ABSTRACT
Pulmonary adenoid cystic carcinoma is a rare and indolent lung malignancy, characterized by a protracted but unpredictable growth behavior. Currently, the treatment of PACC relies on surgery and local radiotherapy. However, treatment options for advanced PACC patients are limited. A larger number of studies demonstrated that advanced PACC patients obtained limited benefit from chemotherapy. Moreover, only a few case reports revealed PACC patients were candidates for target therapy. Therefore, there is an urgent need to develop novel therapies. Due to its rareness, its mutational landscape remains largely elusive. In this study, we performed capture-based ultra-deep sequencing on multiregional surgical specimens obtained from 8 PACC patients using a panel consisting of 295 cancer-related genes. Our data revealed distinctive mutational spectrum of PACC, which differed from non-small cell lung cancer and adenoid cystic carcinomas originated from other anatomical sites. PACC, lacking mutations in a majority of non-small cell lung cancer driver genes, has frequent mutations in genes participating in chromatin remodeling and NOTCH signaling pathway. We also elucidated spatial intra-tumoral heterogeneity, which varied among cases. Most mutations in chromatin remodelers were subclonal. Collectively, our findings elucidated molecular signature associated with PACC and highlighted the potential for epigenetic therapy in this disease.
Keywords: pulmonary adenoid cystic carcinoma, mutational landscape, clonal diversity, chromatin remodeling, NOTCH signaling, epigenetic therapy, Chinese
Introduction
Adenoid cystic carcinoma (ACC), a rare form of adenocarcinoma, primarily arises from the head and neck region, including salivary gland, oropharynx, nasopharynx and paranasal sinuses but can also originate from other anatomical sites such as the lung and breasts.1,2 Pulmonary adenoid cystic carcinoma (PACC), accounting for 0.1–0.5% of primary lung cancers with a 5-year disease-free survival of 50%, shares similar histomorphological features with ACC originated from other sites.3–5 The median age at diagnosis reported by 3 independent studies ranged from 46 to 54, which is younger than the median age reported for other primary lung malignancies.5–7 It is a low-grade tumor characterized by a protracted but unpredictable growth behavior.8,9 Treatment options are limited to surgery and radiation. No effective systemic therapy is available.10,11 Only a few case series or case reports demonstrated efficacies of chemotherapy and targeted therapy, such as EGFR-TKI and ALK-TKI.7,12–14 Surgical resection is the primary treatment for localized disease. However, complete resection is difficult in a significant number of cases due to the central location of the tumor. Radiotherapy is the treatment of choice for partially resected or unresectable tumors.15 Recurrence after complete surgical resection and eventual metastasis are common. The lack of efficacy of chemotherapy agents has been demonstrated by a number of studies,11 thus prompting interests in elucidating new drug targets and developing new regiment.
Due to its rareness, comprehensive genomic profiling of PACC has not been done. Only very few studies have investigated genetic aberrations of PACC, most of them from a single gene or a small panel of genes perspectives.16,17 Huo et al attempted to identify the status of a few major lung cancer-related genes in PACC including KIT, EGFR, BRAF, HRAS, KRAS, PIK3CA, PDGFRA and PTEN and no mutation was detected in any of the above 7 genes.18 Another study utilized immunohistochemistry (IHC), fluorescence in situ hybridization (FISH) and polymerase chain reaction (PCR) to evaluate the mutational status and copy number status of EGFR. This study revealed no EGFR amplification and mutation in exon 18–21 was detected.16 No study has interrogated the mutation spectrum of PACC from a global perspective.
A patient’s tumor is comprised of numerous generically and phenotypically altered competing clones, due to clonal evolution.19 Advances in sequencing techniques have enabled clonal evolution analysis, which revealed intra-tumoral heterogeneity in a number of human malignancies, but not in PACC.20–23 Study investigating localized lung adenocarcinoma revealed limited intra-tumoral heterogeneity, suggesting single biopsy at an appropriate depth is sufficient to reveal a majority of mutations.22 In this study, we investigated multiregional surgical resection samples obtained from 8 patients with localized PACC to interrogate its molecular landscape and intra-tumoral heterogeneity using a panel consisting of 295 cancer-related genes.
Results
Patient characteristics
Of the 8 patients with localized PACC, 4 of them are females; the remaining are males with a median age at diagnosis of 41, ranging from 23 to 62. Seven patients are non-smokers and 1 patient is a current smoker. Six patients had tumor located in the left lung and the remaining two had tumor located in the right lung. All patients received complete resection. The distribution of stage ranged from IB to IIIA and tumor size ranged from 1.2cm to 4.5cm.
Mutation spectrum
In order to interrogate the mutation profile of PACC, we performed capture-based targeted sequencing on multiregional surgical samples of 8 treatment-naïve PACC patients using a panel consisting of 295 cancer-related genes at an average depth of 960x. Multiregional samples were obtained at different regions of the same tumor. We obtained 3 or 4 samples from each patient. Overall, 109 genetic aberrations were identified in tumor tissues, spanning 38 genes from 29 samples, including 70 SNVs and 39 insertions or deletions (INDELs). On average, 4 somatic mutations were identified per sample, corresponding to 5 mutations per megabase, which is lower comparing to NSCLC. We observed substantial inter-tumoral heterogeneity. Fifty-Six mutations were only observed in single patient. The most commonly seen mutations were KAT6A, which occurred in 4/8 patients (Figure 1). KAT6A, a histone acetyltransferase, belongs to a complex which acetylates lysine 9 of histone 3. Chromosomal translocation of this gene has been associated with acute myeloid leukemia.24 Second most frequent occurring mutations included: KMT2D and TET2, which occurred in 3/8 patients. KMT2D is a major mammalian histone H3 lysine 4 mono-methyltransferase. TET2, a DNA demethylase, can also act as a tumor suppressor, whose mutations are commonly seen in hematopoietic malignancies.25 Interestingly, the most frequently mutated genes are epigenetic regulators. It is also worth pointing out that no PACC patients carried mutations in classic driver genes for NSCLC.
Figure 1.
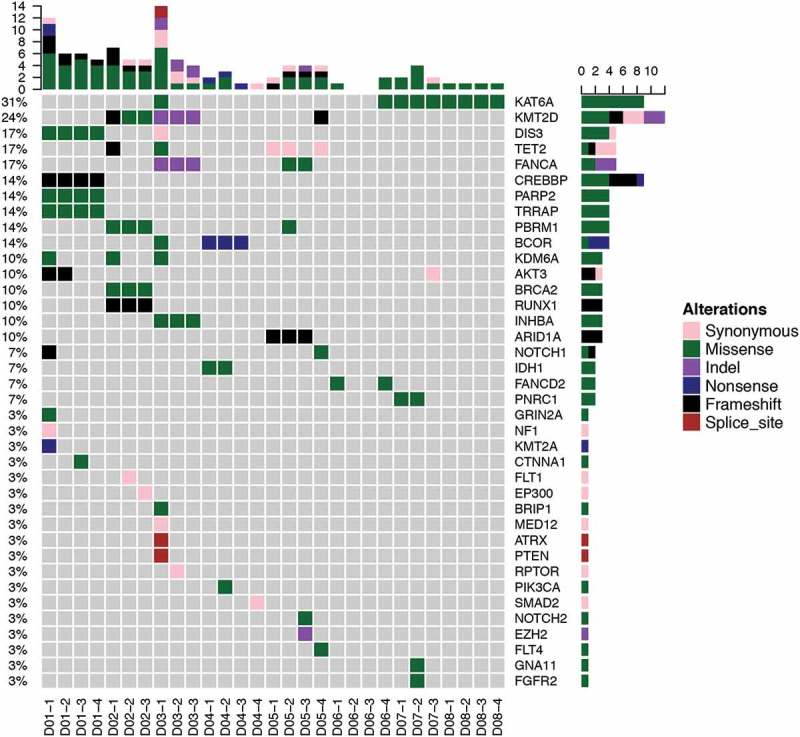
Mutation spectrum. Mutations detected from every sample of PACC patients. Different colors represent different types of mutations. Top bars represent total number of mutations a sample carries; side bars total number of samples with mutation in that particular gene.
PACC has a unique mutational landscape
To further interrogate the molecular profile of PACC, we compared and contrasted the mutational profile of PACC with NSCLC and ACC originated from other anatomical sites. We retrospectively profiled 567 NSCLC samples using the same panel from our archive between January 2013 and December 2017. PACC demonstrated a distinct mutational profile, lacking classic driver mutations associated with NSCLC. In contrast, PACC had frequent mutations in genes participating in chromatin modifications, such as KAT6A, KDM6A, KMT2D, TET2, EP300 and CREBBP (Figure 2). In addition to epigenetic regulators, PACC patients also carried frequent mutations in DIS3, NOTCH1, AKT3, PBRM1, BCOR and FANCA. Next, we compared its mutational profile with salivary gland adenoid cystic carcinoma and breast adenoid cystic carcinoma. Mutational profiles of ACC originated from salivary gland and breasts were retrieved from cBioPortal. Our data revealed limited similarity shared by PACC and ACC originated from other sites. ACC and breast ACC had frequent mutations in NFIB and MYB (Figure 2). Literatures have reported approximately 50% of ACC harbor t(6, 9) translocations, resulting in MYB-NFIB fusion.26,27 In contrast, in our cohort, we observed frequent mutations in epigenetic regulators. Collectively, our data revealed a distinct mutational profile of PACC, which shared limited mutations with NSCLC and ACC originated from salivary gland and breasts.
Figure 2.
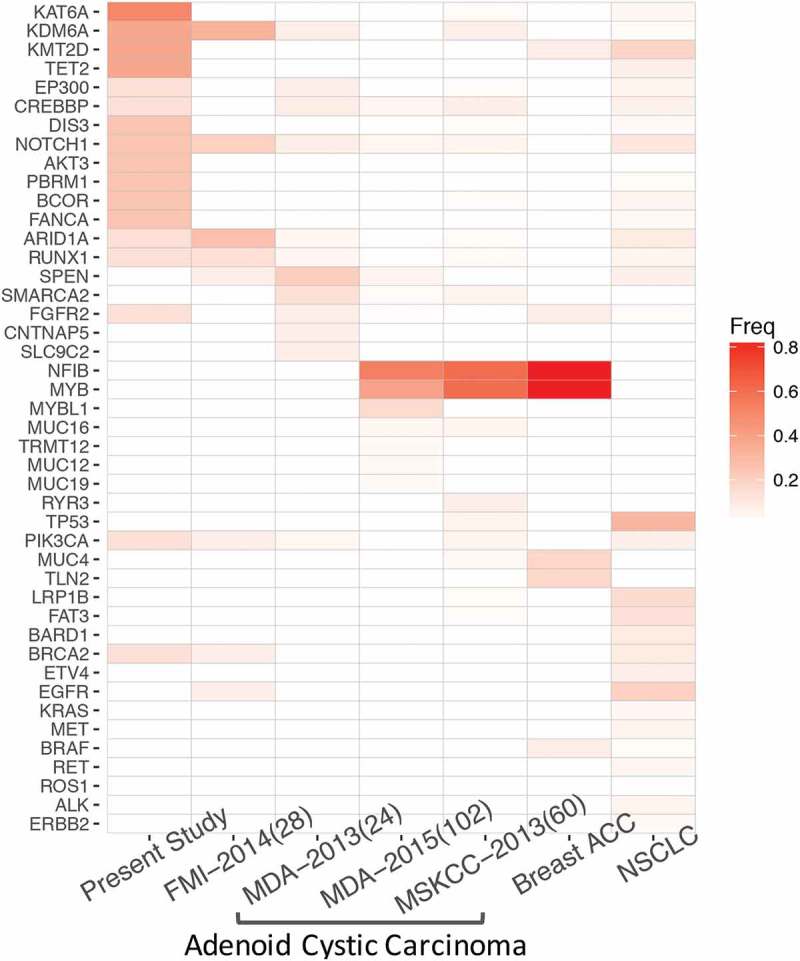
A comparison of mutation spectrum of PACC, ACC and NSCLC. Color gradient represents allelic fraction.
Clonal diversity
Clonal diversity, resulting in variable treatment responses, has been one of the major challenges in treating cancer.28 Comprehensive analysis of multiregional samples aids in understanding intra-tumoral heterogeneity. We obtained 3–4 spatially separated samples from each patient. The number of mutations varied among patients, but no significant associations were derived between total number of mutations and age (p = 0.860), gender (p = 0.886) or tumor size (p = 0.945). Phylogenetic relationship of tumor regions of 6 patient was depicted in Figure 3, where trunk and branches representing ubiquitous and heterogeneous mutations, respectively. Patients D06 and D08 only had 2 and 1 mutation respectively; therefore, no phylogenetic tree was constructed. Intra-tumoral heterogeneity was evident in each tumor studied. Patient D01 demonstrated significant intra-tumoral heterogeneity. Sample D01-1, showed a very different mutational profile, which harbored 10 mutations whereas the other 3 samples obtained from the same tumor harbored 4 or 5 mutations. In this patient, we identified 4 trunk mutations which were shared by all 4 samples, including CREBBP, PARP2, TRRAP and DIS3. In addition, we also identified 1 branch mutations, AKT3 and 6 private mutations that are only carried by a single sample. Interestingly, in patient D04 and D05, no trunk mutation was observed. There is a significant diversity in trunk mutations among the 8 patients. Only KMT2D was identified in more than 1 patient; all other trunk mutations were unique to individual patient. Collectively, our data demonstrate significant intra-tumoral heterogeneity in PACC. Furthermore, classic NSCLC driver mutations29–33 do not drive tumor development in this subset of lung carcinoma.
Figure 3.
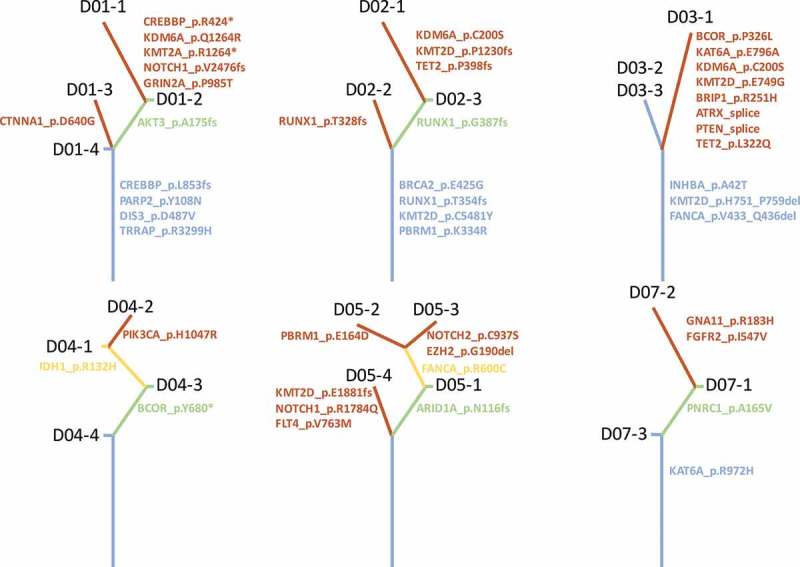
Assessment of intra-tumoral heterogeneity by multiregional sequencing. Each tree represents a patient. Blue, green and brown represent trunk, branch and private mutations, respectively.
Discussion
Due to itc targeted sequencing and identified a few recurring mutations. The most commonly seen mutation was KAT6A, followed by KMT2D and TET2, which play vital roles in chromatin modifications. Their mutations have been frequently identified in both hematological malignancies and solid tumors.25 Interestingly, mutations in TET2 also accumulate with age in otherwise healthy subjects.34 In our cohort, 3 patients carried TET2 mutations, 2 of them were over 60. Their TET2 mutations can potentially be a consequence of aging. PACC demonstrated significant inter-tumoral heterogeneity, evident by approximately 70% of mutations only occurred in single patient.
Furthermore, PACC has a unique genomic profile comparing to NSCLC and other adenoid cystic carcinomas. Common drivers of NSCLC are uncommon events in PACC, in an agreement with a previous study. EGFR, ERBB2, BRAF and ROS1 mutations were not observed in PACC. In contrast, PACC has frequent mutations in NOTCH signaling pathway and epigenetic regulators, most of them participating in catalyzing post-transcriptional modification of histones such as the histone H3K4 methyltransferases: KMT2A/D, histone acetylase (CREBBP and EP300), and the H3K27 methyltransferase EZH2. Many of these mutations are early events (trunk mutations) in disease progression, suggesting they are good therapeutic targets. Patients with mutations in CREBBP or EB300 may benefit from histone deacetylase inhibitors (HDCAi). Efficacies of multiple HDACi have been shown in hematological malignancies. Furthermore, EZH2 inhibitors have also been tested and shown promising results in hematological malignancies.29 Furthermore, the mutational profile of PACC also differs significantly from other ACCs, which has a signature t(6;9) (q22-23; p23-24) chromosomal translocation, occurring in approximately 80% of ACC. Such fusion, resulting in MYB-NFIB fusion and subsequent activation of MYB targets transcription, was not observed in PACC.
Taken together, our study revealed distinctive mutational spectrum of PACC, which differs from NSCLC and ACC originated from other anatomical sites. We also elucidated substantial spatial intra-tumoral heterogeneity.
Materials and methods
Patients and sample preparation
Multiple tissue samples were collected from each patient. Tumor specimens were harvested from primary PACC tumors of patients who underwent surgery at the Xiangya Hospital between January 2013 and December 2017 and reviewed by two independent pathologists. This study was approved by the Institutional Review Board (IRB) of Xiangya Hospital. Written informed content was obtained from every patient.
Tissue DNA extraction
DNA was extracted using QIAamp DNA FFPE tissue kit (Qiagen) according to manufacturer’s instructions. DNA concentration was measured using Qubit dsDNA assay.
NGS library preparation
DNA fragmentation was performed using Covaris M220, followed by end repair, phosphorylation and adaptor ligation. Fragments of size 200–400bp were selected by AMPure beads (Agencourt AMPure XP Kit, Beckman Coulter, California, US) followed by hybridization with capture probes baits, hybrid selection with magnetic beads and PCR amplification. Subsequently, high-sensitivity DNA assay was performed to assess the quality and the size of all fragments.
Capture-based targeted DNA sequencing
Genetic profiles of all tissue samples were assessed by performing capture-based targeted deep sequencing using the OncoScreen panel (Burning Rock Biotech Ltd.), covering 2.02MB of human genomic regions, including all exons and critical introns of 295 genes. DNA quality and size were assessed by high sensitivity DNA assay using a bioanalyzer. All indexed samples were sequenced on a NextSeq 500 (Illumina, Inc., USA) with pair-end reads.
Sequencing data analysis
The sequencing data in the FASTQ format were mapped to the human genome (hg19) using BWA aligner 0.7.10. Local alignment optimization, variant calling and annotation were performed using GATK 3.2, MuTect, and VarScan, respectively. DNA translocation analysis was performed using both Tophat2 and Factera 1.4.3. Gene-level copy number variation was assessed using a t statistic after normalizing reads depth at each region by total reads number and region size, and correcting GC-bias using a LOESS algorithm.
Funding Statement
This investigation was supported by Natural Science Foundation of Hunan Province (NO. 2016JJ3162) and Health Department Foundation of Hunan Province (NO. C2016057).
References
- 1.Bell D, Hanna EY.. Salivary gland cancers: biology and molecular targets for therapy. Curr Oncol Rep. 2012;14(2):166–174. doi: 10.1007/s11912-012-0220-5. [DOI] [PubMed] [Google Scholar]
- 2.Gondivkar SM, Gadbail AR, Chole R, Parikh RV. Adenoid cystic carcinoma: a rare clinical entity and literature review. Oral Oncol. 2011;47(4):231–236. doi: 10.1016/j.oraloncology.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 3.Zhu F, Liu Z, Hou Y, He D, Ge X, Bai C, Jiang L, Li S. Primary salivary gland-type lung cancer: clinicopathological analysis of 88 cases from China. J Thorac Oncol. 2013;8(12):1578–1584. doi: 10.1097/JTO.0b013e3182a7d272. [DOI] [PubMed] [Google Scholar]
- 4.Roden AC, Greipp PT, Knutson DL, Kloft-Nelson SM, Jenkins SM, Marks RS, Aubry MC, García JJ. Histopathologic and cytogenetic features of pulmonary adenoid cystic carcinoma. J Thorac Oncol. 2015;10(11):1570–1575. doi: 10.1097/JTO.0000000000000656. [DOI] [PubMed] [Google Scholar]
- 5.Molina JR, Aubry MC, Lewis JE, Wampfler JA, Williams BA, Midthun DE, Yang P, Cassivi SD. Primary salivary gland-type lung cancer: spectrum of clinical presentation, histopathologic and prognostic factors. Cancer. 2007;110(10):2253–2259. doi: 10.1002/cncr.23048. [DOI] [PubMed] [Google Scholar]
- 6.Hu MM, Hu Y, He JB, Li BL. Primary adenoid cystic carcinoma of the lung: clinicopathological features, treatment and results. Oncol Lett. 2015;9(3):1475–1481. doi: 10.3892/ol.2015.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qing S, Zhou K, Liu X, Li X, Deng F, Ma Y. Primary pulmonary adenoid cystic carcinoma: clinicopathological analyses of 12 cases. Int J Clin Exp Pathol. 2015;8(6):7619–7626. [PMC free article] [PubMed] [Google Scholar]
- 8.Pandey D, Garg PK, Jakhetiya A, Pandey R, Bhoriwal S, Nath D, Kumar S. Surgical experience of primary salivary gland tumors of lung: A case series. Int J Surg. 2015;21:92–96. doi: 10.1016/j.ijsu.2015.06.084. [DOI] [PubMed] [Google Scholar]
- 9.Bennett AK, Mills SE, Wick MR. Salivary-type neoplasms of the breast and lung. Semin Diagn Pathol. 2003;20(4):279–304. [DOI] [PubMed] [Google Scholar]
- 10.Kanematsu T, Yohena T, Uehara T, Ushijima C, Asoh H, Yoshino I, Ichinose Y. Treatment outcome of resected and nonresected primary adenoid cystic carcinoma of the lung. Ann Thorac Cardiovasc Surg. 2002;8(2):74–77. [PubMed] [Google Scholar]
- 11.Dodd RL, Slevin NJ. Salivary gland adenoid cystic carcinoma: a review of chemotherapy and molecular therapies. Oral Oncol. 2006;42(8):759–769. doi: 10.1016/j.oraloncology.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Song Z, Wu W, Zhang Y. Effective treatment with icotinib in primary adenoid cystic carcinoma of the lung with liver metastasis. J Thorac Oncol. 2014;9(9):e67–69. doi: 10.1097/JTO.0000000000000247. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Hau E, Links M, Graham PH. Adenoid cystic carcinoma of the lung: response to tamoxifen after chemoradiation. Asia Pac J Clin Oncol. 2016;12(2):e352–e355. doi: 10.1111/ajco.12184. [DOI] [PubMed] [Google Scholar]
- 14.Shadaba A, Gaze MN, Grant HR. The response of adenoid cystic carcinoma to tamoxifen. J Laryngol Otol. 1997;111(12):1186–1189. [DOI] [PubMed] [Google Scholar]
- 15.Kang DY, Yoon YS, Kim HK, Choi YS, Kim K, Shim YM, Kim J. Primary salivary gland-type lung cancer: surgical outcomes. Lung Cancer. 2011;72(2):250–254. doi: 10.1016/j.lungcan.2010.08.021. [DOI] [PubMed] [Google Scholar]
- 16.Macarenco RS, Uphoff TS, Gilmer HF, Jenkins RB, Thibodeau SN, Lewis JE, Molina JR, Yang P, Aubry M-C. Salivary gland-type lung carcinomas: an EGFR immunohistochemical, molecular genetic, and mutational analysis study. Mod Pathol. 2008;21(9):1168–1175. doi: 10.1038/modpathol.2008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aubry MC, Heinrich MC, Molina J, Lewis JE, Yang P, Cassivi SD, Corless CL. Primary adenoid cystic carcinoma of the lung: absence of KIT mutations. Cancer. 2007;110(11):2507–2510. doi: 10.1002/cncr.23075. [DOI] [PubMed] [Google Scholar]
- 18.Huo Z, Wu H, Li S, Liang Z. Molecular genetic studies on EGFR, KRAS, BRAF, ALK, PIK3CA, PDGFRA, and DDR2 in primary pulmonary adenoid cystic carcinoma. Diagn Pathol. 2015;10:161. doi: 10.1186/s13000-015-0409-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486(7403):395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R, Rosenthal R, et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376(22):2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, Seth S, Chow CW, Cao Y, Gumbs C, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346(6206):256–259. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson K, Lutz C, Van Delft FW, Bateman CM, Guo Y, Colman SM, Kempski H, Moorman AV, Titley I, Swansbury J, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469(7330):356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 24.Gervais C, Murati A, Helias C, Struski S, Eischen A, Lippert E, Tigaud I, Penther D, Bastard C, Mugneret F, et al. Acute myeloid leukaemia with 8p11 (MYST3) rearrangement: an integrated cytologic, cytogenetic and molecular study by the groupe francophone de cytogenetique hematologique. Leukemia. 2008;22(8):1567–1575. doi: 10.1038/leu.2008.128. [DOI] [PubMed] [Google Scholar]
- 25.Dawson MA. The cancer epigenome: concepts, challenges, and therapeutic opportunities. Science. 2017;355(6330):1147–1152. doi: 10.1126/science.aam7304. [DOI] [PubMed] [Google Scholar]
- 26.Mitani Y, Li J, Rao PH, Zhao Y-J, Bell D, Lippman SM, Weber RS, Caulin C, El-Naggar AK. Comprehensive analysis of the MYB-NFIB gene fusion in salivary adenoid cystic carcinoma: incidence, variability, and clinicopathologic significance. Clin Cancer Res. 2010;16(19):4722–4731. doi: 10.1158/1078-0432.CCR-10-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitani Y, Rao PH, Futreal PA, Roberts DB, Stephens PJ, Zhao Y-J, Zhang L, Mitani M, Weber RS, Lippman SM, et al. Novel chromosomal rearrangements and break points at the t(6;9) in salivary adenoid cystic carcinoma: association with MYB-NFIB chimeric fusion, MYB expression, and clinical outcome. Clin Cancer Res. 2011;17(22):7003–7014. doi: 10.1158/1078-0432.CCR-11-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, Wistuba II, Varella-Garcia M, Franklin WA, Aronson SL, Su PF, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311(19):1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia N. An J, Jiang QQ, Li M, Tan J, Hu CP. Analysis of EGFR, EML4-ALK, KRAS, and c-MET mutations in Chinese lung adenocarcinoma patients. Exp Lung Res. 2013;39(8):328–335. doi: 10.3109/01902148.2013.819535. [DOI] [PubMed] [Google Scholar]
- 30.Lu RL, Hu CP, Yang HP, Li YY, Gu QH, Wu L. Biological characteristics and epidermal growth factor receptor tyrosine kinase inhibitors efficacy of EGFR mutation and its subtypes in lung adenocarcinoma. Pathol Oncol Res. 2014;20(2):445–451. doi: 10.1007/s12253-013-9715-0. [DOI] [PubMed] [Google Scholar]
- 31.Xiao D, Lu C, Zhu W, He Q, Li Y, Fu C, Zhou J, Liu S, Tao Y. Comparison of small biopsy specimens and surgical specimens for the detection of EGFR mutations and EML4-ALK in non-small-cell lung cancer. Oncotarget. 2016;7(37):59049–59057. doi: 10.18632/oncotarget.10011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ge L, Shi R. Progress of EGFR-TKI and ALK/ROS1 inhibitors in advanced non-small cell lung cancer. Int J Clin Exp Med. 2015. Jul 15;8(7):10330–10339. [PMC free article] [PubMed] [Google Scholar]
- 33.Tan L, Hu Y, Tao Y, Wang B, Xiao J, Tang Z, Lu T, Tang H. Expression and copy number gains of the RET gene in 631 early and mid stage non-small cell lung cancer cases. Thorac Cancer. 2018 [Epub ahead of print]. doi: 10.1111/1759-7714.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Issa JP. Aging and epigenetic drift: a vicious cycle. J Clin Invest. 2014;124(1):24–29. doi: 10.1172/JCI69735. [DOI] [PMC free article] [PubMed] [Google Scholar]


