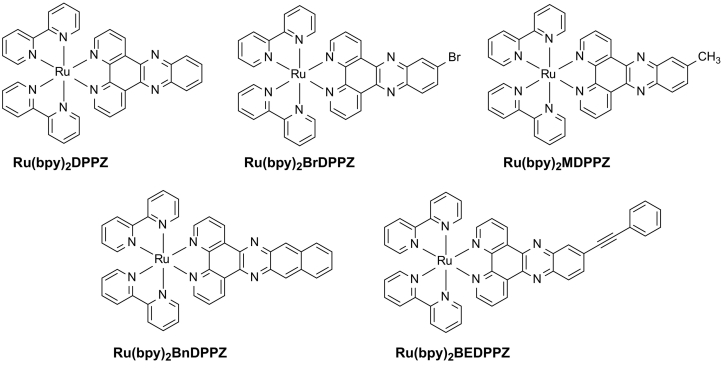
Abstract
Polypyridine Ru(II) complexes have long been deemed to excellent antitumor agents that inhibit the proliferation of breast cancer cells. Nevertheless, their effects on the metastatic potency of breast cancer cells need further research. Herein, a class of polypyridine Ru(II) complexes coordinated with phenazine derivates (DPPZ) ([Ru(bpy)2(DPPZ-R)](ClO4)2, Ru(bpy)2DPPZ: R = -H, Ru(bpy)2BrDPPZ: R = -Br, Ru(bpy)2MDPPZ: R = -CH3, Ru(bpy)2BnDPPZ: R = −acene, Ru(bpy)2BEDPPZ: R = -C ≡ C(C6H5)) was synthesized by introducing different substituent groups to regulate the electron cloud density and planarity of the main ligands. Results indicated that this class of DPPZ-based Ru(II) complexes exhibited promising inhibitory effect against MDA-MB-231 triple-negative breast cancer cells, especially for Ru(bpy)2BEDPPZ, which is comparable with that of cisplatin. In addition, Ru(bpy)2BEDPPZ effectively inhibited the migration and invasion of MDA-MB-231 cells in vitro and suppressed focal adhesion and stress fiber formation. Moreover, it effectively blocked MDA-MB-231 cell metastasis in blood vessels and restrained angiogenesis formation in a zebrafish xenograft breast cancer model. Further studies showed that the mechanisms may involve DNA damage-mediated apoptosis probably due to Ru(bpy)2BEDPPZ, which was enriched in the cell nucleus and induced DNA damage. All these results suggested that the DPPZ-based Ru(II) complexes can act as potent anti-metastasis agents.
Keywords: DPPZ-based Ru(II) complexes, triple-negative breast cancer, angiogenesis inhibitor, metastasis, DNA damage
Graphical Abstract
1. Introduction
Breast cancer is a disease that significantly threatens women's life. The triple-negative breast cancer (TNBC) is considered to be the most dangerous in the different types of breast cancer [1]. It is a common malignant tumor in China and has high degree of malignancy, incidence of recurrence, and metastasis [2]. The site of distant metastasis occurs mainly in the bone, lungs, liver, and brain [3]. Although methods such as endocrine and molecular targeted therapy are used in the treatment of TNBC, they are not effective. Currently, the commonly used treatment for TNBC is chemotherapy [4,5]. Anthracycline-based drugs, paclitaxel, and platinum antitumor drugs mainly target the DNA [[6], [7], [8]]. At the gene level, they affect the function of TNBC cells. However, they have no selectivity against normal cells, causing serious side effects to patients [9]. In TNBC, once metastasis develops in the surrounding tissue, the patient's life is in danger [10]. Thus, developing drugs that can restrain TNBC metastasis and clarifying their mechanism are important [11].
Recently, Ru-based complexes have attracted considerable attention because of their high antitumor activity, low toxicity, and powerful DNA-binding affinity. Two Ru(II) complexes (NAMI-A and KP1019) have entered phase II clinical trials due to their potential antitumor behavior [12]. A number of evidence suggests that Ru(II) complexes have wonderful DNA binding properties [13,14]. DNA is considered as an accepted target for ruthenium complexes. Ru(II) complexes can enter into the cell nucleus and play a key role in biological activity [15]. Barton et al. reported that DPPZ-based Ru(II) complex ([Ru(DIP)2(dppz)]2+) with lipophilic DIP ligand facilitates complex cellular uptake into the cell cytoplasm but has low inhibition against different tumor cells as it cannot enter into the cell nucleus [16]. Moreover, Chao et al. found that a type of Ru(II) complex ([Ru(bpy)(phpy)DPPZ]2+) exhibits high antitumor activity against a series of tumor cell lines through accumulating in the nucleic region to bind DNA [17].
In this study, we designed a class of polypyridine Ru(II)-DPPZ complexes by introducing different substituent groups to regulate the electron cloud density of phenazine ligand. This process may improve molecular planarity and modify the terminal benzene ring to increase lipophilicity, thereby helping complexes enriched into the nucleus to enhance antitumor activity. We described the synthesis and the enhanced potency of this new series of polypyridine Ru(II)-DPPZ complexes to block the metastasis of breast cancer in vitro and in vivo. Further studies showed that this class of Ru(II) complexes may enter into breast cancer cells and localize at the nucleus, which ultimately may induce DNA damage to cause cell apoptosis [18,19]. Therefore, the Ru(II) complexes with low-toxicity can act as potential inhibitors against the metastasis of breast cancer for chemotherapy (Scheme 1).
Scheme 1.
The synthetic route of DPPZ-based Ru(II) complexes.
2. Results and Discussion
2.1. In vitro Tumor Growth Inhibition of the Synthetic Ru(II) Complexes
To better assess the antitumor properties of Ru(II) complexes involving bidentate N-donor ligands with planar aromatic rings, we first evaluated the antiproliferative effects of these complexes on normal and some cancer cell lines (lung A549, breast MCF-7, liver HepG2, triple-negative breast MDA-MB-231, and HaCaT keratinocytes) and compared them with cisplatin. As shown in Table 1, the synthetic Ru(II) complexes emerged acceptable inhibition to various tumor cells after 72 h treatment, especially Ru(bpy)2BEDPPZ, which exhibited better antitumor activity than other complexes. The inhibitory effect (IC50) of Ru(bpy)2BEDPPZ against triple-negative breast MDA-MB-231 cell lines was approximately 17.2 μM, which was better than that of cisplatin (20.9 μM). Its construction is divalent platinum binds to two chlorine atoms and two ammonia molecules and it can bind to DNA and cause cross connection, thus damaging the function of DNA and inhibiting cell mitosis. It is a cellular non-specific drug. However, the ruthenium compound based on a phenazine ligand containing a large aromatic planar structure can better bind to DNA than cisplatin.
Table 1.
Cytotoxic effects of DPPZ-based Ru(II) complexes on human cancer and normal cell lines with the corresponding lipophilicity.
| Comp. | IC50/ μM |
logPo/w | ||||
|---|---|---|---|---|---|---|
| MDA-MB-231 | A549 | MCF-7 | HepG2 | HaCaT | ||
| Ru(bpy)2DPPZ | >100 | >100 | >100 | >100 | >100 | −1.87 |
| Ru(bpy)2BrDPPZ | >100 | 94.1 ± 4.2 | >100 | >100 | >100 | −1.58 |
| Ru(bpy)2MDPPZ | >100 | 93.3 ± 4.1 | 9.7 ± 6.1 | >100 | >100 | −2.17 |
| Ru(bpy)2BnDPPZ | >100 | 32.5 ± 1.6 | >100 | 23.0 ± 1.1 | >100 | −1.65 |
| Ru(bpy)2BEDPPZ | 17.2 ± 0.9 | 20.0 ± 0.7 | 74.9 ± 3.5 | 97.5 ± 4.4 | 47.2 ± 2.6 | −0.87 |
| Cis-platin | 20.9 ± 1.9 | 21.1 ± 0.2 | 86.6 ± 5.9 | 15.5 ± 0.4 | 7.1 ± 0.5 | – |
The electric effect, planarity, and steric hindrance of substituted groups in ligands play key roles in antitumor activity of Ru(II) complexes [20]. Although for the complexes of the electron-donating group, with Ru(bpy)2BnDPPZ modified complexes (with IC50 of 32.5 μM) were better than Ru(bpy)2MDPPZ (with IC50 of 93.3 μM) and Ru(bpy)2BrDPPZ (with IC50 of 94.1 μM) modified one against A549 cells, the best one was Ru(bpy)2BEDPPZ substituted complexes (with IC50 of 20.0 μM). Unlike the IC50 value for different sized planar ligands of complexes to MDA-MB-231 cells, the complexes with high quantity of terminal benzene rings in the ligands had enhanced growth suppression to tumor cells [[21], [22], [23]]. Comprehensive analysis of these results showed that the complexes modified by the electron-donating group can enhance antitumor activities, suggesting that the introduction of a phenylethynyl group in the phenazine ligand can availably improve the growth inhibition of tumor cells by exhibiting certain sensitivity to MDA-MB-231 cells. On the basis of its promising in vitro activity, Ru(bpy)2BEDPPZ was selected for preliminary evaluation in further studies.
2.2. Invasion and Migration of triple-negative breast cells inhibited by Ru(bpy)2BEDPPZ
Considering that metastasis is a severe risk in TNBC treatment, it is important to search for effective drugs that can inhibit it [24]. To investigate the anti-metastatic activity of the complexes, wound healing assay was performed to estimate cell migration and repair ability because the healing degree of scratches can reflect the inhibition effect of drugs on the migration ability of tumor cells. Fig. 1A shows an distinct reduction in the distance of wound closure without drug treatment for 72 h, but wound closure was suppressed with the addition of Ru(bpy)2BEDPPZ. When 2 μM of Ru(bpy)2BEDPPZ was used for treatment, the wound-healing rate was less than half of the control group. When the dose of Ru(bpy)2BEDPPZ was increased to 4 μM, the wound-healing rate further decreased, which suggested that Ru(bpy)2BEDPPZ inhibited migration in a dose-dependent manner.
Fig. 1.
Migration and invasion of MDA-MB-231 cells inhibited by Ru(bpy)2BEDPPZ in vitro. (A) Wound healing assay to evaluate the migration of MDA-MB-231 cells after being treated with Ru(bpy)2BEDPPZ (0, 2 and 4 μM) and DMEM with 10% FBS. Cells were wounded and monitored using a microscope every 24 h. Migration was determined by the rate of cells filling the scratched area.(B) Wound-healing rate of MDA-MB-231 cells induced by Ru(bpy)2BEDPPZ (N = 8). (C) The FITC-gelatin assay to assess the invasion of MDA-MB-231 cells was blocked by Ru(bpy)2BEDPPZ (0 and 5 μM). The number of black holes observed without and with Ru(bpy)2BEDPPZ. (D) The invasive percentage of MDA-MB-231 cells by Ru(bpy)2BEDPPZ (0 and 5 μM). Data were plotted as means ± SEM. Statistical significance was assessed using one-way ANOVA. Values versus the control group: * p < .05.
Moreover, the invasion of MDA-MB-231 cells blocked by Ru(bpy)2BEDPPZ was studied using FITC-gelatin assay. The quantity of black area (degraded area) in the FITC-gelatin showed the invasion capability of tumor cells, because highly invasive MDA-MB-231cells can release matrix metalloproteinases to degrade the FITC-gelatin; that is, high numbers of black area indicated high invasiveness of MDA-MB-231 cells [25]. Fig. 1C shows a high number of black holes in the FITC-gelatin without drug treatment, which suggested that MDA-MB-231 exerted high invasive capability. With 5 μM of Ru(bpy)2BEDPPZ treatment, the invasive capability of MDA-MB-231 cells was markedly suppressed, and the quantity of black holes in FITC-gelatin was decreased. These results suggested that the migration and invasion of MDA-MB-231 cells were effectively inhibited by this class of Ru(II) complexes [26].
The possible mechanism of anti-metastasis by Ru(bpy)2BEDPPZin vitro was studied (Fig. 2). Here, filamentous actin bundles and FA protein points were observed using paxillin [27], a focal adhesion-associated adaptor protein, suggesting cell adhesion ability on the surface [28]. Without treatment with Ru(bpy)2BEDPPZ, a good deal of focal adhesions around the edge of MDA-MB-231 cells were observed. Nevertheless, after Ru(bpy)2BEDPPZ treatment, it is found that a decreased quantity of paxillins in MDA-MB-231 cells with increasing concentration of Ru(bpy)2BEDPPZ, and the cytoskeletal structure became loose. The stress fibers were also inhibited, after Ru(bpy)2BEDPPZ treatment, as represented by F-actin with red fluorescence. The results indicated that Ru(bpy)2BEDPPZ might suppress focal adhesions and stress fibers to inhibit the migration and invasion of MDA-MB-231 cells [29].
Fig. 2.
The influence of Paxillin (green) and F-actin (red) of MDA-MB-231 cells induced by Ru(bpy)2BEDPPZ MDA-MB-231 cells were treatment with Ru(bpy)2BEDPPZ (0, 5 and 10 μM) for 24 h. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.3. Suppression of Breast Cancer Growth and Metastasis in vivo
To detect the anticancer activity of the complexes in vivo, we treated the zebrafish breast cancer model with Ru(bpy)2BEDPPZ. The model was constructed by using transgenic zebrafish (fli1:EGFP), and quantificational breast cancer cells were microinjected into the zebrafish near the subintestinal vessel (SIV) area, in which blood vessels were labeled with green fluorescence and MDA-MB-231 cells with blue fluorescence [30,31]. As shown in Fig. 3, without drug treatment, a spot of blue breast cancer cells was distributed near the SIV area at 0 h. When the time increased to 72 h, an increasing number of MDA-MB-231 cells were observed, and some cells invaded into neighboring tissues and spread into the blood vessels of zebrafish tail, which suggested that MDA-MB-231 cells possessed the ability for infinite proliferation and blood vessel invasion [32]. However, with Ru(bpy)2BEDPPZ (5 μM) treatment for 72 h, the number of MDA-MB-231 cells was notably reduced compared with the control group, which implied that Ru(bpy)2BEDPPZ can effectively suppress the proliferation of TNBC cells in zebrafish. Moreover, scarce MDA-MB-231 cells were found in the blood vessel of zebrafish, indicating that Ru(bpy)2BEDPPZ can suppressed the metastasis of MDA-MB-231 cells in zebrafish xenografts. Moreover, the total area of SIV in the Ru(bpy)2BEDPPZ treatment group zebrafish was obviously less than that in the control group, which indicated that Ru(bpy)2BEDPPZ exhibited a certain inhibitory effect on tumor angiogenesis. The above results suggested that Ru(bpy)2BEDPPZ effectively suppressed the metastasis and proliferation of MDA-MB-231 cells in vivo and can be developed to become a potential candidate to block TNBC metastasis.
Fig. 3.
The proliferation and metastasis of MDA-MB-231 cells in zebrafish xenografts model was inhibited by Ru(bpy)2BEDPPZ Dil-labeled MDA-MB-231 cells (blue) were microinjected into zebrafish embryos, and treatment with Ru(bpy)2BEDPPZ (5 μM) for 72 h. After 48 h, the proliferation and metastasis of the xenografts of MDA-MB-231 cells were imaged under a fluorescence microscope (n = 10/group). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.4. Angiogenesis Inhibited by Ru(bpy)2BEDPPZ in vitro and in vivo
In general, angiogenesis formation act a pivotal part in tumor proliferation and metastasis, some researchers have been studied that the application of Nanoparticles to the inhibition or synergistic treatment tumor through inhibite endothelial tubes formation [33,34], such as nanodiamond, mesoporous silica nanoparticles, etc. [35,36]. Besides, the vascular endothelial growth factor (VEGF) is one of the most critical factor inducing tumor vessel formation [37,38]. As tumor growth requires angiogenesis to provide adequate oxygen and nutrients, inhibiting angiogenesis formation is considered as an important way to overcome breast cancer. Angiogenesis inhibitors are widely used because of their safety and ability to block new blood vessel formation in tumors [39].
Given that tube formation indicates angiogenesis progression, we also demonstrated whether Ru(bpy)2BEDPPZ affects HUVECs' angiogenic effect in vitro by using tube formation assay. As shown in Fig. 4A, without drug interference, HUVECs incubated with VEGF (200 ng/mL) established tubular structures on Matrigel. Then, we treated HUVECs with Ru(bpy)2BEDPPZ in the presence of VEGF and found that the tube formation of these cells was inhibited notably compared with control cells. With increasing Ru(bpy)2BEDPPZ treatment dose, the matrix-induced tube formation of HUVECs was suppressed, indicating its dose-dependent effects [40,41]. Moreover, comprehensive analysis of the tube number, mean node number, and total length of tube suggested that Ru(bpy)2BEDPPZ is responsible for the inhibitory effect of VEGF on the matrix-induced tube formation of HUVECs. These observations indicated that Ru(bpy)2BEDPPZ can suppress the angiogenesis in vitro.
Fig. 4.
The angiogenesis formation inhibited by Ru(bpy)2BEDPPZ in vitro and in vivo. (A)In vitro inhibitory effect of Ru(bpy)2BEDPPZ on the tube formation of HUVECs cells with or without Ru(bpy)2BEDPPZ (0, 5 and 10 μM) for 6 h at 37 °C. The concentration of VEGF is 200 ng/mL. (B) In vivo inhibition of angiogenesis in transgenic zebrafish (fil1:EGFP) with and without Ru(bpy)2BEDPPZ (0, 1.25, 2.5 and 5 μM) at 28.5 °C for 48 h. PTK787 was used as a positive control. (C) Total length of blood vessels of control group (in DMEM containing10%FBS treated group) as 100% (n = 3/group). (D) The mean area of SIVs of control (without Ru(bpy)2BEDPPZ) as 100% were quantified by manual counting at 72 hpf (N = 16). Data were plotted as means ± SEM. Statistical significance was assessed using one-way ANOVA. Values versus the control group: * p < .05.
Furthermore, the inhibitory effect of Ru(bpy)2BEDPPZ against angiogenesis formation was evaluated in zebrafish model. It is found that Ru(bpy)2BEDPPZ equally induces distinct defects in ISV formation. To examine in detail the inhibitory effects of Ru(bpy)2BEDPPZ on ISV formation in vivo, the Tg(fli1a:EGFP) transgenic embryos at 12 h post-fertilization (hpf) were used to treat with various concentrations (1.25, 2.5 and 5.0 μM) of Ru(bpy)2BEDPPZ, owing to GFP expressing under the control of an endothelial-specific promoter at this time point. Given that this moment companies with the construction of the dorsoventral axis, the analysis concentrated only on the inhibition of Ru(bpy)2BEDPPZ on angiogenesis. After treatment with Ru(bpy)2BEDPPZ, ISV was visualized at 48 hpf. In this in vivo angiogenesis model, 5.0 μM of Ru(bpy)2BEDPPZ more effectively inhibited ISV formation than PTK787, which is a commonly used angiogenesis inhibitor. The above results show that angiogenesis formation were inhibited with this type of Ru(II) complex to block breast cancer cell metastasis in vitro and in vivo [42].
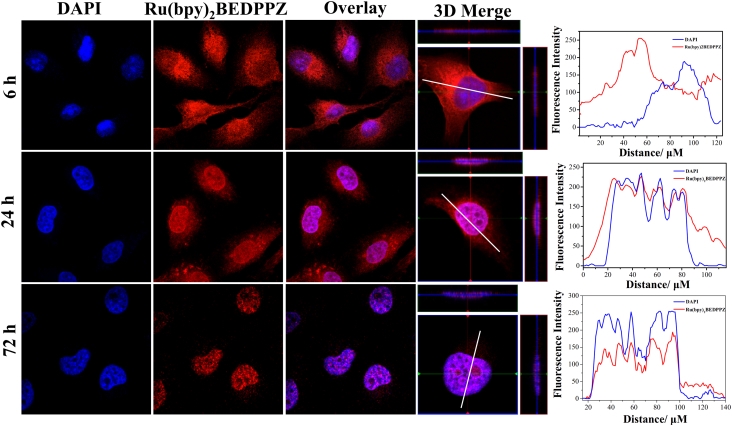
2.5. Ru(bpy)2BEDPPZ Cellular Localization
To investigate the potential antitumor mechanism of the Ru(II) complex, cellular localization of Ru(bpy)2BEDPPZ in MDA-MB-231 cells at different times was investigated, and the results are shown in Fig. 5. The intracellular localization of Ru(bpy)2BEDPPZ with red phosphorescence was slightly distributed in the cell nucleus and primarily enriched in the cell cytoplasm, after treatment for 6 h, which showed a mismatch with DAPI. However, with the increase of time, Ru(bpy)2BEDPPZ entered into the cell nucleus. At 24 h, a small quantity of Ru(bpy)2BEDPPZ was discovered in the cell cytoplasm surrounding the cell nucleus (blue phosphorescence) of the MDA-MB-231 cells. At 72 h, however, Ru(bpy)2BEDPPZ was only gathered in the cell nucleus, but perfectly matched with DAPI. The common target for Ru(II) complex to inhibit tumor cells is DNA. Thus, the localization of Ru(bpy)2BEDPPZ in the cell nucleus may play a key role in inhibiting breast cancer metastasis.
Fig. 5.
Cellular localization of Ru(bpy)2BEDPPZ in MDA-MB-231 cells. Cells were treated with Ru(bpy)2BEDPPZ for 6, 24, 72 h at 37 °C: blue, DAPI; red, ruthenium(II) complexes. [Ru(bpy)2BEDPPZ] = 10 μM. The overlay data were analyzed using Image Pro Plus. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
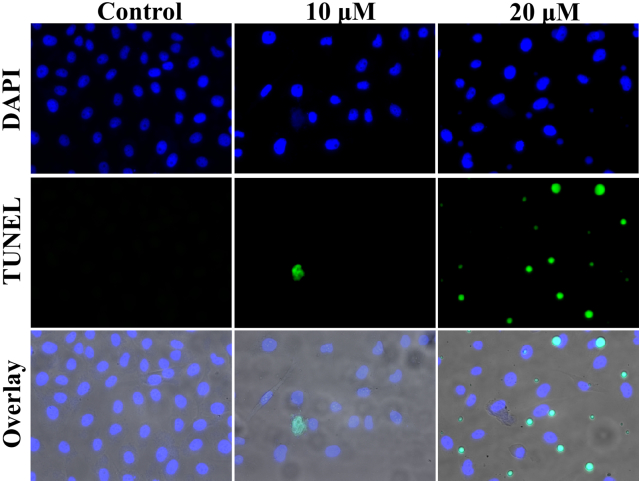
2.6. Growth Inhibition of Ru(bpy)2BEDPPZ by DNA Damage-mediated Apoptosis Induction
As Ru(bpy)2BEDPPZ emitted strong red fluorescence, which would interfere the PI signal, we used TUNEL assay to detect the apoptosis induction properties of 5 in MDA-MB-231 cells. Fig. 6 shows that the number of apoptotic cells was obviously decreased by upon the addition of Ru(bpy)2BEDPPZ. Without drug treatment, a small number of apoptotic cells were observed. However, with 20 μM of Ru(bpy)2BEDPPZ treatment, many apoptotic cells were observed. The apoptotic cells with fragmented DNA can be detected by TUNEL. We posited that Ru(bpy)2BEDPPZ may induce apoptosis of MDA-MB-231 by inducing DNA damage .
Fig. 6.
The apoptosis of MDA-MB-231 cells of Ru(bpy)2BEDPPZ-induced (0, 10 and 20 μM) DNA fragmentation was detected by TUNEL assay and DAPI staining. DAPI, blue; TUNEL, green. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In order to further confirm the DNA damage induced by Ru(bpy)2BEDPPZ, we performed comet assay. Moreover, we also detected the induction of DSBs in MDA-MB-231 cells stained with DSB biomarker γH2AX by neutral comet assays and confocal immunofluorescence assays. As indicated by the statistical increase in olive tail moment, Ru(bpy)2BEDPPZ extremely increased the generation of endogenous DSBs in MDA-MB-231 cells (Fig. 7A). The percentages of γH2AX focal positive cells were obviously increased in MDA-MB-231 cells treated with different concentrations of Ru(bpy)2BEDPPZ (0, 5 and 10 μM). These results indicated that Ru(bpy)2BEDPPZ mainly induced apoptosis of MDA-MB-231 cells through DNA damage, which led to endogenous DSBs [43].
Fig. 7.
(A) DNA damage induced by Ru(bpy)2BEDPPZ as examined by Comet assay. (B) Effects of Ru(bpy)2BEDPPZ on the expression level γH2AX. Cells were treated with Ru(bpy)2BEDPPZ (0, 5 and 10 μM) for 24 h and the length of tail reflects DNA damage in the cells.
3. Conclusions
In this work, we designed and synthesized a class of polypyridine Ru-DPPZ derivatives by microwave-assisted synthesis technology [44,45]. This class of Ru(II) complexes exerted certain inhibition effects against TNBC cells, especially Ru(bpy)2BEDPPZ, which displayed commendable inhibitory activity against the proliferation, migration, and invasion of triple-negative breast cancer MDA-MB-231 cells. The structure-activity relationship analysis of these results suggested that the increased number of aromatic planar rings in ligands can effectively enhance antitumor activity and provided a guiding significance for further optimization of Ru(II) complexes [46,47]. The results of our mechanistic study showed that Ru(bpy)2BEDPPZ was enriched in the cell nucleus to induce DNA damage, leading to tumor cell apoptosis. Further evaluation of in vivo activity indicated that low concentrations of Ru(bpy)2BEDPPZ effectively inhibited the proliferation and metastasis of MDA-MB-231 cells in zebrafish xenografts. In summary, our study showed that the introduction of a phenylethynyl group in Ru(II) complexes can effectively enhance antitumor activity, which provided a guiding significance for optimizing antitumor drugs.
4. Materials and Methods
4.1. Chemicals
All the chemicals and solvents were purchased from commercial vendors. Ruthenium(III) chloride hydrate was obtained from Mitsuwa Chemicals. The ligand dipyrido[3,2-a:2,3′-c]phenazine (DPPZ), 7-bromo-dipyrido[3,2-a:2′,3′-c]phenazine) (7-Br-DPPZ), 6-Methyl-1,2-phenylened- iamine (6-CH3-DPPZ) and Benzo[i]dipyrido[3,2-a:2′,3′-c]phenazine (DPPN) were synthesized by using the similar method in the literature. Using an Anton Paar monowave 300 microwave reactor synthesize these complexes. The 1H NMR and 13C NMR spectra were recorded in a dimethyl-d6 sulfoxide (DMSO-d6) solution detected by a Bruker DRX2500 spectrometer at room temperature. Both Co and Cw were tested by the electronic absorption spectra were recorded with a Shimadzu UV-2550, and the partition coefficient (Po/w) for the complex was calculated according to the equation: Po/w = Ao/Aw. The images of wound-healing assay and comet assay were captured by Leica DMi8 Inverted Microscope (Leica, Germany). Cellular localization and immunofluorescence experiments were performed with an ZEISS LSM 800 Confocal Laser Scanning Microscop (Zeiss, Germany).
4.2. Synthesis and Characterization
4.2.1. Synthesis of [Ru(bpy)2DPPZ](ClO4)2 (Ru(bpy)2DPPZ)
According to the literature, Ru(bpy)2DPPZ was synthesized by using similar metheod but with a few modifications. A mixture of cis-[Ru(bpy)2Cl2]∙2H2O (103.9 mg, 0.2 mmol) and DPPZ (56.4 mg, 0.2 mmol) were heated at reflux for 6 h under N2 atmosphere in 30 mL of 9:1 glycol/water mixture. The solution was cooled to room temperature before the reaction mixture was diluted with 80 mL of water. Then the mixture was filter, and excess sodium perchlorate was added to give a orange solid. Using cooled water and ether to filter and wash the crude products, and purified by flash silica gel column chromatography to afford a red powder, yields 67%. ESI-MS (in CH3CN, m/z): 348.0 ([M-2ClO4−]2+), 795.1 ([M-ClO4−]+). 1H NMR (500 MHz, DMSO-d6): δ 9.64 (dd, J = 8.2, 1.2 Hz, 2H), 8.89 (dd, J = 14.6, 8.2 Hz, 4H), 8.53 (dd, J = 6.5, 3.4 Hz, 2H), 8.25 (ddd, J = 9.3, 6.8, 1 0.3 Hz, 4H), 8.21 (dd, J = 6.6, 3.4 Hz, 2H), 8.15 (td, J = 8.0, 1.3 Hz, 2H), 8.04 (dd, J = 8.2, 5.4 Hz, 2H), 7.85 (d, J = 5.2 Hz, 2H), 7.79 (d, J = 5.4 Hz, 2H), 7.65–7.59 (m, 2H), 7.43–7.38 (m, 2H). 13C NMR (400 MHz, DMSO-d6): δ 155.64 (s), 155.35 (s), 152.28 (s), 150.79 (s), 150.26 (s), 149.19 (s), 140.78 (s), 139.01 (s), 136.94 (s), 136.83 (s), 132.11 (s), 131.45 (s), 129.05 (s), 128.28 (s), 126.78 (s), 123.36 (s), 123.25 (s).
4.2.2. Synthesis of [Ru(bpy)2 7-Br-DPPZ](ClO4)2 (Ru(bpy)2BrDPPZ)
Ru(bpy)2BrDPPZ was synthesized as the same method, but with 7-Br-DPPZ (72 mg, 0.2 mmol), yields 82%. ESI-MS (in CH3CN, m/z): 387.9 ([M-2ClO4−]2+), 875.0 ([M-ClO4−]+). 1H NMR (500 MHz, DMSO-d6): δ 9.56 (ddd, J = 14.6, 8.2, 1.2 Hz, 2H), 8.89 (dd, J = 14.2, 8.3 Hz, 4H), 8.70 (d, J = 2.2 Hz, 1H), 8.44 (d, J = 9.1 Hz, 1H), 8.32–8.20 (m, 5H), 8.16 (tdd, J = 8.1, 2.8, 1.4 Hz, 2H), 8.07–8.00 (m, 2H), 7.85 (d, J = 5.5 Hz, 2H), 7.82–7.77 (m, 2H), 7.64–7.60 (m, 2H), 7.41 (dtd, J = 7.0, 5.7, 1.2 Hz, 2H). 13C NMR (500 MHz, DMSO-d6): δ 157.25 (s), 156.98 (s), 154.25 (s), 154.11 (s), 152.41 (s), 151.90 (s), 151.03 (s), 150.87 (s), 142.72 (s), 141.28 (s), 141.17 (s), 140.91 (s), 138.60 (s), 138.49 (s), 136.13 (s), 133.72 (s), 131.71 (s), 130.46 (s), 130.37 (s), 128.42 (s), 128.27 (s), 128.22 (s), 126.44 (s), 125.03 (s), 124.91 (s).
4.2.3. Synthesis of [Ru(bpy)2(7-CH3-DPPZ)](ClO4)2 (Ru(bpy)2MDPPZ)
Ru(bpy)2MDPPZ was synthesized as the same method, but with 7-CH3-DPPZ (59.2, 0.2 mmol), yields 84%. ESI-MS (in CH3CN, m/z): 354.9 ([M-2ClO4−]2+), 809.0 ([M-ClO4−]+). 1H NMR (500 MHz, DMSO-d6): δ 9.69 (d, J = 8.2 Hz, 1H), 9.62 (d, J = 8.2 Hz, 1H), 8.89 (dt, J = 17.9, 9.0 Hz, 4H), 8.33 (d, J = 8.3 Hz, 1H), 8.27–8.22 (m, 4H), 8.15 (td, J = 8.0, 3.2 Hz, 2H), 8.05 (ddt, J = 12.3, 7.8, 6.3 Hz, 4H), 7.86 (t, J = 5.1 Hz, 2H), 7.79 (dd, J = 10.2, 5.7 Hz, 2H), 7.62 (t, J = 6.6 Hz, 2H), 7.40 (td, J = 13.4, 6.7 Hz, 2H). 13C NMR (126 MHz, DMSO-d6) δ 155.14 (s), 154.86 (s), 151.72 (s), 150.25 (s), 149.79 (s), 148.62 (s), 148.53 (s), 140.54 (s), 139.46 (s), 138.04 (s), 137.21 (s), 136.43 (s), 136.32 (s), 136.00 (s), 131.69 (s), 131.57 (s), 130.76 (s), 130.19 (s), 128.73 (s), 128.48 (s), 127.24 (s), 126.47 (s), 126.28 (s), 126.04 (s), 125.42 (s), 123.66 (s), 122.87 (s), 122.76 (s).
4.2.4. Synthesis of [Ru(bpy)2(DPPN)](ClO4)2 (Ru(bpy)2BnDPPZ)
Ru(bpy)2BnDPPZ was synthesized as the same method, but with DPPN (66.4 mg, 0.2 mmol), yields 51%. ESI-MS (in CH3CN, m/z): 373.0 ([M-2ClO4−]2+)1H NMR (500 MHz, DMSO-d6): δ 9.59 (d, J = 8.1 Hz, 2H), 9.22 (s, 2H), 8.89 (dd, J = 14.3, 8.2 Hz, 4H), 8.45–8.40 (m, 2H), 8.26–8.21 (m,4), 8.17 (t, J = 7.9 Hz, 2H), 8.02 (dd, J = 8.1, 5.4 Hz, 2H), 7.87–7.83 (m, 4H), 7.76 (dd, J = 6.6, 3.1 Hz, 2H), 7.61 (t, J = 14.7, 7.8 Hz, 2H), 7.43 (t, J = 15.3, 8.3 Hz, 2H). 13C NMR (126 MHz, DMSO-d6): δ 141.58 (s), 138.58 (s), 138.48 (s), 138.38 (s), 135.08 (s), 133.72 (s), 131.09 (s), 129.37 (s), 129.07 (s), 128.68 (s), 128.43 (s), 128.32 (s), 128.22 (s), 125.79 (s), 125.00 (s), 124.90 (s).
4.2.5. Synthesis of [Ru(bpy)2(7-BEDPPZ)](ClO4)2 (Ru(bpy)2BEDPPZ)
According to the literatures, the Ru(bpy)2BEDPPZ was synthesized, but with some modifications. A mixture of phenylacetylene (0.09 mL, 0.625 mmol), [Ru(bpy)27-Br-DPPZ](ClO4)2 (122 mg, 0.125 mmol), Pd(PPH3)2Cl2 (3.5 mg, 0.005 mmol) and CuI (2 mg, 0.010 mmol) (15.0 mL) were dissolved in dry Et3N (0.02 mL) under microwaves irradiation at 140 °C for 30 min. The products were purified by using Al2O3 flash column chromatography with CH3CN as the eluent, yield: 48.2%. ESI-MS (in CH3CN, m/z): 398.06 ([M-2ClO4−]2+), 923.12 ([M-ClO4−]+). 1H NMR (500 MHz, DMSO) δ 9.60 (ddd, J = 8.2, 3.3, 1.1 Hz, 2H), 8.94–8.87 (m, 4H), 8.64 (d, J = 1.6 Hz, 1H), 8.52 (d, J = 8.8 Hz, 1H), 8.29 (dd, J = 5.4, 1.0 Hz, 1H), 8.27 (dd, J = 3.6, 1.4 Hz, 2H), 8.24 (d, J = 7.5 Hz, 2H), 8.16 (t, J = 7.8 Hz, 2H), 8.06 (td, J = 8.1, 5.4 Hz, 2H), 7.86 (d, J = 5.4 Hz, 2H), 7.81 (t, J = 5.1 Hz, 2H), 7.74–7.69 (m, 2H), 7.67–7.58 (m, 2H), 7.55–7.51 (m, 3H), 7.42 (t, J = 6.6 Hz, 2H). 13C NMR (126 MHz, DMSO) δ 155.57 (s), 155.30 (s), 152.49 (s), 152.41 (s), 150.79 (s), 150.24 (s), 149.27 (s), 149.20 (s), 140.49 (s), 140.41 (s), 139.78 (s), 139.23 (s), 136.93 (s), 136.83 (s), 133.30 (s), 132.10 (s), 130.62 (s), 128.87 (s), 128.54 (s), 127.79 (s), 126.78 (s), 126.58 (s), 124.99 (s), 123.36 (s), 123.26 (s), 120.31 (s), 92.92 (s), 87.26 (s), 47.41 (s).
4.3. Cell Culture
Human breast cancer MDA-MB-231 and MCF-7 cells, human lung adenocarcinoma A549 cells, hepatocellular carcinoma HepG2 and the immortalized human normal epidermal HaCaT cells were obtained from American Type Culture Collection (ATCC, Manassas, VA). All cell lines were cultured by Dulbecco's modified Eagle medium (DMEM) containing fetal bovine serum (BSA; 10%, HyClone), streptomycin (50 units/mL) and penicillin (100 units/mL).
4.4. MTT Assay
Seeding cells in 96-well tissue culture plates (5 × 103 cells per well 24 h) and treating with different concentration complex (0, 1.56, 3.125, 6.25, 12.5, 25, 50 and 100 μM) for 72 h. Then, MTT stock solution (20 μL/well, 5 mg/mL) was added, companied with incubation for another 4 h. Then, aspirated the medium and replaced with 150 μL/well of DMSO, it was measured at 490 nm using a microplate spectrophotometer (Thermo Scientific Multiskan GO).
4.5. Ru(bpy)2BEDPPZ Wound-Healing Assay
MDA-MB-231 cells (1 × 105 cells/well) were seeded in a 6-well plates with two marked lines on the back. When the cells covered >85% of the culture plate, using a tip (200 μL pipet) to scratch a line on the plate and orthogonal to the mark. Then, treatment with Ru(bpy)2BEDPPZ in different concentrations (0, 2 and 4 μM) in DMEM without FBS and observe migrating cells in the same visual field every 24 h. Based on 10 fields of view for each cell type, the average migration rate and the cell trajectory were computed through Slidebook and Excel software.
4.6. Fluorescein Isothiocyanate (FITC)-Conjugated Gelatin Invasion Assay
According to the manufacturer's instructions (Invitrogen), cells (2 × 105 per well) treating with Ru(bpy)2BEDPPZ (0 and 5 μM) were plated onto the FITC-gelatin-coated coverslips and incubated (37 °C, 24 h). Using laser confocal microscope evaluated and photographed the FITC-gelatin degradation status [48].
4.7. Immunofluorescence
Adding cell suspension culture on 1 cm diameter cover glass which in six-well (Fisher Scientific, Pittsburgh, PA). Then, using 4% formaldehyde solution fixation for 25 min, and cover glass were cultured with these reagents: 5% BSA for 30 min, 1:100 rabbit monoclonal Anti-Paxillin antibody (ab32115) for 60 min, 1:200 Alexa Fluor 488 - conjugated Affinipure Goat Anti-Rabbit IgG(H + L) (proteintech) for 30 min, 1: 100 Rhodamine phalloidin (Amanita phalloides, cytoskeleton) (for F-actin) for 30 min, and stained with DAPI (0.5 μg/mL, 8 min). Using PBS wash two times in each incubation step, then it were transferred onto a microscopic slide, which observed under a laser confocal microscope.
4.8. Tube Formation Assay
Dissolving matrigel (4 °C, 24 h) and it were coated with prechilled 96-well plates (50 μL/well, 37 °C and stay at least 60 min). Then, the unsolid fluid were removed, HUVECs at the density of 3 × 104 were cultured in DMEM with VEGF (200 ng/mL) containing the different concentrations of Ru(bpy)2BEDPPZ (0, 5 and 10 μM) for 6 h. Analyze and capture these images [49].
4.9. Evaluation of the Angiogenic Effect In vivo
24 hpf transgenic Tg(fli1a:EGFP) zebrafish embryos were dechorionated ata petri dishusing protease, then randomly distributed into 6-well microplate, treated with Ru(bpy)2BEDPPZ (0, 1.25, 2.5 and 5 μM). After 48 h treatment at 28.5 °C, using fresh water to wash zebrafish larvae for two times and then transferredinto the 96-well plate, the subintestinal vessel plexus (SIVs) of zebrafish larvae were observed andimaged under the fluorescence microscope. The number of SIVs of each zebrafish was measured via manual counting, average number of SIVs of zebrafish larvae was calculated in each group [50].
4.10. TUNEL Assay
Cells were fixed by using 4% p-formaldehyde in PBS for 30 min and permeabilized simultaneously with 0.3% Triton X-100 in PBS (25 °C, 8 min). Then, adding TUNEL reaction mixture (100 μL/well) into cells for 1 h and under dark condition incubate DAPI (0.5 μg/mL, 37 °C, 8 min). Using fluorescence microscope observe cell morphology.
4.11. Cellular Localization
MDA-MB-231 cells (5 × 104 cells/mL) were incubated with Ru(bpy)2BEDPPZ for 6, 24, 72 h at 37 °C. Then, PBS wash these cells two times, using 4% p-formaldehyde fixed for 30 min, after stained with DAPI (0.5 μg/mL, 8 min). Finally, using confocal laser microscope to observe the cell morphology.
4.12. Single Cell Gel Electrophoresis
MDA-MB-231 cells (5 × 104 cells/mL) were incubated with Ru(bpy)2BEDPPZ (0, 5 and 10 μM) for 72 h. According to 1:7 percentage mix with cell suspension and melted LM agarose. Pipetting the mixture (80 μL) to the slide (Comet Slide TM). Using prechilled lysis solution (4 °C 60 min) immerse the slide before refrigeration for 10 min, and it was wash PBS twice times. Next, using freshly prepared alkaline solution (6 g NaOH, 0.25 mM EDTA (2.5 mL), pH = 13) immerse for 20 min, and it was suffered alkaline solution for electrophoresis (25 V 30 min),when the DNA unwinding. After, using PBS wash the slide twice, DNA was stained with EB after observed under a fluorescence microscope.
4.13. Growth and Metastasis Inhibition of Ru(bpy)2BEDPPZ in vivo
MDA-MB-231 cells were labeled fluorescence by DiI and gathered at a density of 107 cells/mL. Then, cells (50–500 cells) were injected into the perivitelline space near the subintestinal vessels (SIVs) of the transgenic zebrafish (fil1:EGFP) embryos at 48 hpf to establish this zebrafish embryo breast cancer model. Juvenile zebrafish of breast cancer model (48 h old) were incubated in 6-well plates (10 fishes in every well) with 2 mL solutions without or with Ru(bpy)2BEDPPZ (0 and 5 μM) in aquaculture water. The inhibitory effect of Ru(bpy)2BEDPPZ in breast cancer zebra fish were captured pictures every 24 h with a fluorescence microscopy [32,51].
Acknowledgments
Acknowledgments
The authors acknowledge the National Natural Science Foundation of China (Grant Nos. 81572926, 81703349). The China Postdoctoral Science Foundation (Grant No. 2017M610576). The Provincial Major Scientific Research Projects in Universities of Guangdong Province (Grant No. 2014KZDXM053), The Science and Technology Project of Guangdong Province (Grant No. 2014A020212312), The Innovation Projects in Universities of Guangdong Province (2015cxqx151); Tradition Chinese Medicine Bureau of Guangdong Province (20151265), the Innovation Team Projects in Universities of Guangdong Province (2016KCXTD018), the Projects of Guangzhou Key Laboratory of Construction and Application of New Drug Screening Model Systems (No.201805010006) and Key Laboratory of New Drug Discovery and Evaluation of ordinary universities of Guangdong province (No. 2017KSYS002).
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2018.11.010.
Contributor Information
Qiong Wu, Email: wuqiongniu.1113@163.com.
Wenjie Mei, Email: wenjiemei@126.com.
Appendix A. Supplementary Data
Supplementary material
References
- 1.Foulkes W.D., Smith I.E., Reis-Filho J.S. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 2.Dent R., Hanna W.M., Trudeau M., Rawlinson E., Sun P., Narod S.A. Pattern of metastatic spread in triple-negative breast cancer. Breast Cancer Res Treat. 2009;115:423–428. doi: 10.1007/s10549-008-0086-2. [DOI] [PubMed] [Google Scholar]
- 3.DeRose Y.S., Wang G., Lin Y.C., Bernard P.S., Buys S.S., Ebbert M.T. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17:1514–1520. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cleator S., Heller W., Coombes R.C. Triple-negative breast cancer: therapeutic options. Lancet Oncol. 2007;8:235–244. doi: 10.1016/S1470-2045(07)70074-8. [DOI] [PubMed] [Google Scholar]
- 5.F. Andre, C.C. Zielinski, Optimal strategies for the treatment of metastatic triple-negative breast cancer with currently approved agents, Ann Oncol 23 Suppl 6 (2012) vi46–51. [DOI] [PubMed]
- 6.Cardoso F., Durbecq V., Larsimont D., Paesmans M., Leroy J.Y., Rouas G. Correlation between complete response to anthracycline-based chemotherapy and topoisomerase II-alpha gene amplification and protein overexpression in locally advanced/metastatic breast cancer. Int J Oncol. 2004;24:201–209. [PubMed] [Google Scholar]
- 7.Jordan M.A., Wendell K., Gardiner S., Derry W.B., Copp H., Wilson L. Mitotic block induced in HeLa cells by low concentrations of paclitaxel (Taxol) results in abnormal mitotic exit and apoptotic cell death. Cancer Res. 1996;56:816–825. [PubMed] [Google Scholar]
- 8.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 9.Schimmel K.J., Richel D.J., van den Brink R.B., Guchelaar H.J. Cardiotoxicity of cytotoxic drugs. Cancer Treat Rev. 2004;30:181–191. doi: 10.1016/j.ctrv.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Redig A.J., McAllister S.S. Breast cancer as a systemic disease: a view of metastasis. J Intern Med. 2013;274:113–126. doi: 10.1111/joim.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li D., Wang H., Ding Y., Zhang Z., Zheng Z., Dong J. Targeting the NRF-2/RHOA/ROCK signaling pathway with a novel aziridonin, YD0514, to suppress breast cancer progression and lung metastasis. Cancer Lett. 2018;424:97–108. doi: 10.1016/j.canlet.2018.03.029. [DOI] [PubMed] [Google Scholar]
- 12.Hartinger C.G., Zorbas-Seifried S., Jakupec M.A., Kynast B., Zorbas H., Keppler B.K. From bench to bedside--preclinical and early clinical development of the anticancer agent indazolium trans-[tetrachlorobis(1H-indazole)ruthenate(III)] (KP1019 or FFC14A) J Inorg Biochem. 2006;100:891–904. doi: 10.1016/j.jinorgbio.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Hiort C., Lincoln P., Nordén B. DNA binding of .DELTA.- and .LAMBDA.-[Ru(phen)2DPPZ]2+ J Am Chem Soc. 1993;115:3448–3454. [Google Scholar]
- 14.Gao F., Chao H., Zhou F., Yuan Y.X., Peng B., Ji L.N. DNA interactions of a functionalized ruthenium(II) mixed-polypyridyl complex [Ru(bpy)2ppd]2+ J Inorg Biochem. 2006;100:1487–1494. doi: 10.1016/j.jinorgbio.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Gill M.R., Garcia-Lara J., Foster S.J., Smythe C., Battaglia G., Thomas J.A. A ruthenium(II) polypyridyl complex for direct imaging of DNA structure in living cells. Nat Chem. 2009;1:662–667. doi: 10.1038/nchem.406. [DOI] [PubMed] [Google Scholar]
- 16.Puckett C.A., Barton J.K. Methods to explore cellular uptake of ruthenium complexes. J Am Chem Soc. 2007;129:46–47. doi: 10.1021/ja0677564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang H., Zhang P., Yu B., Chen Y., Wang J., Ji L. Targeting nucleus DNA with a cyclometalated dipyridophenazineruthenium(II) complex. J Med Chem. 2014;57:8971–8983. doi: 10.1021/jm501095r. [DOI] [PubMed] [Google Scholar]
- 18.Gill M.R., Thomas J.A. Ruthenium(II) polypyridyl complexes and DNA—from structural probes to cellular imaging and therapeutics. Chem Soc Rev. 2012;41:3179–3192. doi: 10.1039/c2cs15299a. [DOI] [PubMed] [Google Scholar]
- 19.Lord C.J., Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 20.Rajendiran V., Karthik R., Palaniandavar M., Stoeckli-Evans H., Periasamy V.S., Akbarsha M.A. Mixed-ligand copper(II)-phenolate complexes: effect of coligand on enhanced DNA and protein binding, DNA cleavage, and anticancer activity. Inorg Chem. 2007;46:8208–8221. doi: 10.1021/ic700755p. [DOI] [PubMed] [Google Scholar]
- 21.Qian C., Wang J.Q., Song C.L., Wang L.L., Ji L.N., Chao H. The induction of mitochondria-mediated apoptosis in cancer cells by ruthenium(II) asymmetric complexes. Metallomics. 2013;5:844–854. doi: 10.1039/c3mt20270d. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Z., Wu Q., Wu X.H., Sun F.Y., Chen L.M., Chen J.C. Ruthenium(II) complexes as apoptosis inducers by stabilizing c-myc G-quadruplex DNA. Eur J Med Chem. 2014;80:316–324. doi: 10.1016/j.ejmech.2014.04.070. [DOI] [PubMed] [Google Scholar]
- 23.Liu X.W., Li J., Li H., Zheng K.C., Chao H., Ji L.N. Synthesis, characterization, DNA-binding and photocleavage of complexes [Ru(phen)2(6-OH-dppz)]2+ and [Ru(phen)2(6-NO2-dppz)]2+ J Inorg Biochem. 2005;99:2372–2380. doi: 10.1016/j.jinorgbio.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 24.Alonso E.N., Ferronato M.J., Fermento M.E., Gandini N.A., Romero A.L., Guevara J.A. Antitumoral and antimetastatic activity of Maitake D-Fraction in triple-negative breast cancer cells. Oncotarget. 2018;9:23396–23412. doi: 10.18632/oncotarget.25174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Enderling H., Alexander N.R., Clark E.S., Branch K.M., Estrada L., Crooke C. Dependence of invadopodia function on collagen fiber spacing and cross-linking: computational modeling and experimental evidence. Biophys J. 2008;95:2203–2218. doi: 10.1529/biophysj.108.133199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Q., He J., Mei W., Zhang Z., Wu X., Sun F. Arene ruthenium(ii) complex, a potent inhibitor against proliferation, migration and invasion of breast cancer cells, reduces stress fibers, focal adhesions and invadopodia. Metallomics. 2014;6:2204–2212. doi: 10.1039/c4mt00158c. [DOI] [PubMed] [Google Scholar]
- 27.Deakin N.O., Turner C.E. Paxillin comes of age. J Cell Sci. 2008;121:2435–2444. doi: 10.1242/jcs.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaidel-Bar R., Milo R., Kam Z., Geiger B. A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell-matrix adhesions. J Cell Sci. 2007;120:137–148. doi: 10.1242/jcs.03314. [DOI] [PubMed] [Google Scholar]
- 29.Prudent J., Popgeorgiev N., Gadet R., Deygas M., Rimokh R., Gillet G. Mitochondrial Ca(2+) uptake controls actin cytoskeleton dynamics during cell migration. Sci Rep. 2016;6:36570. doi: 10.1038/srep36570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tulotta C., Stefanescu C., Beletkaia E., Bussmann J., Tarbashevich K., Schmidt T. Inhibition of signaling between human CXCR4 and zebrafish ligands by the small molecule IT1t impairs the formation of triple-negative breast cancer early metastases in a zebrafish xenograft model. Dis Model Mech. 2016;9:141–153. doi: 10.1242/dmm.023275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y., Drabsch Y., Pujuguet P., Ren J., van Laar T., Zhang L. Genetic depletion and pharmacological targeting of alphav integrin in breast cancer cells impairs metastasis in zebrafish and mouse xenograft models. Breast Cancer Res. 2015;17:28. doi: 10.1186/s13058-015-0537-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee S.L., Rouhi P., Dahl Jensen L., Zhang D., Ji H., Hauptmann G. Hypoxia-induced pathological angiogenesis mediates tumor cell dissemination, invasion, and metastasis in a zebrafish tumor model. Proc Natl Acad Sci U S A. 2009;106:19485–19490. doi: 10.1073/pnas.0909228106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Setyawati M.I., Tay C.Y., Bay B.H., Leong D.T. Gold nanoparticles induced endothelial leakiness depends on particle size and endothelial cell origin. ACS Nano. 2017;11:5020–5030. doi: 10.1021/acsnano.7b01744. [DOI] [PubMed] [Google Scholar]
- 34.Setyawati M.I., Tay C.Y., Docter D., Stauber R.H., Leong D.T. Understanding and exploiting nanoparticles' intimacy with the blood vessel and blood. Chem Soc Rev. 2015;44:8174–8199. doi: 10.1039/c5cs00499c. [DOI] [PubMed] [Google Scholar]
- 35.Setyawati M.I., Mochalin V.N., Leong D.T. Tuning endothelial permeability with functionalized nanodiamonds. ACS Nano. 2016;10:1170–1181. doi: 10.1021/acsnano.5b06487. [DOI] [PubMed] [Google Scholar]
- 36.Setyawati M.I., Leong D.T. Mesoporous silica nanoparticles as an antitumoral-angiogenesis strategy. ACS Appl Mater Interfaces. 2017;9:6690–6703. doi: 10.1021/acsami.6b12524. [DOI] [PubMed] [Google Scholar]
- 37.Cantarella G., Risuglia N., Dell'eva R., Lempereur L., Albini A., Pennisi G. TRAIL inhibits angiogenesis stimulated by VEGF expression in human glioblastoma cells. Br J Cancer. 2006;94:1428–1435. doi: 10.1038/sj.bjc.6603092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kong W., He L., Richards E.J., Challa S., Xu C.X., Permuth-Wey J. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene. 2014;33:679–689. doi: 10.1038/onc.2012.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Y., Zhang Y., Iwamoto H., Hosaka K., Seki T., Andersson P. Discontinuation of anti-VEGF cancer therapy promotes metastasis through a liver revascularization mechanism. Nat Commun. 2016;7:12680. doi: 10.1038/ncomms12680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paauwe M., Heijkants R.C., Oudt C.H., van Pelt G.W., Cui C., Theuer C.P. Endoglin targeting inhibits tumor angiogenesis and metastatic spread in breast cancer. Oncogene. 2016;35:4069–4079. doi: 10.1038/onc.2015.509. [DOI] [PubMed] [Google Scholar]
- 41.Maji S., Chaudhary P., Akopova I., Nguyen P.M., Hare R.J., Gryczynski I. Exosomal annexin II promotes angiogenesis and breast cancer metastasis. Mol Cancer Res. 2017;15:93–105. doi: 10.1158/1541-7786.MCR-16-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lyu J., Yang E.J., Head S.A., Ai N., Zhang B., Wu C. Pharmacological blockade of cholesterol trafficking by cepharanthine in endothelial cells suppresses angiogenesis and tumor growth. Cancer Lett. 2017;409:91–103. doi: 10.1016/j.canlet.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Su T.T. Cellular responses to DNA damage: one signal, multiple choices. Annu Rev Genet. 2006;40:187–208. doi: 10.1146/annurev.genet.40.110405.090428. [DOI] [PubMed] [Google Scholar]
- 44.Baghbanzadeh M., Carbone L., Cozzoli P.D., Kappe C.O. Microwave-assisted synthesis of colloidal inorganic nanocrystals. Angew Chem Int Ed Engl. 2011;50:11312–11359. doi: 10.1002/anie.201101274. [DOI] [PubMed] [Google Scholar]
- 45.Wu Q., Fan C., Chen T., Liu C., Mei W., Chen S. Microwave-assisted synthesis of arene ruthenium(II) complexes that induce S-phase arrest in cancer cells by DNA damage-mediated p53 phosphorylation. Eur J Med Chem. 2013;63:57–63. doi: 10.1016/j.ejmech.2013.01.037. [DOI] [PubMed] [Google Scholar]
- 46.Meyer E.A., Castellano R.K., Diederich F. Interactions with aromatic rings in chemical and biological recognition. Angew Chem Int Ed Engl. 2003;42:1210–1250. doi: 10.1002/anie.200390319. [DOI] [PubMed] [Google Scholar]
- 47.Liu H.-K., Parkinson J.A., Bella J., Wang F., Sadler P.J. Penetrative DNA intercalation and G-base selectivity of an organometallic tetrahydroanthracene RuII anticancer complex. Chem Sci. 2010;1:258. [Google Scholar]
- 48.Wu Q., Zheng K., Liao S., Ding Y., Li Y., Mei W. Arene Ruthenium(II) Complexes as Low-Toxicity Inhibitor against the Proliferation, Migration, and Invasion of MDA-MB-231 Cells through Binding and Stabilizing c-myc G-Quadruplex DNA. Organometallics. 2016;35:317–326. [Google Scholar]
- 49.Pang X., Yi T., Yi Z., Cho S.G., Qu W., Pinkaew D. Morelloflavone, a biflavonoid, inhibits tumor angiogenesis by targeting rho GTPases and extracellular signal-regulated kinase signaling pathways. Cancer Res. 2009;69:518–525. doi: 10.1158/0008-5472.CAN-08-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lian B.W., Wu Q., Zhang S.Y., Li Y.M., Zhao X.H., Mei W.J. Tissue regeneration promotion effects of phenanthroimidazole derivatives through pro-inflammatory pathway activation. Fish Shellfish Immunol. 2018;80:582–591. doi: 10.1016/j.fsi.2018.06.029. [DOI] [PubMed] [Google Scholar]
- 51.Wang J., Cao Z., Zhang X.M., Nakamura M., Sun M., Hartman J. Novel mechanism of macrophage-mediated metastasis revealed in a zebrafish model of tumor development. Cancer Res. 2015;75:306–315. doi: 10.1158/0008-5472.CAN-14-2819. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material