, Komekado H, & Kikuchi A (2006). Caveolin is necessary for Wnt-3a-dependent internalization of LRP6 and accumulation of β-catenin. Developmental Cell, 11(2), 213–223. 10.1016/j.devcel.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 134.Sheng R, Kim H, Lee H, Xin Y, Chen Y, Tian W, et al. (2014). Cholesterol selectively activates canonical Wnt signalling over non-canonical Wnt signalling. Nature Communications, 5, 4393 10.1038/ncomms5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hofman EG, Ruonala MO, Bader AN, van den Heuvel D, Voortman J, Roovers RC, et al. (2008). EGF induces coalescence of different lipid rafts. Journal of Cell Science, 121 (Pt 15), 2519–2528. 10.1242/jcs.028753. [DOI] [PubMed] [Google Scholar]
- 136.Hofman EG, Bader AN, Gerritsen HC, & van Bergen En Henegouwen PM (2009). EGF induces rapid reorganization of plasma membrane microdomains. Communicative & Integrative Biology, 2(3), 213–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nishioka T, Frohman MA, Matsuda M, & Kiyokawa E (2010). Heterogeneity of phosphatidic acid levels and distribution at the plasma membrane in living cells as visualized by a Foster resonance energy transfer (FRET) biosensor. The Journal of Biological Chemistry, 285(46), 35979–35987. 10.1074/jbc.M110.153007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhao C, Du G, Skowronek K, Frohman MA, & Bar-Sagi D (2007). Phospholipase D2-generated phosphatidic acid couples EGFR stimulation to Ras activation by Sos. Nature Cell Biology, 9(6), 706–712. 10.1038/ncb1594. [DOI] [PubMed] [Google Scholar]
- 139.Zhang F, Wang Z, Lu M, Yonekubo Y, Liang X, Zhang Y, et al. (2014). Temporal production of the signaling lipid phosphatidic acid by phospholipase D2 determines the output of extracellular signal-regulated kinase signaling in cancer cells. Molecular and Cellular Biology, 34(1), 84–95. 10.1128/MCB.00987-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Honda A, Nogami M, Yokozeki T, Yamazaki M, Nakamura H, Watanabe H, et al. (1999). Phosphatidylinositol 4-phosphate 5-kinase alpha is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell, 99(5), 521–532. [DOI] [PubMed] [Google Scholar]
- 141.Michailidis IE, Rusinova R, Georgakopoulos A, Chen Y, Iyengar R, Robakis NK, et al. (2011). Phosphatidylinositol-4, 5-bisphosphate regulates epidermal growth factor receptor activation. Pflügers Archiv, 461(3), 387–397. 10.1007/s00424-010-0904-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ariotti N, Liang H, Xu Y, Zhang Y, Yonekubo Y, Inder K,et al. (2010). Epidermal growth factor receptor activation remodels the plasma membrane lipid environment to induce nanocluster formation. Molecular and Cellular Biology, 30(15), 3795–3804. 10.1128/MCB.01615-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang Y, Gao J, Guo X, Tong T, Shi X, Li L, et al. (2014). Regulation of EGFR nanocluster formation by ionic protein-lipid interaction. Cell Research, 24(8), 959–976. 10.1038/cr.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gureasko J, Kuchment O, Makino DL, Sondermann H, Bar-Sagi D, & Kuriyan J (2010). Role of the histone domain in the autoinhibition and activation of the Ras activator son of sevenless. Proceedings of the National Academy of Sciences of the United States of America, 107(8), 3430–3435. 10.1073/pnas.0913915107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yadav KK, & Bar-Sagi D (2010). Allosteric gating of son of sevenless activity by the histone domain. Proceedings of the National Academy of Sciences of the United States of America, 107(8), 3436–3440. 10.1073/pnas.0914315107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Abankwa D, Hanzal-Bayer M, Ariotti N, Plowman SJ, Gorfe AA, Parton RG, et al. (2008). A novel switch region regulates H-ras membrane orientation and signal output. The EMBO Journal, 27(5), 727–735. 10.1038/emboj.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Abankwa D, Gorfe AA, Inder K, & Hancock JF (2010). Ras membrane orientation and nanodomain localization generate isoform diversity. Proceedings of the National Academy of Sciences of the United States of America, 107(3), 1130–1135. 10.1073/pnas.0903907107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tian T, Harding A, Inder K, Plowman S, Parton RG, & Hancock JF (2007). Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nature Cell Biology, 9(8), 905–914. 10.1038/ncb1615. [DOI] [PubMed] [Google Scholar]
- 149.van Rheenen J, Song X, van Roosmalen W, Cammer M, Chen X, Desmarais V, et al. (2007). EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. The Journal of Cell Biology, 179(6), 1247–1259. 10.1083/jcb.200706206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Roach AN, Wang Z, Wu P, Zhang F, Chan RB, Yonekubo Y, et al. (2012). Phosphatidic acid regulation of PIPKI is critical for actin cytoskeletal reorganization. Journal of Lipid Research, 53(12), 2598–2609. 10.1194/jlr.M028597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pleskot R, Li J, Zarsky V, Potocky M, & Staiger CJ (2013). Regulation of cytoskeletal dynamics by phospholipase D and phosphatidic acid. Trends in Plant Science, 18(9), 496–504. 10.1016/j.tplants.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 152.Wu M, Holowka D, Craighead HG, & Baird B (2004). Visualization of plasma membrane compartmentalization with patterned lipid bilayers. Proceedings of the National Academy of Sciences of the United States of America, 101(38), 13798–13803. 10.1073/pnas.0403835101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pike LJ, Han X, & Gross RW (2005). Epidermal growth factor receptors are localized to lipid rafts that contain a balance of inner and outer leaflet lipids: a shotgun lipidomics study. The Journal of Biological Chemistry, 280(29), 26796–26804. 10.1074/jbc.M503805200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kabouridis PS (2006). Lipid rafts in T cell receptor signalling. Molecular Membrane Biology, 23(1), 49–57. 10.1080/09687860500453673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pierce SK (2002). Lipid rafts and B-cell activation. Nature Reviews. Immunology, 2(2), 96–105. 10.1038/nri726. [DOI] [PubMed] [Google Scholar]
- 156.Dinic J, Riehl A, Adler J, & Parmryd I (2015). The T cell receptor resides in ordered plasma membrane nanodomains that aggregate upon patching of the receptor. Scientific Reports, 5, 10082 10.1038/srep10082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Morino-Koga S, Yano S, Kondo T, Shimauchi Y, Matsuyama S, Okamoto Y, et al. (2013). Insulin receptor activation through its accumulation in lipid rafts by mild electrical stress. Journal of Cellular Physiology, 228(2), 439–446. 10.1002/jcp.24149. [DOI] [PubMed] [Google Scholar]
- 158.Pike LJ (2005). Growth factor receptors, lipid rafts and caveolae: an evolving story. Biochimica et Biophysica Acta, 1746(3), 260–273. 10.1016/j.bbamcr.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 159.Bunnell SC, Hong DI, Kardon JR, Yamazaki T, McGlade CJ, Barr VA, et al. (2002). T cell receptor ligation induces the formation of dynamically regulated signaling assemblies. The Journal of Cell Biology, 158(7), 1263–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Douglass AD, & Vale RD (2005). Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell, 121 (6), 937–950. 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Winter PW, Van Orden AK, Roess DA, & Barisas BG (2012). Actin-dependent clustering of insulin receptors in membrane microdomains. Biochimica et Biophysica Acta, 1818(3), 467–473. 10.1016/j.bbamem.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 162.Dessi S, Batetta B, Pulisci D, Spano O, Anchisi C, Tessitore L, et al. (1994). Cholesterol content in tumor tissues is inversely associated with high-density lipoprotein cholesterol in serum in patients with gastrointestinal cancer. Cancer, 73(2), 253–258. [DOI] [PubMed] [Google Scholar]
- 163.Kolanjiappan K, Ramachandran CR, & Manoharan S (2003). Biochemical changes in tumor tissues of oral cancer patients. Clinical Biochemistry, 36(1), 61–65. [DOI] [PubMed] [Google Scholar]
- 164.Montero J, Morales A, Llacuna L, Lluis JM, Terrones O, Basanez G, et al. (2008). Mitochondrial cholesterol contributes to chemotherapy resistance in hepatocellular carcinoma. Cancer Research, 68(13), 5246–5256. 10.1158/0008-5472.CAN-07-6161. [DOI] [PubMed] [Google Scholar]
- 165.Li YC, Park MJ, Ye SK, Kim CW, & Kim YN (2006). Elevated levels of cholesterol-rich lipid rafts in cancer cells are correlated with apoptosis sensitivity induced by cholesterol-depleting agents. The American Journal of Pathology, 168(4), 1107–1118; quiz 1404-1105. 10.2353/ajpath.2006.050959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Alves AC, Ribeiro D, Nunes C, & Reis S (2016). Biophysics in cancer: the relevance of drug-membrane interaction studies. Biochimica et Biophysica Acta, 1858(9), 2231–2244. 10.1016/j.bbamem.2016.06.025. [DOI] [PubMed] [Google Scholar]
- 167.Lange Y, Swaisgood MH, Ramos BV, & Steck TL (1989). Plasma membranes contain half the phospholipid and 90% of the cholesterol and sphingomyelin in cultured human fibroblasts. The Journal of Biological Chemistry, 264(7), 3786–3793. [PubMed] [Google Scholar]
- 168.Ray TK, Skipski VP, Barclay M, Essner E, & Archibald FM (1969). Lipid composition of rat liver plasma membranes. The Journal of Biological Chemistry, 244(20), 5528–5536. [PubMed] [Google Scholar]
- 169.Das A, Brown MS, Anderson DD, Goldstein JL, & Radhakrishnan A (2014). Three pools of plasma membrane cholesterol and their relation to cholesterol homeostasis. Elife, 3 10.7554/eLife.02882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cancer Genome Atlas Research, N., Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, et al. (2013). The Cancer Genome Atlas Pan-Cancer analysis project. Nature Genetics, 45(10), 1113–1120. 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ribas V, Garcia-Ruiz C, & Fernandez-Checa JC (2016). Mitochondria, cholesterol and cancer cell metabolism. Clinical and Translational Medicine, 5(1), 22 10.1186/s40169-016-0106-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wang B, Rong X, Palladino END, Wang J, Fogelman AM, Martin MG, et al. (2018). Phospholipid remodeling and cholesterol availability regulate intestinal stemness and tumorigenesis. Cell Stem Cell, 22(2), 206–220 e204. 10.1016/j.stem.2017.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kuzu OF, Noory MA, & Robertson GP (2016). The role of cholesterol in cancer. Cancer Research, 76(8), 2063–2070. 10.1158/0008-5472.CAN-15-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Irwin ME, Mueller KL, Bohin N, Ge Y, & Boerner JL (2011). Lipid raft localization of EGFR alters the response of cancer cells to the EGFR tyrosine kinase inhibitor gefitinib. Journal of Cellular Physiology, 226(9), 2316–2328. 10.1002/jcp.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gao J, Wang Y, Cai M, Pan Y, Xu H, Jiang J, et al. (2015). Mechanistic insights into EGFR membrane clustering revealed by super-resolution imaging. Nanoscale, 7(6), 2511–2519. 10.1039/c4nr04962d. [DOI] [PubMed] [Google Scholar]
- 176.Dow LE, O'Rourke KP, Simon J, Tschaharganeh DF, van Es JH, Clevers H, et al. (2015). Apc restoration promotes cellular differentiation and reestablishes crypt homeostasis in colorectal cancer. Cell, 161(7), 1539–1552. 10.1016/j.cell.2015.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Halvey PJ, Zhang B, Coffey RJ, Liebler DC, & Slebos RJ (2012). Proteomic consequences of a single gene mutation in a colorectal cancer model. Journal of Proteome Research, 11(2), 1184–1195. 10.1021/pr2009109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Allott EH, Howard LE, Cooperberg MR, Kane CJ, Aronson WJ, Terris MK, et al. (2014). Serum lipid profile and risk of prostate cancer recurrence: results from the SEARCH database. Cancer Epidemiology, Biomarkers & Prevention, 23(11), 2349–2356. 10.1158/1055-9965.EPI-14-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kitahara CM, Berrington de Gonzalez A, Freedman ND, Huxley R, Mok Y, Jee SH, et al. (2011). Total cholesterol and cancer risk in a large prospective study in Korea. Journal of Clinical Oncology, 29(12), 1592–1598. 10.1200/JCO.2010.31.5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wulaningsih W, Vahdaninia M, Rowley M, Holmberg L, Garmo H, Malmstrom H, et al. (2015). Prediagnostic serum glucose and lipids in relation to survival in breast cancer patients: a competing risk analysis. BMC Cancer, 15, 913 10.1186/s12885-015-1928-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ulmer H, Borena W, Rapp K, Klenk J, Strasak A, Diem G, et al. (2009). Serum triglyceride concentrations and cancer risk in a large cohort study in Austria. British Journal of Cancer, 101(7), 1202–1206. 10.1038/sj.bjc.6605264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yamada K, Araki S, Tamura M, Sakai I, Takahashi Y, Kashihara H, et al. (1998). Relation of serum total cholesterol, serum triglycerides and fasting plasma glucose to colorectal carcinoma in situ. International Journal of Epidemiology, 27(5), 794–798. [DOI] [PubMed] [Google Scholar]
- 183.Jacobs EJ, Newton CC, Thun MJ, & Gapstur SM (2011). Long-term use of cholesterol-lowering drugs and cancer incidence in a large United States cohort. Cancer Research, 71(5), 1763–1771. 10.1158/0008-5472.CAN-10-2953. [DOI] [PubMed] [Google Scholar]
- 184.Murtola TJ, Visvanathan K, Artama M, Vainio H, & Pukkala E (2014). Statin use and breast cancer survival: a nationwide cohort study from Finland. PLoS One, 9(10), e110231 10.1371/journal.pone.0110231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Cardwell CR, Hicks BM, Hughes C, & Murray LJ (2014). Statin use after colorectal cancer diagnosis and survival: a population-based cohort study. Journal of Clinical Oncology, 32(28), 3177–3183. 10.1200/JCO.2013.54.4569. [DOI] [PubMed] [Google Scholar]
- 186.Nielsen SF, Nordestgaard BG, & Bojesen SE (2012). Statin use and reduced cancer-related mortality. The New England Journal of Medicine, 367(19), 1792–1802. 10.1056/NEJMoa1201735. [DOI] [PubMed] [Google Scholar]
- 187.Smith B, & Land H (2012). Anticancer activity of the cholesterol exporter ABCA1 gene. Cell Reports, 2(3), 580–590. 10.1016/j.celrep.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Krycer JR, & Brown AJ (2013). Cholesterol accumulation in prostate cancer: a classic observation from a modern perspective. Biochimica et Biophysica Acta, 1835(2), 219–229. 10.1016/j.bbcan.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 189.Casey SC, Amedei A, Aquilano K, Azmi AS, Benencia F, Bhakta D, et al. (2015). Cancer prevention and therapy through the modulation of the tumor microenvironment. Seminars in Cancer Biology, (35 Suppl), S199–S223. 10.1016/j.semcancer.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kawata S, Takaishi K, Nagase T, Ito N, Matsuda Y, Tamura S, et al. (1990). Increase in the active form of 3-hydroxy-3-methylglutaryl coenzyme A reductase in human hepatocellular carcinoma: possible mechanism for alteration of cholesterol biosynthesis. Cancer Research, 50(11), 3270–3273. [PubMed] [Google Scholar]
- 191.Harwood HJ Jr., Alvarez IM, Noyes WD, & Stacpoole PW (1991). In vivo regulation of human leukocyte 3-hydroxy-3-methylglutaryl coenzyme A reductase: increased enzyme protein concentration and catalytic efficiency in human leukemia and lymphoma. Journal of Lipid Research, 32(8), 1237–1252. [PubMed] [Google Scholar]
- 192.Caruso MG, Notarnicola M, Santillo M, Cavallini A, & Di Leo A (1999). Enhanced 3-hydroxy-3-methyl-glutaryl coenzyme A reductase activity in human colorectal cancer not expressing low density lipoprotein receptor. Anticancer Research, 19(1A), 451–454. [PubMed] [Google Scholar]
- 193.Wong WW, Dimitroulakos J, Minden MD, & Penn LZ (2002). HMG-CoA reductase inhibitors and the malignant cell: the statin family of drugs as triggers of tumor-specific apoptosis. Leukemia, 16(4), 508–519. 10.1038/sj.leu.2402476. [DOI] [PubMed] [Google Scholar]
- 194.Guillaumond F, Bidaut G, Ouaissi M, Servais S, Gouirand V, Olivares O, et al. (2015). Cholesterol uptake disruption, in association with chemotherapy, is a promising combined metabolic therapy for pancreatic adenocarcinoma. Proceedings of the National Academy of Sciences of the United States of America, 112(8), 2473–2478. 10.1073/pnas.1421601112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hilvo M, Denkert C, Lehtinen L, Muller B, Brockmoller S, Seppanen-Laakso T, et al. (2011). Novel theranostic opportunities offered by characterization of altered membrane lipid metabolism in breast cancer progression. Cancer Research, 71(9), 3236–3245. 10.1158/0008-5472.CAN-10-3894. [DOI] [PubMed] [Google Scholar]
- 196.Guenther S, Muirhead LJ, Speller AV, Golf O, Strittmatter N, Ramakrishnan R, et al. (2015). Spatially resolved metabolic phenotyping of breast cancer by desorption electrospray ionization mass spectrometry. Cancer Research, 75(9), 1828–1837. 10.1158/0008-5472.CAN-14-2258. [DOI] [PubMed] [Google Scholar]
- 197.Rysman E, Brusselmans K, Scheys K, Timmermans L, Derua R, Munck S, et al. (2010). De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer Research, 70(20), 8117–8126. 10.1158/0008-5472.CAN-09-3871. [DOI] [PubMed] [Google Scholar]
- 198.Kawashima M, Iwamoto N, Kawaguchi-Sakita N, Sugimoto M, Ueno T, Mikami Y, et al. (2013). High-resolution imaging mass spectrometry reveals detailed spatial distribution of phosphatidylinositols in human breast cancer. Cancer Science, 104(10), 1372–1379. 10.1111/cas.12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Foster DA (2009). Phosphatidic acid signaling to mTOR: signals for the survival of human cancer cells. Biochimica et Biophysica Acta, 1791(9), 949–955. 10.1016/j.bbalip.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Menendez JA, & Lupu R (2007). Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nature Reviews Cancer, 7(10), 763–777. 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 201.Zaidi N, Swinnen JV, & Smans K (2012). ATP-citrate lyase: a key player in cancer metabolism. Cancer Research, 72(15), 3709–3714. 10.1158/0008-5472.CAN-11-4112. [DOI] [PubMed] [Google Scholar]
- 202.Sarker D, Reid AH, Yap TA, & de Bono JS (2009). Targeting the PI3K/AKT pathway for the treatment of prostate cancer. Clinical Cancer Research, 15(15), 4799–4805. 10.1158/1078-0432.CCR-08-0125. [DOI] [PubMed] [Google Scholar]
- 203.Vivanco I, & Sawyers CL (2002). The phosphatidylinositol 3-kinase AKT pathway in human cancer. Nature Reviews Cancer, 2(7), 489–501. 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 204.Podo F, Paris L, Cecchetti S, Spadaro F, Abalsamo L, Ramoni C, et al. (2016). Activation of phosphatidylcholine-specific phospholipase C in breast and ovarian cancer: impact on MRS-detected choline metabolic profile and perspectives for targeted therapy. Frontiers in Oncology, 6, 171 10.3389/fonc.2016.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.El-Mashtoly SF, Yosef HK, Petersen D, Mavarani L, Maghnouj A, Hahn S, et al. (2015). Label-free Raman spectroscopic imaging monitors the integral physiologically relevant drug responses in cancer cells. Analytical Chemistry, 87(14), 7297–7304. 10.1021/acs.analchem.5b01431. [DOI] [PubMed] [Google Scholar]
- 206.Loizides-Mangold U (2013). On the future of mass-spectrometry-based lipidomics. The FEBS Journal, 280(12), 2817–2829. 10.1111/febs.12202. [DOI] [PubMed] [Google Scholar]
- 207.Li J, & Cheng JX (2014). Direct visualization of de novo lipogenesis in single living cells. Scientific Reports, 4, 6807 10.1038/srep06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Le TT, Yue S, & Cheng JX (2010). Shedding new light on lipid biology with coherent anti-Stokes Raman scattering microscopy. Journal of Lipid Research, 51(11), 3091–3102. 10.1194/jlr.R008730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Schieffer D, Naware S, Bakun W, & Bamezai AK (2014). Lipid raft-based membrane order is important for antigen-specific clonal expansion of CD4(+) T lymphocytes. BMC Immunology, 15, 58 10.1186/s12865-014-0058-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.He HT, & Marguet D (2008). T-cell antigen receptor triggering and lipid rafts: a matter of space and time scales. Talking point on the involvement of lipid rafts in T-cell activation. EMBO Reports, 9(6), 525–530. 10.1038/embor.2008.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Zhou Y, Maxwell KN, Sezgin E, Lu M, Liang H, Hancock JF, et al. (2013). Bile acids modulate signaling by functional perturbation of plasma membrane domains. The Journal of Biological Chemistry, 288(50), 35660–35670. 10.1074/jbc.M113.519116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Brown DA, & London E (1998). Structure and origin of ordered lipid domains in biological membranes. The Journal of Membrane Biology, 164(2), 103–114. [DOI] [PubMed] [Google Scholar]
- 213.Bakht O, Pathak P, & London E (2007). Effect of the structure of lipids favoring disordered domain formation on the stability of cholesterol-containing ordered domains (lipid rafts): identification of multiple raft-stabilization mechanisms. Biophysical Journal, 93(12), 4307–4318. 10.1529/biophysj.107.114967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Amaro M, Reina F, Hof M, Eggeling C, & Sezgin E (2017). Laurdan and Di-4-ANEPPDHQ probe different properties of the membrane. Journal of Physics D: Applied Physics, 50(13), 134004 10.1088/1361-6463/aa5dbc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Ramani K, & Balasubramanian SV (2003). Fluorescence properties of Laurdan in cochleate phases. Biochimica et Biophysica Acta, 1618(1), 67–78. [DOI] [PubMed] [Google Scholar]
- 216.Sezgin E, Schneider F, Zilles V, Urbancic I, Garcia E, Waithe D, et al. (2017). Polarity-sensitive probes for superresolution stimulated emission depletion microscopy. Biophysical Journal, 113(6), 1321–1330. 10.1016/j.bpj.2017.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Parasassi T, De Stasio G, d’Ubaldo A, & Gratton E (1990). Phase fluctuation in phospholipid membranes revealed by Laurdan fluorescence. Biophysical Journal, 57(6), 1179–1186. 10.1016/S0006-3495(90)82637-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Gaus K, Zech T, & Harder T (2006). Visualizing membrane microdomains by Laurdan 2-photon microscopy. Molecular Membrane Biology, 23(1), 41–48. 10.1080/09687860500466857. [DOI] [PubMed] [Google Scholar]
- 219.Owen DM, Magenau A, Majumdar A, & Gaus K (2010). Imaging membrane lipid order in whole, living vertebrate organisms. Biophysical Journal, 99(1), L7–L9. 10.1016/j.bpj.2010.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Baumgart T, Hammond AT, Sengupta P, Hess ST, Holowka DA, Baird BA, et al. (2007). Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proceedings of the National Academy of Sciences of the United States of America, 104(9), 3165–3170. 10.1073/pnas.0611357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Sengupta P, Hammond A, Holowka D, & Baird B (2008). Structural determinants for partitioning of lipids and proteins between coexisting fluid phases in giant plasma membrane vesicles. Biochimica et Biophysica Acta, 1778(1), 20–32. 10.1016/j.bbamem.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Owen DM, Oddos S, Kumar S, Davis DM, Neil MA, French PM, et al. (2010). High plasma membrane lipid order imaged at the immunological synapse periphery in live T cells. Molecular Membrane Biology, 27(4–6), 178–189. 10.3109/09687688.2010.495353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Miguel L, Owen DM, Lim C, Liebig C, Evans J, Magee AI, et al. (2011). Primary human CD4+ T cells have diverse levels of membrane lipid order that correlate with their function. Journal of Immunology, 186(6), 3505–3516. 10.4049/jimmunol.1002980. [DOI] [PubMed] [Google Scholar]
- 224.Lingwood D, & Simons K (2010). Lipid rafts as a membraneorganizing principle. Science, 327(5961), 46–50. 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 225.Gowrishankar K, Ghosh S, Saha S C, R., Mayor S, & Rao M (2012). Active remodeling of cortical actin regulates spatiotemporal organization of cell surface molecules. Cell, 149(6), 1353–1367, 10.1016/j.cell.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 226.Ibach J, Radon Y, Gelleri M, Sonntag MH, Brunsveld L, Bastiaens PI, et al. (2015). Single particle tracking reveals that EGFR signaling activity is amplified in Clathrin-coated pits. PLoS One, 10(11), e0143162 10.1371/journal.pone.0143162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Valentine CD, & Haggie PM (2011). Confinement of beta(1)- and beta(2)-adrenergic receptors in the plasma membrane of cardiomyocyte-like H9c2 cells is mediated by selective interactions with PDZ domain and A-kinase anchoring proteins but not caveolae. Molecular Biology of the Cell, 22(16), 2970–2982. 10.1091/mbc.E11-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Jordan S, & Rodgers W (2003). Tcell glycolipid-enriched membrane domains are constitutively assembled as membrane patches that translocate to immune synapses. Journal of Immunology, 171(1), 78–87. [DOI] [PubMed] [Google Scholar]
- 229.Ng XW, Teh C, Korzh V, & Wohland T (2016). The secreted signaling protein Wnt3 is associated with membrane domains in vivo: a SPIM-FCS study. Biophysical Journal, 111(2), 418–429. 10.1016/j.bpj.2016.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Lommerse PH, Snaar-Jagalska BE, Spaink HP, & Schmidt T (2005). Single-molecule diffusion measurements of H-Ras at the plasma membrane of live cells reveal microdomain localization upon activation. Journal of Cell Science, 118(Pt 9), 1799–1809. 10.1242/jcs.02300. [DOI] [PubMed] [Google Scholar]
- 231.Sengupta P, Baird B, & Holowka D (2007). Lipid rafts, fluid/ fluid phase separation, and their relevance to plasma membrane structure and function. Seminars in Cell & Developmental Biology, 18(5), 583–590. 10.1016/j.semcdb.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Haack F, Lemcke H, Ewald R, Rharass T, & Uhrmacher AM (2015). Spatio-temporal model of endogenous ROS and raft-dependent WNT/beta-catenin signaling driving cell fate commitment in human neural progenitor cells. PLoS Computational Biology, 11(3), e1004106 10.1371/journal.pcbi.1004106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Pirruccello J, & Kathiresan S (2010). Genetics of lipid disorders. Current Opinion in Cardiology, 25(3), 238–242. 10.1097/HCO.0b013e328338574d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.De Bont R, & van Larebeke N (2004). Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis, 19(3), 169–185. 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- 235.Llado V, Lopez DJ, Ibarguren M, Alonso M, Soriano JB, Escriba PV, et al. (2014). Regulation of the cancer cell membrane lipid composition by NaCHOleate: effects on cell signaling and therapeutical relevance in glioma. Biochim Biophys Acta, 1838(6), 1619–1627, 10.1016/j.bbamem.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 236.Zalba S, & Ten Hagen TL (2017). Cell membrane modulation as adjuvant in cancer therapy. Cancer Treatment Reviews, 52, 48–57. 10.1016/j.ctrv.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Galeotti T, Borrello S, Minotti G, & Masotti L (1986). Membrane alterations in cancer cells: the role of oxy radicals. Annals of the New York Academy of Sciences, 488, 468–480. [DOI] [PubMed] [Google Scholar]
- 238.Peetla C, Bhave R, Vijayaraghavalu S, Stine A, Kooijman E, & Labhasetwar V (2010). Drug resistance in breast cancer cells: biophysical characterization of and doxorubicin interactions with membrane lipids. Molecular Pharmaceutics, 7(6), 2334–2348. 10.1021/mp100308n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Peetla C, Vijayaraghavalu S, & Labhasetwar V (2013). Biophysics of cell membrane lipids in cancer drug resistance: implications for drug transport and drug delivery with nanoparticles. Advanced Drug Delivery Reviews, 65(13–14), 1686–1698. 10.1016/j.addr.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Niero EL, Rocha-Sales B, Lauand C, Cortez BA, de Souza MM, Rezende-Teixeira P, et al. (2014). The multiple facets of drug resistance: one history, different approaches. Journal of Experimental & Clinical Cancer Research, 33, 37 10.1186/1756-9966-33-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Sherbet GV (1989). Membrane fluidity and cancer metastasis. Experimental Cell Biology, 57(4), 198–205. [DOI] [PubMed] [Google Scholar]
- 242.Sok M, Sentjurc M, Schara M, Stare J, & Rott T (2002). Cell membrane fluidity and prognosis of lung cancer. The Annals of Thoracic Surgery, 73(5), 1567–1571. [DOI] [PubMed] [Google Scholar]
- 243.Campanella R (1992). Membrane lipids modifications in human gliomas of different degree of malignancy. Journal of Neurosurgical Sciences, 36(1), 11–25. [PubMed] [Google Scholar]
- 244.Takahashi K, Hosono M, Sato I, Hata K, Wada T, Yamaguchi K, et al. (2015). Sialidase NEU3 contributes neoplastic potential on colon cancer cells as a key modulator of gangliosides by regulating Wnt signaling. International Journal of Cancer, 137(7), 1560–1573. 10.1002/ijc.29527. [DOI] [PubMed] [Google Scholar]
- 245.Peckys DB, Korf U, & de Jonge N (2015). Local variations of HER2 dimerization in breast cancer cells discovered by correlative fluorescence and liquid electron microscopy. Science Advances, 1(6), e1500165 10.1126/sciadv.1500165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Posada IMD, Lectez B, Sharma M, Oetken-Lindholm C, Yetukuri L, Zhou Y, et al. (2017). Rapalogs can promote cancer cell stemness in vitro in a galectin-1 and H-ras-dependent manner. Oncotarget, 8(27), 44550–44566. 10.18632/oncotarget.17819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Chen J, Liu T, Gao J, Gao L, Zhou L, Cai M, et al. (2016). Variation in carbohydrates between cancer and normal cell membranes revealed by super-resolution fluorescence imaging. Advanced Science (Weinh), 3(12), 1600270 10.1002/advs.201600270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Wu KL, Huang EY, Yeh WL, Hsiao CC, & Kuo CM (2017). Synergistic interaction between galectin-3 and carcinoembryonic antigen promotes colorectal cancer metastasis. Oncotarget, 8(37), 61935–61943. 10.18632/oncotarget.18721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Song S, Ji B, Ramachandran V, Wang H, Hafley M, Logsdon C, et al. (2012). Overexpressed galectin-3 in pancreatic cancer induces cell proliferation and invasion by binding Ras and activating Ras signaling. PLoS One, 7(8), e42699 10.1371/journal.pone.0042699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Liu FT, & Rabinovich GA (2005). Galectins as modulators of tumour progression. Nature Reviews Cancer, 5(1), 29–41. 10.1038/nrc1527. [DOI] [PubMed] [Google Scholar]
- 251.Dennis JW, Nabi IR, & Demetriou M (2009). Metabolism, cell surface organization, and disease. Cell, 139(7), 1229–1241. 10.1016/j.cell.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Ahmad N, Gabius HJ, Andre S, Kaltner H, Sabesan S, Roy R, et al. (2004). Galectin-3 precipitates as a pentamer with synthetic multivalent carbohydrates and forms heterogeneous cross-linked complexes. The Journal of Biological Chemistry, 279(12), 10841–10847. 10.1074/jbc.M312834200. [DOI] [PubMed] [Google Scholar]
- 253.Brewer CF, Miceli MC, & Baum LG (2002). Clusters, bundles, arrays and lattices: novel mechanisms for lectin-saccharide-mediated cellular interactions. Current Opinion in Structural Biology, 12(5), 616–623. [DOI] [PubMed] [Google Scholar]
- 254.Rabinovich GA, Toscano MA, Jackson SS, & Vasta GR (2007). Functions of cell surface galectin-glycoprotein lattices. Current Opinion in Structural Biology, 17(5), 513–520. 10.1016/j.sbi.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Hernandez JD, Nguyen JT, He J, Wang W, Ardman B, Green JM, et al. (2006). Galectin-1 binds different CD43 glycoforms to cluster CD43 and regulate T cell death. Journal of Immunology, 177(8), 5328–5336. [DOI] [PubMed] [Google Scholar]
- 256.Riedl S, Rinner B, Asslaber M, Schaider H, Walzer S, Novak A, et al. (2011). In search of a novel target—phosphatidylserine exposed by non-apoptotic tumor cells and metastases of malignancies with poor treatment efficacy. Biochimica et Biophysica Acta, 1808(11), 2638–2645. 10.1016/j.bbamem.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Birge RB, Boeltz S, Kumar S, Carlson J, Wanderley J, Calianese D, et al. (2016). Phosphatidylserine is a global immunosuppressive signal in efferocytosis, infectious disease, and cancer. Cell Death and Differentiation, 23(6), 962–978. 10.1038/cdd.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Utsugi T, Schroit AJ, Connor J, Bucana CD, & Fidler IJ (1991). Elevated expression of phosphatidylserine in the outer membrane leaflet of human tumor cells and recognition by activated human blood monocytes. Cancer Research, 51(11), 3062–3066. [PubMed] [Google Scholar]
- 259.Wang L, Habib AA, Mintz A, Li KC, & Zhao D (2017). Phosphatidylserine-targeted nanotheranostics for brain tumor imaging and therapeutic potential. Molecular Imaging, 16, 1536012117708722 10.1177/1536012117708722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.He J, Luster TA, & Thorpe PE (2007). Radiation-enhanced vascular targeting of human lung cancers in mice with a monoclonal antibody that binds anionic phospholipids. Clinical Cancer Research, 13(17), 5211–5218. 10.1158/1078-0432.CCR-07-0793. [DOI] [PubMed] [Google Scholar]
- 261.He J, Yin Y, Luster TA, Watkins L, & Thorpe PE (2009). Antiphosphatidylserine antibody combined with irradiation damages tumor blood vessels and induces tumor immunity in a rat model of glioblastoma. Clinical Cancer Research, 15(22), 6871–6880. 10.1158/1078-0432.CCR-09-1499. [DOI] [PubMed] [Google Scholar]
- 262.Huang X, Bennett M, & Thorpe PE (2005). A monoclonal antibody that binds anionic phospholipids on tumor blood vessels enhances the antitumor effect of docetaxel on human breast tumors in mice. Cancer Research, 65(10), 4408–4416. 10.1158/0008-5472.CAN-05-0031. [DOI] [PubMed] [Google Scholar]
- 263.Zhang L, Habib AA, & Zhao D (2016). Phosphatidylserine-targeted liposome for enhanced glioma-selective imaging. Oncotarget, 7(25), 38693–38706, 10.18632/oncotarget.9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Zhou H, Stafford JH, Hallac RR, Zhang L, Huang G, Mason RP, et al. (2014). Phosphatidylserine-targeted molecular imaging of tumor vasculature by magnetic resonance imaging. Journal of Biomedical Nanotechnology, 10(5), 846–855. [DOI] [PubMed] [Google Scholar]
- 265.Zhang L, Zhou H, Belzile O, Thorpe P, & Zhao D (2014). Phosphatidylserine-targeted bimodal liposomal nanoparticles for in vivo imaging of breast cancer in mice. Journal of Controlled Release, 183, 114–123. 10.1016/j.jconrel.2014.03.043. [DOI] [PubMed] [Google Scholar]
- 266.Poyry S, & Vattulainen I (2016). Role of charged lipids in membrane structures—insight given by simulations. Biochimica et Biophysica Acta, 1858(10), 2322–2333. 10.1016/j.bbamem.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 267.van den Bogaart G, Meyenberg K, Risselada HJ, Amin H, Willig KI, Hubrich BE, et al. (2011). Membrane protein sequestering by ionic protein-lipid interactions. Nature, 479(7374), 552–555. 10.1038/nature10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Koldso H, Shorthouse D, Helie J, & Sansom MS (2014). Lipid clustering correlates with membrane curvature as revealed by molecular simulations of complex lipid bilayers. PLoS Computational Biology, 10(10), e1003911 10.1371/journal.pcbi.1003911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Wang J, & Richards DA (2012). Segregation of PIP2 and PIP3 into distinct nanoscale regions within the plasma membrane. Biology Open, 1(9), 857–862. 10.1242/bio.20122071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Honigmann A, van den Bogaart G, Iraheta E, Risselada HJ, Milovanovic D, Mueller V, et al. (2013). Phosphatidylinositol 4, 5-bisphosphate clusters act as molecular beacons for vesicle recruitment. Nature Structural & Molecular Biology, 20(6), 679–686. 10.1038/nsmb.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Coussens LM, Zitvogel L, & Palucka AK (2013). Neutralizing tumor-promoting chronic inflammation: a magic bullet? Science, 339(6117), 286–291. 10.1126/science.1232227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Song M, Wu K, Meyerhardt JA, Ogino S, Wang M, Fuchs CS, et al. (2018). Fiber intake and survival after colorectal cancer diagnosis. JAMA Oncology, 4(1), 71–79. 10.1001/jamaoncol.2017.3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Vargas AJ, & Thompson PA (2012). Diet and nutrient factors in colorectal cancer risk. Nutrition in Clinical Practice, 27(5), 613–623. [DOI] [PubMed] [Google Scholar]
- 274.Chapkin RS, McMurray DN, & Lupton JR (2007). Colon cancer, fatty acids and anti-inflammatory compounds. Current Opinion in Gastroenterology, 23(1), 48–54. 10.1097/MOG.0b013e32801145d7. [DOI] [PubMed] [Google Scholar]
- 275.Shaikh SR, & Edidin M (2008). Polyunsaturated fatty acids and membrane organization: elucidating mechanisms to balance immunotherapy and susceptibility to infection. Chemistry and Physics of Lipids, 153(1), 24–33. 10.1016/j.chemphyslip.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Knapp HR, Hullin F, & Salem N Jr. (1994). Asymmetric incorporation of dietary n-3 fatty acids into membrane aminophospholipids of human erythrocytes. Journal of Lipid Research, 35(7), 1283–1291. [PubMed] [Google Scholar]
- 277.Wassall SR, Brzustowicz MR, Shaikh SR, Cherezov V, Caffrey M, & Stillwell W (2004). Order from disorder, corralling cholesterol with chaotic lipids. The role of polyunsaturated lipids in membrane raft formation. Chemistry and Physics of Lipids, 132(1), 79–88. 10.1016/j.chemphyslip.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 278.Strobel C, Jahreis G, & Kuhnt K (2012). Survey of n-3 and n-6 polyunsaturated fatty acids in fish and fish products. Lipids in Health and Disease, 11, 144 10.1186/1476-511X-11-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Baker EJ, Miles EA, Burdge GC, Yaqoob P, & Calder PC (2016). Metabolism and functional effects of plant-derived omega-3 fatty acids in humans. Progress in Lipid Research, 64, 30–56. 10.1016/j.plipres.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 280.Arterburn LM, Hall EB, & Oken H (2006). Distribution, interconversion, and dose response of n-3 fatty acids in humans. The American Journal of Clinical Nutrition, 83(6 Suppl), 1467S–1476S. [DOI] [PubMed] [Google Scholar]
- 281.Patterson E, Wall R, Fitzgerald GF, Ross RP, & Stanton C (2012). Health implications of high dietary omega-6 polyunsaturated fatty acids. Journal Nutrition Metabolism, 2012, 539426 10.1155/2012/539426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Chapkin RS, Kim W, Lupton JR, & McMurray DN (2009). Dietary docosahexaenoic and eicosapentaenoic acid: emerging mediators of inflammation. Prostaglandins, Leukotrienes, and Essential Fatty Acids, 81(2–3), 187–191. 10.1016/j.plefa.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Cockbain AJ, Toogood GJ, & Hull MA (2012). Omega-3 polyunsaturated fatty acids for the treatment and prevention of colorectal cancer. Gut, 61(1), 135–149. 10.1136/gut.2010.233718. [DOI] [PubMed] [Google Scholar]
- 284.Anti M, Marra G, Armelao F, Bartoli GM, Ficarelli R, Percesepe A, et al. (1992). Effect of omega-3 fatty acids on rectal mucosal cell proliferation in subjects at risk for colon cancer. Gastroenterology, 103(3), 883–891. [DOI] [PubMed] [Google Scholar]
- 285.Anti M, Armelao F, Marra G, Percesepe A, Bartoli GM, Palozza P, et al. (1994). Effects of different doses of fish oil on rectal cell proliferation in patients with sporadic colonic adenomas. Gastroenterology, 107(6), 1709–1718. [DOI] [PubMed] [Google Scholar]
- 286.Bartram HP, Gostner A, Scheppach W, Reddy BS, Rao CV, Dusel G, et al. (1993). Effects of fish oil on rectal cell proliferation, mucosal fatty acids, and prostaglandin E2 release in healthy subjects. Gastroenterology, 105(5), 1317–1322. [DOI] [PubMed] [Google Scholar]
- 287.Hall MN, Chavarro JE, Lee IM, Willett WC, & Ma J (2008). A 22-year prospective study of fish, n-3 fatty acid intake, and colorectal cancer risk in men. Cancer Epidemiology, Biomarkers & Prevention, 17(5), 1136–1143. 10.1158/1055-9965.EPI-07-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Courtney ED, Matthews S, Finlayson C, Di Pierro D, Belluzzi A, Roda E, et al. (2007). Eicosapentaenoic acid (EPA) reduces crypt cell proliferation and increases apoptosis in normal colonic mucosa in subjects with a history of colorectal adenomas. International Journal of Colorectal Disease, 22(7), 765–776. 10.1007/s00384-006-0240-4. [DOI] [PubMed] [Google Scholar]
- 289.Baron JA, Sandler RS, Bresalier RS, Lanas A, Morton DG, Riddell R, et al. (2008). Cardiovascular events associated with rofecoxib: final analysis of the APPROVe trial. Lancet, 372(9651), 1756–1764. 10.1016/S0140-6736(08)61490-7. [DOI] [PubMed] [Google Scholar]
- 290.Graham DJ (2006). COX-2 inhibitors, other NSAIDs, and cardiovascular risk: the seduction of common sense. JAMA, 296(13), 1653–1656. 10.1001/jama.296.13.jed60058. [DOI] [PubMed] [Google Scholar]
- 291.West NJ, Clark SK, Phillips RKS, Hutchinson JM, Leicester RJ, Belluzzi A, et al. (2010). Eicosapentaenoic acid reduces rectal polyp number and size in familial adenomatous polyposis. Gut, 59(7), 918–925. [DOI] [PubMed] [Google Scholar]
- 292.Leitzmann MF, Stampfer MJ, Michaud DS, Augustsson K, Colditz GC, Willett WC, et al. (2004). Dietary intake of n-3 and n-6 fatty acids and the risk of prostate cancer. The American Journal of Clinical Nutrition, 80(1), 204–216. 10.1093/ajcn/80.1.204. [DOI] [PubMed] [Google Scholar]
- 293.Liang P, & Gao M (2017). Fish oil and prostate cancer: effects and clinical relevance. [review]. Cancer Translational Medicine, 3(3), 80–86. 10.4103/ctm.ctm_63_16. [DOI] [Google Scholar]
- 294.Saw CL, Huang Y, & Kong AN (2010). Synergistic antiinflammatory effects of low doses of curcumin in combination with polyunsaturated fatty acids: docosahexaenoic acid or eicosapentaenoic acid. Biochemical Pharmacology, 79(3), 421–430. 10.1016/j.bcp.2009.08.030. [DOI] [PubMed] [Google Scholar]
- 295.Vasudevan A, Yu Y, Banerjee S, Woods J, Farhana L, Rajendra SG, et al. (2014). Omega-3 fatty acid is a potential preventive agent for recurrent colon cancer. Cancer Prevention Research (Philadelphia, Pa.), 7(11), 1138–1148. 10.1158/1940-6207.CAPR-14-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Kawamori T, Lubet R, Steele VE, Kelloff GJ, Kaskey RB, Rao CV, et al. (1999). Chemopreventive effect of curcumin, a naturally occurring anti-inflammatory agent, during the promotion/progression stages of colon cancer. Cancer Research, 59(3), 597–601. [PubMed] [Google Scholar]
- 297.Chen A, Xu J, & Johnson AC (2006). Curcumin inhibits human colon cancer cell growth by suppressing gene expression of epidermal growth factor receptor through reducing the activity of the transcription factor Egr-1. Oncogene, 25(2), 278–287. 10.1038/sj.onc.1209019. [DOI] [PubMed] [Google Scholar]
- 298.Hanai H, Iida T, Takeuchi K, Watanabe F, Maruyama Y, Andoh A, et al. (2006). Curcumin maintenance therapy for ulcerative colitis: randomized, multicenter, double-blind, placebo-controlled trial. Clinical Gastroenterology and Hepatology, 4(12), 1502–1506. 10.1016/j.cgh.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 299.Cruz-Correa M, Shoskes DA, Sanchez P, Zhao R, Hylind LM, Wexner SD, et al. (2006). Combination treatment with curcumin and quercetin of adenomas in familial adenomatous polyposis. Clinical Gastroenterology and Hepatology, 4(8), 1035–1038. 10.1016/j.cgh.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 300.Sharma RA, Euden SA, Platton SL, Cooke DN, Shafayat A, Hewitt HR, et al. (2004). Phase I clinical trial of oral curcumin: biomarkers of systemic activity and compliance. Clinical Cancer Research, 10(20), 6847–6854. 10.1158/1078-0432.CCR-04-0744. [DOI] [PubMed] [Google Scholar]
- 301.Nautiyal J, Banerjee S, Kanwar SS, Yu Y, Patel BB, Sarkar FH, et al. (2011). Curcumin enhances dasatinib-induced inhibition of growth and transformation of colon cancer cells. International Journal of Cancer, 128(4), 951–961, 10.1002/ijc.25410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Hatcher H, Planalp R, Cho J, Torti FM, & Torti SV (2008). Curcumin: from ancient medicine to current clinical trials. Cellular and Molecular Life Sciences, 65(11), 1631–1652. 10.1007/s00018-008-7452-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Park W, Amin AR, Chen ZG, & Shin DM (2013). New perspectives of curcumin in cancer prevention. Cancer Prevention Research (Philadelphia, Pa.), 6(5), 387–400. 10.1158/1940-6207.CAPR-12-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Alrawi SJ, Schiff M, Carroll RE, Dayton M, Gibbs JF, Kulavlat M, et al. (2006). Aberrant crypt foci. Anticancer Research, 26(1A), 107–119. [PubMed] [Google Scholar]
- 305.Carroll RE, Benya RV, Turgeon DK, Vareed S, Neuman M, Rodriguez L, et al. (2011). Phase IIa clinical trial of curcumin for the prevention of colorectal neoplasia. Cancer Prevention Research (Philadelphia, Pa.), 4(3), 354–364. 10.1158/1940-6207.CAPR-10-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Irving GR, Howells LM, Sale S, Kralj-Hans I, Atkin WS, Clark SK, et al. (2013). Prolonged biologically active colonic tissue levels of curcumin achieved after oral administration—a clinical pilot study including assessment of patient acceptability. Cancer Prevention Research (Philadelphia, Pa.), 6(2), 119–128. 10.1158/1940-6207.CAPR-12-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Ariel A, & Serhan CN (2007). Resolvins and protectins in the termination program of acute inflammation. Trends in Immunology, 28(4), 176–183. 10.1016/j.it.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 308.Bazan NG (2007). Omega-3 fatty acids, pro-inflammatory signaling and neuroprotection. Current Opinion in Clinical Nutrition and Metabolic Care, 10(2), 136–141. 10.1097/MCO.0b013e32802b7030. [DOI] [PubMed] [Google Scholar]
- 309.de Urquiza AM, Liu S, Sjoberg M, Zetterstrom RH, Griffiths W, Sjovall J, et al. (2000). Docosahexaenoic acid, a ligand for the retinoid X receptor in mouse brain. Science, 290(5499), 2140–2144. [DOI] [PubMed] [Google Scholar]
- 310.Fan YY, Spencer TE, Wang N, Moyer MP, & Chapkin RS (2003). Chemopreventive n-3 fatty acids activate RXRalpha in colonocytes. Carcinogenesis, 24(9), 1541–1548. 10.1093/carcin/bgg110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Davidson LA, Nguyen DV, Hokanson RM, Callaway ES, Isett RB, Turner ND, et al. (2004). Chemopreventive n-3 polyunsaturated fatty acids reprogram genetic signatures during colon cancer initiation and progression in the rat. Cancer Research, 64(18), 6797–6804. 10.1158/0008-5472.CAN-04-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Jump DB, Botolin D, Wang Y, Xu J, Christian B, & Demeure O (2005). Fatty acid regulation of hepatic gene transcription. The Journal of Nutrition, 135(11), 2503–2506. 10.1093/jn/135.11.2503. [DOI] [PubMed] [Google Scholar]
- 313.Fuentes NR, Kim E, Fan YY, & Chapkin RS (2018). Omega-3 fatty acids, membrane remodeling and cancer prevention. Molecular Aspects of Medicine. 10.1016/j.mam.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Chapkin RS, Akoh CC, & Miller CC (1991). Influence of dietary n-3 fatty acids on macrophage glycerophospholipid molecular species and peptidoleukotriene synthesis. Journal of Lipid Research, 32(7), 1205–1213. [PubMed] [Google Scholar]
- 315.Williams JA, Batten SE, Harris M, Rockett BD, Shaikh SR, Stillwell W, et al. (2012). Docosahexaenoic and eicosapentaenoic acids segregate differently between raft and nonraft domains. Biophysical Journal, 103(2), 228–237. 10.1016/j.bpj.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Levental KR, Lorent JH, Lin X, Skinkle AD, Surma MA, Stockenbojer EA, et al. (2016). Polyunsaturated lipids regulate membrane domain stability by tuning membrane order. Biophysical Journal, 110(8), 1800–1810. 10.1016/j.bpj.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Ma DW, Seo J, Davidson LA, Callaway ES, Fan YY, Lupton JR, et al. (2004). n-3 PUFA alter caveolae lipid composition and resident protein localization in mouse colon. The FASEB Journal, 18(9), 1040–1042. 10.1096/fj.03-1430fje. [DOI] [PubMed] [Google Scholar]
- 318.Seo J, Barhoumi R, Johnson AE, Lupton JR, & Chapkin RS (2006). Docosahexaenoic acid selectively inhibits plasma membrane targeting of lipidated proteins. The FASEB Journal, 20(6), 770–772. 10.1096/fj.05-4683fje. [DOI] [PubMed] [Google Scholar]
- 319.Hou TY, Davidson LA, Kim E, Fan YY, Fuentes NR, Triff K, et al. (2016). Nutrient-gene interaction in colon cancer, from the membrane to cellular physiology. Annual Review of Nutrition, 36, 543–570. 10.1146/annurev-nutr-071715-051039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Stillwell W, & Wassall SR (2003). Docosahexaenoic acid: membrane properties of a unique fatty acid. Chemistry and Physics of Lipids, 126(1), 1–27. [DOI] [PubMed] [Google Scholar]
- 321.Fan YY, Fuentes NR, Hou TY, Barhoumi R, Li XC, Deutz NEP, et al. (2018). Remodelling of primary human CD4+ T cell plasma membrane order by n-3 PUFA. The British Journal of Nutrition, 119(2), 163–175. 10.1017/S0007114517003385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Turk HF, Barhoumi R, & Chapkin RS (2012). Alteration of EGFR spatiotemporal dynamics suppresses signal transduction. PLoS One, 7(6), e39682 10.1371/journal.pone.0039682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Rogers KR, Kikawa KD, Mouradian M, Hernandez K, McKinnon KM, Ahwah SM, et al. (2010). Docosahexaenoic acid alters epidermal growth factor receptor-related signaling by disrupting its lipid raft association. Carcinogenesis, 31(9), 1523–1530. 10.1093/carcin/bgq111. [DOI] [PubMed] [Google Scholar]
- 324.Normanno N, Tejpar S, Morgillo F, De Luca A, Van Cutsem E, & Ciardiello F (2009). Implications for KRAS status and EGFR-targeted therapies in metastatic CRC. Nature Reviews. Clinical Oncology, 6(9), 519–527. 10.1038/nrclinonc.2009.111. [DOI] [PubMed] [Google Scholar]
- 325.Sondermann H, Soisson SM, Boykevisch S, Yang SS, Bar-Sagi D, & Kuriyan J (2004). Structural analysis of autoinhibition in the Ras activator son of sevenless. Cell, 119(3), 393–405. 10.1016/j.cell.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 326.Lee YK, Low-Nam ST, Chung JK, Hansen SD, Lam HYM, Alvarez S, et al. (2017). Mechanism of SOS PR-domain autoinhibition revealed by single-molecule assays on native protein from lysate. Nature Communications, 8, 15061 10.1038/ncomms15061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Hou TY, Monk JM, Fan YY, Barhoumi R, Chen YQ, Rivera GM, et al. (2012). n-3 polyunsaturated fatty acids suppress phosphatidylinositol 4,5-bisphosphate-dependent actin remodelling during CD4+ T-cell activation. The Biochemical Journal, 443(1), 27–37. 10.1042/BJ20111589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Cromm PM, Spiegel J, Grossmann TN, & Waldmann H (2015). Direct modulation of small GTPase activity and function. Angewandte Chemie (International Ed. in English), 54(46), 13516–13537. 10.1002/anie.201504357. [DOI] [PubMed] [Google Scholar]
- 329.Sun Q, Burke JP, Phan J, Burns MC, Olejniczak ET, Waterson AG, et al. (2012). Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angewandte Chemie (International Ed. in English), 51(25), 6140–6143. 10.1002/anie.201201358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Maurer T, Garrenton LS, Oh A, Pitts K, Anderson DJ, Skelton NJ, et al. (2012). Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proceedings of the National Academy of Sciences of the United States of America, 109(14), 5299–5304. 10.1073/pnas.1116510109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Grabocka E, Pylayeva-Gupta Y, Jones MJ, Lubkov V, Yemanaberhan E, Taylor L, et al. (2014). Wild-type H- and N-Ras promote mutant K-Ras-driven tumorigenesis by modulating the DNA damage response. Cancer Cell, 25(2), 243–256. 10.1016/j.ccr.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Jeng HH, Taylor LJ, & Bar-Sagi D (2012). Sos-mediated cross-activation of wild-type Ras by oncogenic Ras is essential for tumorigenesis. Nature Communications, 3, 1168 10.1038/ncomms2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Stephen AG, Esposito D, Bagni RK, & McCormick F (2014). Dragging ras back in the ring. Cancer Cell, 25(3), 272–281. 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 334.Phipps AI, Buchanan DD, Makar KW, Win AK, Baron JA, Lindor NM, et al. (2013). KRAS-mutation status in relation to colorectal cancer survival: the joint impact of correlated tumour markers. British Journal of Cancer, 108(8), 1757–1764. 10.1038/bjc.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Collett ED, Davidson LA, Fan YY, Lupton JR, & Chapkin RS (2001). n-6 and n-3 polyunsaturated fatty acids differentially modulate oncogenic Ras activation in colonocytes. American Journal of Physiology. Cell Physiology, 280(5), C1066–C1075. [DOI] [PubMed] [Google Scholar]
- 336.Fasano E, Serini S, Piccioni E, Toesca A, Monego G, Cittadini AR, et al. (2012). DHA induces apoptosis by altering the expression and cellular location of GRP78 in colon cancer cell lines. Biochimica et Biophysica Acta, 1822(11), 1762–1772. 10.1016/j.bbadis.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 337.Schonberg SA, Lundemo AG, Fladvad T, Holmgren K, Bremseth H, Nilsen A, et al. (2006). Closely related colon cancer cell lines display different sensitivity to polyunsaturated fatty acids, accumulate different lipid classes and downregulate sterol regulatory element-binding protein 1. The FEBS Journal, 273(12), 2749–2765. 10.1111/j.1742-4658.2006.05292.x. [DOI] [PubMed] [Google Scholar]
- 338.Mohammed A, Janakiram NB, Brewer M, Duff A, Lightfoot S, Brush RS, et al. (2012). Endogenous n-3 polyun-saturated fatty acids delay progression of pancreatic ductal adenocarcinoma in Fat-1-p48(Cre/+)-LSL-Kras(G12D/+) mice. Neoplasia (New York, N.Y.), 14(12), 1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Strouch MJ, Ding Y, Salabat MR, Melstrom LG, Adrian K, Quinn C, et al. (2011). A high omega-3 fatty acid diet mitigates murine pancreatic precancer development. The Journal of Surgical Research, 165(1), 75–81. 10.1016/j.jss.2009.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Muratcioglu S, Chavan TS, Freed BC, Jang H, Khavrutskii L, Freed RN, et al. (2015). GTP-dependent K-Ras dimerization. Structure, 23(7), 1325–1335. 10.1016/j.str.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Nan X, Tamguney TM, Collisson EA, Lin LJ, Pitt C, Galeas J, et al. (2015). Ras-GTP dimers activate the mitogen-activated protein kinase (MAPK) pathway. Proceedings of the National Academy of Sciences of the United States of America, 112(26), 7996–8001. 10.1073/pnas.1509123112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Cho KJ, & Hancock JF (2013). Ras nanoclusters: a new drug target? Small GTPases, 4(1), 57–60. 10.4161/sgtp.23145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Chapkin RS, Wang N, Fan YY, Lupton JR, & Prior IA (2008). Docosahexaenoic acid alters the size and distribution of cell surface microdomains. Biochimica et Biophysica Acta, 1778(2), 466–471. 10.1016/j.bbamem.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Kim W, Khan NA, McMurray DN, Prior IA, Wang N, & Chapkin RS (2010). Regulatory activity of polyunsaturated fatty acids in T-cell signaling. Progress in Lipid Research, 49(3), 250–261. 10.1016/j.plipres.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Krausova M, & Korinek V (2014). Wnt signaling in adult intestinal stem cells and cancer. Cellular Signalling, 26(3), 570–579. 10.1016/j.cellsig.2013.11.032. [DOI] [PubMed] [Google Scholar]
- 346.Gregorieff A, & Clevers H (2005). Wnt signaling in the intestinal epithelium: from endoderm to cancer. Genes & Development, 19(8), 877–890. 10.1101/gad.1295405. [DOI] [PubMed] [Google Scholar]
- 347.White BD, Chien AJ, & Dawson DW (2012). Dysregulation of Wnt/beta-catenin signaling in gastrointestinal cancers. Gastroenterology, 142(2), 219–232. 10.1053/j.gastro.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Lucijanic M, Livun A, Tomasovic-Loncaric C, Stoos-Veic T, Pejsa V, Jaksic O, et al. (2016). Canonical Wnt/beta-catenin signaling pathway is dysregulated in patients with primary and secondary myelofibrosis. Clinical Lymphoma, Myeloma & Leukemia, 16(9), 523–526. 10.1016/j.clml.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 349.Guinot A, Oeztuerk-Winder F, & Ventura JJ (2016). miR-17-92/p38alpha dysregulation enhances Wnt signaling and selects Lgr6+ cancer stem-like cells during lung adenocarcinoma progression. Cancer Research, 76(13), 4012–4022. 10.1158/0008-5472.CAN-15-3302. [DOI] [PubMed] [Google Scholar]
- 350.Mitsui Y, Yasumoto H, Nagami T, Hiraki M, Arichi N, Ishikawa N, et al. (2014). Extracellular activation of Wnt signaling through epigenetic dysregulation of Wnt inhibitory factor-1 (Wif-1) is associated with pathogenesis of adrenocortical tumor. Oncotarget, 5(8), 2198–2207. 10.18632/oncotarget.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Wu WK, Wang XJ, Cheng AS, Luo MX, Ng SS, To KF, et al. (2013). Dysregulation and crosstalk of cellular signaling pathways in colon carcinogenesis. Critical Reviews in Oncology/Hematology, 86(3), 251–277. 10.1016/j.critrevonc.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 352.Chan DW, Mak CS, Leung TH, Chan KK, &Ngan HY (2012). Down-regulation of Sox7 is associated with aberrant activation of Wnt/b-catenin signaling in endometrial cancer. Oncotarget, 3(12), 1546–1556, 10.18632/oncotarget.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Gao B (2012). Wnt regulation of planar cell polarity (PCP). Current Topics in Developmental Biology, 101, 263–295. 10.1016/B978-0-12-394592-1.00008-9. [DOI] [PubMed] [Google Scholar]
- 354.Komiya Y, & Habas R (2008). Wnt signal transduction pathways. Organogenesis, 4(2), 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Schepers A, & Clevers H (2012). Wnt signaling, stem cells, and cancer of the gastrointestinal tract. Cold Spring Harbor Perspectives in Biology, 4(4), a007989 10.1101/cshperspect.a007989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Fodde R, & Brabletz T (2007). Wnt/beta-catenin signaling in cancer stemness and malignant behavior. Current Opinion in Cell Biology, 19(2), 150–158. 10.1016/j.ceb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 357.Famili F, Brugman MH, Taskesen E, Naber BE, Fodde R, & Staal FJ (2016). High levels of canonical Wnt signaling lead to loss of stemness and increased differentiation in hematopoietic stem cells. Stem Cell Reports, 6(5), 652–659. 10.1016/j.stemcr.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Oren O, & Smith BD (2016). Eliminating cancer stem cells by targeting embryonic signaling pathways. Stem Cell Reviews. 10.1007/s12015-016-9691-3. [DOI] [PubMed] [Google Scholar]
- 359.Lesko AC, Goss KH, & Prosperi JR (2014). Exploiting APC function as a novel cancer therapy. Current Drug Targets, 15(1), 90–102. [DOI] [PubMed] [Google Scholar]
- 360.Foley PJ, Scheri RP, Smolock CJ, Pippin J, Green DW, & Drebin JA (2008). Targeted suppression of beta-catenin blocks intestinal adenoma formation in APC Min mice. Journal of Gastrointestinal Surgery, 12(8), 1452–1458. 10.1007/s11605-008-0519-6. [DOI] [PubMed] [Google Scholar]
- 361.Lepourcelet M, Chen YN, France DS, Wang H, Crews P, Petersen F, et al. (2004). Small-molecule antagonists of the oncogenic Tcf/beta-catenin protein complex. Cancer Cell, 5(1), 91–102. [DOI] [PubMed] [Google Scholar]
- 362.Emami KH, Nguyen C, Ma H, Kim DH, Jeong KW, Eguchi M, et al. (2004). A small molecule inhibitor of beta-catenin/CREB-binding protein transcription [corrected]. Proceedings of the National Academy of Sciences of the United States of America, 101(34), 12682–12687. 10.1073/pnas.0404875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Inoue K, Fry EA, & Taneja P (2013). Recent progress in mouse models for tumor suppressor genes and its implications in human cancer. Clinical Medicine Insights Oncology, 7, 103–122. 10.4137/CMO.S10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Buchanan FG, Holla V, Katkuri S, Matta P, & DuBois RN (2007). Targeting cyclooxygenase-2 and the epidermal growth factor receptor for the prevention and treatment of intestinal cancer. Cancer Research, 67(19), 9380–9388. 10.1158/0008-5472.CAN-07-0710. [DOI] [PubMed] [Google Scholar]
- 365.Vogelstein B, & Kinzler KW (2012). Winning the war: science parkour. Science Translational Medicine, 4(127), 127ed122. 10.1126/scitranslmed.3004019. [DOI] [PubMed] [Google Scholar]
- 366.Moses H 3rd, Dorsey ER, Matheson DH, & Thier SO (2005). Financial anatomy of biomedical research. JAMA, 294(11), 1333–1342. 10.1001/jama.294.11.1333. [DOI] [PubMed] [Google Scholar]
- 367.Coussens LM, Zitvogel L, & Palucka AK (2013). Neutralizing tumor-promoting chronic inflammation: a magic bullet? Science, 339(6117), 286 10.1126/science.1232227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Ford ES, Bergmann MM, Boeing H, Li C, & Capewell S (2012). Healthy lifestyle behaviors and all-cause mortality among adults in the United States. Preventive Medicine, 55(1), 23–27. 10.1016/j.ypmed.2012.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]