Abstract
Aberrant promoter methylation plays a vital role in colorectal carcinogenesis. However, its role in treatment responses is unclear, especially for metastatic disease. Here, we investigated the association between promoter methylation and treatment outcomes of irinotecan-based chemotherapy in 102 patients with metastatic colorectal cancer. Promoter methylation was examined by methylation-specific polymerase chain reaction for three loci (CHFR, WRN, and SULF2) associated with chemotherapy response and five CpG island methylator phenotype (CIMP)–specific markers (CACNA1G, IGF2, NEUROG1, RUNX3, and SOCS1). Association between CHFR methylation and in vitro sensitivity to irinotecan was also evaluated. Promoter methylation of CHFR, WRN, and SULF2 was identified in 16 (15.7%), 24 (23.5%), and 33 (32.4%) patients, respectively. CIMP status was positive in 22 (21.6%) patients. CHFR methylation was associated with a significantly longer time to progression (TTP) (median: 8.77 vs. 4.43 months, P = .019), with trends favoring higher overall survival (OS) (median: 22.83 vs. 20.17 months, P = .300) and response rates (31.3% vs. 17.4%, P = .300). For patients with unmethylated CHFR, TTP (median: 5.60 vs. 3.53, P = .020) and OS (median: 20.57 vs. 9.23, P = .006) were significantly different according to CIMP status. Colorectal cancer cell lines with CHFR methylation demonstrated increased sensitivity to irinotecan. Both CHFR overexpression and combination with 5-aza-2′-deoxycytidine reversed irinotecan sensitivity in CHFR-methylated cell lines, whereas CHFR knockdown in unmethylated cells restored sensitivity to irinotecan. These data suggest that CHFR methylation may be associated with favorable treatment outcomes of irinotecan-based chemotherapy in patients with metastatic colorectal cancer.
Abbreviations: 5-Aza-CdR, 5-Aza-2′-deoxycytidine; CCLE, the Cancer Cell Line Encyclopedia; CIMP, CpG island methylator phenotype; EGFR, epidermal growth factor receptor; MSP, methylation-specific polymerase chain reaction; OS, overall survival; TCGA, The Cancer Genome Atlas; TTP, time to progression; VEGF, vascular endothelial growth factor
Introduction
Colorectal cancer (CRC) is the third leading cause of cancer-related death worldwide [1]. Since the adoption of anti-EGFR and anti-VEGF(R) antibodies in 2004, the median survival time of patients with metastatic CRC has reached 30 months [2]. However, despite the recent advances in molecular targeted therapy as well as immunotherapy, cytotoxic chemotherapy with fluoropyrimidines and oxaliplatin and/or irinotecan remains the mainstay of therapy and is responsible for the majority of survival gain in metastatic CRC. Therefore, prediction of treatment response or resistance to cytotoxic chemotherapy is a highly significant and clinically relevant issue to further improve the treatment outcomes of patients with metastatic CRC.
Irinotecan, a topoisomerase I inhibitor, is one of the major chemotherapeutic agents for metastatic CRC along with fluoropyrimidines and oxaliplatin. However, only 30%-40% of the patients show an objective response to irinotecan, and there is currently no established biomarker predictive of clinical benefit from irinotecan chemotherapy.
CRC is known to have an abundance of aberrant promoter methylations [3], and methylation status has been studied for potential correlations with treatment outcomes of CRC in this regard [4], [5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15]. Patients with the CpG island methylator phenotype (CIMP), reflecting extensive promoter methylation, were found to benefit from irinotecan chemotherapy in the adjuvant setting. A recent study also demonstrated that the treatment outcomes of metastatic CRC differed significantly according to CIMP status, adding a further prognostic role for CIMP status besides stage II and III CRC [8]. In this study, while CIMP-high metastatic CRC was associated with poorer progression-free survival and overall survival (OS) for oxaliplatin-based first-line chemotherapy, the correlation between CIMP status and treatment outcomes was less prominent for irinotecan-based second-line treatment [8]. As CIMP is one of the key molecular pathways in CRC carcinogenesis and the association between CIMP status and irinotecan response remains unclear, we aimed to further investigate the implications of CIMP in patients with metastatic CRC treated with irinotecan [16].
In addition to CIMP status, silencing by promoter methylation in individual genes has also been suggested to induce resistance or response to irinotecan [15], [17]. Checkpoint with forkhead and ring finger domains (CHFR) encodes the E3 ubiquitin-protein ligase CHFR and has been identified as a mitotic stress checkpoint and tumor suppressor gene. CHFR is frequently inactivated by promoter CpG island methylation in CRC [18], [19], [20]. CHFR methylation was associated with reduced survival in stage II and III CRC [21], [22] and was suggested to be associated with enhanced sensitivity to taxanes in CRC, non–small cell lung cancer, and gastric cancer [23], [24], [25], [26], [27]. Although CHFR methylation has not been directly evaluated in conjunction with irinotecan therapy, recent studies have found that CHFR plays an important role in the early stage of the DNA damage response [28], [29]. As CHFR is highly methylated in CRC and a well-coordinated DNA damage response pathway is required for the repair of irinotecan-induced cellular damage, we postulated that CHFR may be involved in the therapeutic response to irinotecan in addition to taxanes.
Werner syndrome RecQ-like helicase (WRN), known as a tumor suppressor gene with exonuclease function, was reported to be frequently inactivated epigenetically and to correlate with mucinous differentiation in CRC [12], [13], [30]. WRN methylation has been predicted to enhance topoisomerase inhibitor activity by abrogation of its exonuclease function, and the association of WRN methylation and irinotecan response was suggested in a few studies with a relatively small number of patients with colorectal, gastric, and cervical cancers [13], [31], [32].
Heparan sulfate 6-O-endosulfatase gene (SULF2) encodes an oncoprotein with heparin-degrading endosulfatase activity, which activates receptor tyrosine kinases and downstream pathways including MAPK, AKT, and WNT [33]. While the role of SULF2 methylation has been mostly unknown in CRC, SULF2 methylation was associated with irinotecan sensitivity in patients with gastric cancer [32]. In addition, SULF2 silencing increased sensitivity to topoisomerase 1 inhibitors via increased expression of interferon-inducible genes, including ISG15, in non–small cell lung cancer [34].
In this study, we aimed to investigate the association of treatment outcomes for irinotecan-based systemic chemotherapy with methylation in CHFR, WRN, and SULF2 as well as CIMP status in patients with metastatic CRC.
Patients and Methods
Patients and Irinotecan-Based Systemic Chemotherapy
Patients who underwent surgical resection of CRC at the National Cancer Center (NCC), Korea, from 2001 to 2004 were eligible for this retrospective biomarker study if the following criteria were met: pathologically confirmed diagnosis of colorectal adenocarcinoma, age ≥19 years, synchronous or metachronous metastasis, systemic chemotherapy with one of the irinotecan-containing regimens, tumor tissues available at NCC Tumor Bank/Pathology Department, and presence of evaluable lesion(s) before initiation of irinotecan-containing chemotherapy. This study protocol was reviewed and approved by the Institutional Review Board of NCC (IRB No: NCC2014-0075). The study was conducted in accordance with the recommendations of the Declaration of Helsinki for biomedical research involving human subjects.
Patients were treated with one of the following irinotecan-containing chemotherapy regimens: FOLFIRI [irinotecan (180 mg/m2 i.v.), leucovorin (200 mg/m2 i.v. on day 1), and 5-fluorouracil (400 mg/m2 i.v. bolus followed by 2400 mg/m2 continuous i.v. over 46 hours on day 1) every 2 weeks]; XELIRI [irinotecan (250 mg/m2 i.v. on day 1) and capecitabine (1000 mg/m2 p.o. twice a day for 2 weeks) every 3 weeks]; IFL [irinotecan (125 mg/m2 i.v.), leucovorin (20 mg/m2 i.v. on day 1), and 5-fluorouracil (500 mg/m2 i.v. on day 1) weekly for 4 weeks, every 6 weeks]; and irinotecan alone [irinotecan (350 mg/m2 i.v. on day 1) every 3 weeks]. At the physician’s discretion, bevacizumab (5 mg/kg i.v. on day 1 of each cycle of FOLFIRI or 7.5 mg/kg i.v. on day 1 of each cycle of XELIRI) or cetuximab (400 mg/m2 i.v. on day 1 and 250 mg/m2 on day 8 and weekly thereafter) was combined with the cytotoxic chemotherapy. Computed tomography was performed after every four cycles for biweekly regimens and after three cycles for three-weekly regimens during the chemotherapy period or earlier if disease progression was suspected. Disease progression was defined based on the computed tomographic findings.
Methylation Analyses
Analysis of DNA methylation was performed as described previously [35]. Genomic DNA samples from the tumor and adjacent normal tissue were bisulfite-modified using the EZ DNA methylation kit (Zymo Research, Orange, CA) and analyzed for methylation in five CIMP-specific CpG island loci (CACNA1G, IGF2, NEUROG1, RUNX3, SOCS1), as well as CHFR, WRN, and SULF2 using a methylation-specific polymerase chain reaction (MSP) method. Briefly, 1 μg of DNA was denatured using sodium hydroxide, modified by sodium bisulfite, treated again with sodium hydroxide, precipitated with ethanol, and resuspended in water. For each locus, two primer pairs were used for the MSP analysis; the first recognizes and anneals to methylated sequences only, whereas the second set anneals to and amplifies unmethylated alleles. The detailed primer information is provided in Supplementary material 1. The PCR products were then purified using the Wizard DNA purification resin (Promega, Madison, WI). Each PCR product was directly loaded on an 8% acrylamide gel, stained with ethidium bromide, and visualized under UV illumination. CIMP status was considered positive when at least three methylated promoters were identified and as negative when zero to two methylated promoters were identified.
The Cancer Genome Atlas (TCGA) Data Analysis
CHFR DNA methylation (Illumina Infinium HM27 bead array; HM27) and mRNA expression microarray) data from 223 colorectal adenocarcinoma samples from TCGA project were downloaded through cBioPortal (http://www.cbioportal.org; accessed on Jun. 12, 2018).
Cell Lines and Cell Culture
Human CRC cell lines (RKO, HT-29, HCT-116, SNU-81, SW480, DLD-1, SNU-407, CaCo-2, LoVo, SW620, SNU-C4, and SNU-C5) were obtained from the Korea Cell Line Bank and the American Type Culture Collection. Cell lines were grown in DMEM or RPMI-1640 with 10% fetal bovine serum and 1% penicillin-streptomycin at 37°C in a 5% CO2-humidified atmosphere. All cell lines were certified using the GenePrint 10 System (Promega, Madison, WI) by the Omics Core Lab of NCC.
Reagents
Irinotecan HCl trihydrate was purchased from Selleckchem (Houston, TX). The DNA methylation inhibitor 5-aza-2′-deoxycytidine (5-Aza-CdR) was purchased from Sigma-Aldrich (St. Louis, MO). Stock solutions were prepared in dimethyl sulfoxide and stored at −20°C.
Growth Inhibition Assays
A colorimetric assay using the tetrazolium salt MTS was used to assess cell proliferation after treatment with irinotecan. Equivalent numbers of cells (5 × 103 cells/well) were incubated in 0.2 ml culture medium in each well. After 1, 2, and 3 days of culture, 0.1 mg MTS solution (Promega, Madison, WI) was added to each well followed by incubation at 37°C for a further 4 hours. Plates were centrifuged at 450×g for 5 minutes at room temperature, and the medium was removed. Dimethyl sulfoxide (0.15 ml) was added to each well to solubilize the crystals, and the plates were immediately read at 540 nm using a scanning multiwell spectrometer (Bio-Tek instruments Inc., Winooski, VT). The cell proliferation rate was obtained from three biological replicates, and all experiments were performed three times.
Cancer Cell Line Encyclopedia (CCLE) Data Analysis
Pharmacological profiling data for irinotecan was downloaded for 12 colorectal adenocarcinoma cell lines along with the DNA methylation data from the CCLE (http://portal.broadinstitute.org/ccle; accessed on Jun. 13, 2018)
Establishment of CHFR-Overexpressing Cells Using Plasmid DNA Vector
The recombinant plasmid DNA human CHFR (target sequence: 5′-GCGATCGCACGCGT-3′) (RC228526) was purchased from OriGene (Rockville, MD). The recombinant plasmid was then transformed into competent Escherichia coli cells. The bacteria were cultured, and the recombinant plasmids were extracted and purified using PureLink HiPure Plasmid DNA Purification kits (Invitrogen, Carlsbad, CA). HCT-116 and SNU-C5 cells were plated in six-well plates at a density of 3 × 105 cells per well and incubated overnight. Cells were then transfected with the human CHFR vector or a blank control using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Transfection was verified by Western blot analysis.
Small Interfering RNA (siRNA)–Mediated Knockdown of CHFR
siRNA against CHFR and the control sequence were purchased from Qiagen (Chatsworth, CA). The sequence of the CHFR-specific siRNA was 5′-AACCAGAGGTTTGACATGGAA-3′, and AllStars Negative Control siRNA (catalog no. 1027281) was used as the control (nonspecific). siRNA transfection was performed using HiPerFect Transfection Reagent (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Briefly, 12 μl of 20 nM siRNA solution and 20 nM HiPerFect Transfection Reagent was incubated in 100 ml of serum-free RPMI 1640 medium for 10 minutes to facilitate complex formation. The resulting mixture (final concentration 5 nM) was added to SNU-81 and CaCo-2 cells (1 × 106) and incubated in a 60-mm tissue culture dish with 4 ml of RPMI 1640. The cells were then washed at 0, 24, 48, and 72 hours after transfection.
Western Blotting
Briefly, cell homogenates containing equivalent amounts of protein were centrifuged at 4000×g, and the supernatant fractions were subjected to SDS-PAGE. Following electrophoresis, the proteins were transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA) and blocked by incubation for 2 hours at 4°C in 1% Tween 20-TBS buffer containing 1.5% nonfat dry milk (Bio-Rad, Hercules, CA) and 1 mM MgCl2. Membranes were then incubated for 2 hours at room temperature with primary antibodies against CHFR (Santa Cruz bio Technology, Santa Cruz, CA) or β-actin (Cell Signaling Technology, Beverly, MA). Next, the membranes were washed thrice for 15 minutes with blocking solution and incubated with diluted HRP-conjugated secondary antibody (SouthernBiotech, Birmingham, AL) for 1 hour at room temperature. This was followed by washing with blocking solution (thrice for 15 minutes), incubation with WEST-ZOL plus chemiluminescence reagent (iNtRON Biotechnology, Seoul, Korea) for 1 minute, and exposure to film (Kodak Blue XB-1).
Statistical Analyses
The primary aim of the present study was to determine the association of methylation in CHFR, WRN, and SULF2, as well as CIMP status, with time to progression (TTP) of irinotecan-based systemic chemotherapy in patients with metastatic CRC. In addition to TTP, OS and response were analyzed as indicators of treatment outcomes with irinotecan treatment. TTP was defined as the time interval from the date of treatment initiation to the date of disease progression. OS was calculated from the date of treatment initiation to the date of death from any cause. Categorical variables were compared using Pearson’s χ2 test or Fisher’s exact test, and continuous variables were compared using Mann-Whitney test. The Kaplan-Meier method was used for estimating TTP and OS, and comparisons were made using log-rank tests. To adjust for baseline characteristics, we performed multivariate analyses with a Cox proportional hazard model using a forward conditional variable selection method. Age (continuous variable), sex, differentiation (well-differentiated to moderately differentiated vs. poorly differentiated), tumor location (proximal vs. distal), number of metastatic organs (1-2 vs. ≥3), serum carcinoembryonic antigen (CEA) levels (≤5 vs. >5 ng/ml), BRAF mutation status, hMLH1/hMSH2 (proficient vs. deficient), and number of prior systemic chemotherapy lines in the metastatic setting were included as covariates based on previous studies on metastatic CRC [8]. Two-sided P values < .05 were considered statistically significant. Statistical analyses for the clinical study were conducted using R-3.3.4 software, and analyses for in vitro study were performed with GraphPad Prism version 7.0.0 for Windows (GraphPad Software, La Jolla, CA). This study was analyzed and reported according to the Reporting Recommendations for Tumor Marker Prognostic Studies [36].
Results
Patient Characteristics and Irinotecan-Based Chemotherapy
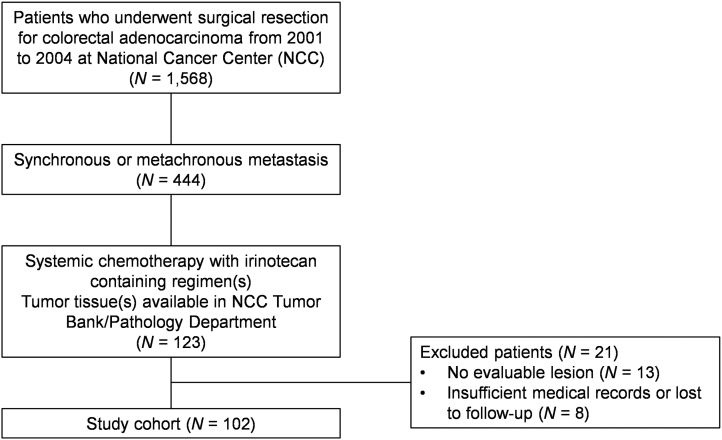
In total, 102 patients were included in this study (Supplementary material 2). The patient characteristics are described in Table 1. According to the inclusion criteria, all patients had undergone resection of the primary tumor. All surgical specimens used for this study were collected from the primary tumors before the initiation of irinotecan treatment and therefore do not reflect the potential effects from subsequent treatments. Twenty-one patients (20.6%) exhibited proximal (cecum to transverse colon) lesions, and 81 (79.4%) exhibited distal lesions. In total, 73 patients (71.6%) presented with synchronous metastatic disease and 29 (28.4%) with metachronous metastasis. Irinotecan-based chemotherapy was administered as first-line therapy in 51 patients (50.0%), second-line in 38 (37.3%), and ≥third-line in 13 patients (12.7%). The irinotecan-containing chemotherapy regimens include FOLFIRI in 44 patients (43.1%), IFL in 15 (14.7%), XELIRI in 21 (20.6%), and irinotecan as a single agent in 22 patients (21.6%). Bevacizumab or cetuximab was added for five and four patients, respectively. The TTP following irinotecan therapy was 5.57 months (95% CI, 3.61-7.52) in the entire population. At the time of last follow-up (March 2018), all 102 patients had died. The median OS after the initiation of irinotecan-based chemotherapy was 20.4 months (95% CI, 18.0-22.8).
Supplementary material 2.
Reporting recommendations for tumour MARKer prognostic studies (REMARK) diagram entailing the study cohort.
Table 1.
Patient Characteristics and CHFR Methylation
| Total (%) | CHFR Methylated (%) | CHFR Unmethylated (%) | P | |
|---|---|---|---|---|
| Total | 102 (100) | 16 (100) | 86 (100) | |
| Age | .550 | |||
| Median (range) | 56 (27-78) | |||
| <65 years | 75 (73.5) | 13 (81.2) | 62 (72.1) | |
| ≥65 years | 27 (26.5) | 3 (18.8) | 24 (27.9) | |
| Sex | >.999 | |||
| Male | 68 (66.7) | 11 (68.8) | 57 (66.3) | |
| Female | 34 (33.3) | 5 (31.2) | 29 (33.7) | |
| Location | .737 | |||
| Right-sided | 21 (20.6) | 4 (25.0) | 17 (19.8) | |
| Left-sided | 81 (79.4) | 12 (75.0) | 69 (80.2) | |
| Histology | .950 | |||
| Nonmucinous | 92 (90.2) | 15 (93.8) | 77 (89.5) | |
| Mucinous | 10 (9.8) | 1 (6.2) | 9 (10.5) | |
| Differentiation | .732 | |||
| Well to moderate | 82 (80.4) | 14 (87.5) | 68 (79.1) | |
| Poor | 20 (19.6) | 2 (12.5) | 18 (20.9) | |
| KRAS/NRAS | .138⁎ | |||
| Wild type | 65 (68.5) | 8 (50.0) | 57 (66.3) | |
| Mutant | 30 (31.6) | 8 (50.0) | 22 (25.6) | |
| BRAF status | .073⁎ | |||
| Wild type | 92 (96.8) | 14 (87.5) | 78 (98.7) | |
| Mutant | 3 (3.2) | 2 (12.5) | 1 (1.3) | |
| hMLH1/hMSH2 | >.999 | |||
| Proficient | 98 (96.1) | 16 (100.0) | 82 (95.3) | |
| Deficient | 4 (3.9) | 0 (0.0) | 4 (4.7) | |
| Presentation | >.999 | |||
| Synchronous | 73 (71.6) | 12 (75.0) | 61 (70.9) | |
| Metachronous | 29 (28.4) | 4 (25.0) | 25 (29.1) | |
| No. metastatic organs | .710 | |||
| 1 | 50 (49.0) | 7 (43.8) | 43 (48.8) | |
| 2 | 39 (38.2) | 6 (37.5) | 33 (38.4) | |
| ≥3 | 14 (13.7) | 3 (18.8) | 11 (12.8) | |
| CEA | .891 | |||
| <5.0 ng/ml | 27 (26.5) | 5 (31.2) | 22 (25.9) | |
| ≥5.0 ng/ml | 74 (72.5) | 11 (68.8) | 63 (74.1) | |
| No. prior treatment(s) | .710 | |||
| 0 | 51 (50.0) | 7 (43.8) | 44 (51.2) | |
| 1 | 38 (37.3) | 6 (37.5) | 32 (37.2) | |
| ≥2 | 13 (12.7) | 3 (18.8) | 10 (11.6) | |
| Irinotecan regimen | .658 | |||
| FOLFIRI | 44 (43.1) | 6 (37.5) | 38 (44.2) | |
| IFL | 15 (14.7) | 4 (25.0) | 11 (12.8) | |
| XELIRI | 21 (20.6) | 3 (18.8) | 18 (20.9) | |
| Irinotecan alone | 22 (21.6) | 3 (18.8) | 19 (22.1) |
P = .024 for KRAS/NRAS/BRAF mutation frequency and CHFR methylation (62.5% vs. 31.2% for CHFR methylated and unmethylated groups, respectively)
CHFR Methylation and Irinotecan Treatment Outcomes
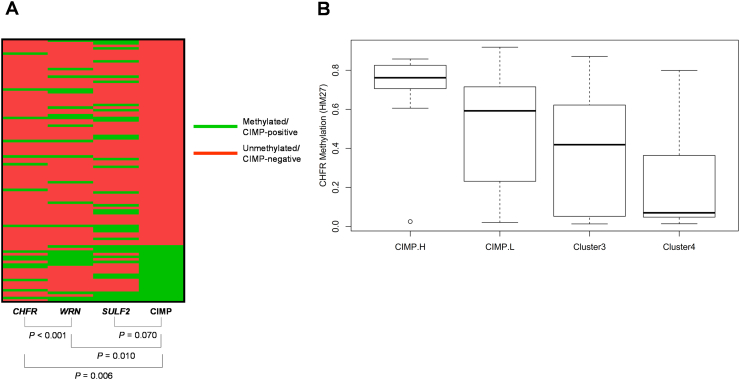
Promoter methylation in CHFR, WRN, and SULF2 was observed in 16 (15.7%), 24 (23.5%), and 33 (32.4%) patients, respectively. Twenty-two (21.6%) patients exhibited at least three methylated CIMP-specific loci and were defined as CIMP-positive. Among the five CIMP-specific loci, NEUROG1 was the most frequently methylated locus (63 patients; 61.8%), followed by CACNA1G (41; 40.2%), IGF2 (28; 27.5%), SOCS1 (16; 15.7%), and RUNX3 (16; 15.7%). In our analysis, CHFR and WRN methylation was significantly associated with CIMP positivity, whereas SULF2 methylation was not (Supplementary Material 3). CHFR methylation was closely associated with the WRN methylation as well. We confirmed a significant increase in CHFR methylation level according to CIMP subtypes using the TCGA dataset (Supplementary material 3). The CHFR-methylated group showed more frequent mutations in KRAS, NRAS, or BRAF compared to the unmethylated group (62.5% vs. 31.2%; P = .024). However, other clinicopathological characteristics including tumor location, BRAF mutation, and MMR status were not associated with methylation of CHFR, WRN, SULF2, or the CIMP status in our analysis (Table 1).
Supplementary material 3.
(A) Correlation between CHFR, WRN, SULF2 methylation, and the CIMP status. P-values are calculated with chi-squared tests. (B) Boxplot depicting CHFR methylation according to CIMP subtypes in TCGA data.
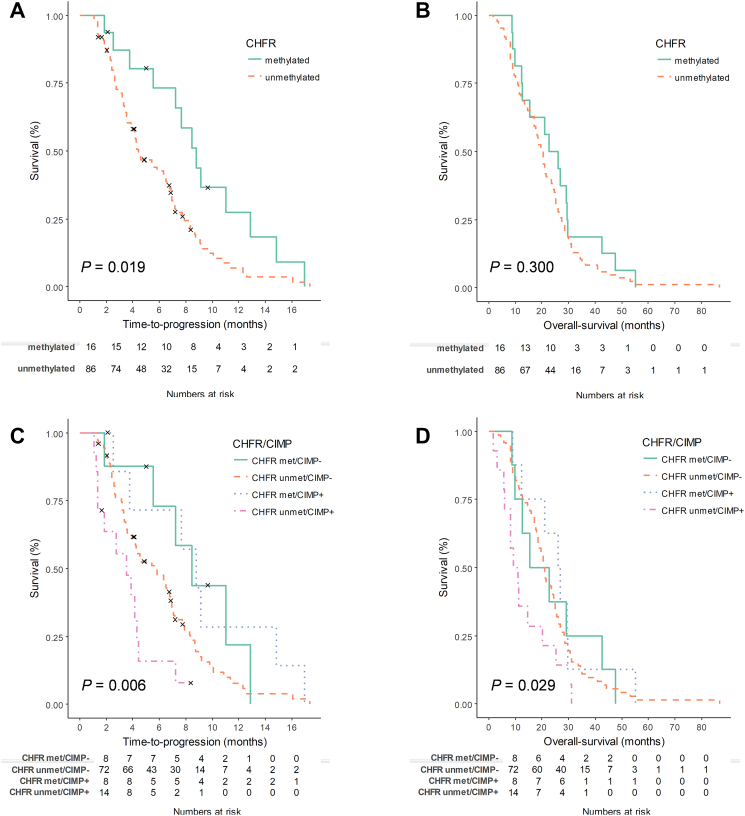
TTP after the initiation of irinotecan-containing chemotherapy was significantly different according to CHFR methylation status (P = .019; Figure 1A). The median TTP was 8.77 months (95% CI, 6.81-10.73) for the CHFR-methylated group and 4.43 months (95% CI, 3.09-5.78) for the unmethylated group. OS showed a trend towards the CHFR-methylated group, but the difference was not significant (P = .300; Figure 1B). The median OS was 22.83 months (95% CI, 13.03-32.63) for the CHFR-methylated group and 20.17 months (95% CI, 17.65-22.68) for the unmethylated group. The response rate was 31.3% vs. 17.4% for patients with methylated and unmethylated CHFR, respectively (P = .278).
Figure 1.
(A-B) Kaplan-Meier curves for TTP and OS of irinotecan treatment according to CHFR methylation. (C-D) Kaplan-Meier curves for TTP and OS of irinotecan treatment according to CHFR methylation and CIMP status.
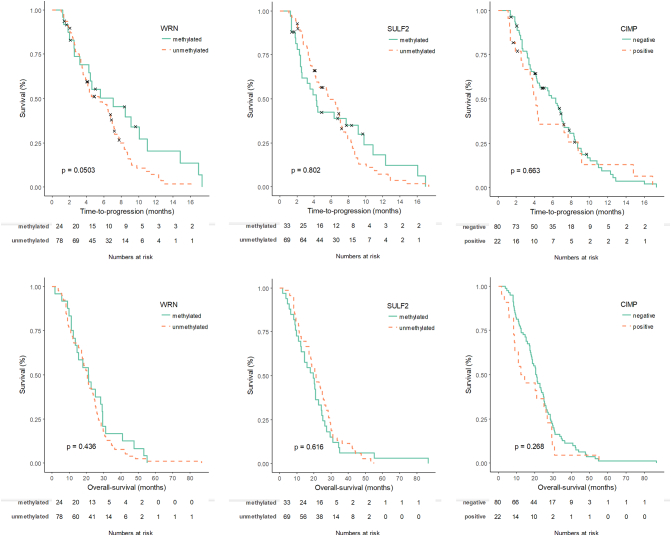
TTP was marginally significant favoring the WRN-methylated group (P = .050; Supplementary material 4). The median TTP was 7.03 months (95% CI, 1.89-12.2) vs. 5.43 (95% CI, 3.32-7.55) for the WRN-methylated and unmethylated groups. There was no difference in OS based on WRN methylation. The treatment outcomes in terms of TTP and OS following irinotecan treatment were not different based on SULF2 methylation or CIMP status (Supplementary Material 4).
Supplementary material 4.
Kaplan-Meier curves for TTP and OS of irinotecan treatment according to WRN/SULF2 methylation and the CIMP status.
In the multivariate analyses for TTP and OS of irinotecan treatment, CHFR methylation was confirmed to be significantly associated with TTP after adjustment for potential confounding factors [adjusted HR, 2.88 (95% CI, 1.50-5.52), P = .001; Table 2], whereas methylation of WRN and SULF2, and the CIMP status were not. Methylation of CHFR, WRN, SULF2, and the CIMP status were not significant factors in the multivariate analysis for OS.
Table 2.
Multivariate Analyses for TTP and OS According to CHFR Status
| TTP |
OS |
||||
|---|---|---|---|---|---|
| Covariates in the Final Model | Adjusted HR (95% CI) | P | Covariates in the Final Model | Adjusted HR (95% CI) | P |
| CHFR unmethylated | 2.88 (1.50-5.52) | .001 | CEA >5.0 ng/ml | 2.33 (1.42-3.80) | .01 |
| No. prior treatments | <.001 | No. prior treatments | .002 | ||
| 1 | 2.78 (1.60-4.86) | <.001 | 1 | 2.78 (1.60-4.86) | <.001 |
| ≥2 | 3.43 (1.69-6.97) | <.001 | ≥2 | 3.43 (1.69-6.97) | <.001 |
| hMLH/hMSH2 deficient | 4.49 (1.02-19.66) | .046 | |||
| Metastatic organs ≥3 | 2.28 (1.19-4.37) | .013 | |||
Multivariate analyses were performed by Cox regression analysis with forward conditional selection method adjusted for age (continuous), sex, differentiation (well to moderately differentiated vs. poorly differentiated), tumor location (right vs. left), metastatic organs (1-2 vs. ≥3), CEA level (≤5 vs. >5.0 ng/ml), BRAF mutation, hMLH1/hMSH2, number of prior treatment(s), promoter methylation, and CIMP status.
As CHFR methylation was associated with CIMP-positive status, the association of CHFR/CIMP status was also analyzed for TTP and OS. TTP and OS were significantly different according to the CHFR/CIMP status (P = .006 for TTP, Figure 1C; P = .029 for OS, Figure 1D). For patients with unmethylated CHFR, TTP and OS were significantly different according to the CIMP status. The median TTP was 5.60 months (95% CI, 3.40-7.81) for the CHFR-unmethylated/CIMP-negative group and 3.53 months (95% CI, 1.60-5.47) for the CHFR-unmethylated/CIMP-positive group (P = .020). The median OS was 20.57 months (95% CI, 18.52-22.61) for the CHFR-unmethylated/CIMP-negative group and 9.23 months (95% CI, 4.28-14.18) for the CHFR-unmethylated/CIMP-positive group (P = .060). In the multivariate analyses for TTP and OS, CHFR/CIMP status was found to be significantly associated with TTP and OS after adjustment for potential confounding factors (Table 3).
Table 3.
Multivariate Analyses for TTP and OS According to CHFR and CIMP Status
| TTP |
OS |
||||
|---|---|---|---|---|---|
| Covariates in the Final Model | Adjusted HR (95% CI) | P | Covariates in the Final Model | Adjusted HR (95% CI) | P |
| CHFR/CIMP status | <.001 | CHFR/CIMP status | <.001 | ||
| CHFR unmethylated/CIMP negative | 2.43 (1.23-4.81) | .011 | CHFR unmethylated/CIMP negative | 3.94 (2.02-7.69) | <.001 |
| CHFR unmethylated/CIMP positive | 6.45 (2.66-15.63) | <.001 | CHFR unmethylated/CIMP positive | 4.72 (2.10-10.64) | <.001 |
| No. prior treatment(s) | <.001 | No. prior treatment(s) | .001 | ||
| 1 | 2.77 (1.58-4.84) | <.001 | 1 | 0.98 (0.59-1.63) | .935 |
| ≥2 | 3.84 (1.87-7.88) | <.001 | ≥2 | 3.42 (1.78-6.60) | <.001 |
| CEA >5.0 ng/ml | 2.80 (1.66-4.72) | <.001 | |||
Multivariate analyses were performed by Cox regression analysis with forward conditional selection method adjusted for age (continuous), sex, differentiation (well to moderately differentiated vs. poorly differentiated), tumor location (right vs. left), metastatic organs (1-2 vs. ≥3), CEA level (≤5 vs. >5.0 ng/ml), BRAF mutation, hMLH1/hMSH2, number of prior treatment(s), and CHFR/CIMP status.
CHFR Methylation and In Vitro Sensitivity to Irinotecan
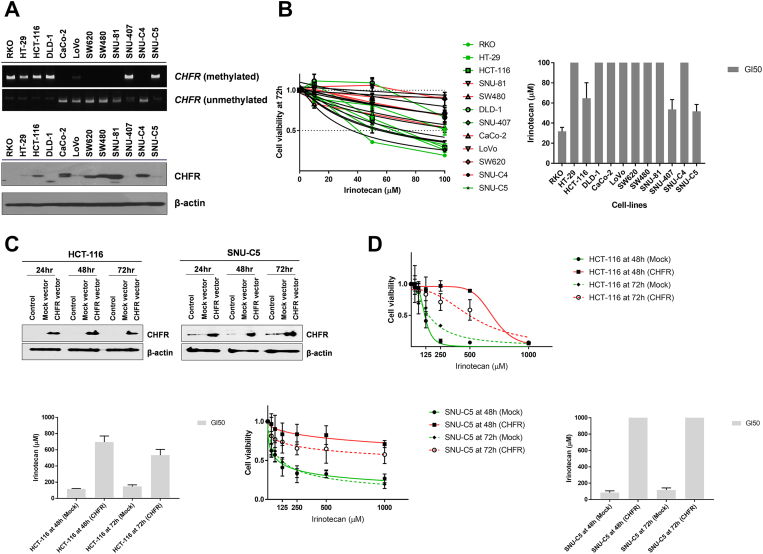
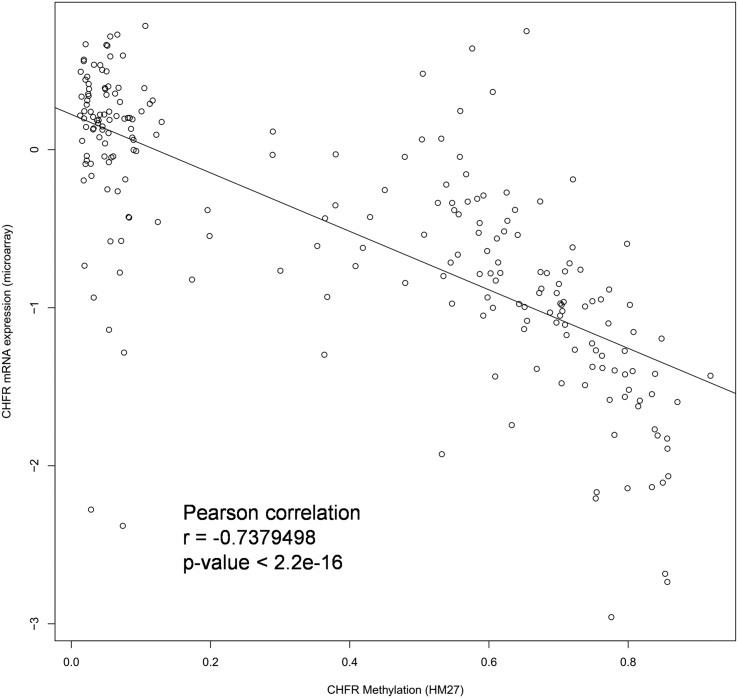
Irinotecan sensitivity in vitro based on CHFR methylation status was evaluated in 12 CRC cell lines. When CHFR promoter methylation status was assessed with MSP, six cell lines (RKO, HT-29, HCT-116, DLD-1, SNU-407, and SNU-C5) showed CHFR methylation, whereas five cell lines (CaCo-2, SW-620, SW-480, SNU-81, SNU-C4) had an unmethylated promoter (Figure 2A). CHFR was partially methylated in LoVo cells. Western blot analysis revealed that protein expression levels of CHFR were inversely correlated with CHFR methylation status (Figure 2A). We confirmed the inverse correlation between CHFR methylation and mRNA expression in clinical samples as well using the TCGA dataset (Pearson correlation coefficient = −0.738, P < 0.001; Supplementary Material 5).
Figure 2.
(A) CHFR promoter methylation in 12 colorectal cancer cell lines as determined by methylation-specific PCR assays (upper panel). CHFR protein expression as determined by Western blotting (lower panel). (B) In vitro sensitivity of 12 colorectal cancer cell lines to increasing concentrations of irinotecan as determined by MTS cell proliferation assays. Cell growth curves (green, CHFR-methylated; red, unmethylated) are based on three independent experiments performed in triplicate (left panel). Nonlinear fit curves for each cell line are shown in black. GI50 values for each cell line are shown as bars representing the mean ± SEM (right panel). (C) Overexpression of CHFR in CHFR-methylated cell lines (HCT-116 and SNU-C5) was confirmed by Western blotting. (D) Irinotecan sensitivity after CHFR overexpression in HCT-116 and SNU-C5. Cell growth curves and GI50 values were determined at 48 and 72 hours after CHFR overexpression following treatment with increasing concentrations of irinotecan. Values are based on three independent experiments performed in triplicate.
Supplementary material 5.
Validation of inverse correlation between CHFR methylation and mRNA expression using TCGA dataset.
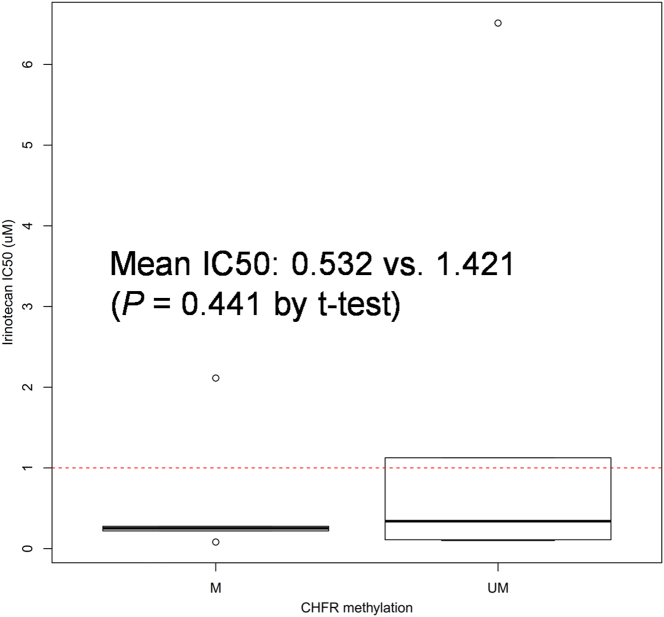
Cell proliferation assay demonstrated that CRC cell lines with CHFR methylation were more sensitive to irinotecan treatment compared to those with unmethylated CHFR (Figure 2B). Notably, all four cell lines with GI50 values <100 μM for irinotecan had methylated CHFR (RKO, SNU-C5, SNU-407, and HCT-116). We also compared the IC50 of 12 CRC cell lines according to CHFR methylation status using CCLE data (Supplementary material 6). Although the mean IC50 was lower in CHFR-methylated cells (n = 6) than in unmethylated cells (n = 6), the difference between groups was not significant (P = .441).
Supplementary material 6.
Boxplot depicting IC50 in CHFR methylated (n = 6) and unmethylated CRC cell-lines (n = 6) in CCLE data.
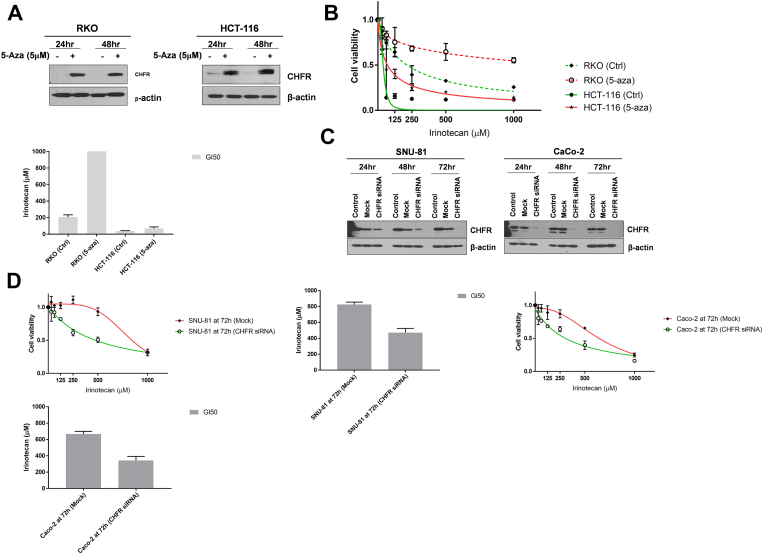
We then investigated the effect of CHFR overexpression on sensitivity to irinotecan in CHFR-methylated HCT-116 and SNU-C5 cells. After CHFR overexpression (Figure 2C), sensitivity to irinotecan measured at 48 and 72 hours was significantly diminished in both cell lines compared to their parental cells (Figure 2D). We also compared the viability of CHFR-methylated cell lines (RKO and HCT-116) with increasing concentrations of irinotecan along with or without 5-Aza-CdR (5 μM), a DNA-demethylating agent (Figure 3, A and B). Both RKO and HCT-116 cells showed CHFR upregulation after 5-Aza-CdR treatment (Figure 3A), and combination with 5-Aza-CdR decreased the growth inhibitory potential of irinotecan in all three cell lines (Figure 3B).
Figure 3.
(A) Western blotting for CHFR in CHFR-methylated cell lines (RKO and HCT-116) at 24 and 48 hours after 5-Aza-CdR treatment at 5 μM. (B) Irinotecan sensitivity in RKO and HCT-116 cells after 5-Aza-CdR treatment. Cell growth curves and GI50 values were plotted at 72 hours after 5-Aza-CdR treatment following treatment with increasing concentrations of irinotecan. (C) Western blotting for CHFR in CHFR-unmethylated cell lines (SNU-81 and CaCo-2) after CHFR knockdown with siRNA at 24, 48, and 72 hours. (D) Irinotecan sensitivity in SNU-81 and CaCo-2 cells after CHFR knockdown. Cell growth curves and GI50 values were plotted at 72 hours after CHFR knockdown following treatment with increasing concentrations of irinotecan. All values are based on three independent experiments performed in triplicate.
We then tested the effect of CHFR knockdown on irinotecan sensitivity in CHFR-unmethylated cell lines (Figure 3, C and D). Both SNU-81 and CaCo-2 cell lines demonstrated loss of CHFR expression at 72 h after siRNA treatment (Figure 3C), and GI50 values were significantly lower in cell lines subjected to CHFR knockdown compared to their parental cells (Figure 3D).
Discussion
In this study, we found that CHFR methylation was predictive of favorable treatment outcomes in terms of TTP in patients with metastatic CRC treated with irinotecan-based systemic chemotherapy. We further expanded our study in vitro and confirmed that CRC cell lines with CHFR methylation were more susceptible to irinotecan compared to those without methylation and that sensitivity to irinotecan could be modulated negatively by CHFR upregulation or positively by downregulating CHFR. In addition, irinotecan treatment outcomes differed according to CIMP status in patients with unmethylated CHFR. To the best of our knowledge, this is the first report on the clinical impact of CHFR solely focused on metastatic CRC and suggests CHFR methylation as a biomarker for irinotecan-based chemotherapy in patients with CRC.
CHFR was methylated in 15.7% of patients with metastatic CRC in our study, and this was lower than in previous studies reporting CHFR methylation in 31%-63% of patients, mostly with stage I-III CRC [18], [19], [21], [22], [37]. When we analyzed the TCGA dataset, CHFR methylation levels did not differ by tumor stage in patients with CRC (P = .511 by ANOVA). Therefore, the lower prevalence of CHFR methylation in this study may be explained by differences in methylation assay or patient selection criteria. The correlation of CHFR with chemotherapy response or resistance has been mostly conducted for taxanes such as paclitaxel or docetaxel [23], [24], [27], [38]. This is because CHFR is known to encode a checkpoint protein that delays entry into metaphase [39]. Hence, the antitumor activity of microtubule inhibitors like taxanes could be boosted in cells with CHFR deficiency. In a previous study, the association of CHFR and chemotherapy response was specific to microtubule inhibitors but not to etoposide, a topoisomerase II inhibitor, or to cisplatin [40]. In a study including 20 patients with CIMP-positive metastatic CRC who received nab-paclitaxel, however, treatment outcomes did not differ according to plasma CHFR methylation status, suggesting that CHFR methylation might not be predictive of taxane sensitivity, at least for patients with CRC. One potential explanation for the association between CHFR methylation and irinotecan response demonstrated in our study comes from the fact that CHFR was first described to function as an E3 ubiquitin ligase, which ubiquitinates and targets proteins for degradation by the S26 proteasome [41]. One of the cellular mechanisms of irinotecan resistance is the repair of irinotecan-induced DNA damage [17]. When the reversible Topo-I-irinotecan-DNA cleavable complex formed by irinotecan treatment collides with the advancing replication fork, Topo-I, the cellular target of irinotecan, is degraded through an ubiquitin/26S proteasome-dependent system, and this facilitates the repair of single-strand breaks, thereby evading irinotecan-induced cellular damage. If the level of CHFR protein, a ubiquitin ligase, is decreased by DNA methylation, ubiquitination of Topo-I by 26S proteasome is impaired, and the subsequent upregulation of Topo-I could again render cancer cells more sensitive to irinotecan-induced cellular damage. This hypothesis is difficult to validate using clinical samples collected at a single time point and is also difficult to demonstrate using public data such as TCGA dataset. In fact, when we correlated CHFR methylation and TOP1 mRNA expression levels using the TCGA dataset, there was no apparent correlation between them (data not shown).
We demonstrated that CHFR methylation was closely associated with CIMP status in our clinical samples as well as in TCGA dataset. Although OS showed a trend favoring the CIMP-negative group (P = .268 for OS after irinotecan treatment) in our cohort, the difference in TTP or OS based on CIMP status was not apparent in the overall population treated with irinotecan. We speculate that the different methodologies used in this study (MSP) and the previous one (MethyLight assay) or differences in patient selection criteria (irinotecan-based chemotherapy) may at least partially contribute to these results. However, in patients with unmethylated CHFR, TTP was poorer in the CIMP-positive group than in the CIMP-negative group, and multivariate analysis confirmed that the combination of CHFR/CIMP was a significant factor for both TTP and OS. Therefore, the significance of CHFR methylation in the treatment response to irinotecan may have weakened the prognostic impact of CIMP in our study cohort.
In our study cohort, WRN was found to be hypermethylated in 23.5% of patients with metastatic CRC, and this rate was lower than the ~40% reported previously [5], [13]. Although DNA hypermethylation in a specific gene has long been regarded to reflect decreased mRNA and protein expression in the corresponding gene, it may not be correlated with repressive gene expression in some cases because of the complex regulation of gene expression besides DNA methylation [42]. Although we included WRN methylation as one of the potential biomarkers in this study, WRN methylation was marginally correlated with TTP, and the correlation was not maintained in a multivariate analysis. Recently, it was reported that WRN methylation was not predictive of good clinical outcomes in 93 patients with metastatic CRC treated with irinotecan and capecitabine [5]. Remarkably, WRN mRNA and protein expression levels were independent of WRN methylation status in the study, which was also confirmed with TCGA data. We suspect that this absence of correlation between WRN methylation and mRNA or protein levels might be the main reason for the lack of correlation in WRN methylation with treatment outcome in our study.
Currently, most of the irinotecan-based systemic chemotherapy is given in combination with anti-EGFR antibodies or anti-VEGF/VEGFR antibodies because combination chemotherapy in both forms have been proven to provide survival benefit in the first-line (cetuximab, panitumumab, bevacizumab) as well as second-line (bevacizumab, ziv-aflibercept, ramucirumab) and third-line settings (cetuximab) when compared to irinotecan-based cytotoxic chemotherapy alone. In our study, we selected patients that had been treated in the era when there was limited access to combination with molecular targeted agents. In fact, most (93/102 patients, 91.2%) of the patients in our study cohort were treated with irinotecan-based systemic chemotherapy alone. Although this selection was not intended and was mainly because there were limitations to the archival tumor tissues collected after 2005 made by domestic legal provisions when this study was planned, we believe that this selection allowed us to better define the pure impact of irinotecan on patient outcomes.
Our study has the following limitations. First, we did not validate our findings in a separate cohort. However, the preclinical observation of our study is fully in agreement with the clinical findings, and the association of CHFR and irinotecan could be mechanistically explainable. Although our findings are not confirmative and are more likely to be hypothesis generating, we believe that these points make our study more generalizable and worthy of further investigation in a larger study cohort. Second, in the absence of another treatment arm, we could not verify that CHFR methylation was predictive or prognostic in nature. However, the changes shown after overexpression and knockdown of CHFR in the preclinical study suggest that CHFR methylation is more likely to be predictive of irinotecan treatment. Third, mainly because of tissue availability, a limited number of patients heterogeneous in their clinical characteristics, including treatment lines, were included. In our study, number of treatment lines was one of the most significant factors in univariate analyses for both TTP and OS, and the association of CHFR methylation or CIMP status with treatment outcomes was more prominent when stratified by number of prior treatment(s). Therefore, the correlation between CHFR methylation and irinotecan sensitivity might have been more evident if we had evaluated a more homogeneous population.
In summary, we report that CHFR is recurrently hypermethylated in metastatic CRC and that methylated CHFR is associated with better treatment outcomes in terms of TTP in patients with metastatic CRC treated with irinotecan-based systemic chemotherapy. Interestingly, in patients with unmethylated CHFR, the CIMP status could further discriminate patient outcomes. The association of CHFR and irinotecan treatment outcomes was confirmed in a preclinical study by overexpressing or knocking down CHFR and investigating the changes in irinotecan sensitivity accordingly. Considering the major importance of irinotecan-based chemotherapy in patients with metastatic CRC and the absence of a biomarker to guide treatment in these patients, we believe that our findings deserve confirmation in a larger patient cohort and could facilitate patient selection for irinotecan chemotherapy.
The following are the supplementary data related to this article.
Sequence information of primers used for the methylation-specific PCR assay.
Declarations of Interest
None.
Acknowledgments
Acknowledgements
We sincerely acknowledge the Omics Core Lab of NCC for supporting the preclinical part of our study.
Funding
This work was supported by the National Cancer Center Korea (grant number NCC-1710880 and 1810950).
Contributor Information
Seung Myung Dong, Email: smdong@ncc.re.kr.
Hee Jin Chang, Email: heejincmd@ncc.re.kr.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Venook AP, Niedzwiecki D, Lenz HJ, Innocenti F, Fruth B, Meyerhardt JA, Schrag D, Greene C, O'Neil BH, Atkins JN. Effect of first-line chemotherapy combined with cetuximab or bevacizumab on overall survival in patients with KRAS wild-type advanced or metastatic colorectal cancer: a randomized clinical trial. JAMA. 2017;317:2392–2401. doi: 10.1001/jama.2017.7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okugawa Y, Grady WM, Goel A. Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology. 2015;149:1204–1225.e12. doi: 10.1053/j.gastro.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crea F, Giovannetti E, Cortesi F, Mey V, Nannizzi S, Gallegos Ruiz MI, Ricciardi S, Del Tacca M, Peters GJ, Danesi R. Epigenetic mechanisms of irinotecan sensitivity in colorectal cancer cell lines. Mol Cancer Ther. 2009;8:1964–1973. doi: 10.1158/1535-7163.MCT-09-0027. [DOI] [PubMed] [Google Scholar]
- 5.Bosch LJ, Luo Y, Lao VV, Snaebjornsson P, Trooskens G, Vlassenbroeck I, Mongera S, Tang W, Welcsh P, Herman JG. WRN promoter CpG island hypermethylation does not predict more favorable outcomes for patients with metastatic colorectal cancer treated with irinotecan-based therapy. Clin Cancer Res. 2016;22:4612–4622. doi: 10.1158/1078-0432.CCR-15-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosch LJW, Trooskens G, Snaebjornsson P, Coupe VMH, Mongera S, Haan JC, Richman SD, Koopman M, Tol J, de Meyer T. Decoy receptor 1 (DCR1) promoter hypermethylation and response to irinotecan in metastatic colorectal cancer. Oncotarget. 2017;8:63140–63154. doi: 10.18632/oncotarget.18702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Meyerhardt JA, Loda M, Giovannucci EL, Fuchs CS. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cha Y, Kim K-J, Han S-W, Rhee YY, Bae JM, Wen X, Cho N-Y, Lee D-W, Lee K-H, Kim T-Y. Adverse prognostic impact of the CpG island methylator phenotype in metastatic colorectal cancer. Br J Cancer. 2016;115:164–171. doi: 10.1038/bjc.2016.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demurtas L, Puzzoni M, Giampieri R, Ziranu P, Pusceddu V, Mandolesi A, Cremolini C, Masi G, Gelsomino F, Antoniotti C. The role of primary tumour sidedness, EGFR gene copy number and EGFR promoter methylation in RAS/BRAF wild-type colorectal cancer patients receiving irinotecan/cetuximab. Br J Cancer. 2017;117:315–321. doi: 10.1038/bjc.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morano F, Corallo S, Niger M, Barault L, Milione M, Berenato R, Moretto R, Randon G, Antista M, Belfiore A. Temozolomide and irinotecan (TEMIRI regimen) as salvage treatment of irinotecan-sensitive advanced colorectal cancer patients bearing MGMT methylation. Ann Oncol. 2018;29:1800–1806. doi: 10.1093/annonc/mdy197. [DOI] [PubMed] [Google Scholar]
- 11.Moutinho C, Martinez-Cardus A, Santos C, Navarro-Perez V, Martinez-Balibrea E, Musulen E, Carmona FJ, Sartore-Bianchi A, Cassingena A, Siena S. Epigenetic inactivation of the BRCA1 interactor SRBC and resistance to oxaliplatin in colorectal cancer. J Natl Cancer Inst. 2014;106 doi: 10.1093/jnci/djt322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogino S, Meyerhardt JA, Kawasaki T, Clark JW, Ryan DP, Kulke MH, Enzinger PC, Wolpin BM, Loda M, Fuchs CS. CpG island methylation, response to combination chemotherapy, and patient survival in advanced microsatellite stable colorectal carcinoma. Virchows Arch. 2007;450:529–537. doi: 10.1007/s00428-007-0398-3. [DOI] [PubMed] [Google Scholar]
- 13.Agrelo R, Cheng WH, Setien F, Ropero S, Espada J, Fraga MF, Herranz M, Paz MF, Sanchez-Cespedes M, Artiga MJ. Epigenetic inactivation of the premature aging Werner syndrome gene in human cancer. Proc Natl Acad Sci U S A. 2006;103:8822–8827. doi: 10.1073/pnas.0600645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Shimodaira H, Soeda H, Komine K, Takahashi H, Ouchi K, Inoue M, Takahashi M, Takahashi S, Ishioka C. CpG island methylator phenotype is associated with the efficacy of sequential oxaliplatin- and irinotecan-based chemotherapy and EGFR-related gene mutation in Japanese patients with metastatic colorectal cancer. Int J Clin Oncol. 2016;21:1091–1101. doi: 10.1007/s10147-016-1017-6. [DOI] [PubMed] [Google Scholar]
- 15.Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet. 2012;13:679–692. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- 16.Bae JM, Kim JH, Kang GH. Molecular subtypes of colorectal cancer and their clinicopathologic features, with an emphasis on the serrated neoplasia pathway. Arch Pathol Lab Med. 2016;140:406–412. doi: 10.5858/arpa.2015-0310-RA. [DOI] [PubMed] [Google Scholar]
- 17.Xu Y, Villalona-Calero MA. Irinotecan: mechanisms of tumor resistance and novel strategies for modulating its activity. Ann Oncol. 2002;13:1841–1851. doi: 10.1093/annonc/mdf337. [DOI] [PubMed] [Google Scholar]
- 18.Toyota M, Sasaki Y, Satoh A, Ogi K, Kikuchi T, Suzuki H, Mita H, Tanaka N, Itoh F, Issa JP. Epigenetic inactivation of CHFR in human tumors. Proc Natl Acad Sci U S A. 2003;100:7818–7823. doi: 10.1073/pnas.1337066100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goel A, Boland CR. Epigenetics of colorectal cancer. Gastroenterology. 2012;143:1442–1460.e1. doi: 10.1053/j.gastro.2012.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Z, Liu J, Jing H, Dong SX, Wu J. The diagnostic and prognostic value of CHFR hypermethylation in colorectal cancer, a meta-analysis and literature review. Oncotarget. 2017;8:89142–89148. doi: 10.18632/oncotarget.19408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cleven AH, Derks S, Draht MX, Smits KM, Melotte V, Van Neste L, Tournier B, Jooste V, Chapusot C, Weijenberg MP. CHFR promoter methylation indicates poor prognosis in stage II microsatellite stable colorectal cancer. Clin Cancer Res. 2014;20:3261–3271. doi: 10.1158/1078-0432.CCR-12-3734. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka M, Chang P, Li Y, Li D, Overman M, Maru DM, Sethi S, Phillips J, Bland GL, Abbruzzese JL. Association of CHFR promoter methylation with disease recurrence in locally advanced colon cancer. Clin Cancer Res. 2011;17:4531–4540. doi: 10.1158/1078-0432.CCR-10-0763. [DOI] [PubMed] [Google Scholar]
- 23.Pelosof L, Yerram SR, Ahuja N, Delmas A, Danilova L, Herman JG, Azad NS. CHFR silencing or microsatellite instability is associated with increased antitumor activity of docetaxel or gemcitabine in colorectal cancer. Int J Cancer. 2014;134:596–605. doi: 10.1002/ijc.28390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Satoh A, Toyota M, Itoh F, Sasaki Y, Suzuki H, Ogi K, Kikuchi T, Mita H, Yamashita T, Kojima T. Epigenetic inactivation of CHFR and sensitivity to microtubule inhibitors in gastric cancer. Cancer Res. 2003;63:8606–8613. [PubMed] [Google Scholar]
- 25.Pillai RN, Brodie SA, Sica GL, Shaojin Y, Li G, Nickleach DC, Yuan L, Varma VA, Bonta D, Herman JG. CHFR protein expression predicts outcomes to taxane-based first line therapy in metastatic NSCLC. Clin Cancer Res. 2013;19:1603–1611. doi: 10.1158/1078-0432.CCR-12-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Privette LM, Petty EM. CHFR: A novel mitotic checkpoint protein and regulator of tumorigenesis. Transl Oncol. 2008;1:57–64. doi: 10.1593/tlo.08109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Yang Y, Lu Y, Herman JG, Brock MV, Zhao P, Guo M. Predictive value of CHFR and MLH1 methylation in human gastric cancer. Gastric Cancer. 2015;18:280–287. doi: 10.1007/s10120-014-0370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kashima L, Idogawa M, Mita H, Shitashige M, Yamada T, Ogi K, Suzuki H, Toyota M, Ariga H, Sasaki Y. CHFR protein regulates mitotic checkpoint by targeting PARP-1 protein for ubiquitination and degradation. J Biol Chem. 2012;287:12975–12984. doi: 10.1074/jbc.M111.321828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu C, Wu J, Paudyal SC, You Z, Yu X. CHFR is important for the first wave of ubiquitination at DNA damage sites. Nucleic Acids Res. 2013;41:1698–1710. doi: 10.1093/nar/gks1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawasaki T, Ohnishi M, Suemoto Y, Kirkner GJ, Liu Z, Yamamoto H, Loda M, Fuchs CS, Ogino S. WRN promoter methylation possibly connects mucinous differentiation, microsatellite instability and CpG island methylator phenotype in colorectal cancer. Mod Pathol. 2007;21:150. doi: 10.1038/modpathol.3800996. [DOI] [PubMed] [Google Scholar]
- 31.Masuda K, Banno K, Yanokura M, Tsuji K, Kobayashi Y, Kisu I, Ueki A, Yamagami W, Nomura H, Tominaga E. Association of epigenetic inactivation of the WRN gene with anticancer drug sensitivity in cervical cancer cells. Oncol Rep. 2012;28:1146–1152. doi: 10.3892/or.2012.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, Xie L, Wang J, Shen J, Liu B. Correlation between the methylation of SULF2 and WRN promoter and the irinotecan chemosensitivity in gastric cancer. BMC Gastroenterol. 2013;13:173. doi: 10.1186/1471-230X-13-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoon S, Lee E-J, Choi J-H, Chung T, Kim DY, Im J-Y, Bae M-H, Kwon J-H, Kim H-H, Kim HC. Recapitulation of pharmacogenomic data reveals that invalidation of SULF2 enhance sorafenib susceptibility in liver cancer. Oncogene. 2018;37:4443–4454. doi: 10.1038/s41388-018-0291-3. [DOI] [PubMed] [Google Scholar]
- 34.Tessema M, Yingling CM, Thomas CL, Klinge DM, Bernauer AM, Liu Y, Dacic S, Siegfried JM, Dahlberg SE, Schiller JH. SULF2 methylation is prognostic for lung cancer survival and increases sensitivity to topoisomerase-I inhibitors via induction of ISG15. Oncogene. 2012;31:4107–4116. doi: 10.1038/onc.2011.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang S, Kim HS, Seo SS, Park SY, Sidransky D, Dong SM. Inverse correlation between RASSF1A hypermethylation, KRAS and BRAF mutations in cervical adenocarcinoma. Gynecol Oncol. 2007;105:662–666. doi: 10.1016/j.ygyno.2007.01.045. [DOI] [PubMed] [Google Scholar]
- 36.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. REporting recommendations for tumour MARKer prognostic studies (REMARK) Br J Cancer. 2005;93:387–391. doi: 10.1038/sj.bjc.6602678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brandes JC, van Engeland M, Wouters KA, Weijenberg MP, Herman JG. CHFR promoter hypermethylation in colon cancer correlates with the microsatellite instability phenotype. Carcinogenesis. 2005;26:1152–1156. doi: 10.1093/carcin/bgi058. [DOI] [PubMed] [Google Scholar]
- 38.Overman M, Adam L, Raghav K, Wang J, Kee B, Fogelman D, Eng C, Vilar E, Shroff R, Dasari A. Phase II study of nab-paclitaxel in refractory small bowel adenocarcinoma and CpG island methylator phenotype (CIMP)-high colorectal cancer. Ann Oncol. 2018;29:139–144. doi: 10.1093/annonc/mdx688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scolnick DM, Halazonetis TD. Chfr defines a mitotic stress checkpoint that delays entry into metaphase. Nature. 2000;406:430–435. doi: 10.1038/35019108. [DOI] [PubMed] [Google Scholar]
- 40.Ogi K, Toyota M, Mita H, Satoh A, Kashima L, Sasaki Y, Suzuki H, Akino K, Nishikawa N, Noguchi M. Small interfering RNA-induced CHFR silencing sensitizes oral squamous cell cancer cells to microtubule inhibitors. Cancer Biol Ther. 2005;4:773–780. doi: 10.4161/cbt.4.7.1896. [DOI] [PubMed] [Google Scholar]
- 41.Derks S, Cleven AH, Melotte V, Smits KM, Brandes JC, Azad N, van Criekinge W, de Bruine AP, Herman JG, van Engeland M. Emerging evidence for CHFR as a cancer biomarker: from tumor biology to precision medicine. Cancer Metastasis Rev. 2014;33:161–171. doi: 10.1007/s10555-013-9462-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ford EE, Grimmer MR, Stolzenburg S, Bogdanovic O, de Mendoza A, Farnham PJ, Blancafort P, Lister R. Frequent lack of repressive capacity of promoter DNA methylation identified through genome-wide epigenomic manipulation. bioRxiv. 2017 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence information of primers used for the methylation-specific PCR assay.