Abstract
Motivation
Orthology inference constitutes a common base of many genome-based studies, as a pre-requisite for annotating new genomes, finding target genes for biotechnological applications and revealing the evolutionary history of life. Although its importance keeps rising with the ever-growing number of sequenced genomes, existing tools are computationally demanding and difficult to employ.
Results
Here, we present SonicParanoid, which is faster than, but comparably accurate to, the well-established tools with a balanced precision-recall trade-off. Furthermore, SonicParanoid substantially relieves the difficulties of orthology inference for those who need to construct and maintain their own genomic datasets.
Availability and implementation
SonicParanoid is available with a GNU GPLv3 license on the Python Package Index and BitBucket. Documentation is available at http://iwasakilab.bs.s.u-tokyo.ac.jp/sonicparanoid.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Due to the recent advancement in DNA sequencing technologies, the number of completely sequenced genomes is growing at an accelerated pace. The accurate inference of orthologous genes encoded on multiple genomes is the key to various analyses based on those datasets (Altenhoff and Dessimoz, 2012). For example, comparative genomics, genome annotation, phylogenomics and the development of genome databases all depend on reliable orthology inference. There are dozens of tools available for orthology inference, including InParanoid (Sonnhammer and Östlund, 2015) and its extension MultiParanoid (Alexeyenko et al., 2006), OrthoMCL (Li, 2003), Hieranoid (Kaduk and Sonnhammer, 2017), OMA (Train et al., 2017), Proteinortho (Lechner et al., 2011), OrthoFinder (Emms and Kelly, 2015) and PANTHER (Mi et al., 2017). InParanoid is one of the oldest and most popular tools and has the best trade-off between specificity and recall (Altenhoff et al., 2016). Here, we present SonicParanoid, which is a fast, accurate and easy-to-use tool for multi-species orthology inference.
2 Materials and methods
SonicParanoid borrows the concepts used in the graph-based algorithm of InParanoid (Remm et al., 2001) because of its reported accuracy (Altenhoff et al., 2016; Chen et al., 2007), but brings changes to the core algorithm and speeds-up and automates the entire process (Supplementary Fig. S1). To reduce the computational time, second-pass alignments and bootstrapping tests are skipped, and MMseqs2 (Steinegger and Söding, 2017) is used instead of legacy-BLAST (Altschul et al., 1997). Furthermore, SonicParanoid adopts a new scoring function and a configurable threshold that take into account sequence-length differences (Supplementary Material and Supplementary Figs S2 and S3). Another difference with the original InParanoid algorithm is in the way that overlapping groups of orthologs are clustered.
In Remm et al. (2001), groups are merged or removed based on comparisons of confidence scores of the grouped orthologs. In contrast, SonicParanoid treats groups as elements of numeric sets, which made the algorithm faster and yet have minimal difference in terms of accuracy (Supplementary Fig. S4). Finally, because the multi-species orthology inference step is fully automated, the users can avoid the cumbersome and error-prone collection of ortholog tables and configuration files creation required by MultiParanoid. Details are described in Supplementary Material.
3 Results and discussion
3.1 Speed evaluation
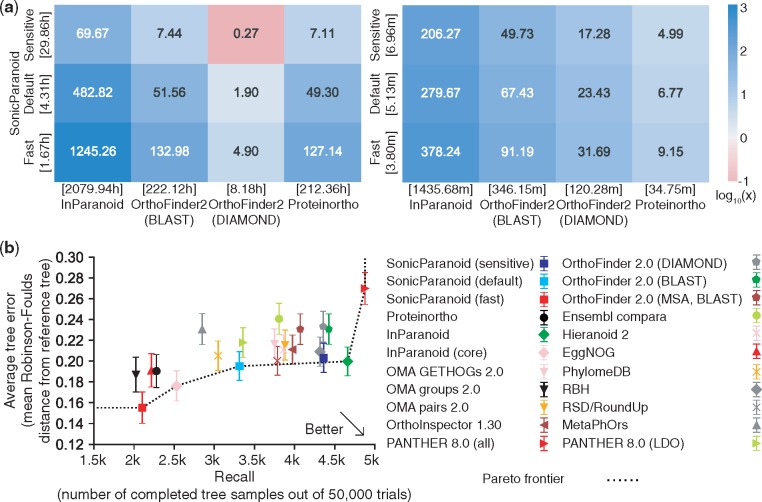
We ran SonicParanoid, InParanoid (ver 4.1), OrthoFinder (ver 2.1.2) and Proteinortho (ver 5.15) on a benchmark proteome dataset provided by the Quest for Orthologs (QfO) consortium (Gabaldón et al., 2009). Details including the hardware and software settings are described in Supplementary Material. Other than InParanoid, OrthoFinder and Proteinortho were selected because they were reported to be fastest among the existing tools (Lechner et al., 2011). When the complete QfO dataset (including 66 proteomes) was processed using eight CPUs, InParanoid, OrthoFinder on BLASTP (Camacho et al., 2009), OrthoFinder on DIAMOND (Buchfink et al., 2015) and Proteinortho required 2079.9, 222.1, 8.2 and 212.4 h, respectively, while SonicParanoid in the default mode reduced the running time to 4.3 h (i.e. 482.8, 51.6, 1.9 and 49.3 times faster, respectively) (Fig. 1a, left). When the execution time was measured without considering the time required for the all-vs-all alignments, SonicParanoid was still faster than the other methods (Fig. 1a, right). This trend was also observed with the eukaryotic and prokaryotic subsets of the QfO dataset as input (Supplementary Figs S5 and S6). We also conducted the scalability analysis of SonicParanoid on large-scale genomic data (Supplementary Fig. S7) as well as memory usage analysis (Supplementary Fig. S8), which show that SonicParanoid is applicable to large genomic datasets.
Fig. 1.
Speed and accuracy of SonicParanoid. (a) Speed-up of SonicParanoid (in the fast, default and sensitive modes) in relation to InParanoid, OrthoFinder2 (on BLASTP and DIAMOND) and Proteinortho on the complete 2011 version of the QfO dataset (left: complete execution, right: orthology inference step only). The numbers on the tiles and their colors indicate the speed-up folds. The numbers in the square brackets represent the execution time in hours (left) or minutes (right). Eight processors were used for each tool. (b) Accuracy of SonicParanoid and other 13 orthology inference tools assessed by the QfO benchmark (generalized species tree discordance test at the last universal common ancestor level). The x-axis represents the numbers of completed tree samples out of 50 000 trials (larger is better), while the y-axis represents the average tree error (smaller is better)
3.2 Accuracy evaluation
The accuracy of SonicParanoid was compared to those of 13 orthology inference methods using the benchmark service from the QfO community (Altenhoff et al., 2016). Details regarding the benchmark datasets construction are described in the Supplementary Material. Overall, the results indicate that SonicParanoid is a good alternative to the existing methods (Fig. 1b and Supplementary Figs S9 and S10). SonicParanoid (especially in the default and sensitive modes) showed accuracy similar to that of InParanoid and near the Pareto frontiers in most tests.
3.3 Usability
To meet various needs from quick assessment to detailed analysis, SonicParanoid provides fast, default and sensitive modes that employ different filtering thresholds for the alignment process. While the default mode would suit most studies, the fast and sensitive modes may be used for evolutionarily close and distant species, respectively (Supplementary Figs S9 and S11).
SonicParanoid only requires users to provide a directory containing the input proteomes to generate ortholog relationship files between proteome pairs and the multi-species ortholog table file (Supplementary Fig. S1). In contrast, InParanoid and MultiParanoid require users to write programs to generate input files for every proteome pair, perform the required InParanoid runs and collect the generated ortholog relationship files for themselves. The entire process is considerably error prone if manually performed, and is difficult to employ for users lacking programing skills.
SonicParanoid also allows seamless addition and deletion of proteomes by reusing the results from previous runs, which is beneficial to users who need to maintain their own orthology databases. Even if the run is interrupted before the computation is completed, for example by power failure, it can be easily resumed without losing previously computed results. Multi-species ortholog tables for subsets of proteomes in a previous run can also be quickly computed.
4 Conclusion
We developed SonicParanoid as a fast, accurate and easy orthology inference tool and evaluated its speed and accuracy using standardized datasets and benchmark. SonicParanoid also has high scalability; it can analyze the 276 proteomes used to build the InParanoid8 orthology database (Sonnhammer and Östlund, 2015) in less than 5 days using only 8 CPUs in the default mode. Considering the ever-growing number of new genomes (Cochrane et al., 2016), we believe the speed, scalability, accuracy and ease-of-use of SonicParanoid will contribute to annotating new genomes, finding target genes in medical and biotechnological applications and revealing the evolutionary history of life.
Supplementary Material
Acknowledgements
We are grateful to Sira Sriswasdi, Martin Frith and all the members of the Iwasaki laboratory for the helpful comments on this study.
Funding
This work was supported by the Japanese Society for the Promotion of Science (Grant Numbers 16H06154, 16H06279, and 17H05834).
Conflict of Interest: none declared.
References
- Alexeyenko A. et al. (2006) Automatic clustering of orthologs and inparalogs shared by multiple proteomes. Bioinformatics, 22, e9–15. [DOI] [PubMed] [Google Scholar]
- Altenhoff A.M. et al. (2016) Standardized benchmarking in the quest for orthologs. Nat. Methods, 13, 425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenhoff A.M., Dessimoz C. (2012) Inferring orthology and paralogy. Methods Mol. Biol., 855, 259–279. [DOI] [PubMed] [Google Scholar]
- Altschul S.F. et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B. et al. (2015) Fast and sensitive protein alignment using DIAMOND. Nat. Methods, 12, 59–60. [DOI] [PubMed] [Google Scholar]
- Camacho C. et al. (2009) BLAST+: architecture and applications. BMC Bioinformatics, 10, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F. et al. (2007) Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS One, 2, e383.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane G. et al. (2016) The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res., 44, D48–D50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms D.M., Kelly S. (2015) OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol., 16, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldón T. et al. (2009) Joining forces in the quest for orthologs. Genome Biol., 10, 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaduk M., Sonnhammer E. (2017) Improved orthology inference with Hieranoid 2. Bioinformatics, 33, 1154–1159. [DOI] [PubMed] [Google Scholar]
- Lechner M. et al. (2011) Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics, 12, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. et al. (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res., 13, 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H. et al. (2017) PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res., 45, D183–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remm M. et al. (2001) Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol., 314, 1041–1052. [DOI] [PubMed] [Google Scholar]
- Sonnhammer E.L.L., Östlund G. (2015) InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res., 43, D234–D239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinegger M., Söding J. (2017) MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol., 35, 1026–1028. [DOI] [PubMed] [Google Scholar]
- Train C.-M. et al. (2017) Orthologous Matrix (OMA) algorithm 2.0: more robust to asymmetric evolutionary rates and more scalable hierarchical orthologous group inference. Bioinformatics, 33, i75–i82. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.