Abstract
Background
Regional distribution of adipose tissue is more important than total amount of body fat in predicting complications associated with obesity. Apolipoprotein B (Apo B) plays a central role in lipid metabolism.
Aim
To investigate the importance of the XbaI polymorphism of Apo B gene (C7673T) as risk factor for visceral obesity and its influence on lipid profile.
Subjects and methods
Total of 122 obese adult females (BMI ⩾ 30 kg/m2): 56 of them with visceral obesity (⩾7 cm by abdominal Ultrasound) and 66 without visceral obesity and 36 age matched non-obese (BMI ⩽ 25 kg/m2) without visceral obesity were studied. Anthropometric assessment, body composition, visceral obesity and lipid profile evaluation were attempted. Genetic analysis of Apo B XbaI was performed using Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP).
Results
Visceral obesity was associated significantly with the presence of the heterozygous (CT) genotype of the XbaI Apo B gene (p < 0.001). Frequency of homozygous (CC) was significantly the least genotype found in females with visceral obesity, while homozygote (TT) genotype was more frequent in those without visceral obesity. T allele (about 70%) was more frequent than C allele (about 30%) in all groups. Significant lowest values of visceral obesity, triglyceride and HDL-C were associated with the presence of (CC) genotype and the highest values were associated with the presence of the heterozygous (CT) genotype; except HDL-C with (TT) genotype.
Conclusions
Study reveals considerable association of Apo B XbaI gene polymorphism with visceral obesity and some lipid profile parameters (TG and HDL-C) among Egyptian females.
Keywords: Visceral obesity, Lipid profile, XbaI, Apo B, Polymorphism, Linkage disequilibrium
1. Introduction
Worldwide obesity has risen to titanic proportions and has become a major worldwide health problem [1]. In longitudinal study published by the Global Burden of Disease Study, the rate of weight gain expanded most rapidly in the Middle East particularly Egypt, which rank among the countries in the world with most overweight people. In Egypt, nearly 70% of the adult population—about 56.5 million people—are considered to be overweight. Over the past three decades, the highest rise in obesity levels among women has been found in Egypt. Being overweight is more than just an esthetic problem; people who are overweight have an increased risk of cardiovascular disease, cancer, diabetes, osteoarthritis, and chronic kidney disease, with most deaths due to cardiovascular problems, including heart attack and stroke [2].
A phenomenon that has received increasing attention is the fact that body shape, and more specifically the regional distribution of adipose tissue, is at least as important, if not more important, than the total amount of body fat in predicting disease-causing complications that have been traditionally associated with obesity. Literature on regional adipose tissue distribution and metabolism has flourished over the past 25 years. They concluded that the proportion of abdominal adipose tissue is a key correlate; and perhaps driver of the health risk associated with overweight and obesity. Visceral obesity has now been established as being part of a complex phenotype including adipose tissue storage dysfunction and ectopic triglyceride accumulation in several sites including the liver [3]. Nazare et al. [4] have reported ethnic differences in risk and prevalence of metabolic diseases including obesity. This may be related to patterns of ethnic-specific body fat distribution.
Dyslipidemias, a group of biochemical disorders, is frequently encountered in obese individuals. The predominant features of dyslipidemias include increased flux of free fatty acids (FFA), raised triglyceride (TG) and low high density lipoprotein cholesterol (HDL-C) levels, a predominance of small dense (atherogenic) low density lipoprotein cholesterol (LDL-C) particles and raised apolipoprotein B (Apo B) values. However, there is a considerable heterogeneity of plasma lipid profile among obese people. The precise cause of this heterogeneity is not entirely clear but has been partly attributed to the degree of visceral adiposity [5].
The Apo B gene is located on the short arm of chromosome (2p23-24) [6], [7]. Several single nucleotide polymorphisms in the Apo B gene have been described, including XbaI [8] and EcoRI [9]. The XbaI polymorphism arises due to a single base variation in exon 26 (at 2488th position ACC → ACT) of the Apo B gene that does not lead to change in amino acid sequence [6]. This polymorphism has been found to be associated with generalized and regional obesity and an increase in various lipoprotein subfractions (total cholesterol [TC], low density lipoprotein cholesterol [LDL-C], and triglycerides [TG]), but the results are conflicting [10].
Because of its central role in lipid transport and metabolism, the examination of the variations of the Apo B gene could help to explain interindividual variation in lipid levels and susceptibility to obesity [11].
1.1. Aim of work
This study has been designed to investigate the importance of the XbaI polymorphism of Apo B gene (C7673T) as risk factor for visceral obesity and its influence on lipid profile among a sample of Egyptian obese females.
2. Subjects and methods
2.1. Subjects
This study was a cross-sectional case control one, consisting of 158 females: 122 obese females (BMI ⩾ 30 kg/m2) and 36 apparently normal healthy ones (BMI ⩽ 25 kg/m2) serving as control group; aged 30–62 years (mean: 49.88 ± 5.6). The obese females were classified into 2 groups: obese with visceral obesity (56) and obese without visceral obesity (66) using cutoff point of 7 cm by abdominal Ultrasound. All subjects enrolled in the study were recruited from outpatient clinic of the “Visceral Obesity Management Unit” at “National Research Centre”; between October 2011 and December 2013. All obese and controls were subjected to a detailed medical history and information was obtained on demographic characteristics concerning history and duration of obesity and medications. Exclusion criteria were: history of smoking, acute or chronic infections, coronary artery disease, congestive heart failure, chronic liver disease, diabetic nephropathy, rheumatic disease and cancer. The participants were informed about the purpose of the study and their permission in the form of written consent was obtained. The protocol was approved by the “Ethics Committee” of the “National Research Centre”. The agreement reference number is 10/119.
Anthropometry assessment: Anthropometric evaluation was performed for all obese and control females. Body weight, height and waist circumference were measured following the recommendations of the International Biological Program [12]. Body weight was determined to the nearest 0.01 kg using a Seca Scale Balance, with the subject wearing minimal clothing and with no shoes. Body height was measured to the nearest 0.1 cm using a Holtain portable Anthropometer. Waist circumference was measured at the level of the umbilicus with the subject standing; with the face directed forward and shoulders relaxed; and breathing normally, using non-stretchable plastic tape to the nearest 0.1 cm. Body mass index (BMI) was calculated as body weight divided by height squared (kg/m2).
Body composition: Each female was also examined by the TANITA Body composition Analyzer. As specified by the manufacturer, the unit was calibrated before testing. The participant stood on the foot board of the device, while she was holding the 2 handles carefully; each by one hand at the same time. By using her sex, age, weight and height approximated to the nearest unit, the percentage body fat (Fat%: an estimate of the fraction of the total body mass that is adipose tissue), fat mass (FM: an estimate of the fraction of the total body weight that is adipose tissue) and fat free mass (FFM: an estimate of the fraction of the total body weight that is not adipose tissue), total body water (TBW) and basal metabolic rate (BMR) were derived.
Ultrasound (US) examination to each female was done to evaluate visceral fat at the umbilicus (USVF) in cm. Intra-abdominal fat thickness measurement was obtained using the “Medison Sonoace X8” ultrasonography equipment. For the visceral fat, a 3.5 MHz transducer was transversely positioned 1 cm above the umbilical scar on the abdominal midline, without exerting any pressure over the abdomen. Visceral fat thickness attempted corresponding to the measurement in centimeters between the internal surface of the abdominal rectus muscle and the posterior aortic wall in the abdominal midline, during expiration. In the current research, we considered visceral fat thickness <3 cm as normal, 3 to <7 as border line and ⩾7 cm with visceral obesity [13]. Then, the obese females were classified into 2 groups: obese with visceral obesity (56) and obese without visceral obesity (66) using cutoff point of 7 cm by Ultrasound. The entire control group had no visceral obesity (visceral fat thickness <3 cm).
2.2. Laboratory investigations
2.2.1. Sample collection
Five milliliters of venous blood samples was withdrawn after 12–14 h overnight fast into two sterile vacutainer (Becton Dickinson, NJ, USA); one containing EDTA for assay Apo B gene variants and the other without additives to separate serum to assay glucose and lipid profile.
2.2.2. Biochemical analysis
Estimation of lipid profile: Plasma levels of triglycerides (TG), high density lipoprotein cholesterol (HDL-C) and total cholesterol (TC) were measured by standardized enzymatic procedures, using kits supplied by Roche Diagnostics (Mannheim, Germany) on the Olympus AU 400 automated clinical chemistry analyzer. Low density lipoprotein cholesterol (LDL-C) was calculated according to formula of Friedwald et al. [14]:
Genotype study of the Apo B XbaI gene polymorphism:
DNA extraction: Genomic DNA was extracted from whole blood using DNA extraction kit (QIAamp DNA Blood Kit; Qiagen). The purity of extracted DNA was checked.
Restriction fragment length polymorphism (PCR-RFLP): The determination of Apo B C7673T polymorphism was done by the Polymerase Chain Reaction–Restriction Fragment Length Polymorphism (PCR-RFLP). The amplification reaction was carried out using primers: 5′-GGAGACTATTCAGAAGCTAA-3′ and 5′-GAAGAGCCTGAAGACTGACT-3′ and the following cycling conditions. Each amplification reaction was performed with 100 ng of genomic DNA; 10 pmol of each primer; 2 μL of 10× buffer solution; 200 μmol/L each of dATP, dCTP, dGTP, and dTTP; and 1 U of Taq polymerase in a total volume of 20 μL. The DNA fragments were generated from initial denaturation at 95 °C for 2.5 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 40 s, and extension at 72 °C for 60 s, with final extension at 72 °C for 5 min. The amplification products were digested with the restriction enzyme XbaI, and the fragments were separated on a 4% agarose gel [15].
2.2.3. Statistical analysis
The data analysis was carried out using the statistical package for social science (SPSS) software version 16 (Chicago, IL, USA). Frequency distribution of the genotypes and alleles was presented as percentage. All numeric variables were expressed as mean ± standard deviation (SD). Chi-square test was used in comparing qualitative data. Comparisons between 3 variables were done using a one way analysis of variance (ANOVA) test. Statistical significance was set at p < 0.05.
3. Results
The study population consisted of 122 obese females; their mean age 49.88 ± 5.6 years; and 36 healthy females as control group with age and sex matching. Comparison between obese and controls revealed that; BMI, waist circumference, total cholesterol, LDL-C and triglycerides levels were significantly higher in obese compared to controls (p < 0.01), while insignificant difference was found for HDL levels.
3.1. Genotyping results
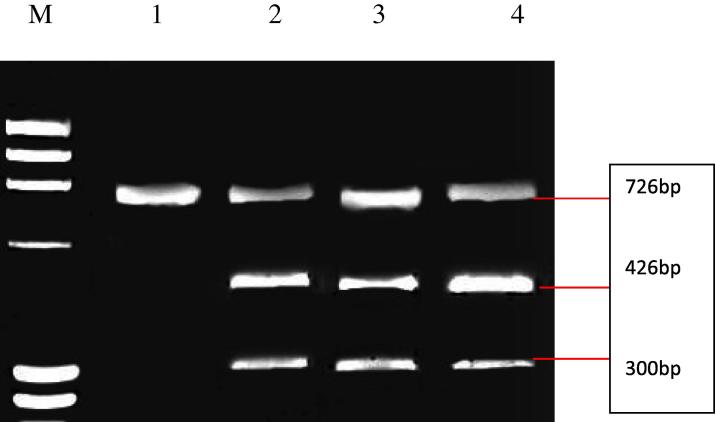
Comparing distribution of the genotypes of XbaI Apo B gene between total obese and normal weight subjects revealed that; the homozygote (TT) genotype was more frequent in both groups (p = 59% and 61.1% respectively) than the other two genotypes (CC and CT), while the homozygote (CC) genotype was the least frequent in both groups (p = 18% and 16.7% respectively) (Table 1). Dividing obese females according to the presence of visceral fat; it was found that visceral obese status was associated significantly with the presence of the heterozygous (CT) genotype of the XbaI Apo B gene; as females carrying (CT) heterozygous represented; 22 out of 56 visceral obese (39.3%), 6 out of 66 (9.1%) obese without visceral fat and 8 out of 36 (22.2%) normal weight subjects; with highly significant differences (p < 0.001). On the contrary the frequency of the homozygous (CC) (the wild type) was significantly the least genotype found in females with visceral obesity (7.1%) than; in females without visceral obesity (27.3%) and in lean subjects (17.7%) (p = 0.002). Moreover, the homozygote (TT) genotype was the most frequent in both obese without visceral obesity and control groups (63.6% and 61.1% respectively) than the obese with visceral obesity (53.6%) (Table 2). Example of Electrophoretic separation of Apo B genotypes (XbaI) among 4 females is presented in Fig. 1.
Table 1.
Genotypes distribution of XbaI Apo B gene in total obese and lean subjects.
| Genotype | Total | Controls | Total obese | p value |
|---|---|---|---|---|
| (N = 158) | (N = 36) | (N = 122) | ||
| N (%) | N (%) | N (%) | ||
| CT | 36 (22.8) | 8 (22.2) | 28 (23) | 0.001 |
| TT | 94 (59.5) | 22 (61.1) | 72 (59) | 0.000 |
| CC | 28 (17.7) | 6 (16.7) | 22 (18) | 0.002 |
N.B.: **p < 0.01 = highly significant differences.
Table 2.
Genotype distribution of XbaI Apo B gene in obese; according to visceral obesity; and lean subjects.
| Genotype | Total | Controls | Obese (N = 122) |
p value | |
|---|---|---|---|---|---|
| Without visceral obesity | With visceral obesity | ||||
| (N = 158) | (N = 36) | (N = 66) | (N = 56) | ||
| N (%) | N (%) | N (%) | N (%) | ||
| CT | 36 (22.8) | 8 (22.2) | 6 (9.1) | 22 (39.3) | 0.002 |
| TT | 94 (59.5) | 22 (61.1) | 42 (63.6) | 30 (53.6) | 0.039 |
| CC | 28 (17.7) | 6 (16.7) | 18 (27.3) | 4 (7.1) | 0.002 |
N.B.: **p < 0.01 = highly significant differences.
Figure 1.
Electrophoretic separation of Apo B genotypes (XbaI). Lane M: ΦX174 Marker. Lane 1: CC genotype (wild-type, homozygous and absence of the restriction site). Lane 2, 3 and 4: CT genotype (heterozygous).
Comparing the allele frequencies of XbaI Apo B gene among the different groups (Table 3, Table 4), revealed that T allele (about 70%) was more frequent than C allele (about 30%) in all groups under study (total sample, females with visceral fat, without visceral fat and in control females). This reflected the higher frequency of T allele than C allele among females with visceral obesity, while the heterozygous (CT) genotype was the most frequent among the females with visceral obesity.
Table 3.
Allele frequency (%) of the XbaI in total obese and lean subjects.
| Allele | Total Frequency (%) | Controls | Total obese | p value |
|---|---|---|---|---|
| (N = 36) (%) | (N = 122) (%) | |||
| C | 30 | 27.8 | 29.5 | 0.000 |
| T | 70 | 72.2 | 70.5 | 0.000 |
N.B.: **p < 0.01 = highly significant differences.
Table 4.
Allele frequency (%) of the XbaI in visceral obese and lean subjects.
| Allele | Total frequency (%) | Controls | Obese |
p value | |
|---|---|---|---|---|---|
| Without visceral obesity | With visceral obesity | ||||
| (N = 36) (%) | (N = 66) (%) | (N = 56) (%) | |||
| C | 30 | 27.8 | 31.8 | 26.8 | 0.019 |
| T | 70 | 72.2 | 68.2 | 73.2 | 0.005 |
N.B.: **p < 0.01 = highly significant differences.
Comparisons of the anthropometric parameters and lipid profile between various genotype of the Apo B XbaI gene revealed statistically significant differences in visceral fat measured by Ultrasound; where the lowest size of visceral obesity among females subjected to this study was associated with the presence of the wild type (CC) genotype and the highest size was associated with the presence of the heterozygous (CT) genotype t(p = 0.017). Concerning lipid profile; the significant lowest level of triglyceride and HDL-C was associated with the presence of the wild type (CC) genotype, and the highest level of triglyceride was associated with the heterozygous (CT) genotype (p = 0.036); and the highest level of HDL-C was associated with the presence of the homozygous (TT) genotype (p = 0.006). However, insignificant statistical differences were detected between these 3 genotypes of Apo B gene regarding BMI, waist circumference, all body composition parameters (Fat%, fat mass, fat free mass, total body water and basal metabolic rate) and the other parameters of lipid profile (total cholesterol and LDL-C) (Table 5).
Table 5.
Comparisons of the anthropometric parameters and lipid profile between the genotype of Apo B XbaI gene.
| Variable | CT (N = 36) |
TT (N = 94) |
CC (N = 28) |
p value | |||
|---|---|---|---|---|---|---|---|
| Mean | ±SD | Mean | ±SD | Mean | ±SD | ||
| Weight (kg) | 98.1500 | 20.43709 | 91.1239 | 23.50827 | 97.7000 | 15.47232 | 0.155 |
| Height (cm) | 159.78 | 5.67255 | 159.16 | 8.91737 | 161.75 | 8.22091 | 0.339 |
| BMI (kg/m2) | 38.2778 | 7.39 | 36.1362 | 8.66 | 37.7357 | 7.93 | 0.354 |
| Waist circumference (cm) | 105.92 | 15.65 | 100.30 | 14.16 | 106.11 | 13.68 | 0.054 |
| US visceral fat (cm) | 6.70 | 2.95 | 5.87 | 1.99 | 5.10 | 1.84 | 0.017∗ |
| Body composition | |||||||
| FAT (%) | 44.29 | 6.56 | 41.25 | 9.70 | 41.74 | 10.99 | 0.269 |
| FM (kg) | 44.64 | 13.75 | 39.17 | 17.15 | 41.70 | 15.04 | 0.229 |
| FFM (kg) | 54.81 | 10.70 | 52.20 | 9.02 | 55.29 | 7.58 | 0.169 |
| TBW | 40.12 | 7.83 | 38.21 | 6.60 | 40.49 | 5.55 | 0.167 |
| BMR (Kcl) | 1713.4 | 329.98 | 1618.7 | 286.76 | 1712.6 | 220.68 | 0.135 |
| Lipid profile | |||||||
| Triglycerides (mg/dl) | 132.65 | 57.26 | 117.68 | 45.19 | 101.23 | 30.24 | 0.036∗ |
| Total cholesterol (mg/dl) | 202.71 | 40.24 | 214.89 | 36.58 | 206.54 | 39.73 | 0.262 |
| HDL-C (mg/dl) | 47.35 | 8.87 | 50.61 | 9.60 | 43.85 | 9.38 | 0.006∗∗ |
| LDL-C (mg/dl) | 128.59 | 40.00 | 140.68 | 37.83 | 143.00 | 36.22 | 0.238 |
N.B.: *p < 0.05 = significant differences. **p < 0.01 = highly significant differences.
4. Discussion
Genetic epidemiological data demonstrate that; in most cases; obesity is a multifactorial disorder with genetic contribution in the range of 40–70%. As obesity is excessive lipid deposition in the fat tissue, there is good reason to study the association between excessive fat mass and genetic polymorphisms of lipid transport proteins. Inherited variations of the apolipoprotein gene have influence circulating lipid concentration. One of the most closely monitored genes which may affect lipid metabolism and may predispose to obesity is the gene for Apo B and XbaI polymorphism [16], [17], [18].
The present study, therefore, attempted to investigate the importance of the XbaI polymorphism of Apo B gene (C7673T) as risk factor for visceral obesity and its influence on lipid profile among a sample of Egyptian obese females.
Regarding XbaI polymorphism of Apo B gene (C7673T) frequency in the current studied sample; both obese and lean, it was obvious that the homozygote (TT) genotype was the most frequent; however the homozygote (CC) genotype was the least frequent.
Rajput-Williams and his colleagues [19], in their analysis reported that; obesity was associated with XbaI that do not change the amino acid sequence. They stated that; these RFLP are likely to be in linkage disequilibrium with nearby functional variation predisposing to obesity. Moreover they postulated that this inherited variation of the apolipoprotein-B gene, influences circulating cholesterol concentration. The Apo B XbaI polymorphism was documented to be associated with obesity and dyslipidemia in North Indian population [20]. Hu et al. [21], support these findings as they postulated that; Apo B XbaI restriction site may serve as potential genetic markers affecting BMI and lipid profiles in children from Guangxi. Among Tunisian population, Apo B restriction fragment length polymorphisms (XbaI) have been found to be associated with variation in lipid levels, obesity and/or coronary artery disease [22]. Sánchez-Cuén and his co-workers [23], agreed with these findings, as they observed a significant association of Apo B XbaI gene polymorphism with obesity and serum lipid levels in Mexican population. While contradictory results were also available; Misra and his colleagues [24] reported that Apo B XbaI gene polymorphism did not associate with obesity in Asian Indian. Srivastava et al. [10] also coincide with this result as they showed that there was an insignificant difference in the genotype and allele frequencies of Apo B XbaI polymorphism between obese and non-obese subjects.
The current study revealed that there were insignificant associations between different genotypes of the Apo B XbaI polymorphism and the studied anthropometric parameters (Weight, Height, BMI, waist circumferences, Fat%, BMR, FM, FFM and TBW) while, the current study demonstrated an influence of XbaI polymorphism of Apo B gene on visceral obesity only. The most important finding emerging from the current study was that the frequency of the heterozygous genotype (CT) was significantly higher in obese with visceral obesity than in the other two groups (lean and obese with no visceral fat), (p = 0.002) while the homozygous wild type genotype (C/C), lacking the XbaI site was more frequent in obese patients without visceral fat (p = 0.017).
Ryo and his colleagues [25] confirmed that regional adipose tissue parameters, especially visceral tissue mass are more closely associated with physiological and pathological processes than total adipose tissue mass.
The genetic determination of lipidemia has been intensively investigated since 1980s. Apo B gene is of particular interest since its protein product plays a crucial role in the formation and clearance of plasma LDL particles. The first papers addressing this topic [26], [27] referred to the T allele as a disadvantageous one. The above authors reported an association between the allele and increased levels of triacylglycerols (TG) and total cholesterol (TC). Talmud et al. [28] demonstrated, in a randomly selected population sample, an association between the (CC) genotype and low TC levels. In a study conducted by Tybjaerg-Hansen et al. [29], the highest levels of LDL-cholesterol were shown in TT homozygote with coronary artery disease (CAD). In hyperlipidemic patients TT homozygotes were found with increased levels of fasting TG [30]. However, the subsequent studies produced opposite results; a group of CC homozygotes showed the highest levels of TC [29] or LDL-cholesterol [31]. Hansen et al. [32] found the highest levels of TC in CC homozygotes whose body mass index (BMI) was <25. A dietary study of Friedlander et al. [33] indicated a more marked increase in blood lipid concentrations in individuals with TT receiving a high-cholesterol diet. The relationship between the XbaI polymorphism of the Apo B gene and serum lipid levels was not documented in the Japanese [34] and Chinese [35] population. This may be due to the much lower cholesterol concentration and a lower incidence of the (T) allele compared to European population.
In the current study, high statistical significant associations of genotypes were shown with; triglyceride concentrations (TG) (p = 0.036) and HDL-C (p = 0.006) levels among the female samples analyzed. The Wild type homozygous genotype was significantly associated with lower TG serum levels (p = 0.036). While there was no association between Apo B gene polymorphism and levels of the other lipid parameters (total cholesterol and LDL). Current study confirms the earlier association between the absence of the XbaI cutting site and the higher TG levels [36].
Hypertriglyceridemia may be the major cause of the other lipid abnormalities since it will lead to delayed clearance of the TG-rich lipoproteins and formation of small dense LDL-C. The mechanism underlying the association between the Apo B XbaI polymorphism and lipid levels is unknown. It could be explained by variation in the synthetic rate of Apo B and Apo B containing lipoproteins, or variation in the removal rate of the Apo B in individuals with different Apo B XbaI genotypes. This effect might be due to the slightly lower LDL catabolic rate in the individuals with the XbaI cutting site. Also, variation in the Apo B gene could influence body fat deposition by affecting the delivery of triglycerides to adipose tissue by Apo B-containing VLDL particles. Other clarification may be; as serum concentration of lipids is highly variable during the day, and the weakness of the association might be due to the fact that serum lipids levels are generally measured in the fasting state during which they reach the lowest levels [37]. Moreover it has been demonstrated that the XbaI polymorphism modifies dietary fat and cholesterol responses in individuals with the XbaI cutting site [38].
There may be several reasons for the controversy or polarity observed in various studies. A simple explanation could be that Apo B XbaI polymorphism exhibits population specific variation, which may be due to gene and environment interactions with variation at the Apo B gene to determine an individual’s plasma lipid and lipoprotein levels. There is general agreement that genetic heterogeneity across races and ethnicities exists in the Apo B gene polymorphisms [39]. Apo B XbaI polymorphism does not lead to changes in the amino acid sequence and cannot be implicated at the structure level. It is possible that some other polymorphism in its vicinity might be present, which is in linkage disequilibrium with Apo B XbaI polymorphism and accountable for the observed association with obesity and lipid levels in other studies. Saha et al. [35], showed a strong disequilibrium between ins/del polymorphism and the XbaI polymorphism of Apo B gene. Moreover, gene–environment interaction may also be responsible for the inconsistency of data due to differences in the diet and lifestyle of populations of various parts of the world [40].
Studies with a larger sample size are necessary and further investigation is needed to confirm or refute the role of this polymorphism and/or other polymorphisms in relation to obesity (visceral obesity). In conclusion, the genetic diagnosis and molecular characterization of Apo B gene mutations will allow a better assessment of response to treatment, prevention and family screening in these subjects.
5. Conclusion
Visceral obesity was associated significantly with the presence of the heterozygous (CT) genotype of the XbaI Apo B gene; in spite of the finding that the homozygote (TT) genotype was the most frequent and the homozygote (CC) genotype was the least frequent. There were significant associations between CT genotype and visceral obesity, triglycerides and HDL-C.
Acknowledgments
We would like to acknowledge our institute “National Research Centre’; Egypt”; without its fund this study could not have been done. We would also like to acknowledge everybody who participated in this study; the employers of our institute who were the participants of this study, the technicians who helped in the laboratory analysis and the doctors who participated in collection of the data. Without their help, this study could not have been completed.
Footnotes
Peer review under responsibility of National Research Center, Egypt.
References
- 1.WHO Media Center, Obesity and Overweight, Fact Sheet No. 311, Updated January 2015.
- 2.Marie N.G., Fleming T., Robinson M., Thomson B., Graetz N., Margono C., Mullany E.C. Lancet. 2014;384(9945):766–781. doi: 10.1016/S0140-6736(14)60460-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tchernof A., Després J. Physiol. Rev. 2013;93:359–404. doi: 10.1152/physrev.00033.2011. [DOI] [PubMed] [Google Scholar]
- 4.Nazare J., Smith J., Borel A., Haffner S., Balkau B., Ross R., Massien C., Alméras N., Després J. Am. J. Clin. Nutr. 2012;96:714–726. doi: 10.3945/ajcn.112.035758. [DOI] [PubMed] [Google Scholar]
- 5.Mooradian A., Haas M., Wehmeier K., Wong N. Obesity (Silver Spring) 2008;16:1152–1160. doi: 10.1038/oby.2008.202. [DOI] [PubMed] [Google Scholar]
- 6.Carlsson P., Darnfors C., Olofsson S.O., Bjursell G. Gene. 1986;49:29–51. doi: 10.1016/0378-1119(86)90383-5. [DOI] [PubMed] [Google Scholar]
- 7.Freedman D.S., Serdula M.K., Percy C.A., Ballew C., White L. J. Nutr. 1997;127:2120S–2127S. doi: 10.1093/jn/127.10.2120S. [DOI] [PubMed] [Google Scholar]
- 8.Priestley L., Knott T., Wallis S., Powell L., Pease R., Brunt H. Nucleic Acids Res. 1985;13:6793. doi: 10.1093/nar/13.18.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang L.S., Miller D.A., Bruns G.A., Breslow J.L. Proc. Natl. Acad. Sci. U.S.A. 1986;83:644–648. doi: 10.1073/pnas.83.3.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srivastava N., Prakash J., Srivastava A., Agarwal C., Pan D., Mittal B. Indian J. Hum. Genet. 2013;19(1):26–31. doi: 10.4103/0971-6866.112880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H., Peng D.Q. Lipids Health Dis. 2011;10:176. doi: 10.1186/1476-511X-10-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiernaux J., Tanner J.M. In: Weiner J.S., Lourie S.A., editors. IBP/Blackwell Scientific Publications; London/Oxford, UK: 1969. (Human Biology: A Guide to Field Methods). [Google Scholar]
- 13.Ribeiro Filho F.F., Faria A.N., Azjen S., Zanella M.T., Ferreira S.R. Obes. Res. 2003;11:1488–1494. doi: 10.1038/oby.2003.199. [DOI] [PubMed] [Google Scholar]
- 14.Friedewald W., Levy R., Fredrickson D. Clin. Chem. 1972;18:499–502. [PubMed] [Google Scholar]
- 15.Zhang L., Zeng Y., Ma M., Yang Q., Hu Z., Du X. Neurol. India. 2009;5:584–588. doi: 10.4103/0028-3886.57805. [DOI] [PubMed] [Google Scholar]
- 16.Freeman D., Griffin B., Holmes A. Arterioscler. Thromb. 1994;14:336–344. doi: 10.1161/01.atv.14.3.336. [DOI] [PubMed] [Google Scholar]
- 17.M. Fiegenbau, Estudo da variabilidade em genes de apolipoproteínas sobre níveis lipídicos e parâmetros de massa e gordura corporal na população de Porto Alegre (Dissertação de Mestrado), Programa de Pós-Graduação em Genética e Biologia Molecular, UFRGS, Porto Alegre, Brazil, 2001.
- 18.Benn M., Nordestgaard B.G., Jensen J.S., Grande P., Sillesen H., Tybjaerg-Hansen A. J. Clin. Endocrinol. Metab. 2005;90:5797–5803. doi: 10.1210/jc.2005-0974. [DOI] [PubMed] [Google Scholar]
- 19.Rajput-Williams J., Knott T., Wallis S., Sweetnam P., Yarnell J., Cox N., Bell G., Miller N., Scott J. Lancet. 1988;2:1442–1446. doi: 10.1016/s0140-6736(88)90930-0. [DOI] [PubMed] [Google Scholar]
- 20.Saha N., Tay J., Heng C., Humphries S. Clin. Genet. 1993;44:113–120. doi: 10.1111/j.1399-0004.1993.tb03861.x. [DOI] [PubMed] [Google Scholar]
- 21.Hu P., Qin Y., Jing C., Lu L., Hu B., Du P. Ann. Hum. Biol. 2009;36(4):411–420. doi: 10.1080/03014460902882475. [DOI] [PubMed] [Google Scholar]
- 22.Kallel A., Feki M., Elasmi M., Souissi M., Sanhaji H., Omar S., Haj Taieb S., Jemaa R., Kaabachi N. Physiol. Res. 2007;56(4):411–417. doi: 10.33549/physiolres.931020. [DOI] [PubMed] [Google Scholar]
- 23.Sánchez-Cuén J., Aguilar-Medina M., Arámbula-Meraz E., Romero-Navarro J., Granados J., Sicairos-Medina L. World J. Gastroenterol. 2010;16:4685–4690. doi: 10.3748/wjg.v16.i37.4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Misra A., Nishanth S., Pasha S., Pandey R., Sethi P., Rawat D. Indian Heart J. 2001;53:177–183. [PubMed] [Google Scholar]
- 25.Ryo M., Kishida K., Nakamura T., Yoshizumi T., Funahashi T., Shimomura L. World J. Radiol. 2014;6:409–416. doi: 10.4329/wjr.v6.i7.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Law A., Wallis S.C., Powell L.M., Pease R.J., Brunt H., Priestley L.M. Lancet. 1986;1:1301–1303. doi: 10.1016/s0140-6736(86)91222-5. [DOI] [PubMed] [Google Scholar]
- 27.Aalto-Setala K., Tikkanen M.J., Taskinen M.R., Nieminen M., Holmberg P., Kontula K. Atherosclerosis. 1988;74:47–54. doi: 10.1016/0021-9150(88)90190-6. [DOI] [PubMed] [Google Scholar]
- 28.Talmud P., Barni N., Kessling A., Carlsson P., Darnfors C., Bjursell G. Atherosclerosis. 1987;67:81–89. doi: 10.1016/0021-9150(87)90267-x. [DOI] [PubMed] [Google Scholar]
- 29.Tybjaerg-Hansen A., Nordestgaard B.G., Gerdes L.U., Humphries S.E. Atherosclerosis. 1991;89(1):69–81. doi: 10.1016/0021-9150(91)90008-q. [DOI] [PubMed] [Google Scholar]
- 30.Jenner K., Sldoll A., Ball M. Atherosclerosis. 1988;69:39–49. doi: 10.1016/0021-9150(88)90287-0. [DOI] [PubMed] [Google Scholar]
- 31.Peacock R., Dunning A., Hamsten A., Tornvall P., Humphries S., Talmud P. Atherosclerosis. 1992;92:151–164. doi: 10.1016/0021-9150(92)90274-k. (See comment in PubMed commons) [DOI] [PubMed] [Google Scholar]
- 32.Hansen P.S., Gerdes L.U., Kllausen I.C., Gregersen N., Faergeman O. Hum. Genet. 1993;91(1):45–50. doi: 10.1007/BF00230221. [DOI] [PubMed] [Google Scholar]
- 33.Friedlander Y., Kaufmann N.A., Cedar H., Weinberg N., Kark J.D. Atherosclerosis. 1993;98:165–177. doi: 10.1016/0021-9150(93)90126-f. [DOI] [PubMed] [Google Scholar]
- 34.Aburatanl H., Murase T., Takaku F., Itoh H., Matsumoto A., Itakura H. N. Engl. J. Med. 1987;317:52. [Google Scholar]
- 35.Saha N., Tay J., Humphries S. Genet. Epidemiol. 1992;9:1–10. doi: 10.1002/gepi.1370090103. [DOI] [PubMed] [Google Scholar]
- 36.Deeb S.S., Failor A., Brown B.G., Brunzell J.D., Albers J.J., Motulsky A.J. Cold Spring Harb. Symp. Quant. Biol. 1986;51:403–409. doi: 10.1101/sqb.1986.051.01.048. [DOI] [PubMed] [Google Scholar]
- 37.Branchi A., Torri A., Berra C., Colombo E., Sommariva D. Intern. Emerg. Med. 2006;1:287–295. doi: 10.1007/BF02934762. [DOI] [PubMed] [Google Scholar]
- 38.Gallegos-Arreola M., Valdez Y., Zúñiga-Corona M., Figuera L., Arnaud-López L., Robles-Cervantes J., González-Ortiz M., Martínez-Abundis E., Puebla-Pérez A., Zúñiga-González G. Asia Pac. J. Clin. Nutr. 2012;21:312–318. [PubMed] [Google Scholar]
- 39.Zhao W., Huang J., Wang L., Li H., Zhang P., Zhao Q., Chen S., Gu F. Biomed. Environ. Sci. 2007;20:260–264. [PubMed] [Google Scholar]
- 40.van Vliet-Ostaptchouk J., Snieder Harold, Lagou Vasiliki. Curr. Nutr. Rep. 2012;1:184–196. doi: 10.1007/s13668-012-0022-2. [DOI] [PMC free article] [PubMed] [Google Scholar]