Abstract
Cutaneous mast cell tumours are one of the most common canine cancers. Approximately 25% of the tumours metastasise. Activating c-kit mutations are present in about 20% of tumours, but metastases occur in the absence of mutations. Tumour metastasis is associated with significantly diminished survival in spite of adjuvant chemotherapy. Available prognostic tests do not reliably predict whether a tumour will metastasise. In this study we compared the global expression profiles of 20 primary cutaneous mast cell tumours that metastasised with those of 20 primary tumours that did not metastasise. The objective was to identify genes associated with mast cell tumour metastatic progression that may represent targets for therapeutic intervention and biomarkers for prediction of tumour metastasis. Canine Gene 1.1 ST Arrays were employed for genome-wide expression analysis of formalin-fixed, paraffin-embedded biopsies of mast cell tumours borne by dogs that either died due to confirmed mast cell tumour metastasis, or were still alive more than 1000 days post-surgery. Decreased gene expression in the metastasising tumours appears to be associated with a loss of cell polarity, reduced cell-cell and cell-ECM adhesion, and increased cell deformability and motility. Dysregulated gene expression may also promote extracellular matrix and base membrane degradation, suppression of cell cycle arrest and apoptosis, and angiogenesis. Down-regulation of gene expression in the metastasising tumours may be achieved at least in part by small nucleolar RNA-derived RNA and microRNA-effected gene silencing. Employing cross-validation, a linear discriminant analysis-based classifier featuring 19 genes that displayed two-fold differences in expression between metastasising and non-metastasising tumours was estimated to classify metastasising and non-metastasising tumours with accuracies of 90–100% and 70–100%, respectively. The differential expression of 9 of the discriminator genes was confirmed by quantitative reverse transcription-PCR.
Introduction
Canine mast cell tumours (MCTs) are neoplastic proliferations which predominantly arise from tissue mast cells in the dermal layer of the skin [1]. MCTs are the most common canine skin tumour [2] with an estimated incidence of 129 per 100,000 dogs. Although most breeds are affected, several breed predispositions have been reported [3]. The majority of MCTs are successfully treated by surgery and/or radiotherapy, but approximately 25% of tumours spread to a regional lymph node, spleen and/or liver, and with local therapy alone death usually follows within 1 year of diagnosis. Dogs that have metastatic disease, or are believed to have a high risk of developing metastatic disease, are often treated with adjunctive chemotherapy. An overall response rate of 47%, with a median response duration of 154 days, has been reported for treatment of measureable MCTs with an often used vinblastine and prednisolone protocol [4], and a median survival time of 1374 days was achieved when an equivalent protocol was utilised post-surgery [5]. However, the side-effects of chemotherapy may include myelosuppression, neutropenia and gastrointestinal disorders.
MCTs are usually classified by histological grade, which is the most important single prognostic factor [1]. Currently, two grading systems are used. The Patnaik grading system is well established and assigns a MCT to one of 3 grades (I, II and III) according to descriptive histological criteria [6]. However, Patnaik grade does not predict metastasis; <10% of grade I, 5–22% of grade II, and >80% of grade III MCTs metastasise [7]. The more recent binary Kiupel histologic grading system [8] utilises more numerical and fewer descriptive criteria and was devised to reduce the discord observed between pathologists applying the Patnaik system. Whilst Kiupel grade is associated with survival time, it also cannot accurately predict metastasis; 37.5% of tumours classified as ‘low grade’ were borne by dogs with distant metastases, whilst 21.9% of MCTs designated as ‘high grade’ were from dogs without distant metastases [9].
Decreased MCT patient survival has been associated with ‘high’ mitotic index [10], increases in the number of Ki-67 positive nuclei [11], argyrophylic nucleolar organizer regions [11] and minichromosome maintenance protein 7 positive cells [12], respectively, and decreased expression of the cell adhesion molecule TSLC1 [13] in cutaneous MCTs. However, although significant differences between the proliferating cell nuclear antigen and argyrophilic nucleolar organizing region counts for metastasising and non-metastasising MCTs were described in a single study [14], neither cell proliferation index has subsequently been shown to be capable of predicting canine cutaneous MCT metastasis [15].
Metastasis is a complex process, each step of which is thought to rely at least in part on cells acquiring specific genetic and/or epigenetic alterations [16] additional to those that drive primary tumour development. The alterations may effect changes in gene expression, and metastasis-associated gene expression signatures have been identified for a number of human tumours [17]. Metastasis-associated gene expression signatures may predict metastasis [18], and several form the basis of routine prognostic tests; e.g. the DecisionDx-UM test [19]. Differential gene expression analysis has also identified biological processes involved in metastasis [20] and candidate metastasis-suppressor [21] and promoting genes [22]. Gene expression analysis of canine cutaneous MCTs was recently performed [23] with the intention of evaluating whether biological behaviour could be predicted on the basis of tumour gene expression profile. However, differential expression analysis compared ‘differentiated’ and ‘undifferentiated’ tumours designated solely on the basis of Kiupel grade [8], with no reference to the presence or absence (or later development) of metastasis.
Clinical management of canine cutaneous MCT would be greatly assisted by the capability to predict tumour metastasis, whilst targeted prevention of metastasis would ultimately represent the most effective life-saving strategy. Elucidation of the molecular genetic contributions to canine cutaneous MCT metastatic progression affords a means of identifying biomarkers of metastasis and potential targets for therapeutic intervention.
In the current study we compared the global gene expression profiles of formalin-fixed, paraffin-embedded biopsies (FFPE) of primary cutaneous MCTs that did and did not metastasise. The aim was to identify genes that are associated with the metastatic progression of cutaneous MCTs, and evaluate the potential for differentiating metastasising and non-metastasising MCTs on the basis of a metastasis-associated gene expression signature.
Materials and methods
Ethics statements
This study was approved by the Animal Health Trust and the University of Liverpool ethics committees, respectively. Informed, written consent was obtained from the owner of each dog whose MCT biopsy was included in this study. A MCT biopsy could be withdrawn from the study at any time. Patient treatment was unaffected by the study.
Tumour samples
Diagnostic histopathology FFPE biopsies of canine primary cutaneous MCTs were collected from dogs treated in the Clinical Oncology departments at the Animal Health Trust Centre for Small Animal Studies and the University of Liverpool Small Animal Teaching Hospital, respectively, between 1997 and 2010. The biopsies were from dogs that were treated for a solitary cutaneous MCT, and for which complete staging information (at the time of initial presentation to the referral hospital), and follow-up information to the time of patient death or a minimum of 1000 days following diagnosis (whichever came first) were available. The occurrence of metastasis was determined by abdominal ultrasound or computed tomography, and cytological/histological examination of a biopsy of one or more regional/draining lymph nodes. For a cytological diagnosis of lymph node metastasis, mast cells had to appear in clusters or sheets, or appear grossly abnormal [14]. MCT biopsies were designated as ‘metastasising’ (M) if they were borne by dogs which died or were euthanased due to MCT metastatic disease <560 days post-surgery/biopsy (regardless of adjuvant chemotherapy, including prednisolone, and/or radiotherapy), and for whom metastasis was confirmed by diagnostic imaging and pathological analysis. Non-metastasising (NM) MCT biopsies were removed from dogs which received no adjuvant therapy (including prednisolone) and were still alive >1000 days post-surgery/biopsy [24], and for whom metastasis was not identified by imaging or pathological analysis.
RNA isolation and purification
Total RNA was isolated from FFPE MCT biopsies using the RecoverAll Total Nucleic Acid Isolation Kit, which incorporates on-column DNase digestion (ThermoFisher Scientific, Paisley, UK). RNA was treated with Heparinase I (Sigma, Gillingham, UK) (10U/μg RNA) in 5mM Tris-HCl (pH7.5), 1mM CaCl2, 4U/μl RNasin Plus RNase Inhibitor (Promega, Southampton, UK) for 3h at 25°C, and subject to further DNase digestion (TURBO DNA-free kit; ThermoFisher Scientific, Paisley, UK). RNA was purified (RNA Clean & Concentrator-5; Zymo Research, Freiburg, Germany) and quantified by RiboGreen fluorometry (Quant-iT RiboGreen RNA Assay Kit, ThermoFisher Scientific, Paisley, UK).
RNA sample selection
The integrity of each FFPE MCT RNA sample was assessed by reverse transcription-quantitative PCR (RT-qPCR) assay of the copy number of a 126bp fragment of a 130–150bp short interspersed nuclear element (SINE) present every 5–8.3kb in the canine genome [25], and shown (by BLAST similarity search against canine mRNA sequences) to occur in the 3’-untranslated region of hundreds of canine mRNAs. cDNA was prepared from 10ng of each total RNA sample using the High-Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific, Paisley, UK), and triplicate PCR assays were performed (PowerUp SYBR Green Master Mix; ThermoFisher Scientific, Paisley, UK) using 1μl aliquots of a 1 in 1.6-fold dilution of each cDNA sample. A quantification cycle (Cq) value was derived for each PCR product using the PCR machine software (StepOne Plus; ThermoFisher Scientific, Paisley, UK), and a geometric mean Cq value calculated for each MCT cDNA sample as a measure of RNA integrity.
Global gene expression profiling
RNA amplification, labelling and microarray hybridisation
Fragmented, biotinylated double-stranded cDNA was prepared from 50ng of each FFPE MCT RNA sample using the SensationPlus FFPE amplification and WT labelling Kit (ThermoFisher Scientific, Paisley, UK), and hybridised (in groups of 4) to a Canine Gene 1.1 ST Array Strip (ThermoFisher Scientific, Paisley, UK). Post-hybridisation washing and staining, and array scanning were performed using the GeneAtlas System Fluidics and Imaging Stations (ThermoFisher Scientific, Paisley, UK), respectively.
Microarray data analysis
Exon-level probe set expression values were generated by quantile normalisation, log2 transformation and signal summarisation, performed using the Robust Multichip Analysis algorithm, implemented within ‘Affymetrix Expression Console Software 1.3’ (ThermoFisher Scientific, Paisley, UK). ‘Outlier arrays’ were considered to be those that had any single sample quality, labelling quality and hybridisation quality metric value ≥2 standard deviations away from the mean of the metric value for all the arrays [26]. Outlier arrays were excluded, and processing of the raw probe-level signal intensity data repeated to generate both quantile normalised and log2-transformed exon and gene-level probe set expression values. Gene-level probe sets (‘Transcript clusters’) with ‘crosshyb_type’ = 1 (unique hybridisation target) and ‘category’ = ‘main’ annotations, and for which at least 1 exon probe set was ‘present’ (detection above background p-value <0.01; [27]) in at least 30% of the tumours in the NM and/or M MCT cohort, were considered to be expressed in the MCTs and were used for subsequent analyses.
Relationships between MCT gene-level expression profiles were visualised by hierarchical clustering (average linkage; similarity metric = Pearson Correlation Coefficient) performed using Cluster [28]. Genes displaying statistically significant differences in expression between M and NM MCTs were identified using a two-tailed t-test for unpaired data. P-values were adjusted by permutation testing [29]. The potential identities of differentially expressed Transcript clusters that represented ‘predicted genes’, or for which no annotation was available, were sought by BLAST similarity search of Transcript cluster sequences against canine and human mRNAs and non-coding RNAs, respectively.
Functional annotation analysis
Over represented functional annotations associated with the differentially expressed genes were identified using DAVID [30] by comparison with the functional annotations attributed to all the ‘crosshyb_type’ = 1 and ‘category’ = ‘main’ annotated Transcript clusters for which at least 1 exon probe set was ‘present’ in at least 30% of the tumours in the NM and/or M MCT cohort.
Reverse transcription-quantitative PCR (RT-qPCR)
The differential expression of selected genes was validated by RT-qPCR. A TaqMan or SYBR Green PCR assay was designed (Beacon Designer, Premier Biosoft; Palo Alto, USA) for each Transcript cluster based upon a unique region within the sequence of the constituent exon probe set that showed the largest statistically significant difference in expression between the M and NM MCTs. Prior to use in RT-qPCR, each MCT cDNA was assayed for the presence of PCR inhibitors (S1 File and S2 Table). Triplicate PCR assays for each preamplified MCT cDNA sample were run on an ABI StepOne Plus PCR machine (ThermoFisher Scientific, Paisley, UK), and a geometric mean Cq value derived. For use as a ‘reference gene’ for normalisation of target gene expression measurements [31], the copy number of a 71bp fragment of a SINE [25] that occurs in the 3’-untranslated region of hundreds of canine mRNAs in each MCT RNA sample was also assayed. MCT Cq values with a standard deviation >0.5 were excluded from further analyses, and genes with a geometric mean Cq of ≥35 were considered not to be expressed. Additional details are included in S1 File and S1 Table.
RT-qPCR data analysis
The geometric mean Cq measures of target gene expression were imported into qbase+ (Biogazelle, Gent, Belgium) and each converted to a relative measure of gene expression (‘Normalised Relative Quantity; NRQ [32]) using a normalisation factor derived from the respective geometric mean canine SINE [25, 31] Cq value. The statistical significance of differences in the expression of genes between M and NM MCTs was determined using a two-tailed t-test for unpaired data performed on log10 transformations of the NRQs.
Class prediction analysis
The optimal classification function for gene expression data-based prediction of ‘metastatic status’ (M or NM) was identified by evaluation of the characteristics of the expression values obtained for the Transcript clusters expressed in the MCTs using the R package SPreFuGED [33], which predicts the performance of representatives of 10 classification functions. Class prediction by Linear Discriminant Analysis was performed using the lda function provided by the R Package MASS [34]. The accuracy of class prediction was estimated through testing by cross-validation. The expression profiles (selected genes) of the MCTs were randomly partitioned into a ‘training data set’ (comprising data for ~90% of the MCTs) and a ‘test data set’ (comprising data for two M and one NM MCT), and the class (M or NM) of the MCTs comprising the test data set predicted. Ten training and test data set combinations were evaluated. The lda function was also run in the ‘leave-one-out cross-validation mode’, whereby the class of each MCT was predicted whilst using the expression data for the remaining (n-1) MCTs as a training data set.
Results
Tumours selected for gene expression profiling
MCT biopsies from 78 dogs were eligible for inclusion in the study. PowerAtlas [35] analysis of Gene Expression Omnibus [36] datasets derived for several human tumours estimated that using 20 tumour samples in each of two ‘outcome groups’ would afford an ‘Estimated Discovery Rate’ (Power) of 73.1–81.7% at the 0.05 significance level. Consequently, the integrity of each MCT RNA was assessed to enable compilation of ‘M’ and ‘NM’ MCT sample groups comprising 20 RNA samples with a similar range of integrities (S3 Table).
Tumours included in differential expression analysis
Sample quality metrics associated with exon-level probe set expression profiles of 40 MCTs were reviewed to identify tumours whose expression profiles differed significantly from the majority of the cohort. Array data for 2 M MCTs and 4 NM MCTs were excluded (S3 and S4 Tables) because for each the ‘percent of probe sets detected above background’ differed by >2 standard deviations from the cohort mean value [26]. Gene-level probe set expression data for 18 M MCTs and 16 NM MCTs was re-processed for further analysis featuring 5,207 Transcript clusters annotated as ‘crosshyb_type’ = ‘1’ and ‘category’ = ‘main’ probe sets, and for which at least 1 exon probe set was ‘present’ in at least 30% of the tumours in the NM and/or M MCT cohort. The raw and processed microarray data has been deposited in the NCBI Gene Expression Omnibus repository (GEO series accession number GSE122590).
The characteristics of the dogs that bore the 18 M MCTs and 16 NM MCTs tumours are detailed in Tables 1 and 2. Nine breeds were represented in the M group and 7 in the NM group, with 5 breeds common to both groups. Multiple representatives of a single breed probably reflect both breed popularity and an increased susceptibility to MCT development [3]; for example, MCTs borne by Labrador Retrievers represent 19% of the NM MCTs and 39% of the M MCTs. Equal proportions of both sexes were represented in the M MCT biopsy group, whilst 69% of the NM MCTs were borne by male dogs. Interestingly, the median age of the dogs affected by M MCTs was 3 years higher than that of the dogs that developed NM tumours. Unsupervised hierarchical clustering of the 34 MCTs on the basis of the expression values of the 20% of Transcript Clusters (1,041) with the highest variance in expression signal, gave no indication of an association between global MCT gene expression profile and breed, sex, or age at diagnosis, respectively (S1 Fig). The mean age of a FFPE NM MCT specimen was 1.6 x higher than that of a FFPE M MCT biopsy, although the FFPE tumour biopsy age does not correlate with tumour RNA integrity (Spearman rank correlation coefficient = 0.13; S3 Table).
Table 1. Dogs bearing metastasising cutaneous mast cell tumours included in differential gene expression analysis.
| Dog ID. | Breed | Sexa | Age at diagnosis (Years) |
|---|---|---|---|
| B1 | Boxer | Fe | 9 |
| CB6 | Cross breed | FeN | 9 |
| CB4 | Cross breed | FeN | 10 |
| CB1 | Cross breed | FeN | 15 |
| CCR1 | Curly Coated Retriever | Fe | 6 |
| D1 | Dogue de Bordeaux | MaN | 2 |
| GS1 | German Shepherd | Ma | 9 |
| GR1 | Golden Retriever | FeN | 8 |
| HV1 | Hungarian Vizsla | MaN | 7 |
| LR10 | Labrador Retriever | FeN | 4 |
| LR1 | Labrador Retriever | FeN | 8 |
| LR2 | Labrador Retriever | Ma | 7 |
| LR5 | Labrador Retriever | Ma | 8 |
| LR9 | Labrador Retriever | Ma | 10 |
| LR8 | Labrador Retriever | MaN | 6 |
| LR3 | Labrador Retriever | MaN | 8 |
| SBT1 | Staffordshire Bull Terrier | FeN | 10 |
| W1 | Whippet | MaN | 11 |
| Mean and standard deviation | 8.17 ± 2.73 | ||
| Median | 8.00 | ||
| Interquartile range | 3.25 |
Table 2. Dogs bearing non-metastasising cutaneous mast cell tumours included in differential gene expression analysis.
| Dog ID. | Breed | Sexa | Age at diagnosis (Years) |
|---|---|---|---|
| B4 | Boxer | Fe | 5 |
| B2 | Boxer | FeN | 4 |
| B3 | Boxer | Ma | 4 |
| CB3 | Cross breed | FeN | 10 |
| CB2 | Cross breed | FeN | 11 |
| CB5 | Cross breed | MaN | 5 |
| ETT1 | English Toy Terrier | Ma | 6 |
| GR2 | Golden Retriever | Ma | 3 |
| GR4 | Golden Retriever | Ma | 7 |
| GR3 | Golden Retriever | MaN | 5 |
| LR4 | Labrador Retriever | Ma | 6 |
| LR6 | Labrador Retriever | MaN | 3 |
| LR7 | Labrador Retriever | MaN | 5 |
| MS1 | Miniature Schnauzer | MaN | 3 |
| SBT2 | Staffordshire Bull Terrier | MaN | 6 |
| W2 | Whippet | Fe | 5 |
| Mean and standard deviation | 5.50 ± 2.21 | ||
| Median | 5.00 | ||
| Interquartile range | 2.00 |
aFe: Female; FeN: Neutered female; Ma: Male; MaN: Neutered male
Genes differentially expressed between M and NM MCTs
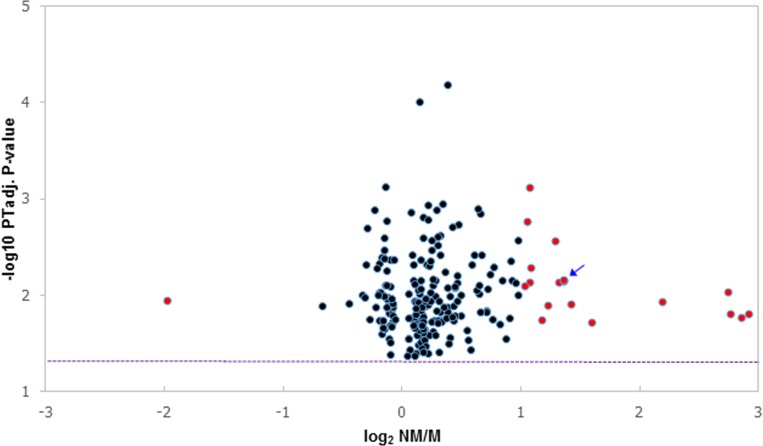
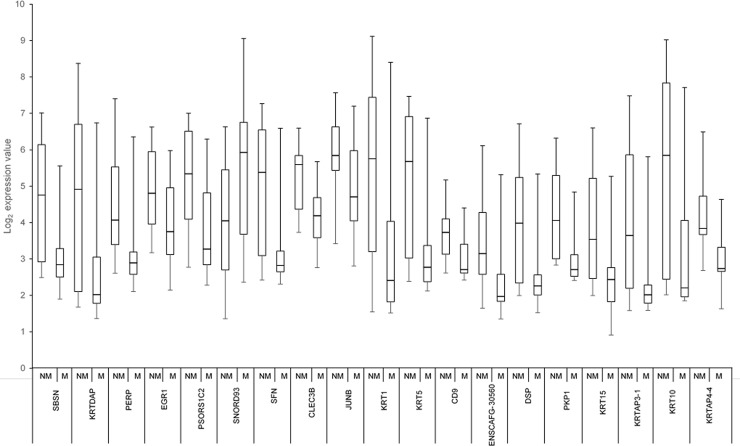
A statistically significant difference in expression (permutation-testing adjusted p-value <0.05) between M and NM MCTs was observed for 218 Transcript clusters; 162 displayed increased expression in the NM MCTs and 56 were expressed at a higher level in the M MCTs (Fig 1). Nineteen genes displayed a >two-fold difference in expression between the M and NM MCTs; 18 of the genes showed increased expression in the NM MCTs (Table 3; Fig 2). Gene annotation was not available for 2 of the Transcript clusters, although each displayed significant sequence similarity to one, or more, mRNAs encoded by a single canine gene. Three genes that exhibited a >two-fold difference in expression are located on each of chromosome 1, 9, and 27, respectively. Two of the genes (SBSN and KRTDAP) are positioned adjacent to each other in a 37.6kb region on chromosome 1 (Table 3). Two chromosomes (CFA14 and CFA31) harbour a higher proportion of the 218 differentially expressed genes than may be expected if their chromosomal distribution only reflected the chromosomal assignments of the genes expressed in the MCTs (S5 Table). Three differentially expressed keratin-associated genes (KRTAP8-1, KRTAP7-1 and KRTAP11-1), which display decreased expression in the M MCTs, lie adjacent to each other within a 69.7kb region on CFA31 (S5 Table).
Fig 1. Genes exhibiting differential expression between metastasising and non-metastasising MCTs.
Expression of 218 genes in 18 metastasising (M) and 16 non-metastasising (NM) MCTs. Each sphere represents an individual gene. The difference in expression between the M and NM MCTs is represented (x-axis) by the log2-transformed fold-change (NM/M). Red spheres denote the 19 genes which exhibit a difference in expression of ≥2.0 (either NM>M, or M>NM). The arrow indicates two genes which cannot be resolved by their x, y co-ordinates. The statistical significance of differences in expression between NM and M MCTs is denoted (y axis) by the minus log10-transformed permutation testing-adjusted unpaired t-test derived p-values. A -log10 PTadj. p-value equivalent to a PTadj. p-value = 0.05 is indicated by the dotted line.
Table 3. Genes displaying >two-fold differences in expression between metastasising and non-metastasising MCTs.
| Gene description (Gene symbol/ID.) | Chromosomal locationb[37] | Fold change (NM/M)d | Adj_p-valuee |
|---|---|---|---|
| TP53 apoptosis effector (PERP) | 1: 30.42 | 2.11 | 0.008 |
| Suprabasin (SBSN) | 1: 117.13c | 2.84 | 0.012 |
| Keratinocyte differentiation-associated protein (KRTDAP) | 1: 117.17c | 7.23 | 0.017 |
| Stratifin (SFN) | 2: 73.27 | 4.71 | 0.012 |
| Plakophilin 1 (PKP1) | 7: 1.92 | 2.45 | 0.003 |
| Keratin 15 (KRT15) | 9: 21.25 | 2.12 | 0.005 |
| Sequence similarity to Keratin-associated protein 4–4 (E-val: 0.0; 470bp; 26%) (KRTAP4-4)a | 9: 21.55 | 2.10 | 0.001 |
| Keratin-associated protein 3–1 (KRTAP3-1) | 9: 21.68 | 2.60 | 0.006 |
| Keratin 10 (KRT10) | 9: 21.86 | 8.50 | 0.014 |
| Early growth response 1 (EGR1) | 11: 26.05 | 2.53 | 0.009 |
| Psoriasis susceptibility 1 candidate 2 (PSORS1C2) | 12: 0.84 | 3.17 | 0.019 |
| Small nucleolar RNA, C/D box 93 (SNORD93) | 14: 36.56 | 0.25 | 0.011 |
| C-type lectin domain family 3, member B (CLEC3B) | 20: 43.32 | 2.57 | 0.007 |
| junB proto-oncogene (JUNB) | 20: 49.36 | 2.25 | 0.018 |
| Epithelial keratin 1 (KRT1) | 27: 2.42 | 7.28 | 0.010 |
| Keratin 5 (KRT5) | 27: 2.57 | 6.81 | 0.013 |
| CD9 antigen (CD9) | 27: 38.74 | 2.07 | 0.002 |
| Sequence similarity to ENSCAFG00000030560 (E-val: 4 x 10−54; 106bp; 60%) (ENSCAFG-30560)a | 31: 38.47 | 2.05 | 0.007 |
| Desmoplakin (DSP) | 35: 7.48 | 2.48 | 0.013 |
aTranscript cluster with no gene annotation. The most significant similarity between the sequence (spliced exons) of the Transcript cluster and a canine mRNA is listed. The significance of the sequence similarity is denoted by the E value and the length of the sequence alignment, and the proportion of the Transcript cluster sequence included in the alignment is stated.
bChromosomal location is denoted by the chromosome name and the gene start base co-ordinate.
cThe genes encoding suprabasin and keratinocyte differentiation-associated protein are adjacent to each on chromosome 1.
dRatio of median gene-level expression values.
ePermutation testing-adjusted t-test p-value
Fig 2. Expression levels of 19 genes that display >two-fold differences in expression between metastasising and non-metastasising MCTs.
The bottom and top of each box denote the expression measurements that encompass the values shared by 25% and 75% of the tumours, respectively. The line within each box represents the median expression value, whilst the vertical lines extending above and below each box indicate the maximum and minimum expression values, respectively. M = metastasising tumour; NM = Non-metastasising MCT.
Functional annotation enrichment analysis
In order to identify biological processes and pathways involved in MCT metastasis, functional annotations over-represented amongst those assigned to the 218 Transcript clusters differentially expressed between the M and NM MCTs were identified by comparison with those attributed to the 5,207 Transcript clusters for which at least 1 exon probe set was ‘present’ in at least 30% of the tumours in the NM and/or M MCT cohort. The frequencies of functional annotations available for 177 of 209 differentially expressed Transcript clusters for which an Ensembl Gene ID [37] could be defined were compared with those available for 4,846 of the ‘present’ Transcript clusters which had an Ensembl Gene ID. Six Gene Ontology Consortium biological processes and two KEGG pathways were enriched amongst the differentially expressed genes (Table 4).
Table 4. Differentially expressed gene-associated enriched functional annotations.
| Gene expression | ||||
|---|---|---|---|---|
| Functional annotationa | Fold enrichmentb | P-valuec | NM > M | M > NM |
| GO BP: 0016337 single organismal cell-cell adhesion | 6.699 | 0.005 | CDSN, DSP, PKP1, SCRIB | |
| GO BP: 0008285 negative regulation of cell proliferation | 3.240 | 0.006 | CD9, ETV3, HDAC4, NF2, SFRP4, SPRY1, STRN, TFAP2A | DNAJA3 |
| GO BP: 0060070 canonical Wnt signaling pathway | 6.029 | 0.008 | BCL9L, MYC, PLPP3, SDC1, SFRP4 | |
| GO BP: 0051496 positive regulation of stress fiber assembly | 7.420 | 0.014 | ARHGEF10, BRAF, EVL, NF2 | |
| GO BP: 0008219 cell death | 12.058 | 0.023 | AXIN1, EMP2, PMP22 | |
| GO BP: 0032060 bleb assembly | 10.336 | 0.031 | EMP1, EMP2, PMP22 | |
| KP cfa04024: cAMP signaling pathway | 2.969 | 0.045 | AFDN, BRAF, CREBBP, FOS | CALM3 |
| KP cfa05210: Colorectal cancer | 4.783 | 0.046 | BRAF, FOS, MYC, PIK3R2 | |
aGO BP: Gene Ontology Biological Process; KP: Kegg Pathway
bFold enrichment—Proportion of 177 differentially expressed genes with the functional annotation/proportion of ~4,846 genes expressed in the MCTs that have the functional annotation.
cP-value: Fisher Exact test p-value (EASE score) modified to reduce false positive results.
Validation of differential expression by RT-qPCR
The expression levels of 9 of the genes which showed >two-fold differences in expression between 18 M and 16 NM MCTs were measured by RT-qPCR assay of the same MCT biopsies (Table 5). The genes selected for validation of differential expression included the only gene that displayed >two-fold increased expression in the M MCTs, the 2 genes represented by Transcript clusters for which gene annotation was not available, and genes representative of different enriched functional annotations (Table 4) and/or associated with different biological processes. Valid gene expression measurements (Cq values) were obtained for fewer than the 34 MCT biopsies assayed because either the Cq was ≥35 or was ‘undetermined’, or the Cq standard deviation for triplicate assays was >0.5. There was a high degree of concordance between the expression levels calculated for individual MCTs (indicated by Spearman rank correlation coefficients in Table 5), and between NM/M fold changes, measured by microarray (exon-level probe set) and RT-qPCR, respectively. For 2 of the genes (EGR1 and KRT10), the differences in expression between the M and NM MCTs was statistically significant. However, statistical significance is affected by both the number of samples and which samples are included in a statistical test. The differences in expression between the NM and M groups attained statistical significance for KRT10 and PERP when the microarray-derived expression measurements for only the MCTs that were included in the statistical analysis of the RT-qPCR generated expression data were analysed (Table 5).
Table 5. Differences in gene expression between M and NM MCTs measured by RT-qPCR.
| RT-qPCR | ||||||
|---|---|---|---|---|---|---|
| Gene symbol/ID. | Exon-level fold changea (NM/M) | No. NM MCTsb | No. M MCTsb | Fold changec (NM/M) | Spearman RCCd | p-valuee (Array) |
| CD9 | 2.25 (1.80) | 12 | 8 | 2.78 | 0.51 | 0.405 (0.090) |
| DSP | 19.08 (1.48) | 7 | 4 | 40.43 | 0.74 | 0.217 (0.152) |
| EGR1 | 3.66 (2.11) | 11 | 14 | 6.05 | 0.69 | 0.028 (0.050) |
| ENSCAFG-30560 | 2.05 (1.43) | 5 | 8 | 7.15 | 0.81 | 0.139 (0.213) |
| KRT10 | 23.43 (43.37) | 11 | 12 | 53.87 | 0.88 | 0.009 (0.017) |
| KRTAP4-4 | 2.10 (2.77) | 7 | 7 | 1.12 | 0.76 | 0.730 (0.208) |
| PERP | 7.38 (4.69) | 11 | 10 | 9.94 | 0.80 | 0.083 (0.041) |
| SBSN | 3.49 (2.62) | 10 | 6 | 14.77 | 0.81 | 0.140 (0.115) |
| SNORD93 | 0.25 (0.39) | 12 | 16 | 0.37 | 0.50 | 0.196 (0.258) |
aFold change differences in expression between 18 M and 16 NM MCTs determined by microarray—Ratio of median expression values for the Exon probe set upon which RT-qPCR assay design was based. In parenthesis are the fold change differences calculated when only the microarray-derived expression values of MCTs that yielded valid Cq values in RT-qPCR assay were considered.
bThe numbers of NM and M MCTs represent the numbers of samples for which valid Cq (Cq <35; Cq SD<0.5) measurements were obtained. ‘Non-valid’ Cq values were attributable to: Cq <35 or ‘undetermined’ and Cq SD>0.5.
cFold change determined by RT-qPCR assay.
dThe Spearman rank correlation coefficient (RCC) indicates the extent of the concordance between the expression values for individual MCT assayed by microarray and RT-qPCR, respectively.
eThe statistical significance of the RT-qPCR measured differences in expression between the NM and M MCTs determined by t-test. The statistical significance of the differences between the microarray-derived gene-level expression values measured for the same MCTs is shown in parenthesis.
Class prediction analysis
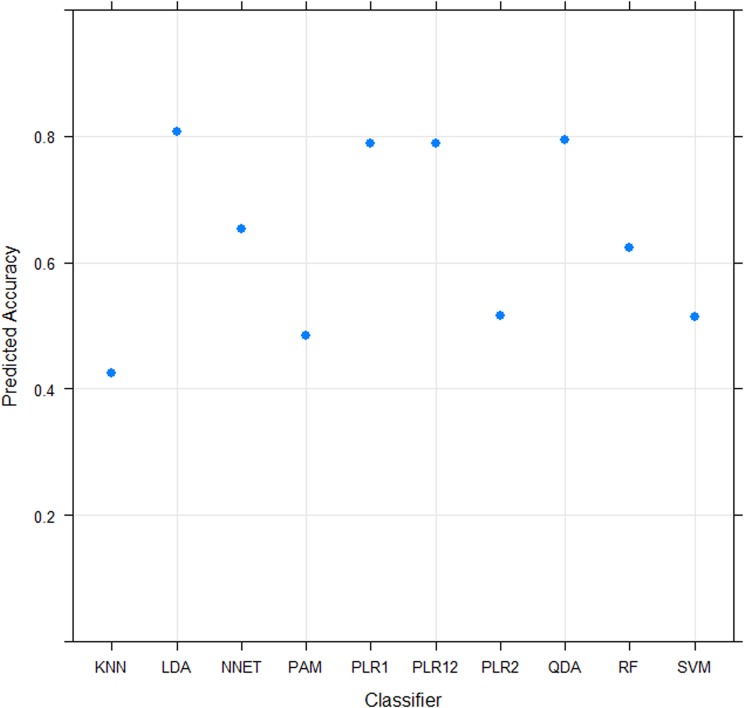
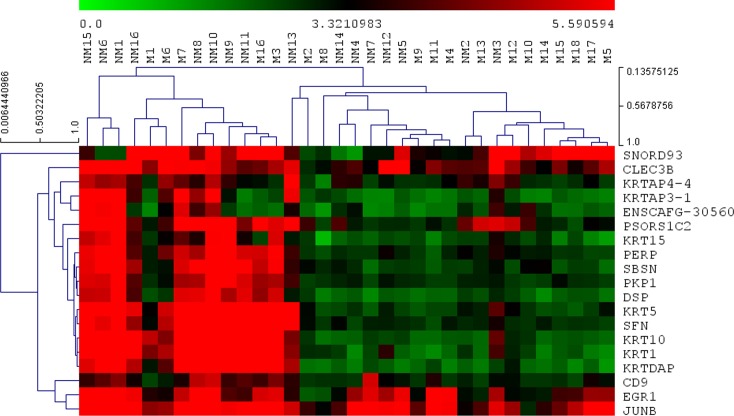
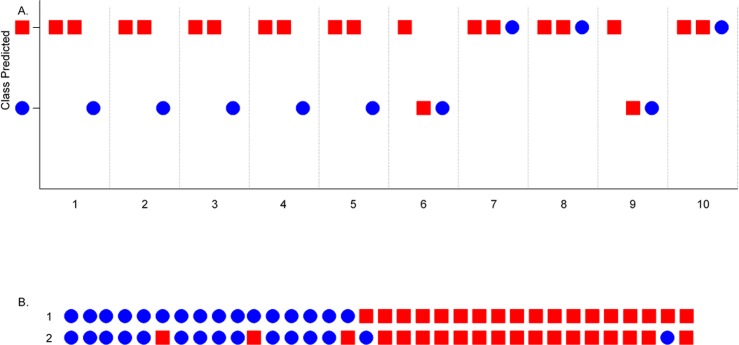
Linear Discriminant Analysis (LDA) was predicted to be the optimal classification function for prediction of ‘metastatic status’ (M or NM) on the basis of the characteristics of the expression values obtained for the 5,207 Transcript clusters ‘present’ in the MCTs (Fig 3). Hierarchical clustering of the 18 M and 16 NM MCTs, on the basis of the microarray-measured expression levels of the 19 genes which displayed >two-fold differences in expression between the M and NM MCTs, separated the MCTs into 2 major groups (Fig 4). The largest group contained 72% of the M MCTs, whilst the NM MCTs were equally divided between the two groups. Although the M and NM MCTs were not partitioned into two groups, class prediction by LDA does not assign a sample to a class on the same basis (i.e. using a measure of the ‘distance’ between 2 samples/pre-created sample groups equal to 1 minus the correlation coefficient) that samples are agglomeratively grouped by hierarchical clustering. Consequently, and as the fold difference in the expression of a gene between classes has been shown to be an effective criterion for ranking genes for use in class prediction [38, 39], the efficacy of using the 19 genes which displayed >two-fold differences in expression between M and NM MCTs in class prediction was evaluated. The performance of the LDA classifier was tested by cross-validation, measuring the accuracy with which 2 M MCTs and 1 NM MCT (randomly selected) were classified (as M or NM) on each of 10 occasions (after the classifier had been trained using the expression values for the remaining 16 M and 15 NM MCTs) (Fig 5). Mean classification accuracies of 90% (M MCTs) and 70% (NM MCTs) were estimated, whilst a median classification accuracy of 100% was achieved for both M and NM MCTs. Evaluating the performance of the classifier in a ‘leave-one-out cross-validation mode’, 88.9% of the 18 M MCTs were correctly classified and 81.3% of the 16 NM MCTs were correctly assigned to the NM class.
Fig 3. Predicted accuracy of classification functions on the basis of the characteristics of the expression values obtained for 5,207 Transcript clusters ‘present’ in the MCTs.
Classification functions evaluated by SPreFuGED [33]: KNN—k-nearest neighbours, LDA—linear discriminant analysis, NNET—feed-forward neural network, PAM—prediction analysis of microarrays, PLR1 - ℓ1ℓ1 penalized logistic regression (Lasso), PLR12 - ℓ1ℓ1 and ℓ2ℓ2 penalized logistic regression (Elastic net), PLR2 - ℓ2ℓ2 penalized logistic regression (Ridge), QDA—quadratic discriminant analysis, RF—Random forest, SVM—support vector machine.
Fig 4. Hierarchical clustering of 18 M and 16 NM MCTs, and 19 genes which display >two-fold differences in expression between M and NM MCTs.
The horizontal colour bar denotes the log2 expression value. The extent of the dissimilarity (equal to 1 minus the Pearson correlation coefficient) between MCTs is indicated by the vertical scale bar in the top right hand corner of the figure, and between genes is indicated by the horizontal scale bar in the top left hand corner of the figure.
Fig 5. Class prediction by Linear Discriminant Analysis.
(A) Results of predicting the classes (M = a square, and NM = a circle) of 3 MCTs (2 M MCTs and 1 NM MCT) on the basis of the expression values of the 19 genes which show statistically significant >two-fold differences in expression between M and NM MCTs. Class prediction was performed on 10 occasions. On each occasion the gene expression data for 34 MCTs (18 M and 16 NM) was randomly divided into a ‘training set’ (90% of the data, comprising 16 M and 15 NM MCTs) and a ‘test set’ (10% of the data, representing 2 M MCTs and 1 NM MCT). The LDA classifier was trained using the training data set and then used to assign the 3 ‘test MCTs’ to either the M or NM class. (B) Results of predicting the class of each of 34 MCTs (16 NM = circles, 18 M = squares) on the basis of the expression values of the 19 genes which show statistically significant >two-fold differences in expression between M and NM MCTs. Over 34 iterations the LDA classifier was trained using the expression data for 33 of the 34 MCTs, and the classification of the remaining MCT predicted. The actual class of each MCT is depicted in row 1 and the predicted class of each MCT is shown in row 2.
Discussion
Mast cell tumours are one of the most common tumours affecting dogs. There is currently no accurate way of predicting whether a tumour is one of the 20–30% of MCTs that will metastasise, and pre-existing micrometastases may not be detected by current imaging modalities. Activating internal tandem duplications (ITDs) in exon 11 of c-kit have been reported in 9% of canine MCTs [40]. The mutations affect the juxtamembrane domain and are associated with a higher histological grade and poor prognosis [40], although they are present in less than 50% of ‘high grade’ MCTs [40, 41]. In the absence of an effective predictive test for MCT metastasis, dogs that bear tumours with unrecognised metastatic potential may not receive adjuvant chemotherapy, whilst dogs erroneously believed to harbour a metastasising tumour may be unnecessarily exposed to the possible side-effects of chemotherapy. The molecular genetic drivers of canine MCT metastasis potentially represent both biomarkers of metastasis and targets for anti-metastasis therapeutics. In the current study we sought to identify ‘across breed’ molecular genetic contributants to canine MCT metastasis by comparing global gene expression in 20 MCTs that metastasised with that in 20 MCTs that did not metastasise. The tumours profiled were borne by 11 breeds (and a variety of cross breeds).
Gene expression associated with mast cell tumour metastasis
Three-quarters of the genes that displayed statistically significant differences in expression between M and NM MCTs showed decreased expression in the M MCTs. This suggests that the ‘balance’ in the molecular genetic contribution to MCT metastasis lies in a reduction of the effect of genes that would otherwise diminish the propensity for metastatic dissemination. Functional annotation enrichment analysis of the differentially expressed genes provides an insight into the biological processes associated with MCT metastasis.
Cell adhesion
The loss of normal cell polarity and adhesion are pivotal to tumour metastasis. Four genes (SCRIB, PKP1, CDSN and DSP) with ‘cell adhesion’ annotation show decreased expression in M MCTs. Failure to maintain the three-dimensional organization of tissues is coincident with disruption of intercellular junctions, loss of cell adhesion and epithelial-mesenchymal transition. The scaffold protein Scribbled Planar Cell Polarity Protein (SCRIB) regulates cell polarity and cell proliferation. In Drosophila, deletion of SCRIB causes loss of apical-basal polarity and in concert with oncogenic Ras activation induces cell proliferation and metastasis [42]. Plakophilin 1 (PKP1), Desmoplankin (DSP) and corneodesmosin (CDSN) are components of the desmosome, intracellular junctions that link cytoskeletal intermediate filaments to the plasma membrane and mediate cell-cell adhesion. Decreased expression of PKP1 in oral squamous cell carcinoma cells increased cell motility [43], and is associated with the metastatic phenotype of several human cancers [44]. Reduced expression of DSP in a number of human primary tumours has been associated with tumour metastasis [45]. DSP has also been identified as a migration suppressor in a mouse model of pancreatic cancer [46], potentially effected by inhibition of β-catenin-dependent Wnt signalling [47].
Canonical Wnt signalling
The role of canonical Wnt signalling in tumour metastasis is tissue-specific, and both activation and inactivation of the pathway have been associated with promoting epithelial-mesenchymal transition [48]. Herein, it is unclear whether activation or inhibition of Wnt signalling promotes MCT metastasis. Decreased expression in the M MCTs of several genes associated with canonical Wnt signalling suggests that diminution of canonical Wnt signalling promotes MCT metastasis. The transcriptional co-activator BCL9L, which promotes β-catenin activity and transcription of Wnt target genes, shows decreased expression in the M MCTs. A similarly reduced level of expression in the M MCTs is noted for the transcription factor, and proto-oncogene, MYC, a target of Wnt signalling. This result is conceptually consistent with the observation that MYC overexpression inhibits cancer cell motility and invasiveness in vitro [49]. SDC1, a cell surface heparan sulphate proteoglycan, promotes canonical Wnt signalling in metastatic melanoma [50]. SDC1 links the cytoskeleton to the extracellular matrix (ECM) and has a role in cell-cell and cell-ECM adhesion, and cell migration. SDC1 expression varies between cancer types, but reduced expression in carcinomas is associated with enhanced cell motility and invasion [51]. PLPP3, a membrane glycoprotein, also shows a decreased level of expression in M MCTs. A major function of PLPP3 is dephosphorylation of extracellular lysophosphatidic acid, a phospholipid with growth factor-like activity that stimulates tumour cell migration and invasion [52]. PLPP3 displays reduced expression in metastasising (versus non-metastasising) sporadic colorectal cancer [53]. Conversely, SFPR4, a member of the secreted frizzled-related family that inhibit Wnt signalling by binding to Wnt proteins or to Frizzled receptors, displays decreased expression in M MCTs.
Negative regulators of cell proliferation
De-regulation of cell proliferation (and a consequent high proliferation rate) is generally associated with tumour aggressiveness. Although the paradigm has been recently challenged, metastatic potential has also been associated with increased resistance/decreased sensitivity to apoptosis [54]. Negative regulators of cell proliferation are enriched amongst the genes exhibiting differential expression between M and NM MCTs. DnaJ Heat Shock Protein Family Member A3 (DNAJA3) is the only one of 9 genes to display increased expression in the M MCTs. DNAJA3 encodes two protein isoforms localised to the mitochondrial matrix which have opposite effects on apoptosis induced by external stimuli. The short isoform suppresses apoptosis [55], and its overexpression has been shown to promote the migration and invasion of non-small cell lung carcinoma cells in vitro [56]. The genes expressed at a lower level in the M MCTs include the transcription factor TFAP2A and transcriptional repressors ETV3, HDAC4, and SPRY1.
Bleb assembly and cell death
The migration of individual tumour cells is facilitated by the formation of bleb plasma membrane protrusions [57], which are initially devoid of the polymeric form of actin. Peripheral myelin protein 22 (PMP22) is an integral membrane protein known to be localised to epithelial and endothelial cell-cell junctions. PMP22 is involved in the linkage of the actin cytoskeleton to the plasma membrane, and overexpression of PMP22 reduces cell growth and motility [58]. Membrane blebbing is also a hallmark of apoptosis and overexpression of PMP22 and the epithelial membrane proteins (EMP) 1 and 2 has been shown to increase cell death in vitro [59]. PMP22, EMP1 and EMP2 had reduced levels of expression in M MCTs.
Stress fibre assembly
Four genes (EVL, BRAF, ARHGEF10 and NF2) annotated as positive regulators of actin stress fibre assembly displayed decreased expression in the M MCTs. Cell softening is necessary for cell invasion and this is achieved through reorganisation of the actin cytoskeletal architecture. Ena/VASP-like (EVL) enhances actin polymerisation and suppresses cell migration [60]. Reduced expression of BRAF in mouse embryonic fibroblasts was associated with a reduction in actin stress fibre content and an increase in cell migration [61]. Rho GTPase guanine nucleotide exchange (ARHGEF10) activates several Rho GTPases promoting actin stress fibre formation [62]. However, transient expression of ARHGEF10 in vitro was associated with the loss of actin stress fibres and the formation of membrane filopodia [63], which facilitate individual tumour cell migration. Neurofibromin 2 (NF2) is thought to encode a protein that links components of the cytoskeletion, including actin, with plasma membrane proteins. NF2 has been shown to stop cell migration by preventing cleavage of the actin-linked transmembrane protein CD44 [64].
cAMP signalling
Altered cyclic nucleotide signalling is a trait of many cancers, although the effect of signalling on cell growth and survival is cancer and cell-type dependent. Five genes associated with cAMP signalling (CALM3, CREBBP, FOS, BRAF, AFDN) show differential expression between M and NM MCTs. Up-regulated in the M MCTs is CALM3, an enzymatic co-factor involved in the regulation of adenyl cyclase (AC) through calcium signalling. Although AC generates cAMP from ATP, its intracellular level is also dependent upon phosphodiesterases. CREBBP binding enhances the transcription factor activity of the cAMP-response element binding protein (CREB) once it is phosphorylated by cAMP-activated protein kinase A. CREB mediates transcription of FOS and JUN, and homodimers of each, or heterodimers of both, form the AP-1 transcription factor complex, which regulates the expression of genes involved in proliferation, apoptosis and cell migration [65]. EPAC, an exchange protein activated by cAMP, activates the GTPase Ras-associated protein 1, which in turn activates BRAF and the adherens junction formation factor (AFDN). AFDN links nectins (transmembrane cell adhesion molecules) at cell-cell junctions to the actin cytoskeleton. The decrease in the expression of both JUNB and FOS observed in the M MCTs is consistent with the decreased expression of CREBBP, and may suppress diminution of proliferation because JUNB is typically a negative regulator of cell proliferation [66]. Down-regulation of JUNB in tumour metastases (relative to primary tumours) is common to many human cancers [17]. Reduced M MCT expression of BRAF and AFDN disrupts cell-cell adhesion favouring cell migration.
Genes displaying two-fold or greater differences in expression between M and NM MCTs
Seven of the 18 genes which show ≥two-fold decreased expression in the metastasising MCTs are keratin genes, or keratin/keratinocyte-associated genes. Keratins are intermediate filaments that form part of the cytoskeleton, and are largely associated with maintaining the mechanical stability and integrity of epithelial cells [67]. Skin epidermal tissue was estimated to constitute 2–3% of the longitudinal MCT biopsy cross-sections from which RNA was isolated for gene expression analysis, and there was no apparent gross difference between the epidermal tissue content of M and NM primary MCT biopsies. However, it is unclear whether differential expression of genes encoding keratin intermediate filaments and epithelial cell-associated desmosomal proteins (DSP, PKP1) reflects differences in epithelial cell (keratinocyte) and/or mast cell gene expression.
Keratins are not detected in canine MCTs by anti-pan cytokeratin immunohistochemistry, which screens for a number of keratins common to many epithelial tissues. However, keratin gene expression is tissue-, differentiation state and functional status-specific [67], and keratin genes have been shown to be expressed in haematopoetic cells [68], as has DSP [69].
In epithelial tumours the down-regulation of specific keratins is believe to alter the cytoskeleton architecture causing increased cellular elasticity and deformability such that cells are better able to permeate through the stroma and migrate away from the primary tumour [70]. It is possible that down-regulation of specific keratin genes in neoplastic mast cells has a similar effect to that deduced for epithelial tumour cells. The altered expression of 8 skin epithelial cell-associated genes (KRT1, KRT5, KRT15, KRTDAP, DSP, PKP1, PERP and SBSN) that displayed >two-fold decreased expression in the M MCTs have previously been associated with the metastasis of a non-epithelial tumour as they were expressed at lower levels in human metastatic cutaneous melanomas than in primary tumours [71]. Down-regulation of KRT15 in tumour metastases (relative to primary tumours) is also common to many solid human cancers [17].
If keratin and desmosomal protein-encoding genes are not expressed in neoplastic canine mast cells, a possibly unlikely alternative proposition is that cytoskeletal reorganisation and reduced adhesiveness of adjacent keratinocytes (potentially neoplastic mast cell-directed) assists neoplastic mast cell cells to escape from the primary tumour. KRT5 and KRT15 are found in keratinocytes occupying the basal layer of the epidermis [67], and their reduced expression in the M MCTs may indicate a loss of basal epithelial cells and/or invasion of the basal layer as an early step in the metastatic cascade. There is increasing appreciation of the role of the tumour tissue microenvironment in facilitating various stages of the metastatic cascade, and evidence that the cells in a primary tumour exploit interactions with surrounding non-malignant cells and the ECM to enable inappropriate growth, local invasion and metastatic dissemination [72]. As a potential precedent, interaction between keratinocytes and cutaneous melanoma cells has been shown to be required for vertical invasion of melanoma cells into the dermis [73].
PERP (TP53 Apoptosis Effector) is a transmembrane 4 desmosomal protein that is involved in maintaining epithelial cell integrity by promoting desmosomal-mediated cell adhesion, but its transcription is also activated by p53 to effect apoptosis [74]. PERP expression is reduced in human primary uveal melanomas that metastasise [75], and PERP has been shown to be down-regulated in murine bone marrow-derived mast cells overexpressing microRNA miR-9, which displays increased expression in ‘biologically high grade MCTs’ [76]. Over-expression of miR-9 enhanced the invasion of mouse malignant mast cell cells in vitro [76].
SBSN (suprabasin) is located in epithelial suprabasal layers and is involved in epidermal differentiation. Both up-regulation [77] and down-regulation [71] of its expression have been associated with tumour metastasis.
SFN (Stratifin or 14-3-3 Sigma) is primarily recognised as a cell cycle check point protein which mediates cycle arrest following DNA damage. However, the identification of SFN-interacting proteins suggests a possible role for SFN in the regulation of cell adhesion, polarity and migration [78]. SFN is frequently silenced by hypermethylation in human cancers, and its decreased expression has been associated with the metastasis of several human cancers [71, 79].
PSORS1C2 (also known as SPR1) is a component of the cross-linked envelope formed on the intracellular side of the cell membrane of terminally differentiated squamous epithelial cells. Reduced PSORS1C2 expression disrupts terminal differentiation and is associated with malignant transformation [80].
CLEC3B encodes a C-type lectin (tetranectin) which is located in the ECM and binds to plasminogen in the presence of plasminogen activators to generate an active protease (plasmin). Plasmin participates in ECM and basement membrane degradation/remodelling, processes key to invasion and metastasis. A reduced serum/plasma CLEC3B level is a biomarker for the metastasis of several human cancers [81].
The role of the transcription factor EGR1 in tumour development and progression is dependent upon the sum of the functions of the genes that it regulates, but it has been shown to up-regulate multiple tumour suppressor genes to inhibit cell growth, proliferation and metastasis [82]. In certain tumour types EGR1 represses transcription of heparanase, which degrades heparan sulphate proteoglycan chains present in the ECM and basement membranes allowing tumour cells to spread and inducing the release of pro-angiogenic chemokines and growth factors [83]. Increased expression of EGR1 in human non-small cell lung carcinomas is associated with up-regulation of KRT18 and reduced lymph node metastasis [84].
CD9 (motility-related protein-1), a member of the transmembrane 4 (tetraspanin) superfamily of cell surface proteins, interacts with intergrin cell adhesion molecules, signalling proteins, and immunoglobulin superfamily members promoting adherence to the ECM and suppressing motility [85]. Decreased expression of CD9 in several human tumours is associated with increased metastatic potential [86]. Canine mast cell tumours are often considered to be an analogue of human gastrointestinal stromal tumours (GISTs) because activating mutations in c-kit occur in both. CD9 expression is recognised as a prognostic marker for gastric GIST [87].
The only gene to show >two-fold increased expression in the MCTs was the non-coding small nucleolar RNA C/D box 93 (SNORD93). Small nucleolar RNAs (snoRNAs) guide sequence-specific post-transcriptional modification of rRNAs and small nuclear RNAs. However, a large proportion of snoRNAs are processed into smaller small nucleolar RNA-derived RNAs (sdRNAs), and a number of C/D box-derived sdRNAs have been shown to suppress gene expression in a manner analogous to microRNAs [88]. SNORD93 has been shown to display increased expression in a metastatic breast cancer cell line [89], and a sdRNA derived from SNORD93 was shown to promote human breast cancer cell invasiveness [90]. A second epigenetic regulator of tumour metastasis, stem-loop pre-microRNA cfa-mir-632, displays increased expression in the M MCTs (M/NM = 1.83). MiR-632 is expressed at high levels in invasive and metastatic human breast cancer cells, and has been shown to down-regulate expression of the heat shock protein DNAJB6 resulting in increased invasive capabilities [91]. MicroRNA-effected gene silencing has been shown to be pivotal in regulating cell adhesion [92].
Differentially expressed genes as targets for anti-metastasis therapeutics
Genes whose altered expression in M MCTs is pro-metastatic may constitute targets for anti-MCT metastasis therapeutics. By way of example, CD9 was the focus of a proof-of-principle study to assess the efficacy of a gene therapy approach to counter lung cancer metastasis. Adenoviral transduction of CD9 in an orthotopic lung cancer model was shown to significantly inhibit lymph node metastasis [93]. Particularly pertinent, is a potential new paradigm in anti-metastatics development that targets actin polymerisation and contractility [94], elements that are integral to both single cell and collective invasion modes of tumour cell migration [57]. Repeated reorganisation of the actin cytoskeleton and the formation of actin-based protrusions are integral to tumour cell migration strategies. In this context, the demonstration herein of the relevance of the altered expression of genes involved in the regulation of actin stress fibre assembly (EVL, BRAF, ARHGEF10 and NF2), and the linkage of the actin cytoskeleton to the plasma membrane (PMP22) and to nectin cell adhesion molecules at cell-cell junctions (AFDN), respectively, to MCT metastasis is significant. If metastasis-promoting down-regulation of gene expression in M MCTs is achieved at least in part though sdRNA/microRNA-effected suppression (as the data obtained in this study suggests), microRNA inhibitors represent a potential therapeutic option [95].
Metastasis-associated gene expression for potential classification of MCTs as metastasising or non-metastasising
The 19 genes that display >two-fold differences in expression between M and NM MCTs collectively represent a cross-breed metastasis-associated gene expression signature that could potentially be used to delineate M and NM MCTs through linear discriminant analysis. Preliminary evaluation, by cross-validation, estimated classification accuracies as 90–100% for M MCTs and 70–100% for NM MCTs. The differential expression of 9 of the 19 genes, including 2 whose identities are currently unconfirmed, was validated by RT-qPCR analysis. If the performance of the discriminator at delineating M from NM MCTs is subsequently validated through trial in further retrospective and prospective studies it would represent a uniquely objective and quantitative tool for predicting canine cutaneous MCT metastasis.
Where they are used, proliferation markers are typically deployed in combination with histological grading to predict the survival of dogs with mast cell tumour. Applying a cut-off score of 1.8, Ki-67 score is a significant predictor of survival of dogs with Patnaik grade II MCTs [11]. However, the effect of inter-operator variability in digital image capture and cell counting is unclear, and ‘poor survival’ may not be associated with metastatic disease.
A previous study sought to identify gene expression markers that are predictive of canine cutaneous MCT behaviour [23]. Gene expression in 13 Kiupel low grade tumours was compared with that in 5 Kiupel high grade tumours. Nearest shrunken centroid classification identified 13 genes that were capable of segregating MCTs into ‘differentiated’ and ‘undifferentiated’ MCT groups, although tumours from dogs that experienced MCT-related death were included in each group. In a subsequent study [96], the gene expression profiles of 40 ‘non-aggressive’ MCTs were compared with those of 7 ‘aggressive’ MCTs. However, aggressive MCTs were not selected due to evidence of distant and/or lymph node metastasis, but based on their histology, and because they were borne by dogs that received systemic treatment and survived for a certain unspecified period of time.
Unbalanced chromosomal abnormalities represent one of the mechanisms by which metastasis-associated changes in gene expression may be effected. Chromosomal grouping (on CFA1 and CFA31, respectively) of genes displaying decreased expression in the M MCTs may be indicative of focal deletions in the M MCTs and/or co-ordinated regulation of transcription. The potential for prognostically-relevant molecular classification of canine MCTs based upon copy number aberrations (CNAs) in MCTs has recently been investigated [97, 98]. CNAs were more frequent in tumours from 6 dogs that died within 6 months of diagnosis (although only 4 of the dogs had confirmed metastasis at diagnosis), and specific gene losses (PTEN and FAS; CFA26) and gains (MAPK3, WNT5B, FGF, FOXM1 and RAD51; CFA27) were associated with a shorter survival time [97]. One of two genes on CFA26 that showed decreased expression in the M MCTs in the present study is located in a ≥1.2Mb CFA26 fragment affected by loss in ~50% of the MCTs from dogs that died within 6 months of diagnosis [97]. A second CNA profiling study identified 4 CNAs that predicted ‘high risk MCTs’ with a sensitivity of 78–94% and specificity of 88–93% [98]. Loss of one copy of CFA5 was reported in ~50% of the ‘high risk MCTs’ [98], and in the present study 15 of 16 differentially expressed genes located on CFA5 show decreased expression in the metastasising MCTs. High risk MCTs were defined as those designated as a Kiupel high grade tumour [8] and/or containing an ITD in c-kit exon 11 [98]. However, since neither the binary grading system nor the presence of an ITD in c-kit exon predicts cutaneous MCT metastasis, the prognostic utility of the proposed 4 CNA-based classification is uncertain.
Limitations
This study featured FFPE biopsies of canine cutaneous MCTs that were surgically removed at first opinion veterinary practices from dogs that were subsequently referred to a specialist veterinary oncology centre. FFPE MCT biopsies were used because it is difficult to collect (in a referral setting) sufficient numbers of fresh (flash frozen or RNAlater-preserved) biopsies, whilst in a primary setting it is more difficult to collate definitive evidence of MCT tumour metastasis. However, global GEP of FFPE tissues using Affymetrix microarrays has been shown to yield biologically authentic and clinically-relevant data [99]. Further retrospective and prospective studies using new, larger cohorts of M and NM MCTs, optimally collected as fresh specimens, will be necessary to validate the capability of the 19-gene LDA classifier at predicting whether a canine cutaneous MCT is a metastasising or non-metastasising tumour. In vitro experimental investigations will ultimately be required to demonstrate if the effects (e.g. on cell adhesion, deformability and motility) on neoplastic canine mast cells of the differences in gene expression (between M and NM MCTs) observed in the current study are as is predicted based on what is known about the function(s) of the genes concerned and (in some cases) data from previous in vitro studies.
Conclusions
Changes in gene expression that mediate metastasis may be temporal, effected by similarly temporal epigenetic regulation, and/or may reflect somatic alterations that become advantageous or are newly acquired in migrating tumour cells. The differences in gene expression displayed by the primary cutaneous mast cell tumours that metastasised (relative to those that did not) appear to reflect the requirements of the initial ‘invasion phase’ of the metastatic cascade. Cell migration is facilitated by loss of cell polarity, reduced cell-cell and cell-ECM adhesion, and cell softening achieved through cytoskeletal reorganisation and disruption of cytoskeleton-plasma membrane links. De-regulation of cell proliferation, and suppression of cell cycle arrest and apoptosis, support invasion, whilst a pro-angiogenic reduction in EGR1 expression promotes intravasation. Some of the genes whose altered expression mediates canine cutaneous MCT metastasis may be potential targets for anti-MCT metastasis therapeutics. This will depend upon the mechanism by which gene expression is altered, and the specificity of the biological function(s) of the genes concerned. Measurement of the expression levels of the 19 genes that display greater than two-fold differences in expression between M and NM primary MCTs may have the potential to form the basis of a test that will predict with a high degree of certainty whether a cutaneous MCT will metastasise. If the performance of the metastasis gene expression signature-associated LDA classifier is validated on an independent MCT cohort it would represent the only test for canine cutaneous MCT metastasis and, as a single assay, an improvement on currently available prognostic indicators for canine cutaneous MCTs.
Supporting information
(PDF)
(A) MCTs labelled according to breed: B—Boxer; CB—Cross breed, CCR—Curly Coated Retriever, D—Dogue de Bordeaux, ETT—English Toy Terrier, GS- German Shepherd, HV—Hungarian Vizsla, LR—Labrador Retriever, MS—Miniature Schnauzer, SBT—Staffordshire Bull Terrier, W—Whippet. (B) MCTs labelled according to sex: Fe—Female, FeN—Neutered female, Ma—Male, MaN—Neutered male. (C) MCTs labelled according to age: Numbers indicate age in years.
(PDF)
(A) RNA integrity assay. (B) Assay for reverse transcription and/or PCR inhibitors. (C) RT-qPCR assays for quantitation of gene expression (optimal probe and primer concentrations, established experimentally, are listed).
(PDF)
(A) Metastasising MCTs. (B) Non-metastasising MCTs.
(PDF)
(A) Metastasising MCTs. (B) Non-metastasising MCTs.
(PDF)
(PDF)
(A) Proportion of genes expressed in MCTs that are located on each chromosome. (B) Positions of differentially expressed genes on chromosome 31.
(PDF)
Acknowledgments
We are indebted to the dog owners who gave us permission to use samples of biopsies of mast cell tumours removed from their dogs in the research study. We are also very grateful to the first opinion veterinary surgeons who contacted the owners of their patients on our behalf, and who provided follow-up clinical information.
Data Availability
The raw and processed microarray data has been deposited in the NCBI Gene Expression Omnibus repository (GEO series accession number GSE122590).
Funding Statement
Funding provided by UK Kennel Club (https://www.thekennelclub.org.uk/). JM is employed by and received salary from Idexx Laboratories. TW is employed by and received salary from Abbey Veterinary Services. RF and TE are employed by and received salary from Finn Pathologists. SB is employed by and received salary from Nationwide Laboratory Services. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Blackwood L, Murphy S, Buracco P, De Vos JP, De Fornel-Thibaud P, Hirschberger J, et al. European consensus document on mast cell tumours in dogs and cats. Vet Comp Oncol. 2012;10:e1–e29. 10.1111/j.1476-5829.2012.00341.x [DOI] [PubMed] [Google Scholar]
- 2.Dobson JM, Samuel S, Milstein H, Rogers K, Wood JL. Canine neoplasia in the UK: estimates of incidence rates from a population of insured dogs. J Small Anim Pract. 2002;43:240–246. [DOI] [PubMed] [Google Scholar]
- 3.Warland J, Dobson J. Breed predispositions in canine mast cell tumour: a single centre experience in the United Kingdom. Vet J. 2013;197:496–498. 10.1016/j.tvjl.2013.02.017 [DOI] [PubMed] [Google Scholar]
- 4.Thamm DH, Mauldin EA, Vail DM. Prednisone and vinblastine chemotherapy for canine mast cell tumor-41 cases (1992–1997). J Vet Intern Med. 1999;13:491–497. [DOI] [PubMed] [Google Scholar]
- 5.Davies DR, Wyatt KM, Jardine JE, Robertson ID, Irwin PJ. Vinblastine and prednisolone as adjunctive therapy for canine cutaneous mast cell tumors. J Am Anim Hosp Assoc. 2004;40:124–130. 10.5326/0400124 [DOI] [PubMed] [Google Scholar]
- 6.Patnaik AK, Ehler WJ, MacEwen EG. Canine cutaneous mast cell tumor: morphologic grading and survival time in 83 dogs. Vet Pathol. 1984;21:469–474. 10.1177/030098588402100503 [DOI] [PubMed] [Google Scholar]
- 7.Welle MM, Bley CR, Howard J, Rüfenacht S. Canine mast cell tumours: a review of the pathogenesis, clinical features, pathology and treatment. Vet Dermatol. 2008;19:321–339. 10.1111/j.1365-3164.2008.00694.x [DOI] [PubMed] [Google Scholar]
- 8.Kiupel M, Webster JD, Bailey KL, Best S, DeLay J, Detrisac CJ, et al. Proposal of a 2-tier histologic grading system for canine cutaneous mast cell tumors to more accurately predict biological behavior. Vet Pathol. 2011; 48:147–155. 10.1177/0300985810386469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stefanello D, Buracco P, Sabattini S, Finotello R, Giudice C, Grieco V, et al. Comparison of 2- and 3-category histologic grading systems for predicting the presence of metastasis at the time of initial evaluation in dogs with cutaneous mast cell tumors: 386 cases (2009–2014). J Am Vet Med Assoc. 2015;246:765–769. 10.2460/javma.246.7.765 [DOI] [PubMed] [Google Scholar]
- 10.Romansik EM, Reilly CM, Kass PH, Moore PF, London CA. Mitotic index is predictive for survival for canine cutaneous mast cell tumors. Vet Pathol. 2007;44:335–341. 10.1354/vp.44-3-335 [DOI] [PubMed] [Google Scholar]
- 11.Scase TJ, Edwards D, Miller J, Henley W, Smith K, Blunden A, et al. Canine mast cell tumors: correlation of apoptosis and proliferation markers with prognosis. J Vet Intern Med. 2006;20:151–158. [DOI] [PubMed] [Google Scholar]
- 12.Berlato D, Stewart J, Newton R, Maglennon GA, Monti P, Flindall A, et al. Evaluation of minichromosome maintenance protein 7 as a prognostic marker in canine cutaneous mast cell tumours. Vet Comp Oncol. 2012;10:135–142. 10.1111/j.1476-5829.2011.00287.x [DOI] [PubMed] [Google Scholar]
- 13.Taylor F, Murphy S, Hoather T, Dobson J, Scase T. TSLC1 tumour-suppressor gene expression in canine mast cell tumours. Vet Comp Oncol. 2010;8:263–272. 10.1111/j.1476-5829.2010.00220.x [DOI] [PubMed] [Google Scholar]
- 14.Simoes JP, Schoning P, Butine M. Prognosis of canine mast cell tumors: a comparison of three methods. Vet Pathol. 1994;31:637–647. 10.1177/030098589403100602 [DOI] [PubMed] [Google Scholar]
- 15.Krick EL, Kiupel M, Durham AC, Thaiwong T, Brown DC, Sorenmo KU. Investigating Associations Between Proliferation Indices, C-kit, and Lymph Node Stage in Canine Mast Cell Tumors. J Am Anim Hosp Assoc. 2017;53:258–264. 10.5326/JAAHA-MS-6265 [DOI] [PubMed] [Google Scholar]
- 16.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell 2011;147(2):275–292. 10.1016/j.cell.2011.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daves MH, Hilsenbeck SG, Ching C Lau CC, and Man T-K. Meta-analysis of multiple microarray datasets reveals a common gene signature of metastasis in solid tumors. BMC Medical Genomics 2011;doi.org/10.1186/1755-8794-4-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Francis P, Namløs HM, Müller C, Edén P, Fernebro J, Berner JM, et al. Diagnostic and prognostic gene expression signatures in 177 soft tissue sarcomas: hypoxia-induced transcription profile signifies metastatic potential. BMC Genomics 2007; 10.1186/1471-2164-8-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Onken MD, Worley LA, Tuscan MD, Harbour JW. An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. J Mol Diagn. 2010;12:461 10.2353/jmoldx.2010.090220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LaTulippe E, Satagopan J, Smith A, Scher H, Scardino P, Reuter V, et al. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–4506. [PubMed] [Google Scholar]
- 21.Ozturk S, Papageorgis P, Wong CK, Lambert AW, Abdolmaleky HM, Thiagalingam A, et al. SDPR functions as a metastasis suppressor in breast cancer by promoting apoptosis. Proc Natl Acad Sci USA. 2016;113:638-6Cancer Cell. 2003. June;3(6):537–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, et al. Genes that mediate breast cancer metastasis to the brain. Nature 2009;459:1005–1009. 10.1038/nature08021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giantin M, Granato A, Baratto C, Marconato L, Vascellari M, Morello EM, et al. Global gene expression analysis of canine cutaneous mast cell tumor: could molecular profiling be useful for subtype classification and prognostication? PLoS One 2014; 10.1371/journal.pone.0095481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krick EL, Billings AP, Shofer FS, Watanabe S, Sorenmo KU. Cytological lymph node evaluation in dogs with mast cell tumours: association with grade and survival. Vet Comp Oncol. 2009;7:130–138. 10.1111/j.1476-5829.2009.00185.x [DOI] [PubMed] [Google Scholar]
- 25.Das M, Chu LL, Ghahremani M, Abrams-Ogg T, Roy MS, Housman D, et al. Characterization of an abundant short interspersed nuclear element (SINE) present in Canis familiaris. Mamm Genome. 1998;9:64–69. [DOI] [PubMed] [Google Scholar]
- 26.QC Metrics for Exon and Gene Design Expression Arrays. A summary based on the Affymetrix Quality Assessment of Exon and Gene Arrays White Paper. Available from: http://static1.1.sqspcdn.com/static/f/1438485/21486054/1359060361517/qc_metrics_exon_gene_qrc.pdf.
- 27.Affymetrix Exon Array Background Correction Revision Date: 2005-09-27, Revision Version: 1.0. Available from: https://assets.thermofisher.com/TFS-Assets/LSG/brochures/exon_background_correction_whitepaper.pdf.
- 28.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 1998;95:14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Camargo A, Azuaje F, Wang H, Zheng H. Permutation—based statistical tests for multiple hypotheses. Source Code Biol Med. 2008; 10.1186/1751-0473-3-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8. Available from: https://david.ncifcrf.gov/. [PubMed]
- 31.Marullo M, Zuccato C, Mariotti C, Lahiri N, Tabrizi SJ, Di Donato S, et al. Expressed Alu repeats as a novel, reliable tool for normalization of real-time quantitative RT-PCR data. Genome Biol. 2010; 10.1186/gb-2010-11-1-r9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hellemans J, Mortier G, De Paepe A, Speleman F and Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biology 2007;doi.org/10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jong VL, Novianti PW, Roes KC, Eijkemans MJ. Selecting a classification function for class prediction with gene expression data. Bioinformatics 2016;32(12):1814–1822. 10.1093/bioinformatics/btw034 [DOI] [PubMed] [Google Scholar]
- 34.Venables WN, Ripley BD. Modern Applied Statistics with S. 4th ed Springer, New York; 2002. R Core Team (2014). [Google Scholar]
- 35.Page GP, Edwards JW, Gadbury GL, Yelisetti P, Wang J, Trivedi P, et al. The PowerAtlas: a power and sample size atlas for microarray experimental design and research. BMC Bioinformatics 2006; 10.1186/1471-2105-7-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/.
- 37.Ensembl Dog genome browser. http://www.ensembl.org/Canis_familiaris/Info/Index.
- 38.Ben-Dor A, Bruhn L, Friedman N, Nachman I, Schummer M, Yakhini Z. Tissue classification with gene expression profiles. J Comput Biol. 2000;7:559–583. 10.1089/106652700750050943 [DOI] [PubMed] [Google Scholar]
- 39.Shi L, Jones WD, Jensen RV, Harris SC, Perkins RG, Goodsaid FM, et al. The balance of reproducibility, sensitivity, and specificity of lists of differentially expressed genes in microarray studies. BMC Bioinformatics 2008;9 Suppl 9:S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zemke D, Yamini B, Yuzbasiyan-Gurkan V. Mutations in the juxtamembrane domain of c-KIT are associated with higher grade mast cell tumors in dogs. Vet Pathol. 2002;39:529–535. 10.1354/vp.39-5-529 [DOI] [PubMed] [Google Scholar]
- 41.Downing S, Chien MB, Kaas PH, Moore PE, London CA. Prevalence and importance of internal tandem duplications in exons 11 and 12 of c-KIT in mast cell tumors of dogs. Am J Vet Res 2002;63,1718–1723. [DOI] [PubMed] [Google Scholar]
- 42.Pagliarini RA, Xu T. A genetic screen in Drosophila for metastatic behavior. Science 2003;302:1227–1231. 10.1126/science.1088474 [DOI] [PubMed] [Google Scholar]
- 43.Sobolik-Delmaire T, Katafiasz D, Keim SA, Mahoney MG, Wahl JK 3rd. Decreased plakophilin-1 expression promotes increased motility in head and neck squamous cell carcinoma cells. Cell Commun Adhes. 2007;14:99–109. 10.1080/15419060701463082 [DOI] [PubMed] [Google Scholar]
- 44.Yang C, Ströbel P, Marx A, Hofmann I. Plakophilin-associated RNA-binding proteins in prostate cancer and their implications in tumor progression and metastasis. Virchows Arch. 2013;463:379–390. 10.1007/s00428-013-1452-y [DOI] [PubMed] [Google Scholar]
- 45.Depondt J, Shabana AH, Florescu-Zorila S, Gehanno P, Forest N. Down-regulation of desmosomal molecules in oral and pharyngeal squamous cell carcinomas as a marker for tumour growth and distant metastasis. Eur J Oral Sci. 1999;107:183–193. [DOI] [PubMed] [Google Scholar]
- 46.Chun MG, Hanahan D. Genetic deletion of the desmosomal component desmoplakin promotes tumor microinvasion in a mouse model of pancreatic neuroendocrine carcinogenesis. PLoS Genet. 2010; 10.1371/journal.pgen.1001120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang L, Chen Y, Cui T, Knösel T, Zhang Q, Albring KF, et al. Desmoplakin acts as a tumor suppressor by inhibition of the Wnt/β-catenin signaling pathway in human lung cancer. Carcinogenesis 2012;33:1863–1870. 10.1093/carcin/bgs226 [DOI] [PubMed] [Google Scholar]
- 48.Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36:1461–1473. 10.1038/onc.2016.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu H, Radisky DC, Yang D, Xu R, Radisky ES, Bissell MJ, et al. MYC suppresses cancer metastasis by direct transcriptional silencing of αv and β3 integrin subunits. Nat Cell Biol. 2012;14:567–574. 10.1038/ncb2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Connell MP, Fiori JL, Kershner EK, Frank BP, Indig FE, Taub DD, et al. Heparan sulfate proteoglycan modulation of Wnt5A signal transduction in metastatic melanoma cells. J Biol Chem. 2009;284:28704–28712. 10.1074/jbc.M109.028498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akl MR, Nagpal P, Ayoub NM, Prabhu SA, Gliksman M, Tai B, et al. Molecular and clinical profiles of syndecan-1 in solid and hematological cancer for prognosis and precision medicine. Oncotarget 2015;6:28693–28715. 10.18632/oncotarget.4981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mills GB, Moolenaar WH. The emerging role of lysophosphatidic acid in cancer. Nature Rev Cancer 2003;3:582–591. [DOI] [PubMed] [Google Scholar]
- 53.Gutiérrez ML, Corchete LA, Sarasquete ME, Del Mar Abad M, Bengoechea O, Fermiñán E, et al. Prognostic impact of a novel gene expression profile classifier for the discrimination between metastatic and non-metastatic primary colorectal cancer tumors. Oncotarget 2017;8:107685–107700. 10.18632/oncotarget.22591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Townson JL, Naumov GN, Chambers AF. The role of apoptosis in tumor progression and metastasis. Curr Mol Med. 2003;3:631–642. [DOI] [PubMed] [Google Scholar]
- 55.Syken J, De-Medina T, Münger K. TID1, a human homolog of the Drosophila tumor suppressor l(2)tid, encodes two mitochondrial modulators of apoptosis with opposing functions. Proc Natl Acad Sci USA 1999;96:8499–8504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang TH, Lin YH, Yang SC, Chang PC, Wang TC, Chen CY. Tid1-S regulates the mitochondrial localization of EGFR in non-small cell lung carcinoma. Oncogenesis 2017; 10.1038/oncsis.2017.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pandya P, Orgaz JL, Sanz-Moreno V. Modes of invasion during tumour dissemination. Mol Oncol. 2017;11:5–27. 10.1002/1878-0261.12019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roux KJ, Amici SA, Fletcher BS, Notterpek L. Modulation of epithelial morphology, monolayer permeability, and cell migration by growth arrest specific 3/peripheral myelin protein 22. Mol Biol Cell. 2005;16:1142–1151. 10.1091/mbc.E04-07-0551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson HL, Wilson SA, Surprenant A, North RA. Epithelial membrane proteins induce membrane blebbing and interact with the P2X7 receptor C terminus. J Biol Chem. 2002;277:34017–34023. 10.1074/jbc.M205120200 [DOI] [PubMed] [Google Scholar]
- 60.Mouneimne G, Hansen SD, Selfors LM, Petrak L, Hickey MM, Gallegos LL, et al. Differential remodeling of actin cytoskeleton architecture by profilin isoforms leads to distinct effects on cell migration and invasion. Cancer Cell. 2012;22:615–630. 10.1016/j.ccr.2012.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pritchard CA, Hayes L, Wojnowski L, Zimmer A, Marais RM, Norman JC. B-Raf acts via the ROCKII/LIMK/cofilin pathway to maintain actin stress fibers in fibroblasts. Mol Cell Biol. 2004;24:5937–5952. 10.1128/MCB.24.13.5937-5952.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fife CM, McCarroll JA, Kavallaris M. Movers and shakers: cell cytoskeleton in cancer metastasis. Br J Pharmacol. 2014;171:5507–5523. 10.1111/bph.12704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Debily MA, Camarca A, Ciullo M, Mayer C, El Marhomy S, Ba I, et al. Expression and molecular characterization of alternative transcripts of the ARHGEF5/TIM oncogene specific for human breast cancer. Hum Mol Genet. 2004;13:323–334. 10.1093/hmg/ddh024 [DOI] [PubMed] [Google Scholar]
- 64.Hartmann M, Parra LM, Ruschel A, Böhme S, Li Y, Morrison H, et al. Tumor Suppressor NF2 Blocks Cellular Migration by Inhibiting Ectodomain Cleavage of CD44. Mol Cancer Res. 2015;13:879–890. 10.1158/1541-7786.MCR-15-0020-T [DOI] [PubMed] [Google Scholar]
- 65.Shaulian E. AP-1-The Jun proteins: Oncogenes or tumor suppressors in disguise? Cell Signal. 2010;22:894–899. 10.1016/j.cellsig.2009.12.008 [DOI] [PubMed] [Google Scholar]
- 66.Jin JY, Ke H, Hall RP, Zhang JY. c-Jun promotes whereas JunB inhibits epidermal neoplasia. J Invest Dermatol. 2011;131:1149–118. 10.1038/jid.2011.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moll R, Divo M, Langbein L. The human keratins: biology and pathology. Histochem Cell Biol. 2008;129:705–33. 10.1007/s00418-008-0435-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Traweek ST, Liu J, Battifora H. Keratin gene expression in non-epithelial tissues. Detection with polymerase chain reaction. Am J Pathol. 1993;142:1111–1118. [PMC free article] [PubMed] [Google Scholar]
- 69.López-Farré AJ, Zamorano-León JJ, Segura A, Mateos-Cáceres PJ, Modrego J, Rodríguez-Sierra P, et al. Plasma desmoplakin I biomarker of vascular recurrence after ischemic stroke. J Neurochem. 2012;121:314–325. 10.1111/j.1471-4159.2012.07683.x [DOI] [PubMed] [Google Scholar]
- 70.Rolli CG, Seufferlein T, Kemkemer R, Spatz JP. Impact of tumor cell cytoskeleton organization on invasiveness and migration: a microchannel-based approach. PLoS One 2010; 10.1371/journal.pone.0008726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang S, Fan W, Wan B, Tu M, Jin F, Liu F, et al. Characterization of long noncoding RNA and messenger RNA signatures in melanoma tumorigenesis and metastasis. PLoS One 2017; 10.1371/journal.pone.0172498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer 2009;9:239–252. 10.1038/nrc2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Golan T, Messer AR, Amitai-Lange A, Melamed Z, Ohana R, Bell RE, et al. Interactions of Melanoma Cells with Distal Keratinocytes Trigger Metastasis via Notch Signaling Inhibition of MITF. Mol Cell. 2015;59:664–676. 10.1016/j.molcel.2015.06.028 [DOI] [PubMed] [Google Scholar]
- 74.Attardi LD. The role of p53-mediated apoptosis as a crucial anti-tumor response to genomic instability: lessons from mouse models. Mutat Res. 2005;569:145–157. 10.1016/j.mrfmmm.2004.04.019 [DOI] [PubMed] [Google Scholar]
- 75.Davies L, Gray D, Spiller D, White MR, Damato B, Grierson I, et al. P53 apoptosis mediator PERP: localization, function and caspase activation in uveal melanoma. J Cell Mol Med. 2009;13:1995–2007. 10.1111/j.1582-4934.2008.00590.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fenger JM, Bear MD, Volinia S, Lin TY, Harrington BK, London CA, et al. Overexpression of miR-9 in mast cells is associated with invasive behavior and spontaneous metastasis. BMC Cancer 2014;14:84 10.1186/1471-2407-14-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu J, Wu G, Li Q, Gong H, Song J, Cao L, et al. Overexpression of Suprabasin is Associated with Proliferation and Tumorigenicity of Esophageal Squamous Cell Carcinoma. Sci Rep. 2016; 10.1038/srep21549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Benzinger A, Muster N, Koch HB, Yates JR 3rd, Hermeking H. Targeted proteomic analysis of 14-3-3 sigma, a p53 effector commonly silenced in cancer. Mol Cell Proteomics. 2005;4:785–795. 10.1074/mcp.M500021-MCP200 [DOI] [PubMed] [Google Scholar]
- 79.Li DJ, Deng G, Xiao ZQ, Yao HX, Li C, Peng F, et al. Identificating 14-3-3 sigma as a lymph node metastasis-related protein in human lung squamous carcinoma. Cancer Lett. 2009;279:65–73. 10.1016/j.canlet.2009.01.028 [DOI] [PubMed] [Google Scholar]
- 80.Hu R, Wu R, Deng J, Lau D. A small proline-rich protein, spr1: specific marker for squamous lung carcinoma. Lung Cancer. 1998;20:25–30. [DOI] [PubMed] [Google Scholar]
- 81.Jensen BA, Clemmensen I. Plasma tetranectin is reduced in cancer and related to metastasia. Cancer 1988;62:869–872. [DOI] [PubMed] [Google Scholar]
- 82.Boone DN, Qi Y, Li Z, Hann SR. Egr1 mediates p53-independent c-Myc-induced apoptosis via a noncanonical ARF-dependent transcriptional mechanism. Proc Natl Acad Sci USA 2011;108:632–637. 10.1073/pnas.1008848108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vlodavsky I, Korner G, Ishai-Michaeli R, Bashkin P, Bar-Shavit R, Fuks Z. Extracellular matrix-resident growth factors and enzymes: possible involvement in tumor metastasis and angiogenesis. Cancer Metastasis Rev. 1990;9:203–226. [DOI] [PubMed] [Google Scholar]
- 84.Zhang H, Chen X, Wang J, Guang W, Han W, Zhang H, et al. EGR1 decreases the malignancy of human non-small cell lung carcinoma by regulating KRT18 expression. Sci Rep. 2014; 10.1038/srep05416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zöller M. Tetraspanins: push and pull in suppressing and promoting metastasis. Nat Rev Cancer 2009;9:40–55. 10.1038/nrc2543 [DOI] [PubMed] [Google Scholar]
- 86.Miyake M, Nakano K, Ieki Y, Adachi M, Huang CL, Itoi S, et al. Motility related protein 1 (MRP-1/CD9) expression: inverse correlation with metastases in breast cancer. Cancer Res. 1995;55:4127–4131. [PubMed] [Google Scholar]
- 87.Setoguchi T, Kikuchi H, Yamamoto M, Baba M, Ohta M, Kamiya K, et al. Microarray analysis identifies versican and CD9 as potent prognostic markers in gastric gastrointestinal stromal tumors. Cancer Sci. 2011;102:883–889. 10.1111/j.1349-7006.2011.01872.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brameier M, Herwig A, Reinhardt R, Walter L, Gruber J. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res. 2011;39:675–686. 10.1093/nar/gkq776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ma R, Zhao H, Patterson D, Borchert G, Xi Y. Oncogenic role of snoRD93 in breast cancer cells. Mol Cancer Res. 2016,14; 10.1158/1557-3125 [DOI] [Google Scholar]
- 90.Patterson DG, Roberts JT, King VM, Houserova D, Barnhill EC, Crucello A, et al. Human snoRNA-93 is processed into a microRNA-like RNA that promotes breast cancer cell invasion. NPJ Breast Cancer 2017; 10.1038/s41523-017-0032-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mitra A, Rostas JW, Dyess DL, Shevde LA, Samant RS. Micro-RNA-632 downregulates DNAJB6 in breast cancer. Lab Invest. 2012;92:1310–1317. 10.1038/labinvest.2012.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Valastyan S, Weinberg RA. Roles for microRNAs in the regulation of cell adhesion molecules. J Cell Sci. 2011;124:999–1006. 10.1242/jcs.081513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Takeda T, Hattori N, Tokuhara T, Nishimura Y, Yokoyama M, Miyake M. Adenoviral transduction of MRP-1/CD9 and KAI1/CD82 inhibits lymph node metastasis in orthotopic lung cancer model. Cancer Res. 2007;67:1744–1749. 10.1158/0008-5472.CAN-06-3090 [DOI] [PubMed] [Google Scholar]
- 94.Gandalovicová A, Rosel D, Fernandes M, Veselý P, Heneberg P, Cermák V, et al. Migrastatics-Anti-metastatic and Anti-invasion Drugs: Promises and Challenges. Trends Cancer 2017;3:391–406. 10.1016/j.trecan.2017.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. 10.1038/nrd.2016.246 [DOI] [PubMed] [Google Scholar]
- 96.Giantin M, Baratto C, Marconato L, Vascellari M, Mutinelli F, Dacasto M, et al. Transcriptomic analysis identified up-regulation of a solute carrier transporter and UDP glucuronosyltransferases in dogs with aggressive cutaneous mast cell tumours. Vet J. 2016;212:36–43. 10.1016/j.tvjl.2016.03.024 [DOI] [PubMed] [Google Scholar]
- 97.Jark PC, Mundin DB, de Carvalho M, Ferioli RB, Anai LA, Marchi FA, et al. Genomic copy number variation associated with clinical outcome in canine cutaneous mast cell tumors. Res Vet Sci. 2017;111:26–30. 10.1016/j.rvsc.2016.11.009 [DOI] [PubMed] [Google Scholar]
- 98.Mochizuki H, Thomas R, Moroff S, Breen M. Genomic profiling of canine mast cell tumors identifies DNA copy number aberrations associated with KIT mutations and high histological grade. Chromosome Res. 2017;25:129–143. 10.1007/s10577-016-9543-7 [DOI] [PubMed] [Google Scholar]
- 99.Linton KM, Hey Y, Saunders E, Jeziorska M, Denton J, Wilson CL, et al. Acquisition of biologically relevant gene expression data by Affymetrix microarray analysis of archival formalin-fixed paraffin-embedded tumours. Br J Cancer 2008;98:1403–1414. 10.1038/sj.bjc.6604316 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(A) MCTs labelled according to breed: B—Boxer; CB—Cross breed, CCR—Curly Coated Retriever, D—Dogue de Bordeaux, ETT—English Toy Terrier, GS- German Shepherd, HV—Hungarian Vizsla, LR—Labrador Retriever, MS—Miniature Schnauzer, SBT—Staffordshire Bull Terrier, W—Whippet. (B) MCTs labelled according to sex: Fe—Female, FeN—Neutered female, Ma—Male, MaN—Neutered male. (C) MCTs labelled according to age: Numbers indicate age in years.
(PDF)
(A) RNA integrity assay. (B) Assay for reverse transcription and/or PCR inhibitors. (C) RT-qPCR assays for quantitation of gene expression (optimal probe and primer concentrations, established experimentally, are listed).
(PDF)
(A) Metastasising MCTs. (B) Non-metastasising MCTs.
(PDF)
(A) Metastasising MCTs. (B) Non-metastasising MCTs.
(PDF)
(PDF)
(A) Proportion of genes expressed in MCTs that are located on each chromosome. (B) Positions of differentially expressed genes on chromosome 31.
(PDF)
Data Availability Statement
The raw and processed microarray data has been deposited in the NCBI Gene Expression Omnibus repository (GEO series accession number GSE122590).