Abstract
Both the social and physical environment shape health, reproduction, and survival across many species, and identifying how these effects manifest at the molecular level has long been a priority in medicine and evolutionary biology. The recent rise of functional genomics has enabled researchers to gain new insights into how environmental inputs shape variation in gene regulation, and consequently, downstream organism-level traits. Here, we discuss recent work on this topic, as well as key knowledge gaps. Research in this area spans a wide range of taxa, but we focus our review on mammalian species because of their close evolutionary proximity to humans and because of their relevance for understanding human health. Improving our understanding of how the environment and the genome are connected promises to shed new light on the mechanisms underlying environmentally-induced disease in humans, as well as the evolution of environmental sensitivity more generally.
Introduction
For decades, evolutionary biologists and medical scientists have asked how environmental variation shapes health, reproduction, and survival. Researchers have long appreciated that the physical environment (e.g., diet, weather conditions, or pathogen exposure) has dramatic effects on fitness in mammalian species. For example, limited resource availability, both during development and later in life, predicts reduced fertility and mortality in humans [1,2] and in several wild mammal populations [3–6]. More recently, it has become clear that social components of the environment can also shape trait variation in ways that are equally profound. In social species, an individual’s position in its social hierarchy (i.e., social status), as well as the degree to which the individual interacts with others (i.e., social connectedness), can predict disease risk and mortality [7–9]. Arguably, the strongest evidence for such social environmental effects comes from our own species [10,11].
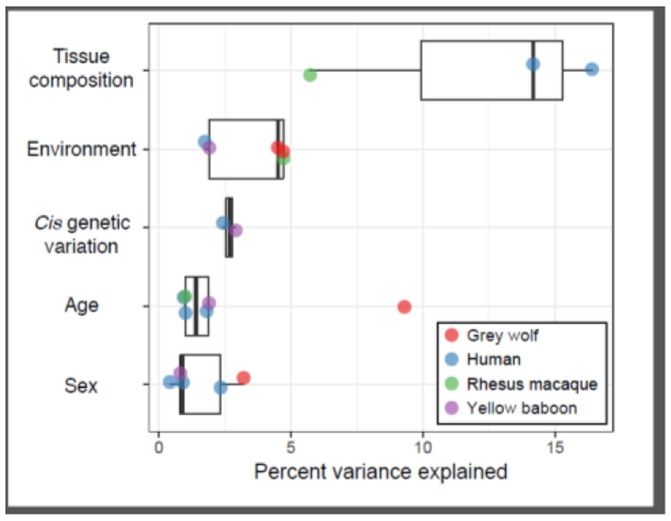
Despite widespread and compelling evidence that environmental challenges affect fitness, we know relatively little about the molecular mechanisms that mediate these relationships, especially at the genome scale. Foundational work in molecular biology, genetics, and neuroscience has identified key molecules and candidate genes involved in sensing and responding to specific environmental inputs [12–14]. However, as new genomic methods have emerged over the last decade, the picture of how environmental variation affects our genomes has widened in scope. Specifically, using functional genomic tools, researchers have started to discover that environmental effects on organism-level traits are often mediated through changes in the way large, coordinated sets of genes are expressed, with environmental effects rivaling other well-known predictors of gene expression variation (e.g., demographic or genetic effects; Figure 1).
Figure 1. The strength of environmental effects on gene regulation is comparable to other well-known predictors of gene regulatory variation.
Using data from several published studies [38,65,83–85], we estimated the mean percent variance in genome-wide gene expression levels explained (PVE) by tissue composition, demographic effects (age and sex), local genetic variation, and a range of social and physical environmental inputs (grey wolf = mange and social status; rhesus macaque = social status; yellow baboon = maternal social connectedness; human = smoking). All studies were conducted in blood-derived samples, and mean PVE was taken from the text, supplementary information, or calculated using publicly available effect size estimates and data files. We note that PVE estimates are strongly influenced by the covariates included in models to detect environmental effects, and by the amount of variation in the environmental variable itself. However, across studies it is clear that environmental effects rival or exceed other widely accepted drivers of gene regulatory variation.
Here, we highlight recent research on how environmental signals affect genome-wide gene regulation, with a focus on select environmental effects that are well-linked to fitness variation and for which functional genomics has begun to provide new mechanistic insight. These examples are not meant as an exhaustive review (for other great reviews of this topic, see: [15–17]), but are instead meant to demonstrate the utility of functional genomics approaches for understanding how environmental variation ‘gets under the skin’ to affect health and survival. In discussing this work, we emphasize recent findings from humans as well as captive and laboratory mammalian models; however, we also identify gaps in our knowledge that could be better addressed using a more diverse set of non-model mammalian systems. Finally, we discuss new ways that functional genomics can be leveraged to understand connections between the environment and fitness, beyond study designs that correlate environmental variation with gene regulatory phenotypes (Box 1). Identifying the mechanistic path from environmental variation to fitness-related traits is important for treating and preventing environmentally-induced disease, and from an evolutionary perspective, for understanding how and when organisms evolve to sense and respond to their surroundings.
Box 1: Emerging questions about the relationship between the environment and fitness that can be answered with genomic tools
As genomic work has progressed, it has become clear that the environment and the genome are intertwined in diverse and important ways, beyond ‘simple’ main effects of environmental variation on gene regulation. Here, we discuss three research areas that explore new types of environment-genome connections – the microbiome, indirect genetic effects, and gene × environment interactions – and address how functional genomic tools can help unravel these emerging links.
The microbiome.
The microbiome (the collection of microbes that live in and on an organism) sits at the interface of the outside environment and internal physiology. Variation in both the physical [68,69] and social environment [70–72] can shape the bacterial communities that exist within an individual, with consequences for physiology, health, and behavior [73,74]. The environmental dependence of the microbiome is thus particularly clear and exciting, and researchers are increasingly turning to functional genomic datasets to understand how the two are mechanistically linked. In particular, researchers are beginning to sequence bacterial RNA (an approach known as ‘metatranscriptomics’) to gain insight into the genes that are actively expressed by the microbes in these complex communities [75]. This approach thus allows researchers to capture information about environmental effects on bacterial function and activity, rather than simple presence/absence.
Indirect genetic effects.
The genotypes of those around you can also influence your behavior and your genome — a concept called indirect genetic effects (IGEs; [76]). For example, using genotype data from over 20,000 parent-offspring pairs, Kong et al. found that genetic variants that exist in parents, but are not transmitted to their children, had an effect on their children’s educational attainment that was ~30% stronger than direct genetic effects [77]. Thus, the parental environment is both profoundly important for educational attainment and affected by parental genotypes — even those genetic variants that are not directly passed on to the offspring. Work in laboratory mice has also found that the impact of IGEs on physiology, behavior, and gene expression can rival or exceed that of direct genetic effects [78,79]. Further, one recent study attempted to map specific loci that affect phenotypic variation in social partners, in an effort to understand the genetic architecture of IGEs ([78] though larger samples will likely be needed to make progress in this area). Moving forward, it will be particularly interesting to expand this work beyond humans and model laboratory systems, and to use functional genomics to trace the mechanistic pathway from genetic variants that exist in social partners to their effects on focal individuals.
Genotype × environment interactions.
There is substantial heterogeneity in how individuals respond to the same environmental challenges. One explanation for this observation is that genetic variation controls each individual’s response. This phenomenon, known as a genotype × environment (G × E) interaction, has captured the attention of evolutionary and health researchers alike, as it has the potential to explain why particular individuals are especially vulnerable (or resilient) to environmental challenges. One effective tool for mapping G × E effects on gene expression is to expose cells from a given individual to contrasting environmental conditions in vitro, and identifying genetic variants that affect gene expression in one condition but not the other. Using this paradigm, researchers have uncovered substantial genetic variation for the way humans respond to bacterial and viral infections, stress hormones, and environmental toxins [80–82]. Importantly, this work has allowed us to begin to understand why responses vary among individuals (e.g., because of ancestry), which genetic variants account for such differences, and how evolutionary processes (e.g., selection or introgression) have established these variants in human populations [17]. As culturing cells from non-model systems is becoming increasingly achievable, it will be important to expand GxE work beyond humans (and eventually, beyond the laboratory). Doing so will allow us to understand whether genetic variation for environmental sensitivity is prevalent across species and environmental contexts; this information will allow us to ask whether the evolutionary processes maintaining genetic variation for plasticity in humans [17] extends to other mammalian and natural systems.
Environmental effects on genome-wide gene regulation
Mammals are able to dynamically respond to changes in their environment by tuning the expression levels of their genes. This ability relies on a diverse set of gene regulatory mechanisms, of which DNA methylation, chromatin accessibility, and histone modifications are the best studied to date. In particular, DNA methylation has received the most attention as a potential molecular mediator of environmental effects on gene expression levels, because of its demonstrated environmental sensitivity as well as the stability of environmentally-induced methylation changes [13,14]. Below, we highlight recent examples of both physical and social environmental components that we have come to recently understand through studies of genome-wide gene expression, as well as the mechanisms that regulate gene expression.
Physical environmental effects on gene regulation
Variation in the physical environment — including what an animal eats, the weather it experiences, and the pathogens it encounters — is intricately connected to physiological change. In mammals, for example, seasonal changes in weather and resource availability can affect reproductive patterns [18], hormone levels [19], and disease risk [20]. It is becoming increasingly clear that these organism-level responses are implemented and maintained at the level of gene regulation. One recent study estimated that at least one quarter of the genes expressed in human blood varied in expression levels across seasons [21]. For instance, winter was associated with heightened expression of proinflammatory genes, which may contribute to higher rates of autoimmune and cardiovascular disease during these months. Interestingly, seasonal expression patterns observed in Europeans were reversed in samples collected from Australians, where the seasons are the opposite of the Northern Hemisphere [21]. Other mammals are less well-studied with respect to seasonal effects on gene regulation, although seasonal expression variation has been documented in the snowshoe hare [22] and some hibernators, including the dwarf lemur [23], and European [24] and Siberian hamsters [25].
Food resource availability, which need not covary with season, also leads to changes in metabolism, life history, and longevity in a range of organisms [1–6]. One of the most dramatic effects is associated not with what you eat, but how much you eat. A 10-40% reduction in caloric intake can lead to a 4-45% increase in lifespan in mammalian species ranging from rodents [26] to rhesus macaques [27]. These dramatic shifts in survival are also reflected in the epigenome of calorically restricted animals: calorically restricted mice and macaques exhibit DNA methylation profiles that mimic those of younger animals [28]. These observations highlight one potential mechanism, delayed epigenetic aging [29], through which caloric restriction may lead to a longer, healthier life. It is important to note, however, that the effects of caloric restriction are not always beneficial. For example, work on the Dutch Hunger Winter, a severe wartime famine, has linked extreme maternal caloric restriction (400-800 calories/day) with an increased risk of insulin resistance, obesity, and cardiovascular disease in offspring conceived during famine [30]. These health consequences are thought to be mediated by changes in DNA methylation that occur near growth and metabolism-related genes in offspring in response to famine [31,32], though the evidence for causal links between in utero caloric restriction, DNA methylation, and metabolic disease has been called into question [33].
In addition to the number of calories consumed, the nutritional content of food resources can alter gene regulation. For example, mice fed a ketogenic diet (characterized by low carbohydrate, moderate protein, and high fat intake) exhibited decreased expression of genes involved in aging-related biological pathways, including insulin signaling and TOR (target of rapamycin) activation; these effects persisted into old age, which may mechanistically explain the link between a ketogenic diet and decreased mortality, improved cognition, and better memory in aging animals [34].
Social environmental effects on gene regulation
For social animals, interactions with conspecifics are one of the most salient components of the environment [35]. In these species, recent evidence points to the importance of gene regulation for translating social environmental variation into variation in reproduction, health, and survival. For instance, social status has been linked to changes in gene expression in species ranging from mice to macaques to humans [36–39]. Specifically, these studies have found evidence that the chronic stress of low social status leads to an increase in the expression of proinflammatory genes both in circulating immune cells [16,38] as well as in brain regions important for memory and cognition [40,41]. In one extreme example of social subordination, the reproductive suppression of subordinate, “non-breeder” naked mole rats is apparent at the level of gene expression in reproductive organs [42]. Subordinate female naked mole rats also exhibit different gene expression profiles in the brain — particularly at genes involved in dopamine metabolism, which is implicated in a range of functions from sexual arousal to aggression to cognition [42].
The effect of the social environment is not limited to social standing. The quality and quantity of social relationships has been shown to alter gene expression as well. For example, in humans, loneliness is associated with increased expression of proinflammatory genes in peripheral blood [16,43] and the brain [44]. Further, a study that experimentally manipulated social status in rhesus macaques found that variation in affiliative relationships partially accounted for the effect of social status on gene expression variation at one third of status-associated genes [38]. Because the gene regulatory effects of social adversity (both low social status and social isolation) are so common across species, and are often concentrated in proinflammatory or innate immune genes, this signature is sometimes called the ‘conserved transcriptional response to adversity’ (CTRA; [45]).
Open questions about environmental effects on gene regulation
While progress has been made toward understanding how gene expression connects environmental insults with fitness-related traits, several key gaps emerge from the summaries provided above. First, it is relatively rare for researchers to investigate both environmentally responsive gene expression patterns and their underlying gene regulatory mechanisms in a single system. Studies that have done so have almost universally focused on DNA methylation [31,46–49] (but see [50] for a study showing effects of social status on chromatin accessibility and gene expression). However, there is no reason to think other regulatory mechanisms are not equally environmentally responsive. For example, human dendritic cells can remodel their transcription factor binding, chromatin, histone, and DNA methylation profiles over the course of a few hours in response to bacterial infection [51,52]. To date, however, the degree to which these same mechanisms also respond dynamically to social and other physical environmental stimuli remains largely unknown, as does the degree to which environmentally-associated variation in gene regulatory mechanisms are causal to changes in gene expression and organisms-level phenotypes [33]. Studies that utilize time course experiments [52], Mendelian Randomization or other causal inference tools [33,53,54], and manipulation of epigenetic marks [55,56] promise to help further our understanding of the causal chain linking variation in the environment with variation in gene regulatory mechanisms, gene expression, and ultimately fitness.
A second open question is to what degree are the same environmentally-responsive genes affected across similar contexts, tissues, or species? While some progress has been made in identifying proinflammatory and innate immune genes as common targets of social adversity in mammalian blood [45], this work is so far limited to captive animals and humans, and more work is needed to understand the generality of this framework. It is possible that there is a conserved molecular “toolkit” that underlies shared gene regulatory responses to environmental variation [57]. However, because of the limited taxonomic diversity of gene regulatory work, little progress has been made in understanding how or why a given set of genes evolve to sense and respond to environmental variation.
Finally, we do not yet understand when in the lifespan animals are most sensitive to social or physical environmental inputs. Mounting evidence suggests that some environmental stressors encountered early in life may permanently alter gene regulatory programs, and thus go on to shape physiology and health across the lifecourse. For example, a groundbreaking study in laboratory mice found that maternal diet during pregnancy influenced offspring methylation near the agouti gene, which in turn affected agouti gene expression, fur color, body mass, and susceptibility to diabetes later in life [12,58]. More recently, these findings have been extended to a diverse set of early life environmental contexts [31,47,59,60]. However, evidence that gene expression programs dynamically react to the current environment has also been growing in parallel [38,61], leaving open the question of which environments, genes, and organisms are governed by ‘early embedding’ of environmental variation into the genome versus dynamic responses. Addressing this gap is essential for understanding when in the lifespan environmental improvements or interventions are likely to have the biggest impact.
Mammalian systems inform our understanding of evolution and human health
As models for human health, studies of mammals can help us to understand the most salient features of an individual’s environment, including how they mechanistically alter health and survival. Remaining challenges include (i) understanding the gene regulatory mechanisms that causally link environmental effects to gene expression variation and, ultimately health and survival; (ii) identifying the specific genes that are most strongly affected by environmental challenges; and (iii) understanding how the answers to (i) and (ii) vary across the lifecourse. Addressing these challenges will require multiple approaches, including longitudinal, prospective work in humans as well as studies of animal models. While work in humans can struggle with retrospective reporting of environmental circumstances, as well as confounded relationships between key environmental components (e.g., social adversity can be associated with reduced access to healthcare and poor nutrition [62]). These issues can be circumvented in animal models. Further, in captive situations, experimental manipulations can identify causal links between the environment and gene regulation — experiments that would be difficult or impossible to conduct in humans.
However, it is important to point out that certain questions will be difficult to answer in the handful of popular mammalian models researchers have focused on to date, namely laboratory rodents and captive primates.Though powerful, these systems (i) do not capture the full range of physical and social environmental variation that exists in nature, (ii) typically focus on isolating the effects of one environmental variable, and may therefore miss key interactions among different environmental components [38,63,64], and (iii) are not sufficient to support comparative work on the evolution of environmentally sensitive gene regulatory programs. Thus, without expanding genomic work to a larger set of environments and species (including wild populations), we will remain unable to understand how genomic plasticity evolves in response to the complex environmental variation observed in nature. While a few steps have been taken in this direction (e.g., [23,46,54,65]), more needs to be done. Fortunately, there is a rich history of studying environmental effects in natural systems, and modern genomic tools are rapidly becoming more streamlined and optimized for samples collected in the wild or in species without existing genomic tools [66,67]. Thus, non-model, natural systems appear ripe for molecular analysis of well-understood environment-fitness relationships.
Acknowledgements
We thank Jenny Tung and Rachel Johnston for their thoughtful feedback on an earlier version of the manuscript. This work was supported by National Science Foundation (SBE-1723237) and National Institutes of Health (R00-AG051764), and a postdoctoral fellowship to AJL from the Helen Hay Whitney Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Moore SE, Cole TJ, Poskitt EM, Sonko BJ, Whitehead RG, McGregor I a., Prentice a. M: Season of birth predicts mortality in rural Gambia. Nature 1997, 388:434. [DOI] [PubMed] [Google Scholar]
- 2.Rickard IJ, Holopainen J, Helama S, Helle S, Russell AF, Lummaa V: Food availability at birth limited reproductive success in historical humans. Ecology 2010, 91:3515–3525. [DOI] [PubMed] [Google Scholar]
- 3.Gaillard JM, Festa-Bianchet M, Delorme D, Jorgenson J: Body mass and individual fitness in female ungulates: bigger is not always better. Proc Biol Sci 2000, 267:471–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beehner JC, Onderdonk DA, Alberts SC, Altmann J: The ecology of conception and pregnancy failure in wild baboons. Behav Ecol 2006, 17:741–750. [Google Scholar]
- 5.Hamel S, Gaillard J-M, Festa-Bianchet M, Côté SD: Individual quality, early-life conditions, and reproductive success in contrasted populations of large herbivores. Ecology 2009, 90:1981–1995. [DOI] [PubMed] [Google Scholar]
- 6.Altmann S: Diets of yearling female primates (Papio cynocephalus) predict lifetime fitness. Proc Natl Acad Sci U S A 1991, 88:420–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holt-Lunstad J, Smith TB, Layton JB: Social relationships and mortality risk: a meta-analytic review. PLoS Med 2010, 7:e1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chetty R, Stepner M, Abraham S, Lin S, Scuderi B, Turner N, Bergeron A, Cutler D: The Association Between Income and Life Expectancy in the United States, 2001-2014. JAMA 2016, 315:1750–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silk JB: The adaptive value of sociality in mammalian groups. Philos Trans R Soc Lond B Biol Sci 2007, 362:539–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holt-Lunstad J, Smith TB, Layton JB: Social relationships and mortality risk: a meta-analytic review. PLoS Med 2010, 7:e1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chetty R, Stepner M, Abraham S, Lin S, Scuderi B, Turner N, Bergeron A, Cutler D: The Association Between Income and Life Expectancy in the United States, 2001-2014. JAMA 2016, 315:1750–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolff GL, Kodell RL, Moore SR, Cooney CA: Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J 1998, 12:949–957. [PubMed] [Google Scholar]
- 13.Jaenisch R, Bird A: Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 2003, 33 Suppl:245–254. [DOI] [PubMed] [Google Scholar]
- 14.Jirtle RL, Skinner MK: Environmental epigenomics and disease susceptibility. Nat Rev Genet 2007, 8:253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tung J, Gilad Y: Social environmental effects on gene regulation. Cell Mol Life Sci 2013, 70:4323–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cole SW: Human Social Genomics. PLoS Genet 2014, 10:e1004601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanz J, Randolph HE, Barreiro LB: Genetic and evolutionary determinants of human population variation in immune responses. Curr Opin Genet Dev 2018, 53:28–35. [DOI] [PubMed] [Google Scholar]
- 18.Holekamp KE, Szykman M, Boydston EE, Smale L: Association of seasonal reproductive patterns with changing food availability in an equatorial carnivore, the spotted hyaena (Crocuta crocuta). Reproduction 1999, 116:87–93. [DOI] [PubMed] [Google Scholar]
- 19.Gesquiere LR, Onyango PO, Alberts SC, Altmann J: Endocrinology of year-round reproduction in a highly seasonal habitat: environmental variability in testosterone and glucocorticoids in baboon males. Am J Phys Anthropol 2011, 144:169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson RJ: Seasonal immune function and sickness responses. Trends Immunol 2004, 25:187–192. [DOI] [PubMed] [Google Scholar]
- 21.Castro Dopico X, Evangelou M, Ferreira RC, Guo H, Pekalski ML, Smyth DJ, Cooper N, Burren OS, Fulford AJ, Hennig BJ, et al. : Widespread seasonal gene expression reveals annual differences in human immunity and physiology. Nat Commun 2015, 6:7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferreira MS, Alves PC, Callahan CM, Marques JP, Mills LS, Good JM, Melo-Ferreira J: The transcriptional landscape of seasonal coat colour moult in the snowshoe hare. Mol Ecol 2017, 26:4173–4185. [DOI] [PubMed] [Google Scholar]
- 23.Faherty SL, Villanueva-Cañas JL, Blanco MB, Albà MM, Yoder AD: Transcriptomics in the wild: Hibernation physiology in free-ranging dwarf lemurs. Mol Ecol 2018, 27:709–722. [DOI] [PubMed] [Google Scholar]
- 24.Sáenz de Miera C, Monecke S, Bartzen-Sprauer J, Laran-Chich M-P, Pévet P, Hazlerigg DG, Simonneaux V: A circannual clock drives expression of genes central for seasonal reproduction. Curr Biol 2014, 24:1500–1506. [DOI] [PubMed] [Google Scholar]
- 25.Petri I, Diedrich V, Wilson D, Fernández-Calleja J, Herwig A, Steinlechner S, Barrett P: Orchestration of gene expression across the seasons: Hypothalamic gene expression in natural photoperiod throughout the year in the Siberian hamster. Sci Rep 2016, 6:29689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swindell WR: Dietary restriction in rats and mice: a meta-analysis and review of the evidence for genotype-dependent effects on lifespan. Ageing Res Rev 2012, 11:254–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mattison JA, Colman RJ, Beasley TM, Allison DB, Kemnitz JW, Roth GS, Ingram DK, Weindruch R, de Cabo R, Anderson RM: Caloric restriction improves health and survival of rhesus monkeys. Nat Commun 2017, 8:14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maegawa S, Lu Y, Tahara T, Lee JT, Madzo J, Liang S, Jelinek J, Colman RJ, Issa J-PJ: Caloric restriction delays age-related methylation drift. Nat Commun 2017, 8:539. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Aging-associated DNA methylation (DNAm) changes are conserved across mice, rhesus macaques, and humans, and the rate of this “epigenetic drift” correlated with differences in lifespan across these three species. Further, a caloric restriction, which extends the lifespan of these mammals, also slowed the rate of drift in rhesus macaques and mice. These findings suggests that epigenetic drift may affect lifespan and that dietary interventions increase longevity by slowing epigenetic drift.
- 29.Horvath S, Raj K: DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet 2018, 19:371–384. [DOI] [PubMed] [Google Scholar]
- 30.Roseboom T, de Rooij S, Painter R: The Dutch famine and its long-term consequences for adult health. Early Hum Dev 2006, 82:485–491. [DOI] [PubMed] [Google Scholar]
- 31.Tobi EW, Goeman JJ, Monajemi R, Gu H, Putter H, Zhang Y, Slieker RC, Stok AP, Thijssen PE, Muller F, et al. : DNA methylation signatures link prenatal famine exposure to growth and metabolism. Nat Commun 2014, 5:5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tobi EW, Slieker RC, Luijk R, Dekkers KF, Stein AD, Xu KM, Biobank-based Integrative Omics Studies Consortium, Slagboom PE, van Zwet EW, Lumey LH, et al. : DNA methylation as a mediator of the association between prenatal adversity and risk factors for metabolic disease in adulthood. Sci Adv 2018, 4:eaao4364. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Using whole blood samples from 1) individuals conceived during the Dutch Hunger Winter and 2) their siblings conceived during non-famine periods (as controls), Tobi et al. asked whether DNA methylation (DNAm) mediated the association between in utero famine exposure and metabolic health. The authors used formal mediation to show that DNAm at 6 loci explain ~80% of the relationship between famine exposure and altered triglyceride levels, providing evidence that DNAm is a key mechanisms linking environmental signals and health in humans.
- 33.Richmond R, Relton C, Davey Smith G: What evidence is required to suggest that DNA methylation mediates the association between prenatal famine exposure and adulthood disease? 2018, [Google Scholar]; • In their response to Tobi et al., Richmond and colleagues question the claim that the effects of famine exposure on metabolic health are causally mediated by DNA methylation (DNAm). In particular, they highlight that the ‘natural experiment’ study design of Tobi et al. cannot rule out confounding or reverse causation (e.g., famine exposure could increase triglyceride levels, which in turn alter DNA methylation). Using a recently developed tool for causal inference, Mendelian Randomization, the authors show that the mediating relationship uncovered by Tobi et al. is likely much weaker (or undetectable) relative to initial claims.
- 34.Newman JC, Covarrubias AJ, Zhao M, Yu X, Gut P, Ng C-P, Huang Y, Haidar S, Verdin E: Ketogenic Diet Reduces Midlife Mortality and Improves Memory in Aging Mice. Cell Metab 2017, 26:547–557.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The authors found that putting mice on a cyclical ketogenic diet, which mimics some of metabolic components of caloric restriction, reduced weight gain and mid-life mortality while improving memory and overall health. These consequences appear to be mediated by changes in metabolism associated gene expression in the liver and kidney; further, a key transcription factor, PPARα, appears to play a crucial role in this regulatory cascade. Thus, targeting the PPARα pathway may help to induce some of the beneficial outcomes of a ketogenic diet.
- 35.Sapolsky RM: The influence of social hierarchy on primate health. Science 2005, 308:648–652. [DOI] [PubMed] [Google Scholar]
- 36.Hinwood M, Tynan RJ, Day TA, Walker FR: Repeated social defeat selectively increases δFosB expression and histone H3 acetylation in the infralimbic medial prefrontal cortex. Cereb Cortex 2011, 21:262–271. [DOI] [PubMed] [Google Scholar]
- 37.Runcie DE, Wiedmann RT, Archie EA, Altmann J, Wray GA, Alberts SC, Tung J: Social environment influences the relationship between genotype and gene expression in wild baboons. Philos Trans R Soc Lond B Biol Sci 2013, 368:20120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snyder-Mackler N, Sanz J, Kohn JN, Brinkworth JF, Morrow S, Shaver AO, Grenier J-C, Pique-Regi R, Johnson ZP, Wilson ME, et al. : Social status alters immune regulation and response to infection in macaques. Science 2016, 354:1041–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The authors experimentally manipulated the social status of female rhesus macaques and found 1) a strong proinflammatory gene expression signature of social adversity (low social status) in peripheral immune cells and 2) an effect of social status on an individual’s response to bacterial infection. Importantly, the authors found that the gene regulatory consequences of social adversity were reversible, suggesting large-scale plasticity in gene expression at social adversity-associated loci.
- 39.Lehmann ML, Weigel TK, Elkahloun AG, Herkenham M: Chronic social defeat reduces myelination in the mouse medial prefrontal cortex. Sci Rep 2017, 7:46548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKim DB, Niraula A, Tarr AJ, Wohleb ES, Sheridan JF, Godbout JP: Neuroinflammatory Dynamics Underlie Memory Impairments after Repeated Social Defeat. J Neurosci 2016, 36:2590–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niraula A, Wang Y, Godbout JP, Sheridan JF: Corticosterone Production during Repeated Social Defeat Causes Monocyte Mobilization from the Bone Marrow, Glucocorticoid Resistance, and Neurovascular Adhesion Molecule Expression. J Neurosci 2018, 38:2328–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mulugeta E, Marion-Poll L, Gentien D, Ganswindt SB, Ganswindt A, Bennett NC, Blackburn EH, Faulkes CG, Heard E: Molecular insights into the pathways underlying naked mole-rat eusociality. 2017, doi: 10.1101/209932. [DOI] [Google Scholar]
- 43.Cole SW, Levine ME, Arevalo JMG, Ma J, Weir DR, Crimmins EM: Loneliness, eudaimonia, and the human conserved transcriptional response to adversity. Psychoneuroendocrinology 2015, 62:11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Canli T, Yu L, Yu X, Zhao H, Fleischman D, Wilson RS, De Jager PL, Bennett DA: Loneliness 5 years ante-mortem is associated with disease-related differential gene expression in postmortem dorsolateral prefrontal cortex. Transl Psychiatry 2018, 8:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cole SW: Social regulation of human gene expression: mechanisms and implications for public health. Am J Public Health 2013, 103 Suppl 1 :S84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lea AJ, Altmann J, Alberts SC, Tung J: Resource base influences genome-wide DNA methylation levels in wild baboons (Papio cynocephalus). Mol Ecol 2016, 25:1681–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Lea et al. profiled genome-wide, whole-blood DNA methylation (DNAm) levels in two groups of wild baboons exposed to different diets. They identified >1000 sites that were differentially methylated between the two groups, and provided statistical and experimental evidence that these differences affect gene expression levels at metabolism-related genes. These findings provide some of the first support for a role for DNA methylation in mediating ecological effects on phenotypic traits in wild animals.
- 47.McDade TW, Ryan C, Jones MJ, Maclsaac JL, Morin AM, Meyer JM, Borja JB, Miller GE, Kobor MS, Kuzawa CW: Social and physical environments early in development predict DNA methylation of inflammatory genes in young adulthood. Proc Natl Acad Sci U S A 2017, 114:7611–7616. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This study of a large, longitudinal cohort in the Philippines found that early life adversity was associated with DNA methylation (DNAm) levels at 10 loci in adult blood cells. Adversity-associated loci were located near genes involved in inflammation and, in a few cases, predicted variation in inflammatory biomarkers. The results suggest that changes in DNAm might provide a mechanistic link between early life adversity and negative health outcomes in adulthood.
- 48.Dunn EC, Soare TW, Simpkin AJ, Suderman MJ, Zhu Y, Klengel T, Smith ADAC, Ressler K, Relton CL: Sensitive periods for the effect of childhood adversity on DNA methylation: Results from a prospective, longitudinal study. 2018, doi: 10.1101/271122. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Dunn and colleagues found that the timing of early life adversity exposure was important for predicting patterns of DNA methylation (DNAm) in blood cells. Specifically, adversity experienced during infancy affected DNAm levels at many loci — comprising 80% of the loci associated with adversity experienced any time during development. These results suggest that infancy is a particularly sensitive period in which early life adversity can become biologically embedded in the developing immune system.
- 49.Lam LL, Emberly E, Fraser HB, Neumann SM, Chen E, Miller GE, Kobor MS: Factors underlying variable DNA methylation in a human community cohort. Proceedings of the National Academy of Sciences 2012, 109:17253–17260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Snyder-Mackler N, Sanz J, Kohn JN, Voyles TN, Pique-Regi R, Wilson ME, Barreiro LB, Tung J: Social status alters chromatin accessibility and the gene regulatory response to glucocorticoid stimulation in rhesus macaques. 2018, doi: 10.1101/365049. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Using an experimental manipulation of social status in rhesus macaques, Snyder-Mackler et al revealed that status alters immune cell gene expression and chromatin accessibility; that dexamethasone stimulation attenuates the status-related effects; and that low status animals may have less accessible DNA at glucocorticoid response-associated transcription factors. Together, these results demonstrate that social status affects multiple aspects of immune cell gene regulation, including chromatin accessibility; and that the cellular context (e.g., environmental stimulations) can alter the strength of this association.
- 51.Pacis A, Tailleux L, Lambourne J, Yotova V, Dumaine A, Luca F, Grenier J-C, Hansen KD, Gicquel B, Yu M, et al. : Bacterial Infection Remodels the DNA Methylation Landscape of Human Dendritic Cells. Genome Res 2015, doi: 10.1101/gr.192005.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pacis A, Mailhot-Leonard F, Tailleux L, Randolph HE, Yotova V, Dumaine A, Grenier J-C, Barreiro LB: Gene activation precedes DNA demethylation in response to infection in human dendritic cells. 2018, doi: 10.1101/358531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davey Smith G, Smith GD, Ebrahim S: “Mendelian randomization”: can genetic epidemiology contribute to understanding environmental determinants of disease?*. Int J Epidemiol 2003, 32:1–22. [DOI] [PubMed] [Google Scholar]
- 54.Lea AJ, Akinyi MY, Nyakundi R, Mareri P, Nyundo F, Kariuki T, Alberts S, Archie E, Tung J: Dominance rank-associated immune gene expression is widespread, sex-specific, and a precursor to high social status in wild male baboons. 2018, doi: 10.1101/366021. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Using genome-wide gene expression data collected from wild baboons, Lea et al. find that social status (i.e., dominance rank) is strongly associated with immune-related gene expression in males, but not females. The authors further leverage Mendelian Randomization to show that rank-related expression patterns are precursors, not consequences, of high status in males, in support of the idea that physiological condition determines who attains high rank. This work provides the first test of the relationship between social status and gene regulation in wild primates, as well as insights into the causal nature of this link.
- 55.Lea AJ, Vockley CM, Johnston RA, Del Carpio CA, Barreiro LB, Reddy TE, Tung J: Genome-wide quantification of the effects of DNA methylation on human gene regulation. 2017, doi: 10.1101/146829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ford EE, Grimmer MR, Stolzenburg S, Bogdanovic O, de Mendoza A, Farnham PJ, Blancafort P, Lister R: Frequent lack of repressive capacity of promoter DNA methylation identified through genome-wide epigenomic manipulation. 2017, doi: 10.1101/170506. [DOI] [Google Scholar]
- 57.Rittschof CC, Robinson GE: Behavioral Genetic Toolkits: Toward the Evolutionary Origins of Complex Phenotypes. Curr Top Dev Biol 2016, 119:157–204. [DOI] [PubMed] [Google Scholar]
- 58.Waterland RA, Jirtle RL: Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 2003, 23:5293–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waterland R a., Kellermayer R, Laritsky E, Rayco-Solon P, Harris RA, Travisano M, Zhang W, Torskaya MS, Zhang J, Shen L, et al. : Season of conception in rural gambia affects DNA methylation at putative human metastable epialleles. PLoS Genet 2010, 6:e1001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lea AJ, Archie EA, Tung J, Alberts SC: Developmental plasticity: bridging research in evolution and human health. Evolution, Medicine, and Public Health 2018, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Levine ME, Crimmins EM, Weir DR, Cole SW: Contemporaneous Social Environment and the Architecture of Late-Life Gene Expression Profiles. Am J Epidemiol 2017, 186:503–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paula Braveman LG: The Social Determinants of Health: It’s Time to Consider the Causes of the Causes. Public Health Rep 2014, 129:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen E, Miller GE, Kobor MS, Cole SW: Maternal warmth buffers the effects of low early-life socioeconomic status on pro-inflammatory signaling in adulthood. Mol Psychiatry 2011, 16:729–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lea AJ, Altmann J, Alberts SC, Tung J: Developmental constraints in a wild primate. Am Nat 2015, 185:809–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Charruau P, Johnston RA, Stahler DR, Lea A, Snyder-Mackler N, Smith DW, vonHoldt BM, Cole SW, Tung J, Wayne RK: Pervasive Effects of Aging on Gene Expression in Wild Wolves. Mol Biol Evol 2016, 33:1967–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bengston SE, Dahan RA, Donaldson Z, Phelps SM, van Oers K, Sih A, Bell AM: Genomic tools for behavioural ecologists to understand repeatable individual differences in behaviour. Nat Ecol Evol 2018, doi: 10.1038/s41559-017-0411-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lea AJ, Vilgalys TP, Durst PAP, Tung J: Maximizing ecological and evolutionary insight in bisulfite sequencing data sets. Nat Ecol Evol 2017, 1:1074–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. : Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505:559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smits SA, Leach J, Sonnenburg ED, Gonzalez CG, Lichtman JS, Reid G, Knight R, Manjurano A, Changalucha J, Elias JE, et al. : Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science 2017, 357:802–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, Scott NM, Gibbons SM, Larsen P, Shogan BD, Weiss S, et al. : Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 2014, 345:1048–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tung J, Barreiro LB, Burns MB, Grenier J-C, Lynch J, Grieneisen LE, Altmann J, Alberts SC, Blekhman R, Archie EA: Social networks predict gut microbiome composition in wild baboons. Elife 2015, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moeller AH, Foerster S, Wilson ML, Pusey AE, Hahn BH, Ochman H: Social behavior shapes the chimpanzee pan-microbiome. Sci Adv 2016, 2:e1500997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lynch SV, Pedersen O: The Human Intestinal Microbiome in Health and Disease. N Engl J Med 2016, 375:2369–2379. [DOI] [PubMed] [Google Scholar]
- 74.Dinan TG, Cryan JF: Brain–gut–microbiota axis — mood, metabolism and behaviour. Nat Rev Gastroenterol Hepatol 2017, 14:69–70. [DOI] [PubMed] [Google Scholar]
- 75.Franzosa EA, Hsu T, Sirota-Madi A, Shafquat A, Abu-Ali G, Morgan XC, Huttenhower C: Sequencing and beyond: integrating molecular “omics” for microbial community profiling. Nat Rev Microbiol 2015, 13:360–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moore AJ, Brodie ED, Wolf JB: Interacting Phenotypes and the Evolutionary Process: I. Direct and Indirect Genetic Effects of Social Interactions. Evolution 1997, 51:1352. [DOI] [PubMed] [Google Scholar]
- 77.Kong A, Thorleifsson G, Frigge ML, Vilhjalmsson BJ, Young Al, Thorgeirsson TE, Benonisdottir S, Oddsson A, Halldorsson BV, Masson G, et al. : The nature of nurture: Effects of parental genotypes. Science 2018, 359:424–428. [DOI] [PubMed] [Google Scholar]
- 78.Baud A, Casale FP, Nicod J, Stegle O: Genome-wide association study of social genetic effects on 170 phenotypes in laboratory mice. 2018, doi: 10.1101/302349. [DOI] [Google Scholar]
- 79.Baud A, Mulligan MK, Casale FP, Ingels JF, Bohl CJ, Callebert J, Launay J-M, Krohn J, Legarra A, Williams RW, et al. : Genetic Variation in the Social Environment Contributes to Health and Disease. PLoS Genet 2017, 13:e1006498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moyerbrailean GA, Richards AL, Kurtz D, Kalita CA, Davis GO, Harvey CT, Alazizi A, Watza D, Sorokin Y, Hauff N, et al. : High-throughput allele-specific expression across 250 environmental conditions. Genome Res 2016, 26:1627–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maranville JC, Luca F, Richards AL, Wen X, Witonsky DB, Baxter S, Stephens M, Di Rienzo A: Interactions between glucocorticoid treatment and cis-regulatory polymorphisms contribute to cellular response phenotypes. PLoS Genet 2011, 7:e1002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nédélec Y, Sanz J, Baharian G, Tung J, Yotova V, Barreiro Correspondence LB, Szpiech ZA, Pacis A, Dumaine A, Grenier J, et al. : Genetic Ancestry and Natural Selection Drive Population Differences in Immune Responses to Pathogens. Cell 2016, 167:113. [DOI] [PubMed] [Google Scholar]
- 83.Penninx BWJH, Beekman ATF, Smit JH, Zitman FG, Nolen WA, Spinhoven P, Cuijpers P, De Jong PJ, Van Marwijk HWJ, Assendelft WJJ, et al. : The Netherlands Study of Depression and Anxiety (NESDA): Rationale, objectives and methods. Int J Methods Psychiatr Res 2008, 17:121–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tung J, Zhou X, Alberts SC, Stephens M, Gilad Y: The genetic architecture of gene expression levels in wild baboons. Elife 2015, 4:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hoffman GE, Schadt EE: variancePartition: interpreting drivers of variation in complex gene expression studies. BMC Bioinformatics 2016, 17:483. [DOI] [PMC free article] [PubMed] [Google Scholar]