Abstract
There exist some patients who face recurrent total fertilization failure during assisted reproduction treatment, but the pathological mechanism underlying is elusive. Here, by using sc-RNA-seq method, the transcriptome profiles of ten abnormally fertilized zygotes were assessed, including five zygotes from one patient with recurrent Poly-PN zygotes, and five zygotes from a patient with pronuclear fusion failure. Four zygotes with three pronuclear (Tri-PN) were collected from four different patients as controls. After that, we identified 951 and 1697 significantly differentially expressed genes (SDEGs) in Poly-PN and PN arrest zygotes, respectively as compared with the control group. KEGG analyses indicated down regulated genes in the Poly-PN group included oocyte meiosis related genes, such as PPP2R1B, YWHAZ, MAD2L1, SPDYC, SKP1 and CDC27, together with genes associated with RNA processing, such as SF3B1, LOC645691, MAGOHB, PHF5A, PRPF18, DDX5, THOC1 and BAT1. In contrast, down regulated genes in the PN arrest group, included cell cycle genes, such as E2F4, DBF4, YWHAB, SKP2, CDC23, SMC3, CDC25A, CCND3, BUB1B, MDM2, CCNA2 and CDC7, together with homologous recombination related genes, such as NBN, XRCC3, SHFM1, RAD54B and RAD51. Thus, our work provides a better understanding of transcriptome profiles underlying RTFF, although it based on a limited number of patients.
Introduction
Despite nearly forty years of scientific and clinical advance in the field of assisted reproduction, there still exist some rarely patients, even though rarely occur, who have to face recurrent total fertilization failure (RTFF) without any visual precautionary indicator1–3, even some of them could be rescued by assisted oocyte activation4. Therefore, elucidating the internal mechanism of fertilization failure is of great importance for these patients. However, until now, the relevant etiological analysis was often restricted to morphology during IVF, such as immature oocytes5, improper meiosis6, zygotes with abnormal pronuclei7, and di-pronuclear zygote failing mitotic cleavage8. Due to small amount of material available, deciphering mechanisms underlying these defects remain technical challenging.
Recent technical advances in single-cell sequencing open a new era for exploring the biological state of a single cell at both the DNA and RNA levels for studying variations in genome9,10, transcriptome, and epigenome11 separately or in parallel12. Originally adopted by Surani’s research team13, this approach has been applied successfully in discriminating cell types14–19, elucidating regulatory circuits20, and investigating tumor heterogeneity21,22. In reproductive biology fields, this technique has been used for screening transcriptome of tissues23 and germline cells at different stages24–29. The single cell sequencing technique has great potential in clinical implication30–32, especially in the diagnosis for clarifying the molecular mechanisms of fertilization failure at a single cell resolution. So the aim of this work was to characterize the pathological changes of human zygotes with RTFF at the transcriptional level.
Results
Clinical treatment history of the RTFF patients
As clinical treatment history shown (Table 1), one patient experienced two stimulated cycles under different procedures with 4 and 5 Poly-PN fertilized eggs after ICSI treatment, respectively. Another patient had all zygotes with PN arrest, with 18, 7 and 9 matured oocytes retrieved separately in three cycles although three different ovarian stimulation procedures were employed each time. Moreover, There were no significant differences in serum levels of FSH, LH, E2 and progesterone at baseline and trigger day in patients with Poly-PN, PN arrest, and the control groups (Supplementary Fig. 1), indicating that the observed defects in the zygotes were more likely associated with oocyte original molecular defects rather than ovarian stimulation protocol.
Table 1.
Embryonic developmental consequence of the patients with RTFF during assisted reproduction treatment.
| Clinical cycles | Patient with recurred Poly-PN zygotes | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Oocyte | Oocyte | Fertilization | |||||||
| MII | MI | GV | Abnormal | 0PN | Poly-PN | ||||
| 1st | HMG+MPA+EE | ICSI | 11 | 5 | 3 | 2 | 1 | 1 | 4 |
| 2nd | HMG+MPA+CC | ICSI | 7 | 6 | 0 | 1 | 0 | 1 | 5 |
| Patient with recurred PN-arrest zygotes | |||||||||
| Oocyte | Fertilization | ||||||||
| Total | MII | MI | GV | Abnormal | 2PN | PN arrest | |||
| 1st | Short protocol | IVF/ICSI | 19 | 9/9 | 0 | 0 | 1 | 9/9 | 18 |
| 2nd | HMG + MPA | ICSI | 8 | 7 | 0 | 0 | 1 | 7 | 7 |
| 3rd | HMG + CC | ICSI | 10 | 9 | 0 | 1 | 0 | 9 | 9 |
Transcriptome profiles in Poly-PN and PN-arrest zygotes
It is crucial for oocyte to accumulate indispensable mRNAs to ensure its later use for fertilization and subsequent cell division before the zygotic gene activation33. As the scarce of the oocytes for RTFF patients, it was difficult to collect enough donated oocytes for our study. Therefore, we investigate the transcriptome profile using the unfertilized oocyte after clinical treatment. The procedure of our work was shown in Fig. 1. After sequencing using Illumina HiSeq 2,500 sequencer, we obtain about 142 million clean reads, of which 116 million clean reads mapped to human genome reference. On average, 15,058, 14,995 and 17,713 genes (FPKM ≥ 0.1), 10,471, 10,451 and 10,289 genes (FPKM ≥ 1) or 4,539, 4,410 and 3,630 genes (FPKM ≥ 10) were acquired in Tri-PN, Poly-PN and PN arrest groups, respectively (Fig. 2a). These results were consistent with the data from a previous study24, implying that our technology has the similar sensitivity and coverage.
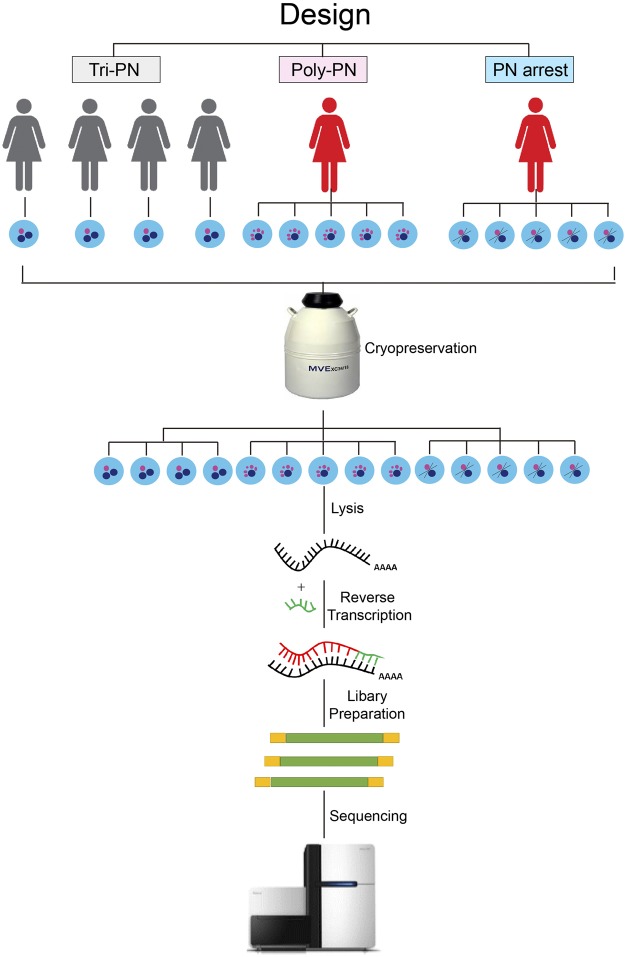
Figure 1.
Overview of the scRNA-seq experimental design.
Figure 2.
Transcriptome profile of the Poly-PN and PN-arrest zygotes. (a) The number of reference transcripts averagely found in each sample in different groups based on clustering of expression patterns for 14 single cell samples from Poly-PN, PN arrest and Control groups; (c,d) Scatterplot showing the number of genes up regulated (red) and down regulated (blue) in Poly PN (c) and PN arrest zygotes (d) separately compared with control group.
To compare the global transcriptome profiles of unfertilized eggs or zygotes among different groups, we analyzed data by hierarchical clustering, and the results indicated that 14 zygotes from 3 groups were clustered into corresponding groups and separated from each other (Fig. 2b). Four Tri-PN zygotes from different patients have the similar transcriptome profile in spite of the heterogeneity in patient source. Interestingly, we also found that five PN arrest zygotes clustered closer with four Tri-PN zygotes, but away from the five Poly-PN zygotes. This finding indicated that the underlying mechanisms was quite different between the Poly-PN and PN arrest group.
To clarify underlying mechanisms, we analyzed major differences of expression profile among different groups. To rule out technical errors causing artifacts of gene expression, all reference genes with average FPKM > 0.5 in any of three groups were used for subsequent analysis. According to the criteria of fold change > 2 or < 0.5 and P < 0.001, 951 (227 up regulated and 724 down regulated) and 1,697 genes (205 up regulated and 1,492 down regulated) were found to be significantly differentially expressed genes (SDEGs) in Poly-PN and PN arrest zygotes, respectively, as compared with the control group (Fig. 2c,d and Supplementary Tables 1 and 2).
The SDEGs are involved in different biological processes between Poly-PN and PN arrest zygotes
In order to clarify the biological function of these differential expressed genes, the KEGG analyses were applied to SDEGs of Poly-PN and PN arrest zygotes, separately. As the results shown (Table 2), for the up regulated genes, there was a enrichment of genes whose products are related to RNA processing and translation, such as RNA splicing (P-value = 1.90 × 10−2 for Poly-PN) or Ribosome biogenesis (P-value = 5.80 × 10−9 for Poly-PN and 1.10 × 10−84 for PN arrest), together with energy consuming related items, such as Huntington’s disease (P-value = 6.40 × 10−3 for Poly-PN), Parkinson’s disease (P-value = 1.20 × 10−3 for PN arrest) and Oxidative phosphorylation (P-value = 5.20 × 10−5 for PN arrest). Genes involved in Wnt signaling pathway (P-value = 1.00 × 10−2 for Poly-PN), Notch signaling pathway (P-value = 2.00 × 10−2 for Poly-PN) and some other signaling pathways in cancer were also enriched (Table 2 and Supplementary Tables 3 and 4). For down regulated genes, the Poly-PN specific down regulated genes were mainly enriched in Oocyte meiosis (P-value = 3.60 × 10−2), Spliceosome (P-value = 4.00 × 10−3), Pyrimidine metabolism (P-value = 1.60 × 10−2), Citrate cycle (P-value = 3.80 × 10−3) (Table 3 and Supplementary Table 5). Whereas, the PN arrest specific down regulated genes mainly belonged to Cell cycle (P-value = 7.60 × 10−3), Homologous recombination (P-value = 2.40 × 10−2) and Amino sugar or nucleotide sugar metabolism (P-value = 4.60 × 10−2) (Table 3 and Supplementary Table 6). Furthermore, The SDEGs down regulated overlapped in both of this two groups were mainly related to Basal transcription factors (P-value = 3.10 × 10−2 for Poly-PN and 4.20 × 10−3 for PN arrest), Ubiquitin mediated proteolysis (P-value = 4.70 × 10−2 for Poly-PN and 7.70 × 10−3 for PN arrest) and Glycan biosynthesis (P-value = 2.00 × 10−2 for Poly-PN and 3.10 × 10−2 for PN arrest).
Table 2.
KEGG Signaling pathways enrichment of SDEGs up regulated in Poly-PN and PN arrest groups separately compared with controls.
| Items | Count | % | P-Value | Genes | |
|---|---|---|---|---|---|
| SDEGs (Poly-PN/Control) | Ribosome | 12 | 5.5 | 5.80E-09 | RPSA, RPL23, RPL9, RPL35, RPL3, RPS9, RPL7A, RPS11, RPS20, RPS4X, RPS8, RPS24 |
| Huntington’s disease | 8 | 3.6 | 6.40E-03 | POLR2F, TAF4, EP300, NDUFB8, NDUFA6, CREBBP, UQCRQ, ATP5J | |
| Wnt signaling pathway | 7 | 3.2 | 1.00E-02 | PRKCA, CTBP1, TCF7, EP300, CREBBP, FRAT1, FRAT2 | |
| Pathway in cancer | 10 | 4.5 | 1.90E-02 | PRKCA, CTBP1, TCF7, EP300, RALBP1, CREBBP, FOXO1, LAMC1, IKBKB, LAMB1 | |
| Spliceosome | 6 | 2.7 | 1.90E-02 | CHERP, PCBP1, LSM7, SNRPD2, SNRPG, SF3B2 | |
| Notch signaling pathway | 4 | 1.8 | 2.00E-02 | CTBP1, EP300, PSEN2, CREBBP | |
| Prostate cancer | 5 | 2.3 | 2.40E-02 | TCF7, EP300, CREBBP, FOXO1, IKBKB | |
| SDEGs (PN arrest/Control) | Ribosome | 56 | 28.1 | 1.10E-84 | RPL36A, RPL19, RPL13, RPLP2, RPS2, RPS3, RPS3A, RPLP0, RPLP1, RPL10, RPL12, RPS27A, RPS4X, RPS18, RPS19, RPL41, RPS16, RPS17, RPS15, RPS12, RPS13, RPS11, UBA52, RPL35, RPS15A, RPL36, RPL37, RPL38, RPS25, RPL30, RPS27, RPS28, RPL32, RPS29, RPL31, RPL8, RPL3, RPL10A, RPL7A, RPL4, RPS20, RPS21, RPS23, RPS24, RPL26, RPS9, RPL27, RPL23A, RPL24, RPS5, RPL28, RPL29, RPL23, RPL13A, RPL37A |
| Oxidative phosphorylation | 11 | 5.5 | 5.20E-05 | ATP5D, NDUFA2, NDUFB10, NDUFB8, NDUFB9, NDUFA6, COX6A1, ATP5L, COX6C, NDUFA11, NDUFB2 | |
| Parkinson’s disease | 9 | 4.5 | 1.20E-03 | ATP5D, NDUFA2, NDUFB10, NDUFB8, NDUFB9, NDUFA6, COX6A1, COX6C, NDUFB2 | |
| Alzheimer’s disease | 9 | 4.5 | 5.50E-03 | ATP5D, NDUFA2, NDUFB10, NDUFB8, NDUFB9, NDUFA6, COX6A1, COX6C, NDUFB2 | |
| Huntington’s disease | 9 | 4.5 | 9.90E-03 | ATP5D, NDUFA2, NDUFB10, NDUFB8, NDUFB9, NDUFA6, COX6A1, COX6C, NDUFB2 |
Table 3.
KEGG Signaling pathways enrichment of SDEGs down regulated in Poly-PN and PN arrest groups separately compared with controls.
| Items | Count | % | P-Value | Genes | |
|---|---|---|---|---|---|
| SDEGs (Poly-PN/Control) | Citrate cycle (TCA cycle) | 6 | 0.9 | 0.0038 | LOC642502, DLST, SUCLG1, DLD, PDHA2, FH |
| Spliceosome | 12 | 1.7 | 0.004 | SFRS7, SF3B1, LOC645691, MAGOHB, PHF5A, HNRNPC, PRPF18, DDX5, SF3B4, PRPF38A, THOC1, BAT1 | |
| Pyrimidine metabolism | 9 | 1.3 | 0.016 | POLR2H, UMPS, POLR2E, RRM2, PNPT1, DCK, ZNRD1, POLR2D, DUT | |
| N-Glycan biosynthesis | 6 | 0.9 | 0.02 | TUSC3, ALG3, DPM1, ALG6, MGAT5, ALG13 | |
| Basal transcription factors | 5 | 0.7 | 0.031 | TAF9B, GTF2A1L, GTF2B, TBPL1, GTF2H1 | |
| Oocyte meiosis | 9 | 1.3 | 0.036 | PPP2R1B, YWHAZ, MAD2L1, CCNB2, PPP2CA, FBXO5, SKP1, CDC27, SPDYC | |
| Ubiquitin mediated proteolysis | 10 | 1.4 | 0.047 | UBE2N, UBE2E3, UBE2D3, UBA3, UBE2W, PIAS1, SKP1, UBOX 5, TRAF6, CDC27 | |
| SDEGs (PN arrest/Control) | Basal transcription factors | 8 | 0.6 | 4.20E-03 | TAF11, GTF2E1, GTF2I, TAF4B, GTF2F2, TAF9B, GTF2B, GTF2H1 |
| Cell cycle | 16 | 1.1 | 7.60E-03 | CDC7, E2F4, DBF4, YWHAB, SKP2, CDC23, ANAPC10, SMC3, CDC25A, ORC2L, CCND3, ORC4L, BUB1B, ORC5L, MDM2, CCNA2 | |
| Ubiquitin mediated proteolysis | 17 | 1.2 | 7.70E-03 | UBE2G1, SKP2, CDC23, UBE2F, ANAPC10, UBE2H, UBE2C, BIRC2, BRCA1, UBE2E3, TRIM32, UBA3, UBE2W, MDM2, PIAS2, RCHY1, TRAF6 | |
| Proteasome | 8 | 0.6 | 2.10E-02 | PSMA1, PSMC6, PSMD12, PSMA4, PSMC1, SHFM1, PSMA7, PSMD7 | |
| Homologous recombination | 6 | 0.4 | 2.40E-02 | NBN, XRCC3, RAD51L1, SHFM1, RAD54B, RAD51 | |
| O-Glycan biosynthesis | 6 | 0.4 | 3.10E-02 | GALNT3, GALNT1, C1GALT1C1, GALNT11, GCNT1, C1GALT1 | |
| Amino sugar and nucleotide sugar metabolism | 7 | 0.5 | 4.60E-02 | GNPDA1, GNPDA2, HEXB, UGDH, NAGK, FPGT, UGP2 |
Among the total 1,956 down regulated genes in different annotations, there were about 464 (23.7%) genes specifically down regulated in Poly PN, 1,233 (63.0%) genes specifically down regulated in PN arrest and only 259 (13.3%) genes down regulated overlap in both of this two groups (Fig. 3a). The Poly-PN specific down regulated genes included oocyte meiosis related genes such as Protein Phosphatase (PPP2R1B), YWHAZ, MAD2L1, SPDYC, SKP1 and CDC27 (Fig. 3b). Certain genes associated with RNA processing, such as those encoding splicing factor genes SF3B1, LOC645691, MAGOHB, PHF5A, PRPF18, DDX5, THOC1 and BAT1 were also down regulated (Fig. 3c), perhaps contributing to the fertilization failure in Poly-PN group. In contrast, the PN arrest specific down regulated genes, such as E2F4, DBF4, YWHAB, SKP2, CDC23, SMC3, CDC25A, CCND3, BUB1B, MDM2, CCNA2, CDC7 were involved in Cell cycle (Fig. 3d) and NBN, XRCC3, SHFM1, RAD54B, RAD51 were Homologous recombination related genes (Fig. 3e). These results implied that the Poly-PN might have defects during oocyte meiosis, whereas defects of PN arrest zygotes were involved in Cell cycle and Homologous recombination.
Figure 3.
Relative expression levels of SDEGs specifically expressed in Poly-PN and or PN arrest zygotes. (a) The Venn diagram of down regulated SDEGs in Poly-PN and PN arrest groups separately compared with those in control group; (b–e) Scatterplot showing the relative expression levels of the particularly interesting SDEGs specifically down regulated in Poly-PN or PN arrest zygotes. P < 0.001 was indicated significantly different.
Functional validation of the selected genes by gene knock down
We randomly chose two of these meiosis related genes (PPP2CA and SKP1) and validated their function in mice oocyte, the results indicated that both PPP2CA and SKP1 knock down did not show any difference with the corresponding control group in either oocyte maturation or fertilization (Fig. 4a,b). In order to clarify the mechanism underlying, we analyzed available published single cell RNA-Seq data sets corresponding to fertilization process of human and mice, focusing on the 43 selected genes, which enriched in Meiosis, Spliceosome, Cell cycle and Homologous recombination items separately. As the results shown (Fig. 4c), human and mice have very different expression patterns of the selected genes during their fertilization process. Thus, more clinical cases, but not the mice, might be an ideal model for validation of the function of these selected genes in future.
Figure 4.
(a,b) Validation of two meiosis ralated genes in mice oocytes using gene knock down experiment. (c) Comparative expression of these identified genes during fertilization in human (bottom layer) and mice (up layer), respectively. HR, Homologous recombination.
Discussion
Some patients have oocytes incapable of completing the whole process of fertilization, including defective sperm entry, oocyte activation, pronuclear formation or fusion34, as well as some failure in mitotic division35. In this work, we profiled the transcriptome of the Poly-PN and PN arrest zygotes from two patients with RTFF, and found Poly-PN zygotes showed defects in Meiosis and RNA processing and PN arrest zygotes had defects in Cell cycle and DNA homologous recombination.
For meiosis, oocytes need to undergo meiotic DNA replication and homologous chromosomes segregation, and then arrest in metaphase of meiosis II awaiting fertilization33. After sperm penetration, oocyte resumes meiosis and segregates sister chromatids and completes the meiosis II. So the differentially expressed genes in Poly-PN might play critical roles in this biological process. For example, the subunit of the SCF E3 ubiquitin ligase (SKP1) has been reported to be important in the progression of recombination during oocyte meiosis36. The APC core subunit (CDC27) and the checkpoint protein (MAD2) play critical roles in segregating sister chromatids during oocyte meiosis37. Similarly, some other genes down regulated in Poly-PN zygotes, such as Protein Phosphatase (PPP2R1B), YWHAZ and SPDYC were also in associated with meiosis38,39.
Furthermore, during oocyte maturation, it also needs to accumulate sufficient maternal RNA to ensure oocyte maturation, fertilization and subsequently embryo development until the embryonic genome is activated40. So certain RNA processing genes identified in Poly-PN zygotes, such as those encoding splicing factor genes THOC141, SF3B142, LOC645691, exon junction complex core component related gene (MAGOHB)43, PHD finger-like domain-containing protein 5 A (PHF5A)44, some pre-mRNA processing factor 18 related gene (PRPF18)45 and RNA helicases related genes (DDX5 and BAT1)46,47, are involved in regulating RNA secondary structure and pre-mRNA splicing, which might be responsible for RNA maturation during oocyte meiosis.
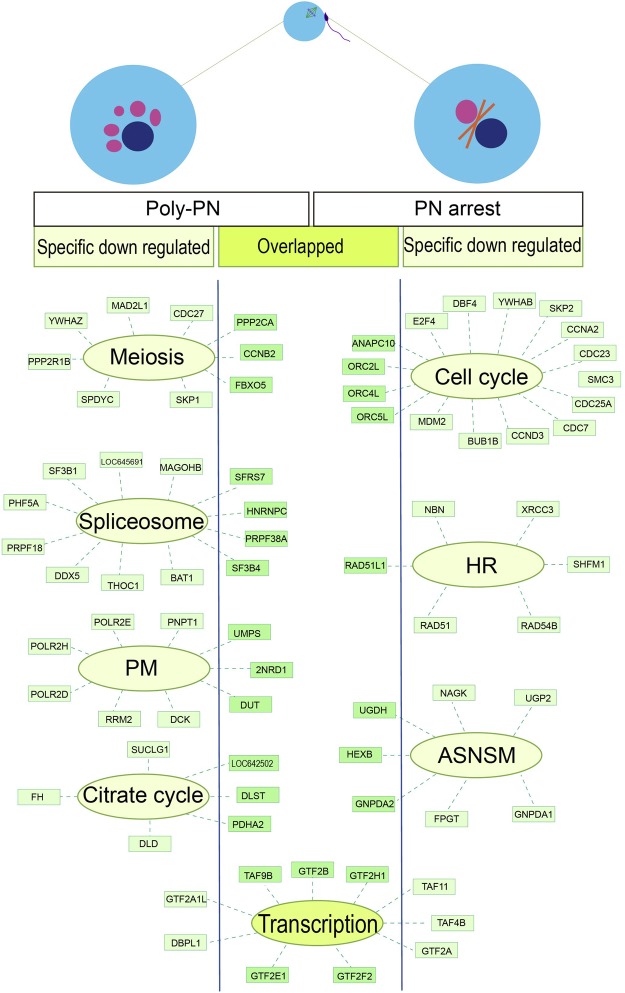
Upon fertilization, the zygotes undergoes chromatin remodeling, genomes reprogramming or DNA repairing, and the cell cycle machinery must be switched from meiotic to mitotic chromosome segregation48. Our results indicated that the PN arrest specific down regulated genes mainly related to these biological process. For example, the cyclin associated kinase (CCNA2 and CCND3) were required for sister chromatid segregation49, and structural maintenance and segregation of chromosome proteins (SMC3 and BUB1B) have been reported to be in associated with developmental potential of human pre-implantation zygotes50. Cell cycle related genes (CDC7, CDC23 and CDC25A) and some other genes including DBF4, YWHAB, SKP2 and MDM2 were also found to be significantly down regulated in PN arrest group. Moreover, some other genes specifically down-regulated in PN arrest zygotes, including check point proteins codon genes (RAD54B and RAD51)51,52, DNA repair related genes (XRCC3)53, chromosome integrity maintenance genes (NBN)54,55 and SHFM1, all of which were involved in key proteins for homologous recombination. In addition, we also found some genes down regulated overlap for both Poly-PN and PN arrest groups and these genes in different annotations were classified according to the expression specification and illustrated in a model (Fig. 5).
Figure 5.
Module visualization of Poly-PN and PN arrest specifically down regulated SDEGs. Module visualization of Poly-PN and PN arrest specifically down regulated SDEGs. Down regulated SDEGs were shown Poly-PN specifically (left panel) PN arrest specifically (right panel) and genes down regulated overlap in both of this two groups (middle panel), PM, Pyrimidine metabolism; HR, Homologous recombination; ASNSM, Amino sugar or nucleotide sugar metabolism.
Taken together, our work found Poly-PN have some problems in oocyte meiosis and RNA processing, whereas PN arrest showed defects during mitosis cell cycle or homologous recombination during meiosis and this could provide new targets for therapeutic intervention by modulating these corresponding signaling pathways in the future. Remarkably, as the scarce of the RTFF patients, we could not collect enough oocyte samples for single cell RNA sequencing. So more clinical cases need to be collected and further verification need to be performed in the future.
Methods
Ethics statement
All procedures were approved by the Research Ethics Committee of Shanghai Jiao tong University School of Medicine and informed consent was obtained from participants at IVF center of the Ninth people’s hospital. We confirmed that all patients have written informed consent for the use of their zygotes for this research. Animals were maintained at 23 °C in a 12-h (7:00–19:00) light and 12-h (19:00–7:00) dark schedule, and all experimental procedures were performed in accordance with Institutional Animal Care and Use Committee guidelines of Shanghai Jiao Tong University School of Medicine.
Patients, ovarian stimulation, oocyte retrieval, and the IVF/ICSI procedure
For all patients in our study, five types of ovarian stimulation protocols were used: (1) Human Menotrophins Gonadotrophin (hmG, Lizhu Pharmaceutical Trading Co.) co-treated with Medroxyprogesterone acetate (MPA, Shanghai Sine Pharmaceutical Ltd.) (hMG + MPA); (2) Human Menotrophins Gonadotrophin co-treated with Clomifene Citrate (CC, Medochemie Ltd.) (hMG + CC); (3) Human Menotrophins Gonadotrophin co-treated with Medroxyprogesterone acetate and Clomifene Citrate (hMG + MPA + CC); (4) Human Menotrophins Gonadotrophin co-treated with Medroxyprogesterone acetate and ethinyl estradiol (EE, Shanghai Sine Pharmaceutical Ltd.) (hMG + MPA + EE) and (5) Short protocol, in which patients were administered with GnRHa daily beginning on menstrual cycle day 2 and with hMG daily beginning on menstrual cycle day 3. Follicle growth was monitored by ultrasound examination. Serum FSH, LH, E2, and progesterone concentrations were measured serially using the chemiluminescence (Abbott Biologicals B.V.) method on the same days as the ultrasound exams. Human Chorionic Gonadotrophin (hCG, Lizhu Pharmaceutical Trading Co.) at a 1000–5000 IU dose was administered when the dominant follicles reached 18 mm in diameter. Cumulus oocyte complexes were recovered transvaginally with ultrasound guidance 34–36 hours post hCG. After retrieval, oocytes were maintained in human tubal fluid (HTF; Irvine Scientific) medium plus 10% synthetic serum substitute (SSS; Irvine Scientific) for about 2 hours before In vitro fertilization (IVF)/Intracytoplasmic sperm injection (ICSI).
For ICSI treatment, the cumulus oophorus were removed mechanically from oocytes with denuding pipettes in solution with 80IU hyaluronidase (Sigma) followed by injection. For IVF treatment, cumulus oocyte complexes were inseminated with about 0.3–0.5 × 106/ml motile spermatozoa in HTF medium and the cumulus oophorus were removed 18 hours later. Fertilized eggs from both IVF and ICSI groups were cultured in 20 μl continuous single culture medium (CSC: Irvine Scientic: USA) individually under oil and incubated at 37 °C humidified atmosphere under 5% CO2, 5% O2, and 90% N2 for pre-implantation culture. As a policy of our center, fertilization was assessed by the presence of two pronuclei 16–18 hours post insemination, followed by confirming the embryonic development 66–68 hours post insemination. The zygotes with more than three tiny pronuclei following the ICSI procedure were recognized as Poly-PN zygotes. The zygotes with normal pronuclei but failed to fuse until 66–68 hours post fertilization were name as PN-arrest zygotes. Tri-PN zygotes from four different IVF patients were used as controls. All samples above were collected and vitrified using Cryotip method and then stored in liquid nitrogen until subsequent experimental treatment.
Preparation and quality control of single-cell cDNAs
The method for RNA extraction was carried out as described previously56. Briefly, after thawing, each zygote was washed twice and transferred into lysate buffer. Then the reverse transcription reaction was performed directly on whole cell lysate using SuperScript II reverse transcriptase (Life Technologies). We performed 15 cycles of PCR to amplify cDNA and the PCR product was purified by using AMPure XP beads (Beckman Coulter). Agilent high-sensitivity DNA chip kit on a BioAnalyzer (Agilent Technologies) was used for checking the quality of cDNAs according the size distribution to ensure cDNAs contained few short fragments (<500 bp) and showed peak sizes between 1.5 kb–2 kb.
RNA-Seq library construction and sequencing
According to the manual of TruePrep DNA Library Prep Kit V2 for Illumina (Vazyme Biotech), the quality of RNA-Seq sequencing library was checked by using Agilent high-sensitivity DNA chip. The libraries showing the peak around 300 bp was chosen for high-throughput sequencing on the Illumina HiSeq 2500 platform using the dual index sequencing strategy with single-end reads length of 50 bp.
Bioinformatics process for sequencing data
Individual sample from different zygotes has its own unique barcode sequence and could be separated from clean data. We used Tophat v2.0.957 to assemble the reads into NCBI build 37 hg19 genome and used Cufflinks v2.1.158 to calculate gene expression level. Clustering was used to process hierarchical clustering using Euclidean distance metric in the R packages59. Gene expression levels were measured by using fragment per kilobase of exon per million mapped reads (FPKM). To rule out technical errors and increase the power to detect biological function, all reference genes with average FPKM > 0.5 in any of three groups and the criterion of P < 0.001 or P < 0.01 together with FC (fold change) >2 or <0.5 were used to identify differentially expressed genes for subsequent biological analysis using ArrayTrackTM software (FDA’s own bioinformatics and genomics tool, http://www.fda.gov/ScienceResearch/BioinformaticsTools/Arraytrack/default.htm).
KEGG pathway analysis
Database for Annotation Visualization and Integrated Discovery (DAVID V6.7; https://david.ncifcrf.gov/) was used to perform KEGG pathway analysis60,61.
ShRNA design and in vitro transcription
For short hairpin RNA (shRNA) design, we selected an siRNA-target sequence on the NCBI RNAi database for each targeted genes, and the forward and reverse primers for each gene (SKP1 F: ATAGGGGGCT GCAAACTACT TAGACATTTC AAGAGA ATGT CTAAGTAGTT TGCAGCCTTT TTTG; SKP1 R: GATCCAAAAA AGGCTGCAAA CTACTTAGAC ATTCTCTTGA AATGTCTAAG TAGTTTGCAG CCCC; PPP2CA F: ATAGGGTGGA ACTTGACGAC ACTCTTATTC AAGAGATAAG AGTGTCGTCA AGTTCCATTT TTTGPPP2CA R: GATCCAAAAA ATGGAACTTG ACGACACTCT TATCTCTTGA ATAAGAGTGT CGTCAAGTTC CACC) were annealed and cloned into a T7 promoter containing vector pcDNA3.1(+) using Bsa1 and BamH1 restriction enzyme site, shRNA was transcribed in vitro from linearized pcDNA3.1-shRNA plasmid using MEGA short script T7 kit (Life Technology) and purified using MEGA clear kit (Life Technology) and mixed in RNase-free water at the concentration of 50 ng/μl for subsequent use.
Oocyte microinjection, parthenogenetic activation and development assessment
Female mice aged 6–8 weeks were induced to superovulate by i.p. injection of 10 IU of pregnant mare’s gonadotrophin (PMSG) (Ningbo Hormone Products Co.). Cumulus oocyte complexes (COCs) were collected at 46 h post PMSG. For COCs retrieval, the ovaries were removed immediately and put into 4 ml HTF medium plus 10% SSS (Irvine Scientific) and 0.2 mM IBMX (Sigma). The COCs were released into this medium by puncturing ovaries with a 27 g needle. The cumulus cells were released mechanically using mouth pipette and only those with normal morphologies were used for RNA injection.
All injected oocytes were cultured for maturation in a CO2 incubator for 16 hours for maturation before activation. The activation medium used was KSOM (Millipore) supplemented with 10 mM SrCl2. After being washed twice in activation medium, oocytes were incubated first in activation medium for 2.5 hours and then in activation medium without SrCl2 for 3.5 hours at 37 °C in a humidified atmosphere with 5% CO2, 5% O2, and 90% N2. Both the activation medium and KSOM for subsequent short culture of oocytes were supplemented with 5 μg/mL cytochalasin B. Six hours after the onset of activation, the fertilization rate was assessed by count pronuclear formation.
Statistical analysis
Serum hormone data were analyzed by GraphPad Prism software using two-way repeated measures ANOVA. Bonferroni post tests were used for pairwise comparisons. **P < 0.01; ***P < 0.001. Data for relative expression levels in Poly-PN and PN arrest zygotes were separately compared with control were analyzed using a two-tailed, unpaired Student’s t test. P < 0.001 indicated as significantly different. The maturation and fertilization rate of the oocytes between gene knock down and control group were analyzed by using Chi-Squared Test.
Electronic supplementary material
Acknowledgements
We thank the voluntary research participants and all the doctors and embryologists in our center. We also thank Qiang Wu (SJTU) for technique assistance for RNA sequencing and Aaron J. Hsueh (Stanford) for critical reading of manuscript. This work was supported by grants from the National Natural Science Foundation of China (Grant No. 31200825, 81571397 and 81571486), the Fundamental Research Funds for the Central Universities (17JCYB12), Shanghai Committee of Science and Technology, China (Grant No. 16411963800) and Shanghai Three-year Plan on Promoting TCM Development, China (Grant No. ZY3-LCPT-2-2006).
Author Contributions
L.S., Y.X.Z. and Y.P.K. designed the study; L.S., Q.F.L. and L.H.S. collected the samples; Y.X.Z., L.L.J. and J.Z. constructed the library; L.S. and H.B.W. performed the RNAi experiment; L.S., Y.X.Z. and H.S. analyzed the data; L.S. and Y.P.K. supervised the study; L.S. wrote the manuscripts.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lun Suo and Yu xiao Zhou contributed equally.
Contributor Information
Lun Suo, Email: suoyunfei123@gmail.com.
Yan ping Kuang, Email: kuangyanp@126.com.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-36275-6.
References
- 1.Flaherty SP, Payne D, Matthews CD. Fertilization failures and abnormal fertilization after intracytoplasmic sperm injection. Hum Reprod. 1998;13(Suppl 1):155–164. doi: 10.1093/humrep/13.suppl_1.155. [DOI] [PubMed] [Google Scholar]
- 2.Mahutte NG, Arici A. Failed fertilization: is it predictable? Current opinion in obstetrics & gynecology. 2003;15:211–218. doi: 10.1097/00001703-200306000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Combelles CM, et al. Diagnosing cellular defects in an unexplained case of total fertilization failure. Hum Reprod. 2010;25:1666–1671. doi: 10.1093/humrep/deq064. [DOI] [PubMed] [Google Scholar]
- 4.Kuentz P, et al. Assisted oocyte activation overcomes fertilization failure in globozoospermic patients regardless of the DPY19L2 status. Hum Reprod. 2013;28:1054–1061. doi: 10.1093/humrep/det005. [DOI] [PubMed] [Google Scholar]
- 5.Thornton MH, Francis MM, Paulson RJ. Immature oocyte retrieval: lessons from unstimulated IVF cycles. Fertility and sterility. 1998;70:647–650. doi: 10.1016/S0015-0282(98)00274-X. [DOI] [PubMed] [Google Scholar]
- 6.Filges I, et al. Recurrent triploidy due to a failure to complete maternal meiosis II: whole-exome sequencing reveals candidate variants. Molecular human reproduction. 2015;21:339–346. doi: 10.1093/molehr/gau112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenbusch BE, Schneider M. Recurrent failure of pronucleus formation after intracytoplasmic sperm injection. Arch Gynecol Obstet. 1999;262:185–188. doi: 10.1007/s004040050247. [DOI] [PubMed] [Google Scholar]
- 8.Rawe VY, Olmedo SB, Nodar FN, Ponzio R, Sutovsky P. Abnormal assembly of annulate lamellae and nuclear pore complexes coincides with fertilization arrest at the pronuclear stage of human zygotic development. Hum Reprod. 2003;18:576–582. doi: 10.1093/humrep/deg114. [DOI] [PubMed] [Google Scholar]
- 9.Lu S, et al. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science. 2012;338:1627–1630. doi: 10.1126/science.1229112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan L, et al. Live births after simultaneous avoidance of monogenic diseases and chromosome abnormality by next-generation sequencing with linkage analyses. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:15964–15969. doi: 10.1073/pnas.1523297113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angermueller C, et al. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nature methods. 2016;13:229–232. doi: 10.1038/nmeth.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macaulay IC, et al. Separation and parallel sequencing of the genomes and transcriptomes of single cells using G&T-seq. Nature protocols. 2016;11:2081–2103. doi: 10.1038/nprot.2016.138. [DOI] [PubMed] [Google Scholar]
- 13.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nature methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 14.Poulin JF, Tasic B, Hjerling-Leffler J, Trimarchi JM, Awatramani R. Disentangling neural cell diversity using single-cell transcriptomics. Nature neuroscience. 2016;19:1131–1141. doi: 10.1038/nn.4366. [DOI] [PubMed] [Google Scholar]
- 15.Zeisel A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 16.Li J, et al. Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types. EMBO reports. 2016;17:178–187. doi: 10.15252/embr.201540946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Treutlein B, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawlor N, et al. Single-cell transcriptomes identify human islet cell signatures and reveal cell-type-specific expression changes in type 2 diabetes. Genome research. 2017;27:208–222. doi: 10.1101/gr.212720.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han X, et al. Mapping the Mouse Cell Atlas by Microwell-Seq. Cell. 2018;172:1091–1107 e1017. doi: 10.1016/j.cell.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Jaitin DA, et al. Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell. 2016;167:1883–1896 e1815. doi: 10.1016/j.cell.2016.11.039. [DOI] [PubMed] [Google Scholar]
- 21.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng C, et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell. 2017;169:1342–1356 e1316. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 23.Krjutskov K, et al. Single-cell transcriptome analysis of endometrial tissue. Hum Reprod. 2016;31:844–853. doi: 10.1093/humrep/dew008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue Z, et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature. 2013;500:593–597. doi: 10.1038/nature12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan L, et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nature structural & molecular biology. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- 26.Petropoulos S, et al. Single-Cell RNA-Seq Reveals Lineage and X Chromosome Dynamics in Human Preimplantation Embryos. Cell. 2016;167:285. doi: 10.1016/j.cell.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blakeley P, et al. Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development. 2015;142:3151–3165. doi: 10.1242/dev.123547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi J, et al. Dynamic transcriptional symmetry-breaking in pre-implantation mammalian embryo development revealed by single-cell RNA-seq. Development. 2015;142:3468–3477. doi: 10.1242/dev.123950. [DOI] [PubMed] [Google Scholar]
- 29.Liu W, et al. Identification of key factors conquering developmental arrest of somatic cell cloned embryos by combining embryo biopsy and single-cell sequencing. Cell discovery. 2016;2:16010. doi: 10.1038/celldisc.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Q, et al. Single-cell analysis of differences in transcriptomic profiles of oocytes and cumulus cells at GV, MI, MII stages from PCOS patients. Scientific reports. 2016;6:39638. doi: 10.1038/srep39638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang R, et al. RNA-Seq-Based Transcriptome Analysis of Changes in Gene Expression Linked to Human Pregnancy Outcome After In Vitro Fertilization-Embryo Transfer. Reprod Sci. 2016;23:134–145. doi: 10.1177/1933719115597766. [DOI] [PubMed] [Google Scholar]
- 32.Fragouli E, Lalioti MD, Wells D. The transcriptome of follicular cells: biological insights and clinical implications for the treatment of infertility. Human reproduction update. 2014;20:1–11. doi: 10.1093/humupd/dmt044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clift D, Schuh M. Restarting life: fertilization and the transition from meiosis to mitosis. Nature reviews. Molecular cell biology. 2013;14:549–562. doi: 10.1038/nrm3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kort DH, et al. Human embryos commonly form abnormal nuclei during development: a mechanism of DNA damage, embryonic aneuploidy, and developmental arrest. Hum Reprod. 2016;31:312–323. doi: 10.1093/humrep/dev281. [DOI] [PubMed] [Google Scholar]
- 35.Azzarello A, Hoest T, Mikkelsen AL. The impact of pronuclei morphology and dynamicity on live birth outcome after time-lapse culture. Hum Reprod. 2012;27:2649–2657. doi: 10.1093/humrep/des210. [DOI] [PubMed] [Google Scholar]
- 36.McLoud JD, Yang M. The conserved function of skp1 in meiosis. Frontiers in genetics. 2012;3:179. doi: 10.3389/fgene.2012.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peter M, et al. The APC is dispensable for first meiotic anaphase in Xenopus oocytes. Nature cell biology. 2001;3:83–87. doi: 10.1038/35050607. [DOI] [PubMed] [Google Scholar]
- 38.Hu MW, et al. Scaffold subunit Aalpha of PP2A is essential for female meiosis and fertility in mice. Biology of reproduction. 2014;91:19. doi: 10.1095/biolreprod.114.120220. [DOI] [PubMed] [Google Scholar]
- 39.Meng J, et al. The role of 14-3-3epsilon interaction with phosphorylated Cdc25B at its Ser321 in the release of the mouse oocyte from prophase I arrest. PloS one. 2013;8:e53633. doi: 10.1371/journal.pone.0053633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L, Lu X, Dean J. The maternal to zygotic transition in mammals. Molecular aspects of medicine. 2013;34:919–938. doi: 10.1016/j.mam.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, et al. Thoc1 deficiency compromises gene expression necessary for normal testis development in the mouse. Molecular and cellular biology. 2009;29:2794–2803. doi: 10.1128/MCB.01633-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Golas MM, Sander B, Will CL, Luhrmann R, Stark H. Molecular architecture of the multiprotein splicing factor SF3b. Science. 2003;300:980–984. doi: 10.1126/science.1084155. [DOI] [PubMed] [Google Scholar]
- 43.Singh KK, Wachsmuth L, Kulozik AE, Gehring NH. Two mammalian MAGOH genes contribute to exon junction complex composition and nonsense-mediated decay. RNA biology. 2013;10:1291–1298. doi: 10.4161/rna.25827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Will CL, et al. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. The EMBO journal. 2002;21:4978–4988. doi: 10.1093/emboj/cdf480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minakuchi, M. et al. Pre-mRNA Processing Factor Prp18 Is a Stimulatory Factor of Influenza Virus RNA Synthesis and Possesses Nucleoprotein Chaperone Activity. Journal of virology91 (2017). [DOI] [PMC free article] [PubMed]
- 46.Gonzalez-Duarte RJ, et al. The expression of RNA helicase DDX5 is transcriptionally upregulated by calcitriol through a vitamin D response element in the proximal promoter in SiHa cervical cells. Molecular and cellular biochemistry. 2015;410:65–73. doi: 10.1007/s11010-015-2538-4. [DOI] [PubMed] [Google Scholar]
- 47.Peelman LJ, et al. The BAT1 gene in the MHC encodes an evolutionarily conserved putative nuclear RNA helicase of the DEAD family. Genomics. 1995;26:210–218. doi: 10.1016/0888-7543(95)80203-X. [DOI] [PubMed] [Google Scholar]
- 48.Ladstatter S, Tachibana-Konwalski K. A Surveillance Mechanism Ensures Repair of DNA Lesions during Zygotic Reprogramming. Cell. 2016;167:1774–1787 e1713. doi: 10.1016/j.cell.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Touati SA, et al. Cyclin A2 is required for sister chromatid segregation, but not separase control, in mouse oocyte meiosis. Cell reports. 2012;2:1077–1087. doi: 10.1016/j.celrep.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 50.Yanez LZ, Han J, Behr BB, Reijo Pera RA, Camarillo DB. Human oocyte developmental potential is predicted by mechanical properties within hours after fertilization. Nature communications. 2016;7:10809. doi: 10.1038/ncomms10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang AT, et al. A Dominant Mutation in Human RAD51 Reveals Its Function in DNA Interstrand Crosslink Repair Independent of Homologous Recombination. Molecular cell. 2015;59:478–490. doi: 10.1016/j.molcel.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanaka K, Kagawa W, Kinebuchi T, Kurumizaka H, Miyagawa K. Human Rad54B is a double-stranded DNA-dependent ATPase and has biochemical properties different from its structural homolog in yeast, Tid1/Rdh54. Nucleic acids research. 2002;30:1346–1353. doi: 10.1093/nar/30.6.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kurumizaka H, et al. Homologous-pairing activity of the human DNA-repair proteins Xrcc3.Rad51C. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5538–5543. doi: 10.1073/pnas.091603098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carney JP, et al. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/S0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 55.Varon R, et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/S0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- 56.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nature protocols. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 57.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deng W, Wang Y, Liu Z, Cheng H, Xue Y. HemI: a toolkit for illustrating heatmaps. PloS one. 2014;9:e111988. doi: 10.1371/journal.pone.0111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.