Significance
Over 50 years ago, Dan Janzen proposed an integrative framework relating latitudinal differences in climate variability to elevational trends in species diversity. We show that tropical species in three independent insect clades have (i) narrower thermal breadths, (ii) decreased dispersal and higher population structure, and (iii) higher cryptic diversity and speciation rates. This research tests all of the key predictions of Janzen’s hypothesis in related taxa. Our work advances the understanding of how climate variability shapes global diversity patterns, moving beyond simple correlations, to mechanistic links between climate, local adaptation, dispersal, and montane species richness.
Keywords: climate variability, thermal tolerance, elevation gradient, speciation, cryptic diversity
Abstract
Species richness is greatest in the tropics, and much of this diversity is concentrated in mountains. Janzen proposed that reduced seasonal temperature variation selects for narrower thermal tolerances and limited dispersal along tropical elevation gradients [Janzen DH (1967) Am Nat 101:233–249]. These locally adapted traits should, in turn, promote reproductive isolation and higher speciation rates in tropical mountains compared with temperate ones. Here, we show that tropical and temperate montane stream insects have diverged in thermal tolerance and dispersal capacity, two key traits that are drivers of isolation in montane populations. Tropical species in each of three insect clades have markedly narrower thermal tolerances and lower dispersal than temperate species, resulting in significantly greater population divergence, higher cryptic diversity, higher tropical speciation rates, and greater accumulation of species over time. Our study also indicates that tropical montane species, with narrower thermal tolerance and reduced dispersal ability, will be especially vulnerable to rapid climate change.
Tropical montane regions are among the most biodiverse ecosystems on earth, with higher species richness and endemism than temperate mountains (1–3). Numerous hypotheses have been proposed to explain these global patterns of biodiversity, yet few provide a mechanistic explanation for why tropical mountains generate or harbor more species than mountains at higher latitudes (2, 3). The Climate Variability Hypothesis (CVH) postulates that seasonal climatic variation increases with latitude, selecting for species with broader thermal tolerances and higher dispersal capacities (4–6). In 1967, Janzen (7) extended the CVH to elevation gradients, hypothesizing that reduced seasonality produces greater thermal stratification along tropical mountains, selecting for narrower thermal tolerances, and therefore limiting dispersal across tropical elevation gradients (8). Over time, dispersal limitation should result in population divergence and eventually speciation (9, 10), and if sustained over longer time periods, higher species richness in tropical mountains (2).
Janzen’s 1967 framework is compelling because it provides a mechanistic explanation for how climatic variation shapes thermal physiology and dispersal and, ultimately, higher rates of speciation in tropical mountains (10). Several studies have confirmed some of Janzen’s predictions: many tropical species have narrower thermal tolerances (11–13), exhibit greater population genetic differentiation (7, 14), occupy narrower elevation ranges (15, 16), and have greater species turnover along elevation gradients (17) than their temperate counterparts. However, a definitive test of the mechanisms linking climate variability and tropical montane diversity has remained elusive because it requires the collection and integration of large and diverse datasets of physiological traits, gene flow, species diversity, and speciation rates for related montane taxa from both tropical and temperate regions (2, 10).
Here, we quantify thermal tolerance and dispersal traits of tropical and temperate species in three distinct stream insect orders—mayflies (Ephemeroptera), stoneflies (Plecoptera), and caddisflies (Trichoptera)—and examine their role in promoting higher speciation rates in tropical (Ecuadorian Andes) versus temperate (Colorado Rockies) montane streams. First, we characterized the thermal tolerance of 35 tropical and 27 temperate phylogenetically paired taxa to test if thermal breadth covaries with climate variability across latitude (13). Second, we estimated effective dispersal of four tropical and four temperate taxa along each elevation gradient to test if gene flow is reduced and population genetic divergence is higher in tropical mountains. Third, we quantified differences in regional taxonomic richness and cryptic diversity based on analyses of 20 tropical and 26 temperate stream insect communities to test if they are higher in the tropics. The integration of these empirical measurements provides a unique test of the link between climate variability and trait-mediated diversification rates in tropical and temperate mountains (Dataset S1).
Results and Discussion
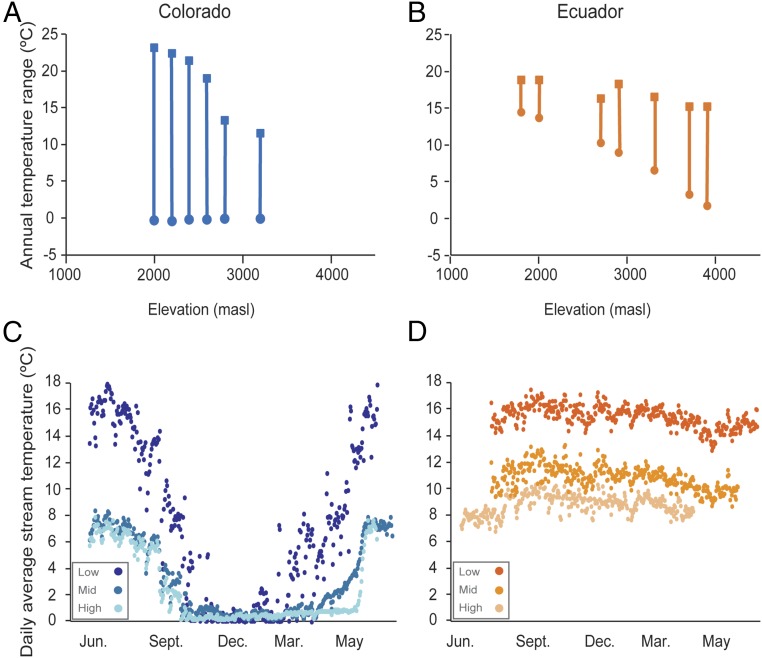
We found that stream temperature variation and the degree of overlap in temperature across elevation were greater in Colorado than in Ecuador, confirming the climatic context for regional differences in selection and diversification (13, 18) (Fig. 1). As predicted, tropical species, defined first by using morphology and then delimited by DNA barcoding (hereafter referred to as “barcoded species”), had narrower thermal breadths than their temperate counterparts (Fig. 2A). Specifically, when acclimated to a common temperature, temperate insects from a given elevation had on average a higher critical thermal maximum (CTMAX) and a lower critical thermal minimum (CTMIN) than phylogenetically paired tropical insects at the same elevation (Fig. 2A and SI Appendix, Table S1) (13). The broader thermal breadth (i.e., the difference between CTMAX and CTMIN) in temperate barcoded species is most pronounced at low elevations because temperate barcoded species exhibit significantly lower CTMIN values than tropical ones, despite being acclimated to the same temperature (Fig. 2A and SI Appendix, Table S1) (13).
Fig. 1.
Variation in annual temperature overlap and stream temperature profiles for temperate and tropical streams. Annual maximum (squares) and minimum (circles) temperatures show that temperate streams (A) have greater annual temperature variation than tropical streams (B). Temperature profiles (daily average temperature over 1 y) at low, mid-, and high elevation streams show that temperate streams (C) show higher seasonal variance than tropical ones (D) at all elevations.
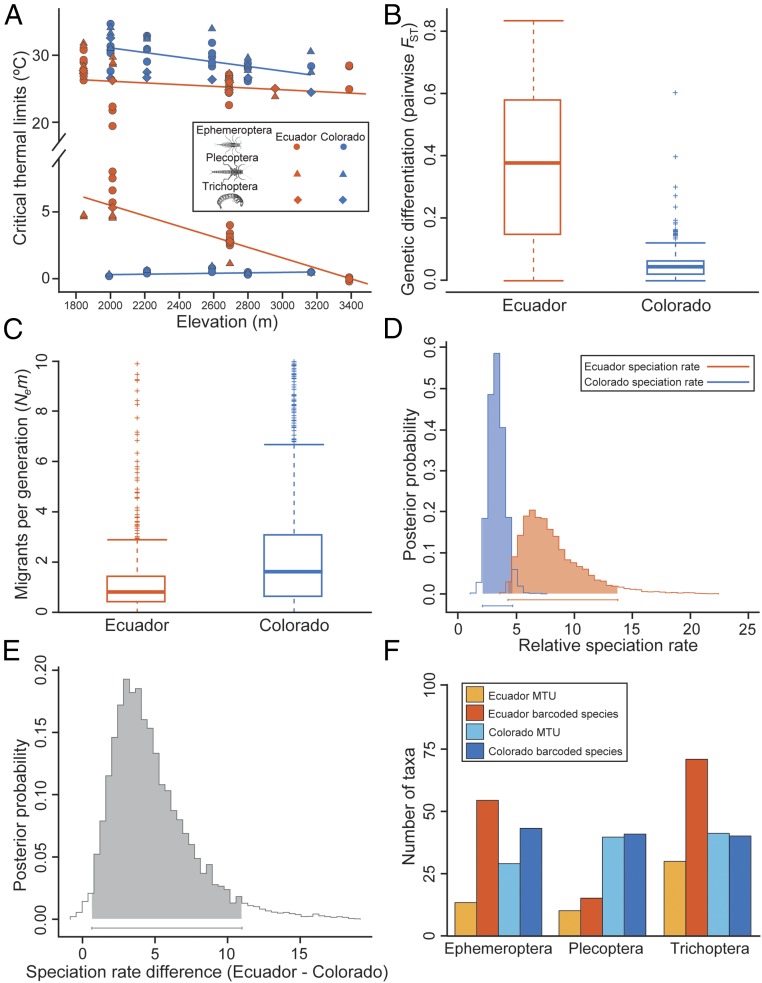
Fig. 2.
Variation in physiological and dispersal traits leading to higher speciation rates in tropical stream insects. (A) Critical thermal limits as a function of elevation. In tropical barcoded species (orange), CTMAX temperatures were on average lower and CTMIN temperatures were higher compared with temperate relatives (blue) across elevations. Thus, thermal breadths are narrower in tropical insects at all elevations. (B and C) Dispersal, quantified as pairwise population FST and the effective number of migrants (Nem) per generation, indicates more extensive movement and higher levels of genetic connectivity among higher latitude sites in Colorado. (D and E) Bayesian parameter estimation for the full BiSSE model shows that speciation rates are higher in stream insect communities in the Ecuadorian Andes than in the Colorado Rockies. (F) The number of barcoded species compared with those identified with morphology alone (MTUs) was higher in Ecuador compared with Colorado, confirming higher cryptic diversity in the tropics.
We next characterized fine-scale gene flow along elevation gradients for four tropical and four temperate species complexes paired to represent all three insect clades. For gene flow analyses, we used morphological taxonomic units (MTUs)—i.e., species identified based on morphology alone—that often comprise species complexes. Both in Ecuador and in Colorado, the focal MTUs are known to harbor divergent lineages and/or undescribed diversity across their ranges. We chose MTUs for gene flow analyses because our goal was to quantify how much genetic differentiation and gene flow occur along the entire extent of the elevational gradient, and thus across boundaries of diversifying lineages and potential cryptic species in each region. We used double-digest restriction-site associated DNA sequencing (ddRADseq) (19) to genotype hundreds of individuals (316–990) at hundreds to thousands of SNP loci (419–4,544) (SI Appendix, Table S2). Clustering of genotypes into genetic demes revealed that genetic structure was lower in all MTUs from Colorado (mean = 2.75 demes, SD = 1.5) than in those from Ecuador (mean = 11.5 demes, SD = 3.9; SI Appendix, Fig. S1). Estimates of pairwise population genetic differentiation (FST) and effective migration (Nem) confirmed overall lower levels of dispersal and population connectivity along the tropical gradient. The ranges of FST values between populations along the elevation gradient were lower in Colorado, indicating higher levels of interbreeding among temperate populations (Fig. 2B), and this finding was concordant with higher estimated migration rates among populations in Colorado than in Ecuador (P < 0.001) (Fig. 2C and SI Appendix, Fig. S2).
To be comparable, estimates of gene flow among populations in different regions must account for landscape features. We applied a linear mixed-effects model to quantify the effects of geographic distance and elevation on population genetic differentiation at each latitude. The best-fit model indicated significant interactions between latitude and geographic (Euclidean) distance (F1,1154 = 1.86; P = 0.012) and latitude and elevation difference (F1,1154 = 33.20; P < 0.001), confirming that elevation and geographic distance have greater effects on genetic differentiation among tropical stream insect populations compared with temperate ones (SI Appendix, Table S3). Combined, our population genomic results show extensive population structure and reduced dispersal rates in tropical taxa in each of the three clades, resulting from higher isolation-by-distance and isolation-by-elevation in tropical mountains.
To compare regional taxonomic richness, we surveyed all taxa in the three focal clades along our tropical and temperate elevation gradients and quantified the number of MTUs (11,433 specimens examined: Ecuador—4,511 and Colorado—6,922) and barcoded species (3,980 specimens barcoded: Ecuador—1,495; Colorado—2,485) (9) (Dataset S2). The difference between MTU and barcoded species at a survey site is a measure of cryptic diversity. Our surveys revealed higher MTU richness in Colorado than in Ecuador, but this pattern was reversed in two of the insect orders when taking cryptic diversity into account. Barcoded species richness was higher in Ecuador than in Colorado for Ephemeroptera (54 vs. 41 barcoded species) and Trichoptera (71 vs. 35 barcoded species), but not Plecoptera (14 vs. 36 barcoded species; Fig. 2F). This result underscores the importance of quantifying cryptic diversity in studies of richness, especially when comparing regions where species discovery and taxonomic knowledge are very different (20).
To test whether ancestral location (temperate or tropical) could explain among-clade differences in barcoded species diversity, we estimated Bayesian supertree phylogenies for the barcoded species sampled in our regional taxonomic surveys and inferred the location of the common ancestor for barcoded species in each clade (SI Appendix, Fig. S3). We found that the Trichoptera barcoded species in our study most likely had a tropical common ancestor, whereas the ancestors of Ephemeroptera and Plecoptera were temperate (SI Appendix, Fig. S3). Thus, the Trichoptera barcoded species that we sampled have had a longer history in the tropics, followed by the Ephemeroptera. In contrast, the Plecoptera barcoded species have had the shortest time to diversify in the tropics (21–24). Despite these differences in biogeographic history, we observed consistently larger increases in the number of barcoded species vs. MTUs in Ecuador compared with Colorado (Ephemeroptera: +350% vs. +40%; Plecoptera: +56% vs. +3%; Trichoptera: +137% vs. −3%; Fig. 2F), indicating higher levels of cryptic diversity in the tropics, irrespective of the time for tropical speciation in each clade.
Finally, to test for latitudinal differences in speciation rates, we used binary state speciation and extinction models (BiSSE) (25). For the dataset including barcoded species for all three clades combined, the full BiSSE model with parameters estimated using Markov Chain Monte Carlo (MCMC) inferred a higher speciation rate for Ecuador compared with Colorado (Fig. 2D); the 95% credibility interval for the difference in speciation rates (Ecuador–Colorado) was positive and did not include zero (Fig. 2E and SI Appendix, Fig. S4). For barcoded species in each insect order, we also fit full BiSSE models to estimate parameter distributions using maximum likelihood and 20,000 post burn-in trees that were used to create the clade supertrees. We then compared rates across latitude by determining latitudinal differences in parameter estimates and examining the 95% credibility interval for these rate differences. For individual clades, we found consistent trends toward higher speciation rates in Ecuador compared with Colorado, with stronger effects for the Trichoptera and Plecoptera than the Ephemeroptera (SI Appendix, Fig. S5). Because these diversification analyses are based on a phylogeny of barcoded species, the estimates of speciation are not biased by regional differences in historical taxonomic effort and cryptic diversity; they show that the rate of origination of barcoded species is higher in the tropics independently of the number of barcoded species in each MTU in each region. Overall, our results demonstrate narrower thermal breadths, less gene flow, higher population divergences, higher cryptic diversity, and higher speciation rates in the tropics.
Empirical support for the CVH has been equivocal (2, 10), likely because most studies use range size as a proxy for dispersal capacity and latitude as a proxy for climatic variability, thereby disregarding important variance in organismal traits and local environmental fluctuations within latitudes (15, 26). Range sizes of organisms are a consequence of individual species’ responses to many environmental and biotic factors, as well as to historical contingencies (1, 27), and thus are imprecise proxies for selection on thermal tolerance and dispersal, the phenotypic traits that contribute directly to diversification (8). Our study shows that quantifying these key organismal traits provides a more mechanistic understanding of how populations diverge across environmental (selection) gradients (28, 29). We do not claim that other biotic factors, such as species competition and host–pathogen dynamics, are unimportant in determining the boundaries of species’ ranges; many studies clearly show that these factors are significant (30, 31). However, in the case of strong abiotic gradients, such as those found in mountains, the effects of climatic variability on local adaptation and dispersal are likely major drivers of diversification. Analyses of these organismal traits reveal commonalities and idiosyncrasies in rates of diversification among taxa, and they also have the potential to predict species’ responses to changing climate (32).
Our integrative approach provides the strongest evidence to date that reduced climate variability favors narrow thermal tolerance and reduced dispersal across elevational gradients, which in turn increase speciation rates. We were able to test the CVH by linking microevolutionary processes at the population genomic level to macroevolutionary patterns of species diversification, an approach that may be the key to testing the generality of the CVH in other taxonomic groups (2). Our study demonstrates the insights possible when integrating data on processes operating at different temporal scales—from local thermal adaptation, to ongoing gene flow among adjacent populations, to estimates of speciation that occurred in the distant past. For example, the magnitude of genetic divergence among populations within a species could be an indicator of the potential for speciation (2) because the likelihood of reproductive isolation increases with genetic distance (33). In fact, surveys in vertebrates show that the magnitude of population genetic differentiation within species parallels the species-richness gradients observed at a global scale (7, 34), a pattern expected if most incipient species reach species status. Our findings confirm that within-species processes of population divergence are indeed mirrored in among-species patterns of cryptic diversity and speciation rates at different latitudes. Furthermore, our results underscore how a focus on species richness alone can fail to reveal important aspects of global patterns in biodiversity. For example, we found Plecoptera species richness to be lower in the Andes compared with the Rockies; however, this recently arrived group is also speciating at a higher rate in the tropics, which may eventually produce greater species diversity.
Finally, our study has important implications for predicting the vulnerability of species to climate change. Global warming is projected to increase seasonal variability and alter daily maximum and minimum temperatures (35, 36), with potentially negative consequences for species’ persistence and future diversification (37). Tropical ectotherms persist close to the edge of their evolved thermal tolerances (38), and thus, paradoxically, the same traits that promote high speciation and diversity are the ones that render them especially vulnerable to rapid changes in thermal environments. Characterizing species in terms of traits that are directly linked to global warming (e.g., thermal tolerance) or that represent potential adaptive capacity to adjust (e.g., dispersal ability) provides a powerful framework for predicting evolutionary responses, species range shifts (39), and ecological changes (40) in natural communities across geographic gradients of projected climate change.
Methods
Study Area and Collection.
To characterize temperature profiles at different elevations for streams in both regions, we measured stream temperature using Solinst temperature/level loggers in the Poudre River drainage in Colorado and HOBO level loggers in the Papallacta River drainage in Ecuador over a 12-mo period between 2013 and 2015. Loggers recorded temperature at 15-min intervals each day. We calculated daily average temperatures over the course of a year for each stream to characterize variation in stream temperature.
We collected aquatic insect samples from small (first–third order), minimally impacted montane streams at ∼200-m elevation intervals from 1,556 to 3,478 meters above sea level (m asl) in Colorado (Cache la Poudre basin in the Front Range of the Rocky Mountains) (9) and 1,664–4,248 m asl in Ecuador (Napo basin in the Oriental Andean Cordillera) (41). To collect insects, we used D-frame nets with 500-µm mesh and searched under available substrates. From each sample, we separated all aquatic larvae of Ephemeroptera (mayflies), Plecoptera (stoneflies), and Trichoptera (caddisflies). For estimates of regional diversity, we supplemented larval collections with adults collected from streamside vegetation. For measurements of gene flow, we subsampled phylogenetically paired species complexes with broad elevational distributions and sufficient sample sizes across sites (Dataset S1 and SI Appendix, Table S2). All specimens used for genetic analyses were field-preserved in 100% ethanol.
Measuring Latitudinal Differences in Thermal Tolerance.
To test one of the primary predictions of the CVH, we measured the CTMAX and CTMIN temperatures of temperate and tropical barcoded species from various elevations (13). The CVH predicts that tropical species should have narrower thermal breadths, which was calculated as the difference between CTMAX and CTMIN. To account for the large seasonal differences at different latitudes, we conducted experiments in the Colorado Rockies between June and August, when stream temperatures at a given elevation were similar to those in the Ecuadorian Andes. We paired streams at the different latitudes by elevation and acclimated barcoded species from those streams for 48 h at the same temperature (SI Appendix, Table S1). For the experiments, we ramped temperature by 0.3 °C/min until we observed a loss of righting response, at which point individuals were placed in a recovery tank; only individuals that survived the treatment were included in the analysis (13). A critical aspect of this experimental design is that by acclimating barcoded species from similar elevations, but different latitudes, at the same acclimation temperature, we were able to control for environmental effects on measures of thermal breadth. Indeed, we could not acclimate species from all elevations to a single acclimation temperature because attempts to acclimate low elevation tropical species to cooler temperatures or high elevation tropical species to warmer temperatures resulted in obvious signs of thermal stress or mortality (13). To statistically control for evolutionary history while comparing thermal breadth values across latitude and thermal breadth across streams of different temperatures, we used phylogenetic generalized least squares regression (42) fit with an Ornstein–Uhlenbeck model (43, 44) of trait evolution. We also calculated mean critical limits for barcoded species in each taxonomic order and for each elevation for the temperate and tropical comparisons (SI Appendix, Table S1).
Genomic Sequence Data Collection.
SNP genotyping was performed using ddRADseq (19). We performed sample demultiplexing, de novo assembly, and SNP calling for each MTU in Stacks v 1.19 (45) using previously published parameters (46). An initial set of SNPs was filtered to remove loci with minor allele frequencies <0.01 for the demographic analyses in ∂a∂i (47) (SI Appendix, Table S2). For frequency-based analyses, a more stringent filter was applied to remove loci with minor allele frequencies <0.05 and those that were typed in <60% of individuals for a given MTU. We excluded loci in linkage disequilibrium.
Restriction-site associated DNA sequencing tag sequences, SNP data, and detailed collection localities for all eight focal MTUs are available in the Dryad Digital Repository (https://doi.org/10.5061/dryad.m728c47).
Testing for Latitudinal Differences in Gene Flow and Dispersal Rates.
For each MTU, we computed chord distances (Dc) among sampling sites with the dist.genpop function in the R adegenet package (48). We then identified the number of genetic clusters in the data by performing successive K-means clustering, increasing the number of clusters (K) in each model with the find.clusters function. We used Bayesian Information Criterion (BIC) to assess the models for K values ranging from 1 to 35, depending on the MTU. The curve of BIC statistics for each K was smoothed using a Lowess approach to select the best-fit model (SI Appendix, Fig. S1).
We estimated migration rates among sites along the elevation gradients for each MTU to test the hypothesis that gene flow among sites is more restricted in the tropics. Effective migration rates (Nem) between population pairs were inferred in the program ∂a∂i (47), which uses a diffusion approximation approach on the folded 2D allele frequency spectrum (AFS) estimated over all SNPs. Provided with a parameterized demographic model, ∂a∂i calculates the expected AFS for that model under random genetic drift and then maximizes the similarity to the observed AFS using maximum likelihood. Because little information is known about the actual demographic histories of the individual populations, we assumed a simple population split model for all pairwise analyses, with an ancestral population of constant effective size N0 that splits at time Ts into two subpopulations of constant effective size N1 and N2, linked by constant symmetric migration occurring at rate m after the split (SI Appendix, Fig. S2). ∂a∂i specifies migration rates in units of Nem, where Ne is the inferred effective population size of the ancestral population (given by N0 in our model). While this simple split model is an oversimplification of the true demographic history of any given population pair, the migration rates inferred within each MTU, when compared over many different pairs, provide information about the relative magnitude of migration for temperate and tropical pairs within each clade. We calculated effective migration rates among populations for MTUs in each clade independently (SI Appendix, Fig. S2) and for all MTUs pooled by region (Fig. 2C). Finally, we estimated pairwise population divergences (FST) within each species complex using adegenet (Fig. 2B).
To determine the extent to which subpopulation differentiation was influenced by the landscape, and if geographic distance or elevation differences among sites disproportionately affected genetic differentiation at different latitudes, we used linear mixed-effects models that accounted for the correlated nature of pairwise genetic distance measurements (49, 50). Mixed-effects models were fit using the lme and gls functions in the R package nlme, and correlation matrices were generated using the corMLPE function (https://github.com/nspope/corMLPE). The centered geographic distances and elevational differences among sites were the independent variables, and Dc was the dependent variable. Taxonomic membership of each MTU was nested within latitude and included as a random effect (SI Appendix, Table S3).
Determining Regional Taxonomic Richness and Cryptic Diversity.
At each of 20 tropical and 26 temperate streams, we determined the total number of MTUs present in each of our three focal clades and then used DNA barcoding to assign individuals to barcoded species (9) (Dataset S2). We identified specimens morphologically to the lowest level possible using available literature (51–54). For each MTU identified at each site, we then DNA-barcoded (55) up to 10 specimens using standard protocols from the Canadian Center for DNA Barcoding (56, 57). We analyzed a total of 3,980 sequences including 3,052 previously published records (9, 58) and 928 new specimens for this study. We used refined single-linkage clustering to delimit groups of specimens into barcoded species (59).
To determine relative levels of cryptic diversity at tropical and temperate sites, we quantified the increase in the number of barcoded species versus the number of MTUs originally identified in each clade. To compare latitudinal differences in regional taxonomic richness, we used the total number of barcoded species in Ecuador and Colorado, thus avoiding the underestimation of species richness in less-studied tropical faunas (9). Final identifications of all barcoded species are available in Dataset S2. Specimens, locality, and sequence data have been accessioned in the Barcode of Life Database (www.boldsystems.org/index.php/Public_SearchTerms?query=DS-TTADDD) (60) and in GenBank (MH838053–MH841890).
Historical Context for Latitudinal Differences in Species Richness.
We estimated a Bayesian phylogeny for the barcoded species included in our study. We estimated a tree constrained according to well-supported higher-level taxonomy groupings for the three clades (Dataset S3). In determining constraints, we applied a set of decision rules in cases of conflicting published phylogenetic relationships (9, 61). Each barcoded species is one tip in the phylogeny; thus we randomly chose among the longest available sequences for each barcoded species and aligned them in MAFFT v.7 (62) using strategy G-INS-i with offset value 0.1 and all other options set as default. Once aligned, we used jModelTest2 (63, 64) to select the best-fit nucleotide-substitution model based on the Akaike Information Criterion, which was GTR + Γ. We then conducted six runs each with 100,000,000 generations of Bayesian MCMC in BEAST v.2.3.2 (65). We modeled lineage-specific substitution rates using a relaxed clock with log-normally distributed rates (66–68) and diversification using a birth–death tree prior (69–71). For each run, we plotted the -ln likelihood scores against generation time in Tracer v 1.6 (72) and examined the effective sample sizes for parameters to ensure that all six analyses reached stationarity. We combined the trees from each run in LogCombiner v 2.3.2 (65), discarded the first 25% of generations as burn-in, and resampled every 22,500 trees. We summarized a total of 20,000 post burn-in trees in TreeAnnotator v 2.3.2 to create the maximum clade credibility (MCC) supertree. We then collapsed clades with <0.95 posterior probability on the MCC supertree. This tree was subsequently divided into three subtrees for independent clade analyses.
For independent clade analyses, we used stochastic character mapping (73) to infer the most probable ancestral location and the relative historical presence of each clade in the tropics. We applied pattern-based stochastic character mapping (73) as opposed to other events-based methods because all of the taxa detected in our study are endemic to single continents (North or South America). We used maximum likelihood to fit equal-rates and all-rates-different transition matrices and compared them using a likelihood-ratio test. After determining the best-fit model, we used the R package phytools (74) to simulate 10,000 stochastic character maps for ancestral state coded as “tropical” or “temperate.” The results of stochastic mappings were summarized as pie charts indicating the posterior probability of the location of each ancestral node (SI Appendix, Fig. S3).
Latitudinal Differences in Diversification.
For barcoded species from all three clades, together and separately, we tested for latitudinal differences in macroevolutionary rates using BiSSE (25) implemented in the R package diversitree (75). Although geographic state speciation and extinction models (GeoSSE) are ideal for testing for character-dependent diversification based on biogeography (76), we used BiSSE rather than GeoSSE because the latter is not suitable for datasets containing species endemic to single continents or for trees containing polytomies (77, 78). For the analysis of all barcoded species together, we used a Bayesian MCMC version of BiSSE designed to handle polytomies to fit the full model. We then calculated and examined the 95% credibility interval for latitudinal differences (Ecuador–Colorado) between parameter estimates. Lack of inclusion of zero in the 95% credibility interval indicated a significant difference in parameter estimates for Ecuador versus Colorado. For analyses of barcoded species in each clade, we determined the maximum-likelihood parameter estimates for the full BiSSE model for each of the 20,000 post burn-in trees summarized to create our supertrees and calculated the 95% credibility interval for latitudinal differences (Ecuador–Colorado) between parameter estimates.
Supplementary Material
Acknowledgments
We thank D. G. Gannon, José Schreckinger, Maja Celiscak, and numerous student interns for field and laboratory assistance, Amy McMahon for insect illustrations, and Andrea Landeira-Dabarca for stream temperature data. This research was supported by National Science Foundation (NSF) Dimensions of Biodiversity Grants DEB-1046408, DEB-1045960, and DEB-1045991; NSF Graduate Research Fellowships (to B.A.G. and A.A.S.); and the Atkinson Center for a Sustainable Future. Collection permits and field site access were granted by the US Department of Interior National Park Service (Permit ROMO-2011-SCI-0042); the Continental Divide Research Learning Center, US Department of Agriculture Forest Service (Authorization ID: CAN440); Ministerio de Ambiente de Ecuador (Permits 56-IC-FAU/FLO-DPN/MA and MAE-DNB-CM-2015-0017); the Cayambe-Coca Ecological Reserve; and the residents of Oyacachi.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Restriction-site associated DNA sequences, SNP data, and detailed collection localities for all eight focal morphological taxonomic units are available in the Dryad Digital Repository, https://datadryad.org/, (doi:10.5061/dryad.m728c47). Specimens, locality, and sequence data have been accessioned in the Barcode of Life Database (DOI:dx.doi.org/10.5883/DS-TTADDD) and in GenBank (MH838053–MH841890).
See Commentary on page 12337.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809326115/-/DCSupplemental.
References
- 1.Graham CH, et al. The origin and maintenance of montane diversity: Integrating evolutionary and ecological processes. Ecography. 2014;37:711–719. [Google Scholar]
- 2.Mittelbach GG, et al. Evolution and the latitudinal diversity gradient: Speciation, extinction and biogeography. Ecol Lett. 2007;10:315–331. doi: 10.1111/j.1461-0248.2007.01020.x. [DOI] [PubMed] [Google Scholar]
- 3.Fine PVA. Ecological and evolutionary drivers of geographic variation in species diversity. Annu Rev Ecol Evol Syst. 2015;46:369–392. [Google Scholar]
- 4.Stevens GC. The latitudinal gradient in geographical range: How so many species coexist in the tropics. Am Nat. 1989;133:240–256. [Google Scholar]
- 5.Dobzhansky T. Evolution in the tropics. Am Sci. 1950;38:209–221. [Google Scholar]
- 6.Gaston KJ, Chown SL. Elevation and climatic tolerance: A test using dung beetles. Oikos. 1999;86:584–590. [Google Scholar]
- 7.Janzen DH. Why mountain passes are higher in the tropics. Am Nat. 1967;101:233–249. [Google Scholar]
- 8.Martin PR, McKay JK. Latitudinal variation in genetic divergence of populations and the potential for future speciation. Evolution. 2004;58:938–945. doi: 10.1111/j.0014-3820.2004.tb00428.x. [DOI] [PubMed] [Google Scholar]
- 9.Gill BA, et al. Cryptic species diversity reveals biogeographic support for the ‘mountain passes are higher in the tropics’ hypothesis. Proc Biol Sci. 2016;283:20160553. doi: 10.1098/rspb.2016.0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghalambor CK, Huey RB, Martin PR, Tewksbury JJ, Wang G. Are mountain passes higher in the tropics? Janzen’s hypothesis revisited. Integr Comp Biol. 2006;46:5–17. doi: 10.1093/icb/icj003. [DOI] [PubMed] [Google Scholar]
- 11.Addo-Bediako A, Chown SL, Gaston KJ. Thermal tolerance, climatic variability and latitude. Proc Biol Sci. 2000;267:739–745. doi: 10.1098/rspb.2000.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sunday JM, Bates AE, Dulvy NK. Global analysis of thermal tolerance and latitude in ectotherms. Proc Biol Sci. 2011;278:1823–1830. doi: 10.1098/rspb.2010.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shah AA, et al. Climate variability predicts thermal limits of aquatic insects across elevation and latitude. Funct Ecol. 2017;31:2118–2127. [Google Scholar]
- 14.Eo SH, Wares JP, Carroll JP. Population divergence in plant species reflects latitudinal biodiversity gradients. Biol Lett. 2008;4:382–384. doi: 10.1098/rsbl.2008.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan W-P, et al. Seasonal and daily climate variation have opposite effects on species elevational range size. Science. 2016;351:1437–1439. doi: 10.1126/science.aab4119. [DOI] [PubMed] [Google Scholar]
- 16.McCain CM. Vertebrate range sizes indicate that mountains may be ‘higher’ in the tropics. Ecol Lett. 2009;12:550–560. doi: 10.1111/j.1461-0248.2009.01308.x. [DOI] [PubMed] [Google Scholar]
- 17.Huey RB. Latitudinal pattern of between-altitude faunal similarity: Mountains might be ‘higher’ in the tropics. Am Nat. 1978;112:225–229. [Google Scholar]
- 18.Zuloaga J, Kerr JT. Over the top: Do thermal barriers along elevation gradients limit biotic similarity? Ecography. 2017;40:478–486. [Google Scholar]
- 19.Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS One. 2012;7:e37135. doi: 10.1371/journal.pone.0037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith MA, et al. Extreme diversity of tropical parasitoid wasps exposed by iterative integration of natural history, DNA barcoding, morphology, and collections. Proc Natl Acad Sci USA. 2008;105:12359–12364. doi: 10.1073/pnas.0805319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zwick P. Phylogenetic system and zoogeography of the Plecoptera. Annu Rev Entomol. 2000;45:709–746. doi: 10.1146/annurev.ento.45.1.709. [DOI] [PubMed] [Google Scholar]
- 22.Savage HM. Biogeographic classification of the Neotropical Leptophlebiidae (Ephemeroptera) based upon geological centers of ancestral origin and ecology. Stud Neotrop Fauna Environ. 1987;22:199–222. [Google Scholar]
- 23.Malm T, Johanson KA, Wahlberg N. The evolutionary history of Trichoptera (Insecta): A case of successful adaptation to life in freshwater. Syst Entomol. 2013;38:459–473. [Google Scholar]
- 24.Dijkstra K-DB, Monaghan MT, Pauls SU. Freshwater biodiversity and aquatic insect diversification. Annu Rev Entomol. 2014;59:143–163. doi: 10.1146/annurev-ento-011613-161958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maddison WP, Midford PE, Otto SP. Estimating a binary character’s effect on speciation and extinction. Syst Biol. 2007;56:701–710. doi: 10.1080/10635150701607033. [DOI] [PubMed] [Google Scholar]
- 26.Cadena CD, et al. Latitude, elevational climatic zonation and speciation in New World vertebrates. Proc Biol Sci. 2012;279:194–201. doi: 10.1098/rspb.2011.0720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jankowski JE, Londoño GA, Robinson SK, Chappell MA. Exploring the role of physiology and biotic interactions in determining elevational ranges of tropical animals. Ecography. 2013;36:1–12. [Google Scholar]
- 28.Zamudio KR, Bell RC, Mason NA. Phenotypes in phylogeography: Species’ traits, environmental variation, and vertebrate diversification. Proc Natl Acad Sci USA. 2016;113:8041–8048. doi: 10.1073/pnas.1602237113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith BT, et al. The drivers of tropical speciation. Nature. 2014;515:406–409. doi: 10.1038/nature13687. [DOI] [PubMed] [Google Scholar]
- 30.Sexton JP, McIntyre PJ, Angert AL, Rice KJ. Evolution and ecology of species range limits. Annu Rev Ecol Evol Syst. 2009;40:415–436. [Google Scholar]
- 31.Brown JH, Stevens GC, Kaufman DM. The geographic range: Size, shape, boundaries, and internal structure. Annu Rev Ecol Syst. 1996;27:597–623. [Google Scholar]
- 32.Sunday JM, Bates AE, Dulvy NK. Thermal tolerance and the global redistribution of animals. Nat Clim Chang. 2012;2:686–690. [Google Scholar]
- 33.Coyne JA, Orr HA. Patterns of speciation in Drosophila. Evolution. 1989;43:362–381. doi: 10.1111/j.1558-5646.1989.tb04233.x. [DOI] [PubMed] [Google Scholar]
- 34.Bernatchez L, Wilson CC. Comparative phylogeography of Nearctic and Palearctic fishes. Mol Ecol. 1998;7:431–452. [Google Scholar]
- 35.Wang G, Dillon ME. Recent geographic convergence in diurnal and annual temperature cycling flattens global thermal profiles. Nat Clim Chang. 2014;4:988–992. [Google Scholar]
- 36.Karl TR, et al. Global warming: Evidence for asymmetric diurnal temperature change. Geophys Res Lett. 1991;18:2253–2256. [Google Scholar]
- 37.Vasseur DA, et al. Increased temperature variation poses a greater risk to species than climate warming. Proc Biol Sci. 2014;281:20132612. doi: 10.1098/rspb.2013.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deutsch CA, et al. Impacts of climate warming on terrestrial ectotherms across latitude. Proc Natl Acad Sci USA. 2008;105:6668–6672. doi: 10.1073/pnas.0709472105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kearney M, Porter W. Mechanistic niche modelling: Combining physiological and spatial data to predict species’ ranges. Ecol Lett. 2009;12:334–350. doi: 10.1111/j.1461-0248.2008.01277.x. [DOI] [PubMed] [Google Scholar]
- 40.Pyne MI, Poff NL. Vulnerability of stream community composition and function to projected thermal warming and hydrologic change across ecoregions in the western United States. Glob Change Biol. 2017;23:77–93. doi: 10.1111/gcb.13437. [DOI] [PubMed] [Google Scholar]
- 41.Lessman J. Freshwater vertebrate and invertebrate diversity patterns in an Andean-Amazon basin: Implications for conservation efforts. Neotrop Biodivers. 2016;2:99–114. [Google Scholar]
- 42.Grafen A. The phylogenetic regression. Philos Trans R Soc Lond B Biol Sci. 1989;326:119–157. doi: 10.1098/rstb.1989.0106. [DOI] [PubMed] [Google Scholar]
- 43.Butler MA, King AA. Phylogenetic comparative analysis: A modeling approach for adaptive evolution. Am Nat. 2004;164:683–695. doi: 10.1086/426002. [DOI] [PubMed] [Google Scholar]
- 44.Hansen TF. Stabilizing selection and the comparative analysis of adaptation. Evolution. 1997;51:1341–1351. doi: 10.1111/j.1558-5646.1997.tb01457.x. [DOI] [PubMed] [Google Scholar]
- 45.Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. Stacks: An analysis tool set for population genomics. Mol Ecol. 2013;22:3124–3140. doi: 10.1111/mec.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Polato NR, et al. Genetic diversity and gene flow decline with elevation in montane mayflies. Heredity (Edinb) 2017;119:107–116. doi: 10.1038/hdy.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009;5:e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jombart T. adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- 49.Clarke RT, Rothery P, Raybould AF. Confidence limits for regression relationships between distance matrices: Estimating gene flow with distance. J Agric Biol Environ Stat. 2002;7:361–372. [Google Scholar]
- 50.Van Strien MJ, Keller D, Holderegger R. A new analytical approach to landscape genetic modelling: Least-cost transect analysis and linear mixed models. Mol Ecol. 2012;21:4010–4023. doi: 10.1111/j.1365-294X.2012.05687.x. [DOI] [PubMed] [Google Scholar]
- 51.Domínguez E, Fernández H. 2009. Macroinvertebrados Bentónicos Sudamericanos: Sistemática y Biología [South American Benthic Macroinvertebrates: Systematics and Biology] (Fundación Miguel Lillo Tucumán, Argentina)
- 52.Merritt RW, Cummins KW, Berg MB. An Introduction to the Aquatic Insects of North America. Kendall Hunt Publishing; Dubuque, IA: 2008. [Google Scholar]
- 53.Domínguez E, Molineri C, Pescador ML, Hubbard MD, Nieto C. 2006. Ephemeroptera of South America. Aquatic Biodiversity in Latin America, eds Adis J, Arias JR, Rueda-Delgado G, Wantzen KM (Pensoft, Sofia, Bulgaria), Vol 2.
- 54.Stark B, Froehlich PC, del Carmen Zuñiga ML. 2009. South American Stoneflies (Plecoptera). Aquatic Biodiversity in Latin America, eds Adis J, Arias JR, Rueda-Delgado G, Wantzen KM (Pensoft, Sofia, Bulgaria), Vol 5.
- 55.Hebert PDN, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc Biol Sci. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hajibabaei M, et al. Critical factors for assembling a high volume of DNA barcodes. Philos Trans R Soc Lond B Biol Sci. 2005;360:1959–1967. doi: 10.1098/rstb.2005.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ivanova NV, DeWaard JR, Hebert PDN. An inexpensive, automation-friendly protocol for recovering high-quality DNA. Mol Ecol Notes. 2006;6:998–1002. [Google Scholar]
- 58.Gill BA, et al. Morphological taxonomy, DNA barcoding, and species diversity in southern Rocky Mountain headwater streams. Freshw Sci. 2014;33:288–301. [Google Scholar]
- 59.Ratnasingham S, Hebert PDN. A DNA-based registry for all animal species: The barcode index number (BIN) system. PLoS One. 2013;8:e66213. doi: 10.1371/journal.pone.0066213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ratnasingham S, Hebert PDN. bold: The barcode of life data system ( www.barcodinglife.org) Mol Ecol Notes. 2007;7:355–364. doi: 10.1111/j.1471-8286.2007.01678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poff NL, et al. Functional trait niches of North American lotic insects: Traits-based ecological applications in light of phylogenetic relationships. J North Am Benthol Soc. 2006;25:730–755. [Google Scholar]
- 62.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 64.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bouckaert R, et al. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2014;10:e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thorne JL, Kishino H, Painter IS. Estimating the rate of evolution of the rate of molecular evolution. Mol Biol Evol. 1998;15:1647–1657. doi: 10.1093/oxfordjournals.molbev.a025892. [DOI] [PubMed] [Google Scholar]
- 67.Thorne JL, Kishino H. Divergence time and evolutionary rate estimation with multilocus data. Syst Biol. 2002;51:689–702. doi: 10.1080/10635150290102456. [DOI] [PubMed] [Google Scholar]
- 68.Kishino H, Thorne JL, Bruno WJ. Performance of a divergence time estimation method under a probabilistic model of rate evolution. Mol Biol Evol. 2001;18:352–361. doi: 10.1093/oxfordjournals.molbev.a003811. [DOI] [PubMed] [Google Scholar]
- 69.Yang Z, Rannala B. Bayesian phylogenetic inference using DNA sequences: A Markov chain Monte Carlo method. Mol Biol Evol. 1997;14:717–724. doi: 10.1093/oxfordjournals.molbev.a025811. [DOI] [PubMed] [Google Scholar]
- 70.Popovic L. Asymptotic genealogy of a critical branching process. Ann Appl Probab. 2004;14:2120–2148. [Google Scholar]
- 71.Gernhard T. The conditioned reconstructed process. J Theor Biol. 2008;253:769–778. doi: 10.1016/j.jtbi.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 72.Rambaut A, Suchard MA, Xie D, Drummond AJ. 2014 Tracer v1.6. Available at beast.community. Accessed August 01, 2016.
- 73.Huelsenbeck JP, Nielsen R, Bollback JP. Stochastic mapping of morphological characters. Syst Biol. 2003;52:131–158. doi: 10.1080/10635150390192780. [DOI] [PubMed] [Google Scholar]
- 74.Revell LJ. Phytools: An R package for phylogenetic comparative biology (and other things) Methods Ecol Evol. 2011;3:217–223. [Google Scholar]
- 75.Fitzjohn RG. Diversitree: Comparative phylogenetic analyses of diversification in R. Methods Ecol Evol. 2012;3:1084–1092. [Google Scholar]
- 76.Goldberg EE, Lancaster LT, Ree RH. Phylogenetic inference of reciprocal effects between geographic range evolution and diversification. Syst Biol. 2011;60:451–465. doi: 10.1093/sysbio/syr046. [DOI] [PubMed] [Google Scholar]
- 77.Parada A, D’Elía G, Palma RE. The influence of ecological and geographical context in the radiation of Neotropical sigmodontine rodents. BMC Evol Biol. 2015;15:172. doi: 10.1186/s12862-015-0440-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Givnish TJ, et al. Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc Biol Sci. 2015;282:20151553. doi: 10.1098/rspb.2015.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.