Significance
Colwellia psychrerythraea 34H is a cold-adapted marine bacterium that represents a genus and species cosmopolitan to the cold ocean. To our knowledge, metabolic flux studies of an obligate psychrophile like 34H are not available. We characterized the physiology and metabolism of 34H, at normal-growth temperature of 4 °C and upper-stress condition of room temperature (23 °C), by integrating metabolic flux studies (tracking 13C-labeled compounds) and genetic analyses (transcriptomics). Results from these system-level analyses reveal unique metabolic features under cold salty conditions, which broaden our understanding of microbial ecology in the cold ocean, currently vulnerable to global warming. Specific findings have relevance to bioremediation of pollutants from the petroleum industry, increasingly active in polar seas, and to biomanufacturing of cold-adapted enzymes.
Keywords: marine psychrophile, metabolic flux, ED pathway, short-chain fatty acids, gluconeogensis
Abstract
Colwellia psychrerythraea 34H is a model psychrophilic bacterium found in the cold ocean—polar sediments, sea ice, and the deep sea. Although the genomes of such psychrophiles have been sequenced, their metabolic strategies at low temperature have not been quantified. We measured the metabolic fluxes and gene expression of 34H at 4 °C (the mean global-ocean temperature and a normal-growth temperature for 34H), making comparative analyses at room temperature (above its upper-growth temperature of 18 °C) and with mesophilic Escherichia coli. When grown at 4 °C, 34H utilized multiple carbon substrates without catabolite repression or overflow byproducts; its anaplerotic pathways increased flux network flexibility and enabled CO2 fixation. In glucose-only medium, the Entner–Doudoroff (ED) pathway was the primary glycolytic route; in lactate-only medium, gluconeogenesis and the glyoxylate shunt became active. In comparison, E. coli, cold stressed at 4 °C, had rapid glycolytic fluxes but no biomass synthesis. At their respective normal-growth temperatures, intracellular concentrations of TCA cycle metabolites (α-ketoglutarate, succinate, malate) were 4–17 times higher in 34H than in E. coli, while levels of energy molecules (ATP, NADH, NADPH) were 10- to 100-fold lower. Experiments with E. coli mutants supported the thermodynamic advantage of the ED pathway at cold temperature. Heat-stressed 34H at room temperature (2 hours) revealed significant down-regulation of genes associated with glycolytic enzymes and flagella, while 24 hours at room temperature caused irreversible cellular damage. We suggest that marine heterotrophic bacteria in general may rely upon simplified metabolic strategies to overcome thermodynamic constraints and thrive in the cold ocean.
Many bioprocesses rely on the innate metabolic strengths of nonmodel environmental microorganisms. From this perspective, microbiologists are exploring new microbes from extreme environments for advantageous characteristics (1). Advances in sequencing techniques, genomic analyses, and metabolic analyses have aided species characterization by providing the functional output of microorganisms and revealing their genotype-to-phenotype regulations. For some environments, the functional metabolisms of representative species (mainly thermophiles and mesophiles) have been characterized in detail (2–5). For cold environments, few studies have addressed the metabolic pathways of cold-adapted microorganisms (psychrophiles) directly, the emphasis being on genomic and diversity analyses instead (6–8). However, most of Earth’s biosphere is cold (>75% volumetrically is below 5 °C), particularly the microbially dominated ocean with its mean temperature of 4 °C (9).
In this study, Colwellia psychrerythraea 34H (henceforth, 34H), a psychrophilic extremophile (unable to grow above 18 °C) of ecological importance in the cold ocean, was used as a model species for investigating the metabolism of cold-adapted microbial life. This Gram-negative, heterotrophic gammaproteobacterium, originally isolated from arctic marine sediments (10), is considered cosmopolitan to polar regions and the cold deep sea (11, 12). Although ice adapted in many ways (11), 34H also represents the cold dark ocean in its lack of proteorhodopsins, photoactive proteins used by many bacteria in the surface ocean to generate supplemental energy (13). Genomic studies of 34H and other strains of C. psychrerythraea have revealed many metabolic pathways that are important in the geochemical cycling of nutrients in cold marine environments (14), including those for hydrocarbon degradation and denitrification (14, 15). For example, an amplified genome of C. psychrerythraea, present in the Colwellia-rich microbial community that responded at depth to the Deepwater Horizon oil spill (12, 16, 17), has the potential to degrade ethane and propane (components of natural gas), suggesting a possible role in bioremediation of cold environments. Additionally, 34H has the capacity to produce industrially relevant compounds, such as polyhdyroxyalkanoate and polyamides (14) and unique cryoprotectants (18).
Able to grow at temperatures as low as −12 °C (19), 34H has been used to study cold-adapted proteins (20) and enzymes (21), extracellular polysaccharide substances (18, 22), and motility and chemohalotaxis at subzero temperatures (23, 24). Recently, investigation of cold-adapted enzymes has increased in importance due to the potential economic and ecological advantages they can provide over their higher-temperature-requiring counterparts in industrial processes (25). Previous studies of cold-adapted microbes have revealed a variety of molecular adaptations that allow their activity and survival under extreme conditions, including elevated amounts of unsaturated fatty acids in the membrane (keeping it from rigidifying in the cold), an overall higher content of polar amino acids (affecting protein thermolability), enzymes with high specific activity at low temperatures (10, 19, 25), and antifreeze molecules (18, 22). However, to our knowledge, a comprehensive metabolic characterization of a psychrophilic extremophile is not available. With climate change (26, 27) and concerns about pollutants from resource-extraction industries, which continue to develop in the deep sea and newly ice-free Arctic regions, microbial metabolic responses in cold environments is an emerging frontier. Using 34H as a model marine psychrophile, this research elucidates physiological functions, biomass composition, and functional metabolism by quantifying metabolites, using 13C-metabolic flux analyses (13C-MFA), and sequencing transcriptomes [RNA-sequencing (RNA-Seq)] under normal-growth cold temperature (4 °C) and thermally stressful room temperature (above its upper-growth temperature of 18 °C). Dynamic 13C-labeling techniques (28) and metabolic modeling (29) in particular have been used to decipher cellular metabolism, responses to genetic perturbation (28), and innate pathway regulations (30, 31) in other organisms, but have not been applied to a psychrophilic organism. By comparing 34H responses not only to different temperatures but also to responses of mesophilic Escherichia coli [which also contains the Entner–Doudoroff (ED) and Embden–Meyerhof–Parnas (EMP) pathways], insights into the distinctive metabolic functioning of an obligate psychrophile emerge.
Results
Growth Characteristics and Biomass Composition.
The first step toward applying dynamic 13C labeling to 34H was to quantify its growth physiology at 4 °C in a defined minimal medium of artificial seawater (ASW) supplemented with glucose (1 g⋅L−1 and trace vitamins). Under these conditions, the specific growth rate was 0.008 h−1 for a doubling time of about 3.6 d (SI Appendix, Fig. S1A). In ASW medium with lactate (1 g⋅L−1) instead, the rate was faster (0.011 h−1, doubling time of 2.6 d; SI Appendix, Fig. S2A), though in complex rich medium, the doubling time is ∼5 h. The specific glucose uptake rate, determined by measuring glucose concentration in the minimal ASW medium over time with an enzymatic assay, was ∼2.3 mg⋅h−1 (SI Appendix, Fig. S1A). Unlike heterotrophic mesophiles (e.g., E. coli and Shewanella species), acetate was not secreted by 34H to a detectable level during glucose or lactate metabolism. The biomass composition of 34H was quantified experimentally (SI Appendix, Fig. S1 B–D) during growth on glucose. A significant portion of the dry weight biomass was ash (20%), presumably due to its required growth in a sea-salt medium. On an ash-free biomass basis, the content was found to be comparable to previously reported E. coli measurements (32): 53% protein and 15% lipids. The RNA and DNA contents were calculated based on the protein:RNA and RNA:DNA ratios found in E. coli (33, 34), resulting in 20% RNA and 3% DNA. Although the protein profile was comparable to that of E. coli (SI Appendix, Fig. S1C), the percentage of polar and charged amino acid residues in 34H was higher (51% vs. 47%). The lipid composition revealed a large component of unsaturated (56%) and short chain (12%, containing 12 carbons or less) fatty acids (SI Appendix, Fig. S1D), both of which comprised a larger percentage of the composition than in E. coli (32).
Steady-State 13C-Fingerprinting and -MFA.
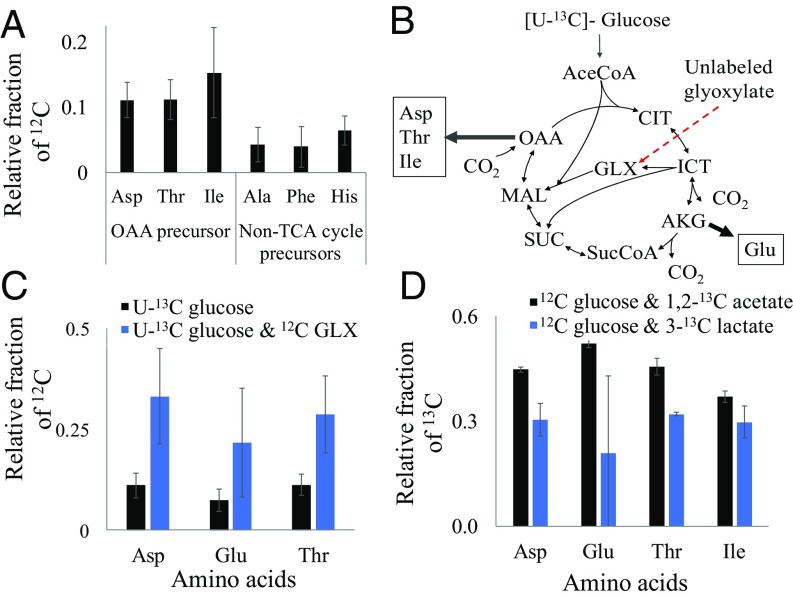
Next, the metabolism of 34H was investigated using 13C-fingerprinting and 13C-MFA. The experimentally determined biomass composition was used to construct a biomass equation (SI Appendix, Table S1, reaction 74) describing the drain of metabolite precursors to make cellular components, following a previously reported method (35). Then, the annotated genome from the Kyoto Encyclopedia of Genes and Genomes database (kegg.jp) was used to construct a simplified core metabolic network using WUFlux software (36) (full model in SI Appendix, Table S1), supplemented with the previously determined biomass equation. The metabolic network was similar to that of E. coli, with the exception of the anaplerotic reactions which are important for replenishing TCA cycle metabolites. The 34H genome shows genes that potentially encode enzymes for the anaplerotic reactions [phosphoenolpyruvate (PEP)+CO2 <=> oxaloacetate (OAA) and pyruvate (PYR)+CO2 <=> OAA or malate] annotated as PEP decarboxylase (cps4595), malate dehydrogenase (cps0331 and cps4262), and oxaloacetate decarboxylase (cps1048∼1050). We considered the possibility that these enzymes may have plasticity for both forward and reverse reactions, and thus investigated the ability of 34H to incorporate atmospheric 12CO2 into its TCA cycle metabolites after growing with U-13C (fully labeled) glucose. The resulting significant (P value = 0.0294) dilution (>10%) of labeled carbons in OAA cycle-derived amino acids relative to amino acids from non-TCA cycle precursors (<5% dilution) suggests that 34H incorporates CO2 into its TCA cycle metabolites via anaplerotic reactions; these reactions were included in the metabolic network (Fig. 1A).
Fig. 1.
12C- and 13C-fingerprinting experiments with 34H revealing its ability to incorporate CO2 into TCA cycle metabolites, bypass the lower half of the TCA cycle (glyoxylate shunt), and utilize multiple substrates simultaneously (no glucose catabolic repression). (A) Labeling profiles of amino acids derived from OAA in cultures grown with U-13C glucose. (B) Entry points for the incorporation of glucose and glyoxylate into the TCA cycle (indicated by red arrow). See SI Appendix for abbreviations for metabolites. (C) Labeling profiles of proteinogenic amino acids from the TCA cycle in cultures grown with only U-13C glucose and with U-13C glucose and 12C glyoxylate. (D) Labeling profiles of amino acids derived from the TCA cycle in cultures grown on unlabeled glucose supplemented with 1,2-13C acetate or 3-13C lactate. Error bars indicate SD of the mean (n = 2).
13C-fingerprinting was performed to probe the activity of the glyoxylate shunt, which bypasses the lower half of the TCA cycle (Fig. 1B). Cultures were grown in the presence of U-13C glucose and unlabeled glyoxylate, and the amino acid dilution of labeled carbons was determined. Again, a significant (P value <0.01) amount of unlabeled carbon (∼25%) was observed in the TCA cycle-derived amino acids from cells grown on U-13C glucose and unlabeled glyoxylate relative to the cells grown on only U-13C glucose (∼10%). This degree of unlabeled carbon indicates that the glyoxylate shunt is functional. We also investigated effects of glucose catabolic repression. Cultures were grown in the presence of two carbon substrates, one labeled (U-13C acetate or 3-13C lactate) and one unlabeled (glucose), and were harvested during exponential growth phase before the glucose in the culture was exhausted. Analysis showed that 34H amino acids had a significant amount of labeling under both the labeled acetate (∼50% labeling) and lactate (∼30% labeling) growth conditions compared with the natural abundance of labeled carbon (1%), indicating that the microbe is capable of coutilizing glucose with other substrates (Fig. 1).
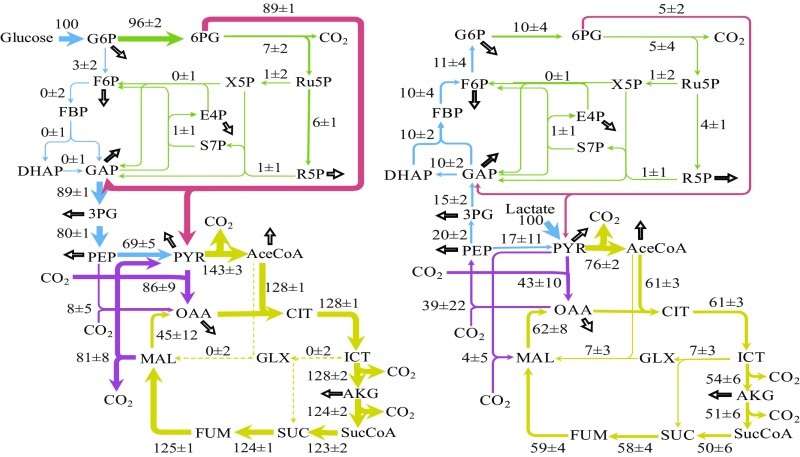
Using our constructed metabolic network, 13C-MFA studies were conducted to quantify the central metabolic pathways of 34H when grown on 1,2-13C glucose or 3-13C lactate. The resulting flux maps are presented in Fig. 2. MFA on glucose-grown 34H cells revealed that the ED pathway is the primary route of glycolysis. Relative to the glucose uptake rate, ∼89% of the flux passes through the ED pathway. Flux through the decarboxylate step of the pentose phosphate (PP) pathway is equivalent to 7% of the glucose uptake rate. The upper portion of the EMP pathway (glucose-6-phosphate to glyceraldehyde-3-phosphate) is estimated to be relatively inactive, with little to no flux. The MFA results confirmed a highly functional TCA cycle in 34H operating with CO2 fixation reactions (i.e., the PYR shunt and PEP carboxylase) during glucose metabolism.
Fig. 2.
Resulting metabolic flux map when 34H was grown at 4 °C on 1,2-13C glucose (Left) or 3-13C lactate (Right). Net fluxes are shown for reversible reactions. Metabolic pathways are indicated by colors: red, ED pathway; blue, EMP pathway; yellow, TCA cycle; green, PP pathway; and purple, anaplerotic reactions. Thicker arrows represent higher flux rates; dashed arrows represent lowest flux rates. Hollow black arrows indicate metabolite drainage for biomass formation. See SI Appendix, Table S1 for full flux values, confidence intervals, and abbreviations for metabolites.
In parallel, MFA on lactate-grown cells showed that 10% flux relative to the lactate uptake rate proceeded through gluconeogenesis. Most of the incoming lactate flux was converted to acetyl-CoA, and then biomass without acetate secretion, providing a metabolic route for more rapid growth on lactate than glucose (Fig. 2). The PEP carboxykinase is an active starting point for gluconeogenesis. The PP pathway maintained its small amount of flux under both conditions, suggesting that only a limited amount of PP pathway flux is necessary for cell growth. A difference between growth on glucose vs. lactate was the activation of the glyoxylate shunt during growth on lactate.
Pool Size and Dynamic Labeling of Central Metabolites.
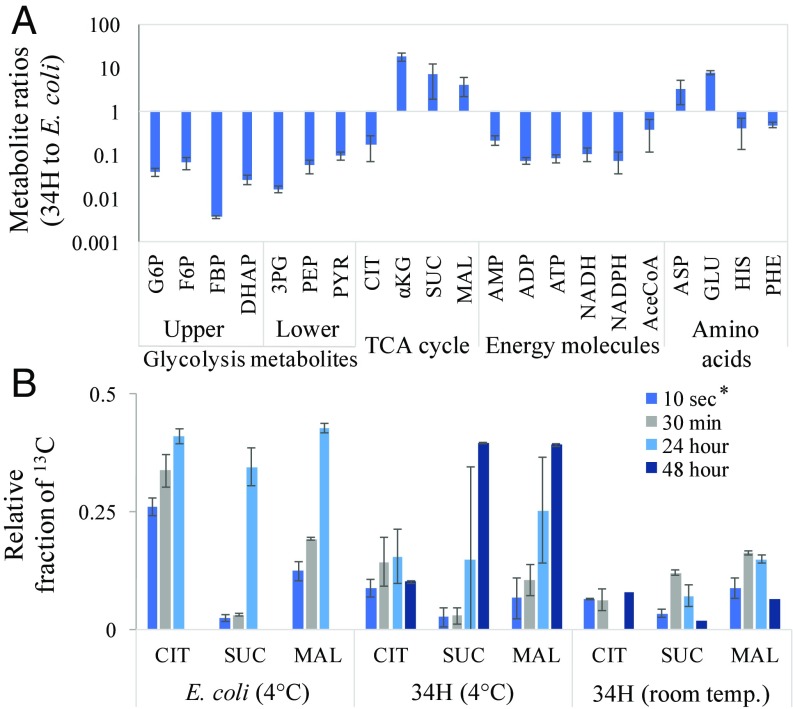
The metabolic pool size (i.e., concentration of intracellular metabolites) of 34H was compared with that of E. coli during exponential growth at their respective normal-growth temperatures (4 °C and 37 °C) using the isotopic ratio method previously described (37). The analysis (Fig. 3A) indicated that, except for citrate, 34H has a much higher (4–17 times higher) pool size of TCA cycle metabolites (αKG, succinate, malate) and TCA cycle-derived amino acids. The pool sizes of non-TCA cycle-derived amino acids and acetyl-CoA were approximately 2-fold smaller in 34H, while the EMP pathway metabolites were 10- to 50-fold smaller. The small pool sizes of the EMP pathway metabolites are in agreement with the MFA model showing that 34H does not utilize the EMP pathway. The pool sizes of energy molecules (ATP, NADH, and NADPH) were 10- to 100-fold smaller in 34H and the relative energy charge was lower than that found in E. coli. These small pool sizes of energy molecules suggest that the growth of 34H may be inherently energy limited.
Fig. 3.
Isotopic-labeling ratio methods and dynamic-labeling experiments with 34H revealing its metabolic pool sizes and metabolic behavior compared with E. coli. (A) Ratio of normal-growth 34H and E. coli metabolite pool sizes (Log10 scale). (B) Comparison of metabolic-labeling responses of TCA metabolites in cold-stressed (4 °C, following acclimation at 4 °C for 1 h) E. coli, normal-growth (4 °C) 34H, and heat-stressed (room temperature, following acclimation for 1 h at room temperature) 34H in media pulsed with U-13C glucose. E. coli was not sampled at 48 h; citrate was not detected in 34H room-temperature samples at 24 h. For 10-s data (asterisk), cell metabolism may have been active during the 5-min centrifugation (0 °C) step. Error bars indicate SD of the mean (n = 2, except for 34H room temperature where n = 4).
A dynamic labeling approach was then applied to investigate 34H metabolism at both 4 °C and room temperature. E. coli metabolism at 4 °C was used as a benchmark comparison in lieu of any available dynamic labeling experiments on cold-adapted bacteria. Cultures were grown in unlabeled media (34H at 4 °C and E. coli at 37 °C), and exponentially growing cells were placed at room temperature (34H) or 4 °C (E. coli) for 1 h. To examine glucose catabolism, cultures were then pulsed with concentrated U-13C glucose, followed by liquid chromatography-mass spectrometry (LC-MS) analysis of downstream metabolite labeling. Despite no growth of E. coli after equilibration at 4 °C, its upper glycolysis pathway was still highly active and intermediates became labeled within minutes after the pulse (SI Appendix, Fig. S3 and Dataset S1). 34H upper glycolytic metabolites (i.e., F6P) were labeled more slowly due to low activity of the EMP. During incubation at 4 °C, the TCA cycle in both E. coli and 34H was labeled much more slowly than glycolysis (Fig. 3). Additionally, labeling enrichment of succinate in 34H was initially much less than malate and citrate, indicating the presence of metabolic rate-limiting steps in the lower branch of the TCA cycle. At room temperature, 34H catabolism appeared to be strongly inhibited. TCA cycle metabolite labeling was reduced compared with normal-temperature growth (Fig. 3B), indicating reduction in the synthesis of biomass precursors. Glycolytic labeling was detected for 24 h (SI Appendix, Fig. S3 and Dataset S1), but labeling signatures in all metabolites became less visible thereafter (due to cell damage and metabolite degradation), relative to normal-growth temperature (4 °C; Fig. 3B and SI Appendix, Fig. S3), and no energy molecules (ATP, ADP, AMP) were detected at 48 h. Cultures exposed to room temperature for over 24 h did not recover at 4 °C in complex medium (SI Appendix, Fig. S3C), indicating irreversible cell damage.
RNA-Seq of C. psychrerythraea 34H.
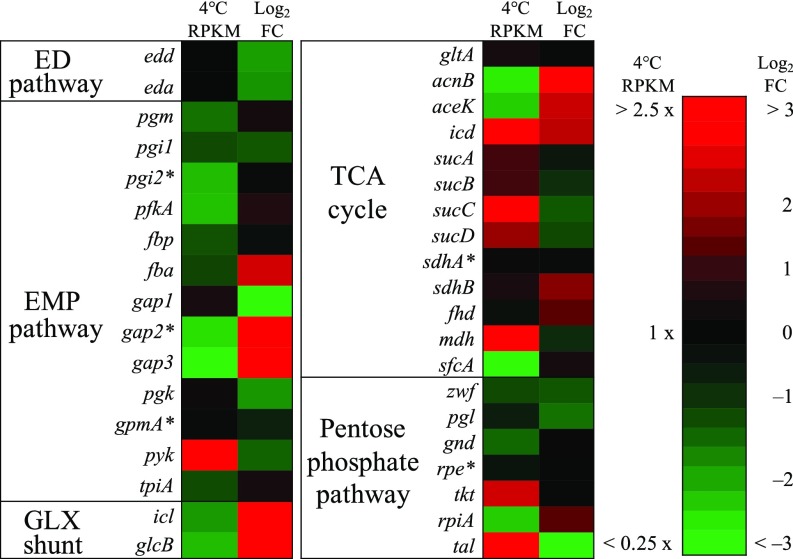
To provide additional insights into the metabolic pathways of 34H, RNA-Seq was performed (Dataset S2). As a qualitative comparison of gene expression level within samples, reads per kilobase million (RPKM) were normalized to the edd gene from the ED pathway. At 4 °C the ED pathway genes (edd and eda) had similar RPKM readings and were two of the more highly expressed genes (Fig. 4). The ED pathway expression levels were much higher than the upper portion of the EMP pathway. This high expression level supports MFA results showing that the ED pathway is the main glycolysis route. After exposure to elevated temperature (23 °C, too warm for growth) for 2 h, RNA-Seq showed a significant down-regulation of the ED pathway, an up-regulation of the upper portion of the TCA cycle and glyoxylate shunt, and a general down-regulation of the PP pathway (Fig. 4). Although 34H remains motile and displays chemotaxis for a similar period of time at temperatures as warm as 27 °C (26), most flagella-associated genes were down-regulated at the elevated temperature (SI Appendix, Fig. S4). No significant down-regulation of methyl-accepting chemotaxis proteins, however, or for choline and glycine/betaine expression was detected (SI Appendix, Fig. S4 and Dataset S2). The down-regulation of catabolism together with the results of dynamic labeling provide evidence that 34H ceases nutrient uptake and biosynthesis when stressed by elevated temperature, even as it maintains osmoregulation and may continue to search for nutrients.
Fig. 4.
Transcriptomic expression data of 34H at normal-growth conditions and differential expression data after exposure to temperature-stressed conditions as determined by RNA-Seq. Left column represents RPKM expression levels normalized to gene edd-1 (ED pathway). Right column represents Log2(fold change) (Log2FC) of differential gene expression from 4 °C to 23 °C. Scale for RPKM expression and Log2FC presented on the far Right. An asterisk next to gene names indicates data with Log2FC Benjamini–Hochberg false discovery rate (FDR)-adjusted P values <0.05. See SI Appendix, Fig. S4 and Dataset S2 for full gene expression details.
Discussion
This physiological study provides molecular-level insights (summarized in SI Appendix, Fig. S5) into the metabolism of a cold-adapted marine bacterium, C. psychrerythraea 34H, which may serve as a model for similar heterotrophic microorganisms in the extensive cold regions of the ocean. At the molecular level, the finding that 34H actively produces high amounts of unsaturated and short chain fatty acids helps to explain how it maintains the fluidity of its lipid membrane bilayer at cold temperatures (38). The ready microbial production of short chain fatty acids is also of industrial interest, as they can be used as drop-in jet fuel. The larger percentage of polar amino acids relative to mesophilic E. coli likely contributes to unique protein and supramolecular interactions in the cold marine environment and confirms, in live culture, earlier proteome predictions based on the 34H genome (14). The finding that 34H biomass contains a relatively high percentage of ash may reflect a role for inorganic salts in maintaining osmotic pressure or cryoprotection, supplementing its known use of organic compatible solutes (39) and extracellular polysaccharides under osmotic stress (22).
A key finding at the metabolic level is the demonstration that 34H, which contains both the EMP and ED pathways, favors the use of the ED pathway. In general, the EMP pathway generates more ATP than the ED pathway, facilitating biomass production; indeed, the use of the EMP pathway is ubiquitous in terrestrial microorganisms (40). This ubiquity differs strikingly, however, from the marine bacteria examined in a recent study, where most strains (all Bacteroidetes, all grown at 30 °C) relied on the ED pathway for glucose catabolism (41). To gain additional insight into this difference, we revisited previous thermodynamic analyses of the ED and EMP pathways (40, 42) from a cold marine perspective. The previous models calculated the thermodynamic data based on average concentrations of 1 mM for each metabolite, as well as low ionic strength and room temperature. Because marine microbes experience higher (seawater) salt levels, higher pH, and lower temperatures, the equilibrator calculator (43) was used to estimate the ∆G of each enzyme step in the two pathways at 0.67 M (ionic strength of ASW) and pH of 7.5 (in the seawater range of 7.5–8.5). The Keq obtained for these conditions was then used to estimate the effect of decreasing the temperature to 4 °C (Methods). The total ∆G of each pathway was determined by summing each individual step. The use of more realistic marine conditions led to an increase in thermodynamic favorability at 4 °C that was larger for the ED pathway (∆G of −36 kJ⋅mol−1) than the EMP pathway (∆G of −8 kJ⋅mol−1) (SI Appendix, Fig. S6), thus identifying a potential driving force for use of the ED pathway in the cold.
A second possible driving force for preferential use of the ED pathway in 34H is that it allows for simultaneous uptake of diverse carbon sources (44), a potentially important strategy in nutrient-poor areas of the cold ocean. In contrast, model microorganisms that utilize the EMP pathway, like E. coli, often experience catabolic repression. Directing glycolytic flux through the ED pathway rather than the EMP pathway may be a key strategy for avoiding glucose catabolic repression (44). The ED pathway also requires fewer enzymes (40), saving cellular resources by reducing the amount of protein needed for catabolism. Importantly, low temperature and high salt will slow both mRNA translation and metabolite diffusion. Thus, fewer enzyme steps and metabolite transport barriers among cascade reactions benefit an organism in the cold ocean. To further explore the potential advantages of the ED pathway at low temperature, we applied dynamic labeling to two E. coli mutants at 4 °C (mutational approaches with 34H have not yet been achieved): (i) a ∆ptsG mutant, where ptsG is a key gene in the glucose phosphotransferase system (PTS); and (ii) a ∆ptsG mutant overexpressing the ED pathway. As the PTS is a highly regulated glucose phosphorylation system (absent in 34H) tied to catabolic repression, these ∆ptsG E. coli mutants could be used to obtain insights into ED and EMP pathway utilization at cold temperature. The ∆ptsG + ED mutant showed faster 13C enrichment in glycolytic intermediates than the ∆ptsG mutant (53 ± 2% vs. 33 ± 3% in dihydroxyacetone phosphate [DHAP] labeled for 5 s following a U-13C glucose pulse; SI Appendix, Fig. S7), supporting the advantage of the ED pathway for glucose catabolism in the cold.
Recent reviews (45, 46) of cold-adapted bacteria, covering comparative transcriptomic and proteomic studies across a range of temperatures, further support the EMP pathway as unfavorable in cold marine environments. For many of the species examined, the EMP pathway was down-regulated at temperatures <10 °C, while alternative carbon pathways (such as the glyoxylate and methyglyoxal pathways) were activated (45). Although the cold-adapted terrestrial bacterium Planococcus halocryophilus (isolated from Arctic permafrost) does encode an ED pathway gene (2-dehydro-3-deoxyphosphogluconate aldolase, BBI08_12080), the full function of its ED pathway has not been studied; the ED pathway may represent a distinctive metabolic feature for the survival and successful competition of psychrophilic bacteria in the cold marine environment.
Compared with E. coli’s faster metabolism, 34H shows flexibility in its flux network, as well as the capacity to use multiple carbon sources and employ anaplerotic pathways to regulate its TCA cycle. It also demonstrates some unique metabolic regulations in the cold. It accumulated larger intracellular pools of central metabolites at normal-growth temperature (e.g., TCA cycle metabolites, including glutamate, which are known osmoprotectants) (47), but smaller pools of glycolytic intermediates and intracellular energy metabolites (e.g., ATP, NADH, NADPH); it did not accumulate the overflow byproduct acetate, possibly due to its slower metabolism. At a warmer stressful room temperature, 34H still survives and engages in chemotaxis in the short term (24), but our transcriptomic and dynamic labeling data show that its catabolic pathways and the majority of flagella-associated genes become significantly down-regulated. Psychrophilic 34H clearly has genetic mechanisms that shut down metabolism under longer-term high-temperature stress (24), which may lead to irreparable cell damage, leaving it with only short-term survival strategies in warm waters.
Materials and Methods
Chemicals and Growth Conditions.
Difco Marine Broth 2216 was purchased from Becton, Dickinson and Company. All other chemicals and 13C-labeled substrates were purchased from Sigma-Aldrich. 34H was grown in half-organic-strength Marine Broth 2216 or in ASW (per liter: 24 g NaCl, 7 g MgSO4•7H2O, 0.7 g KCl, 5.3 g MgCl2•6H2O, 1.3 g TAPSO buffer, 2 g NH4Cl, pH 7.6) (26) medium supplemented with 1 g⋅L−1 glucose, lactate, or acetate and, as vitamins enhanced growth in this minimal medium, 5 mL⋅L−1 of 100× RPMI 1640 Vitamin Mix in Dulbecco’s phosphate buffer solution (Sigma-Aldrich). Cell cultures were grown in 125-mL glass flasks at 4 °C with an agitation rate of 50 rpm. E. coli cells were grown in Luria–Bertani liquid medium or M9 minimal media [per liter: 200 mL 5× M9 salts (Sigma-Aldrich), 2 mL 1 M MgSO4, 100 μL 1 M CaCl2] supplemented with 1 g⋅L−1 glucose. Growth was monitored by measuring OD600 using a UV-visible spectrophotometer.
Biomass Composition Analysis.
The lipid composition was performed by Microbial ID, Inc. For glycogen measurement, 5 mg of dried biomass was digested with 1 mL of 30% KOH at 100 °C for 20 min. Contents were then cooled and glycogen was precipitated by adding 1 mL of 95% ethanol and boiling the sample in a hot water bath. Ten milliliters of a 0.2% anthrone reagent solution and 5 mL of ddH2O were used to dissolve the precipitate. Absorption at 680 nm was measured against a series of standards. Protein and amino acid composition analysis was performed by the Molecular Structure Facility at University of California Davis (msf.ucdavis.edu/). The ash measurement was obtained from heating dried biomass in a Lindberg Blue Muffle Furnace (Thermo Fisher Scientific) to 600 °C for 24 h. DNA and RNA were estimated using the protein:DNA and DNA:RNA ratios from E. coli (33, 34).
13C Cultures and Dynamic Labeling Experiments.
A stock culture of 34H was inoculated into half-organic-strength marine broth medium. Cells were grown to an OD600 of 0.40 and then inoculated at a ratio of 0.5 vol/vol% into ASW medium containing the appropriate 13C carbon substrates ([1-13C] glucose, [1,2-13C] glucose, [U-13C] glucose, [U-13C] glucose, unlabeled glyoxalate, unlabeled glucose, [1,2-13C] acetate, or [3-13C] lactate). Cells were subcultured in the same medium at an inoculation ratio of 0.5 vol/vol% to minimize carryover of unlabeled carbon. Biomass samples for fingerprinting experiments and MFA were harvested from exponentially growing cells and stored at −20 °C until analysis. For dynamic labeling, cultures grown with unlabeled glucose were pulsed with a concentrated [U-13C] glucose solution to obtain a final concentration of 1 g⋅L−1 of labeled glucose. The heat-stressed dynamic labeled 34H cultures were moved from 4 °C to room temperature (23 °C) at an OD600 of 0.35 and allowed to acclimate for 1 h before pulsing labeled glucose. E. coli cultures were inoculated from Luria–Bertani liquid plates into 5 mL Luria–Bertani liquid media and grown at 37 °C overnight. M9 minimal medium was inoculated at 0.3 vol/vol%. At an OD600 of ∼0.8, E. coli cultures were moved to a 4 °C room and acclimated for 1 h (temperature of the culture monitored with a thermometer) before pulsing with labeled glucose (final labeled glucose concentration of 1 g⋅L−1).
Comparison of Metabolite Concentrations.
Metabolite concentrations in 34H were analyzed using an approach based on isotope ratios. Specifically, E. coli was grown with U-13C glucose (1g⋅L−1) and 13C-bicarbonate (1g⋅L−1) at 37 °C, while 34H was cultured with unlabeled medium at 4 °C. For metabolite quantification, fully labeled E. coli cells served as internal standards that were mixed with 34H cultures. The mixed biomass was quenched to ∼0 °C (temperature monitored with a thermometer) using a liquid nitrogen bath before metabolite extraction for LC-MS analysis. The resulting isotopic ratio of each metabolite (13C labeled vs. unlabeled) was normalized by biomass and labeling enrichment of E. coli to obtain the relative 34H metabolite pool sizes by benchmarking them against E. coli.
GC-MS Analysis.
The proteinogenic amino acids were hydrolyzed using 6 N HCl at 100 °C. Samples were centrifuged at 9,500 × g for 5 min to pellet cell debris. The supernatant was transferred to a new tube and dried by filtered air for 8 h. The amino acids were derivatized with N-(tert-butyldimethylsilyl)-N-methyl-trifluoroacetamide containing an equal volume of the solvent, tetrahydrofuran (29), and then analyzed for their 13C mass fraction using GC-MS. The m/z ions [M-57]+ or [M-15]+, and [M-159]+ or [M-85]+ were used for isotopic tracing. The [M-15]+ fragment was used for leucine and isoleucine because their [M-57]+ fragment overlaps with other mass peaks. The mass isotopomer distributions obtained were then corrected for natural stable isotopes using a previously described algorithm (48). For the anaplerotic pathway P value, the labeling data of the TCA cycle-derived and non-TCA cycle-derived amino acids were compared using a Student’s t test. Similarly, the P value between the glyoxylate shunt and labeled-13C glucose was obtained using labeling data of the TCA cycle-derived amino acids.
LC-MS Analysis.
At the specified time points, pulsed cultures were quick quenched using liquid N2, centrifuged at 5,000 × g for 5 min at 0 °C, and washed with 0.9% NaCl before being repelleted. Samples were then prepared as previously described (35) and run according to published protocols (35, 49). Full details are provided in SI Appendix, Methods.
MFA.
MFA was performed with the free software, WUFlux that we developed (36). Briefly, the built-in MATLAB function “fmincon” was employed for nonlinear optimization, which minimizes the sum of squared residuals (SSR) between experimentally and computationally determined data weighted by measured variances. The Monte Carlo method was used to determine the 95% confidence intervals of the central metabolic pathways, and the χ2 test was applied to determine the goodness of fit.
RNA-Sequencing and Data Analysis.
Transcriptomic data from 34H were obtained under normal-growth conditions at 4 °C and after 2 h of exposure to 23 °C. Exponentially growing cells were pelleted and immediately frozen using liquid nitrogen. RNA was extracted using a Quick-RNA Miniprep Kit (Zymo). RNA profiling (library preparation, sequencing, and data analysis) was performed by the Genome Technology Access Center at Washington University (https://gtac.wustl.edu/) as previously described (29). Biological triplicates were sampled at each condition.
Thermodynamic Calculations.
The equilibrator calculator was used for initial estimation with the ionic strength as an input (43). The change in Gibb’s free energy formula (SI Appendix, Methods) was used to estimate the change of free energy from standard temperature to 4 °C.
Analysis of Engineered ED Pathway in E. coli.
The ∆ptsG and ∆ptsG + ED E. coli mutants were generated as reported previously (44). Briefly, the ∆ptsG strain was obtained from the Keio Collection (Coli Genetic Stock Center) and transformed with the plasmid carrying the ED pathway genes (edd and eda). The strains were grown in M9 minimal medium supplemented with 2 g⋅L−1 unlabeled glucose in shaking flasks (200 rpm, 37 °C). Exponentially growing cultures were transferred to 4 °C and allowed to equilibrate for 2 h (200 rpm) before being pulsed with [U-13C] glucose for a final concentration of 4 g⋅L−1. Cells were harvested at specified time points after the pulse, and the metabolites were extracted using the chloroform–methanol method and analyzed on LC-MS as previously described. All time points were completed in duplicate.
Compliance with Ethical Standards.
This article does not contain any studies with human participants or animals performed by any of the authors.
Supplementary Material
Acknowledgments
We thank Whitney Hollingshead for preparing cell cultures and the Genome Technology Access Center (Department of Genetics, Washington University School of Medicine) for RNA-Seq analysis. This work was also part of the Department of Energy (DOE) Joint BioEnergy Institute (www.jbei.org) supported by the DOE Office of Science, Office of Biological and Environmental Research, through Contract DE-AC02-05CH11231 between the Lawrence Berkeley National Laboratory and the DOE. The Donald Danforth Plant Science Center QTRAP LC-MS/MS acquisition was supported by National Science Foundation (NSF) Grant DBI-1427621. J.J.C. was supported by NSF Grant DBI-1356669 and J.W.D by the Karl M. Banse Professorship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807804115/-/DCSupplemental.
References
- 1.Ramond JB, Makhalanyane TP, Tuffin MI, Cowan DA. Normalization of environmental metagenomic DNA enhances the discovery of under-represented microbial community members. Lett Appl Microbiol. 2015;60:359–366. doi: 10.1111/lam.12380. [DOI] [PubMed] [Google Scholar]
- 2.Olson DG, et al. Glycolysis without pyruvate kinase in Clostridium thermocellum. Metab Eng. 2017;39:169–180. doi: 10.1016/j.ymben.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 3.McKinlay JB, et al. Non-growing Rhodopseudomonas palustris increases the hydrogen gas yield from acetate by shifting from the glyoxylate shunt to the tricarboxylic acid cycle. J Biol Chem. 2014;289:1960–1970. doi: 10.1074/jbc.M113.527515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cordova LT, Cipolla RM, Swarup A, Long CP, Antoniewicz MR. 13C metabolic flux analysis of three divergent extremely thermophilic bacteria: Geobacillus sp. LC300, Thermus thermophilus HB8, and Rhodothermus marinus DSM 4252. Metab Eng. 2017;44:182–190. doi: 10.1016/j.ymben.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jazmin LJ, et al. Isotopically nonstationary 13C flux analysis of cyanobacterial isobutyraldehyde production. Metab Eng. 2017;42:9–18. doi: 10.1016/j.ymben.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowman JP. Genomic analysis of psychrophilic prokaryotes. In: Margesin R, Schinner F, Marx JC, Gerday C, editors. Psychrophiles: From Biodiversity to Biotechnology. Springer; Berlin: 2008. pp. 265–284. [Google Scholar]
- 7.Allen MA, et al. The genome sequence of the psychrophilic archaeon, Methanococcoides burtonii: The role of genome evolution in cold adaptation. ISME J. 2009;3:1012–1035. doi: 10.1038/ismej.2009.45. [DOI] [PubMed] [Google Scholar]
- 8.Murray AE, Grzymski JJ. Diversity and genomics of Antarctic marine micro-organisms. Philos Trans R Soc Lond B Biol Sci. 2007;362:2259–2271. doi: 10.1098/rstb.2006.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pickard GL, Emery WJ. Descriptive Physical Oceanography: An Introduction. 5th Ed Pergamon Press; New York: 1990. [Google Scholar]
- 10.Huston AL, Krieger-Brockett BB, Deming JW. Remarkably low temperature optima for extracellular enzyme activity from Arctic bacteria and sea ice. Environ Microbiol. 2000;2:383–388. doi: 10.1046/j.1462-2920.2000.00118.x. [DOI] [PubMed] [Google Scholar]
- 11.Boetius A, Anesio AM, Deming JW, Mikucki JA, Rapp JZ. Microbial ecology of the cryosphere: Sea ice and glacial habitats. Nat Rev Microbiol. 2015;13:677–690. doi: 10.1038/nrmicro3522. [DOI] [PubMed] [Google Scholar]
- 12.Techtmann SM, et al. Colwellia psychrerythraea strains from distant deep sea basins show adaptation to local conditions. Front Environ Sci. 2016;4:33. [Google Scholar]
- 13.Béjà O, Spudich EN, Spudich JL, Leclerc M, DeLong EF. Proteorhodopsin phototrophy in the ocean. Nature. 2001;411:786–789. doi: 10.1038/35081051. [DOI] [PubMed] [Google Scholar]
- 14.Methé BA, et al. The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc Natl Acad Sci USA. 2005;102:10913–10918. doi: 10.1073/pnas.0504766102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mason OU, Han J, Woyke T, Jansson JK. Single-cell genomics reveals features of a Colwellia species that was dominant during the Deepwater Horizon oil spill. Front Microbiol. 2014;5:332. doi: 10.3389/fmicb.2014.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valentine DL, et al. Propane respiration jump-starts microbial response to a deep oil spill. Science. 2010;330:208–211. doi: 10.1126/science.1196830. [DOI] [PubMed] [Google Scholar]
- 17.Redmond MC, Valentine DL. Natural gas and temperature structured a microbial community response to the Deepwater Horizon oil spill. Proc Natl Acad Sci USA. 2012;109:20292–20297. doi: 10.1073/pnas.1108756108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carillo S, et al. A unique capsular polysaccharide structure from the psychrophilic marine bacterium Colwellia psychrerythraea 34H that mimics antifreeze (glyco)proteins. J Am Chem Soc. 2015;137:179–189. doi: 10.1021/ja5075954. [DOI] [PubMed] [Google Scholar]
- 19.Llyd EW, Deming JW. Characterization of a cold-active bacteriophage on two psychrophilic marine hosts. Aquat Microb Ecol. 2006;45:15–29. [Google Scholar]
- 20.Yamauchi S, Ueda Y, Matsumoto M, Inoue U, Hayashi H. Distinct features of protein folding by the GroEL system from a psychrophilic bacterium, Colwellia psychrerythraea 34H. Extremophiles. 2012;16:871–882. doi: 10.1007/s00792-012-0483-7. [DOI] [PubMed] [Google Scholar]
- 21.Huston AL, Methe B, Deming JW. Purification, characterization, and sequencing of an extracellular cold-active aminopeptidase produced by marine psychrophile Colwellia psychrerythraea strain 34H. Appl Environ Microbiol. 2004;70:3321–3328. doi: 10.1128/AEM.70.6.3321-3328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marx JG, Carpenter SD, Deming JW. Production of cryoprotectant extracellular polysaccharide substances (EPS) by the marine psychrophilic bacterium Colwellia psychrerythraea strain 34H under extreme conditions. Can J Microbiol. 2009;55:63–72. doi: 10.1139/W08-130. [DOI] [PubMed] [Google Scholar]
- 23.Junge K, Eicken H, Deming JW. Motility of Colwellia psychrerythraea strain 34H at subzero temperatures. Appl Environ Microbiol. 2003;69:4282–4284. doi: 10.1128/AEM.69.7.4282-4284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Showalter GM, Deming JW. Low-temperature chemotaxis, halotaxis and chemohalotaxis by the psychrophilic marine bacterium Colwellia psychrerythraea 34H. Environ Microbiol Rep. 2018;10:92–101. doi: 10.1111/1758-2229.12610. [DOI] [PubMed] [Google Scholar]
- 25.Kavitha M. Cold active lipases—An update. Front Life Sci. 2016;9:226–238. [Google Scholar]
- 26.Smetacek V, Nicol S. Polar ocean ecosystems in a changing world. Nature. 2005;437:362–368. doi: 10.1038/nature04161. [DOI] [PubMed] [Google Scholar]
- 27.Colangelo-Lillis JR, Deming JW. Genomic analysis of cold-active Colwelliaphage 9A and psychrophilic phage-host interactions. Extremophiles. 2013;17:99–114. doi: 10.1007/s00792-012-0497-1. [DOI] [PubMed] [Google Scholar]
- 28.Tang JK, You L, Blankenship RE, Tang YJ. Recent advances in mapping environmental microbial metabolisms through 13C isotopic fingerprints. J R Soc Interface. 2012;9:2767–2780. doi: 10.1098/rsif.2012.0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Au J, Choi J, Jones SW, Venkataramanan KP, Antoniewicz MR. Parallel labeling experiments validate Clostridium acetobutylicum metabolic network model for (13)C metabolic flux analysis. Metab Eng. 2014;26:23–33. doi: 10.1016/j.ymben.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 30.Haverkorn van Rijsewijk BR, Nanchen A, Nallet S, Kleijn RJ, Sauer U. Large-scale 13C-flux analysis reveals distinct transcriptional control of respiratory and fermentative metabolism in Escherichia coli. Mol Syst Biol. 2011;7:477. doi: 10.1038/msb.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishii N, et al. Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science. 2007;316:593–597. doi: 10.1126/science.1132067. [DOI] [PubMed] [Google Scholar]
- 32.Neidhardt FC, Ingraham JL, Schaechter M. 1990. Physiology of the Bacterial Cell: A Molecular Approach (Sinauer Associates, Sunderland, MA), p xii, 506 p.
- 33.Pramanik J, Keasling JD. Stoichiometric model of Escherichia coli metabolism: Incorporation of growth-rate dependent biomass composition and mechanistic energy requirements. Biotechnol Bioeng. 1997;56:398–421. doi: 10.1002/(SICI)1097-0290(19971120)56:4<398::AID-BIT6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 34.Pramanik J, Keasling JD. Effect of Escherichia coli biomass composition on central metabolic fluxes predicted by a stoichiometric model. Biotechnol Bioeng. 1998;60:230–238. doi: 10.1002/(sici)1097-0290(19981020)60:2<230::aid-bit10>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 35.Abernathy MH, et al. Deciphering cyanobacterial phenotypes for fast photoautotrophic growth via isotopically nonstationary metabolic flux analysis. Biotechnol Biofuels. 2017;10:273. doi: 10.1186/s13068-017-0958-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He L, Wu SG, Zhang M, Chen Y, Tang YJ. WUFlux: An open-source platform for 13C metabolic flux analysis of bacterial metabolism. BMC Bioinformatics. 2016;17:444. doi: 10.1186/s12859-016-1314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wan N, et al. Cyanobacterial carbon metabolism: Fluxome plasticity and oxygen dependence. Biotechnol Bioeng. 2017;114:1593–1602. doi: 10.1002/bit.26287. [DOI] [PubMed] [Google Scholar]
- 38.Casanueva A, Tuffin M, Cary C, Cowan DA. Molecular adaptations to psychrophily: The impact of ‘omic’ technologies. Trends Microbiol. 2010;18:374–381. doi: 10.1016/j.tim.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 39.Firth E, Carpenter SD, Sorensen HL, Collins RE, Deming JW. Bacterial use of choline to tolerate salinity shifts in sea-ice brines. Elem Sci Anth. 2016;4:000120. [Google Scholar]
- 40.Flamholz A, Noor E, Bar-Even A, Liebermeister W, Milo R. Glycolytic strategy as a tradeoff between energy yield and protein cost. Proc Natl Acad Sci USA. 2013;110:10039–10044. doi: 10.1073/pnas.1215283110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klingner A, et al. Large-Scale 13C flux profiling reveals conservation of the Entner-Doudoroff pathway as a glycolytic strategy among marine bacteria that use glucose. Appl Environ Microbiol. 2015;81:2408–2422. doi: 10.1128/AEM.03157-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noor E, et al. Pathway thermodynamics highlights kinetic obstacles in central metabolism. PLOS Comput Biol. 2014;10:e1003483. doi: 10.1371/journal.pcbi.1003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noor E, et al. An integrated open framework for thermodynamics of reactions that combines accuracy and coverage. Bioinformatics. 2012;28:2037–2044. doi: 10.1093/bioinformatics/bts317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abernathy MH, et al. Comparative studies of glycolytic pathways and channeling under in vitro and in vivo modes. AIChE J. July 30, 2018 doi: 10.1002/aic.16367. [DOI] [Google Scholar]
- 45.Tribelli PM, López NI. Reporting key features in cold-adapted bacteria. Life (Basel) 2018;8:E8. doi: 10.3390/life8010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raymond-Bouchard I, Whyte LG. In: From Transcriptomes to Metatranscriptomes: Cold Adaption and Active Metabolisms of Psychrophiles from Cold Environments. Psychrophiles from Biodiversity to Biotechnology. Margesin R, editor. Springer International Publishing; New York: 2017. pp. 437–457. [Google Scholar]
- 47.Sévin DC, Stählin JN, Pollak GR, Kuehne A, Sauer U. Global metabolic responses to salt stress in fifteen species. PLoS One. 2016;11:e0148888. doi: 10.1371/journal.pone.0148888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wahl SA, Dauner M, Wiechert W. New tools for mass isotopomer data evaluation in (13)C flux analysis: Mass isotope correction, data consistency checking, and precursor relationships. Biotechnol Bioeng. 2004;85:259–268. doi: 10.1002/bit.10909. [DOI] [PubMed] [Google Scholar]
- 49.Ma F, Jazmin LJ, Young JD, Allen DK. Isotopically nonstationary 13C flux analysis of changes in Arabidopsis thaliana leaf metabolism due to high light acclimation. Proc Natl Acad Sci USA. 2014;111:16967–16972. doi: 10.1073/pnas.1319485111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.