Abstract
How cells respond to a myriad of stimuli with finite signaling machinery is central to immunology. In naive T cells, the inherent effect of ligand strength on activation pathways and endpoints remains controversial, confounded by environmental fluctuations and intercellular variability within populations. Here, we study how ligand potency affects CD8+ T cell activation in vitro using genome-wide RNA, multi-dimensional protein and functional measurements in single cells. Our data reveal that strong ligands drive more efficient and uniform activation than weak ligands, but all activated cells are fully cytolytic. Importantly, activation follows the same transcriptional pathways, regardless of ligand potency. Thus, stimulation strength does not intrinsically dictate T cell activation route or phenotype; instead it controls how rapidly and simultaneously cells initiate activation, allowing limited machinery to elicit wide-ranging responses.
Cytotoxic T lymphocytes (CTLs) are critical for immune defense against tumors and viral pathogens. These programmed killer cells develop from naive CD8+ T cells over a multi-day activation process that depends on the activating stimulus as well as the surrounding immunological environment. T cell activation begins when the T cell receptor (TCR) on a naive CD8+ T cell recognizes peptide-class I major histocompatibility complexes (pMHC) on an antigen presenting cell (APC)1,2. If the TCR-pMHC interaction is strong enough to overcome negative feedback loops that control basal T cell signaling, additional signaling components are recruited to the plasma membrane. This initiates multiple coordinated enzymatic cascades that regulate integrin affinity, cytoskeletal rearrangements, metabolic changes and transcription factor activity, which are essential for T cell proliferation and effector differentiation.
Altering the affinity of the TCR-pMHC interaction can dramatically affect immunological outcomes in both thymic selection and T cell activation in the periphery3,4. In thymocytes, TCR-pMHC affinity determines whether cells undergo positive or negative selection5–7. In the periphery, different studies have found T cell responses to be graded or thresholded with respect to ligand affinity, with stronger ligands generally inducing a larger T cell response8–13. In vitro studies have observed that reducing ligand affinity can decrease readouts of naive CD8+ T cell activation including calcium fluxes, kinase and transcription factor activity, surface receptor and cytokine expression, and proliferation5,11,13–23. Similarly, changing ligand concentration or costimulatory signals can affect T cell activation11,16,21,24–28. However, T cell activation assays differ between studies, with some reported as analog and others digital with respect to ligand affinity3,4,17,18,21,23,27,29,30. Additionally, reports conflict over whether ligand potency controls response magnitude13,15,16,20, speed19, frequency17,18, or a combination thereof21,22,30. A substantial challenge in investigating early T cell activation is that populations of identically stimulated T cells exhibit extensive heterogeneity. Previous studies were limited in the number of markers that could be examined in individual cells or required pooled populations for genome-wide measurements. Thus, it has been difficult to identify the intrinsic mechanism by which ligand potency affects the integrated organization of gene expression changes within an individual cell. Single-cell technologies now allow individual cells to be interrogated at the molecular level in an unbiased, genome-wide manner, enabling us to address this question.
Here we employ five single-cell methods to measure the genome-wide transcriptomes and targeted proteomic and functional profiles of naive CD8+ T cells activated in a controlled in vitro environment with peptide ligands of various potencies. We find that reducing ligand potency decreases the rate with which single T cells initiate transcriptional activation, driving increased heterogeneity in the pool of cells. However, we observe that all cells that activate, regardless of the potency of their primary stimulus, can achieve the same spectrum of effector protein profiles, are capable of cytolytic granule release, and reach this activated state using the same transcriptional pathway. Thus, we show that stimulation strength dictates how rapidly and uniformly cells respond.
Results
Distinct transcriptional phases of early T cell activation
We first sought to characterize transcriptomic changes during the initial six hours of naive CD8+ T cell activation by single-cell RNA-sequencing (scRNA-seq). We used the OT-I transgenic TCR model to facilitate strict control over the timing of antigen encounter and the strength of the activating stimulus. Cells were isolated from OT-I TCR transgenic Rag1-deficient mice, in which all peripheral CD8+ T cells recognize the ovalbumin peptide SIINFEKL presented by the MHC class I molecule H-2Kb. Ex vivo CD8+ OT-I T cells were stimulated with pure peptide for self-presentation to avoid contamination by antigen-presenting-cell RNA. By staining cells for surface protein expression before sorting into lysis buffer for scRNA-seq, we simultaneously measured expression of four surface proteins and quantified genome-wide mRNA in the same cells. We assayed one protein that is downregulated with activation, CD62L (L-selectin, encoded by Sell), and three proteins upregulated with activation, CD69, CD25 (encoded by Il2ra) and CD44.
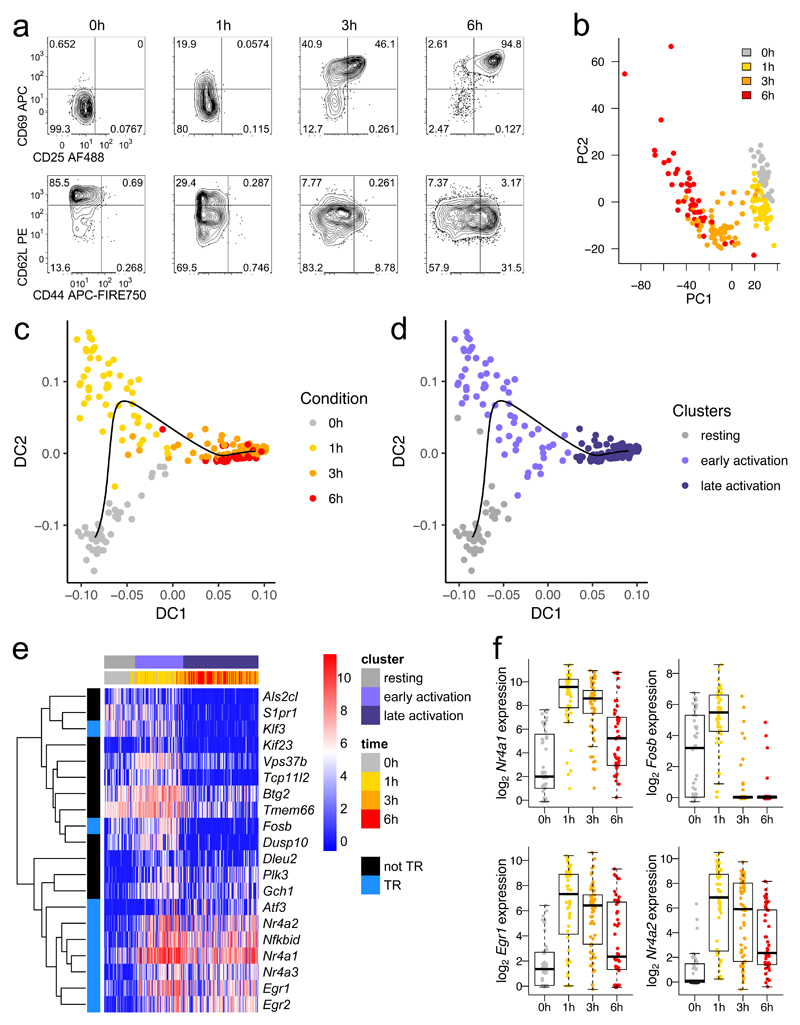
We first profiled a time course of strong stimulation by sequencing the transcriptomes of individual OT-I CD8+ T cells after 0, 1, 3 and 6 hours of activation with 1 μM of the high potency OT-I TCR cognate peptide SIINFEKL (N4) in the presence of IL-2 (Supplementary Fig. 1a and Methods). 87% of cells passed quality control filtering, resulting in 44-64 cells per condition. Protein profiles revealed a rapid drop in CD62L expression, followed by sequential increases in CD69, CD25 and CD44 expression (Fig. 1a). Surface protein measurements and principal components analysis applied to the mRNA data confirmed that, despite a uniform activation stimulus, these cells were heterogeneous during the first 6 hours of activation (Fig. 1a,b). Because of this diversity, simply grouping the cells by sampling time cannot achieve the fine resolution required to understand coordination of gene expression. Instead, we took advantage of the heterogeneity within each stimulation condition to order all cells by their progressively changing transcriptional profiles, referring to the progress in ‘pseudotime’ instead of real time. We used a diffusion pseudotime method31, a computational approach which finds the most direct path through the observed cell states based upon each cell’s gene expression profile. Fitting a diffusion pseudotime trajectory to the single-cell transcriptomic data, we created a fine-grained map of activation (Fig. 1c, Supplementary Fig. 1b).
Fig. 1. A burst of transcriptional regulatory machinery characterizes early T cell activation.
a, OT-I CD8+ T cells were stimulated with high potency ovalbumin peptide (N4) for 0, 1, 3, or 6 hours before sorting for scRNA-seq by FACS. Protein expression flow cytometry measurements are representative of at least 2 independent experiments. b, Principal components analysis of scRNA-seq of cells sorted in a. c, Diffusion pseudotime analysis of sequenced cells: cells are plotted by diffusion components (DCs) 1 and 2, with a black line delineating the pseudotime trajectory. d, Plot in c is colored by clusters in diffusion pseudotime. e, The top 20 genes transcriptionally upregulated in the early activation cluster versus the resting and late activation clusters are depicted in a heatmap with genes clustered by Pearson correlation. Blue (TR) indicates transcriptional regulatory genes. f, Expression of selected transcription factors from e is shown with a box plot indicating the median, boxed interquartile range, and whiskers extending to the most extreme point up to 1.5 x the interquartile range. (b-f) n = 44 cells for 0h, 51 for 1h, 64 for 3h, and 46 for 6h.
The distribution along pseudotime of all cells from the various time points revealed three clusters of resting, early activated and late activated cells (Fig. 1d, Supplementary Fig. 1c,d). We performed a differential expression analysis to compare each activated cluster to the resting cells. Cells in the early stages of activation primarily increased expression of genes involved in immune and regulatory processes, whereas cells in the later stages of activation showed strong upregulation of genes involved in metabolic and biosynthetic functions (Supplementary Table 1), which are critical for T cell effector differentiation32,33. We then examined genes differentially expressed in the early activation cluster compared to both the resting and late activation clusters to identify transient early expression changes. Filtering for differentially expressed genes uniquely upregulated in early activation revealed an enrichment for genes that encode transcriptional regulators (enrichment p value 1.8 x 10-9, Fig. 1e, Supplementary Fig. 1e). In particular, the early response NR4A family orphan nuclear receptors, including Nr4a1 (Nur77) whose expression has been found to reflect TCR signaling activity27,34–36, were most highly expressed 1 hour after activation (Fig. 1f). Additionally, early growth response factors (Egr1 and Egr2) and the AP-1 transcription factor subunit Fosb followed similar expression patterns. Thus, we identified two distinct phases of the activation response.
Ligand potency controls response rate of CD8+ T cells
To determine how TCR stimulation strength might affect this early response, we selected two transcription factors characteristic of the early activation profile, Nr4a1 and Fosb, and examined their expression by RNA flow cytometry while modulating TCR signaling strength. We stimulated cells with each of four peptides with previously characterized potency for stimulating the OT-I TCR5,37: the cognate SIINFEKL peptide (N4); 2 variants with the amino acid changes T4 and G4, in decreasing order of potency; and NP68, which does not bind the OT-I TCR. The same concentration (1 μM) of each peptide was used, which should saturate MHC occupancy5,11. We cultured all cells in equivalent concentrations of exogenous IL-2, as this cytokine is required for T cells to enter the cell cycle and proliferate. Given that IL-2 production varies with stimulation strength33, addition of exogenous IL-2 enabled us to identify intrinsic changes in gene activation that were dependent solely on stimulation strength (Supplementary Fig. 2a,b).
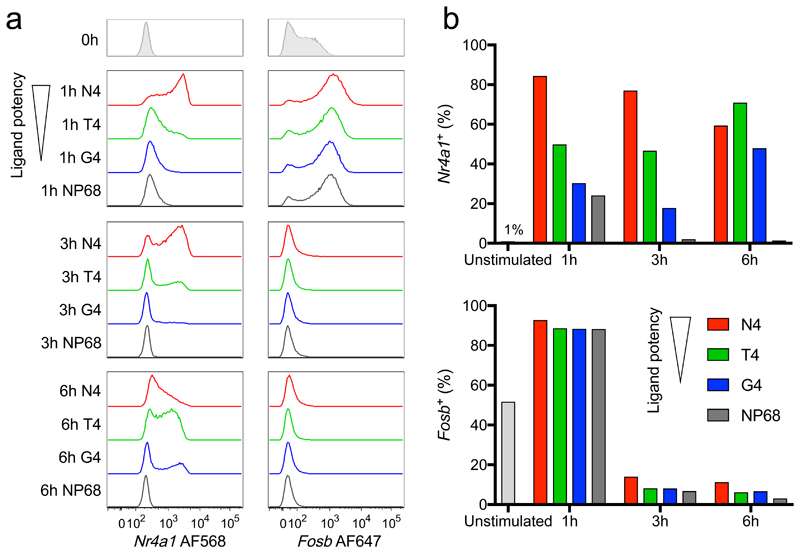
RNA hybridization was verified through examination of the control gene Rpl39 (Supplementary Fig. 2c,d). Nr4a1 induction after 1 hour was dependent on ligand strength, with higher potency ligands driving a larger percentage of cells expressing Nr4a1 mRNA, in accordance with previous studies27,34 (Fig. 2). In cells stimulated with the most potent ligand (N4), this percentage gradually declined by 6 hours, but in cells stimulated with the reduced potency ligands (T4 and G4, respectively), the percentage of Nr4a1+ cells increased between 3 and 6 hours, suggesting a delayed peak in the population response. Cells stimulated with the null peptide NP68 upregulated Nr4a1 to a small extent after 1 hour, suggesting a combination of TCR-dependent and -independent effects on induction. In contrast, Fosb was strongly upregulated at 1 hour and rapidly returned to baseline by 3 hours, regardless of the peptide stimulus. These results indicate that within the immediate early burst of transcription factor expression, certain genes exemplified by Nr4a1 respond primarily to TCR stimulation, while another set of regulators including Fosb are likely driven by TCR-independent factors acting in the first hour of tissue culture. Our observation that ligands of lower potency result in reduced immediate and delayed maximum expression of Nr4a1 suggests that stimulation strength either alters the rate with which cells embark on a universal transcriptional activation pathway or controls the utilization (or coordination) of different activation pathways.
Fig. 2. Early response genes can be TCR-dependent or TCR-independent.
a, OT-I CD8+ T cells were stimulated with high potency N4 peptide, reduced potency ligands (T4 or G4) or a non-binding control peptide (NP68) for the indicated times before examination of Nr4a1 and Fosb expression by RNA flow cytometry. Samples were gated on live cells in which the control gene Rpl39 was detected. b, Plots depict the percentage of cells detected expressing each transcription factor. Results (a, b) are representative of 3 independent experiments.
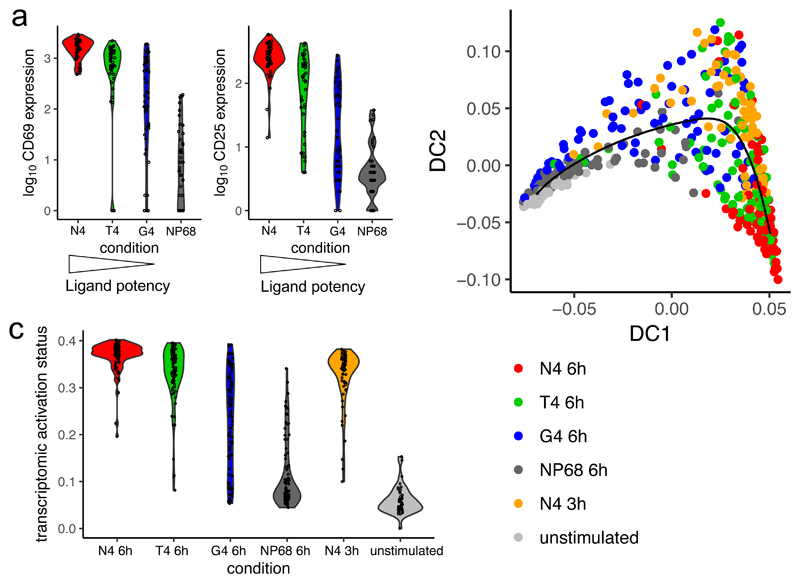
To test these possibilities, we performed scRNA-seq on OT-I CD8+ T cells stimulated with the same ligands for 6 hours. Protein profiling revealed that reducing ligand potency increased heterogeneity in protein markers of early activation (Fig. 3a, Supplementary Fig. 3a). To determine whether ligand potency controls transcriptional activation pathways, we combined data from cells stimulated for 6 hours with all ligands and the most potent ligand (N4) stimulation time course. 93% of cells in this combined data set passed quality control filtering, leaving 44-94 cells per condition. We excluded cells cultured for only 1 hour to avoid the immediate TCR-independent effects described above. Using diffusion pseudotime analysis, we fitted a trajectory to the cells and found that it tracked activation status (Fig. 3b). We observed that cells stimulated with medium (T4) and low (G4) potency ligands did not follow a different activation trajectory from those stimulated with the strongest ligand (N4). This indicated that all ligands promote the same major transcriptional changes, including upregulation of biosynthetic and metabolic machinery. As with protein expression in the early hours of activation, reducing ligand potency resulted in greater heterogeneity with respect to progress along the activation trajectory (Fig. 3c). Cells activated by weaker ligands were not universally less activated, with a proportion of cells achieving activation comparable to cells stimulated with the highest potency ligand. This indicates that stimulation strength controls the probability of a cell activating at any given moment, regulating the rate with which cells initiate transcriptional activation rather than the speed with which they progress once activation is initiated. When measurements are summarized across the whole T cell population, reduction in activation efficiency would appear as a reduction in the magnitude or fraction of cells responding, which have been described in numerous studies (exemplified by references15,17,18,20,21,30). Only through genome-wide single-cell experiments could we distinguish the effect of response rate from differences in the percentage, speed of progression, or phenotype of responding cells.
Fig. 3. Ligand potency determines activation rate.
OT-I CD8+ T cells were stimulated for 6 hours with the various peptide ligands or for 3 hours with high affinity (N4) peptide for scRNA-seq. a, Violin plots depict the distribution of surface protein expression measurements in sequenced cells stimulated for 6 hours; n = 45 cells for N4, 44 for T4, 48 for G4, and 47 for NP68. A second independent experiment is depicted in Supplementary Fig. 3a. b, An activation trajectory was fit to the transcriptomic data by diffusion pseudotime analysis; n = 91 cells for N4 6h, 91 for T4 6h, 94 for G4 6h, 93 for NP68 6h, 64 for N4 3h, and 44 for unstimulated. Plot depicts combined data from two independent scRNA-seq experiments. c, Distribution of cells along the pseudotime transcriptomic activation trajectory fit in b separated by stimulation condition.
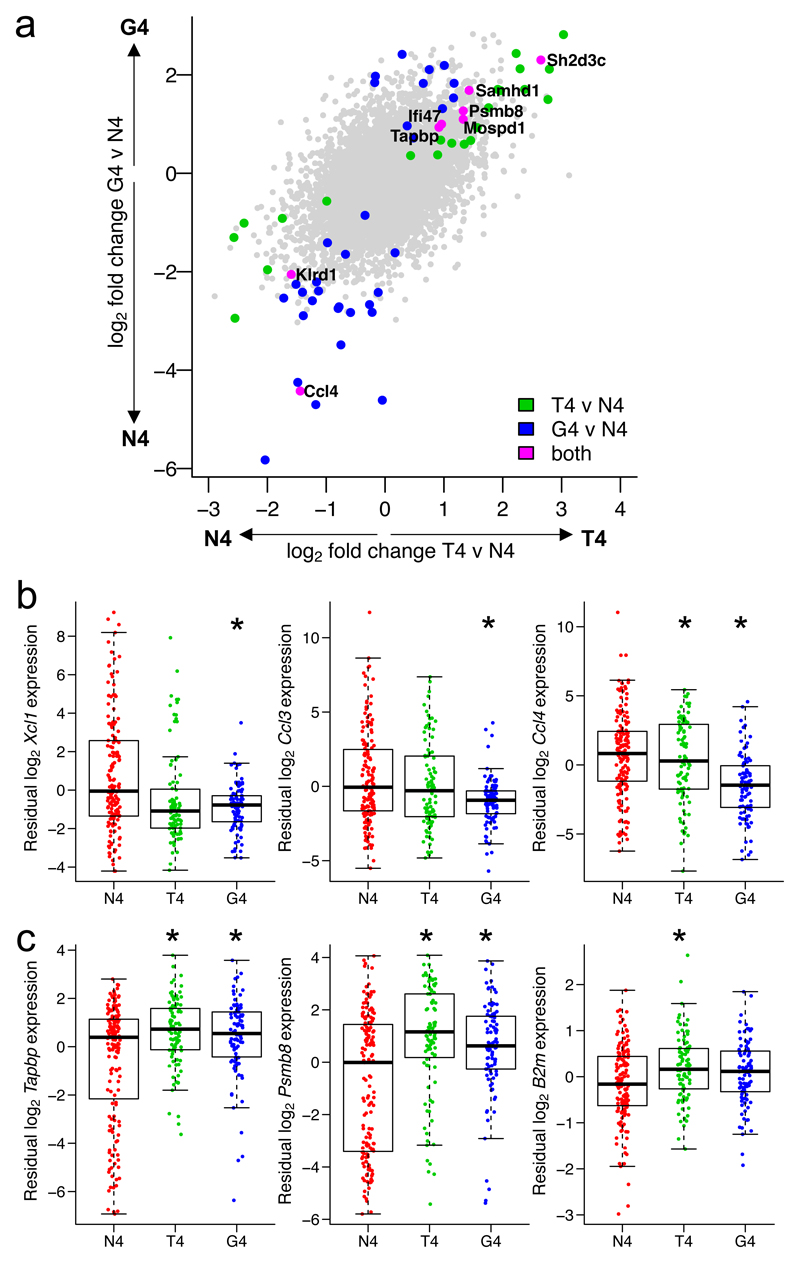
Although we found the main activation trajectory to be shared between cells stimulated by high or reduced potency ligands, it remained possible that stimulation strength might cause subtle deviations. Extensive heterogeneity between cells makes it challenging to directly compare cells at the same activation stage after stimulation with different ligands. Even sorting cells based on binary measures of activation status14,21,30 might fail to account for heterogeneity within the activated populations. A benefit of our single-cell approach is that we can take advantage of the heterogeneity within stimulated populations to control for differences in activation state while comparing cells stimulated with various ligands. CD69 surface protein expression was strongly correlated with transcriptomic activation status, demonstrating activation along our transcriptomic trajectory. CD69 expression could therefore be used as an independently measured feature to control for activation state in a differential RNA expression analysis between cells treated with the various ligands (Supplementary Fig. 3b). This allowed us to ask the question: given the activation state defined by CD69 expression, what is differentially expressed between stimulation conditions? Our analysis revealed only a small number of genes expressed at different levels, confirming that cells activated by different ligands share a transcriptional response (G4 vs N4, 41 of 8854 genes, 0.46%; and T4 vs N4, 30 of 8854 genes, 0.34% of genes differentially expressed). Expression of a small subset of genes was directly impacted by ligand potency (Fig. 4a, Supplementary Tables 2,3). This included downregulated expression of genes encoding chemokines such as Xcl1, Ccl3 and Ccl4, particularly in cells stimulated with the low potency (G4) ligand, and upregulated expression of genes encoding proteins with roles in class I antigen presentation such as Psmb8, Tapbp and B2m, particularly in cells stimulated with the medium potency (T4) ligand (Fig. 4b,c, Supplementary Fig. 3c,d). Although many genes, including Ccl3 and Ccl4 chemokines, have been previously observed to exhibit altered expression with weak ligand stimulation15,28, here we demonstrate that this occurs for only a small subset of genes when we account for the activation status of the cells. Of particular importance, genes directing the metabolic and biosynthetic programs of T cell activation were not enriched in our differentially expressed gene lists, suggesting that all activated cells, regardless of ligand potency, can mount these energetic effector differentiation processes. Likewise, early transcription of Gzmb, encoding the important cytolytic effector molecule Granzyme B, did not significantly differ between stimulation conditions when accounting for cellular activation status (Supplementary Fig. 3e). Our results indicate that while there are specific genes involved in leukocyte recruitment and endogenous peptide presentation whose expression depends on ligand strength, the overwhelming majority of genes expressed in early activation depend on activation status and not the potency of the stimulus.
Fig. 4. Differential expression between cells activated by ligands of different potencies accounting for activation status.
a, Log2-fold changes from a differential expression analysis between T4 (medium potency) and N4 (high potency), or G4 (low potency) and N4, peptide stimuli in Fig. 3, accounting for each cell’s activation status as defined by log10 CD69 surface protein expression. Plot depicts all 8854 genes tested; differentially expressed genes (FDR < 0.05) for T4 versus N4 are shown in green; G4 versus N4, blue; intersection of T4 versus N4 and G4 versus N4, magenta and labelled. b, Residual expression of selected chemokine genes in scRNA-seq data after accounting for CD69 surface protein expression, plotted by stimulation condition. c, As b for selected genes associated with class I antigen presentation. (b-c) Box plots show the median, boxed interquartile range, and whiskers extending to the most extreme point up to 1.5 x the interquartile range; n = 155 cells for N4, 91 for T4, 94 for G4. * FDR < 0.05; statistical test described in Methods and statistics detailed in Supplementary Table 2.
Activated T cells achieve the same spectrum of effector phenotypes regardless of stimulation potency
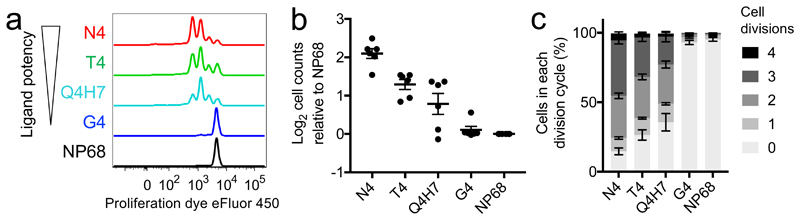
Having observed that, regardless of the primary stimulus, activated T cells progress along the same transcriptomic activation trajectory, we next examined whether cells exhibited phenotypic differences associated with stimulation strength during the first two days of effector T cell differentiation. Because OTI T cells can undergo extensive death when activated with soluble peptide for multiple days, we instead used peptide-pulsed APCs to activate naive T cells for two days (Supplementary Fig. 4a). In this system, we observed negligible activation after two days by the low potency (G4) peptide (Fig. 5a). This is in contrast to pure peptide stimulation with low potency ligand, which, consistent with our observations of transcriptional activity at 6 hours, drove full activation at two days (Supplementary Fig. 4b). Ex vivo CD8+ OT-I T cells were stimulated in IL-2-supplemented media with autologous T-depleted splenocytes (cells remaining after CD8+ T cell isolation) pulsed with one of the 4 peptides used in our transcriptomic studies or Q4H7 peptide, whose potency for stimulating OT-I T cells lies between that of T4 and G4 peptides5 (medium-low potency). After 2 days of activation, the majority of T cells stimulated with the three strongest ligands (N4, T4, Q4H7) were proliferating (Fig. 5a). The proportion of cells in later division cycles and the total number of T cells were associated with stimulation strength (Fig. 5b,c), in agreement with previous observations suggesting an increased percentage of proliferating cells, shorter time to first division, or greater proliferation with stronger TCR-pMHC interactions11,12,14,16,19,21,27. However, we noted that the most proliferative cells in each condition that stimulated proliferation had all undergone 4 divisions, indicating that the few cells that began proliferating immediately after stimulation with weaker ligands were not slower to divide. Although it has been previously reported that stimulation strength affects the proportion of proliferating cells and the time to first division22, our results now show that weaker ligands do not cause a universal delay in proliferation, but rather reduce the number of cells that activate immediately. Together with our results from early activation transcriptomic analysis, we conclude that ligand potency determines the rate with which an individual cell initiates activation and thus the heterogeneity of activation across the population.
Fig. 5. Cell number but not maximal division number is associated with ligand potency.
a, OT-I CD8+ T cells were stimulated with autologous T-depleted splenocytes pulsed with various peptides. After 2 days, proliferation was measured by flow cytometry. Results are representative of 10 separate mice in 7 independent experiments. b, Cells activated as in a were counted and normalized to the null peptide (NP68) condition. Plot depicts counts from 6 separate mice in 5 independent experiments; line indicates mean and error bars denote SEM. c, Mean percentages of cells in each division cycle were quantified from the 10 separate mice in 7 independent experiments described in a; error bars depict SEM.
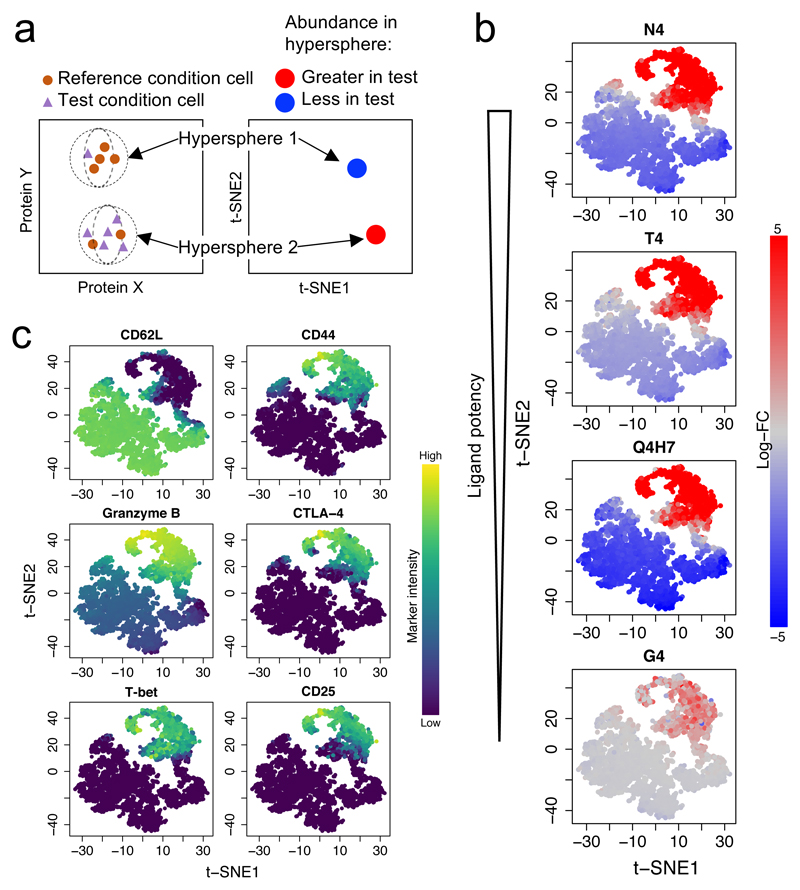
Proliferation represents only one component of effector differentiation, and expression of cytokines, cytotoxic mediators, and costimulatory or co-inhibitory receptors can control the phenotype of differentiated CTLs. In order to examine effector phenotypes without making prior assumptions about which proteins would be co-expressed, we used mass cytometry to simultaneously measure 19 surface and intracellular proteins related to T cell differentiation and effector function. After two days of stimulation with peptide-pulsed autologous T-depleted splenocytes, we tested for subpopulations of cells that changed in abundance39 under stimulation with any ovalbumin peptide variant compared to the null peptide NP68. The high-dimensional space defined by the 19 measured proteins was divided into phenotypic hyperspheres and the abundance of cells from each condition was quantified within each hypersphere (Fig. 6a). We identified phenotypic hyperspheres that differed significantly in abundance in any peptide stimulation condition (Fig. 6b). Examination of protein expression profiles that defined each phenotype revealed that, similar to its proliferation profile, the least potent ligand (G4) stimulated only a very small proportion of cells to transition from a CD44loCD62Lhi naive phenotype to a CD44hiCD62Llo effector phenotype (Fig. 6c, Supplementary Fig. 5). In contrast, stronger ligands (N4, T4, and Q4H7) drove increased abundance of effector populations co-expressing many effector-associated proteins including the high affinity IL-2 receptor subunit CD25, the cytotoxic mediator Granzyme B, the coinhibitory receptor CTLA-4, and the transcription factor T-bet that promotes effector CD8+ T cell differentiation. This effector phenotype was present in all activated populations, regardless of the potency of the primary stimulus, and was validated for a subset of proteins by flow cytometry (Supplementary Fig. 6a,b). Granzyme B expression levels in Granzyme B+ cells were marginally reduced with the highest potency stimulus (N4) (Supplementary Fig. 6c,d), as observed previously in vivo8, but in contrast to that previous study, we did not observe a decrease in the percentage of Granzyme B+ cells with strong stimulation (Supplementary Fig. 6b). Of note, IFN-γ was not expressed in all effector T cells after two days of stimulation, instead emerging in a small subset of cells (up to 20%) primarily under stimulation with ligands of medium/medium-low potency (T4 or Q4H7) (Supplementary Fig. 6e-g). Divergence of IFN-γ expression from other effector phenotypes was noted in an earlier study14. However, its infrequent expression in effector cells and disconnect from other effector protein phenotypes suggests that IFN-γ is not representative of cytolytic effector cell differentiation in this system.
Fig. 6. Effector T cells exhibit the same spectrum of phenotypes, regardless of primary stimulation strength.
a, A schematic of differential abundance testing for mass cytometry data using cydar software demonstrates how cells are assigned to hyperspheres and how the abundance of cells within each hypersphere is compared between conditions. b, OT-I CD8+ T cells from two separate mice were stimulated as in Fig. 5 for 2 days before profiling activation markers by mass cytometry. Analysis identified subpopulations of cells that changed in abundance with any stimulus. Plots depict a t-SNE visualization of the significantly differentially abundant hyperspheres (FDR < 0.05, 5138 of 5160 tested), colored by their log2 fold-change in abundance between NP68 (null peptide) stimulation and the indicated ovalbumin peptide variant (statistical test described in Methods). c, The plot from b is colored by marker intensity for selected proteins.
We verified effector protein expression phenotypes using wild-type splenocytes as APCs (Supplementary Fig. 4c,d). Under wild-type APC activation, G4 stimulation activated a small population of cells. These activated cells behaved similarly to those activated by other primary stimuli. Together, our phenotypic profiling supports a model in which ligand potency determines the rate with which cells embark on effector differentiation but not the spectrum of effector phenotypes they can achieve.
All differentiated effector cells are cytolytic
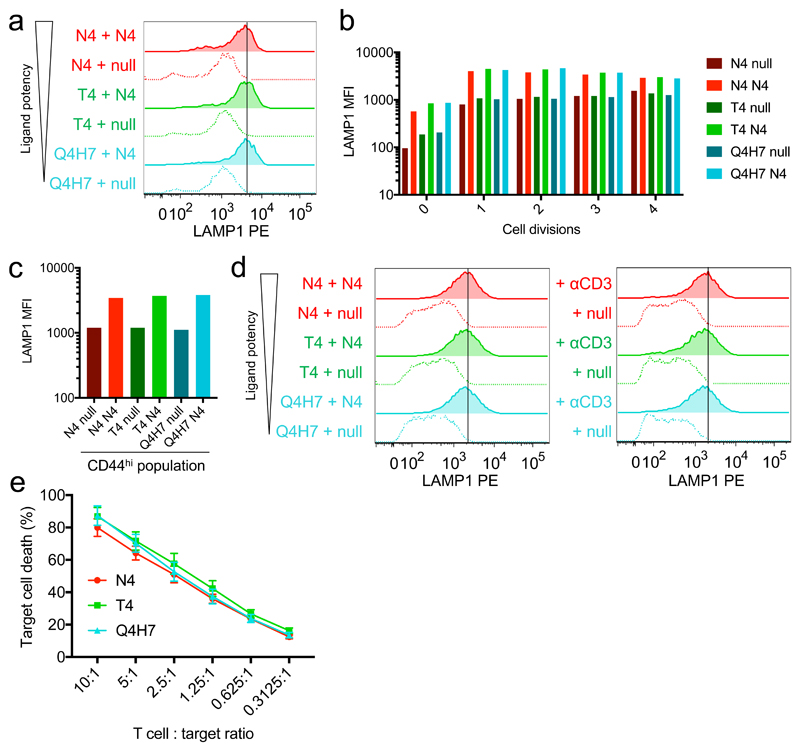
The main effector function of CTLs is the targeted release of cytolytic granules upon encountering an antigen-presenting target cell. Cytolytic granules carry the lysosome-associated transmembrane protein LAMP1, which enables measurement of degranulation in individual cells via the quantity of LAMP1 transiently trafficked to the cell surface during antigen challenge. To test how the TCR-pMHC affinity of a cell’s primary stimulus might affect its degranulation upon subsequent antigen encounter, we assayed degranulation of CD8+ T cells activated with ligands of different potencies in response to target cell challenge. We first looked at the ability of OTI T cells activated for 2 days with peptide-pulsed APCs to respond to a second stimulus. Co-culture with target cells presenting the cognate N4 peptide drove an increase in LAMP1 detected on the cell surface in all effector populations (Fig. 7a). Subsetting cells based on division number or CD44 expression revealed that cells in the same activation state exhibited similar degranulation profiles upon challenge, regardless of their primary stimulus (Fig. 7b,c). Again, use of wild-type APCs for primary stimulation yielded similar results (Supplementary Fig. 7a-c). After a week in culture, CTLs induced by ligands that drove substantial proliferation and effector differentiation (N4, T4, or Q4H7) still demonstrated strong degranulation capacity (Fig. 7d). Although we observed a slight dependence of degranulation on the potency of the primary stimulus (Supplementary Fig. 7d,e), this trend was absent in a target cell killing assay (Fig. 7e). It was previously demonstrated that only proliferating cells were competent in cytolytic activity14. Our results demonstrate that the converse of this relationship is true by showing that all tested ligands that induce proliferation can produce cytolytically competent CTLs.
Fig. 7. All activated CD8+ T cells can degranulate regardless of the affinity of their primary stimulus.
a, OTI CD8+ T cells activated for 2 days as in Fig. 5 and 6 were challenged with high potency ovalbumin peptide (N4)-pulsed EL4 cells for 3 hours. Degranulation was quantified by adding anti-LAMP1 PE to the cell culture medium to measure lysosomal trafficking by flow cytometry. b, Cells in a were gated based on their proliferation dye intensity to identify those that had divided 0, 1, 2, 3, or 4 times before comparing LAMP1 median fluorescence intensity (MFI). c, As in b, CD44hi cells were gated before comparing LAMP1 MFI. Bar plots (b-c) depict mean of two technical replicates in one representative experiment. Results (a-b) are representative of 6 separate mice from 5 independent experiments and (c) 5 separate mice from 4 independent experiments. d, T cells stimulated as in a were cultured for 7 days before a 3-hour challenge with ovalbumin peptide (N4)-pulsed EL4 cells (left) or plate-bound anti-CD3ε (right). Results are representative of 8 and 5 separate mice examined in 6 and 4 independent experiments, respectively. e, T cells activated as in a were cultured until day 8 and were tested for their ability to kill N4-pulsed EL4 cells by LDH release assay. Plot depicts combined data from 5 separate mice in 3 independent experiments; mean plotted with error bars representing SEM.
Discussion
Previous studies have led to conflicting models of graded and threshold activation responses to ligand potency, including T cell response speed, magnitude or proportion of responding cells5,11,13,14,17–22,30. We resolve these different interpretations with an unbiased approach revealing that ligand potency determines the rate with which cells initiate transcriptional activation. Support for our model can be found in studies over the last decade that have looked at specific parts of the T cell response: the time to start of proliferation but not proliferation speed was previously found to be associated with signal strength22; likewise, low potency Listeria monocytogenes infection resulted in curtailed expansion of antigen-specific T cells but complete secondary responses upon rechallenge9; and ligand potency was shown to determine the average delay in starting proliferation but not CD69 expression in activated cells19,21. We demonstrate that these previous results can all be attributed to signal strength-dependent modulation of the rate of response via shared activation machinery.
Several in vivo reports have claimed that primary stimulation strength can alter CD8+ T cell differentiation fate12,15,23. However, it has been difficult to tease apart the impact of ligand potency from that of antigen presentation, costimulatory context, and other cells responding in vivo. The duration of T cell-APC interactions influences the magnitude or phenotype of T cell responses in different contexts12,36,40,41. In addition, cells stimulated with reduced potency ligands take longer on average to accumulate phosphorylated signaling mediators downstream of the TCR19,42 and may require more sustained antigen presentation14. Furthermore, the length of time a T cell remains conjugated to an APC8,12,43, its motility10,38 and retention time in lymphoid organs in vivo8,9 are associated with ligand affinity, and affinity-dependent interaction time is also associated coinhibitory receptor expression43. Therefore, a T cell’s environment will change with ligand affinity, which will likely impact in vivo cell fate and not reflect the cell-intrinsic capacity to respond. Finally, autocrine and paracrine availability of the cytokine IL-2 modulates T cell activation and proliferation11,14,16,24,27,28,44 through PI3K and Myc activity18,26,33,45, and weak stimulation can result in insufficient IL-2 to support a proliferative response11,16,27,33,46. However, IL-2 secreted by activated CD4+ and CD8+ T cells would likely be expressed in the lymphoid tissue microenvironment in a physiological, polyclonal response, and cells stimulated by weak ligands can co-opt IL-2 from co-cultured cells stimulated by strong ligands26. Variations between experimental protocols pertaining to these factors have left the field unclear as to whether cell-intrinsic ligand strength-dependent T cell phenotypes exist. We demonstrate that in a controlled cellular environment, all activated cells can achieve an effector phenotype via shared transcriptional machinery.
Previous in vivo cytotoxicity studies have demonstrated reduced total killing by CTLs induced by low potency ligands8,9,12. However, two reports noted that, early after activation, low potency stimulation resulted in greater cytolytic capacity per cell compared to high potency stimulation8,9. Strongly stimulated T cells were instead retained in the periarteriolar lymphoid sheaths of the spleen9 or in the interfollicular regions of the lymph node in a CXCR3-dependent manner8, reducing their availability for killing. In our data, RNA expression of Cxcr3 exhibited a non-significant trend toward downregulation in G4-stimulated cells after accounting for activation status at 6 hours (Supplementary Table 2), and genes encoding other chemokines and receptors were upregulated with high potency stimulation, suggesting additional potential mechanisms. In contrast, in an ex vivo killing assay, CTLs generated in vivo with high or low potency ligands in the presence of exogenous IL2 exhibited comparable cytotoxicity28. In our controlled in vitro system, we find that all T cells that achieve an effector phenotype are cytolytically competent, suggesting that large divergences observed in vivo are not intrinsic to the T cell response to TCR stimulation.
Ligand potency can affect TCR-induced signaling events upstream of transcription including calcium fluxes47,48, TCR-coreceptor interaction42, post-translational modifications of signaling cascade components5,13,17,19,29,42, and transcription factor nuclear translocation13. Such signaling mechanisms may drive the altered rate of transcription initiation that we have identified49. It is interesting to consider what might cause specific cells to be the first to respond, particularly to low potency stimulation. CD8 and SHP-1 (encoded by Ptpn6) protein expression levels and markers of metabolic activity are associated with response propensity in mature CTL50 and naive T cells21, respectively. Unfortunately, due to the destructive nature of the single-cell sequencing technologies, cells cannot be comprehensively profiled both before and after activation. Hypothesis-driven sorting experiments may be fruitful in identifying additional markers of poised cells, but ideally a genome-wide screening experiment would be required to understand all contributing factors.
Our results emphasize the importance of using single-cell approaches to measure highly heterogeneous systems and identify rate-based responses46. These methods have allowed us to examine the coordination of mRNA and protein phenotypes in individual cells to answer a fundamental question in T cell biology: how does stimulation strength control the cell-intrinsic naive T cell response? We demonstrate that the primary effect of ligand affinity is to control the rate with which naive T cells initiate activation, not to lower effector gene expression within activated cells or drive the use of alternative transcriptional pathways. Additional environmental cues dependent on the nature of the antigen and other responding immune cell populations can adjust the ultimate phenotype of each clonal T cell response, but none of these characteristics is intrinsically determined by TCR signal strength. Indeed, the fact that heterogeneous, low efficiency T cell activation responses can achieve full effector phenotype and function via induction of the same genes that mediate synchronous responses to high potency ligands would seem fundamental to maintaining flexibility and sensitivity in the immune system. Using response rate to modulate clonal population size according to ligand strength makes use of common transcriptional machinery while allowing high affinity clones to become the most prevalent and suppressing low affinity clones with a near-zero activation rate. This model provides a mechanism by which peripheral CD8+ T cells can generate a full CTL response to a vast range of peptide antigens with a finite set of rearranged receptors and signaling components.
Methods
Mice
Mice were bred and housed in the University of Cambridge CBS facility. The genotype of OT-I Rag-deficient mice (OT-I Rag1tm1Bal on a C57BL/6 background) was confirmed prior to study. The wild-type C57BL/6N mouse line was obtained from the Wellcome Trust Sanger Institute Mouse Genetics Project (Sanger MGP).
Cell culture
The murine lymphoblast EL4 cell line, originally from the Sir William Dunn School of Pathology Cell Bank, Oxford, was maintained in DMEM (Gibco) supplemented with 10% fetal bovine serum (Biosera) and penicillin-streptomycin (Sigma) and tested negative for mycoplasma.
Dissected spleens from OT-I Rag1-deficient mice were homogenized through a 70 μm filter. CD8+ T cells were isolated using the CD8a+ T Cell Isolation Kit, mouse (Miltenyi). T cells were stimulated either with addition of purified peptide or with pulsed APCs. T-depleted OT-I splenocytes (the positive fraction from CD8a+ T cell separation) were used as APCs. Results were confirmed using wild-type splenocytes treated with RBC lysis buffer (eBioscience) as APCs. APCs were irradiated with ~3000 rad and pulsed for 2 hours with peptide before washing and co-culture with isolated T cells. OT-I APCs were cultured at a 5:1 ratio and wild-type APCs at a 2.5:1 ratio with T cells. Where indicated, T cells were stained with proliferation dye eFluor450 (eBioscience) according to the manufacturer’s instructions before culture. Cells were cultured in media composed of complete RPMI 1640 medium (Gibco), 10% fetal bovine serum (Biosera), penicillin-streptomycin (Sigma), sodium pyruvate (Gibco), L-glutamine (Sigma), β-mercaptoethanol (Gibco), and, unless indicated otherwise, 20 ng/mL (≥ 100 U/mL) murine IL-2 (Peprotech). The following peptides were used for stimulation at 1 μM unless otherwise indicated: SIINFEKL (N4), SIITFEKL (T4), SIIQFEHL (Q4H7), SIIGFEKL (G4), and ASNENMDAM (NP68) (Cambridge Bioscience). Although 1 μM is a higher concentration than required for maximal activation of OT-I cells with pure N4 peptide11,17,33, we sought to keep this variable constant across all ligands including G4 for which 1 μM was required to maximize induction of CD69 expression19. We observed no inhibitory effect of excess N4 peptide on early activation protein phenotypes (Supplementary Fig. 1a).
The rate of activation in response to the lowest potency ligand G4 differed between pure peptide and peptide-pulsed APC antigen presentation systems. Under pure peptide stimulation, G4 peptide was capable of activating transcription in approximately half of cells after 6 hours and by two days had activated proliferation in the whole population (Fig. 2,3 and Supplementary Fig. 4b). In contrast, G4-pulsed APCs induced negligible proliferation at two days (Fig. 5 and Supplementary Fig. 4c). These differences may be due to concentration and chronicity of antigen presentation, such that changing the antigen presenting system shifts the rate with which each ligand activates T cells but does not affect the ordering of these rates or the observation that all stimulation conditions capable of driving T cell activation use similar machinery. Our model in which ligand potency intrinsically determines relative activation rate is consistent with all of these observations.
Sorting for single-cell RNA-seq
Isolated naive OT-I T cells were stained with proliferation dye. 105 cells per well were stimulated with 1 μM peptide in 200 μL media in 96-well round-bottom plates. Cells were washed in PBS, blocked with FcR blocking antibody (clone 93, Biolegend) and stained with Zombie Aqua (Biolegend), anti-CD8a eVolve 655 (clone 53-6.7, eBioscience), anti-CD44 APC-eFluor780 or APC-FIRE750 (clone IM7, eBioscience or Biolegend, respectively), anti-CD62L PE (clone MEL-14, eBioscience), anti-CD25 Alexa Fluor 488 (clone PC61.5, eBioscience), and anti-CD69 APC (clone H1.2F3, eBioscience). Anti-CD154 PE-Cy7 (clone MR1, Biolegend) and anti-CD71 PerCP-Cy5.5 (clone R17217, Biolegend) were also included in one experiment, but expression levels were largely invariant and therefore not included in the analysis. Cells were sorted on a BD Influx (BD Biosciences), selecting live cells that expressed CD8 and that had not proliferated (Supplementary Fig. 8a). We did not see proliferation at the time points used for scRNA-seq. Expression of all surface proteins was recorded for each indexed cell. Two scRNA-seq experiments were performed: experiment 1 consisted of 2 96-well plates of cells stimulated with the various peptides for 6 hours; experiment 2 consisted of 4 96-well plates of cells stimulated with N4, T4, G4, or NP68 for 6 hours, with N4 for 3 hours, with N4 for 1 hour, or unstimulated. Within each experiment, cells from all conditions were included on each plate to avoid confounding of technical effects between plates. In each experiment, at least one well was left blank (without a cell) to monitor background levels of ambient RNA.
Single-cell RNA-seq
Sequencing library preparation was adapted from Picelli et al51 with the following modifications. Cells were sorted into 96-well plates holding 4 μL of lysis buffer composed of 2.3 U SUPERase In RNase inhibitor (Thermo Fisher Scientific), 0.11 % (v/v) Triton X-100 (Sigma), 12.5 mM DTT (Thermo Fisher Scientific), and 2.5 mM dNTP mix (Thermo Fisher Scientific). 1 μL annealing mix containing ERCC RNA Spike-In Mix (Thermo Fisher Scientific; final dilution 1 in 3 x 107) and 10 μM oligo-dT30VN (Sigma) was added to each well before reverse transcription with SuperScript II (Invitrogen). cDNA amplification was performed with 23 PCR cycles and the resulting PCR products purified with Ampure XP Beads (Agencourt) at a volume ratio of 0.7:1 beads:DNA. Libraries were prepared using the Nextera XT DNA Sample Preparation Kit and indexes from the Nextera XT Index Kit v2 Set A and Nextera XT Index Kit v2 Set D (Illumina). Each plate of libraries was pooled, cleaned with Ampure XP beads, and quantified using KAPA Library Quantification Kit (Roche). Equimolar quantities of libraries from each plate were combined for sequencing. Single-end 50bp sequencing was performed using a HiSeq 4000 (Illumina). Experiment 1 was sequenced in a single lane. Experiment 2 was sequenced on two lanes and the data merged.
Single-cell RNA-seq alignment and QC
Sequencing reads were aligned to the mm10 genome and ERCC transcripts using subread v 1.5.152 in RNA-seq mode with unique mapping. The number of reads mapping to exonic regions of each gene were counted using featureCounts53 (implemented in Rsubread v1.20.6) with a minimum quality score threshold of 10 and annotation from Ensembl GRCm38 version 82. Each sequencing run was aligned and counted separately. For cells sequenced twice, counts were summed.
Data were organized using the scran and scater Bioconductor packages54,55. Cells were filtered as follows for quality control: exclusion of samples where FACS had recorded multiple sorted cells; exclusion of cells with low numbers of mapped reads or genes detected (threshold defined as 3 MADs below the median log10-value); exclusion of cells with a high percentage of reads mapped to ERCC spike-in controls or mitochondrial genes (3 MADs above the median value); exclusion of cells with high or low ratio of ERCC spike-in controls to endogenous genes (3 MADs from the median difference of log2-values). All filters were applied on the whole data set and the union of excluded cells were removed. After filtering, experiment 1 retained 184 of 191 sorted cells, and experiment 2 retained 344 of 376 sorted cells. Blank wells were automatically excluded through the filtering steps.
Single-cell RNA-seq analysis
Within each sequencing run, genes were filtered for those detected in at least 5% of cells and those with a mean count of at least 1. Data was normalized using size factors calculated from ERCC spike-in controls to account for variation in total mRNA56. Log2-transformation was performed on the normalized expression values, after adding a pseudocount of 1. Batch effects between 96-well plates were removed by applying ComBat57 in the sva Bioconductor package to the log-expression values, including stimulation condition as a preserved covariate. For analyses of the combined data, genes were required to meet filtering criteria in both sequencing data sets before following the same normalization and batch correction procedure on the combined data.
Non-branching diffusion pseudotime analysis was performed using a modified version of the dpt package31 to create a diffusion map of the cells and find the most probable activation trajectory. Analysis was performed using the first 50 scaled principal components and a Euclidean distance metric. The starting point of the fit trajectory, at pseudotime 0, is known as the root cell. We required that the root cell be from the unstimulated condition. In the situation in which a root cell was not automatically identified from this condition, pseudotime was fit to only the subset of cells from the most and least stimulated conditions to find a root cell, and this cell was subsequently used as root in the full analysis. Clustering along pseudotime was performed using the classInt package for 1-dimensional class interval selection using the “jenks” method. To identify the most biologically variable genes among unstimulated cells, we first used the trendVar function of the scran Bioconductor package54 to fit a parametric curve followed by loess smoothing (span = 0.95) to the variance versus the mean log-expression of the spike-in transcripts. Biological variances of endogenous genes were then obtained by decomposing the variance using the decomposeVar function. Heatmaps were generated using the pheatmap R package.
Differential expression analyses were performed by GLM in edgeR58,59 with a likelihood ratio test, using effective library sizes derived from the spike-in size factors and blocking on the plate of origin for each cell. We performed differential expression analysis between stimuli of varying affinities while controlling for activation status by additionally blocking on a spline basis matrix (4 degrees of freedom) for log10 CD69 protein expression within each sequencing run to allow for non-linear trends in protein expression.
GO category enrichment of differentially expressed genes (FDR < 0.05) was performed using the goana function from the limma Bioconductor package. The significance threshold for enrichment was set at FDR < 0.05 (Benjamini Hochberg procedure). Transcriptional regulatory gene enrichment was calculated by hypergeometric test of enrichment for transcription factor genes (annotated in TcoF-DB v260) within genes significantly upregulated (log-FC > 0, FDR < 0.05) in the early activation cluster compared with resting and late activation clusters. The gene universe for this analysis consisted of all genes tested for differential expression.
RNA flow cytometry
Control genes for RNA flow cytometry were determined from scRNA-seq data. We began with the sequencing run containing both the high potency ligand (N4) time course and reduced potency ligand stimulations for 6 hours (experiment 2). Genes were filtered for those detected in at least 80% of cells in each condition. Gene variances were calculated from the spike-in-normalized, log-transformed reads for each plate separately and averaged. We then performed an ANOVA test on the expression of each gene with respect to condition using the full spike-in-normalized, log-transformed, batch-corrected data set. Genes were filtered based on an average variance less than 2 and an ANOVA p value greater than 0.01. The top non-pseudogenes in this list were Slc34a2 and Rpl39. Examining these genes in scRNAseq experiment 1 (consisting of only 6-hour stimulated samples) revealed that Slc34a2 maintained a lack of bias by stimulation condition, and Rpl39 expression was slighlty different between conditions, significant by ANOVA without multiple testing correction. Both genes remained strongly expressed across cells. A PrimeFlow expression assay (Thermo Fisher Scientific) with detectable signal was not available for Slc34a2, so Rpl39 was used as a control gene to check that cells were permeabilized and probes hybridized and amplified.
Isolated naive OT-I T cells were stimulated with 1 μM peptide for 0-6 hours in complete media as in the scRNA-seq experiment. After stimulation, cells were immediately moved to ice before RNA flow cytometry was performed using the PrimeFlow system (Thermo Fisher Scientific). Cell surfaces were stained with Zombie Aqua Fixable Viability Kit (Biolegend) before fixation and permeabilization. Cells were stained with Nr4a1 AF568, Fosb AF647, and Rpl39 AF488. Data were acquired on a BD LSRFortessa and analyzed in FlowJo (Supplementary Fig. 8b).
Mass cytometry
Cells from two mice were stimulated separately with irradiated autologous T-depleted splenocytes pulsed with 1 μM N4, T4, Q4H7, G4 or NP68 peptide. For each condition, two million isolated naive OT-I T cells were mixed with 10 million peptide-pulsed APCs in 4 mL media in 6-well plates. Antibodies for mass cytometry are listed in Supplementary Table 4. 155Gd-anti-CD44 (clone IM7) and 160Gd-anti-PE (clone PE001) were conjugated from Biolegend MaxPar Ready antibodies using the Maxpar Antibody Labeling Kit (Fluidigm) according to the manufacturer’s instructions with 83 and 74 μg antibody, respectively. All antibodies were directly conjugated to metals except for CD62L-APC, CD11a-FITC, and LAMP1-PE, which were then probed with metal-conjugated anti-APC, anti-FITC, and anti-PE antibodies, respectively. Staining for mass cytometry was performed using sequential MaxPar reagent kits (Fluidigm) in the following steps. Live cells were stained with 0.25 μM Cell ID Cisplatin for 5 minutes at room temperature. Three million cells from each sample were barcoded using the Cell-ID 20-Plex Pd Barcoding Kit and pooled for staining. For surface staining, cells were blocked with 1 μg FcR blocking antibody (clone 93, Biolegend) per million cells before staining with primary followed by secondary antibodies, all in MaxPar Cell Staining Buffer. For intracellular staining, cells were fixed and permeabilized with the Maxpar Nuclear Antigen Staining Buffer Set. Cells were again incubated with FcR blocking reagent before staining with primary and secondary intracellular antibodies, all in Nuclear Antigen Staining Perm. After washing with Nuclear Antigen Staining Perm and Cell Staining Buffer, cells were stained overnight in 125 nM Cell-ID Intercalator-Ir in Maxpar Fix and Perm Buffer before analyzing on a Helios CyTOF system (Fluidigm) using Fluidigm CyTOF Software to collect and process the data. We note that intracellular IFN-γ measurements were made without addition of a Golgi-interrupting compound to avoid effects on other intracellular components measured.
Mass cytometry data analysis
Mass cytometry data was analyzed using the cydar Bioconductor package39. Cells were pooled across samples and mass-labelled marker values were transformed using the Logicle transformation61. We imposed the following filters sequentially: removal of cells acquired during a clog in the Helios machine, removal of normalization beads, inclusion of singlet cells labelled with Ir191 and Ir193 DNA markers, removal of dead cells labelled with high levels of Pt195, removal of cells expressing extremely low levels of the hematopoietic marker CD45 (5 MAD filter), removal of non-T cells marked by extremely low expression of TCRb and CD8a (6 MAD and 5 MAD filters, respectively, because surface TCRb and CD8a expression can drop dramatically with internalization upon activation). Cells were assigned to hyperspheres with a radius defined from a per-marker log-intensity tolerance of 0.5, and differential abundance was analyzed using the edgeR Bioconductor package with a quasi-likelihood GLM fit62 including the mouse of origin as a blocking factor for each sample. Significant hyperspheres were identified by analysis of deviance to detect those with differential abundance in any condition, controlling the spatial FDR at 5%. A t-SNE plot was generated using the Rtsne package63 with a perplexity value of 50.
Flow cytometry
FC receptors were blocked with anti-mouse CD16/32 (clone 93, Biolegend) before staining. Dead cells were identified using the Zombie Aqua Fixable Viability Kit (Biolegend). Cells were stained with the following antibody clones: anti-mouse CD8a (clone 53-6.7, Biolegend), anti-mouse/human CD44 (clone IM7, Biolegend), anti-mouse CD62L (clone MEL-14, eBioscience), anti-mouse/human Granzyme B (clone GB11, Biolegend), anti-mouse CTLA4 (clone UC1-4B9, Biolegend), anti-mouse CD25 (clone PC61.5, eBioscience), and anti-mouse IFN-γ (clone XMG1.2, Biolegend). Intracellular staining of Granzyme B, CTLA-4, and IFN-γ was performed using the Foxp3/Transcription Factor Staining Buffer Set (eBioscience). To count cells, 123count eBeads (eBioscience) were added to flow cytometry tubes immediately before flow cytometer acquisition. Data was acquired on a BD LSRFortessa and analyzed in FlowJo. Cells were gated for size, single cells, living cells, and CD8+ cells before examination of proliferation curves (Supplementary Fig. 8c). Cells were further gated on proliferation dye+ cells to exclude any CD8+ T cells in the APC fraction before quantification of division numbers and examination of surface and intracellular proteins. Statistical analyses of results from separate mice were performed using GraphPad Prism software.
Degranulation assay
Activated T cells were assayed for degranulation upon challenge with ovalbumin (N4)-pulsed EL4 cells or antibody-based TCR stimulation. Cells were stained with proliferation dye eFluor 450 (eBioscience) before stimulation to measure this parameter in degranulating cells. For cellular challenge, EL4 cells were pulsed with 1 μM of the highest potency N4 peptide for 1 hour and washed. Un-pulsed EL4 cells were used as a control. T cell stimulation cultures were mixed 1:1 with EL4 cells in media supplemented with anti-mouse LAMP1 PE (clone eBio1D4B, eBioscience, 2 μg/mL). For antibody-based challenge, CTLs were cultured on plates coated with 1 μM anti-CD3ε (clone 145-2C11, BD Biosciences) for 1 hour at 37 °C. After 3 hours, cells were stained on ice. FC receptors were blocked with anti-mouse CD16/32 (clone 93, Biolegend) and cells were stained with the Zombie Aqua Fixable Viability Kit (Biolegend) and antibodies to CD8a (53-6.7, Biolegend) and CD44 (IM7, Biolegend). Data were acquired immediately on a BD LSRFortessa and analyzed in FlowJo. Cells were gated for size, single cells, living cells, and CD8+ cells before examination of LAMP1 surface trafficking (Supplementary Fig. 8d,e). For samples activated for 2 days before degranulation testing, cells were also gated on proliferation dye+ cells to exclude any T cells in the residual irradiated APC population where necessary. Where possible, samples were run in duplicate and measurements averaged. Statistical analyses of results from separate mice were performed using GraphPad Prism software.
Killing assay
After 8 days of activation, activated T cells were assayed for their ability to kill ovalbumin (N4)-pulsed EL4 cells. Cells were plated at T:EL4 cell ratios ranging from 10:1 to 0.3125:1 in round-bottom 96-well plates in RPMI without phenol red (Gibco), supplemented with 2% fetal bovine serum. EL4 cells without ovalbumin pulse were used as a control. Maximum death was estimated by adding lysis buffer to the same number of EL4 cells. All conditions were performed in duplicate. Cells were co-cultured for 3 hours and EL4 cell death assessed by lactate dehydrogenase (LDH) release using the CytoTox 96 Non-Radioactive Cytotoxicity Assay (Promega). Absorbances at 490nm were read on a VersaMax microplate reader (Molecular Devices) using SoftmaxPro 5.4.1 software. Changes in absorbance between wells containing pulsed and un-pulsed EL4 wells were calculated and compared to maximum EL4 death. Statistical analyses of results from separate mice were performed using GraphPad Prism software.
Supplementary Material
A differential expression analysis was performed on scRNA-seq data between cells in the early activation cluster and the resting naive T cell cluster, and between cells in the late activation cluster and the resting cluster from Fig. 1. Differentially expressed genes were tested for Gene Ontology (GO) category enrichment. Enriched categories (FDR < 0.05) are reported.
Results from the sc-RNAseq differential expression analysis between cells stimulated with peptides of different potencies (n = 155 cells for N4, 91 for T4, 94 for G4), controlling for activation status by CD69 protein expression depicted in Fig. 4.
A differential expression analysis was performed on scRNA-seq data between peptides of different potencies, controlling for CD69 expression as described in Fig. 4 and Supplementary Table 2. Differentially expressed genes were tested for Gene Ontology (GO) category enrichment. Enriched categories (FDR < 0.05) are reported.
Antibodies used for mass cytometry experiment.
Acknowledgements
This work was funded by an MRC Skills Development Fellowship to ACR (MR/P014178/1); the Wellcome Trust, grants [103930] and [100140] to GMG; Cancer Research UK, core funding to JCM [A17197]; EMBL, core funding to JCM; the University of Cambridge; and Hutchison Whampoa Limited. Single cell collection and analysis was supported through MRC Clinical Research Infrastructure funds for the Cambridge Single Cell Facility (MR/M008975/1). WWYL and BG were supported by Bloodwise (12029) and Cancer Research UK (C1163/A12765 and C1163/A21762). This research was supported by the CIMR Flow Cytometry Core Facility. In particular, we wish to thank R. Schulte and C. Cossetti for their advice and support in cell sorting. We would also like to thank the CRUK-CI Flow Cytometry core, particularly M. Strzelecki and R. Grenfell, and Genomics core for their resources and assistance. We thank the Wellcome Trust Sanger Institute Mouse Genetics Project (Sanger MGP) and its funders for providing the wild-type C57BL/6 mouse line; funding information may be found at www.sanger.ac.uk/mouseportal. We thank C. Gawden-Bone, J. Warland, A. Denton and G. Frazer for critical reading of the manuscript.
Footnotes
Ethics statement
This research has been regulated under the Animals (Scientific Procedures) Act 1986 Amendment Regulations 2012 following ethical review by the University of Cambridge Animal Welfare and Ethical Review Body (AWERB).
Data availability statement
scRNA-seq data are available in the ArrayExpress repository, accession number E-MTAB-6051. Mass cytometry data are available in the Cytobank repository, accession number 66456. The remaining data that support the findings of this study are available from the corresponding authors upon request.
Code availability statement
Analysis code for scRNA-seq and mass cytometry data is available at https://github.com/MarioniLab/SingleCellAPL2018.
Reporting summary
Additional information can be found in the Life Sciences Reporting Summary included with this article.
Author contributions
ACR, JCM and GMG designed the study and wrote the manuscript. ACR carried out the experiments and analyses under the supervision of JCM and GMG. ATLL designed analytical pipelines and software and advised analyses. WWYL and BG advised and supervised, respectively, cell sorting and library preparation for scRNA-seq, which were optimized by WWYL. All authors edited and approved the final manuscript.
Competing interests:
The authors declare no competing interests.
References
- 1.Brownlie RJ, Zamoyska R. T cell receptor signalling networks: branched, diversified and bounded. Nat Rev Immunol. 2013;13:257–269. doi: 10.1038/nri3403. [DOI] [PubMed] [Google Scholar]
- 2.Cantrell D. Signaling in lymphocyte activation. Cold Spring Harb Perspect Biol. 2015;7 doi: 10.1101/cshperspect.a018788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conley JM, Gallagher MP, Berg LJ. T Cells and Gene Regulation: The Switching On and Turning Up of Genes after T Cell Receptor Stimulation in CD8 T Cells. Front Immunol. 2016;7:76. doi: 10.3389/fimmu.2016.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zikherman J, Au-Yeung B. The role of T cell receptor signaling thresholds in guiding T cell fate decisions. Curr Opin Immunol. 2015;33:43–48. doi: 10.1016/j.coi.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daniels MA, et al. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature. 2006;444:724–729. doi: 10.1038/nature05269. [DOI] [PubMed] [Google Scholar]
- 6.Hogquist KA, et al. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 7.Fu G, et al. Themis sets the signal threshold for positive and negative selection in T-cell development. Nature. 2013;504:441–445. doi: 10.1038/nature12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ozga AJ, et al. pMHC affinity controls duration of CD8+ T cell-DC interactions and imprints timing of effector differentiation versus expansion. J Exp Med. 2016;213:2811–2829. doi: 10.1084/jem.20160206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zehn D, Lee SY, Bevan MJ. Complete but curtailed T-cell response to very low-affinity antigen. Nature. 2009;458:211–214. doi: 10.1038/nature07657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skokos D, et al. Peptide-MHC potency governs dynamic interactions between T cells and dendritic cells in lymph nodes. Nat Immunol. 2007;8:835–844. doi: 10.1038/ni1490. [DOI] [PubMed] [Google Scholar]
- 11.Denton AE, et al. Affinity thresholds for naive CD8+ CTL activation by peptides and engineered influenza A viruses. J Immunol. 2011;187:5733–5744. doi: 10.4049/jimmunol.1003937. [DOI] [PubMed] [Google Scholar]
- 12.King CG, et al. T cell affinity regulates asymmetric division, effector cell differentiation, and tissue pathology. Immunity. 2012;37:709–720. doi: 10.1016/j.immuni.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmer E, Drobek A, Stepanek O. Opposing effects of actin signaling and LFA-1 on establishing the affinity threshold for inducing effector T-cell responses in mice. Eur J Immunol. 2016;46:1887–1901. doi: 10.1002/eji.201545909. [DOI] [PubMed] [Google Scholar]
- 14.Auphan-Anezin N, Verdeil G, Schmitt-Verhulst AM. Distinct thresholds for CD8 T cell activation lead to functional heterogeneity: CD8 T cell priming can occur independently of cell division. J Immunol. 2003;170:2442–2448. doi: 10.4049/jimmunol.170.5.2442. [DOI] [PubMed] [Google Scholar]
- 15.Man K, et al. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat Immunol. 2013;14:1155–1165. doi: 10.1038/ni.2710. [DOI] [PubMed] [Google Scholar]
- 16.Marchingo JM, et al. T cell signaling. Antigen affinity, costimulation, and cytokine inputs sum linearly to amplify T cell expansion. Science. 2014;346:1123–1127. doi: 10.1126/science.1260044. [DOI] [PubMed] [Google Scholar]
- 17.Navarro MN, Feijoo-Carnero C, Arandilla AG, Trost M, Cantrell DA. Protein kinase D2 is a digital amplifier of T cell receptor-stimulated diacylglycerol signaling in naive CD8(+) T cells. Sci Signal. 2014;7:ra99. doi: 10.1126/scisignal.2005477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Preston GC, et al. Single cell tuning of Myc expression by antigen receptor signal strength and interleukin-2 in T lymphocytes. EMBO J. 2015;34:2008–2024. doi: 10.15252/embj.201490252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosette C, et al. The impact of duration versus extent of TCR occupancy on T cell activation: a revision of the kinetic proofreading model. Immunity. 2001;15:59–70. doi: 10.1016/s1074-7613(01)00173-x. [DOI] [PubMed] [Google Scholar]
- 20.Yao S, et al. Interferon regulatory factor 4 sustains CD8(+) T cell expansion and effector differentiation. Immunity. 2013;39:833–845. doi: 10.1016/j.immuni.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balyan R, et al. Modulation of Naive CD8 T Cell Response Features by Ligand Density, Affinity, and Continued Signaling via Internalized TCRs. J Immunol. 2017;198:1823–1837. doi: 10.4049/jimmunol.1600083. [DOI] [PubMed] [Google Scholar]
- 22.Hommel M, Hodgkin PD. TCR affinity promotes CD8+ T cell expansion by regulating survival. J Immunol. 2007;179:2250–2260. doi: 10.4049/jimmunol.179.4.2250. [DOI] [PubMed] [Google Scholar]
- 23.Nayar R, et al. Graded levels of IRF4 regulate CD8+ T cell differentiation and expansion, but not attrition, in response to acute virus infection. J Immunol. 2014;192:5881–5893. doi: 10.4049/jimmunol.1303187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchingo JM, et al. T-cell stimuli independently sum to regulate an inherited clonal division fate. Nat Commun. 2016;7:13540. doi: 10.1038/ncomms13540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Gisbergen KP, et al. The costimulatory molecule CD27 maintains clonally diverse CD8(+) T cell responses of low antigen affinity to protect against viral variants. Immunity. 2011;35:97–108. doi: 10.1016/j.immuni.2011.04.020. [DOI] [PubMed] [Google Scholar]
- 26.Voisinne G, et al. T Cells Integrate Local and Global Cues to Discriminate between Structurally Similar Antigens. Cell Rep. 2015;11:1208–1219. doi: 10.1016/j.celrep.2015.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Au-Yeung BB, et al. IL-2 Modulates the TCR Signaling Threshold for CD8 but Not CD4 T Cell Proliferation on a Single-Cell Level. J Immunol. 2017;198:2445–2456. doi: 10.4049/jimmunol.1601453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verdeil G, Puthier D, Nguyen C, Schmitt-Verhulst AM, Auphan-Anezin N. STAT5-mediated signals sustain a TCR-initiated gene expression program toward differentiation of CD8 T cell effectors. J Immunol. 2006;176:4834–4842. doi: 10.4049/jimmunol.176.8.4834. [DOI] [PubMed] [Google Scholar]
- 29.Altan-Bonnet G, Germain RN. Modeling T cell antigen discrimination based on feedback control of digital ERK responses. PLoS Biol. 2005;3:e356. doi: 10.1371/journal.pbio.0030356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allison KA, et al. Affinity and dose of TCR engagement yield proportional enhancer and gene activity in CD4+ T cells. Elife. 2016;5 doi: 10.7554/eLife.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haghverdi L, Buttner M, Wolf FA, Buettner F, Theis FJ. Diffusion pseudotime robustly reconstructs lineage branching. Nat Methods. 2016;13:845–848. doi: 10.1038/nmeth.3971. [DOI] [PubMed] [Google Scholar]
- 32.Pollizzi KN, Powell JD. Integrating canonical and metabolic signalling programmes in the regulation of T cell responses. Nat Rev Immunol. 2014;14:435–446. doi: 10.1038/nri3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan TCJ, et al. Suboptimal T-cell receptor signaling compromises protein translation, ribosome biogenesis, and proliferation of mouse CD8 T cells. Proc Natl Acad Sci U S A. 2017;114:E6117–E6126. doi: 10.1073/pnas.1700939114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moran AE, et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashouri JF, Weiss A. Endogenous Nur77 Is a Specific Indicator of Antigen Receptor Signaling in Human T and B Cells. J Immunol. 2017;198:657–668. doi: 10.4049/jimmunol.1601301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Au-Yeung BB, et al. A sharp T-cell antigen receptor signaling threshold for T-cell proliferation. Proc Natl Acad Sci U S A. 2014;111:E3679–3688. doi: 10.1073/pnas.1413726111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alam SM, et al. Qualitative and quantitative differences in T cell receptor binding of agonist and antagonist ligands. Immunity. 1999;10:227–237. doi: 10.1016/s1074-7613(00)80023-0. [DOI] [PubMed] [Google Scholar]
- 38.Moreau HD, et al. Dynamic in situ cytometry uncovers T cell receptor signaling during immunological synapses and kinapses in vivo. Immunity. 2012;37:351–363. doi: 10.1016/j.immuni.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 39.Lun ATL, Richard AC, Marioni JC. Testing for differential abundance in mass cytometry data. Nat Methods. 2017 doi: 10.1038/nmeth.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prlic M, Hernandez-Hoyos G, Bevan MJ. Duration of the initial TCR stimulus controls the magnitude but not functionality of the CD8+ T cell response. J Exp Med. 2006;203:2135–2143. doi: 10.1084/jem.20060928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Stipdonk MJ, et al. Dynamic programming of CD8+ T lymphocyte responses. Nat Immunol. 2003;4:361–365. doi: 10.1038/ni912. [DOI] [PubMed] [Google Scholar]
- 42.Yachi PP, Ampudia J, Zal T, Gascoigne NR. Altered peptide ligands induce delayed CD8-T cell receptor interaction--a role for CD8 in distinguishing antigen quality. Immunity. 2006;25:203–211. doi: 10.1016/j.immuni.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 43.Zahm CD, Colluru VT, McNeel DG. Vaccination with High-Affinity Epitopes Impairs Antitumor Efficacy by Increasing PD-1 Expression on CD8+ T Cells. Cancer Immunol Res. 2017;5:630–641. doi: 10.1158/2326-6066.CIR-16-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pipkin ME, et al. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 2010;32:79–90. doi: 10.1016/j.immuni.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heinzel S, et al. A Myc-dependent division timer complements a cell-death timer to regulate T cell and B cell responses. Nat Immunol. 2017;18:96–103. doi: 10.1038/ni.3598. [DOI] [PubMed] [Google Scholar]
- 46.Tkach KE, et al. T cells translate individual, quantal activation into collective, analog cytokine responses via time-integrated feedbacks. Elife. 2014;3:e01944. doi: 10.7554/eLife.01944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen JL, et al. Ca2+ release from the endoplasmic reticulum of NY-ESO-1-specific T cells is modulated by the affinity of TCR and by the use of the CD8 coreceptor. J Immunol. 2010;184:1829–1839. doi: 10.4049/jimmunol.0902103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le Borgne M, et al. Real-Time Analysis of Calcium Signals during the Early Phase of T Cell Activation Using a Genetically Encoded Calcium Biosensor. J Immunol. 2016;196:1471–1479. doi: 10.4049/jimmunol.1502414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mayya V, Dustin ML. What Scales the T Cell Response? Trends Immunol. 2016;37:513–522. doi: 10.1016/j.it.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 50.Feinerman O, Veiga J, Dorfman JR, Germain RN, Altan-Bonnet G. Variability and robustness in T cell activation from regulated heterogeneity in protein levels. Science. 2008;321:1081–1084. doi: 10.1126/science.1158013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 52.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 54.Lun AT, McCarthy DJ, Marioni JC. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 2016;5:2122. doi: 10.12688/f1000research.9501.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCarthy DJ, Campbell KR, Lun AT, Wills QF. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017;33:1179–1186. doi: 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lun ATL, Calero-Nieto FJ, Haim-Vilmovsky L, Gottgens B, Marioni JC. Assessing the reliability of spike-in normalization for analyses of single-cell RNA sequencing data. Genome Res. 2017;27:1795–1806. doi: 10.1101/gr.222877.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 58.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmeier S, Alam T, Essack M, Bajic VB. TcoF-DB v2: update of the database of human and mouse transcription co-factors and transcription factor interactions. Nucleic Acids Res. 2017;45:D145–D150. doi: 10.1093/nar/gkw1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parks DR, Roederer M, Moore WA. A new "Logicle" display method avoids deceptive effects of logarithmic scaling for low signals and compensated data. Cytometry A. 2006;69:541–551. doi: 10.1002/cyto.a.20258. [DOI] [PubMed] [Google Scholar]
- 62.Lun AT, Chen Y, Smyth GK. It's DE-licious: A Recipe for Differential Expression Analyses of RNA-seq Experiments Using Quasi-Likelihood Methods in edgeR. Methods Mol Biol. 2016;1418:391–416. doi: 10.1007/978-1-4939-3578-9_19. [DOI] [PubMed] [Google Scholar]
- 63.van der Maaten LJP. Accelerating t-SNE using tree-based algorithms. J Mach Learn Res. 2014;15:3221–3245. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A differential expression analysis was performed on scRNA-seq data between cells in the early activation cluster and the resting naive T cell cluster, and between cells in the late activation cluster and the resting cluster from Fig. 1. Differentially expressed genes were tested for Gene Ontology (GO) category enrichment. Enriched categories (FDR < 0.05) are reported.
Results from the sc-RNAseq differential expression analysis between cells stimulated with peptides of different potencies (n = 155 cells for N4, 91 for T4, 94 for G4), controlling for activation status by CD69 protein expression depicted in Fig. 4.
A differential expression analysis was performed on scRNA-seq data between peptides of different potencies, controlling for CD69 expression as described in Fig. 4 and Supplementary Table 2. Differentially expressed genes were tested for Gene Ontology (GO) category enrichment. Enriched categories (FDR < 0.05) are reported.
Antibodies used for mass cytometry experiment.