Abstract
Posttranscriptional modifications of rRNA occur in the nucleolus where rRNA modification guide RNAs, or snoRNAs, concentrate. On the other hand, scaRNAs, the modification guide RNAs for spliceosomal snRNAs, concentrate in the Cajal body (CB). It is generally assumed, therefore, that snRNAs must accumulate in CBs to be modified by scaRNAs. Here we demonstrate that the evidence for the latter postulate is not consistent. In the nucleus, scaRNA localization is not limited to CBs. Furthermore, canonical scaRNAs can modify rRNAs. We suggest that the conventional view that scaRNAs function only in the CB needs revision.
Keywords: Cajal body, RNA modification, scaRNA, snoRNA
INTRODUCTION
Site-specific positioning of the most abundant posttranscriptional RNA modifications, such as pseudouridylation and 2′-O-methylation, is usually mediated by specialized small RNAs called guide RNAs. The majority of these guide RNAs concentrate in the nucleolus and are responsible for modification of ribosomal RNAs (rRNAs). Therefore, they are designated small nucleolar RNAs, or snoRNAs (Maxwell and Fournier 1995; Smith and Steitz 1997; Decatur and Fournier 2003). Early studies of rRNA modification demonstrated that most if not all of the 2′-O-methylation of rRNAs occurs on the nascent 45S precursor (for reviews, see Maden 1990; Decatur and Fournier 2003). Thus, the nucleolus is the place where newly transcribed rRNAs meet matching modification guide RNAs and become modified cotranscriptionally.
Spliceosomal U snRNAs represent another class of heavily modified RNAs. Their guide RNAs, with one exception (U6 snRNA 2′-O-methylation), are often concentrated in a separate nuclear compartment, the Cajal body (CB). Consequently, these modification guide RNAs are named small CB-specific RNAs, or scaRNAs. CB-localization elements (CAB-box and G.U/U.G wobble stem) and their association with the WDR79 protein are essential for targeting scaRNAs to CBs (Richard et al. 2003; Tycowski et al. 2009; Deryusheva and Gall 2013; Marnef et al. 2014). CBs are found in various eukaryotic species and are usually identified by the marker protein coilin. They are also characterized by a high concentration of spliceosomal snRNAs and core components of their modification machinery, such as the methyltransferase fibrillarin and the pseudouridine synthase dyskerin. The fact that modification guide RNPs and their cognate substrate RNAs both accumulate to high concentration in the CB led to the hypothesis that snRNA modification itself occurs there. In support of this hypothesis, fragments of U2 and U5 snRNAs became modified when they were specifically targeted to CBs, whereas fragments of rRNA and U6 snRNA became modified when they were targeted to nucleoli (Ganot et al. 1999; Jády et al. 2003).
However, the evidence that snRNA modification occurs only in CBs has never been strong. Most notably, CBs are not universal cell components, being absent from many normal cells, including stem cells (Liu et al. 2009). Furthermore, snRNA modification is unaffected in the coilin mutant of Drosophila, which lacks CBs entirely (Deryusheva and Gall 2009). Here we describe additional observations that question the exclusive roles of the CB and scaRNAs in posttranscriptional modification of spliceosomal snRNAs.
OBSERVATIONS AND CONCLUSIONS
Subtle structural changes make substrate RNA unmodifiable
Key evidence that snRNA modifications occur in CBs came from an experimental setup in which test RNA fragments were inserted into scaRNA U87/SCARNA5 for targeting to CBs (Jády et al. 2003). In our study of Drosophila scaRNA modifications, we adapted U87-based constructs to express stable artificial substrate RNAs in a yeast cell system (Deryusheva and Gall 2013). When we tested the ability of Drosophila scaRNA: ΨU2–55 to modify 28S rRNA, we accidentally created a construct with a motif specifically recognized by the yeast pseudouridine synthase Pus4 (Fig. 1A, underlined in top sequence). The presence of this accidentally created motif resulted in the positioning of an extra pseudouridine outside the test 28S rRNA sequence inserted in the U87 backbone RNA (Fig. 1B). To avoid any confusion, we mutated the Pus4p-modified uridine to cytosine. Unexpectedly, this new substrate RNA became unmodifiable, although a 1-nt mutation should not alter the substrate-guide RNA base-pairing (Fig. 1C). When adenine replaced the same uridine, the scaRNA:ΨU2–55 target position became efficiently modified again (Fig. 1D). Importantly, the expression level of the exogenous guide RNA scaRNA:ΨU2–55 was similar in all experiments, as verified by northern blots. Thus, minor alterations in secondary structure of a substrate RNA dramatically change the accessibility of RNA target sequences to the modification machinery (Fig. 1B′–D′).
FIGURE 1.
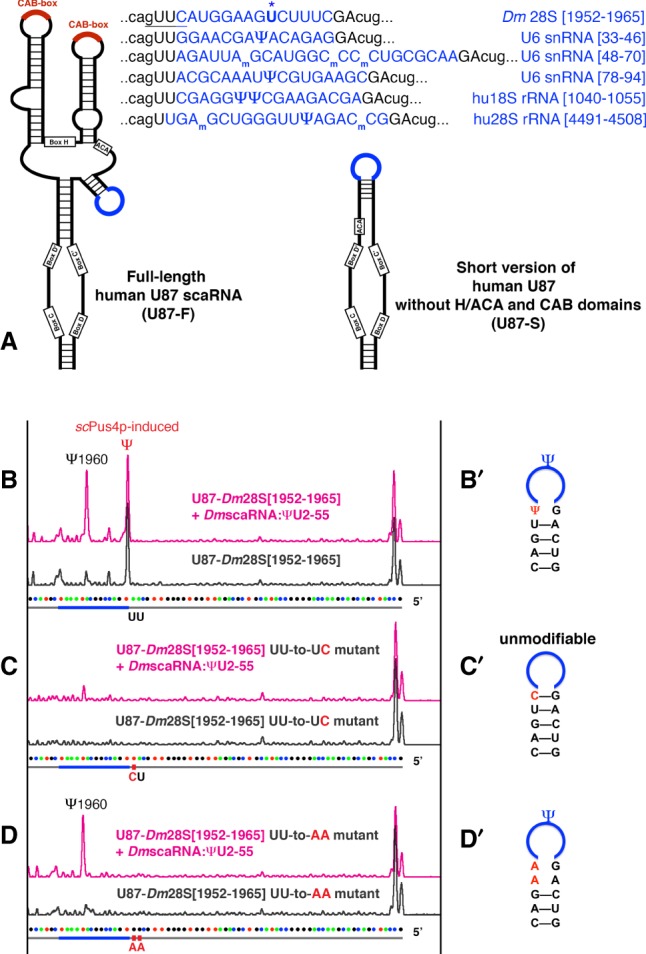
Structural alterations in substrate RNA can change modification efficiency. (A) Schematic drawing of two types of artificial substrate constructs used in this study. Highlighted in blue are fragments of Drosophila 28S, human 18S and 28S rRNAs, and U6 snRNA inserted into human U87 scaRNA. The yeast Pus4p-specific motif is underlined in the U87-Dm28S [1952–1965] substrate sequence; the tested target uridine is boldfaced and marked with a star (top row). (B–D) Modification of U87-Dm28S [1952–1965] substrate RNA mediated by Drosophila scaRNA:ΨU2–55 in yeast. (B′–D′) Predicted secondary structure at the site of substrate insertion. Induced pseudouridylation and point mutations are depicted. The U-to-C point mutation shown in C′ stabilizes the stem at the substrate loop and inhibits positioning of Ψ1960, as shown in C.
These observations prompted us to pursue an alternative explanation for negative results in RNA-targeting experiments.
Fragments of U6 snRNA and rRNAs become modified whether targeted to Cajal bodies or to nucleoli
Early experiments from the Kiss laboratory (Jády et al. 2003) demonstrated that fragments of U6 snRNA were not modified when targeted to CBs. This result supported the idea that typical snoRNAs in the nucleolus acted as guide RNAs for 2′-O-methylation of U6 snRNA (Tycowski et al. 1998; Ganot et al. 1999). Later, U6 pseudouridylation guide RNAs were identified and found concentrated in CBs (Tycowski et al. 2009; Deryusheva and Gall 2013). From these findings, one would expect that 2′-O-methylation of U6 snRNA occurs in the nucleolus and pseudouridylation in the CB. Yet, neither type of modification had been detected when U6 snRNA fragments were targeted to CBs (Jády et al. 2003).
To resolve these contradictory findings, we generated similar U87 constructs containing fragments of U6 snRNA (Fig. 1A). Because minor structural alterations can make substrate RNA unmodifiable, we first tested all constructs in a yeast cell system to be sure that they could be modified appropriately (data not shown). We then transfected HeLa cells. Full-length U87 scaRNA with inserted fragments of U6 snRNA concentrated in CBs as expected (Fig. 2A). However, in contrast to previously reported results (Jády et al. 2003), the inserted sequences became modified at positions specific to U6 snRNA, both pseudouridylation and 2′-O-methylation (Fig. 2B). Furthermore, rRNA fragments inserted into U87 scaRNA and targeted to CBs were also modified (Fig. 2B). Importantly, when fragments of U6 snRNA were inserted into U87 lacking the H/ACA domain and CB-localization signals (Fig. 1A, U87-S), they also became properly modified (Fig. 2B, blue trace). We should emphasize that the analysis is complicated by the fact that transiently expressed exogenous RNAs often show mislocalization within the transfected cells. In our experiments, we verified the concentration of substrate RNAs in nuclear bodies using fluorescent in situ hybridization (FISH). No more than 5% of the cells showed a weak signal in nucleoli in addition to accumulation in CBs, when they were transfected with full-length U87 scaRNA (Fig. 2A, middle panel). This minor mislocalized fraction of test substrate RNA cannot explain the very high efficiency of modification observed: Reverse transcription was terminated at modified positions, and few full-length fragments were detected (Fig. 2B, red traces). These observations demonstrate that targeting of RNA to a particular nuclear compartment is not essential for proper posttranscriptional modification.
FIGURE 2.
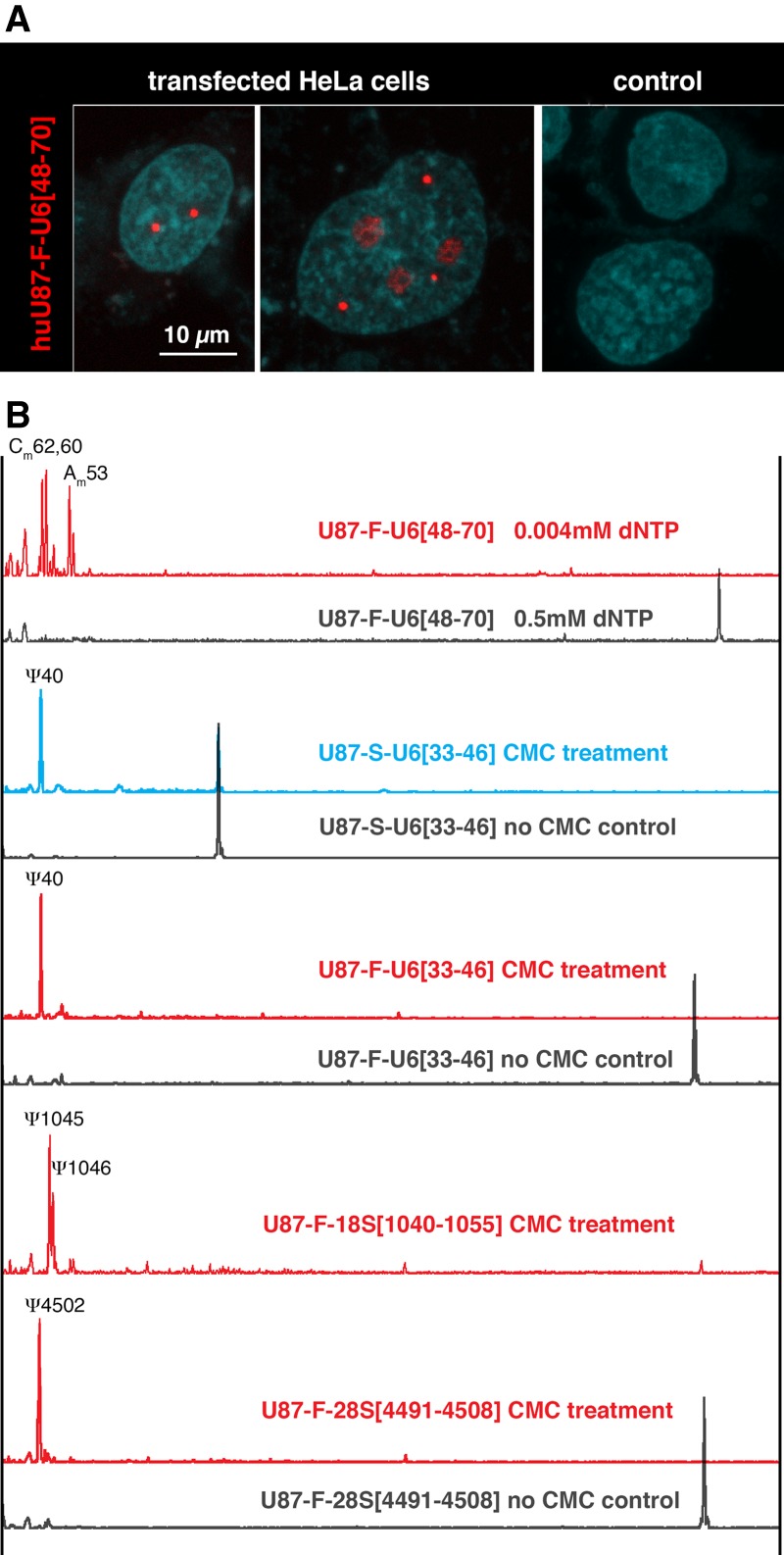
(A) A full-length human U87 scaRNA that contains a U6 snRNA insertion (U87-F-U6[48–70]) concentrates in CBs. (Left panel) Bright red FISH signals are detected in CBs of transfected HeLa cells after hybridization with a Cy3-labeled U87 antisense probe. (Middle panel) A few cells have a weak signal in the nucleoli. (Right panel) Control nontransfected cells show no detectable signal. Similar FISH patterns were observed when HeLa cells were transfected with U87-F constructs containing fragments of 18S and 28S rRNAs (not shown). (B) Fragments of rRNAs and U6 snRNA are properly modified when they are targeted to CBs (red traces). Modification also occurs when the fragments are inserted into a U87-S construct that lacks the H/ACA domain, which contains the CAB-box (blue trace). In each case, black traces are controls.
A canonical scaRNA can modify rRNA
Since the early discovery of dual guide RNAs for modification of U6 snRNA and 28S rRNA (Tycowski et al. 1998), several scaRNAs were predicted to mediate modification of both spliceosomal U snRNAs and rRNAs (Yuan et al. 2003; Deryusheva and Gall 2013). In fact, two canonical scaRNAs in Drosophila, scaRNA:ΨU1–6 and scaRNA:ΨU2–55, modified substrate RNAs corresponding to 18S and 28S rRNA sequences, when they were tested in yeast cells (Deryusheva and Gall 2013). Nevertheless, endogenous Drosophila rRNAs were not modified at the predicted target positions. One could argue that modification guide RNAs behave differently when expressed in yeast; for instance, yeast snoRNA snR81 normally modifies both U2 snRNA and 25S rRNA (Ma et al. 2005). Furthermore, the pseudouridylation pocket in snR81 that is specific to 25S rRNA mediates the formation of a stress-inducible pseudouridine in U2 snRNA (Wu et al. 2011). Thus, while it was evident that scaRNAs can modify rRNAs in yeast, it remained an open question whether the same is true in higher eukaryotes.
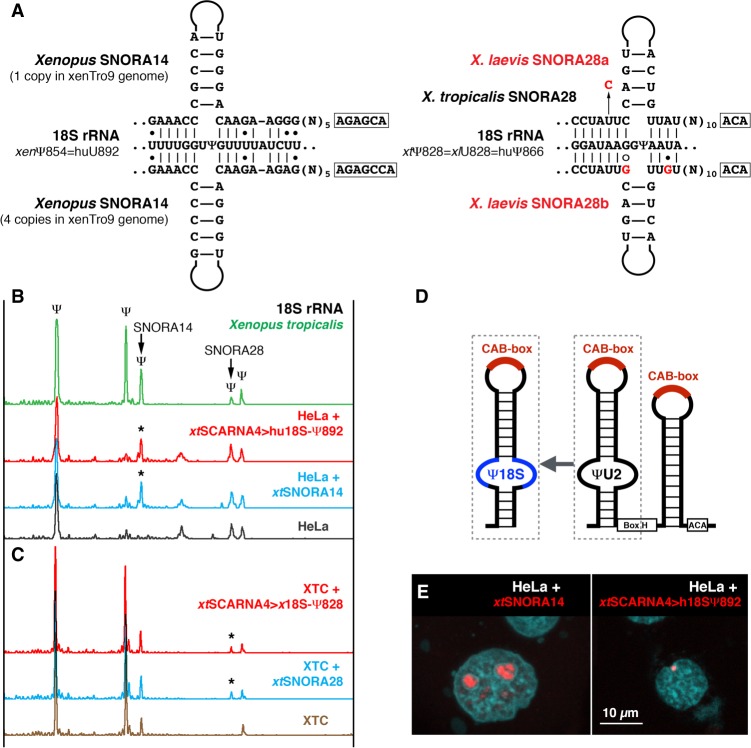
To address this question, we developed an assay using vertebrate cell lines. In our bioinformatics analysis of vertebrate snoRNAs, we identified amphibian and zebrafish SNORA14 as a potential guide RNA for 18S rRNA pseudouridylation at a position equivalent to position 892 in human 18S rRNA (Fig. 3A, left scheme). We showed pseudouridylation of this position in zebrafish and four amphibian species: axolotl, newt, Xenopus tropicalis (Fig. 3B, top green trace), and X. laevis (Fig. 3C), but no pseudouridylation in lizard, chicken, mouse, and human (Fig. 3B, bottom gray trace). We expressed Xenopus SNORA14 in HeLa cells and observed pseudouridylation of human 18S rRNA at position 892 (Fig. 3B, blue trace, star). This is an example of a typical snoRNA (Fig. 3E) that modifies rRNA at the predicted position. Then we generated an artificial guide RNA for human 18S-892 by antisense element replacement in X. tropicalis SCARNA4, which normally modifies U2 snRNA at position 41 (Fig. 3D). When expressed in HeLa cells, this chimeric scaRNA, xtSCARNA4>hu18S-Ψ892, concentrates in CBs as expected (Fig. 3E) and, despite the CB localization, induces modification of human 18S rRNA (Fig. 3B, red trace, star). Thus, these experiments demonstrate (i) that Xenopus SNORA14 is a genuine guide RNA for the tested position, (ii) that human 18S rRNA is promiscuous for pseudouridylation at this position, and more importantly (iii) that both snoRNAs and typical scaRNAs can modify rRNA.
FIGURE 3.
Typical scaRNAs can modify 18S rRNA. (A) Postulated base-pairing of Xenopus snoRNAs, SNORA14, and SNORA28, with 18S rRNA. Differences between X. tropicalis and X. laevis SNORA28 are highlighted in red. (B) 18S rRNA from X. tropicalis is pseudouridylated by SNORA28 at position 828 and by SNORA14 at position 854 (top green trace). In HeLa cells, the position corresponding to the Xenopus SNORA14 target is not modified (bottom gray trace). When Xenopus SNORA14 was expressed in HeLa cells, position 892 (equivalent to position 854 in Xenopus) became pseudouridylated (blue trace, star). Human 18S-892 guide RNA made from Xenopus SCARNA4 (xtSCARNA4>hu18S-Ψ892) also induced the pseudouridylation of 18S rRNA in HeLa cells (red trace, star). (C) In contrast to human and X. tropicalis, 18S rRNA in X. laevis is not pseudouridylated at position 828 (bottom brown trace). Expression of X. tropicalis SNORA28 in XTC cells induced pseudouridylation of this position (blue trace, star). This pseudouridine was also induced by the chimeric scaRNA xtSCARNA4>x18S-Ψ828 (red trace, star). (D) Schematic representation of 18S guide RNA generated from scaRNA. (E) In transfected HeLa cells, FISH with Cy3-labeled antisense RNA probes shows that xtSNORA14 localizes in nucleoli (left panel, red signal) and xtSCARNA4>hu18SΨ892 localizes in CBs (right panel, red signal).
To confirm this observation in other cells, we took advantage of differences in 18S rRNA modification between X. laevis and X. tropicalis. In X. laevis, position 828 in 18S rRNA is not pseudouridylated (Fig. 3C, bottom brown trace); this position is equivalent to position 866 in human and is known as a highly conserved pseudouridine across different vertebrate species, including X. tropicalis (Fig. 3B). Coinciding with the observed modification pattern, the corresponding guide RNA SNORA28 is mutated in X. laevis (Fig. 3A, right scheme). We transfected the X. laevis cell line XTC with constructs to express either X. tropicalis SNORA28 or chimeric scaRNA xtSCARNA4>xl18S-Ψ828. In both cases, X. laevis 18S rRNA became modified at position 828 (Fig. 3C, blue and red traces, stars). These experiments clearly demonstrate that scaRNA modification activity is not limited to spliceosomal snRNAs.
In fact, neither snoRNAs nor scaRNAs are limited to specific compartments. A recent study from Karla Neugebauer's laboratory demonstrated that various scaRNAs, when injected into cultured HeLa cells, concentrate in nucleoli in addition to CBs; similarly, snoRNAs, while obviously concentrated in nucleoli, were not excluded from CBs (Machyna et al. 2014). scaRNAs in Xenopus oocytes concentrate in CB-like nuclear organelles called pearls and in numerous extrachromosomal nucleoli; the removal of the CB-localization signal from a scaRNA did not change this localization (Nizami and Gall 2012). An earlier study of Xenopus oocytes (Yu et al. 2001) suggested that spliceosomal snRNAs concentrate in nucleoli and that post-transcriptional modification takes place there. In these experiments, however, the accumulation of U snRNAs was demonstrated on nuclear spreads. In such preparations, the nucleoplasm is not retained on the slides (Yu et al. 2001; Gerbi and Lange 2002; Gerbi et al. 2003). If RNA distribution is analyzed in whole oocyte nuclei isolated in mineral oil, no enrichment of U2 snRNA is observed in nucleoli (Fig. 4A), whereas scaRNA concentration there is evidently high (Fig. 4B). It is worth noting that even this observation of scaRNA concentration in nucleoli is misleading. In the case of fluorescently labeled scaRNAs injected into Xenopus oocytes, one can easily estimate that ∼98% of the nuclear signal is dispersed throughout the nucleoplasm, despite the presence of hundreds of extremely bright nucleoli (Fig. 4B). Thus, scaRNAs do normally concentrate in CBs, but they are also dispersed in the nucleoplasm and can re-localize to another nuclear compartment, especially in the absence of canonical CBs.
FIGURE 4.
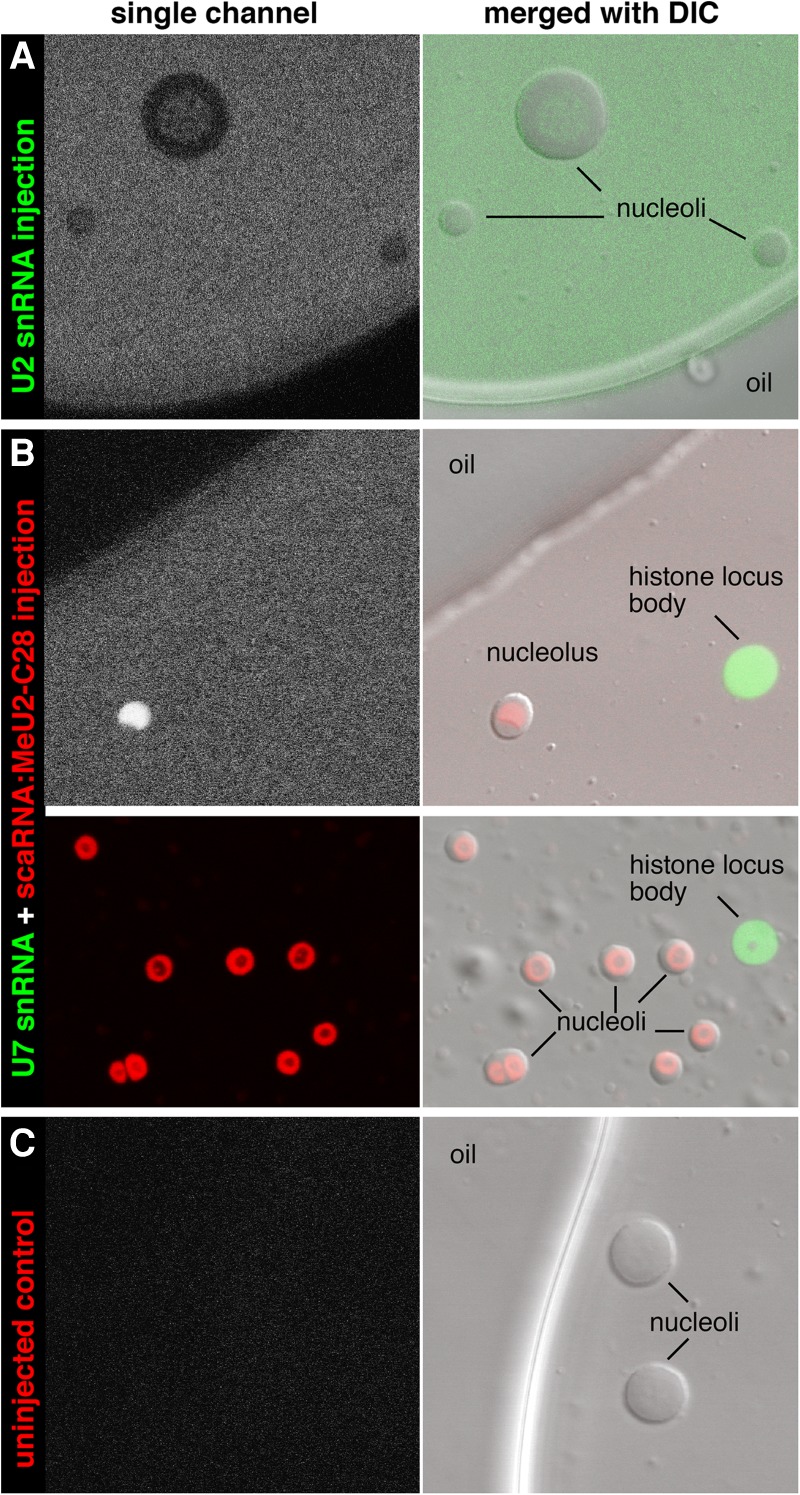
Distribution of fluorescently labeled snRNAs and scaRNA in X. laevis oocyte nuclei. RNAs were injected into the cytoplasm of intact oocytes. Eighteen hours later, nuclei were isolated under mineral oil, which preserves the nucleoplasm in which the organelles are suspended. (A) Oocytes injected with fluorescein-U2 snRNA. U2 snRNA (green) is not enriched in nucleoli relative to the nucleoplasm. (B) Oocytes injected with a mixture of fluorescein-U7 snRNA and in vitro-transcribed Drosophila scaRNA:MeU2-C28 labeled with Cy5. U7 snRNA (green) concentrates in histone locus bodies, whereas the scaRNA (red) concentrates in nucleoli. (C) Uninjected control nuclei do not fluoresce.
Final remarks
Normally scaRNAs concentrate in CBs, but their nuclear localization is not restricted to CBs and CBs are not required for scaRNA activity. Furthermore, scaRNA guide activity is not limited to spliceosomal RNAs. Similarly, snoRNAs can be found outside the nucleolus. scaRNPs contain at least one component not present in snoRNPs, the WDR79 protein, which is bound to the CAB-box. Yet neither WDR79 nor the CAB-box are essential for scaRNP guide activity (Deryusheva and Gall 2009). In summary, there is no sharp distinction between snoRNAs and scaRNAs. Their substrate specificities overlap as well as their localization within the nucleus.
MATERIALS AND METHODS
Expression of exogenous guide and substrate RNAs
Constructs were made in which fragments of U6 snRNA, 18S and 28S rRNAs were inserted in the human U87/SCARNA5 coding sequence (schematic representation shown in Fig. 1A); these chimeric U87 sequences themselves replaced intronic snoRNA snR18 in the yeast EFB1 gene. The resultant DNA fragments were cloned into pCS2 and p426Gal1 vectors for expression in vertebrate cell lines and in the yeast Saccharomyces cerevisiae, respectively. X. tropicalis SNORA14, SNORA28, and SCARNA4 occur within introns; these introns, along with their flanking exons, were amplified from genomic DNA and cloned into pCS2 vector. PCR-based mutagenesis was used to replace antisense element sequences in xtSCARNA4. Yeast expression constructs for these guide RNAs were generated as previously described (Deryusheva and Gall 2018).
Human HeLa and X. laevis XTC cells were transfected with the listed constructs using ViaFect and FuGene HD transfection reagents (Promega), respectively. Yeast expression constructs were introduced into BY4741 “wild-type” yeast strain using the standard lithium acetate transformation method. RNA was extracted from cultured transfected and control cells using TRIzol reagent and Direct-zol RNA MiniPrep kit (Zymo Research). Yeast RNA was extracted using the hot acid phenol method. Expression of exogenous RNAs was verified by northern blot analysis as previously described (Deryusheva and Gall 2013).
RNA modification analysis
Modifications of human and Xenopus 18S rRNA and artificial substrate RNAs inserted in the human U87 backbone were analyzed using a fluorescent primer extension method as previously described (Deryusheva and Gall 2009). CMC treatment of test RNA was used to map pseudouridines. A low concentration of dNTP in the reaction mix was used for 2′-O-methylation mapping. 6-FAM-labeled oligonucleotides were the following: SD339 (3′-terminus of human U87) AGGTCTCAGATTGAAAACTTGAGATCA and SD493 (1002–1033 nt of human 18S or 964–995 nt of Xenopus 18S rRNA) CGTTCTTGATTAATGAAAACGTTCTTGGCAAA.
Because RNA modification analyses based on reverse transcription are only semiquantitative, each sample was tested in duplicate or triplicate and separated on capillary columns in serial dilutions. All experiments were performed in parallel. Expression levels of exogenous guide and substrate RNAs were verified by northern blots, and their nuclear localization was determined by FISH.
Fluorescent in situ hybridization (FISH)
Transfected and control mock-transfected HeLa and XTC cells were grown on 4-well culture slides. The slides were rinsed in phosphate-buffered saline (PBS) and fixed in 4% formaldehyde/PBS for 10–15 min. The fixed cells were permeabilized in 0.3% Triton X-100 in PBS for 30 min followed by 70% methanol overnight. Slides were then either rehydrated and used for FISH experiments or stored refrigerated in 70% methanol until use. Cy3-labeled antisense RNA probes were made by in vitro transcription reactions using DNA constructs or PCR fragments as templates. Hybridization was performed overnight at 42°C. Slides were washed in 0.2× SSC three times for 10 min, stained with DAPI, and mounted in a Mowiol/DABCO antifade solution. Images were captured using a laser-scanning confocal microscope (Leica SP5) with a 63× (NA 1.40) planapochromatic objective.
Injections into Xenopus oocytes
In vitro transcribed fluorescently labeled RNAs (fluorescein-U2 snRNA, fluorescein-U7 snRNA, Cy5-DmscaRNA:MeU2-C28) were injected into X. laevis oocytes using a Nanoject microinjector (Drummond). The injection procedure and amount of injected RNAs were exactly the same as in the in vitro modification assay with unlabeled U2 snRNA and its guide scaRNA:MeU2-C28 (Deryusheva and Gall 2009). After overnight incubation, injected and control Xenopus oocyte nuclei were isolated under mineral oil. While still in oil, nuclei were transferred onto slides, gently covered with a coverslip, and analyzed by confocal microscopy. All manipulations with Xenopus were performed according to approved IACUC protocols.
ACKNOWLEDGMENTS
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health (grant number R01 GM33397 to J.G.G.). J.G.G. is American Cancer Society Professor of Developmental Genetics.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.068593.118.
REFERENCES
- Decatur WA, Fournier MJ. 2003. RNA-guided nucleotide modification of ribosomal and other RNAs. J Biol Chem 278: 695–698. 10.1074/jbc.R200023200 [DOI] [PubMed] [Google Scholar]
- Deryusheva S, Gall JG. 2009. Small Cajal body-specific RNAs of Drosophila function in the absence of Cajal bodies. Mol Biol Cell 20: 5250–5259. 10.1091/mbc.e09-09-0777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deryusheva S, Gall JG. 2013. Novel small Cajal body-specific RNAs identified in Drosophila: probing guide RNA function. RNA 19: 1802–1814. 10.1261/rna.042028.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deryusheva S, Gall JG. 2018. Orchestrated positioning of post-transcriptional modifications at the branch point recognition region of U2 snRNA. RNA 24: 30–42. 10.1261/rna.063842.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot P, Jády BE, Bortolin ML, Darzacq X, Kiss T. 1999. Nucleolar factors direct the 2′-O-ribose methylation and pseudouridylation of U6 spliceosomal RNA. Mol Cell Biol 19: 6906–6917. 10.1128/MCB.19.10.6906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbi SA, Lange TS. 2002. All small nuclear RNAs (snRNAs) of the [U4/U6.U5] Tri-snRNP localize to nucleoli; Identification of the nucleolar localization element of U6 snRNA. Mol Biol Cell 13: 3123–3137. 10.1091/mbc.01-12-0596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbi SA, Borovjagin AV, Odreman FE, Lange TS. 2003. U4 snRNA nucleolar localization requires the NHPX/15.5-kD protein binding site but not Sm protein or U6 snRNA association. J Cell Biol 162: 821–832. 10.1083/jcb.200301071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády BE, Darzacq X, Tucker KE, Matera AG, Bertrand E, Kiss T. 2003. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J 22: 1878–1888. 10.1093/emboj/cdg187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JL, Wu Z, Nizami Z, Deryusheva S, Rajendra TK, Gao H, Beumer KJ, Carroll D, Matera AG, Gall JG. 2009. Coilin is essential for Cajal body organization in Drosophila melanogaster. Mol Biol Cell 20: 1661–1670. 10.1091/mbc.e08-05-0525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Yang C, Alexandrov A, Grayhack EJ, Behm-Ansmant I, Yu YT. 2005. Pseudouridylation of yeast U2 snRNA is catalyzed by either an RNA-guided or RNA-independent mechanism. EMBO J 24: 2403–2413. 10.1038/sj.emboj.7600718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machyna M, Kehr S, Straube K, Kappei D, Buchholz F, Butter F, Ule J, Hertel J, Stadler PF, Neugebauer KM. 2014. The coilin interactome identifies hundreds of small noncoding RNAs that traffic through Cajal bodies. Mol Cell 56: 389–399. 10.1016/j.molcel.2014.10.004 [DOI] [PubMed] [Google Scholar]
- Maden BE. 1990. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol 39: 241–303. 10.1016/S0079-6603(08)60629-7 [DOI] [PubMed] [Google Scholar]
- Marnef A, Richard P, Pinzón N, Kiss T. 2014. Targeting vertebrate intron-encoded box C/D 2′-O-methylation guide RNAs into the Cajal body. Nucleic Acids Res 42: 6616–6629. 10.1093/nar/gku287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell ES, Fournier MJ. 1995. The small nucleolar RNAs. Annu Rev Biochem 35: 897–934. 10.1146/annurev.bi.64.070195.004341 [DOI] [PubMed] [Google Scholar]
- Nizami ZF, Gall JG. 2012. Pearls are novel Cajal body-like structures in the Xenopus germinal vesicle that are dependent on RNA pol III transcription. Chromosome Res 20: 953–969. 10.1007/s10577-012-9320-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Darzacq X, Bertrand E, Jády BE, Verheggen C, Kiss T. 2003. A common sequence motif determines the Cajal body-specific localisation of box H/ACA scaRNAs. EMBO J 22: 4283–4293. 10.1093/emboj/cdg394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CM, Steitz JA. 1997. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell 89: 669–672. 10.1016/S0092-8674(00)80247-0 [DOI] [PubMed] [Google Scholar]
- Tycowski KT, You ZH, Graham PJ, Steitz JA. 1998. Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol Cell 2: 629–638. 10.1016/S1097-2765(00)80161-6 [DOI] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Kukoyi A, Steitz JA. 2009. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell 34: 47–57. 10.1016/j.molcel.2009.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Xiao M, Yang C, Yu YT. 2011. U2 snRNA is inducibly pseudouridylated at novel sites by Pus7p and snR81 RNP. EMBO J 30: 79–89. 10.1038/emboj.2010.316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y-T, Shu MD, Narayanan A, Terns RM, Terns MP, Steitz JA. 2001. Internal modification of U2 small nuclear (sn)RNA occurs in nucleoli of Xenopus oocytes. J Cell Biol 152: 1279–1288. 10.1083/jcb.152.6.1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan G, Klämbt C, Bachellerie JP, Brosius J, Hüttenhofer A. 2003. RNomics in Drosophila melanogaster: identification of 66 candidates for novel non-messenger RNAs. Nucleic Acids Res 31: 2495–2507. 10.1093/nar/gkg361 [DOI] [PMC free article] [PubMed] [Google Scholar]