Abstract
Polymerization induces hydrolysis of ATP bound to actin, followed by γ-phosphate release, which helps advance the disassembly of actin filaments into ADP-G-actin. Mechanical understanding of this correlation between actin assembly and ATP hydrolysis has been an object of intensive studies in biochemistry and structural biology for many decades. Although actin polymerization and depolymerization occur only at either the barbed or pointed ends and the kinetic and equilibrium properties are substantially different from each other, characterizing their properties is difficult to do by bulk assays, as these assays report the average of all actin filaments in solution and are therefore not able to discern the properties of individual actin filaments. Biochemical studies of actin polymerization and hydrolysis were hampered by these inherent properties of actin filaments. Total internal reflection fluorescence (TIRF) microscopy overcame this problem by observing single actin filaments. With TIRF, we now know not only that each end has distinct properties, but also that the rate of γ-phosphate release is much faster from the terminals than from the interior of actin filaments. The rate of γ-phosphate release from actin filament ends is even more accelerated when latrunculin A is bound. These findings highlight the importance of resolving structural differences between actin molecules in the interior of the filament and those at either filament end. This review provides a history of observing actin filaments under light microscopy, an overview of dynamic properties of ATP hydrolysis at the end of actin filament, and structural views of γ-phosphate release.
Keywords: TIRF, Single actin filament observation, ATP hydrolysis, Phosphate (Pi) release
Introduction
All eukaryotic cells need cytoskeletons to support three-dimensional shapes and maintain various dynamic cell motilities. Actin is a cytoskeletal filament protein well known for its role in generating forces that propel cell membrane protrusions at the leading edge of cells, such as filopodia and lamellipodia, by its polymerization (Blanchoin et al. 2014). Polymerization dynamics of actin in vitro has been researched for 70 years and interactions with many actin-binding proteins have been discovered. From the beginning of the study of treadmilling (Wegner 1976), there was an inconsistent wide gap between the slow intrinsic rates of actin polymerization and depolymerization, and the much faster velocity of actin-driven cell protrusion. In an effort to bridge the gap, some accessory proteins like formins have been discovered that can accelerate polymerization (Kovar and Pollard 2004), whereas proteins such as ADF/cofilin and the gelsolin family promote disassembly (De La Cruz et al. 2015; Kinosian et al. 1998). Investigations into the fine-tuning of actin polymerization dynamics, which is able to produce forces and change velocities under various cellular functions, were greatly aided with the improvement in technology allowing the observation of single actin filament elongation and depolymerization in real time. In this review, we describe actin polymerization and depolymerization dynamics with ATP hydrolysis and structural fluctuations that were elucidated through direct observations of single actin filaments and structural analyses.
Paradigm shift of actin polymerization research was triggered by TIRF microscopy development
An early stage of actin filament observation under light microscopy was performed by using dark-field microscopy in 1980 (Nagashima and Asakura 1980). Instead of labeling actin with a fluorescent dye, actin filaments were mixed with skeletal muscle myosin II and diluted to 24 nM by a phosphate-buffered solution. Decoration of actin filaments with heavy meromyosin (HMM) enhanced the mass of the actin filaments, so that acto-myosin filaments were able to be visualized (Fig. 1a). By measuring the thermal bending motions of actin filaments over time, it was confirmed that actin filaments are semi-flexible polymers. In 1984, as epifluorescence light microscopy was commercially available, single actin filaments labeled with fluorescent phalloidin were able to be visualized without myosin (Fig, 1b) (Honda et al. 1986). Brownian motion observation of actin filaments over time revealed that actin filaments became more flexible when interacting with myosin, while tropomyosin-bound actin filaments became rigid (Yanagida et al. 1984). However, the direct measurement of polymerization and depolymerization of individual actin filaments was missed under these conditions because of the following two reasons; first, to measure the intrinsic elongation rate, actin must be polymerized in conditions with concentrations of actin monomers above a certain concentration, called the critical concentration. However, too many actin filaments were nucleated above this concentration; thus, individual actin filaments were indistinguishable because actin filaments overlapped with each other. Second, phalloidin prevents actin depolymerization. Indeed, this is why phalloidin is widely used to fix the actin cytoskeleton. In 2001, the elongation rate was determined by observing single actin filaments under an epifluorescence microscope (Ishiwata et al. 2001). It was performed by mixing rhodamine phalloidin and non-labeled actin while below the critical concentration. It was a unique approach, although depolymerization was suppressed. In vitro, phalloidin minimizes the density of actin filaments and phalloidin-bound actin filaments have a twofold longer persistence length than bare actin filaments (Isambert et al. 1995). These issues were solved as a result of progress in single-molecule imaging techniques. The combination of TIRF (total internal reflection fluorescence) microscopy and a highly sensitive camera together with an image intensifier such as II-SIT (image intensifier and silicon intensified target) allowed us to visualize individual actin filaments located near the glass surface (Fujiwara et al. 2002). Barbed ends grow much faster than pointed ends (Fig. 1c, arrowheads). Single actin filament observation with actin alone revealed that the elongation rates of individual actin filaments are uniform. Although actin filament elongation rate at the barbed end is reduced when increasing the ratio of fluorescence-labeled actin to unlabeled actin monomers (Amann and Pollard 2001), the intrinsic elongation rate, as estimated from the intercept of the elongation rate vs. label ratio, is 7.4~12/μM/s, regardless of which conjugated fluorescent dye is used (Fujiwara et al. 2002; Kuhn and Pollard 2005). The measured elongation rates were comparable to previous research performed by using EM of 11.6/μM/s (Pollard 1986). Although elongation from the barbed end is easily detected because of its fast rate, the elongation from the pointed end tends to be buried in noise, because it has a slow elongation rate and high critical concentration.
Fig. 1.
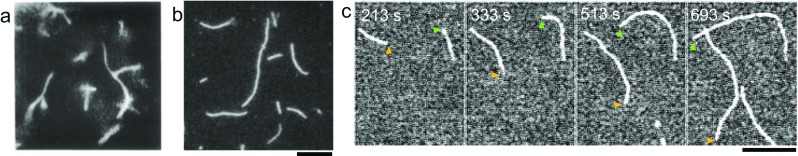
History of visualizing actin filaments under light microscopy. a Non-fluorescent dye conjugated actin filaments were visualized by dark-field light microscopy. Actin filaments were decorated by HMM. Image was taken by a high-sensitivity television camera with × 1500 magnification. Image a is from Nagashima and Asakura (1980). b FITC-labeled actin filaments (50 nM, 85% label) in the presence of 0.1 mM phalloidin monitored by using epi fluorescence microscope. The bar represents 10 μm. c Time course image of elongation of 15% Alexa488-lysine-labeled Mg-ATP-actin of 1 μM monitored by TIRF microscope with EMCCD. Scale bar is 10 μm. Arrowheads indicate the fast-growing end of each filament and time was indicated in each panel (t = 0, actin was mixed with the polymerization buffer)
Observation of individual actin filaments without phalloidin suggested that the length of actin filaments were fluctuated at steady state (Fujiwara et al. 2002). One-dimensional Brownian motion of the filament length at around the critical concentration of actin proposed a unique mechanism, that this fluctuation may be caused by the association and dissociation of small actin clusters consisting of six subunits at filament ends. Similar behavior was also observed in Kuhn and Pollard in 2005 (Kuhn and Pollard 2005) by using TIRF microscopy with an EMCCD (electron-multiplying charge-coupled device). With these experiments, actin biochemistry was able to be more carefully quantified. A three-color detection system showed the regulatory mechanism of the barbed end of single actin filaments with mDia1 and capping protein (CP) (Bombardier et al. 2015). Solution exchanges in microfluidic chambers combined with TIRF microscopy showed that actin depolymerization is accelerated by profilin (Jégou et al. 2011). These approaches applied to future studies will answer even more regulatory mechanisms of the cytoskeleton at the level of the single filament.
Single actin filament observation revealed that the on and off rates of Pi are different between the interior and terminal subunits of actin filaments
ATP-, ADP-Pi-, and ADP-actin can all associate and dissociate at both barbed and pointed ends of filaments, although the majority of actin monomers bind ATP in living cells (Rosenblatt et al. 1995). Once an ATP-actin monomer is polymerized, its conformation changes (Oda et al. 2009) and ATP is hydrolyzed irreversibly at the rate of 0.3/s (Blanchoin and Pollard 2002). Including the above, solution experiments also estimated that γ-phosphate dissociates slowly from actin at ~ 0.001–0.006/s (Blanchoin and Pollard 1999; Carlier and Pantaloni 1986, 1988; Melki et al. 1996), and the dissociation of γ-phosphate occurs at random locations along an actin filament (Blanchoin and Pollard 1999). In 2011, Jégou et al. visually confirmed that actin filaments that were polymerized from ATP-actin are depolymerized at a gradually increasing rate until it reaches the rate of ADP-actin (Jégou et al. 2011). This observation confirmed that γ-phosphate release occurs at random locations. These kinetic rates show that ATP hydrolysis is crucial as a timekeeper for the aging of an actin filament, in particular that the slow γ-phosphate release from the filament is important to extend the lifetime of each actin filament. However, this slow and random dissociation of γ-phosphate from actin filaments raised a question for treadmilling; a few γ-phosphates remaining in old regions of actin filaments may be enough to pause the treadmilling by inhibiting depolymerization. In fact, the concentration of inorganic phosphate (Pi) in a cell (Burt et al. 1977) is high enough to bind Pi to actin filaments, as shown in vitro where Pi binding to filamentous actin easily occurs at similar concentrations (Kd = 1.5 mM) (Fujiwara et al. 2007), suggesting that treadmilling may be suppressed in a cell. Single actin filament observation by TIRF microscopy solved this question by showing that both association and dissociation rates of Pi to the end of actin filaments are much faster than the rates in internal locations (Fig. 2a (Fujiwara et al. 2007)). The estimated rates of γ-phosphate release from ends are very rapid at 2/sec, which is over 650 times faster than the γ-phosphate release from the internal sections of an actin filament. The progressively accelerated depolymerization of actin filaments that was polymerized from Mg-ATP-actin (Jégou et al. 2011) suggests that rapid γ-phosphate release from the terminal ends of actin filaments is influenced by both the surrounding environment and the nucleotide state of neighboring subunits (Korn et al. 1987). While it is still unclear how many actin subunits from the terminal ends cooperatively dissociate Pi rapidly, this rapid Pi release allows actin filaments to depolymerize, which promotes treadmilling. The rapid γ-phosphate dissociation, which is followed by actin depolymerization, was not detected until single actin filament observation was performed. Rapid γ-phosphate dissociation from the terminal ends explains how treadmilling can occur in physiological conditions containing several millimolars of Pi without any actin-associated proteins.
Fig. 2.
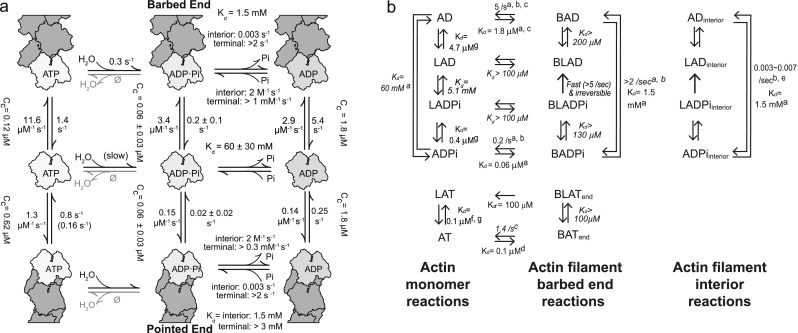
Rates of polymerization and depolymerization of actin in each nucleotide state. a Kinetic and thermodynamic parameters of actin polymerization at the barbed- and pointed-end polymerization with bound ATP (left top, middle and bottom), ADP-Pi (center top, middle and bottom), or ADP (right top, middle and bottom). Numbers are kinetic rates of each reaction (arranged from Fujiwara et al. 2007). Kds and Cc indicate equilibrium dissociation constants of reactions and the critical concentration, respectively. Two association and dissociation rates of Pi binding are shown at both ends, because of the loose binding of Pi at the terminal actin subunits. b Thermodynamic reactions of actin in the presence of latrunculin A. Abbreviations: A, actin; L, Latrunculin A; T, ATP; D, ADP: Pi, inorganic phosphate; B, barbed end. Numbers next to arrows are dissociation rate or equilibrium dissociation constants (Kd). (Left) Monomeric actin reactions. (Middle) Reactions on actin filament barbed ends. (Right) Reactions of interior subunits in actin filaments. Parameters shown in italics were calculated using detailed balance. Superscripts indicate references: a, Fujiwara et al. (2002)); b, Jégou et al. (2011)); c, Pollard (1986)); d, Kuhn and Pollard (2005)); e, Blanchoin and Pollard (1999)); f, Coué et al. (1987)); g Fujiwara et al. (2018))
Recently, TIRF observation was used to investigate how latrunculin A inhibits actin polymerization. In addition to actin monomer sequestering function, latrunculin A increased the dissociation rate from both ends at the rate of ADP-actin subunits, which is the maximum intrinsic depolymerization rate (Fig. 2b (Fujiwara et al. 2018)). This fast depolymerization mechanism is highly possibly because latrunculin A accelerates γ-phosphate release from terminal subunits of actin filaments. This finding confirms that γ-phosphate loosely binds to actin subunits at terminal subunits. Thus, the γ-phosphate dissociates easily and is also affected by additional factors like latrunculin A. When actin depolymerization rate is estimated under various rates of phosphate release at barbed end in a cell, the depolymerization rate is increased up to 60% if latrunculin A accelerates γ-phosphate release to 5/s, or more than doubles when γ-phosphate is released at 20/s.
Exactly how many actin subunits near terminal ends bind γ-phosphate is unclear, as it is also unclear that ADP-Pi-actin subunits at the ends may take the same conformation from the bulk of the ADP-Pi-actin subunits in the middle of filaments. Ratios and rates of ADP-Pi-actin binding may be changed with surrounding conditions, and these kinetic rates can be determined by solution assays. However, the functional mechanism of actin as a biopolymer requires structural information to fully understand actin polymerization dynamics. For example, latrunculin A favors monomeric actin rather than filamentous actin-binding ADP-Pi (Fujiwara et al. 2018). This suggests that the latrunculin A binding manner varies based upon the structures of monomeric and filamentous actin. EM data of actin structure at pointed ends supports this hypothesis (Narita et al. 2011), by showing different conformations of actin at filament terminals and interiors (Oda et al. 2009). While there is agreement that the γ-phosphate release at terminal ends of actin filaments is fast, the known ATP hydrolysis initiated from actin polymerization and the known conformational actin dynamics do not fully explain this mechanism. In the next paragraph, we are going to describe γ-phosphate binding from a structural viewpoint.
Structural view of filament ends and Pi regulation
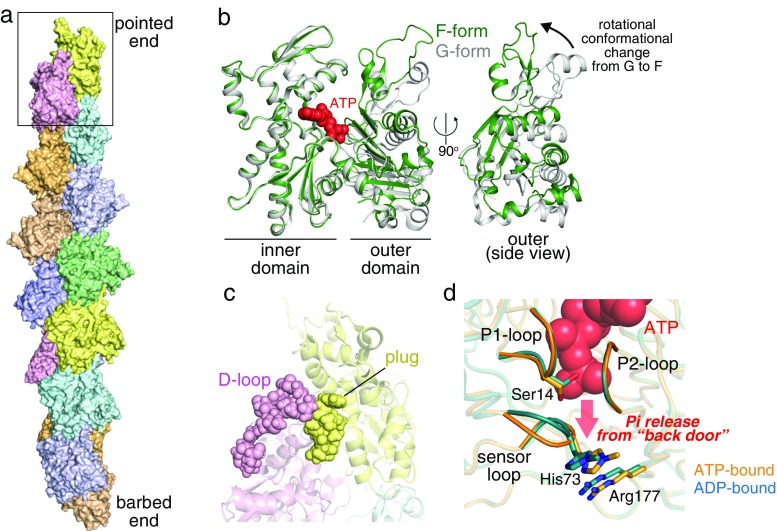
Structurally, an actin molecule consists of two domains, an inner and an outer domain (Kabsch et al. 1990), that undergo a rotational movement relative to each other upon polymerization (Fig. 3b) (Oda et al. 2009). Because ATP is located between the two major domains, its hydrolysis is likely to be triggered by a structural rearrangement around the nucleotide induced by the G- to F-conformational change (Merino et al. 2018; Oda et al. 2009). Owing to the recent technology progress of cryo-EM, the structure of the actin filament has been described at near-atomic resolutions (Fig. 3a) (Mentes et al. 2018; Pospich et al. 2017; von der Ecken et al. 2015; Tanaka et al. 2018). In 2018, a study suggested that, based on filament structures with different nucleotide states, ATP hydrolysis and the following γ-phosphate release mainly affect the conformation of the DNase I binding loop (D-loop), a critical region mediating the longitudinal subunit interaction, thereby modulating the filament stability (Merino et al. 2018).
Fig. 3.
Actin structures. a Structure of F-actin (PDB: 2zwh (Oda et al. 2009)). Top, pointed end (marked by a box; enlarged in c); bottom, barbed end. b Conformational change of actin from monomeric form (G-form) (gray, PDB: 1j6z (Otterbein et al. 2001)) to filamentous form (F-form) (green). Front (left) and side (right) views. In the side view, inner domains are omitted for clarity. c The pointed-end structure (PDB: 2y83). An extra link is formed between the plug of the pointed-end subunit and D-loop of the penultimate subunit (shown in spheres). d A close-up view of the nucleotide-binding site, shown as the back side of actin is oriented to the bottom. AMPPNP-bound (orange, PDB: 1nwk (Graceffa and Dominguez 2003)) and ADP-bound (cyan, 1j6z) G-form actin structures are superimposed. P1, P2, and sensor loops are highlighted. Residues proposed for involvement in controlling the Pi release are shown in stick models
It is reasonable to assume that, compared to filament interior subunits, end-exposed subunits adopt different conformations because they have fewer neighbors. Currently, a few studies have addressed the filament end structures, even including terminal structures capped by other proteins (Narita et al. 2011, 2006; Rao et al. 2014; Urnavicius et al. 2015). Among these, a cryo-EM structure of non-capped pointed-end (23-Å resolution) revealed that there is an extra link between the two pointed-end subunits that may have a functional relevance in regard to its significantly slow dynamics (Fig. 3c) (Narita et al. 2011). Elucidation of the filament end structures by single-particle cryo-EM analysis has been hampered by difficulty to collect large numbers of the end images required for a robust averaging process. Because the assembly and disassembly occur exclusively at the ends, direct structural information of the filament terminals, but not averaged structures of subunits along the filament, would be valuable to understand the kinetic differences between the two ends that have been established by biochemical studies.
ADP-G-actin seldomly incorporates Pi from the solution (Kd = 60 mM) (Fujiwara et al. 2007). This can be explained by structural differences in the active site between ATP- and ADP-G-actins (Fig. 3d). Although the overall structures are nearly identical in both nucleotide states, there is a slight difference in the P1 loop, one of the two loops that clamp the phosphate tail of the nucleotides (Graceffa and Dominguez 2003; Otterbein et al. 2001; Rould et al. 2006). In ADP-G-actin, a side chain of Ser14 on the P1 loop flips to a space that is occupied by γ-phosphate in ATP-G-actin. This local change in the active site is accompanied by a structural alteration of the sensor loop carrying methylated His73. These conformational rearrangements may act as a structural barrier in ADP-G-actin for Pi uptake. On the contrary, ADP-bound subunits in the filaments, especially those located near barbed ends, accept Pi more readily than ADP-G-actin (Kd = 1.5–3 mM) (Carlier and Pantaloni 1988; Fujiwara et al. 2007). This suggests that, in the filaments, Pi release does not induce a significant conformational change in the active site (particularly at the γ-phosphate site). A more likely explanation is that the conformation of subunits in the filament is designed to be stable when it binds γ-phosphate so that ADP-F-actin easily incorporates Pi. This is in line with biochemical data showing that ADP-Pi-F-actin is most stable during the actin polymerization cycle (Fujiwara et al. 2007). Near-atomic resolution cryo-EM F-actin structures appear to support this notion (Merino et al. 2018), although higher resolution information is required for a detailed comparison of the active site structures. Interestingly, the pointed ends exhibit a weak affinity for Pi (Fujiwara et al. 2007), implying that, upon Pi release, subunits located at this less-dynamic end may undergo a substantial conformational change in the nucleotide-binding pocket, just as occurs in G-actin.
Once actin is polymerized, the γ-phosphate bond is broken from the other two sequential phosphates in an ATP’s tail as a result of hydrolysis. In actin filaments, this phosphate tail of ATP locates as its γ-phosphate faces the filament axis side. It is therefore likely that Pi produced by the hydrolysis escapes from the subunit at the backside of the molecule (Fig. 3d). This back door Pi release pathway was proposed based on a molecular dynamics simulation (Wriggers and Schulten 1999). This study assumed that His73, Arg177, and some residues on the P1 and P2 loops play a key role in controlling the timing of Pi release. Interestingly, these residues are exposed to the solution particularly at the barbed-end subunits, potentially explaining the high sensitivity for Pi of this dynamic end.
Concluding remarks
To understand actin-based cell motility, the intrinsic kinetic rates for the polymerization and depolymerization reactions including the coordinated conformations of monomeric and filamentous actin at each nucleotide state are necessary. As the light microscope observation technique is developed, ATP hydrolysis and molecular dynamics of each actin subunit in different locations on one filament may be deciphered. By adding more functions such as microfluidic system, and other spatial and temporal manipulation systems including structural information, the dynamic mechanisms of actin with its binding proteins will be determined as a biomolecular machine. Structural dynamics is crucial information to understand the mechanical properties of actin polymerization with ATP hydrolysis. Unveiling structural differences between actin molecules along an actin filament should answer how individual biopolymers change their disposition of response and maintain the cell cytoskeleton.
Acknowledgments
We thank all editors (Enrique De La Cruz, Laurent Blanchoin, and Mike Ostap) who helped editing this review. We are also grateful to Shin’ichi Ishiwata and Timothy Day for the careful reading and editing.
Funding
This study was funded by the JSPS KAKENHI Grant Number 17K07373 and Toyota Physical and Chemical Research Institute, 41-1, Yokomichi, Nagakute, Aichi 480-1192, Japan.
Conflict of interest
Ikuko Fujiwara declares that she has no conflict of interest. Shuichi Takeda declares that he has no conflict of interest. Toshiro Oda declares that he has no conflict of interest. Hajime Honda declares that he has no conflict of interest. Akihiro Narita declares that he has no conflict of interest. Yuichiro Maéda declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Contributor Information
Ikuko Fujiwara, Email: fujiwara.ikuko@nitech.ac.jp.
Shuichi Takeda, Email: takeda.shuichi@f.mbox.nagoya-u.ac.jp.
References
- Amann KJ, Pollard TD. Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy. Proc Natl Acad Sci U S A. 2001;98:15009–15013. doi: 10.1073/pnas.211556398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD. Mechanism of interaction of Acanthamoeba actophorin (ADF/Cofilin) with actin filaments. J Biol Chem. 1999;274:15538–15546. doi: 10.1074/jbc.274.22.15538. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD. Hydrolysis of ATP by polymerized actin depends on the bound divalent cation but not profilin. Biochemistry. 2002;41:597–602. doi: 10.1021/bi011214b. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev. 2014;94:235–263. doi: 10.1152/physrev.00018.2013. [DOI] [PubMed] [Google Scholar]
- Bombardier JP, Eskin JA, Jaiswal R, Correa IR, Jr, Xu MQ, Goode BL, Gelles J. Single-molecule visualization of a formin-capping protein ‘decision complex’ at the actin filament barbed end. Nat Commun. 2015;6:8707. doi: 10.1038/ncomms9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt CT, Glonek T, Barany M. Analysis of living tissue by phosphorus-31 magnetic resonance. Science. 1977;195:145–149. doi: 10.1126/science.188132. [DOI] [PubMed] [Google Scholar]
- Carlier MF, Pantaloni D. Direct evidence for ADP-Pi-F-actin as the major intermediate in ATP-actin polymerization. Rate of dissociation of Pi from actin filaments. Biochemistry. 1986;25:7789–7792. doi: 10.1021/bi00372a001. [DOI] [PubMed] [Google Scholar]
- Carlier MF, Pantaloni D. Binding of phosphate to F-ADP-actin and role of F-ADP-Pi-actin in ATP-actin polymerization. J Biol Chem. 1988;263:817–825. [PubMed] [Google Scholar]
- Coué M, Brenner SL, Spector I, Korn ED. Inhibition of actin polymerization by latrunculin A. FEBS Lett. 1987;213:316–318. doi: 10.1016/0014-5793(87)81513-2. [DOI] [PubMed] [Google Scholar]
- De La Cruz EM, Martiel JL, Blanchoin L. Mechanical heterogeneity favors fragmentation of strained actin filaments. Biophys J. 2015;108:2270–2281. doi: 10.1016/j.bpj.2015.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara I, Takahashi S, Tadakuma H, Funatsu T, Ishiwata S. Microscopic analysis of polymerization dynamics with individual actin filaments. Nat Cell Biol. 2002;4:666–673. doi: 10.1038/ncb841. [DOI] [PubMed] [Google Scholar]
- Fujiwara I, Vavylonis D, Pollard TD. Polymerization kinetics of ADP- and ADP-Pi-actin determined by fluorescence microscopy. Proc Natl Acad Sci U S A. 2007;104:8827–8832. doi: 10.1073/pnas.0702510104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara I, Zweifel M, Courtemanche N, Pollard TD. Latrunculin A accelerates actin filament depolymerization in addition to sequestering actin monomers. Curr Biol. 2018;28:3183–3192. doi: 10.1016/j.cub.2018.07.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graceffa P, Dominguez R. Crystal structure of monomeric actin in the ATP state. Structural basis of nucleotide-dependent actin dynamics. J Biol Chem. 2003;278:34172–34180. doi: 10.1074/jbc.M303689200. [DOI] [PubMed] [Google Scholar]
- Honda H, Nagashima H, Asakura S. Directional movement of F-actin in vitro. J Mol Biol. 1986;191:131–133. doi: 10.1016/0022-2836(86)90428-6. [DOI] [PubMed] [Google Scholar]
- Isambert H, Venier P, Maggs AC, Fattoum A, Kassab R, Pantaloni D, Carlier MF. Flexibility of actin filaments derived from thermal fluctuations. Effect of bound nucleotide, phalloidin, and muscle regulatory proteins. J Biol Chem. 1995;270:11437–11444. doi: 10.1074/jbc.270.19.11437. [DOI] [PubMed] [Google Scholar]
- Ishiwata S, Tadashige J, Masui I, Nishizaka T, Kinosita K., Jr . Microscopic analysis of polymerization and fragmentation of individual actin filaments. In: dos Remedios CG, Thomas DD, editors. Molecular Interactions of Actin: Actin Structure and Actin-binding Proteins. Heidelberg: Springer Verlag; 2001. pp. 79–94. [DOI] [PubMed] [Google Scholar]
- Jégou A, Niedermayer T, Orbán J, Didry D, Lipowsky R, Carlier MF, Romet-Lemonne G. Individual actin filaments in a microfluidic flow reveal the mechanism of ATP hydrolysis and give insight into the properties of profilin. PLoS Biol. 2011;9:e1001161. doi: 10.1371/journal.pbio.1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Atomic structure of the actin: DNase I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- Kinosian HJ, Newman J, Lincoln B, Selden LA, Gershman LC, Estes JE. Ca2+ regulation of gelsolin activity: binding and severing of F-actin. Biophys J. 1998;75:3101–3109. doi: 10.1016/S0006-3495(98)77751-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn ED, Carlier MF, Pantaloni D. Actin polymerization and ATP hydrolysis. Science. 1987;238:638–644. doi: 10.1126/science.3672117. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci U S A. 2004;101:14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JR, Pollard TD. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys J. 2005;88:1387–1402. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melki R, Fievez S, Carlier MF. Continuous monitoring of Pi release following nucleotide hydrolysis in actin or tubulin assembly using 2-amino-6-mercapto-7-methylpurine ribonucleoside and purine-nucleoside phosphorylase as an enzyme-linked assay. Biochemistry. 1996;35:12038–12045. doi: 10.1021/bi961325o. [DOI] [PubMed] [Google Scholar]
- Mentes A, et al. High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing. Proc Natl Acad Sci U S A. 2018;115:1292–1297. doi: 10.1073/pnas.1718316115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino F, et al. Structural transitions of F-actin upon ATP hydrolysis at near-atomic resolution revealed by cryo-EM. Nat Struct Mol Biol. 2018;25:528–537. doi: 10.1038/s41594-018-0074-0. [DOI] [PubMed] [Google Scholar]
- Nagashima H, Asakura S. Dark-field light microscopic study of the flexibility of F-actin complexes. J Mol Biol. 1980;136:169–182. doi: 10.1016/0022-2836(80)90311-3. [DOI] [PubMed] [Google Scholar]
- Narita A, Takeda S, Yamashita A, Maeda Y. Structural basis of actin filament capping at the barbed-end: a cryo-electron microscopy study. EMBO J. 2006;25:5626–5633. doi: 10.1038/sj.emboj.7601395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita A, Oda T, Maeda Y. Structural basis for the slow dynamics of the actin filament pointed end. EMBO J. 2011;30:1230–1237. doi: 10.1038/emboj.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T, Iwasa M, Aihara T, Maéda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- Otterbein LR, Graceffa P, Dominguez R. The crystal structure of uncomplexed actin in the ADP state. Science. 2001;293:708–711. doi: 10.1126/science.1059700. [DOI] [PubMed] [Google Scholar]
- Pollard TD. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J Cell Biol. 1986;103:2747–2754. doi: 10.1083/jcb.103.6.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospich S, Kumpula EP, von der Ecken J, Vahokoski J, Kursula I, Raunser S. Near-atomic structure of jasplakinolide-stabilized malaria parasite F-actin reveals the structural basis of filament instability. Proc Natl Acad Sci U S A. 2017;114:10636–10641. doi: 10.1073/pnas.1707506114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao JN, Madasu Y, Dominguez R. Mechanism of actin filament pointed-end capping by tropomodulin. Science. 2014;345:463–467. doi: 10.1126/science.1256159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt J, Peluso P, Mitchison TJ. The bulk of unpolymerized actin in Xenopus egg extracts is ATP-bound. Mol Biol Cell. 1995;6:227–236. doi: 10.1091/mbc.6.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rould MA, Wan Q, Joel PB, Lowey S, Trybus KM. Crystal structures of expressed non-polymerizable monomeric actin in the ADP and ATP states. J Biol Chem. 2006;281:31909–31919. doi: 10.1074/jbc.M601973200. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Takeda S, Mitsuoka K, Oda T, Kimura-Sakiyama C, Maéda Y, Narita A. Structural basis for cofilin binding and actin filament disassembly. Nat Commun. 2018;9:1860. doi: 10.1038/s41467-018-04290-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnavicius L, et al. The structure of the dynactin complex and its interaction with dynein. Science. 2015;347:1441–1446. doi: 10.1126/science.aaa4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Ecken J, Müller M, Lehman W, Manstein DJ, Penczek PA, Raunser S. Structure of the F-actin-tropomyosin complex. Nature. 2015;519:114–117. doi: 10.1038/nature14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner A. Head to tail polymerization of actin. J Mol Biol. 1976;108:139–150. doi: 10.1016/S0022-2836(76)80100-3. [DOI] [PubMed] [Google Scholar]
- Wriggers W, Schulten K. Investigating a back door mechanism of actin phosphate release by steered molecular dynamics. Proteins. 1999;35:262–273. doi: 10.1002/(SICI)1097-0134(19990501)35:2<262::AID-PROT11>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Yanagida T, Nakase M, Nishiyama K, Oosawa F. Direct observation of motion of single F-actin filaments in the presence of myosin. Nature. 1984;307:58–60. doi: 10.1038/307058a0. [DOI] [PubMed] [Google Scholar]