Abstract
Different techniques were adopted for molecular characterization of several indigenous strains of Bacillus thuringiensis (Bt) previously isolated from Egyptian soil samples. These isolates show different toxicity levels against neonate larvae of both insect species; Spodoptera littoralis (Biosduval); and Helicoverpa armigera (Hübner). The parasporal crystals among the most potent isolates contained polypeptides of about 127 and 130 kDa. PCR screening for genes encoding different Cry genes was performed. The Cry 1 gene is the most abundant in these isolates (83.33%) among tested Cry-type genes, followed by Cry 1 gene subfamilies (Cry 1B and Cry 1C) with percentage of 38.88% and 77.77%, respectively. The tested isolates showed the presence of Cry 2A(a,b) gene, but not all of these isolates were positive for Cry 2 gene (55.55%). Only 27.77% and 16.66% of the tested isolates harbor Cry 4 and Cry 3 genes, respectively. All strains were negative in PCR assays for the Vip 3Aa1 gene. Moreover, DNA fingerprinting using RAPD-PCR was performed to detect the genetic similarities and dissimilarities among the different isolates and standard strains. Assessment of Bt diversity based on the combined analysis of their protein and RAPD-PCR banding patterns was performed. This study demonstrates that Bt strains isolated from Egyptian soil samples can be distinguished and identified on the basis of the distribution of Cry-type genes and RAPD fingerprints.
Keywords: Bacillus thuringiensis, Bioassay, Molecular characterization, SDS-PAGE, Cry genes, RAPD-PC
1. Introduction
Bacillus thuringiensis (Bt) is widely distributed in natural soils as reported by many authors e.g. [1], [2], [3], [4], [5], [6], [7], [8], [9]. [8], [10] studied the distribution of Bt in the soils of 22 governorates in Egypt searching for new indigenous isolates with potential activity against some lepidopterous insects.
The present study aims to determine the diversity and the genetic variability of Bt isolates from Egyptian soils and their potency against some lepidopterous insects. The choice of the selected isolates was based on their potential activity against the two lepidopterous insects S. littoralis and H. armigera.
PCR is a highly sensitive and rapid method to detect and identify the Cry genes, predict insecticidal activities of Bt strains, determine the distribution of Cry genes, and detect new genes. Several Bt strain collections have been described in the literature e.g. [9], [11], [12], [13], [14], [15], [16], [17], [18], [19]. There is a considerable genetic diversity among Bt strains, most of which are naturally multigenic, harboring between 2 and 12 Cry genes, which are generally located on low copy number, high molecular weight (>30 kDa) plasmids. Some Cry genes are also located on chromosomes [20]. The presence of conserved blocks of homologous DNA among different Cry genes suggests recombination between chromosomal and plasmid-borne Cry genes. The Cry genes are flanked by transposable elements, which may be involved in Cry gene amplification and diversification [21].
2. Materials and methods
A soil sampler was used to collect the samples of soil from the fields to a depth of 15 cm. The collection sites had no history of the treatment with Bt The samples were stored in sealed sterile vials at 4 °C until analyzed. Eighteen Bt isolates, from soil samples collected from different Egyptian governorates and three standard strains, namely Agerin®, Dipel® 2X and HD-635 were used in this study. The choice of the isolates was based on their potential activity against S. littoralis (Biosduval) and H. armigera (Hübner). For culturing the isolates obtained, the method described by [22] was adopted. Small quantities of B. thuringiensis can easily be recovered by the lactose-acetone co-precipitation procedure of [23]. Different larval instars of each species were collected as field strains from tomato fields. Colonies were established in the laboratory and reared for a minimum of one generation to eliminate parasitoids and pathogens and to obtain sufficient numbers at the selected age-classes of larval development. Laboratory colonies of both species were established on a semi-artificial diet according to [24].
2.1. Bioassay
For bioassay, the spore-crystal complexes of forty isolates were evaluated for their potential activity against S. littoralis and H. armigera. The standard bioassay procedures were followed according to [25] and they were carried out using neonate larvae of each species. The potential activity of the isolates was determined in relation to different standard strains. HD-635 was used as a standard for S. littoralis since 1989 [26]. Both of Agerin® and Dipel® 2X were selected as standards for H. armigera.
The diet-incorporation bioassay method was adopted. In this method, the Bt preparation is mixed into agar-based diet at 55 °C. After the diet cools and solidifies, the test insects were transferred into the treated diet containers covered with perforated lids. The containers were incubated for a suitable period of time.
All bioassays were done on the neonate larvae of the target insects. Screening of different Bt isolates preparations was made at 500 μg/ml. One droplet of (0.04%) Tween 60 was added to obtain a homogenous solution and mixed with 100 ml diet. The assay procedure proposed by [24] was adopted. The potential activity of the different isolates was evaluated using 50 healthy neonate larvae of each insect species and for each treatment. The same number was used as a control. The larvae were separated into 5 replicates (10 larvae/replicate) for each treatment. The percentage of larval mortalities was reported after 7 days from treatment and corrected according to [27]
T = No. of dead larvae in treated replicates.
C = No. of dead larvae in control replicates.
Six serial dilutions of each of the tested isolates were prepared (500, 250, 125, 62.5, 31.25 and 15.62 μg/ml). Ten ml from each concentration was mixed with 100 ml diet. The median lethal concentrations LC50 of potent isolates were computed through probit analysis within 95% confidence limits using Propan program. The potency of each isolate in relation to standard was calculated as follows:
2.2. Molecular characterization of Bt Isolates
Three molecular approaches were used to characterize the Bt isolates with reference to the three tested standards, these approaches are:
-
2.2.1.
Protein analysis using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) [28].
-
2.2.2.
Polymerase chain reaction analyses were performed to identify the existence of Cry genes. The primers used, their sequence, the spectrum of the genes and the sizes of the PCR products are shown in Table 1. The isolation of the total genomic DNA from the isolates was carried out according to [29]. Amplification was performed in 20-μl reaction mixtures (containing of 1.0 μl of 10 mM dNTPs, 2.5 μl of 10 X Buffer, containing MgCl2, 2.5 μl of 50-ng/ μl template DNA, 0.5 μl of Taq polymerase, 0.5 μl of 50 pmol from each primer and 12.5 μl sterile distilled water) in a Thermal Cycler, beginning at 95 °C for 5 min and followed by 34 cycle program consisting of a denaturation step at 94 °C for 30 s, an annealing at primer specific temperature (Table 1) for 50 s and an extension step at 72 °C for 75 s; followed by a final extension step at 72 °C for 7 min. PCR products were size fractionated by electrophoresis in 1X% agarose in the presence of ethidium bromide. Characteristic DNA banding patterns were readily and reproducibly visualized using a gel documentation system and the resulting bands were scored using the Labimage program (Kapelan Bio-Imaging, Leipzig, Germany).
-
2.2.3.
Randomly Amplified Polymorphic DNA (RAPD) markers are produced by performing PCR reactions using random primers and can be used to distinguish between closely related organisms [30], [31], [32], [33]. RAPD technology was applied to total DNA extracted from single Bt colonies of selected strains. Twenty 10-mer primers were used (OPA-01, OPA-02, OPA-03, OPA-04, OPA-05, OPA-07, OPA-09, OPA-15, OPA-20, OPB-02, OPB-03, OPB-04, OPB-18, OPG-01, OPG-02, OPG-03, OPG-04, OPZ-01, OPZ-03, OPZ-06). DNA amplification was performed in 25-μl reaction consisting of 2.0 μl of 10 mM dNTPs, 5.0 μl of10 X Buffer, containing MgCl2, 2.0 μl of 50-ng/ μl template DNA, 0.2 μl of Taq polymerase, 2.0 μl of 50 pmol from each primer and 13.8 μl sterile distilled water. Amplifications were performed in a Thermal Cycler (Biometra, Germany), beginning at 92 °C for 5 min, followed by a 45 cycle reaction start denaturation step at 24 °C for 45 s, annealing step at 36 °C for 45 s and an extension step at 72 °C for 90 s. The reaction was completed with final extension step at 72 °C for 10 min. PCR products were size fractionated by electrophoresis in 1X% agarose in the presence of ethidium bromide. Characteristic DNA banding patterns were readily and reproducibly visualized using a gel documentation system and the resulting bands were scored using the Labimage program. The isolates were clustered based on the matrix of genetic dissimilarities (Euclidean distance). Scoring matrix was created based on the RAPD banding patterns and these data were then analyzed using the unweighted pair-group method with arithmetic averages (UPGMA) algorithm with SPSS (Version 16, SPSS Inc., USA) and Statistica (Version 8, StatSoft Inc., Tulsa, Oklahoma, 2007) software packages.
Table 1.
Primers sequences used in PCR screening of the tested Cry-genes.
| Primer | Sequence 5′ → 3′ | Product size | Annealing temp. (°C) | References |
|---|---|---|---|---|
| Cry1,Un1(d) | CATGATTCATGCGGCAGATAAAC | 274–277 bp | 55 | [12] |
| Cry1,Un1(r) | TTGTGACACTTCTGCTTCCCATT | |||
| Cry1B CJ8 | CTTCATCACGATGGAGTAA | 367 bp | 46 | [51] |
| Cry1B CJ9 | CATAATTTGGTCGTTCTGTT | |||
| Cry1c CJ10 | AAAGATCTGGAACACCTTT | 130 bp | 45 | [51] |
| Cry1c CJ11 | CAAACTCTAAATCCTTTCAC | |||
| Cry2,Un2(d) | GTTATTCTTAATGCAGATGAATGGG | 689–701 bp | 55 | [12] |
| Cry2,Un2(r) | CGGATAAAATAATCTGGGAAATAGT | |||
| Cry2Aa,b(d) | CAG ATA CCC TTG CTC GTG TAA | 1072 bp | 54 | [52] |
| Cry2Aa,b(r) | ATA GGC CCG TGC TCC ACC AGG | |||
| Cry3,Un3(d) | CGTTATCGCAGAGAGATGACATTAAC | 589–604 bp | 55 | [12] |
| Cry3,Un3(r) | CATCTGTTGTTTCTGGAGGCAAT | |||
| Cry4,Un4(d) | GCATATGATGTAGCGAAACAAGCC | 439 bp | 60 | [12] |
| Cry4,Un4(r) | GCGTGACATACCCATTTCCAGGTCC | |||
| Vip3Aa1(d) | ATGAACAAGAATAATACTAAATTAAGC | Varied according to subspecies | 54 | [53] |
| Vip3Aa1(r) | GGTCGACTTACTTAATAGAGACATCG |
3. Results
The selection of Bt isolates was based on their potential activity against the target insects. The tested isolates have varying potencies against S. littoralis and H. armigera. The potency of the tested isolates as shown in Table 2 indicates that some of them were highly toxic against neonate larvae of H. armigera (S.Sinai 2, Qena 1, Qena 2, Sohag 1, Asut 1, Tosk 1, Tosk 2); causing 80–96% larval mortality; four isolates (Isma 1, Port 1, N.Sinai 2, S.Sinai 3) resulted in 63–70% mortality; while the isolates R.Sea 1, Matr 1, S.Sinai 1 and Asut 2 gave 50–58% larval mortality. None of the isolates caused mortality for S. littoralis above 50%. Subsequently, the LC50 for the isolates which show high toxicity levels (⩾80% mortality) against H. armigera are illustrated in Table 3.
Table 2.
Potency of Bt isolates obtained from Egyptian soils against neonate larvae of target insects at 500 μg/ml after 7 days.
| Governorate | Isolate code | % of larval mortality of Bt isolates after 7 days, at 500 μg/ml against neonate larvae of |
|
|---|---|---|---|
| S. littoralis | H. armigera | ||
| North Sinai | N.Sinai 1 | 20 | 30 |
| N.Sinai 2 | 50 | 70 | |
| N.Sinai 3 | 32 | 40 | |
| Port Said | Port 1 | 48 | 74 |
| Matruh | Matr 1 | 28 | 58 |
| South Sinai | S.Sinai 1 | 24 | 50 |
| S.Sinai 2 | 45 | 80 | |
| S.Sinai 3 | 32 | 67 | |
| Ismailia | Isma 1 | 50 | 67 |
| Red Sea | R.Sea 1 | 30 | 52 |
| Asyut | Asut 1 | 45 | 82 |
| Asut 2 | 29 | 50 | |
| Sohag | Sohg 1 | 40 | 92 |
| Qena | Qena 1 | 47 | 95 |
| Qena 2 | 40 | 86 | |
| Qena 3 | 25 | 40 | |
| Aswan | Tosh 1 | 40 | 90 |
| Tosh 2 | 36 | 88 | |
Table 3.
LC50 and LC90 (μg/ml) at Confidence limits (95%) of potent Bt isolates obtained from soil samples against neonate larvae of H. armigera.
| Isolate | LC50 (Confidence limits at 95%) | LC90 (Confidence limits at 95%) | Slope ± S.E. | Potency in relation to Dipel® 2X (IU/mg) | Potency in relation to Agerin® (IU/mg) |
|---|---|---|---|---|---|
| S.Sinai 2 | 157.4 (123.3–208.9) | 1105.8 (683.4–2303) | 1.51 ± 0.18 | 37912.0 | 40723.8 |
| Qena 1 | 86.9 (63.4–118.8) | 620.9 (420.2–1032.3) | 1.14 ± 0.16 | 68668.8 | 73761.8 |
| Qena 2 | 128.7 (102.6–164.7) | 782.4 (518.4–1434.3) | 1.64 ± 0.18 | 46380.4 | 49820.3 |
| Sohag 1 | 102.3 (81.2–129.7) | 694.2 (455.7–1296.9) | 1.61 ± 0.18 | 58362.9 | 62691.5 |
| Asut 1 | 140.6 (111.5–182.1) | 892.8 (578–1706. 5) | 1.59 ± 0.18 | 42458.6 | 45607.6 |
| Tosk 1 | 101.4 (79.9–129.9) | 640.7 (430.8–1145.3) | 1.53 ± 0.17 | 58849.7 | 63214. |
| Tosk.2 | 109.8 (87.2–139.9) | 695.6 (463.1–1265.6) | 1.59 ± 0.18 | 54371.8 | 58404.4 |
| Dipel® 2X | 186.5 (141.8–262.1) | 1638 (1913.7–4166.3) | 1.36 ± 0.17 | 32,000 | 32,000 |
| Agerin ® | 200.4 (152–284.1) | 1732.9 (959.9–4468.4) | 1.37 ± 0.17 | 32,000 | 32,000 |
3.1. Protein analysis
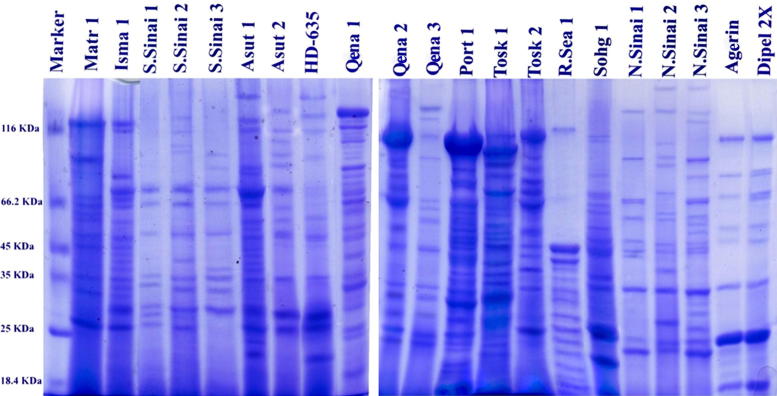
Electrophoretic patterns of different isolates and standards reveal bands with different molecular weights that ranged from 197 to 21 KDa (Fig. 1). Although, each isolate has a unique protein pattern, there are shared bands among different isolates and standards. The most shared bands among the tested isolates and the standard HD-635 have molecular weight of 25.5 KDa. Shared bands were detected among the most potent isolates e.g. 130 KDa that was detected in the isolates Tosk 1, Tosk 2, Qena 1, and Sohg1, also 127 KDa, 124 KDa, 100 KDa, 98 KDa etc were detected with other isolates.
Figure 1.
Protein pattern of different Bt isolates.
3.2. Specific PCR
Different isolates were analyzed for the presence of different crystal genes (Cry genes) to explore the differences in gene profile between the isolates and to determine if the differences in gene profiles correlate with differences in the insecticidal activities. PCR reactions were made using specific primers of Cry genes (Cry 1, Cry 1B, Cry 1C, Cry 2, Cry 2A(a,b), Cry 3, Cry 4 and Vip 3Aa1) encoding for toxins that are highly specific for different insect pests and yielded reproducible polymorphic bands of PCR products (Table 4).
Table 4.
Polymorphic bands detected among different Bt isolates using specific primers for Cry-type genes.
| B. t. isolates | Molecular weight of polymorphic bands (bp) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Cry Cry 1 | Cry 1B | Cry 1C | Cry 2 | Cry 2Aa,b | Cry 3 | Cry 4 | Vip 3Aa1 | |
| Isma 1 | 375 | – | 130 | – | 340 | – | – | – |
| Tosk 1 | 274 | 367 | 1050, 620, 130 | 701 | 1072, 340 | 780, 604, 340, 220 | 670, 525, 400, 290 | – |
| Tosk 2 | 274 | 367 | 1050, 620, 130 | 701 | 1072, 340 | 780, 604, 340, 220 | 439, 290 | – |
| Port 1 | 360, 274 | – | 300, 130 | – | 340 | – | – | – |
| Dipel® 2X | 274 | 367 | 1050, 620, 130 | 701 | 1072 | – | 480 | – |
| Agrin® | 274 | 367 | 1050, 620, 130 | 701 | 1072 | – | 439 | – |
| Sohg 1 | 360, 150 | – | 690, 130 | – | 960, 320 | – | – | – |
| Matr 1 | 1000, 360 | 367 | 620, 475, 180 | 701 | 1072, 340, 200 | – | – | – |
| Qena 1 | 375, 274 | 1210, 680,520 | 1050, 620, 250 | 870, 510 | 1000, 500, 320, 180 | – | – | – |
| Qena 2 | 274 | 367 | 1050, 620, 130 | 701 | 1072 | 780, 604, 340, 220 | – | – |
| Qena3 | – | – | – | – | – | – | – | – |
| R.Sea 1 | – | – | 130 | – | – | – | – | – |
| Asut 1 | 500, 360, 230 | – | 600, 220 | – | – | – | – | – |
| Asut 2 | 200 | – | 220 | – | – | – | – | – |
| S.Sinai 1 | 277 | – | – | 701 | – | – | – | – |
| S.Sinai 2 | 1000, 750, 500, 360, 277 | 720 | 1050, 620, 250 | – | – | – | 290 | – |
| S.S.inai 3 | 277 | 367 | 1050, 620, 130 | – | 1072 | – | 290 | – |
| N.Sinai 1 | – | – | – | – | – | – | – | – |
| N.Sinai 2 | 1000, 600, 480, 360, 230 | – | 1350, 1050 | 701 | – | – | 439 | – |
| N.S.inai 3 | 200 | – | – | – | – | – | – | – |
| HD-635 | 277 | – | – | – | 1072 | – | – | – |
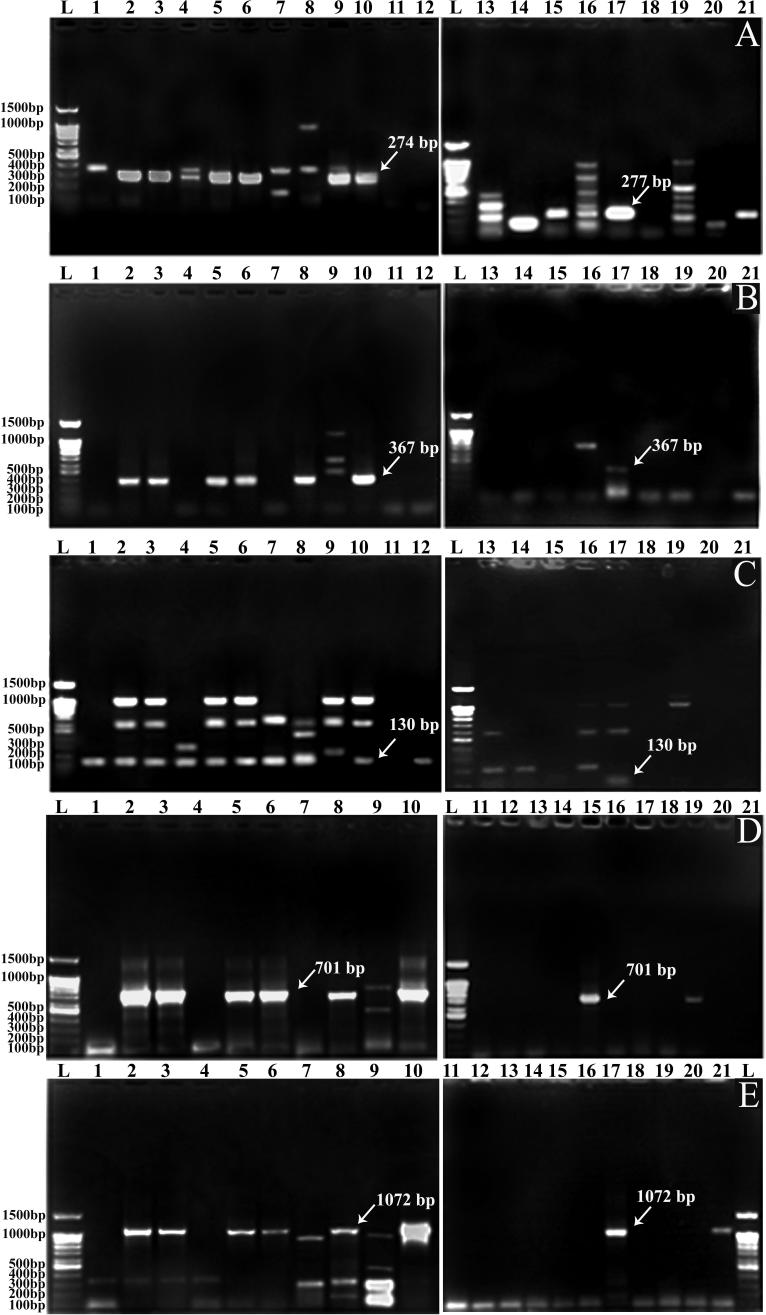
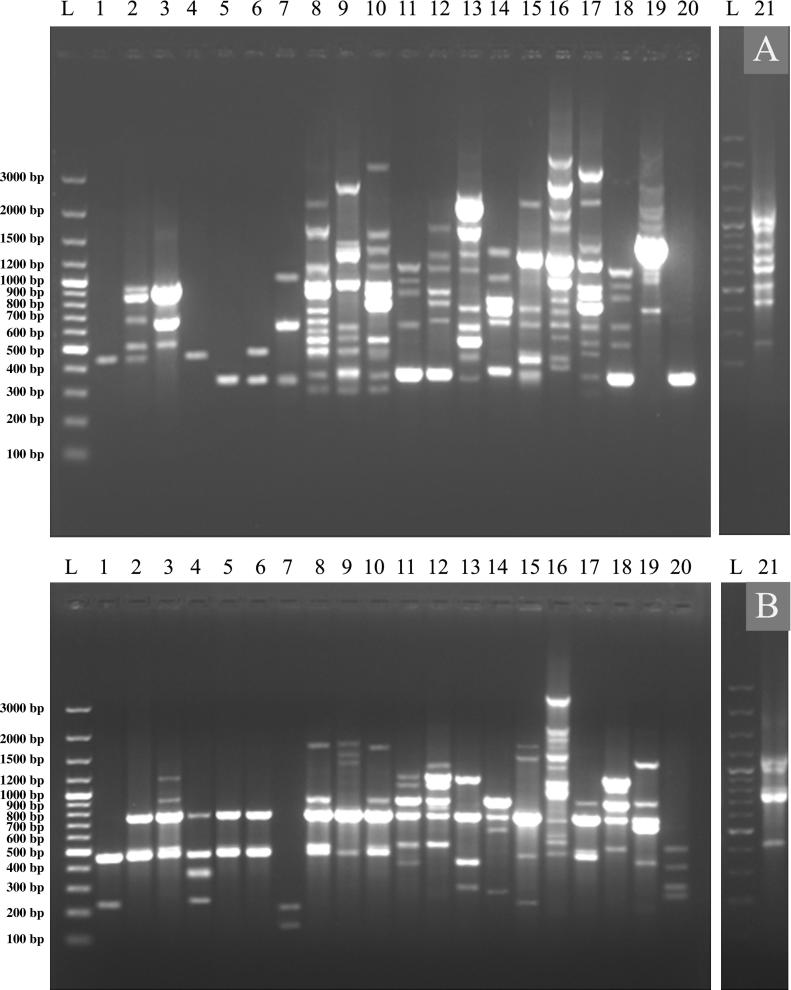
For example, the PCR amplifications using universal primers for Cry 1 genes are illustrated in Fig.2A. Fifteen isolates were shown to contain this gene. Nine isolates produced PCR products of the expected size (274–277 bp). Three isolates were negative for the presence of Cry 1 gene (Qena 3, R.Sea 1 and N.Sinai 1 isolates). However, some isolates produced bands of different sizes, larger or smaller than the expected size (Isma 1, Sohg 1, Matr 5, Asut 1, Asut 2, N.Sinai 2 and N.Sinai 3 isolate). The most interesting observation was the detection of the expected band, in addition to extra unique band(s) among the tested isolates. For example, Port 1 and Qena 1 isolates yielded additional bands at 360 bp and 375 bp, respectively. On the other hand, six isolates produced bands of different sizes from the expected band size; these isolates were, Isma 1, Sohg 1, Matr 5, Asut 1, Asut 2, N.Sinai 2 and N.Sinai 3. The presence or absence of Cry 1 subfamilies in the tested Bt strains was also determined. Two different subgroups of Cry 1 gene (Cry 1B and Cry 1C) were examined (Fig. 2B and C, respectively). Data revealed that seven isolates harbor the Cry 1B gene and fourteen isolates harbor the Cry 1C gene.
Figure 2.
PCR amplification profile of Cry-type genes using universal specific primers for different native and standard Bt isolates (Lane 1: 100 bp SibEnzyme DNA ladder; Lane 1–21: Isma 1, Tosk 1, Tosk 2, Port 1, Dipel® 2X, Agerin®, Sohg 1, Matr1, Qena 1, Qena 2, Qena 3, R.Sea 1, Asut 1, Asut 2, S.Sinai 1, S.Sinai 2, S.Sinai 3, N.Sinai 1, N.Sinai 2, N.Sinai 3 and HD-635, respectively). A: Cry 1 gene, B: Cry 1B gene, C: Cry 1C gene, D: Cry 2 gene and E: Cry 2A (a,b) gene.
The distribution of Cry-2 family and subfamily (Cry 2A(a,b)) genes among different indigenous Bt isolates was also tested. It appears that seven isolates harbor the Cry 2 gene and ten isolates harbor the Cry 2A(a,b) gene (Fig. 2D and E). On the other hand, three isolates harbor the Cry 3 gene; and 5 isolates harbor the Cry 4 gene.
Concerning the Cry-type genes of the standard Bt strains used in this study, Dipel® 2X and Agerin®; it is clear that they contain all tested Cry-type genes except Cry 3 and Vip 3Aa1. However, the standard HD-635 harbors only two types of Cry-genes, namely; Cry 1 and Cry 2A(a,b). The Cry 1 family and subfamily (Cry 1C) were the most persistent among the tested and standard isolates. In contrast, all isolates and standard strains are free from the presence of vegetative insecticidal protein (Vip)-like sequences.
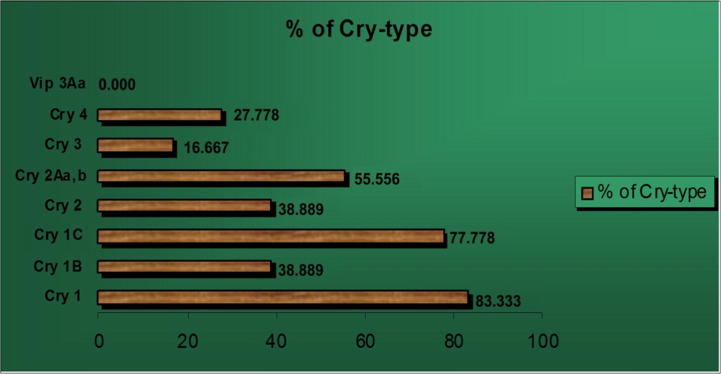
The frequency of each Cry-type gene among the tested isolates is shown in Fig. 3. Among the tested Cry-type genes, Cry 1 family and subfamilies (Cry 1B and Cry 1C) genes were the most abundant at 83.33%, 38.88%. and 77.77%, respectively. Moreover, the frequency of the Cry 2 gene was 38.88%. Interestingly, not all the isolates that contained Cry 2 yielded PCR products using primers for the Cry 2A(a,b) gene (S.Sinai 1 and N.Sinai 2 isolates). 55.55% of the tested isolates contained the Cry 2A(a,b) gene. On the other hand, the least frequent Cry-type genes were Cry 3 gene, which was detected in only 16.66% of the tested isolates. The Cry 4 gene was detected in 27.77% of the isolates. Finally, none of the isolates had any evidence for the Vip 3Aa1 gene.
Figure 3.
The percentages of presence of Cry-type genes using specific primers among different indigenous Bt isolates.
3.3. RAPD-PCR analysis
RAPD analysis was performed to measure genetic difference among sample and standard isolates. Out of the twenty pre-selected random primers, only two (OPA-20 and OPB-18) revealed reproducible polymorphisms. From the data, the percentage of polymorphism was 130 and this was recorded for OPA-20 primer Also, in the case of OPB-18 primer, the percentage of polymorphism was 25.263%. The richest PCR amplification patterns as detected using OPA-20 are shown in Fig.4A. In general, some isolates have uniquely characteristic bands; For example 1235 bp for S.Sinai 1 isolate; while 3625, 2568, 1985, 1719, 836, 400 bp were detected only for S.Sinai 2 isolate; 3100 bp for S.Sinai 3 isolate; three bands having 2266, 1490 and 1290 bp were detected for Qena 1 isolate; two bands having 3534, 794 bp for Qena 2 isolate; three bands having 2035, 1327 and 1137 bp were detected for Asut 1 isolate; 383 bp was detected only for Asut 2 isolate; 529 and 264 bp were detected only for N. Sinai 2 isolate and finally a unique band at 1308 bp was clearly detected for R.Sea 1 isolate.
Figure 4.
Agarose gel electrophoresis of RAPD-PCR products of different Bt isolates using OPA-20 (A) and OPA-18 (B). Lane 1: 100 bp Fermantas DNA ladder; Lane 1–21: Isma 1, Tosk 1, Tosk 2, Port 1, Dipel® 2X, Agerin®, Sohg 1, Matr1, Qena 1, Qena 2, Qena 3, R.Sea 1, Asut 1, Asut 2, S.Sinai 1, S.Sinai 2, S.Sinai 3, N.Sinai 1, N.Sinai 2, N.Sinai 3 and HD-635, respectively.
Also, common polymorphic bands among different isolates and reference ones were detected using OPB-18 primer (Fig.4B). For example, amplicon of 792 bp was common for the three reference isolates in addition to Port 1, Matr 1, Qena 1, Qena 2, Qena 3, Asut 1, Asut 2 and R.Sea 1 isolates. Among the tested isolates, S.Sinai 2 isolate was highly differentiated from the other ones by amplification of six unique polymorphic bands having, 3320, 2140, 1940, 1387, 608 and 565 bp. In addition, there are some unique characteristic polymorphic bands distributed among different isolates. For instance, polymorphic bands of 1880, 1645 and 1480 bp were peculiar for Qena 1 isolate; 375 and 250 bp for Port 1 isolate; 1838 and 536 bp for Matr 1 isolate; 463 bp for Isma 1 isolate; 154 bp for Sohg 1 isolate; 1240 bp for Qena 3 isolate; 869 bp for R.Sea 1 isolate; 1200 bp for Asut 1 isolate; 286 bp for Asut 2 isolate; and 890 bp for N.Sinai 1 isolate. Also, two polymorphic bands having 410 and 272 bp are characteristic for N.Sinai 3.
3.4. Assessment of Bt diversity based on combined analysis of their protein and RAPD-PCR banding patterns
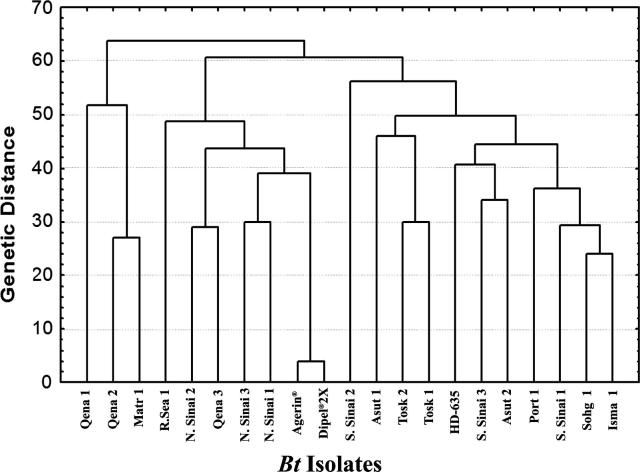
The dissimilarity distance values among the isolates based on combined analysis of protein pattern and RAPD-PCR are calculated. The distance values were found to range from 4.00 (most related) to 58.00 (distantly related). The highest genetic distance value (58.00) was observed between the Qena 1 and S.Sinai 2 isolates. However, the lowest genetic distance was recorded among the Agerin® and Dipel® 2X at 4.00, followed by genetic distance of 20.00 between Sohg 1 isolate and both Agerin® and Dipel® 2X standards.
Diversity analysis of the twenty-one isolates (Fig. 5) showed high levels of genetic diversity; these were subdivided into three main clusters. Cut off point at 60 dissimilarity points were fixed as maximum dissimilarity. Cluster I involved Qena 1, Qena 2 and Matr 1 isolates. Cluster II was divided into two main subclusters; the first contained N.Sinai 2 isolate; and the 2nd subcluster involved two groups. The first group contained Qena 3 and R.Sea 1 isolates. The 2nd group was divided into two subgroups, one containing N.Sinai 1 and N.Sinai 3 isolates; whereas the other one contained the standards (Agerin® and Dipel® 2X). The third cluster was the largest having isolates (S.Sinai 1, S.Sinai 2, S.Sinai 3, Asut 1, Asut 2, Tosk 1, Tosk 2, Isma 1, Port 1, Sohg 1 and HD-635). The salient features of this tree indicate that both Agerin® and Dipel® 2X are genetically very close, with high genetic similarity, clustering in one subgroup. So, it may be concluded that both Agerin® and Dipel® 2X are genetically very close and may be of the same genetic origin.
Figure 5.
Phylogenetic tree based on combined protein and RAPD-PCR.
4. Discussion
In this study, different approaches (SDS-PAGE, specific PCR using different Cry genes and RAPD-PCR) have been used to characterize the Bt isolates and to explain the variability in their potency. Protein analysis indicated that it is possible to distinguish between different isolates based on protein banding patterns and in most cases there is a positive linkage between the molecular weights of the major components of the crystal proteins and potential activities of the isolate. For instance, shared bands among the most potent isolates, such as 130 KDa was detected in Tosk 1, Tosk 2, Qena 1, and Sohg 1 isolates (shared with third reference strain; HD-635); 127 KDa for Qena 2, Asut 1 and S.Sinai 2 isolates (shared with the reference strains; Agerin® and Dipel® 2X); 124 KDa for Qena 1 and Sohg 1 isolates; 100 KDa for Tosk 1, Qena 1 and S.Sinai 2 isolates; isolates; 98 KDa for Tosk 2, Qena 1, Qena 2 and S.Sinai 2 isolates; 79 KDa for Tosk 1, Tosk 2 isolates (shared with the reference strains; Agerin® and Dipel® 2X); 76 KDa for Qena 1 and Sohg 1 isolates; 71 KDa for Sohg 1, Qena 1 Qena 2 and Asut 1 isolates (shared with third reference strain; HD-635); and finally, low molecular weight protein band at 49 KDa for Qena 1, Qena 2, Asut 1, Tosk 1 and Tosk 2 isolates whereas, the isolates which show moderate potential activity against neonate larvae of H. armigera shared with one or more bands having molecular weight of 120, 100, 98, 79 and 71 KDa. It seems that, Bt with protoxin that ranged between 49 and 79 KDa as low molecular weight proteins and high molecular weight proteins of 130–98 KDa would be potent against H. armigera. The relation between the potential activity of the isolates and their protein pattern was investigated by many authors e.g. [10], [18], [34], [35], [36], [37].
Moreover, results obtained from PCR analysis of Cry genes revealed that the most common Cry genes found in the tested Bt isolates are Cry 1 and Cry 1 subfamily genes, followed by Cry 2 and its subfamily genes. In this concern, many authors reported that the most common Cry genes found in nature are those within the Cry 1 gene and/or Cry 1 subfamily combinations [9], [38], [39], [40], [41], [42], [43]. The Cry 1 and Cry 1 subfamily linkage may be explained by their common location on the same replicon [44]. On the other hand, [45] found that the most abundant gene was Cry 2, followed by Cry 1 gene family. Although, most isolates produced a positive single band at 274–277 bp and this was obtained with universal primers for Cry 1 gene, extra band/bands were amplified. Similar findings were reported [11], [46] who suggested that isolates containing novel Cry genes may give PCR products different in size relative to the standard or may completely lack PCR products. [47] suggested that this could be due to the fact that two or three cry genes might be positioned next to each other, forming an operon. In this case, priming of an oligonucleotide primer to the neighbor cry gene may produce an extra PCR product larger than expected size. On the other hand, the absence of the PCR product in some isolates was detected. Coinciding with this, [12], [13] found that 89 out of 215 field-collected strains of Bt did not produce PCR products with universal primers. The third approach for molecular characterization (RAPD-PCR) showed to be simple, useful, fast, and informative and it is unique in that no information concerning the sequence is needed. It gives the opportunity to get information about the biodiversity in a group of isolates. To investigate the diversity among the tested Bt isolates and other standard formulations, two reproducible and characteristic RAPD patterns were produced using OPA-20 primer and OPB-18 primer random primers. In this context, many authors used RAPD analysis to characterize different Bt isolates [48], [49], [50].
5. Conclusions
It appears that Bt strains can be distinguished and identified on the basis of molecular characterization techniques such as SDS-PAGE, specific PCR of Cry-type genes and RAPD-PCR fingerprint. Genetic variability was obvious among different isolates and thus causing variation in their potency against the target lepidopterous insects. The combination of classical and molecular methods makes it more feasible to group the Bt isolates, than if the classical methods are used.
Acknowledgements
The authors wish to express their gratitude to the National Research Centre, Cairo, for providing and financial support of this study.
Footnotes
Peer review under responsibility of National Research Center, Egypt.
Contributor Information
H.S. Salama, Email: hsarsalama@hotmail.com.
N.M. Abd El-Ghany, Email: nesreen_nrc@yahoo.com.
M.M. Saker, Email: sakrmahmoud@yahoo.com.
References
- 1.Salama H.S., Foda M., Zaki F., Ragaei M. Angew. Zool. 1986;3:257–265. [Google Scholar]
- 2.Ohba M., Aizawa K. J. Invertebr. Pathol. 1986;47:277–282. [Google Scholar]
- 3.Ohba M., Aizawa K. J. Invertebr. Pathol. 1986;47:12–20. [Google Scholar]
- 4.Martin A.W., Travers R.S. Appl. Environ. Microb. 1989;55:2437–2442. doi: 10.1128/aem.55.10.2437-2442.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morris O.N., Converse V., Kanagaratnam P., Cote J.C. Can. Entomol. 1998;130(4):515–537. [Google Scholar]
- 6.Chen F.C., Tsai M.C., Peng C.H., Chak K.F. Curr. Microbiol. 2004;48:270–275. doi: 10.1007/s00284-003-4195-1. [DOI] [PubMed] [Google Scholar]
- 7.Zhao Y.J., Wei W., Li R.P., Yuan S., Dai Y.J., Yao J. J. Microbiol. 2009;28(4):43–46. [Google Scholar]
- 8.Salama H.S., Saker M., Salama M., El-Banna A., Ragaei M., Abd El-Ghany N.M. Arch. Phytopathol. Pfl. 2012;45(7):856–868. [Google Scholar]
- 9.Abd El-Ghany N.M., Abdel Ghany E.M., Salama H.S. Biopest. Int. 2015;11(1) [Google Scholar]
- 10.Merdan A., Salama H.S., Labib E., Ragaei M., Abd El-Ghany N.M. Arch. Phytopathol. Pfl. 2010;43(12):1165–1176. [Google Scholar]
- 11.Carozzi N.B., Kramer V.C., Warren Evola G.W.S., Koziel M.G. Appl. Environ. Microb. 1991;57:3057–3061. doi: 10.1128/aem.57.11.3057-3061.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ben-Dov E., Zaritsky A., Dahan E., Barak Z., Sinai R., Manasherob R., Khamraev A., Troitskaya E., Dubitsky A., Berezina N., Margalith Y. Appl. Environ. Microb. 1997;63:4883–4890. doi: 10.1128/aem.63.12.4883-4890.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ben-Dov E., Wang Q., Zaritsky A., Manasherob R., Barak Z., Schneider B., Khamraev A., Baizhanov M., Glupov V., Margalith Y. Appl. Environ. Microb. 1999;65:3714–3716. doi: 10.1128/aem.65.8.3714-3716.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi S.K., Shin B.S., Kong E.M., Rho H.M., Park S.H. Curr. Microbiol. 2000;41:65–69. doi: 10.1007/s002840010093. [DOI] [PubMed] [Google Scholar]
- 15.Wang J., Boets A., Rie J.V., Ren G. J. Invertebr. Pathol. 2003;82:63–71. doi: 10.1016/s0022-2011(02)00202-1. [DOI] [PubMed] [Google Scholar]
- 16.Guidelli-Thuler A.M., Sena J.A.D., Abreu I.L., Davolos C.C., Alves S.B., Polanczyk R.A., Valicente F.H., Lemos M.V.F. Arquivos do Instituto Biologico (Sao Paulo) 2008;75(4):405–414. [Google Scholar]
- 17.Álvarez A., Virla E.G., Pera L.M. M.D. Baigorí. Biotechnol. Lett. 2009;31:1899–1903. doi: 10.1007/s10529-009-0091-5. [DOI] [PubMed] [Google Scholar]
- 18.Zheng A., Zhu J., Tan F., Guan P., Yu X., Wang S., Deng Q., Li S., Liu H., Li P. Ann. Microbiol. 2010;60(1):129–134. [Google Scholar]
- 19.Saker M., Salama H.S., Ragaei M., Abd El-Ghany N.M. Arch. Phytopathol. Pfl. 2012;45(1):110–125. [Google Scholar]
- 20.Carlson C.R., Kolsto A.B. J. Bacteriol. 1993;175:1053–1060. doi: 10.1128/jb.175.4.1053-1060.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lereclus D., Mahillon J., Menou G., Lecadet M.M. Mol. Gen. Genet. 1986;204:52–57. doi: 10.1007/BF00330186. [DOI] [PubMed] [Google Scholar]
- 22.Morris O.N., Converse V., Kanagaratnam P. J. Invertebr. Pathol. 1996;67:129–136. doi: 10.1006/jipa.1997.4667. [DOI] [PubMed] [Google Scholar]
- 23.Dulmage H.T., Correa J.A., Martinez A.J. J. Invertebr. Pathol. 1970;15:15–20. doi: 10.1016/0022-2011(70)90093-5. [DOI] [PubMed] [Google Scholar]
- 24.Khalifa A., Salama H.S., El-Sharaby A. Zeitschrift für angewandte Entomologie (J. Appl. Entomol.) 1973;73:129–132. [Google Scholar]
- 25.Dulmage H.T. Bull. Entomol. Soc. Am. 1973;19(4):200–202. [Google Scholar]
- 26.Salama H.S., Foda M.S., Sharaby A. Trop. Pest Manage. 1989;35:326–330. [Google Scholar]
- 27.Abbott W.S. J. Econ. Entomol. 1925;18:265–267. [Google Scholar]
- 28.Laemmli U.K. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Beard C.E., Ranasinghe C., Akhurst R.J. Lett. Appl. Microbiol. 2001;33:241–245. doi: 10.1046/j.1472-765x.2001.00982.x. [DOI] [PubMed] [Google Scholar]
- 30.Welsh J., McClelland M. Nucl. Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams S., Friedrich L., Dincher S., Carozzi N., Kessmann H., Ward E., Ryals J. Biotechnology. 1993;7:194–200. [Google Scholar]
- 32.Bassam B.J., Caetano-Anollẻs G., Gresshoff P.M. Appl. Microbiol. Biotechnol. 1992;38:70–76. doi: 10.1007/BF00169422. [DOI] [PubMed] [Google Scholar]
- 33.Hadrys H., Balick M., Schierwater B. Mol. Ecol. 1992;1:55–63. doi: 10.1111/j.1365-294x.1992.tb00155.x. [DOI] [PubMed] [Google Scholar]
- 34.Chak K.F., Ellar D.J. J. Gen. Microbiol. 1987;133:2921–2931. doi: 10.1099/00221287-133-10-2921. [DOI] [PubMed] [Google Scholar]
- 35.Höfte H., Whitely H.R. Microbiol. Rev. 1989;53:242–255. doi: 10.1128/mr.53.2.242-255.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haider M.Z., Mahmood S. J. Basic Microb. 1990;30:251–258. doi: 10.1002/jobm.3620300406. [DOI] [PubMed] [Google Scholar]
- 37.Itoua-Apoyolo C., Drif L., Vassal J.M., De Barjac H., Bossy J.P., Leclant F., Frutos R. Appl. Environ. Microb. 1995;61(12):4343–4347. doi: 10.1128/aem.61.12.4343-4347.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bravo A., Sarabia S., Lopez L., Ontiveros H., Abarca C., Ortiz A., Ortiz M., Lina L., Villalobos F.J., Pena G. Appl. Environ. Microb. 1998;64:4965–4972. doi: 10.1128/aem.64.12.4965-4972.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valicente F.H., Barreto M.R., de Vasconcelos M.J.V., de Figueiredo J.E.F., Paiva E. Anais da Sociedade Entomologica do Brasil. 2000;29(1):147–153. [Google Scholar]
- 40.Martinez C., Caballero P. J. Appl. Microbiol. 2000;92(4):745–752. doi: 10.1046/j.1365-2672.2002.01579.x. [DOI] [PubMed] [Google Scholar]
- 41.Porcar M., Juárez-Pérez V. FEMS Microbiol. Rev. 2003;26:419–432. doi: 10.1111/j.1574-6976.2003.tb00624.x. [DOI] [PubMed] [Google Scholar]
- 42.Zhu J., Tan F.R., Tang J., Li Y.Y., Zheng A.P., Li P. Ann. Microbiol. 2009;59(1):1–8. [Google Scholar]
- 43.Wu C.B., Guan Y., Qiu J.J., Huang Z.M., Zhang A.H. J. Econ. Animal. 2009;13(2):72–76. [Google Scholar]
- 44.Sanchis V., Lereclus D., Menou G., Chaufaux J., Lecadet M.M. Mol. Microbiol. 1988;2:393–404. doi: 10.1111/j.1365-2958.1988.tb00044.x. [DOI] [PubMed] [Google Scholar]
- 45.Arrieta G., Hernández A., Espinoza A.M. Rev. Biol. Trop. 2004;52(3):757–764. [PubMed] [Google Scholar]
- 46.Kuo W.S., Chak K.F. Appl. Environ. Microb. 1996;62:1369–1377. doi: 10.1128/aem.62.4.1369-1377.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown K.L., Whiteley H.R. J. Bacteriol. 1992;174:549–557. doi: 10.1128/jb.174.2.549-557.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brousseau R., Saintonge A., Préfontaine G., Masson L., Cabana J. Appl. Environ. Microb. 1993;59(1):114–119. doi: 10.1128/aem.59.1.114-119.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansen B.M., Damgaard P.H., Eilenberg J., Pedersen J.C. J. Invertebr. Pathol. 1998;71:106–114. doi: 10.1006/jipa.1997.4712. [DOI] [PubMed] [Google Scholar]
- 50.Yaradoni S.N., Krishnaraj P.U., Ussuf K.K., Kuruvinashetti M.S. J. Plant Biol. 2003;30(1):7–13. Society for Plant physiology and Biochemistry, New Delhi, India. [Google Scholar]
- 51.Cerón J., Covarrubias L., Quintero R., Ortíz A., Ortíz M., Aranda E., Lina L., Bravo A. Appl. Environ. Microb. 1994;60:353–356. doi: 10.1128/aem.60.1.353-356.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Asano S., Bando H., Iizuka T. J. Ser. Sci. Japan. 1993;62:223–227. [Google Scholar]
- 53.Ritu B., Dalal M., Panguluri S.K., Jagadish B., Mandaokar A.D., Singh A.K., Kumar P.A. FEMS Microbiol. Lett. 2005;243:467–472. doi: 10.1016/j.femsle.2005.01.011. [DOI] [PubMed] [Google Scholar]