Abstract
Background
Influenza virus infections are a major public health concern worldwide. Conventional treatments against the disease are designed to target viral proteins. However, the emergence of viral variants carrying drug-resistant mutations can outpace the development of pathogen-targeting antivirals. Diphyllin and bafilomycin are potent vacuolar ATPase (V-ATPase) inhibitors previously shown to have broad-spectrum antiviral activity. However, their poor water solubility and potential off-target effect limit their clinical application.
Methods
In this study, we report that nanoparticle encapsulation of diphyllin and bafilomycin improves the drugs’ anti-influenza applicability.
Results
Using PEG-PLGA diblock copolymers, sub-200 nm diphyllin and bafilomycin nanoparticles were prepared, with encapsulation efficiency of 42% and 100%, respectively. The drug-loaded nanoparticles have sustained drug release kinetics beyond 72 hours and facilitate intracellular drug delivery to two different influenza virus-permissive cell lines. As compared to free drugs, the nanoparticulate V-ATPase inhibitors exhibited lower cytotoxicity and greater in vitro antiviral activity, improving the therapeutic index of diphyllin and bafilomycin by approximately 3 and 5-fold, respectively. In a mouse model of sublethal influenza challenge, treatment with diphyllin nanoparticles resulted in reduced body weight loss and viral titer in the lungs. In addition, following a lethal influenza viral challenge, diphyllin nanoparticle treatment conferred a survival advantage of 33%.
Conclusions
These results demonstrate the potential of the nanoparticulate V-ATPase inhibitors for host-targeted treatment against influenza.
Keywords: influenza virus, vacuolar ATPase inhibitor, diphyllin, bafilomycin, nanoparticles
Introduction
Influenza viruses belong to the family Orthomyxoviridae and can be categorized into four major types: A, B, C, and D.1,2 Influenza A and B viruses that routinely spread in people cause seasonal flu epidemics each year. Influenza viruses inflict millions of infection cases in human and animals every year, and effective antivirals are an essential countermeasure against the disease. Amantadine is the first synthetic compound that inhibits influenza virus replication; the compound and its derivatives inhibit matrix-2 ion channels to block the migration of H+ ions into the interior of the virus particles, a process critical for virus uncoating to occur.3 In recent years, however, influenza virus resistance to these compounds has been widely reported.4,5 Another class of antiviral agent is neuraminidase (NA) inhibitors, which include oseltamivir, zanamivir, and peramivir. These antiviral agents inhibit viral NA activity, which plays an important role in early influenza infection of the human airway epithelium and in virus budding.6 While oseltamivir is currently the most common commercial anti-influenza drug, resistance against NA inhibitors has been observed.5,7 On the contrary, several genome-wide screens have identified host factors essential for influenza virus replication.8–10 As an alternative to the aforementioned pathogen-targeted antivirals, growing efforts are devoted to blocking or promoting host factors to fight influenza viruses.11 By modulating host factors involved in viral replications, these host-targeted antiviral strategies may be less susceptible to strain variations and mutations as they do not exert a selective pressure on the target pathogen.
Among host factors that can be targeted for antiviral treatments, vacuolar ATPases (V-ATPases) are a promising target for intercepting virus entry into host cells. V-ATPases are ubiquitous proton pumps located in the endomembrane system of all eukaryotic cells.12 Among viral threats such as influenza viruses, flaviviruses, vaccinia viruses, bornaviruses, rhabdoviruses, and coronaviruses, V-ATPase-mediated endosomal acidification is an essential cellular process for viral entry.13–17 Inhibition of V-ATPase-mediated endosomal acidification may thus pave ways to new antiviral treatments with broad applicability and low susceptibility to drug-resistant mutation. Several V-ATPase inhibitors have been studied, among which plecomacrolide bafilomycin is the first discovered and perhaps the most notable example.18 While these compounds have shown antiviral potentials, their clinical application is thwarted by toxicity concerns.19–21 In addition, V-ATPase inhibitors are often poorly water soluble, which presents further drug delivery challenges. Previously, we showed that diphyllin, a new class of the V-ATPase inhibitor,12 is effective in blocking influenza virus infection,22 and its nanoformulation showed improved safety and effectiveness in inhibiting the feline coronavirus.23 Toward improving V-ATPase inhibitors for influenza treatment, we herein prepare diphyllin-loaded polymeric nanoparticles comprised of poly(ethylene glycol)-block-poly(lactide-co-glycolide) (PEG-PLGA) and examined its efficacy against influenza virus in vitro and in vivo. In parallel, we assessed the applicability of nanoparticle-mediated delivery to the commonly studied bafilomycin. The particular nanocarrier was chosen as PLGA-based polymeric nanoparticles that have been broadly adopted for enhancing the delivery of hydrophobic drugs.24 The biodegradable polymer is also widely used in FDA-approved products and is therefore readily translatable.25 The material has also been shown to induce little innate immune activation.26 In this study, the physicochemical properties and drug release kinetics of both diphyllin and bafilomycin-loaded nanoparticles were examined. Cellular uptake of the nanoparticles was assessed in two different influenza virus-permissive cell lines, including MH-S and ARPE-19 (adult retinal pigment epithelial cell line-19). We further demonstrated that the nanoparticulate V-ATPase inhibitors were less cytotoxic than the free drug compounds and reduced influenza viral infection in vitro. Using diphyllin nanoparticles, we also showed that the host-targeted treatment can be applied to multiple strains of influenza viruses and successfully reduced disease severity in a mouse model of influenza virus infection, highlighting the potential of nanoparticulate V-ATPase inhibitors for antiviral development.
Materials and methods
Cells and viruses
MDCK (Madin – Darby canine kidney) cells were maintained in the DMEM supplemented with 10% FBS and 1% penicillin/streptomycin/amphotericin B (PSA). MH-S (murine alveolar macrophages [AMs]) cells were maintained in ATCC-formulated RPMI-1640 medium supplemented with 2-mercaptoethanol to a final concentration of 0.05 mM, FBS to a final concentration of 10%, and 1% PSA. For influenza virus infection in MDCK and MH-S cells, infection medium (RPMI containing 0.75% BSA, 0.05 mM 2-mercaptoethanol, 1% PSA, and 2 µg/mL L-1-p-Tosylamino-2-phenylethyl chloromethyl ketone-treated trypsin) was used. ARPE-19 cells were maintained in the DMEM/F12 supplemented with 10% FBS and 1% PSA. All the cells were purchased from the Bioresource Collection and Research Center (BCRC) (Hsinchu, Taiwan) and cultured at 37°C and 5% CO2. All the reagents for cell culture were purchased from Invitrogen. Influenza A virus (A/Puerto Rico/8/1934(H1N1)) was kindly provided by Professor Shin-Ru Shih in Chang Gung University. A/HKx31(H3N2) was kindly provided by Professor Hung-Chih Yang in National Taiwan University. Viruses were propagated in the allantoic cavity of 10-day-old embryonated chicken eggs. The viral titer was determined as below.
Preparation of V-ATPase inhibitor-loaded nanoparticles
Diphyllin (Sigma-Aldrich Co., St Louis, MO, USA) or bafilomycin (LC Laboratory, Woburn, MA, USA) was encapsulated into a PEG-functionalized PLGA nanoparticle using an oil-in-water emulsion technique. Briefly, 0.5 mg of diphyllin or bafilomycin was dissolved with 20 mg of PEG-PLGA diblock copolymers (Sigma #764760) in 1 mL of chloroform (Honeywell International Inc., Morris Plains, NJ, USA). The solution was subsequently added to 5 mL of 10 mM phosphate buffer (pH 8.0) and dispersed using a probe sonicator for 1 minute 30 seconds with 50% power to form the single emulsion. Following the sonication, the emulsion was placed inside a fume hood under gentle stirring (180–220 rpm) to evaporate the chloroform. Following 3 hours of solvent evaporation, the resulting nanoparticles were collected and washed in 10 mL of the phosphate buffer under ultracentrifugation 60,000 g, 25 minutes. Then, the nanoparticles were resuspended in 10% sucrose for subsequent characterization. Empty PEG-PLGA nanoparticles were also prepared as a control for this study. For preparing fluorescently labeled nanoparticles, 0.01 mg of a DiD fluorescent dye (1,1′-Dioctadecyl-3,3,3′,3′-Tetramethylindodicarbocyanine, 4-Chlorobenzenesulfonate Salt) (633 nm/647 nm) (Thermo Fisher Scientific, Waltham, MA, USA; #D7757) was added to the solvent phase before the single emulsion process.
Transmission electron microscopy (TEM) and dynamic light scattering (DLS)
For TEM observation, negative staining was applied. First, 10 µL nanoparticles was added to glow-discharged grid and the droplet was placed on the grid for 30 seconds. Then, 10 drops of ddH2O was applied to the grid to wash away the excessive nanoparticles. Subsequently, the grid was stained with 10 µL of 1% uranyl acetate for 30 seconds and the staining was subsequently removed by wicking with a piece of filter paper. Nanoparticle size and zeta potential were determined by DLS using Malvern Zetasizer Nano ZS (Malvern Instruments, Malvern, UK) following the manufacturer’s instructions. Each measurement was made with 1 mL of the nanoparticles at 1 mg/mL in water in a folded capillary zeta cell (Malvern Panalytical, DTS1070).
High-performance liquid chromatography (HPLC)
The content of diphyllin or bafilomycin in the PEG-PLGA nanoparticles was quantified by HPLC (Agilent Technologies, Santa Clara, CA, USA; 1,100 series). Briefly, 200 µL of nanoparticles loaded with diphyllin or bafilomycin was dissolved with 800 µL of acetone, which was then air-dried at 60°C. The samples were then dissolved in mobile phase solvent (diphyllin: 40% acetonitrile/bafilomycin: 87% ace-tonitrile) and filtered prior to HPLC. Ascentis C18 column was equipped in mobile phase. The optimal detection wavelength was 254 nm, and the column temperature was 25°C. Serially diluted samples of diphyllin (0.125, 0.03125, 0.0078125, and 0.001953125 mg/mL) or bafilomycin (0.5, 0.25, 0.125, 0.0625, and 0.03125 mg/mL) dissolved in the mobile phase solvent were prepared as standards for the HPLC analysis.
Drug release studies
The drug release kinetics were determined in either phosphate buffer (1.16 mM Na2HPO4, 38.9 mM NaH2PO4, pH 7.4) or acetate buffer (0.33% acetate acid, 1.31% sodium acetate, pH 5.0). A set of dialysis tubes (Slide-A-Lyzer MINI Dialysis Device) were loaded with 50 µL drug-loaded nanoparticles containing 0.05 mg of diphyllin or bafilomycin. The tubes were then placed in a tank of phosphate buffer or acetate buffer. The drug-loaded nanoparticles were harvested from the dialysis tubes at 30 minutes, 1 hour, 2 hours, 4 hours, 6 hours, 23 hours, 48 hours, and 72 hours. The nanoparticle samples were then dissolved with acetone, and the drug contents were analyzed by HPLC.
Cellular uptake studies
MH-S cells and ARPE-19 cells were seeded in 8-well chamber slides (3×104/well) 1 day prior to the experiment. These cells were treated with DiD-labeled diphyllin or bafilomycin nanoparticles (1 mg/mL) in the culture media. After 24 hours of incubation at 37°C, culture supernatant was removed and the cells were washed with PBS buffer. Untreated cells were used as a control. The cells were fixed using 10% formalin (V/V) in water. Cellular nuclei were stained with DAPI, and fluorescence microscopy (Olympus Corporation, Tokyo, Japan; IX83) was used to examine cellular uptake of the fluorescent nanoparticles.
In vitro cytotoxicity evaluation of free drugs and drug-loaded nanoparticles
For evaluating 50% cytotoxic concentration (CC50), MH-S cells were grown on a 96-well clear polystyrene microplate at a density of 20,000 cells per well 1 day prior to the experiment. Free drugs or drug-loaded nanoparticles were twofold serially diluted in cell media and added to the cell monolayer in four replicates. After 72 hours of incubation, 5 µL of alamarBlue reagent (Thermo Fisher Scientific) was added to each well to evaluate the cellular viability. Absorbance values were measured at 570 nm with a reference wavelength of 610 nm. The CC50 of free drugs or drug-loaded nanoparticles was analyzed using Prism (GraphPad Software, Inc., La Jolla, CA, USA).
Cell cytopathic effect (CPE) inhibition assay
The CPE inhibition assay was used to determine the IC50 of free drug or drug-loaded nanoparticles (the concentration of drug showed 50% inhibition of virus-induced CPE) against influenza virus H1N1. Briefly, MH-S cells were seeded on 96-well microplates (10,000 cells/well) 1 day before the experiment. Free drug or drug-loaded nanoparticles in infection medium were added to MH-S cells 1 hour before infection. After 1 hour of incubation, cells were infected with influenza virus H1N1 at a multiplicity of infection (MOI) of 1. After infection, viruses were removed and the free drug or drug-loaded nanoparticles in cell medium were added back to the cells. After 24 hours, cellular viability was examined by an alamarBlue assay. The IC50 of free drugs or drug-loaded nanoparticles were determined using Prism (GraphPad).
Assessment of antiviral activity of free drug and drug-loaded nanoparticles
To evaluate the activity of free drug or drug nanoparticles in blocking the viral replication, MH-S cell cultures were used. The cells were pretreated 1 hour with free drugs or drug-loaded nanoparticles and then infected with influenza virus (H1N1, MOI=1; H3N2, MOI=5) for 1 hour. After infection, viruses were removed and the cells were washed. Subsequently, fresh cell media containing drug-loaded nanoparticles were added to the cells. After 24 hours of incubation, the supernatant was harvested to assess the viral titer.
Evaluation of antiviral efficacy of diphyllin nanoparticles in vivo
Anti-influenza efficacy study was performed in mice. Groups of six 6-week-old female BALB/c mice were purchased from the National Laboratory Animal Center, Taipei, Taiwan. Mice were infected with a nonlethal dose [100,000 PFU (plaque-forming unit), equal to 0.5 LD50] or a lethal dose (250,000 PFU, equal to 1.3 LD50) of influenza H1N1 virus via intranasal administration under light anesthesia with Zoletil 50 (18.75 mg/kg). Right after the virus infection, mice were intravenously injected with diphyllin nanoparticles containing 10 µg of diphyllin for 1 or 3 days. Mice receiving empty PEG-PLGA nanoparticles were set as a control group. Following the virus infection, mice were monitored daily for weight change and survival. For the nonlethal dose infection experiment, two mice of each group were sacrificed on day 5 postviral infection. Lungs of the mice were harvested and homogenated in the infection medium, and the viral load was determined by the quantitative RT-PCR (qRT-PCR) described below. Animal experiments in this study were conducted in accordance with the guidelines of the institutional animal care and use committee (IACUC) of the National Taiwan University (Approval number: NTU105-EL-00181).
Influenza virus titration
For tissue culture infective dose (TCID50) assays, MDCK cells were seeded in 96-well microplates (2×104 cells/well) 1 day before the experiment. First, the growth media were removed and the cells were washed with PBS buffer. The samples were tenfold serially diluted with infection medium, and 100 µL of the diluted samples was added to the cells and incubated at 37°C for 1 hour. After the infection, media were removed and the cells were washed. Subsequently, 100 µL of the fresh virus media was added into each well. After 72 hours of incubation, the supernatant was harvested to perform the direct hemagglutination assay spot assay with 1% chicken RBCs, and the viral titers were determined with the Reed and Muench method.27 For plaque assays, MDCK cells were seeded in a 6-well plate to obtain 80% confluency (8×105 cells/well) 1 day before experiments. First, the growth medium was removed and the cells were washed with PBS buffer. The samples were tenfold serially diluted with infection medium, and 200 µL of the diluted samples was added to the cells and then incubated at 37°C for 1 hour under gentle shaking. After the infection, culture media were removed and cells were washed with PBS. Two milliliters of 0.3% agarose in infection medium was then added to each well. After 72 hours of incubation, the cells were fixed with 10% formalin for 30 minutes. The agarose overlay was removed with running water. The cells were stained with 1% crystal violet at room temperature for 15 minutes. Once dry, the viral plaques were counted, and the viral titers were determined. For viral RNA quantification in the lung of the mice, a qRT-PCR was performed as previously described28 with some modifications. Briefly, viral RNA was extracted using a viral nucleic acid extraction kit (Geneaid Biotech Ltd., Taipei, Taiwan). cDNA was synthesized using the QuantiNova Reverse Transcription Kit (Qiagen NV, Venlo, the Netherlands) following manufacturer’s instructions. For qPCR, the 20 µL reaction mixture was prepared by mixing 500 nM of primers M1-F and primer M1-R, and 250 nM of FAM-labeled probe (Integrated DNA Technologies), 10 µL of 2× PrimeTime Gene Expression MasterMix (Integrated DNA Technologies), and 2 µL of cDNA. The cycling protocol in a thermal cycler consisted of 3 minutes of preheating at 95°C and followed by 45 cycles of 15 seconds of denaturation at 95°C, 1 minute of primer annealing and extension at 60°C (CFX Connect, Bio-Rad Laboratories Inc., Hercules, CA, USA). Two microliters of plasmid DNA containing various copy numbers of the matrix protein gene of influenza virus was used for standard curve construction in qRT-PCR for viral load quantification. Two replicates were performed for each samples or standards.
Statistical analyses
Data were analyzed by unpaired t-test or ANOVA followed by the Dunnett’s multiple comparison tests using Prism (Graph-Pad). P-value <0.05 is considered statistically significant.
Results
Characterization of diphyllin- and bafilomycin-loaded nanoparticles
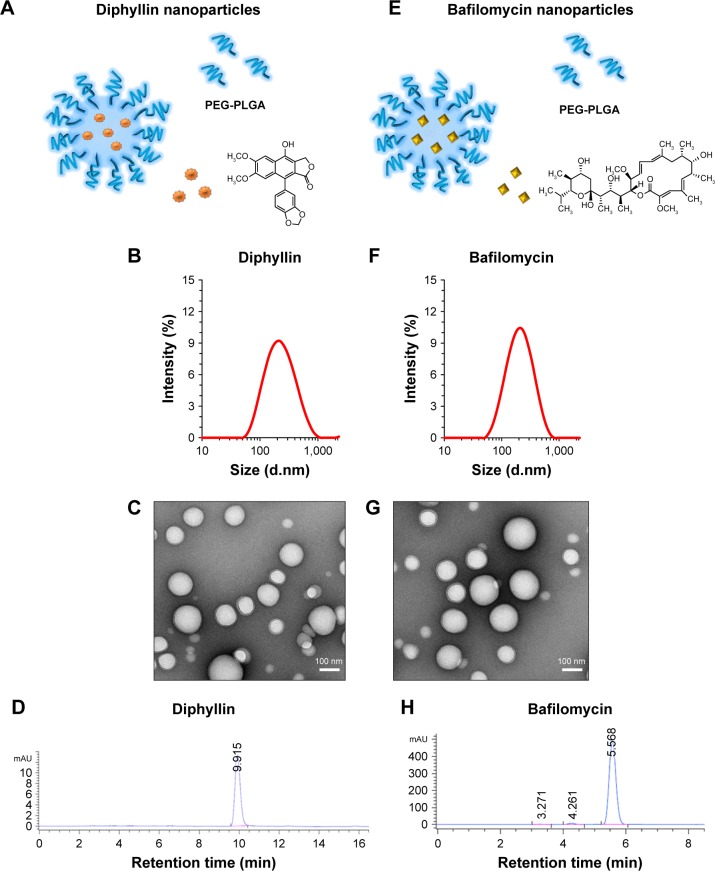
The composition of diphyllin nanoparticles is illustrated in Figure 1A. Using DLS, the diphyllin-loaded nanoparticles in this study showed a Z-average diameter of 178 nm (Figure 1B). The resulting diphyllin nanoparticles showed a uniformly spherical shape as visualized by TEM (Figure 1C). The quantification of diphyllin loading in PEG-PLGA nanoparticles was determined by HPLC. The standard curve was constructed with serially diluted samples of diphyllin (0.125, 0.03125, 0.0078125, and 0.001953125 mg/mL) that separately corresponded to each area value 2,140.27, 587.06, 154.80, and 35.19 (mAU*second) (Figure S1A) and the peak detection of diphyllin appeared at ~9.9 minutes (Figure 1D). With the reference of standard curve, the content of diphyllin in nanoparticles for each batch was determined. In this study, the averaged loading efficiency of diphyllin was 42%.
Figure 1.
Characterization of drug-loaded nanoparticles.
Notes: Illustrations of (A) diphyllin-loaded PEG-PLGA nanoparticles and (E) bafilomycin-loaded PEG-PLGA nanoparticles. Size measurements of (B) diphyllin nanoparticle and (F) bafilomycin nanoparticles were obtained by DLS. (C) Diphyllin nanoparticles and (G) bafilomycin nanoparticles were visualized under TEM following negative staining with uranyl acetate. Scale bars = 100 nm. Encapsulation of (D) diphyllin and (H) bafilomycin were confirmed by HPLC.
Abbreviations: PEG-PLGA, poly(ethyleneglycol)-block-poly(lactide-co-glycolide); DLS, dynamic light scattering; TEM, transmission electron microscopy; HPLC, high-performance liquid chromatography.
The schematic presentation of bafilomycin-loaded nanoparticles is illustrated in Figure 1E. Using DLS, the bafilomycin-loaded nanoparticles used in this study had a Z-average diameter of 197 nm (Figure 1F). The resulting bafilomcycin-loaded nanoparticles showed a uniformly spherical shape as visualized by TEM (Figure 1G). The quantification of bafilomycin loading in PEG-PLGA nanoparticles was determined by HPLC. The standard curve was constructed with serially diluted samples of bafilomycin (0.5, 0.25, 0.125, 0.0625, and 0.03125 mg/mL) that separately corresponded to each area value 7,270.52, 3,652.66, 1,751.47, 901.46, and 421.81 (mAU*second) (Figure S1B). The peak detection of bafilomycin appeared at ~5.5 minutes (Figure 1H). With the reference of standard curve, the content of bafilomycin in nanoparticles for each batch was determined. In this study, the loading efficiency of bafilomycin was ~100%. Both nanoparticulate V-ATPase inhibitors showed similar physicochemical properties. Interestingly, bafilomycin encapsulation was much more efficient than diphyllin, which may be attributed to bafilomycin’s lower water solubility.
Release kinetics of drug-loaded nanoparticles
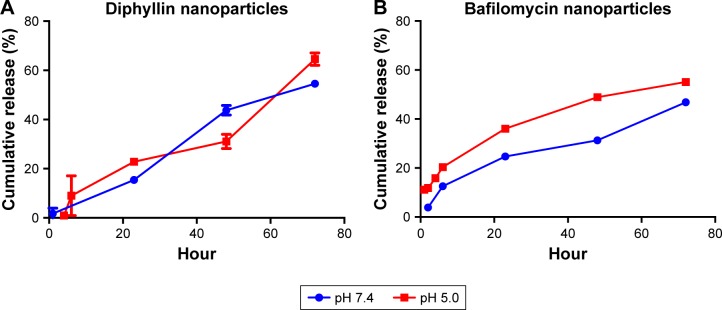
This drug release study was performed in phosphate buffer (pH 7.4) and acetate buffer (pH 5.0) to simulate the extracellular and endosomal condition, respectively. In diphyllin nanoparticles, little diphyllin was released from the nanoparticles in the first 6 hours. After 23 hours, 22% of the diphyllin was released in the acidic buffer and 15% of diphyllin was released in the phosphate buffer. After 72 hours, 64% of diphyllin was released in the acidic buffer and 54% of diphyllin was released in the phosphate buffer (Figure 2A). Overall, diphyllin release from the nanoparticles was largely similar between the two pH values during the observation period. For bafilomycin-loaded nanoparticles, 20% of bafilomycin was released in the acidic buffer and 12% of bafilomycin was released in the phosphate buffer in the first 6 hours. After 23 hours, 36% of bafilomycin was released in the acidic buffer and 24% of bafilomycin was released in the phosphate buffer. After 72 hours, 55% of bafilomycin was released in the acidic buffer and 43% of bafilomycin was released in the phosphate buffer (Figure 2B). Bafilomycin release was accelerated in pH 5.0 compared with pH 7.4 overall. Both diphyllin and bafilomycin nanoparticles exhibited sustained drug release profiles for over 72 hours.
Figure 2.
Release kinetics of drug-loaded nanoparticles.
Notes: Release kinetics of (A) diphyllin nanoparticles and (B) bafilomycin nanoparticles were assessed in a phosphate buffer (pH 7.4) and an acidic buffer (pH 5.0) at 37°C. HPLC was used to measure the drug content following dialysis at various time points. Data in the plot represent the mean ± SEM out of three test replicates.
Abbreviations: HPLC, high-performance liquid chromatography; SEM, standard error of the mean.
Assessment of cellular uptake of nanoparticles
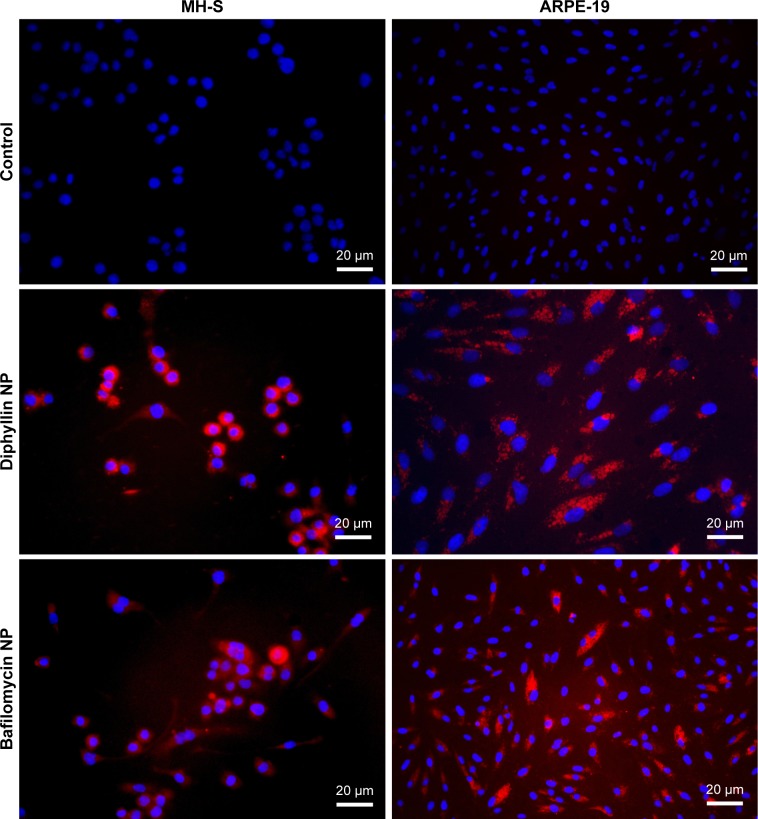
To verify that the nanocarriers can efficiently deliver their drug payload intracellularly, MH-S cells and ARPE-19 cells were treated with fluorescently DiD-labeled diphyllin and bafilomycin nanoparticles. The fluorescence signal was examined at 400× magnification under a fluorescence microscope. The nucleus stained by DAPI was shown in blue, and the nanoparticles were shown in red. While untreated control cells revealed no fluorescence, both MH-S and ARPE-19 cells took up nanoparticles efficiently (Figure 3), indicating successful intracellular drug delivery by the nanoparticles.
Figure 3.
Cellular uptake of drug-loaded nanoparticles.
Notes: MH-S cells and ARPE-19 cells were incubated with 1 mg/mL of either diphyllin nanoparticles (diphyllin NP) or bafilomycin nanoparticles (bafilomycin NP) loaded with a fluorescent DiD dye for 24 hours. Fluorescence images validate nanoparticles uptake by the cells. Cellular nuclei (DAPI) were imaged in blue. Scale bars = 20 µm.
Abbreviations: ARPE, adult retinal pigment epithelium; NP, nanoparticle; DiD, 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine,4-chlorobenzenesulfonate salt.
CC50 of free drugs and drug-loaded nanoparticles
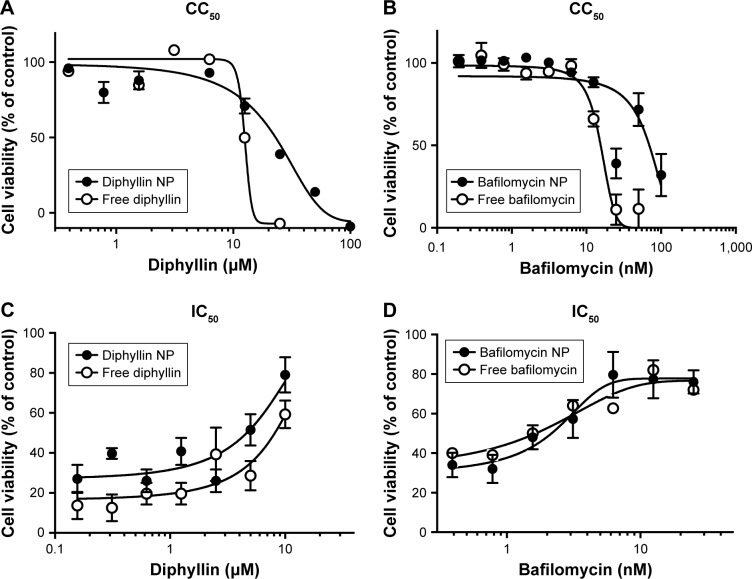
To assess the safety of the drug-loaded nanoparticles, a standard alamarBlue assay was used to determine the cytotoxicity of free drugs and drug-loaded nanoparticles in MH-S cells. Following a 3-day incubation, the CC50 values of free diphyllin and diphyllin nanoparticles were 12.5 µM and 21.89 µM, respectively (Figure 4A). The improved safety following nanoparticle encapsulation was also observed with bafilomycin. A fourfold reduction in cytotoxicity was observed with the nanoformulation as the CC50 values of free bafilomycin and bafilomycin nanoparticles were 15.54 nM and 69.6 nM, respectively (Figure 4B). These results indicate improved safety achieved by the nanoparticulate diphyllin and bafilomycin, which may facilitate sustained release and enhance drug localization in the endolysosomal compartment for reduced side effects.
Figure 4.
CC50 and IC50 of free and nanoparticulate V-ATPase inhibitors.
Notes: (A) Free diphyllin or diphyllin NP was added to MH-S cells and incubated for 72 hours. An alamarBlue assay was performed and cell viability was normalized to the value of untreated medium controls (100%). (B) Free bafilomycin or bafilomycin nanoparticles (bafilomycin NP) were added to MH-S cells and incubated for 72 hours. An alamarBlue assay was performed and cell viability was normalized to the value of untreated medium controls (100%). (C) Free diphyllin or diphyllin NP was added to MH-S cells and incubated for 1 hour. Cells were then infected with influenza virus H1N1 at an MOI of 1 for another hour. Viruses were removed and the free diphyllin or diphyllin NP was added back to the cells. Twenty-four hours later, cellular viability was examined by an alamarBlue assay, and cell viability was normalized to the value of uninfected cell controls (100%). (D) Free bafilomycin or bafilomycin NP was added to MH-S cells and incubated for 1 hour. Cells were then infected with influenza virus H1N1 at an MOI of 1 for another hour. Viruses were removed and the free bafilomycin or bafilomycin NP was added back to the cells. Twenty-four hours later, cellular viability was examined by an alamarBlue assay, and cell viability was normalized to the value of uninfected cell controls (100%). Data in the plot represent mean ± SEM out of four test replicates.
Abbreviations: CC50, 50% cytotoxic concentration; IC50, 50% inhibitory concentration; V-ATPase, vacuolar ATPase; NP, nanoparticle; MOI, multiplicity of infection; SEM, standard error of the mean.
IC50 of free drug and drug-loaded nanoparticles
The IC50 of free drug and drug-loaded against influenza virus H1N1 was determined by the CPE inhibition assay in MH-S cells. The results showed that the IC50 of free diphyllin was 8.149 µM, whereas that of the diphyllin nanoparticles was 4.640 µM (Figure 4C), demonstrating improved antiviral activity with the nanoformulation. Calculation of therapeutic index (TI = CC50/IC50) yielded values of 1.53 and 4.72 for the free diphyllin and diphyllin nanoparticles, respectively. Similarly, the antiviral activity of free bafilomycin and bafilomycin nanoparticles against influenza virus H1N1 was also compared in MH-S cells. The results showed that the IC50 of free bafilomycin was 2.280 nM, whereas that of the bafilomycin nanoparticles was 2.046 nM (Figure 4D). The benefit of nanoparticle delivery was less pronounced with bafilomycin, which may be attributed to the strong potency of the compound. Calculation of TI yielded values of 6.82 and 34.02 for the free bafilomycin and bafilomycin nanoparticles, respectively. Compared with free drugs, the nanoparticulate V-ATPase inhibitors exhibited lower cytotoxicity and greater antiviral activity, improving the therapeutic index of diphyllin and bafilomycin by approximately three- and fivefold, respectively.
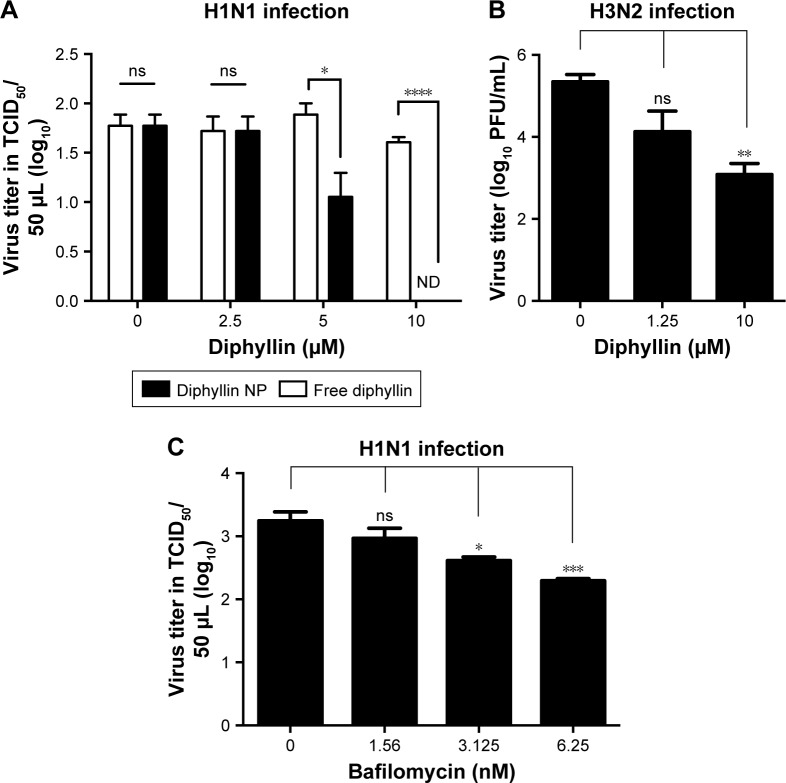
Inhibition of the replication of influenza virus
To further compare and assess the viral inhibitory effect of free or nanoparticulate V-ATPase inhibitors, the yield of progeny virus upon nanoparticulate diphyllin or bafilomycin treatment in MH-S cells was examined. The results showed that diphyllin nanoparticles induced dose-dependent inhibition against the H1N1 infection. The group treated with 5 and 10 µM diphyllin nanoparticles had a significant decrease in viral TCID50 titer compared with the untreated and the free diphyllin-treated groups (Figure 5A), demonstrating that the nanoparticle-mediated delivery could enhance the anti-influenza activity of the V-ATPase inhibitor. We further examined the nanoparticle’s activity against an H3N2 influenza virus to assess its potential as a broad-spectrum antiviral. As shown in Figure 5B, the mean viral PFU titer of the group treated with 10 µM diphyllin nanoparticles was 1.6×103 PFU/mL, whereas the mean virus titer of the untreated (medium only) group was 2.7×105 PFU/mL, validating that the diphyllin nanoparticle can be applied against multiple strains of influenza viruses.
Figure 5.
Antiviral activity of free and nanoparticulate V-ATPase inhibitors.
Notes: (A) MH-S cells were pretreated with free diphyllin or diphyllin nanoparticles (diphyllin NP) and were infected with H1N1 virus at an MOI of 1. Following 24 hours of incubation, the supernatants were harvested to assess the viral TCID50 titers. Viral titers between free diphyllin and diphyllin NP-treated groups were compared by unpaired t-test. (B) MH-S cells were pretreated with diphyllin nanoparticles and were infected with H3N2 virus at an MOI of 5. Following 24 hours of incubation, the supernatants were harvested to assess virus titers by plaque assay. (C) MH-S cells were pretreated with bafilomycin nanoparticles and were infected with H1N1 at an MOI of 1. Following 24 hours of incubation, the supernatants were harvested to assess the viral TCID50 titers. Viral titers among drug-treated groups and the untreated control group were compared by one-way ANOVA followed by Dunnett’s multiple comparisons test (ns: nonsignificant, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001). Data in the plot represent mean ± SEM out of three replicates.
Abbreviations: V-ATPase, vacuolar ATPase; NP, nanoparticle; MOI, multiplicity of infection; TCID50, 50% tissue culture infective dose; SEM, standard error of the mean.
In parallel, the anti-influenza activity for the bafilomycin nanoparticles was also assessed. Similarly, the MH-S cells were pretreated with bafilomycin nanoparticles in various concentrations and then infected with influenza virus H1N1. After infection, the virus was removed and bafilomycin nanoparticles were added to cells. After 24 hours of incubation, the supernatant was harvested to determine the viral titer by TCID50. The results showed that the bafilomycin nanoparticles exhibited dose-dependent inhibition against H1N1 virus infection. The viral titer in the group treated with 6.25 nM bafilomycin was 102.29 TCID50/50 µL and the group treated with 3.125 nM bafilomycin was 102.61 TCID50/50 µL. These groups significantly reduced viral titer compared with the medium control group (103.24 TCID50/50 µL) (Figure 5C), demonstrating that the nanoparticle delivery strategy can be applied to different types of V-ATPase inhibitors for antiviral applications.
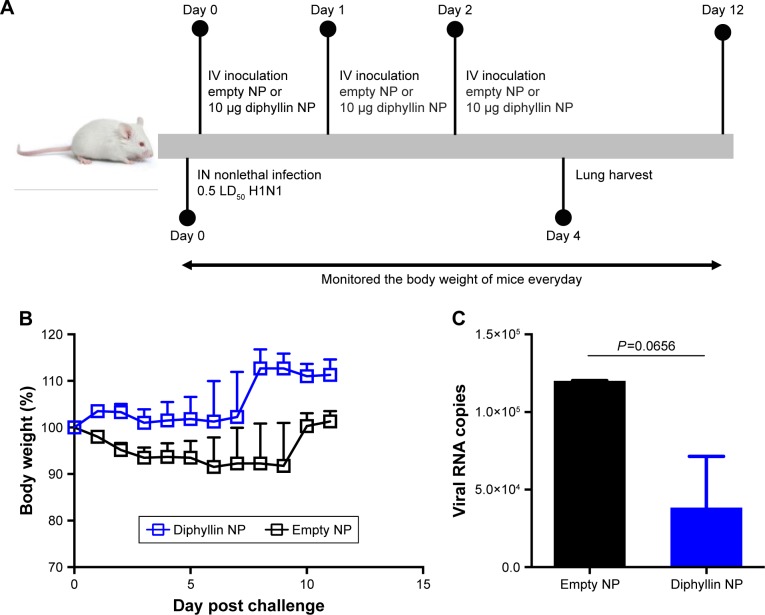
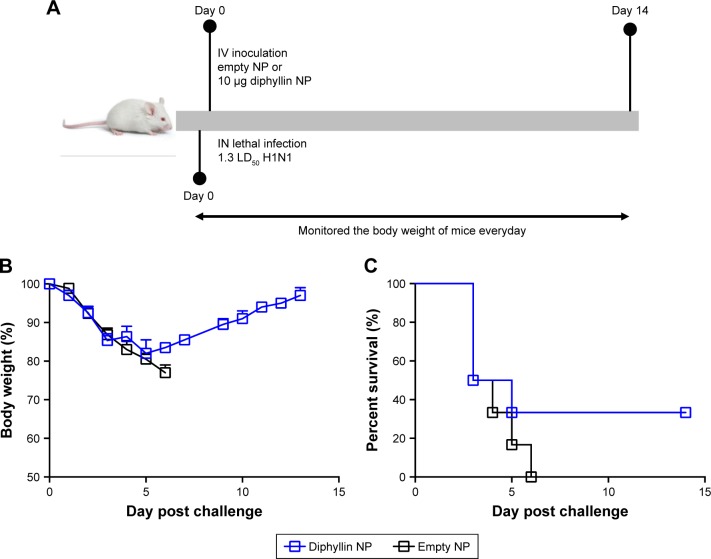
Antiviral efficacy of diphyllin nanoparticles in vivo
As the diphyllin nanoparticles were previously shown to be well tolerated in mice,23 we further examined the applicability of the nanoparticulate V-ATPase inhibitors for influenza treatment in vivo, a mouse model of influenza infection was used. Mice were inoculated intranasally with a nonlethal dose of H1N1 virus and intravenously treated with diphyllin nanoparticles equivalent to 10 µg of diphyllin or empty nanoparticles of equivalent polymer content daily for 3 days (Figure 6A). The mice were subsequently monitored for weight loss and lung viral load. In the study, the control mice progressively lost weight for 9 days. In contrast, mice treated with diphyllin nanoparticles have reduced body weight loss (Figure 6B). The lungs of mice were harvested for viral RNA assessment on day 5 postinfection. Although statistical significance was not reached, diphyllin nanoparticle treatment alleviated the viral load in the lungs of mice (Figure 6C). We further evaluated the efficacy of diphyllin nanoparticles in a lethal infection of H1N1 virus (Figure 7A). Compared with the control group, mice receiving single treatment of diphyllin nanoparticles showed reduced weight loss (Figure 7B) and improved survival rate (33%, 2/6) upon termination of the study (Figure 7C). In contrast, all the empty nanoparticle-receiving mice succumbed to virus infection within 6 days postinfection. These in vivo data collectively suggest that the diphyllin nanoparticles are able to reduce disease severity and enhance the survival outcome, mirroring the anti-influenza activity we observed with in vitro investigations.
Figure 6.
Antiviral efficacy of diphyllin nanoparticles in vivo in a nonlethal dose viral infection.
Notes: (A) Schematic presentation of the mice treatment schedule. Mice (6 mice/group) were inoculated intranasally with 100,000 PFU (0.5 LD50) of H1N1 and intravenously treated with nanoparticles loaded with 10 µg of diphyllin nanoparticles or empty nanoparticles of equivalent polymer content daily for 3 days. (B) The body weight of the mice was monitored in both groups. (C) Viral load in lungs of mice in both groups on day 5 postinfection. Two groups were compared by unpaired t-test. Data in the plot represent the mean ± SEM.
Abbreviations: NP, nanoparticle; IV, intravenous; IN, intranasal; PFU, plaque-forming unit; LD50, 50% lethal dose; SEM, standard error of the mean.
Figure 7.
Antiviral efficacy of diphyllin nanoparticles in vivo in a lethal dose viral infection.
Notes: (A) Schematic presentation of the mice treatment schedule. Mice (6 mice/group) were inoculated intranasally with 250,000 PFU (1.3 LD50) of H1N1 and intravenously treated with nanoparticles loaded with 10 µg of diphyllin nanoparticles or empty nanoparticles of equivalent polymer content daily for 1 day. (B) The body weight of the mice was monitored in both groups. (C) The survival rate of the mice was evaluated in both groups.
Abbreviations: NP, nanoparticle; IV, intravenous; IN, intranasal; PFU, plaque-forming unit; LD50, 50% lethal dose.
Discussion
Because of the emergence of drug-resistant viral strains, more and more anti-influenza studies begin to focus on host-targeted therapies. In our study, we utilized two V-ATPase inhibitors, diphyllin and bafilomycin, to interfere with endosome acidification, which is a critical process for influenza virus replication. Previous studies have also relied on the inhibition of endosome acidification to fight against influenza virus infection,19,20,29 and they have used alternative V-ATPase inhibitor saliphenylhalamide (SaliPhe) and several older V-ATPase inhibitors such as concanamycin A and archazolid B to study their efficacy against influenza A virus in vitro. All the aforementioned V-ATPase inhibitors have been shown to inhibit endosomal acidification and influenza virus replication in vitro, demonstrating broad-spectrum treatment potentials.
Drug delivery and drug toxicity are two critical factors that can undermine the therapeutic potential of a candidate compound. To overcome these challenges, nanoparticulate drug delivery system offers several advantages in terms of improving drug delivery and reducing toxicity compared with the traditional forms of drugs. In our study, diphyllin and bafilomycin are hydrophobic compounds that possess cytotoxicity concerns. To improve the applicability of these compounds, we adopted a PEG-PLGA-based polymeric nanoparticle system, which readily encapsulates water-insoluble molecules in its hydrophobic polymeric core. The nanocarriers can then facilitate delivery of diphyllin and bafilomycin in the absence of organic solvents, steadying the drugs’ pharmacokinetics through PEG-mediated long circulation and sustained drug release.30 Upon nanoparticle encapsulation, the water solubility of the drugs is improved and the cytotoxicity reduced. Moreover, diphyllin and bafilomycin showed a sustained release profile, which is advantageous toward improving the drugs’ pharmacokinetics. In our previous report, we showed that diphyllin nanoparticles could enhance the compound’s efficacy against the feline coronavirus,23 and this study further verified the nanocarriers’ effect against influenza virus. Other studies have shown similar benefits upon nanoparticle encapsulation of antivirals. For instance, SaliPhe, a poorly soluble compound, has been encapsulated into porous silicon nanoparticles. SaliPhe-loaded porous silicon nanoparticles improved the compound’s water solubility and reduced its side effect. However, the SaliPhe nanoparticles’ in vivo efficacy remains to be studied.29 In another example, HIV-I integrase strand transfer inhibitor Dolutegravir was encapsulated into nanoparticles by a polymer p407. After nanoparticle encapsulation, this drug could better target macrophages and attenuate viral replication and spread of virus to CD4+ T cells.31 These results echo our present finding that nanoparticulate drug delivery system can be a useful tool for improving the delivery of V-ATPase inhibitors.
AMs are susceptible to influenza virus infection,32,33 and they play a protective role of immunity in the disease.34 Depletion of AMs during influenza infection promotes secondary bacterial superinfections, thereby increasing severity of the disease.35 MH-S cells, an AM cell line derived from BALB/c mice, retain many of the properties of AMs36 and has been used as an in vitro model for influenza studies.37 In our data, two nanoparticulate V-ATPase inhibitors exhibited potent anti-influenza activity in AMs in vitro, and the effectiveness in other respiratory epithelial cells such as A549, Calu-3, RPMI 2650 cells, or a recently developed well-differentiated airway epithelium culture system38 are of interest for future investigation. Beyond respiratory tract tissues, several ocular epithelial cells have been shown to support productive replication of both avian and human influenza viruses in vitro or ex vivo,29,39–42 and influenza virus has been isolated from the eye of infected birds.43 In this study, ARPE-19 cells, a retinal epithelial cell line, were shown to uptake the drug nanoparticles efficiently, suggesting the potential application of V-ATPase-targeting treatment for the viral infection in the eyes. Further studies may be extended to validate the antiviral effectiveness of diphyllin or bafilomycin nanoparticles in this cell.
A major advantage behind antiviral approaches based on interference or promotion of host factors is that they can have broad-spectrum antiviral activities and are less susceptible to drug resistance mutations. As numerous cellular pathways are essential for viral replication, host-directed therapeutics have the potential to reduce viral replication and diminish harmful inflammatory responses.11,44 Several host-targeted antivirals are currently under investigation in late-phase clinical trials.45 In addition to V-ATPase, inhibitors of other host factors may be contemplated for improving the design of antiviral nanoparticles. Given the recent rise in immunomodulatory nanoparticle development,46,47 antiviral nanoparticles may also incorporate innate immune modulators to further enhance treatment potency. This study, for the first time, demonstrates the therapeutic promise of nanoparticulate V-ATPase inhibitors in a mouse model of influenza virus infection. The use of diphyllin nanoparticle reduced the severity of influenza and improved the survival outcome in a mouse model of infection, and further investigation of its prophylactic or therapeutic efficacy is warranted. For future studies, drug dosing and administration route can be further optimized. As the nasal epithelium serves as the portal of entry for the influenza virus,2 a local intranasal administration of the antiviral nanoparticles may be considered. Moreover, an accumulated line of evidence shows that drug combinations significantly improve the anti-influenza efficacy for many existing treatments.48–50 We previously showed that combinations of diphyllin and oseltamivir or of diphyllin and amantadine demonstrated enhanced antiviral effect and cell protection.22 Several randomized controlled trials are set to test the safety and efficacy of various combinations of influenza antivirals.51 In a recent example, administration of protectin D1, a host-targeted therapeutic candidate, showed 100% protection against a lethal influenza viral challenge when combined with the peramivir, a pathogen-targeted antiviral drug, while treatment with protectin D1 or peramivir alone provided only partial protection (30%).52 In light of these successful combinatorial treatment, simultaneous delivery of multiple synergistic antiviral drugs by a single nanocarrier may be designed under the versatile nanotechnology platform. Furthermore, targeting specificity of host-directed therapeutics may be enhanced by surface modifications of drug-loaded nanoparticles.53 For instance, influenza virus targeting by nanoparticles was recently demonstrated with a cell membrane-coated nanoparticle.54 The strategy may be applied to further facilitate antiviral drug targeting. Future studies are warranted to further validate and improve the nanoparticle-based antiviral treatments.
Conclusion
In summary, we have successfully constructed and characterized diphyllin-loaded nanoparticles and bafilomycin-loaded nanoparticles. The nanoparticles achieved a sustained release beyond 72 hours and were efficiently taken up by multiple types of cell lines. In cell-based assays, nanoformulated diphyllin or bafilomycin revealed enhanced safety and viral inhibitory activities than free drugs. Moreover, diphyllin-loaded nanoparticles showed broad-spectrum activity against two subtypes of virus infections, H1N1 and H3N2. In selecting diphyllin for further in vivo studies, diphyllin-loaded nanoparticles were well tolerated in mice and reduced the weight loss and lung viral load after H1N1 influenza virus infection. The nanoparticles also improved mice survival following a lethal influenza viral challenge. Collectively, this work highlights nanoformulated V-ATPase inhibitors as potential host-targeted treatments against influenza. Continued development of the nanoformulations is warranted.
Supplementary material
HPLC analysis of diphyllin and bafilomycin.
Notes: (A) The standard curve was constructed with serially diluted samples of diphyllin (0.125, 0.03125, 0.0078125, and 0.001953125 mg/mL). (B) The standard curve was constructed with serially diluted samples of bafilomycin (0.5, 0.25, 0.125, 0.0625, and 0.03125 mg/mL).
Abbreviation: HPLC, high-performance liquid chromatography.
Acknowledgments
This work was supported by National Taiwan University (107L104315 to H-WC), Ministry of Science and Technology of Taiwan (MOST-106-2923-B-002-004-MY3 to H-WC), Infectious Diseases Research and Education Center (to Y-TC), and the Career Development Award from Academia Sinica (to C-MJH).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bouvier NM, Palese P. The biology of influenza viruses. Vaccine. 2008;26(Suppl 4):D49–D53. doi: 10.1016/j.vaccine.2008.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dou D, Revol R, Östbye H, Wang H, Daniels R. Influenza A virus cell entry, replication, virion assembly and movement. Front Immunol. 2018;9:1581. doi: 10.3389/fimmu.2018.01581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies WL, Grunert RR, Haff RF, et al. Antiviral activity of 1-adamantanamine (amantadine) Science. 1964;144(3620):862–863. doi: 10.1126/science.144.3620.862. [DOI] [PubMed] [Google Scholar]
- 4.Dong G, Peng C, Luo J, et al. Adamantane-resistant influenza a viruses in the world (1902–2013): frequency and distribution of M2 gene mutations. PLoS One. 2015;10(3):e0119115. doi: 10.1371/journal.pone.0119115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yen HL. Current and novel antiviral strategies for influenza infection. Curr Opin Virol. 2016;18:126–134. doi: 10.1016/j.coviro.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Cohen M, Zhang XQ, Senaati HP, et al. Influenza A penetrates host mucus by cleaving sialic acids with neuraminidase. Virol J. 2013;10:321. doi: 10.1186/1743-422X-10-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hurt AC. The epidemiology and spread of drug resistant human influenza viruses. Curr Opin Virol. 2014;8:22–29. doi: 10.1016/j.coviro.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 8.König R, Stertz S, Zhou Y, et al. Human host factors required for influenza virus replication. Nature. 2010;463(7282):813–817. doi: 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han J, Perez JT, Chen C, et al. Genome-wide CRISPR/Cas9 screen identifies host factors essential for influenza virus replication. Cell Rep. 2018;23(2):596–607. doi: 10.1016/j.celrep.2018.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hao L, Sakurai A, Watanabe T, et al. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature. 2008;454(7206):890–893. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baillie JK, Digard P. Influenza – time to target the host? N Engl J Med. 2013;369(2):191–193. doi: 10.1056/NEJMcibr1304414. [DOI] [PubMed] [Google Scholar]
- 12.Huss M, Wieczorek H. Inhibitors of V-ATPases: old and new players. J Exp Biol. 2009;212(Pt 3):341–346. doi: 10.1242/jeb.024067. [DOI] [PubMed] [Google Scholar]
- 13.Pierson TC, Diamond MS. Degrees of maturity: the complex structure and biology of flaviviruses. Curr Opin Virol. 2012;2(2):168–175. doi: 10.1016/j.coviro.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Townsley AC, Weisberg AS, Wagenaar TR, Moss B. Vaccinia virus entry into cells via a low-pH-dependent endosomal pathway. J Virol. 2006;80(18):8899–8908. doi: 10.1128/JVI.01053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albertini AA, Baquero E, Ferlin A, Gaudin Y. Molecular and cellular aspects of rhabdovirus entry. Viruses. 2012;4(1):117–139. doi: 10.3390/v4010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez-Dunia D, Cubitt B, de la Torre JC. Mechanism of Borna disease virus entry into cells. J Virol. 1998;72(1):783–788. doi: 10.1128/jvi.72.1.783-788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bowman EJ, Siebers A, Altendorf K. Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc Natl Acad Sci U S A. 1988;85(21):7972–7976. doi: 10.1073/pnas.85.21.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ochiai H, Sakai S, Hirabayashi T, Shimizu Y, Terasawa K. Inhibitory effect of bafilomycin A1, a specific inhibitor of vacuolar-type proton pump, on the growth of influenza A and B viruses in MDCK cells. Antiviral Res. 1995;27(4):425–430. doi: 10.1016/0166-3542(95)00040-s. [DOI] [PubMed] [Google Scholar]
- 20.Müller KH, Kainov DE, El Bakkouri K, et al. The proton translocation domain of cellular vacuolar ATPase provides a target for the treatment of influenza A virus infections. Br J Pharmacol. 2011;164(2):344–357. doi: 10.1111/j.1476-5381.2011.01346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeganeh B, Ghavami S, Kroeker AL, et al. Suppression of influenza A virus replication in human lung epithelial cells by noncytotoxic concentrations bafilomycin A1. Am J Physiol Lung Cell Mol Physiol. 2015;308(3):L270–L286. doi: 10.1152/ajplung.00011.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen HW, Cheng JX, Liu MT, et al. Inhibitory and combinatorial effect of diphyllin, a v-ATPase blocker, on influenza viruses. Antiviral Res. 2013;99(3):371–382. doi: 10.1016/j.antiviral.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu CJ, Chang WS, Fang ZS, et al. Nanoparticulate vacuolar ATPase blocker exhibits potent host-targeted antiviral activity against feline coronavirus. Sci Rep. 2017;7(1):13043. doi: 10.1038/s41598-017-13316-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makadia HK, Siegel SJ. Poly lactic-co-glycolic acid (PLGA) as biodegradable controlled drug delivery carrier. Polymers (Basel) 2011;3(3):1377–1397. doi: 10.3390/polym3031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hrkach J, Von Hoff D, Mukkaram Ali M, et al. Preclinical development and clinical translation of a PSMA-targeted docetaxel nanoparticle with a differentiated pharmacological profile. Sci Transl Med. 2012;4(128):128ra39. doi: 10.1126/scitranslmed.3003651. [DOI] [PubMed] [Google Scholar]
- 26.Roberts RA, Shen T, Allen IC, Hasan W, DeSimone JM, Ting JP. Analysis of the murine immune response to pulmonary delivery of precisely fabricated nano- and microscale particles. PLoS One. 2013;8(4):e62115. doi: 10.1371/journal.pone.0062115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. Am J Epidemiol. 1938;27(3):493–497. [Google Scholar]
- 28.Ward CL, Dempsey MH, Ring CJ, et al. Design and performance testing of quantitative real time PCR assays for influenza A and B viral load measurement. J Clin Virol. 2004;29(3):179–188. doi: 10.1016/S1386-6532(03)00122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bimbo LM, Denisova OV, Mäkilä E, et al. Inhibition of influenza A virus infection in vitro by saliphenylhalamide-loaded porous silicon nanoparticles. ACS Nano. 2013;7(8):6884–6893. doi: 10.1021/nn402062f. [DOI] [PubMed] [Google Scholar]
- 30.Hu CM, Fang RH, Luk BT, Zhang L. Polymeric nanotherapeutics: clinical development and advances in stealth functionalization strategies. Nanoscale. 2014;6(1):65–75. doi: 10.1039/c3nr05444f. [DOI] [PubMed] [Google Scholar]
- 31.Sillman B, Bade AN, Dash PK, et al. Creation of a long-acting nano-formulated dolutegravir. Nat Commun. 2018;9(1):443. doi: 10.1038/s41467-018-02885-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Powe JR, Castleman WL. Canine influenza virus replicates in alveolar macrophages and induces TNF-alpha. Vet Pathol. 2009;46(6):1187–1196. doi: 10.1354/vp.08-VP-0229-P-FL. [DOI] [PubMed] [Google Scholar]
- 33.Rodgers BC, Mims CA. Influenza virus replication in human alveolar macrophages. J Med Virol. 1982;9(3):177–184. doi: 10.1002/jmv.1890090304. [DOI] [PubMed] [Google Scholar]
- 34.He W, Chen CJ, Mullarkey CE, et al. Alveolar macrophages are critical for broadly-reactive antibody-mediated protection against influenza A virus in mice. Nat Commun. 2017;8(1):846. doi: 10.1038/s41467-017-00928-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghoneim HE, Thomas PG, McCullers JA. Depletion of alveolar macrophages during influenza infection facilitates bacterial superinfections. J Immunol. 2013;191(3):1250–1259. doi: 10.4049/jimmunol.1300014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mbawuike IN, Herscowitz HB. MH-S, a murine alveolar macrophage cell line: morphological, cytochemical, and functional characteristics. J Leukoc Biol. 1989;46(2):119–127. doi: 10.1002/jlb.46.2.119. [DOI] [PubMed] [Google Scholar]
- 37.Zhang R, Ai X, Duan Y, et al. Kaempferol ameliorates H9N2 swine influenza virus-induced acute lung injury by inactivation of TLR4/MyD88-mediated NF-κB and MAPK signaling pathways. Biomed Pharmacother. 2017;89:660–672. doi: 10.1016/j.biopha.2017.02.081. [DOI] [PubMed] [Google Scholar]
- 38.Wu NH, Yang W, Beineke A, et al. The differentiated airway epithelium infected by influenza viruses maintains the barrier function despite a dramatic loss of ciliated cells. Sci Rep. 2016;6:39668. doi: 10.1038/srep39668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan RW, Kang SS, Yen HL, et al. Tissue tropism of swine influenza viruses and reassortants in ex vivo cultures of the human respiratory tract and conjunctiva. J Virol. 2011;85(22):11581–11587. doi: 10.1128/JVI.05662-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michaelis M, Geiler J, Klassert D, Doerr HW, Cinatl J., Jr Infection of human retinal pigment epithelial cells with influenza A viruses. Invest Ophthalmol Vis Sci. 2009;50(11):5419–5425. doi: 10.1167/iovs.09-3752. [DOI] [PubMed] [Google Scholar]
- 41.Creager HM, Kumar A, Zeng H, Maines TR, Tumpey TM, Belser JA. Infection and replication of influenza virus at the ocular surface. J Virol. 2018;92(7):pii: e02192-17. doi: 10.1128/JVI.02192-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Belser JA, Lash RR, Garg S, Tumpey TM, Maines TR. The eyes have it: influenza virus infection beyond the respiratory tract. Lancet Infect Dis. 2018;18(7):e220–e227. doi: 10.1016/S1473-3099(18)30102-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lenny BJ, Shanmuganatham K, Sonnberg S, et al. Replication capacity of avian influenza A(H9N2) virus in pet birds and mammals, Bangladesh. Emerg Infect Dis. 2015;21(12):2174–2177. doi: 10.3201/eid2112.151152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ludwig S. Disruption of virus-host cell interactions and cell signaling pathways as an anti-viral approach against influenza virus infections. Biol Chem. 2011;392(10):837–847. doi: 10.1515/BC.2011.121. [DOI] [PubMed] [Google Scholar]
- 45.Koszalka P, Tilmanis D, Hurt AC. Influenza antivirals currently in late-phase clinical trial. Influenza Other Respir Viruses. 2017;11(3):240–246. doi: 10.1111/irv.12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin LC, Chattopadhyay S, Lin JC, Hu CJ. Advances and opportunities in nanoparticle- and nanomaterial-based vaccines against bacterial infections. Adv Healthc Mater. 2018;7(13):e1701395. doi: 10.1002/adhm.201701395. [DOI] [PubMed] [Google Scholar]
- 47.Chattopadhyay S, Chen JY, Chen HW, Hu CJ. Nanoparticle vaccines adopting virus-like features for enhanced immune potentiation. Nanotheranostics. 2017;1(3):244–260. doi: 10.7150/ntno.19796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen JT, Smee DF, Barnard DL, et al. Efficacy of combined therapy with amantadine, oseltamivir, and ribavirin in vivo against susceptible and amantadine-resistant influenza A viruses. PLoS One. 2012;7(1):e31006. doi: 10.1371/journal.pone.0031006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haasbach E, Hartmayer C, Planz O. Combination of MEK inhibitors and oseltamivir leads to synergistic antiviral effects after influenza A virus infection in vitro. Antiviral Res. 2013;98(2):319–324. doi: 10.1016/j.antiviral.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 50.Belardo G, Cenciarelli O, La Frazia S, Rossignol JF, Santoro MG. Synergistic effect of nitazoxanide with neuraminidase inhibitors against influenza A viruses in vitro. Antimicrob Agents Chemother. 2015;59(2):1061–1069. doi: 10.1128/AAC.03947-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dunning J, Baillie JK, Cao B, Hayden FG, International Severe Acute Respiratory and Emerging Infection Consortium (ISARIC) Antiviral combinations for severe influenza. Lancet Infect Dis. 2014;14(12):1259–1270. doi: 10.1016/S1473-3099(14)70821-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morita M, Kuba K, Ichikawa A, et al. The lipid mediator protectin D1 inhibits influenza virus replication and improves severe influenza. Cell. 2013;153(1):112–125. doi: 10.1016/j.cell.2013.02.027. [DOI] [PubMed] [Google Scholar]
- 53.Hu CM, Zhang L. Nanoparticle-based combination therapy toward overcoming drug resistance in cancer. Biochem Pharmacol. 2012;83(8):1104–1111. doi: 10.1016/j.bcp.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 54.Chen HW, Fang ZS, Chen YT, et al. Targeting and enrichment of viral pathogen by cell membrane cloaked magnetic nanoparticles for enhanced detection. ACS Appl Mater Interfaces. 2017;9(46):39953–39961. doi: 10.1021/acsami.7b09931. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HPLC analysis of diphyllin and bafilomycin.
Notes: (A) The standard curve was constructed with serially diluted samples of diphyllin (0.125, 0.03125, 0.0078125, and 0.001953125 mg/mL). (B) The standard curve was constructed with serially diluted samples of bafilomycin (0.5, 0.25, 0.125, 0.0625, and 0.03125 mg/mL).
Abbreviation: HPLC, high-performance liquid chromatography.