Graphical abstract
Keywords: Hepatitis, Autoimmune, Variants, Paediatrics, Cytokines, Tumour necrosis factor
Highlights
-
•
CARD10 rs6000782 and TNF rs1799724 SNPs are associated with pAIH.
-
•
Carriers of minor alleles of both variants are at a higher risk of pAIH.
-
•
The mutant genotype of rs6000782 increases serum levels of NFκB-p65 and TNF-α.
-
•
High levels of cytokines are coupled with elevated AST and ALT and prolonged INR.
-
•
No CARD10 rs6000782 and TNF rs1799724 variants predispose towards cirrhosis.
Abstract
Although the pathogenesis of paediatric-onset autoimmune hepatitis (pAIH) remains incompletely understood, genetic variants and environmental factors are known to be involved. Caspase recruitment domain family member 10 (CARD10) is a scaffold protein that participates in a complex pathway activating nuclear factor kappa-B (NFκB) and tumour necrosis factor alpha (TNF-α). This study aimed to investigate the association of CARD10 rs6000782 (g.37928186A > C) and TNF gene promoter rs1799724 (c.-1037C > T) variants with pAIH susceptibility in a cohort of Egyptian children. The research was also extended to assess the relationship of these variants with levels of NFκB-p65 and TNF-α. Fifty-six pAIH patients and 44 age- and sex-matched healthy controls were included. Variant genotyping was performed by polymerase chain reaction (PCR). Serum NFκB-p65 and TNF-α levels were measured using enzyme-linked immunosorbent assays (ELISAs). rs6000782 C and rs1799724 T alleles, separate or in combination, were significantly increased in pAIH patients compared to controls. Serum levels of NFκB-p65 and TNF-α were higher in pAIH differentiating both groups. Moreover, the recessive model of rs6000782 revealed a significant association with the levels of both NFκB-p65 and TNF-α. In conclusion, rs6000782 and rs1799724 variants are potential genetic risk factors for pAIH predisposition, with the former affecting NFκB-p65 and TNF-α levels. Overall, the inflammatory cascade was associated with the degree of liver cell destruction. Clinically, screening and genetic counselling are recommended for relatives of pAIH patients.
Introduction
Autoimmune hepatitis is an idiopathic, relatively rare, chronic, progressive self-perpetuating inflammatory disorder of the liver that may lead to cirrhosis and hepatocellular carcinoma [1]. The disorder is characterized by female predominance and the presence of elevated serum levels of transaminases, autoantibodies, and immunoglobulins [2]. The prevalence and clinical expression of AIH varies greatly according to age, race, and ethnicity [3], [4], [5]. Among the scant literature discussing the onset of paediatric-onset autoimmune hepatitis (pAIH), a recent Chinese study reported an incidence of 0.4 per 100,000 children [6]. Overall, the pathogenesis of AIH remains incompletely understood; however, genetics, represented by single-nucleotide polymorphisms (SNPs), and environmental factors are involved [7]. The most investigated genes are those located within the human leukocyte antigen (HLA) region, though evaluation of the non-HLA genetics of AIH may provide novel targets for diagnosis, treatment, and prevention [8].
The caspase recruitment domain family member 10 gene (CARD10 [MIM: 607209]), located 12,643 bp downstream in the 22q13.1 region, encodes the CARD10 protein, formerly known as CARD-containing membrane-associated guanylate kinase (MAGUK) protein 3. CARD10 is activated via stimulation of G protein-coupled receptors by lysophosphatidic acid and angiotensin II [9]. CARD10 is a scaffold protein in the CARD10/b-cell CLL lymphoma 10 (BCL10)/mucosa-associated lymphoid tissue (MALT1) pathway, which induces pro-inflammatory nuclear factor kappa-B (NFκB) activation and is widely expressed in a variety of non-hematopoietic tissues, including hepatocytes [10]. NFκB-p65 is a subunit of the NFκB complex and plays a crucial role in inflammatory and immune responses [11]. The SNP rs6000782 (g.37532179A > C), which has been mapped to the CARD10 gene, was found in a genome-wide association study (GWAS) to be associated with AIH [12]. Nonetheless, a lack of this association was reported in a Japanese population comprising a replication cohort [13].
Tumour necrosis factor alpha (TNF-α) is a pleiotropic, potent cytokine that is mainly involved in promoting strong inflammatory and immune responses [14], and TNF-α overproduction may predispose towards autoimmunity [15]. The gene encoding TNF-α is located within the Class III region of the human major histocompatibility complex on chromosome 6 (6p21.33) [16]. Several variants have been identified in the promoter region of the tumour necrosis factor gene (TNF [MIM: 191160]), with the potential to cause changes within regulatory sites and thus affect the regulation and/or function of TNF-α production [17], [18]. Overexpression of TNF-α in AIH patients’ sera has been reported, with the level directly correlating with prognosis [19]. In addition, the TNF promoter polymorphism rs1799724 (c.-1037C > T) has been associated with autoimmune diseases, such as ankylosing spondylitis and Crohn’s disease [20], [21] and with susceptibility to hepatitis B virus infection in some populations [22].
Nonetheless, no data are available to date concerning the relationships of CARD10 gene variations and pAIH or AIH in a Middle Eastern population. Furthermore, TNF (c.-1037C > T) has not been studied in relation to AIH. Therefore, the current study aimed to investigate the association of CARD10 rs6000782 and TNF rs1799724 with pAIH susceptibility in a cohort of Egyptian children. Relationships between these SNPs, clinical and biochemical data and levels of NFκB-p65 and TNF-α were also assessed.
Patient and methods
Study population
This prospective case-control study was carried out on 56 Egyptian children diagnosed with AIH [males = 15 (26.79%) and females = 41 (73.21%); average age 8.36 ± 3.44 (mean ± SD), range 4–17 years]. All patients were recruited from the outpatient clinics of the hepatology units at Kasr El-Aini and New Children Pediatric Hospitals of Cairo University between July 2016 and July 2017.
All pAIH patients were diagnosed based on history, physical examination, liver biochemical profile, autoantibodies, ultrasound findings and scoring systems according to simplified diagnostic criteria of the international autoimmune hepatitis group [23]. Percutaneous liver biopsy was performed for those without contraindications (n = 41), revealing the typical hallmarks of AIH, including interface hepatitis, hepatocellular rosette formation and piecemeal necrosis in 73% and findings compatible with AIH in 27%. A score of 6 was obtained in 20 patients, and 36 patients had a score equal to or greater than 7. Liver cirrhosis was diagnosed based on clinical and ultrasound findings, coagulopathy, and decreased serum albumin together with liver biopsy whenever not contraindicated.
Patients with associated chronic viral liver disease (hepatitis B virus (HBV) and hepatitis C virus (HCV)), overlap syndrome, or other autoimmune diseases and those who had undergone liver transplantation were excluded. A group of 44 age- and sex-matched children (14 males (31.8%), 30 females (68.2%); age 8.93 ± 2.6) were recruited as a control group. The controls were negative for autoantibodies and had no familial history of autoimmune disease.
The study’s protocol and all procedures performed were approved by the Research Ethics Committee for experimental and clinical studies at the Faculty of Pharmacy of Cairo University in Cairo, Egypt (approval number: BC1748). Written informed consent was obtained from the guardians of all subjects in addition to the personal consent of children older than 7 years in accordance with the ethical standard laid down in the 1975 Helsinki declaration as revised in 2008.
Blood sampling and laboratory tests
Baseline venous blood samples (10 mL) were collected by well-trained nurses. Complete laboratory analysis was performed under aseptic conditions. Six millilitres of blood was allowed to clot and then centrifuged at 1500g for 10 min to separate the serum, which was then divided into four aliquots and stored at −20 °C until further use for laboratory analysis. Laboratory tests included liver biochemical profile, i.e., aspartate aminotransferase (AST), alanine aminotransferase (ALT), total bilirubin, and albumin levels, which were analysed spectrophotometrically using the automated chemical AU680 Analyzer (Beckman Coulter, Inc. supplied by BM-Egypt, Cairo, Egypt). Immunoglobulin G (IgG) levels were determined by means of nephelometry using a BN ProSpec System provided by Siemens-Egypt (Cairo, Egypt). Levels of antinuclear antibodies (ANA), smooth muscle antibodies (SMA), and antibodies against liver kidney microsomes (anti-LKM) were measured using immunofluorescence.
The remaining 4 mL of blood was collected in vacutainer tubes containing ethylenediaminetetraacetic acid (EDTA) and stored at −80 °C for DNA extraction.
DNA extraction and genotyping
Genomic DNA extraction from whole blood was performed using a QIAamp DNA blood Mini kit (Qiagen, CA, USA supplied by Clinilab, Cairo, Egypt); yield and purity were measured using a Q5000 UV-Vis Spectrophotometer (Quawell, China provided by Matrix Scientific Trade Co., Giza, Egypt). Genotyping was performed using Applied Biosystem Step One™ Real-Time PCR System with TaqMan allelic discrimination assays using predesigned primer/probe sets for CARD10 rs6000782 (A/C) [NC_000022.10: g.37928186A > C, TaqMan SNP Genotyping Assays ID: C__30428160_10] and TNF-α -857C/T [NM_000594.3:c.-1037C > T, dbSNP ID: rs1799724, TaqMan SNP Genotyping Assays ID C__11918223_10] (Applied Biosystems, CA, USA supplied by Clinilab, Cairo, Egypt).
The PCR reaction mixture consisted of the following: 12.5 µL TaqMan Genotyping Master Mix (2x), 1.25 µL 20x working SNP genotyping assay, genomic DNA: 20/DNA concentration in µL, and DNase-free water was added to reach a total volume of 25 µL. Sample denaturation and enzyme activation were performed at 95 °C for 10 min, followed by 50 cycles of PCR amplification at 95 °C for 15 s and 60 °C for 90 s; allelic discrimination plate reading and analysis were performed using Sequence Detection System (SDS) software (Applied Biosystems, CA, USA supplied by Clinilab, Cairo, Egypt). VIC dye and FAM dye were used for allele discrimination.
Quantitative detection of serum NFκB-p65 and TNF-α by ELISA
Serum NFκB-p65 and TNF-α levels were measured using enzyme-linked immunosorbent assays (ELISAs) according to the manufacturers’ instructions. NFκB-p65 was assayed using a human NFκB-p65 ELISA kit (Glory Science Co., Ltd., Del Rio, TX, USA) and TNF-α using an AssayMax Human TNF-alpha ELISA kit (Assaypro, MO, USA); the kits were supplied by Leader Trade Co., Cairo, Egypt. The results of serum NFκB-p65 and TNF-α levels are expressed as ng/L and pg/mL, respectively.
Statistical analysis
All statistical calculations were performed using GraphPad Prism 6.0 (GraphPad Software, CA, USA provided by https://www.graphpad.com) for Microsoft Windows. Values are reported as the mean ± standard deviation (SD) or number (percentage) when appropriate. Prior to association analyses, deviation from Hardy-Weinberg equilibrium (HWE) was tested for each polymorphism using the chi-squared (χ2) test, which was also applied for comparing categorical data. Fisher’s exact test was employed when the expected frequency was less than five. Continuous normally distributed variables were compared using Student’s t-test or one-way analysis of variance (ANOVA); for non-parametric data, Mann Whitney or Kruskal Wallis tests were used when appropriate. Correlations were determined by Pearson and Spearman correlations. Odds ratios (ORs) with 95% confidence intervals were calculated. P values < 0.05 were considered statistically significant.
Results
Demographic and biochemical profile
The demographic and laboratory data of the 56 pAIH patients and 44 healthy subjects enrolled in this study are provided in Table 1. All diagnosed cases were sporadic. The results show high statistical significance of biochemical profile parameters among the two groups (P < 0.0001), with wide variability within the cases and female predominance. Most cases had ANA and SMA titres ≥ 1:40 (71.43% and 92.86%, respectively). Four of the 5 patients with anti-LKM antibody positivity also showed both ANA and SMA titres ≥ 1:40, and a concomitant speckled ANA pattern was observed for one patient.
Table 1.
Baseline characteristics of pAIH patients and controls.
| Characteristics | pAIH n = 56 (%) | Controls n = 44 (%) | P value |
|---|---|---|---|
| Demographic data | |||
| Female | 41 (73.21) | 30 (68.18) | 0.659 |
| Male | 15 (26.79) | 14 (31.82) | |
| Age, years, mean ± SD | 8.36 ± 3.44 | 8.93 ± 2.6 | 0.1 |
| Laboratory investigations | |||
| AST, (IU/L), mean ± SD | 600.36 ± 424.44 | 19.14 ± 7.26 | <0.0001* |
| ALT, (IU/L), mean ± SD | 543.71 ± 382.01 | 16.66 ± 7.22 | <0.0001* |
| Total bilirubin, (mg/dL), mean ± SD | 4.92 ± 3.71 | 0.39 ± 0.23 | <0.0001* |
| Albumin, (g/dL), mean ± SD | 3.32 ± 0.68 | 4.511 ± 0.5 | <0.0001* |
| INR, mean ± SD | 1.78 ± 0.67 | 1.03 ± 0.085 | <0.0001* |
| IgG, (mg/dL), mean ± SD | 3019 ± 1353 | 958.8 ± 139.8 | <0.0001* |
| ANA ≥ 1:40, n (%) | 40 (71.43) | 0 (0) | <0.0001* |
| SMA ≥ 1:40, n (%) | 52 (92.86) | 0 (0) | <0.0001* |
| Anti-LKM ≥ 1:40, n (%) | 5 (8.93) | 0 (0) | 0.065 |
| Clinical data | |||
| Cirrhosis, n (%) | 23 (41.07) | – | – |
| Hypersplenism, n (%) | 10 (17.86) | – | – |
Values are expressed as the mean ± standard deviation (SD) or number n (%). pAIH, paediatric-onset autoimmune hepatitis; AST, aspartate aminotransferase; ALT, alanine aminotransferase; INR, international normalized ratio; IgG, immunoglobulin G; ANA, antinuclear antibodies; SMA, smooth muscle antibodies; anti-LKM, antibodies against liver kidney microsomes. Categorical data were analysed using Fisher’s exact test. Continuous data were analysed using an unpaired Student’s t-test or the Mann Whitney test after testing for normality.
Indicates statistical significance (P < 0.05).
HWE
As shown in Table 2, the genotype distributions of the CARD10 rs6000782 and TNF rs1799724 polymorphisms agreed with those expected for Hardy–Weinberg equilibrium at P = 0.13 and P = 0.64, respectively, in patients and P = 0.11 and P = 0.26, respectively, in controls.
Table 2.
Hardy–Weinberg equilibrium for rs6000782 and rs1799724 in pAIH patients and controls.
| Genotype | Observed frequency | Expected frequencya | P value b | ||
|---|---|---|---|---|---|
| rs6000782 | cases | AA | 55.36 | 52.3 | X2 = 2.323 P = 0.127 |
| AC | 33.93 | 40.0 | |||
| CC | 10.71 | 7.7 | |||
| controls | AA | 72.73 | 74.6 | X2 = 2.492 P = 0.114 |
|
| AC | 27.27 | 23.6 | |||
| CC | 0 | 1.9 | |||
| rs1799724 | cases | CC | 55.36 | 56.3 | X2 = 0.225 P = 0.635 |
| CT | 39.28 | 37.5 | |||
| TT | 5.36 | 6.3 | |||
| controls | CC | 79.55 | 80.6 | X2 = 1.297 P = 0.255 |
|
| CT | 20.45 | 18.4 | |||
| TT | 0 | 1.0 | |||
Results are presented as percentages.
Expected genotype frequencies based on observed allele frequencies and assuming Hardy–Weinberg equilibrium.
P values were calculated using the χ2 test for Hardy–Weinberg equilibrium at individual loci.
Genotyping
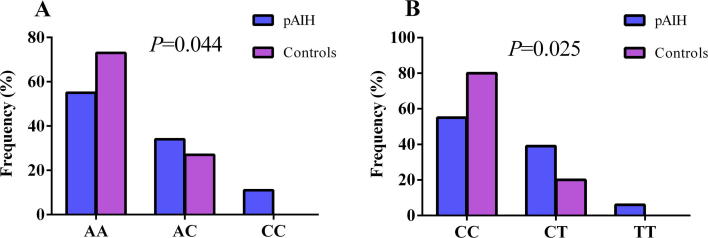
The genotype frequencies for rs6000782 and rs1799724 according to the full genotype model are described in Fig. 1. Other genotypic models, including additive, dominant, recessive, and allelic models, were compared in cases and controls, as shown in Table 3. For both SNPs, statistical significance between both groups was found in the full genotype and allelic models.
Fig. 1.
Genotype distribution according to the full genotype model of (A) CARD10 rs6000782 and (B) TNF rs1799724 in pAIH patients and controls. P was calculated using the χ2 test. Statistical significance was considered at P < 0.05.
Table 3.
Genotype and allele frequency distribution of rs6000782 and rs1799724 in pAIH patients and controls.
| Genotypes and alleles | pAIH n = 56 (%) |
Controls n = 44 (%) | OR | 95% CI | P value | |
|---|---|---|---|---|---|---|
| rs6000782 A/C | AA | 31 (55.36) | 32 (72.73) | Ref | ||
| AC | 19 (33.93) | 12 (27.27) | 1.63 | 0.68–3.92 | 0.283 | |
| CC | 6 (10.71) | 0 (0) | – | – | 0.027* | |
| AC + CC | 25 (44.64) | 12 (27.27) | 2.15 | 0.92–5.02 | 0.096 | |
| AA + AC | 50 (89.29) | 44 (100) | Ref | |||
| CC | 6 (10.71) | 0 (0) | – | – | 0.033* | |
| Allele A | 81 (72.3) | 76 (86.4) | Ref | |||
| Allele C | 31 (27.7) | 12 (13.6) | 2.42 | 1.16–5.06 | 0.023* | |
| rs1799724 C/T | CC | 31 (55.36) | 35 (79.55) | Ref | ||
| CT | 22 (39.28) | 9 (20.45) | 2.76 | 1.11–6.88 | 0.031* | |
| TT | 3 (5.36) | 0 (0) | – | – | 0.114 | |
| CT + TT | 25 (44.64) | 9 (20.45) | 3.14 | 1.27–7.73 | 0.019* | |
| CC + CT | 53 (94.64) | 44 (100) | Ref | |||
| TT | 3 (5.36) | 0 (0) | – | – | 0.253 | |
| Allele C | 84 (75) | 79 (89.77) | Ref | |||
| Allele T | 28 (25) | 9 (10.23) | 2.93 | 1.30–6.59 | 0.010* | |
Values are expressed as numbers (percentages). CI, confidence interval; Ref, reference.
Indicates statistical significance (P < 0.05).
For rs6000782, the minor homozygote CC genotype was significantly more abundant among the cases. The recessive model (AA + AC vs CC) also showed a significant difference (P = 0.03). Conversely, the rs1799724 heterozygous CT genotype was significantly less common in controls. The common homozygous CC genotype in the dominant model also showed statistical significance when compared to the CT + TT genotypes.
The joint effect of the studied gene polymorphisms was examined in patients with pAIH compared to the control group (Table 4). The results revealed that the combination of the homozygous minor allele (CC) of the first SNP with the heterozygous genotype (CT) of the latter greatly predisposed patients towards pAIH. In addition, the presence of rs6000782 and rs1799724 CT haplotypes carried a high risk of pAIH occurrence.
Table 4.
Haplotype and joint analysis of rs6000782 (A/C) and rs1799724 (C/T) polymorphisms in pAIH patients compared to controls.
| pAIH, n (%) | Controls, n (%) | P | OR (CI) | |
|---|---|---|---|---|
| Combined genotypes | ||||
| AA + CC | 19 (33.93) | 27 (61.36) | Ref | |
| AA + CT | 11 (19.64) | 5 (11.36) | 0.083 | 3.13 (0.93–10.5) |
| AA + TT | 1 (1.79) | 0 (0) | 0.426 | – |
| AC + CC | 12 (21.43) | 8 (18.18) | 0.189 | 2.1 (0.73–6.2) |
| AC + CT | 6 (10.71) | 4 (9.09) | 0.315 | 2.1 (0.53–8.6) |
| AC + TT | 1 (1.79) | 0 (0) | 0.426 | – |
| CC + CC | 0 (0) | 0 (0) | – | – |
| CC + CT | 5 (8.93) | 0 (0) | 0.018* | – |
| CC + TT | 1 (1.79) | 0 (0) | 0.426 | – |
| Combined alleles | ||||
| AC | 128 (57.14) | 138 (78.41) | Ref | |
| AT | 34 (15.18) | 14 (7.96) | 0.005* | 2.62 (1.34–5.1) |
| CC | 40 (17.86) | 20 (11.36) | 0.010* | 2.16 (1.20–3.88) |
| CT | 22 (9.82) | 4 (2.27) | <0.001* | 5.93 (1.99–17.7) |
Values are expressed in numbers. Ref, reference. pAIH, n = 56, healthy controls, n = 44.
Indicates statistical significance (P < 0.05).
NFκB-p65 and TNF-α
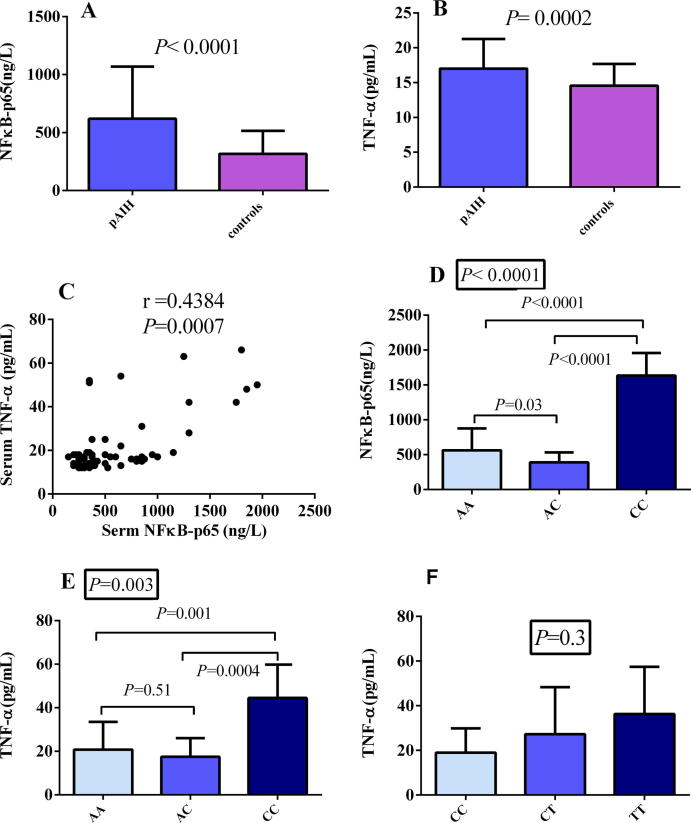
Levels of both cytokines were significantly elevated among the cases in comparison to the controls (Fig. 2A and B). Moreover, each correlated strongly with the other, as shown in Fig. 2C (P = 0.0007). Upon comparing the serum levels of both cytokines to rs6000782 genotypes of the studied patients, the effect of the homozygous minor allele (CC) was evident wherein the levels of NFκB-p65 and TNF-α were significantly higher than those of AA (Fig. 2D and E). The recessive model of this SNP showed the same results, with P < 0.0001 for NFκB-p65 and P = 0.0003 for TNF-α.
Fig. 2.
Serum levels of NFκB-p65 and TNF-α. (A) NFκB-p65 in cases and controls. (B) TNF-α among cases and controls. (C) Correlation of NFκB-p65 and TNF-α levels. (D) NFκB-p65 among different rs6000782 genotypes. (E) TNF-α among different rs6000782 genotypes. (F) TNF-α among different rs1799724 genotypes. Values are expressed as the mean ± standard deviation (SD). P values in boxes were calculated by ANOVA. Statistical significance at P < 0.05.
TNF-α levels were higher in the rs1799724 TT group than in the other genotype groups (Fig. 2F). However, the increased levels did not show statistical significance across the three genotypes in any of the studied models.
Relationship of studied variants and cytokines with clinical and biochemical profiles of patients
Table 5 shows the association of the rs6000782 CC genotype with elevated ALT, international normalized ratio (INR), and IgG levels compared to those of the AA genotype. In addition, the recessive model (not shown in the table) confirmed the association with the three abovementioned parameters. In the current study, a lack of association between rs1799724 in all models and the different parameters studied was found.
Table 5.
Association of rs6000782 and rs1799724 genotypes with characteristics of pAIH patients.
| Parameter | rs6000782 |
rs1799724 |
||||||
|---|---|---|---|---|---|---|---|---|
| (n=) | AA (31) |
AC (19) |
P | CC (6) |
P | CC (31) |
CT + TT (25) |
P |
| Sex (F/M) | 23/8 | 12/7 | 0.53 | 6/0 | 0.31 | 22/9 | 19/6 | 0.77 |
| Age (years) | 8.1 ± 3.1 | 9.3 ± 4.2 | 0.25 | 6.8 ± 1.1 | 0.31 | 8.9 ± 3.8 | 7.7 ± 2.9 | 0.22 |
| AST (IU/L) | 550 ± 414 | 561 ± 381 | 0.73 | 986 ± 478 | 0.09 | 582 ± 453 | 622 ± 394 | 0.73 |
| ALT (IU/L) | 454 ± 276 | 525 ± 382 | 0.56 | 1,066 ± 497 | 0.02* | 498 ± 343 | 601 ± 426 | 0.58 |
| T. bilirubin (mg/dL) | 4.5 ± 3.2 | 4.5 ± 3.6 | 0.96 | 8.1 ± 5.5 | 0.09 | 4.9 ± 3.7 | 5 ± 3.8 | 0.75 |
| Albumin (g/dL) | 3.3 ± 0.7 | 3.5 ± 0.6 | 0.21 | 2.8 ± 0.8 | 0.16 | 3.4 ± 0.6 | 3.2 ± 0.8 | 0.15 |
| INR | 1.6 ± 0.5 | 1.8 ± 0.7 | 0.43 | 2.5 ± 0.9 | 0.01* | 1.7 ± 0.6 | 1.9 ± 0.7 | 0.15 |
| IgG (mg/dL) | 2,894 ± 1,226 | 2,788 ± 1,109 | 0.57 | 4,399 ± 2,028 | 0.03* | 2,795 ± 1,298 | 3,298 ± 1,394 | 0.06 |
| ANA ≥ 1:40, n | 22 | 13 | 1 | 5 | 1 | 26 | 14 | 0.05 |
| SMA ≥ 1:40, n | 30 | 16 | 0.15 | 6 | 1 | 30 | 22 | 0.31 |
| Anti-LKM ≥ 1:40, n | 4 | 1 | 0.64 | 0 | 1 | 3 | 2 | 1 |
| Cirrhosis, n | 12 | 7 | 1 | 4 | 0.37 | 11 | 12 | 0.42 |
Values are expressed as the mean ± standard deviation (SD) or number n (%). pAIH, paediatric-onset autoimmune hepatitis; AST, aspartate aminotransferase; ALT, alanine aminotransferase; INR, international normalized ratio; IgG, immunoglobulin G; ANA, antinuclear antibodies; SMA, smooth muscle antibodies; anti-LKM, antibodies against liver kidney microsomes. Categorical data were analysed using Fisher’s exact test. Continuous data were analysed using an unpaired Student’s t-test or the Mann Whitney test after testing for normality.
Indicates statistical significance (P < 0.05).
NFκB-p65 was positively correlated with the necroinflammatory markers AST (r = 0.304 and P = 0.023) and ALT (r = 0.331 and P = 0.013), though no correlation was found with the synthetic function of both cytokines (Table 6). Furthermore, no associations between the studied cytokines and sex, autoantibody or IgG level, or cirrhosis were observed.
Table 6.
Correlations between NFκB-p65 and TNF-α serum levels and biochemical parameters of pAIH patients.
| NFκB-p65 |
TNF-α |
|||
|---|---|---|---|---|
| r | P value | r | P value | |
| AST | 0.304 | 0.023* | 0.203 | 0.135 |
| ALT | 0.331 | 0.013* | 0.114 | 0.402 |
| Total bilirubin | 0.184 | 0.176 | 0.155 | 0.253 |
| Albumin | −0.223 | 0.098 | −0.089 | 0.517 |
| INR | 0.251 | 0.063 | 0.201 | 0.138 |
NFκB-p65, nuclear factor kappa B-p65; TNF-α, tumour necrosis factor alpha; r, correlation coefficient; AST, aspartate aminotransferase; ALT, alanine aminotransferase; INR, international normalized ratio.
Indicates statistical significance (P < 0.05).
Discussion
Polymorphisms in cytokine genes affect their levels and consequently the balance between type 1 and 2 T-helper cells. This imbalance predisposes individuals towards many autoimmune diseases, including AIH [24]. Although AIH has classically been sub-categorized into types 1 and 2 according to autoantibody profiles, recent studies highlight the lack of genetic, clinical, biochemical and histological bases for this categorization [25]. Thus, the research presented herein intended to examine the effect of SNPs rs6000782 and rs1799724 on the serum levels of NFκB-p65 and TNF-α, the occurrence of pAIH, and the clinical and biochemical presentation.
Of note, the CARD10 rs6000782 allele frequencies among the healthy subjects included in this study were intermediate compared to those reported for other populations, namely, the Egyptian control children in the present study showed A and C allele frequencies of 86.4% and 13.6%, respectively. Based on data from the HapMap project, version 2010–08_phase II + III, the allele A frequencies were 96.7% and 95.6% in European and Japanese populations, respectively; in contrast, a Nigerian population showed an allele A frequency of only 63.3%. These differences may be attributed to the fact that modern Egyptians are descendants of mixed ancestries due to historical genetic admixture and racial mixing. Regardless, the abovementioned discrepancy was not observed for TNF rs1799724, with controls exhibiting a C allele frequency of 89.77%, comparable to European, Japanese, and Nigerian populations, of 93.3%, 82.2%, and 96.7%, respectively. It should be noted that in this study, the genotype frequencies of both cases and controls were in HWE, ensuring no genotyping deviation, batch effects or population stratification.
In the present investigation, CARD10 rs6000782 proved to be significantly associated with pAIH, with a higher frequency of the mutant homozygous CC genotype and the recessive model (CC vs. AA + AC) among cases compared to controls; this was also observed for the C allele frequency. The earliest GWAS for AIH was carried out by de Boer and colleagues using the gene sequencing method; among the findings was the novel reporting of an association of the CARD10 rs6000782 C allele with AIH in German and Dutch populations [12]. However, this relationship did not meet the criteria of P < 5 × 10−8 required to declare genome-wide significance [26]. Failure to confirm these results using the PCR-restriction fragment length polymorphism technique was reported in a replication study involving a Japanese population by Migita et al. [13]. A possible explanation for these discrepant results is the ethnic variations and/or environmental risk factors among different populations.
This study is the first to investigate the functional role of CARD10 rs6000782 and its relationship to the levels of certain cytokine. The CC genotype was associated with significantly higher levels of both NFκB-p65 and TNF-α compared to the AA genotype. This may be explained by the fact that CARD10 is required for G-protein-coupled receptor, protein kinase C and epidermal growth factor receptor-induced NFκB activation [27], [28], with consequent production of proinflammatory cytokines, such as TNF-α in lymphocytes, hepatocytes and hepatic stellate cells [29]. The results elaborate on the finding of a strong positive correlation between NFκB-p65 and TNF-α levels, and this report thus confirms the hypothesis of an altered cytokine profile in patients with pAIH. The rs6000782 SNP with an activated inflammatory cascade was reflected as a positive association between the mutant CC genotype, the degree of liver cell destruction (AST and ALT elevation) and deterioration in synthetic function (prolonged INR). Overall, the positive correlations between NFκB-p65 and AST and ALT levels support these findings.
To the best of the authors’ knowledge, this study provides the first evidence of an association between TNF rs1799724 and increased pAIH susceptibility. The present study reveals that compared with the CC genotype, the rs1799724 CT genotype is a risk factor for pAIH. Despite conflicting data regarding rs17999724 and autoimmune diseases, the findings agree with meta-analyses of diseases such as ankylosing spondylitis [20], Crohn’s disease [21] and vitiligo [18] using additive, dominant, and allele models. In contrast, a lack of this association was documented for idiopathic nephrotic syndrome [30] and rheumatoid arthritis [31]. These contradictory results might be explained by differences in ethnic origin or disease presentation or in number of individuals studied.
Although the specific role of TNF-α in AIH pathogenesis has not yet been completely explained, using an experimental model, Gatselis et al. concluded that TNF-α is essential for the induction of AIH via upregulation of hepatic C-C motif chemokine ligand 20 expression, promoting the migration of dysregulated splenic T-cells [32]. This confirms that pAIH patients show significantly higher levels of TNF-α than do controls, in agreement with the results of Akberova et al. [33]. It is important to highlight that TNF-α promoter polymorphisms are reported to be involved in modulating the expression, and consequently, the level of TNF-α [17].
An attempt to identify an association between rs1799724 and TNF-α levels revealed a tendency for higher levels in the additive model (CT vs. CC) and the dominant model (CT + TT vs CC); however, this was not statistically significant, which may be explained by the wide spectrum of disease activity among patients at diagnosis. This explanation is extended to support both the lack of association of the dominant model and the lack of correlation of TNF-α levels on one hand to the different studied biochemical parameters on the other hand. A lack of such correlation was also reported in a cohort of AIH adults [33].
This study’s results emphasize that the combined presence of these two variants in the studied paediatric population played a central role in pAIH susceptibility, confirming that this disease is predisposed by the combination of low-penetrance susceptibility alleles. The two gene combinations indicate that by modulating cytokine levels, these SNPs are potential genetic susceptibility markers. Such results confirm the heterogeneous nature and complex immunopathogenesis of AIH and may provide a better understanding of key molecular effectors in this disease, which may aid the development of more specific and patient-tailored immunotherapies [34].
Study limitations
The relatively small number of patients is a limitation of this study. However, there is an overall scarcity of data in the literature concerning AIH in children compared to adults. Extending the investigation of the studied associations to larger sample sets and ethnically different populations is warranted to validate that the aforementioned findings would be replicated in other groups. Nevertheless, the data reported herein may have important clinical implications regarding family studies and screening as well as genetic counselling for individuals related to pAIH patients.
Conclusions
The CARD10 rs6000782 CC genotype and TNF rs1799824 CT heterozygous genotype are associated with AIH in paediatric patients. Combined carriers of CARD10 rs6000782 (C) and TNF rs1799824 (T) alleles are at a higher risk of pAIH. Furthermore, the data suggest that the CC genotype of CARD10 rs6000782 is associated with increased serum levels of both NFκB-p65 and TNF-α, as reflected biochemically in an elevation of serum transaminases and reduced synthetic liver function.
Acknowledgments
Acknowledgments
The authors sincerely acknowledge the staff members of the hepatology outpatient clinic at New Children Pediatrics Hospital of Cairo University. This work was partially supported by the Research Service of the Faculty of Pharmacy, Cairo University. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest
The authors have no conflicts of interest to declare.
Footnotes
Peer review under responsibility of Cairo University.
References
- 1.Manns M.P., Lohse A.W., Vergani D. Autoimmune hepatitis – update 2015. J Hepatol. 2015;62(S1):S100–S111. doi: 10.1016/j.jhep.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Tang J., Zhou C., Zhang Z.J., Sen Zheng S. Association of polymorphisms in non-classic MHC genes with susceptibility to autoimmune hepatitis. Hepatobiliary Pancreat Dis Int. 2012;11(2):125–131. doi: 10.1016/s1499-3872(12)60136-2. [DOI] [PubMed] [Google Scholar]
- 3.Lohse A., Chazouilleres O., Daleko G., Drenth J., Heneghan M., Hofer H. EASL clinical practice guidelines: autoimmune hepatitis. J Hepatol. 2015;63(4):971–1004. doi: 10.1016/j.jhep.2015.06.030. [DOI] [PubMed] [Google Scholar]
- 4.Van Gerven N.M.F., Verwer B.J., Witte B.I., Van Erpecum K.J., Van Buuren H.R., Maijers I. Epidemiology and clinical characteristics of autoimmune hepatitis in the Netherlands. Scand J Gastroenterol. 2014;49(10):1245–1254. doi: 10.3109/00365521.2014.946083. [DOI] [PubMed] [Google Scholar]
- 5.Delgado J.-S., Vodonos A., Malnick S., Kriger O., Wilkof-Segev R., Delgado B. Autoimmune hepatitis in southern Israel: a 15-year multicenter study. J Dig Dis. 2013;14(11):611–618. doi: 10.1111/1751-2980.12085. [DOI] [PubMed] [Google Scholar]
- 6.Li X., Chen H., Cai Y., Zhang P., Chen Z. Association of STAT4 and PTPN22 polymorphisms and their interactions with type-1 autoimmune hepatitis susceptibility in Chinese Han children. Oncotarget. 2017;8(37):60933–60940. doi: 10.18632/oncotarget.17458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Webb G.J., Hirschfield G.M. Genetics of autoimmune liver disease: a brief summary for clinicians. Dig Dis. 2014;32(5):e1–e6. doi: 10.1159/000366174. [DOI] [PubMed] [Google Scholar]
- 8.Webb G.J., Hirschfield G.M., Krawitt E.L., Gershwin M.E. Cellular and molecular mechanisms of autoimmune hepatitis. Annu Rev Pathol Mech Dis. 2018;13(1):247–292. doi: 10.1146/annurev-pathol-020117-043534. [DOI] [PubMed] [Google Scholar]
- 9.D’Andrea E.L., Ferravante A., Scudiero I., Zotti T., Reale C., Pizzulo M. The dishevelled, EGL-10 and pleckstrin (DEP) domain-containing protein DEPDC7 binds to CARMA2 and CARMA3 proteins, and regulates NF-kB activation. PLoS ONE. 2014;9(12):1–14. doi: 10.1371/journal.pone.0116062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blonska M., Lin X. NF-κB signaling pathways regulated by CARMA family of scaffold proteins. Cell Res. 2011;21(1):55–70. doi: 10.1038/cr.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nolan G.P., Ghosh S., Liou H.C., Tempst P., Baltimore D. DNA binding and I kappa B inhibition of the cloned p65 subunit of NF-kappa B, a rel-related polypeptide. Cell Av. 1991;64(5):961–969. doi: 10.1016/0092-8674(91)90320-x. [DOI] [PubMed] [Google Scholar]
- 12.De Boer Y.S., Van Gerven N.M.F., Zwiers A., Verwer B.J., Van Hoek B., Van Erpecum K.J. Genome-wide association study identifies variants associated with autoimmune hepatitis type 1. Gastroenterology. 2014;147(2) doi: 10.1053/j.gastro.2014.04.022. 443–452.e5. [DOI] [PubMed] [Google Scholar]
- 13.Migita K., Jiuchi Y., Furukawa H., Nakamura M., Komori A., Yasunami M. Lack of association between the CARD10 rs6000782 polymorphism and type 1 autoimmune hepatitis in a Japanese population. BMC Res Notes. 2015;8(1):4–8. doi: 10.1186/s13104-015-1733-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newton K., Dixit V.M. Signaling in innate immunity and inflammation. Cold Spring Harb Perspect Biol. 2012;4(3):1–19. doi: 10.1101/cshperspect.a006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim E.Y., Moudgil K.D. Immunomodulation of autoimmune arthritis by pro-inflammatory cytokines. Cytokine. 2017;98(April):87–96. doi: 10.1016/j.cyto.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang B.-B., Liu X.-Z., Sun J., Yin Y.-W., Sun Q.-Q. Association between TNF α gene polymorphisms and the risk of Duodenal Ulcer: a meta-analysis. PLoS ONE. 2013;8(2) doi: 10.1371/journal.pone.0057167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El-Tahan R.R., Ghoneim A.M., El-Mashad N. TNF-α gene polymorphisms and expression. Springerplus. 2016;5(1) doi: 10.1186/s40064-016-3197-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laddha N.C., Dwivedi M., Begum R. Increased Tumor Necrosis Factor (TNF)-α and its promoter polymorphisms correlate with disease progression and higher susceptibility towards Vitiligo. PLoS ONE. 2012;7(12) doi: 10.1371/journal.pone.0052298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li S., Huang X., Zhong H., Chen Z., Peng Q., Deng Y. Tumour necrosis factor alpha (TNF-α) genetic polymorphisms and the risk of autoimmune liver disease: a meta-analysis. J Genet. 2013;92(3):617–628. [PubMed] [Google Scholar]
- 20.Li Y., Tang H.B., Bian J., Bin Li B, Gong T.F. Genetic association between TNF-α −857 C/T polymorphism and ankylosing spondylitis susceptibility: evidence from a meta-analysis. Springerplus. 2016;5(1) doi: 10.1186/s40064-016-3603-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao Y.Q., Dong S.Q., Gao M. Association between TNF-α rs1799724 and rs1800629 polymorphisms and the risk of Crohn’s disease. Genet Mol Res. 2015;14(4):15811–15821. doi: 10.4238/2015.December.1.33. [DOI] [PubMed] [Google Scholar]
- 22.Xia Q., Zhou L.F., Liu D., Chen Z., Chen F. Relationship between TNF-<alpha> gene promoter polymorphisms and outcomes of hepatitis B virus infections: a meta-analysis. PLoS ONE. 2011;6(5) doi: 10.1371/journal.pone.0019606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hennes E.M., Zeniya M., Czaja A.J., Parés A., Dalekos G.N., Krawitt E.L. Simplified criteria for the diagnosis of autoimmune hepatitis. Hepatology. 2008;48(1):169–176. doi: 10.1002/hep.22322. [DOI] [PubMed] [Google Scholar]
- 24.Akkoc T. Re: The role of Th1/Th2 cells and associated cytokines in autoimmune hepatitis. Turkish J Gastroenterol. 2017;28(2):115–116. doi: 10.5152/tjg.2017.14021. [DOI] [PubMed] [Google Scholar]
- 25.Muratori P., Lalanne C., Fabbri A., Cassani F., Lenzi M., Muratori L. Type 1 and type 2 autoimmune hepatitis in adults share the same clinical phenotype. Aliment Pharmacol Ther. 2015;41(12):1281–1287. doi: 10.1111/apt.13210. [DOI] [PubMed] [Google Scholar]
- 26.Nicolae D.L. Testing untyped alleles (TUNA)-applications to genome-wide association studies. Genet Epidemiol. 2006;30(8):718–727. doi: 10.1002/gepi.20182. [DOI] [PubMed] [Google Scholar]
- 27.Jiang T., Grabiner B., Zhu Y., Jiang C., Li H., You Y. CARMA3 is crucial for EGFR-induced activation of NF-κB and tumor progression. Cancer Res. 2011;71(6):2183–2192. doi: 10.1158/0008-5472.CAN-10-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grabiner B.C., Blonska M., Lin P.C., You Y., Wang D., Sun J. CARMA3 deficiency abrogates G protein-coupled receptor-induced NF-κB activation. Genes Dev. 2007;21(8):984–996. doi: 10.1101/gad.1502507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McAllister-Lucas L.M., Ruland J., Siu K., Jin X., Gu S., Kim D.S.L. CARMA3/Bcl10/MALT1-dependent NF-kappaB activation mediates angiotensin II-responsive inflammatory signaling in nonimmune cells. Proc Natl Acad Sci USA. 2007;104(1):139–144. doi: 10.1073/pnas.0601947103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tieranu I., Dutescu M.I., Bara C., Tieranu C.G., Balgradean M., Popa O.M. Preliminary study regarding the association between tumor necrosis factor alpha gene polymorphisms and childhood idiopathic nephrotic syndrome in Romanian pediatric patients. Maedica (Buchar) 2017;12(3):164–168. [PMC free article] [PubMed] [Google Scholar]
- 31.Sadaf T., John P., Bhatti A., Jahangir S., Kiani A.K., Gill F.A. Lack of tumor necrosis factor alpha gene polymorphism -857c/t (rs1799724) association in Pakistani rheumatoid arthritis patients. Int J Rheum Dis. 2016;19(11):1119–1125. doi: 10.1111/1756-185X.12857. [DOI] [PubMed] [Google Scholar]
- 32.Iwamoto S., Kido M., Aoki N., Nishiura H., Maruoka R., Ikeda A. TNF-α is essential in the induction of fatal autoimmune hepatitis in mice through upregulation of hepatic CCL20 expression. Clin Immunol. 2013;146(1):15–25. doi: 10.1016/j.clim.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 33.Akberova D., Kiassov A.P., Abdulganieva D. Serum cytokine levels and their relation to clinical features in patients with autoimmune liver diseases. J Immunol Res. 2017;2017(Il):1–7. doi: 10.1155/2017/9829436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cassim S., Bilodeau M., Vincent C., Lapierre P. Novel immunotherapies for autoimmune hepatitis. Front Pediatr. 2017;5(January):1–8. doi: 10.3389/fped.2017.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]